Abstract
Osteoporosis (OP) is an age-related disease, and osteoporotic fracture is one of the major causes of disability and mortality in elderly patients (>70 years old). As the pathogenesis and molecular mechanism of OP remain unclear, the identification of disease biomarkers is important for guiding research and providing therapeutic targets. In the present study, core genes and microRNAs (miRNAs) associated with OP were identified. Differentially expressed genes (DEGs) between human mesenchymal stem cell specimens from normal osseous tissues and OP tissues were detected using the GEO2R tool of the Gene Expression Omnibus database and Morpheus. Network topological parameters were determined using NetworkAnalyzer. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were performed using the Database for Annotation, Visualization and Integrated Discovery, and ClueGO. Cytoscape with the Search Tool for the Retrieval of Interacting Genes and Molecular Complex Detection plug-in was used to visualize protein-protein interactions (PPIs). Additionally, miRNA-gene regulatory modules were predicted using CyTargetLinker in order to guide future research. In total, 915 DEGs were identified, including 774 upregulated and 141 downregulated genes. Enriched GO terms and pathways were determined, including ‘nervous system development’, ‘regulation of molecular function’, ‘glutamatergic synapse pathway’ and ‘pathways in cancer’. The node degrees of DEGs followed power-law distributions. A PPI network with 541 nodes and 1,431 edges was obtained. Overall, 3 important modules were identified from the PPI network. The following 10 genes were identified as core genes based on high degrees of connectivity: Albumin, PH domain leucine-rich repeat-containing protein phosphatase 2 (PHLPP2), DNA topoisomerase 2-α, kininogen 1 (KNG1), interleukin 2 (IL2), leucine-rich repeats and guanylate kinase domain containing, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit γ (PIK3CG), leptin, transferrin and RNA polymerase II subunit A (POLR2A). Additionally, 15 miRNA-target interactions were obtained using CyTargetLinker. Overall, 7 miRNAs co-regulated IL2, 3 regulated PHLPP2, 3 regulated KNG1, 1 regulated PIK3CG and 1 modulated POLR2A. These results indicate potential biomarkers in the pathogenesis of OP and therapeutic targets.
Keywords: osteoporosis, bioinformatics analysis, topological parameter, protein-protein interaction, core gene, microRNA
Introduction
Osteoporosis (OP) is a major public health issue with a gradually increasing incidence, from 14.94% prior to 2008 to 27.96% in the period between 2012 and 2015 in China (1,2). Nearly 27.5 million individuals in Europe were affected by OP in 2010 (3). Furthermore, the diagnosis of OP is difficult until the occurrence of bone fractures. Accordingly, research on the pathogenesis and molecular mechanism of OP is required in order to identify biomarkers and therapeutic targets.
Mutations in genes associated with OP have been detected. For example, cystatin A (expressed by the CSTA gene) is essential for epidermal development and maintenance (4). CSTA interacts with various genes, including one encoding a tyrosine kinase binding protein. These genes are associated with the immune regulation of osteoclasts (5). Additionally, a member of the fibroblast growth factors (FGFs) family, FGF21, is an essential endocrine hormone that regulates glucose and lipid metabolism (6). FGF21 affects bone development; it is inversely associated with regional bone mass density (BMD) (7). However, few studies have evaluated the core genes involved in OP using a bioinformatics approach. Although previous studies have identified several potential genes and proteins associated with OP, topological analyses are required in order to characterize the complex underlying networks. Furthermore, few studies have explored the microRNA (miRNA/miR)-gene regulatory networks to generate novel premises for OP research.
In the present study, the gene expression dataset GSE35956 was selected from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo) (8), and the GEO2R online analysis software (https://www.ncbi.nlm.nih.gov/geo/geo2r/) was used to uncover differentially expressed genes (DEGs). Using these loci, a protein-protein interaction (PPI) network was obtained and a network topological analysis was performed in order to identify core genes with high degrees of connectivity. In addition, the functions of the DEGs and 3 central modules were analyzed, including analyses of over-represented biological processes (BPs), molecular functions (MFs), cellular components (CCs), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (9,10). miRNAs that regulate these core genes were predicted, providing a basis for further studies.
Materials and methods
Data
The gene expression dataset GSE35956 was obtained from the publicly accessible GEO database. GSE35956 was obtained through rigorous scientific design and the data can be analyzed with high quality. The selection criteria were as follows: i) Entry type: Series; ii) Organism: Homo sapiens; and iii) Experiment type: Expression profiling by array. The dataset includes human mesenchymal stem cell (hMSC) samples from 5 middle-aged donors without any indication of the syndrome (age range, 42–67 years; mean age, 57.6 years; sex, 4 female and 1 male) and 5 patients (age range, 79–94 years; mean age, 86.2 years; sex, female) suffering from primary OP (hMSC-OP) (11). Non-osteoporotic donors with total hip arthroplasty due to osteoarthritis and/or hip dysplasia were selected and MSCs from the bone marrow of the donors were obtained. MSCs of OP donors were isolated from femoral heads following low-energy fracture of the femoral neck. Benisch et al (11) used the Significance Analysis of Microarrays software (http://statweb.stanford.edu/~tibs/SAM/) to compare gene expression patterns of 2 groups of hMSC populations. In the present study, a separate analysis of 2 groups was performed and each group contained the 5 aforementioned samples. The study was based on the GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array; Affymetrix; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The series matrix file of GSE35956 was downloaded from the GEO database.
Data processing for DEG identification
GEO2R and Morpheus (https://software.broadinstitute.org/morpheus/) were used to identify DEGs among experimental samples (12). GEO2R offers a convenient interface enabling sophisticated R-based analyses of GEO data and is useful for the identification and evaluation of DEGs (8). P<0.05 and |logFC|≥2 were set as the thresholds for DEG detection.
Gene Ontology (GO) and KEGG pathway analyses of DEGs
GO analyses are used to annotate genes or gene products and to determine biological characteristics of high-throughput genome or transcriptome data (10). KEGG is a group of databases for various biological data, including genomes and biological pathways. The Database for Annotation, Visualization and Integrated Discovery v6.8 (DAVID; http://david.ncifcrf.gov/) is a net-based online bioinformatics resource with tools for the functional interpretation of large-scale gene or protein datasets (13). The Functional Annotation Tool of DAVID was used and then the upregulated and downregulated DEGs were inserted into the tool for GO and KEGG analysis, respectively. In particular, Homo sapiens were required to be selected in order to limit the annotation of the species, thereby generating a summary of gene-species mapping. Finally, the data was downloaded and P<0.05 was used to indicate a statistically significant difference.
Construction of functional annotation maps
In order to evaluate the biological functions of the proteins in the PPI networks based on upregulated, downregulated and total DEGs, the ClueGO plug-in v2.5.0 was used to identify over-represented BP terms for protein members in the network (14). ClueGO integrates GO terms into a PPI network and creates a functional annotation map that represents the associations between terms. The κ score was set to 0.4, indicating the resemblance of GO terms for associated genes. In addition, KEGG pathways were used to combine associated genes with corresponding pathways and P<0.05 indicated a significant difference.
Network topological parameters
Network topological analyses were used for the comparison and characterization of complex networks. NetworkAnalyzer v2.7 (Department of Computational Biology and Applied Algorithmics at the Max Planck Institute for Informatics, Saarbrücken, Germany), which is part of Cytoscape v3.6.0, was used to analyze network topological parameters (15). NetworkAnalyzer only fits points with positive coordinate values and provides the association between the given data points and the corresponding points on the fitted curve. This coefficient provides the ratio of variability in a dataset, which is described by a fitted linear model. To determine whether two random variables satisfy the linear association, the correlation coefficient can be calculated with the linear regression model and the least squares method. Consequently, the R2 value is calculated on logarithmized data, where the power-law curve: y=a xb is transformed into linear model: ln y=ln a + b ln x, where × and y are positive random variables, and a and b are constants >0. The power law distribution appears as a straight line with a slope of a power exponent. This linear association is the basis for judging whether a random variable satisfies a power law in a given instance, and the power-law distribution of node degree was the main parameter used to evaluate the network topology.
PPI network and module analysis
The Search Tool for the Retrieval of Interacting Genes (STRING) v10.5 (https://string-db.org/) is an online tool designed to evaluate PPI information (16) and this was used to detect potential associations among the DEGs. The results were input into Cytoscape v3.6.0 (http://www.cytoscape.org/) to map the associations among the DEGs (17). A confidence score ≥0.4 and a maximum number of interactors of 0 were set as criteria. Molecular Complex Detection (MCODE) v1.5.1 (Bader Lab, University of Toronto, Toronto, Ontario, Canada) was used to evaluate the modules in the PPI network in Cytoscape (18). The definition of false discovery rate (FDR) is the expected value of the number of false rejects as a percentage of all rejected null hypotheses. The P-value is generally performed using a Student's t-test or χ2 test for the analysis of differential gene expression, and the false positive test can be performed on the P-value using the Benjamini FDR check method (19). In other words, the second screening of DEGs can be performed by the FDR significance parameter. The screening conditions were as follows: Degree, 2; node score, 0.2; k-core, 2; and maximum depth, 100. A KEGG pathway enrichment analysis for DEGs in the module was performed using the Functional Annotation Tool based on the DAVID for these cluster genes and annotation summary results are depicted as a Functional Annotation Chart in Fig. 1. The count threshold was set to 2, which means the minimum number of genes for the corresponding term is 2. Additionally, Homo sapiens were required to be selected to limit the annotation of the species. P<0.05 indicated a significant difference.
