Tissue homeostasis is sustained by stem cell self-renewal and differentiation. How stem cells coordinately differentiate into multiple cell types is largely unclear. Recent studies underline the heterogeneity among stem cells or common progenitors, suggesting coordination occurs at the stem cell/progenitor level1–4. Here, by tracking and manipulating the same stem cells and their progeny at the single-cell level in live mice, we uncover an unanticipated flexibility of homeostatic stem cell differentiation in hair follicles. Though stem cells have been shown to be flexible upon injury, we demonstrate that hair germ stem cells at the single-cell level can flexibly establish all the differentiation lineages even in uninjured conditions. Furthermore, stem cell derived hair progenitors in the structure called matrix, previously thought to be unipotent, flexibly change differentiation outcomes as a consequence of unexpected dynamic relocation. Finally, the flexible cell fate determination mechanism maintains normal differentiation and tissue architecture against ectopic differentiation stimulus induced by Wnt activation. This work provides a model of continually fate channeling and late commitment of stem cells to achieve coordinated differentiation and robust tissue architecture.
Classical view of stem cell differentiation assumes that stem cells are uniformly multipotent, and they stereotypically produce diverse differentiated cells through lineage-restricted progenitors in a stepwise manner5. This model is challenged by recent studies in hematopoietic system, which highlight the heterogeneity within stem cell or common progenitor pools by employing single-cell analyses and clonal lineage tracking approaches1, 2, 4. The heterogeneous stem cells often differ in their differentiation behaviors based on their intrinsic properties such as epigenetic configuration4. However, stem cells/progenitors might still display flexibility on their differentiation paths, since stem cells have been shown to be equipotent in intestinal epithelium homeostasis6, 7, and lineage commitment appears to be a continuum during human steady-state hematopoiesis8. Though stem cells can certainly adopt flexibility under tissue injury9, it is still unclear how flexible stem cells/progenitors differentiate during homeostasis, and if flexible, how far into the process of differentiation this flexibility would still be retained.
Skin hair follicle represents an excellent model to spatiotemporally interrogate the differentiation process during homeostasis due to the multiple differentiated lineages generated by the stem cells during each hair cycle, as well as the well-characterized differentiated cell identities and tissue anatomy10, 11. During the resting phase of hair cycle, stem cells reside in the lower portion of hair follicles, where they are organized into two compartments, the bulge and hair germ, with distinct functional contributions to hair growth (Fig. 1a)12, 13. Specifically, the hair germ stem cells have been shown to give rise to differentiated cells in the following hair growth phase3, 14. At the beginning of a growth phase, the hair germ stem cells undergo oriented divisions and downward extension to generate progenitors that are organized along the basement membrane around the mesenchymal dermal papilla, within a compartment called the matrix (Fig. 1a)3, 15, 16. It has been shown that the matrix progenitors divide asymmetrically to renew their pool while producing distinct cell-types that differentiate upwards along the inner length of the follicle3, 15, 17. Additionally, the progenitor cells in the matrix are thought to be unipotent and molecularly distinct based on single-cell RNA-seq and classical lineage tracing analysis3, 18. Current models posit that the position a progenitor occupies around the mesenchyme dictates a specific differentiated cell type3, 15, 19. Like other tissues, stem cells in the hair follicle can acquire plasticity of fate determination under injury conditions9. However, it remains unclear, during homeostasis, how the hair germ stem cells diversify into distinct lineage-restricted matrix progenitors and establish the upward differentiation trajectories. One previous lineage tracing study showed heterogeneity within hair follicle stem cells regarding the number of lineages they generate, though it was unclear what accounts for the heterogeneous behaviors20. Another recent work uncovered spatial heterogeneity of molecular signatures within the stem cell population through single-cell sequencing3. These together suggest that the hair germ stem cells might be heterogeneously primed for differentiation lineage establishment. Testing this hypothesis requires fate tracking of the same stem cells within these heterogeneous pools during the differentiation process within the same animals.
Fig 1. Stem cells are spatially primed for differentiation lineage establishment in hair follicle.
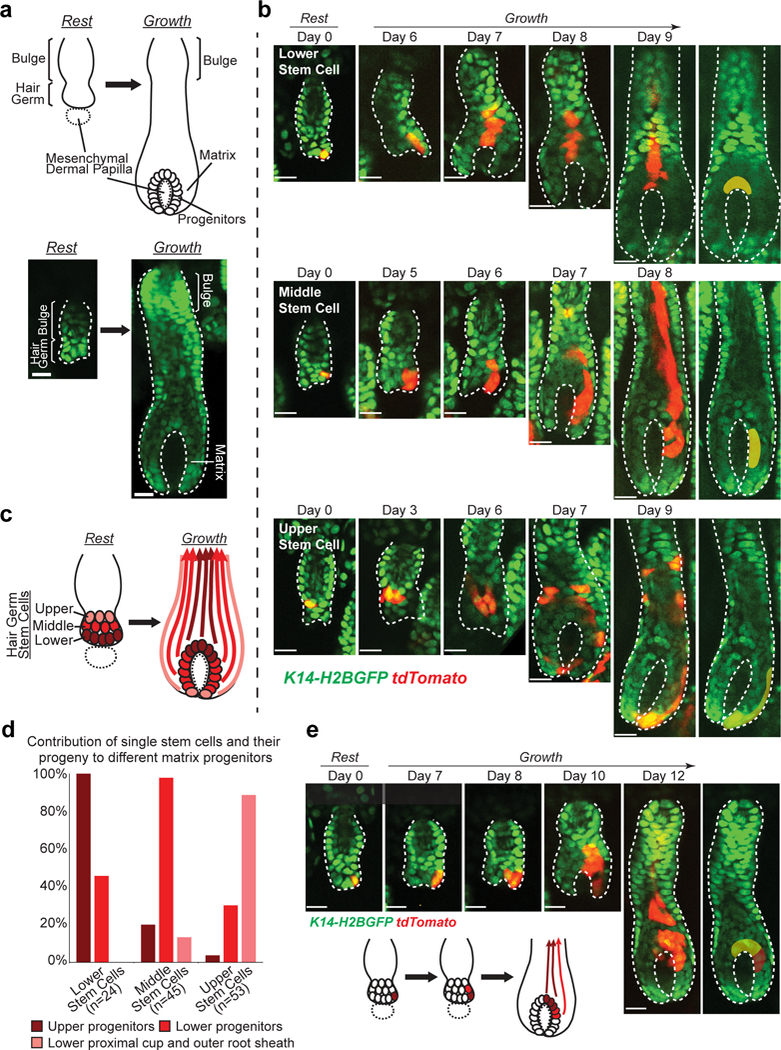
a, Schematic and two-photon images of resting and growing hair follicles. b, Representative examples of tracked lineages from single stem cells located at different positions of resting hair follicles, showing distinct contributions of the spatially organized hair germ stem cells to the generation of the matrix progenitor subgroups and their upward moving differentiated lineages. Subgroups of progenitors arising from single stem cells are pseudo-colored. Images representative of 122 hair follicles from 7 mice. c, A schematic showing spatially reversed organization of the hair germ stem cells into the matrix progenitor subgroups during hair follicle growth. Dotted circle indicates mesenchymal dermal papilla. Solid line outlines the epithelium. d, Frequency of generating certain subgroup of matrix progenitors by single hair germ stem cells at different initial positions. Column shows the percentage of hair follicles with a single stem cell labeled at a given position that generated a certain subgroup of progenitors. Frequency is calculated based on the lineage tracing of 24 single lower stem cells, 45 single middle stem cells and 53 single upper stem cells from 7mice. Note that single stem cells can sometimes contribute to multiple subgroups of progenitors. Source data are provided in Supplementary Table 1. e, A representative example of a single hair germ stem cell that generates both upper and lower matrix progenitor subgroups through expansion across positions in the early stage. Subgroups of progenitors are differentially pseudo-colored. Epithelial nuclei were marked by K14-H2BGFP (green in a, b and e). Lgr5-CreER and R26-flox-STOP-tdTomato (red in b and e) were used to induce stem cell labeling. Hair follicle epithelium is outlined by dashed lines (a, b and e). The tracking was typically performed from Telogen to Anagen IIIc/IV. Scale bars, 20 μm.
