Abstract
In the past 4 years, because of the advent of new cameras, many ribosome structures have been solved by cryoelectron microscopy (cryo-EM) at high, often near-atomic resolution, bringing new mechanistic insights into the processes of translation initiation, peptide elongation, termination, and recycling. Thus, cryo-EM has joined X-ray crystallography as a powerful technique in structural studies of translation. The significance of this new development is that structures of ribosomes in complex with their functional binding partners can now be determined to high resolution in multiple states as they perform their work. The aim of this article is to provide an overview of these new studies and assess the contributions they have made toward an understanding of translation and translational control.
To understand translation and translational control, an understanding of structure as well as dynamics of the various processes is required. The ability to see interactions at the functional sites in atomic detail, facilitated by much of the recent work, has made a large difference in this respect. One mechanism can be singled out that is ubiquitous, with different ramifications in initiation, translocation, decoding, termination, and recycling; in each of these steps of translation, the action of a GTPase is required, and typically the engagement of a factor creates an atomic constellation that triggers guanosine triphosphate (GTP) hydrolysis, followed by changes in both the factor and the ribosome.
There are tantalizing questions on how this action unfolds in each case, and the reason so much is still unanswered is that it occurs quite rapidly, and that its structural characterization therefore requires some kind of interference, such as the use of a nonhydrolyzable GTP analog or mutation at a key site, each time invoking questions about the authenticity of the state trapped. Thus, the nature of the decisive powerstroke that brings us from point A to point B in the processive machinery is uncertain.
Among all steps of translation, eukaryotic initiation is probably the least understood because of the multitude of steps and the complexity of the scanning mechanism leading to the placement of the AUG codon at the P site of the small subunit. Next there come the variants of translation initiation through the viral internal ribosome entry site (IRES) mechanism; here it is necessary to understand how the IRES is able to “push the right buttons” on the ribosome to take over control. Another highlight of this review is the recent progress in the structural characterization of ribosomes of parasitic protozoans; here, the reason little is known thus far is related to the prevalence of the diseases that they cause in underdeveloped countries and the politics of science funding.
As translation of the genetic code into polypeptides proceeds through its main phases of initiation, peptide elongation, termination, and recycling, the ribosome undergoes numerous conformational changes and ligand binding/unbinding events. For a long time, steps toward the goal of obtaining information on the structural basis of these events, critical for understanding the mechanistic basis of biological function, have been handicapped by shortcomings of the two main structural visualization techniques, X-ray crystallography and single-particle cryoelectron microscopy (cryo-EM). Single-particle cryo-EM, recently highlighted by the 2017 Nobel Prize in Chemistry, is the visualization of molecules in a thin layer of vitreous ice following rapid plunge-freezing of the sample into a cryogen, or a freezing agent, at liquid-nitrogen temperature (McDowall et al. 1983). Raw data collected on the electron microscope comes in the form of two-dimensional “snapshots” called micrographs, each of which include a few to over 100 copies of a biological molecule. Because molecules are randomly oriented, all view directions are sampled when a sufficient number of micrographs are taken. As the electron dose is spread over a large number of molecules, radiation damage is avoided or strongly reduced. Raw data are subjected to a specialized series of computational techniques, during which individual copies of the specimen are identified and the two-dimensional view represented by each copy is assigned a viewing angle, and a three-dimensional reconstruction of the Coulomb potential distribution (colloquially called “density map”) can ultimately be computed.
While X-ray crystallography, starting with publications in 2000 (Ban et al. 2000; Schluenzen et al. 2000; Wimberly et al. 2000), has been the source of many high-resolution atomic models of the ribosome structure, it is intrinsically limited as a technique by the conformational selection taking place during the formation of the crystal, such that many interesting functional states will escape visualization. Conversely, states that are only transiently visited may receive inordinate attention because they happen to be captured in crystals diffracting to high resolution. Cryo-EM, on the other hand, apart from the fact that it does not require crystals, has the advantage that it is capable of displaying all states coexisting in the sample without restriction. The much lower sample volume required is another benefit. However, until recently, this technique has been limited in resolution to 5–6 Ångströms (Å, 10−10 meters) for asymmetric molecules (e.g., Hashem et al. 2013a) because of the low quality of the recording medium. This situation has been radically changed with the commercial introduction, in 2012, of direct electron detecting cameras (see McMullan et al. 2009), which allow near-atomic resolution to be reached. For the ribosome field, this has been a game changer as it is now possible to study ligand-binding events (e.g., with transfer RNA [tRNA] or various translation factors) in minute detail. Even ab initio structural modeling is now possible over large stretches of the reconstructed density map; magnesium ions can be located; and ribosomal RNA (rRNA) modifications can be directly identified by the positions of extra density in the map (see Fischer et al. 2015; Liu et al. 2016). This greatly expanded potential of cryo-EM has led an increasing number of crystallographers to adopt the new technique, helped by the fact that both techniques of structure research share many mathematical concepts and modeling tools.
The aim of this article is to give an overview of the recent gain in fundamental knowledge on ribosome structure and function, now that cryo-EM has joined X-ray crystallography as a powerful alternative technique of high-resolution structure research. However, the ensuing large proliferation of structures precludes an exhaustive coverage, and some areas receive particular attention.
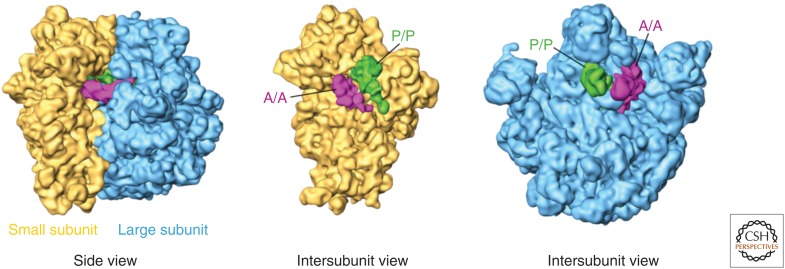
Readers unfamiliar with the ribosome may benefit from a brief introduction to its structure (Fig. 1). In all organisms, the ribosome is a 2- to 4.5-megadalton (MDa) structure made up of a small subunit (SSU) and a large subunit (LSU). Both subunits are composed of rRNA surrounded by many smaller ribosomal proteins (r-proteins). From a side view, the solvent side of the SSU appears relatively flat, whereas that of the LSU is rounded. A view of the SSU’s intersubunit side displays its head region, complete with a leftward-facing beak, above a thin neck region that separates the head from the body of the subunit. The body features a large platform on the right and a smaller shoulder on the left, narrowing somewhat toward a pointed left foot and right foot in eukaryotes, or to a single spur in prokaryotes. Meanwhile, the solvent side of the large subunit appears roughly round with the central protuberance at the top. The messenger RNA (mRNA) passes through the mRNA channel created by the groove between the head and the body of the SSU at its intersubunit face. The mRNA and tRNAs both pass through the three adjacent A (aminoacyl), P (peptidyl), and E (exit) sites, which are located on both the SSU and LSU intersubunit faces.
Figure 1.
General features of ribosome structure and positions of transfer RNAs (tRNAs). The large subunit appears in blue and the small subunit in yellow. A-site (A/A) tRNA is pink, and P-site (P/P) tRNA is green. Note that the E site is to the right of the P site as shown from the intersubunit view of the small subunit and simultaneously to the left of the P site as shown from the view of the large subunit. A hybrid-state A/P tRNA would appear as an occupied A site on the small subunit and an occupied P site on the large subunit; in the same way, a P/E tRNA occupies the P site on the small subunit and the E site on the large subunit. (From Agirrezabala et al. 2008; adapted, with permission, from Elsevier © 2008.)
EUBACTERIAL TRANSLATION AND TRANSLATIONAL CONTROL
In previous studies, before 2013, cryo-EM reconstructions could only be used to study large conformational changes such as intersubunit rotation or subunit head swivel, at intermediate (∼5–9 Å) resolutions. At high (2.5–3.5 Å) resolutions readily attainable now, where domain motions and molecular contacts are much better defined, insights have been gained into key events during translation, and into diverse mechanisms of translational control. Additionally, the capability of single-particle cryo-EM to visualize multiple states in a single sample has particularly benefited the understanding of multistep processes such as mRNA-tRNA translocation.
