Abstract
Coding and noncoding RNAs serve a crucial role in tumorigenesis. Circulating RNAs have been recognized as a novel category of biomarkers for a variety of physiological and pathological conditions. To identify plasma RNA biomarkers for gastric cancer (GC), a genome-wide transcriptome analysis using GeneChip® Human Transcriptome Array, which contains probe sets covering exons of ~67500 coding and noncoding transcripts of annotated genes, was performed to screen for the RNAs that exhibited differential expression in the plasma samples of patients with GC and controls. The expression levels of 6 candidate RNAs, including regulator of G-protein signaling 18 (RGS18), integral membrane protein 2B, pro-platelet basic protein (PPBP), nucleosome assembly protein1-like 1, n324674 and ENST00000442382 were assessed in the plasma samples of 81 patients with GC and 77 healthy participants using reverse transcription-quantitative polymerase chain reaction. Furthermore, the expression levels of RGS18 and PPBP mRNAs were indicated to be significantly differentially expressed (P<0.0001) in an independent panel of plasma samples of 36 patients with GC compared with 34 healthy participants. The potential association of RGS18 and PPBP mRNA expression levels with clinicopathological features was subsequently analyzed. Receiver operating characteristic analysis indicated that the combination of these 2 mRNAs with an area under curve <0.812 was an improved indicator for gastric cancer compared with respective individual levels. The results of the present study indicate that RGS18 and PPBP mRNA expression was significantly downregulated in the plasma of patients with GC, and the combination of these 2 mRNAs may be a useful diagnostic or prognostic marker for GC.
Keywords: plasma, RNAs, gastric cancer, biomarker, microarray, regulator of G-protein signalling 18, pro-platelet basic protein
Introduction
Gastric cancer (GC) is the third leading cause of cancer-associated mortality, particularly in East Asia, Eastern Europe and South America (1–3) and the 5-year overall survival rate remains <25% (4). Therefore, identification of molecular biomarkers, which contribute to the early stratification of patients with poor prognosis, would aid in selecting the most effective and appropriate therapeutic strategy (3–5). However, numerous GC-associated biomarkers, involving carcinoembryonic antigen (CEA), carbohydrate antigen 19-9, carbohydrate antigen 72-4 (CA72-4) and carbohydrate antigens 125, frequently used for diagnosis, prognosis prediction and monitoring of postoperative recurrence (6,7) are not sensitive or specific for the detection of GC, particularly in early diagnosis. Therefore, identification of novel biomarkers for GC diagnosis have been a major focus of recent investigations (8–10).
A previous study indicated that the release of nucleic acids into the blood was associated with apoptosis and necrosis of cancer cells, including thyroid cancer, nasopharyngeal carcinoma and lung cancer, in the tumor microenvironment. In addition, the study reported that secretion has also been indicated to be a potential source of cell-free nucleic acids (11). Therefore, the detection of cell-free (cf) RNAs in plasma or serum may serve as a ‘liquid biopsy’, which would be useful for numerous diagnostic applications and would reduce the requirement for tumor tissue biopsies (11,12). A variety of extracellular RNAs have been detected and used as diagnostic and prognostic indicators for early diagnosis and disease outcomes (11–13). In contrast to miRNA, complete mRNA and long non-coding RNA (lncRNA) molecules have been reported to be limited in the plasma (14). Plasma mRNAs and lncRNAs exist mainly in the form of RNA fragments. Therefore, it is difficult to screen potential plasma RNA markers using traditional lncRNA or mRNA microarray with a single probe mapped to one gene. GeneChip® Human Transcriptome Array 2.0 (HTA2.0) contains >6.0 million distinct probes, covering coding and non-coding transcripts, and was designed with ~10 probes per exon, therefore ensuring that complete and accurate expression data was obtained in the present study, even for fragmented RNA molecules.
Therefore, in the present study, the Human Transcriptome Array 2.0 was used to screen candidate RNAs that were differentially expressed between plasma samples from healthy participants and patients with GC. The differentially expressed RNA molecules were validated in a training set and assessed in the plasma samples of 81 patients with GC and 77 healthy participants using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The optimal candidate RNA biomarkers were isolated in a test set of 36 patients with GC and 34 healthy participants and the association between expression levels and patient clinicopathological data was determined to assess the diagnostic value of these biomarkers in patients with GC. The results of the present study demonstrated that certain RNAs, including G-protein signaling 18 (RGS18) and pro-platelet basic protein (PPBP), may be significantly downregulated in the plasmas of patients with GC. RGS18 has been reported to have a combined and cell-specific role in regulating the function of cancer cells, along with the progression of various types of cancer (15). PPBP has been reported to stimulate various cellular processes, including regulating the growth and metastasis of cholangiocarcinoma (16). The findings of the present study indicate that a combination of RGS18 and PPBP mRNAs may be used as a diagnostic or prognostic marker of GC.
