Abstract
Lung cancer is one of the leading causes of cancer-associated mortality throughout the world. The prognosis of the disease depends on many factors including the stage and type of cancer. Many studies have identified various microRNAs (miRNAs) that affect the prognosis of lung cancer. In order to systemically analyze the available clinical data, the present study performed a meta-analysis to examine all evidence on the potential role of miRNAs as novel predictors of survival in lung cancer. Literature published in English prior to February 1st, 2018 was searched through PubMed to review all of the associations between individual miRNAs and groups of miRNAs with the prognosis of lung cancer. Data was extracted using standard forms and pooled odds ratios with 95% confidence intervals (CIs) were calculated. A total of 15 eligible studies were included in the meta-analysis. These represented 1,753 lung cancer patients and 20 miRNAs. A total of 8 downregulated miRNAs were associated with poorer overall survival (OS) [hazard ratio (HR)=0.59, 95% CI: 0.47–0.75, P<1×10−4], while 10 upregulated miRNAs were associated with poorer OS (HR=1.76, 95% CI: 1.31–2.35, P<1×10−4). Additionally, low miRNA expression was associated with lymph node metastasis [LNM; relative risk (RR)=0.53, 95% CI: 0.46–0.61, P<1×10−4]. The expression of miRNAs was not associated with lung cancer stage (RR=1.07, 95% CI: 0.94–1.22, P=0.23). Expression levels of different miRNAs were associated with the OS and LNM of patients with lung cancer. These miRNAs may be applied as potential prognostic markers in lung cancer.
Keywords: microRNAs, lung cancer, prognosis, meta-analysis
Introduction
The incidence and mortality of lung cancer is on the rise worldwide (1) and is the leading cause of death among men and the second leading cause of cancer death among women all over the world (2). In China, it is estimated that more than 4 million cases of cancer and nearly 3 million cancer deaths occur (3). Most lung cancer patients were diagnosed as at advanced stages and were clinically inoperable. Conventional treatments including surgery, chemotherapy and radiotherapy for lung cancer have proven to be ineffective for late stages of lung cancer. The 5-year survival rate of lung cancer is poor (4). Early diagnosis and accurate prognosis analysis are important to improve the survival rate of lung cancer patients. Discovery of microRNAs (miRNAs/miRs) has opened up a new horizon for predicting the prognosis of lung cancer. Many cancer-associated miRNAs can predict the prognosis of lung cancer and can be targeted for treatment (5), but the results from individual experiments of the miRNAs are still inconsistent.
miRNAs are short noncoding RNAs that consist of 21–23 nucleotides in length, which participate in the process of translational repression or degradation of mRNA (6). Emerging studies showed that the expression levels of miRNAs in cancer were different from that in normal tissues. Besides, the specific expression signatures were correlated with prognosis of cancer, such as breast cancer, colorectal cancer and gastric cancer. According to the existing research results, a growing number of researchers found that different genes were related to the prognosis of lung cancer. miRNAs may play a key role in the pathogenesis of lung cancer and can be targets for potential therapeutics for the disease (7,8). For example, it was reported that high levels of miR-211 were associated with poor survival in human non-small cell lung cancer (NSCLC) patients. miR-211 promotes tumor proliferation and invasion via regulating MxA expression in NSCLC, which suggested that manipulation of the level of miR-211 may provide a novel therapy for NSCLC patients in the future (9). miR-197 expression was associated with tumor size and identified as a novel independent predictor of unfavorable prognosis for NSCLC patients (10). miR-638 levels were associated with the survival of NSCLC patients and may be also considered a potential independent predictor for NSCLC prognosis (11). The associations between miRNAs and lung cancers were reported in many meta-analyses (12–14), but the evaluation of the prognostic role of miRNAs in lung cancer has not been fully analyzed. In this report, we systematically performed a meta-analysis of miRNAs profiling with cancer stage, LNM and OS rate of the lung cancer patient.
Materials and methods
Study selection and inclusion criteria
Literature in PubMed database was comprehensively searched (last updated search being February 1st, 2018). The publications were identified with a combination of the key words: miRNA, miRNAs, Micro RNA, RNA Micro, miRNA, Primary MicroRNA, Primary miRNA, pri-miRNA, pri miRNA, small temporal RNA, small stRNA, small temporal RNA, pre-miRNA, pre miRNA, lung neoplasms, lung neoplasia, lung neoplasias, lung tumors, lung benign neoplasms, lung benign neoplasm, lung benign malignancy, lung malignancies, lung cancer and lung cancers.
To be included in the meta-analysis, studies had to meet the following criteria: i) The subjects of studies were lung cancer patients diagnosed through pathology or cytology; ii) the studies aimed to investigate the relationship between miRNAs and lung cancer patients; iii) patients were grouped according to expression levels of miRNAs, which were measured in primary tumor tissues or adjacent normal tissue; iv) related clinical pathological characteristics were shown, such as tumor stages of cancers (T), lymph node metastasis (LNM), or distant metastasis (DM); v) available data contained information about the prognostic value of patients with survival outcomes, such as overall survival (OS)/recurrence-free survival (RFS)/event-free survival(EFS)/distant metastases-free survival (DMFS)/progression-free survival (PFS); vi) data contained hazard ratio (HR) or relative risk (RR) and corresponding 95% CI, directly or indirectly; and vii) full-text paper was available.
Studies were excluded if one of the following existed: i) Duplicate publications; ii) non-human study or non-clinical study or animal study or non-English study; iii) reviews, case reports, letters, editorials, and expert opinions; and iv) studies without available data or no complete text.
Data extraction
For the eligible studies, data was extracted independently by two investigators (DY and SY). Disagreements were resolved by discussion with another investigator (WX or YZ). The following data was extracted: i) Publication information: The first author's name, the year of publication and the country of origin; ii) patients' characteristic information: Number of participants, type of lung cancer, type of miRNAs, clinical tumor stage, and follow-up duration; iii) miRNA detection information: Tissue sample, method and cut-off values; iv) prognosis information: The number of patients with lymph nodes metastasis and different tumor stage; and v) survival analysis and multivariate analysis: ORs of miRNAs for LNM, HRs of miRNAs for OS and corresponding 95% Cl and P-values. This data was either obtained from the original article directly with sufficient data or via Engauge Digitizer version 4.1 to extract when only Kaplan-Meier curves were available.
Two investigators (YZ and WX) used the Newcastle-Ottawa Scale (NOS) to independently assess the quality of all the included studies. There were three major sections (selection, comparability and outcome) in NOS. The selection consisted of adequacy information of case definition, number of case in the research and representative miRNAs. The comparability contained miRNA exposure, detection method and cut-off values. The outcome included assessment results and adequate follow-up time. The highest score of NOS criteria was 8 with the lowest score being 0. The higher the score on this test is indicative of a better quality and a study with an NOS score equal to or more than 5 is considered to be of good quality. Finally, we used Begg's test to evaluate potential publication bias. The result pattern was not significantly impacted by removing a single study each time.
