Abstract
Gastrointestinal cancer is one of the most common causes of mortality globally. The present study examined the influence of cytokine genetic polymorphisms [interleukin (IL)-1B C-31T, IL-1RN VNTR, IL-6 C-634G, IL-8 T-251A, IL-10 T-819C and IL-10 A-1082G] on clinical outcomes in patients with gastrointestinal cancer in palliative care. A total of 59 patients with gastrointestinal cancer who were admitted to Iga City General Hospital were analyzed. Genotyping was conducted using a polymerase chain reaction with confronting two-pair primers. Patients with at least one IL-1RN 2 allele demonstrated a significantly better survival (P=0.0275) while those with IL-6−634 G/G demonstrated a worse survival (P=0.0024). Multivariate analyses using the Cox proportional hazard model revealed that those with at least one IL-1RN 2 allele, IL-6−634 G/G or IL-10−1082 A/G had a significantly elevated adjusted hazard ratio of 9.20 (P=0.014), 41.01 (P=0.001) or 6.49 (P=0.046), respectively, compared with those with each homozygous wild-type polymorphism. In addition, the evaluation of weight loss by genotype revealed the potential influence of IL-10 T-819C genotype (P=0.072). IL-1RN, IL-6 and IL-10 polymorphisms were associated with the survival of patients with gastrointestinal cancer, suggesting the clinical feasibility of genetic testing in patients with gastrointestinal cancer in palliative care.
Keywords: cytokines, single nucleotide polymorphisms, cancer palliative care
Introduction
Gastrointestinal cancer is one of the most common causes of death worldwide. Many patients remain incurable mainly because the disease is detected late, thus requiring palliative medical care. Some of the most important factors to be considered are patients' nutritional status including weight loss, muscle wasting known as sarcopenia, and inflammation (1). Systemic inflammation that results from the tumor existence and progression has been shown to play key roles in these adverse effects, and recent studies have suggested that some genetic polymorphisms involved in immune or inflammatory processes may have an influence on patient outcomes, such as weight loss or survival, through the modulation of these pathways (2,3). Our research group has reported the essential roles of interleukin (IL)-1B, IL-1RN, IL-6, IL-8 and IL-10 in human gastrointestinal cancers, underscoring the importance of these cytokines in the etiology of gastrointestinal cancers and subsequent systemic reactions (4–6). One rural hospital in central Japan, Iga General Hospital in the town of Ninja, provides palliative care including nutritional intervention for such patients with gastrointestinal cancers, and this hospital has accumulated considerable amounts of clinical data since the establishment of an outpatient clinic for cancer chemotherapy and palliative care in 2011. Here, we analyzed the body composition of almost all of the cancer patients who visited this clinic monthly, using the body composition measurement machine, InBody® (https://inbodyusa.com/). Our aim was to evaluate the patients' health conditions including nutritional status to provide more effective palliative care, including nutritional interventions. In this study, we used clinical data from Iga General Hospital to examine the influence of genetic cytokine polymorphisms [IL-1B C-31T, IL-1RN variable number tandem repeats (VNTR), IL-6 C-634G, IL-8 T-251A, IL-10 T-819C, and IL-10 A-1082G] on clinical outcomes of the gastrointestinal cancer patients, to determine how to establish personalized palliative care for gastrointestinal cancer patients based on genetic information.
Materials and methods
Patients and samples
Data from 59 patients with gastrointestinal cancers (consisting of esophageal, gastric, colorectal, biliary, and pancreatic cancers) who visited the outpatient clinic for cancer chemotherapy and palliative care at Iga General Hospital (Iga, Japan) were analyzed. All of the patients underwent palliative chemotherapy (53 patients after surgery, 6 patients who did not undergo surgery) together with the body composition measurement including total body weight, skeletal muscle weight, fat weight, and water weight each time they visited this clinic, which was approximately once a month. All of the patients agreed to provide their clinical data for analyses and their blood for DNA testing after written informed consent. Patients' weight loss was evaluated as a weight loss of >5% body weight (WL5) or that of >10% body weight (WL10) within 6 months after recruitment (which was equal to the time they underwent surgery or the initial course of chemotherapy). This study was approved by the Institutional Review Board of Nagoya University Graduate School of Medicine (Nagoya, Japan; approval no. 2013-0220-10).
DNA sample preparation and genotyping
DNA was extracted from the buffy coat using the Qiagen DNeasy mini kit (Qiagen, Hilden, Germany). Genotyping for IL-1B C-31T, IL-1RN VNTR, IL-6 C-634G, IL-8 T-251A, and IL-10 T-819C polymorphisms was conducted using polymerase chain reaction with confronting two-pair primers (PCR-CTPP) (7), and that for the IL-10 A-1082G polymorphism was conducted using the ABI PRISM 7300 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The sequences of the primers used are the same as previously described (8–10).