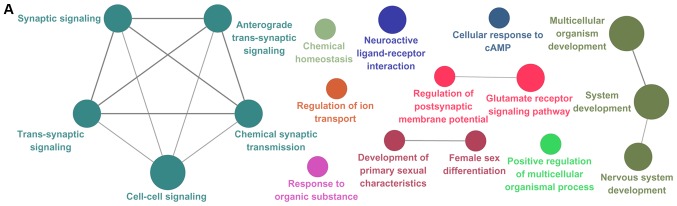
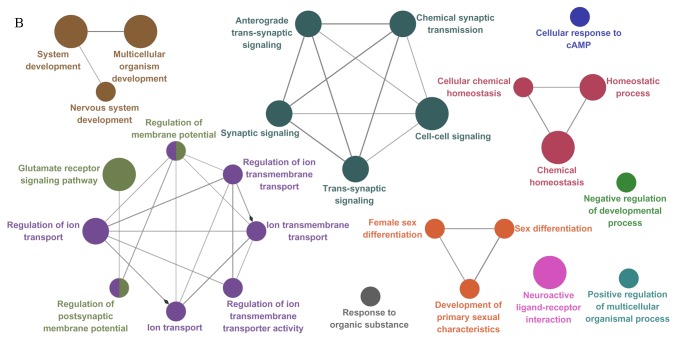
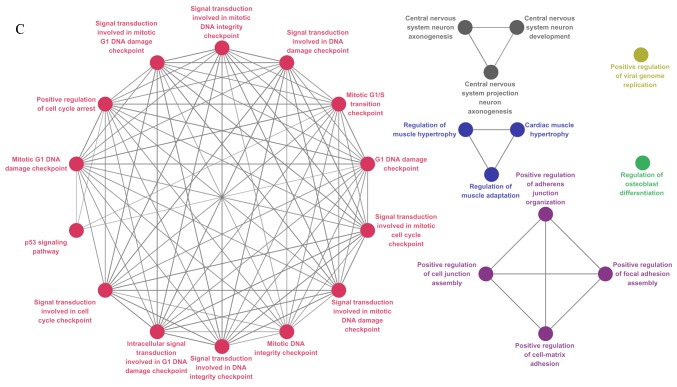
Figure 1.
Functional annotation map of PPI subnetworks for (A) all DEGs, (B) upregulated DEGs and (C) downregulated DEGs. The GO biological process terms for total, upregulated and downregulated DEGs included in the PPI subnetworks are represented in (A), (B) and (C), respectively. GO terms are presented as nodes. PPI, protein-protein interactions; DEG, differentially expressed gene; GO, Gene Ontology.
Prediction of miRNA-gene regulatory modules
CyTargetLinker v3.0.1 (https://projects.bigcat.unimaas.nl/cytargetlinker/) was utilized to predict miRNA-target interactions (MTIs) for the genes, which were presented in a graphical manner using an extension of the network (20), with the network established by the core genes can be extended to be associated with miRNAs. A regulatory interaction network (RegIN) is a network containing regulatory interactions, often derived from online interaction databases (20). To generate a RegIN using CyTargetLinker in Cytoscape, Homo sapiens MTIs were obtained from the experimentally validated database miRTarBase v4.4, which includes 20,942 MTIs, and from 2 predicted miRNA databases, MicroCosm v5.0, with 541,039 MTIs, and TargetScan v6.2, with 511,040 MTIs (http://projects.bigcat.unimaas.nl/cytargetlinker/regins). Each regulatory interaction comprises 2 nodes, a source (regulatory component) and a target biomolecule, connected by a single directed edge. In the present study, the top 10 hub genes were selected for the extension of the network using CyTargetLinker.
Results
Identification of DEGs
The gene expression profiles of 10 samples in the gene expression dataset GSE35956, including 5 OP and 5 control samples, were analyzed. A total of 915 DEGs between the OP and control samples were identified using GEO2R and Morpheus, including 774 upregulated and 141 downregulated genes.
GO function and KEGG pathway enrichment analysis
GO and KEGG pathway enrichment analyses were performed using DAVID and the ClueGO plug-in in order to gain a comprehensive understanding of the functions of the DEGs. The results of the GO analysis indicated that upregulated and downregulated genes were enriched for various BP terms, which are listed in Table I. For upregulated DEGs these included ‘nervous system development’, ‘homeostatic process’, ‘cell-cell signaling’, ‘chemical homeostasis’ and ‘cellular chemical homeostasis’, and for downregulated DEGs they included ‘regulation of MF’, ‘single-organism organelle organization’, ‘cytoskeleton organization’, ‘regulation of GTPase activity’ and ‘actin cytoskeleton organization’. In the MF category, the upregulated genes were enriched for ‘RNA polymerase II transcription factor activity, sequence-specific DNA binding’, ‘channel activity’, ‘passive transmembrane transporter activity’, ‘ligand-gated ion channel activity’ and ‘ligand-gated channel activity’, while the downregulated genes were enriched for ‘nucleotide binding’, ‘nucleoside phosphate binding’, ‘purine ribonucleoside triphosphate binding’, ‘purine ribonucleoside binding’ and ‘extracellular matrix structural constituent’. A CC analysis further demonstrated that the upregulated genes were enriched for ‘synapse’, ‘synapse part’, ‘synaptic membrane’, ‘postsynaptic membrane’ and ‘excitatory synapse’, while the downregulated genes were enriched for ‘extracellular matrix’, ‘perinuclear region of cytoplasm’, ‘proteinaceous extracellular matrix’, ‘cell leading edge’ and ‘ruffle’.
Table I.
Gene Ontology analysis of differentially expressed genes associated with osteoporosisa.
| Expression | Category | Term | Countb | %c | P-value |
|---|---|---|---|---|---|
| Upregulated | GOTERM_BP_FAT | GO:0007399~nervous system development | 106 | 13.73 | 4.20×10−6 |
| GOTERM_BP_FAT | GO:0042592~homeostatic process | 88 | 11.40 | 1.06×10−6 | |
| GOTERM_BP_FAT | GO:0007267~cell-cell signaling | 85 | 11.01 | 2.52×10−7 | |
| GOTERM_BP_FAT | GO:0048878~chemical homeostasis | 66 | 8.55 | 1.42×10−7 | |
| GOTERM_BP_FAT | GO:0055082~cellular chemical homeostasis | 47 | 6.09 | 3.77×10−6 | |
| GOTERM_MF_FAT | GO:0000981~RNA polymerase II transcription factor activity, sequence-specific DNA binding | 43 | 5.57 | 1.96×10−5 | |
| GOTERM_MF_FAT | GO:0015267~channel activity | 31 | 4.02 | 3.65×10−4 | |
| GOTERM_MF_FAT | GO:0022803~passive transmembrane transporter activity | 31 | 4.02 | 3.80×10−4 | |
| GOTERM_MF_FAT | GO:0015276~ligand-gated ion channel activity | 16 | 2.07 | 6.49×10−5 | |
| GOTERM_MF_FAT | GO:0022834~ligand-gated channel activity | 16 | 2.07 | 6.49×10−5 | |
| GOTERM_CC_FAT | GO:0045202~synapse | 51 | 6.61 | 7.19×10−7 | |
| GOTERM_CC_FAT | GO:0044456~synapse part | 42 | 5.44 | 4.81×10−6 | |
| GOTERM_CC_FAT | GO:0097060~synaptic membrane | 29 | 3.76 | 6.53×10−8 | |
| GOTERM_CC_FAT | GO:0045211~postsynaptic membrane | 22 | 2.85 | 4.28×10−6 | |
| GOTERM_CC_FAT | GO:0060076~excitatory synapse | 20 | 2.59 | 1.79×10−5 | |
| Downregulated | GOTERM_BP_FAT | GO:0065009~regulation of molecular function | 34 | 24.29 | 0.001 |
| GOTERM_BP_FAT | GO:1902589~single-organism organelle organization | 23 | 16.43 | 0.001 | |
| GOTERM_BP_FAT | GO:0007010~cytoskeleton organization | 18 | 12.86 | 0.002 | |
| GOTERM_BP_FAT | GO:0043087~regulation of GTPase activity | 14 | 10.00 | 9.63×10−4 | |
| GOTERM_BP_FAT | GO:0030036~actin cytoskeleton organization | 12 | 8.57 | 0.002 | |
| GOTERM_MF_FAT | GO:0000166~nucleotide binding | 29 | 20.71 | 0.003 | |
| GOTERM_MF_FAT | GO:1901265~nucleoside phosphate binding | 29 | 20.71 | 0.003 | |
| GOTERM_MF_FAT | GO:0035639~purine ribonucleoside triphosphate binding | 24 | 17.14 | 0.004 | |
| GOTERM_MF_FAT | GO:0032550~purine ribonucleoside binding | 24 | 17.14 | 0.004 | |
| GOTERM_MF_FAT | GO:0005201~extracellular matrix structural constituent | 5 | 3.57 | 0.003 | |
| GOTERM_CC_FAT | GO:0031012~extracellular matrix | 10 | 7.14 | 0.015 | |
| GOTERM_CC_FAT | GO:0048471~perinuclear region of cytoplasm | 10 | 7.14 | 0.044 | |
| GOTERM_CC_FAT | GO:0005578~proteinaceous extracellular matrix | 8 | 5.71 | 0.017 | |
| GOTERM_CC_FAT | GO:0031252~cell leading edge | 7 | 5.00 | 0.046 | |
| GOTERM_CC_FAT | GO:0001726~ruffle | 6 | 4.29 | 0.006 |
The top 5 terms were selected according to P-value.