Statistical analysis
The STATA software version 12.0 (StataCorp LP, College Station, TX, USA) was used for the following statistical analysis and to generate a forest plot to show the content of this statistical analysis. I2 statistics and P-values were used for investigating the heterogeneity among combined studies. The heterogeneity was regarded as significant when the I2 value >50% or a P-value <0.05 for Q test, while I2 values <50% or P-value >0.05 indicated that there was no significant heterogeneity, indicating that a random-effects model had been used to test heterogeneity.
When evaluating the association between miRNAs, prognosis, LNM and T, we used both HR/RR and 95% CI. Sensitivity analyses were used to test the effect of each study on pooled results. The potential publication bias was performed by Begg's test and funnel plots were used to show the results. P<0.05 was considered to indicate a statistically significant difference. The biased risks of all the included studies were according to the basis for assessing the internal validity of the prognostic study and the recommendations on the biomarker research report (15–17).
Results
Characteristics of eligible studies, publication bias and sensitivity analysis
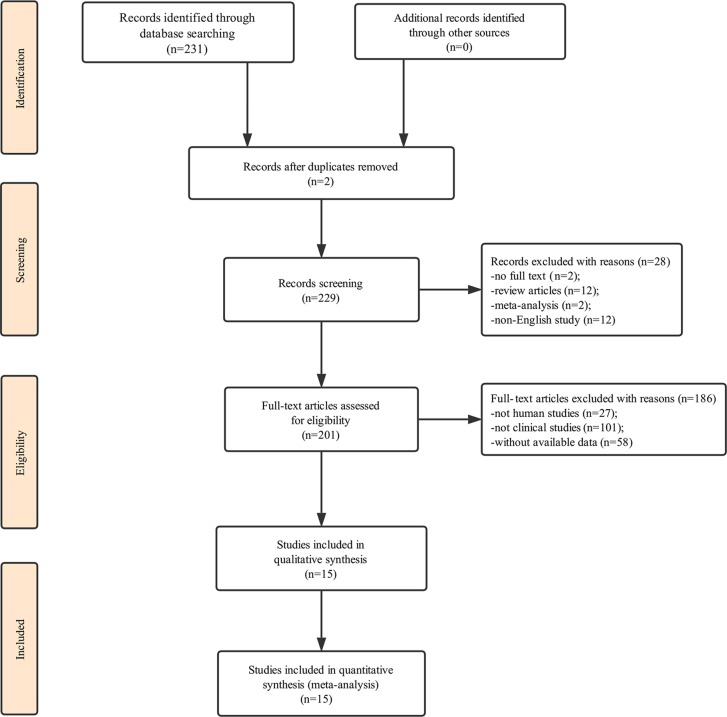
Our initial search of the database identified 231 publications. Two duplicate studies were excluded. We first carefully reviewed the titles and abstracts of each article, a total of 28 of them were excluded due to not containing full text, review articles, meta-analysis or not being in English. We then performed a detailed evaluation of each full text and 186 of them were further excluded due to being animal experiments, irrelevant clinical studies or without data available; therefore, only 15 articles were eligible for the final analysis (10,11,18–30). The process of study selection is presented in Fig. 1 and the main characteristics of the included articles are summarized in Table I.
Figure 1.
A flow diagram demonstrating the study selection process. Initially, a total of 231 records were identified on the database. Duplicate records were excluded to make sure each paper was only represented once within our set of records. A total of 28 records were excluded after reading the titles or the abstracts as these studies were reviews, meta-analyses, studies that were not in English or the full text was not available. After full-text reading of the studies that were not on human subjects, not clinical studies or without available data another 186 records were excluded. This process led to the identification of 15 studies eligible for systematic review.
Table I.
Characteristics of studies in the present meta-analysis.
| A, Study information | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| miRNA expression | |||||||||||
| Author, year | Country | miRNA | Total no. | Sample tissue | Detection method | Cut-off | Expression status | High expression | Low expression | Refs. | |
| Mavridis et al, 2015 | Greece | 197 | 124 | Frozen tissue | qPCR | X-tile algorithm | UR | 49 | 75 | (10) | |
| Wang et al, 2015 | P.R. China | 638 | 189 | Serum | qPCR | X-tile algorithm | DR | 112 | 77 | (11) | |
| Chen et al, 2013 | P.R. China | 146α | 101 | Tissue | qPCR | Median | DR | 0 | 101 | (18) | |
| Chen et al, 2017 | P.R. China | 148α | 159 | Tissue | qPCR | Median | DR | 73 | 86 | (19) | |
| Cui et al, 2013 | P.R. China | 125b | 260 | Serum | qPCR | Median | UR | 260 | 0 | (20) | |
| Guo et al, 2015 | P.R. China | 204 | 126 | Plasma | qPCR | Median | DR | 66 | 60 | (21) | |
| Li et al, 2015 | P.R. China | 148α | 73 | Frozen tissue | qPCR | Mean | DR | 31 | 42 | (22) | |
| Liu et al, 2012 | P.R. China | 21 | 70 | Frozen tissue | qPCR | Mean | UR | 48 | 22 | (23) | |
| 141 | 70 | 49 | 21 | ||||||||
| 200c | 70 | 38 | 32 | ||||||||
| Nadal et al, 2014 | USA | 411 | 24 | Frozen tissue | qPCR | Median | UR | 24 | 0 | (24) | |
| 370 | 33 | Frozen tissue | qPCR | Median | UR | 33 | 0 | ||||
| 376α | 34 | Frozen tissue | qPCR | Median | UR | 34 | 0 | ||||
| Skrzypski et al, 2014 | Poland | 192 | 223 | Frozen tissue | qPCR | Median | UR | 90 | 133 | (25) | |
| 662 | 223 | Frozen tissue | qPCR | Median | UR | 90 | 133 | ||||
| Sun et al, 2013 | P.R. China | 150 | 90 | Frozen tissue | qPCR | Median | UR | 0 | 90 | (26) | |
| 3940-5p | 90 | Frozen tissue | qPCR | Median | 0 | 90 | |||||
| Võsa et al, 2011 | Estonia | 374α | 36 | Tissue | qPCR | Median | DR | 18 | 18 | (27) | |
| Wang et al, 2016 | P.R. China | 148b | 39 | Frozen tissue | qPCR | Median | DR | 18 | 21 | (28) | |
| Jiang et al, 2016 | P.R. China | 26b | 154 | Frozen tissue | qPCR | Median | DR | 77 | 77 | (29) | |
| Rothschild et al, 2012 | Bern | 381 | 18 | Tissue | qPCR | Median | DR | 3 | 15 | (30) | |
| B, Statistics | |||||||||||
| miRNA expression | |||||||||||
