Abstract
Hepatitis E virus (HEV) possesses many of the features of other positive-stranded RNA viruses but also adds HEV-specific nuances, making its virus–host interactions unique. Slow virus replication kinetics and fastidious growth conditions, coupled with the historical lack of an efficient cell culture system to propagate the virus, have left many gaps in our understanding of its structure and replication cycle. Recent advances in culturing selected strains of HEV and resolving the 3D structure of the viral capsid are filling in knowledge gaps, but HEV remains an extremely understudied pathogen. Many steps in the HEV life cycle and many aspects of HEV pathogenesis remain unknown, such as the host and viral factors that determine cross-species infection, the HEV-specific receptor(s) on host cells, what determines HEV chronicity and the ability to replicate in extrahepatic sites, and what regulates processing of the open reading frame 1 (ORF1) nonstructural polyprotein.
Hepatitis E virus (HEV) is one of the leading causes of acute viral hepatitis worldwide. In developing countries with poor sanitation conditions, an estimated 20 million infections occur annually with approximately 3.3 million acute HEV cases, resulting in about 56,600 deaths per year (Lozano et al. 2012; Rein et al. 2012). In industrialized countries, clinical cases of HEV tend to be more sporadic, with zoonotic transmission via direct contact with infected animals or consumption of contaminated animal meats being the most likely routes of infection. Domestic animals, including pigs, deer, and rabbits, harbor the zoonotic genotype (gt)3 HEV, which can contaminate commercial meat products bound for human consumption (Feagins et al. 2007; Cossaboom et al. 2011, 2016). The true incidence of HEV infection within the United States is unknown owing to the lack of Food and Drug Administration (FDA)–approved diagnostics for testing patients. Further complicating diagnosis, the incubation period of hepatitis E following exposure to the virus ranges from 2 to 10 weeks. The symptoms of acute hepatitis E are indistinguishable from other types of acute viral hepatitis, which include mild fever, anorexia, nausea, and vomiting lasting for days. Less common symptoms include abdominal pain, skin rash, or joint pain. In addition, patients can develop jaundice with dark urine and hepatomegaly. In rare instances, fulminant hepatitis may also occur (Goel and Aggarwal 2016). The rate of fulminant hepatitis greatly increases in HEV-infected women who are in their second or third trimester of pregnancy, resulting in acute liver failure, fetal loss, and mortality rates as high as 25%. More recently, chronic HEV infections have become a significant clinical problem in immunosuppressed populations, especially in solid organ transplant recipients (Kamar et al. 2011, 2013; Kamar and Izopet 2014; Fang and Han 2017), which could allow the virus to acquire mutations that could potentially enhance pathogenicity and transmissibility (van Tong et al. 2016).
CLASSIFICATION
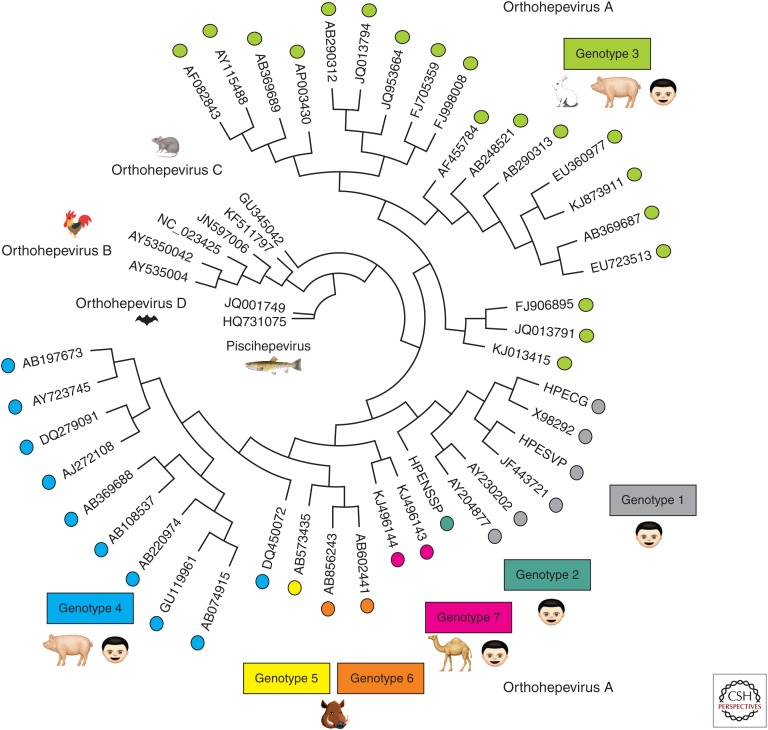
HEV is a single-stranded positive-sense RNA virus classified within the Hepeviridae family, of which there are two genera based on sequence divergence: Orthohepevirus and Piscihepevirus. The genus Piscihepevirus contains a single member known as the cutthroat trout virus (Batts et al. 2011). The genus Orthohepevirus includes avian and mammalian HEVs and are subdivided into four distinct species, Orthohepevirus A–D. These species are classified based on host range and sequence identities (Fig. 1). The species Orthohepevirus A includes at least seven HEV genotypes (HEV-1 to HEV-7), which collectively infect humans and a number of other animal species, including pigs, boars, deer, mongooses, rabbits, and camels (Smith et al. 2016). gt1 to 4 infect humans, with gt1 and gt2 being restricted to humans and gt3 and gt4 being zoonotic, infecting both humans and several other animal