Abstract
Hodgkin's lymphoma (HL) is a common hematologic tumor, and the incidence is increasing. At present, it is considered that miRNAs are closely related to HL. Substantial attention has been paid to the effects of miRNA on the pathophysiological process of HL. This study was focused on the potential role of miR-24-3p in HL by targeting DEDD. The reverse transcription-quantitative PCR (RT-qPCR) results demonstrated that miR-24-3p expression was highly elevated and DEDD expression reduced inversely in HL tissues compared to adjacent tissues. According to the results of CKK-8 assays, miR-24-3p was able to accelerate HL cell proliferation. In addition, the results of the Transwell assays also indicated that miR-24-3p promoted the invasion and migration abilities of HL cells. Moreover, the results demonstrated that miR-24-3p inhibited DEDD expression. Hence, the present study revealed that miR-24-3p could accelerate HL development through inhibiting DEDD.
Keywords: microRNA-24-3p, Hodgkin's lymphoma, proliferation, migration, invasion, DEDD
Introduction
Hodgkin's lymphoma (HL) is a common lymphatic tumor, and the incidence rate by year of HL new cases range from three to four per 100,000 western individuals (1). HL is an malignant tumor derived from B cells (2). As it is hard to diagnose advanced HL, and the pathophysiological mechanisms of HL remain unknown, approximately 20% of the newly diagnosed cases die from HL (3). Consequently, more effective treatments to increase HL survival rates need to be developed. At the same time, understanding of the mechanisms of HL development is required. Studies of factors associated with HL, such as genes and miRNAs, are also important.
miRNAs are 19–23 nucleotides in length. As a subgroup of non-coding miRNAs, they play a role in regulating post-transcriptionally via targeting mRNA 3′UTR, causing their degradation or suppressing mRNA translation (4–6). An increasing number of studies have emerged, providing evidence that various miRNAs can exert oncogenic or tumor suppressive functions (7,8). For example, miR-148a suppresses migration and invasion of breast cancer (9). miRNA-29b suppresses tumor angiogenesis, invasion, and metastasis (10). miR-24-3p was proven to be involved in multiple kinds of tumors, including HL (11), head and neck squamous cell cancer (12), and bladder carcinoma (13). However, the function of miR-24-3p in mediating HL invasion and migration remains largely elusive.
The death effector domain-containing protein (DEDD) has a relevance to different kinds of physiological processes, including cell mitosis, cycle and apoptosis (14). Studies have shown that DEDD may be used as a potential therapeutic target and prognostic marker to cure metastasis of carcinoma (15,16). However, the effects of DEDD on regulating HL have not yet been reported. To better understand the anti-metastatic functions and mechanisms of DEDD in HL, the present study investigated the DEDD expression levels in HL tissues and cell lines. At the same time, the correlation between miR-24-3p and DEDD in HL was analyzed.
Materials and methods
Cell cultures
HL cell lines [L1236 (CSC-C0538, Creative Bioarray) and L428 (ws101345, ATCC)] were cultured in RPMI-1640 medium (Cellgro; Corning, Inc., Corning, NY, USA) in an atmosphere with 5% CO2 at 37°C. The medium contained penicillin/streptomycin, 5% L428, 10% L1236 as well as 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Germinal center (GC)-B cells were sorted from tonsil tissue samples of three HL donors aged between 3 and 10 years. Two of the three GC-B cells were purified >98% from human tonsil tissues on the basis of the CD20+lgD−CD38+ expression, as previously described. The third sample was magnetic-activated cell sorting purified >95% based on expression of IgD−CD138−CD3−CD10+. Written informed consents were obtained for the use of the tonsil samples from the parents of the children.
Tissue specimens
We collected the HL patients' tonsil tissue samples and matched normal tissues from the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China) between 2015 and 2017. The obtained tissues were snap-frozen at −80°C. The parents of the HL patients provided written informed consent. The present study obtained approval from the Ethics Committee of the First Affiliated Hospital of Zhengzhou University.
