Abstract
Recent studies have described fungal communities in indoor environments using gene sequencing-based approaches. In this study, dust-borne fungal communities were elucidated from a water-damaged office building located in the northeastern region of the United States using internal transcribed spacer (ITS) rRNA gene sequencing. Genomic DNA was extracted from 5 mg of floor dust derived from 22 samples collected from either the lower floors (n = 8) or a top floor (n = 14) of the office building. ITS gene sequencing resolved a total of 933 ITS sequences and was clustered into 216 fungal operational taxonomic units (OTUs). Analysis of fungal OTUs at the 97% similarity threshold showed a difference between the lower and top floors that was marginally significant (p = 0.049). Species richness and diversity indices were reduced in the lower floor samples compared to the top floor samples and there was a high degree of compositional dissimilarity within and between the two different areas within the building. Fungal OTUs were placed in the phyla Ascomycota (55%), Basidiomycota (41%), Zygomycota (3%), Glomeromycota (0.4%), Chytridiomycota (0.3%), and unassigned fungi (0.5%). The Ascomycota classes with the highest relative abundances included the Dothideomycetes (30%) and Eurotiomycetes (16%). The Basidiomycota consisted of the classes Ustilaginomycetes (14%), Tremellomycetes (11%), and Agaricomycetes (8%). Sequence reads derived from the plant pathogen Ustilago syntherismae were the most abundant in the analysis as were obligate Basidiomycota yeast species that accounted for 12% and 11% of fungal ITS sequences, respectively. ITS gene sequencing provides additional insight into the diversity of fungal OTUs. These data further highlight the contribution of fungi placed in the phylum Basidiomycota, obligate yeasts, as well as xerophilic species that are typically not resolved using traditional culture methods.
Keywords: Exposure assessment, fungus, gene sequencing, occupational, office building
Introduction
Exposure to fungal bioaerosols within office buildings has become a significant public health concern. Recent meta-analyses and consensus documents have identified associations between fungal exposure and some respiratory morbidities;[1–3] however, methods to assess fungal exposure lack standardization and have been restricted to the examination of viable or nonviable bioaerosols. A priori taxa selection, observer identification bias, and difficulties associated with the differentiation of fungal from non-fungal particles are limiting variables related to these methodological approaches. Recent advances in chemical or immunoassay detection platforms have also enabled the quantification of (1→3)-β-D-glucan, ergosterol, secondary microbial volatile organic compounds, secondary fungal metabolites or high molecular weight proteins, such as allergens. Due to limitations associated with each of these methodological approaches, consensus threshold levels for fungal contaminants in indoor and occupational environments have not been established.
During the last two decades, molecular-based exposure assessment methods utilizing broad range gene targets have been increasingly employed to identify microorganisms,[4] in particular fungal bioaerosols, in a variety of indoor, outdoor, and occupational environments.[5–11] Based on the amplification of variable regions within internal transcribed spacer region of 18S ribosomal DNA (ITS), it has been possible to detect fungal species. Methods employed have included quantitative polymerase chain reaction,[12] and more recently, metagenomic approaches such as denaturing gradient gel electrophoresis,[13] Sanger sequencing,[7,14,15] or next generation sequencing.[8] Studies that have utilized these approaches have provided new insight into the distribution of previously overlooked fungal species, differences in fungal diversity metrics, and associations with asthma and asthma development.[16–18]
In the U.S., few sequencing studies have evaluated indoor fungal diversity within an occupational setting, such as an office building with documented water damage. In 2006, a State Department of Health and the National Institute for Occupational Safety and Health (NIOSH) conducted an environmental survey in an office building located in the northeastern region of the U.S. following a cluster of reported sarcoidosis and asthma-like cases among employees.[19] Assessment of microbial contaminants in the survey included viable culture methods and (1→3)-β-D-glucan analysis.[20] In this study, we extend the initial microbial assessment and analyze a subset of the 120 original floor dust samples by ITS rRNA gene sequencing analysis. The aim of using this methodological approach is to build upon the initial viable culture dataset and provide a comprehensive analysis of the species richness, diversity, and relative abundance of fungal communities present in the office building dust samples.
Methods
Building conditions and dust sample collection
In 2006, a State Department of Health, in collaboration with NIOSH, conducted a health and environmental survey of a cluster of sarcoidosis cases in a State Office building located in the northeastern region of the U.S.[19] The building, that was constructed in 1978 (old building), and a three-story annex (new annex) added in 1991, had a history of water infiltration and microbial contamination. The building housed 136 employees at the time of the survey.[19] The environmental survey was conducted due to employee indoor air quality concerns and included the collection of 120-floor dust samples from various locations within the building as previously described.[20] To build upon the original viable culture datasets and to further elucidate the fungal communities within the office building, a subset of samples (n = 22) for which dust aliquots were available and stored at −80°C were evaluated by ITS rRNA gene sequencing analysis. Sample extracts from two locations within the office building defined as the lower floors (ground and second floors; n = 8) and top floor (third floor; n = 14) were analyzed to examine whether there were microbial community structure differences within different areas of the office building.