Statistical analysis
The χ2 test was used to compare the frequencies in the contingency tables. A Kaplan-Meier curve together with the log-rank test and the Cox proportional hazard model were used for the evaluation of survival-time analyses. The unconditional logistic regression model adjusted for sex and age was used to estimate the risk of weight loss. In addition, to evaluate the effect of measured genotypes on body composition of the participating patients, the β-values for the slope of each body composition element [i.e., the skeletal muscle weight (kg) and extra-cellular water [in proportion to the total body weight)] per allele model, which was the additive model, were calculated using linear regression. All P-values were two-sided, and P<0.05 was considered to indicate a statistically significant difference.
Results
Patient characteristics
Patient characteristics are summarized in Table I. A total of 59 patients with gastrointestinal cancers in palliative care participated in the study, in whom colorectal cancer patients (67.8%) and patients with Union for International Cancer Control (UICC) clinical stage IV (49.2%) predominated, and polymorphisms of IL-1B C-31T, IL-1RN VNTR, IL-6 C-634G, IL-8 T-251A, IL-10 T-819C and IL-10 A-1082G were successfully genotyped for all the patients.
Table I.
Characteristics of the study subjects.
| Variables | n (or years) | |||||||
|---|---|---|---|---|---|---|---|---|
| Age [years (sd)] | 69.1 (8.4) | |||||||
| Sex [n (%)] | ||||||||
| Male | 37 (62.7) | |||||||
| Female | 22 (37.3) | |||||||
| Cancer type [n (%)] | ||||||||
| Esophageal | 2 (3.4) | |||||||
| Stomach | 11 (18.6) | |||||||
| Colorectal | 40 (67.8) | |||||||
| Pancreatic | 5 (8.5) | |||||||
| Billiary | 1 (1.7) | |||||||
| UICC stage [n (%)] | ||||||||
| I | 5 (8.5) | |||||||
| II | 10 (16.9) | |||||||
| III | 15 (25.4) | |||||||
| IV | 29 (49.2) | |||||||
| Genotype frequency | ||||||||
| IL-1B C-31T (rs1143627) | ||||||||
| C/C | 17 (28.8) | |||||||
| C/T | 30 (50.9) | |||||||
| T/T | 12 (20.3) | |||||||
| IL-1RN VNTR | ||||||||
| 2/2 | 1 (1.7) | |||||||
| 2/5 | 3 (5.1) | |||||||
| 5/5 | 53 (89.8) | |||||||
| 5/6 | 2 (3.4) | |||||||
| IL-6 C-634G (rs1800796) | ||||||||
| C/C | 36 (61.0) | |||||||
| C/G | 20 (33.9) | |||||||
| G/G | 3 (5.1) | |||||||
| IL-8 T-251A (rs4073) | ||||||||
| T/T | 23 (39.0) | |||||||
| A/T | 28 (47.4) | |||||||
| A/A | 8 (13.6) | |||||||
| IL-10 T-819C (rs3021097) | ||||||||
| T/T | 26 (44.1) | |||||||
| C/T | 27 (45.7) | |||||||
| C/C | 6 (10.2) | |||||||
| IL-10 A-1082G (rs1800896) | ||||||||
| A/A | 52 (88.1) | |||||||
| A/G | 7 (11.9) | |||||||
| G/G | 0 (0.0) | |||||||
IL, interleukin; UICC, Union for International Cancer Control.
Correlation between cytokine polymorphisms and weight loss
Table II shows the frequency of weight loss stratified by genotype for each cytokine polymorphism. Although a significant increase in the risk of 10% weight loss (WL10) was observed in patients with an increased number of C alleles with the IL-10 T-819C polymorphism (per-allele OR=3.09, P=0.046, by the sex- and age-adjusted additive model), none of the other cytokine polymorphisms showed statistical significance. Additionally, further analyses of change in the body composition by genotype revealed a significant reduction in skeletal muscle weight by the increasing number of the long-repeat (L) alleles of the IL-1RN VNTR polymorphism (P=0.001). Additionally, there was a significant increase in the weight proportion of extra-cellular water (ECW) with an increasing number of C alleles in the IL-10 T-819C polymorphism (P=0.040; Table III).
Table II.
Risk of weight loss by cytokine genotype.