Cell transfection
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Carlsbad, CA, USA) was used to transfect miR-24-3p mimics or inhibitor as well as overexpression vector of DEDD into HL cells. Plasmids were transfected into HL cell lines by X-tremeGENE HP DNA Transfection Reagent (Roche Diagnostics, Basel, Switzerland).
Reverse transcription-quantitative PCR (RT-qPCR)
The TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to isolate the total RNA from HL tissues, adjacent normal tissues and cultured L1236 and L428 cells respectively according to the manufacturer's protocol. The reverse transcription reaction was then conducted to synthesize cDNAs using M-MLV Reverse Transcriptase reagent kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA). RT-qPCR was conducted by Bio-Rad CFX96™ Real-Time PCR Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, USA) with the SYBR-Green kit (Takara Bio, Inc., Otsu, Japan) using specific primers (Table I). The conditions for PCR were as follows: 95°C for 5 min, 40 cycles of denaturation at 95°C (15 sec), 50°C (30 sec) and 72°C (30 sec). The miR-24-3p expression was normalized to U6 while the DEDD expression was normalized to GAPDH. The 2−ΔΔCq method (17) was used to calculate the relative expression levels of genes.
Table I.
Primer sequences for RT-qPCR.
| Primers | Sequences |
|---|---|
| miRNA-24-3p | F 5′-AACACACCTATTCAAGGATTCA-3′ |
| R 5′-CCCATTCAGCAGGAACAGAAA-3′ | |
| U6 | F 5′-CTCGCTTCGGCAGCACA-3′ |
| R 5′-AACGCTTCACGAATTTGCGT-3′ | |
| DEDD | F 5′-TCCCCAGCCCTCTAAAACAG-3′ |
| R 5′-CCGCAGTCTGATGTCACATG-3′ | |
| GAPDH | F 5′-TGCACCACCAACTGCTTAGC-3′ |
| R 5′-GGCATGGACTGTGGTCATGAG-3′ |
RT-qPCR, reverse transcription-quantitative PCR; U6: snRNA, small nuclear RNA; DEDD: death effector domain-containing protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; F, forward; R, reverse.
Western blotting
HL cells were lysed by chilled RIPA lysis buffer (9803; Cell Signaling Technology, Inc., Danvers, MA, USA) which contained phenylmethylsulfonyl fluoride protease inhibitor. The lysates were put onto ice for half an hour, then centrifuged at 14,000 × g at 4°C for 15 min. The BCA Protein Assay kit (Thermo Fisher Scientific, Inc.) was applied to detect the protein concentration according to the manufacturer's protocol. A total of 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate the proteins and then the separated proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane which was incubated at 4°C overnight with primary antibodies diluted in 5% milk. The primary antibodies were as follows: rabbit polyclonal antibodies against DEDD (dilution, 1:1,000; cat. no. ab203655; Abcam, Cambridge, MA, USA) and GAPDH (dilution, 1:1,000, cat. no. ab37168; Abcam). A secondary incubation step was performed with appropriate secondary antibody for 1 h at room temperature. The proteins were detected by a chemiluminescence method. GAPDH (Abcam) was used as an internal loading control.
Cell Counting kit-8 (CCK-8)
The cell proliferation ability of HL cells was observed using CCK-8 (Beyotime Institute of Biotechnology, Haimen, China). HL cells with different transfections were seeded and cultured in 96-well plates for 0, 12, 24, 48 and 72 h, respectively, adding 10 µl CCK-8 reagent into the 96-well plates. At 2 h after being cultured at 37°C, the absorbance of HL cells in different groups at 450 nm was detected by a microplate reader (BioTeke, Winooski, VT, USA).
Transwell assays
The migration and invasion abilities of the treated HL cell lines were assessed by Transwell assays. For the migration assay, the treated cells (1×105 cells/well) were seeded in the top chambers, and RPMI-1640 medium containing 20% FBS was seeded into the bottom chambers, being incubated at 37°C for 24 h with 5% CO2. After that, the migratory cells were fixed, stained and counted. The difference between the migration and invasion assay was that Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) was added into the wells.
