Abstract
Previous studies indicate that miR-149 could both inhibit and promote the development of human cancer depending on the tumor type. GIT1 was found to play an important role in regulating cell migration. However, the specific function of miR-149-5p and GIT1 in the progression of medullary thyroid carcinoma (MTC) remains unknown. The purpose of this study was to confirm the function of miR-149-5p in MTC and explore its downstream regulation. Moreover, miR-149-5p level in MTC was detected via RT-quantitative PCR (RT-qPCR). GIT1 expression levels were assessed by RT-qPCR and western blot analysis. The cell proliferation and invasion were detected through MTT or Transwell assay respectively. In addition, miR-149-5p was identified to directly target GIT1 in MTC via dual luciferase assay. The results suggested that miR-149-5p level was obviously declined in MTC. Functionally, miR-149-5p overexpression inhibited proliferation and invasion. Moreover, miR-149-5p directly targeted GIT1 and was negatively associated with its expression in MTC. Conversely, GIT1 expression was obviously increased in MTC. GIT1 overexpression partially reversed the inhibitory action of miR-149-5p in MTC. miR-149-5p suppressed the proliferation and invasion of MTC cells through targeting GIT1, which would create new therapeutic avenues for MTC treatment.
Keywords: medullary thyroid carcinoma, miR-149-5p, invasion, proliferation, GIT1
Introduction
Medullary thyroid carcinoma (MTC) is a malignant tumor that originates from the paracytic cells of the thyroid follicle, accounting for approximately 3–8% of thyroid cancer (1). Clinical malignant degree of MTC is higher than that of papillary thyroid carcinoma. MTC is more prone to lymph node metastasis which results in more difficulty in treatment (2). MTC is divided into sporadic entity and hereditary: 75–80% is sporadic (sMTC) and 20–25% is hereditary (hMTC) (3). Moreover, because MTC is not sensitive to radiotherapy, surgery is the preferred method for the treatment of MTC (4). Moreover, early diagnosis is more important for patients with MTC, because MTC is a moderate malignancy. Therefore, finding new markers for early diagnosis of MTC is important for improving its survival rate.
Recently, microRNA (miRNA) was demonstrated to regulate the progression of human cancer through repressing or activating the corresponding gene expression (5). Especially, many miRNAs have been reported to be significantly associated with human metastatic MTC (6). For instance, miR-21 has been identified as an overexpressed marker in MTC (7). miR-182 was found to promote cell invasion by linking to RET oncogene in MTC (8). Inversely, miR-129-5p was reported to regulate cell growth, migration and apoptosis in MTC by repressing RET (9). In previous studies, miR-149 was identified as a suppressive miRNA in various human cancers, such as non-small cell lung cancer (10), bladder carcinoma (11) and gastric carcinoma (12). On the contrary, the upregulation of miR-149-5p was identified in acute myeloid leukemia and glioma (13,14). However, the specific function of miR-149-5p in MTC still remains unknown.
G protein-coupled receptor kinase interacting ArfGAP 1 (GIT1) is a multifunctional scaffold protein which could activate cell formation and spread (15). In recent years, GIT1 was identified to promote cell migration and invasion in breast cancer (16). Moreover, GIT1 was also found to regulate the proliferation and EMT of non-small cell lung cancer cells regulated by miR-138 (17). Additionally, it was reported that GIT1 promoted osteoblastic proliferation and differentiation (18). Importantly, Chan et al reported that miR-149/GIT1 axis repressed integrin signaling and metastasis in breast cancer (19). However, the function of miR-149/GIT1 axis in MTC is still unclear.
In this study, we mainly investigated the dysregulated expression levels of miR-149-5p and GIT1 in MTC. At the same time, their effects on cell proliferation and invasion were explored in MTC. Furthermore, we clarified the interaction between miR-149-5p and GIT1 in MTC. Our findings may create new therapeutic avenues for MTC treatment.
Materials and methods
Clinical tissues
Thiry-six paired surgical tumor specimens and adjacent tissue samples were obtained from the Sir Run Run Shaw Hospital, College of Medicine, Zhejiang University (Hangzhou, China) between February 2016 and March 2017 after receiving written informed consent. None of the patients received treatment prior to the operation. Human tissue was frozen in liquid nitrogen and then stored at −80°C for further experiment. This experiment was approved by the Institutional Ethics Committee of Sir Run Run Shaw Hospital, College of Medicine, Zhejiang University.
