Abstract
Artemisinin-based combination therapies (ACTs) have substantially reduced worldwide malaria burden and deaths. But malaria parasites have become resistant to artemisinins. Prior studies suggested two different molecular pathways of artemisinin-resistance. Here we unify recent findings into a single model, where elevation of a lipid, phosphatidylinositol-3- phosphate (PI3P) results in vesicle expansion that increases the engagement with the unfolded protein response (UPR). Vesicle expansion (rather than increasing individual genetic determinants of the UPR) efficiently induces artemisinin resistance likely by promoting ‘proteostasis’ (protein translation coupled to proper protein folding and vesicular remodeling) to mitigate artemisinin-induced proteopathy (death from global abnormal protein-toxicity). Vesicular amplification engages the host red cell, suggesting that artemisinin resistant malaria may also persist by taking advantage of host niches and escaping the immune response.
Introduction
World-wide levels of malaria have substantially decreased over the last decade. Notably, twenty-one countries are expected to have eliminated the disease by 2020 [1]. Yet a substantial burden remains. In 2016, 445 000 deaths and 216 million new cases were caused by Plasmodium falciparum, the most virulent of human malaria parasites [1]. Artemisinin-based combination therapies (ACTs) are frontline, fast acting drugs that have been key to reducing the malaria burden and deaths. However, P. falciparum resistance to artemisinins and ACTs has emerged, threatening global malaria control and elimination.
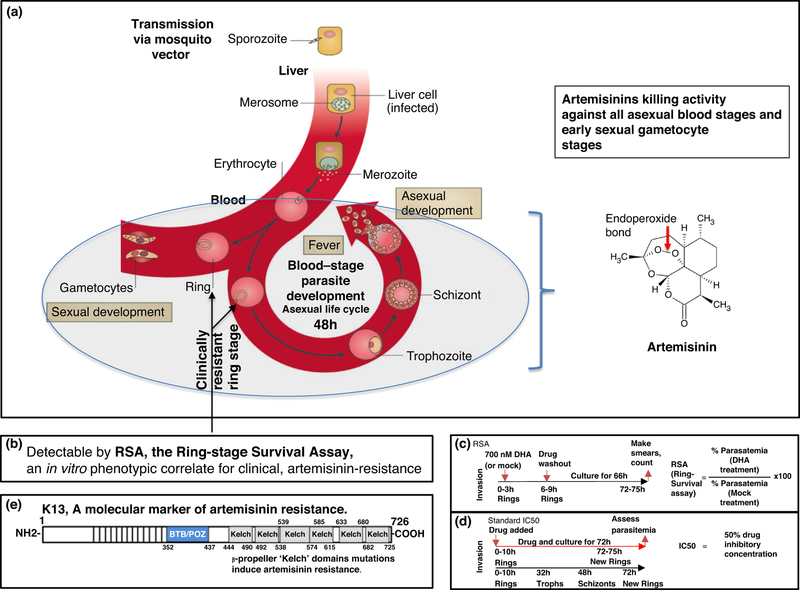
P. falciparum infection in humans is initiated when an infected mosquito bites to release sporozoites that infect liver cells. In the liver, sporozoites develop into merozoites which emerge into the blood stream to invade erythrocytes (red blood cells) and develop through distinct ring, trophozoite and schizont stages over a 48 hour asexual life cycle (Figure 1a). The blood stages of infection cause all of the symptoms and pathologies of malaria (and are thus targeted by all antimalarials in current use). Parasite killing by artemisinins depends on cleavage of their endoperoxide bond (Figure 1a). Proteomic studies indicate that artemisinins alkylate hundreds of proteins [2●●,3●●] suggesting they may kill by inducing ‘proteopathy’ or global degeneration of the parasite’s (proteinaceous) cytoplasm. Resistance to artemisinins observed in the clinic, is seen in ring parasites that show ‘delayed clearance’ from the circulation, in response to direct administration of drug to patients [4,5]. Trophozoite and schizont stages that sequester in tissues are not found in circulation and hence not detected in blood smears used to monitor parasites [6].
Figure 1:
Life cycle of Plasmodium falciparum and measures of artemisinin drug resistance. (a) (Adapted from Delves, M., Scheurer, C, et al. 2012 [48]). P. falciparum malaria infection in humans is initiated when an infected mosquito bites and releases sporozoites that infect liver cells and develop into merozoites. These merozoites emerge into the blood and invade erythrocytes (red blood cells), where they develop over a 48-hour asexual life cycle through morphologically defined ring, trophozoite and schizont stages. Schizont lysis leads to the release of daughter merozoites (which coincides with fever), to initiate a new asexual life cycle. A small subpopulation of ring-form parasites develops into sexual gametocyte stages which are taken up by the mosquito during a blood meal. The killing activity of artemisinins (against asexual and early sexual gametocyte stages) is dependent on the cleavage of their endoperoxide bond. Clinical resistance to artemisinins is seen at the parasite ring stage. (b) Clinical artemisinin resistance is measured in vitro by the Ring-stage Survival Assay (RSA). (c) In the RSA, ring-stage parasites 0–3 hour, are treated with maximal concentrations (700 nM) of dihydroartemisinin (DHA, the active form of all artemisinins) seen in plasma for 6 hours, after which the drug is washed out, mimicking pharmacological exposure seen in patients. Parasites are subsequently allowed to progress for another 66 hours, after which parasitemia is determined. The RSA value is calculated as shown. (d) The standard IC50 assay is carried out by exposing ring, trophozoite and schizont stages to continuous drug treatment over 72 hours. (e) Plasmodium falciparum K13 (PfKelch13) is a causal molecular marker of artemisinin resistance. It contains a single BTB domain (common to all Broad-complex, Tramtrack and Bric-a-bric domain (BTB) family proteins), which is present at the amino terminus, followed by multiple copies of β-propeller ‘Kelch’ repeats. K13 contains six β-propeller Kelch domains, mutations in which induce artemisinin resistance.
