Abstract
Mutations of telomerase reverse transcriptase (TERT) and the α thalassemia/mental retardation syndrome X-linked (ATRX) genes have been the subject of numerous studies on the classification and prognosis of glioma. However, the association between TERT and ATRX in World Health Organization (WHO) grade II to IV glioma remains unclear. The present study utilized Sanger sequencing and immunohistochemical methods to detect the expression of the TERT promoter region, ATRX mutations and proliferation marker protein Ki-67 (Ki-67) protein expression in 179 cases of glioma. The current study analyzed these variables and their association with clinicopathological characteristics to further basic research and provide a theoretical basis for the clinical diagnosis and treatment of this type of tumor (1). The results demonstrated that TERT promoter mutations were negatively associated with ATRX. Additionally, Ki-67 protein expression in TERT wild-type samples was higher compared with samples with ATRX deletion. Overall, the results demonstrated, for the first time to the best of the authors' knowledge, that TERT promoter mutations are negatively associated with ATRX expression in WHO grade II to IV gliomas. These findings provide a theoretical basis for further basic research and may improve clinical diagnosis and treatment of glioma in the future.
Keywords: telomerase reverse transcriptase promoter, α thalassemia/mental retardation syndrome X-linked, isocitrate dehydrogenase, mutation, proliferation marker protein Ki-67, glioma
Introduction
Mutations in telomerase reverse transcriptase (TERT) and the α thalassemia/mental retardation syndrome X-linked (ATRX) gene are the subject of numerous studies on the classification and prognosis of glioma (1). A telomere is a nucleoprotein complex containing hundreds of nucleotide repeats that exists at the end of all eukaryotic chromosomes and gradually shortens with each round of mitosis (2). Telomerase is a reverse transcriptase that uses its own RNA as a template to add nucleotides to telomeres (3). Cancer cells characteristically acquire the infinite capability to divide by maintaining telomere length through the sustained expression of telomerase, or in the absence of telomerase, by an alternative lengthening of telomeres (ALT) mechanism (3). Previous studies have demonstrated that mutations in the core region of the TERT promoter in glioma and various other tumors lead to increased telomerase activity (2,4–7). In addition, various tumors with alternative telomeres exhibit ATRX gene mutations, with an incidence of ~75% in grade II–III glioma (2). Therefore, these mutations may be important to glioma formation and development.
The increasing number of reports on the mechanism of telomere maintenance in glioma has been crucial for disease treatment and patient prognosis (4,5,7–15). However, to the best of the authors' knowledge, no studies have assessed the distribution and importance of ATRX alterations and TERT promoter mutations in glioma. The aim of the present study was therefore to assess ATRX and TERT promoter mutations in glioma and to analyze their role in the proliferative activity and WHO pathological grades of gliomas, thus furthering research and providing a theoretical basis for the clinical diagnosis, and treatment of glioma.
Materials and methods
General information
Tumor tissues were obtained from August 2016 to June, 2017 at Tianjin Huanhu Hospital (Tianjin, China). To ensure the accuracy of pathological diagnosis, the human glioma genome database in China, (Chinese Glioma Genome Atlas, CGGA, http://www.cgga.org.cn/), was used, and >80% of the tissue was tumor tissue. All of the tissues were diagnosed by an experienced pathologist. The present study was approved by the Medical Ethics Committee of Tianjin Huanshu Hospital and all patients provided signed informed consent. The inclusion criteria were as follows: i) Specimens must be pathologically diagnosed with grade II–IV glioma using the 2016 version of the World Health Organization (WHO) central nervous system tumor classification (16); and ii) The pathology library had complete and adequate paraffin specimens. Samples were excluded if tissues were of poor quality and if they were classified as WHO grade I (low grade) glioma. Following screening, 179 specimens were obtained, which included 38 WHO grade II glioma samples, 43 grade III samples and 98 grade IV samples (108 males and 71 females; age range, 8–84 years; median age, 48.5 years).
Immunohistochemistry
ATRX immunohistochemistry of polyclonal rabbit antibody (dilution 1:400; product code HPA001906; Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) was performed using an automated immunostainer (Leica Bond III; Leica Microsystems GmbH, Wetzlar, Germany) and standard protocols (10). Evaluation was performed by two observers simultaneously on a multi-headed microscope and scoring was done in consensus. Only nuclear staining was considered for evaluation. Cases with more than 10% positive tumor cells were scored as positive. Endothelial cells, cortical neurons and infiltrating inflammatory cells were generally positive and served as internal positive controls. Cases with negative tumors cells in which vessel cells and neurons were not stained were not evaluated and not considered for further statistical evaluation. In cases with inhomogeneous immunoreaction, areas with highest staining were scored. Regarding the analysis of Ki-67, the following methods were used in this study: Immunohistochemical staining was conducted and the proportion of the malignant cells staining positive for the nuclear antigen Ki-67 was evaluated in a quantitative and visual way using light microscopes (magnification, ×400). The Ki-67 percentage score is defined as the percentage of positively stained tumor cells among the total number of malignant cells assessed. Withal only the positivity is of interest independent of the intensity of coloration. To ensure quality assurance of the staining, positive control tissues are completed. A Ki-67 cut-off point of 27.2% was defined mean of Ki-67 expression (Table I) The complete specimen is investigated and checked for immunostaining tumor cell nuclei. Scoring is conducted considering the whole tumor section and not only limited to the hot spots of the carcinoma or to the most evident positive parts within the invasive segment or the front of necrosis.