To understand how the hair germ stem cells generate matrix progenitors and establish differentiation lineages, we tracked the same stem cells in live mice through the intravital imaging approach previously established in the lab14, 16. Specifically, to track stem cell contribution to the differentiation lineages in the hair growth phase (also called Anagen), we labeled single stem cells in the resting (or Telogen) hair follicles with low induction of an inducible Cre-dependent fluorescent reporter (Lgr5-CreER; R26-flox-STOP-tdTomato). K14-H2BGFP (histone H2B fused with green fluorescent protein (GFP) and driven by the Keratin 14 promoter) was used as a general epithelial fluorescent marker as previously described (Fig. 1b)16, 21. As we tracked the hair germ stem cells over time, we found that their initial positions, with respect to the bottom of the resting hair follicle, predicted which subgroups of matrix progenitors they generated (Fig. 1b-d). Interestingly, the organization of the matrix progenitors was inversed when compared to the one of the stem cells (Fig. 1c). This suggests that stem cells located at different positions are primed to establish differentiation lineages, which is consistent with a recent model of spatial heterogeneity of the resting hair germ stem cells3. Specifically, we showed that the lower stem cells generated the upper matrix progenitor subgroup that is known to differentiate into the inner hair lineages (medulla and cortex); the middle stem cells generated the lower matrix progenitor subgroup which is known to differentiate into the outer hair lineages (cuticle and inner root sheath); while the most upper stem cells produced the outer root sheath (ORS), the undifferentiated layer covering the outside of the growing hair follicles, as well as the cells at the bottom called lower proximal cup (LPC) (Fig. 1b-d). Interestingly however, we also found that some stem cells could generate differentiated lineages across multiple subgroups, probably due to the early expansion of the stem cells into multiple positions (Fig. 1d, e).
It is known that, during early growth phase, hair follicle epithelium expands downwards to encapsulate the underlying mesenchyme (Supplementary Video 1)10. Our data makes us wonder how this morphogenetic process is regulated to achieve the spatially reversed organization of stem cells into matrix progenitors shown above. To understand that, we analyzed the stem cell behaviors during the early hair follicle growth stages (Anagen I-II). Close revisits of the genetically labeled single stem cells showed that most stem cells underwent oriented divisions, which is consistent with our previous observations (Fig. 2a)16. As hair follicles start to grow, the stem cells in the hair germ amplify to produce suprabasal cells (Supplementary Video 2). During those early stem cell amplification stage, we found progeny of the middle and upper stem cells largely stayed basal (Fig. 2a,c,d). In contrast, progeny of the lower stem cells were often observed being displaced into the suprabasal layer (Fig. 2a,c,d). These early suprabasal cells are likely to differentiate into K79+ canal cells and companion layer cells identified previously22, 23, though future work will be needed to address this. In addition, lower stem cells frequently underwent cell death shown by both lineage tracing and time-lapse analyses (Fig. 2a,b,d and Supplementary Video 3). Together, these spatially regulated stem cell behaviors resulted in differential expansion rates in the basal layer at different positions (Fig. 2c,e and Supplementary Video 4). As a result of this process, only the higher stem cells expand downwards to generate the lower matrix progenitors leading to spatially reversed organization of the stem cell progeny (Fig. 1c).
Fig 2. Spatially regulated stem cell behaviors lead to spatially reversed organization of stem cell progeny.
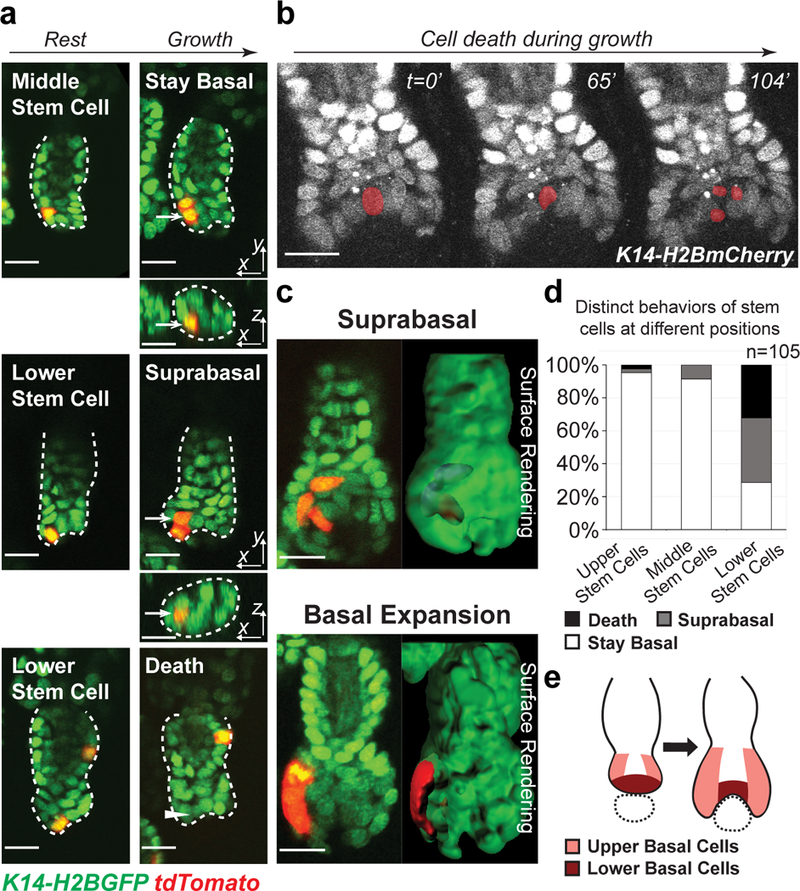
a, Representative examples of lineage tracing of single hair germ stem cells at different positions in early hair follicle growth phase, showing progeny of middle stem cells stayed basal, while progeny of lower stem cells were displaced into the suprabasal layer or underwent cell death. Arrows indicate the stem cell progeny. Arrowhead indicates where the dead stem cell was. Hair follicle epithelium is outlined by dashed lines. The tracking was typically performed from Telogen to Anagen I. Images representative of 105 hair follicles from 6 mice. b, Time-lapse frames showing stem cell death at lower position. The dying cell is pseudo-colored. Images representative of 5 mice. Two independent time-lapses are provided in Supplementary Video 3. c, Single z-planes and 3D surface rendering of hair follicles during lineage tracing showing suprabasal displacement (blue) of cells deriving from lower stem cell (up) and basal expansion of cells deriving from upper stem cell (down). d, Frequency of hair germ stem cells at different positions undergoing cell death, suprabasal displacement and basal expansion. 100% stacked column shows the percentage of hair follicles with a single stem cell labeled at a given position that performed a certain behavior. n=105 hair follicles in 6 mice. e, A schematic showing distinct behaviors of hair germ stem cells at different positions lead to differential basal expansion and mesenchyme encapsulation. Dotted circle indicates mesenchymal dermal papilla. Epithelial nuclei were marked by K14-H2BGFP (green in a and c) or K14-H2BmCherry (white in b). Lgr5-CreER and R26-flox-STOP-tdTomato (red in a and c) were used to induce stem cell labeling. Scale bars, 20 μm.