Count is the number of enriched genes for each term. GO, Gene Ontology; BP, biological process; MF, molecular function; CC, cellular component.
Represents the percentage of genes contained in each term to the upregulated or downregulated genes.
In addition, 6 KEGG pathways were identified, as listed in Table II, involving the ‘glutamatergic synapse’, ‘adrenergic signaling in cardiomyocytes’, ‘neuroactive ligand-receptor interaction’, ‘Rap1 signaling pathway’ and ‘cAMP signaling pathway’ for upregulated DEGs, and ‘pathways in cancer’ for downregulated DEGs.
Table II.
Kyoto Encyclopedia of Genes and Genomes pathway analysis of DEGs associated with osteoporosisa.
| Category of DEGs | Term | Countb | %c | P-value | Genes |
|---|---|---|---|---|---|
| Upregulated | hsa04080:Neuroactive ligand-receptor interaction | 28 | 3.63 | 2.34×10−7 | AVPR2, MCHR1, GRIK2, NPY2R, GRIK3, GABRB1, F2RL1, GRIK5, HCRTR1, LTB4R, CNR1, P2RY1, CHRNA4, GRID1, GABRG3, GABRA2, PTGER3, NPBWR2, LEP, P2RY10, ADRB1, GRM2, PRLR, GRM8, CHRM1, P2RX2, GHSR, TSHR |
| hsa04015:Rap1 signaling pathway | 19 | 2.46 | 1.45×10−4 | PIK3CG, ADCY1, MAGI2, PGF, EFNA2, SIPA1L2, KITLG, FGF23, RGS14, PRKCB, CNR1, P2RY1, RASGRP2, CALML6, RAPGEF3, ANGPT2, RAPGEF1, AKT2, ITGA2B | |
| hsa04024:cAMP signaling pathway | 16 | 2.07 | 0.002 | PIK3CG, HCN2, ADCY1, PTGER3, CFTR, CNGB1, CNGB3, ADRB1, CHRM1, CAMK2D, CALML6, RAPGEF3, GHSR, CAMK2A, TSHR, AKT2 | |
| hsa04724:Glutamatergic synapse | 11 | 1.42 | 0.004 | SLC17A8, SLC1A2, ADCY1, DLGAP1, GRM2, GRM8, GRIK2, GRIK3, GRIK5, SHANK3, PRKCB | |
| hsa04261:Adrenergic signaling in cardiomyocytes | 11 | 1.42 | 0.020 | PIK3CG, ADCY1, ADRB1, CACNG8, PPP1R1A, CACNB1, CAMK2D, CALML6, RAPGEF3, CAMK2A, AKT2 | |
| Downregulated | hsa05200:Pathways in cancer | 7 | 55.00 | 0.030 | FGFR2, LAMA4, WNT4, HSP90B1, APC2, PIAS2, CDK2 |
The top 5 terms were selected for upregulated DEGs and one term was selected for downregulated DEGs according to P-values.
Count is the number of enriched genes for each term. DEGs, differentially expressed genes.
Represents the percentage of genes contained in each term to the upregulated or downregulated genes.
The results of the ClueGO analysis demonstrated that upregulated and downregulated DEGs were particularly enriched for BP terms, including ‘multicellular organism development’, ‘system development’, ‘response to organic substance’, ‘nervous system development’ and ‘cell-cell signaling’ for upregulated DEGs, and ‘regulation of osteoblast differentiation’, ‘mitotic DNA integrity checkpoint’, ‘central nervous system neuron development’, ‘mitotic G1/S transition checkpoint’ and ‘mitotic G1 DNA damage checkpoint’ for downregulated DEGs (Table III). KEGG pathways involved in ‘neuroactive ligand-receptor interaction’ and the ‘p53 signaling pathway’ for upregulated and downregulated DEGs, respectively, were identified.
Table III.
Summary of ClueGO analysis of DEGs for biological processes associated with osteoporosisa.
| DEG expression | GO ID | GO term | Genes, nb | %c | P-value | Genes |
|---|---|---|---|---|---|---|
| Upregulated | GO:0007275 | Multicellular organism | 220 | 4.14 | 5.03×10−7 | ADAMTS9, ADCY1, AKT2, ALG5, AMOT, ANGPT2, ANKRD54, ANPEP, APLP1, APOH, AQP1, ARHGAP26, ASB4, ASCL2, ASNS, ATCAY, ATN1, AVPR2, AXIN2, BCAN, BMP7, BMP8B, BRSK1, BRSK2, CAMK2A, CAMK2D, CBX2, CCK, CCR4, CD24, CD74, CHRM1, CNGB1, CNR1, CNTN4, COL10A1, CPB2, CUX2, CYP19A1, DACH1, DCHS2, DDC, DMBX1, DNAH11, DRAXIN, DYRK1B, EFNA2, EGR2, EMX1, EN2, EPHA4, ESRRB, EZH1, F2RL1, FEZF2, FGF23, FOXE1, FOXG1, FOXO1, FOXP2, FZD10, GABRB1, GAL, GHSR, GJA5, GJB6, GPM6B, GPRC5B, HDAC10, HES4, HID1, HIPK1, HMGA2, HNF4A, HOOK1, HPRT1, HSPB6, HTN3, IBSP, IKZF1, IKZF3, IL2, IRF4, ITGA2B, JMJD6, JPH1, KCNIP2, KITLG, KLHL17, KLHL32, KLKB1, KMT2B, KRT40, LEP, LFNG, LHX2, LHX6, LHX8, LPAL2, LRRC7, LSAMP, LST1, LTBP4, LYL1, MAB21L2, MAGI2, MARK4, MCF2, MEF2D, MEX3C, MINK1, MMP11, MOBP, MOV10L1, MT1G, MYCBPAP, MYH7B, MYO3B, MYO7A, NDRG2, NEUROG2, NFATC4, NME8, NOBOX, NPAS1, NPPC, NPY2R, NR0B1, NR2F6, NRP2, NRXN1, NTRK3, OBSL1, OLFM1, P2RX2, P2RY1, PAK3, PCDH15, PCDH8, PCDHGC5, PCSK6, PCYT1B, PECAM1, PGF, PHOSPHO1, PIK3CG, PKP2, PLCG2, POLB, PPARGC1A, PPARGC1B, PPDPF, PRKCB, PRLR, PSG1, PTPRB, RAB11FIP4, RAD21L1, RAPGEF1, RAPGEF3, RARRES2, RAX, RFX4, RGS14, RIMS1, RIPPLY2, SCEL, SCN3B, SCRT1, SDK2, SEMA4D, SEMA6B, SERPINA5, SETDB2, SEZ6L, SF1, SFTPD, SH3TC2, SHANK3, SHOX, SIAH3, SIN3A, SLC17A8, SLC1A2, SLC8A3, SMIM6, SOST, SOX10, SOX2, SOX8, SPATA19, ST8SIA2, STMN4, STOX1, TAF10, TBX3, TBX6, TCF21, TEF, TERT, TEX11, THPO, TIE1, TNFRSF11A, TP63, TRIB1, TRIM54, TRPC6, TRPM1, TSHR, TSSK1B, TTPA, TUB, UBE4B, WDR72, WT1, XIRP2, ZFP36L1, ZFP36L2, ZFP42 |