Table I.
Associations between various factors and TERT status.
| TERT mutations | ||||
|---|---|---|---|---|
| Variable | Total number | Mut | WT | P-value |
| Number | 179 | 97 | 82 | |
| Age (years) | ||||
| ≤48.5 | 71 | 46 | 25 | 0.038 |
| >48.5 | 108 | 72 | 36 | |
| Sex | ||||
| Male | 108 | 58 | 50 | 0.253 |
| Female | 71 | 31 | 40 | |
| WHO grade | ||||
| II | 38 | 17 | 21 | 0.015 |
| III | 43 | 26 | 17 | |
| IV | 98 | 54 | 44 | |
| IDH | ||||
| Mut | 73 | 35 | 38 | 0.066 |
| WT | 106 | 62 | 44 | |
| ATRX | ||||
| Expression | 128 | 86 | 37 | 0.001 |
| Loss | 50 | 11 | 39 | |
| Ki-67 expression (%) | ||||
| ≤27.12a | 88 | 45 | 43 | 0.167 |
| >27.12 | 91 | 53 | 38 | |
| Recurrence (n) | ||||
| Recurrence | 8 | 5 | 3 | 0.442 |
| Non-recurrence | 180 | 93 | 87 | |
Mean of Ki-67 expression is 27.12%. TERT, telomerase reverse transcriptase; WHO, world health organization; IDH, isocitrate dehydrogenase; ATRX, α thalassemia/mental retardation syndrome X-linked; Mut, mutation; WT, wild-type.
Sanger sequencing
A pyrosequencing assay was designed to examine the two reported mutation hot spots in the TERT promoter. They result in a cytidine to thymidine transition at position 1,295,228 or 1,295,250 on the reverse strand of chromosome 5. These mutations are termed C228T and C250T, respectively. The following primer pair for polymerase chain reaction (PCR) was designed to amplify a region including both sites; forward, GTCCTGCCCCTTCACCTT (biotinylated at the 5′ end) and reverse, GCACCTCGCGGTAGTGG (biotinylated at the 5′ end). The primers for pyrosequencing were designed immediately upstream of the hotspot as follows: TGTAAAACGACGGCCAGT (biotinylated at the 5′ end), CAGGAAACAGCTATGACC (biotinylated at the 5′ end). The mutation hotspots at codon 132 of IDH1 and codon 140 of IDH2 were also screened using pyrosequencing. The following oligonucleotide primers were used for amplification; IDH1 forward, ATGAGAAGAGGGTTGAGGAGTT (biotinylated at the 5′ end) and reverse, GCAAAATCACATTATTGCCAAC (biotinylated at the 5′ end); IDH2 forward, GCTGCAGTGGGACCACTATT (biotinylated at the 5′ end) and reverse, CTAGGCGAGGAGCTCCAGT (biotinylated at the 5′ end). The primers for pyrosequencing were designed immediately upstream of the hotspot as follows: For IDH1, IDH2: TGTAAAACGACGGCCAGT (biotinylated at the 5′ end), CAGGAAACAGCTATGACC (biotinylated at the 5′ end). The pyrosequencing assays were designed to detect all known mutations at IDH1 R132 or IDH2 R140. Templates for Sanger sequencing were prepared by amplifying genomic DNA with the same primer pair as for pyrosequencing without biotinylation of the reverse primer. Cycle sequencing was carried out using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol with the forward PCR primer as a sequencing primer.
Statistical methods
SPSS 19.0 statistical software (IBM Corp., Armonk, NY, USA) was used for data analysis. The association between the indexes was statistically analyzed using logistic regression, the Fisher exact test and Logistic Regression. One-way analysis of variance was used to analyze Ki-67 expression data with Fisher's Least Significant Difference post-hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
TERT promoter mutations are positively associated with age and WHO grade, but negatively associated with ATRX mutations
In the present study, the Logistic Regression was utilized to assess the effects of age, sex, WHO classification, IDH mutation, ATRX loss and Ki-67 protein expression on TERT promoter mutations (Table I). The results demonstrated that sex, IDH mutation and Ki-67 protein expression did not significantly affect TERT promoter mutations. However, age, WHO grade and ATRX loss were significantly associated. Logistic regression was then utilized to asses age, WHO grade and ATRX loss in TERT promoter mutations. The three variables included in the model (age, WHO grade and ATRX loss) were all determined to be associated with TERT mutation status (Table II). The probability of mutation in the TERT promoter in ATRX wild-type samples was 20.40 (1/0.049) times higher than that in ATRX mutants. Furthermore, the TERT mutation probability increased by 4.9% for each additional year of age (Table II). In addition, TERT promoter mutations were associated with WHO grade. Using WHO grade IV as a reference variable, the odds ratio of TERT promoter mutation was 7.447 and 8.211 for WHO grades II and III, respectively (Table II).