Materials and methods
Tissue and blood samples
Peripheral blood samples were collected from 81 patients with GC and 77 healthy participants (Table I) at the Chinese People's Liberation Army General Hospital (Beijing, China) and 36 patients with GC and 34 healthy participants (Table II) at the China-Japan Union Hospital of Jilin University (Changchun, China). GC tissues and paired para-tumorous tissues were collected from 26 patients (Table III), who underwent surgery for GC at the Chinese PLA General Hospital (Beijing, China). None of the patients were administered radiotherapy or chemotherapy treatment prior to the operation. All tissue specimens and blood samples were immediately frozen in liquid nitrogen and stored at −80°C. All samples were staged in accordance with the Tumor-Node-Metastasis (TNM) classification and the criteria of the Union for International Cancer Control (UICC) and the tumor grade was assessed in accordance to the UICC criteria (17,18). Written informed consent was obtained from all patients. The Ethics Committee of the Chinese PLA General Hospital (Beijing, China) and the China-Japan Union Hospital of Jilin University (Changchun, China) approved the use of samples for the present study.
Table I.
Basic characteristics of enrolled individuals in the training set.
| Control group (n=77) | GC group (n=81) | |
|---|---|---|
| Clinical parameter | Number of cases | Number of cases |
| Age (years) | ||
| ≤60 | 37 (48.1%) | 42 (51.9%) |
| >60 | 40 (51.9%) | 39 (48.1%) |
| Mean age | 61 | 60 |
| Age range | 36–84 | 36–84 |
| Sex | ||
| Male | 49 (63.6%) | 64 (79.0%) |
| Female | 28 (36.4%) | 17 (21.0%) |
| Differentiateda | ||
| PDACb | 56 | |
| MDAC | 20 | |
| WDAC | 5 | |
| Invasion depth | ||
| T1 | 10 | |
| T2 | 19 | |
| T3 | 7 | |
| T4 | 37 | |
| Uncategorized | 8 | |
| Regional lymph nodes | ||
| N0 | 25 | |
| N1 | 17 | |
| N2 | 11 | |
| N3 | 11 | |
| Uncategorized | 17 | |
| Distant metastasis | ||
| Yes | 10 | |
| No | 63 | |
| Uncategorized | 8 | |
| TNM stagingc | ||
| I | 19 | |
| II–IV | 52 | |
| Size (cm3) | ||
| ≥4 | 42 | |
| <4 | 24 | |
| Uncategorized | 15 |
World Health Organization classification of tumors of the digestive system (17).
Mucinous carcinoma and poorly differentiated adenocarcinoma were classified into a group due to the analogously poor prognosis.
American Joint Committee on Cancer staging system (18). GC, gastric cancer; TNM, Tumor-Node-Metastasis; PDAC, poorly-differentiated adenocarcinoma; MDAC, moderately-differentiated adenocarcinoma; WDAC, well-differentiated adenocarcinoma.
Table II.
Basic characteristics of enrolled individuals in testing set.
| Control group (n=34) | GC group (n=36) | |
|---|---|---|
| Clinical parameter | Number of cases | Number of cases |
| Age (years) | ||
| ≤60 | 19 (55.9%) | 11 (30.6%) |
| >60 | 15 (44.1%) | 25 (69.4%) |
| Mean age | 59 | 58 |
| Age range | 36–78 | 36–83 |
| Sex | ||
| Male | 16 (47.1%) | 20 (55.6%) |
| Female | 18 (52.9) | 16 (44.4%) |
| Differentiateda | ||
| PDACb | 6 | |
| MDAC | 12 | |
| WDAC | 1 | |
| Invasion depth | ||
| T1-T2 | 2 | |
| T3-T4 | 7 | |
| Regional lymph nodes | ||
| N0 | 3 | |
| N1-N3 | 6 | |
| Distant metastasis | ||
| Yes | 1 | |
| No | 8 | |
| TNM stagingc | ||
| I | 2 | |
| II–IV | 7 | |
| Size (cm3) | ||
| ≥4 | 16 | |
| <4 | 4 |
World Health Organization classification of tumors of the digestive system (17).
Mucinous carcinoma and poorly differentiated adenocarcinoma were classified into a group due to the analogously poor prognosis.
American Joint Committee on Cancer staging system (18). GC, gastric cancer; TNM, Tumor-Node-Metastasis; PDAC, poorly-differentiated adenocarcinoma; MDAC, moderately-differentiated adenocarcinoma; WDAC, well-differentiated adenocarcinoma.
Table III.
The clinical parameters of patients with GC and GC tumors.