The parasite gene pfkelch13 (K13) is the primary marker of artemisinin resistance as defined both clinically and in vitro using the Ring-stage Survival Assay (RSA; [6] Figure 1b). In the RSA, parasites are treated at maximal concentrations of artemisinins achieved in plasma (~700 nM) for 6 hours to mimic pharmacological drug exposure seen in patients (Figure 1c). Notably, only the earliest stages of rings (0–3 hour in the life cycle) manifest artemisinin resistance, while later stages (including 9– 12 hour and 18–21 hour rings) do not [6]. Moreover, in vivo clinical artemisinin resistance cannot be gauged in a standard P. falciparum IC50 assay (Figure 1d, carried out over 72 hours of continuous drug pressure for effects on parasite proliferation through the trophozoite/schizont stages).
Based on its sequence and structure, K13 is predicted to a substrate adapter of an E3 ligase [7] (Figure 1e). Its mammalian orthologues (the best characterized of which is Keap1 [7]) target binding, ubiquitination and proteosomal degradation of select substrates, keeping their levels low to maintain proper cellular homeostasis. Mutations in the b-propeller ‘Kelch’ domain diminish targeting and raise substrate levels in a cell. In cancer, K13 orthologues confer resistance to drugs that kill by inducing ‘proteopathy’ in tumors [8]. In P. falciparum, two major K13 effector mechanisms have been proposed to overcome artemisinin-induced proteopathy and death. They are namely, first, proteostatic dysregulation of parasite phosphatidylinositol-3- kinase resulting in elevation of parasite PI3P [9] and second, upregulation of parasite oxidative stress and protein damage pathways via the unfolded protein response (UPR) [10]. In this review we discuss the most recent advances and convergence of mechanisms in a unified model of K13-dependent and independent states of artemisinin resistance, with implications for resistance to partner drugs as well as new emergent antimalarials. We conclude by deliberating on how understanding mechanisms of resistance sheds critical insights into challenges that need to be overcome to eliminate parasites from regions with high and low endemicity and rapidly changing rates of transmission of malaria in the world.
Pathways of artemisinin resistance
The identification of K13 mutations of artemisinin resistance in P. falciparum malaria galvanized the identification of underlying mechanisms. Comparative, global transcriptomic profiling of over a thousand clinical strains led to the identification of molecular signatures for the resistant K13 mutant parasites in this set, and implicated upregulation of parasite oxidative stress and protein damage pathways via the UPR, as a mechanism of resistance [10]. Concurrent independent studies using both clinical and laboratory isolates suggested K13 acted as an adapter of PfPI3K in an E3 ligase complex to control protein levels of PfPI3K, in complete absence of changes in pfpi3k gene transcript levels [9]. Wild type K13 protein bound PfPI3K catalyzed kinase ubiquitinylation and degradation by the proteasome, while K13 mutants failed to bind, increasing levels of PfPI3K and elevating its lipid product PI3P. Moreover, elevation of PI3P alone efficiently conferred artemisinin resistance [9]. Yet the function of PI3P elevation and its connection to the UPR remained elusive.
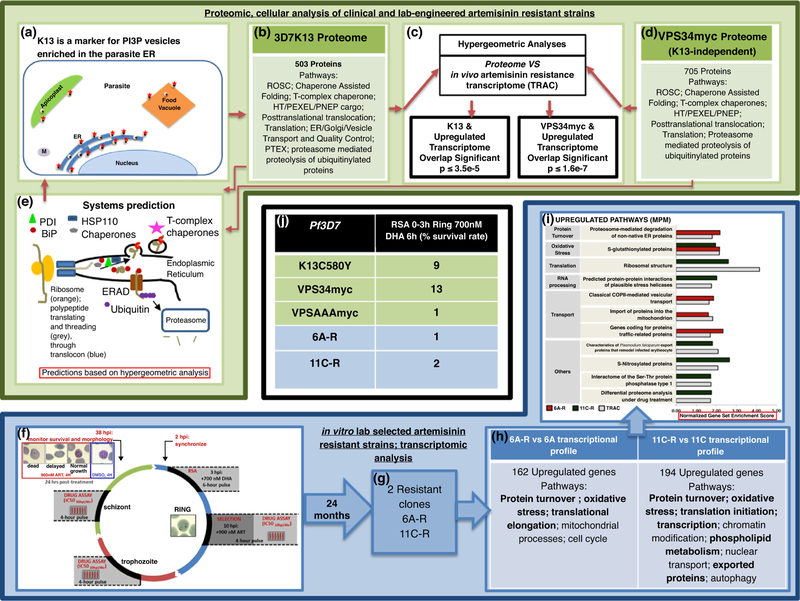
Two recent studies have directly shed additional light on these pathways, their relatedness and function in resistance [11●●,12●●]. At the cellular level, high resolution immunoelectron microscopy established that K13 localizes to PI3P vesicles, serving as a marker for these vesicles in the parasite (shown in a schematic in Figure 2a; [11●●]). K13-PI3P vesicles reside predominantly in the parasite ER (and at lower levels in the food vacuole and the apicoplast). The major artemisinin resistance mutation K13C580Y quantitatively increased PI3P- vesicles. Biochemical isolation of these vesicles and their proteomic analyses enabled determining their constituent properties and as well as comparisons with the in vivo resistance clinical transcriptome. As summarized in Figure 2b, the K13/PI3P vesicle proteome was enriched in multiple proteostasis systems of protein export, quality-control and folding in the parasite ER and cytoplasm including oxidative protein damage, stress and UPR. Hypergeometric analyses (a discrete probability distribution to predict success of outcomes) suggested significant overlap of the vesicle proteome with upregulated (but not downregulated) genes of the in vivo artemisinin resistance transcriptome (Figure 2c) reinforcing the association between vesicle amplification and resistance. Synthetic elevation of PI3P (by transgenic expression of human PI3K called VPS34) in absence of K13 mutation, conferred greater than a log increase in artemisinin resistance as measured by RSA and amplified vesicle proteomic signatures of ER proteostasis systems (Figure 2d and e). The proteomes significantly overlapped with adaptive responses to oxidative stress and UPR associated with in vivo, clinical artemisinin resistance (Figure 2c). Together, these data suggested that expansion of homeostatic ER vesiculation is a major determinant of artemisinin resistance and likely to be a primary trigger for amplification of pathways of parasite oxidative stress and protein damage pathways via the UPR.