| Polymorphism | WL5 | WL5 (−) | Crude OR | aOR | WL10 | WL10 (−) | Crude OR | aOR |
|---|---|---|---|---|---|---|---|---|
| IL-1B C-31T (rs1143627) | ||||||||
| C/C | 11 | 4 | 1 | 1 | 4 | 11 | 1 | 1 |
| C/T | 17 | 9 | 0.69 | 0.76 | 11 | 15 | 2.02 | 2.07 |
| (0.17–2.79) | (0.18–3.19) | (0.51–8.05) | (0.50–8.53) | |||||
| T/T | 4 | 6 | 0.24 | 0.20 | 4 | 6 | 1.83 | 1.73 |
| (0.04–1.33) | (0.03–1.19) | (0.33–10.10) | (0.31–9.78) | |||||
| IL-1RN VNTR | ||||||||
| L/L | 31 | 17 | 1 | 1 | 18 | 30 | 1 | 1 |
| L/2+2/2a | 1 | 2 | 0.27 | 0.27 | 1 | 2 | 0.83 | 0.87 |
| (0.02–3.25) | (0.02–3.29) | (0.07–9.86) | (0.07–10.45) | |||||
| IL-6 C-634G (rs1800796) | ||||||||
| C/C | 19 | 12 | 1 | 1 | 12 | 19 | 1 | 1 |
| C/G | 11 | 6 | 1.16 | 1.12 | 5 | 12 | 0.66 | 0.69 |
| (0.34–3.96) | (0.32–3.97) | (0.19–2.35) | (0.19–2.54) | |||||
| G/G | 2 | 1 | 1.26 | 1.11 | 2 | 1 | 3.17 | 3.32 |
| (0.10–15.49) | (0.08–14.89) | (0.26–38.84) | (0.25–44.45) | |||||
| IL-8 T-251A (rs4073) | ||||||||
| T/T | 15 | 5 | 1 | 1 | 10 | 10 | 1 | 1 |
| A/T | 14 | 11 | 0.42 | 0.40 | 7 | 18 | 0.39 | 0.30 |
| (0.12–1.53) | (0.10–1.60) | (0.11–1.34) | (0.08–1.20) | |||||
| A/A | 3 | 3 | 0.33 | 0.33 | 2 | 4 | 0.50 | 0.46 |
| (0.05–2.21) | (0.05–2.26) | (0.07–3.38) | (0.06–3.29) | |||||
| IL-10 T-819C (rs3021097) | ||||||||
| T/T | 11 | 10 | 1 | 1 | 5 | 16 | 1 | 1 |
| C/T | 19 | 8 | 2.16 | 2.48 | 12 | 15 | 2.56 | 3.24 |
| (0.66–7.10) | (0.71–8.69) | (0.73–9.01) | (0.84–12.43) | |||||
| C/C | 2 | 1 | 1.82 | 2.08 | 2 | 1 | 6.40 | 8.73 |
| (0.14–23.25) | (0.15–28.05) | (0.47–86.34) | (0.60–127.38) P for trend=0.046 | |||||
| C/T+C/C | 21 | 9 | 2.12 | 2.44 | 14 | 16 | 2.80 | 3.55 |
| (0.67–6.76) | (0.72–8.30) | (0.82–9.62) | (0.94–13.35) P=0.061 | |||||
| IL-10 A-1082G (rs1800896) | ||||||||
| A/A | 30 | 15 | 1 | 1 | 17 | 28 | 1 | 1 |
| A/G | 2 | 4 | 0.25 | 0.22 | 2 | 4 | 0.82 | 0.75 |
| (0.04–1.52) | (0.04–1.43) | (0.14–4.99) | (0.12–4.65) | |||||
WL5 (10), indicates the number of subjects with weight loss more than 5% (or 10%); WL5 (10) (−), indicates those with no weight loss; OR, odds ratio (95% confidence interval in the parenthesis); aOR, adjusted odds ratio (adjusted for age and sex); IL, interleukin.
Those with L/2 and 2/2 genotypes were combined together because of the small number of the subjects in each stratum. Data for weight loss (body weights at two time-points; baseline and 6 months later) was available in 51 subjects.
Table III.
Change in body composition by genotypes.
| Polymorphism | β for the slope of skeletal musclea,b | P | β for the slope of ECWa,b | P |
|---|---|---|---|---|
| IL-1B C-31T (rs1143627) | 0.172 (−0.100, 0.445) | 0.211 | 0.0077 (−0.0963, 0.1116) | 0.883 |
| IL-1RN VNTR | −0.681 (−1.064, −0.299) | 0.001 | 0.0208 (−0.1385, 0.1801) | 0.795 |
| IL-6 C-634G (rs1800796) | −0.040 (−0.367, 0.288) | 0.809 | 0.0007 (−0.0051, 0.0019) | 0.245 |
| IL-8 T-251A (rs4073) | −0.175 (−0.458, 0.107) | 0.219 | 0.0059 (−0.1019, 0.1136) | 0.913 |
| IL-10 T-819C (rs3021097) | −0.117 (−0.410, 0.176) | 0.427 | 0.1119 (−0.0051, 0.2187) | 0.040 |
| IL-10 A-1082G (rs1800896) | −0.267 (−0.858, 0.325) | 0.370 | 0.0036 (−0.2204, 0.2277) | 0.974 |
β-values for the slope of each body composition element per allele (additive) model were calculated using linear regression; ECW, extra-cellular water (in proportion to the total body weight).
Monthly body composition data was available in 58 subjects.
