Abstract
Hepatocellular carcinoma (HCC) is a heterogeneous malignancy type with limited approaches for treatment. Additionally, inappropriate immune therapy indicates that the understanding the underlying mechanism of HCC is necessary. The aim of the present study was to investigate the influence of a novel circular RNA (circRNA), circRNA of AR-suppressed PABPC1 91 bp (CircARSP91), on immune surveillance induced by natural killer (NK) cells. An in vitro cell cytotoxicity assay was performed to determine the cytotoxicity of NK cells against HCC cells. A specific plasmid for circRNA overexpression was used to establish stable cell lines. Additionally, samples from patients with HCC were analyzed to determine the association between the present in vitro data and those of clinical settings. CircARSP91 could increase the susceptibility of HCC cells to NK cell cytotoxicity. Following screening multiple factors that could influence the activation of NK cells, it was determined that such a phenotype may be caused by upregulating UL16 binding protein 1 (ULBP1) expression in HCC cells at the mRNA and protein levels. Additionally, the data generated from patient samples significantly support a positive association between CircARSP91 and ULBP1. In conclusion, CircARSP91 could enhance innate immune surveillance by strengthening the cytotoxicity of NK cells, implying that circRNA may serve a role in tumor immunity.
Keywords: circular RNA, ULBP1, natural killer cell, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the second leading cause of cancer-associated mortalities globally and accounts for ~80% of all primary liver cancer cases in 2012 (1). The leading risk factor for HCC is cirrhosis arising from viral hepatitis, alcoholic liver disease and non-alcoholic fatty liver disease (2). Thus far, only a limited portion of patients with HCC globally qualify for primary resection or liver transplantation as a HCC treatment, while the remaining patients have no efficient treatment to prolong survival time (3–5). Sorafenib is the only drug approved by the Food and Drug Administration in USA for advanced HCC, but with unsatisfactory effects (6,7).
Such a dilemma causes researchers to continue the search for novel therapeutics that could also improve the early diagnosis rate of HCC (8). However, an improved understanding of HCC initiation and progression is the precondition for future progress in developing novel treatments (9). Recently, circular RNAs (circRNAs) have emerged as novel molecules that mediate tumor biological behavior and function through multiple mechanisms (10–13). However, circRNAs in HCC are only mentioned by a limited number of publications (12–14). Additionally, the notable sex disparity observed in HCC initiation and prognosis is always considered as an important phenotype (15).
Recently, a study reported a functioning circRNA determined in HCC and termed it circRNA of AR-suppressed PABPC1 91 bp (CircARSP91; hsa_circ_0085154) (16). According to their report, CircARSP91 could suppress HCC proliferation with no effect on metastasis. Notably, it was determined in the present study that CircARSP91 in tumor cells could alter the susceptibility to the cytotoxicity of natural killer (NK) cells in HCC. Additionally, the best of our knowledge, no publication has illustrated circRNA function in tumor immune surveillance. During mechanism dissection, it was successfully demonstrated that CircARSP91 may enhance the cytotoxicity of NK cells against HCC via upregulating UL16-binding protein-1 (ULBP1), a killer cell lectin similar to the receptor K1 (NKG2D) ligand that mediates the interaction between tumor cells and NK cells (17).
Materials and methods
Cell culture
MHCC-97h and SK-Hep1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 1% glutamine, and 1% penicillin/streptomycin. NK-92MI cells were maintained in α-minimum essential medium (Invitrogen; Thermo Fisher Scientific, Inc.) with 0.2 mM inositol, 0.1 mM 2-mercaptoethanol, 0.02 mM folic acid, horse serum (Gibco; Thermo Fisher Scientific, Inc.) to a final concentration of 12.5% and FBS to a final concentration of 12.5%, based on American Type Culture Collection (ATCC) guidelines (18). The SK-Hep1 and NK-92MI cells were obtained from ATCC (Manassas, VA, USA), and MHCC-97 h cells were provided by Professor Zheng Shusen of Zhejiang University, as produced in a previous study (19).
In vitro transfection and infection
The overexpression plasmid for CircARSP91, pCDH-CircARSP91 and its empty vector, pCDH, were provided by Professor Cai Xiujun produced as previous described (16). The pCDH-CircARSP91 and empty vector pCDH (#72265; Addgene, Inc., Cambridge, MA, USA) could express green fluorescence protein as a marker for infection. Cells (SK-Hep1 and MHCC-97H) stably expressing CircARSP91 were established based on a previous report (18). Briefly, 293T cells (ATCC) were transfected with pCDH (or pCDH-CircARSP91), psPAX2 (#12260; Addgene, Inc.) packaging plasmid and pMD2.G (#12259; Addgene, Inc.) envelope plasmid for 48 h to obtain the lentivirus supernatant, which was frozen at −80°C for later infection to produce stable polyclonal populations. Small interfering RNAs (siRNAs) targeting CircARSP91 were designed and generated by Guangzhou RiboBio Co., Ltd. (Guangzhou, China). For the siRNA interference assays, cells (SK-Hep1 and MHCC-97H) were transfected using the Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) reverse transfection protocol, according to the manufacturer's protocols (10 µg siRNA transfected for 48 h prior to the subsequent experiments). The control group for the knocking down experiments used scramble sequence. The sequences of siRNA were: si-CircARSP91 #1, 5′-CAGACCTCATCACTCAGAA-3′; si-circARSP91 #2, 5′-CCAGACCTCATCACTCAGA-3′; si-circARSP91 #3, 5′-CCTCATCACTCAGAACCGT-3′; and si-scramble, 5′-CCGAAGGCCGTGTCCCGAG-3′.