Table II.
Logistic regression analysis of factors associated with TERT.
| 95% CI of EXP (B) | |||||||
|---|---|---|---|---|---|---|---|
| Factor | B | S.E. | df | Sig. | Exp (B) | Minimum | Upper limit |
| Age | 0.049 | 0.024 | 1 | 0.038 | 1.050 | 1.003 | 1.100 |
| WHO grade | 2 | 0.015 | |||||
| WHO II | 2.008 | 0.883 | 1 | 0.023 | 7.447 | 1.319 | 42.034 |
| WHO III | 2.106 | 0.827 | 1 | 0.011 | 8.211 | 1.623 | 41.542 |
| ATRX | −3.007 | 0.927 | 1 | 0.001 | 0.049 | 0.008 | 0.304 |
TERT, telomerase reverse transcriptase; WHO, world health organization; ATRX, α thalassemia/mental retardation syndrome X-linked; B, regression coefficient; S.E., standard error; Sig., significance; df, degree of freedom; exp, odd ratio.
ATRX mutations are positively associated with age and IDH mutations
The Logistic Regression was also used to assess the effects of age, sex, WHO classification, IDH mutation, TERT mutation and Ki-67 protein expression on ATRX status (Table III). The results indicated that sex, WHO classification and Ki-67 expression did not significantly affect ATRX stats. However, age and IDH mutation were determined to be significantly associated. The associations between age, IDH mutation and ATRX loss was further assessed using logistic regression. The two variables included in the model (age and IDH mutation) were determined to be statistically significant (Table IV). The probability of ATRX increased by 8.8% for each additional year of age. Additionally, the probability of ATRX mutation in IDH mutant samples was 14.03 times higher than that in IDH wild-type samples.
Table III.
The association between various factors and ATRX status.
| Variable | Total number | Loss | Expression | P-value |
|---|---|---|---|---|
| Number | 179 | 50 | 123 | |
| Age | ||||
| ≤48.5 | 71 | 39 | 32 | 0.005 |
| >48.5 | 108 | 17 | 91 | |
| Sex | ||||
| Male | 108 | 31 | 77 | 0.739 |
| Female | 71 | 25 | 46 | |
| WHO grade | ||||
| II | 38 | 21 | 17 | 0.507 |
| III | 43 | 24 | 19 | |
| IV | 98 | 71 | 27 | |
| IDH | ||||
| Mut | 73 | 34 | 39 | 0.003 |
| WT | 106 | 22 | 84 | |
| TERT | ||||
| Mut | 97 | 11 | 86 | 0.001 |
| WT | 82 | 45 | 37 | |
| Ki-67 expression index | ||||
| ≤27.12 | 87 | 36 | 51 | 0.131 |
| >27.12 | 91 | 20 | 71 | |
| Recurrence (n) | ||||
| Recurrence | 8 | 4 | 4 | 0.135 |
| Non-recurrence | 171 | 52 | 119 | |
ATRX, α thalassemia/mental retardation syndrome X-linked; WHO, world health organization; IDH, isocitrate dehydrogenase; TERT, telomerase reverse transcriptase; Ki-67, proliferation marker protein Ki-67.
Table IV.
Logistic regression analysis of various factors with ATRX.
| 95% CI of EXP (B) | |||||||
|---|---|---|---|---|---|---|---|
| B | S.E. | df | Sig. | Exp (B) | Minimum | Upperlimit | |
| Age | 0.085 | 0.030 | 1 | 0.005 | 1.088 | 1.026 | 1.154 |
| IDH | 2.641 | 0.888 | 1 | 0.003 | 14.033 | 2.464 | 79.927 |
ATRX, α thalassemia/mental retardation syndrome X-linked; IDH, isocitrate dehydrogenase; B, regression coefficient; S.E., standard error; Sig, significance; df, degree of freedom; exp, odd ratio.