Correlation between cytokine polymorphisms and patients' survival
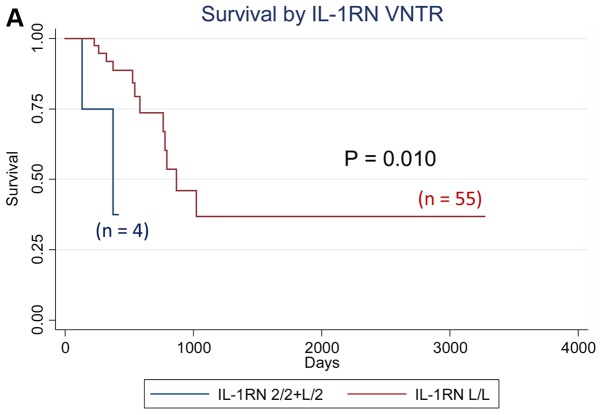
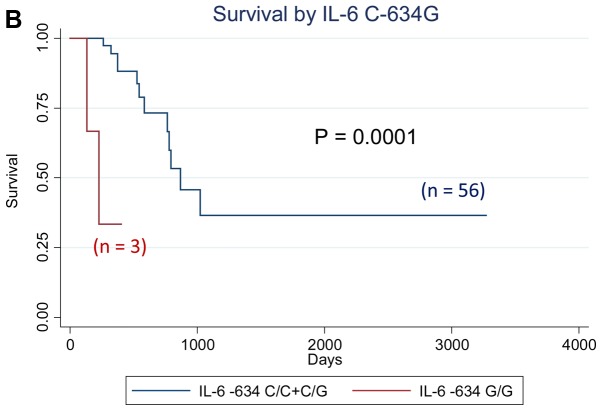
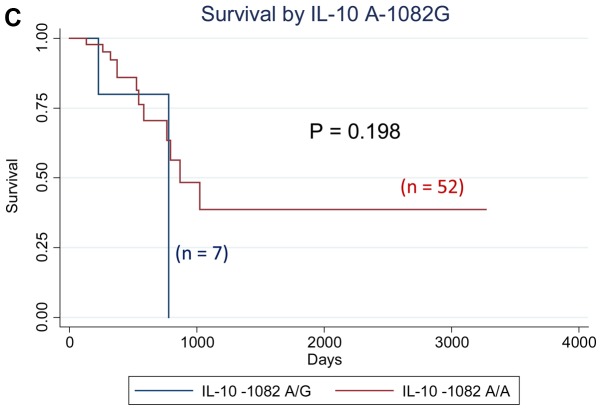
We analyzed the effect of all cytokine polymorphisms examined on patient survival using the Kaplan-Meier method. Those with at least one IL-1RN VNTR 2 allele showed significantly worse survival (P=0.010, logrank test), whereas those with the IL-6−634 G/G genotype showed significantly worse survival (P=0.0001), as shown in Fig. 1. Further multivariate analyses using the Cox proportional hazard model revealed that the IL-10−1082 A/G genotype was an independent prognostic factor for overall survival in patients with gastrointestinal cancer, with the adjusted hazard ratio (aHR) of 6.48 (95% CI: 1.03–40.54, P=0.046) when adjusted for sex, age, and clinical stage, whereas the aHR for the IL-1 RN VNTR [aHR=8.84 (95% CI: 1.48–52.85, P=0.017) for subjects with 2 allele vs. L/L genotype] and that for the IL-6 C-634G [aHR=39.15 (4.25–360.67, P=0.001) for subjects with G/G genotype vs. others] also had significantly elevated aHRs (Table IV). The effects of the IL-1RN VNTR, IL-6 C-634G, and IL-10 A-1082G polymorphisms on the survival of patients with colorectal cancer, which is the most frequent type of cancer, were in the same direction as those of all the patients in this study (data not shown).
Figure 1.
Survival curve by cytokine genotypes. (A) Survival by IL-1RN VNTR; (B) Survival by IL-6 C-634G; (C) Survival by IL-10 A-1082G. P-values indicate Log-rank P-values. IL, interleukin.
Table IV.
Patient prognosis by cytokine genotypes.