| GO:0048731 | System development | 203 | 4.28 | 1.94×10−7 | ADAMTS9, ADCY1, AKT2, AMOT, ANGPT2, ANKRD54, ANPEP, APLP1, APOH, AQP1, ARHGAP26, ASB4, ASCL2, ASNS, ATCAY, ATN1, AVPR2, AXIN2, BCAN, BMP7, BMP8B, BRSK1, BRSK2, CAMK2A, CAMK2D, CCK, CCR4, CD24, CD74, CHRM1, CNGB1, CNR1, CNTN4, COL10A1, CPB2, CUX2, CYP19A1, DCHS2, DMBX1, DNAH11, DRAXIN, DYRK1B, EFNA2, EGR2, EMX1, EN2, EPHA4, ESRRB, EZH1, F2RL1, FEZF2, FGF23, FOXE1, FOXG1, FOXO1, FOXP2, FZD10, GABRB1, GAL, GHSR, GJA5, GJB6, GPM6B, GPRC5B, HDAC10, HES4, HID1, HIPK1, HMGA2, HPRT1, HSPB6, HTN3, IBSP, IKZF1, IKZF3, IL2, IRF4, ITGA2B, JMJD6, JPH1, KCNIP2, KITLG, KLHL17, KLHL32, KLKB1, KMT2B, KRT40, LEP, LFNG, LHX2, LHX6, LHX8, LPAL2, LRRC7, LSAMP, LST1, LYL1, MAB21L2, MAGI2, MARK4, MCF2, MEF2D, MEX3C, MINK1, MOBP, MT1G, MYH7B, MYO3B, MYO7A, NDRG2, NEUROG2, NFATC4, NOBOX, NPAS1, NPPC, NPY2R, NR0B1, NR2F6, NRP2, NRXN1, NTRK3, OBSL1, OLFM1, P2RX2, P2RY1, PAK3, PCDH15, PCDH8, PCDHGC5, PCSK6, PCYT1B, PECAM1, PGF, PHOSPHO1, PIK3CG, PKP2, PLCG2, POLB, PPARGC1A, PPARGC1B, PPDPF, PRKCB, PRLR, PSG1, PTPRB, RAB11FIP4, RAD21L1, RAPGEF1, RAPGEF3, RARRES2, RAX, RFX4, RGS14, RIMS1, RIPPLY2, SCEL, SCN3B, SCRT1, SDK2, SEMA4D, SEMA6B, SERPINA5, SETDB2, SEZ6L, SF1, SFTPD, SH3TC2, SHANK3, SHOX, SIN3A, SLC17A8, SLC1A2, SLC8A3, SMIM6, SOX10, SOX2, SOX8, ST8SIA2, STMN4, STOX1, TAF10, TBX3, TBX6, TCF21, TERT, TEX11, THPO, TIE1, TNFRSF11A, TP63, TRIB1, TRPC6, TRPM1, TSHR, TTPA, TUB, UBE4B, WDR72, WT1, XIRP2, ZFP36L1, ZFP36L2, ZFP42 | |
| GO:0010033 | Response to organic substance | 138 | 4.34 | <0.001 | ABCA2, ABCC2, ADCY1, AIM2, AKT2, ANGPT2, APLP1, AQP1, AQP4, ASNS, ATP6V0A4, AVPR2, BMP7, BMP8B, BRSK2, CACNB1, CAMK2A, CAMK2D, CCDC3, CCL23, CCR4, CD24, CD6, CD74, CFTR, CGN, CHRM1, CHRNA4, CNR1, CPB2, CRLF2, CSF2RA, CUX2, CXCL5, DBH, DDC, EGR2, ELANE, EPHA4, ESRRB, F2RL1, FABP4, FGF23, FOXO1, FOXP2, FZD10, GABRB1, GAL, GBP5, GHSR, GJB6, GPD1, GRIK5, HCN2, HCRTR1, HFE2, HID1, HIPK1, HNF4A, HPRT1, IBSP, IL2, IL22RA2, IL37, IRF4, ITIH4, KANK2, KHSRP, LATS2, LEP, LMO3, LPAL2, LRRC19, LRRC3, LTBP4, MAGI2, MAP2K7, MAPK4, MCF2, MEFV, MT1G, MTHFR, MTSS1L, NFATC4, NPPC, NR0B1, NR2F6, NRP2, NRXN1, NTRK3, P2RX2, P2RY1, PAK3, PAQR9, PCSK6, PELO, PGF, PID1, PIK3CG, PLCG2, POLB, POLR2A, PPARGC1A, PPARGC1B, PRKCB, PRLR, PSG1, RAPGEF1, RAPGEF3, RARRES2, RERG, RIPOR1, RNASE7, RNF31, RXRB, SIN3A, SLC11A1, SLC1A2, SLC8A3, SORBS1, SOST, SOX10, SOX2, SOX30, TAT, TCF21, TERT, THPO, TIE1, TNFRSF11A, TP63, TRIB1, TSHR, TUB, UBE4B, WT1, ZFP36L1, ZFP36L2 | |
| GO:0007399 | Nervous system development | 109 | 4.61 | <0.001 | ADCY1, AKT2, APLP1, AQP1, ARHGAP26, ASCL2, ATCAY, ATN1, AVPR2, BCAN, BMP7, BRSK1, BRSK2, CAMK2A, CAMK2D, CCK, CCR4, CHRM1, CNGB1, CNR1, CNTN4, CUX2, DMBX1, DRAXIN, EFNA2, EGR2, EMX1, EN2, EPHA4, EZH1, FEZF2, FOXG1, FOXP2, FZD10, GABRB1, GAL, GHSR, GPM6B, GPRC5B, HDAC10, HES4, HIPK1, HPRT1, IL2, KCNIP2, KLHL17, LEP, LHX2, LHX6, LHX8, LRRC7, LSAMP, LST1, MAB21L2, MAGI2, MARK4, MCF2, MEF2D, MINK1, MOBP, MYO7A, NDRG2, NEUROG2, NFATC4, NPAS1, NPPC, NR0B1, NR2F6, NRP2, NRXN1, NTRK3, OBSL1, OLFM1, P2RY1, PAK3, PCDH15, PCDH8, PCDHGC5, PPARGC1A, RAPGEF1, RAX, RFX4, RGS14, RIMS1, SCN3B, SCRT1, SDK2, SEMA4D, SEMA6B, SEZ6L, SH3TC2, SHANK3, SLC17A8, SLC1A2, SLC8A3, SOX10, SOX2, SOX8, ST8SIA2, STMN4, TBX3, TBX6, TERT, TP63, TRPC6, TRPM1, TSHR, UBE4B, ZFP36L1 | |
| GO:0007267 | Cell-cell signaling | 94 | 5.64 | 1.18× 10−8 | ADRB1, AMER3, ANPEP, AQP1, AXIN2, BRSK1, BRSK2, CACNB1, CADPS, CAMK2A, CCL23, CD24, CFTR, CHRM1, CHRNA4, CNR1, CNTN4, CUX2, CXCL5, DBH, DLGAP1, DRAXIN, EFNA2, EGR2, EPHA4, FGF23, FOXO1, FZD10, GAL, GHSR, GJA5, GPRC5B, GRID1, GRIK2, GRIK3, GRIK5, GRM2, GRM8, HCN2, HCRTR1, HMGA2, HNF4A, IL2, JPH3, KCNIP2, LATS2, LEP, LTBP4, MAGI2, MCHR1, MINK1, MYCBPAP, NDRG2, NETO1, NFATC4, NPBWR2, NPPC, NPTX2, NPY2R, NRXN1, P2RX2, P2RY1, PANX2, PCDH15, PCDH8, PCDHGC5, PGF, PKP2, PLCG2, RAPGEF1, RAPGEF3, RASL10B, RGS14, RIMS1, SCEL, SCN3B, SHANK3, SHISA9, SIRPG, SLC1A2, SLC5A7, SLC8A3, SORCS3, SOST, SOX10, SOX2, STXBP5L, SYNPO, TBX3, TERT, TNFRSF11A, TP63, TSHR, UNC13C | |
| Downregulated | GO:0045667 | Regulation of osteoblast differentiation | 5 | 4.42 | 0.001 | FGFR2, IL6ST, PIAS2, PRKD1, WNT4 |
| GO:0044774 | Mitotic DNA integrity checkpoint | 5 | 4.31 | 0.001 | CDK2, CNOT7, PLK3, TOP2A, TP73 | |
| GO:0021954 | Central nervous system neuron development | 4 | 5.41 | 0.001 | ADARB1, FGFR2, NFIB, SZT2 | |
| GO:0044819 | Mitotic G1/S transition checkpoint | 4 | 5.19 | 0.002 | CDK2, CNOT7, PLK3, TP73 | |
| GO:0031571 | Mitotic G1 DNA damage checkpoint | 4 | 5.19 | 0.002 | CDK2, CNOT7, PLK3, TP73 |
The top 5 terms were selected according to P-value.
Number of enriched genes for each term. GO, Gene Ontology; DEGs, differentially expressed genes; BP, biological process.
Represents the percentage of genes contained in each term to the upregulated or downregulated genes.
Functional annotation map of PPI subnetworks
ClueGO provided a functional annotation map for PPI subnetworks, in which protein members are enriched corresponding to their GO terms and pathways (Fig. 1). Regulation of osteoblast differentiation was linked to 5 genes. Among these, FGF receptor 2, interleukin 6 signal transducer and Wnt family member 4 genes were also associated with a number of BP terms, including ‘central nervous system projection neuron axonogenesis’, ‘cardiac muscle hypertrophy’, ‘positive regulation of focal adhesion assembly biological process’ and ‘pathways in cancer’.
Analyses of network topological properties
The node degrees for the total upregulated and downregulated DEGs generally obeyed power-law distributions, as demonstrated in Fig. 2. Supplementary topological parameters, including the clustering coefficient, network centralization and network density, are listed in Table IV.
Figure 2.