Genomic DNA extraction
Genomic DNA (gDNA) was extracted from dust samples (n = 22) using the High Pure PCR Template Kit (Roche), as previously described.[15] Dust (5 mg) was added to a bead beater tube containing 300 mg of glass beads (212–300 μm; Sigma, St. Louis, MO) followed by 350 μl of the kit tissue lysis buffer. The tubes were vigorously vortexed followed by 30 sec in a bead beater at high speed (BioSpec Products, Bartlesville, OK). The gDNA extracts were centrifuged two times at 20,000 × g for 1 min. The super-natants were transferred to new microcentrifuge tubes, 20 μl of CelLytic B Cell Lysis Reagent (Sigma) was added, and then incubated at 37°C for 30 min. The kit’s binding buffer (200 μL) and 40 μl of proteinase K solution were added, and the tubes incubated for 10 min at 70°C. The samples were then mixed with 100 μl of isopropanol, washed, and eluted as recommended by the manufacturer (Roche). The eluate was then aliquoted into a labeled 1.5 ml Eppendorf tube and stored at −20°C. An extraction reagent blank was additionally included in the analysis as a control.
PCR amplification, cloning, and Sanger sequencing
Fungal rDNA was amplified using polymerase chain reaction (PCR) amplification with the primer pair Fun18Sf (forward primer: 5′-TTGCTCTTCAACGAGGAAT-3′) and ITS4 (reverse primer: 5′-TCCTCCGCTTATTGATATGC-3′) using a modification of the method presented by Pitkaranta.[15] Briefly, three replicate PCR reactions were prepared for each sample using 33.3 μl PCR grade water (Sigma), 5 μl 10X PCR buffer (Invitrogen, Carlsbad, CA), 1.5 μl 50 mM MgCl2, 1 μl 10 mm deoxynucleoside triphosphate, 0.5 μl 50 mM betaine, 2.5 μl dimethyl sulfoxide, 0.5 μl 20 μm forward (Fun18Sf) and reverse (ITS4) primer, 0.2 μl of Platinum TAQ polymerase (Invitrogen), and 5 μL of template DNA for a final volume of 50 μl. PCR was performed using an Eppendorf Mastercycler Pro (Hamburg, Germany) using the following conditions: initial denaturation at 95°C for 3 min; 6 cycles of denaturation at 96°C for 30 sec, annealing at 50°C for 45 sec, primer extension at 72°C for 3 min and then followed by a further 20 cycles of denaturation at 96°C for 30 sec, acnnealing at 50°C for 45 sec, primer extension at 72°C for 1 min; and final extension at 72°C for 10 min. The primers amplify an approximately 700–1000 base pair fragment that includes the 3′ end of the 18S gene (240 bp), the ITS region, and the 5′ end of the 28S gene (~50 bp) as previously described.[14] Triplicate PCR reactions were combined for each sample and purified using a QIAquick PCR purification kit following the methods recommended by the manufacturer (Qiagen, Valencia, CA). Purified PCR products (8 μl) were then analyzed on a 1% agarose gel as previously described.[15,21] Reagent negative controls that were prepared in parallel to samples did not reveal amplification of contaminant fungal DNA.
Fungal ITS amplicons were cloned into the pDRIVE vector using a PCR cloning kit (Qiagen) according to manufacturer’s instructions. Ligated plasmids were then transformed into TOP10 chemically competent E. coli (Invitrogen), as previously described.[15,21] The transformants were spread onto Luria-Bertani (LB) agar plates containing 100 mg/ml ampicillin and a top layer of X-gal and then incubated overnight at 37°C. Inactivation of the lacZ gene resulted in the production of white colonies (positive) that contained the ITS amplicon insert. Forty-eight positive colonies per dust sample were randomly selected (except four samples with low DNA yield) and cultured in 1.5 ml LB media containing 100 mg/ml ampicillin overnight at 37°C. Glycerol stocks were then prepared in 96-well plates and sent to Genewiz, Inc. (South Plainfield, NJ) for Sanger sequencing analysis as previously described.[15,21]
Plasmid DNA ITS regions were sequenced in both directions using primers T7 and SP6 allowing for sequencing of the entire ITS region. Sequence chromatogram files (.ab1) were downloaded, trimmed, and forward and reverse reads assembled using Geneious R7 Software (Biomatters Ltd, Auckland, New Zealand) before downstream analysis.[15] A preliminary quality filtering step evaluated assembled sequences for the presence of primers, low-quality sequences, chimeras, and short nucleotide read lengths. Sequences that could not be trimmed due to poor sequence quality or could not assemble did not pass filtering and were omitted from the analysis.
