Abstract
To investigate the prospective roles and the clinicopathological application of microRNA-21-5p (miR-21-5p) in hepatocellular carcinoma (HCC), the present review is based on 24 studies and bioinformatics investigation. Firstly, HCC-associated miR-21-5p data were aggregated from literature databases and two public genomic data repositories, including the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA). Potential target genes of miR-21-5p in HCC were identified using TCGA and GEO, Natural Language Processing and 14 online software packages. The oncogenic roles of these target genes was probed for understanding using Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analysis. Hub genes were further investigated by protein-protein interaction network (PPI) analysis. Comprehensive meta-analysis, including 10 microarrays from GEO datasets, 13 literature studies and TCGA-based RNA sequencing data, indicated a reliable diagnostic capacity of miR-21-5p [area under the curve (AUC), 0.887; sensitivity, 0.78% and specificity, 0.79%]. The healthy control group (AUC, 0.926; sensitivity, 0.87% and specificity, 0.82%) demonstrated high diagnostic capacity of miR-21-5p compared with the chronic hepatitis B infection group (AUC, 0.904; sensitivity, 0.75% and specificity, 0.84%). A total of 10 significant enrichment pathways were indicated by KEGG analysis, with cytokine-cytokine receptor interaction exhibiting the highest score. A total of 5 genes, hepatocyte growth factor, forkhead box O1 (FOXO1), thrombospondin 1, estrogen receptor 1 (ESR1) and C-X-C motif chemokine ligand 12 were selected from 39 overlapping genes, according to the PPI network. Target genes were assembled in GO terms associated with ‘response to chemical stimulus’, ‘cell surface’ and ‘growth factor binding’. In particular, low expression of FOXO1 and ESR1 was associated with miR-21-5p expression. In conclusion, upregulated expression of miR-21-5p may be a functional regulator of the metabolism or apoptosis in HCC and a novel tumor marker for the early diagnosis of HCC.
Keywords: hepatocellular carcinoma, miR-21-5p, target genes, diagnostic value, bioinformatics enrichment analysis
Introduction
As the second leading cause of cancer-associated mortality, hepatocellular carcinoma (HCC) is one of the most prevalent types of cancer worldwide (1). The incidence rates of HCC have increased over the past two decades, with >40,000 cases occurring in 2017 in the United States alone (2). The majority of patients with HCC do not receive early treatment and have a poor long-term overall survival (OS) rate, due to malignant features, such as late-stage presentation, metastasis, migration and the lack of HCC symptoms and specific biomarkers (3). Therefore, the diagnosis of HCC prior to reaching the advanced stage of the disease is imperative, in order to achieve improved treatment outcomes for patients. Currently, α-fetoprotein (AFP) is most widely used diagnostic serological marker for HCC (4). However, the expression of AFP is not specific to HCC, since it is also expressed in patients with chronic hepatitis B infection without HCC (5). Nevertheless, AFP-positivity is prevalently manifested in HCC tumors of large size, in the middle to advanced Tumor-Node-Metastasis stage and of high pathological grade compared with AFP-negative status. Therefore, the accuracy of serum AFP for the detection of early-stage HCC is limited (6,7). Molecular targeted drugs have become clinically available, however, only a limited group of patients benefit from this treatment, due to their high price, unclear efficacy and variation in patient response (8,9). Therefore, a more precise marker is urgently required for the early detection of HCC and the development of novel therapeutic approaches.
Intracellular communication, gene dysregulation and environmental factors have been indicated to be involved in the extremely complex underlying mechanisms of occurrence, advancement and metastasis of HCC (10,11). From a molecular perspective, it has been indicated that multiple genes and cellular pathways take part in the origin and development of HCC (12,13). MicroRNAs (miRNAs) are small non-coding endogenous RNAs, ~22 nucleotides in length, serving important roles in proliferation, differentiation, apoptosis, invasion and metastasis, as well as numerous other cellular biological processes (14,15). It has been reported that numerous miRNAs exhibit different expression levels in various types of cancer, indicating their future clinical diagnostic value (16). Among them, miRNA-21 (miR-21) has been indicated to be overexpressed in mammalian cells and has been extensively examined in numerous types of cancer, including esophageal cancer, lung adenocarcinoma, pancreatic adenocarcinoma and tongue squamous cell carcinoma (17–21). A number of studies have reported the diagnostic value of elevated expression of miR-21 in HCC compared with healthy controls or patients with chronic hepatitis B infection (22–25).
High-throughput technologies, including microarrays and RNA sequencing, have served a crucial role in global gene expression research. The application of gene chip-based gene expression profiles has generated extensive information and provided a theoretical insight into the carcinogenesis of HCC. The Gene Expression Omnibus (GEO), as well as The Cancer Genome Atlas (TCGA), are public data portals regarding numerous cancer and non-cancerous samples, providing an unprecedented source of tumor-associated information for the identification of novel biomarkers. The development of natural language processing (NLP) has also been significant, focusing on the interactions between natural languages and computers and containing a large amount of laboratory and clinical data. The underlying biological mechanism of hub genes can be analyzed through Gene Ontology (GO) functional annotation, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment and protein-protein interaction (PPI) network analysis. To the best of our knowledge, the limitation of previous studies regarding miRNA in HCC is their sole focus on serum, plasma, blood or tissue samples (26,27).
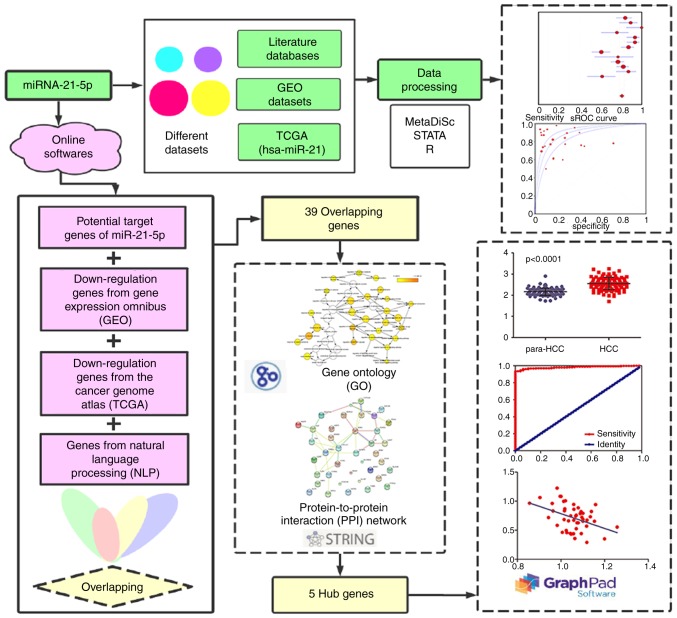
In the present study, a comprehensive analysis was performed by combining literature studies, KEGG analysis, the GEO database, online software platforms and NLP. In addition, functional annotation and signaling pathway analyses of possible genes were performed with GO enrichment analysis, KEGG analysis and PPI networks. Subsequently, genes with the highest PPI scores were further investigated (Fig. 1). The aim of the present study was to further understand the underlying pathogenic mechanism and potential clinical value of miR-21-5p in HCC, in order to provide precise diagnostic insights for HCC.
Figure 1.
Flow chart of the study protocol. HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas; GEO, Gene Expression Omnibus; GO, Gene Ontology; PPI, protein-protein interaction network; NLP, Natural Language Processing; sROC, summary receiver operating characteristic.
Materials and methods
Meta-analysis of the literature regarding the clinical role of miR-21-5p
Data acquisition
HCC-associated miR-21-5p data were collected from 14 databases: PubMed (http://www.ncbi.nlm.nih.gov/pubmed), Embase (www.elsevier.com/solutions/embase-biomedical-research), EBSCO (http://search.ebscohost.com), Wiley Online Library (http://onlinelibrary.wiley.com), Science Direct (https://www.sciencedirect.com/), Web of Science (http://webofknowledge.com/WOS), Cochrane Central Register of Controlled Trials (http://cochranelibrary-wiley.com/cochranelibrary/search?searchRow.searchOptions.searchProducts=clinicalTrialsDoi), Ovid (http://gateway.ovid.com/), LILACS (http://lilacs.bvsalud.org/en/), Google Scholar (scholar.google.com.cn/), Chinese Chong Qing VIP (http://qikan.cqvip.com/), China National Knowledge Infrastructure (CNKI) (http://www.cnki.com.cn), Chinese Wan Fang database (http://med.wanfangdata.com.cn/) and China Biology Medicine disc (http://www.sinomed.ac.cn/). The literature searches of the present study were restricted to human studies. In order to acquire all relevant studies, the references of review papers and other relevant studies were also manually searched. Literature sources were retrieved from July 9 to September 8 2017 without restrictions on language. As miR-21-5p has been also referred to as miR-21, the key words and medical subject headings (MeSH) included the following: (miR-21 OR miRNA-21 OR microRNA-21 OR miR21 OR miRNA21 OR microRNA21 OR ‘miR 21’ OR ‘miRNA-21’ OR ‘microRNA 21’ OR miR-21-5p OR miR-21-5p OR microRNA-21-5p) AND (malignan* OR cancer OR tumor OR tumour OR neoplas* OR carcinoma) AND (hepatocellular OR liver OR hepatic OR HCC).
Inclusion and exclusion criteria
A study was incorporated when it matched the filter criteria: i) An interpretation of the expression level of miR-21-5p in patients with HCC; ii) number of cases was reported in the study; iii) histopathological examination was used for diagnosis, iv) values of expression available directly or indirectly. Additionally, the following conditions caused the exclusion of a study: i) Duplicate publications; ii) failure to specify the control groups; iii) non-human subjects or, iv) lack of original data, such as review, case report or conference note.
Collection and analysis of GEO data
GEO series (GSE) studies were gathered from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The information obtained included the expression profile of miR-21-5p, the fold-change value in HCC and the type of control sample. There was no restriction on the specific type of control group. Cell line assays or assays not considering expression were excluded. The following information was recorded for GSE chips: Main contributor (first author), publishing year, country, sample type, experiment type, the platform of the GSE chips and the number of patients with HCC and control patients. True positive (TP), false positive (FP), false negative (FN), true negative (TN) rates and area under the curve (AUC) were also recorded. All the expression values of miR-21-5p from GEO data were log2 scaled. The level of expression of miR-21-5p between patients with HCC and controls was compared using GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA).