| Sample | Sex | Age (years) | Differentiateda | TNM stagingb | Depth of invasion | LN metastasis |
|---|---|---|---|---|---|---|
| 1 | Male | 58 | MDACc | IIIB | T3 | N2 |
| 2 | Male | 74 | PDAC | II | T3 | N0 |
| 3 | Female | 79 | PDAC | III | T3 | N2 |
| 4 | Female | 76 | PDAC | IIIB | T3 | N2 |
| 5 | Male | 73 | MDAC | IV | T3 | N3 |
| 6 | Male | 87 | MDAC | IA | T1 | N0 |
| 7 | Female | 71 | MDAC | IV | T4 | N2 |
| 8 | Male | 73 | MDAC | II | T2 | N1 |
| 9 | Male | 57 | PDAC | IB | T2 | N0 |
| 10 | Female | 34 | PDAC | IA | T1 | N0 |
| 11 | Male | 75 | PDAC | IIIB | T3 | N2 |
| 12 | Male | 54 | MDAC | IB | T2 | N0 |
| 13 | Female | 70 | PDAC | IV | T3 | N3 |
| 14 | Male | 62 | MDAC | II | T2 | N1 |
| 15 | Female | 67 | MDAC | IV | T4 | N2 |
| 16 | Male | 26 | MDAC | IIIA | T3 | N1 |
| 17 | Female | 45 | MDAC | IIIB | T3 | N2 |
| 18 | Male | 59 | PDAC | IV | T3 | N3a |
| 19 | Male | 50 | MDAC | II | T2 | N1 |
| 20 | Male | 52 | MDAC | IV | T4a | N3b |
| 21 | Female | 42 | MDAC | II | T3 | N0 |
| 22 | Female | 67 | MDAC | IIIB | T3 | N2 |
| 23 | Female | 41 | PDAC | IV | T4a | N3b |
| 24 | Male | 53 | MDAC | IIIB | T3 | N2 |
| 25 | Male | 46 | PDAC | IV | T4a | N3b |
| 26 | Female | 34 | PDAC | IB | T2 | N0 |
Mean age, 59 years; age range, 26–87 years.
World Health Organization classification of tumors of the digestive system (17).
The American Joint Committee on Cancer staging system (18).
Mucinous carcinoma and poorly differentiated adenocarcinoma were classified into a group due to the analogously poor prognosis. TNM, Tumor-Node-Metastasis; LN, lymph node; PDAC, poorly-differentiated adenocarcinoma; MDAC, moderately-differentiated adenocarcinoma; WDAC, well-differentiated adenocarcinoma.
Microarray analysis
Cell-free RNA was extracted from the mixed plasma samples of patients with GC and healthy participants (n=20, individual blood samples) using the miRNeasy Serum/Plasma kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's protocol, and treated with RNase-Free DNase set (Qiagen GmbH), according to the manufacturer's protocol. The sample preparation and microarray hybridization were performed by Shanghai Bohao Biotechnology Co., Ltd. (Shanghai, China). Total RNA was amplified, labelled and purified using an Affymetrix WT PLUS reagent kit (Affymetrix; Thermo Fisher Scientific Inc., Waltham, MA, USA) to obtain labelled cDNA. Samples were hybridized to a GeneChip® Human Transcriptome Array 2.0 (Affymetrix; Thermo Fisher Scientific Inc.), according to the manufacturer's protocol. Arrays were scanned using the GeneChip® Scanner 3000 (Affymetrix; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Command Console software 4.10 (Affymetrix; Thermo Fisher Scientific Inc.) was used with default settings to control the scanner and summarize probe cell intensity data. Raw data were then normalized using Expression Console software 4.10 (Affymetrix; Thermo Fisher Scientific, Inc.).
Total RNA preparation and RT-qPCR
Total RNA was extracted from tissue samples using TRIzol reagent (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), according to the manufacturer's protocol. Plasma cfRNA was extracted from 200 µl plasma using the RNeasy Serum/Plasma kit (Qiagen GmbH), according to the manufacturer's protocols. cDNA was synthesized and amplified using ImProm-II™ reverse transcriptase (Promega Corporation, Madison, WI, USA), according to the manufacturer's protocol. The primer sequences are indicated in Table IV. qPCR was subsequently performed using the SYBR qPCR mix (Takara Biotechnology Co., Ltd., Tokyo, Japan) with an Mx 3000p instrument (Agilent Technologies Inc., Santa Clara, CA, USA), according to the manufacturer's protocols. Amplification of the cDNA was confirmed using melting curve analysis. cDNA was reverse transcribed from 20 µl plasma cfRNA and the following thermocycling conditions were used for the qPCR: Initial denaturation at 95°C for 5 min; 45 cycles of denaturation at 95°C for 10 sec and annealing and extending at 60°C for 20 sec. The relative expression level of each RNA was quantified using the 2−∆∆Cq method (19) with the 18S rRNA gene as the endogenous control for data normalization, since its expression level does not significantly differ in the plasma samples of patients with GC and healthy participants (20,21).
Table IV.
Sequences of primers used in the present study.
| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| RGS18 | TGGACTAGAGGCTTTTAC | ATTTGTTGAGGTCCCTTG |
| ITM2B | TATTCAGAAACGTGAAGC | CTTGACTGTTCAAGAAC |
| PPBP | TGAGACAGAATGAAACAC | AGGTGATGAATCTGCTG |
| NAP1L1 | TGGCCAGCATCTGAGAAC | CACGATGAACCTATTCTG |
| n324674 | TGGATCACCTGAGGTCAG | GGGTTCAAGCAATTCTCC |
| ENST00000442382 | TTCAGCATGGCTGGGAAG | CTCTGTCCTGCTGCCTTG |
| 18S rRNA | GTAACCCGTTGAACCCCATT | CCATCCAATCGGTAGTAGCG |
RGS18, regulator of G protein signaling 18; ITM2B, integral membrane protein 2B; PPBP, pro-platelet basic protein; NAP1L1, nucleosome assembly protein 1 like 1.