TERT wild-type and ATRX wild-type tumors exhibit high Ki-67 protein expression; however the wild-type TERT promoter with ATRX mutations exhibits low Ki-67 protein expression
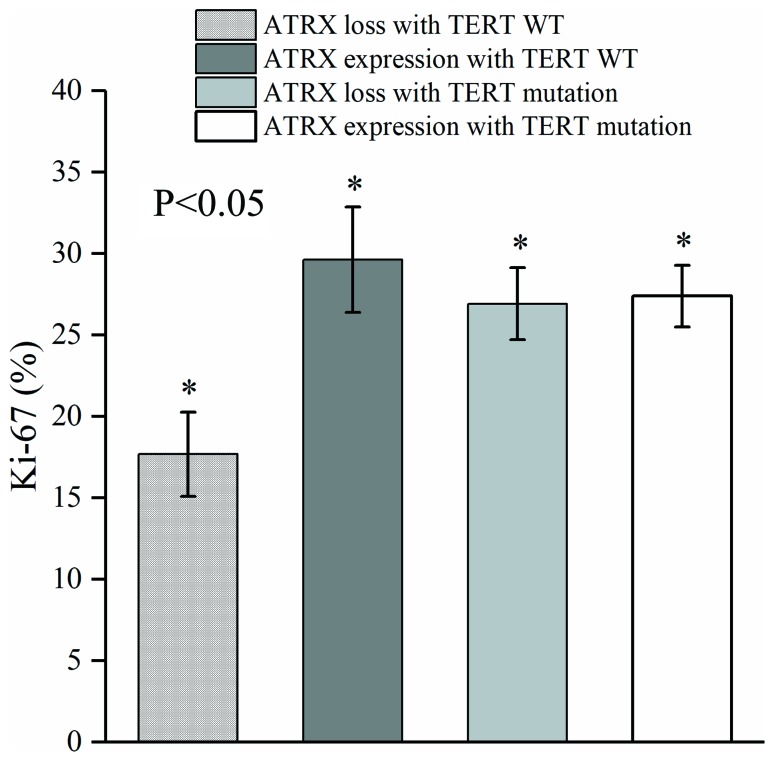
Karsy et al (17) have identified ATRX and TERT mutations in anaplastic gliomas. To assess the association of ATRX and TERT mutations with WHO grade II–IV gliomas, tissues in these genes were collected (Table V). In addition, ATRX mutations were added to TERT promoter mutations to observe their association with Ki-67 protein expression (Fig. 1). Difference of measurement data was compared with single factor analysis of variance (P=0.018). The results demonstrated that TERT promoter mutations were significantly different in ATRX mutant and wild-type populations. In the TERT wild-type group, Ki-67 protein was least abundant in patients that were ATRX-deficient, and Ki-67 expression was highest in patients with ATRX expression (Fig. 2).
Table V.
The association between ATRX and TERT status.
| WHO grade | ATRX loss/retention (%) | TERT loss/WT (%) | ATRX loss and TERT WT | ATRX loss and TERT loss | ATRX retention and TERT WT | ATRX retention and TERT loss |
|---|---|---|---|---|---|---|
| II | 21/17 (44.74) | 21/17 (44.74) | 17 | 0 | 4 | 17 |
| III | 22/19 (44.18) | 17/26 (60.47) | 12 | 3 | 5 | 23 |
| IV | 68/27 (27.55) | 44/54 (55.10) | 16 | 8 | 28 | 46 |
ATRX, α thalassemia/mental retardation syndrome X-linked; TERT, telomerase reverse transcriptase; WHO, world health organization; WT, wild-type.
Figure 1.
Difference between ATRX mutants and wild-type populations. In the wild-type TERT group, Ki-67 protein was least abundant in ATRX-deficient patients. In the four groups, Ki-67 expression was highest in patients without ATRX deletion. *P<0.05. TERT, telomerase reverse transcriptase; ATRX, α thalassemia/mental retardation syndrome X-linked; WT, wild-type; Ki-67, proliferation marker protein Ki-67.
Figure 2.
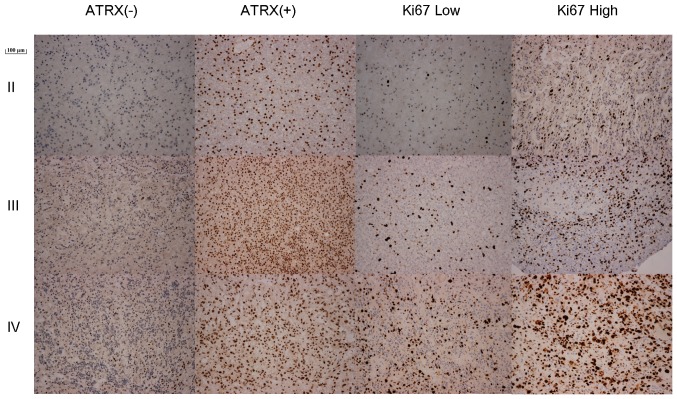
ATRX and Ki-67 staining in II–IV grade glioma (magnification, ×400). The dark brown stain indicates ATRX wild-type (+) and the dark blue stain indicate ATRX mutant-type (−). ATRX, α thalassemia/mental retardation syndrome X-linked; Ki-67, proliferation marker protein Ki-67.