The spatially distinct progenitor contributions shown above, together with the previously demonstrated heterogeneous molecular signatures suggest that hair follicle stem cells may have pre-determined cell fates based on their initial positions3. However, the observation that at lower frequency stem cells can produce a wider range of lineages (Fig. 1d) suggests that, while primed, stem cells may still have a certain degree of flexibility in differentiation. Stem cells are known to be plastic during tissue repair across a number of tissues including the hair follicle9. Yet, whether stem cell differentiation in the hair follicle follows flexible principles under uninjured conditions needs to be tested. If stem cells are not flexible to differentiate, we would predict that dramatically reducing the number of stem cells able to contribute to tissue growth will compromise the formation of matrix progenitors as well as significantly delay tissue growth. Alternatively, even few stem cells capable of contributing to tissue growth will properly generate the matrix and sustain hair follicle growth (Fig. 3a). To test this hypothesis, we developed a genetic system that allows us to block the contribution of most stem cells to hair follicle growth by inhibiting their proliferation and track the remaining ones for their ability to generate the matrix during these uninjured settings. Through this genetic approach, we can 1) mark all epithelial cells with an mCherry reporter (H2BmCherry), and 2) overexpress the cell cycle inhibitor Cdkn1b (also known as p27)24 specifically in hair follicle stem cells, and label these non-proliferating cells with a GFP reporter (H2BGFP) (Lgr5-CreER; R26-flox-STOP-tTA; tetO-Cdkn1b; pTRE-H2BGFP; K14-H2BmCherry) (Fig. 3a). By a high induction of Cre activity, we blocked the proliferation of most stem cells (H2BGFP+;H2BmCherry+) and tracked the remaining H2BGFP-;H2BmCherry+ stem cells to see whether these few proliferating stem cells can generate the entire matrix, or fail to compensate (Fig. 3a, Supplementary Video 5 and Supplementary Fig. 1a). Intriguingly, even a few (down to one) proliferating stem cells were capable to generate the entire matrix progenitor population (Fig. 3b). More surprisingly, they sustained hair follicle growth as fast as the control (Fig. 3b). Additionally, the initial positions of non-inhibited stem cells did not lead to difference in hair follicle growth rate (Fig. 3c and Supplementary Fig. 1b). These data suggest that even if stem cells are spatially primed, they are flexible to establish a diverse set of differentiation lineages as well as actively maintain a normal rate of hair follicle growth during homeostasis.
Fig 3. Stem cells are fully potent to establish all the differentiation lineages in the hair follicle.
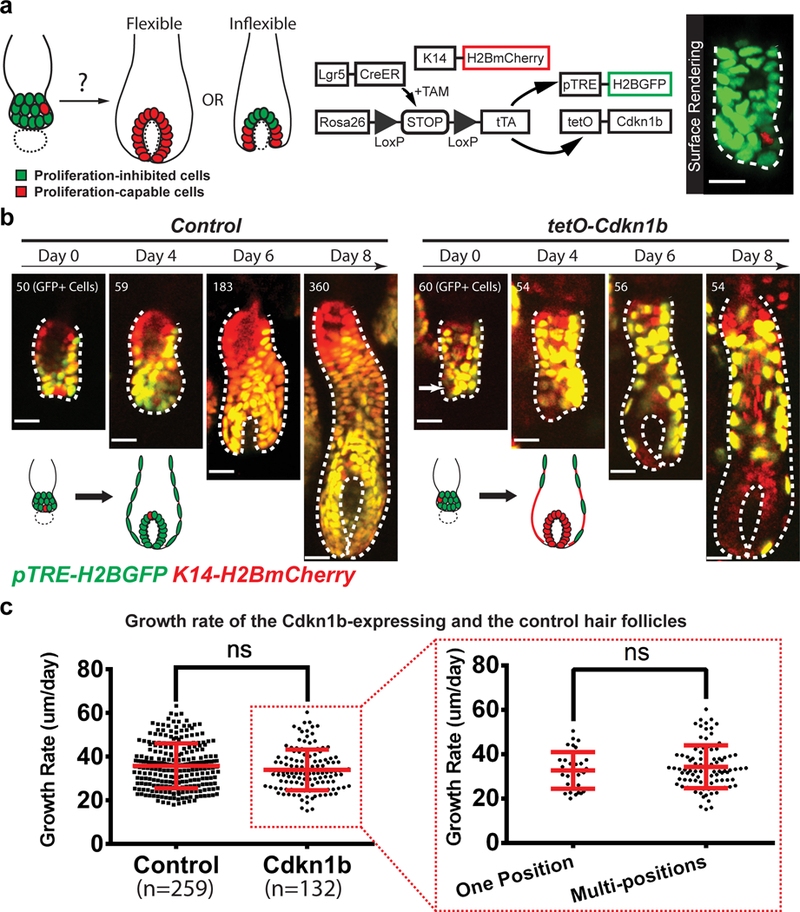
a, Schematic diagrams showing the approach for testing the flexibility of hair follicle stem cells in differentiation through inhibiting proliferation of most stem cells. Dotted circle indicates mesenchymal dermal papilla. 3D surface rendering of a hair follicle (right) showing proliferation inhibition and labeling of all (green) but one (red) hair germ cells. b, Representative examples and schematic showing when most stem cells are impaired to proliferate, only few non-inhibited stem cells (arrow) can flexibly generate the entire matrix during hair follicle growth. Note that the GFP+ cells in the Cdkn1b-overexpressing hair follicle (right) failed to expand in contrast to the control (left). Epithelial nuclei were marked by K14-H2BmCherry (red). Cdkn1b-overexpressing cells and the control cells were labeled by H2BGFP (green). The tracking was typically performed from Telogen to Anagen IIIc. Images representative of 259 hair follicles from 4 control mice and 132 hair follicles from 3 Cdkn1b overexpression mice. c, No significant growth rate difference between the Cdkn1b-overexpressing hair follicles and the controls, nor between hair follicles carrying non-inhibited stem cells at one position versus multi-positions. Plot shows the mean±SD (n=259 hair follicles in 4 control mice, n=132 hair follicles in 3 Cdkn1b overexpression mice, n=34 hair follicles carrying non-inhibited stem cells at one position, n=98 hair follicles carrying non-inhibited stem cells at multi-positions). ns, not significant, p=0.0820 and 0.3939 for the two graphs. Two-sided unpaired t-test was used to calculate p value. Statistical source data are provided in Supplementary Table 1. Hair follicle epithelium is outlined by dashed line (a and b). Scale bars, 20 μm.