Statistical analyses
Statistical analyses were performed using GraphPad Prism 5.0 (GraphPad Software Inc., La Jolla, CA, USA) and SPSS 17.0 (SPSS, Inc., Chicago, IL, USA). Student's t-tests were used to evaluate differences in the RNA expression levels of tissue and plasma samples from patients with GC and healthy controls. The specificity, sensitivity and area under the curve (AUC) of each RNA sample was determined using receiver operating characteristic (ROC) curve analysis. Using the binary outcome of patients with GC or healthy control samples as dependent variables, a logistic regression model was established using the stepwise model selection method. All statistical tests were 2-tailed and P<0.05 was considered to indicate a statistically significant difference.
Results
Plasma RNA screening and data analysis
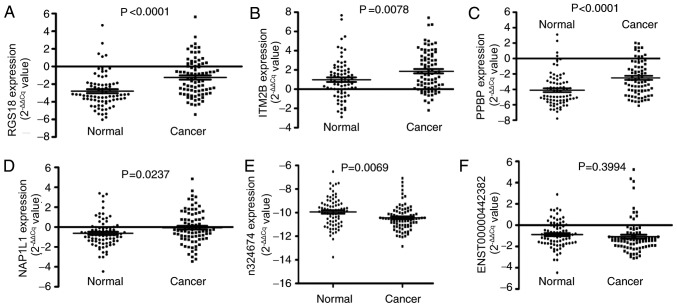
To investigate whether aberrantly expressed RNAs are present in the plasma of patients with GC, 2 pools of plasma specimens were generated from the GC group (n=16) and the control group (n=16). Circulating RNAs in the 2 plasma pools were analyzed using microarray [GeneChip® Human Transcriptome Array 2.0 (Affymetrix; Thermo Fisher Scientific Inc.)]. Numerous significantly differentially expressed RNAs between the 2 pools were detected, including 716 RNAs with a fold-change ≥2. Among these, 577 RNAs were upregulated and 139 RNAs were downregulated in patients with GC compared with the control group. There were 333 RNAs with a fold-change ≥3, including 272 RNAs that were upregulated and 61 RNAs that were downregulated in patients with GC compared with the control group (Fig. 1). Based on the expression levels of probe set regions and integrated transcripts, 19 RNA molecules were selected for verification in the 2 plasma pools using RT-qPCR. It was indicated that the expression levels of 2 non-coding RNAs (n324674 and ENST00000442382) were significantly elevated, while a total of 4 mRNAs, regulator of G-protein signaling 18 (RGS18), integral membrane protein 2B (ITM2B), pro-platelet basic protein (PPBP) and nucleosome assembly protein1-like 1 (NAP1L1), were reduced in the plasma of patients with GC (Figs. 2 and 4). Therefore, the expression levels of these 6 RNAs were further verified in the large-scale training sample of patients with GC.
Figure 1.
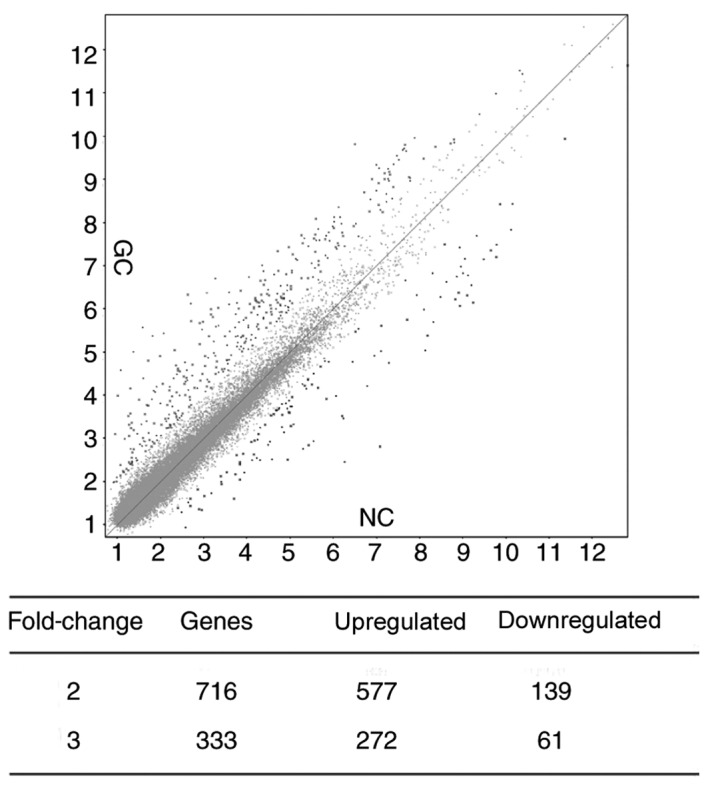
Scatter plot analysis of the transcriptome array. NC, negative control; GC, gastric cancer.
Figure 2.
Large-scale training plasma samples of patients with GC (n=81) and healthy participants (n=77) of candidate RNAs. (A) Scatter plots of plasma levels of regulator of G-protein signaling 18. (B) Scatter plots of plasma levels of integral membrane protein 2B. (C) Scatter plots of plasma levels of pro-platelet basic protein. (D) Scatter plots of plasma levels of nucleosome assembly protein1-like 1. (E) Scatter plots of plasma levels of n324674. (F) Scatter plots of plasma levels of ENST00000442382. Expression levels of RNAs (2−ΔΔCq scale y-axis) were normalized to that of the 18S rRNA gene. RGS18, regulator of G-protein signaling 18; ITM2B, integral membrane protein 2B; PPBP, pro-platelet basic protein; NAP1L1, nucleosome assembly protein1-like 1.