| Author, year | High with T2-4/3-4 | Low with T2-4/3-4 | High with LNM | Low with LNM | Survival analysis | Multivariate analysis | HR statistic | HR (95% CI) | Follow-up, months | Refs. | |
| Mavridis et al, 2015 | 30 | 38 | OS | YES | SC | 1.91 (1.75–3.19) | 84 | (10) | |||
| Wang et al, 2015 | 73 | 51 | 87 | 70 | OS | YES | SC | 0.61 (0.35–1.06) | 60 | (11) | |
| Chen et al, 2013 | 0 | 73 | 0 | 66 | OS | Unreported | SC | 0.56 (0.21–1.51) | 60 | (18) | |
| Chen et al, 2017 | 58 | 70 | 27 | 48 | OS | YES | SC | 0.85 (0.54–1.35) | 120 | (19) | |
| Cui et al, 2013 | 260 | 0 | OS | YES | Rep | 2.03 (1.14–3.05) | 25 | (20) | |||
| Guo et al, 2015 | 0 | 49 | OS | YES | SC | 0.54 (0.31–0.94) | 60 | (21) | |||
| DFS | 0.78 (0.44–1.36) | ||||||||||
| Li et al, 2015 | 0 | 37 | 9 | 30 | NA | Unreported | NA | (22) | |||
| Liu et al, 2012 | 34 | 0 | 32 | 0 | OS | YES | SC | 1.31 (0.42–3.03) | 24 | (23) | |
| 34 | 0 | 32 | 0 | 1.02 (0.37–2.82) | |||||||
| 34 | 0 | 32 | 0 | 1.28 (0.04–2.53) | |||||||
| Nadal et al, 2014 | 5 | 0 | OS | YES | Rep | 3.37 (1.42–8.0) | 60 | (24) | |||
| DFS | 3.32 (1.61–6.88) | ||||||||||
| 16 | 0 | OS | YES | Rep | 3.37 (1.42–8.0) | 60 | |||||
| DFS | 3.32 (1.61–6.88) | ||||||||||
| 22 | 0 | OS | YES | Rep | 3.37 (1.42–8.0) | 60 | |||||
| DFS | 3.32 (1.61–6.88) | ||||||||||
| Skrzypski et al, 2014 | 17 | 24 | OS | YES | SC | 1.24 (1.06–1.45) | 72 | (25) | |||
| DMFS | 1.12 (1.01–1.24) | ||||||||||
| 17 | 24 | OS | YES | SC | 2.58 (1.52–4.37) | 72 | |||||
| DMFS | 1.23 (1.09–1.38) | ||||||||||
| Sun et al, 2013 | 0 | 52 | 0 | 34 | NA | YES | NA | (26) | |||
| 0 | 52 | 0 | 34 | ||||||||
| Võsa et al, 2011 | OS | YES | SC | 0.98 (0.16–5.88) | 84 | (27) | |||||
| Wang et al, 2016 | 8 | 19 | 9 | 7 | OS | YES | SC | 0.86 (0.31–2.38) | 60 | (28) | |
| Jiang et al, 2016 | 27 | 43 | 18 | 51 | OS | YES | SC | 0.61 (0.26–1.44) | 60 | (29) | |
| Rothschild et al, 2012 | OS | Unreported | SC | 0.37 (0.25–0.66) | 35 | (30) | |||||
| EFS | 0.48 (0.31–0.93) | ||||||||||
‘Total no.’ refers to the number of studies included in the meta-analysis of each microRNA; HR, hazard ratio; 95 % CI; 95% confidence interval; I2 is a measure of between-study heterogeneity; LNM, lymph node metastasis; T, tumor stage of cancers; OS, overall survival; RFS, recurrence-free survival; EFS, event-free survival; DFS, disease-free survival; DMFS, distant metastases-free survival; NA, not available; qPCR, quantitative polymerase chain reaction; SC, survival curve; DR, downregulation; UR, upregulation.
All of the included studies are considered as high quality, as their NOS score was equal to or more than 5, while five of them scored 8 in the quality assessment. Quality assessment of eligible studies is shown in Table II.
Table II.
Quality assessment of eligible studies (NOS).
| Selection | Comparability | Outcome | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Author, year | Adequacy of case definition | No. of cases | Representativeness of the cases | Ascertainment of exposure | Ascertainment of detection method | Ascertainment of cut-off | Assessment of outcome | Adequate follow-up | Total | Refs. |
| Mavridis et al, 2015 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | (10) |
| Wang et al, 2015 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | (11) |
| Chen et al, 2013 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | (18) |
| Chen et al, 2017 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | (19) |
| Cui et al, 2013 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 7 | (20) |
| Guo et al, 2015 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 5 | (21) |
| Li et al, 2015 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 5 | (22) |
| Liu et al, 2012 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 7 | (23) |
| Nadal et al, 2014 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | (24) |
| Skrzypski et al, 2014 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | (25) |
| Sun et al, 2013 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 6 | (26) |
| Võsa et al, 2011 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | (27) |
| Wang et al, 2016 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | (28) |
| Jiang et al, 2016 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | (29) |
| Rothschild et al, 2012 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | (30) |
NOS, Newcastle-Ottawa Scale.
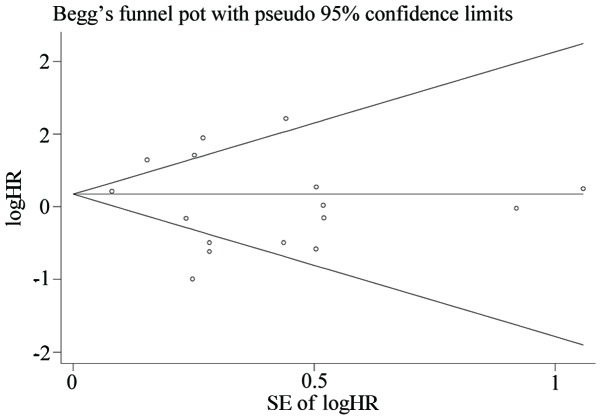
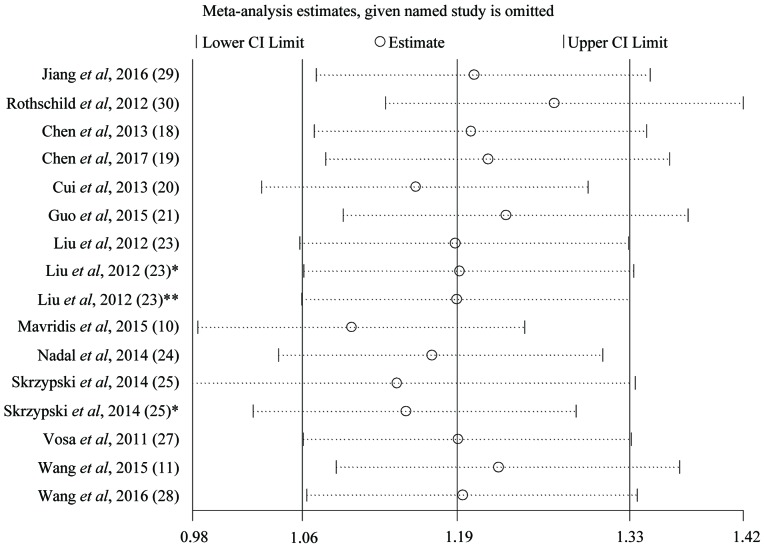
We used Begg's test and funnel plots to evaluate potential publication bias. The shape of the funnel plot of the OS group was symmetrical (Begg's test, t=−0.78, P=0.447), indicating that no significant publication bias was observed by the Begg's test (Fig. 2). Sensitivity analysis of the publications in the OS group is presented in Fig. 3. The results pattern was not significantly impacted by removing a single study each time.