| Polymorphism | Comparison groups | Log-rank P | Model | Crude HR | aHR-1 | aHR-2 |
|---|---|---|---|---|---|---|
| IL-1B C-31T (rs1143627) | All 3 genotypes | 0.916 | Additive | 1.03 | 0.89 | 0.74 |
| (0.53–2.00) | (0.39–2.03) | (0.33–1.65) | ||||
| vt. hetero+vt. homo vs. wt. homo | 0.792 | Dominant | 1.16 | 1.02 | 0.93 | |
| (0.39–3.44) | (0.29–3.54) | (0.27–3.21) | ||||
| vt. homo vs. others | 0.889 | Recessive | 0.91 | 0.65 | 0.34 | |
| (0.25–3.30) | (0.14–3.04) | (0.06–1.83) | ||||
| IL-1RN VNTRa | All 3 genotypes | <0.0001 | Additive | 8.29 | 8.28 | 8.88 |
| (2.05–33.47) | (2.05–33.51) | (2.07–38.14) | ||||
| vt. hetero+vt. homo vs. wt. homo | 0.010 | Dominant | 6.83 | 6.61 | 8.84 | |
| (1.25–37.36) | (1.19–36.71) | (1.48–52.85) | ||||
| vt. homo vs. others | <0.0001 | Recessive | – | – | – | |
| IL-6 C-634G (rs1800796) | All 3 genotypes | 0.0005 | Additive | 2.61 | 2.71 | 3.05 |
| (1.03–6.58) | (0.99–7.40) | (1.07–8.67) | ||||
| vt. hetero+vt. homo vs. wt. homo | 0.206 | Dominant | 1.96 | 1.92 | 1.97 | |
| (0.68–5.65) | (0.64–5.73) | (0.66–5.90) | ||||
| vt. homo vs. others | 0.0001 | Recessive | 13.31 | 15.01 | 39.15 | |
| (2.40–73.84) | (2.22–101.41) | (4.25–360.67) | ||||
| IL-8 T-251A (rs4073) | All 3 genotypes | 0.869 | Additive | 0.84 | 0.77 | 0.97 |
| (0.37–1.87) | (0.34–1.77) | (0.40–2.39) | ||||
| vt. hetero+vt. homo vs. wt. homo | 0.798 | Dominant | 0.87 | 0.83 | 0.94 | |
| (0.30–2.50) | (0.29–2.43) | (0.32–2.78) | ||||
| vt. homo vs. others | 0.601 | Recessive | 0.58 | 0.43 | 1.07 | |
| (0.08–4.49) | (0.05–3.77) | (0.11–10.23) | ||||
| IL-10 T-819C (rs3021097) | All 3 genotypes | 0.541 | Additive | 1.10 | 1.07 | 1.06 |
| (0.52–2.36) | (0.49–2.34) | (0.45–2.50) | ||||
| vt. hetero+vt. homo vs. wt. homo | 0.478 | Dominant | 1.49 | 1.50 | 1.24 | |
| (0.49–4.47) | (0.48–4.66) | (0.39–4.00) | ||||
| vt. homo vs. others | 0.578 | Recessive | 0.57 | 0.50 | 0.74 | |
| (0.07–4.34) | (0.06–3.96) | (0.09–5.87) | ||||
| IL-10 A-1082G | A/G vs. A/A | 0.198 | – | 2.66 | 2.98 | 6.48 |
| (rs1800896) | (0.57–12.50) | (0.60–14.73) | (1.03–40.54) |
HR, hazard ratio; aHR, adjusted hazard ratio (aHR-1, adjusted for sex and age; aHR-2, adjusted for age, sex and clinical stage [stage 4]).
For IL-1RN VNTR, L/L, L/2, and 2/2 genotypes were defined as wt. homo, vt. hetero and vt. homo, respectively, where L allele includes 5- and 6-repeat allele.
Discussion
The present study examined the influence of several cytokine polymorphisms on the clinical outcome of gastrointestinal cancer patients, and revealed that polymorphisms in IL-1RN and IL-6 genes have a significant impact on patient survival, based on data obtained from a single institution. The findings obtained were consistent with the previous reports (11,12), which underscored the potential clinical importance of sequence variations of these cytokines in gastrointestinal cancer patients' medical care. IL-1RN encodes the antagonist against IL-1α and IL-1β, and is well-known to play crucial roles in controlling the balance of various proinflammatory processes such as in central nervous system events in the human brain, in the pathogenesis of skin psoriasis, or in the risk of gastrointestinal cancer (13–15). IL-6 is also known to play key roles in tumor-induced inflammation and various subsequent responses including the acute phase responses leading to elevated CRP levels and tumor growth in gastrointestinal cancer (16).