Figure 4.
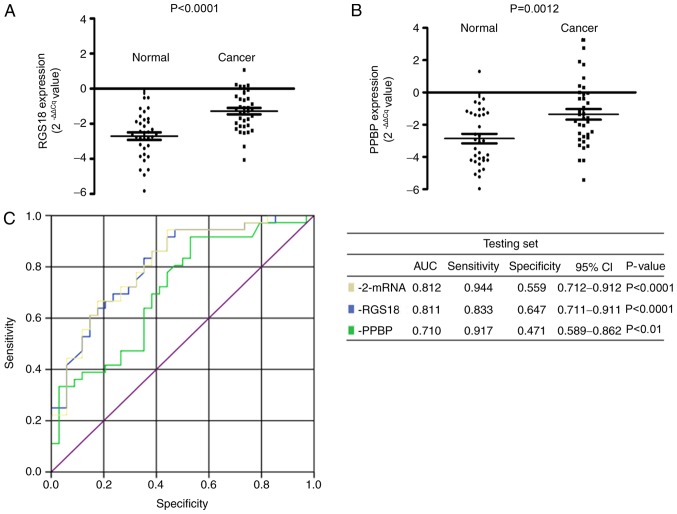
Validation of RNAs in plasma samples of the testing set (n=70). (A) Scatter plot of plasma levels of RGS18 expression in patients with gastric cancer (n=36) and healthy participants (n=34). (B) Scatter plot of plasma levels of PPBP expression in patients with gastric cancer (n=36) and healthy participants (n=34). Expression levels of RNAs (2−ΔΔCq scale y-axis) are normalized to that of 18S rRNA. (C) Receiver operating characteristic curve analysis of plasma RNAs: RGS18 and PPBP. RGS18, regulator of G-protein signaling 18; PPBP, pro-platelet basic protein; CI, confidence interval; AUC, area under the curve.
Large-scale validation in plasma samples
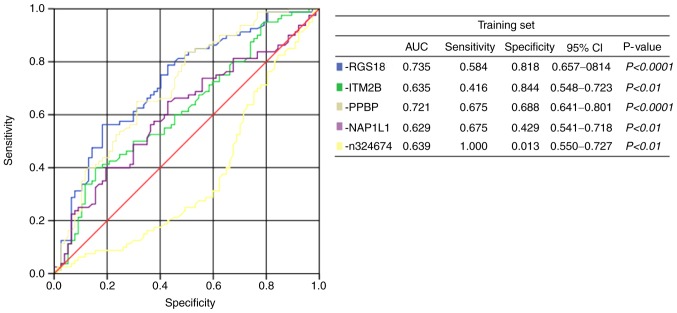
In the large-scale training sample of patients with GC (n=81) and healthy participants (n=77), the expression level of n324674 (P =0.0069) was indicated to be increased in the plasma of patients with GC compared with that of healthy participants. However, the expression level of RGS18 (P<0.0001), ITM2B (P=0.0078), PPBP (P<0.0001) and NAP1L1 (P=0.0237) were reduced in the plasma of patients with GC (Fig. 2). To evaluate whether plasma levels of the 5 candidate RNAs could be used as diagnostic markers for GC, ROC curve analyses were performed. It was indicated that the plasma levels of RGS18, ITM2B, PPBP, NAP1L1 and n324674 discriminated patients with GC from healthy participants, with AUCs of the ROC curves of 0.735 [95% confidence interval (CI), 0.657–0.814], 0.635 (95% CI, 0.548–0.723), 0.721 (95% CI, 0.641–0.801), 0.629 (95% CI, 0.541–0.718) and 0.639 (95% CI, 0.550–0.727), respectively (Fig. 3).
Figure 3.
Receiver operating characteristic curve analysis of plasma RNAs: RGS18, ITM2B, PPBP, NAP1L1 and n324674. RGS18, regulator of G-protein signaling 18; ITM2B, integral membrane protein 2B; PPBP, pro-platelet basic protein; NAP1L1, nucleosome assembly protein1-like 1; CI, confidence interval, AUC, area under the curve.
RGS18 and PPBP, the most significantly differentially expressed RNAs, were subsequently selected for further validation in an independent testing set of patients with GC (n=36) and healthy participants (n=34) (Table II). The results confirmed that the expression levels of RGS18 (P<0.0001) and PPBP (P=0.0012) were reduced in the plasma samples from patients with GC compared with those from healthy participants (Fig. 4). In addition, the AUC value of the 2-mRNA panel was 0.812, higher than that of the individual mRNAs. The association between various clinicopathological features (age, sex, tumor size, histological differentiation, invasion depth, regional lymph nodes, distant metastasis and TNM stage) and expression levels of RGS18 and PPBP in GC plasma samples (n=117), including the training (n=81) and testing sets (n=36), was subsequently investigated. It was indicated that PPBP expression was reduced in female patients with GC compared with male patients with GC (P=0.0328; Fig. 5).
Figure 5.