Figure 2:
Mechanisms of artemisinin resistance. (a–e) Proteomic and cellular studies of clinical and lab-engineered artemisinin resistance. (Summarized from Bhattacharjee et al. 2018 [11●●]). (a) K13 (red star) is a marker for PI3P (black dots) in vesicles (yellow) enriched in the parasite ER (dark blue). K13-PI3P vesicles are also observed in the food vacuole (orange) and apicoplast (green). These vesicles were not enriched at the parasite mitochondria (M), nucleus or plasma membrane. (b) Biochemical isolation followed by proteomic analysis of the 3D7K13 vesicle revealed several parasite pathways of proteostasis systems enriched in reactive oxidative stress complex (ROSC), Chaperone Assisted Folding; T-complex chaperone; HT/PEXEL/PNEP (host-targeting/plasmodium export element/PEXEL-negative exported proteins) cargo; posttranslational translocation; translation; ER/Golgi/vesicle transport and quality control; plasmodium translocon of exported proteins (PTEX); proteasome mediated proteolysis of ubiquitinylated proteins. (c) Hypergeometric analyses revealed significant overlap between the K13-vesicle proteome and upregulated (but not downregulated) genes of the in vivo artemisinin resistant transcriptome (TRAC) [10]. (d) Synthetic resistance by elevation of PI3P induced independent of K13 mutation (using VPS34myc) yielded vesicular proteomes enriched in pathways of ROSC, Chaperone Assisted Folding, T- complex chaperones, HT/PEXEL/PNEP, Posttranslational translocation, Translation, Proteasome mediated proteolysis of ubiquitinylated proteins. Hypergeometric analyses revealed significant overlap between the synthetic PI3P (VPS34myc) proteome and upregulated (but not downregulated) genes of the in vivo artemisinin resistant transcriptome (TRAC; c). (e) Vesicular K13 and synthetic PI3P proteomes predict a system of vesicular proteostasis containing oxidative stress responses to protein damage, the UPR and ERAD (ER-associated degradation) pathway including (but not limited to) key ER proteins such as PDI (protein disulfide isomerase), BiP (heat shock protein 70). This model is validated by localization of K13 to vesicles (panel a). (f–i). Transcriptomic studies of in vitro lab selected artemisinin resistance (summarized from Rocamora et al., 2018 [12●●]). (f) In vitro selection of P. falciparum clones after long term exposure to artemisinins. The 3D7 strain was exposed at mid ring (10 hour+) to short (4 hour) pulses of a clinically relevant dose (900 nM) of artemisinins continuously for two years. (g) Two in vitro-selected resistant clones 6A-R and 11C-R were generated. (h) Their transcriptional profiling showed enhancement of adaptive responses against oxidative stress and protein damage (shown in bold). (i) Normalized Gene Set Enrichment Analysis (GSEA) Score was used to depict overlap between significantly upregulated functionalities of the in vitro lab selected resistant lines and in vivo artemisinin resistant transcriptome [10]. (j) Ring-stage survival assay (RSA) values associated of indicated strains reported by Bhattacharjee et al., 2018 [11●●], Mbengue et al., 2015 [9], Rocamora et al., 2018 [12●●].
In the second study [12●●], in vitro-selected artemisinin resistant parasite lines were derived from a laboratory strain (3D7) of P. falciparum exposed to multiple short pulses of artemisinins during the asexual cycle and grown long term (for 24 months). Two isolated clones without mutation in K13 (Figure 2f–g, clones 6A-R and 11C-R) showed changes of 1.7–2.5 fold increase in artemisinin resistance in a standard IC50 assay (measured over 72 hours) but with low RSA values of 1% (the cut off for resistance) and 2%. Their gene expression profiles showed some enhancement of adaptive responses against oxidative stress and protein damage (Figure 2h) but failed to show significant intersection to the in vivo transcriptome (further supporting K13 is associated with clinical artemisinin resistance). A normalized gene set enrichment score (GSEA) was used to capture links in the gene sets of the in vitro selected transcriptome to clinical artemisinin resistance at the transcriptional level (Figure 2i). In a modified in vitro assay where 10 hour parasites were pulsed for 4 hours (thought to mimic in vivo cycles of drug pressure), the in vitro-selected strains showed higher tolerance for artemisinin than parental counterparts. Trans expression of two top hit genes PfTrx1 and PfSpp2 (respectively involved in cellular protection and protein translation) in the parent 3D7 strain, showed a small increase in resistance when measured in 10 hour rings pulsed for 4 hours but did not contribute resistance measurable by RSA (Figure 2j).