Cell culture
The human MTC cell lines TT, MZ-CRC-1 and NThy-ori 3.1 human primary thyroid epithelial cells were used for this experiment. All the cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were seeded in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS) and cultured at 37°C with 5% CO2.
Cell transfection
The miR-149-5p mimic (5′-UCUGGCUCCGUGUCUUCACUCCC-3′) or mimic-NC (5′-UUCUCCGAACGUGUCACGUTT-3′), miR-149-5p inhibitor (5′-GGGAGUGAAGACACGGAGCCAGA-3′), or inhibitor-NC (5′-CAGUACUUUUGUGUAGUACAA-3′); GIT1 siRNA (si-GIT1, 5′-GUGCCAAUAUGAGCUCAGUTT-3′ and 5′-AGUGAGCUCAUAUUGGCACTT-3′) and its negative control (5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′) were purchased from RiboBio (Guangzhou, China) and then they were transferred into TT cells with Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to manufactures' protocols. Then they were further incubated for 48 h at 37°C in an incubator.
RT-quantitative PCR (RT-qPCR)
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was applied for extracting total RNA containing miRNA to quantitate miR-149-5p expression in MTC tissues and cell lines. RT-qPCR was carried out through the SYBR Green Master Mix (Roche Molecular Diagnostics, Pleasanton, CA, USA) on 7900HT Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). U6 and GAPDH were used as control for miR-149-5p and GIT1. The cycling conditions for RT-qPCR were as follows: 5 min at 95°C, followed by 40 cycles of 95°C for 30 sec and 60°C for 45 sec. The primers were designed as follows: miR-149, 5′-GGCTCTGGCTCCGTGTCTT-3′ and 5′-CAGTGCAGGGTCCGAGGTATT-3′; U6, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and 5′-CGCTTCACGAATTTGCGTGTCAT-3′; and GAPDH, 5′-GCCTTCCGTGTCCCCACTGC-3′ and 5′-CAATGCCAGCCCCAGCGTCA-3′. Its expression was calculated using the 2−∆∆cq method (20).
Luciferase activity assay
TargetScan (http://www.targetscan.org/) predicted biological targets of miRNAs by searching for the presence of conserved 8mer, 7mer, and 6mer sites that match the seed region of each miRNA (21). The wild or mutant type of 3′-UTR of GIT1 was inserted into the pGL3 luciferase vector (Promega Corporation, Madison, WI, USA) for luciferase reporter experiments. Then, wild- or mutant-type of 3′-UTR of GIT1 and miR-149-5p mimics were transfected into TT cells. Subsequently, the Dual Luciferase Assay (Promega Corporation) was applied to analyze luciferase activity.
MTT assay for cell proliferation
The MTT (3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2H-tetrazolium bromide) assay was applied to measure cell proliferation. Cells (4×103/well) were seeded onto 96-well plates in medium. The cells containing miR-149-5p mimic or inhibitor were incubated for 24, 48, 72 or 96 h. After incubation, the cells added with MTT (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) were incubated for 4 h at 37°C. The absorbance at 570 nm (OD=570 nm) was detected with a spectrophotometer (Bio-Rad, Hercules, CA, USA).
Cell invasion assay
Cell invasion was assessed by Transwell assay. Transwells were coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) to study cell invasion. Cells (2×105) were planted into the upper chambers (8 µm pore size; Corning Incorporated, Corning, NY, USA) and medium with 20% FBS was put into the lower chamber. Then the cells were incubated at 37°C with 5% CO2 for 18 h. Then the invasive cells on the lower surface were fixed with 4% PFA and stained with 0.1% crystal violet. Cells were counted under a light microscope. (Nikon, Missisauga, ON, Canada).
Western blot analysis
The protein samples were obtained using RIPA lysis buffer. Proteins were separated through a 10% SDS-PAGE and incubated with 5% non-fat milk in PVDF membranes at room temperature. Next we incubated the membranes overnight at 4°C with primary anti-GIT1 antibody (dilution, 1:1,000; rabbit polyclonal; cat. no. ab153958; Abcam, Cambridge, MA, USA), primary anti-GAPDH antibody (dilution, 1:1,000; rabbit monoclonal; cat. no. ab9485; Abcam) and subsequently incubated with secondary goat anti-rabbit IgG H&L (HRP) antibody (dilution, 1:2,000; cat. no. ab6721; Abcam). Then, protein expression levels were measured by ECL (Pierce; Thermo Fisher Scientific, Inc.).