species. HEV-5 and HEV-6 have only been identified in wild boars thus far (Smith et al. 2014). HEV-7 from camelids has recently been reported to infect humans and has caused chronic disease in a liver transplant patient (Lee et al. 2016). Orthohepevirus B species was originally known as the avian HEV and primarily infects chickens, causing hepatitis-splenomegaly syndrome with decreased egg production (Haqshenas et al. 2001). More recently, members of the Orthohepevirus B species have also been detected in wild birds (Reuter et al. 2016; Zhang et al. 2017). Avian HEV does not appear to be able to infect mammals (Yugo et al. 2014). The Orthohepevirus C species consists of isolates from rat, greater bandicoot, Asian musk shrew, ferret, and mink, but also is not thought to infect humans. The Orthohepevirus D species consists of isolates found in bats (Smith et al. 2015). The taxonomy of the Hepeviridae family will continue to evolve as more genetically divergent strains are identified from an expanding host of animal species.
Figure 1.
A phylogenetic tree encompassing all of the newly International Committee for Taxonomy of Viruses (ICTV)–recognized reference strains of the family Hepeviridae. All recently proposed reference strains for hepatitis E virus (HEV) (Smith et al. 2014, 2016) were aligned and placed into a circular phylogenetic tree using Geneious software v10.2.2 by Biomatters (see geneious.com). Sequence alignment parameters included global alignment with free end gaps with a cost matrix of 65% similarity. The tree was generated using the Tamura-Nei genetic distance model with a neighbor joining tree build method. The more divergent strains of HEV are located toward the center (Piscihepevirus) and the more conserved strains are located around the circumference (Orthohepevirus A strains). The Orthohepevirus A strains are color-coded to match their respective genotype. Cartoons depict which host species the viruses are known to infect. Orthohepevirus C are also able to infect ferrets, mink, Asian musk shrew, and greater bandicoot (Smith et al. 2014). Orthohepevirus B are reported to infect turkeys, little egrets (Reuter et al. 2016), and wild birds (Zhang et al. 2017).
PROPERTIES OF VIRIONS AND VIRION STRUCTURE
The HEV virion is a small (320–340 Å) particle (Balayan et al. 1983) with a T = 3 icosohedral capsid lattice comprised of 180 copies of the capsid protein (Xing et al. 1999, 2010; Guu et al. 2009). Crystal structures revealed three functional domains named S (shell), M (middle), and P (protruding). The S domain is the most conserved region among HEV genotypes (Zhai et al. 2006) and forms the icosahedral shell serving as the base for the M and P domains. The S domain crystal structure reveals a jelly-roll fold common to many small RNA viruses (Rossmann and Johnson 1989). The P domain is hypothesized to be the binding site for the cellular receptor and is recognized by neutralizing antibodies (He et al. 2008). The P domain is further divided into the P1 region, which has threefold protrusions, and the P2 region, which has twofold spikes. Both P regions possess β-barrel folds and potential polysaccharide-binding sites that are postulated to help in cell–receptor binding and/or capsid disassembly (Guu et al. 2009). The M domain interacts strongly with both the S and P domains, and likely contributes to virion particle stability (Xing et al. 2010). The amino-terminal region of the capsid protein appears to be critical for forming the T = 3 icosahedral structure, as capsid protein expression products lacking the amino-terminal 111 amino acids assemble into virus-like particles (VLPs) with a T = 1 symmetry that are smaller than the native virions. In addition, the carboxy-terminal region of the capsid, although not involved in virion morphology, is required for virus replication. Virus particles lacking the 52 carboxy-terminal amino acids showed defective RNA encapsidation and had less stable virions (Shiota et al. 2013).