Figure 2.
Begg's funnel plot of OS group. The meta-analysis for microRNA expression levels and OS in patients with lung cancer was analyzed using a funnel plot because it exceeded the pre-requisite of 10 studies. The y-axis represents the SE (logHR), which served as a measure of precision, where the higher the SE, the less precise the study. The HR has been plotted along the x-axis. The pattern of the study was symmetric which indicated that the bias of the study was small. SE, standard error; logHR, log hazard ratio; OS, overall survival.
Figure 3.
Sensitivity analysis of the publications in the overall survival group. Liu et al, 2012 (23) represents the data of miR-21. Liu et al, 2012 (23)* represents the data of miR-141. Liu et al, 2012 (23)** represents the data of miR-200c. Skrzypski et al, 2014 (25) represents the data of miR-192. Skrzypski et al, 2014 (25)* represents the data of miR-662. miR, microRNA; CI, confidence interval.
Association between expression levels of miRNAs and LNM in lung cancer
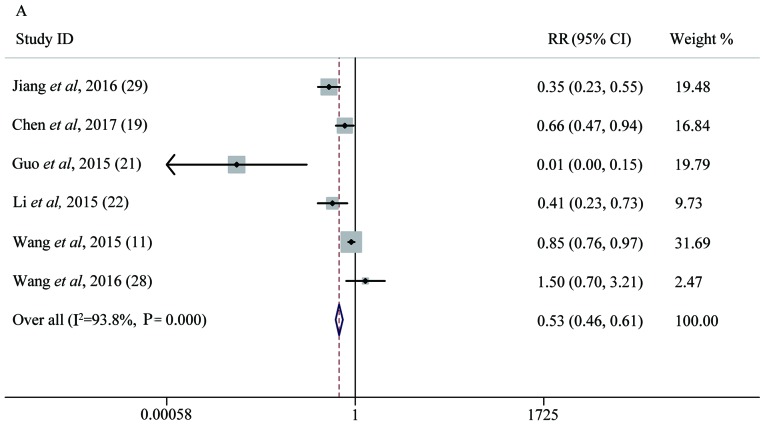
Six studies with 740 lung cancer patients were included to assess the association between the expression levels of different miRNAs and LNM. Five miRNAs were evaluated in this group, including miR-26b, miR-148α, miR-204, miR-638 and miR-148b. There was significant heterogeneity between the studies included (P<1×10−4, I2=93.8%). The random-effects model was applied and miR-148α was investigated in two studies (19,22). There were 150 patients who became LNM among 377 patients with high miRNA expression, with a percentage of 39.8%; but patients with low miRNA expression were more likely to become LNM as the percentage was 71.8%. Analysis showed that the pooled RR was 0.53 (95% CI: 0.46–0.61, P<1×10−4), which indicated that low expression of miRNAs was predictive of LNM (Fig. 4A).
Figure 4.
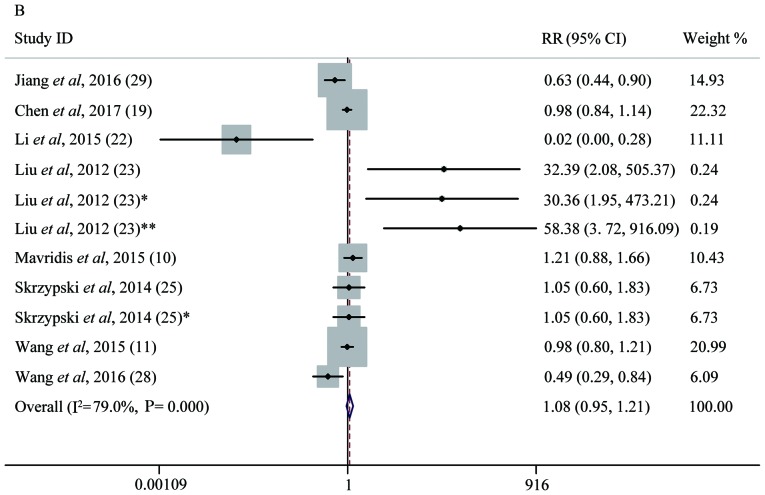
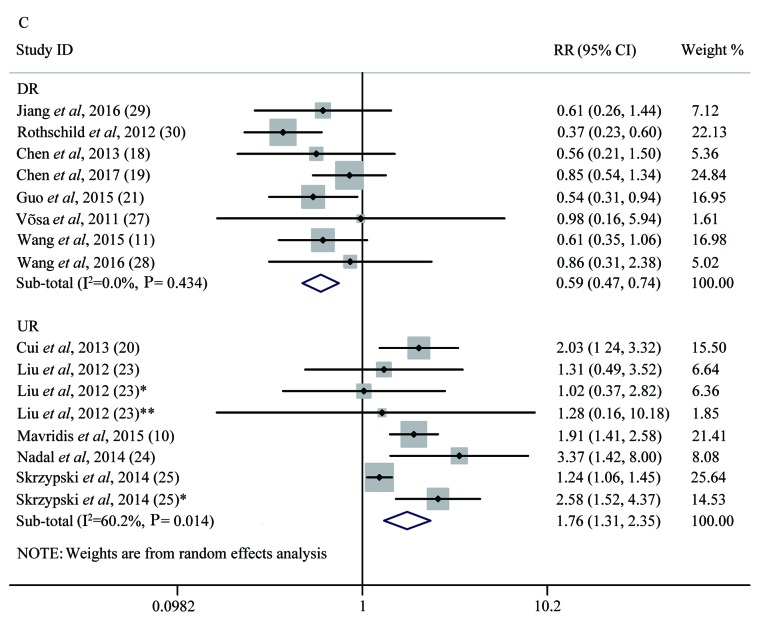
Forest plot analysis. (A) Forest plot of the LNM group. (B) Forest plot of the T group. (C) Forest plot of the OS group. Each plot A-C corresponds to the meta-analysis of a different group: (A) LNM, (B) T, and (C) OS. The effect size for the estimate of each study was presented as a grey square proportional in size to the weight of that study. The confidence interval around that effect size was presented as a horizontal line. The vertical line across these estimates represents HR=1 and any horizontal line crossing this vertical line represents a non-statistically significant result. The summary effect size was presented as a rhombus, the center of which represents the summary effect size and the width of which represents its CI. All meta-analyzed groups, apart from the T group (plot B), were statistically significantly associated with microRNAs. LNM, lymph node metastasis; RR, relative risk; CI, confidence interval; T, tumor stages of cancer; OS, overall survival; DR, downregulated; UR, upregulated.