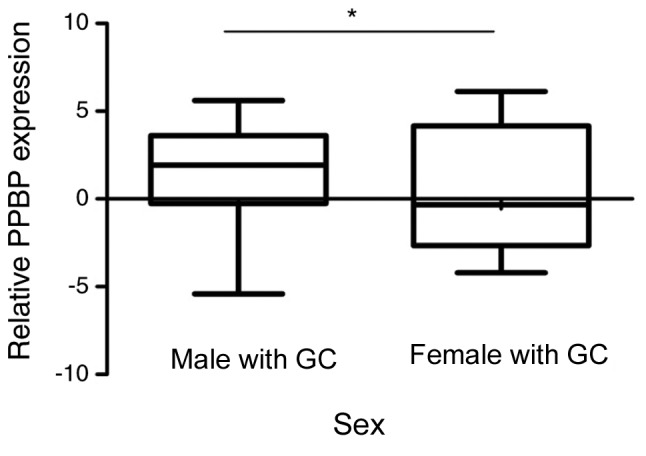
Relative PPBP expression in gastric cancer plasma samples and its clinical significance. PPBP expression was significantly decreased in female patients with GC compared with male patients with GC. *P<0.05. PPBP, pro-platelet basic protein
Detection of RNA expression in GC tissues
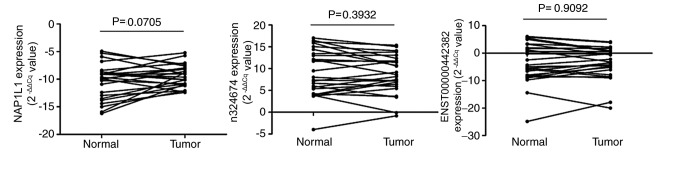
The levels of the 6 candidate RNAs in 26 GC tissue samples and paired adjacent histologically normal tissues were assessed. RGS18, ITM2B and PPBP expression could not be detected using RT-qPCR in GC tissues, whereas the expression of NAP1L1, n324674 and ENST00000442382 was detected. However, their expression patterns in the tissues were different from those in plasma samples. NAP1L1 was downregulated in 9 tumor tissues compared with levels in normal tissues, while n324674 was upregulated in 11 tissues and ENST00000442382 was upregulated in 12 tissues (Fig. 6). According to these results, it was speculated that these plasma RNAs may not derive directly from gastric carcinoma tissues.
Figure 6.
Relative expression levels of NAP1L1, n324674 and ENST00000442382 in 26 gastric cancer and paired normal gastric tissue samples. No significant differences between normal and tumor tissues were observed according to paired t-tests. NAP1L1, nucleosome assembly protein1-like.
Discussion
GC is the third leading cause of cancer-associated mortality worldwide (1–3). GC tumor development and progression is a multi-step processes, involving numerous genetic and epigenetic alterations. However, only a few oncogenes and tumor suppressor genes have been previously identified as being involved in gastric carcinogenesis (22–25). Previous research has reported the use of biomarkers for early detection and prediction of chemotherapeutic sensitivity and prognosis in patients with various types of cancer (11). Among the various approaches reported in previous research, blood-based testing has been indicated as the ideal method for identifying biomarkers in cancer, due to its simplicity and minimal invasiveness (26). However, conventional plasma biomarkers, including CEA and CA72-4, lack sufficient sensitivity and specificity (6,7). Similar to other molecules, cfRNA has been stably detected in plasma or serum samples (27–29). Previous studies have indicated that plasma mRNAs and lncRNAs may be protected by exosome encapsulation and complex formation with proteins, similar to plasma microRNAs (27,30,31). Genome-wide screening approaches have provided opportunities to develop novel diagnostic or prognostic markers and to identify novel therapeutic targets (32).
In the present study, the Human Transcriptome Array was used to identify RNAs that are differentially expressed in plasma samples from patients with GC and healthy participants. The results indicated that the expression levels of RGS18, ITM2B, PPBP and NAP1L1 were significantly decreased in GC plasma samples compared with healthy participant plasma samples, whereas those of n324674 were significantly increased, confirming the differential expression of the aforementioned RNAs in 81 patients with GC and 77 healthy participants. The association between the plasma levels of these 2 RNAs in 117 patients with GC (training and testing sets) and various clinicopathological factors was analyzed. The results indicated that the differential expression of PPBP between patients with GC and healthy participants was more significant in females, as females with GC exhibited decreased expression levels of PPBP compared with male patients with GC. In terms of the utility of these RNAs as biomarkers, it was indicated that plasma levels of RGS18 and PPBP discriminated patients with GC from healthy participants with a combined AUC of 0.812.
Among the 6 candidate RNAs, n324674 and ENST00000442382 were lncRNAs. Numerous lncRNAs have been reported to be associated with disease, particularly cancerous diseases (33). lncRNAs, including H19, imprinted maternally expressed transcript (non-protein coding) (H19), hepatocellular carcinoma upregulated lncRNA, colon cancer associated transcript 1 and run-related transcription factor 1, have been reported to serve functional roles in GC (23,34,35). H19 has also been reported to circulate in the plasmas of patients with GC (24). Therefore, n324674 may be a novel lncRNA associated with GC.