Luciferase reporter assays
The treated HL cells were co-transfected with luciferase reporter plasmids which contained wild-type or mutant 3′UTRs of DEDD and miR-24-3p by Lipofectamine 2000. At 48 h after transfection, the Dual-Luciferase Reporter Assay kit (Promega Corp., Madison, WI, USA) was used to perform the luciferase reporter assays.
Statistical analysis
All the above experiments were performed 3 times. The statistical analysis was evaluated by the GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA, USA) together with SPSS 18.0 version (SPSS, Inc. Chicago, IL, USA). Students t test and one way ANOVA followed by Tukeys post hoc test were used to analyze two or multiple groups, respectively. Statistically significant difference was set at P<0.05.
Results
miR-24-3p expression is upregulated and DEDD expression is downregulated in HL
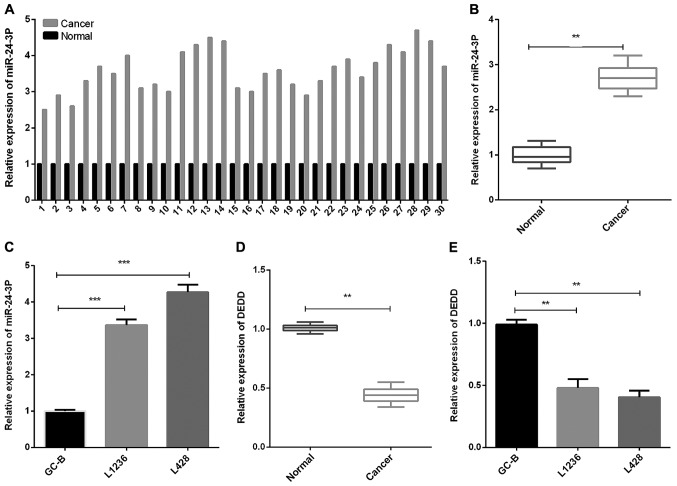
To evaluate miR-24-3p and DEDD expression levels in HL, we collected HL tissues and matched normal tissues from 30 HL patients, and then measured DEDD expression as well as miR-24-3p expression. The results of RT-qPCR revealed that the miR-24-3p expression in HL tissues was significantly increased in contrast with that in the normal control tonsil tissues (Fig. 1A and B, P<0.01). Moreover, we found that the miR-24-3p expression level in GC-B cells was decreased compared with that in HL cells (L1236 and L428) (Fig. 1C, P<0.001). The DEDD expression in HL tissues was significantly decreased compared to normal tonsil tissues (Fig. 1D, P<0.01). Furthermore, similar results were found in GC-B and HL cells (L1236 and L428) (Fig. 1E, P<0.01).
Figure 1.
miR-24-3p expression is elevated and DEDD expression is reduced in HL tissues and cell lines. (A) The miR-24-3p expression in HL tissues and matched normal tissues of 30 HL patients were detected by RT-qPCR. (B) RT- qPCR was performed to measure the miR-24-3p expression in HL tissues (n=30) and matched normal tissues (n=30). (C) miR-24-3p expression in HL L1236 and L428 cells was measured by RT-qPCR. (D) The DEDD expression in human HL tissues (n=30) and matched normal tissues (n=30) was detected by RT-qPCR. (E) The DEDD expression was measured using RT-qPCR in HL cells. **P<0.01, ***P<0.001. HL, Hodgkin's lymphoma; RT-qPCR, reverse transcription-quantitative PCR.