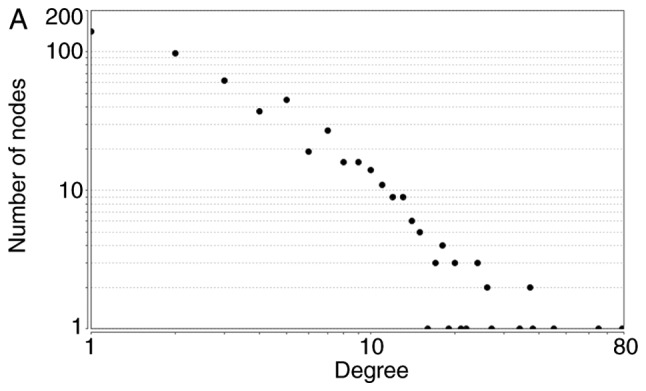
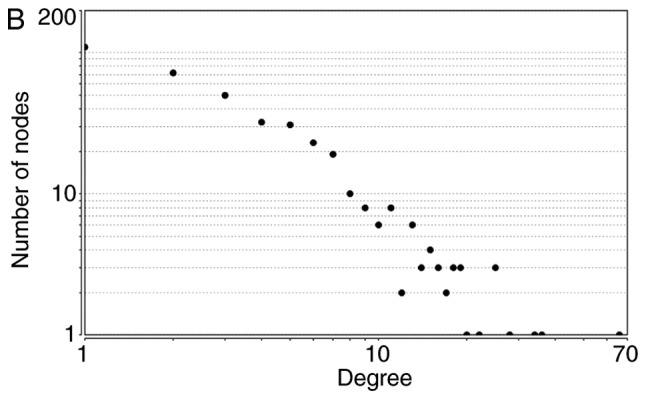
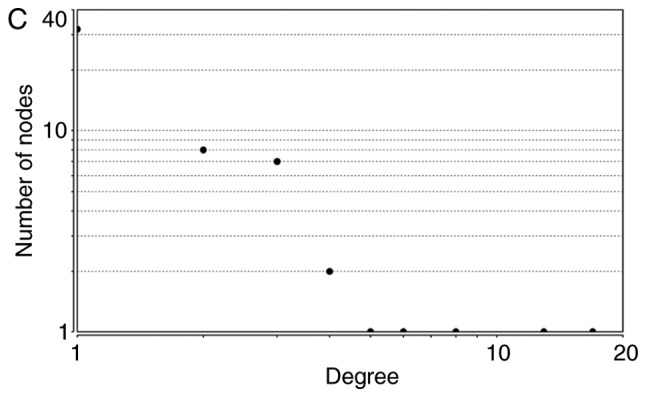
Power-law distributions of node degrees for (A) the total, (B) the upregulated and (C) the downregulated PPI subnetworks. Degree is a parameter used to describe the number of interacting partner proteins. As the number of linked nodes increases, the average degree decreases. PPI, protein-protein interactions.
Table IV.
Topological parameters for the total, the upregulated and the downregulated PPI subnetworks.
| PPI subnetwork | y=axb | R2 | Correlation coefficient | Clustering coefficient | Network centralization | Network density |
|---|---|---|---|---|---|---|
| Total | y=256.19x−1.478 | 0.863 | 0.931 | 0.249 | 0.137 | 0.010 |
| Upregulated | y=191.96x−1.462 | 0.901 | 0.939 | 0.256 | 0.153 | 0.012 |
| Downregulated | y=18.646x−1.291 | 0.809 | 0.984 | 0.142 | 0.287 | 0.044 |
R2 is a measure of the fit quality, with a range of 0–1 (a higher value indicates a better fit). The correlation between observations and the corresponding points on the fitted curve was determined. Clustering coefficient denotes an average of the clustering coefficients of all nodes in the network. Network centralization is a connectivity distribution index. Network density is the normalized value of the average number of neighbors. PPI, protein-protein interaction.
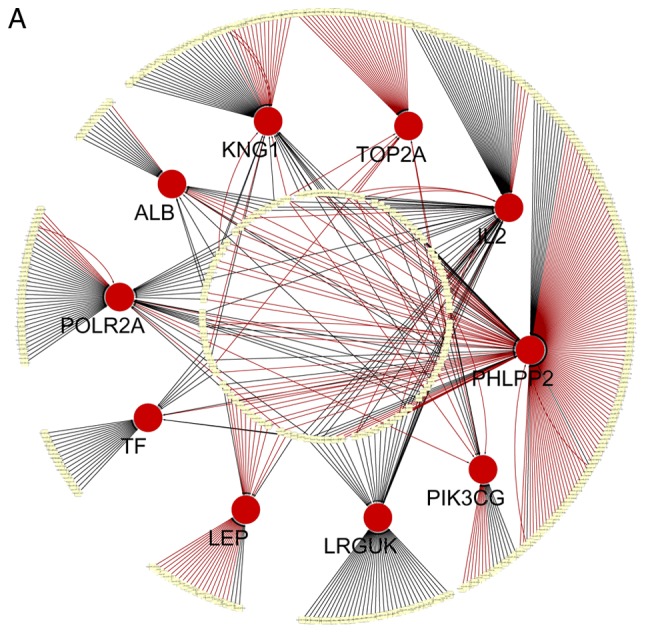
Core genes and modules in the PPI network
The top 10 core genes with the highest node degrees were determined using Cytoscape and the STRING database. These were albumin (ALB), PH domain leucine-rich repeat-containing protein phosphatase 2 (PHLPP2), DNA topoisomerase 2-α (TOP2A), kininogen 1 (KNG1), interleukin 2 (IL2), leucine-rich repeats and guanylate kinase domain containing (LRGUK), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit γ (PIK3CG), leptin (LEP), transferrin (TF) and RNA polymerase II subunit A (POLR2A). Using an MCODE analysis, several significant modules were identified, including 541 nodes and 1431 edges. The 3 modules with the lowest P-values were selected to evaluate enriched pathways (Fig. 3 and Table V).
Figure 3.
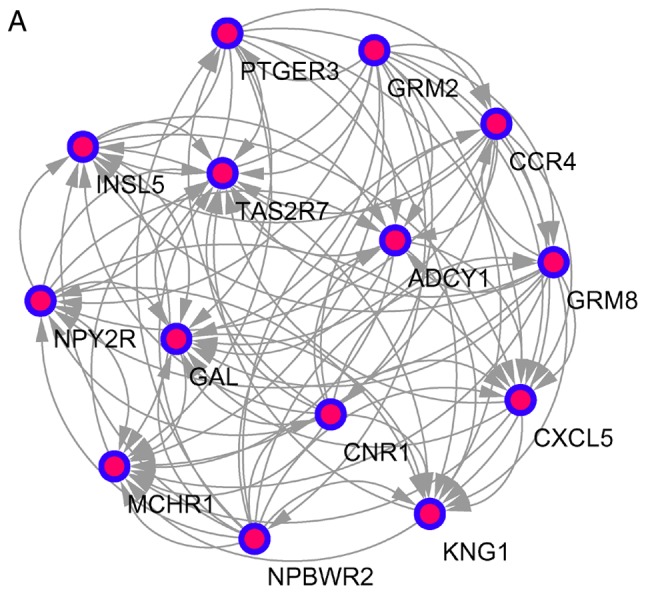
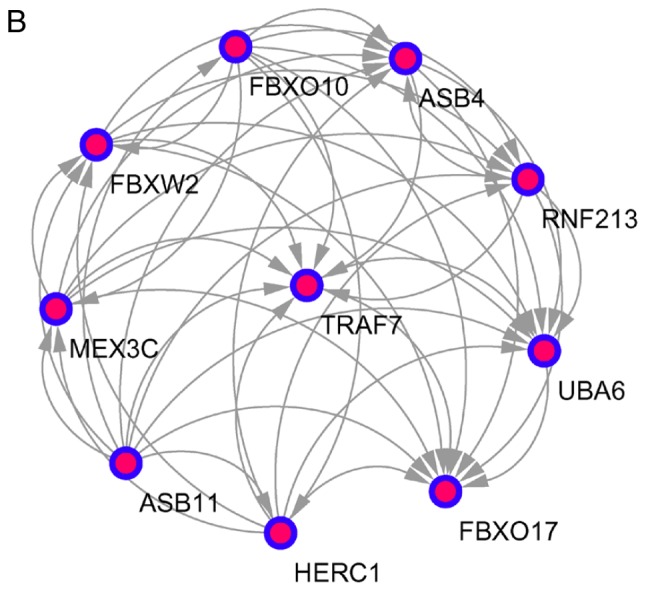
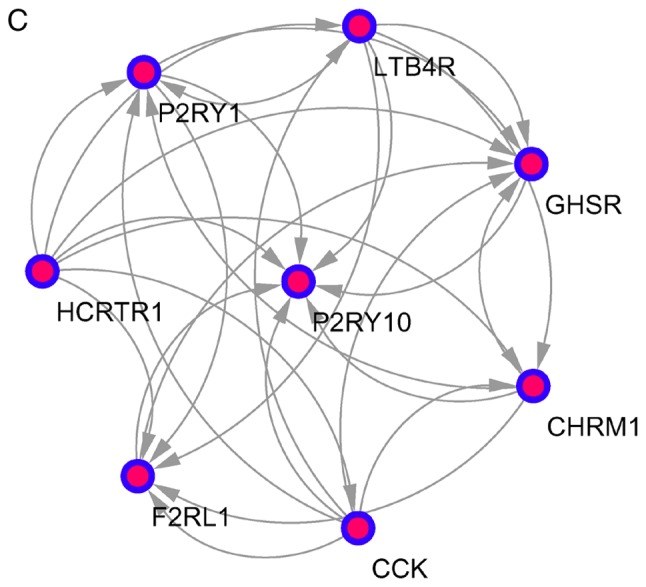
Top 3 modules from the protein-protein interactions network. (A) Module A, 14 nodes and 91 edges; (B) module B, 10 nodes and 45 edges; and (C) module C, 8 nodes and 28 edges.