Our findings that stem cells are flexible to differentiate cause us to ask how far into the process of differentiation this flexibility would still be retained. To this end, we captured matrix dynamics in later hair follicle growth stages (from Anagen III to VI) with more challenging deeper imaging and performed longer revisits. To identify distinct differentiated cell types, we took advantage of their previously well-characterized morphologies as well as their positions with respect to the hair follicle architecture, and confirmed by whole mount staining with molecular differentiation markers (Fig. 4a)3, 15. Unexpectedly, when we tracked lower stem cells into late hair growth stages, we found the descendent top matrix progenitors, which proliferated in early stages, all stopped self-renewing and underwent terminal differentiation (Fig. 4b). This observation implies that the matrix progenitors need constant replenishment at the top position. To begin to interrogate how the replenishment occurs at the population level, we labeled and tracked the lower part of the matrix by utilizing the light-inducible lineage tracing approach we have previously established (K14-H2BPAmCherry; K14-actinGFP)25. Revisits of the same hair follicles over time showed extension of the labeling from the lower to the top part of the matrix, suggesting either relocation or expansion of the lower matrix progenitors (Supplementary Fig. 2). To gain resolution at the single cell level, we tracked individual lower progenitors along with their upward differentiating progeny by using Hopx-CreER26 in combination with R26-flox-STOP-tdTomato and K14-H2BGFP. Surprisingly, those matrix progenitors at the epithelial-mesenchymal interface were not fixed in their positions. Instead, they were continuously relocated upwards along the basement membrane, resulting into the generation of distinct types of differentiated cells that corresponded to their newly acquired positions (Fig. 4c,g, Supplementary Fig. 4c). When they reached the top position, they stopped self-renewal as shown above (Fig. 4b). We also observed that this dynamic “cellular flow” was fueled by the outer root sheath (ORS) cells, which moved into the matrix through lower proximal cup to generate new progenitors (Fig.4d). The progressive cell relocation was also captured by lineage tracing through traditional tissue section analysis (Supplementary Fig. 3). Concomitantly, the upper ORS cells, which were often derived from the lower bulge stem cells, kept expanding and moving downwards to replenish the cells that were relocated into the matrix (Supplementary Fig. 4a,b). Together, while these data show that matrix progenitor cells use positional information to generate specific differentiated cell types in line with previous finding, they demonstrate the unanticipated ability of matrix progenitors to continually change positions around the mesenchyme. Therefore, matrix progenitors are not unipotent, but rather are competent to give rise to multiple differentiated lineages. Furthermore, the matrix progeny were observed, upon detachment from the basement membrane, to still be able to give rise to distinct lineages than the ones on the main differentiation trajectory, both towards outer and inner cell fates (Fig. 4e,f, Supplementary Fig. 4c). These data establish a previously unappreciated temporal framework where cell fate commitment occurs later than previously thought so that progenitors and their progeny can flexibly differentiate on demand (Fig. 4g).
Fig 4. Hair progenitors undergo dynamic relocation and change differentiation outcomes.
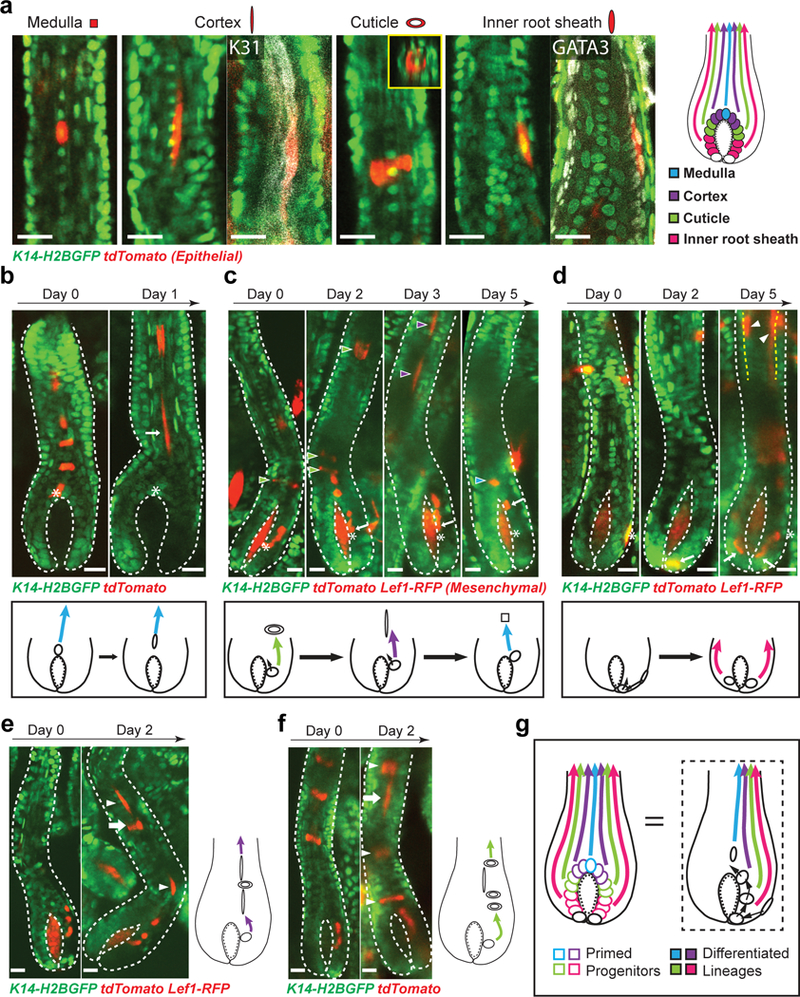
a, Morphologies and positions of differentiated hair lineages. Whole mount staining of cortex marker (K31, white) and inner root sheath (IRS) marker (GATA3, white) confirms the molecular signature of differentiated cells. XZ view of the labeled cuticle cell showing its distinctive ring shape (inset). Images representative of 20 mice. b, Lineage tracing of matrix progenitors and schematic show top progenitors stop self-renewal. Images representative of 44 hair follicles from 11 mice. c, Lineage tracing of matrix progenitors and schematic show lower progenitors are continually relocated upwards and change differentiation outcomes (arrowheads) corresponding to their new positions. Arrowheads with different colors indicate distinct differentiation lineages produced at each time