Discussion
Telomeres consist of hundreds of nucleotide repeats that are present at the end of all eukaryotic chromosomes as a nucleoprotein complex (18). These gradually shorten in length following each round of mitosis, which eventually leads to aging (19). The gradual depletion of telomeres has been defined as one of the signs of biological senescence (20). Telomerase is a reverse transcriptase that uses its own RNA as a template to add nucleotides to telomeres (6). Cancer cells characteristically acquire the infinite capability to divide by maintaining telomere length through the sustained expression of telomerase or via the ALT mechanism (3). The human TERT gene encodes a telomerase catalytic subunit, which maintains telomeres by increasing their length (21). TERT is located on chromosome 5p15.33 and the promoter region of this gene is considered the most important regulatory element for its expression (22). At present, there are known inhibitors of TERT transcription, including cellular tumor antigen p53 (p53), which downregulates TERT transcription in a E3 ubiquitin-protein ligase SP1-dependent manner (23). Therefore, the deletion of p53 can activate the ALT pathway (24). TERT mutations are most commonly observed in medulloblastomas and gliomas, with TERT promoter mutations being identified in ~80% of all primary glioblastoma (7). The results of the present study indicated that TERT promoter mutations were 44.74% (17/38) in WHO grade II gliomas, 60.47% (18/43) in WHO grade III gliomas and 55.10% (42/98) in WHO grade IV gliomas, which are lower than those reported in previous literature (6,25). However, the data of the present study also revealed that TERT promoter mutation was associated with WHO classification, indicating that TERT promoter detection may be used for glioma prognosis, prediction and molecular typing.
ATRX is located on the Xq21.1 chromosome and encodes a 280 kDa nucleoprotein, which is involved in numerous cellular functions, including DNA recombination, repair and transcriptional regulation (26). Human ATRX mutations lead to the development of thalassemia, mental retardation, α-thalassemia X-linked mental retardation and other genetic conditions (27). ATRX mutations are present in at least 15 types of human tumors, including neuroblastoma, osteosarcoma and pancreatic neuroendocrine tumors (28). However, the role of ATRX in tumorigenesis remains to be elucidated. The ATRX protein may serve an important epigenetic role as it can be deposited on heterochromatin and telomeric DNA (29). In numerous different types of tumor, ALT activation is associated with the loss of function of the ATP-dependent helicase ATRX or its interacting partner (histone H3.3) (30). Similar to TERT promoter mutations, ATRX mutations are also associated with mutations in the tumor suppressor gene p53 (31); ATRX regulates the upstream pathways of cancer by modulating p53 (32). In the current study, the probability of mutations in the TERT promoter region in ATRX mutants samples was less than that in ATRX wild-type which the Exp (B) is 0.049 (Table II). Due to the association between ATRX deletion and ALT phenotype, another mechanism of telomere retention in tumor cells involves TERT (29). TERT promoter point mutations result in an increase of telomerase expression (33). Certain studies have demonstrated that TERT mutations are present in gliomas (2,4–7,17,33). Therefore, the present study hypothesized that alterations in ATRX and mutations in the TERT promoter region of glioma tissues may be mutually exclusive due to functional redundancy. Furthermore, a previous study demonstrated that ATRX and TERT mutations are mutually exclusive in anaplastic gliomas (7). Following this, the current study tested glioma tissues for mutations within these genes. In tissues with ATRX loss and wild-type TERT, 17 cases of WHO grade II, 12 cases of WHO grade III and 16 cases of WHO grade IV gliomas were identified (Table V). Although previous studies have demonstrated that ATRX loss and TERT promoter mutations in glioma may be mutually exclusive due to functional redundancy (17). Our results, which were 3 and 8 parents with grade III and IV gliomas, respectively, who had both mutations (Table V), suggest that contrary to previous conclusions in WHO grade II–IV glioma. However, it was also suggested that either mutation may contribute to the formation of glioma.
In glioma, molecular markers have independent prognostic significance (34). ATRX mutations are associated with alternative lengthening of telomeres in gliomas and tumor progression (15). The present study revealed that ATRX mutations were closely associated with IDH mutations in the present study. ATRX serves an important role in the regulation of chromatin remodeling (35). It is associated with IDH mutations and it may exhibit different epigenetic features (36). It is closely associated with gliomas of the CpG island methylation phenotype (36). The genetic differences between ATRX expression and non-transcription factors may be the basis of different clinical course in glioma.
The status of IDH in glioma may be predicted by ATRX sequencing (10). ATRX mutations/ALT phenotypes in gliomas can be observed by immunohistochemical staining (29). ATRX mutations are also associated with alternative lengthening of telomeres in gliomas and tumor progression; however, wild-type ATRX does not necessarily exclude the presence of IDH mutations (11,17,37). In the current study, almost all WHO II glioma were not associated with ATRX deletion. In one study on glioma, none of the 12 patients with IDH mutations exhibited ATRX deletions (38). Therefore, the current study hypothesizes that IDH and ATRX should be analyzed simultaneously in these tumors to reliably classify gliomas.
The results of the current study determined that mutations in the TERT promoter and ATRX gene were associated with age. This may be due to patients with low-grade glioma exhibiting high levels of DNA methylation, resulting in increased TERT expression (39). The prolongation and maintenance of telomere length in the tumor cells of young patients also may be more dependent on ALT mechanisms and the epigenetic regulation of telomerase (40). In addition, low-grade glioma cells originate from cells that maintain a strong ability to differentiate (41). These telomerases are in an activated state, such that mutation of the TERT promoter or ATRX is not required to achieve telomerase activation (3).