Detailed structural studies have focused on two pivotal stages of the polypeptide elongation cycle (see reviews by Voorhees and Ramakrishnan 2013; Rodnina and Wintermeyer 2016): elongation factor G (EF-G)-assisted mRNA-tRNA translocation and tRNA selection/decoding. As fast and highly dynamic events involving a multitude of domain motions, these are very hard to capture by cryo-EM because of the high degree of ensuing heterogeneity. Particular difficulties are posed by structural states that differ in structural constellations (e.g., base flipping) in a local region only, whereas remaining virtually unchanged elsewhere. In these cases, a technique called local classification can still be used, by singling out the region of interest through imposition of an appropriate mask in the application of the classification algorithm (Penczek et al. 2006). In a variant of the technique introduced by Scheres (2016), the projected average density is subtracted from the data, thereby increasing the sensitivity of the local classification.
Research on the structural basis of mRNA-tRNA translocation was recently reviewed by Ling and Ermolenko (2016) and Frank (2017). Despite a long history of structural studies, the role of EF-G as it interacts with the ribosome during mRNA-tRNA translocation is still not well understood, even though there is basic agreement on the factor’s catalytic action leading to a large (104- to 106-fold) acceleration of the translocation rate (Katunin et al. 2002). Brownian intersubunit motion of the ribosome has been observed in the absence of factors (Agirrezabala et al. 2008, 2012; Cornish et al. 2008; Julián et al. 2008), and the role of EF-G as providing a “pawl” in the promotion of translocation is generally accepted, implying the existence of some type of power stroke. Specifically, this pawl is manifest in the tip of EF-G’s domain IV, which disrupts the codon–anticodon interaction (Frank et al. 2007; Taylor et al. 2007), a notion that has received renewed support in a recent mutation study (Liu et al. 2014).
Intermediate states in factor-free intersubunit rotation (Fischer et al. 2010; Agirrezabala et al. 2012), also observed by X-ray crystallography (Zhang et al. 2009; Tourigny et al. 2013; Zhou et al. 2014), reflect a trajectory across the ribosome’s “metastable” free-energy landscape accessible through ambient thermal energy (Munro et al. 2009). Brilot et al. (2013) used cryo-EM to determine the structure of the 70S ribosome bound with EF-G, trapped in the pre-translocation state using the antibiotic viomycin. The structure shows a normally transient state where the A-site tRNA is still bound to the ribosome as well. A surprising discovery of the Steitz group made by X-ray crystallography (Lin et al. 2015) is a large conformational change of EF-G that apparently occurs on the binding of this factor to the ribosome. It is still unclear how this result fits into the overall picture of translocation. In their cryo-EM study, Li et al. (2015) were able to trap EF-G on the pre-translocational ribosome in two different binding configurations, one bound to the rotated and the other to the unrotated ribosome, making use of the mutation H94A. The results of the study allowed the investigators to formulate necessary and sufficient conditions for GTPase activation. Accordingly, stabilization of the factor by contacts with protein S12 and the L11-lobe is required, a condition that is only fulfilled in the rotated configuration.
In the area of tRNA selection, the most interesting question is the discrimination between cognate and near-cognate codon–anticodon pairing, essential for the fidelity of translation. Here it is clear from previous work (Ogle et al. 2001) that discrimination is achieved as a result of at least two successive steps of probing the short codon–anticodon helix with the help of three bases: A1492, A1493, and G650. It has been a challenge to image the ribosome-A/T-tRNA complex for a programmed ribosome and bring order into the sequence of events. Here a comparison between two cryo-EM studies, Agirrezabala et al. (2011) and Loveland et al. (2017) shows the progress recently achieved in the examination of the near-cognate case in a striking way, as the former did not achieve resolution sufficient to delineate the positions of the bases with certainty, whereas the latter shows the complex at close to 3 Å resolution in multiple states of intermediate engagement. In passing, it should also be noted that Fischer et al. (2015), working with cryo-EM, achieved 2.9 Å resolution with the cognate aminoacylated tRNA-EF-Tu•GDP-ribosome complex stabilized with kirromycin, a big step forward from the previous reconstruction of the same complex by Villa et al. (2009), while shedding no new light upon dynamics of tRNA selection. The main gain of the study was the identification and enumeration of rRNA modifications.
It is increasingly recognized that structural snapshots are not sufficient to understand translocation (or any of the steps of translation, for that matter) and that information on real-time dynamics is required. A whole series of recent studies uses single-molecule fluorescence resonance energy transfer (FRET) to study the timing of binding, domain motions on EF-G, inter-subunit rotation, small subunit head swivel, and mRNA-tRNA translocation with respect to the small subunit (Chen et al. 2011, 2014, 2016; Adio et al. 2015; Salsi et al. 2015; Sharma et al. 2016; Wasserman et al. 2016; Kim and Tinoco 2017). A detailed model for the sequence of events inferred from structural and single-molecule FRET (smFRET) studies is described by Wasserman et al. (2016). A new method called single-molecule polarized total internal reflection fluorescence (polTIRF) microscopy provides information not just on distance but on relative orientation between domains. Application of this method to the ribosome indeed allowed the investigators to observe a power stroke of domain IV of EF-G during translocation (Chen et al. 2016).
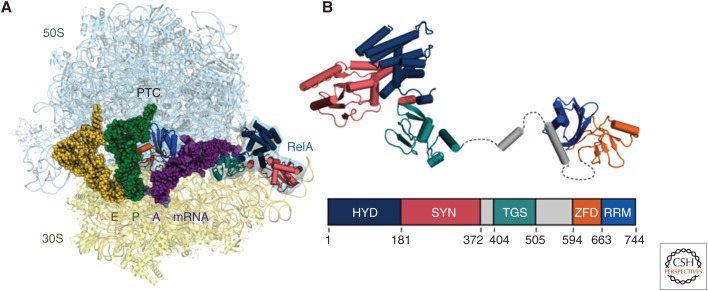
Important inroads have also been made into the understanding of translational control in eubacteria involving the binding of specific protein factors, an advance helped by the improvement in spatial resolution. Here the emerging common theme is the existence of very specific mechanisms for structural recognition of an abnormal state of the translation process, often going hand-in-hand with stress conditions in the cell, which requires intervention and rescue. EttA is a newly found ATP-binding cassette (ABC) transporter protein, which is found to bind at the ribosomal E site and suppresses translation when the energy supply in the cell falls below a critical level (Boel et al. 2014; Chen et al. 2014). EttA has a “feeler” domain that is apparently able to sense the presence or absence of a peptide on the P-site tRNA and controls the action of the factor through a mechanism of conformational signaling accordingly. RelA is a protein factor active in the stringent response, a response to conditions of amino acid starvation. In Escherichia coli, RelA synthesizes alarmones on binding to the ribosome and encountering a deacylated tRNA at the A site (Agirrezabala et al. 2013). Recent cryo-EM studies have uncovered the detailed molecular mechanism, which involves sensing the absence of the aminoacyl group on the CCA end of the strongly distorted, A/T-shaped A-site tRNA (Fig. 2) (Brown et al. 2016; Loveland et al. 2016). BipA is a GTPase closely related to EF-G and engaged at the same factor-binding site of the ribosome, but only if a tRNA is bound at the A site. Curiously, the ribosome was found in the rotated position (Kumar et al. 2015), again implying specific recognition of a local conformational anomaly. BipA is known to be essential to survival at low temperature, nutrient depletion, and various other stress conditions, but the molecular mechanism of its action is as yet unknown. Finally, ArfA is a factor that recruits release factor RF2 on encountering a ribosome containing truncated mRNA, a condition characterized by the presence of a peptidyl-tRNA in the P site and an empty A site. Thus ArfA rescues translation in bacteria lacking the trans-translation mechanism. In their recent work, Demo et al. (2017) were able to elucidate the structure of the ArfA-RF2-bound ribosome and describe its likely mechanism of action. Interestingly, this capability adds to the versatility of RF2, the protein factor already known from its classical role as a class I release factor in stop codon recognition and, more recently, in translation quality control (Zaher and Green 2009).
Figure 2.