Fungal OTU and diversity analysis
All trimmed and assembled sequences were then analyzed to determine operational taxonomic units (OTUs). Using MOTHUR v1.32.1 software,[22] unique sequences within the complete dataset were searched to identify and group identical sequences. Clustering of unique sequences was performed using ClustalW Alignment algorithm with a 97% similarity threshold. Fasta files that contained trimmed representative sequences of each clustered OTU group were prepared using MOTHUR v1.32.1 software.[22] The representative sequences of each clustered OTU were then queried against The National Center for Biotechnology Information (NCBI) using the Basic Local Alignment Tool (BLAST), as previously described.[15] OTU identification and abundance data was prepared for each analyzed sample. Identification to species level required a >97% identity score, otherwise, identifications were made using the following guidelines: ≤97% but >95% for genus level, ≤95% but >90% for family level, ≤90% but >85% for order level, ≤85% but >80% for class level, and ≤80% but >77% for phylum level. Fungal OTUs unable to be placed at these taxonomic levels were identified as “unidentified fungi.”
For richness analyses, OTU abundance datasets at the 97% similarity threshold were used to generate rarefaction curves and 95% confidence intervals in EstimateS v9.1[23] for lower and top floor samples. Extrapolated curves were additionally calculated by augmenting the empirical sample set by a factor of 3X using EstimateS v9.1.[23] OTUs defined at a 97% similarity threshold were also used to estimate richness and diversity indices including Chao2, the Shannon diversity index and the Simpson index of diversity for lower and top floor dust samples according to the equations presented by Magurran.[24] The Bray-Curtis dissimilarity coefficient, a beta diversity measure, was also calculated to evaluate sample dissimilarity within and between lower and top floor samples as previously described.[24,25]
Statistical analysis
Statistical analysis and graphical presentation (OTU abundance, rarefaction curves, and relative abundance of fungal community distribution) of OTUs and diversity measures captured from the evaluation of lower and top floor dust samples was completed using Sigmaplot v12.5 (Systat Software Inc, San Jose, CA). The data were log transformed before analysis and used to run a one-way analysis of variance (ANOVA) to measure statistical significance at α = 0.05.
Results
Using the universal fungal primer pair Fun18Sf/ITS4, DNA was amplified from all extracted dust samples. After filtering, 933 ITS clones were assembled from the Sanger sequencing datasets. The DNA sequences were clustered into 270 individual OTUs following 97% similarity cutoff analysis.[22] Fungal OTUs (n = 216) derived from 765 ITS sequence reads accounted for 80% of all identified OTUs and were placed in 5 fungal divisions. Additional OTUs were placed in the Viridiplantae (n = 51) and Protozoa (n = 3). The fifteen most abundant OTUs = accounted for 59% of all sequence reads.
The number of fungal OTUs at the 97% similarity threshold is shown in Figure 1A. For all samples, a mean of 17.68 fungal OTUs per sample was identified following the clustering of unique sequences (Figure 1A). Analysis of the number of OTUs showed a difference between the lower and top floors that was marginally significant (p = 0.049). The marginal difference in lower and top floor OTUs was also reflected in the estimates of species richness and diversity shown in Table 1 as well as the rarefaction curves presented in Figure 1B. Rarefaction analysis showed lower estimated species richness in the lower floor samples compared to the top floor samples. However, richness may have been underestimated as the rarefaction curves did not reach a horizontal asymptote for either sample group. Other calculated diversity estimates including Chao2, Shannon, and Simpson indices demonstrated lower species richness and diversity in the lower floor samples compared to top floor samples (Table 1). The Bray-Curtis dissimilarity coefficient scores for lower (0.91) and top floors (0.75) as well as between floors (0.88) showed a compositional dissimilarity between the fungi identified in samples derived from the two sampling areas within the building.
Figure 1.
Operational Taxonomic Unit (OTUs) analysis of samples derived from the State Office Building lower floor and top floor samples. OTUs were defined at 97% similarity threshold. (A) Bar graph representing the mean number of fungal OTUs identified for all, lower floor and top floor samples. Error bars represent standard deviation. (B) Rarefaction analysis (reference samples solid shapes) of fungi for lower floors (denoted by a solid triangle) and top floor (denoted by a solid circle) samples. Hollow symbols with dotted lines represent extrapolation up to three times the sample size. Grey error bars represent 95% confidence interval.