Collection and clinical parameter analysis of TCGA data
TCGA miR-seq data was downloaded from Firebrowse (http://firebrowse.org/), for the entry of liver HCC (‘LIHC’) on April 17th, 2017. Subsequently, the data was manipulated by SPSS 21.0 (IBM Corp., Armonk, NY, USA). Clinical parameters, including age, sex, grade, stage, vascular invasion, history of primary risk factors, hepatitis B virus infection and histological type, were visualized using GraphPad Prism 5.0.
Comprehensive meta-analysis based on studies from literature, GEO and TCGA
Consistent data elements for the integrated meta-analysis were extracted. Following the input of accuracy data, including TP, FP, FN and TN, of each study into MetaDiSc 1.4, the integrated value of sensitivity (SEN), specificity (SPE), positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR) and AUC was calculated (28). Receiver operator characteristic (ROC) curve was applied to evaluate the overall diagnostic accuracy. AUC was calculated for the combined pool of comprehensive data. Depending on the use of a healthy control group or chronic hepatitis B infection control group from the extracted studies, two additional analyses were performed in addition to the meta-analysis of the literature.
Subgroup analyses
In order to evaluate the diagnostic capacity of miR-21-5p from different experimental methods and sample types and figure out the heterogeneity, four subgroup analyses were performed. Diagnostic efficiency of each subgroup was performed with MetaDiSc 1.4.
Potential target gene collection and bioinformatics investigation
Collection of potential target genes
Genes were selected when identified by >3 of the 14 online software packages (miRanda, mirbridge, miRWalk, Microt4, mirTarbase, PITA, miRDB, RNA22, miRNAMap, miRMap, RNAhybrid, PicTar, Targetscan and PolymiRTS). Overlapping genes were identified by the combination of the predicted genes from 14 online software packages, TCGA, GEO and NLP. Overlapping genes were considered to be possible target genes of miR-21-5p.
Functional annotation
GO functional annotation were performed and visualized by Bingo plug-in units from Cytoscape 3.5.0 (Cytoscape, Seattle, WA, USA) (29). GO pathway enrichment analysis consists of three components: i) Biological process (BP); ii) molecular function (MF), and iii) cellular component (CC). A BP term with P<0.005 was considered to be statistically significant. P<0.05 was denoted as the cut-off criteria for CC and MF terms. The functional annotation data were extracted from the downloaded files on August 31th, 2018.
PPI network construction and signal pathway analyses
A bioinformatics database called Search Tool for the Retrieval of Interacting Genes/Proteins (STRING 10.5; http://www.string-db.org) was applied to establish a gene interaction network and excavate notable pathways associated with the potential target genes of miR-21-5p (30). KEGG pathways were determined from the Analysis module and the complete PPI network was drawn on August 31th, 2018. Each internal node in the PPI map represents one potential gene, while the edges between genes indicate a regulatory connection between the genes. These connections can be vividly synthesized and scored. The higher the score, the closer the association between the genes. Genes with high scores may occupy a critical position in the network or act as hub genes. A false discovery rate (FDR) <0.05 value was regarded as the cut-off criteria for pathways by KEGG pathway analysis.
Diagnostic values of hub genes
In order to investigate the association between the five hub genes and the progression of HCC, expression level in para-HCC non-cancerous tissues and HCC tissues (data from TCGA) was compared by single-sample t-test and ROC analysis. The degree of diagnostic value was assessed by the AUC of each gene. Correlation analysis of the hub genes and miR-21-5p expression was further established using Pearson correlation coefficient. GraphPad Prism software was utilized to generate scatter diagrams and visualize associations.
Statistical analysis
Cochran Q-test and the inconsistency index (I2) was calculated to evaluate the heterogeneity. I2 >50% and/or P<0.05 was considered to indicate that heterogeneity existed in the study and the random-effects model was accepted. Otherwise, a fixed-effects model was used for statistical purposes (31,32). The area under the sROC curve was used to evaluate the diagnostic value of miR-21-5p in HCC. The diagnosis information from the literature data and comprehensive data were recorded separately. The diagnostic efficiency of each meta-analysis was performed with MetaDiSc 1.4. P<0.05 was considered to indicate a statistically significant difference. The potential publication bias was investigated with Deeks funnel plot asymmetry test.
Results
Meta-analysis of the literature regarding the clinical role of miR-21-5p
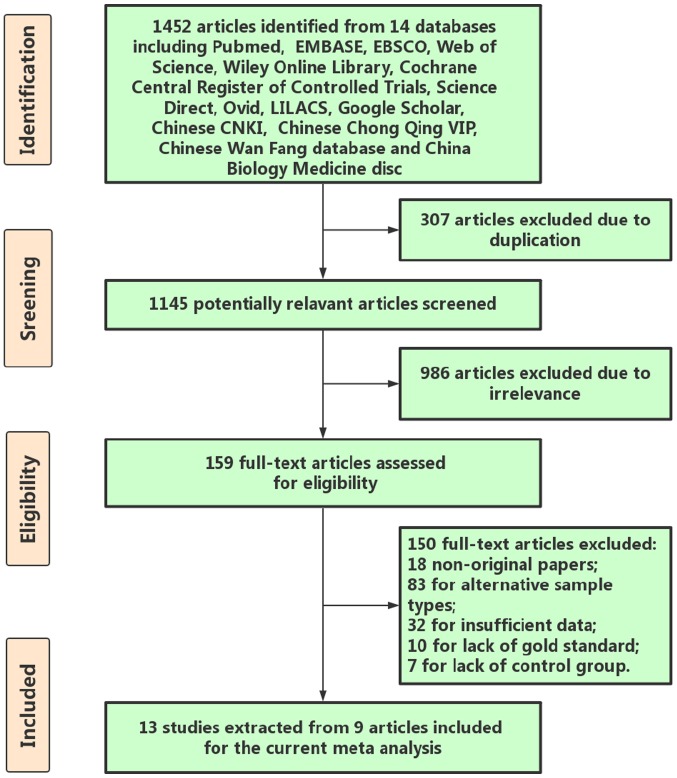
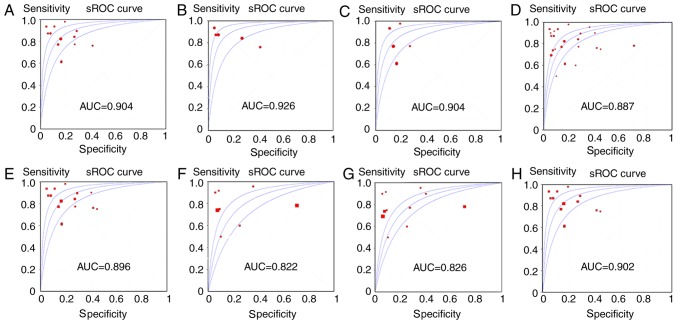
By retrieving and complying with the inclusion and exclusion criteria, 13 eligible studies were incorporated into the meta-analysis of the literature (33–39) (Table I). The flowchart indicated in Fig. 2 illustrates the selection and retrieval process. A total of 7 studies were performed in China, 3 in Egypt and 3 in Japan. The literature data were entered into MetaDiSc 1.4 and it was indicated that there was significant heterogeneity in the pooled estimates of SEN, SPE, PLR, NLR, and DOR, P<0.05. Therefore, the random-effects model was used. The pooled sensitivity and specificity were 0.81 (95% confidence interval; (CI), 0.79–0.83) and 0.82 (95% CI, 0.80–0.85). The forest plots also revealed that the pooled PLR, NLR and DOR were 4.56 (95% CI, 3.29–6.31), 0.21 (95% CI, 0.15–0.30) and 24.56 (95% CI, 13.85–43.54), respectively. The sROC demonstrated an AUC of 0.904 from the 13 studies for the literature meta-analysis (Table II; Fig. 3A). Consequently, the SEN, SPE, PLR, NLR and DOR of the healthy control group were 0.87 (95% CI, 0.83–0.90), 0.82 (95% CI, 0.77–0.86), 6.62 (95% CI, 2.48–15.63), 0.17 (95% CI, 0.10–0.29) and 39.09 (95% CI, 9.98–154.03), respectively (Table II). Correspondingly, the SEN, SPE, PLR, NLR and DOR of the chronic hepatitis B patient group were 0.75 (95% CI, 0.71–0.79), 0.84 (95% CI, 0.78–0.88), 4.36 (95% CI, 3.23–5.88), 0.27 (95% CI, 0.16–0.45) and 18.66 (95% CI, 7.89–44.15), respectively (Table II). The sROC demonstrated an AUC of 0.926 and 0.904 from the healthy control group and chronic hepatitis B patient group (Table II; Fig. 3B and C).
Table I.
