Abstract
Single nucleotide polymorphisms (SNPs) in microRNA may affect its expression and regulation of target genes, which may consequently alter individual susceptibility to cancer. In this study we aimed to investigate associations between miR-122 polymorphisms and hepatocellular carcinoma (HCC) in a southern Chinese population. Three selected SNPs in miR-122 (rs9966765, rs1135519, and rs17669) were genotyped in 1050 HCC patients and 1079 cancer-free controls using Sequenom MassARRAY platform and the associations of the three SNPs and HCC risk were evaluated. We found that individuals with the rs1135519 CC genotypes had a significant increased risk of HCC than those with TT genotypes (adjusted OR=2.71, 95% CI=1.15-6.36, and P=0.022), while the rs9966765 CC genotypes showed a borderline significant association with increased risk of HCC when compared with the GG genotypes (adjusted OR=2.38, 95% CI=0.99-5.75, and P=0.052). There was also a significant increased risk of HCC when combining risk genotypes of these loci, i.e., rs1135519 CC and rs9966765 CC. Compared with the low-risk group (0 risk genotype), the high risk group (1-2 risk genotypes) had significantly increased risk of HCC (OR=1.61, 95% CI=1.05-2.44, and P=0.028). Further genotype-expression analysis revealed that cases carrying the CC genotype of rs1135519 had lower levels of miR-122 expression than those with the TT genotype. Our results suggest that SNP of rs1135519 modulates miR-122 expression and contributes to the genetic susceptibility of HCC, either independently or together with rs9966765 in miR-122. Further well-designed studies with lager sample sizes are needed to confirm our findings.
1. Introduction
Hepatocellular carcinoma (HCC) is the second most common malignancies worldwide, with an estimated 782,500 new cases and 745,500 deaths reported annually, of which more than 50% are from China [1]. As the major risk factors, the relationships of hepatitis B/C virus (HBV/HCV) infection, tobacco use, and alcohol abuse and HCC have been well established [2, 3]. However, the exact mechanism of developing HCC is still unclearly. Recently, germline genetic variations, such as single nucleotide polymorphisms (SNPs), have been reported to be associated with HCC, which may help to uncover the molecular mechanism of development of HCC [4, 5].
MicroRNAs (miRNAs) are a class of small noncoding single-stranded RNAs that function as negative gene regulators by cleavage target mRNA or inhibiting its translation through binding to its 3'-untranslated region (UTR) [6, 7]. It is estimated that over 1,800 precursor miRNAs have been identified in the human genome, targeting 60% of the whole genes [8, 9]. MiRNAs are well conserved in eukaryotic organisms and considered to be a vital component of genetic regulation [10, 11]. Emerging studies have shown that miRNAs are involved in diverse biological processes, including development, differentiation, cell growth, and apoptosis [12]. Additionally, aberrant expression of miRNAs has been implicated in a wide variety of human cancers, including HCC [13].
MiR-122 is a highly abundant liver-specific miRNA that plays a pivotal role in liver development and hepatic function regulation [14, 15]. MiR-122 acts as a tumor suppressor against HCC by binding to the target genes involved in various biological processes in HCC, including cell proliferation, apoptosis, and angiogenesis [16, 17]. Recently, it has been also shown that downregulation of miR-122 is correlated with metastasis and poor prognosis of HCC [18, 19]. Many studies have demonstrated that SNPs in miRNA can alter their expression level and may contribute to the susceptibility to cancer [20–22]. Given the important role of miR-122 in liver pathology, we hypothesized that germline genetic variants within miR-122 would influence susceptibility to HCC. To test this hypothesis, we conducted a case-control study to investigate the association between SNPs in miR-122 and the risk of HCC in a Chinese population.
2. Materials and Methods
2.1. Ethics Statement
This study was conducted in accordance with the approved guidelines by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University, and informed consent was obtained from all patients. And the Ethics Committee of the First Affiliated Hospital of Guilin Medical University approves that this study has complied with the Declaration of Helsinki.