According to the WHO classification of tumors of the 2016 World Health Organization Classification of Tumors of the Central Nervous System, in small specimens, such as those obtained from a stereotactic biopsy, a single case of mitosis may suggest significant proliferative activity. Ki-67 labelling may be helpful to illustrate this (16). Therefore, Ki-67 was used to evaluate proliferation in the present study. The Ki-67 protein, which encodes two isoforms with molecular weights of 345 and 395 kDa, was originally discovered by Scholzen and Gerdes in 2000 (42). The expression of the Ki-67 protein is associated with the proliferative activity of the internal cell population in malignant tumors and is a marker of tumor invasiveness (43). The role of Ki-67 in the determination of prognosis has been previously studied, such that it is considered a potential marker for tumors of the central nervous system (44). The present study did not identify a statistically significant association between ATRX mutations and Ki-67 expression in the TERT promoter mutation group. However, when ATRX mutations were added to the TERT promoter mutations, it was demonstrated that TERT promoter mutations were significantly different between ATRX mutants and wild-type populations (Fig. 2). Difference of measurement data was compared with single factor analysis of variance (P=0.018). In the wild-type TERT group, Ki-67 was least abundant in the ATRX-deficient patients. Furthermore, Ki-67 expression was highest in the four groups without ATRX deletion. Due to the fact that the Ki-67 promoter possesses three Sp1 binding sites and that p53 inhibits Sp1 transcription (45), p53 may inhibit Ki-67 expression via a p53-Sp1-dependent pathway. One possibility is that the p53-binding motif affects the transcriptional inhibition of the Ki67 promoter (42). P53 may also change the interaction at the Sp1 binding site of the Ki67 promoter (42). ATRX mutations are associated with mutations in the tumor suppressor gene p53 (38). Therefore, the results of the present study demonstrated that the expression of Ki-67 was lowest in the wild-type TERT/ATRX deletion group. Ki-67 expression reflects the rate of tumor proliferation (16,42). Many factors are adjusted in tumor proliferation, including the second mitochondria-derived activator of caspases (Smac) (46), DNA replication licensing factor MCM7 (47), p53 (48), B-cell lymphoma 2 (49) proliferating cell nuclear antigen (50) and cluster of differentiation 105 (49). The highest expression of Ki-67 was observed in wild-type TERT/ATRX-deficient patients, which may indicate an association with the aforementioned regulatory mechanisms. Further studies are required to demonstrate these connections. To the best of the authors' knowledge, the present study assessed ATRX and the TERT promoter mutations together for the first time, dividing them into four subtypes based on two markers. The molecular classification of these mutations may better predict patient prognosis and guide clinical treatment strategies in the future.
However, the present study was limited as statistical methods were only utilized to determine the association among TERT, ATRX and other associated indicators, with the aim of solving clinical problems, ranging from tumor proliferation, recurrence and metastasis. The current study only assesses significant statistical information with regard to stage II to IV glioma tumors and TERT and ATRX analysis. Further experiments are therefore required to assess its internal associations, such as signal transduction pathway, interactions between proteins and epigenetic differences.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JL and XZ contributed to the writing of this manuscript, while MS, YH, XY and YF contributed the conception design and editing of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Medical Ethics Committee of Tianjin Huanshu Hospital and all patients provided written informed consent.
Patient consent for publication
Study participants provided their consent for the publication of any data/associated images.
Competing interests
The authors declare that there have no competing interests.
References
- 1.Nandakumar P, Mansouri A, Das S. The role of ATRX in glioma biology. Front Oncol. 2017;7:236. doi: 10.3389/fonc.2017.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuan Y, Qi C, Maling G, Xiang W, Yanhui L, Ruofei L, Yunhe M, Jiewen L, Qing M. TERT mutation in glioma: Frequency, prognosis and risk. J Clin Neurosci. 2016;26:57–62. doi: 10.1016/j.jocn.2015.05.066. [DOI] [PubMed] [Google Scholar]