In vitro cytotoxicity assays
In vitro cytotoxicity assays were performed using NK-92MI cells and HCC tumor cell lines (SK-Hep1 and MHCC-97H). The tumor cells were seeded at a density of 60% in humidified incubator at 37°C overnight. Subsequently, tumor cells were washed three times with phosphate buffered saline (PBS) and labeled with carboxyfluorescein succinimidyl ester (CFSE; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), according to manufacturer's protocols (20). Following the washing steps (three times with PBS), NK-92MI cells were added in and co-cultured with target cells (SK-Hep1 and MHCC-97H) at effector cell:target cell (E:T) ratios of 5:1 for 4 h at 37°C in an atmosphere containing 5% CO2 in DMEM supplemented with 10% FBS as previously described (21,22). A culture of target cells alone was used as a negative control. Each experimental condition was performed in triplicate. The cells were then washed three times with PBS containing 0.5% bovine serum album (Miltenyi Biotec, Inc., San Diego, CA, USA) and stained with propidium iodide (PI) (Beyotime Institute of Biotechnology, Haimen, China), according to the manufacturer's protocol, prior to data acquisition on a BD LSRII flow cytometer (BD Biosciences, Franklin Lakes, CA, USA). The data were analyzed using FlowJo 10.0 (FlowJo LLC, Ashland, OR, USA). The target cells were identified as CFSE-positive (+), and the effector cells were identified as CFSE-negative (−). The dead target cells were identified as CFSE+/PI+. Spontaneous death was defined as the proportion of dead target cells cultured alone (negative control), and this value was subtracted from the proportion of dead target cells co-cultured with effector cells. Each cytotoxicity assay was repeated in at least three independent experiments. Cytotoxicity was calculated as % cytotoxicity =100×[no. of CFSE+/PI+ cells]/[(no. of CFSE+ cells)+(no. of CFSE+/PI+ cells)].
ELISA
Conditioned media (CM) collected from the interaction between HCC cells and NK-92MI cells were evaluated using a granzyme B ELISA kit (BMS2027; Thermo Fisher Scientific, Inc.). The standard curve was produced to determine the granzyme B concentration. In brief, the CM was collected 2 h later after adding NK-92MI cells to HCC cells (SK-Hep1 and MHCC-97H). The control groups had no NK-92MI cells added. Subsequently, the CM was incubated with an antibody-coated plate (from the ELISA kit) for further detection. All the procedures were performed according to the manufacturer's protocols.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). A total of 1 µg total RNA was subjected to reverse transcription using Superscript III transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.). RT-qPCR was conducted using a Bio-Rad CFX96 system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) with SYBR® Green (Invitrogen; Thermo Fisher Scientific, Inc.) to determine the mRNA expression level of a gene of interest. The expression levels were normalized to that of GAPDH RNA (forward, 5′-TGTGGGCATCAATGGATTTGG-3′ and reverse, 5′-ACACCATGTATTCCGGGTCAAT-3′). The primers used are as follows: Major histocompatibility complex class I polypeptide-related sequence A (MIC)-A, forward, 5′-CTTCAGAGTCATTGGCAGACAT-3′, and reverse, 5′-TGTGGTCACTCGTCCCAACT-3′; MIC-B, forward, 5′-TCTTCGTTACAACCTCATGGTG-3′, and reverse, 5′-TCCCAGGTCTTAGCTCCCAG-3′; ULBP1, forward, 5′-TAAGTCCAGACCTGAACCACA-3′, and reverse, 5′-TCCACCACGTCTCTTAGTGTT-3′; ULBP2, forward, 5′-AGCAACTGCGTGACATTCAG-3′, and reverse, 5′-GCCATCCTATACAGTCTCCCA-3′; ULBP3, forward, 5′-TCTATGGGTCACCTAGAAGAGC-3′, and reverse, 5′-TCCACTGGGTGTGAAATCCTC-3′; B7-H6, forward, 5′-CTTTTATTCCCAACCCCTCAACA-3′, and reverse, 5′-CACATCGGTACTCTCCTGCTT-3′; interleukin (IL)-12a, forward, 5′-CCTTGCACTTCTGAAGAGATTGA-3′, and reverse, 5′-ACAGGGCCATCATAAAAGAGGT-3′; IL-12b, forward, 5′-ACCCTGACCATCCAAGTCAAA-3′, and reverse, 5′-TTGGCCTCGCATCTTAGAAAG-3′; IL-15, forward, 5′-TTGGGAACCATAGATTTGTGCAG-3′, and reverse, 5′-GGGTGAACATCACTTTCCGTAT-3′; poliovirus receptor (PVR), forward, 5′-TGGAGGTGACGCATGTGTC-3′, and reverse, 5′-GTTTGGACTCCGAATAGCTGG-3′; Poliovirus receptor-related 2, forward, 5′-GGATGTGCGAGTTCAAGTGCT-3′, and reverse 5′-TGGGACCCATCTTAGGGTGG-3′; and CircARSP91, forward, 5′-GGTGCCAGACCTCATCACTC-3′, and reverse 5′-GAGCAGTCCAGCGAGGACTT-3′. The thermocycling conditions were: 95°C for 3 min, and then 95°C for 10 sec and 55°C for 30 sec for 40 cycles, followed by storage at 4°C. Normalization was performed using the 2−ΔΔq method (23).
Western blot analysis
Cells were lysed in radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology), and proteins were determined by the BCA method (Pierce™ Microplate BCA Protein assay kit, Thermo, Fisher Scientific, Inc.). A total of 60 µg were separated on 10% SDS-PAGE, followed by transferring to polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). Following blocking the membranes (5% non-fat milk in tris-buffered saline with 0.05% Tween-20 at room temperature for 1 h), they were incubated with appropriate dilutions of specific primary antibodies (4°C for overnight) and then were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (room temperature for 1 h). Subsequently, they were visualized using the enhanced chemiluminescent system (Thermo Fisher Scientific, Inc.). The anti-GAPDH (1:1,000; cat no. 6c5) antibody was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Additionally, the anti-ULBP1 (1:1,000; cat no. ab176566), ULBP2 (1:1,000; cat no. ab173286) and ULBP3 (1:1,000; cat no. ab89931) antibodies were purchased from Abcam (Cambridge, UK). All the HRP-conjugated secondary antibodies (1:1,000; cat nos. A0239 and A0258) were purchased from Beyotime Institute of Biotechnology.
