Abstract
Previously, certain experiments have suggested that density-regulated re-initiation and release factor (DENR) could serve important roles in cancer, however, to the best of our knowledge, a comprehensive analysis of DENR and its association with cancer patient survival is lacking. The aim of the current study was to investigate the expression of DENR in multiple tumour types and to evaluate the effects of DENR on survival in malignancies. Sample expression profiles were downloaded from the Gene Expression Omnibus database. Association between DENR expression and clinicopathological features was analysed by Chi-square tests. The effects of DENR on survival were evaluated by Kaplan-Meier analysis. The results of the current study demonstrate that DENR expression was upregulated in nine cancer types. High DENR expression indicated poor prognosis of patients. The results of the present study demonstrated that DENR is highly expressed in multiple tumour types and may be used as a potential prognostic marker and therapeutic target.
Keywords: cancer biomarkers, poor prognosis, density-regulated re-initiation and release factor
Introduction
The density-regulated re-initiation and release factor (DENR) gene has been mapped to the 12q24 region of chromosome 12 in humans (1). DENR encodes a 198 amino acid protein. Deyo et al (1) identified that DENR encodes a novel protein, the expression of which is upregulated in cells cultured at high density, in comparison with those grown at low density. Cell-cell contact is similarly important to immune system function and malignant tumour metastasis (2,3). Certain studies have suggested that alterations in cell density lead to changes in the biology of a cell, including modulated expression of differentiation properties of LLC-PK1 cells (4) and gene transcriptional activity, including MYC proto-oncogene, BHLH transcription factor (5). One potential initiating mechanism for DENR increase is cell contact itself. Reinert et al (6) revealed novel actions of MCT1, the monocarboxylate transporter 1 gene located at chromosome Xq22-24 (7), whose protein product is comprised of an SUI1 domain involving the translation initiation factor, eukaryotic initiation factor 2 (eIF2) (8). Reinert et al (6) identified that MCT1 is a cap-binding protein that recruits and interacts with DENR and subsequently, modifies the mRNA translational profile. Using a yeast two-hybrid method, MCT1 was demonstrated to interact with DENR involving the SUI1 domain, which may play a role in the pathogenesis of human breast carcinoma (6–9). Skabkin et al (10) studied the interactions of Ligatin and MCT-1/DENR and identified related proteins that singly (Ligatin) or jointly (MCT-1 and DENR) elevate efficient eIF2-independent recruitment of Met-tRNA to 40S/mRNA complexes, and demonstrated that MCT-1/DENR could elevate the release of mRNA. A report by Schleich et al (11) suggested that DENR affects the translation of mRNAs involved in cell proliferation. Recently, a study identified that DENR was highly associated with glioma susceptibility (12). Although, to the best of our knowledge, the specific role of DENR has not been elucidated in humans, these studies suggest an important role in cancer. However, a comprehensive analysis of DENR expression and its relation to the survival of cancer patients is lacking.
The current study evaluated the expression of DENR in normal and cancerous tissues in humans. In addition, the current study analysed the significance of DENR in cancer prognosis using publicly available datasets of gene expression coupled with clinical outcomes.
Materials and methods
Data collection
The expression status of DENR in human healthy and tumour tissues was studied using the GEO dataset (http://www.ncbi.nlm.nih.gov/geo/), which included microarray-based experiments measuring mRNA expression. The primary filtering criteria were set to ‘Cancer vs. Normal Analysis’ and ‘Differential Analysis’, and the raw data. (CEL format) of each dataset were obtained online. The following dataset were included in the present study: GSE15852 (13); GSE8671 (14); GSE13911 (15); GSE10072 (16); GSE6344 (17); GSE14520 (18); GSE15471 (19); GSE9844 (20) and GSE5563 (21). A standard data normalisation process was used for all datasets. The platform filtering criterion was set to ‘Affymetrix U133’ to minimise platform variation when detecting differences in DENR expression between cancer types. The Affymetrix U133 platform includes three types of arrays: Human Genome U133A 2.0, Human Genome U133A&B and Human Genome U133 Plus 2.0 array. These arrays differed by the number of probe sets on the chip, but all included DENR. The association between DENR expression and clinicopathological features was analysed, and the relationship between DENR and survival was calculated using the GEO dataset and the TCGA (https://cancergenome.nih.gov/). The following clinicopathological features and survival dataset were included in the present study: GSE15459 (22); GSE30219 (23); GSE27020 (24); and breast, kidney, rectal, pancreatic cancer primary data from TCGA (25). Finally, the expression profiles and clinical information for 3,389 tissue samples from nine common cancer types were downloaded, including breast, tongue, gastric, colorectal, lung, renal, pancreatic and vulvar cancers and hepatocellular carcinoma.