Information and clinical data on the 24 included studies.
| Accession | First author | Publication year | Country | Experiment type | Platform | Sample type | Number of patients with HCC | Control patients | TP | FP | FN | TN | AUC | (Refs.) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PMID:21229610 | Xu et al (a) | 2011 | China | RT-qPCR | Serum | 101 | 89 | 85 | 24 | 16 | 65 | 0.870 | (33) | |
| PMID:22403344 | Liu et al (c) | 2012 | China | RT-qPCR | Serum | 57 | 59 | 51 | 17 | 6 | 42 | 0.865 | (34) | |
| PMID:26302751 | Amr et al (b) | 2015 | Egypt | RT-qPCR | Serum | 23 | 17 | 23 | 3 | 0 | 14 | 0.943 | (35) | |
| PMID:26669589 | Zhuang et al (a) | 2015 | China | RT-qPCR | Serum | 52 | 43 | 35 | 17 | 11 | 24 | 0.621 | (36) | |
| PMID:27113935 | El-Tawdi et al (a) | 2016 | Egypt | RT-qPCR | Serum | 78 | 42 | 73 | 2 | 5 | 40 | 0.957 | (14) | |
| PMID:27113935 | El-Tawdi et al (b) | 2016 | Egypt | RT-qPCR | Serum | 78 | 36 | 73 | 4 | 5 | 32 | 0.938 | (14) | |
| PMID:21749846 | Tomimaru et al (a) | 2012 | Japan | RT-qPCR | Plasma | 126 | 50 | 110 | 4 | 16 | 46 | 0.953 | (37) | |
| PMID:21749846 | Tomimaru et al (b) | 2012 | Japan | RT-qPCR | Plasma | 126 | 30 | 77 | 5 | 49 | 25 | 0.773 | (37) | |
| PMID:21283620 | Mizuguchi et al (b) | 2011 | Japan | RT-qPCR | Tissue | 22 | 22 | 17 | 6 | 5 | 16 | 0.780 | (38) | |
| PMID:28477010 | Guo et al (b) | 2017 | China | RT-qPCR | Serum | 175 | 64 | 135 | 9 | 40 | 55 | 0.789 | (24) | |
| PMID:28477010 | Guo et al (c) | 2017 | China | RT-qPCR | Serum | 175 | 278 | 144 | 45 | 31 | 233 | 0.849 | (24) | |
| CNKI | Qin et al (a) | 2013 | China | RT-qPCR | Plasma | 55 | 50 | 48 | 3 | 7 | 47 | 0.956 | (39) | |
| CNKI | Qin et al (b) | 2014 | China | RT-qPCR | Plasma | 55 | 60 | 34 | 10 | 21 | 50 | 0.771 | (39) | |
| GSE22058 | Burchard et al | 2010 | USA | Non-coding RNA profiling by array | GPL10457 | Tissue | 96 | 96 | 71 | 7 | 25 | 89 | 0.877 | (40) |
| GSE10694 | Li et al | 2008 | China | Non-coding RNA profiling by array | GPL6542 | Tissue | 78 | 88 | 61 | 62 | 17 | 26 | 0.506 | (41) |
| GSE41874 | Morita et al | 2013 | Japan | Non-coding RNA profiling by array | GPL7722 | Tissue | 6 | 4 | 3 | 0 | 3 | 4 | 0.625 | – |
| GSE21279 | Hou et al | 2010 | China | Non-coding RNA profiling by array | GPL9052 | Tissue | 4 | 11 | 3 | 1 | 1 | 10 | 0.841 | (42) |
| GSE57555 | Murakami et al | 2015 | Japan | Non-coding RNA profiling by array | GPL16699 | Tissue | 5 | 16 | 3 | 4 | 2 | 12 | 0.625 | (43) |
| GSE67882 | Ghosh et al | 2015 | India | Non-coding RNA profiling by array | GPL10850 | Tissue | 4 | 8 | 4 | 0 | 0 | 8 | 1.000 | – |
| GSE69580 | Hung et al | 2015 | China | Non-coding RNA profiling by array | GPL10850 | Tissue | 5 | 5 | 5 | 0 | 0 | 5 | 1.000 | – |
| GSE12717 | Su et al | 2008 | China | Non-coding RNA profiling by array | GPL7274 | Tissue | 10 | 6 | 10 | 2 | 0 | 4 | 0.800 | (44) |
| GSE54751 | Zhao et al | 2014 | USA | Expression profiling by RT-qPCR | GPL18262 | Tissue | 10 | 10 | 9 | 4 | 1 | 6 | 0.710 | (45) |
| GSE50013 | Shen et al | 2013 | USA | Expression profiling by RT-qPCR | GPL15497 | Plasma | 12 | 8 | 9 | 4 | 3 | 5 | 0.615 | (46) |
| TCGA | Tissue | 375 | 50 | 260 | 3 | 115 | 47 | 0.840 |
GO, Gene Ontology; HCC, hepatocellular carcinoma; AUC, area under the curve; TP, true positive; FP, false positive; FN, false negative; TN, true negative; RT-qPCR, reverse transcriptase-quantitative polymerase chain reaction, (a), healthy control group; (b), control group with chronic hepatitis B infection; (c), non-cancerous control group (healthy and chronic hepatitis B carriers); TCGA, The Cancer Genome Atlas.
Figure 2.
Flow chart indicating the identification of qualified studies for miR-21-5p as a diagnostic marker for HCC. HCC, hepatocellular carcinoma.
Table II.
Summary estimates of diagnostic criteria and the 95% confidence intervals.
| Analysis | Literature studies | Control group: Healthy | Control group: CHB | Integrated studies |
|---|---|---|---|---|
| Sensitivity (95% CI) | 0.81 (0.79–0.83) | 0.87 (0.83–0.90) | 0.75 (0.71–0.79) | 0.78 (0.76–0.80) |
| Specificity (95% CI) | 0.82 (0.80–0.85) | 0.82 (0.77–0.86) | 0.84 (0.78–0.88) | 0.79 (0.77–0.82) |
| Positive LR (95% CI) | 4.56 (3.29–6.31) | 6.22 (2.48–15.63) | 4.36 (3.23–5.88) | 4.46 (2.91–6.84) |
| Negative LR (95% CI) | 0.21 (0.15–0.30) | 0.17 (0.10–0.29) | 0.27 (0.16–0.45) | 0.26 (0.20–0.33) |
| DOR (95% CI) | 24.56 (13.85–43.54) | 39.09 (9.92–154.03) | 18.66 (7.89–44.15) | 20.17 (11.65–34.92) |
| AUC | 0.904 | 0.926 | 0.904 | 0.887 |
CI, confidence interval; LR, likelihood ratio; DOR, diagnostic odds ratio; AUC, area under the curve; CHB, chronic hepatitis B infection.
Figure 3.
The sROC analysis of miR-21-5p expression. (A) Healthy control group. (B) Chronic hepatitis B control group. (C) Integrated studies. (D) RT-qPCR subgroup. (E) Array subgroup. (F) Tissue subgroup. (G) Serum subgroup. (H) Plasma subgroup. sROC, summary receiver operating characteristic; AUC, area underthe curve; RT-qPCR, reverse transcriptase-quantitative polymerase chain reaction; miR, microRNA.
Collection and analysis of GEO data
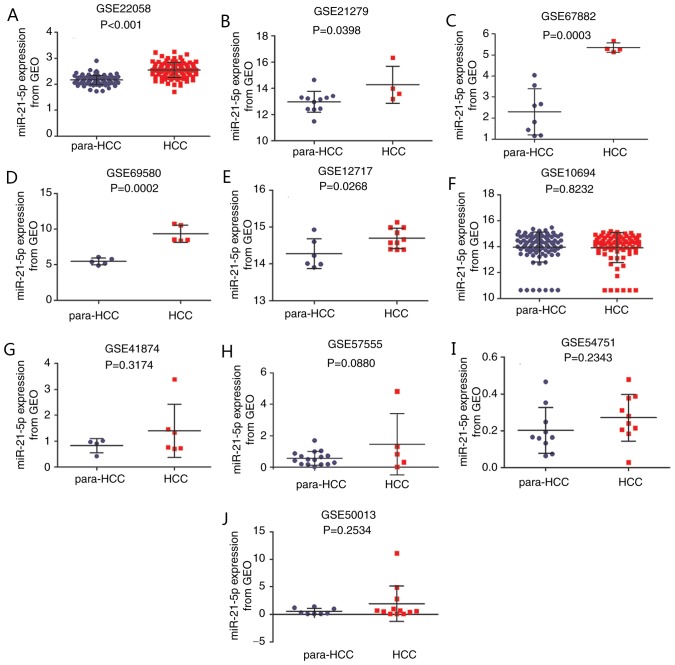
Gene expression in HCC samples and corresponding adjacent para-tumorous liver tissues were compared. A total of 10 GSE chips from GEO datasets were available for the comprehensive meta-analysis (40–46) (Table I). A total of 8 chips were sequenced by array, while 2 chips were analyzed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). With the exception of one chip that used serum, the samples for the other 9 chips were tissue samples. Expression differed among the chips. Significant differences were observed in 5 GSE chips, which indicated increased expression of miR-21-5p in HCC tissues compared with the corresponding adjacent non-cancerous tissues (P<0.05). The results were as follows: 2.55±0.0299 vs. 2.16±0.0176 (GSE22058; P<0.0001; Fig. 4A), 14.28±0.7054 vs. 12.97±0.2419 (GSE21279; P=0.0398; Fig. 4B), 5.34±0.1105 vs. 2.30±0.3856 (GSE67882; P=0.0003; Fig. 4C), 9.35±0.5412 vs. 5.46±0.2113 (GSE69580, P=0.0002, Fig. 4D) and 14.70±0.0877 vs. 14.28±0.1655 (GSE12717; P=0.0268; Fig. 4E). No statistically significant difference between HCC tissues and the adjacent non-cancerous tissues was indicated in the other 5 GSE chips. The results for these chips were as follows: 13.93±0.1315 vs. 13.97±0.1231 (GSE10694; P=0.8232; Fig. 4F), 1.40±0.4191 vs. 0.83±0.1376 (GSE41874; P=0.3174; Fig. 4G), 1.46±0.8684 vs. 0.57±0.1082 (GSE57555; P=0.0880; Fig. 4H), 0.27±0.0401 vs. 0.20±0.0394 (GSE54751; P=0.2343; Fig. 4I) and 1.917±0.9283 vs. 0.55±0.1955 (GSE50013; P=0.2534; Fig. 4J).
Figure 4.
The expression level of miR-21-5p in HCC tissues and adjacent non-cancerous tissues from GEO data. (A) GSE22058. (B) GSE21279. (C) GSE67882. (D) GSE69580. (E) GSE12717. (F) GSE10694. (G) GSE41874. (H) GSE57555. (I) GSE54751. (J) GSE50013. HCC, hepatocellular carcinoma; GEO, Gene Expression Omnibus; miR, microRNA.