Mechanism of artemisinin resistance
Proteostasis mechanisms of artemisinin resistance are more complex than amplification/mutation of a transporter or enzyme seen in the case of resistance to other antimalarials [13–15]. Consequently, multiple approaches and measurements have been used to assess artemisinin resistance in laboratory studies. Nonetheless, since the RSA provides a gold standard of in vitro artemisinin resistance that has been validated against in vivo resistance, it provides a robust index to simplify comparisons between findings across multiple studies. Since variations may arise due to differences in strain background, we used 3D7 strain data to compare the effect of K13C580Y mutation (responsible 80% of resistant parasites in SE Asia) as well as proposed determinants of K13-independent resistance. As shown in panel 2j, transgenic elevation of parasite PI3P (via expression of VPS34) in absence of K13 mutation, induced greater than a log change of resistance in the RSA and comparable to levels induced by K13C580Y. Moreover, expression of VPS34myc amplified a proteome that significantly overlapped with upregulated genes of the in vivo clinical transcriptome (Figure 2d). In vitro chemo-selected resistant parasites also independent of K13 showed much lower levels of resistance (RSAs 1–2) and some association with the in vivo clinical artemisinin resistance transcriptome. But transgenic expression of the high value candidate genes of the oxidative stress and protein damage, did not result in an RSA survival rate of 2% (Figure 2j; although they may show a small contribution in a less stringent resistance assay).
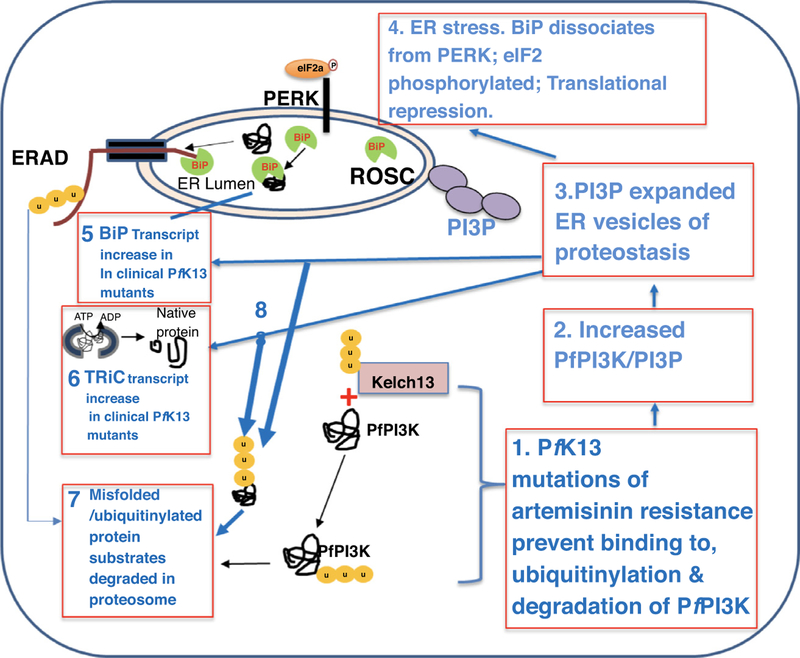
In sum, amplification of PI3P vesiculation as modeled in Figure 3 (steps 1–3) remains the stronger predictor of artemisinin resistance. Its transcriptional imprint maybe upregulation of ER vesiculation, parasite oxidative stress and protein damage pathways via the UPR. Individual components of adaptive responses against oxidative stress and protein damage may act in concert within PI3P proteostatic vesicles or in addition to them, to confer resistance (Figure 3, steps 4–6). Therefore, although in vitro-selected artemisinin-resistant parasites were not analyzed for levels of PfPI3K-protein or PI3P-lipid vesicles [12●●], their transcriptional profiles support the model in Figure 3, that expansion of homeostatic ER vesiculation is a major determinant of artemisinin resistance. The model in Figure 3 also explains additional findings from recent studies that PERK (also known as PK4; step 4) in the presence of artemisinins, phosphorylates eukaryotic initiation factor-2a leading to translation arrest and the induction of latency [16●●], providing a high level of proof that that Plasmodium manifests at least one (of 3) well- defined effector arms of the UPR of eukaryotes in artemisinin resistance.