point. Images representative of 149 hair follicles from 17 mice. d, Lineage tracing of outer root sheath (ORS) cells and schematic show ORS cells move into the matrix and produce differentiated lineages (arrowheads). Images representative of 49 hair follicles from 8 mice. Yellow dashed lines indicate the interface between ORS and inner lineages. e, An example of concurrent multi-lineage differentiation of matrix progenitors and schematic showing a single progenitor produced an outer differentiated cell (cuticle cell, arrow) among cortex cells (arrowhead). f, An example of concurrent multi-lineage differentiation of matrix progenitors and schematic showing a single progenitor produced an inner differentiated cell (cortex cell, arrow) among cuticle cells (arrowhead). g, A model of primed progenitors with distinct molecular signatures undergo dynamic relocation and change differentiation outcomes. Epithelial nuclei were marked by K14-H2BGFP (green in a, b, c, d, e and f). Hopx-CreER (a, b, c, e and f) or Lgr5-CreER (b, c, d) in combination with R26-flox-STOP-tdTomato (red) was used to induce cell labeling. The tracking was performed from Telogen (b) or Anagen IIIc (b, c, d, e and f) to Anagen VI. Hair follicle epithelium is outlined by dashed lines (b, c, d, e and f). Mesenchymal dermal papilla was labeled by Lef1-RFP in c, d and e. Asterisk and arrow indicate the original and current progenitor cell position respectively in b, c and d. Scale bars, 20 μm.
So far, we have shown that the hair follicle matrix represents a system that can flexibly determine cell fates as well as refresh the germinative population through dynamic cellular flow. We hypothesize that these features could buffer against aberrant differentiation cues leading to robust orchestration of the differentiation lineages and overall maintenance of hair architecture. To test that we sought to ectopically activate Wnt signaling, a signal pathway that is known to function in hair differentiation27, 28, by inducing a β-catenin gain-of-function (βCatGOF) mutation within the matrix. We and others previously also showed that the induction of the βCatGOF mutation in hair follicle stem cells caused ectopic growth and differentiation (Supplementary Fig. 5a)29–32. To specifically induce the βCatGOF mutation in a subset of the matrix progenitors, we utilized Shh-CreER33 in combination with the Cre-inducible βCatGOF allele (β-cateninflox(Ex3))34. As shown by both mRNA expression and downstream Wnt signaling reporter (TCF-H2BGFP)35, we successfully expressed the βCatGOF mutant allele and activated Wnt in ectopic positions within the matrix (Fig. 5a-c). Intriguingly, the hair follicles that expressed β-catenin mutant matrix cells (labelled with K14-H2BGFP and R26-flox-STOP-tdTomato reporter) for multiple days still differentiated properly, based on both cell morphologies, as well as molecular marker expression shown by whole mount staining (Fig. 5d). We reasoned that the observed normal tissue architecture in the face of ectopic Wnt activation might be explained by either elimination of the βCatGOF mutant cells or integration of them within the normal hair differentiation process. To test these hypotheses, we sought to track βCatGOF mutant cells by combining the permanent tdTomato labelling with the TCF-H2BGFP reporter, which shows the initial ectopic Wnt activation (Fig. 5e). Revisits showed that the tdTomato+;H2BGFPHi βCatGOF mutant progenitors were captured to integrate into the normal differentiation process over time, therefore excluding an elimination scenario at the population level (Fig. 5e). The mutant cells were gradually relocated into the top matrix area, where Wnt signal is high, through a “cellular flow” mechanism (Fig. 5e and Supplementary Fig. 5a). The progeny of the mutant cells still obeyed the position-dependent rule of differentiation, suggesting a corrected cell fate (Fig. 5e). To more directly track the mutant cells, we generated a mouse strain carrying an inducible GFP-tagged βCatGOF mutant allele (pTRE-dNβCatGFP) (Supplementary Fig. 5b). The directly tagged βCatGOF mutant cells induced in the matrix also differentiated normally and underwent relocation (Supplementary Fig. 5c). These data support a model whereby flexible cell fate determination ensures robust differentiation and tissue architecture.
Fig 5. Robust differentiation is maintained upon ectopic differentiation stimulus in the matrix progenitors.