Association between miRNA levels and lung cancer T stages
Seven studies containing 1,031 patients were included to evaluate the relationship between miRNA level and tumor T stage. We divided T1 into a low T stage group and T2-4 into high a T stage group, according to T stage level (T1/2/3/4). Significant heterogeneity was found among the studies (P<1×10−4, I2=79%) and a random-effects model study showed a pooled RR of 1.07 with a 95% CI of 0.94–1.22 (P=0.23), indicating that the expression of miRNAs might not be a direct predictor of T stage (Fig. 4B).
Association between the expression of miRNAs and OS of lung cancer patients
Thirteen studies of 1,536 lung cancer patients were included to assess the correlation between different miRNA expression levels and OS. Subgroup analysis was performed on the basis of the expression of miRNAs. Eight studies of 768 lung cancer patients reported the relationship between downregulated expression of different miRNAs and OS, including miRNA-26b, miRNA-381, miRNA-146α, miRNA-148α, miRNA-204, miRNA-374α, miRNA-638 and miRNA-148b. There was no significant heterogeneity among the combined studies (P=0.434, I2=0%) by applying the random effects model (31). We found that low expression of these miRNAs was associated with shorter OS of lung cancer patients, according to the pooled HR of 0.59 with a 95% CI of 0.47–0.75, (P<1×10−4). High expression of 8 miRNAs mentioned above increased the likelihood of survival. Five studies of 768 lung cancer patients reported the relationship between upregulated expression of different miRNAs and OS, including miRNA-125b, miRNA-21, miRNA-141, miRNA-200c, miRNA-197, miRNA-41, miRNA-370, miRNA-376α, miRNA-192 and miRNA-662. There was significant heterogeneity among the combined studies (P=0.014, I2=60.2%). High expression of these 10 miRNAs was associated with poorer OS of lung cancer patients, according to the pooled HR of 1.76 with a 95% CI of 1.31–2.35 (P<1×10−4) (Fig. 4C). Only 2 studies included, had studied the association between the expression of miRNAs and disease-free survival (DFS) of lung cancers, while only 1 study accessed the association between the expression of miRNAs and DMFS of lung cancer patients. Since there were less than three of each type of study there was no test for association. All meta-analysis results are shown in Table III.
Table III.
Results of meta-analysis.
| Outcomes | No. of studies | No. of patients | HR/OR/RR (95% CI) | P-value | Heterogeneity I2 (%) | Publication bias P-value |
|---|---|---|---|---|---|---|
| LNM | 6 | 740 | 0.53 (0.46–0.61) | <1×10−4 | 93.8 | 0.145 |
| T | 7 | 1,031 | 1.07 (0.94–1.22) | 0.23 | 79.0 | 0.527 |
| OS (DR) | 8 | 768 | 0.59 (0.47–0.75) | <1×10−4 | 0.0 | 0.447 |
| OS (UR) | 5 | 768 | 1.76 (1.31–2.35) | <1×10−4 | 60.2 | 0.447 |
CI, confidence interval; HR, hazard ratio; OR, odds ratio; RR, relative risk; LNM, lymph node metastasis; T, tumor state of cancers; OS, overall survival; DR, downregulation; UR, upregulation.
Discussion
Lung cancer is the most commonly diagnosed cancers among men and women. It is also the most commonly diagnosed cancer and the leading cause of cancer death in men aged 75 years or older (3). Although early diagnosis, targeting therapy and immunotherapy treatment of lung cancer have developed rapidly in recent years, a vast majority of lung cancers are still diagnosed at a late stage (32) and have poor prognosis. The 5-year survival rate is only approximately 10% and is one of the lowest among cancer patients (33). Finding new biomarkers can not only improve early diagnosis but also provide new targeting means after prognosis analysis. miRNAs have emerged as new regulators of cancer genomes and accumulating studies have found that many miRNAs are associated with the prognosis of lung cancer, especially NSCLC (34,35).
On the one hand, there are few studies focus on the relationship between the same miRNA and lung cancer, so it is impossible for us to discuss. A meta-analysis is a statistical analysis that combines the results of multiple scientific studies. In our meta-analysis, we examined all evidences on the potential role of various miRNAs in lung cancer in order to systemically analyze the available clinical data. On the other hand, the targets and mechanism of miRNAs in lung cancer are still unclear, summarizing all the similar researches can provide us a direction in clinical work and improve our comprehension of potential intricate biological mechanism. We analyzed and evaluated the association between expression levels of multiple miRNAs and clinical prognosis for lung cancer patients, which laid a foundation for the diagnosis, prognosis evaluation and targeted therapy of lung cancer.
We found that high expression of miR-125b, miR-21, miR-141, miR-200c, miR-197, miR-41, miR-370, miR-376α, miR-192 and miR-662 resulted in a lower level of survival among patients. We also found that low levels of expression of miR-26b, miR-381, miR-146α, miR-148α, miR-204, miR-374α, miR-638 and miR-148b also resulted in a lower level of survival among patients. Our analysis showed that high expression of miRNAs was correlated with LNM in lung cancer. We could not observe the relationships between the expression of various miRNAs and tumor stages, DMS or DMFS, which may be due to the samples size of this study.