Figure 3:
Model unifying proteostasis in ER and cytoplasm of Plasmodium falciparum in mechanisms of artemisinin resistance. Pathways of protein quality control in the ER and the cytoplasm stimulate concerted mechanisms of protein translation, translocation, vesicular export and additional chaperone functions to enable proteostasis and thereby restore proper folding of proteins and their function in a cell. A model is proposed for proteostasis pathways to rescue Plasmodium falciparum parasites from artemisinin-induced protein damage, proteopathy and death. In artemisinin-resistant parasites, K13 mutations prevent binding, ubiquitinylation and degradation of PfPI3K (step 1, [9]). This leads to an increase in the kinase and thereby its lipid product, PI3P (step 2) causing expansion of homeostatic PI3P-vesicles of proteostasis from the ER (step 3, [11●●]) that may underlie a mechanism of autophagy. In addition, these vesicles contain BiP, a key component of the ROSC. BiP is usually bound to UPR transmembrane receptors, but under conditions of stress disassociates from the membrane and binds to misfolded proteins, shifting equilibrium away from and activating UPR receptors. In P. falciparum, the UPR receptor, ER transmembrane sensor protein-kinase R (PKR)-like ER kinase (PERK, also known as PK4) has been shown to phosphorylate elongation initiation factor 2a (eIF2a), leading to translational repression, and a reduction of general protein synthesis in artemisinin resistant parasites (step 4, [16●●]). Increase in BiP transcript levels in vivo artemisinin resistance (step 5) [10], may reflect a response linked to steps 4 and 3. The T-complex protein 1 (TCP1) ring complex (TRiC) chaperone transcript also increased in in vivo artemisinin resistance [10] may enable misfolded proteins in the cytoplasm to become properly folded (step 6) appears associated with step 3 [11●●]. In the ER-associated degradation (ERAD) pathway misfolded proteins bound and unfolded by BiP (step 5), are translocated to the cytoplasm, ubiquitinylated and degraded in the proteasome (through the ubiquitin-26S proteasome pathway (step 7). Misfolded proteins in the cytoplasm that are not rescued by TRiC maybe ubiquitinylated and targeted for degradation in the proteasome (step 8, [32]).
In higher eukaryotes, PI3P expansion in the ER stimulates autophagy to efficiently remove misfolded/toxic protein aggregates [17–19] as well as unconventional autophagic secretion [20]. Longitudinal genomic surveillance studies have identified artemisinin resistance associated malarial gene loci [21●,22●,23●] with multiple genes including several belonging to phosphoinositide pathways and autophagic pathways. A vesicular/autophagic resistancemechanism may be effective against new drugs that induce toxicity through protein aggregates or ‘proteopathy’ [24●●,25] and may present many targets such as plasmepsins also detected in the vesicle proteome [11●●], to facilitate resistance to partner drugs such as recently reported in multi drug resistant malaria [26,27]. However, a caveat to the ‘autophagy hypothesis’ is that PI3P vesicles in K13 mutants contain single membranes lacking luminal staining, while auto-phagosomes are double membraned and contain cellular components targeted for degradation. ER autophagy remains poorly understood in malaria parasites [28,29] and its mechanistic analyses need further study in artemisinin resistance.
Artemisinin resistance mechanism of widespread parasite remodeling and host engagement
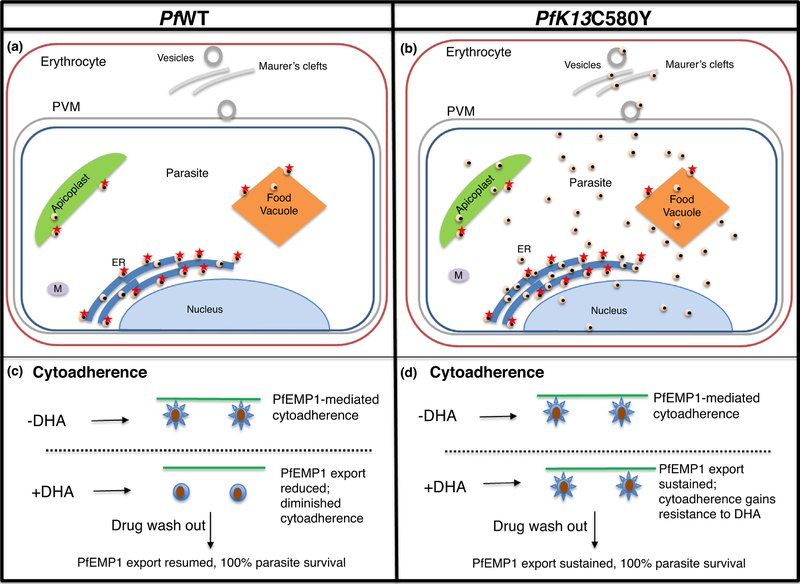
An unexpected feature of PI3P vesicles is that upon amplification they reach all destinations in mutant parasite (Figure 4a and b). This may explain complexity of artemisinin resistance as well as how a single K13-proteo- static determinant may reflect selective pressure in multiple organellar systems and hundreds of parasite genes implicated in resistance [2●●,3●●,30–36]. Since PI3P elevation is sufficient to induce resistance, expansion of PI3P vesicles may also account for K13-independent resistance [37●]. Another intriguing feature of PI3P vesicle expansion in resistant parasites is that the lipid also targets the red cell (Figure 4b) suggesting host factors may modulate artemisinin resistance [11●●]. Consistent with findings that PI3P is detected in intra-erythrocytic vesicles and Maurer’s clefts utilized in parasite protein export (Figure 4b), K13 mutations regulate the dynamics of export of PfEMP1 (the major parasite virulence determinant) to the host red cell. The K13 vesicle-proteome contains host-targeted protein translocation export (‘PTEX’) machinery [11●●,38,39] and several cargo with relevant export signals [40–42]. Whether all exported cargo utilize this pathway is not known: export to the host cell is complex and there may be steps of export selectivity that remain poorly understood [43–45]. Nonetheless dissemination of PI3P vesicles of proteostasis may also provide mechanisms to mitigate artemisinin damage to the host red cell. Finally, PfEMP1 export and cytoad- herence are inhibited by dihydroartemisinin in wild type parasites but both processes become resistant to drug in K13 mutants (Figure 4c, d, [11●●]). This suggests that in addition to protecting the infected red cell from proteopathy, K13 resistance mutations may provide better adherence to host receptors even in presence of drug. This may contribute to immune protection by helping the parasite avoid the spleen at both high and low parasite burdens. Additionally, K13 mutations may facilitate persistence of resistant parasites seen at low and asymptomatic parasitemias despite mass drug administration [46,47]. Since artemisinins and ACTs remain critical tools for malaria elimination, it will be important to understand how resistance enhances survival mechanisms within the parasite as well as the parasite’s engagement with its host.