The remaining RNAs are mRNAs. PPBP has been reported to stimulate various cellular processes, including DNA synthesis, mitosis, glycolysis and intracellular cyclic adenosine monophosphate accumulation (16). NAP1L1 participates in DNA replication and may serve a role in modulating chromatin formation and regulating proliferation (36). The underlying mechanisms of PPBP and NAP1L1 in GC require further investigation. RGS proteins have been reported to interact with and negatively regulate G protein activation, and the RGS gene has been indicated to regulate platelet aggregation, haemostasis and thrombosis (37). The ITM2B gene has been reported to be a target of BCL6 repression in lymphomas and neurodegenerative diseases and ITM2B protein generated by alternative splicing has been reported to induce apoptosis in hematopoietic cell lines (38).
Previous studies have demonstrated that the levels of particular RNAs in body fluids were inconsistent with their corresponding levels in tissues (39). It has been reported that one reason for this is that cfRNAs may be transferred to body fluids by exosomes (40,41). Previous studies have reported that certain RNAs can be enriched in exosomes and selectively released from healthy and malignant cells (42). In the present study, RGS18, ITM2B and PPBP could not be detected in GC tissues by RT-qPCR and the expression patterns of NAP1L1, n324674 and ENST00000442382 in GC tissues were not consistent with those in plasma samples. This unexpected result suggests that these RNAs in plasma may derive from other tissues and not GC tissues.
In conclusion, the results of the present study demonstrate that certain RNAs, including RGS18 and PPBP, were significantly downregulated in the plasma of patients with GC compared with healthy controls. These findings imply that a combination of these 2 mRNAs could be used as a diagnostic or prognostic marker for GC.
Acknowledgements
Not applicable.
Funding
The present study was supported partially by the National Key Research and Development Program of China (grant no. 2016YFC1303600) and the Chinese National Natural Science Foundation Projects (grant nos. 31370760, 91540202 and 31470782).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
HF and XZ conceived the project and contributed to the study design. CS, HL and ZP performed the data collection. CS, HL and DK analyzed and interpreted the data. HL, CS and HF wrote the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from all patients. The Ethics Committee of the Chinese PLA General Hospital (Beijing, China) and the China-Japan Union Hospital of Jilin University (Changchun, China) approved the use of samples for the present study.
Patient consent for publication
Written informed consent for the publication of any associated data was obtained from all the patient, or parent, guardian or next of kin (in case of deceased patients).
Competing interests
The authors declare that they have no competing interests.
References
- 1.Stewart BW, Wild C, editors. World cancer report. WHO Press; 2014. International Agency for Research on Cancer and World Health Organization. [Google Scholar]
- 2.Wu HH, Lin WC, Tsai KW. Advances in molecular biomarkers for gastric cancer: miRNAs as emerging novel cancer markers. Exp Rev Mol Med. 2014;16:e1. doi: 10.1017/erm.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu Z, Ye G, Zhang X, Xiao B, Guo J. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer. 2014;120:3320–3328. doi: 10.1002/cncr.28882. [DOI] [PubMed] [Google Scholar]
- 4.Durães C, Almeida GM, Seruca R, Oliveira C, Carneiro F. Biomarkers for gastric cancer: Prognostic, predictive or targets of therapy? Virchows Arch. 2014;464:367–378. doi: 10.1007/s00428-013-1533-y. [DOI] [PubMed] [Google Scholar]
- 5.Fassan M, Baffa R, Kiss A. Advanced precancerous lesions within the GI tract: The molecular background. Best Pract Res Clin Gastroenterol. 2013;27:159–169. doi: 10.1016/j.bpg.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Nakane Y, Okamura S, Akehira K, Boku T, Okusa T, Tanaka K, Hioki K. Correlation of preoperative carcinoembryonic antigen levels and prognosis of gastric cancer patients. Cancer. 1994;73:2703–2708. doi: 10.1002/1097-0142(19940601)73:11<2703::AID-CNCR2820731109>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 7.Marrelli D, Pinto E, De Stefano A, Farnetani M, Garosi L, Roviello F. Clinical utility of CEA CA 19-9 and CA 72-4 in the follow-up of patients with resectable gastric cancer. Am J Surg. 2001;181:16–19. doi: 10.1016/S0002-9610(00)00549-3. [DOI] [PubMed] [Google Scholar]
- 8.Feng F, Tian Y, Xu G, Liu Z, Liu S, Zheng G, Guo M, Lian X, Fan D, Zhang H. Diagnostic and prognostic value of CEA CA19-9, AFP and CA125 for early gastric cancer. BMC Cancer. 2017;17:737. doi: 10.1186/s12885-017-3738-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuoka T, Yashiro M. Biomarkers of gastric cancer: Current topics and future perspective. World J Gastroenterol. 2018;24:2818–2832. doi: 10.3748/wjg.v24.i26.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L, Wang B, Ye G, Xiao B, Guo J. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol. 2015;36:2007–2012. doi: 10.1007/s13277-014-2807-y. [DOI] [PubMed] [Google Scholar]