RelA in action on the ribosome, as revealed by cryoelectron microscopy (cryo-EM). (A) RelA monitors the CCA end of an incoming putative aminoacyl-transfer RNA (tRNA) and binds to the ribosome on recognizing the absence of an amino acid. (B) Model of ribosome-bound form of RelA oriented from amino to carboxyl terminus, with the domain organization shown below; hydrolase (HYD), synthetase (SYN), TGS (threonyl-tRNA synthetase, GTPase and SpoT), Zinc-finger (ZFD) and RNA recognition motif (RRM) domains. Flexible elements between ordered RelA domains are shown in dashed lines. (From Brown et al. 2016; adapted, with permission, from Springer Nature © 2016.)
EUKARYOTIC RIBOSOME STRUCTURE AND FUNCTION
In the past few years, studies of ribosome structure for eukaryotes have led to a better appreciation of their diversity, particularly because of the great variation in rRNA expansion segments and the addition of a variety of eukaryotic-specific proteins. As we describe below, special cases are the ribosomes of parasitic protozoans, which for the first time reached resolutions that bring them within reach of drug design, as resolutions in the 2.5 to 3.5 Å range allow bound drug molecules to be directly located and visualized.
Yeast and Higher Eukaryotes
The first X-ray structures of eukaryotic ribosomes, denoted as 80S ribosomes, each comprising one 40S small subunit and one 60S large subunit, were from Tetrahymena thermophila (a 40S•eIF1 complex [Rabl et al. 2011] and a 60S•eIF6 complex [Klinge et al. 2011]) and yeast (80S [Ben-Shem et al. 2011]), and were reviewed by Wilson and Doudna Cate (2012) and Klinge et al. (2012). Specific progress with the ribosome structure of yeast was discussed by Yusupova and Yusupov (2014). Yusupov’s group also published 16 X-ray structures of the drug-bound yeast ribosome (de Loubresse et al. 2014; Yusupova and Yusupov 2017). 80S ribosomes from a growing number of species were solved by cryo-EM in various states of translation, at close to atomic resolutions: canine (Voorhees and Hegde 2016), porcine (Voorhees et al. 2013, 2014), and human (Khatter et al. 2015).
A number of studies focused on translation mediated by IRESs from viruses on ribosomes from yeast (Fernández et al. 2014; Abeyrathne et al. 2016; Murray et al. 2016), rabbit (Hashem et al. 2013c; Yamamoto et al. 2014, 2015; Muhs et al. 2015), and human (Quade et al. 2015). Besides adding to our rapidly growing structural knowledge base, these reconstructions offer novel insights into the way the eukaryotic ribosome is hijacked by viral mRNA. These works are reviewed in further detail below.
Parasitic Protozoans
In recent cryo-EM studies, several structures of ribosomes from a number of protozoan parasites have been solved at near-atomic resolutions (2.5 to 3 Å), elucidating their unique features, which are distinct from those of other eukaryotes. The resulting structures provide a basis for future functional studies of the translational machinery of protozoan parasites and have established the ribosome as a promising drug target against these human pathogens. These include a number of trypanosomatids and Plasmodium falciparum, all causing debilitating and often-fatal diseases. Current treatments against these parasites have poor efficacy, high toxicity, and increasing levels of drug resistance (Andrews et al. 2014). The importance of these parasites in epidemic diseases warrants a special highlight in our review.
Among pathogenic protozoans, the trypanosomatids, Trypanosoma cruzi, Trypanosoma brucei, and Leishmania spp., are a unique family causing insect-borne diseases: Chagas disease, African trypanosomiasis (sleeping sickness), and Leishmaniasis, respectively. Altogether, an estimated 37 million people are infected with these parasites worldwide. The first cryo-EM structures of ribosomes from this family (for T. cruzi at 12 Å [Gao et al. 2005] and for T. brucei at 5.5 Å [Hashem et al. 2013a]) provided a first glimpse and overview of their architecture. More detailed insights have only recently emerged with the advent of the new direct-detector technology. Cryo-EM reconstructions of native 60S subunits from T. cruzi at 2.5 Å (Liu et al. 2016, 2017) and Leishmania donovani at 2.8 Å (Shalev-Benami et al. 2016), as well as 80S ribosomes of L. donovani at 2.9 Å (Zhang et al. 2016) have revealed the ribosome structure in unprecedented detail, making it possible to discern rRNA nucleotides, amino acid side chains, rRNA modifications, solvent molecules, and metal ions. As a result, atomic models are now available for the ribosomes of all three species.
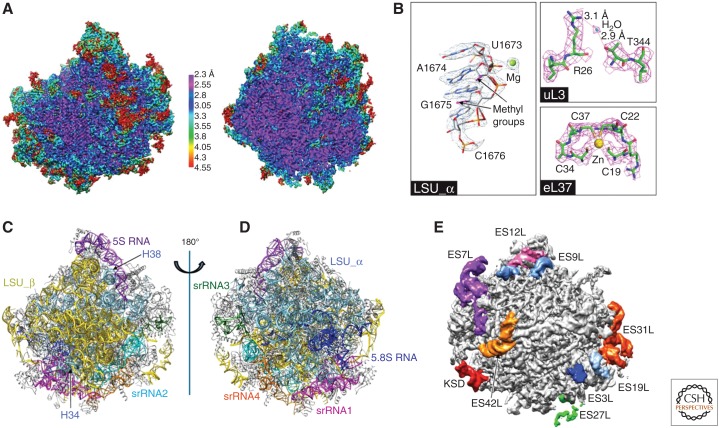
A very striking feature of trypanosomatid ribosomes is the fragmentation of the 28S rRNA in the large subunit, discovered by use of nucleic acid electrophoresis (Campbell et al. 1987). Following the terminology of Hashem et al. (2013a), we refer to the six fragments as LSU-α, LSU-β, and srRNA1 through srRNA4. The two largest pieces, LSU-α and LSU-β, are located on the solvent and interface side, respectively, and provide the scaffold for the large subunit. Locations of these rRNA components and the unique ways they are knitted together in the mature ribosome were described in various depths (Fig. 3) (Hashem et al. 2013a; Liu et al. 2016; Shalev-Benami et al. 2016; Zhang et al. 2016). The srRNA1 piece is at the bottom of the large subunit, as viewed from the solvent side, opposite the 5S rRNA. It lies close to the 5.8S rRNA, without being associated with it. The remaining three pieces srRNA2-4 are in close contact with one another and situated mainly on the left side of the large subunit under the P stalk (solvent-side view). Strikingly, despite the fragmented nature of the rRNA, the ribosomal functional cores adopt the complete 3D architecture that is common to other eukaryotic ribosomes. Although the exact mechanism of rRNA fragmentation in trypanosomatids is unknown, the high-resolution structures of the T. cruzi and L. donovani 60S large ribosomal subunit suggest that cleavage occurs during or postassembly (Liu et al. 2016; Shalev-Benami et al. 2016). Further, these structures contain clues on how these rRNA fragments are most likely assembled (Liu et al. 2016). In fact, this “archeological” approach to the interpretation of structure has resulted in a proposal for the chronology of biosynthesis of trypanosomatid ribosomes (Liu et al. 2016).
Figure 3.
Structure of the large ribosomal subunit of Trypanosoma cruzi. (A) Sharpened cryoelectron microscopy (cryo-EM) map of the 60S subunit, colored by local resolution and viewed from the subunit interface. (Left) Surface view. (Right) Central cutaway view. (B) Selected views of density for ribosomal RNA (rRNA) and proteins, with associated ions and water molecule. (C,D) rRNA architecture of the large subunit: interface (C) and solvent (D) view. (E) Some rRNA expansion segments in the unsharpened map of a large subunit, viewed from the solvent side. (From Liu et al. 2016; adapted, with permission, from the National Academy of Sciences in conjunction with Creative Commons licensing.)
Compared with other eukaryotic ribosomes, in particular with the yeast ribosome, the cytosolic ribosome in trypanosomatids shows a set of trypanosome-specific components both in RNAs and in proteins. In the large subunit, a number of rRNA kinetoplastid-specific expansion segments (ESs) are located in the periphery and are unusually large. Many r-proteins have carboxy-terminal or amino-terminal extensions. Most remarkably, the r-protein eL19 contains an extremely long carboxy-terminal extension, doubling its size compared to its yeast counterpart. In addition, kinetoplastid ribosomes lack the r-protein eL41, resulting in the absence of the eukaryote/archaea-specific intersubunit bridges eB12 and eB14. In contrast to the large differences seen in the large subunit rRNA, the small subunit is more conserved.