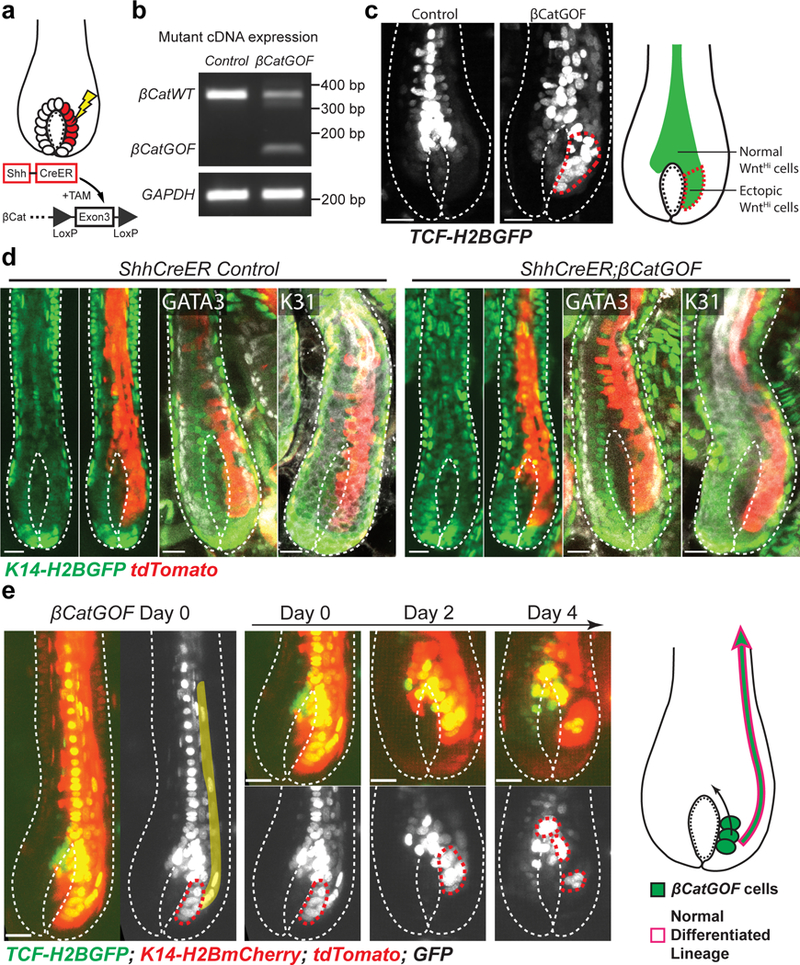
a, A schematic of inducing β-catenin gain-of-function (βCatGOF) mutation in the matrix by Shh-CreER. b, RT-PCR showing expression of βCatGOF allele in total ear skin samples after induction. Unprocessed original scans are provided in Supplementary Fig. 6.c, Live imaging of Wnt reporter (TCF-H2BGFP, white) mice showing induction of βCatGOF mutation (right) causes ectopic Wnt activation in the lower matrix (red dashed line) when compared with the control (left). Images were typically captured on Anagen IV-VI hair follicles. Images representative of 101 hair follicles from 9 mice. d, Live imaging and whole mount staining of differentiation markers (GATA3 and K31, white) showing hair follicles carrying βCatGOF mutation for multiple days have normal differentiation. Epithelial nuclei were marked by K14-H2BGFP (green). R26-flox-STOP-tdTomato (red) was used to identify the mutant cells. Images captured on Anagen IV-VI hair follicles. Images representative of 113 hair follicles from 3 control mice and 222 hair follicles from 4 βCatGOF mice. e, Representative images showing, after induction in the matrix, βCatGOF mutant cells (red dashed lines) were gradually relocated into the top matrix area and sent progeny upwards to differentiate normally (pseudo-colored). Epithelial nuclei were marked by K14-H2BmCherry (nuclei, red). TCF-H2BGFP (green or white) and R26-flox-STOP-tdTomato (cytoplasm, brighter red) were used to identify and track the mutant cells. The tracking was performed from Anagen V to VI. Images representative of 101 hair follicles from 9 mice. Hair follicle epithelium is outlined by dashed lines (c, d and e). Scale bars, 20 μm.
Tissue resident stem cells need to generate multiple differentiation cell types in a coordinated fashion to sustain tissue function in homeostasis. While the orchestration might be achieved at any step during stem cell differentiation, current thinking has focused on coordination occurring at the stem cell/common progenitor level, as recent studies highlight the heterogeneity within stem cells or common progenitors through single-cell analysis and clonal lineage tracing1–4. However, the inability to track the same stem cells over time in a spatial context during their differentiation has hampered to obtain insights into this fundamental question. Here, by spatiotemporally tracking and manipulating stem cells in live mice, we have shown that stem cell fate determination is a flexible process during homeostatic hair follicle growth. Although we found that stem cells appeared to be spatially primed for generating progenitors and differentiation lineages, they still could flexibly contribute to any lineage on demand. The flexibility is even retained in the hair matrix progenitors that were previously thought unipotent, as evidenced by the changing lineages they produce both with and without relocation. These together support a model that fate commitment happens later than previously anticipated, while stem cells and progenitors are continually tuned towards certain fates to meet the tissue requirement. The distinct stem cells’/progenitors’ molecular signatures captured by previous single-cell sequencing study3 may indicate primed states that can still be overridden by environmental signals. Other fast turnover tissues, such as intestine, might use the same strategy to maintain robust differentiation while facing high-level cellular dynamics.
Furthermore, with the ability to revisit the same cells in the same tissue, we also captured the unforeseen cellular dynamics during hair follicle growth. There appear to be a directional “cellular flow” that starts from the outer root sheath (ORS) and continues within the matrix progenitor layer, to refresh the germinative population and to refill the top progenitors that stop self-renewal. Similar phenomenon was observed in transplanted chimeric vibrissal follicles, where implanted cells move along the ORS into the matrix36. Since the matrix might compile mutant cells derived from the stem cells close to the skin surface, the “cellular flow” offers a way to flush the abnormal cells out of the matrix and keep the normal hair follicle function. Consistent with that, βCatGOF mutant cells, which can cause aberrant growth when induced in the hair follicle stem cells, are relocated within the matrix and integrated into normal differentiation lineages with no outgrowth emergence. Previous studies also showed that inducing oncogenes in the matrix progenitors could not cause tumor formation37. These together suggest that hair follicle matrix represents a robust system, which can tolerate/utilize various mutant cells and maintain stable differentiation.
Methods
Mice
K14-H2BGFP21, pTRE-H2BGFP21, K14-actinGFP38, Lef1-RFP39 and K14-Cre40 mice were obtained from E. Fuchs. β-cateninflox(Ex3)34 mice were obtained from M.Taketo. K14-H2BmCherry41 and K14-H2BPAmCherry25 mice were generated in the lab and described previously. Lgr5-CreER42, Hopx-CreER26, Shh-CreER43, R26-flox-STOP-tdTomato44, R26-flox-STOP-tTA45, tetO-Cdkn1b24, TCF-H2BGFP35 and mTmG46 mice were obtained from The Jackson Laboratory. The pTRE-dNβCatGFP mice were generated by the Yale Transgenic Facility. Specifically, the fragment of N-terminal truncated β-catenin and GFP fusion47 was amplified from the pCAG-delta90GFP plasmid (Addgene #26645) and inserted into the pL3-TRE-MCS-polyA-2L vector (Addgene #11719) by using the SacII and NotI cloning sites. The 4.7kb fragment between XhoI and FseI sties of the resulting construct was injected to make the transgenic mouse line. All the mice were bred to a mixed albino background.