miR-24-3p accelerates HL cell proliferation
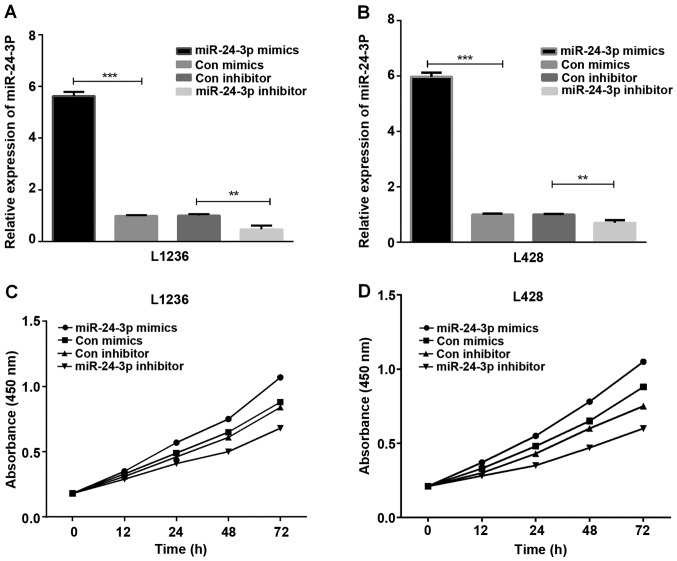
The HL cells (L1236 and L428) transfected with mimics or inhibitor of miR-24-3p were used for detecting the role of miR-24-3p in HL cell proliferation. The results of RT-qPCR assays demonstrated that the miR-24-3p mimic expression was high in L1236 and L428 cells (Fig. 2A and B, P<0.001 and <0.01). Subsequently, the CCK-8 assay was conducted to observe HL cell proliferation ability, and the results demonstrated that miR-24-3p promoted L1236 cell (Fig. 2C) and L428 cell (Fig. 2D) proliferation.
Figure 2.
miR-24-3p accelerates HL cell proliferation. (A and B) The miR-24-3p expression in transfected (A) L1236 and (B) L428 cells were measured using RT-qPCR. (C and D) The CCK-8 assay was conducted to investigate proliferation of transfected (C) L1236 and (D) L428 cells at 0, 12, 24, 48 and 72 h. **P<0.01, ***P<0.001. HL, Hodgkin's lymphoma; RT-qPCR, reverse transcription-quantitative PCR; CCK-8, Cell Counting kit-8.
miR-24-3p accelerates HL cell invasion and migration
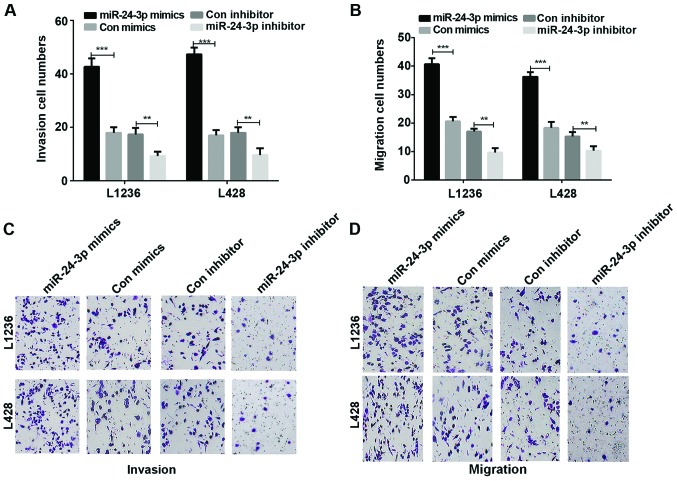
We studied cell invasion and migration abilities in HL cells which were transfected with mimics or inhibitor of miR-24-3p. The results showed that there was a significant rise in the invasion of L136 and L428 cells transfected with mimics of miR-24-3p in contrast with the HL cells transfected with control mimics (P<0.001). On the contrary, the Transwell results demonstrated a significant decrease in the invasion of L136 and L428 cells transfected with inhibitor of miR-24-3p compared to the control group (Fig. 3A and C, P<0.001 and <0.01). In addition, according to the Transwell assays, the results also demonstrated that miR-24-3p promoted HL migration ability (Fig. 3B and D, P<0.001 and <0.01).
Figure 3.
miR-24-3p accelerates HL cell invasion and migration. (A) The invasion and (B) the migration cell numbers of L1236 and L428 were counted. (C) Cell invasion was detected by the Transwell assay in transfected L1236 and L428 cells. (D) The Transwell assay was conducted to detect cell migration in transfected L1236 and L428 cells. **P<0.01, ***P<0.001. HL, Hodgkin's lymphoma.