Statistical analysis
The obtained data are shown as the mean ± SD. Statistical analysis was analyzed with SPSS.19 (IBM Corp., Armonk, NY, USA) and GraphPad Prism 6.0. The correlation of miR-149-5p with clinicopathological characteristics of MTC was calculated through the Chi-square test. Differences among multiple groups were calculated according to the analysis of variance (ANOVA) with Tukey-Kramer post hoc test. The overall survival was analyzed by Kaplan-Meier method and the log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
The dysregulated expression levels of miR-149-5p and GIT1 examined in MTC
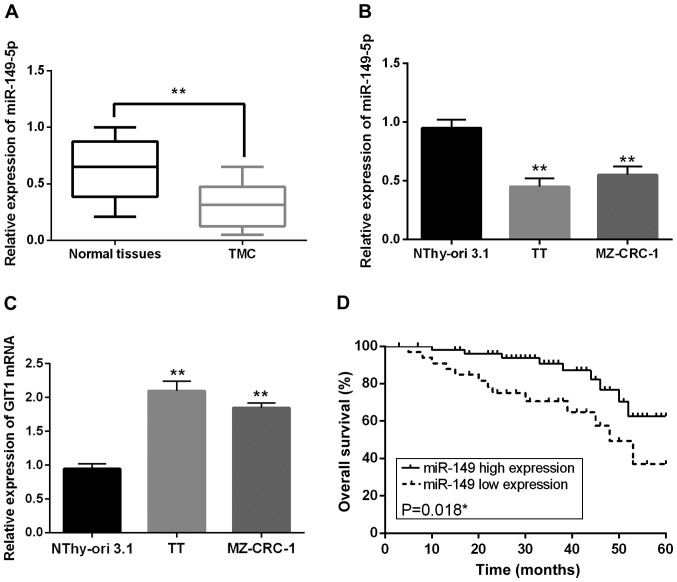
Primarily, the expression levels of miR-149-5p and GIT1 were identified in MTC via RT-qPCR assay. Then decreased expression of miR-149-5p was observed in MTC tissues compared with the normal tissues (Fig. 1A). Consistent with the above result, downregulation of miR-149-5p was also identified in TT and MZ-CRC-1 cell lines in comparison with NThy-ori 3.1 cells (Fig. 1B). Surprisingly, upregulation of GIT1 was detected in TT and MZ-CRC-1 cells compared with the control (Fig. 1C). Thus, we considered that the dysregulated expression levels of miR-149-5p and GIT1 might be associated with tumorigenesis of MTC. Next, the correlation between miR-149-5p level and the clinicopathological characteristics was analyzed in MTC patients. As shown in Table I, decreased miR-149-5p expression was found to be obviously correlated with distant metastases (P=0.013) and TNM stage (P=0.043). Furthermore, survival analysis indicated that low miR-149-5p expression was notably related to shorter overall survival of MTC patients (P=0.0018; Fig. 1D).
Figure 1.
miR-149-5p and GIT1 levels are changed in MTC. (A) The expression levels of miR-149-5p in MTC tissues and normal tissues were detected via RT-qPCR. (B) The miR-149-5p expression in TT, MZ-CRC-1 and NThy-ori 3.1 normal thyroid epithelial cells (control). (C) The GIT1 expression in TT, MZ-CRC-1 and NThy-ori 3.1 normal thyroid epithelial cells (control). (D) Kaplan-Meier overall survival curves of MTC patients. Low miR-149-5p expression was notably related to shorter overall survival of MTC patients. **P<0.01.
Table I.
Relationship between miR-149-5p expression and the clinicopathological characteristics of MTC patients.
| miR-149-5p | ||||
|---|---|---|---|---|
| Characteristics | Cases | High | Low | P-value |
| Sex | 0.655 | |||
| Male | 16 | 6 | 10 | |
| Female | 20 | 5 | 15 | |
| Tumor size | 0.222 | |||
| >5 cm | 19 | 7 | 12 | |
| ≤5 cm | 17 | 5 | 12 | |
| Distant metastases | 0.043a | |||
| Present | 11 | 4 | 7 | |
| Absent | 25 | 10 | 15 | |
| Lymph node metastasis | 0.086 | |||
| Present | 13 | 5 | 8 | |
| Absent | 23 | 10 | 13 | |
| TNM stage | 0.013a | |||
| I/II | 21 | 8 | 13 | |
| III/IV | 15 | 6 | 9 | |
Statistical analyses were performed by the χ2 test.