Collection of clinicopathological characteristics from TCGA
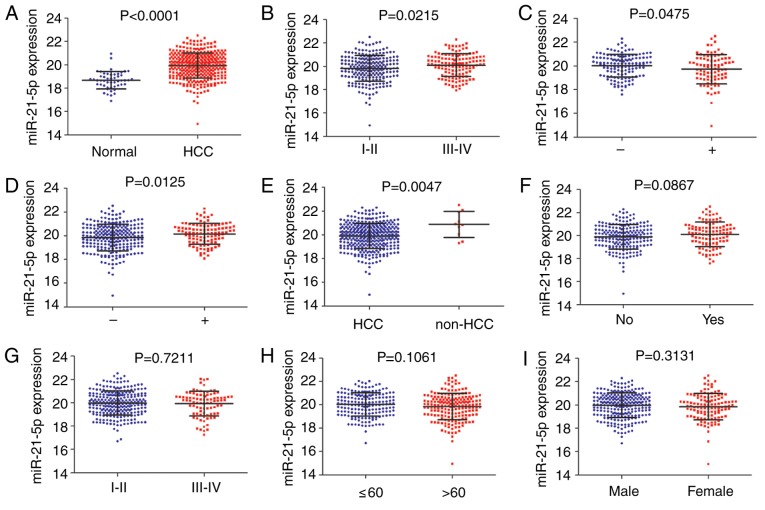
A total of 9 clinicopathological characteristics were associated with miR-21-5p expression level. Significant overexpression of miR-21-5p was observed in HCC tissues (19.95±1.0672), compared with the non-cancerous tissues (18.67±0.7504; P<0.0001; Fig. 5A). Increased expression of miR-21-5p was identified for advanced tumor grade stage III–IV (20.10±0.0840) compared with early tumor grade stage I–II (19.84±0.0735) (P=0.0215; Fig. 5B). miR-21-5p differential expression was associated with history of primary risk factors. Factors, hepatitis B virus (HBV)-infected, alcohol consumption and nonalcoholic fatty liver disease, defined ‘positive’, and ‘negative’ was defined as no history of risk factors. Significant differences were observed between positive (19.73±0.1328) and negative (20.02±0.0811) (P=0.0475; Fig. 5C). HBV-infected patients exhibited a significantly increased expression level (20.15±0.0865) compared with HBV-negative patients (19.84±0.0723) (P=0.0125; Fig. 5D). Increased expression level of miR-21-5p was identified in HCC tumors (20.88±0.3447) compared with other histological types of liver such as hepatocholangiocarcinoma and fibrolamellar carcinoma (19.91±0.0565) (P=0.0047; Fig. 5E). No significant differences in miR-21-5p expression level were associated with vascular invasion, stage, age or sex (Fig. 5F-I).
Figure 5.
The expression level of miR-21-5p according to clinicopathological parameters in HCC from TCGA data. (A) Normal control and HCC. (B) Grade. (C) History of primary risk factors. (D) Hepatitis B virus infection. (E) Histological type. (F) Vascular invasion. (G) Stage. (H) Age. (I) Sex. HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas; miR, microRNA.
Integrated diagnostic value
A total of 24 studies, including 10 microarrays from GEO datasets, 13 literature studies and TCGA-based RNA sequencing data were included for the comprehensive meta-analysis. Significant heterogeneity was identified in all five pooled effects (SEN, SPE, PLR, NLR and DOR) by the Q and I2 tests (all I2 >50% and P<0.05). Accordingly, a random-effects model was employed. As presented in Table II, the overall SEN, SPE, PLR, NLR and DOR of the studies were 0.78 (95% CI, 0.76–0.80), 0.79 (95% CI, 0.77–0.82), 4.46 (95% CI, 2.91–6.84), 0.26 (95% CI, 0.20–0.33) and 20.17 (95% CI, 11.65–34.92), respectively. The AUC value of integrated meta-analysis was 0.887 (Fig. 3D).
Subgroup analyses
Subgroup analyses based on experiment type (RT-qPCR or array) and sample type (tissue or serum/plasma) were conducted. For the experiment type subgroups, the SEN, SPE, PLR, NLR and DOR of the RT-qPCR subgroup were 0.81 (95% CI, 0.79–0.83), 0.82 (95% CI, 0.79–0.84), 4.12 (95% CI, 3.02–5.61), 0.22 (95% CI, 0.15–0.30) and 22.00 (95% CI, 12.73–38.01), respectively. The corresponding 5 values in the array subgroup were 0.77 (95% CI, 0.71–0.83), 0.68 (95% CI, 0.61–0.74), 4.57 (95% CI, 1.30–16.06), 0.39 (95% CI, 0.24–0.64) and 15.70 (95% CI, 3.32–74.29) (Table III). The values of SEN, SPE and DOR were markedly increased in the RT-qPCR subgroup compared with the array subgroup. For the subgroup analysis of different sample types, the SEN, SPE, PLR, NLR and DOR of the tissue subgroup were 0.73 (95% CI, 0.69–0.76), 0.72 (95% CI, 0.67–0.77), 4.35 (95% CI, 1.66–11.39), 0.36 (95% CI, 0.28–0.47) and 15.24 (95% CI, 5.00–45.45). The SEN, SPE, PLR, NLR and DOR in the serum/plasma subgroup were 0.81 (95% CI, 0.79–0.83), 0.82 (95% CI, 0.79–0.85), 4.44 (95% CI, 3.16–6.25), 0.21 (95% CI, 0.15–0.30) and 23.99 (95% CI, 13.27–43.37) (Table III). The value of SEN, SPE, PLR and DOR were increased in the serum/plasma subgroup compared with the tissue subgroup. AUC values from the sROC of the RT-qPCR subgroup, array subgroup, tissue subgroup and serum/plasma subgroup were 0.896, 0.822, 0.826 and 0.902, respectively (Table III; Fig. 3). Nevertheless, no publication bias existed in the meta-analysis according to Deeks test. (P=0.877; Fig. 6).
Table III.
Summary estimates of diagnostic criteria and the 95% confidence intervals of four subgroups.
| Analysis | RT-qPCR subgroup | Non-coding RNA profiling by array subgroup | Tissue subgroup | Serum/plasma subgroup |
|---|---|---|---|---|
| Sensitivity (95% CI) | 0.81 (0.79–0.83) | 0.77 (0.71–0.83) | 0.73 (0.69–0.76) | 0.81 (0.7869–0.83) |
| Specificity (95% CI) | 0.82 (0.79–0.84) | 0.68 (0.61–0.74) | 0.72 (0.67–0.77) | 0.82 (0.79–0.85) |
| Positive LR (95% CI) | 4.12 (3.02–5.61) | 4.57 (1.30–16.06) | 4.35 (1.66–11.39) | 4.44 (3.16–6.25) |
| Negative LR (95% CI) | 0.22 (0.15–0.30) | 0.39 (0.24–0.64) | 0.36 (0.28–0.47) | 0.21 (0.15–0.30) |
| DOR (95% CI) | 22.00 (12.73–38.01) | 15.70 (3.32–74.29) | 15.24 (5.00–46.45) | 23.99 (13.27–43.37) |
| AUC | 0.896 | 0.822 | 0.826 | 0.902 |
CI, confidence interval; LR, likelihood ratio; DOR, diagnostic odds ratio; AUC, area under the curve; RT-qPCR, reverse transcriptase-quantitative polymerase chain reaction.
Figure 6.
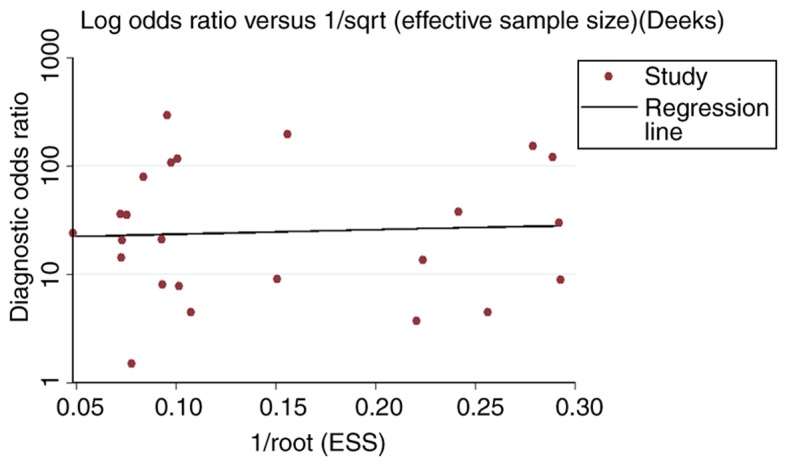
The publication bias of miR-21-5p, based on the comprehensive meta-analysis. ESS, effective sample size; miR, microRNA.
Potential target gene collection and bioinformatics analysis
Collection of prospective target genes
A total of 48,446 target genes were identified from 14 prediction software packages and genes had to be identified ≥3 times to be regarded as potential target genes of miR-21-5p. Reduced-expression genes assembled from TCGA and GEO and genes selected from NLP were integrated to identify overlapping genes. Out of the 10,911 potential target genes, 1,123 reduced-expression genes from TCGA, 2,956 reduced-expression genes from GEO and 1,800 genes were analyzed for intersection. This resulted in 39 target genes attained for the following bioinformatics analyses.