- 3.Heidenreich B, Rachakonda PS, Hemminki K, Kumar R. TERT promoter mutations in cancer development. Curr Opin Genet Dev. 2014;24:30–37. doi: 10.1016/j.gde.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Walsh KM, Codd V, Smirnov IV, Rice T, Decker PA, Hansen HM, Kollmeyer T, Kosel ML, Molinaro AM, McCoy LS, et al. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet. 2014;46:731–735. doi: 10.1038/ng.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arita H, Narita Y, Fukushima S, Tateishi K, Matsushita Y, Yoshida A, Miyakita Y, Ohno M, Collins VP, Kawahara N, et al. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol. 2013;126:267–276. doi: 10.1007/s00401-013-1141-6. [DOI] [PubMed] [Google Scholar]
- 6.Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, Pekmezci M, Rice T, Kosel ML, Smirnov IV, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, Friedman AH, Friedman H, Gallia GL, Giovanella BC, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci USA. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heaphy CM, Subhawong AP, Hong SM, Goggins MG, Montgomery EA, Gabrielson E, Netto GJ, Epstein JI, Lotan TL, Westra WH, et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol. 2011;179:1608–1615. doi: 10.1016/j.ajpath.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hewer E, Vajtai I, Dettmer MS, Berezowska S, Vassella E. Combined ATRX/IDH1 immunohistochemistry predicts genotype of oligoastrocytomas. Histopathology. 2016;68:272–278. doi: 10.1111/his.12743. [DOI] [PubMed] [Google Scholar]
- 11.Jiao Y, Killela PJ, Reitman ZJ, Rasheed AB, Heaphy CM, de Wilde RF, Rodriguez FJ, Rosemberg S, Oba-Shinjo SM, Nagahashi Marie SK, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kannan K, Inagaki A, Silber J, Gorovets D, Zhang J, Kastenhuber ER, Heguy A, Petrini JH, Chan TA, Huse JT. Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget. 2012;3:1194–1203. doi: 10.18632/oncotarget.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JC, Jeng YM, Liau JY, Tsai JH, Hsu HH, Yang CY. Alternative lengthening of telomeres and loss of ATRX are frequent events in pleomorphic and dedifferentiated liposarcomas. Mod Pathol. 2015;28:1064–1073. doi: 10.1038/modpathol.2015.67. [DOI] [PubMed] [Google Scholar]
- 14.Liu XY, Gerges N, Korshunov A, Sabha N, Khuong-Quang DA, Fontebasso AM, Fleming A, Hadjadj D, Schwartzentruber J, Majewski J, et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012;124:615–625. doi: 10.1007/s00401-012-1031-3. [DOI] [PubMed] [Google Scholar]
- 15.Wiestler B, Capper D, Holland-Letz T, Korshunov A, von Deimling A, Pfister SM, Platten M, Weller M, Wick W. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013;126:443–451. doi: 10.1007/s00401-013-1156-z. [DOI] [PubMed] [Google Scholar]
- 16.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 17.Karsy M, Guan J, Cohen AL, Jensen RL, Colman H. New molecular considerations for glioma: IDH, ATRX, BRAF, TERT, H3 K27M. Curr Neurol Neurosci Rep. 2017;17:19. doi: 10.1007/s11910-017-0722-5. [DOI] [PubMed] [Google Scholar]
- 18.Armanios M. Telomeres and age-related disease: How telomere biology informs clinical paradigms. J Clin Invest. 2013;123:996–1002. doi: 10.1172/JCI66370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez FJ, Scheithauer BW, Burger PC, Jenkins S, Giannini C. Anaplasia in pilocytic astrocytoma predicts aggressive behavior. Am J Surg Pathol. 2010;34:147–160. doi: 10.1097/PAS.0b013e3181c75238. [DOI] [PubMed] [Google Scholar]
- 20.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Podlevsky JD, Chen JJ. It all comes together at the ends: Telomerase structure, function, and biogenesis. Mutat Res. 2012;730:3–11. doi: 10.1016/j.mrfmmm.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 23.Kanaya T, Kyo S, Hamada K, Takakura M, Kitagawa Y, Harada H, Inoue M. Adenoviral expression of p53 represses telomerase activity through down-regulation of human telomerase reverse transcriptase transcription. Clin Cancer Res. 2000;6:1239–1247. [PubMed] [Google Scholar]
- 24.Farooqi AS, Dagg RA, Choi LM, Shay JW, Reynolds CP, Lau LM. Alternative lengthening of telomeres in neuroblastoma cell lines is associated with a lack of MYCN genomic amplification and with p53 pathway aberrations. J Neurooncol. 2014;119:17–26. doi: 10.1007/s11060-014-1456-8. [DOI] [PubMed] [Google Scholar]
- 25.Venneti S, Huse JT. The evolving molecular genetics of low-grade glioma. Adv Anat Pathol. 2015;22:94–101. doi: 10.1097/PAP.0000000000000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibbons RJ, Wada T, Fisher CA, Malik N, Mitson MJ, Steensma DP, Fryer A, Goudie DR, Krantz ID, Traeger-Synodinos J. Mutations in the chromatin-associated protein ATRX. Hum Mutat. 2008;29:796–802. doi: 10.1002/humu.20734. [DOI] [PubMed] [Google Scholar]
- 27.Gibbons RJ, Pellagatti A, Garrick D, Wood WG, Malik N, Ayyub H, Langford C, Boultwood J, Wainscoat JS, Higgs DR. Identification of acquired somatic mutations in the gene encoding chromatin-remodeling factor ATRX in the alpha-thalassemia myelodysplasia syndrome (ATMDS) Nat Genet. 2003;34:446–449. doi: 10.1038/ng1213. [DOI] [PubMed] [Google Scholar]