Table 1.
Fungal species richness and diversity indices calculated for lower and top floor samples.
| Chao-2 | Shannon Diversity Indexa | Simpson Index of Diversityb | Bray-Curtis Dissimilarity Coefficientc | ||
|---|---|---|---|---|---|
| Lower Floors | 163 | 2.06 (0.78) | 0.84 (0.16) | 0.91 (0.12) | 0.88 (0.11) |
| Top Floor | 382.55 | 2.62 (0.35) | 0.92 (0.04) | 0.75 (0.13) | |
Shannon diversity index represents the mean and standard deviation in brackets for each sample group.
Simpson index of diversity represents the mean and standard deviation in brackets for each sample group.
Bray-Curtis dissimilarity coefficient represents the mean and standard deviation calculated for within and between lower and top floor samples.
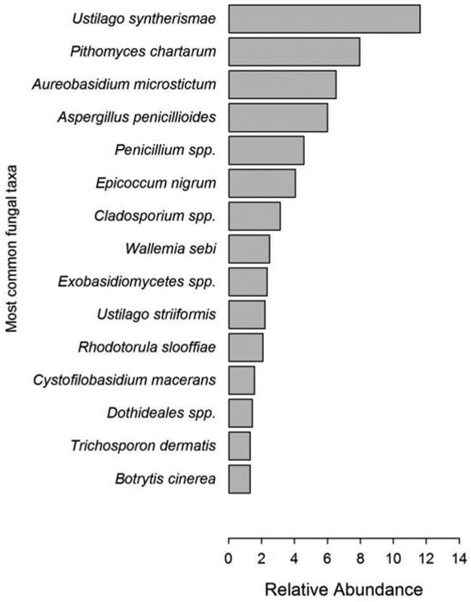
The phylum Ascomycota was the most prevalent fungal phylum encountered in the dust samples and accounted for 55% of all identified fungal sequences (Figure 2). Figure 3 shows the relative abundance of Ascomycota classes that were elucidated in the Sanger sequencing analysis. The class Dothideomycetes (30%) had the highest relative abundance and consisted of fungal sequences placed in the orders Pleosporales (17%), Dothideales (8%), and Capnodiales (4%) (Figure 3). The most common OTUs identified within the order Pleosporales included Pithomyces chartarum (8%) and Epicoccum nigrum (4%) (Figure 4), whereas Alternaria alternata accounted for less than 1% (data not shown). The most common Dothideales and Capnodiales OTUs identified in the analysis included Aureobasidium microstictum (8%) and Cladosporium spp. (3%), respectively (Figures 3 and 4). The class Eurotiomycetes (16%) was the second most common Ascomycota class (Figure 3) and primarily consisted of fungal OTUs placed in the order Eurotiales (14%) (Figure 3), such as Aspergillus penicillioides (6%), and Penicillium species (5%) (Figure 4). Other identified species placed in the Eurotiales included A. fumigatus (0.8%), A. halophilicus (0.5%), A. versicolor (0.5%), and A. ustus (0.1%) (data not shown).
Figure 2.
Relative abundance of the fungal phyla identified in the analysis of 22 floor dust samples. The x-axis represents the percentage of 765 fungal sequences.
Figure 3.
Rank order of the ten most abundant fungal classes and orders identified in the analysis of twenty-two-floor dust samples. Y-axes represent the rank order of relative abundance of sequences placed in Ascomycota class, Basidiomycota class, Ascomycota order, and Basidiomycota order. Not included in this figure are the Chytridiomycota and Glomeromycota that were identified to phylum level. The Zygomycota was composed of species placed in one Order, the Mucorales. The x-axis represents the percentage of 765 fungal sequences.
Figure 4.
The relative abundance of identified fungal OTUs (sequences with 97% similarity) that accounted for greater than 1% of all fungal clone libraries (n = 765). Not included in this figure is the Basidiomycota genus Cryptococcus that accounted for 4.31% of total fungal sequences.
The phylum Basidiomycota accounted for 41% of the fungal sequences identified in the dust sample analysis (Figure 2). The Ustilaginomycetes (14%) was the most common Basidiomycota class (Figure 3) and included the fungal OTUs Ustilago syntherismae (12%) and U. striiformis (2%) (Figure 4). Figure 3 also shows the Basidiomycota class Tremellomycetes (11%) was predominated by obligate yeast species placed in the orders Tremellales (8%) and Cystofilobasidiales (2%) (Figure 3). The order Tremellales was composed of sequence reads derived from 14 Cryptococcus species (4%) and 2 Trichosporon species (Figure 4). Other common Basidiomycota included the Agaricomycetes (8%), Wallemia sebi (2%), Exobasidiomycetes (2%), and Sporidioboles species (2%) (Figures 3 and 4). Other identified Basidiomycota included the Pucciniales (1%), Sterigmatomyces halophilus (0.3%), Malasseziales (0.3%), and unassigned Basidiomycota (0.3%) (data not shown).