2.2. Information and Sample Collection of Study Subjects
The present hospital-based case-control study included a total of 1050 HCC patients and 1079 cancer-free controls, which were consecutively recruited from First Affiliated Hospital of Guangxi Medical University and Affiliated Cancer Hospital of Guangxi Medical University from June 2007 to April 2011. Among the subjects, 589 HCC cases and 597 controls were collected between June 2007 and January 2010 as reported in our earlier studies [23]. Hence, an additional 461 HCC cases and 482 controls were recruited from February 2010 to April 2011. Altogether, a total of 1050 HCC cases and 1079 controls were used in the present study. All of the HCC patients were newly diagnosed and histologically confirmed. Those who had a prior history of other cancers, metastasized cancers, or previous radiotherapy or chemotherapy before the recruitment were excluded. The cancer-free controls were recruited from the Department of Orthopedics and Ophthalmology in the same period and were frequency matched to the cases by age (±5 years) and sex, without genetic relationships with the HCC patients. After signing an informed consent form, information including demographic data, history of tobacco and alcohol consumption and chronic HBV infection were obtained from each subject through face-to-face interviews conducted by trained investigators. Ever smokers were defined as persons who had smoked more than 100 cigarettes in their lifetime; ever drinkers were defined as persons who had used alcohol at least once a week for more than one year. HBV infection was defined as positive for HBV surface antigen (HBsAg). 5 mL of peripheral blood was collected for serology test and molecular genetic analysis. Thirty-two HCC cases with the CC or TT genotypes of rs1135519 were selected from cases with one of these two genotypes. Tumor tissues for these selected cases were collected from patients who had undergone surgery in the First Affiliated Hospital of Guilin Medical University.
2.3. SNPs Selection and Genotyping
The NCBI dbSNP database were used to selected common SNPs in 2kb upstream and downstream of miR-122 gene, with the minor allele frequency (MAF) > 0.05 in the CHB population (Chinese Han in Beijing). And the pairwise linkage disequilibrium (LD) had an r2 threshold of 0.8 on the NIEHS data-base (https://snpinfo.niehs.nih.gov/). Finally, rs9966765, rs1135519, and rs17669 in miR-122 were selected for further study. DNA was extracted from peripheral blood by phenol–chloroform extraction and stored at -80°C. The three selected common SNPs in miR-122 were genotyped by using the Agena MassARRAY genotyping system (Agena, San Diego, CA) according to the manufacturer's instructions. Each PCR reaction mixture contained 10 ng of genomic DNA, 0.5 μL 10×PCR Buffer, 0.4 μL 25 mM MgCl 2, 0.1 μL25 mM dNTPs, 1 μL0.5 uM primer Mix, and 0.2 μL 5 U/μL Hot Star Taq polymerase. Reaction was performed at 94°C for 15 min, followed by 45 cycles at 94°C for 20 s, 56°C for 30 s, and 72°C for 1 min, with a final incubation at 72°C for 3 min. The extension reactions were performed at 94°C for 30 s and then 94°C for 5 s, followed by 40 cycles at 52°C for 5 s, 5 cycles at 80°C for 5 s, with a final incubation at 72°C for 3min. Purified extension reaction products were spotted onto a 384-well Spectro CHIPs and measured by using the platform MALDI-TOF mass spectrometry within the Agena MassARRAY system. Genotype calling was performed and analyzed by using the MassARRAY Typer software version 4.0. All SNPs were successfully genotyped with high call rates (>94%). For quality control, we randomly selected 10% of the samples for repeating genotyping and found the consistent rate was 100%.