Data pre-processing and representation
Samples were defined as either carcinoma or normal. Datasets from the nine cancer types were normalised by the RMA algorithm (26) using R statistical 3.1 software (https://www.r-project.org/). Expression values of DENR were dichotomised into low- and high-expression groups using the median as a cut-off value.
Data analysis
Data were analysed using R statistical 3.1 software. Paired or unpaired t-tests were used to analyse expression differences between carcinoma and normal groups. Associations between gene expression and clinicopathological characteristics were evaluated by Chi-square test. Overall survival and recurrence-free survival were evaluated by Kaplan-Meier analysis and a log-rank test. GSEA was used to predict the gene sets modulated by DENR. Expression differences were analysed between normal and carcinoma tissues by paired or unpaired t-tests with threshold values of P<0.05. P<0.05 was considered to indicate a statistically significant difference. Bars indicate the median and the interquartile range of the dataset analysis in the present study.
Results
DENR upregulation in multiple cancers
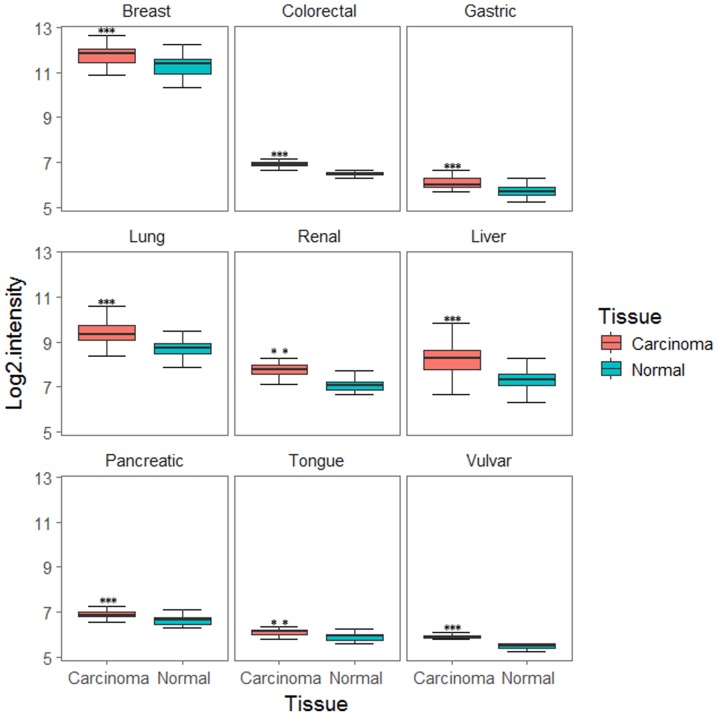
To examine DENR expression in human cancers the tumour expression profiles were downloaded and normalised by RMA. As demonstrated in Fig. 1, DENR expression was significantly increased in the following cancer tissues compared with normal tissues: Breast cancer (43 cancer tissues, 43 normal tissues; GSE15852; P=1.591×10−05), colorectal cancer (32 cancer tissues, 32 normal tissues; GSE8671; P=9.991×10−13), gastric cancer (31 cancer tissues, 31 normal tissues; GSE13911; P=1.179×10−06), lung cancer (58 cancer tissues, 49 normal tissues; GSE10072; P=2.116×10−11), renal cancer (10 cancer tissues, 10 normal tissues; GSE6344; P=0.002771), hepatocellular carcinoma (247 cancer tissues, 239 normal tissues; GSE14520; P<2.2×10−16), pancreatic cancer (39 cancer tissues, 39 normal tissues; GSE15471; P=2.249×10−05), tongue cancer (26 cancer tissues, 12 normal tissues; GSE9844; P=0.006865) and vulvar cancer (9 cancer tissues, 10 normal tissues; GSE5563; P=3.844×10−07).