Fungal sequences were also placed in 3 additional fungal divisions, including the Zygomycota (3%), Glomeromycota (0.4%), and Chytridiomycota (0.3%) (Figure 2). Fungal sequences that could not be identified using reference ITS sequences in the NCBI database were placed into a group entitled unidentified fungi that accounted for 0.5% of the analyzed fungal sequences.
Discussion
Clone library sequencing of full-length ITS regions is an exposure assessment tool increasingly used to elucidate dust-borne fungal taxa. Data derived from these studies have shown clone libraries to be predominantly composed of fungi placed in the phylum Ascomycota but have also highlighted the relative abundance of additional fungal phyla that may not be cultured on standard nutrient medium or identified using microscopic methods of analysis.[9,14,15,26] The Basidiomycota have emerged from these studies to be more prevalent in the environment than previously estimated using traditional methods.[9,15] In this study, we build upon viable culture data collected from an office building that had a history of water infiltration and microbial contamination and demonstrate the contribution of the Basidiomycota, as well as other previously overlooked dust-borne fungal populations.[19]
Culture-dependent assessment of fungi in the office building elucidated 48 Ascomycota species and two Basidiomycota species.[20] In comparison, clone libraries contained a much broader diversity of fungal DNA and included 765 ITS sequences placed in 216 fungal OTUs. These results revealed an approximately 4-fold increase in OTUs compared to cultured species; a finding that is consistent with previously published molecular profiling studies.[15,26] The Basidiomycota accounted for 41% of clone libraries and included 92 OTUs; a finding that is 46-fold higher compared to the species identified in the original culture analysis.[20] Compared to previous Sanger sequencing studies conducted within our laboratory, the prevalence of Basidiomycota was also up to 5-fold higher in dust from our study building compared to dust derived from homes in the mid-west of the U.S.[15] Fungal populations can vary between geographic environments(27), and diversity has been described to be higher in temperate compared to tropical regions,[28] as well as in environments with water infiltration.[10,29] Although the abundance of the Basidiomycota can be as high as 64% in the outdoor environment,[9] these results further demonstrate the abundance of the Basidiomycota within an indoor occupational environment that has been overlooked using traditional methods of fungal analysis.
Evaluation of the number of OTUs, species richness, and diversity indices revealed lower floor samples to have a marginally reduced number of OTUs compared to samples derived from the top floor. Although the lower floor samples consisted of a smaller sample size that reduces the ability to detect statistical differences reliably, we cannot exclude additional limiting variables not assessed in the present study. Anthropogenic disturbance, outdoor sourced fungi, and fungi associated with water damaged building materials (leaking pipes or moisture intrusion through the building envelope) are additional sources of variability that can alter community structures. Calculation of the Bray-Curtis dissimilarity confirmed this variability as coefficient scores showed a compositional dissimilarity within and between fungi identified in lower and top floor sampling locations. Compared to next generation sequencing approaches, Sanger sequencing of clone libraries produced rarefaction curves that were approaching the horizontal asymptote. Although this result demonstrates that additional samples may have further resolved increased fungal richness, this methodological approach improves the elucidation of viable and nonviable fungal species in dust samples.
The Agaricomycetes is a Basidiomycota class that has been identified to be abundant in previous sequencing studies[9,26,30,31] and accounted for 8% of fungal sequences in the current survey. The Ustilagomycetes, in particular, the genus Ustilago, had the highest proportion of all clone libraries identified in the phylum Basidiomycota (14%). The fungal species, U. syntherismae and U. striiformis, are plant pathogens that cause loose smut of Digitaria species (crabgrass) or stripe smut of grass, respectively. The prevalence of these two Ustilago species in the analysis may be due to passive microbial transport where workers functioned as secondary emitters of Ustilago biomass that had either been tracked in on footwear or emitted from the clothing of the worker.[29,32,33] In addition, Ustilago species were not identified in the original culture analysis but have been previously identified in sequencing studies of the indoor environment often in lower relative abundances.[29] The identification of Ustilago species in this analysis represents an additional burden of respirable spores and other fungal components that could have been encountered by workers within this office building.