Figure 4:
Expansion of PI3P vesicles by the major K13C580Y mutation of artemisinin resistance. (a) In Plasmodium falciparum wild type (WT) cells, PI3P vesicles (black in yellow spheres) with associated K13 (red star) are seen at parasite ER, apicoplast and food vacuole. (b) In the major mutation of artemisinin resistance K13C580Y, PI3P vesicles are amplified exported from the ER and disseminated in all organelles throughout the parasite as well as the erythrocyte, where PI3P is detected on vesicles and ‘Maurer’s clefts’ known to mediate the export of virulence determinants such as a major adhesin family (PfEMP1) to infected host cell surface. (c) Export of PfEMP1 (blue spikes) and cytoadherence of infected erythrocytes (blue circles with brown spheres) to host receptors (green line) are diminished by dihydroartemisinin (DHA) which is known to reduce levels of PI3P in artemisinin sensitive parasites [9,11●●]. (d) Export of PfEMP1 and cytoadherence are not diminished by DHA in artemisinin resistant parasites that show elevation in PI3P vesicles in the parasite and erythrocyte [11●●].
Conclusions
On the heels of the identification of K13 resistance marker, different technological approaches of transcriptomics (revealing associative UPR responses) versus functional and genetic studies (showing requirement of the lipid PI3P), suggested two distinct processes of artemisinin resistance. However, subsequent work using proteomics as an important bridging technology, suggests that both function in the same pathway. Furthermore, individual ER-UPR components are involved in artemisinin induced latency, but do not induce significant levels of resistance. Other candidate genes have not been tested. However, expansion of PI3P vesicles efficiently induces a log increase in artemisinin resistance. Vesicle formation and turnover are complex processes that may remodel both the parasite and red cell, suggesting that understanding their functions may provide insights about parasite mechanisms to survive exposure to artemisinins as well as host pathogenesis.
Acknowledgements
The authors apologize to colleagues whose work could not be cited owing to space limitation. We thank members of the Haldar lab for insightful discussion. Work in the authors’ laboratories was supported by the US National Institutes of Health (R01 HL069630 and HL130330).
Footnotes
Conflict of interest statement
Nothing declared.
Papers of particular interest, published within the period of review, have been highlighted as
● of special interest
●● of outstanding interest
References and recommended reading
- 1.WHO: World Malaria Report 2017 2017.
- 2.Wang J, Zhang C-J, Chia WN, Loh CCY, Li Z, Lee YM, He Y, Yuan L-X, Lim TK, Liu M et al. : Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum. Nat Commun 2015, 6.●● First of two studies showing artemisinins kill by targeting hundreds of proteins suggesting parasite death is caused by proteopathy.
- 3.Ismail HM, Barton V, Phanchana M, Charoensutthivarakul S, Wong MHL, Hemingway J, Biagini GA, O’neill PM, Ward SA, Chibale K et al. : Artemisinin activity-based probes identify multiple molecular targets within the asexual stage of the malaria parasites Plasmodium falciparum 3D7. PNAS 2016, 113.●● First of two studies showing artemisinins kill by targeting hundreds of proteins suggesting parasite death is caused by proteopathy.
- 4.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM: Evidence of artemisinin-resistant malaria in Western Cambodia. N Engl J Med 2008, 359. [DOI] [PubMed] [Google Scholar]
- 5.Dondorp AM, Nosten F, Yi P, Das D, Hanpithakpong W, Lee SJ, Ringwald P, Imwong M, Chotivanich K et al. : Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 2009, 361:455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, Lim P, Mao S, Sopha C, Sam B et al. : Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug response studies. Lancet Infect Dis 2013, 13:1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta VA, Beggs AH: Kelch proteins: emerging roles in skeletal muscle development and diseases. Skelet Muscle 2014. 10.1186/2044-5040-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikesitch N, Ling SCW: Molecular mechanisms in multiple myeloma drug resistance. J Clin Pathol 2016, 69:97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mbengue A, Bhattacharjee S, Pandharkar T, Liu H, Estiu G, Stahelin RV, Rizk SS, Njimoh DL, Ryan Y, Chotivanich K et al. : A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature 2015, 520:683–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mok S, Ashley EA, Ferreira PE, Zhu L, Lin Z, Yeo T, Chotivanich K, Imwong M, Pukrittayakamee S, Dhorda M et al. : Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science 2015, 347:431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhattacharjee S, Coppens I, Mbengue A, Suresh N, Ghorbal M,Slouka Z, Safeukui I, Tang H-Y, Speicher DW, Stahelin RV et al. : Remodeling of the malaria parasite and host human red cell by vesicle amplification that induces artemisinin resistance. Blood 2018. 10.1182/blood-2017-11-814665.●● This study revealed an ER PI3P-vesicular system causing artemisinin resistance modifies malaria parasite and host red cells.