Figure 1.
Expression of DENR in multiple cancer types. These data were extracted from the Gene Expression Omnibus database. DENR was upregulated in breast, gastric, colorectal, renal, lung, liver, pancreatic, tongue and vulvar cancers. **P<0.01, ***P<0.001 vs. normal, by paired or unpaired t-test. DENR, density-regulated re-initiation and release factor.
Associations between DENR expression and clinicopathological features of patients with nine common cancer types
The data summarised in Table I reveal significant associations between DENR expression and clinicopathological features of patients with multiple cancer types, including hepatocellular carcinoma (primary data from GSE14520), gastric cancer (primary data from GSE15459), lung cancer (primary data from GSE30219), breast cancer (primary data from TCGA), kidney cancer (primary data from TCGA), rectal cancer (primary data from TCGA) and pancreatic cancer (primary data from TCGA). Associations between DENR expression and clinicopathological features for head and neck cancer (primary data from GSE9844) including, age, gender and lymph node metastases were not significant. In addition, data involving clinicopathological features for vulvar cancer in public databases were not available.
Table I.
Correlation between DENR expression and clinicopathological features of patients with nine common cancer types.
| DENR expression | ||||||
|---|---|---|---|---|---|---|
| Type of cancer | Characteristic | Case (n) | High (n) | Low (n) | χ2 value | P-value |
| Hepatocellular carcinoma (primary data from GSE14520) | AFP (ng/ml−1) | |||||
| >300 | 100 | 69 | 31 | 25.407 | <0.001 | |
| ≤300 | 118 | 41 | 77 | |||
| AJCC TNM stage | ||||||
| I | 93 | 41 | 52 | 6.005 | 0.049 | |
| II | 77 | 37 | 40 | |||
| III | 39 | 32 | 17 | |||
| CLIP stage | ||||||
| 0 | 97 | 33 | 64 | 18.308 | <0.001 | |
| 1 | 74 | 47 | 27 | |||
| 2–5 | 48 | 30 | 18 | |||
| Gastric cancer (primary data from GSE15459) | Lauren classification | |||||
| Diffuse | 75 | 29 | 46 | 6.348 | 0.041 | |
| Mixed | 99 | 57 | 42 | |||
| Intestinal | 18 | 10 | 8 | |||
| Subtype | ||||||
| Invasive | 51 | 28 | 23 | 55.099 | <0.001 | |
| Metabolic | 40 | 2 | 28 | |||
| Unstable | 70 | 54 | 16 | |||
| Proliferative | 31 | 12 | 19 | |||
| Lung cancer (primary data from GSE30219) | AJCC T stage | |||||
| I | 166 | 68 | 98 | 11.694 | 0.008 | |
| II | 69 | 42 | 27 | |||
| III | 31 | 20 | 11 | |||
| IV | 21 | 12 | 9 | |||
| Lymph node metastasis | ||||||
| Positive | 93 | 63 | 30 | 16.878 | <0.001 | |
| Negative | 208 | 83 | 115 | |||
| Breast invasive carcinoma (primary data from TCGA) | Age/year | |||||
| ≤65 | 759 | 357 | 402 | 8.461 | 0.004 | |
| >65 | 319 | 181 | 138 | |||
| AJCC T stage | ||||||
| T1 | 279 | 126 | 153 | 13.487 | 0.004 | |
| T2 | 633 | 340 | 293 | |||
| T3 | 138 | 56 | 82 | |||
| T4 | 40 | 25 | 15 | |||
| AJCC N stage | ||||||
| N0 | 515 | 253 | 262 | 9.324 | 0.025 | |
| N1 | 361 | 173 | 188 | |||
| N2 | 120 | 75 | 45 | |||
| N3 | 77 | 34 | 43 | |||
| Kidney chromophobe carcinoma (primary data from TCGA) | AJCC TNM stage | |||||
| I–III | 60 | 27 | 33 | 6.600 | 0.010 | |
| IV | 6 | 6 | 0 | |||