- 11.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 12.Viereck J, Thum T. Circulating noncoding RNAs as biomarkers of cardiovascular disease and injury. Circ Res. 2017;120:381–399. doi: 10.1161/CIRCRESAHA.116.308434. [DOI] [PubMed] [Google Scholar]
- 13.Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516. doi: 10.1038/srep11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsui NB, Ng EK, Lo YM. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin Chem. 2002;48:1647–1653. [PubMed] [Google Scholar]
- 15.Sethakorn N, Dulin NO. RGS expression in cancer: Oncomining the cancer microarray data. J Recept Signal Transduct Res. 2013;33:166–171. doi: 10.3109/10799893.2013.773450. [DOI] [PubMed] [Google Scholar]
- 16.Guo Q, Jian Z, Jia B, Chang L. CXCL7 promotes proliferation and invasion of cholangiocarcinoma cells. Oncol Rep. 2017;37:1114–1122. doi: 10.3892/or.2016.5312. [DOI] [PubMed] [Google Scholar]
- 17.Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO classifcation of tumours of the digestive system. Fourth Edition. World Health Organization. 2010;3:417. [Google Scholar]
- 18.Washington K. 7th edition of the AJCC cancer staging manual: Stomach. Ann Surg Oncol. 2010;17:3077–3079. doi: 10.1245/s10434-010-1362-z. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.March-Villalba JA, Martínez-Jabaloyas JM, Herrero MJ, Santamaria J, Aliño SF, Dasi F. Cell-free circulating plasma hTERT mRNA is a useful marker for prostate cancer diagnosis and is associated with poor prognosis tumor characteristics. PLoS One. 2012;7:e43470. doi: 10.1371/journal.pone.0043470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ke D, Li H, Zhang Y, An Y, Fu H, Fang X, Zheng X. The combination of circulating long noncoding RNAs AK001058, INHBA-AS1, MIR4435-2HG, and CEBPA-AS1 fragments in plasma serve as diagnostic markers for gastric cancer. Oncotarget. 2017;8:21516–21525. doi: 10.18632/oncotarget.15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 23.Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, Fang G. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437–445. doi: 10.1007/s00432-012-1324-x. [DOI] [PubMed] [Google Scholar]
- 24.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T, et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013;33:3185–3193. [PubMed] [Google Scholar]
- 27.Tani N, Ichikawa D, Ikoma D, Tomita H, Sai S, Ikoma H, Okamoto K, Ochiai T, Ueda Y, Otsuji E, et al. Circulating cell-free mRNA in plasma as a tumor marker for patients with primary and recurrent gastric cancer. Anticancer Res. 2007;27:1207–1212. [PubMed] [Google Scholar]
- 28.Zhou H, Xu W, Qian H, Yin Q, Zhu W, Yan Y. Circulating RNA as a novel tumor marker: An in vitro study of the origins and characteristics of extracellular RNA. Cancer Lett. 2008;259:50–60. doi: 10.1016/j.canlet.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 29.Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174–1179. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iguchi H, Kosaka N, Ochiya T. Secretory microRNAs as a versatile communication tool. Commun Integr Biol. 2010;3:478–481. doi: 10.4161/cib.3.5.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Hu Z, Wang W, Ba Y, Ma L, Zhang C, Wang C, Ren Z, Zhao Y, Wu S, et al. Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int J Cancer. 2012;130:1620–1628. doi: 10.1002/ijc.26177. [DOI] [PubMed] [Google Scholar]
- 33.Gutschner T, Diederichs S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448:315–322. doi: 10.1016/j.bbrc.2013.12.126. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: A clinical and in vitro investigation. Oncol Rep. 2014;31:358–364. doi: 10.3892/or.2013.2850. [DOI] [PubMed] [Google Scholar]
- 36.Yan Y, Yin P, Gong H, Xue Y, Zhang G, Fang B, Chen Z, Li Y, Yang C, Huang Z, et al. Nucleosome assembly protein 1-like 1 (Nap1l1) regulates the proliferation of murine induced pluripotent stem cells. Cell Physiol Biochem. 2016;38:340–350. doi: 10.1159/000438634. [DOI] [PubMed] [Google Scholar]
- 37.Alshbool FZ, Karim ZA, Vemana HP, Conlon C, Lin OA, Khasawneh FT. The regulator of G-protein signaling 18 regulates platelet aggregation, hemostasis and thrombosis. Biochem Biophys Res Commun. 2015;462:378–382. doi: 10.1016/j.bbrc.2015.04.143. [DOI] [PubMed] [Google Scholar]
- 38.Baron BW, Baron RM, Baron JM. The ITM2B (BRI2) gene is a target of BCL6 repression: Implications for lymphomas and neurodegenerative diseases. Biochim Biophys Acta. 2015;1852:742–748. doi: 10.1016/j.bbadis.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui L, Zhang X, Ye G, Zheng T, Song H, Deng H, Xiao B, Xia T, Yu X, Le Y, Guo J. Gastric juice MicroRNAs as potential biomarkers for the screening of gastric cancer. Cancer. 2013;119:1618–1626. doi: 10.1002/cncr.27903. [DOI] [PubMed] [Google Scholar]
- 40.Fellouse FA. Stromal exosomes allow cancer cell autoactivation. Med Sci (Paris) 2014;30:405–407. doi: 10.1051/medsci/20143004014. (In French) [DOI] [PubMed] [Google Scholar]
- 41.Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MÁ, Bernad A, Sánchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gezer U, Özgür E, Cetinkaya M, Isin M, Dalay N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int. 2014;38:1076–1079. doi: 10.1002/cbin.10301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.