Patient selection and immunohistochemistry (IHC) staining
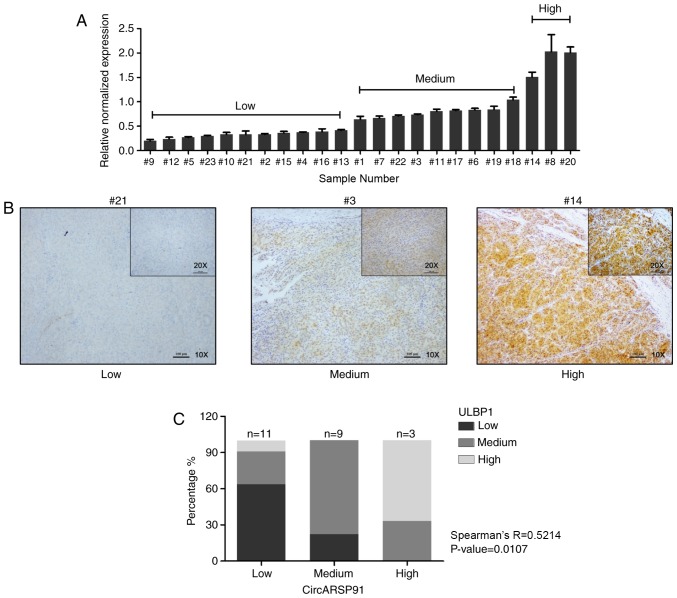
A total of 23 samples from patients with HCC were randomly selected (age range, 47 to 72 years old with a mean age of 61 years; 18 male and 5 female patients), which were collected at Shengzhou People's Hospital (Shengzhou, China) from September 1, 2014, to October 1, 2016. The present study was approved by the Research Ethics Committee of Shengzhou People's Hospital with written informed patient consent. The pathology records were reviewed to identify samples with confirmed HCC. The IHC slides of all 23 patients used for ULBP1 scoring were reviewed by two pathologists in a double-blind manner. The staining results were measured semi-quantitatively on a scale of low, medium and high expression. Staining was scored as follows: Low, faint, barely discernable cytoplasmic staining for ULBP1 in tumor cells; medium, moderate, smooth cytoplasmic staining of the tumor cells with moderate intensity for ULBP1; and high, apparent granular, dark-brown staining in the cytoplasm for ULBP1. Representative examples of low, medium and high expression IHC staining for ULBP1 are demonstrated in Fig. 4.
Figure 4.
Positive correlation between CircARSP91 and ULBP1 in samples from patients with HCC. (A) Total RNA was extracted from 23 human HCC samples collected at Shengzhou People's Hospital (Shengzhou, China) and reverse transcription-quantitative polymerase chain reaction was performed to detect the expression level of CircARSP91. Subsequently, the results were divided into three groups based on their relative normalized expression (different from the grouping for immunohistochemistry): Low, ≤0.5; medium, 0.5–1.0; and high, >1.0. (B) Representative images for ULBP1 immunohistochemistry staining in human HCC samples. (C) Spearman's correlation analysis for CircARSP91 and ULBP1 based on the stained clinical samples. R=0.5214, P=0.0107. The data depicted are the mean ± standard deviation. CircARSP91, circRNA of AR-suppressed PABPC1 91 bp; ULBP1, UL16-binding protein-1; HCC, hepatocellular carcinoma.
IHC staining for ULBP1 was performed using a standard streptavidin-biotin-peroxidase immunostaining procedure (16). The antibody used for anti-ULBP1 was identical to that used in western blotting, but with a dilution of 1:100. The cutoff was set as 5%, according to previous publication (24). Appropriate positive (normal human liver tissue) and negative controls (without primary antibody incubation) were used in each case.
Nude mouse liver orthotopic tumor model
A total of 12 male 6–8-week-old nude mice were purchased from Vital River Laboratory Animal Technology Co. Ltd., (Beijing, China). Mice were housed in a temperature at 27°C, 50% relative humidity, alternately exposed to light for 10 h and without light for 14 h pathogen-free environment. All mice had free access to food and water. The mice were randomly divided into two groups. The MHCC97H vector and MHCC97H pCDH-ARSP91 cells were engineered to express the luciferase gene (PCDNA3.0-luciferase; cat no. #21375; Addgene, Inc.) by stable transfection as previously described (18,22), and the positive stable clones were selected with G418 and expanded in culture. The cells were resuspended in PBS with extracellular matrix Matrigel at a 1:1 ratio. Each group was co-injected with vector or pCDH-ARSP91 cells (MHCC-97H) and NK-92MI cells (E:T ratio=1:5) into the left lobe of the liver as previously described (18,22). A tumor cell concentration of 5×107 cells/ml in a 120-µl volume was injected into each mouse. Following injection, pressure was applied to the injection spot with a cotton bud for 3 min to prevent bleeding. Subsequently, the abdominal cavity was closed with a suture. Each group contained one mouse that succumbed following injection surgery due to liver bleeding. Additionally, the necessary procedure was performed to minimize the pain of the mouse based on 3R principles (25), including conducting the surgery under general anesthesia status and sacrificing the mouse appropriately. The interaction between tumor cells and NK cells should last 3 days (18,22). Following the cytotoxicity function of co-injected NK cells, the remaining quantity of tumor cells may be different resulting in a differing tumor size, based on the expression level of circ-ARSP91 of the HCC cells. Tumor formation and size were tracked using a Fluorescent Imager (IVIS Spectrum; PerkinElmer, Hopkinton, MA, USA; www.perkinelmer.com) starting 2 weeks after tumor injection and using a mouse tail vein injection of 150 mg/kg luciferin.