| Rectal adenocarcinoma (primary data from TCGA) | AJCC T stage | |||||
| T1 | 4 | 3 | 1 | 9.423 | 0.024 | |
| T2 | 13 | 9 | 4 | |||
| T3 | 65 | 34 | 31 | |||
| T4 | 10 | 1 | 9 | |||
| Pancreatic adenocarcinoma (primary data from TCGA) | Sex | |||||
| Male | 98 | 60 | 38 | 10.989 | <0.001 | |
| Female | 80 | 29 | 51 | |||
| Head and neck carcinoma and vulvar carcinoma | Clinicopathological feature data for head and neck cancer and vulvar cancer in public databases were either non-significant or not available. | |||||
Data analysed by Chi-square test. DENR, density-regulated re-initiation and release factor; AFP, α-fetoprotein; AJCC, American Joint Committee on Cancer; CLIP, Cancer of the Liver Italian Program; TNM, tumour-node-metastases; T, tumour; N, lymph node; n, number of samples; TCGA, The Cancer Genome Atlas.
The results presented in Table I demonstrate that increased DENR expression was significantly associated with more aggressive phenotypes. This was measured by the American Joint Committee on Cancer (AJCC) classification of malignant tumours including T, N, M and TNM stages (27). DENR expression was significantly higher in advanced AJCC tumour stages than in early tumour stages in multiple cancers including hepatocellular cancer, lung cancer, breast cancer, kidney cancer and rectal cancer. DENR expression was significantly elevated in lymph node metastases compared to that in non-lymph node metastases in lung cancer. High DENR expression was positively associated with histological tumour subtype and Lauren classification (27) in gastric cancer. DENR expression was significantly increased in high α-fetoprotein (AFP) tumours compared with low AFP tumours in hepatocellular cancer.
Association of DENR expression with survival outcome in gastric cancer, hepatocellular carcinoma, lung cancer, kidney cancer and laryngeal cancer
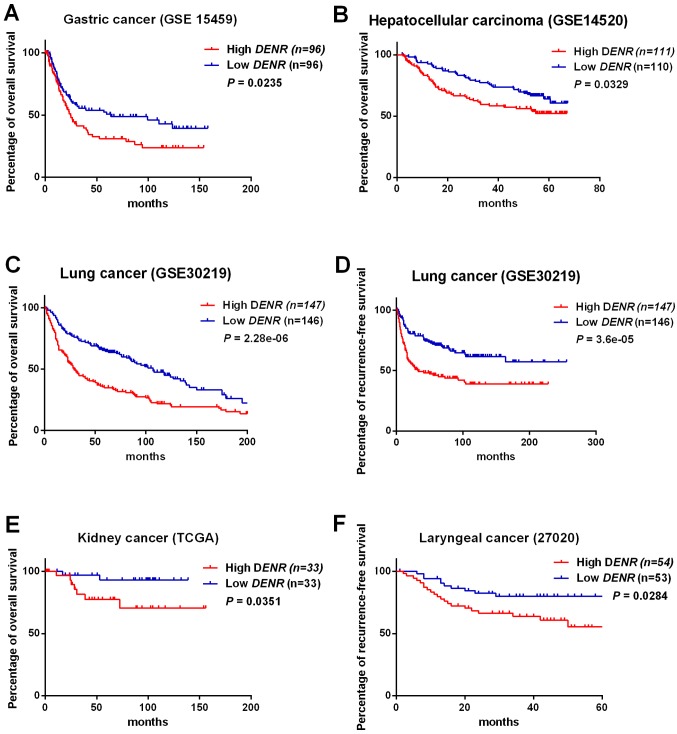
DENR expression (low DENR, n=96; high DENR, n=96) in gastric cancer tissues (primary data from GSE15459) was significantly associated with reduced overall survival (χ2=5.1 and P=0.0235 in log-rank test; Fig. 2A). Expression of DENR (low DENR, n=110; high DENR, n=111) in hepatocellular carcinoma tissues (primary data from GSE14520) was significantly associated with worse overall survival (χ2=4.6 and P=0.0329 in log-rank test; Fig. 2B). In lung cancer tissues (primary data from GSE30219), DENR expression (low DENR, n=146; high DENR, n=147) was significantly associated with poor overall survival (χ2=22.3 and P=2.28×10−06 in log-rank test; Fig. 2C). For lung cancer (primary data from GSE30219), expression of DENR (low DENR, n=146; high DENR, n=147) was significantly associated with diminished recurrence-free survival (χ2=17.1 and P=3.6×10−05 in log-rank test; Fig. 2D). DENR expression (low DENR, n=33; high DENR, n=33) in kidney cancer tissues (primary data from TCGA) was significantly associated with poor overall survival (χ2=4.4 and P=0.0351 in log-rank test; Fig. 2E). DENR levels (low DENR, n=53; high DENR, n=54) in laryngeal cancer tissues (primary data from GSE27020) were significantly associated with reduced recurrence-free survival (χ2=4.8 and P=0.0284 in log-rank test; Fig. 2F).