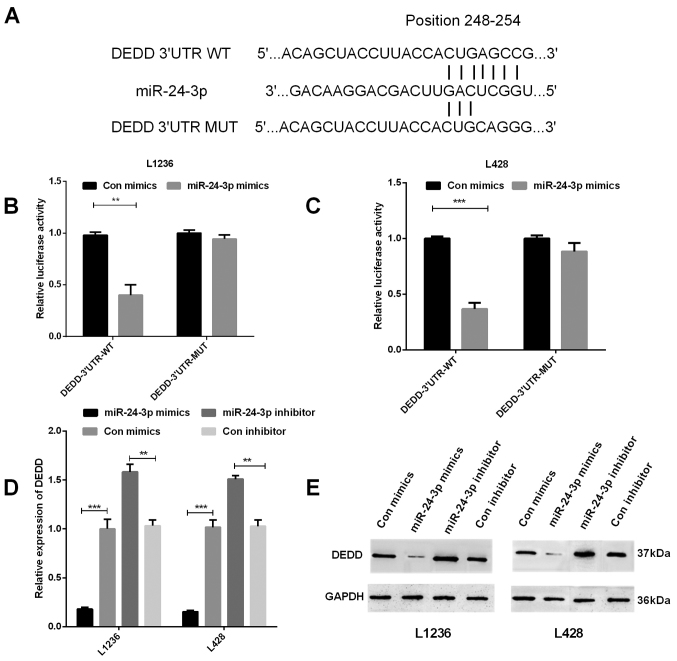
miR-24-3p suppresses DEDD gene transcription in HL by targeting its 3′UTR
To investigate whether DEDD expression had relevance to miR-24-3p expression and to better understand the function of miR-24-3p in HL, TargetScan was used to find the target sites in the DEDD sequence of miR-24-3p (Fig. 4A). We measured the DEDD 3′UTR luciferase activities by performing luciferase reporter gene assays. We co-transfected mimics of miR-24-3p and wild-type DEDD 3′UTR vector or mutant DEDD 3′UTR vector into HL cells to observe whether DEDD was the target of miR-24-3p, and then detected the role of miR-24-3p in regulating the mRNA and protein expression of DEDD. The results showed that there was a significant decrease of fluorescence activity in both L1236 (P<0.01) and L428 (P<0.001) cells co-transfected with the wild-type DEDD 3′UTR vector and miR-24-3p in contrast with the control group, however, between cells co-transfected with the mutant DEDD 3′UTR vector and miR-24-3p and the control group, there was no significant difference (Fig. 4B and C). The results of RT-qPCR and western blotting both demonstrated that miR-24-3p could inhibit DEDD expression in L1236 and L428 cells (Fig. 4D and E, P<0.001 and <0.01).
Figure 4.
The gene transcription of DEDD in HL was inhibited by miR-24-3p. (A) TargetScan was applied to predict the target site of miR-24-3p in the DEDD sequence. (B and C) The luciferase reporter gene assays were performed to detect the fluorescence activities of the DEDD 3′UTR in (B) L1236 and (C) L428 cells which were co-transfected with wild-type DEDD 3′UTR or mutational type DEDD 3′UTR and miR-24-3p, respectively. (D) The DEDD mRNA expression in transfected L1236 and L428 cells was determined using RT-qPCR. (E) The DEDD protein expression was evaluated in transfected L1236 and L428 cells by western blotting. **P<0.01, ***P<0.001. HL, Hodgkin's lymphoma; RT-qPCR, reverse transcription-quantitative PCR.