Table V.
Enriched pathways for modules A-Ca.
| Modules | Terms | P-value | FDR | Genes |
|---|---|---|---|---|
| A | Neuroactive ligand-receptor interaction | 1.53×10−6 | 0.001 | MCHR1, PTGER3, GRM2, GRM8, CNR1, NPY2R, NPBWR2 |
| B | Ubiquitin mediated proteolysis | 0.021 | 2.096 | UBA6, HERC1 |
| C | Neuroactive ligand-receptor interaction | 3.94×10−9 | 2.36×10−6 | HCRTR1, P2RY10, LTB4R, CHRM1, P2RY1, F2RL1, GHSR |
One term was selected for each module according to P-value. FDR, false discovery rate.
Identification of miRNA-gene regulatory modules
CyTargetLinker was used to predict miRNA-gene interactions for the 10 core genes. In total, 305 predicted MTIs were identified using the MicroCosm database and 277 predicted MTIs were identified using the TargetScan database, including 456 nodes and 582 edges. Additionally, setting a threshold functionality of 2, 15 MTIs were found from the MicroCosm and TargetScan databases. The genes and miRNAs are shown in Fig. 4. Specifically, 7 miRNAs that co-regulate IL2, 3 miRNAs that regulate PHLPP2, 3 miRNAs that regulate KNG1, 1 miRNA that regulates PIK3CG and 1 miRNA that modulates POLR2A were detected (Table VI).
Figure 4.
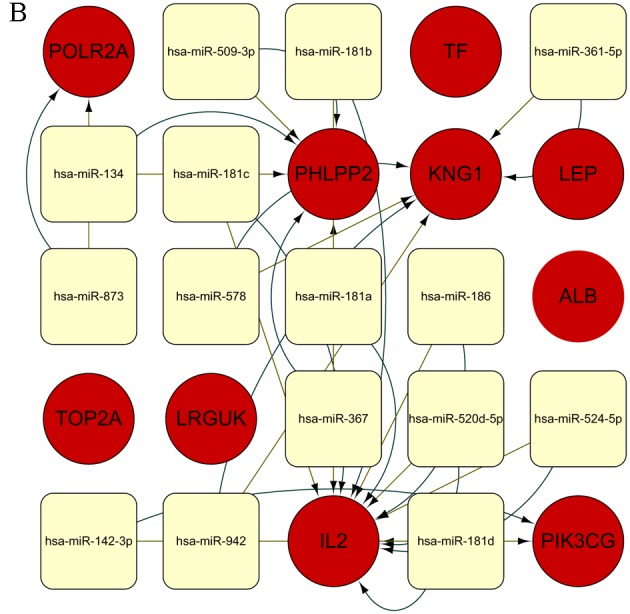
miRNA regulation of genes. (A) Extended regulatory network of miRNAs and target genes. The MTIs obtained from the MicroCosm database (n=305) and TargetScan database (n=277) are indicated in black and red, respectively. The overlap threshold function was set to display only MTIs present in 2 regulatory interaction networks. (B) Defined number of predicted miRNAs and target genes. Genes and miRNAs are denoted as red circles and yellow rounded rectangles, respectively. Additionally, green edges represent MTIs from the database TargetScan v6.2, and yellow edges represent MTIs from the database MicroCosm v5.0. The names of the genes and miRNAs are displayed on the nodes. miRNA, microRNA; MTI, microRNA-target interaction.
Table VI.
A list of 5 hub genes and their predicted miRNAs from the CyTargetLinker extension network analysisa.
| Gene | miRNAs |
|---|---|
| IL2 | hsa-miR-524-5p, hsa-miR-520d-5p, hsa-miR-181b, hsa-miR-181a, hsa-miR-181d, hsa-miR-181c, hsa-miR-186 |
| PHLPP2 | hsa-miR-509-3p, hsa-miR-134, hsa-miR-367 |
| KNG1 | hsa-miR-361-5p, hsa-miR-578, hsa-miR-942 |
| PIK3CG | hsa-miR-142-3p |
| POLR2A | hsa-miR-873 |
These were selected among 10 hub genes with a threshold of 2 regulatory interaction networks. The miRNA-target interactions were obtained from the MicroCosm database (305) and TargetScan database (277). miRNA/miR, microRNA; IL2, interleukin 2; PHLPP2, PH domain leucine-rich repeat-containing protein phosphatase 2; KNG1, kininogen 1; PIK3CG, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit γ; POLR2A, RNA polymerase II subunit A.
Discussion
OP has become a pervasive public health problem; it is characterized by decreased bone strength and an increased risk of fractures (21). The pathogenesis of OP involves an imbalance between osteoclast resorption and osteoblast bone formation. MSCs have multiple differentiation potential, and they can directly participate in bone formation by phenotypic differentiation into osteoblasts and indirectly via paracrine effects. During the process of aging, MSCs in bone marrow transform into other types of cells, including adipocytes and osteoblasts, leading to OP (22). Owing to the high prevalence and gravity of OP, understanding its pathogenic and molecular mechanisms is urgent for drug development and treatment. Microarray and high-throughput sequencing approaches have been used extensively to predict therapeutic targets for diseases, including OP. In the present study, the gene expression dataset GSE35956 was obtained from the GEO database to explore core genes and predict regulatory miRNAs involved in OP. Specifically, 5 hMSC specimens from normal osseous tissues and 5 hMSC specimens from OP samples were included and 915 DEGs were identified, including 774 upregulated and 141 downregulated genes.
Subsequently, network topological properties were analyzed to distinguish between the predicted network and a random network. A node degree approximating a power-law distribution is a standard characteristic of scale-free networks, and the PPI network in this study obeyed this rule (23,24). The 3 subnetworks were true complex biological networks characterized as scale-free, suggesting that the PPI network is reliable and robust.
OP-associated DEGs identified in the present study are not necessarily biologically meaningful. A number of effective methods, including GO and KEGG analyses, can be used to determine the importance of DEGs, with the goal of clarifying the roles of individual molecules in BPs. Accordingly, GO and KEGG pathway analyses were performed using the DEGs in the present study. Upregulated DEGs were primarily associated with ‘nervous system development’, ‘multicellular organism development’, ‘RNA polymerase II transcription factor activity’, ‘sequence-specific DNA binding and synapse’, while downregulated DEGs were associated with the ‘regulation of molecular function’, ‘regulation of osteoblast differentiation’, ‘nucleotide binding’ and ‘extracellular matrix’. Based on the KEGG pathway analysis, upregulated DEGs were enriched for ‘neuroactive ligand-receptor interaction’, ‘Rap1 signaling pathway’, ‘cAMP signaling pathway’, ‘glutamatergic synapse’ and ‘adrenergic signaling in cardiomyocytes’, while the downregulated DEGs were involved in the ‘p53 signaling pathway’.
Based on an analysis of the PPI network, the following core genes were identified: ALB, PHLPP2, TOP2A, KNG1, IL2, LRGUK, PIK3CG, LEP, TF and POLR2A. Among these, ALB had the highest degree of connectivity. These hub genes, particularly ALB, TOP2A, KNG1, IL2, PIK3CG, LEP and TF, were enriched for homeostatic processes and the regulation of apoptotic processes. A recent study has reported that homeostatic processes are imbalanced in postmenopausal osteoporotic women (25). In addition, triggering apoptotic processes could promote osteoclast apoptosis, thus alleviating OP (26). It could be hypothesized that the hub genes are associated with the development of OP. ALB and its product, which has a good binding capacity for hormones, is involved in various BPs, including antioxidant activity, the regulation of apoptotic processes and the cellular response to starvation. The precise function of ALB and its role in OP are unclear. However, a previous study found that the expression of ALB differed significantly between a normal group and an OP group (P<0.05), and others have linked oxidative status with bone alterations (27–29). PHLPP2 is involved in the regulation of protein kinase B (Akt) and protein kinase C signaling, and mediates dephosphorylation in the C-terminal domain hydrophobic motif of members of the AGC serine/threonine protein kinase family. The protein is also involved in signal transduction and hippocampus development. PHLPP2 exhibits nuclear localization and is expressed in numerous tissue types, including the small intestine, brain, bone marrow and ovaries (30). PHLPP inhibitors may relieve mechanical pain and slow cartilage degradation in osteoarthritic joints (31). Therefore, PHLPP2 has a potential regulatory role in OP and is a target for future research. TOP2A, which encodes a DNA topoisomerase, is associated with various processes, including chromosome condensation (32). Mutations in TOP2A are associated with the development of drug resistance, including etoposide and doxorubicin (33–35). Exposure to atmospheric oxygen promotes TOP2A expression in mouse MSCs, leading to oxidative stress, reduced cell viability and the inhibition of cell proliferation (36). KNG1 exhibits alternative splicing to produce 2 distinct proteins: High- and low-molecular-weight kininogen. KNG1 also factors in the G-protein-coupled receptor signaling pathway and the inflammatory response. Zhang et al (37) revealed that KNG1 is significantly associated with the complement and coagulation cascade pathway in Kashin-Beck disease. Further research is required to clarify the role of KNG1 in OP. IL2 proteins are involved in cytokine secretion and are vital for the proliferation of T and B lymphocytes. Targeted knock-outs of similar genes in mice cause ulcerative diseases, suggesting that IL2 serves an important role in antigen-stimulated immune responses (38). The present study revealed that IL2 is associated with the positive regulation of tissue remodeling, including bone remodeling, but little is known about the effects and mechanisms of IL2 in OP. LRGUK comprises 3 domains: A leucine-rich repeat, a guanylate kinase-like domain and an unnamed domain (39). LRGUK exhibits biased expression in the testis, bone marrow and lungs, and functions in ATP-binding and kinase activity. Studies on LRGUK are rare. One previous study demonstrated that LRGUK is associated with type 2 diabetes and fasting glucose levels (40). PIK3CG phosphorylates inositol lipids and is involved in the immune response process; it serves a vital role by recruiting pleckstrin homology domain-containing proteins to the membrane, including Akt1, and by activating signaling cascades involved in cell growth, proliferation and differentiation (41). Additionally, PIK3CG regulates bone homeostasis by modulating osteoclastogenesis and bone homeostasis (42), suggesting that this gene is linked to OP. LEP encodes a protein secreted by white adipocytes and serves an important role in energy balance regulation. This protein is also involved in endocrine function and in the regulation of immune and inflammatory responses. Tariq et al (43) found that body weight and body mass index impact BMD, whereas serum leptin is not associated with BMD. Zheng et al (44) identified that LEP overexpression in bone marrow stromal cells can stimulate periodontal regeneration in osteoporotic conditions. The association between LEP and OP is unclear, and should be a focus of future studies. TF encodes an extracellular or secreted glycoprotein with an N-terminal transmembrane domain and a short cysteine-rich cytoplasmic loop prior to the unique C-terminal ends (45). The protein is involved in cellular iron ion homeostasis, post-translational protein modification and the regulation of protein stability. The putative iron sensor TF receptor 2 can bind to iron-loaded TF in the bloodstream, which increases the expression of hepcidin by stimulating the bone morphogenetic protein signaling pathway (46). POLR2A provides a platform for transcription, mRNA processing and chromosome remodeling (47). This gene encodes an essential subunit of RNA polymerase II and is ubiquitously expressed in the testis, skin and bone marrow, and is involved in numerous physiological processes, including the regulation of gene silencing by miRNA and of RNA splicing (48,49). However, the biological functions of POLR2A in OP remain unknown.