P. falciparum, an apicomplexan parasite transmitted by the bite of an infected female mosquito (Anopheles spp.), is the causative agent of the most severe form of human malaria. Malaria constitutes an immense global public health burden; in 2015, it caused an estimated 214 million clinical episodes and 438,000 deaths (www.who.int/mediacentre/factsheets/fs094/en). Using cryo-EM, the parasite-specific features and structural dynamics of the P. falciparum 80S (Pf80S) ribosome were recently characterized (Wong et al. 2014, 2017; Sun et al. 2015). In the first of these studies, Wong et al. (2014) solved the structure of the Pf80S ribosome bound to emetine, a eukaryotic protein synthesis inhibitor, at a resolution of 3.2 Å. The Pf80S ribosome displays extensive differences in the lengths of its rRNA expansion segments and protein extensions in comparison to the human ribosome.
As for the r-proteins, 15 of these are extended in P. falciparum when compared to yeast ribosomes, which are commonly used as a reference against other eukaryotic ribosomes. Importantly, these extensions result in unique interactions not observed in the ribosomes of the human host (Wong et al. 2014; Sun et al. 2015). For instance, r-protein eL41 has a 14-residue amino-terminal extension that reaches into a pocket formed by 18S rRNA, bridging the large and small subunits. An additional small bridge is formed between the platform of the small subunit and the region around the L1 stalk via interactions between the carboxy-terminal helix extension of eL8 and the carboxy-terminal helix eS1. Stabilizing interactions are also observed near the L1 and P stalks, likely with functional implications on translation in P. falciparum. Sun et al. (2015), with a sample purified from a schizont-stage P. falciparum cell extract, obtained five reconstructions showing the ribosome either empty or in four different tRNA-binding states, two of which (an 80S with tRNAs in the P/P and E/E sites, and a rotated 80S bound by hybrid-state A/P and P/E tRNAs) are recognizable as part of the canonical elongation cycle. This study is an example for the capability of single-particle cryo-EM to provide multiple functionally relevant structures from the same sample.
Altogether, the newly determined structures of ribosomes from human parasites provide a platform for functional studies of parasite-specific features (rRNA expansion segments, r-protein extensions, dynamics, assembly, etc.) and the development of new therapeutics. Future studies are expected to explore the binding of translation initiation, elongation, and release factors in these parasites, as they are incompletely understood. Thus, there is vast potential for high-resolution cryo-EM to characterize the functions of parasite-specific elements as well as complexes to guide the design of drugs targeting their translational machinery.
Canonical Eukaryotic Translation Initiation
Eukaryotic translation initiation occurs in four stages: 43S preinitiation complex formation, mRNA attachment, scanning and start codon recognition, and large ribosomal subunit joining. This process requires the concerted action of over a dozen protein factors at different times, with concomitant conformational changes in those factors, the ribosome, tRNA, and mRNA (Fig. 4).
Figure 4.
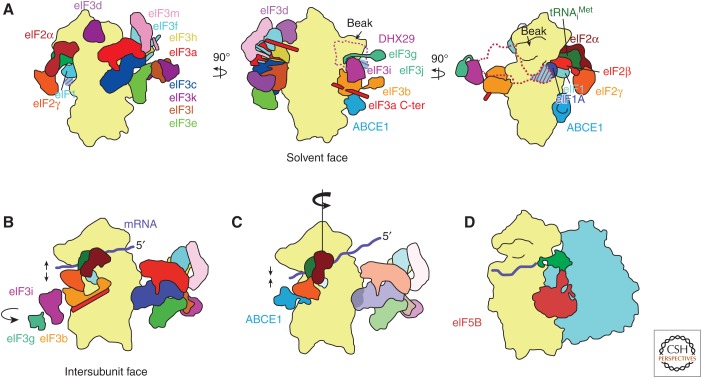
A model of eukaryotic translation initiation. (Note that eukaryotic initiation factor [eIF]4 subunits and eIF5 are not shown.) (A) 43S complex. Factors eIF1, eIF1A, ternary complex, and eIF3 bind to the 40S subunit. DHX29 is required only for initiation on highly structured messenger RNAs (mRNAs) in mammals, and is shown as a dotted outline. The binding site of the substoichiometric subunit eIF3j overlaps with part of DHX29’s binding site; the area of overlap is indicated with diagonal lines. The mammalian homolog of factor eIF3 is shown; yeast eIF3 comprises only core subunits a and c and peripheral subunits b, i, g, and j. Factor ABCE1 likely binds to the 40S subunit interface, at a conserved GTPase binding site. (B) 48S-open complex. mRNA is delivered to the complex, and start codon scanning begins. The latch between rRNA helices 18 and 24 on the 40S subunit opens to allow mRNA entry. Subunits eIF3b, i, and g move from the solvent face to the intersubunit face of the 40S subunit, likely on mRNA binding. (C) 48S-closed complex. Scanning concludes on start codon recognition in the 40S subunit P site. The 40S subunit head rotates toward the solvent side of the subunit, aiding the closure of the latch. Subunits eIF3b, i, and g have dissociated from the 40S subunit. Factor ABCE1 binds the 40S subunit interface. Initiator tRNA is accommodated fully in the P site, caused, in part, by eIF2β’s releasing its contacts with eIF1 and eIF1A in favor of contacting the 40S subunit head, moving the ternary complex toward the 40S subunit head away from the body, whereas the anticodon remains securely in the P site. Factors eIF1 (shown transparently) and the carboxy-terminal tail (CTT) of eIF1A (not shown) begin to dissociate. Factor eIF5 (not shown) catalyzes GTP hydrolysis on eIF2, after which both factors will dissociate. (D) 80S complex. Factor eIF5B catalyzes 60S subunit joining. Upon subunit joining, eIF5B will dissociate with eIF1A (not shown), leaving an elongation-competent 80S complex. (Figure based on data in des Georges et al. 2015 and Simonetti et al. 2016.)
In the field of eukaryotic initiation, structural knowledge has lagged far behind the knowledge in other steps of translation. This has been the result mainly of the low stability and structural variability of the various complexes engaged in preinitiation and their pathway toward assembly. The resulting compositional and conformational heterogeneity of such complexes poses challenges in visualization. A breakthrough was achieved with the discovery that DHX29, a newly discovered initiation factor (Pisareva et al. 2008; Parsyan et al. 2009), stabilizes the preinitiation complex to some extent, sufficient for visualization by cryo-EM (Hashem et al. 2013b). It is worth noting, however, that only 4% of the data showed the complete complex (with the exception of eIF1), and that all other classes presented complexes where some of the factors were missing. It is only because of the power of today’s classification methods that such a small portion of the data can be identified. The reason for the low yield of intact complexes is unknown, but it may be related to the low binding affinity of some of the factors and to the forces that act on the specimen during the preparation of the EM grid (Glaeser 2016). This low yield explains the relatively low resolution of the reconstruction. Nevertheless, in this work, the structure and position of eIF3 on the ribosome were well defined for the first time.
43S Preinitiation Complex
Helicase DHX29, first visualized by Hashem et al. (2013b) as part of the 43S preinitiation complex, is required to translate highly structured mRNAs bound to the mammalian 43S complex (Pisareva et al. 2008). DHX29 is positioned on the 40S subunit shoulder near the mRNA entry channel latch, consistent with a role in unwinding double-stranded stems of incoming mRNA. Additionally, the resolution of this 43S complex, 11.6Å, was sufficient to clarify the orientation of the eIF3 core octamer bound to the 40S subunit, which differed significantly from the previous understanding of its orientation (Siridechadilok et al. 2005); in its actual orientation, the eIF3 core contacts much less of the 40S subunit than previously thought.
des Georges et al. (2015) presented the first set of cryo-EM reconstructions that collectively show all but eIF3j of the 13 subunits of mammalian eIF3 in the context of the 43S complex. The PCI/MPN octameric core is attached to the platform of the 40S subunit via subunits eIF3a and eIF3c; the eIF3 core in mammals differs substantially from that in yeast, which comprises only subunits eIF3a and eIF3c. In a separate reconstruction, the investigators resolved peripheral subunits eIF3bgi on the solvent side of the 40S subunit near the shoulder. In a third reconstruction, a density seen previously (Hashem et al. 2013b) at low resolution is now visible in des Georges et al. (2015) at intermediate resolution and tentatively assigned to eIF3d, the first time a cryo-EM density is attributed to this peripheral subunit.