2.4. Quantitative Real-Time Reverse Transcriptase-Polymerase Chain Reaction Assay
miR-122 expression was measured by reverse transcription-PCR according to the TaqMan microRNA Assay protocol (Bio Miao Biological Technology (Beijing) Co.,Ltd.). RNAs were extracted with TRIZOL reagent (Invitrogen, CA, USA) following the manufacturer's instructions were reverse-transcribed using a PrimeScript II 1st Strand cDNA Synthesis Kit ( TAKARA). The expression level of mature miR-122 was examined using SYBR FAST qPCR Kit Master Mix(2×) Universal (KAPA Biosystems), and the comparative Ct method comparing to the transcription level of U6 RNA was used to calculate the expression levels.
2.5. Statistical Analysis
The differences of distributions of selected variables and genotypes between cases and controls were assessed using χ2 test. Hardy-Weinberg equilibrium analysis was assessed by a goodness of fit χ2 test. The associations between miR-122 SNPs and the risk of HCC were estimated by calculating the odds ratios (ORs) and 95% confidence intervals (95%CIs) with unconditional multivariate logistic regression model. All statistical tests were two-tailed. P values below 0.05 were regarded as indicating statistical significance. All analyses were performed using SPSS software, version 13.0 (SPSS Institute, Chicago, IL).
3. Result
3.1. Characteristics of the Study Population
The selected characteristics of 1050 HCC cases and 1079 cancer-free controls included in this study are shown in Table 1. There were no statistical differences in the distribution of age and sex between cases and controls (P=0.132 and 0.618, respectively). However, cases were more likely to be smokers (P<0.001), drinkers (P<0.001), and HBV carriers (P<0.001) than the controls.
Table 1.
Frequency distributions of the clinical features between HCC cases (n=1050) and controls (n=1079) [n(%)].
| Variables | Cases | Controls | P a |
|---|---|---|---|
| Age | 0.053 | ||
| <48 | 463 (44.10) | 521 (48.29) | |
| ≥48 | 587 (49.00) | 558 (51.71) | |
| Sex | 0.618 | ||
| Male | 917 (87.33) | 950 (88.04) | |
| Female | 133 (12.67) | 129 (11.96) | |
| Smoking | <0.001 | ||
| Never | 663 (63.14) | 908 (84.15) | |
| Ever | 387 (36.86) | 171 (15.85) | |
| Drinking | <0.001 | ||
| Never | 698 (66.48) | 937 (86.84) | |
| Ever | 352 (33.52) | 142 (13.16) | |
| HBV infection | <0.001 | ||
| No | 176 (16.76) | 981 (90.92) | |
| Yes | 874 (83.24) | 98 (9.08) |
a P-value for Chi-square test.
3.2. Association between miR-122 Polymorphisms and the Risk of HCC
The primary information of the selected SNPs is listed in Table 2. The observed genotype frequencies for the SNPs of miR-122 among the control subjects were all in agreement with HWE (P=0.544 for rs9966765, P=0.447 for rs1135519, and P=0.525 for rs17669, respectively). The calling rates were all above 94%. The distributions of genotype frequencies for these three SNPs in the cases and controls are summarized in Table 3. The frequencies of rs1135519 TT, TC, and CC genotypes were 68.65%, 28.37%, and 2.98% in HCC cases while they were 73.14%, 25.09%, and 1.77% in the controls, respectively. There were statistically significant difference in the distribution of rs1135519 genotypes between cases and controls (P =0.033). However, no significant difference in the genotype frequencies of rs9966765 or rs17669 was found (P = 0.071 for rs9966765 and P = 0.112 for rs17669, respectively).
Table 2.
The information of the miR-122 SNPs.
| SNP | Location | Base change | Genotypes | MAF a | P HWE b | Genotyping rate (%) |
|---|---|---|---|---|---|---|
| rs9966765 | 5' near gene | G > C | GG CG CC | 0.142 | 0.544 | 94.9 |
| rs1135519 | 5' near gene | T > C | TT CT CC | 0.144 | 0.447 | 94.9 |
| rs17669 | 3' near gene | A > G | AA GA GG | 0.143 | 0.525 | 94.6 |
a Minor allele frequency;
bP-value for Hardy-Weinberg equilibrium tests.