A module analysis of the PPI network demonstrated that OP is linked to neuroactive ligand-receptor interactions and ubiquitin-mediated proteolysis. The neuroactive ligand-receptor interaction pathway can be activated by targeting the cell membrane G protein coupled receptor, which is involved in signal transduction from the extracellular to the intracellular components, including glycine, serine and threonine (50,51). Neuroactive steroids, which influence the modulation of the γ-aminobutyric acid receptor, are hormones that act as regulators of neurotransmitter receptors to either enhance or suppress neuronal activity (52). Moreover, ubiquitin-mediated proteolysis has been reported to serve a key role in MSC osteogenic or adipogenic differentiation (53) and the ubiquitin-mediated proteolysis pathway serves a critical role in various processes, including the cell cycle, cellular response to stress, DNA repair and immune regulation (54). Therefore, combined with the present results, these data indicate that neuroactive ligand-receptor interactions and ubiquitin-mediated proteolysis may serve key roles in the progression of OP, and are potential targets for drug development and treatment.
In the present study, an integrated miRNA-gene analysis of core genes associated with OP was performed. An overlap threshold of 2 was set to identify only interactions supported by 2 MTI databases, and to ensure credible and accurate results. Overall, 15 predicted miRNAs and their target genes were screened. IL2 was regulated by miRNAs hsa-miR-524-5p, hsa-miR-520d-5p, hsa-miR-181b, hsa-miR-181a, hsa-miR-181d, hsa-miR-181c and hsa-miR-186. These miRNAs were enriched for biological function terms, including ‘molecular function’, ‘enzyme regulator activity’ and ‘immune system process’. PHLPP2 was associated with miRNAs hsa-miR-509-3p, hsa-miR-134 and hsa-miR-367, which are mainly associated with organelles. miRNA hsa-miR-361-5p was observed to modulate KNG1, along with hsa-miR-942 and hsa-miR-578. These are involved in blood coagulation and platelet activation. Furthermore, hsa-miR-142-3p may serve a vital role in OP by regulating PIK3CG, which is involved in the estrogen and B cell receptor signaling pathways. miRNA hsa-miR-873, which is associated with metabolic pathways, co-regulates POLR2A. The present results indicate that a series of miRNAs simultaneously regulate the same gene, as observed for IL2, PHLPP2 and KNG1. Further studies are required to verify these interactions.
In conclusion, the results of the present study improve our understanding of the progression of OP based on an in-depth bioinformatics analysis of DEGs. In total, 915 DEGs and 10 core genes (ALB, PHLPP2, TOP2A, KNG1, IL2, LRGUK, PIK3CG, LEP, TF and POLR2A) involved in OP were identified. Additionally, miRNAs associated with target genes were identified; these may be crucial for the initiation and progression of OP. However, further experimental studies are required in order to determine the precise roles of these genes and miRNAs. The present study provides a useful set of genes and miRNAs for future studies on the molecular mechanisms of OP and for the determination of therapeutic targets.
Acknowledgements
Not applicable.
Funding
The National Natural Science Foundation of China (grant no. 81573874) provided financial support for the conduct of the study.
Availability of data and materials
The datasets generated and analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository (https://www.ncbi.nlm.nih.gov/geo/) (8).
Authors' contributions
QLF, YC and FT designed the study. YC, SMY, FT and FXL analyzed and interpreted the data. YC, FT and QLF were major contributors in the writing of the manuscript. All authors read and approved the submitted manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Body JJ, Bergmann P, Boonen S, Boutsen Y, Devogelaer JP, Goemaere S, Kaufman JM, Rozenberg S, Reginster JY. Evidence-based guidelines for the pharmacological treatment of postmenopausal osteoporosis: A consensus document by the Belgian Bone Club. Osteoporos Int. 2010;21:1657–1680. doi: 10.1007/s00198-010-1223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen P, Li Z, Hu Y. Prevalence of osteoporosis in China: A meta-analysis and systematic review. BMC Public Health. 2016;16:1039. doi: 10.1186/s12889-016-3712-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Svedbom A, Hernlund E, Ivergård M, Compston J, Cooper C, Stenmark J, McCloskey E, Jönsson B, Kanis JEU Review Panel of IOF, corp-author. Osteoporosis in the European Union: A compendium of country-specific reports. Arch Osteoporos. 2013;8:137. doi: 10.1007/s11657-013-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma M, Chen X, Lu L, Yuan F, Zeng W, Luo S, Yin F, Cai J. Identification of crucial genes related to postmenopausal osteoporosis using gene expression profiling. Aging Clin Exp Res. 2016;28:1067–1074. doi: 10.1007/s40520-015-0509-y. [DOI] [PubMed] [Google Scholar]
- 5.Takayanagi H. Osteoimmunology: Shared mechanisms and crosstalk between the immune and bone systems. Nat Rev Immunol. 2007;7:292–304. doi: 10.1038/nri2062. [DOI] [PubMed] [Google Scholar]
- 6.Li X, Ge H, Weiszmann J, Hecht R, Li Y, Véniant M, Xu J, Wu X, Lindberg R, Li Y. Inhibition of lipolysis may contribute to the acute regulation of plasma FFA and glucose by FGF21 in ob/ob mice. FEBS Lett. 2009;583:3230–3234. doi: 10.1016/j.febslet.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 7.Hao RH, Gao JL, Li M, Huang W, Zhu DL, Thynn HN, Dong SS, Guo Y. Association between fibroblast growth factor 21 and bone mineral density in adults. Endocrine. 2018 doi: 10.1007/s12020-017-1507-y. [DOI] [PubMed] [Google Scholar]
- 8.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2012;41:D991–D995. doi: 10.1093/nar/gks1193. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benisch P, Schilling T, Klein-Hitpass L, Frey S, Seefried L, Raaijmakers N, Krug M, Regensburger M, Zeck S, Schinke T, et al. The transcriptional profile of mesenchymal stem cell populations in primary osteoporosis is distinct and shows overexpression of osteogenic inhibitors. PLoS One. 2012;7:e45142. doi: 10.1371/journal.pone.0045142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venet D, Detours V, Bersini H. A measure of the signal-to-noise ratio of microarray samples and studies using gene correlations. PLoS One. 2012;7:e51013. doi: 10.1371/journal.pone.0051013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 14.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doncheva N, Assenov Y, Domingues F, Albrecht M. Topological analysis and interactive visualization of biological networks and protein structures. Nat Protoc. 2012;7:670–685. doi: 10.1038/nprot.2012.004. [DOI] [PubMed] [Google Scholar]
- 16.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Roy Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 20.Kutmon M, Kelder T, Mandaviya P, Evelo CT, Coort SL. CyTargetLinker: A Cytoscape app to integrate regulatory interactions in network analysis. PLoS One. 2013;8:e82160. doi: 10.1371/journal.pone.0082160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tella SH, Gallagher JC. Prevention and treatment of postmenopausal osteoporosis. J Steroid Biochem Mol Biol. 2014;142:155–170. doi: 10.1016/j.jsbmb.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho SS, Vollmer NL, Refaat MI, Jeon O, Alsberg E, Lee MA, Leach JK. Bone morphogenetic protein-2 promotes human mesenchymal stem cell survival and resultant bone formation when entrapped in photocrosslinked alginate hydrogels. Adv Healthc Mater. 2016;5:2501–2509. doi: 10.1002/adhm.201600461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- 24.Wu B, Xie J, Du Z, Wu J, Zhang P, Xu L, Li E. PPI network analysis of mRNA expression profile of ezrin knockdown in esophageal squamous cell carcinoma. Biomed Res Int. 2014;2014:651954. doi: 10.1155/2014/651954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korkmaz V, Kurdoglu Z, Alisik M, Turgut E, Sezgın OO, Korkmaz H, Ergun Y, Erel O. Thiol/disulfide homeostasis in postmenopausal osteoporosis. J Endocrinol Invest. 2017;40:431–435. doi: 10.1007/s40618-016-0585-7. [DOI] [PubMed] [Google Scholar]