The mice were sacrificed after 8 weeks by cervical dislocation, and the liver tumors were isolated for weighing. Prior to weighing, the isolated tumors were maintained in PBS. All animal studies were approved by the Ethics Committee of Experimental Animals of Zhejiang University (Shengzhou, China) and performed under the supervision and guidelines of the Zhejiang University (http://www.lac.zju.edu.cn/).
Statistical analysis
The data are expressed as the means ± standard deviations from ≥3 independent experiments. Statistical analyses involved an unpaired t-test, one-way analysis of variance (with the Student Newman-Keul's method used for the post-hoc test) and Spearman's correlation with SPSS 17.0 (SPSS Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Establishment of HCC cell lines with stably overexpressed or knocked down CircARSP91
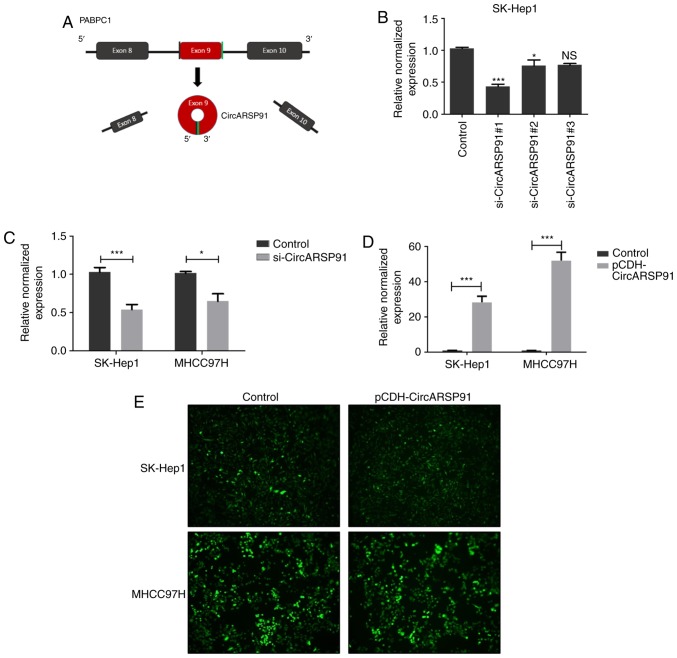
To assess the function of CircARSP91 on the biological behavior of HCC, HCC cells with a stable expression of CircARSP91 were established based on a previous report (18). The formation of CircARSP91 was performed in a previous tudy by the circularization of exon 9 of PABPC1, indicating that the total length of CircARSP91 was small (91 bp), compared with other reported constructs, including circRNA sponge for miR-7 (1,485 bp; Fig. 1A) (10). A total of 3 knockdown siRNA constructs were designed based on the junction site of CircARSP91, and the knockdown efficiency was assessed (Fig. 1B). Subsequently, si-CircARSP91 #1 was selected for subsequent experiments, and stable CircARSP91 knockdown was performed in SK-Hep1 and MHCC97H cells (Fig. 1C). These two cell lines have a moderate endogenous expression level of circARSP91 (qPCR Cq values of ~25 for SK-Hep1 and 27 for MHCC97H, with GAPDH having a value of ~19; data not shown). Additionally, CircARSP91 overexpression was performed in the identical cell lines successfully using a lentivirus system (Fig. 1D). The transfection efficiency of the lentivirus was evaluated by the fluorescence intensity (Fig. 1E). These results demonstrate the successful establishment of overexpressed and knockdown cells.
Figure 1.
Establishment of stable CircARSP91 overexpressing and knockdown cell lines. (A) Schematic diagram for Circ-ARSP91 formation. (B) The knockdown efficiency was evaluated in SK-Hep1 cells for si-CircARSP91. (C) Stable CircARSP91 knockdown hepatocellular carcinoma cell lines were established in SK-Hep1 and MHCC97H cells. The knockdown efficiency was evaluated by RT-qPCR. Overexpression efficiency of CircARSP91 in SK-Hep1 and MHCC97H cells was assessed by (D) RT-qPCR and (E) green fluorescence. The data depicted are the mean ± standard deviation. ***P<0.001 and *P<0.05 vs. the control. CircARSP91, circRNA of AR-suppressed PABPC1 91 bp; NS, not significant; si-CircARSP91, small interfering-CircARSP91; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
CircARSP91 increases the susceptibility of HCC cells to NK cell cytotoxicity