Subunit eIF3j was finally visualized by Aylett et al. (2015) as part of a yeast 40S•eIF1•eIF1A• eIF3 complex. These investigators enriched for complete complexes using a lysine cross-linking technique that tethered components to one another (Erzberger et al. 2014). Subunit eIF3j is a substoichiometric, nonessential component of eIF3, conserved between yeast and mammals (Valásek et al. 2001; see Browning et al. 2001); in the 40S•eIF1•eIF1A•eIF3 complex, eIF3j is situated at the mRNA entry channel on the 40S subunit shoulder at the intersubunit face, near the mRNA entry channel (Fraser et al. 2007). This binding site overlaps with that of DHX29, indicating that their binding associations in the 43S complex are mutually exclusive. Subunit eIF3j contacts eIF1A and, because eIF1 and eIF1A bind cooperatively (Maag and Lorsch 2003), the cryo-EM study by Aylett et al. (2015) is the first evidence that links binding of eIF3 to that of eIF1 and eIF1A. eIF3 makes relatively few contacts with the 40S subunit, so its interactions with eIF1 and eIF1A are particularly important.
Also visible for the first time in a cryo-EM reconstruction is the helical, spectrin-like carboxy-terminal domain of subunit eIF3a, which extends along the solvent side of the 40S subunit, from the eIF3ac subunit core situated on the platform to the peripheral eIF3bgi subunit module near the shoulder.
48S Translation Initiation Complex
For some time, it was difficult to directly assess structural differences between the stages of scanning and start codon recognition during initiation, because the two were thought to differ only by relatively small conformational changes and by start codon placement. Hussain et al. (2014) broke ground in this area, using a tRNA variant to stabilize start codon recognition to yield a reconstruction of a partial yeast 48S complex (py48S), containing the 40S subunit, eIF1, eIF1A, ternary complex (i.e., eIF2•GTP-Met-tRNAiMet or nonhydrolyzable GTP analog), and mRNA, with partial putative eIF5 occupancy. The 4 Å structure, which represents an initiation complex just after start codon recognition, presents initiator tRNA, eIF1, and the amino-terminal tail (NTT) of eIF1A together for the first time. According to this structural mapping, the initiator tRNA interacts with the mRNA start codon in the P site; the eIF1A NTT is ordered and stabilizes the codon–anticodon duplex; and eIF1 is seen in a novel conformation in which its two β-hairpins have shifted to avoid a clash with the tRNA in its current close proximity to the mRNA. Additionally, the mRNA entry channel latch between h18 on the 40S subunit head and h34 on its body (Spahn et al. 2001) is in a closed state, as expected on start codon recognition. Furthermore, the reconstruction reveals a 13° clockwise swivel of the 40S subunit head, with the beak having moved toward the solvent face of the subunit, about the axis of h28, the “neck” between the 40S subunit head and body. This swivel motion brings h29 of the 18S rRNA from the 40S subunit head into contact with the anticodon stem-loop (ASL) of initiator tRNA, thereby stabilizing the tRNA in its start codon-bound state.
Llácer et al. (2015) built on this result by introducing at 6 Å the first open-state py48S complex, omitting eIF5 and using an mRNA variant to stabilize a scanning complex (with 40S subunit, eIF1, eIF1A, ternary complex, mRNA, and parts of eIF3 visible) in addition to another py48S-closed complex at 4.9 Å similar to the one in Hussain et al. (2014) (with the same components as the open complex in addition to eIF5). Here “open” and “closed” refer to the state of the mRNA entry channel latch. The 40S head tilts away from the body to open the latch during scanning, which permits mRNA movement through the now widened channel between the 40S subunit head and neck; the latch closes after start codon recognition, to “lock” the mRNA in position.
Specifically, Llácer et al. (2015) showed that on start codon recognition, h28 is compressed, bringing the 40S subunit head closer to the body to close the latch. The 40S subunit body component of the P site is brought into contact with the initiator tRNA’s ASL, helping to accommodate tRNAi fully in the P site. Concomitantly, subunit eIF2β releases its contacts with eIF1 and eIF1A in favor of contacting the 40S subunit head. As a result, the bulk of the ternary complex moves toward the 40S subunit head and away from the body, whereas the ASL is stationary in the 40S subunit P site (Llácer et al. 2015; Simonetti et al. 2016). In conjunction with rearrangements of other initiation factors, including eIF1, eIF1A, and eIF2α, the 40S subunit head positions the initiator tRNA in the P site in a closed, scanning-arrested state.
Simonetti et al. (2016), via a late-stage mammalian 48S subunit reconstruction (containing the 40S subunit, ternary complex, mRNA, eIF3i, and eIF3g), reinterpreted Llácer et al.’s results by swapping the attributions of eIF3b and eIF3i, both of which bear WD40 domains of similar size. The reassigned factor eIF3b appears on the intersubunit face of the 40S subunit in both open and closed complexes, which suggests that it has moved from the 40S subunit solvent face before scanning and start codon recognition, and likely on mRNA attachment. The relocated eIF3b contacts eIF2γ, maintaining the ternary complex in a scanning-competent conformation, as opposed to the 40S head-proximal conformation mentioned above on start codon recognition. A follow-up analysis of the same 48S subunit reconstruction (Mancera-Martínez et al. 2017) indicates that eIF3i and eIF3g likely relocate with eIF3b from the 40S subunit solvent face to the intersubunit face as a module, although whether eIF3i+g remain bound to the 40S subunit is unclear. Importantly, the investigators additionally reassign a density on the 40S subunit intersubunit face to the NTPase ABCE1, positing that ABCE1 binds the interface on start codon recognition, after dissociation of the eIF3b (along with i+g). Moreover, ABCE1 may cycle on and off the 40S subunit throughout ribosome recycling and subsequent translation initiation, possibly acting as an anti-association factor and a gatekeeper to multiple steps of translation initiation.
Fernández et al. (2013) visualized a complex representing the result of the final step of eukaryotic initiation, eIF5B-mediated subunit joining. Their 6.6 Å structure of factor eIF5B in complex with the yeast 80S ribosome and the initiator aminoacylated tRNA in the P/I state—a conformation in the P site with the large-subunit end bound to an initiation factor—was achieved by optimizing the sample in a stepwise fashion in response to shortcomings of early reconstructions of the complex, because of partial occupancy of some components and tRNA heterogeneity. The resultant reconstruction shows that eIF5B binds tightly to ribosomal protein eL40 at the base of the P stalk, such that there would be a steric clash if eL40 were ubiquitinated. In fact, as eL40 is ubiquitinated through the late stages of 60S subunit maturation (Fernández-Pevida et al. 2012), eIF5B promotes subunit joining strictly with mature 60S subunits. Furthermore, some aspects of this complex that are conserved in bacteria—the P/I conformation of the initiator tRNA, salient intersubunit rotation, and the general conformation of ribosome-bound eIF5B—corroborate an earlier low-resolution cryo-EM reconstruction in bacteria (Allen et al. 2005) that featured eIF5B’s prokaryotic homolog IF2. This suggests that eIF5B’s role in initiation is universally conserved, which would explain why eIF5B is required even when most initiation factors are not during many types of noncanonical 5′-cap-independent initiation.
With all these pieces in place, it is possible now to put a composite model together for the stepwise progression of canonical translation initiation in eukaryotes (Fig. 4).
IRES-Mediated Initiation and Translocation
Cryo-EM has advanced the understanding of noncanonical translation initiation via IRESs. In viruses, IRESs are classified into four types, according in part to the subset of initiation factors that they require. Type 3 IRESs, which require very few initiation factors, and type 4 IRESs, which require none, depart the most from canonical requirements, and recent cryo-EM studies have helped to elucidate the mechanisms of types 3 and 4 IRES function (Kwan and Thompson 2018) and of IRES-bound ribosome behavior.