Functional annotation and signal pathway analyses
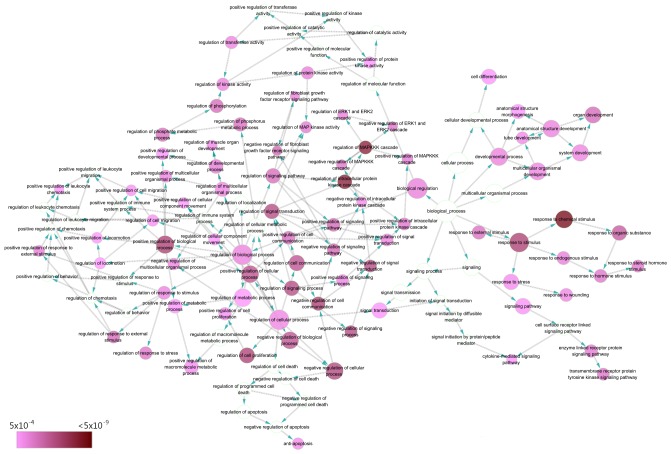
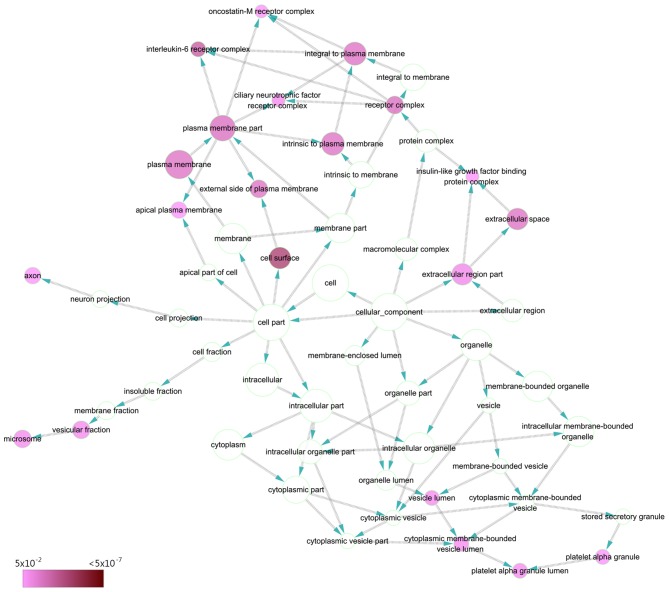
The GO annotation system in DAVID identified the 39 target genes significantly involved in the following biological processes: ‘response to chemical stimulus’ (GO:0042221; P=2.08×10−11), ‘regulation of MAPKKK cascade’ (GO:0043408; P=1.68×10−10) and ‘regulation of intracellular protein kinase cascade’ (GO:0010627; P=2.39×10−10) (Table IV; Fig. 7). As for cellular components, target genes were concentrated in terms of the ‘cell surface’ (GO:0009986; P=9.04×10−7), ‘interleukin-6 receptor complex’ (GO:0005896; P=1.57×10−5) and ‘plasma membrane part’ (GO:0044459; P=8.66×10−5) (Table IV; Fig. 8). The ten significant GO terms were exhibited for BP, CC and MF.
Table IV.
GO functional annotation of the target genes of miR-21-5p.
| GO ID | GO term | Category | Count | P-value | Correlation P-value |
|---|---|---|---|---|---|
| GO:0042221 | Response to chemical stimulus | BP | 20 | 2.08E-11 | 3.0689E-08 |
| GO:0043408 | Regulation of MAPKKK cascade | BP | 9 | 1.68E-10 | 1.173E-07 |
| GO:0010627 | Regulation of intracellular protein kinase cascade | BP | 11 | 2.39E-10 | 1.173E-07 |
| GO:0048518 | Positive regulation of biological process | BP | 21 | 4.33E-09 | 1.2905E-06 |
| GO:0050896 | Response to stimulus | BP | 26 | 4.92E-09 | 1.2905E-06 |
| GO:0010648 | Negative regulation of cell communication | BP | 10 | 5.26E-09 | 1.2905E-06 |
| GO:0042127 | Regulation of cell proliferation | BP | 14 | 6.66E-09 | 1.4006E-06 |
| GO:0048523 | Negative regulation of cellular process | BP | 19 | 1.09E-08 | 0.000002015 |
| GO:0009968 | Negative regulation of signal transduction | BP | 7 | 1.77E-08 | 0.000002835 |
| GO:0023057 | Negative regulation of signaling process | BP | 7 | 1.99E-08 | 0.000002835 |
| GO:0009986 | Cell surface | CC | 8 | 9.04E-07 | 0.00010939 |
| GO:0005896 | Interleukin-6 receptor complex | CC | 2 | 1.57E-05 | 0.00095048 |
| GO:0044459 | Plasma membrane part | CC | 14 | 8.66E-05 | 0.0034849 |
| GO:0043235 | Receptor complex | CC | 4 | 1.15E-04 | 0.0034849 |
| GO:0005886 | Plasma membrane | CC | 19 | 2.19E-04 | 0.0040511 |
| GO:0005615 | Extracellular space | CC | 8 | 2.57E-04 | 0.0040511 |
| GO:0005887 | Integral to plasma membrane | CC | 10 | 2.61E-04 | 0.0040511 |
| GO:0009897 | External side of plasma membrane | CC | 4 | 2.78E-04 | 0.0040511 |
| GO:0031226 | Intrinsic to plasma membrane | CC | 10 | 3.01E-04 | 0.0040511 |
| GO:0044421 | Extracellular region part | CC | 8 | 1.60E-03 | 0.019331 |
| GO:0019838 | Growth factor binding | MF | 6 | 2.10E-07 | 0.000056609 |
| GO:0034875 | Caffeine oxidase activity | MF | 2 | 3.53E-05 | 0.0031751 |
| GO:0033695 | Oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor | MF | 2 | 3.53E-05 | 0.0031751 |
| GO:0016725 | Oxidoreductase activity, acting on CH or CH2 groups | MF | 2 | 2.62E-04 | 0.017694 |
| GO:0005496 | Steroid binding | MF | 3 | 5.66E-04 | 0.023457 |
| GO:0051117 | ATPase binding | MF | 2 | 6.07E-04 | 0.023457 |
| GO:0008083 | Growth factor activity | MF | 4 | 6.09E-04 | 0.023457 |
| GO:0060089 | Molecular transducer activity | MF | 13 | 1.17E-03 | 0.023457 |
| GO:0004871 | Signal transducer activity | MF | 13 | 1.17E-03 | 0.023457 |
| GO:0005125 | Cytokine activity | MF | 4 | 1.35E-03 | 0.023457 |
GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function.
Figure 7.
Directed acyclic graph of pathways identified by GO analysis from the perspective of BP. Each node represents a specific GO term of a BP (P<0.005). BP, biological process; GO, Gene Ontology.
Figure 8.
Directed acyclic graph of pathways identified by GO analysis from the perspective of CC. Each node represents a specific GO term of a CC (P<0.05). CC, cellular component; GO, Gene Ontology.
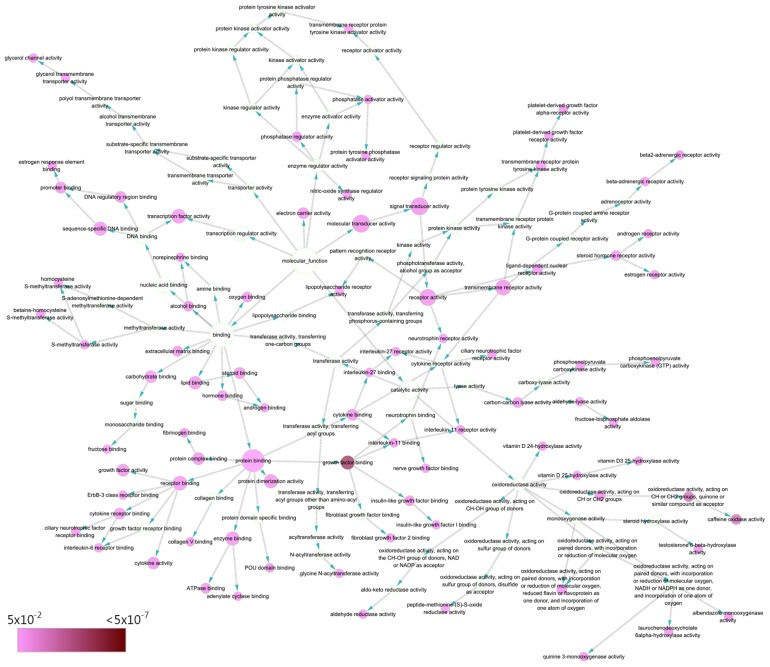
Genes were prominently accumulated in 3 molecular functions, including ‘growth factor binding’ (GO:0019838; P=2.10×10−7), ‘caffeine oxidase activity’ (GO:0034875; P=3.53×10−5) and ‘oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor’ (GO:0033695; P=3.53×10−5 (Table IV; Fig. 9). Additionally, five pathways took precedence in KEGG pathway analysis (FDR<0.01): ‘Cytokine-cytokine receptor interaction’ (hsa04060; FDR=0.00262), ‘Rap1 signaling pathway’ (hsa04015; FDR=0.00393), ‘PI3K-Akt signaling pathway’ (hsa04151; FDR=0.00393), ‘Malaria’ (hsa05144; FDR=0.00659) and ‘MicroRNAs in cancer’ (hsa05206; FDR=0.00894) (Table V).
Figure 9.
Directed acyclic graph of pathways identified by GO analysis from the perspective of MF. Each node represents a specific GO term of a MF (P<0.05). NADH, nicotinamide adenine dinucleotide; NADPH, nicotinamide adenine dinucleotide phosphate; ATP, adenosine triphosphate; MF, molecular function; GO, gene ontology.
Table V.
KEGG pathway analysis of the target genes of miR-21-5p.
| KEGG ID | Name | Count | FDR | Gene symbol |
|---|---|---|---|---|
| hsa04060 | Cytokine-cytokine receptor interaction | 6 | 0.00262 | CCR1, CXCL12, HGF, IL6ST, NGFR, PDGFRA |
| hsa04015 | Rap1 signaling pathway | 5 | 0.00393 | HGF, NGFR, PDGFRA, TEK, THBS1 |
| hsa04151 | PI3K-Akt signaling pathway | 6 | 0.00393 | HGF, NGFR, PDGFRA, TEK, THBS1, TLR4 |
| hsa05144 | Malaria | 3 | 0.00659 | HGF, THBS1, TLR4 |
| hsa05206 | MicroRNAs in cancer | 4 | 0.00894 | PDGFRA, SPRY2, THBS1, ZEB2 |
| hsa05202 | Transcriptional misregulation in cancer | 4 | 0.0126 | DUSP6, FOXO1, IGFBP3, NGFR |
| hsa05215 | Prostate cancer | 3 | 0.0204 | AR, FOXO1, PDGFRA |
| hsa05323 | Rheumatoid arthritis | 3 | 0.0204 | CXCL12, TEK, TLR4 |
| hsa04014 | Ras signaling pathway | 4 | 0.0225 | HGF, NGFR, PDGFRA, TEK |
| hsa05205 | Proteoglycans in cancer | 4 | 0.0225 | ESR1, HGF, THBS1, TLR4 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; FDR, false discovery rate.