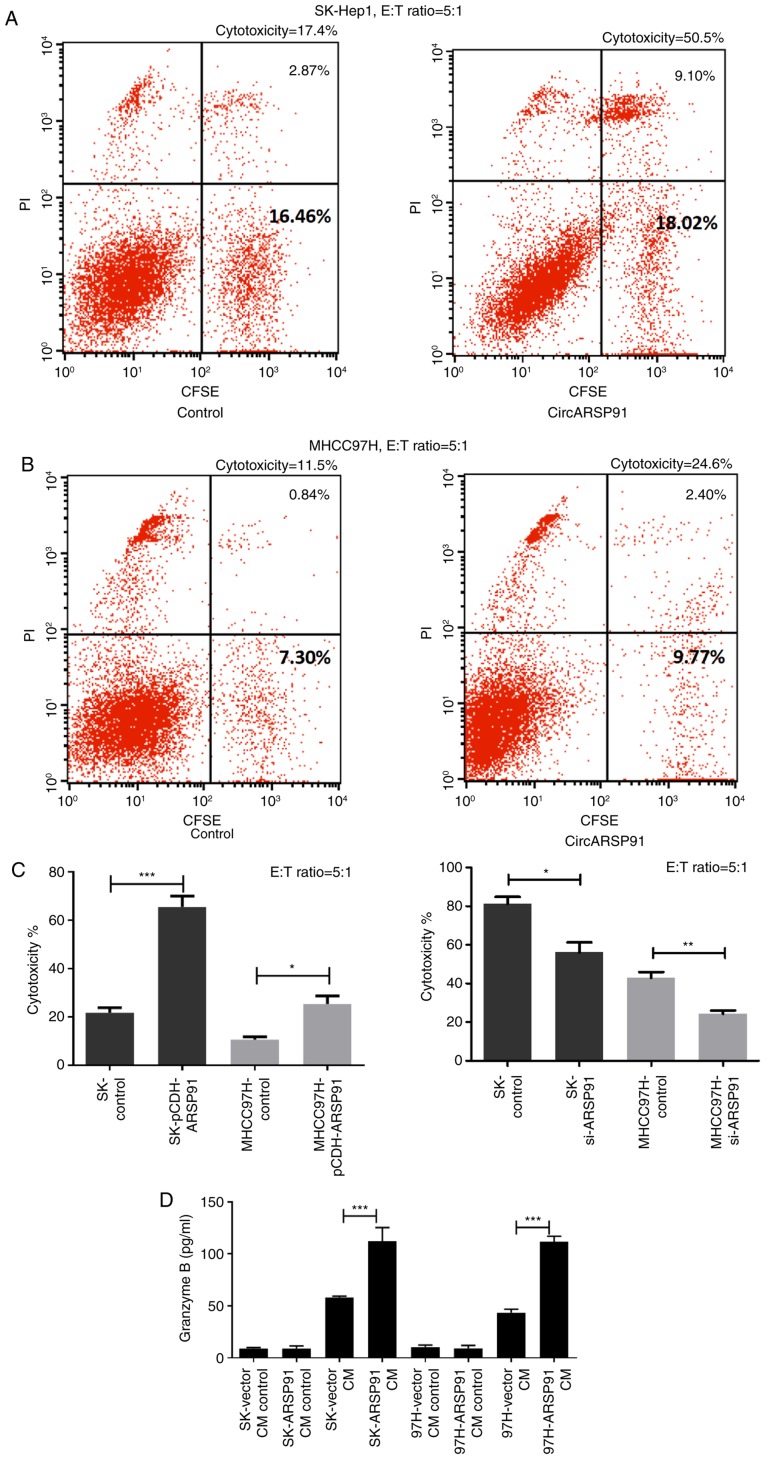
Following acquiring HCC cells that modulate the expression of CircARSP91, the cytotoxicity of NK cells was assessed against them. In brief, HCC cells were stained with CFSE first, followed by co-culturing stained HCC cells with NK-92MI cells at an E:T ratio of 5:1 for 4–8 h. Thereafter, PI staining was performed to localize the damaged tumor cells, and the final detection was performed using flow cytometry. Cytotoxicity was calculated as % cytotoxicity=100×[no. of CFSE+/PI+ cells]/[(no. of CFSE positive cells)+(no. of CFSE+/PI+ cells)] (Fig. 2A and B). Notably, it was determined that CircARSP91 could upregulate the susceptibility of HCC cells to NK cell cytotoxicity, indicating that the overexpression of CircARSP91 caused HCC cells to become increasingly vulnerable to NK cells while the knockdown of circARSP91 assisted tumor cells in escaping from the immune surveillance of NK cells (Fig. 2C). Additionally, since increased granzyme B release resulted in improved activated status of NK cells, the CM from the cytotoxicity assay was collected and the granzyme B concentration was detected among different groups. The results indicated that CircARSP91 could significantly enhance NK-92MI cell activation (Fig. 2D).
Figure 2.
CircARSP91 of HCC enhances the cytotoxicity of natural killer cells against tumor cells. (A) The SK-Hep1 cells were labelled with CFSE and NK-92MI cells were applied to tumor cells for 4 h at an E:T ratio of 5:1. Subsequently, PI staining was used to detect dead cells and all the cells were collected for flow cytometry analysis. The right upper quadrant depicts CFSE-positive and PI-positive cells, and the right lower quadrant depicts CFSE-positive cells alone. The percentages in the image are for the right upper quadrant (2.87, and 9.10 0.84 and 2.40%) or all of the right quadrant (16.46, 18.02, 7.30 and 9.77%). The cytotoxicity was calculated as % cytotoxicity=100×[no. of CFSE+/PI+ cells]/[(no. of CFSE positive cells) + (no. of CFSE+/PI+ cells)]. The spontaneous death of target cells was excluded in the final results. (B) A similar procedure was performed on MHCC-97H cells and the percentages in the image are for the right upper quadrant (0.84 and 2.40%) or all of the right quadrant (7.30 and 9.77%). (C) Quantification results for cytotoxicity calculation. The left panel depicts the assay performed with CircARSP91 overexpressing HCC cells, and the right panel depicts the assay performed with si-CircARSP91 cells. (D) CM was also collected to evaluate granzyme B release from NK-92MI cells. CM was collected 2 h after adding NK-92MI cells to HCC cells. The control groups had no NK-92MI cells added. The data depicted are the mean ± standard deviation. ***P<0.001, **P<0.01, *P<0.05. E:T, effector cell:target cell; CFSE, carboxyfluorescein succinimidyl ester; PI, propidium iodide; HCC, hepatocellular carcinoma; CircARSP91, circRNA of AR-suppressed PABPC1 91 bp; si-ARSP91, small interfering-CircARSP91; CM, conditioned media.