There is an increasing amount of evidence that genetic factors play major roles in the regulation of cytokine production, such as for IL-1RN and IL-6 (9,17,18). Carriers of the IL-1RN 2 polymorphism have been shown to have higher IL-1RN circulating levels than non-carriers (19,20), while some other experimental reports suggest the association of the IL-1RN polymorphism with the amount of IL-1B production, but the directions of the effect seem inconsistent (7). The IL-1RN VNTR polymorphism is consisted of two to six tandem repeats of 86-bp conserved sequence, which is located in the putative protein binding sites and thus may influence gene expression (15,21). Considering that the genes for IL-1RN and IL-1B are both located on chromosome 2q14 within a 360-kb region, it is possible that the genetic variation of IL-1RN VNTR may affect IL-1B levels through linkage disequilibrium on the genome. The present study could not reproduce the previously reported association of IL-1RN VNTR 2 allele with a better prognosis in gastrointestinal cancer patients, which might be explained by the modulation of the IL-1B levels that results from the IL-1RN VNTR polymorphism, or by random error because of the relatively small sample size and disease heterogeneity. The significant trend of reduced skeletal muscle weight associated with a higher number of IL-1RN VNTR L alleles, however, is consistent with a previous report (11), considering the lower IL-1RN circulating levels in L allele carriers and the subsequent exacerbated inflammation induced by IL-1B. For the IL-6 polymorphism, the IL-6−634 G allele is reported to result in higher plasma IL-6 levels, and is associated with a higher risk of cachexia (12). The finding of a worse prognosis in those with the IL-6−634 G/G genotype is consistent with a previous report (12), and is regarded as biologically plausible considering that higher IL-6 levels that result from the IL-6−634 G/G genotype may exacerbate systemic inflammation, thereby leading to the worse prognosis in gastrointestinal cancer patients under palliative care. The association of the presence of the IL-10−819 C allele with advanced stages of colorectal cancers, and with possible weight loss and an increase in ECW, may suggest possible important roles of this variation in the IL-10 gene in the etiology of colorectal cancer patients. Previous studies have shown that IL-10 exerts its anti-inflammatory effects independent of IL-10R, PIK3, or p70S6 kinase (22). Considering that the T allele of the IL-10 T-819C polymorphism was associated with a higher IL-10 expression, these associations of the IL-10−819 C allele with possible weight loss in gastrointestinal cancer patients might be explained by the reduced IL-10 expression as a result of this polymorphism (23). The G allele of IL-10 A-1082G polymorphism was reportedly associated with worse prognosis in patients with gastrointestinal cancers (24). The IL-10 A-1082G polymorphism lies within the promoter region of the IL-10 gene, where the G/G genotype of this polymorphism was shown to be associated with increased levels of IL-10 production (25). Our results in this study verified the clinical impact of the IL-10−1082 G allele on gastrointestinal cancer patient prognosis, suggesting the potential prognostic role of this IL-10 SNP in gastrointestinal cancer patients receiving palliative care. There also exist several other genetic polymorphisms reportedly associated with the risk of cachexia or weight loss in cancer patients to date, such as those in P-selectin (SELP), leptin receptor (LEPR), tumor necrosis factor (TNF), lymphotoxin alpha (LTA) or tumor necrosis factor receptor superfamily member 1A (TNFRSF1A) genes, which might possibly have biological interactions with the genes investigated in the present study, and would become potential candidates for future investigations as well (26,27).
With regard to technical aspects, a strength of this study design is the frequent monitoring of the patients with detailed data, such as body composition, body weight, and laboratory data, which was conducted at least once to twice per month. The limitations, however, would be the smaller sample size, which should be increased in follow-up studies. The present study result may also suggest that genetic testing of gastrointestinal patients may be useful for predicting patients' need for supportive care such as nutritional intervention or possible molecular therapies, such as recombinant IL-1RN (11). It has been reported that elevated resting energy expenditure in several types of cancer patients is associated with the presence of a systemic inflammatory response, which may lead to a worse clinical outcomes in patients (25). Nutritional interventions such as administration of eicosapentaenoic acid (EPA) have been shown to be effective in partially controlling the systemic inflammation in these cancer patients (28). To examine this possibility, further studies taking into consideration the efficacy of nutritional interventions based on the genetic information are possible future research directions.
In conclusion, this is the first study that showed genetic polymorphisms can be used to predict gastrointestinal cancer patient survival. We also confirmed the previously reported roles of IL-RN VNTR and IL-6 C-634G polymorphisms in predicting survival, suggesting the potential feasibility of genetic testing in gastrointestinal cancer patients with palliative care. The sample size of the present study is limited, and the findings should be verified with larger number of patients, to confirm the possible establishment of personalized palliative care for gastrointestinal cancer patients.
Acknowledgements
The authors would like to thank Ms. Yoko Mitsuda and Ms. Keiko Shibata (Nagoya University, Nagoya, Japan) for their technical assistance.
Funding
The present study was supported in part by Grants-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science and Technology, JSPS KAKENHI Grant Number JP (grant no. 25460745).
Availability of data and materials
The datasets used and/or analyzed in the present study are available from the corresponding author on reasonable request.
Authors' contributions
AH, YO, YS and CM contributed to the entire study design, data collection and analyses, and reviewed the final version of the paper critically. YM, KO, KT, RN, YI, HS, HU and MT provided the data from the outpatient clinic to the analysts, provided clinical interpretation of the results, and reviewed the paper critically. SM and AO contributed to the collection of clinical data and genotyping of the patient samples, and reviewed the paper. YT and DCM provided an interpretation of the results from the viewpoint of expert surgical scientists and reviewed the final version of the paper.
Ethical approval and consent to participate
All of the patients agreed to provide their clinical data for analyses and their blood for DNA testing after written informed consent. This study was approved by the Institutional Review Board of Nagoya University Graduate School of Medicine (approval no. 2013-0220-10).