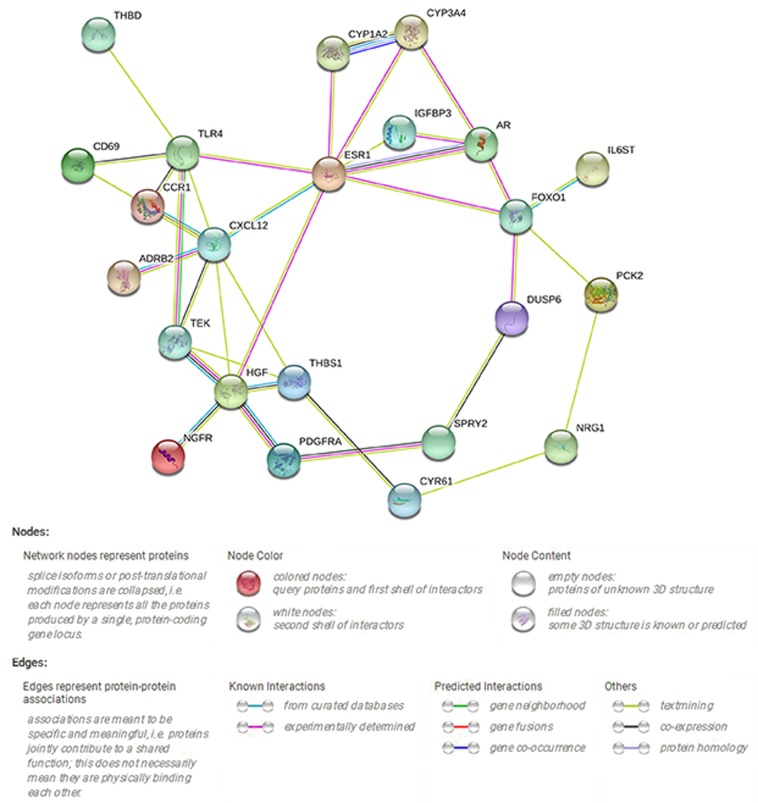
PPI network construction
The PPI network indicating the interactions between 39 target genes of miR-21-5p is demonstrated by 39 nodes and 36 edges in Fig. 10. A total of 5 hub genes were identified according to their scores, including HGF, FOXO1, THBS1, ESR1 and CXCL12.
Figure 10.
The protein-protein interaction network of the target genes of miR-21-5p.
Diagnostic values of the 5 hub genes
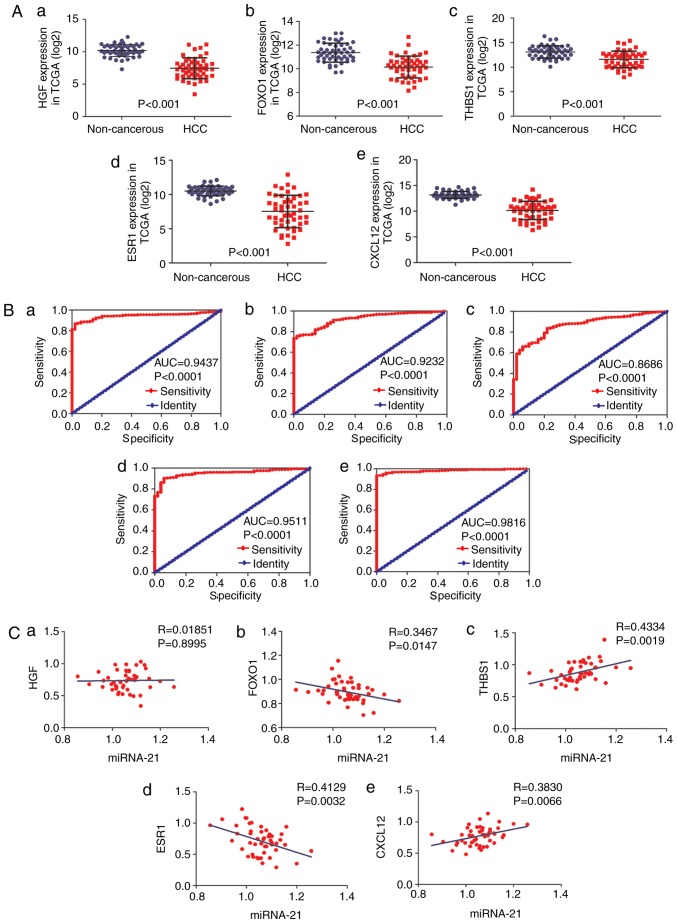
Significantly increased expression of HGF was observed in normal controls (10.19±0.13) compared with patients with HCC (7.45±0.23) (P<0.001; Fig. 11Aa). Similarly, the FOXO1 expression level was significantly increased in normal controls (11.36±0.11) compared with patients with HCC (10.15±0.13) (P<0.001; Fig. 11Ab). Patients with HCC had a significantly decreased THBS1 expression level (11.57±0.24) compared with healthy controls (13.09±0.18) (P<0.001; Fig. 11Ac). Expression of ESR1 was significantly increased in normal controls (10.51±0.10) compared with patients with HCC (7.52±0.33) (P<0.001; Fig. 11Ad). Significantly increased CXCL12 expression level was observed in normal controls (13.18±0.10) compared with patients with HCC (10.15±0.25) P<0.001; Fig. 11Ae). The AUCs of HGF, FOXO1, THBS1, ESR1 and CXCL12 were 0.9437 (95% CI, 0.9220–0.9654; P<0.0001; Fig. 11Ba), 0.9232 (95% CI, 0.8951–0.9513; P<0.0001; Fig. 11Bb), 0.8686 (95% CI, 0.8258–0.9114; P<0.0001; Fig. 11Bc), 0.9511 (95% CI, 0.9299–0.9724; P<0.0001; Fig. 11Bd) and 0.9816 (95% CI, 0.9705–0.9927; P<0.0001; Fig. 11Be), respectively. The results of correlation analysis between the hub genes (HGF, FOXO1, THBS1, ESR1 and CXCL12) and miR-21-5p were as follows: (R=0.01851; P=0.08995; Fig. 11Ca), (R=−0.3467; P=0.0147; Fig. 11Cb), (R=0.4334; P=0.0019; Fig. 11Cc), (R=−0.4129; P=0.0032; Fig. 11Cd), (R=0.3830; P=0.0066; Fig. 11Ce), respectively.
Figure 11.
Expression levels, ROC curves and correlation analysis of the 5 hub genes. (A) Expression levels of (a) HGF, (b) FOXO1, (c) THBS1, (d) ESR1 and (e) CXCL12 in HCC tissues and non-cancerous tissues from TCGA data. (B) ROC curves of (a) HGF, (b) FOXO1, (c) THBS1, (d) ESR1 and (e) CXCL12 in HCC from TCGA data. (C) Correlation analysis of (a) HGF, (b) FOXO1, (c) THBS1, (d) ESR1 and (e) CXCL12 and miR-21-5p from TCGA data. HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas; ROC, receiver operating characteristic; HGF, hepatocyte growth factor; FOXO1, forkhead box 1; THBS1, thrombospondin 1; ESR1, estrogen receptor 1; CXCL12, C-X-C motif chemokine ligand 12.
Discussion
The pathogenesis of HCC has been extensively investigated and there is a consensus that miRNAs may act as vital diagnostic markers for the detection of multiple types of malignancy. In particular, miR-21-5p, located on chromosome 17q23.1, has been reported to be implicated in cancer diagnosis and prognosis (47,48). Numerous previous studies have investigated the underlying functional mechanism of miR-21-5p in numerous types of tumor. Wu et al (49), Zeng et al (50), Qu et al (51) and Markou et al (52) have investigated miR-2-5p in colorectal cancer, gastric cancer, pancreatic cancer and non-small cell lung cancer, respectively. Previous studies have also reported a decrease in tumor-cell proliferation, migration and invasion following the knockdown of miR-21-5p expression in HCC cells (53,54). Tomimaru et al demonstrated that the ROC analysis of plasma miR-21 yielded an AUC of 0.953 (87.3% sensitivity and 92.0% specificity) for differentiating HCC from healthy participants. In addition, when distinguishing between liver cancer and chronic hepatitis, plasma miR-21 yielded an AUC of 0.773 (83.3% specificity and 61.1% sensitivity) (37). This meta-analysis of integrated studies from literature, GEO and TCGA demonstrated appreciable diagnostic significance of miR-21-5p for HCC. Studies based on RT-qPCR and serum/plasma have been identified to have improved diagnostic value compared with the array and tissue studies. Furthermore, the detection of miR-21 expression from serum/plasma is non-invasive, making it more applicable in a clinical setting.
In previous studies, miR-21-5p diagnostic ability has been reported, due to an increased level of expression being correlated with HBV infection, advanced tumor grade, history of risk factors and advanced pathological stage. HBV infection is considered to be a major risk factor for hepatocarcinogenesis (55). However, the underlying mechanism of miRNAs with HBV-associated HCC requires further investigation. Xie et al have identified certain factors contributing to HCC, including long-term HBV infection, high levels of HBV replication, HBV genotype, specific HBV variants, HBV integration and HBV coding proteins (56). Xie et al (57) have summarized the change in expression of numerous miRNAs in HBV infection and have demonstrated the upregulation of miR-21 expression in HBV-associated HCC. In the present study, a more accurate diagnostic effect of miR-21-5p was demonstrated in distinguishing patients with HCC from a healthy population (AUC=0.926) compared with patients from chronic HBV (AUC=0.904). The result further verified the elevated expression of miR-21-5p in HBV-associated HCC.