CircARSP91 enhances the cytotoxicity of NK cells via upregulating ULBP1 in HCC
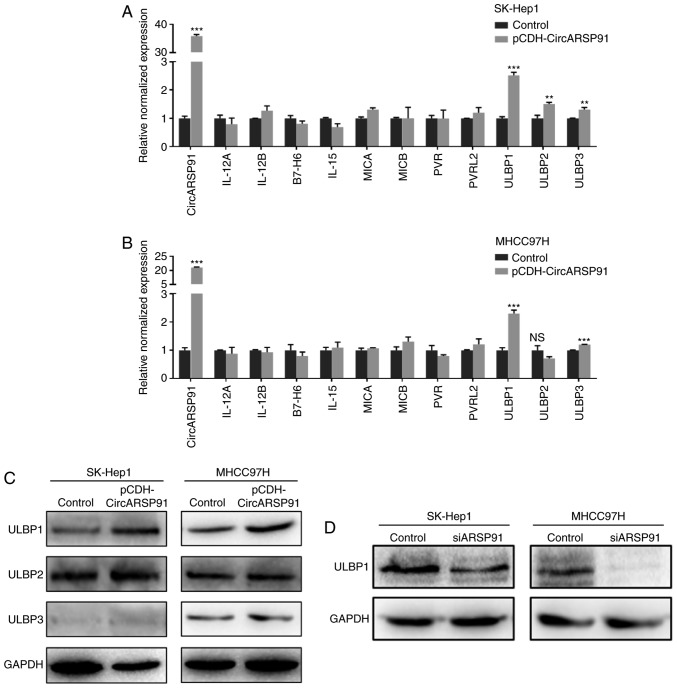
Previous reports reported the association between AR and the cytotoxicity of NK cells in HCC by screening molecules that could influence the interaction between NK cells and HCC tumor cells (18,22). Subsequently, similar screening was performed using RT-qPCR and it was determined that the expression of ULBPs was significantly increased following overexpressing CircARSP91 in the HCC cell lines (Fig. 3A and B). However, when it was attempted to verify the mRNA level results by immunoblotting, only the ULBP1 level was demonstrated to be increased by CircARSP91 (Fig. 3C). Additionally, the knockdown data indicated a consistent trend (Fig. 3D).
Figure 3.
CircARSP91 increases ULBP1 expression in HCC cells. (A) RT-qPCR was applied to screen tumor-associated NK cell molecules following overexpressing CircARSP91 in SK-Hep1 cells. (B) A similar screening was performed on MHCC-97H cells. The results narrowed down promising NK cell activation-associated molecules to the ULBP family, indicating that CircARSP91 may boost the ULBP mRNA level in HCC cells. (C) Western blotting was performed using ULBP-specific antibodies and it was determined that ULBP1 could be upregulated by CircARSP91 in SK-Hep1 and MHCC97H cells, while ULBP2 and ULBP3 failed to be consistent with the RT-qPCR results. (D) Knocking down CircARSP91 in HCC cells lines could decrease ULBP1 expression. The data depicted are the mean ± standard deviation. ***P<0.001, **P<0.01 and *P<0.05 vs. the control. HCC, hepatocellular carcinoma; CircARSP91, circRNA of AR-suppressed PABPC1 91 bp; si-ARSP91, small interfering-CircARSP91; ULBP1, UL16-binding protein-1; NK, natural killer; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; IL, interleukin; MICA, major histocompatibility complex class I polypeptide-related sequence A; PVR, poliovirus receptor; PVRL2, Poliovirus receptor-related 2; NS, not significant.
ULBP1 expression is positively correlated with CircARSP91 in human HCC samples
To further investigate the correlation between ULBP1 and CircARSP91, 23 human HCC samples were collected from Shengzhou People's Hospital from September 1, 2014, to October 1, 2016. Subsequently, the total RNA was extracted from these samples and the CircARSP91 expression level was detected, which could be divided into three grades, low, medium and high expression (Fig. 4A). Additionally, IHC staining was performed in the same samples to determine the ULBP1 level, and the representative images are depicted in Fig. 4B. Thereafter, Spearman's correlation was performed, and a positive correlation (R=0.4252, P=0.0383) was revealed (Fig. 4C).
The CircARSP91 expression level enhances the cytotoxicity of NK-92MI cells in an orthotopic HCC mouse model
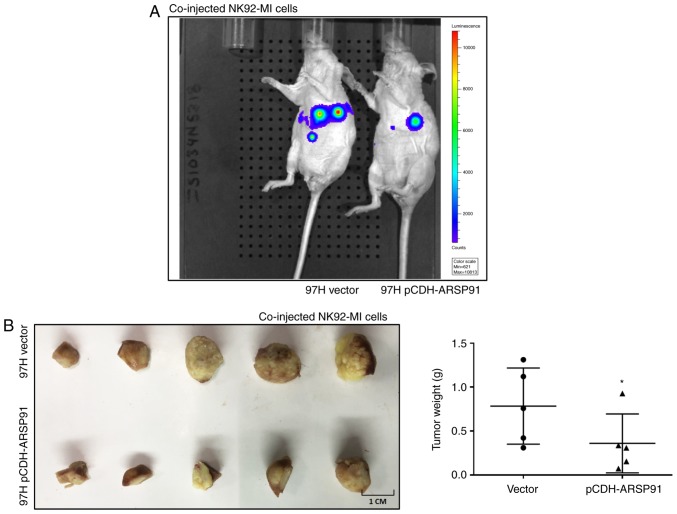
To confirm the aforementioned in vitro cell line data in vivo, an orthotopic xenograft nude mouse model was developed, as previously described (18). In brief, the HCC cell line MHCC97H with or without overexpressed CircARSP91 were co-injected with NK-92MI cells at an E:T ratio of 5:1 into the left lobes of nude mouse livers. The tumor formation and size were tracked using an IVIS detection system. A total of 8 weeks after injection, the IVIS images revealed that the tumor size of the pCDH-ARSP91 group was notably smaller, compared with the control group, implying that CircARSP91 caused tumor cells to be more vulnerable to the cytotoxicity of NK cells (Fig. 5A). Since a large amount of NK cells was injected and the effect of these NK cells would be fast and strong (18), the phenotype generated should not be due to endogenous NK cells. Following sacrificing the mice, the results from the tumor weight measurements indicated a consistent conclusion (Fig. 5B). However, only one mouse exhibited metastasis, which makes the comparison of metastasis status between the two groups unpractical.
Figure 5.