- 12.Rocamora F, Zhu L, Liong KY, Dondorp A, Miotto O, Mok S,Bozdech Z: Oxidative stress and protein damage responses mediate artemisinin resistance in malaria parasites. PLOS Pathog 2018, 14:e1006930.●● Transcriptomic analysis ofin vitro lab strains selected long term for artemisinin resistance.
- 13.Mixson-Hayden T, Jain V, McCollum AM, Poe A, Nagpal AC, Dash AP, Stiles JK, Udhayakumar V, Singh N: Evidence of selective sweeps in genes conferring resistance to chloroquine and pyrimethamine in Plasmodium falciparum isolates in India. Antimicrob Agents Chemother 2010, 54:997–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gesase S, Gosling RD, Hashim R, Ord R, Naldoo I, Madebe R, Mosha JF, Joho A, Mandia V, Mrema H et al. : High resistance of Plasmodium falciparum to sulphadoxine/pyrimethamine in Northern Tanzania and the emergence of DHPS resistance mutation at codon 581. PLoS ONE 2009, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdul-Ghani R, Farag HF, Allam AF: Sulfadoxine-pyrimethamine resistance in Plasmodium falciparum: a zoomed image at the molecular level within a geographic context. Acta Trop 2013, 125:163–190. [DOI] [PubMed] [Google Scholar]
- 16.Zhang M, Gallego-Delgado J, Fernandez-Arias C, Waters NC,Rodriguez A, Tsuji M, Wek RC, Nussenzweig V, Sullivan WJ: Inhibiting the Plasmodium eIF2α kinase PK4 prevents artemisinin-induced latency. Cell Host Microbe 2017, 22 766–776.e4.●● This study showed artemisinin resistance induces latency (promoting survival) through phosphorylation of Plasmodium eukaryotic initiation factor 2a (eIF2a).
- 17.Marat AL, Haucke V: Phosphatidylinositol 3-phosphates—at the interface between cell signalling and membrane traffic. EMBO J 2016, 35:561–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nascimbeni AC, Codogno P, Morel E: Phosphatidylinositol-3- phosphate in the regulation of autophagy membrane dynamics. FEBS J 2017, 284:1267–1278. [DOI] [PubMed] [Google Scholar]
- 19.Kaminska J, Rzepnikowska W, Polak A, Flis K, Soczewka P, Bala K, Sienko M, Grynberg M, Kaliszewski P, Urbanek A et al. : Phosphatidylinositol-3-phosphate regulates response of cells to proteotoxic stress. Int J Biochem Cell Biol 2016, 79:494–504. [DOI] [PubMed] [Google Scholar]
- 20.Ponpuak M, Mandell MA, Kimura T, Chauhan S, Cleyrat C, Deretic V: Secretory autophagy. Curr Opin Cell Biol 2015, 35:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Cabrera M, Yang J, Yuan L, Gupta B, Liang X,Kemirembe K, Shrestha S, Brashear A, Li X et al. : Genome-wide association analysis identifies genetic loci associated with resistance to multiple antimalarials in Plasmodium falciparum from China–Myanmar border. Sci Rep 2016, 6:1–12.● GWAS study identifying locus on chromosome 10 containing autophagy related protein 18 (ATG18) was associated with decreased sensitivities to artemisinins.
- 22.Dwivedi A, Reynes C, Kuehn A, Roche DB, Khim N, Hebrard M,Milanesi S, Rivals E, Frutos R, Menard D et al. : Functional analysis of Plasmodium falciparum subpopulations associated with artemisinin resistance in Cambodia. Malar J 2017, 16:1–17.● Genomic assessment of emergence of parasite genes and associated pathways linked to artemisnin resistance in Cambodian parasite isolates.
- 23.Cerqueira GC, Cheeseman IH, Schaffner SF, Nair S, McDew-White M, Phyo AP, Ashley EA, Melnikov A, Rogov P, Birren BW et al. : Longitudinal genomic surveillance of Plasmodium falciparum malaria parasites reveals complex genomic architecture of emerging artemisinin resistance. Genome Biol 2017, 18:1–13.● Longitudinal genomic surveillance study showing greatest temporal changes at SNPs in K13 followed by phosphatidylinositol-4-kinase, over the time course of emergence of artemisinin resistance in Thailand.
- 24.Straimer J, Gnädig NF, Stokes BH, Ehrenberger M, Crane AA, Fidock DA: Plasmodium falciparum K13 mutations differentially impact ozonide susceptibility and parasite fitness in vitro. MBio 2017, 8:1–12.●● This study showed that K13 mutants of artemisinin resistance display differential resistance to ozonides that (like artemsinins) also activate to kill upon cleavage of an endoperoxide bond.