GENOME STRUCTURE
Genome Organization
The mammalian HEV genome is a single-stranded, positive-sense RNA ∼7200 nucleotides in length, whereas the avian HEV genome is ∼6650 nucleotides long (Huang et al. 2004). HEV genome RNA possesses a 5′ 7-methylguanosine cap structure followed by a short 5′ untranslated region (UTR) of 26 nucleotides, three major open reading frames ([ORFs]: ORF1, ORF2, and ORF3), and a 3′UTR (Fig. 2) (Zhang et al. 2001). ORF1 encodes the viral nonstructural polyprotein. There is an RNA structural element in the upstream region of ORF1 that binds to the capsid protein and is thought to be the RNA packaging signal (Surjit et al. 2004). Overlapping the X and helicase (Hel) domains in gt1 HEV is a small frameshifted ORF4 that is controlled by an internal ribosome entry site (IRES)-like RNA structure that directs translation of ORF4 under endoplasmic reticulum (ER) stress. There is a junction region between ORF1 and ORF3 containing cis-active elements thought to control the expression of a subgenomic bicistronic messenger RNA (mRNA) (Fig. 2) (Graff et al. 2006; Huang et al. 2007). Both the sequence and the stem-loop structure in the junction region play important roles in HEV replication (Cao et al. 2010). This bicistronic mRNA encodes the ORF3 multifunctional protein (vp13), and the ORF2 capsid protein. ORF3 substantially overlaps the 5′ coding sequence of ORF2 in a separate reading frame, but neither ORF3 nor ORF2 overlaps ORF1. Following ORF2 is the 3′UTR, which terminates in a 3′ polyadenylated tail. A 3′ cis-active element is found within the 3′UTR, which overlaps the carboxy-terminal sequence of the ORF2, and is necessary for viral RNA replication by binding to the RNA-dependent RNA polymerase (RdRp) domain of the ORF1 polyprotein (Agrawal et al. 2001).
Figure 2.
Genomic organization of hepatitis E virus (HEV). The genome of HEV is ∼7.2 kb in length and is composed of a single-stranded positive-sense RNA molecule. The genome contains a 7-methylguanosine RNA cap at the 5′ end and is polyadenylated at the 3′ terminus. There are three conserved open reading frames (ORFs) found in all known HEV strains: ORF1, ORF2, and ORF3. ORF1 encodes the nonstructural polyproteins with putative functional domains including methyltransferase (Met), Y domain, papain-like cysteine protease (PCP), hypervariable region (HVR), helicase (Hel), and RNA-dependent RNA polymerase (RdRp). ORF2 encodes the capsid structural protein. ORF3 encodes a multifunctional phosphoprotein (also known as VP13). ORF2 and ORF3 proteins are translated from a bicistronic subgenomic RNA 2.2 kb in length. In addition to these three ORFs, the genotype (gt)1 HEV encodes an ORF4, which generates a protein from an internal ribosome entry site (IRES)-like element in response to endoplasmic reticulum (ER) stress. This ORF4 protein is a viral replication enhancer. A number of RNA structural elements within the HEV genome contribute to RNA packaging (5′ RNA stem loops), translation of ORF2, ORF3 (junction region CREs), and ORF4 (IRES-like element), and in binding of the genomic RNA to the RdRp (3′ CRE). Positions and lengths of ORFs and indicated features are based on the prototypical gt1 HEV (Sar55 strain) sequence (GenBank accession number AF444002.1).