The role of DEDD in regulating miR-24-3p effects in HL cell migration and invasion
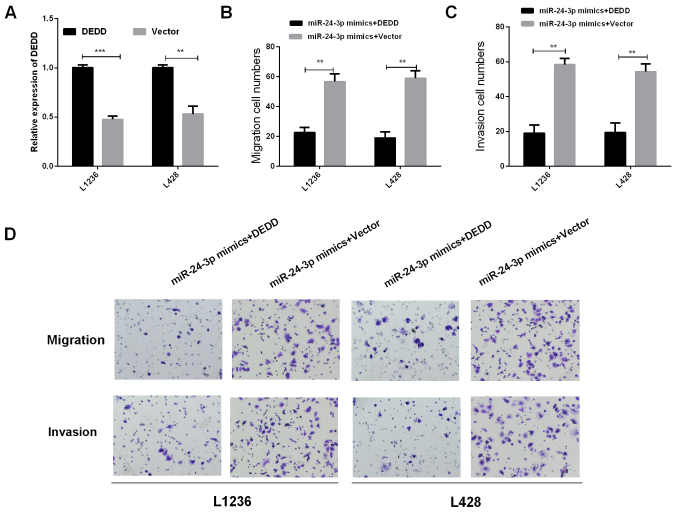
We investigated whether DEDD was required in regulating miR-24-3p functions of promoting HL cell invasion and migration. Overexpression vector of DEDD and miR-24-3p mimics were co-transfected into L1236 and L428 cells using RT-qPCR to detect the DEDD mRNA expression, the results are shown in Fig. 5A. Then, we detected the migration and invasion in HL cells which were co-transfected with overexpression vector of DEDD and miR-24-3p mimics using Transwell assay. The results revealed that the overexpression of DEDD markedly reversed miR-24-3p-mediated promotion of cell migration and invasion in HL cells (Fig. 5B-D, P<0.01). Collectively, these data suggested that DEDD may reverse partial function of miR-24-3p in HL cells.
Figure 5.
DEDD may regulate miR-24-3p function in HL cell invasion and migration. (A) The DEDD mRNA expression in HL cells transfected with overexpression vector of DEDD using RT-qPCR. (B) The migration and (C) the invasion cell numbers of L1236 and L428 co-transfected with overexpression vector of DEDD and miR-24-3p mimics were counted. (D) Transwell assay showed the cell migration and invasion abilities in HL cells co-transfected with overexpression vector of DEDD and miR-24-3p mimics. **P<0.01, ***P<0.001. HL, Hodgkin's lymphoma; RT-qPCR, reverse transcription-quantitative PCR.
Discussion
HL is a particular B-cell malignancy, accounting for a large percentage of lymphomas (18). Despite significant advances in the treatment of HL, existing treatments fail to cure 10–20% of HL patients. Similarly, 10–20% of HL patients may be over-treated. So, reliable predictive biomarkers for HL patients who need intensive therapy remain a challenge. Such biomarkers could also provide insight into the biology of HL.
Several studies have indicated that different kinds of miRNAs are involved in different kinds of biological activities, for instance, cell proliferation, migration, metastasis and inflammation (19,20). Multiple studies have found that many different miRNAs play an important role in the development of HL. For example, miR-124a methylation is associated with aggressive HL disease (21); miR-374b can suppress cell proliferation and promote cell apoptosis by inhibiting AKT1 and Wnt-16 in T-cell lymphoblastic lymphoma (22); miR-9 methylation is a common event in HL and participated in HL pathogenesis (23); overexpression of miR-155 and inhibition of its target NIAM enhance cell growth in HL (24).
miR-24-3p is one of the most studied miRNAs in different kinds of tumors. Over the past decade, increasing evidence has indicated that dysregulation of miRNAs makes contributions to various aspects of tumorigenesis process. miR-24-3p is a master regulator in gene regulation and is related to diverse human disease (25). Several other studies also revealed an oncogenic role for miR-24-3p in other types of cancer, for instance, miR-24-3p has been proved to promote glioma cell proliferation by targeting MXI1 (26); miR-24-3p represses small cell lung carcinoma VP16-DDP chemoresistance through ATG4A (27). However, as far as we know, there have been no previous studies which report the mechanism of miR-24-3p in HL. To explore the functional relevance of miR-24-3p in HL, we compared its expression level in HL tissues to that in paracarcinoma tissues. The results demonstrated that the miR-24-3p expression level in HL tissues was higher than that in paracarcinoma tissues. In addition, it also indicated that miR-24-3p could promote HL cell proliferation, invasion and migration through DEDD.