- 28.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pickett HA, Reddel RR. Molecular mechanisms of activity and derepression of alternative lengthening of telomeres. Nat Struct Mol Biol. 2015;22:875–880. doi: 10.1038/nsmb.3106. [DOI] [PubMed] [Google Scholar]
- 31.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tönjes M, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 32.Elsässer SJ, Allis CD, Lewis PW. Cancer. New epigenetic drivers of cancers. Science. 2011;331:1145–1146. doi: 10.1126/science.1203280. [DOI] [PubMed] [Google Scholar]
- 33.Pekmezci M, Rice T, Molinaro AM, Walsh KM, Decker PA, Hansen H, Sicotte H, Kollmeyer TM, McCoy LS, Sarkar G, et al. Adult infiltrating gliomas with WHO 2016 integrated diagnosis: Additional prognostic roles of ATRX and TERT. Acta Neuropathol. 2017;133:1001–1016. doi: 10.1007/s00401-017-1690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Bent MJ, Brandes AA, Taphoorn MJ, Kros JM, Kouwenhoven MC, Delattre JY, Bernsen HJ, Frenay M, Tijssen CC, Grisold W, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: Long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31:344–350. doi: 10.1200/JCO.2012.43.2229. [DOI] [PubMed] [Google Scholar]
- 35.Bassett AR, Cooper SE, Ragab A, Travers AA. The chromatin remodelling factor dATRX is involved in heterochromatin formation. PLoS One. 2008;3:e2099. doi: 10.1371/journal.pone.0002099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai J, Zhu P, Zhang C, Li Q, Wang Z, Li G, Wang G, Yang P, Li J, Han B, et al. Detection of ATRX and IDH1-R132H immunohistochemistry in the progression of 211 paired gliomas. Oncotarget. 2016;7:16384–16395. doi: 10.18632/oncotarget.7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leeper HE, Caron AA, Decker PA, Jenkins RB, Lachance DH, Giannini C. IDH mutation, 1p19q codeletion and ATRX loss in WHO grade II gliomas. Oncotarget. 2015;6:30295–30305. doi: 10.18632/oncotarget.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castelo-Branco P, Choufani S, Mack S, Gallagher D, Zhang C, Lipman T, Zhukova N, Walker EJ, Martin D, Merino D, et al. Methylation of the TERT promoter and risk stratification of childhood brain tumours: An integrative genomic and molecular study. Lancet Oncol. 2013;14:534–542. doi: 10.1016/S1470-2045(13)70110-4. [DOI] [PubMed] [Google Scholar]
- 40.Koelsche C, Sahm F, Capper D, Reuss D, Sturm D, Jones DT, Kool M, Northcott PA, Wiestler B, Böhmer K, et al. Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta Neuropathol. 2013;126:907–915. doi: 10.1007/s00401-013-1195-5. [DOI] [PubMed] [Google Scholar]
- 41.Taphoorn MJ. Neurocognitive sequelae in the treatment of low-grade gliomas. Semin Oncol. 2003;30:45–48. doi: 10.1053/j.seminoncol.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 42.Scholzen T, Gerdes J. The Ki-67 protein: From the known and the unknown. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 43.Klöppel G, Perren A, Heitz PU. The gastroenteropancreatic neuroendocrine cell system and its tumors: The WHO classification. Ann N Y Acad Sci. 2004;1014:13–27. doi: 10.1196/annals.1294.002. [DOI] [PubMed] [Google Scholar]
- 44.Ishihara M, Mukai H, Nagai S, Onozawa M, Nihei K, Shimada T, Wada N. Retrospective analysis of risk factors for central nervous system metastases in operable breast cancer: Effects of biologic subtype and Ki67 overexpression on survival. Oncology. 2013;84:135–140. doi: 10.1159/000345321. [DOI] [PubMed] [Google Scholar]
- 45.Nakano T, Ohno T, Ishikawa H, Suzuki Y, Takahashi T. Current advancement in radiation therapy for uterine cervical cancer. J Radiat Res. 2010;51:1–8. doi: 10.1269/jrr.09132. [DOI] [PubMed] [Google Scholar]
- 46.Hu HY, Liu H, Zhang JW, Hu K, Lin Y. Clinical significance of Smac and Ki-67 expression in pancreatic cancer. Hepatogastroenterology. 2012;59:2640–2643. doi: 10.5754/hge12071. [DOI] [PubMed] [Google Scholar]
- 47.Liu YZ, Jiang YY, Hao JJ, Lu SS, Zhang TT, Shang L, Cao J, Song X, Wang BS, Cai Y, et al. Prognostic significance of MCM7 expression in the bronchial brushings of patients with non-small cell lung cancer (NSCLC) Lung Cancer. 2012;77:176–182. doi: 10.1016/j.lungcan.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Golmohammadi R, Pejhan A. The prognostic value of the P53 protein and the Ki67 marker in breast cancer patients. J Pak Med Assoc. 2012;62:871–875. [PubMed] [Google Scholar]
- 49.Boonyaphiphat P, Pruegsanusak K, Thongsuksai P. The prognostic value of p53, Bcl-2 and Bax expression in laryngeal cancer. J Med Assoc Thai. 2012;95:1317–1320. [PubMed] [Google Scholar]
- 50.Ben-Izhak O, Bar-Chana M, Sussman L, Dobiner V, Sandbank J, Cagnano M, Cohen H, Sabo E. Ki67 antigen and PCNA proliferation markers predict survival in anorectal malignant melanoma. Histopathology. 2002;41:519–525. doi: 10.1046/j.1365-2559.2002.01444.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.