P<0.05 was considered significant.
miR-149-5p inhibits cell proliferation and invasion in MTC
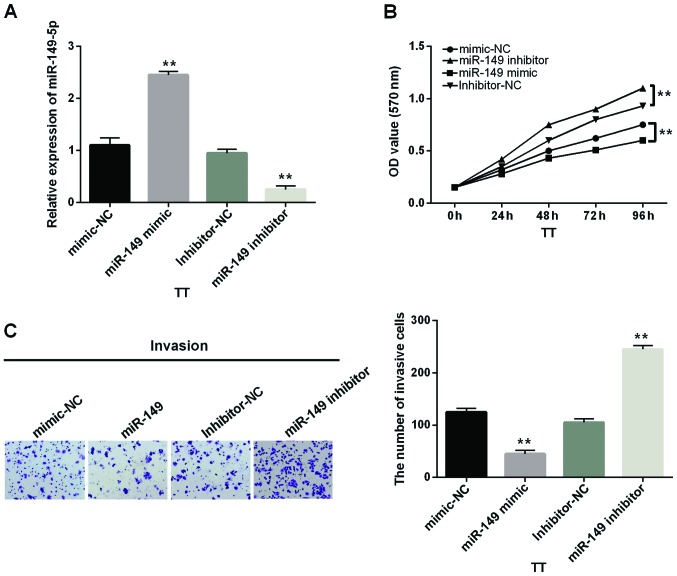
Next, we transfected miR-149-5p mimics or inhibitor into TT cells to explore its function in MTC. The transfection efficiency was measured by RT-qPCR (Fig. 2A). Functionally, the cell proliferation was distinctly suppressed by miR-149-5p mimics whereas enhanced by miR-149-5p inhibitor (Fig. 2B). In addition, the role of miR-149-5p for the invasion of MTC cells was investigated. Similarly to the result of cell proliferation, miR-149-5p mimics repressed cell invasion while miR-149-5p inhibitors promoted cell invasion in MTC cells (Fig. 2C). According to these results, miR-149-5p was verified as a suppressive miRNA in human MTC.
Figure 2.
miR-149-5p inhibits proliferation and invasion of MTC cells. (A) miR-149-5p expression was examined in TT cells containing miR-149-5p mimics or inhibitor via RT-qPCR. (B) Cell proliferation was measured in cells containing miR-149-5p mimics or inhibitor via MTT. (C) Cell invasion was measured in cells with miR-149-5p mimics or inhibitor via Transwell analysis **P<0.01.
GIT1 was confirmed to be a direct target of miR-149-5p in MTC
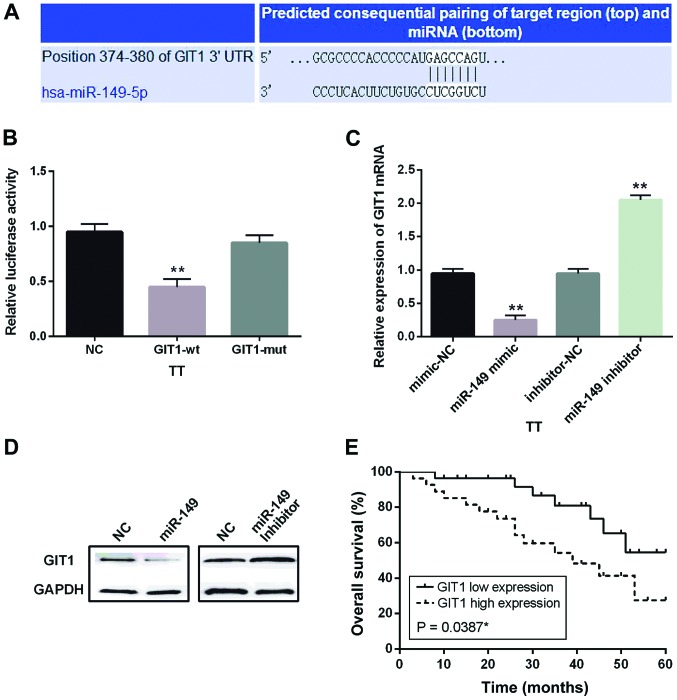
Subsequently, the target genes of miR-149-5p were searched in TargetScan to further analyze its regulatory pathway in MTC. Based on the prediction, we selected GIT1 as the target of miR-149-5p (Fig. 3A). Then we confirmed this prediction by luciferase reporter assays. As we predicted, the luciferase activity in cells harbored wild-type GIT1 and miR-149-5p mimic was lower than that in the control group. However, luciferase activity in cells harboring mutant-type GIT1 and miR-149-5p mimics had almost no change (Fig. 3B). Furthermore, we examined the mRNA and protein expression levels of GIT1 in TT cells containing miR-149-5p mimics or inhibitors to verify the interaction between miR-149-5p and GIT1. The results indicated that miR-149-5p mimics impeded GIT1 expression and miR-149-5p inhibitors promoted its expression (Fig. 3C and D). Taken together, miR-149-5p was verified to directly target GIT1 and negatively associated with its expression in MTC. Besides, Kaplan-Meier survival analysis was performed to analyze the correlation between GIT1 and prognosis of MTC patients. The results showed that high GIT1 expression was related to shorter OS of MTC patients (P=0.0387; Fig. 3E).