miRNAs can regulate growth factors, target tumor suppression genes and other transcriptional factors to promote or inhibit the proliferation of cancer in humans. miR-125b may be associated with TGF-β to stimulate cancer growth and the potential activation of TGF-β with the possibility of playing a role in promoting cancer (36). miR-21 is one of the most commonly observed aberrant miRNAs in human cancers and a large scale miRNA analysis of 540 samples from six different types of solid tumors showed that miR-21 was the only miRNA that was upregulated in all cancer types (37). miR-21 is believed to target many tumor suppressors to regulate epithelial-mesenchymal transition, invasion and metastasis in lung cancer (38). There is evidence that miR-141 promoted the proliferation of non-small cell lung cancer cells by regulating the expression of PH domain leucine-rich-repeats protein phosphatase 1 and 2 (PHLPP1 and PHLPP2) (39). miR-200c was found to exert tumor-suppressive effects for NSCLC through the suppression of USP25 expression (40). miRNA-197 acts as an oncogene downregulating p53 and FUS1 tumor suppressor gene expression (41,42). It was also shown to be associated with brain metastasis in EGFR-mutant lung cancer (43). miR-370 was reported to inhibit the progression of non-small cell lung cancer by downregulating the oncogene TRAF4 (44). miR-376α could suppress the proliferation and invasion of non-small-cell lung cancer by targeting c-Myc (45). miR-192 was found to regulate chemo-resistance of lung adenocarcinoma for gemcitabine and cisplatin combined therapy by targeting Bcl-2 (46). miR-662 may provide an alternative mechanism of downregulation of a tumor suppressor gene GDF10 that belongs to the TGF-β family ligands (25). miR-26b was shown to function as a critical regulator of tumor progression and carcinogenesis because it acts as either a tumor suppressor or an oncogene in various cancers (29). miR-381 targeted IGF-1R to deactivate the protein kinase B (AKT) and extracellular signal-regulated kinase (ERK) signaling pathways (47). Overexpression of miR-146α significantly enhanced cell apoptosis, inhibiting cell viability and motility in vitro and in vivo and miR-146α could specially degrade the mRNA of cyclin J in the development of acquired drug resistance to DDP-based chemotherapy in NSCLC cells (48). Overexpression of miRNA-148α inhibited Wnt-1 protein expression in cancer cells. Knocking down Wnt-1 by siRNA had a similar effect to that of miRNA-148α overexpression on cell migration and invasion in lung cancer cells (19). miR-148b was reported to regulate radio-resistance of lung cancer cells by modulating the level of MLH1 expression (49). miRNA-204 suppressed human non-small cell lung cancer by targeting activating transcription factor 2 (ATF2) (50). miR-374α had very significantly affected pathways of cell migration (27). Downregulation of miRNA-638 promotes angiogenesis and growth of hepatocellular carcinoma by targeting VEGF (51).
Though the sample types were different from each other in the publications included in our analysis, we put them together for some reasons. Firstly, all the studies included in the paper used qPCR, allowing comparisons to be made between them. Secondly, it was reported that serum miRNAs have potential clinical value as tumor markers for screening NSCLC. Serum expression levels of miR-125b and miR-22 in NSCLC patients were significantly higher than those with benign lung diseases and those in the healthy controls (52). It also reported that serum and plasma could be better mediums to measure miRNA compared to sputum and whole blood. When combining CT scanning with miRNA measured in serum or plasma, the sensitivity value increased (35). Last but not least, there is a continued need for the development of minimally-invasive, cost-effective and easy methods to diagnose lung cancer at an earlier, curable stage. The miRNA levels in plasma, serum and other biological fluids could be optimistic and promising markers in the future.
Several high-quality studies arouse us great interest before we drafted our study. Similar publications have been published looking into the value of miRs in lung cancer (53–55). Since the topic has been hot in recent research, we performed this analysis. Different from previous studies, we further evaluated the relationship between expression levels of multiple miRNAs and cancer stage and LNM for lung cancer patients. In our meta-analysis, we summarized and reviewed all the similar researches and identified that a few miRNAs could serve as a novel predictor for the prognosis of NSCLC. It also provided very useful resources for new therapeutic targets for the management of lung cancer in the future. However, we also understood that there had a number of limitations in our research. First, it was only based on the results of one database (PubMed) and some relevant studies might have been missed. Second, only 15 articles were included in this analysis and all of the cancer types were NSCLC. Third, though the detection method of miRNAs in all studies analyzed were reverse transcription qPCR, the kits, analytical methods and positive results evaluation criteria might have had variations which may have led to different experimental results. Fourth, the search language in this study was limited to English and other languages such as Chinese were not included, which might lead to an inevitable bias. Fifth, although no significant publication bias was found in the included studies, it still could not be completely avoided as positive results are more likely to be accepted while negative results are more likely to be rejected or not published. Sixth, the sample types were different from each other in the publications included in the analysis. These results were combined and therefore the overall determined conclusion of the study might not be widely applicable due to the inclusion of these different samples.
All in all, we have examined a substantial number of prognostic data about the association of various miRNAs and the survival of lung cancer patients. Our meta-analysis identified that a few miRNAs could serve as a novel predictor of prognosis in NSCLC. Larger studies are required to further confirm our observation and understanding of the roles of these miRNAs and their targets in cancer cells that can bring new therapeutic methods to NSCLC lung cancer treatment.
Acknowledgements
The authors would like thank their colleagues at the Department of Thoracic Medicine of Shenzhen Second People's Hospital (Guangdong, China) for their assistance during data extraction and analysis.