Figure 2.
Association of DENR expression with patient prognosis in gastric cancer, hepatocellular carcinoma, lung cancer, kidney cancer and laryngeal cancer. The association was investigated by Kaplan-Meier analysis and log-rank test. Overall survival of (A) patients with gastric cancer (data from GSE15459) and (B) patients with hepatocellular carcinoma (data from GSE14520). (C) Overall survival and (D) recurrence-free survival of patients with lung cancer (data from GSE 30219). (E) Overall survival of patients with kidney cancer (data from TCGA) and (F) recurrence-free survival of patients with laryngeal cancer (data from GSE27020). DENR, density-regulated re-initiation and release factor; GSE, gene set enrichment.
In summary, these results demonstrate that high DENR expression is indicative of a poor prognosis for patients with hepatocellular carcinoma, gastric cancer, kidney cancer, laryngeal cancer and lung cancer.
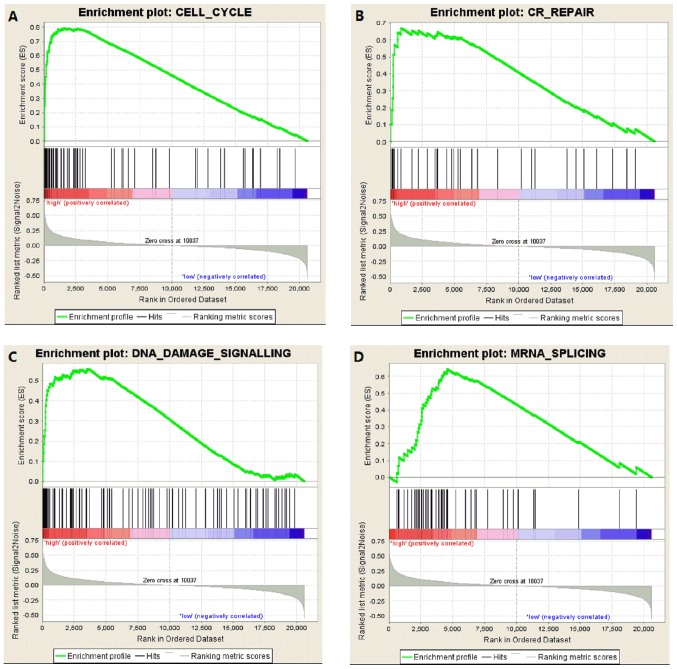
GSEA for lung cancer tissues with high DENR expression
Fig. 3 demonstrates the results of GSEA for lung cancer tissues with high DENR expression (primary data from GSE30219). The curated gene set was used as an example dataset. The results revealed that enriched gene sets were associated with CELL_CYCLE [P<0.001, false discovery rate (FDR)=0.068, enrichment score (ES)=0.793; Fig. 3A], CR_REPAIR (cancer-related DNA repair) (P=0.012, FDR=0.071, ES=0.670; Fig. 3B), DNA_DAMAGE_SIGNALLING (P=0.002, FDR=0.056, ES=0.558; Fig. 3C) and MRNA_SPLICING (P<0.001, FDR=0.003, ES=0.644; Fig. 3D). These findings indicated that DENR regulates cell cycle, cancer-related DNA repair, DNA damage signalling and mRNA splicing activities.