Type 3 IRESs, such as the classical swine fever virus (CSFV) IRES, bind to the 40S subunit in a way that clashes with the mammalian eIF3 core’s ribosomal contacts, eS1, eS26, and eS27. Hashem et al. (2013c) observed that the IRES resolves the clash by displacing the eIF3 core entirely from the ribosome and instead binding eIF3 via the IRES’s own domain III, holding eIF3 in a position 55 Å shifted and 60° rotated from its canonical position on the 40S subunit platform.
Initiation with the type 3 IRES appears to rely on 40S subunit rolling, a movement usually confined to the elongation stage during canonical translation. Specific to eukaryotes, rolling is a ∼6° rotation about the longest axis of the 40S subunit, orthogonal to intersubunit rotation (Budkevich et al. 2011) with the beak moving closer to the 60S subunit. Canonically, rolling occurs in the early pre-translocation state where it serves to shift an incoming tRNA in the A/T conformation, in which the 3′ acceptor end of the tRNA is not yet situated in the 60S A site, into the A/A conformation, which is fully accommodated in the A site. Reverse rolling takes place concurrently with intersubunit reverse rotation to yield the posttranslocation complex (Budkevich et al. 2014). However, Yamamoto et al. (2014) showed that rolling occurs in the final steps of hepatitis C virus (HCV) IRES-mediated initiation. The subunit-joining GTPase eIF5B recruits the 60S subunit to a 40S•HCV-IRES•Met-tRNAiMet complex in the rolled state, yielding a pre-translocation-like 80S complex presented at 8.2 Å. eIF5B•GTP hydrolysis induces reverse rolling as eIF5B begins to disengage from the complex; this posttranslocation-like state is captured in an 8.6 Å reconstruction. Yamamoto et al. propose that subsequent eIF5B dissociation yields an elongation-competent complex. Thus, the HCV IRES uses canonical-elongation-like rolling to complete initiation.
Fernández et al. (2014) obtained two reconstructions of a model type 4 IRES, the cricket paralysis virus (CrPV) intergenic IRES, bound to a yeast 80S ribosome in rotated and nonrotated states, at sufficiently high resolution (3.7 Å and 3.8 Å, respectively) to build an atomic model of the full-length IRES. They took advantage of the fact that the 80S ribosome from the yeast Kluyveromyces lactis is stable (Rodicio and Heinisch 2013) and does not aggregate at pH 6, and used it to build complexes with the CrPV IRES RNA, as RNA and especially RNA pseudoknots, which are found in the CrPV IRES, are stable at pH 6 (Li and Breaker 1999; Nixon and Giedroc 2000). Importantly, the resultant high-resolution structures revealed that the CrPV IRES binds the ribosome with its start codon-initiator tRNA mimic (Costantino et al. 2008), called pseudoknot I (PKI), in the A site rather than in the P site as the canonical start codon-initiator tRNA complex does; the 80S ribosome•CrPV IRES assembly therefore requires translocation to occur before elongation proceeds, and represents a pre-translocation complex rather than an initiation complex. In fact, the CrPV IRES-bound 80S ribosome is in equilibrium between rotated and nonrotated states (Fernández et al. 2014).
Type 3 and 4 IRESs both bind between the head and body of the 40S subunit in a way that constrains the flexibility of the 40S head position, tilting the head away from the body and thereby forcing the latch into its open position (Quade et al. 2015; Yamamoto et al. 2015; Murray et al. 2016). In this way, the IRESs mimic the induction of head tilt and latch opening by factors eIF1, eIF1A, and eIF3 during canonical initiation (Aylett et al. 2015; Llácer et al. 2015). In the case of the HCV IRES, the model type 3 IRES head tilt of about 17° persists until P-site tRNA binding occurs, which takes place after 60S subunit joining in HCV IRES-driven initiation (Yamamoto et al. 2015). The incoming tRNA displaces the HCV IRES domain II from its position as a wedge between the 40S subunit head and body, and domain II undergoes a 55° rotation resulting in contact with the 28S rRNA on the 60S subunit. In type 4 IRES-driven initiation, stem loops IV and V of the IRES insert themselves between the head and body of the 40S subunit (Fernández et al. 2014; Koh et al. 2014).
During eEF2-dependent translocation, the type 4 IRES must shift through the intersubunit space as a single flexible body. Two groups (Muhs et al. 2015; Murray et al. 2016) elucidated individual mid- and posttranslocation states, respectively, of ribosome-bound type 4 CrPV IRES. In a tour-de-force of cryo-EM data collection and classification, Abeyrathne et al. (2016) captured the entire arc of IRES translocational motion by five high-resolution (3.5 Å to 4.2 Å) cryo-EM reconstructions of the type 4 IRES from Taura syndrome virus (TSV) bound to the 80S ribosome, which was bound in turn by eEF2. The TSV IRES, in an extended conformation before translocation, compresses and then extends again in a stepwise fashion, whereas its PK1 moves from the A site to the P site. Concerted intersubunit rotation and 40S head swivel accompany the movement of the IRES. As the ribosome proceeds from the rotated state (3° more rotated than when eEF2 is absent [Murray et al. 2016]) to the nonrotated state, 40S subunit head swivel progresses from moderate (12°) to high (17°) to minimal (1°). The swiveling action toward the 60S subunit appears to align the 40S subunit head A-site region with the body P-site region to permit the IRES PK1 to advance from the A site to the P site (Abeyrathne et al. 2016) in addition to a previously known role in which swiveling permits P-site tRNA’s shift to the E site (Spahn et al. 2004; Schuwirth et al. 2005).
CONCLUSION
When compared to the knowledge of only 4 years ago, the structural information available on ribosomes from eubacteria and eukaryotes is now much richer and more detailed. Especially, the addition of information on multiple states and dynamics obtained by single-particle cryo-EM has helped to broaden the functional knowledge base in eubacterial and eukaryotic translation. As we have shown, there are beautiful examples for the elucidation of detailed mechanisms in the recent structural literature on translational control and tRNA selection in eukaryotes, and IRES-mediated initiation and translocation in eukaryotes. In all, we can look forward to a much-enriched integration of biochemical, structural, and single-molecule FRET data relating to translation than previously thought possible.