Table 3.
Association between SNPs in miR-122 and HCC risk [n (%)].
| Genotypes | Cases a | Controls a | OR (95% CI) b | P b |
|---|---|---|---|---|
| rs9966765 | ||||
| GG | 725 (69.38) | 788 (73.58) | 1 | |
| CG | 291 (27.85) | 264 (24.65) | 1.08 (0.80-1.46) | 0.630 |
| CC | 29 (2.78) | 19 (1.77) | 2.38 (0.99-5.75) | 0.052 |
| rs1135519 | ||||
| TT | 714 (68.65) | 786 (73.14) | 1 | |
| CT | 295 (28.37) | 270 (25.09) | 1.09 (0.80-1.47) | 0.563 |
| CC | 31 (2.98) | 19 (1.77) | 2.71 (1.15-6.36) | 0.022 |
| rs17669 | ||||
| AA | 725 (69.85) | 789 (73.46) | 1 | |
| AG | 285 (27.45) | 266 (24.77) | 1.17 (0.96-1.42) | 0.123 |
| GG | 28 (2.70) | 19 (1.77) | 1.60 (0.89-2.90) | 0.117 |
a The numbers were not the same for each SNP due to different calling rates.
b Adjusted for age, sex, smoking, drinking, and HBV infection in logistic regression model.
After adjusting for age, sex, smoking, drinking, and HBV infection, we found that rs1135519 CC genotype was significantly associated with an increased risk of HCC when compared with TT genotype (adjusted OR=2.71, 95%CI=1.15-6.36, and P=0.022). We also found rs9966765 CC genotype had a borderline significant association with increased risk of HCC when compared with GG genotype (adjusted OR=2.38, 95% CI=0.99-5.75, and P=0.052). Although AG and GG genotypes of rs17669 tended to increased HCC risk (adjusted OR=1.17, 95% CI=0.96-1.42, P=0.123 for AG, OR=1.60, 95% CI =0.89-2.90, and P=0.117 for GG) when compared with AA genotype, however, it showed a nonsignificant association (Table 3).
3.3. Association Analysis of the Combined Risk Genotypes in miR-122 and HCC Risk
Considering a possible combined effect of the two SNPs, i.e., rs9966765 and rs1135519, on risk of HCC, we conducted combined analysis by using the number of the putative risk genotypes (i.e., rs9966765 CC and rs1135519 CC). As shown in Table 4, the combined risk genotypes were found to be associated with increased risk of HCC. The frequencies of 1-2 risk genotypes in HCC patients (2.9%) were higher than that in controls (1.8%), which showed statistical significance (P<0.05). Individuals with 1-2 risk genotypes had a significantly increased risk of HCC when compared with those harboring 0 risk genotypes after adjustment for age, sex, smoking, drinking, and HBV infection (adjusted OR =1.61, 95% CI=1.05-2.44, and P=0.028).
Table 4.
Association of number of risk genotypes and HCC risk [n(%)].
| Number of risk genotype a | Case | Control | OR (95% CI) b | P b |
|---|---|---|---|---|
| 0 | 1009 (97.1) | 1050 (98.2) | 1 | |
| 1-2 | 30 (2.9) | 19 (1.8) | 1.61 (1.05–2.44) | 0.028 |
a Risk genotypes used were rs9966765 CC and rs1135519 CC.
b Adjusted for age, sex, smoking, drinking status and HBV infection.
3.4. Correlation between rs1135519 Genotypes and the Expression Levels of miR-122
To explore functional relevance of rs1135519 genotypes, we conducted genotype-phenotype correlation analysis between rs1135519 genotypes and miR-122 expression level in 32 HCC tissues. Compared with individuals carrying TT genotype (n=28), rs1135519 CC genotype carriers (n=4) had lower levels of miR-122 expression (∆Ctmean=10.13±0.75 for CC vs. ∆Ctmean=11.21±0.88 for TT, P=0.013, Figure 1), suggesting that rs1135519 SNP could modulate the miR-122 expression.