Figure 3.
GSEA for lung cancer tissues with high DENR expression (data from GSE30219). GSEA revealed that the enriched gene sets were associated with (A) cell cycle, (B) CR_REPAIR (cancer DNA related repair), (C) DNA damage signalling and (D) mRNA splicing. GSEA, gene set enrichment analysis; DENR, density-regulated re-initiation and release factor.
Discussion
In the current study, it has been demonstrated that DENR expression is upregulated in different cancer tissues compared with that observed in normal tissues. Deyo et al (1) demonstrated that the DENR gene maps to human chromosome 12. Skabkin et al (28) revealed that the release of tRNA and mRNA is associated with eukaryotic translation initiation factor 1 (eIF1)/eIF1A, Ligatin or MCT-1/DENR. Furthermore, Schleich et al (11) demonstrated that the regulation of the translation of a specific set of mRNAs is associated with DENR. The results of GSEA for lung cancer tissues with high DENR expression identified in the current study established that enriched gene sets were associated with the MRNA_SPLICING categorisation. These findings indicated that DENR regulates mRNA splicing. A previous study disclosed that cells harbour a previously unappreciated translational control system that involves DENR and exerts a key role in tissue development and supporting cellular proliferation (11). The current study demonstrates that DENR is associated with cell cycle, CR_REPAIR (cancer-related DNA repair), DNA damage signalling, disrupted DNA replication, DNA repair and cell cycle progression, which promotes uncontrolled proliferation of tumour cells (29). Similarly, the current study identified that elevated DENR expression was significantly associated with advanced AJCC tumour stages in multiple cancer types including hepatocellular cancer, lung cancer, breast cancer, kidney cancer and rectal cancer.
Advanced tumour stage, tumour size and lymph node metastasis are each indicative of poor prognosis (30,31). High DENR expression was associated with these disadvantageous features in the current study and was also associated with poor overall survival or recurrence-free survival in hepatocellular carcinoma, gastric cancer, kidney cancer, laryngeal cancer and lung cancer. In hepatocellular carcinoma, DENR expression was significantly increased in high AFP tumours compared with low AFP tumours. A recent study suggested that AFP is associated with malignant hepatic tumours (32). The current study revealed high DENR expression was positively associated with the histological tumour invasive subtype and metabolic subtype in gastric cancer. A previous study demonstrated that invasive subtype is associated with malignant gastric cancer and poor prognosis (33). The current study revealed that DENR expression in gastric cancer tissues is significantly associated with overall survival. High DENR expression is indicative of poor prognosis for patients with gastric cancer.
In conclusion, the current study demonstrated that DENR is highly expressed in multiple cancer types and is associated with poor survival of patients with hepatocellular carcinoma, gastric cancer, kidney cancer, laryngeal cancer and lung cancer. Elevated expression of DENR may contribute to tumourigenesis and affect clinical outcome.
Acknowledgements
The present study was performed in accordance with The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) publication policy, as outlined in TCGA data portal (http://cancergenome.nih.gov) and GEO data portal (http://www.ncbi.nlm.nih.gov/geo/).
Funding
Not applicable.
Availability of data and materials
The results of the present study are in whole or part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/) and the GEO dataset (http://www.ncbi.nlm.nih.gov/geo/).