- 25.O’Neill PM, Amewu RK, Charman SA, Sabbani S, Gnä dig NF, Straimer J, Fidock DA, Shore ER, Roberts NL, Wong MHL et al. : A tetraoxane-based antimalarial drug candidate that overcomes PfK13-C580Y dependent artemisinin resistance. Nat Commun 2017, 8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witkowski B, Duru V, Khim N, Ross LS, Saintpierre B, Beghain J, Chy S, Kim S, Ke S, Kloeung N et al. : A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype–genotype association study. Lancet Infect Dis 2017, 17:174–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, Almagro-Garcia J, Neal AT, Sreng S, Suon S et al. : Genetic markers associated with dihydroartemisinin–piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype–phenotype association study. Lancet Infect Dis 2017, 17:164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitamura K, Kishi-Itakura C, Tsuboi T, Sato S, Kita K, Ohta N, Mizushima N: Autophagy-related Atg8 localizes to the apicoplast of the human malaria parasite Plasmodium falciparum. PLoS ONE 2012. 10.1371/journal.pone.0042977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cervantes S, Bunnik EM, Saraf A, Conner CM, Escalante A, Sardiu ME, Ponts N, Prudhomme J, Florens L, Le Roch KG: The multifunctional autophagy pathway in human malaria parasite, Plasmodium falciparum. Autophagy 2014, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miotto O, Amato R, Ashley EA, MacInnis B, Almagro-Garcia J, Amaratunga C, Lim P, Mead D, Oyola SO, Dhorda M et al. : Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet 2015, 47:226–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miotto O, Almagro-Garcia J, Manske M, Macinnis B, Campino S, Rockett KA, Amaratunga C, Lim P, Suon S, Sreng S et al. : Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet 2013, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dogovski C, Xie SC, Burgio G, Bridgford J, Mok S, McCaw JM, Chotivanich K, Kenny S, Gnä dig N, Straimer J et al. : Targeting the cell stress response of Plasmodium falciparum to overcome artemisinin resistance. PLoS Biol 2015, 13:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Huang L, Li J, Fan Q, Long Y, Li Y, Zhou B: Artemisinin directly targets malarial mitochondria through its specific mitochondrial activation. PLoS ONE 2010, 5:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohrbach P, Sanchez CP, Hayton K, Friedrich O, Patel J, Sidhu ABS, Ferdig MT, Fidock DA, Lanzer M: Genetic linkage of pfmdr1 with food vacuolar solute import in Plasmodium falciparum. EMBO J 2006, 25:3000–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eckstein-Ludwing U, Webb RJ, van Goethem IDA, East JM, Lee AG, Kimura M, O’Neill PM, Bray PG, Ward SA, Krishna S: Artemisinins target the SERCA of Plasmodium falciparum. Nature 2003, 424:957–961. [DOI] [PubMed] [Google Scholar]
- 36.Bhisutthibhan J, Pan X, Hossler A, Walker DJ, Charles A, Carlton J, Dame JB, Meshnick SR, Hossler P, Yowell C: The Plasmodium falciparum translationally controlled tumor protein homolog and its reaction with the antimalarial drug artemisinin. J Biol Chem 1998, 273:16192–16198. [DOI] [PubMed] [Google Scholar]
- 37.Mukherjee A, Bopp S, Magistrado P, Wong W, Daniels R,Demas A, Schaffner S, Amaratunga C, Lim P, Dhorda M et al. : Artemisinin resistance without pfkelch13 mutations in Plasmodium falciparum isolates from Cambodia. Malar J 2017, 16:1–12.● First report and establishment of clinical strains resistant to artemisinin in the absence of K13 mutations.
- 38.Elsworth B, Matthews K, Nie CQ, Kalanon M, Charnaud SC, Sanders PR, Chisholm S, Counihan N, Shaw PJ, Pino P et al. : PTEX is an essential nexus for protein export in malaria parasites. Nature 2014. 10.1038/nature13555. [DOI] [PubMed] [Google Scholar]
- 39.Beck JR, Muralidharan V, Oksman A, Goldberg DE: PTEX component HSP101 mediates export of diverse malaria effectors into host erythrocytes. Nature 2014, 511:592–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hiller NL, Bhattacharjee S, Ooij C, Van Liolios K, Harrison T, Lopezestran C, Haldar K: A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science 2004, 306:1934–1937. [DOI] [PubMed] [Google Scholar]
- 41.Marti M, Baum J, Rug M, Tilley L, Cowman AF: Signal-mediated export of proteins from the malaria parasite to the host erythrocyte. J Cell Biol 2005, 171:587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spielmann T, Gilberger TW: Protein export in malaria parasites: do multiple export motifs add up to multiple export pathways? Trends Parasitol 2010, 26:6–10. [DOI] [PubMed] [Google Scholar]
- 43.Tarr SJ, Cryar A, Thalassinos K, Haldar K, Osborne AR: The C- terminal portion of the cleaved HT motif is necessary and sufficient to mediate export of proteins from the malaria parasite into its host cell. Mol Microbiol 2013, 87:835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Koning-Ward TF, Dixon MW, Tilley L, Gilson PR: Plasmodium species: master renovators of their host cells. Nat Rev Microbiol 2016. 10.1038/nrmicro.2016.79. [DOI] [PubMed] [Google Scholar]
- 45.Haldar K: Protein trafficking in apicomplexan parasites: crossing the vacuolar Rubicon. Curr Opin Microbiol 2016. 10.1016/j.mib.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phommasone K, Adhikari B, Henriques G, Pongvongsa T, Phongmany P, Von Seidlein L, White NJ, Day NPJ, Dondorp AM, Newton PN et al. : Asymptomatic Plasmodium infections in 18 villages of southern Savannakhet Province, Lao PDR (Laos). Malar J 2016, 15:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nyunt MH, Shein T, Zaw NN, Han SS, Muh F, Lee SK, Han JH, Thant KZ, Han ET, Kyaw MP: Molecular evidence of drug resistance in asymptomatic malaria infections, Myanmar, 2015. Emerg Infect Dis 2017, 23:517–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delves M, Plouffe D, Scheurer C, Meister S, Wittlin S, Winzeler EA, Sinden RE, Leroy D: The activities of current antimalarial drugs on the life cycle stages of Plasmodium: a comparative study with human and rodent parasites. PLoS Med 2012, 9. [DOI] [PMC free article] [PubMed]