Patient consent for publication
All study participants approved the publication of the present study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Toiyama Y, Miki C, Inoue Y, Tanaka K, Mohri Y, Kusunoki M. Evaluation of an inflammation-based prognostic score for the identification of patients requiring postoperative adjuvant chemotherapy for stage II colorectal cancer. Exp Ther Med. 2011;2:95–101. doi: 10.3892/etm.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan BH, Fearon KC. Cytokine gene polymorphisms and susceptibility to cachexia. Curr Opin Support Palliat Care. 2010;4:243–248. doi: 10.1097/SPC.0b013e32833e4a5d. [DOI] [PubMed] [Google Scholar]
- 3.Tan BH, Ross JA, Kaasa S, Skorpen F, Fearon KC. European Palliative Care Research Collaborative: Identification of possible genetic polymorphisms involved in cancer cachexia: A systematic review. J Genet. 2011;90:165–177. doi: 10.1007/s12041-011-0027-4. [DOI] [PubMed] [Google Scholar]
- 4.Okugawa Y, Miki C, Toiyama Y, Yasuda H, Yokoe T, Saigusa S, Hiro J, Tanaka K, Inoue Y, Kusunoki M. Loss of tumoral expression of soluble IL-6 receptor is associated with disease progression in colorectal cancer. Br J Cancer. 2010;103:787–795. doi: 10.1038/sj.bjc.6605827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toiyama Y, Miki C, Inoue Y, Minobe S, Urano H, Kusunoki M. Loss of tissue expression of interleukin-10 promotes the disease progression of colorectal carcinoma. Surg Today. 2010;40:46–53. doi: 10.1007/s00595-009-4016-7. [DOI] [PubMed] [Google Scholar]
- 6.Konishi N, Miki C, Yoshida T, Tanaka K, Toiyama Y, Kusunoki M. Interleukin-1 receptor antagonist inhibits the expression of vascular endothelial growth factor in colorectal carcinoma. Oncology. 2005;68:138–145. doi: 10.1159/000086768. [DOI] [PubMed] [Google Scholar]
- 7.Hamajima N, Saito T, Matsuo K, Kozaki K, Takahashi T, Tajima K. Polymerase chain reaction with confronting two-pair primers for polymorphism genotyping. Jpn J Cancer Res. 2000;91:865–868. doi: 10.1111/j.1349-7006.2000.tb01026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamajima N, Matsuo K, Saito T, Tajima K, Okuma K, Yamao K, Tominaga S. Interleukin 1 polymorphisms, lifestyle factors and Helicobacter pylori infection. Jpn J Cancer Res. 2001;92:383–389. doi: 10.1111/j.1349-7006.2001.tb01106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suma S, Naito M, Wakai K, Sasakabe T, Hattori Y, Okada R, Kawai S, Hishida A, Morita E, Nakagawa H, et al. Effects of IL6 C-634G polymorphism on tooth loss and their interaction with smoking habits. Oral Dis. 2015;21:807–813. doi: 10.1111/odi.12352. [DOI] [PubMed] [Google Scholar]
- 10.Hamajima N, Katsuda N, Matsuo K, Saito T, Hirose K, Inoue M, Zaki TT, Tajima K, Tominaga S. High anti-Helicobacter pylori antibody seropositivity associated with the combination of IL-8-251TT and IL-10-819TT genotypes. Helicobacter. 2003;8:105–110. doi: 10.1046/j.1523-5378.2003.00130.x. [DOI] [PubMed] [Google Scholar]
- 11.Graziano F, Ruzzo A, Santini D, Humar B, Tonini G, Catalano V, Berardi R, Pizzagalli F, Arduini F, Bearzi I, et al. Prognostic role of interleukin-1beta gene and interleukin-1 receptor antagonist gene polymorphisms in patients with advanced gastric cancer. J Clin Oncol. 2005;23:2339–2345. doi: 10.1200/JCO.2005.01.0819. [DOI] [PubMed] [Google Scholar]
- 12.Zhang D, Zhou Y, Wu L, Wang S, Zheng H, Yu B, Li J. Association of IL-6 gene polymorphisms with cachexia susceptibility and survival time of patients with pancreatic cancer. Ann Clin Lab Sci. 2008;38:113–119. [PubMed] [Google Scholar]
- 13.Carter DB, Deibel MR, Jr, Dunn CJ, Tomich CS, Laborde AL, Slightom JL, Berger AE, Bienkowski MJ, Sun FF, McEwan RN, et al. Purification, cloning, expression and biological characterization of an interleukin-1 receptor antagonist protein. Nature. 1990;344:633–638. doi: 10.1038/344633a0. [DOI] [PubMed] [Google Scholar]
- 14.Arend WP. Interleukin 1 receptor antagonist. A new member of the interleukin 1 family. J Clin Invest. 1991;88:1445–1451. doi: 10.1172/JCI115453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue H, Lin B, Ni P, Xu H, Huang G. Interleukin-1B and interleukin-1 RN polymorphisms and gastric carcinoma risk: A meta-analysis. J Gastroenterol Hepatol. 2010;25:1604–1617. doi: 10.1111/j.1440-1746.2010.06428.x. [DOI] [PubMed] [Google Scholar]