Figure 3.
GIT1 was confirmed to be a direct target of miR-149-5p in MTC. (A) The binding sites of miR-149-5p on the 3′-UTR of GIT1. (B) Luciferase reporter assay. (C and D) The mRNA and protein expression levels of GIT1 were analyzed in cells containing miR-149-5p mimics or inhibitor. (E) Kaplan-Meier overall survival curves of MTC patients. High GIT1 expression was related to shorter OS of MTC patients. *P<0.05, **P<0.01.
GIT1 regulates cell proliferation and invasion in MTC
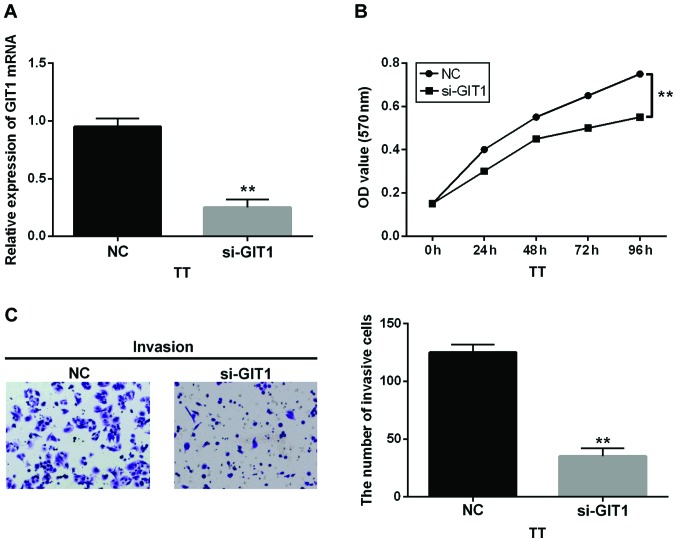
Immediately following the above findings, we transfected GIT1 siRNA into TT cells to explore its specific function in MTC. Then the GIT1 expression was identified in transfected cells (Fig. 4A). Interestingly, the si-GIT1 had the same effect as miR-149-5p overexpression in MTC which inhibited cell proliferation in MTC cells (Fig. 4B). Similarly, si-GIT1 also suppressed the invasion of MTC cells (Fig. 4C). Therefore, we inferred that GIT1 might function as an oncogene in human MTC.
Figure 4.
GIT1 regulates the proliferation and invasion of MTC cells. (A) The mRNA expression of GIT1 was measured in cells containing GIT1 siRNA. (B) The proliferation in cells containing si-GIT1. (C) Cell invasion analysis of TT cells with si-GIT1 **P<0.01.