Authors' contributions
DW and SZ conceived and designed the study. Data were curated and analyzed by DW, SZ, LW and PZ. Data collection and software analysis were performed by DW, LW, CR and MW. All authors contributed to the writing of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Deyo JE, Chiao PJ, Tainsky MA. Drp, a novel protein expressed at high cell density but not during growth arrest. DNA Cell Biol. 1998;17:437–447. doi: 10.1089/dna.1998.17.437. [DOI] [PubMed] [Google Scholar]
- 2.Kehry MR, Hodgkin PD. B-cell activation by helper T-cell membranes. Crit Rev Immunol. 1994;14:221–238. doi: 10.1615/CritRevImmunol.v14.i3-4.20. [DOI] [PubMed] [Google Scholar]
- 3.Bussemakers MJ, Schalken JA. The role of cell adhesion molecules and proteases in tumor invasion and metastasis. World J Urol. 1996;14:151–156. doi: 10.1007/BF00186894. [DOI] [PubMed] [Google Scholar]
- 4.Amsler K. Role of cell density/cell-cell contact, and growth state in expression of differentiated properties by the LLC-PK1 cell. J Cell Physiol. 1994;159:331–339. doi: 10.1002/jcp.1041590216. [DOI] [PubMed] [Google Scholar]
- 5.Lee LA, Resar LM, Dang CV. Cell density and paradoxical transcriptional properties of c-Myc and Max in cultured mouse fibroblasts. J Clin Invest. 1995;95:900–904. doi: 10.1172/JCI117741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinert LS, Shi B, Nandi S, Mazan-Mamczarz K, Vitolo M, Bachman KE, He H, Gartenhaus RB. MCT-1 protein interacts with the cap complex and modulates messenger RNA translational profiles. Cancer Res. 2006;66:8994–9001. doi: 10.1158/0008-5472.CAN-06-1999. [DOI] [PubMed] [Google Scholar]
- 7.Prosniak M, Dierov J, Okami K, Tilton B, Jameson B, Sawaya BE, Gartenhaus RB. A novel candidate oncogene, MCT-1, is involved in cell cycle progression. Cancer Res. 1998;58:4233–4237. [PubMed] [Google Scholar]
- 8.Kasperaitis MA, Voorma HO, Thomas AA. The amino acid sequence of eukaryotic translation initiation factor 1 and its similarity to yeast initiation factor SUI1. FEBS Lett. 1995;365:47–50. doi: 10.1016/0014-5793(95)00427-B. [DOI] [PubMed] [Google Scholar]
- 9.Levenson AS, Thurn KE, Simons LA, Veliceasa D, Jarrett J, Osipo C, Jordan VC, Volpert OV, Satcher RL, Jr, Gartenhaus RB. MCT-1 oncogene contributes to increased in vivo tumorigenicity of MCF7 cells by promotion of angiogenesis and inhibition of apoptosis. Cancer Res. 2005;65:10651–10656. doi: 10.1158/0008-5472.CAN-05-0845. [DOI] [PubMed] [Google Scholar]
- 10.Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schleich S, Strassburger K, Janiesch PC, Koledachkina T, Miller KK, Haneke K, Cheng YS, Kuechler K, Stoecklin G, Duncan KE, Teleman AA. DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature. 2014;512:208–212. doi: 10.1038/nature13401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Truvé K, Dickinson P, Xiong A, York D, Jayashankar K, Pielberg G, Koltookian M, Murén E, Fuxelius HH, Weishaupt H, et al. Utilizing the dog genome in the search for novel candidate genes involved in glioma development-genome wide association mapping followed by targeted massive parallel sequencing identifies a strongly associated locus. PLoS Genet. 2016;12:e1006000. doi: 10.1371/journal.pgen.1006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pau Ni IB, Zakaria Z, Muhammad R, Abdullah N, Ibrahim N, Aina Emran N, Hisham Abdullah N, Syed Hussain SN. Gene expression patterns distinguish breast carcinomas from normal breast tissues: The Malaysian context. Pathol Res Pract. 2010;206:223–228. doi: 10.1016/j.prp.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Sabates-Bellver J, Van der Flier LG, de Palo M, Cattaneo E, Maake C, Rehrauer H, Laczko E, Kurowski MA, Bujnicki JM, Menigatti M, et al. Transcriptome profile of human colorectal adenomas. Mol Cancer Res. 2007;5:1263–1275. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- 15.D'Errico M, de Rinaldis E, Blasi MF, Viti V, Falchetti M, Calcagnile A, Sera F, Saieva C, Ottini L, Palli D, et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer. 2009;45:461–469. doi: 10.1016/j.ejca.