The contribution of obligate yeasts associated with the skin, such as Candida, Trichosporon, Malassezia, and Cryptococcus species, have been elucidated in previous sequencing studies and reported to account for as much as 29.6% of all fungal sequences in indoor environments.[17,26,27,34] In the dust samples, endogenous fungal flora, including Malassezia, Rhodotorula, Trichosporon, and Cryptococcus, were also prevalent and accounted for 22% of Basidiomycota clone libraries. Nonpathogenic Cryptococcus species were particularly abundant in the clone library analysis, a finding that confirms previous sequencing studies that have identified Cryptococcus species to be prevalent in indoor air,[15,35] outdoor air,[30] and building floor dust.[17] Recent next generation sequencing studies have also identified positive associations between Cryptococcus species and mild asthma within a pediatric asthma cohort[18] and low Cryptococcus diversity was a risk factor for increased asthma in children.[17] The current study further highlights additional yeast species that were overlooked in the initial viable culture analysis.
The phylum Ascomycota accounted for 55% of fungal clone libraries in our study, a finding consistent with previous analyses,[15,26] although the prevalence of this phylum can be as high as 92% in some indoor environments.[15] The Dothideomycetes was the most prominent Ascomycota class identified and accounted for 30% of all fungal clone libraries. The abundance of this class is similar to previously reported sequencing studies of the indoor built environment[27] and a water damaged Finnish office building.[26] The class was predominated by the order Pleosporales, which includes allergenically important species that share Alt a 1 allergen orthologues.[36,37] Interestingly, the spectrum and relative abundance of Pleosporalean fungi identified in the dust of our study building was similar to that identified in dust derived from children’s homes participating in the Kansas City Safe and Healthy Homes Program.[15] Other prominent Ascomycota orders identified in the analysis included the Eurotiales, Dothidiales, and Capnodiales that commonly grow within the phylloplane. These findings are consistent with sequenced datasets derived from a water-damaged office building or flooded indoor environments.[10,26]
Compared to traditional analytical methods for fungi, sequencing fungal DNA represents a platform that enables an objective approach to identifying microorganisms, in particular, fungal species that were not capable of growth on nutrient media.[4] An example of the benefits of this approach includes the identification of fungi placed in the Basidiomycota, obligate yeasts, as well as xerophilic species that require specific culture conditions, such as A. halophilicus.[38] Although the species identified in the clone libraries corresponded to the original viable culture data,[20] these results further demonstrate several challenges associated with identifying fungal ITS sequences.[39] Short amplicons within Cladosporium and Penicillium species and homology within Aspergillus ITS sequences are limiting factors that challenge taxonomic placement.[40] Extraction biases may also limit the ability to amplify DNA from species that produce amerospores, such as Aspergillus and Penicillium species, and may account for the smaller proportion of clone libraries that were represented by these genera.[21] Other limiting variables that may have influenced the results of this analysis include, small sample size, discriminating viable from non-viable fungi, carpet age, variations in sample collection techniques, amplification biases of the selected primer set,[41,42] variable copy number of rDNA,[43] as well as ITS paralogues.[43] These variables require further evaluation so future studies can utilize standardized methods of extraction and amplification that will enable improved intersurvey comparisons.
Conclusions
Sequencing-based approaches have provided unique insight into the richness, diversity, and relative abundance of dust-borne fungi that could not be classified or were overlooked using traditional methods of assessment.[4] In this study, we built upon an existing culture-dependent dataset and demonstrated a 4-fold increase in identified OTUs in dust samples compared to culture-dependent approaches. Evaluation of OTUs, species richness, and diversity indices showed marginal differences between lower and top floors although there was dissimilarity between the clustered fungal sequences. An increased relative abundance of Basidiomycota fungi placed in the order Ustilaginales was additionally observed. Predominant species included U. syntherismae and U. striiformis, as well as obligate fungal yeasts. Important fungal aeroallergens in the Ascomycota order Pleosporales that share Alt a 1 allergen orthologues were additionally elucidated in the analysis. These sequencing data are among the first to report fungal richness and diversity derived from dust samples within a U.S. Office building and highlight a variety of indoor and outdoor sourced fungi that were not captured using culture-dependent methods.
Acknowledgments
Funding
This study was supported in part by an interagency agreement between NIOSH and NIEHS (AES12007001-1-0-6) as a collaborative National Toxicology Program research activity.
Disclaimer
The findings and the conclusions in this article are those of the authors and do not necessarily represent the views of the National Institute for Occupational Safety and Health.
Footnotes
The authors declare no conflict of interest.