GIT1 overexpression partially reversed the inhibitory action of miR-149-5p in MTC
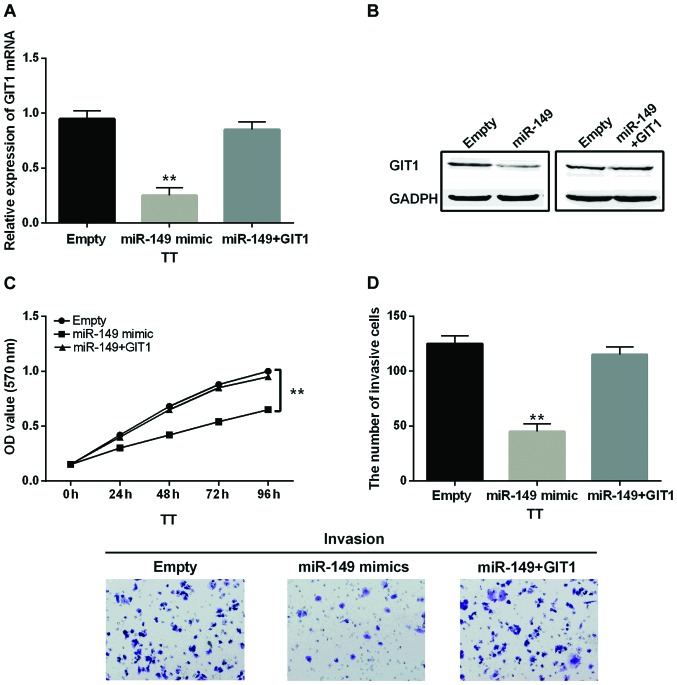
Finally, miR-149-5p mimics and GIT1 vector were co-transfected into TT cells to investigate how GIT1 interacted with miR-149-5p in MTC. There was almost no difference in mRNA and protein expression levels of GIT1 in cells with miR-149-5p mimics and GIT1 vector compared with the control group (Fig. 5A and B). It reflected that the GIT1 vector restored the reduction of GIT1 expression induced by miR-149-5p mimics. Simultaneously, the inhibitory effect of miR-149-5p on cell proliferation was also reversed by upregulation of GIT1 in MTC (Fig. 5C). Similarly, the inhibitory effect of miR-149-5p on cell invasion was restored by GIT1 overexpression in MTC as well (Fig. 5D). As a result, we considered that GIT1 overexpression partially reversed the inhibitory action of miR-149-5p in MTC.
Figure 5.
GIT1 overexpression partially reversed the inhibitory action of miR-149-5p in MTC. (A and B) The mRNA and protein expression levels of GIT1 were measured in cells containing GIT1 vector and miR-149-5p. (C) The cell proliferation in cells containing GIT1 vector and miR-149-5p. (D) The cell invasion in cells containing GIT1 vector and miR-149-5p **P<0.01.
Discussion
In this study, the function of miR-149-5p and its downstream regulation were investigated in MTC. The results suggested that miR-149-5p expression was obviously declined in MTC. Moreover, miR-149-5p overexpression was found to inhibit the proliferation and invasion of MTC cells. Furthermore, miR-149-5p was confirmed to directly target GIT1 and negatively associated with its expression in MTC. Contrary to miR-149-5p, GIT1 expression was distinctly increased in MTC. GIT1 overexpression partially reversed the inhibitory action of miR-149-5p in MTC.
Recently, many miRNAs have been reported to regulate tumorigenesis and progression of human cancer (22). The dysregulated expression of miRNAs could be identified as a feature of tumorigenesis (23). Especially, miR-149 could both inhibit and promote the development of human cancer depending on the tumor type. For instance, miR-149 had been demonstrated to be upregulated in nasopharyngeal carcinoma and participated in its progression (24). The same carcinogenic effect for miR-149 was also found in melanoma which miR-149 upregulation generated apoptotic resistance (25). On the contrary, miR-149 was reported to suppress cell migration and invasion through inhibiting FOXM1 in colorectal cancer (26). Luo et al demonstrated that miR-149 repressed cell metastasis via regulating PPM1F in hepatocellular carcinoma (27). Consistent with the above studies, we also found that miR-149-5p was downregulated in MTC and it had inhibitory effect on the proliferation and invasion of MTC cells as well.
GIT1 was predicted and verified as a direct target gene of miR-149-5p to explore the downstream regulation of miR-149-5p in MTC. Previous studies had demonstrated that GIT1 was a direct target gene of miR-195 (28) and miR-491 (29) in chondrocytes and hepatocellular carcinoma. GIT1 was able to act as a biomarker for the tumorigenesis of oral squamous cell carcinoma (30). Importantly, miR-149 had been reported to target GIT1 in breast cancer (19). Thus, this study was carried out to confirm whether there was a similar relationship among miR-149 and GIT1 in MTC. As expected, miR-149-5p was found to directly target GIT1 and negatively related to its expression. Besides that, we also proposed that GIT1 overexpression could restore the inhibitory action of miR-149-5p in MTC.