The present study provides evidence that miR-24-3p is involved in regulating HL cell proliferation, invasion and migration. At the same time, miR-24-3p expression was increased and that of DEDD expression was decreased in HL tissues. Moreover, miR-24-3p was able to suppress the gene transcription of DEDD. Hence, the present study demonstrated that miR-24-3p promoted HL progression via suppressing DEDD. Thus, it is suggested that miR-24-3p may be a key potential therapeutic target for the treatment of HL.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
JW drafted the manuscript. JW, KY and XLv were mainly devoted to cell culture. JW, QY and MS performed RT-qPCR. XLi and HS were responsible for western blotting. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China). Signed informed consent was obtained from the parents of the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Gibcus JH, Tan LP, Harms G, Schakel RN, de Jong D, Blokzijl T, Möller P, Poppema S, Kroesen BJ, van den Berg A. Hodgkin lymphoma cell lines are characterized by a specific miRNA expression profile. Neoplasia. 2009;11:167–176. doi: 10.1593/neo.08980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vardiman JW. The World Health Organization (WHO) classification of tumors of the hematopoietic and lymphoid tissues: An overview with emphasis on the myeloid neoplasms. Chem Biol Interact. 2010;184:16–20. doi: 10.1016/j.cbi.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Paydas S, Acikalin A, Ergin M, Celik H, Yavuz B, Tanriverdi K. Micro-RNA (miRNA) profile in Hodgkin lymphoma: Association between clinical and pathological variables. Med Oncol. 2016;33:34. doi: 10.1007/s12032-016-0749-5. [DOI] [PubMed] [Google Scholar]
- 4.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: Progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lima CR, Gomes CC, Santos MF. Role of microRNAs in endocrine cancer metastasis. Mol Cell Endocrinol. 2017;456:62–75. doi: 10.1016/j.mce.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 8.Hemmatzadeh M, Mohammadi H, Karimi M, Musavishenas MH, Baradaran B. Differential role of microRNAs in the pathogenesis and treatment of esophageal cancer. Biomed Pharmacother. 2016;82:509–519. doi: 10.1016/j.biopha.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Jiang Q, He M, Ma MT, Wu HZ, Yu ZJ, Guan S, Jiang LY, Wang Y, Zheng DD, Jin F, et al. MicroRNA-148a inhibits breast cancer migration and invasion by directly targeting WNT-1. Oncol Rep. 2016;35:1425–1432. doi: 10.3892/or.2015.4502. [DOI] [PubMed] [Google Scholar]
- 10.Fang JH, Zhou HC, Zeng C, Yang J, Liu Y, Huang X, Zhang JP, Guan XY, Zhuang SM. MicroRNA-29b suppresses tumor angiogenesis, invasion, and metastasis by regulating matrix metalloproteinase 2 expression. Hepatology. 2011;54:1729–1740. doi: 10.1002/hep.24577. [DOI] [PubMed] [Google Scholar]
- 11.Yuan Y, Kluiver J, Koerts J, de Jong D, Rutgers B, Abdul Razak FR, Terpstra M, Plaat BE, Nolte IM, Diepstra A, et al. miR-24-3p is overexpressed in Hodgkin lymphoma and protects Hodgkin and Reed-Sternberg cells from apoptosis. Am J Pathol. 2017;187:1343–1355. doi: 10.1016/j.ajpath.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 12.Sun X, Xiao D, Xu T, Yuan Y. miRNA-24-3p promotes cell proliferation and regulates chemosensitivity in head and neck squamous cell carcinoma by targeting CHD5. Future Oncol. 2016;12:2701–2712. doi: 10.2217/fon-2016-0179. [DOI] [PubMed] [Google Scholar]