Figure 1.
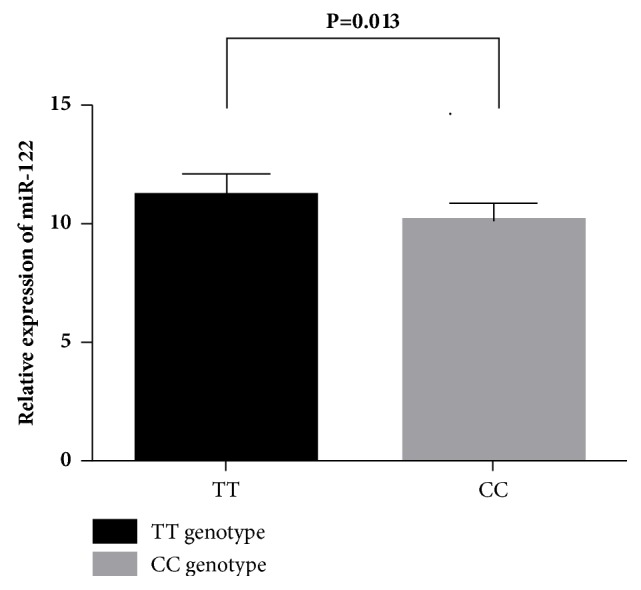
The relative expression of miR-122 in HCC tissues with rs1135519 CC and TT genotypes. P value is from t test.
4. Discussion
In this case-control study, we investigated the associations between three selected SNPs (i.e., rs9966765, rs1135519, and rs17669) in miR-122 and risk of HCC by using 1050 HCC patients and 1079 cancer-free controls in a Chinese population. We found that polymorphisms of rs1135519 modulates miR-122 expression and contributes to the genetic susceptibility of HCC, either independently or together with rs9966765 in miR-122.
As liver-specific miRNA, miR-122 has been reported to be down-regulated and associated with development, metastasis, recurrence, and poor prognosis of HCC [18, 19, 24]. Reduced levels of miR-122 in HCC may result in chromosomal instability through deregulation of cyclin G1 or p53-dependent pathways [25, 26]. Functional studies demonstrated that miR-122 could suppress cell proliferation and induces cell apoptosis in HCC by directly targeting Wnt/beta-catenin pathway, while the activation of oncogene c-Myc may induce transcriptional repression of miR-122 in HCC [27, 28]. Studies in mouse models using modified antisense miR-122 showed that its depletion compromised the liver function and reduced cholesterol level by targeting expression of genes involved in cholesterol biosynthesis that was found to facilitate cystogenesis and hepatocarcinogenesis [29, 30]. Thus, it was speculated that special SNPs in miR-122 gene might affect its expression and subsequently alter the risk of HCC. In this study, the CC genotype of rs1135519 was correlated with lower expression of miR-122, which was consistent with its risk effect on HCC.
Polymorphisms in miRNA with potential functions in its expression may influence the individual susceptibility to cancers, which have been well illustrated. Xu et al. [31] reported that HCC risk of rs2910164 GG genotype in miR-146a was 2-times of CC genotype among male (OR=2.02, 95% CI=1.06–3.85, and P=0.034). Further investigation disclosed that GG genotype conferred a higher expression level of mature miR-146a. Qi et al. [32] showed that the A to G base change of rs999885 in the promoter region of miR-106b-25 cluster may provide an increased risk for HCC in HBV persistent carriers by altering the expression of the miR-106b-25 cluster. Our previous research also showed that miR-199a rs74723057 polymorphism combined with MET rs1621 may influence susceptibility to HCC [33]. To date, only one published study had reported the effect of genetic variants in miR-122 on risk of HCC. Liu et al. [34] found that the C to A base change of rs4309483 located in the regulatory regions of miR-122 was associated with decreased expression of miR-122 and thus showed an increased risk of HCC in a case-control study, which was consistent with our study.