The CircARSP91 expression level enhances the cytotoxicity of NK-92MI cells in an orthotopic HCC mouse model. (A) The orthotopic HCC tumor mouse model was monitored with an IVIS image system prior to sacrifice at 8 weeks after co-injection. Representative images are depicted between the MHCC97H vector and MHCC97H pCDH-ARSP91. (B) At 8 weeks after injection, the mice were sacrificed to isolate the tumor from the liver (left panel). The tumors were stored in PBS prior to weighing, and the weight measurements indicate enhanced cytotoxicity of natural killer cells in the pCDH-ARSP91 group (right panel). The data depicted are the mean ± standard deviation. *P<0.05. HCC, hepatocellular carcinoma; CircARSP91, circRNA of AR-suppressed PABPC1 91 bp; pCDH-ARSP91, overexpression of pCDH-CircARSP91.
Discussion
In the present study, a novel function of CircARSP91 that could enhance the cytotoxicity of NK cells against HCC tumor cells was elaborated. Such a phenotype may be due to one of the NKG2D ligands, ULBP1, which could be upregulated by CircARSP91 at the mRNA and protein levels. Additionally, when human HCC samples were used to perform IHC staining and RT-qPCR, a positive association was identified between CircARSP91 and ULBP1. A detailed mechanism dissection is still required to gain a more comprehensive explanation of how CircARSP91 regulates ULBP1. As for the direct function of CircARSP91 in HCC tumor cells, it was considered that this would not interfere with the observation of the cytotoxicity of NK cells. The cytotoxicity was measured based on the membrane damage and cell lysis while CircARSP91 would not influence these aspects according to the present observation during cell culture.
The function of circRNAs has been illustrated based on their ability to act as a miRNA ‘sponge’, blocking miRNA regulation of target genes (10,26). However, a previous study on CircARSP91 reported that CircARSP91 may not function as a miRNA sponge due to it being predicted to bind tumor suppressors instead (16). Furthermore, an increasing number of researchers have regarded the sponging function of circRNAs as exceptional due to the majority of circRNAs not having sufficient miRNA-binding sites to function as sponges (27,28). Recently, other molecular mechanisms of gene regulation by circRNAs have been reported (29). For example, circRNAs, including circular poly(A) binding protein nuclear 1, can act as competitors with their cognate mRNA for RNA binding proteins and can modulate the rate of translation of target mRNAs (30). Additionally, a circRNA derived from the SEPALLATA3 gene was demonstrated to interact with its cognate DNA, and affect the recruitment of splicing factors to the nascent transcript and influence splicing by exon-skipping (31). In the present study, the mRNA and protein of ULBP1 were upregulated by CircARSP91, and it was considered that CircARSP91 may interact with the ULBP1 promoter region and recruit RNA polymerase II to enhance the expression of the ULBP1 gene, as previously reported (32), or that CircARSP91 could indirectly modulate ULBP1 by other mechanisms.
ULBP1, an NKG2D ligand, could activate receptors expressed by NK cells primarily involved in the elimination of transformed and infected cells (33). Researchers have reported that numerous drugs (like cisplatin and trichostatin A) could influence the status of ULBPs by enhancing the cytotoxicity of NK cells (22,34). However an increasing number of investigations have tried to apply NK cell-associated therapy against cancer (35,36). Therefore, it was considered that with accumulated evidence NK cell-associated therapy could progress toward the next stage for patients with cancer in the near future. In the present study, the association between non-coding RNA and NK cells was determined for a novel circRNA, CircARSP91, which could boost the expression of ULBP1 and assist NK cells in identifying target tumor cells. To the best of our knowledge, this circRNA-associated immune system phenomenon in HCC has not been published elsewhere. Notably, this data could provide valuable information for the future development of associated therapeutics to improve the control of liver cancer.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CircARSP91
circRNA of AR-suppressed PABPC1 91 bp
- NK cell
natural killer cell
- E:T
effector cell:target cell
- ULBP1
UL16 binding protein 1
- HCC
hepatocellular carcinoma
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
QY designed the experiments and prepared the manuscript. Data collection and experiments were performed by YM and CZ. BZ and HY analyzed the data and all authors contributed to the revision of the manuscript.
Ethics approval and consent to participate
All animal studies were approved by the Ethics Committee of Experimental Animals of Zhejiang University (Shengzhou, China) and performed under the supervision and guidelines of the Zhejiang University. The present study was approved by the Research Ethics Committee of Shengzhou People's Hospital.
Patient consent for publication
All the patients' samples used in this manuscript was approved by the Research Ethics Committee of Shengzhou People's Hospital with patient consent for publication.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Park JW, Chen M, Colombo M, Roberts LR, Schwartz M, Chen PJ, Kudo M, Johnson P, Wagner S, Orsini LS, Sherman M. Global patterns of hepatocellular carcinoma management from diagnosis to death: The BRIDGE Study. Liver Int. 2015;35:2155–2166. doi: 10.1111/liv.12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohammed MS, Ferrier G, Abouda G, Corless L. Socio-economic deprivation significantly impacts clinical management and survival in hepatocellular carcinoma. J Hepatol. 2018;68:623–625. doi: 10.1016/j.jhep.2017.09.031. [DOI] [PubMed] [Google Scholar]