- 16.Fearon KC, Glass DJ, Guttridge DC. Cancer cachexia: Mediators, signaling and metabolic pathways. Cell Metab. 2012;16:153–166. doi: 10.1016/j.cmet.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 17.Furuta T, El-Omar EM, Xiao F, Shirai N, Takashima M, Sugimura H. Interleukin 1beta polymorphisms increase risk of hypochlorhydria and atrophic gastritis and reduce risk of duodenal ulcer recurrence in Japan. Gastroenterology. 2002;123:92–105. doi: 10.1053/gast.2002.34156. [DOI] [PubMed] [Google Scholar]
- 18.Ferrari SL, Ahn-Luong L, Garnero P, Humphries SE, Greenspan SL. Two promoter polymorphisms regulating interleukin-6 gene expression are associated with circulating levels of C-reactive protein and markers of bone resorption in postmenopausal women. J Clin Endocrinol Metab. 2003;88:255–259. doi: 10.1210/jc.2002-020092. [DOI] [PubMed] [Google Scholar]
- 19.Hurme M, Santtila S. IL-1 receptor antagonist (IL-1Ra) plasma levels are co-ordinately regulated by both IL-1Ra and IL-1beta genes. Eur J Immunol. 1998;28:2598–2602. doi: 10.1002/(SICI)1521-4141(199808)28:08<2598::AID-IMMU2598>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 20.Vamvakopoulos J, Green C, Metcalfe S. Genetic control of IL-1beta bioactivity through differential regulation of the IL-1 receptor antagonist. Eur J Immunol. 2002;32:2988–2996. doi: 10.1002/1521-4141(2002010)32:10<2988::AID-IMMU2988>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 21.Vamvakopoulos JE, Taylor CJ, Morris-Stiff GJ, Green C, Metcalfe S. The interleukin-1 receptor antagonist gene: A single-copy variant of the intron 2 variable number tandem repeat (VNTR) polymorphism. Eur J Immunogenet. 2002;29:337–340. doi: 10.1046/j.1365-2370.2002.00319.x. [DOI] [PubMed] [Google Scholar]
- 22.Hamajima N, Katsuda N, Matsuo K, Saito T, Hirose K, Inoue M, Zaki TT, Tajima K, Tominaga S. High anti-Helicobacter pylori antibody seropositivity associated with the combination of IL-8-251TT and IL-10-819TT genotypes. Helicobacter. 2003;8:105–110. doi: 10.1046/j.1523-5378.2003.00130.x. [DOI] [PubMed] [Google Scholar]
- 23.Crawley JB, Williams LM, Mander T, Brennan FM, Foxwell BM. Interleukin-10 stimulation of phosphatidylinositol 3-kinase and p70 S6 kinase is required for the proliferative but not the antiinflammatory effects of the cytokine. J Biol Chem. 1996;271:16357–16362. doi: 10.1074/jbc.271.27.16357. [DOI] [PubMed] [Google Scholar]
- 24.Deans DA, Tan BH, Ross JA, Rose-Zerilli M, Wigmore SJ, Howell WM, Grimble RF, Fearon KC. Cancer cachexia is associated with the IL10-1082 gene promoter polymorphism in patients with gastroesophageal malignancy. Am J Clin Nutr. 2009;89:1164–1172. doi: 10.3945/ajcn.2008.27025. [DOI] [PubMed] [Google Scholar]
- 25.McMillan DC. An inflammation-based prognostic score and its role in the nutrition-based management of patients with cancer. Proc Nutr Soc. 2008;67:257–262. doi: 10.1017/S0029665108007131. [DOI] [PubMed] [Google Scholar]
- 26.Tan BH, Ross JA, Kaasa S, Skorpen F, Fearon KC. European Palliative Care Research Collaborative: Identification of possible genetic polymorphisms involved in cancer cachexia: A systematic review. J Genet. 2011;90:165–177. doi: 10.1007/s12041-011-0027-4. [DOI] [PubMed] [Google Scholar]
- 27.Tan BH, Fladvad T, Braun TP, Vigano A, Strasser F, Deans DA, Skipworth RJ, Solheim TS, Damaraju S, Ross JA, et al. P-selectin genotype is associated with the development of cancer cachexia. EMBO Mol Med. 2012;4:462–471. doi: 10.1002/emmm.201200231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ries A, Trottenberg P, Elsner F, Stiel S, Haugen D, Kaasa S, Radbruch L. A systematic review on the role of fish oil for the treatment of cachexia in advanced cancer: An EPCRC cachexia guidelines project. Palliat Med. 2012;26:294–304. doi: 10.1177/0269216311418709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed in the present study are available from the corresponding author on reasonable request.