Several potential limitations in the current study should be considered. Firstly, as a hospital-based case-control study, the selection bias was unavoidable. To reduce the potential section bias, we had applied a rigorous design in selecting cases and controls by frequency matching with age and sex. Secondly, the sample size may limit the statistical power of our study, especially for subgroup analysis, although it had more than 2000. As a result, our results still need further validation. Finally, the precise molecular mechanisms underlying that how the SNP altering miR-122 expression need to be illustrated in future work.
In conclusion, we identified two genetic variants in miR-122 (rs9966765 and rs1135519), which may individually or jointly modulate the risk of HCC. Our results suggest the potential role the two SNPs in regulation of miR-122 expression and development of HCC. Further larger and well-designed studies with diverse populations and functional assays are warranted to validate our findings.
Acknowledgments
This study was supported by the grant from National Natural Science Foundation of China (NSFC, 81660567, 81460516) and Guangxi Natural Science Foundation Project for Returnees (2015 GXNSFCB 139007).
Contributor Information
Songqing He, Email: dr_hesongqing@163.com.
Hongping Yu, Email: yhp268@163.com.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
Chunhua Bei and Shun Liu contributed equally to this work.
References
- 1.Torre L. A., Bray F., Siegel R. L., Ferlay J., Lortet-Tieulent J. Global cancer statistics, 2012. CA: A Cancer Journal for Clinicians. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Dickinson J., Wun Y., Wong S. Modelling death rates for carriers of hepatitis B. Epidemiology and Infection. 2002;128(01) doi: 10.1017/S0950268801006410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Serag H. B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142(6):1264–1273. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farazi P. A., DePinho R. A. Hepatocellular carcinoma pathogenesis: from genes to environment. Nature Reviews Cancer. 2006;6(9):674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 5.Qiu M., Liu Y., Yu X., Qin L., Bei C., Zeng X. polymorphisms and the risk of hepatocellular carcinoma. Tumor Biology. 2016;37:3863–3870. doi: 10.1007/s13277-015-4222-4. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.Sun K., Lai E. C. Adult-specific functions of animal microRNAs. Nature Reviews Genetics. 2013;14(8):535–548. doi: 10.1038/nrg3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutvágner G., Zamore P. D. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297(5589):2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 9.Wienholds E., Kloosterman W. P., Miska E., et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309(5732):310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 10.Flynt A. S., Lai E. C. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nature Reviews Genetics. 2008;9(11):831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hobert O. miRNAs Play a Tune. Cell. 2007;131(1):22–24. doi: 10.1016/j.cell.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 12.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Croce C. M. Causes and consequences of microRNA dysregulation in cancer. Nature Reviews Genetics. 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morita K., Taketomi A., Shirabe K., et al. Clinical significance and potential of hepatic microRNA-122 expression in hepatitis C. Liver International. 2011;31(4):474–484. doi: 10.1111/j.1478-3231.2010.02433.x. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y., Shen A., Rider P. J. A liver-specific microRNA binds to a highly conserved RNA sequence of hepatitis B virus and negatively regulates viral gene expression and replication. The FASEB Journal. 2011;25(12):4511–4521. doi: 10.1096/fj.11-187781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin C. J., Gong H. Y., Tseng H. C., Wang W. L., Wu J. L. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochemical and Biophysical Research Communications. 2008;375(3):315–320. doi: 10.1016/j.bbrc.2008.07.154. [DOI] [PubMed] [Google Scholar]