- 13.Yu G, Jia Z, Dou Z. miR-24-3p regulates bladder cancer cell proliferation, migration, invasion and autophagy by targeting DEDD. Oncol Rep. 2017;37:1123–1131. doi: 10.3892/or.2016.5326. [DOI] [PubMed] [Google Scholar]
- 14.Hua F, Xue JF, Lü XX, Hu ZW. DEDD decreases Smad3 activity, promotes tumor cell apoptosis and inhibits proliferation. Yao Xue Xue Bao. 2013;48:680–685. (In Chinese) [PubMed] [Google Scholar]
- 15.Carstens JL, Lovisa S, Kalluri R. Microenvironment-dependent cues trigger miRNA-regulated feedback loop to facilitate the EMT/MET switch. J Clin Invest. 2014;124:1458–1460. doi: 10.1172/JCI75239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lv Q, Hua F, Hu ZW. DEDD, a novel tumor repressor, reverses epithelial-mesenchymal transition by activating selective autophagy. Autophagy. 2012;8:1675–1676. doi: 10.4161/auto.21438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Wang F, Xie X, Yang X, Jiang G, Gu J. The influence of marital status on the survival of patients with Hodgkin lymphoma. Oncotarget. 2017;8:51016–51023. doi: 10.18632/oncotarget.16879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Paris PL, Chen J, Ngo V, Yao H, Frazier ML, Killary AM, Liu CG, Liang H, Mathy C, et al. Next generation sequencing of pancreatic cyst fluid microRNAs from low grade-benign and high grade-invasive lesions. Cancer Lett. 2015;356:404–409. doi: 10.1016/j.canlet.2014.09.029. (2 Pt B) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Leva G, Croce CM. The role of microRNAs in the tumorigenesis of ovarian cancer. Front Oncol. 2013;3:153. doi: 10.3389/fonc.2013.00153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Ben Dhiab M, Ziadi S, Ksiaa F, Louhichi T, Ben Gacem R, Ben Zineb A, Amara K, Hachana M, Trimeche M. Methylation of miR124a-1, miR124a-2, and miR124a-3 in Hodgkin lymphoma. Tumour Biol. 2015;36:1963–1971. doi: 10.1007/s13277-014-2802-3. [DOI] [PubMed] [Google Scholar]
- 22.Qian D, Chen K, Deng H, Rao H, Huang H, Liao Y, Sun X, Lu S, Yuan Z, Xie D, et al. MicroRNA-374b suppresses proliferation and promotes apoptosis in T-cell lymphoblastic lymphoma by repressing AKT1 and Wnt-16. Clin Cancer Res. 2015;21:4881–4891. doi: 10.1158/1078-0432.CCR-14-2947. [DOI] [PubMed] [Google Scholar]
- 23.Ben Dhiab M, Ziadi S, Louhichi T, Ben Gacem R, Ksiaa F, Trimeche M. Investigation of miR9-1, miR9-2 and miR9-3 methylation in Hodgkin lymphoma. Pathobiology. 2015;82:195–202. doi: 10.1159/000432402. [DOI] [PubMed] [Google Scholar]
- 24.Slezak-Prochazka I, Kluiver J, de Jong D, Smigielska-Czepiel K, Kortman G, Winkle M, Rutgers B, Koerts J, Visser L, Diepstra A, et al. Inhibition of the miR-155 target NIAM phenocopies the growth promoting effect of miR-155 in B-cell lymphoma. Oncotarget. 2016;7:2391–2400. doi: 10.18632/oncotarget.6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chhabra R, Dubey R, Saini N. Cooperative and individualistic functions of the microRNAs in the miR-23a~27a~24-2 cluster and its implication in human diseases. Mol Cancer. 2010;9:232. doi: 10.1186/1476-4598-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu W, Liu M, Peng X, Zhou P, Zhou J, Xu K, Xu H, Jiang S. miR-24-3p and miR-27a-3p promote cell proliferation in glioma cells via cooperative regulation of MXI1. Int J Oncol. 2013;42:757–766. doi: 10.3892/ijo.2012.1742. [DOI] [PubMed] [Google Scholar]
- 27.Pan B, Chen Y, Song H, Xu Y, Wang R, Chen L. Mir-24-3p downregulation contributes to VP16-DDP resistance in small-cell lung cancer by targeting ATG4A. Oncotarget. 2015;6:317–331. doi: 10.18632/oncotarget.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.