ORF1
ORF1 is the largest ORF in the HEV genome with ∼5082 nucleotides that can be translated directly from the viral genome to produce a polyprotein containing the viral nonstructural proteins (Tsarev et al. 1992; Emerson et al. 2001). The functional domains within ORF1 include: methyltransferase (Met), Y domain, papain-like cysteine protease (PCP), hypervariable region (HVR) or proline-rich region (PRR), X domain, Hel, and the RdRp mentioned above (Fig. 2). Although ORF1 appears to be related to Rubi-like viruses such as Rubivirus, Betatetravirus, or Benyvirus, whether the ORF1 polyprotein requires processing for functionality remains debatable (Ansari et al. 2000; Ropp et al. 2000; Sehgal et al. 2006; Suppiah et al. 2011; Perttila et al. 2013). An initial study in which ORF1 was expressed via a vaccinia virus system showed cleavage by the PCP (Ropp et al. 2000), but the same group later showed that ORF1 processing was deficient in HEK293T cells (Suppiah et al. 2011). When ORF1 was expressed in Escherichia coli and cell-free systems based on HepG2 cell lysates, no processing was detected (Ansari et al. 2000). Another study using a cell-free translation system showed the appearance of two ORF1 products not dependent on the presence of the PCP domain, but failed to show these products in radiolabeled human cells (Perttila et al. 2013). However, additional studies have shown that transfection of HepG2 cells with RNA derived from an infectious complementary DNA (cDNA) clone of HEV produced cleaved products corresponding to the sizes of Met, Hel, and RdRp (Panda et al. 2000). When expressed via a baculovirus expression system in insect cells, processing appears to occur and could be inhibited by E-64d, a cysteine protease inhibitor. More recently, an E. coli–derived PCP was shown to process ORF1 in vitro (Paliwal et al. 2014) and expression of an HEV replicon in a subclone of Huh7 human liver cells (S10-3) resulted in cleavage products (Parvez 2013). Taken together, these data suggest that ORF1 is likely processed in a manner that is fastidious—requiring specific concentrations of ORF1 polyprotein, the correct cellular conditions, and potentially viral-specific contributions to properly function.
Methyltransferase
The HEV Met domain lies within amino acids 60–240 (Koonin et al. 1992; Emerson et al. 2001; Zhang et al. 2001), and appears similar to that of the family Bromoviridae within the α-like supergroup of RNA viruses (van der Poel et al. 2001). The predicted HEV Met domain contains an invariant histidine residue, an aspartic acid-X-X-arginine amino acid motif, and an invariant tyrosine amino acid, as would be expected from known methyltransferase motifs I, II, and IV, respectively (Rozanov et al. 1992). Additionally, expression of ORF1 cDNA in insect cells produces a 110 kDa protein and a putative proteolytic product of 80 kDa. The 110 kDa protein possesses guanine-7-methyltransferase and guanylyl transferase activity in in vitro assays (Magden et al. 2001).
Y Domain
The Y domain spans amino acids 216 to 442 but its function remains elusive. Sequence analysis of a number of the alphavirus-like superfamily of single-stranded positive-sense RNA viruses suggests that the Y domain is an extension of the methyltransferase carboxy-terminal (core) region (Ahola and Karlin 2015). The HEV Y domain possesses a potential palmitoylation site and a conserved α-helical segment. Amino acids Cys-336, Cys-337, and Trp-413 were found to be universally conserved among HEV strains and critical for viral RNA replication (Parvez 2017). It was suggested that these conserved residues in the Y domain may be involved in targeting of the viral protein to membranes of the replication complex. Disruption of secondary RNA structures without altering protein-coding sequences also resulted in replication defects, thus suggesting that RNA structure within the Y domain is important for virus replication (Parvez 2017).
Papain-Like Cysteine Protease
The functions of the PCP domain in HEV ORF1 protein processing and the HEV life cycle remains controversial (Fig. 3). What is known thus far is that the PCP domain of HEV has some sequence similarity to the protease domain of rubella virus (Koonin et al. 1992). In rubella virus, the PCP proteolytically processes the nonstructural proteins (NSPs) both in cis and trans, and is susceptible to mutation of Cys-1152 within the catalytic domain, which inhibits NSP processing (Marr et al. 1994; Liang and Gillam 2000). HEV ORF1 expressed via a vaccinia virus system was not affected by mutation of Cys-483 within the putative catalytic domain (Ropp et al. 2000). More recently, it was shown that mutation of cysteine residues at 457, 459, 471, 472, 481, 483, and histidine residues at 443, 497, and 590 within the PCP domain abolished HEV RNA replication in a gt1 HEV-based replicon system in Huh7 cells (Parvez 2013). However, these mutants were not directly tested for ORF1 processing ability. In addition, PCP expressed in an E. coli system has the ability to cleave in vitro–translated ORF1 protein, and this processing was sensitive to treatment with protease inhibitors, suggesting that PCP is a bona fide chymotrypsin-like protease (Paliwal et al. 2014). Further validation of PCP protease activity is necessary to definitively understand its role in the HEV life cycle.