A total of 5 hub genes (HGF, FOXO1, THBS1, ESR1 and CXCL12) were identified by PPI network construction. It should be noted that FOXO1 and ESR1 were negatively correlated with miR-21-5p. Forkhead box O1 (FOXO1 or FKHR) has been reported to be the target gene of miR-21 in various types of tumor, including large B-cell lymphoma, pancreatic ductal adenocarcinoma and glioblastoma (58–60). The aforementioned studies have identified that overexpression of miR-21 decreased the level of FOXO1. Dong et al have claimed that FOXO1 inhibits the invasion and metastasis of HCC (61). However, the specific function of FOXO1 in HCC has not yet been determined. The role of estrogen receptor 1 (ESR1) in breast cancer has been investigated, but also its clinical relevance in prostate, endometrial and other types of cancer (62). The expression of ESR1 can predict the grade and stage of non-muscle-invasive bladder carcinoma have been reported (63). Dou et al have stated that methylation of ESR1 in HBV-associated HCC may be affected by HBV (64). A study by Hishida et al (65) considered ESR1 as a candidate tumor suppressor gene in HCC. THBS1 may serve a role as an inhibitor of tumor growth, cell migration and neovascularization in lung cancer (66). From the functional annotation analysis in the present study, the hub genes participated in various processes, including ‘response to chemical stimulus’, ‘cell surface’ and ‘growth factor binding’. It was speculated that miR-21-5p expression may be involved in metabolism or apoptotic processes of HCC.
While the present study provides additional evidence supporting the use of miR-21-5p to diagnose HCC, there are limitations that should be acknowledged. Firstly, significant heterogeneity was unavoidable. As noted in the meta-analysis, samples from serum or plasma and the use of RT-qPCR were more precise in diagnosing HCC compared with studies using tissues or the array method, which indicated that the sample type and experiment type may influence the accuracy of diagnosis. Secondly, variation in sample size, sex ratio and age may also have contributed to heterogeneity. Thirdly, miR-21-5p expression levels in patients with HCC from different geographic locations and times may also have an effect. In addition, the results were based solely on data from a range of databases. Therefore, verification experiments to reveal the underlying molecular mechanisms of miR-21-5p are required to confirm the results of the present study, particularly the pathway and functional analysis of miR-21-5p.
In conclusion, an accumulation of data from GEO datasets, TCGA, NLP and literature databases was performed to describe the application potential of miR-21-5p in HCC. Using bioinformatics methods, including pathway enrichment analyses, functional annotation by GO and correlation analysis, the underlying molecular mechanism of miR-21-5p was investigated. Subgroup analyses for different types of experiment and sample were also conducted, in order to compare the diagnostic capacity and identify sources of heterogeneity. FOXO1 and ESR1 were negatively correlated with upregulation of miR-21-5p expression. The highest enriched pathway (cytokine-cytokine receptor interaction) requires further investigation. Prospective studies with large cohorts are required to verify the present study's findings, and to clarify the underlying molecular mechanism of miR-21-5p in HCC.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- HCC
hepatocellular carcinoma
- TCGA
The Cancer Genome Atlas
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PPI
protein-protein interaction network
- NLP
Natural Language Processing
- TP
true positive
- FP
false positive
- TN
true negative
- FN
false negative
- SEN
sensitivity
- SPE
specificity
- PLR
positive likelihood ratio
- NLR
negative likelihood ratio
- DOR
diagnostic odds ratio
- ROC
receiver operating characteristic
- AUC
area under the curve
Funding
The present study was partly supported by the Fund of National Natural Science Foundation of China (grant no. NSFC81260222 and NSFC81060202) and the Innovation Project of Guangxi Graduate Education (grant no. YCSW2017105).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
XZZ and YD conceived and designed the study, acquired data, interpreted the results and drafted the manuscript. HY and GC contributed to the acquisition of funding and support. XZZ and YD analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wallace MC, Preen D, Jeffrey GP, Adams LA. The evolving epidemiology of hepatocellular carcinoma: A global perspective. Expert Rev Gastroenterol Hepatol. 2015;9:765–779. doi: 10.1586/17474124.2015.1028363. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Corvalan AH. Early diagnosis of hepatocellular carcinoma by microRNAs: Shining a light from the genome's ‘dark matter’. Dig Dis Sci. 2012;57:2737–2739. doi: 10.1007/s10620-012-2384-0. [DOI] [PubMed] [Google Scholar]
- 4.Omata M, Cheng A, Kokudo N, Kudo M, Lee JM, Jia J, Tateishi R, Han KH, Chawla YK, Shiina S, et al. Asia-Pacific clinical practice guidelines on the management of hepatocellular carcinoma: A 2017 update. Hepatol Int. 2017;11:317–370. doi: 10.1007/s12072-017-9799-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang N, Feng J, Li ZR, Ming KH, Lei XX, Xu BL. Evaluation of serum α-fetoprotein levels during different infection phases of CHB patients. Clin Lab. 2018;64:43–49. doi: 10.7754/Clin.Lab.2017.170526. [DOI] [PubMed] [Google Scholar]
- 6.Bai DS, Zhang C, Chen P, Jin SJ, Jiang GQ. The prognostic correlation of AFP level at diagnosis with pathological grade, progression, and survival of patients with hepatocellular carcinoma. Sci Rep. 2017;7:12870. doi: 10.1038/s41598-017-12834-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniele B, Bencivenga A, Megna AS, Tinessa V. Alpha-fetoprotein and ultrasonography screening for hepatocellular carcinoma. Gastroenterology. 2004;127(Suppl 1):S108–S112. doi: 10.1053/j.gastro.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 8.Marks EI, Yee NS. Molecular genetics and targeted therapy in hepatocellular carcinoma. Curr Cancer Drug Targets. 2016;16:53–70. doi: 10.2174/1568009615666150916092903. [DOI] [PubMed] [Google Scholar]
- 9.Akhtar B, Muhammad F, Sharif A, Akhtar M, Majeed W. Diverse signaling pathways and current status of molecular targeted treatments for hepatocellular carcinoma. Crit Rev Eukaryot Gene Expr. 2017;27:373–385. doi: 10.1615/CritRevEukaryotGeneExpr.2017021006. [DOI] [PubMed] [Google Scholar]
- 10.Singh A, Kumar R, Pandey A. Hepatocellular carcinoma: Causes, mechanism of progression and biomarkers. Curr Chem Genom Transl Med. 2018;12:9–26. doi: 10.2174/2213988501812010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klungboonkrong V, Das D, McLennan G. Molecular mechanisms and targets of therapy for hepatocellular carcinoma. J Vasc Interv Radiol. 2017;28:949–955. doi: 10.1016/j.jvir.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Herceg Z, Paliwal A. Epigenetic mechanisms in hepatocellular carcinoma: How environmental factors influence the epigenome. Mutat Res. 2011;727:55–61. doi: 10.1016/j.mrrev.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Aigner A. MicroRNAs (miRNAs) in cancer invasion and metastasis: Therapeutic approaches based on metastasis-related miRNAs. J Mol Med. 2011;89:445–457. doi: 10.1007/s00109-010-0716-0. [DOI] [PubMed] [Google Scholar]
- 14.El-Tawdi AH, Matboli M, Shehata HH, Tash F, El-Khazragy N, Azazy Ael-S, Abdel-Rahman O. Evaluation of circulatory RNA-Based biomarker panel in hepatocellular carcinoma. Mol Diagn Ther. 2016;20:265–277. doi: 10.1007/s40291-016-0200-9. [DOI] [PubMed] [Google Scholar]
- 15.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 16.Backes C, Meese E, Keller A. Specific miRNA disease biomarkers in blood, serum and plasma: Challenges and prospects. Mol Diagn Ther. 2016;20:509–518. doi: 10.1007/s40291-016-0221-4. [DOI] [PubMed] [Google Scholar]
- 17.Wurster DH. Sex-chromosome translocations and karyotypes in bovid tribes. Cytogenetics. 1972;11:197–207. doi: 10.1159/000130189. [DOI] [PubMed] [Google Scholar]
- 18.Fendereski M, Zia MF, Shafiee M, Safari F, Saneie MH, Tavassoli M. MicroRNA-196a as a potential diagnostic biomarker for esophageal squamous cell carcinoma. Cancer Invest. 2017;35:78–84. doi: 10.1080/07357907.2016.1254228. [DOI] [PubMed] [Google Scholar]
- 19.Du C, Zheng J, Lu X, Wang Y. Downregulation of miR-18a or miR-328 inhibits the invasion and migration of lung adenocarcinoma A549 cells. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2016;32:1051–1054. (In Chinese) [PubMed] [Google Scholar]
- 20.Cheng RF, Wang J, Zhang JY, Sun L, Zhao YR, Qiu ZQ, Sun BC, Sun Y. MicroRNA-506 is up-regulated in the development of pancreatic ductal adenocarcinoma and is associated with attenuated disease progression. Chin J Cancer. 2016;35:64. doi: 10.1186/s40880-016-0128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun L, Liang J, Wang Q, Li Z, Du Y, Xu X. MicroRNA-137 suppresses tongue squamous carcinoma cell proliferation, migration and invasion. Cell Prolif. 2016;49:628–635. doi: 10.1111/cpr.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan X, Wang ZX, Wang R. MicroRNA-21: A novel therapeutic target in human cancer. Cancer Biol Ther. 2010;10:1224–1232. doi: 10.4161/cbt.10.12.14252. [DOI] [PubMed] [Google Scholar]