Funding
The present study was supported by a grant from the Shenzhen Municipal Science and Technology Innovation Committee (grant no. JCY20170412170814624).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
DY and SY extracted and assessed that data from eligible studies. WX and YZ interpreted the data for miRNA expression levels and prognosis in NSCLC. LW and MZ assessed and revised the data and manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Li D, Song L, Wen Z, Li X, Jie J, Wang Y, Peng L. Strong evidence for LncRNA ZNRD1-AS1, and its functional Cis-eQTL locus contributing more to the susceptibility of lung cancer. Oncotarget. 2016;7:35813–35817. doi: 10.18632/oncotarget.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castro D, Moreira M, Gouveia AM, Pozza DH, De Mello RA. microRNAs in lung cancer. Oncotarget. 2017;8:81679–81685. doi: 10.18632/oncotarget.20955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aghanoori MR, Mirzaei B, Tavallaei M. miRNA molecular profiles in human medical conditions: Connecting lung cancer and lung development phenomena. Asian Pac J Cancer Prev. 2014;15:9557–9565. doi: 10.7314/APJCP.2014.15.22.9557. [DOI] [PubMed] [Google Scholar]
- 7.Kozomara A, Griffiths-Jones S. miR base: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong Y, Chen Z, Guo S, Liao X, Xie H, Zheng Y, Cai B, Huang P, Liu Y, Zhou Q, et al. TUG1, SPRY4-IT1, and HULC as valuable prognostic biomarkers of survival in cancer: A PRISMA-compliant meta-analysis. Medicine (Baltimore) 2017;96:e8583. doi: 10.1097/MD.0000000000008583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang M, Shi J, Peng N, He S. MicroRNA-211 promotes non-small-cell lung cancer proliferation and invasion by targeting MxA. OncoTargets Ther. 2017;10:5667–5675. doi: 10.2147/OTT.S143084. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Mavridis K, Gueugnon F, Petit-Courty A, Courty Y, Barascu A, Guyetant S, Scorilas A. The oncomiR miR-197 is a novel prognostic indicator for non-small cell lung cancer patients. Br J Cancer. 2015;112:1527–1535. doi: 10.1038/bjc.2015.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F, Lou JF, Cao Y, Shi XH, Wang P, Xu J, Xie EF, Xu T, Sun RH, Rao JY, et al. miR-638 is a new biomarker for outcome prediction of non-small cell lung cancer patients receiving chemotherapy. Exp Mol Med. 2015;47:e162. doi: 10.1038/emm.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Võsa U, Vooder T, Kolde R, Vilo J, Metspalu A, Annilo T. Meta-analysis of microRNA expression in lung cancer. Int J Cancer. 2013;132:2884–2893. doi: 10.1002/ijc.27981. [DOI] [PubMed] [Google Scholar]
- 13.Guan P, Yin Z, Li X, Wu W, Zhou B. Meta-analysis of human lung cancer microRNA expression profiling studies comparing cancer tissues with normal tissues. J Exp Clin Cancer Res. 2012;31:54. doi: 10.1186/1756-9966-31-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang Y. An expression meta-analysis of predicted microRNA targets identifies a diagnostic signature for lung cancer. BMC Med Genomics. 2008;1:61. doi: 10.1186/1755-8794-1-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Y, Ge Y, Wang Y, Ma G, Wang X, Liu H, Wang M, Zhang Z, Chu H. The association of rs710886 in lncRNA PCAT1 with bladder cancer risk in a Chinese population. Gene. 2017;627:226–232. doi: 10.1016/j.gene.2017.06.021. [DOI] [PubMed] [Google Scholar]
- 16.Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting recommendations for tumor marker prognostic studies (REMARK): Explanation and elaboration. PLoS Med. 2012;9:e1001216. doi: 10.1371/journal.pmed.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouwmeester W, Zuithoff NP, Mallett S, Geerlings MI, Vergouwe Y, Steyerberg EW, Altman DG, Moons KG. Reporting and methods in clinical prediction research: A systematic review. PLoS Med. 2012;9:1–12. doi: 10.1371/journal.pmed.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P, Dewaele A, Sadones J, Geers C, De Grève J. miR-146α inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS One. 2013;8:e60317. doi: 10.1371/journal.pone.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Min L, Ren C, Xu X, Yang J, Sun X, Wang T, Wang F, Sun C, Zhang X. miRNA-148α serves as a prognostic factor and suppresses migration and invasion through Wnt1 in non-small cell lung cancer. PLoS One. 2017;12:e0171751. doi: 10.1371/journal.pone.0171751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui EH, Li HJ, Hua F, Wang B, Mao W, Feng XR, Li JY, Wang X. Serum microRNA 125b as a diagnostic or prognostic biomarker for advanced NSCLC patients receiving cisplatin-based chemotherapy. Acta Pharmacol Sin. 2013;34:309–313. doi: 10.1038/aps.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo W, Zhang Y, Zhang Y, Shi Y, Xi J, Fan H, Xu S. Decreased expression of miR-204 in plasma is associated with a poor prognosis in patients with non-small cell lung cancer. Int J Mol Med. 2015;36:1720–1726. doi: 10.3892/ijmm.2015.2388. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Yu T, Cao J, Liu L, Liu Y, Kong HW, Zhu MX, Lin HC, Chu DD, Yao M, et al. MicroRNA-148α suppresses invasion and metastasis of human non-small-cell lung cancer. Cell Physiol Biochem. 2015;37:1847–1856. doi: 10.1159/000438546. [DOI] [PubMed] [Google Scholar]
- 23.Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ, Wang YK, Zeng F, Zhou JH, Zhang YK. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol. 2012;29:618–626. doi: 10.1007/s12032-011-9923-y. [DOI] [PubMed] [Google Scholar]
- 24.Nadal E, Zhong J, Lin J, Reddy RM, Ramnath N, Orringer MB, Chang AC, Beer DG, Chen G. A microRNA cluster at 14q32 drives aggressive lung adenocarcinoma. Clin Cancer Res. 2014;20:3107–3117. doi: 10.1158/1078-0432.CCR-13-3348. [DOI] [PubMed] [Google Scholar]
- 25.Skrzypski M, Czapiewski P, Goryca K, Jassem E, Wyrwicz L, Pawłowski R, Rzyman W, Biernat W, Jassem J. Prognostic value of microRNA expression in operable non-small cell lung cancer patients. Br J Cancer. 2014;110:991–1000. doi: 10.1038/bjc.2013.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Y, Su B, Zhang P, Xie H, Zheng H, Xu Y, Du Q, Zeng H, Zhou X, Chen C, et al. Expression of miR-150 and miR-3940-5p is reduced in non-small cell lung carcinoma and correlates with clinicopathological features. Oncol Rep. 2013;29:704–712. doi: 10.3892/or.2012.2152. [DOI] [PubMed] [Google Scholar]
- 27.Võsa U, Vooder T, Kolde R, Fischer K, Välk K, Tõnisson N, Roosipuu R, Vilo J, Metspalu A, Annilo T. Identification of miR-374α as a prognostic marker for survival in patients with early-stage nonsmall cell lung cancer. Genes Chromosomes Cancer. 2011;50:812–822. doi: 10.1002/gcc.20902. [DOI] [PubMed] [Google Scholar]