- 4.Mancuso A, Maringhini A. Management of hepatocellular carcinoma recurrence after liver transplant is far from perfect. Am J Surg. 2018;216:389–390. doi: 10.1016/j.amjsurg.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 5.European Association For The Study Of The Liver; European Organisation For Research And Treatment Of Cancer: EASL-EORTC clinical practice guidelines: Management of hepatocellular carcinoma. J Hepatol. 2012;56:908–943. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Bruix J, Raoul JL, Sherman M, Mazzaferro V, Bolondi L, Craxi A, Galle PR, Santoro A, Beaugrand M, Sangiovanni A, et al. Efficacy and safety of sorafenib in patients with advanced hepatocellular carcinoma: Subanalyses of a phase III trial. J Hepatol. 2012;57:821–829. doi: 10.1016/j.jhep.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Parikh ND, Marshall VD, Singal AG, Nathan H, Lok AS, Balkrishnan R, Shahinian V. Survival and cost-effectiveness of sorafenib therapy in advanced hepatocellular carcinoma: An analysis of the SEER-Medicare database. Hepatology. 2017;65:122–133. doi: 10.1002/hep.28881. [DOI] [PubMed] [Google Scholar]
- 8.Fu J, Wang H. Precision diagnosis and treatment of liver cancer in China. Cancer Lett. 2018;412:283–288. doi: 10.1016/j.canlet.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Sia D, Villanueva A, Friedman SL, Llovet JM. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology. 2017;152:745–761. doi: 10.1053/j.gastro.2016.11.048. [DOI] [PubMed] [Google Scholar]
- 10.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 11.Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Patton JG, Zhang B. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6:37982. doi: 10.1038/srep37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, Yang BB. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell death and differ. 2017:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Li Y, Zheng Q, He J, Chen B, Lyu D, Zheng B, Xu Y, Long Z, Zhou Y, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, Fan J, Liu L, Qin W. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 15.Ma WL, Lai HC, Yeh S, Cai X, Chang C. Androgen receptor roles in hepatocellular carcinoma, fatty liver, cirrhosis and hepatitis. Endocr Relat Cancer. 2014;21:R165–R182. doi: 10.1530/ERC-13-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi L, Yan P, Liang Y, Sun Y, Shen J, Zhou S, Lin H, Liang X, Cai X. Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis. 2017;8:e3171. doi: 10.1038/cddis.2017.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soriani A, Zingoni A, Cerboni C, Iannitto ML, Ricciardi MR, Di Gialleonardo V, Cippitelli M, Fionda C, Petrucci MT, Guarini A, et al. ATM-ATR-dependent up-regulation of DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic agents results in enhanced NK-cell susceptibility and is associated with a senescent phenotype. Blood. 2009;113:3503–3511. doi: 10.1182/blood-2008-08-173914. [DOI] [PubMed] [Google Scholar]
- 18.Shi L, Lin H, Li G, Jin RA, Xu J, Sun Y, Ma WL, Yeh S, Cai X, Chang C. Targeting androgen receptor (AR)→IL12A signal enhances efficacy of sorafenib plus NK cells immunotherapy to better suppress HCC progression. Mol Cancer Ther. 2016;15:731–742. doi: 10.1158/1535-7163.MCT-15-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding S, Chen G, Zhang W, Xing C, Xu X, Xie H, Lu A, Chen K, Guo H, Ren Z, et al. MRC-5 fibroblast-conditioned medium influences multiple pathways regulating invasion, migration, proliferation, and apoptosis in hepatocellular carcinoma. J Transl Med. 2015;13:237. doi: 10.1186/s12967-015-0588-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krzywinska E, Kantari-Mimoun C, Kerdiles Y, Sobecki M, Isagawa T, Gotthardt D, Castells M, Haubold J, Millien C, Viel T, et al. Loss of HIF-1α in natural killer cells inhibits tumour growth by stimulating non-productive angiogenesis. Nat Commun. 2017;8:1597. doi: 10.1038/s41467-017-01599-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Y, Kuang DM, Pan WD, Wan YL, Lao XM, Wang D, Li XF, Zheng L. Monocyte/macrophage-elicited natural killer cell dysfunction in hepatocellular carcinoma is mediated by CD48/2B4 interactions. Hepatology. 2013;57:1107–1116. doi: 10.1002/hep.26192. [DOI] [PubMed] [Google Scholar]
- 22.Shi L, Lin H, Li G, Sun Y, Shen J, Xu J, Lin C, Yeh S, Cai X, Chang C. Cisplatin enhances NK cells immunotherapy efficacy to suppress HCC progression via altering the androgen receptor (AR)-ULBP2 signals. Cancer Lett. 2016;373:45–56. doi: 10.1016/j.canlet.2016.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Cho EY, Chang MH, Choi YL, Lee JE, Nam SJ, Yang JH, Park YH, Ahn JS, Im YH. Potential candidate biomarkers for heterogeneity in triple-negative breast cancer (TNBC) Cancer Chemother Pharmacol. 2011;68:753–761. doi: 10.1007/s00280-010-1548-x. [DOI] [PubMed] [Google Scholar]
- 25.Törnqvist E, Annas A, Granath B, Jalkesten E, Cotgreave I, Öberg M. Strategic focus on 3R principles reveals major reductions in the use of animals in pharmaceutical toxicity testing. PLoS One. 2014;9:e101638. doi: 10.1371/journal.pone.0101638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haque S, Harries LW. Circular RNAs (circRNAs) in health and disease. Genes (Basel) 2017;8(pii):E353. doi: 10.3390/genes8120353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, Gorospe M. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G, Cildir G, Jourdain A, Tergaonkar V, Schmid M, Zubieta C, Conn SJ. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. 2017;3:17053. doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
- 32.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 33.Huergo-Zapico L, Acebes-Huerta A, López-Soto A, Villa-Álvarez M, Gonzalez-Rodriguez AP, Gonzalez S. Molecular bases for the regulation of NKG2D ligands in cancer. Front Immunol. 2014;5:106. doi: 10.3389/fimmu.2014.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin S, Kim M, Lee SJ, Park KS, Lee CH. Trichostatin A sensitizes hepatocellular carcinoma cells to enhanced NK cell-mediated killing by regulating immune-related genes. Cancer Genomics Proteomics. 2017;14:349–362. doi: 10.21873/cgp.20045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu T, Cui T, Peng L, Kong S, Zou J, Tian X. The anti-hepatocellular carcinoma activity of Mel-P15 is mediated by natural killer cells. Oncol Lett. 2017;14:6901–6906. doi: 10.3892/ol.2017.7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu L, Li XJ, Kalimuthu S, Gangadaran P, Lee HW, Oh JM, Baek SH, Jeong SY, Lee SW, Lee J, Ahn BC. Natural killer cell (NK-92MI)-based therapy for pulmonary metastasis of anaplastic thyroid cancer in a nude mouse model. Front Immunol. 2017;8:816. doi: 10.3389/fimmu.2017.00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.