References
- [1].WHO: WHO Guidelines for Indoor Air Quality: Dampness and Mould. Geneva: WHO Regional Office for Europe, 2009. [PubMed] [Google Scholar]
- [2].Institute of Medicine of the National Academies: Damp indoor spaces and health. Washington, DC: The National Academies Press, 2004. [PubMed] [Google Scholar]
- [3].Mendell MJ, Mirer AG, Cheung K, Tong M, and Douwes J: Respiratory and allergic health effects of dampness, mold, and dampness-related agents: A review of the epidemiologic evidence. Environ. Health Perspect. 119(6):748–756 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Petti CA: Detection and identification of microorganisms by gene amplification and sequencing. Clin. Infect. Dis 44(8):1108–1114 (2007). [DOI] [PubMed] [Google Scholar]
- [5].Scott JA, Summerbell RC, and Green BJ: Detection of indoor fungi bioaerosols In Fundamentals of Mold Growth in Indoor Environments and Strategies for Healthy Living, Adnan O and Samson RA (eds.), pp. 353–373. Amsterdam, The Netherlands: Wageningen Academic Publishers, 2011. [Google Scholar]
- [6].Summerbell RC, Green BJ, Coor D, and Scott JA: Molecular Methods for Bioaerosol Characterization In Microorganisms in Home and Indoor Work Environments: Diversity, Health Impacts, Investigation and Control, Second Edition, Flannigan B, Samson RA and Miller JD (eds.), pp. 247–265. Boca Raton, FL: CRC Press, Taylor and Francis Group, 2011. [Google Scholar]
- [7].Pashley CH, Fairs A, Free RC, and Wardlaw AJ: DNA analysis of outdoor air reveals a high degree of fungal diversity, temporal variability, and genera not seen by spore morphology. Fungal. Biol 116(2):214–224 (2012). [DOI] [PubMed] [Google Scholar]
- [8].Madsen AM, Zervas A, Tendal K, and Nielsen JL: Microbial diversity in bioaerosol samples causing ODTS compared to reference bioaerosol samples as measured using Illumina sequencing and MALDI-TOF. Environ. Res 140:255–267 (2015). [DOI] [PubMed] [Google Scholar]
- [9].Frohlich-Nowoisky J, Pickersgill DA, Despres VR, and Poschl U: High diversity of fungi in air particulate matter. Proc. Natl. Acad. Sci. U S A 106(31):12814–12819 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Emerson JB, Keady PB, Brewer TE, et al. : Impacts of flood damage on airborne bacteria and fungi in homes after the 2013 Colorado Front Range flood. Environ. Sci. Technol 49(5):2675–2684 (2015). [DOI] [PubMed] [Google Scholar]
- [11].Fierer N, Liu Z, Rodriguez-Hernandez M, Knight R, Henn M, and Hernandez MT: Short-term temporal variability in airborne bacterial and fungal populations. Appl. Environ. Microbiol 74(1):200–207 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Vesper S, McKinstry C, Haugland R, et al. : Development of an Environmental Relative Moldiness index for US homes. J. Occup. Environ. Med 49(8):829–833 (2007). [DOI] [PubMed] [Google Scholar]
- [13].Johansson E, Reponen T, Meller J, Vesper S, and Yadav J: Association of Streptomyces community composition determined by PCR-denaturing gradient gel electrophoresis with indoor mold status. Environ. Monit. Assess 186(12):8773–8783 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pitkaranta M, Meklin T, Hyvarinen A, et al. : Analysis of fungal flora in indoor dust by ribosomal DNA sequence analysis, quantitative PCR, and culture. Appl. Environ. Microbiol 74(1):233–244 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rittenour WR, Ciaccio CE, Barnes CS, et al. : Internal transcribed spacer rRNA gene sequencing analysis of fungal diversity in Kansas City indoor environments. Environ. Sci. Process. Impacts 16(1):33–43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Reponen T, Lockey J, Bernstein DI, et al. : Infant origins of childhood asthma associated with specific molds. J. Allergy Clin. Immunol 130(3):639–644.e635 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dannemiller KC, Mendell MJ, Macher JM, et al. : Next-generation DNA sequencing reveals that low fungal diversity in house dust is associated with childhood asthma development. Indoor Air 24(3):236–247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dannemiller KC, Gent JF, Leaderer BP, and Peccia J: Indoor microbial communities: Influence on asthma severity in atopic and nonatopic children. J. Allergy Clin. Immunol (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Laney AS, Cragin LA, Blevins LZ, et al. : Sarcoidosis, asthma, and asthma-like symptoms among occupants of a historically water-damaged office building. Indoor Air 19(1):83–90 (2009). [DOI] [PubMed] [Google Scholar]