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Abeyrathne PD, Koh CS, Grant T, Grigorieff N, Korostelev AA. 2016. Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome. eLife 5: e14874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adio S, Senyushkina T, Peske F, Fischer N, Wintermeyer W, Rodnina MV. 2015. Fluctuations between multiple EF-G-induced chimeric tRNA states during translocation on the ribosome. Nat Commun 6: 7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Lei J, Brunelle JL, Ortiz-meoz RF, Green R, Frank J. 2008. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell 32: 190–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Schreiner E, Trabuco LG, Lei J, Ortiz-Meoz RF, Schulten K, Green R, Frank J. 2011. Structural insights into cognate vs. near-cognate discrimination during decoding. EMBO J 30: 1497–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Liao HY, Schreiner E, Fu J, Ortiz-Meoz RF, Schulten K, Green R, Frank J. 2012. Structural characterization of mRNA-tRNA translocation intermediates. Proc Natl Acad Sci 109: 6094–6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Fernandez IS, Kelley AC, Carton DG, Ramakrishnan V, Valle M. 2013. The ribosome triggers the stringent response by RelA via a highly distorted tRNA. EMBO Rep 14: 811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. 2005. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121: 703–712. [DOI] [PubMed] [Google Scholar]
- Andrews KT, Fisher G, Skinner-Adams TS. 2014. Drug repurposing and human parasitic protozoan diseases. Int J Parasitol Drugs Drug Resist 4: 95–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylett CHS, Boehringer D, Erzberger JP, Schaefer T, Nenad Ban N. 2015. Structure of a Yeast 40S–eIF1–eIF1A–eIF3–eIF3j initiation complex. Nat Struct Mol Biol 22: 269–271. [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen JJ, Moore PB, Steitz TA. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, de Loubresse NG, Melnikov S, Jenner L, Yusupova G, Yusupov M. 2011. The structure of the Eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529. [DOI] [PubMed] [Google Scholar]
- Boel G, Smith PC, Ning W, Englander MT, Chen B, Hashem Y, Testa AJ, Fischer JJ, Wieden HJ, Frank J, et al. 2014. The ABC-F protein EttA gates ribosome entry into the translation elongation cycle. Nat Struct Mol Biol 21: 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N. 2013. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc Natl Acad Sci 110: 20994–20999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Fernández IS, Gordiyenko Y, Ramakrishnan V. 2016. Ribosome-dependent activation of stringent control. Nature 534: 277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning KS, Gallie DR, Hershey JW, Hinnebusch AG, Maitra U, Merrick WC, Norbury C. 2001. Unified nomenclature for the subunits of eukaryotic initiation factor 3. Trends Biochem Sci 26: 284. [DOI] [PubMed] [Google Scholar]
- Budkevich T, Giesebrecht J, Altman RB, Munro JB, Mielke T, Nierhaus KH, Blanchard SC, Spahn CMT. 2011. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol Cell 44: 214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budkevich TV, Giesebrecht J, Behrmann E, Loerke J, Ramrath DJF, Mielke T, Ismer J, Hildebrand PW, Tung C-S, Nierhaus KH, et al. 2014. Regulation of the mammalian elongation cycle by subunit rolling: A eukaryotic-specific ribosome rearrangement. Cell 158: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DA, Kubo K, Graham Clark C, Boothroyd JC. 1987. Precise identification of cleavage sites involved in the unusual processing of trypanosome ribosomal RNA. J Mol Biol 196: 113–124. [DOI] [PubMed] [Google Scholar]
- Chen C, Stevens B, Kaur J, Cabral D, Liu H, Wang Y, Zhang H, Rosenblum G, Smilansky Z, Goldman YE, et al. 2011. Single-molecule fluorescence measurements of ribosomal translocation dynamics. Mol Cell 42: 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Boel G, Hashem Y, Ning W, Fei J, Wang C, Gonzalez RL Jr, Hunt JF, Frank J. 2014. EttA regulates translation by binding the ribosomal E site and restricting ribosome-tRNA dynamics. Nat Struct Mol Biol 21: 152–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Cui X, Beausang JF, Zhang H, Farrell I, Cooperman BS, Goldman YE. 2016. Elongation factor G initiates translocation through a power stroke. Proc Natl Acad 113: 7515–7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Noller HF, Ha T. 2008. Spontaneous intersubunit rotation in single ribosomes. Mol Cell 30: 578–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. 2008. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat Struct Mol Biol 15: 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Loubresse NG, Prokhorova I, Holtkamp W, Rodnina MV, Yusupova G, Yusupov M. 2014. Structural basis for the inhibition of the eukaryotic ribosome. Nature 513: 517–522. [DOI] [PubMed] [Google Scholar]
- Demo G, Svidritskiy E, Madireddy R, Diaz-Avalos R, Grant T, Grigorieff N, Sousa D, Korostelev AA. 2017. Mechanism of ribosome rescue by ArfA and RF2. eLife 10.7554/eLife.23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- des Georges A, Dhote V, Kuhn L, Hellen CU, Pestova TV, Frank J, Hashem Y. 2015. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature 525: 491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Stengel F, Pellarin R, Zhang S, Schaefer T, Aylett CHS, Cimermančič P, Boehringer D, Sali A, Aebersold R, et al. 2014. Molecular architecture of the 40S•eIF1•eIF3 translation initiation complex. Cell 158: 1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández IS, Bai XC, Hussain T, Kelley AC, Lorsch JR, Ramakrishnan V, Scheres SH. 2013. Molecular architecture of a eukaryotic translational initiation complex. Science 342: 1240585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández IS, Bai XC, Murshudov G, Scheres SH, Ramakrishnan V. 2014. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell 157: 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Pevida A, Rodríguez-Galán O, Díaz-Quintana A, Kressler D, de la Cruz J. 2012. Yeast ribosomal protein L40 assembles late into precursor 60S ribosomes and is required for their cytoplasmic maturation. J Biol Chem 287: 38390–383407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. 2010. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature 466: 329–333. [DOI] [PubMed] [Google Scholar]
- Fischer N, Neumann P, Konevega AL, Bock LV, Ficner R, Rodnina MV, Stark H. 2015. Structure of the E. coli ribosome–EF-Tu complex at <3 Å resolution by Cs-corrected cryo-EM. Nature 520: 567–570. [DOI] [PubMed] [Google Scholar]
- Frank J. 2017. The translation elongation cycle—Capturing multiple states by cryo-electron microscopy. Philos Trans R Soc Lond B Biol Sci 372: 20160180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. 2007. The process of mRNA-tRNA translocation. Proc Natl Acad Sci 104: 19671–19678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CS, Berry KE, Hershey JW, Doudna JA. 2007. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol Cell 26: 811–819. [DOI] [PubMed] [Google Scholar]
- Gao H, Ayub MJ, Levin MJ, Frank J. 2005. The structure of the 80S ribosome from Trypanosoma cruzi reveals unique rRNA components. Proc Natl Acad Sci 102: 10206–10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaeser RM. 2016. How good can cryo-EM become? Nat Methods 13: 28–32. [DOI] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Fu J, Buss SN, Jossinet F, Jobe A, Zhang Q, Liao HY, Grassucci RA, Bajaj C, et al. 2013a. High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome. Nature 494: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HL, Grassucci RA, Hellen CUT, Pestova TV, Frank J. 2013b. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell 153: 1108–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU, Frank J. 2013c. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature 503: 539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain T, Llácer JL, Fernández IS, Munoz A, Martin-Marcos P, Savva CG, Lorsch JR, Hinnebusch AG, Ramakrishnan V. 2014. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell 159: 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julián P, Koneveg AL, Scheres SHW, Lázaro M, Gil D, Wintermeyer W, Rodnina MV, Valle M. 2008. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci 105: 16924–16927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katunin VI, Savelsbergh A, Rodnina MV, Wintermeyer W. 2002. Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry 41: 12806–12812. [DOI] [PubMed] [Google Scholar]
- Khatter H, Myasnikov AG, Natchiar SK. 2015. Structure of the human 80S ribosome. Nature 520: 640–645. [DOI] [PubMed] [Google Scholar]
- Kim HK, Tinoco I Jr. 2017. EF-G catalyzed translocation dynamics in the presence of ribosomal frameshifting stimulatory signals. Nucl Acids Res 45: 2865–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. 2011. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334: 941–948. [DOI] [PubMed] [Google Scholar]
- Klinge S, Voigts-Hoffmann M, Leibundgut M, Ban N. 2012. Atomic structures of the eukaryotic ribosome. Trends Biochem Sci 37: 189–198. [DOI] [PubMed] [Google Scholar]
- Koh CS, Brilot AF, Grigorieff N, Korostelev AA. 2014. Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center. Proc Natl Acad Sci 111: 9139–9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Chen Y, Ero R, Ahmed T, Tan J, Li Z, Wong ASW, Bhushan S, Gao YG. 2015. Structure of BipA in GTP form bound to the ratcheted ribosome. Proc Natl Acad Sci 112: 10944–10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Kwan T, Thompson SR. 2018. Noncanonical translation initiation in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Breaker RR. 1999. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J Am Chem Soc 121: 5364–5372. [Google Scholar]
- Li W, Liu Z, Koripella RK, Langlois R, Sanyal S, Frank J. 2015. Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G. Sci Adv 1: e1500169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Gagnon MG, Bulkley D, Steitz TA. 2015. Conformational changes of elongation factor G on the ribosome during tRNA translocation. Cell 160: 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, Ermolenko DN. 2016. Structural insights into ribosome translocation. WIREs RNA 7: 620–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Song G, Zhang D, Zhang D, Li Z, Lyu Z, Dong J, Achenbach J, Gong W, Zhao XS, et al. 2014. EF-G catalyzes tRNA translocation by disrupting interactions between decoding center and codon–anticodon duplex. Nat Struct Mol Biol 21: 817–824. [DOI] [PubMed] [Google Scholar]