- 17.Ma L., Liu J., Shen J., et al. Expression of miR-122 mediated by adenoviral vector induces apoptosis and cell cycle arrest of cancer cells. Cancer Biology & Therapy. 2014;9(7):554–561. doi: 10.4161/cbt.9.7.11267. [DOI] [PubMed] [Google Scholar]
- 18.Yin S., Fan Y., Zhang H. Differential TGFβ pathway targeting by miR-122 in humans and mice affects liver cancer metastasis. Nature Communications. 2016;7, article 11012 doi: 10.1038/ncomms11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thakral S., Ghoshal K. miR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Current Gene Therapy. 2015;15(2):142–150. doi: 10.2174/1566523214666141224095610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Y., Liu L., Liu J. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. International Journal of Cancer. 2011;128(2):412–417. doi: 10.1002/ijc.25342. [DOI] [PubMed] [Google Scholar]
- 21.Ryan B. M., Robles A. I., Harris C. C. Genetic variation in microRNA networks: the implications for cancer research. Nature Reviews Cancer. 2010;10(6):389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu F., Yang L., Zhang L., et al. Polymorphism in mature microRNA-608 sequence is associated with an increased risk of nasopharyngeal carcinoma. Gene. 2015;565(2):180–186. doi: 10.1016/j.gene.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 23.Zeng X., Liu S., Yu H., et al. DNA repair capacity, DNA-strand break repair gene polymorphisms, and the incidence of hepatocellular carcinoma in southwestern Guangxi of China. DNA and Cell Biology. 2012;31(8):1384–1391. doi: 10.1089/dna.2012.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L., Zhang Z., Wang F. The efficacy of miRNA122, a novel therapeutic target, for predicting the progression of hepatocellular carcinoma (HCC) Cellular & Molecular Immunology. 2012;9(2):103–104. doi: 10.1038/cmi.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gramantieri L., Ferracin M., Fornari F., et al. Cyclin G1 is a target of miR-122a, a MicroRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Research. 2007;67(13):6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 26.Xu J., Zhu X., Wu L., et al. MicroRNA-122 suppresses cell proliferation and induces cell apoptosis in hepatocellular carcinoma by directly targeting Wnt/β-catenin pathway. Liver International. 2012;32(5):752–760. doi: 10.1111/j.1478-3231.2011.02750.x. [DOI] [PubMed] [Google Scholar]
- 27.Wang B., Hsu S.-H., Wang X., et al. Reciprocal regulation of microRNA-122 and c-Myc in hepatocellular cancer: Role of E2F1 and transcription factor dimerization partner 2. Hepatology. 2014;59(2):555–566. doi: 10.1002/hep.26712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S., Qiu L., Yan X., et al. Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G 1-modulated P53 activity. Hepatology. 2012;55(3):730–741. doi: 10.1002/hep.24809. [DOI] [PubMed] [Google Scholar]
- 29.Tsai W.-C., Hsu S.-D., Hsu C.-S., et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. The Journal of Clinical Investigation. 2012;122(8):2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krützfeldt J., Rajewsky N., Braich R., et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 31.Xu T., Zhu Y., Wei Q., et al. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 2008;29(11):2126–2131. doi: 10.1093/carcin/bgn195. [DOI] [PubMed] [Google Scholar]
- 32.Qi F., Huang M., Pan Y., et al. A genetic variant in the promoter region of miR-106b-25 cluster predict clinical outcome of HBV-related hepatocellular carcinoma in Chinese. PLoS ONE. 2014;9(1):p. e85394. doi: 10.1371/journal.pone.0085394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q., Yu X., Li Q., et al. Association between miR-199a rs74723057 and MET rs1621 polymorphisms and the risk of hepatocellular carcinoma. Oncotarget. 2016;7(48):79365–79371. doi: 10.18632/oncotarget.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y., Xie K., Wen J., Deng M., Li J., Hu Z. A genetic variant in microRNA-122 regulatory region confers risk for chronic hepatitis B virus infection and hepatocellular carcinoma in Han Chinese. Journal of Medical Virology. 2014;86(10):1669–1674. doi: 10.1002/jmv.23996. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