- 23.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 24.Guo X, Lv X, Lv X, Ma Y, Chen L, Chen Y. Circulating miR-21 serves as a serum biomarker for hepatocellular carcinoma and correlated with distant metastasis. Oncotarget. 2017;8:44050–44058. doi: 10.18632/oncotarget.17211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eteriia GP. Characteristics of suture materials for bronchoplasty. Vestn Khir Im I I Grek. 1971;107:103–108. (In Russian) [PubMed] [Google Scholar]
- 26.Wang X, Zhang J, Zhou L, Lu P, Zheng ZG, Sun W, Wang JL, Yang XS, Li XL, Xia N, et al. Significance of serum microRNA-21 in diagnosis of hepatocellular carcinoma (HCC): Clinical analyses of patients and an HCC rat model. Int J Clin Exp Pathol. 2015;8:1466–1478. [PMC free article] [PubMed] [Google Scholar]
- 27.Wen Y, Han J, Chen J, Dong J, Xia Y, Liu J, Jiang Y, Dai J, Lu J, Jin G, et al. Plasma miRNAs as early biomarkers for detecting hepatocellular carcinoma. Int J Cancer. 2015;137:1679–1690. doi: 10.1002/ijc.29544. [DOI] [PubMed] [Google Scholar]
- 28.Zamora J, Abraira V, Muriel A, Khan K, Coomarasamy A. Meta-DiSc: A software for meta-analysis of test accuracy data. BMC Med Res Methodol. 2006;6:31. doi: 10.1186/1471-2288-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson D, White IR, Thompson SG. Extending DerSimonian and Laird's methodology to perform multivariate random effects meta-analyses. Stat Med. 2010;29:1282–1297. doi: 10.1002/sim.3602. [DOI] [PubMed] [Google Scholar]
- 32.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58:882–893. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 33.Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, Huang L, Li H, Tan W, Wang C, Lin D. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136–142. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- 34.Liu AM, Yao TJ, Wang W, Wong KF, Lee NP, Fan ST, Poon RT, Gao C, Luk JM. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: A retrospective cohort study. BMJ Open. 2012;2:e000825. doi: 10.1136/bmjopen-2012-000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amr K, Ezzat W, Elhosary Y, Hegazy A, Fahim H, Kamel R. The potential role of miRNAs 21 and 199-a in early diagnosis of hepatocellular carcinoma. Gene. 2016;575:66–70. doi: 10.1016/j.gene.2015.08.038. [DOI] [PubMed] [Google Scholar]
- 36.Zhuang C, Jiang W, Huang D, Xu L, Yang Q, Zheng L, Wang X, Hu L. Serum miR-21, miR-26a and miR-101 as potential biomarkers of hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2016;40:386–396. doi: 10.1016/j.clinre.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, Marubashi S, Tanemura M, Tomokuni A, Takemasa I, Umeshita K, et al. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol. 2012;56:167–175. doi: 10.1016/j.jhep.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 38.Mizuguchi Y, Mishima T, Yokomuro S, Arima Y, Kawahigashi Y, Shigehara K, Kanda T, Yoshida H, Uchida E, Tajiri T, Takizawa T. Sequencing and bioinformatics-based analyses of the microRNA transcriptome in hepatitis B-related hepatocellular carcinoma. PLoS ONE. 2011;6:e15304. doi: 10.1371/journal.pone.0015304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin Z, Zhu X, Huang Y. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. Sichuan Med J. 2013;34:1463–1465. [Google Scholar]
- 40.Burchard J, Zhang C, Liu AM, Poon RT, Lee NP, Wong KF, Sham PC, Lam BY, Ferguson MD, Tokiwa G, et al. microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol Syst Biol. 2010;6:402. doi: 10.1038/msb.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W, Xie L, He X, Li J, Tu K, Wei L, Wu J, Guo Y, Ma X, Zhang P, et al. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int J Cancer. 2008;123:1616–1622. doi: 10.1002/ijc.23693. [DOI] [PubMed] [Google Scholar]
- 42.Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, Qin L, Wu X, Zheng Y, Yang Y, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer cell. 2011;19:232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 43.Murakami Y, Kubo S, Tamori A, Itami S, Kawamura E, Iwaisako K, Ikeda K, Kawada N, Ochiya T, Taguchi YH. Comprehensive analysis of transcriptome and metabolome analysis in intrahepatic cholangiocarcinoma and hepatocellular carcinoma. Sci Rep. 2015;5:16294. doi: 10.1038/srep16294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 45.Zhao L, Zhang Y. miR-342-3p affects hepatocellular carcinoma cell proliferation via regulating NF-kappaB pathway. Biochem Biophys Res Commun. 2015;457:370–377. doi: 10.1016/j.bbrc.2014.12.119. [DOI] [PubMed] [Google Scholar]
- 46.Shen J, Wang A, Wang Q, Gurvich I, Siegel AB, Remotti H, Santella RM. Exploration of genome-wide circulating microRNA in hepatocellular carcinoma: MiR-483-5p as a potential biomarker. Cancer Epidemiol Biomarkers Prev. 2013;22:2364–2373. doi: 10.1158/1055-9965.EPI-13-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clancy C, Joyce MR, Kerin MJ. The use of circulating microRNAs as diagnostic biomarkers in colorectal cancer. Cancer Biomark. 2015;15:103–113. doi: 10.3233/CBM-140456. [DOI] [PubMed] [Google Scholar]
- 48.Labib H, Elantouny N, Ibrahim N, Alnagar A. Upregulation of microRNA-21 is a poor prognostic marker in patients with childhood B cell acute lymphoblastic leukemia. Hematology. 2017;22:392–397. doi: 10.1080/10245332.2017.1292204. [DOI] [PubMed] [Google Scholar]
- 49.Wu Y, Song Y, Xiong Y, Wang X, Xu K, Han B, Bai Y, Li L, Zhang Y, Zhou L. MicroRNA-21 (Mir-21) promotes cell growth and invasion by repressing tumor suppressor PTEN in colorectal cancer. Cell Physiol Biochem. 2017;43:945–958. doi: 10.1159/000481648. [DOI] [PubMed] [Google Scholar]
- 50.Zeng Z, Wang J, Zhao L, Hu P, Zhang H, Tang X, He D, Tang S, Zeng Z. Potential role of microRNA-21 in the diagnosis of gastric cancer: A meta-analysis. PLoS One. 2013;8:e73278. doi: 10.1371/journal.pone.0073278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qu K, Zhang X, Lin T, Liu T, Wang Z, Liu S, Zhou L, Wei J, Chang H, Li K, et al. Circulating miRNA-21-5p as a diagnostic biomarker for pancreatic cancer: Evidence from comprehensive miRNA expression profiling analysis and clinical validation. Sci Rep. 2017;7:1692. doi: 10.1038/s41598-017-01904-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54:1696–1704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- 53.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng YH, Tsao CJ. Emerging role of microRNA-21 in cancer. Biomed Rep. 2016;5:395–402. doi: 10.3892/br.2016.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang HI, Yeh SH, Chen PJ, Iloeje UH, Jen CL, Su J, Wang LY, Lu SN, You SL, Chen DS, et al. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst. 2008;100:1134–1143. doi: 10.1093/jnci/djn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie Y. Hepatitis B virus-associated hepatocellular carcinoma. Adv Exp Med Biol. 2017;1018:11–21. doi: 10.1007/978-981-10-5765-6_2. [DOI] [PubMed] [Google Scholar]
- 57.Xie K, Zhang Y, Liu J, Zeng Y, Wu H. MicroRNAs associated with HBV infection and HBV-related HCC. Theranostics. 2014;4:1176–1192. doi: 10.7150/thno.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song W, Wang L, Wang L, Li Q. Interplay of miR-21 and FoxO1 modulates growth of pancreatic ductal adenocarcinoma. Tumour Biol. 2015;36:4741–4745. doi: 10.1007/s13277-015-3123-x. [DOI] [PubMed] [Google Scholar]
- 59.Lei BX, Liu ZH, Li ZJ, Li C, Deng YF. miR-21 induces cell proliferation and suppresses the chemosensitivity in glioblastoma cells via downregulation of FOXO1. Int J Clin Exp Med. 2014;7:2060–2066. [PMC free article] [PubMed] [Google Scholar]
- 60.Go H, Jang JY, Kim PJ, Kim YG, Nam SJ, Paik JH, Kim TM, Heo DS, Kim CW, Jeon YK. MicroRNA-21 plays an oncogenic role by targeting FOXO1 and activating the PI3K/AKT pathway in diffuse large B-cell lymphoma. Oncotarget. 2015;6:15035–15049. doi: 10.18632/oncotarget.3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong T, Zhang Y, Chen Y, Liu P, An T, Zhang J, Yang H, Zhu W, Yang X. FOXO1 inhibits the invasion and metastasis of hepatocellular carcinoma by reversing ZEB2-induced epithelial-mesenchymal transition. Oncotarget. 2017;8:1703–1713. doi: 10.18632/oncotarget.13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun H, Hou J, Shi W, Zhang L. Estrogen receptor 1 (ESR1) genetic variations in cancer risk: A systematic review and meta-analysis. Clin Res Hepatol Gastroenterol. 2015;39:127–135. doi: 10.1016/j.clinre.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 63.Breyer J, Wirtz R, Laible M, Schlombs K, Erben P, Kriegmair MC, Stoehr R, Eidt S, Denzinger S, Burger M, et al. ESR1, ERBB2, and Ki67 mRNA expression predicts stage and grade of non-muscle-invasive bladder carcinoma (NMIBC) Virchows Arch. 2016;469:547–552. doi: 10.1007/s00428-016-2002-1. [DOI] [PubMed] [Google Scholar]
- 64.Dou CY, Fan YC, Cao CJ, Yang Y, Wang K. Sera DNA methylation of CDH1, DNMT3b and ESR1 promoters as biomarker for the early diagnosis of hepatitis B virus-related hepatocellular carcinoma. Dig Dis Sci. 2016;61:1130–1138. doi: 10.1007/s10620-015-3975-3. [DOI] [PubMed] [Google Scholar]
- 65.Hishida M, Nomoto S, Inokawa Y, Hayashi M, Kanda M, Okamura Y, Nishikawa Y, Tanaka C, Kobayashi D, Yamada S, et al. Estrogen receptor 1 gene as a tumor suppressor gene in hepatocellular carcinoma detected by triple-combination array analysis. Int J Oncol. 2013;43:88–94. doi: 10.3892/ijo.2013.1951. [DOI] [PubMed] [Google Scholar]
- 66.Weng TY, Wang CY, Hung YH, Chen WC, Chen YL, Lai MD. Differential expression pattern of THBS1 and THBS2 in lung cancer: Clinical outcome and a systematic-analysis of microarray databases. PLoS One. 2016;11:e0161007. doi: 10.1371/journal.pone.0161007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.