In conclusion, miR-149-5p suppressed the proliferation and invasion of MTC cells through regulating GIT1. Moreover, downregulation of miR-149-5p and upregulation of GIT1 were found to be related to the poor prognosis of MTC patients. Therefore, we considered that restoring miR-149-5p expression by silencing GIT1 could create new therapeutic avenues for MTC treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XY contributed significantly to analysis and wrote the manuscript. XC contributed to the conception of the study. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Sir Run Run Shaw Hospital, College of Medicine, Zhejiang University (Hangzhou, China). Signed informed consents were obtained from the patients or guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Leboulleux S, Baudin E, Travagli JP, Schlumberger M. Medullary thyroid carcinoma. Clin Endocrinol (Oxf) 2004;61:299–310. doi: 10.1111/j.1365-2265.2004.02037.x. [DOI] [PubMed] [Google Scholar]
- 2.Santarpia L, Ye L, Gagel RF. Beyond RET: Potential therapeutic approaches for advanced and metastatic medullary thyroid carcinoma. J Intern Med. 2009;266:99–113. doi: 10.1111/j.1365-2796.2009.02112.x. [DOI] [PubMed] [Google Scholar]
- 3.Wells SA, Jr, Asa SL, Dralle H, Elisei R, Evans DB, Gagel RF, Lee N, Machens A, Moley JF, Pacini F, et al. American Thyroid Association Guidelines Task Force on Medullary Thyroid Carcinoma, corp-author. Revised American Thyroid Association guidelines for the management of medullary thyroid carcinoma. Thyroid. 2015;25:567–610. doi: 10.1089/thy.2014.0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drosten M, Pützer BM. Mechanisms of disease: Cancer targeting and the impact of oncogenic RET for medullary thyroid carcinoma therapy. Nat Clin Pract Oncol. 2006;3:564–574. doi: 10.1038/ncponc0610. [DOI] [PubMed] [Google Scholar]
- 5.Iorio MV, Croce CM. Causes and consequences of microRNA dysregulation. Cancer J. 2012;18:215–222. doi: 10.1097/PPO.0b013e318250c001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santarpia L, Calin GA, Adam L, Ye L, Fusco A, Giunti S, Thaller C, Paladini L, Zhang X, Jimenez C, et al. A miRNA signature associated with human metastatic medullary thyroid carcinoma. Endocr Relat Cancer. 2013;20:809–823. doi: 10.1530/ERC-13-0357. [DOI] [PubMed] [Google Scholar]
- 7.Chu YH, Hardin H, Schneider DF, Chen H, Lloyd RV. MicroRNA-21 and long non-coding RNA MALAT1 are overexpressed markers in medullary thyroid carcinoma. Exp Mol Pathol. 2017;103:229–236. doi: 10.1016/j.yexmp.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Spitschak A, Meier C, Kowtharapu B, Engelmann D, Pützer BM. MiR-182 promotes cancer invasion by linking RET oncogene activated NF-κB to loss of the HES1/Notch1 regulatory circuit. Mol Cancer. 2017;16:24. doi: 10.1186/s12943-016-0563-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duan L, Hao X, Liu Z, Zhang Y, Zhang G. MiR-129-5p is down-regulated and involved in the growth, apoptosis and migration of medullary thyroid carcinoma cells through targeting RET. FEBS Lett. 2014;588:1644–1651. doi: 10.1016/j.febslet.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Ke Y, Zhao W, Xiong J, Cao R. miR-149 inhibits non-small-cell lung cancer cells EMT by targeting FOXM1. Biochem Res Int. 2013;2013:506731. doi: 10.1155/2013/506731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang D, Du G, Xu A, Xi X, Li D. Expression of miR-149-3p inhibits proliferation, migration, and invasion of bladder cancer by targeting S100A4. Am J Cancer Res. 2017;7:2209–2219. [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Zheng X, Zhang Z, Zhou J, Zhao G, Yang J, Xia L, Wang R, Cai X, Hu H, et al. MicroRNA-149 inhibits proliferation and cell cycle progression through the targeting of ZBTB2 in human gastric cancer. PLoS One. 2012;7:e41693. doi: 10.1371/journal.pone.0041693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian P, Yan L. Inhibition of MicroRNA-149-5p induces apoptosis of acute myeloid leukemia cell line THP-1 by targeting fas ligand (FASLG) Med Sci Monit. 