- 28.Wang R, Ye F, Zhen Q, Song T, Tan G, Chu W, Zhang Y, Lv B, Zhao X, Liu J. MicroRNA-148b is a potential prognostic biomarker and predictor of response to radiotherapy in non-small-cell lung cancer. J Physiol Biochem. 2016;72:337–343. doi: 10.1007/s13105-016-0485-5. [DOI] [PubMed] [Google Scholar]
- 29.Jiang LP, Zhu ZT, He CY. Expression of miRNA-26b in the diagnosis and prognosis of patients with non-small-cell lung cancer. Future Oncol. 2016;12:1105–1115. doi: 10.2217/fon.16.21. [DOI] [PubMed] [Google Scholar]
- 30.Rothschild SI, Tschan MP, Jaggi R, Fey MF, Gugger M, Gautschi O. microRNA-381 represses ID1 and is deregulated in lung adenocarcinoma. J Thorac Oncol. 2012;7:1069–1077. doi: 10.1097/JTO.0b013e31824fe976. [DOI] [PubMed] [Google Scholar]
- 31.Al Khalaf MM, Thalib L, Doi SA. Combining heterogenous studies using the random-effects model is a mistake and leads to inconclusive meta-analyses. J Clin Epidemiol. 2011;64:119–123. doi: 10.1016/j.jclinepi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 32.Sittka A, Schmeck B. MicroRNAs in the lung. Adv Exp Med Biol. 2013;774:121–134. doi: 10.1007/978-94-007-5590-1_7. [DOI] [PubMed] [Google Scholar]
- 33.Wang T, Nelson RA, Bogardus A, Grannis FW., Jr Five-year lung cancer survival: Which advanced stage nonsmall cell lung cancer patients attain long-term survival? Cancer. 2010;116:1518–1525. doi: 10.1002/cncr.24871. [DOI] [PubMed] [Google Scholar]
- 34.Mizuno K, Mataki H, Seki N, Kumamoto T, Kamikawaji K, Inoue H. MicroRNAs in non-small cell lung cancer and idiopathic pulmonary fibrosis. J Hum Genet. 2017;62:57–65. doi: 10.1038/jhg.2016.98. [DOI] [PubMed] [Google Scholar]
- 35.Gyoba J, Shan S, Roa W, Bédard EL. Diagnosing lung cancers through examination of micro-RNA biomarkers in blood, plasma, serum and sputum: A review and summary of current literature. Int J Mol Sci. 2016;17:494. doi: 10.3390/ijms17040494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Zhang Y, Fu Y, Zhang J, Yin L, Pu Y, Liang G. MicroRNA-125b may function as an oncogene in lung cancer cells. Mol Med Rep. 2015;11:3880–3887. doi: 10.3892/mmr.2014.3142. [DOI] [PubMed] [Google Scholar]
- 37.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markou A, Zavridou M, Lianidou ES. miRNA-21 as a novel therapeutic target in lung cancer. Lung Cancer (Auckl) 2016;7:19–27. doi: 10.2147/LCTT.S60341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mei Z, He Y, Feng J, Shi J, Du Y, Qian L, Huang Q, Jie Z. MicroRNA-141 promotes the proliferation of non-small cell lung cancer cells by regulating expression of PHLPP1 and PHLPP2. FEBS Lett. 2014;588:3055–3061. doi: 10.1016/j.febslet.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 40.Li J, Tan Q, Yan M, Liu L, Lin H, Zhao F, Bao G, Kong H, Ge C, Zhang F, et al. miRNA-200c inhibits invasion and metastasis of human non-small cell lung cancer by directly targeting ubiquitin specific peptidase 25. Mol Cancer. 2014;13:166. doi: 10.1186/1476-4598-13-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fiori ME, Barbini C, Haas TL, Marroncelli N, Patrizii M, Biffoni M, De Maria R. Antitumor effect of miR-197 targeting in p53 wild-type lung cancer. Cell Death Differ. 2014;21:774–782. doi: 10.1038/cdd.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du L, Schageman JJ, Subauste MC, Saber B, Hammond SM, Prudkin L, Wistuba II, Ji L, Roth JA, Minna JD, et al. miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol Cancer Res. 2009;7:1234–1243. doi: 10.1158/1541-7786.MCR-08-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Remon J, Alvarez-Berdugo D, Majem M, Moran T, Reguart N, Lianes P. miRNA-197 and miRNA-184 are associated with brain metastasis in EGFR-mutant lung cancers. Clin Trans Oncol. 2016;18:153–159. doi: 10.1007/s12094-015-1347-2. [DOI] [PubMed] [Google Scholar]
- 44.Chen T, Gao F, Feng S, Yang T, Chen M. MicroRNA-370 inhibits the progression of non-small cell lung cancer by downregulating oncogene TRAF4. Oncol Rep. 2015;34:461–468. doi: 10.3892/or.2015.3978. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Cong W, Wu G, Ju X, Li Z, Duan X, Wang X, Gao H. miR-376α suppresses the proliferation and invasion of non-small-cell lung cancer by targeting c-Myc. Cell Biol Int. 2018;42:25–33. doi: 10.1002/cbin.10828. [DOI] [PubMed] [Google Scholar]
- 46.Cao J, He Y, Liu HQ, Wang SB, Zhao BC, Cheng YS. MicroRNA 192 regulates chemo-resistance of lung adenocarcinoma for gemcitabine and cisplatin combined therapy by targeting Bcl-2. Int J Clin Exp Med. 2015;8:12397–12403. [PMC free article] [PubMed] [Google Scholar]
- 47.Tu C, Wang F, Wan J. MicroRNA-381 inhibits cell proliferation and invasion in endometrial carcinoma by targeting the IGF-1R. Mol Med Rep. 2018;17:4090–4098. doi: 10.3892/mmr.2017.8288. [DOI] [PubMed] [Google Scholar]
- 48.Shi L, Xu Z, Wu G, Chen X, Huang Y, Wang Y, Jiang W, Ke B. Up-regulation of miR-146α increases the sensitivity of non-small cell lung cancer to DDP by downregulating cyclin J. BMC Cancer. 2017;17:138. doi: 10.1186/s12885-017-3132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhai G, Li G, Xu B, Jia T, Sun Y, Zheng J, Li J. miRNA-148b regulates radioresistance in non-small lung cancer cells via regulation of MutL homologue 1. Biosci Rep. 2016;36:e00354. doi: 10.1042/BSR20150300. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Zhang S, Gao L, Thakur A, Shi P, Liu F, Feng J, Wang T, Liang Y, Liu JJ, Chen M, et al. miRNA-204 suppresses human non-small cell lung cancer by targeting ATF2. Tumour Biol. 2016;37:11177–11186. doi: 10.1007/s13277-016-4906-4. [DOI] [PubMed] [Google Scholar]
- 51.Cheng J, Chen Y, Zhao P, Liu X, Dong J, Li J, Huang C, Wu R, Lv Y. Downregulation of miRNA-638 promotes angiogenesis and growth of hepatocellular carcinoma by targeting VEGF. Oncotarget. 2016;7:30702–30711. doi: 10.18632/oncotarget.8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi GL, Chen Y, Sun Y, Yin YJ, Song CX. Significance of serum microRNAs in the auxiliary diagnosis of non-small cell lung cancer. Clin Lab. 2017;63:133–140. doi: 10.7754/Clin.Lab.2016.160710. [DOI] [PubMed] [Google Scholar]
- 53.Zhan B, Lu D, Luo P, Wang B. Prognostic value of expression of microRNAs in non-small cell lung cancer: A systematic review and meta-analysis. Clin Lab. 2016;62:2203–2211. doi: 10.7754/Clin.Lab.2016.160426. [DOI] [PubMed] [Google Scholar]
- 54.Yu N, Zhang Q, Liu Q, Yang J, Zhang S. A meta-analysis: microRNAs' prognostic function in patients with nonsmall cell lung cancer. Cancer Med. 2017;6:2098–2105. doi: 10.1002/cam4.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang M, Li X, Quan X, Li Xi, Zhou B. Clinically correlated microRNAs in the diagnosis of non-small cell lung cancer: A systematic review and meta-analysis. Biomed Res Int. 2018;2018:5930951. doi: 10.1155/2018/5930951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.