To induce single stem cell labeling, Lgr5-CreER; R26-flox-STOP-tdTomato; K14-H2BGFP mice were given a single dose of tamoxifen (1 μg/g in corn oil) around postnatal day 17 by intraperitoneal injection. To induce single matrix progenitor cell labeling, Hopx-CreER; R26-flox-STOP-tdTomato; K14-H2BGFP mice were administered a single dose of 500 μg tamoxifen between postnatal day 24 and 26. To induce outer root sheath cell labeling, Lgr5-CreER; R26-flox-STOP-tdTomato; K14-H2BGFP mice were administered a single dose of 100 μg tamoxifen between postnatal day 24 and 27. In some mice, Lef1-RFP was used to visualize mesenchymal dermal papilla. To induce Cdkn1b and pTRE-H2BGFP expression in most stem cells, Lgr5-CreER; R26-flox-STOP-tTA; tetO-Cdkn1b; pTRE-H2BGFP; K14-H2BmCherry mice were given two or three injections of 2 mg tamoxifen between postnatal day 17 and 19. To induce βCatGOF mutation in the matrix, Shh-CreER; β-cateninflox(Ex3); R26-flox-STOP-tdTomato; K14-H2BGFP or Shh-CreER; β-cateninflox(Ex3); TCF-H2BGFP; R26-flox-STOP-tdTomato; K14-H2BmCherry mice were given three injections of 2 mg tamoxifen at postnatal day 24, 25 and 26. To induce dNβCatGFP expression in the matrix, Shh-CreER; R26-flox-STOP-tTA; pTRE-dNβCatGFP; K14-H2BmCherry mice were given one injection of 2 mg tamoxifen on postnatal day 25. All time courses of in vivo imaging began at least 3 days after induction. Since the timing of the hair cycle is slightly variable between individual mice, in some cases, we kept imaging and monitored the hair cycle until the right stages before analyzing data. In those cases, the day of the desired starting hair stages is referred to as “Day 0” in the figures. Revisits were typically performed every one or two days. The length of the revisits depends on the goals of the specific experiments and is indicated in the figures and legends. Mice from experimental and control groups were randomly selected for live imaging experiments. No blinding was done. Each “n” stands for a tracking course of multiple time points for lineage tracing experiments. Hair follicle stages were judged according to the previous literature9. All studies and procedures involving animal subjects were approved by the Institutional Animal Care and Use Committee at Yale School of Medicine and conducted in accordance with the approved animal handling protocol #11303. This study is compliant with all relevant ethical regulations regarding animal research.
In vivo imaging
Imaging procedures were similar to those previously described13, 16. Mice were anaesthetized with intraperitoneal injection of 5μl/g of ketamine/xylazine cocktail mix (15mg/ml/ and 1mg/ml, respectively, in PBS). Anaesthesia was maintained throughout the course of the experiment with vaporized isoflurane delivered by a nose cone (1% in oxygen and air). Mice were placed on a warming pad during imaging. The ear was mounted on a custom-made stage and a glass coverslip was placed directly against it. A LaVision TriM Scope II (LaVision Biotec) microscope equipped with a Chameleon Vision II (Coherent) two-photon laser (using 940 or 990 nm for live imaging, 880 nm for whole-mounts) and a Chameleon Discovery (Coherent) two-photon laser (using 1120 nm for live imaging and whole-mounts) was used to acquire z-stacks of 90–200 μm in 2–3 μm steps through either a Zeiss 20x/1.0 or a Nikon 40x/1.15 water immersion objective. Optical sections were scanned with a field of view of 0.09 or 0.25 mm2. For imaging large areas, multiple tiles (up to 36) of optical fields were captured using a motorized stage. To activate photo-activatable H2BmCherry (H2BPAmCherry), a 405 nm laser was used to scan the target area for 1 min at 10% laser power. Patterns of hair follicle clusters were used as landmarks for revisiting the same skin area. For time-lapse recordings, serial optical sections were obtained at 13min intervals.
Image analysis
Raw image stacks were imported into Fiji (ImageJ, NIH) or Imaris (BitPlane) for analysis. Tiles of optical fields were stitched in Fiji. All the analyses are based on the 3D view of the hair follicles. In the case of progenitor lineage tracing, progenitors and their progeny were distinguished based on their 3D positions in relation to the mesenchymal surface. In the case of multiple labelled progenitors, we only analyzed hair follicles containing labeled progenitors that were located away from each other. This allowed us to clearly resolve those individual progenitors along with their progeny based on the distinct trajectories of hair differentiation lineages which are characterized by distinct curvatures and specific positions in the epithelial concentric layers15. Additionally, progenitor relocation was judged separately, and scored as occurring when at least one progenitor had relocated. Due to the similar morphologies of the inner root sheath cuticle and the hair shaft cuticle, we refer to both of them as “cuticle” to increase rigor. Similarly, we refer to all the three inner root sheath cell types as “inner root sheath”. Selected optical planes or z-projections of sequential optical sections were used to assemble figures. Hair follicle growth rate was calculated based on the length measurement through Fiji. 3D surface rendering of selected hair follicle cells was performed in Imaris.
Immunofluorescence
For whole mount staining, ear skins were dissected and fixed in 4% paraformaldehyde in PBS for 1 hr at room temperature (RT) then overnight at 4 ºC. Fixed tissues were cut into small pieces and incubated in the blocking solution (5% normal goat serum, 1% bovine serum albumin and 2% Triton X-100 in PBS) for 6–8 hrs at RT. Blocked tissues were incubated with primary antibodies diluted in the blocking solution for ~66 hrs at RT. Tissues were then washed in PBS with 2% Triton X-100 for 6–8 hrs and incubated with secondary antibodies diluted in the blocking solution for ~66 hrs at RT. After staining, tissues were washed in PBS with 2% Triton X-100 for 6–8 hrs and mounted in VECTASHIELD Antifade Mounting Medium for fluorescence (VECTOR). For tissue section analysis, back skins were dissected and either directly embedded in OCT (Tissue Tek), or fixed with 4% paraformaldehyde in PBS for 1 hr at 4 ºC before embedding to preserve the tdTomato signal. Frozen OCT blocks were sectioned at 30 μm to have more 3D information. For BrdU staining, mice were intraperitoneally injected once with 50 μg/g BrdU. Back skins were embedded in OCT 6 hrs after injection. Immunofluorescence staining on the sections were described previously (Science 2014). Primary antibodies used were as follows: guinea pig anti-K31 (1:50, Progen, GP-hHa1), rabbit anti-GATA3 (1:250, Abcam, ab199428), chicken anti-GFP (1:1000, Abcam, ab13970), rabbit anti-Ki67 (1:200, Abcam, ab15580), rat anti-BrdU (1:200, Abcam, ab6326). Secondary antibodies used were as follows: goat anti-guinea pig Alexa633, goat anti-rabbit Alexa633, goat anti-rat Alexa633 and goat anti-chicken Alexa488 (Thermo Fisher).
RT-PCR
Total RNA was isolated from ear skin with an RNeasy Fibrous Tissue Mini Kit (Qiagen) according to the manufacturer’s instructions. cDNA was then made using Superscript III First-Strand Synthesis kit (Thermo Fisher). PCR was then performed to detect the βCatGOF expression with primers: Fwd: 5’-TGAAGCTCAGCGCACAG-3’, and Rev: 5’-CATGCCCTCATCTAGCGTC-3’, which binds exon2 and exon4 of β-catenin gene respectively. GAPDH was used as the internal control.
Statistics and reproducibility.
Statistical calculations were performed using Prism 6 (GraphPad). Two-sided unpaired t-test was used to determine the significance between two groups. A p value of <0.05 was considered significant; precise p values can be found in the figure legends. No statistical method was used to predetermine sample size. Mouse numbers represent the biological replicates. Sample size and replicates are indicated in the figure legends. All the experiments were repeated at least three times.