- 26.Lee EJ, Kim JL, Kim YH, Kang MK, Gong JH, Kang YH. Phloretin promotes osteoclast apoptosis in murine macrophages and inhibits estrogen deficiency-induced osteoporosis in mice. Phytomedicine. 2014;21:1208–1215. doi: 10.1016/j.phymed.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 27.Huang C, Li S. Association of blood neutrophil lymphocyte ratio in the patients with postmenopausal osteoporosis. Pak J Med Sci. 2016;32:762–765. doi: 10.12669/pjms.323.10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spilmont M, Léotoing L, Davicco MJ, Lebecque P, Mercier S, Miot-Noirault E, Pilet P, Rios L, Wittrant Y, Coxam V. Pomegranate and its derivatives can improve bone health through decreased inflammation and oxidative stress in an animal model of postmenopausal osteoporosis. Eur J Nutr. 2014;53:1155–1164. doi: 10.1007/s00394-013-0615-6. [DOI] [PubMed] [Google Scholar]
- 29.Wauquier F, Leotoing L, Coxam V, Guicheux J, Wittrant Y. Oxidative stress in bone remodelling and disease. Trends Mol Med. 2009;15:468–477. doi: 10.1016/j.molmed.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang SM, Feigenson M, Begun DL, Shull LC, Culley KL, Otero M, Goldring MB, Ta LE, Kakar S, Bradley EW, Westendorf JJ. Phlpp inhibitors block pain and cartilage degradation associated with osteoarthritis. J Orthop Res. 2018;36:1487–1497. doi: 10.1002/jor.23781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madabhushi R. The Roles of DNA Topoisomerase IIβ in Transcription. Int J Mol Sci. 2018;19(pii):E1917. doi: 10.3390/ijms19071917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lang AJ, Mirski SE, Cummings HJ, Yu Q, Gerlach JH, Cole SP. Structural organization of the human TOP2A and TOP2B genes. Gene. 1998;221:255–266. doi: 10.1016/S0378-1119(98)00468-5. [DOI] [PubMed] [Google Scholar]
- 34.Kaplan E, Gündüz U. Expression analysis of TOP2A, MSH2 and MLH1 genes in MCF7 cells at different levels of etoposide resistance. Biomed Pharmacother. 2012;66:29–35. doi: 10.1016/j.biopha.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Raguz S, Adams C, Masrour N, Rasul S, Papoutsoglou P, Hu Y, Cazzanelli G, Zhou Y, Patel N, Coombes C, et al. Loss of O6-methylguanine-DNA methyltransferase confers collateral sensitivity to carmustine in topoisomerase II-mediated doxorubicin resistant triple negative breast cancer cells. Biochem Pharmacol. 2013;85:186–196. doi: 10.1016/j.bcp.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 36.Boregowda SV, Krishnappa V, Chambers JW, Lograsso PV, Lai WT, Ortiz LA, Phinney DG. Atmospheric oxygen inhibits growth and differentiation of marrow-derived mouse mesenchymal stem cells via a p53-dependent mechanism: Implications for long-term culture expansion. Stem Cells. 2012;30:975–987. doi: 10.1002/stem.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang F, Wen Y, Guo X, Zhang Y, Wang S, Yang T, Shen H, Chen X, Tan L, Tian Q, Deng HW. Genome-wide pathway-based association study implicates complement system in the development of Kashin-Beck disease in Han Chinese. Bone. 2015;71:36–41. doi: 10.1016/j.bone.2014.09.025. [DOI] [PubMed] [Google Scholar]
- 38.Li J, Hou Y, Ding X, Hou W, Song B, Zeng Y. Overexpression, purification, molecular characterization and the effect on tumor growth of ribosomal protein L22 from the Giant Panda (Ailuropoda melanoleuca) Mol Biol Rep. 2014;41:3529–3539. doi: 10.1007/s11033-014-3217-3. [DOI] [PubMed] [Google Scholar]
- 39.Okuda H, DeBoer K, O'Connor AE, Merriner DJ, Jamsai D, O'Bryan MK. LRGUK1 is part of a multiprotein complex required for manchette function and male fertility. FASEB J. 2017;31:1141–1152. doi: 10.1096/fj.201600909R. [DOI] [PubMed] [Google Scholar]
- 40.Laramie JM, Wilk JB, Williamson SL, Nagle MW, Latourelle JC, Tobin JE, Province MA, Borecki IB, Myers RH. Polymorphisms near EXOC4 and LRGUK on chromosome 7q32 are associated with Type 2 Diabetes and fasting glucose; the NHLBI Family Heart Study. BMC Med Genet. 2008;9:46. doi: 10.1186/1471-2350-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jean S, Kiger AA. Classes of phosphoinositide 3-kinases at a glance. J Cell Sci. 2014;127:923–928. doi: 10.1242/jcs.093773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang H, Chang W, Hurley M, Vignery A, Wu D. Important roles of PI3Kgamma in osteoclastogenesis and bone homeostasis. Proc Natl Acad Sci USA. 2010;107:12901–12906. doi: 10.1073/pnas.1001499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tariq S, Baig M, Tariq S, Shahzad M. Association of serum leptin with bone mineral density in postmenopausal osteoporotic females. Gynecol Endocrinol. 2017;33:287–291. doi: 10.1080/09513590.2016.1261103. [DOI] [PubMed] [Google Scholar]
- 44.Zheng B, Jiang J, Chen Y, Lin M, Du Z, Xiao Y, Luo K, Yan F. Leptin overexpression in bone marrow stromal cells promotes periodontal regeneration in a rat model of osteoporosis. J Periodontol. 2017;88:808–818. doi: 10.1902/jop.2017.170042. [DOI] [PubMed] [Google Scholar]
- 45.Ramsey J, Mukhopadhyay S. Disentangling the frames, the state of research on the alphavirus 6K and TF proteins. Viruses. 2017;9(pii):E228. doi: 10.3390/v9080228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Worthen C, Enns C. The role of hepatic transferrin receptor 2 in the regulation of iron homeostasis in the body. Front Pharmacol. 2014;5:34. doi: 10.3389/fphar.2014.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao DY, Gish G, Braunschweig U, Li Y, Ni Z, Schmitges FW, Zhong G, Liu K, Li W, Moffat J, et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature. 2016;529:48–53. doi: 10.1038/nature16469. [DOI] [PubMed] [Google Scholar]
- 48.Hui-Ying X, Da-Hong Z, Li-Juan J, Xiao-Jie L. Anticancer opportunity created by loss of tumor suppressor genes. Technol Cancer Res Treat. 2016;15:729–731. doi: 10.1177/1533034615604798. [DOI] [PubMed] [Google Scholar]
- 49.David CJ, Boyne AR, Millhouse SR, Manley JL. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes Dev. 2011;25:972–983. doi: 10.1101/gad.2038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lauss M, Kriegner A, Vierlinger K, Noehammer C. Characterization of the drugged human genome. Pharmacogenomics. 2007;8:1063–1073. doi: 10.2217/14622416.8.8.1063. [DOI] [PubMed] [Google Scholar]
- 51.Bhattarai UR, Katuwal Bhattarai M, Li F, Wang D. Insights into the temporal gene expression pattern in lymantria dispar larvae during the baculovirus induced hyperactive stage. Virol Sin. 2018 Jul 25; doi: 10.1007/s12250-018-0046-x. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J, Cheng J, Zhang C, Li X. Cardioprotection effects of sevoflurane by regulating the pathway of neuroactive ligand-receptor interaction in patients undergoing coronary artery bypass graft surgery. Comput Math Methods Med. 2017;2017:3618213. doi: 10.1155/2017/3618213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhuang Q, Mao W, Xu P, Li H, Sun Z, Li S, Qiu G, Li J, Zhang J. Identification of differential genes expression profiles and pathways of bone marrow mesenchymal stem cells of adolescent idiopathic scoliosis patients by microarray and integrated gene network analysis. Spine (Phila Pa 1976) 2016;41:840–855. doi: 10.1097/BRS.0000000000001394. [DOI] [PubMed] [Google Scholar]
- 54.Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: Biological regulation via destruction. Bioessays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository (https://www.ncbi.nlm.nih.gov/geo/) (8).