2016;22:5116–5123. doi: 10.12659/MSM.899114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen X, Li J, Liao W, Wang J, Chen H, Yao Y, Liu H, Ding K. microRNA-149 targets caspase-2 in glioma progression. Oncotarget. 2016;7:26388–26399. doi: 10.18632/oncotarget.8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manabe R, Kovalenko M, Webb DJ, Horwitz AR. GIT1 functions in a motile, multi-molecular signaling complex that regulates protrusive activity and cell migration. J Cell Sci. 2002;115:1497–1510. doi: 10.1242/jcs.115.7.1497. [DOI] [PubMed] [Google Scholar]
- 16.Tao WY, Wang CY, Sun YH, Su YH, Pang D, Zhang GQ. MicroRNA-34c suppresses breast cancer migration and invasion by targeting GIT1. J Cancer. 2016;7:1653–1662. doi: 10.7150/jca.14762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Wang Q, Wen R, Liang J, Zhong X, Yang W, Su D, Tang J. MiR-138 inhibits cell proliferation and reverses epithelial-mesenchymal transition in non-small cell lung cancer cells by targeting GIT1 and SEMA4C. J Cell Mol Med. 2015;19:2793–2805. doi: 10.1111/jcmm.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tu XM, Gu YL, Ren GQ. miR-125a-3p targetedly regulates GIT1 expression to inhibit osteoblastic proliferation and differentiation. Exp Ther Med. 2016;12:4099–4106. doi: 10.3892/etm.2016.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan SH, Huang WC, Chang JW, Chang KJ, Kuo WH, Wang MY, Lin KY, Uen YH, Hou MF, Lin CM, et al. MicroRNA-149 targets GIT1 to suppress integrin signaling and breast cancer metastasis. Oncogene. 2014;33:4496–4507. doi: 10.1038/onc.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Volinia S, Visone R, Galasso M, Rossi E, Croce CM. Identification of microRNA activity by Targets' Reverse EXpression. Bioinformatics. 2010;26:91–97. doi: 10.1093/bioinformatics/btp598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 23.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo Z, Zhang L, Li Z, Li X, Li G, Yu H, Jiang C, Dai Y, Guo X, Xiang J, et al. An in silico analysis of dynamic changes in microRNA expression profiles in stepwise development of nasopharyngeal carcinoma. BMC Med Genomics. 2012;5:3. doi: 10.1186/1755-8794-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin L, Hu WL, Jiang CC, Wang JX, Han CC, Chu P, Zhang LJ, Thorne RF, Wilmott J, Scolyer RA, et al. MicroRNA-149*, a p53-responsive microRNA, functions as an oncogenic regulator in human melanoma. Proc Natl Acad Sci USA. 2011;108:15840–15845. doi: 10.1073/pnas.1019312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu K, Liu X, Mao X, Xue L, Wang R, Chen L, Chu X. MicroRNA-149 suppresses colorectal cancer cell migration and invasion by directly targeting forkhead box transcription factor FOXM1. Cell Physiol Biochem. 2015;35:499–515. doi: 10.1159/000369715. [DOI] [PubMed] [Google Scholar]
- 27.Luo G, Chao YL, Tang B, Li BS, Xiao YF, Xie R, Wang SM, Wu YY, Dong H, Liu XD, et al. miR-149 represses metastasis of hepatocellular carcinoma by targeting actin-regulatory proteins PPM1F. Oncotarget. 2015;6:37808–37823. doi: 10.18632/oncotarget.5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu YL, Rong XX, Wen LT, Zhu GX, Qian MQ. miR-195 inhibits the proliferation and migration of chondrocytes by targeting GIT1. Mol Med Rep. 2017;15:194–200. doi: 10.3892/mmr.2016.5982. [DOI] [PubMed] [Google Scholar]
- 29.Yang X, Ye J, Yan H, Tang Z, Shen J, Zhang J, Yang L. MiR-491 attenuates cancer stem cell-like properties of hepatocellular carcinoma by inhibition of GIT-1/NF-κB-mediated EMT. Tumour Biol. 2016;37:201–209. doi: 10.1007/s13277-015-3687-5. [DOI] [PubMed] [Google Scholar]
- 30.Huang WC, Chan SH, Jang TH, Chang JW, Ko YC, Yen TC, Chiang SL, Chiang WF, Shieh TY, Liao CT, et al. miRNA-491-5p and GIT1 serve as modulators and biomarkers for oral squamous cell carcinoma invasion and metastasis. Cancer Res. 2014;74:751–764. doi: 10.1158/0008-5472.CAN-13-1297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.