Supplementary Material
Acknowledgments
We thank J. Moore and E. Legué for feedback on the manuscript and other Greco lab members for helpful discussion. We thank E. Fuchs for the K14-H2BGFP, pTRE-H2BGFP, K14-actinGFP and K14-Cre mice, and M.Taketo for the β-cateninflox(Ex3) mice. We thank the Yale Transgenic Facility for generating the pTRE-dNβCatGFP mice. This work is supported by The New York Stem Cell Foundation, The Edward Mallinckrodt, Jr. Foundation, the Glenn Foundation for Medical Research, the HHMI Scholar award, and the National Institute of Arthritis and Musculoskeletal and Skin Disease (NIAMS), NIH, grants no. 2R01AR063663–06A1, no. 1R01AR072668–01 and no. 5R01AR067755–02. V.G. is a New York Stem Cell Foundation Robertson Investigator and HHMI Scholar. T.X. was supported by The James Hudson Brown - Alexander Brown Coxe Postdoctoral Fellowships.
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
Data availability
Statistics source data for Fig. 3c and Supplementary Fig. 1b have been provided as Supplementary Table 1. Additional images for Fig. 5 have been deposited at the Figshare with DOI: 10.6084/m9.figshare.7170422. All the data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Paul F et al. Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell 163, 1663–1677 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Notta F et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science 351, aab2116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang H, Adam RC, Ge Y, Hua ZL & Fuchs E Epithelial-Mesenchymal Micro-niches Govern Stem Cell Lineage Choices. Cell 169, 483–496 e413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu VW et al. Epigenetic Memory Underlies Cell-Autonomous Heterogeneous Behavior of Hematopoietic Stem Cells. Cell 168, 944–945 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao MP, Seita J & Weissman IL Establishment of a normal hematopoietic and leukemia stem cell hierarchy. Cold Spring Harb Symp Quant Biol 73, 439–449 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Snippert HJ et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Grun D et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525, 251–255 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Velten L et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat Cell Biol 19, 271–281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanpain C & Fuchs E Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science 344, 1242281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller-Rover S et al. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J Invest Dermatol 117, 3–15 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Xin T, Greco V & Myung P Hardwiring Stem Cell Communication through Tissue Structure. Cell 164, 1212–1225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greco V et al. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell 4, 155–169 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito M, Kizawa K, Hamada K & Cotsarelis G Hair follicle stem cells in the lower bulge form the secondary germ, a biochemically distinct but functionally equivalent progenitor cell population, at the termination of catagen. Differentiation 72, 548–557 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Rompolas P, Mesa KR & Greco V Spatial organization within a niche as a determinant of stem-cell fate. Nature 502, 513–518 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Legue E & Nicolas JF Hair follicle renewal: organization of stem cells in the matrix and the role of stereotyped lineages and behaviors. Development 132, 4143–4154 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Rompolas P et al. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature 487, 496–499 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seldin L, Muroyama A & Lechler T NuMA-microtubule interactions are critical for spindle orientation and the morphogenesis of diverse epidermal structures. Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang AB, Zhang YV & Tumbar T Gata6 promotes hair follicle progenitor cell renewal by genome maintenance during proliferation. EMBO J 36, 61–78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sequeira I & Nicolas JF Redefining the structure of the hair follicle by 3D clonal analysis. Development 139, 3741–3751 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Legue E, Sequeira I & Nicolas JF Hair follicle renewal: authentic morphogenesis that depends on a complex progression of stem cell lineages. Development 137, 569–577 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Tumbar T et al. Defining the epithelial stem cell niche in skin. Science 303, 359–363 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veniaminova NA et al. Keratin 79 identifies a novel population of migratory epithelial cells that initiates hair canal morphogenesis and regeneration. Development 140, 4870–4880 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mesler AL, Veniaminova NA, Lull MV & Wong SY Hair Follicle Terminal Differentiation Is Orchestrated by Distinct Early and Late Matrix Progenitors. Cell Rep 19, 809–821 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pruitt SC, Freeland A, Rusiniak ME, Kunnev D & Cady GK Cdkn1b overexpression in adult mice alters the balance between genome and tissue ageing. Nat Commun 4, 2626 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rompolas P et al. Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science 352, 1471–1474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeda N et al. Hopx expression defines a subset of multipotent hair follicle stem cells and a progenitor population primed to give rise to K6+ niche cells. Development 140, 1655–1664 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi YS et al. Distinct functions for Wnt/beta-catenin in hair follicle stem cell proliferation and survival and interfollicular epidermal homeostasis. Cell Stem Cell 13, 720–733 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kandyba E & Kobielak K Wnt7b is an important intrinsic regulator of hair follicle stem cell homeostasis and hair follicle cycling. Stem Cells (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gat U, DasGupta R, Degenstein L & Fuchs E De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell 95, 605–614 (1998). [DOI] [PubMed] [Google Scholar]
- 30.Lo Celso C, Prowse DM & Watt FM Transient activation of beta-catenin signalling in adult mouse epidermis is sufficient to induce new hair follicles but continuous activation is required to maintain hair follicle tumours. Development 131, 1787–1799 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Brown S et al. Correction of aberrant growth preserves tissue homeostasis. Nature 548, 334–337 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deschene ER et al. beta-Catenin activation regulates tissue growth non-cell autonomously in the hair stem cell niche. Science 343, 1353–1356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levy V, Lindon C, Harfe BD & Morgan BA Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Dev Cell 9, 855–861 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Harada N et al. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J 18, 5931–5942 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrer-Vaquer A et al. A sensitive and bright single-cell resolution live imaging reporter of Wnt/ss-catenin signaling in the mouse. BMC Dev Biol 10, 121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oshima H, Rochat A, Kedzia C, Kobayashi K & Barrandon Y Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell 104, 233–245 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Blanpain C Tracing the cellular origin of cancer. Nat Cell Biol 15, 126–134 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Vaezi A, Bauer C, Vasioukhin V & Fuchs E Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Dev Cell 3, 367–381 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Rendl M, Lewis L & Fuchs E Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol 3, e331 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vasioukhin V, Degenstein L, Wise B & Fuchs E The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci U S A 96, 8551–8556 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mesa KR et al. Niche-induced cell death and epithelial phagocytosis regulate hair follicle stem cell pool. Nature 522, 94–97 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barker N et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Harfe BD et al. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517–528 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Madisen L et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L et al. Restricted expression of mutant SOD1 in spinal motor neurons and interneurons induces motor neuron pathology. Neurobiol Dis 29, 400–408 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Muzumdar MD, Tasic B, Miyamichi K, Li L & Luo L A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Chenn A & Walsh CA Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297, 365–369 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