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 16.Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tun HW, Marlow LA, von Roemeling CA, Cooper SJ, Kreinest P, Wu K, Luxon BA, Sinha M, Anastasiadis PZ, Copland JA. Pathway signature and cellular differentiation in clear cell renal cell carcinoma. PLoS One. 2010;5:e10696. doi: 10.1371/journal.pone.0010696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX, Wang XW. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55:2016–2027. [PubMed] [Google Scholar]
- 20.Ye H, Yu T, Temam S, Ziober BL, Wang J, Schwartz JL, Mao L, Wong DT, Zhou X. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9:69. doi: 10.1186/1471-2164-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santegoets LA, Seters MV, Helmerhorst TJ, Heijmans-Antonissen C, Hanifi-Moghaddam P, Ewing PC, van Ijcken WF, van der Spek PJ, van der Meijden WI, Blok LJ. HPV related VIN: Highly proliferative and diminished responsiveness to extracellular signals. Int J Cancer. 2007;121:759–766. doi: 10.1002/ijc.22769. [DOI] [PubMed] [Google Scholar]
- 22.Ooi CH, Ivanova T, Wu J, Lee M, Tan IB, Tao J, Ward L, Koo JH, Gopalakrishnan V, Zhu Y, et al. Oncogenic pathway combinations predict clinical prognosis in gastric cancer. PLoS Genet. 2009;5:e1000676. doi: 10.1371/journal.pgen.1000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rousseaux S, Debernardi A, Jacquiau B, Vitte AL, Vesin A, Nagy-Mignotte H, Moro-Sibilot D, Brichon PY, Lantuejoul S, Hainaut P, et al. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med. 2013;5:186ra66. doi: 10.1126/scitranslmed.3005723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fountzilas E, Kotoula V, Angouridakis N, Karasmanis I, Wirtz RM, Eleftheraki AG, Veltrup E, Markou K, Nikolaou A, Pectasides D, Fountzilas G. Identification and validation of a multigene predictor of recurrence in primary laryngeal cancer. PLoS One. 2013;8:e70429. doi: 10.1371/journal.pone.0070429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomczak K, Czerwińska P, Wiznerowicz M. The cancer genome atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19A:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 27.Washington K. 7th edition of the AJCC cancer staging manual: Stomach. Ann Surg Oncol. 2010;17:3077–3079. doi: 10.1245/s10434-010-1362-z. [DOI] [PubMed] [Google Scholar]
- 28.Skabkin MA, Skabkina OV, Hellen CU, Pestova TV. Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol Cell. 2013;51:249–264. doi: 10.1016/j.molcel.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 30.Carter CL, Allen C, Henson DE. Relation of tumor size, lymph node status, and survival in 24,740 breast cancer cases. Cancer. 1989;63:181–187. doi: 10.1002/1097-0142(19890101)63:1<181::AID-CNCR2820630129>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 31.Hanrahan EO, Gonzalez-Angulo AM, Giordano SH, Rouzier R, Broglio KR, Hortobagyi GN, Valero V. Overall survival and cause-specific mortality of patients with stage T1a, bN0M0 breast carcinoma. J Clin Oncol. 2007;25:4952–4960. doi: 10.1200/JCO.2006.08.0499. [DOI] [PubMed] [Google Scholar]
- 32.Ryu T, Takami Y, Wada Y, Tateishi M, Matsushima H, Mikagi K, Saitsu H. Double- and triple-positive tumor markers predict early recurrence and poor survival in patients with hepatocellular carcinoma within the Milan criteria and Child-Pugh class A. J Gastrointest Surg. 2017;21:957–966. doi: 10.1007/s11605-017-3394-1. [DOI] [PubMed] [Google Scholar]
- 33.Rugge M, Meggio A, Pennelli G, Piscioli F, Giacomelli L, De Pretis G, Graham DY. Gastritis staging in clinical practice: The OLGA staging system. Gut. 2007;56:631–636. doi: 10.1136/gut.2006.106666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The results of the present study are in whole or part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/) and the GEO dataset (http://www.ncbi.nlm.nih.gov/geo/).