- [20].Park JH, Cox-Ganser JM, White SK, et al. : Bacteria in a water-damaged building: associations of actinomycetes and nontuberculous mycobacteria with respiratory health in occupants. Indoor Air (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rittenour WR, Park JH, Cox-Ganser JM, Beezhold DH, and Green BJ: Comparison of DNA extraction methodologies used for assessing fungal diversity via ITS sequencing. J. Environ. Monit 14(3):766–774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Schloss PD, Westcott SL, Ryabin T, et al. : Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol 75(23):7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Colwell RK, Chao A, Gotelli NJ, et al. : Models and estimators linking individual-based and sample-based rarefaction, extrapolation and comparison of assemblages. J. Plant Ecol 5(1):3–21 (2012). [Google Scholar]
- [24].Magurran AE: Measuring Biological Diversity. Oxford: Blackwell Publishing, 2004. [Google Scholar]
- [25].Legendre P, and Legendre LFJ: Numerical Ecology. Elsevier, 2012. [Google Scholar]
- [26].Pitkaranta M, Meklin T, Hyvarinen A, et al. : Molecular profiling of fungal communities in moisture damaged buildings before and after remediation–a comparison of culture-dependent and culture-independent methods. BMC Microbiol. 11:235 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yamamoto N, Hospodsky D, Dannemiller KC, Nazaroff WW, and Peccia J: Indoor emissions as a primary source of airborne allergenic fungal particles in classrooms. Environ. Sci. Technol 49(8):5098–5106 (2015). [DOI] [PubMed] [Google Scholar]
- [28].Amend AS, Seifert KA, Samson R, and Bruns TD: Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. Proc. Natl. Acad. Sci. USA 107(31):13748–13753 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dannemiller KC, Gent JF, Leaderer BP, and Peccia J: Influence of housing characteristics on bacterial and fungal communities in homes of asthmatic children. Indoor Air 26(2):179–192 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yamamoto N, Bibby K, Qian J, et al. : Particle-size distributions and seasonal diversity of allergenic and pathogenic fungi in outdoor air. ISME J. 6(10):1801–1811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Shin SK, Kim J, Ha SM, et al. : Metagenomic insights into the bioaerosols in the indoor and outdoor environments of childcare facilities. PLoS One 10(5):e0126960 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Adams RI, Bhangar S, Pasut W, et al. : Chamber bioaerosol study: outdoor air and human occupants as sources of indoor airborne microbes. PLoS One 10(5):e0128022 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pasanen AL, Kalliokoski P, Pasanen P, Salmi T, and Tossavainen A: Fungi carried from farmers’ work into farm homes. Am Ind Hyg Assoc J 50(12): 631–633 (1989). [DOI] [PubMed] [Google Scholar]
- [34].Findley K, Oh J, Yang J, et al. : Topographic diversity of fungal and bacterial communities in human skin. Nature 498(7454):367–370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Adams RI, Miletto M, Taylor JW, and Bruns TD: Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J. 7(7):1262–1273 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Saenz-de-Santamaria M, Postigo I, Gutierrez-Rodriguez A, et al. : The major allergen of Alternaria alternata (Alt a 1) is expressed in other members of the Pleosporaceae family. Mycoses 49(2):91–95 (2006). [DOI] [PubMed] [Google Scholar]
- [37].Hong SG, Cramer RA, Lawrence CB, and Pryor BM: Alt a 1 allergen homologs from Alternaria and related taxa: analysis of phylogenetic content and secondary structure. Fungal Genet. Biol 42(2):119–129 (2005). [DOI] [PubMed] [Google Scholar]
- [38].Micheluz A, Manente S, Tigini V, et al. : The extreme environment of a library: Xerophilic fungi inhabiting indoor niches. Int. Biodeter. Biodegrad 99:1–7 (2015). [Google Scholar]
- [39].Schoch CL, Seifert KA, Huhndorf S, et al. : Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 109(16):6241–6246 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Schubert K, Groenewald JZ, Braun U, et al. : Biodiversity in the Cladosporium herbarum complex (Davidiellaceae, Capnodiales), with standardisation of methods for Cladosporium taxonomy and diagnostics. Stud. Mycol 58:105–156 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Toju H, Tanabe AS, Yamamoto S, and Sato H: High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS One 7(7):e40863 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ihrmark K, Bodeker IT, Cruz-Martinez K, et al. : New primers to amplify the fungal ITS2 region–evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol 82(3):666–677 (2012). [DOI] [PubMed] [Google Scholar]
- [43].Vetrovsky T, Kolarik M, Zifcakova L, Zelenka T, and Baldrian P: The rpb2 gene represents a viable alternative molecular marker for the analysis of environmental fungal communities. Mol. Ecol. Resour 16(2):388–401 (2016). [DOI] [PubMed] [Google Scholar]