- Liu Z, Gutierrez-Vargas C, Wei J, Grassucci RA, Ramesh M, Espina N, Sun M, Tutuncuoglu B, Madison-Antenucci S, Woolford JL, et al. 2016. Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit. Proc Natl Acad Sci 113: 12174–12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Gutierrez-Vargas C, Wei J, Grassucci RA, Sun M, Espina N, Madison-Antenucci S, Tong L, Frank J. 2017. Determination of the ribosome structure to a resolution of 2.5 Å by single-particle cryo-EM. Prot Sci 26: 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llácer JL, Hussain T, Marler L, Aitken CE, Thakur A, Lorsch JR, Hinnebusch AG, Ramakrishnan V. 2015. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol Cell 59: 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland AB, Bah E, Madireddy R, Zhang Y, Brilot AF, Grigorieff N, Korostelev AA. 2016. Ribosome•RelA structures reveal the mechanism of stringent response activation. eLife 10.7554/eLife.17029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland AB, Demo G, Grigorieff N, Korostelev AA. 2017. Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature 546: 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D, Lorsch JR. 2003. Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J Mol Biol 330: 917–924. [DOI] [PubMed] [Google Scholar]
- Mancera-Martínez E, Querido J, Valasek LS, Simonetti A, Hashem Y. 2017. ABCE1: A special factor that orchestrates translation at the crossroad between recycling and initiation. RNA Biol 10.1080/15476286.2016.1269993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall AW, Chang JJ, Freeman R, Lepault J, Walter CA, Dubochet J. 1983. Electron microscopy of frozen hydrated sections of vitreous ice and vitrified biological samples. J Microsc 131: 1–9. [DOI] [PubMed] [Google Scholar]
- McMullan G, Chen S, Henderson R, Faruqi A. 2009. Detective quantum efficiency of electron area detectors in electron microscopy. Ultramicroscopy 109: 1126–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhs M, Hilal T, Mielke T, Skabkin MA, Sanbonmatsu KY, Pestova TV, Spahn CMT. 2015. Cryo-EM of ribosomal 80S complexes with termination factors reveals the translocated cricket paralysis virus IRES. Mol Cell 57: 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Sanbonmatsu KY, Spahn CMT, Blanchard SC. 2009. Navigating the ribosome’s metastable energy landscape. Trends Biochem Sci 34: 390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J, Savva CG, Shin BS, Dever TE, Ramakrishnan V, Fernández IS. 2016. Structural characterization of ribosome recruitment and translocation by type IV IRES. eLife 10.7554/eLife.13567.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon PL, Giedroc DP. 2000. Energetics of a strongly pH dependent RNA tertiary structure in a frameshifting pseudoknot. J Mol Biol 296: 659–671. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902. [DOI] [PubMed] [Google Scholar]
- Parsyan A, Shahbaziana D, Martineaua Y, Petroulakisa E, Alaina T, Larssona O, Mathonneta G, Tettweilera G, Hellen CU, Pestova TV, et al. 2009. The helicase protein DHX29 promotes translation initiation, cell proliferation, and tumorigenesis. Proc Natl Acad Sci 106: 22217–22222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penczek PA, Frank J, Spahn CM. 2006. A method of focused classification, based on the bootstrap 3D variance analysis, and its application to EF-G-dependent translocation. J Struct Biol 154: 184–194. [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. 2008. Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell 135: 1237–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quade N, Boehringer D, Leibundgut M, van den Heuvel J, Ban N. 2015. Cryo-EM structure of Hepatitis C virus IRES bound to the human ribosome at 3.9 Å resolution. Nat Commun 6: 8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. 2011. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331: 730–736. [DOI] [PubMed] [Google Scholar]
- Rodicio R, Heinisch JJ. 2013. Yeast on the milky way: Genetics, physiology and biotechnology of Kluyveromyces lactis. Yeast 30: 165–177. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W. 2016. Protein elongation, co-translational folding and targeting. J Mol Biol 428: 2165–2185. [DOI] [PubMed] [Google Scholar]
- Salsi E, Farah E, Netter Z, Dann J, Ermolenko DN. 2015. Movement of elongation factor G between compact and extended conformations. J Mol Biol 427: 454–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. 2016. Processing of structurally heterogeneous cryo-EM data in RELION. Methods Enzymol 579: 125–157. [DOI] [PubMed] [Google Scholar]
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon H, Franceschi F, et al. 2000. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102: 615–623. [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310: 827–834. [DOI] [PubMed] [Google Scholar]
- Simonetti A, Brito Querido J, Myasnikov AG, Mancera-Martínez E, Renaud A, Kuhn L, Hashem Y. 2016. eIF3 peripheral subunits rearrangement after mRNA binding and start-codon recognition. Mol Cell 63: 206–217. [DOI] [PubMed] [Google Scholar]
- Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. 2005. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310: 1513–1515. [DOI] [PubMed] [Google Scholar]
- Shalev-Benami M, Zhang Y, Matzov D, Halfon Y, Zackay A, Rozenberg H, Zimmerman E, Bashan A, Jaffe CL, Yonath A, et al. 2016. 2.8 Å cryo-EM structure of the large ribosomal subunit from the eukaryotic parasite Leishmania. Cell Rep 16: 288–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma H, Adio S, Senyushkina T, Belardinelli R, Peske F, Rodnina MV. 2016. Kinetics of spontaneous and EF-G-accelerated rotation of ribosomal subunits. Cell Rep 16: 2187–2196. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291: 1959–1962. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jørgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JPG, Frank J. 2004. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J 23: 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Li W, Blomqvist K, Das S, Hashem Y, Dvorin JD, Frank J. 2015. Dynamical features of the Plasmodium falciparum ribosome during translation. Nucl Acids Res 43: 10515–10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DJ, Nilsson J, Merrill AR, Andersen GR, Nissen P, Frank J. 2007. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J 26: 2421–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourigny DS, Fernandez IS, Kelley AC, Ramakrishnan V. 2013. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science 340: 1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valásek L, Phan L, Schoenfeld LW, Valásková V, Hinnebusch AG. 2001. Related eIF3 subunits TIF32 and HCR1 interact with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J 20: 891–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, Frank J. 2009. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc Natl Acad Sci 106: 1063–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Hegde RS. 2016. Structure of the Sec61 channel opened by a signal sequence. Science 351: 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Ramakrishnan V. 2013. Structural basis of the translational elongation cycle. Annu Rev Biochem 82: 203–236. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Fernández IS, Scheres SHW, Hegde RS. 2013. Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell 157: 1632–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Fernandez IS, Scheres SHW, Hegde RS. 2014. Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell 157: 1632–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman MR, Alejo, JL, Altman, RB, Blanchard, SC. 2016. Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat Struct Mol Biol 23: 333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN, Doudna Cate JH. 2012. The structure and function of the Eukaryotic ribosome. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339. [DOI] [PubMed] [Google Scholar]
- Wong W, Bai X, Brown A, Fernandez IS, Hanssen E, Condron M, Tan YH, Baum J, Scheres SHW. 2014. Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine. eLife 3: e03080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W, Bai XC, Sleebs BE, Triglia T, Brown A, Thompson JK, Jackson KE, Hanssen E, Marapana DS, Fernandez IS, et al. 2017. Mefloquine targets the Plasmodium falciparum 80S ribosome to inhibit protein synthesis. Nat Microbiol 2: 17031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Unbehaun A, Loerke J, Behrmann E, Collier M, Bürger J, Mielke T, Spahn CM. 2014. Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA. Nat Struct Mol Biol 21: 721–727. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Collier M, Loerke J, Ismer J, Schmidt A, Hilal T, Sprink T, Yamamoto K, Mielke T, Bürger J, et al. 2015. Molecular architecture of the ribosome-bound hepatitis C virus internal ribosomal entry site RNA. EMBO J 34: 3042–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupova G, Yusupov M. 2014. High-resolution structure of the eukaryotic 80S ribosome. Ann Rev Biochem 83: 467–486. [DOI] [PubMed] [Google Scholar]
- Yusupova G, Yusupov M. 2017. Crystal structure of eukaryotic ribosome and its complexes with inhibitors. Philos Trans R Soc Lond B Biol Sci 372: 20160184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R. 2009. Quality control by the ribosome following peptide bond formation. Nature 457: 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Dunkle J, Cate JHD. 2009. Structures of the ribosome in intermediate states of ratcheting. Science 325: 1014–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lai M, Chang W, Yu I, Ding K, Mrazek J, Ng HL, Yang OO, Maslov DA, Zhou ZH. 2016. Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes. Nat Commun 7: 13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, Noller HF. 2014. How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science 345: 1188–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]