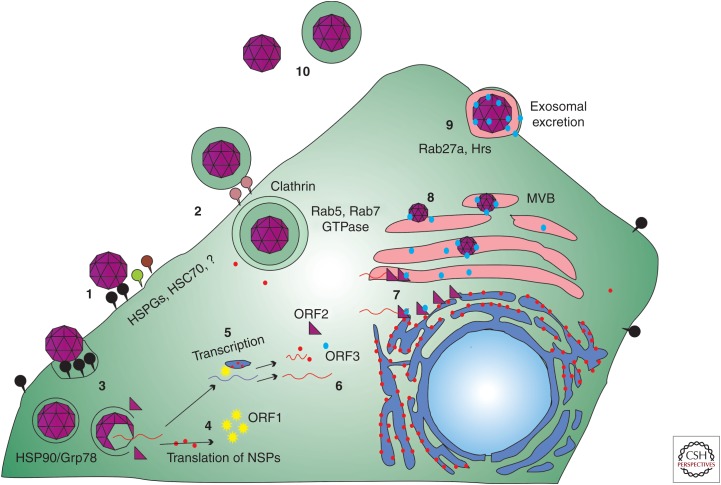
Figure 3.
Life cycle of hepatitis E virus (HEV). (1) Nonenveloped HEV virions bind to cellular receptors. Heparin sulfate proteoglycans (HSPGs) and heat shock cognate protein 70 (HSC70) are thought to be the attachment receptors for HEV but more research is required to determine the entry receptors for HEV. Once bound to the cell, virions are taken into the cell through a dynamin-2, clathrin, and cholesterol-dependent process. (2) Quasi-enveloped HEV virions are capable of entering the cell via a clathrin-mediated pathway. This entry pathway appears to be dependent on Rab 5 and Rab7 GTPases and, unlike the unenveloped HEV virions, also requires lysosome function before uncoating within the host cell. (3) Heat shock proteins including HSP90 and Grp78 have been implicated in transporting virions to sites of genome release. (4) Once released into the cytoplasm, the positive-strand genomic RNA serves as messenger RNA (mRNA) for the translation of the open reading frame (ORF)1 nonstructural polyprotein. (5) Whether or not processing occurs for the ORF1 polyprotein remains debatable, but the viral nonstructural proteins within ORF1 transcribe the positive-sense RNA genome into a negative-strand intermediate. This step occurs at either the rough endoplasmic reticulum (RER) or RER-derived membrane vesicles and involves host proteins, including elongation initiation factors, and viral proteins, including ORF4 for genotype (gt)1 HEV replication. The subgenomic bicistronic mRNA is generated from the negative-stranded RNA and serves as messenger for translation of ORF2 and ORF3 proteins. (6) Host ribosomes translate the newly synthesized RNAs into more nonstructural proteins and the ORF2 and ORF3 proteins. (7) There are a number of unknown steps to virus assembly. ORF2 binds to viral genomic RNA and begins to multimerize. ORF3 also binds to ORF2 during transition to the plasma membrane. (8) Particles become membrane-associated likely by budding into intracellular vesicles via interaction of the ORF3 protein with host vacuolar sorting proteins such as tumor suppressor gene 101. These enveloped particles retain the trans-Golgi network protein 2 protein and CD63 on their lipid surface until particle release. (9) The plasma membrane serves as the final site for release of enveloped virions via the exosomal release pathway and is sensitive to disruption of Rab27a and Hrs. (10) At some point during transit through the host digestive system, the viral envelope is lost. HEV released into the bloodstream appears to retain its quasi-envelope, which is thought to mask the capsid shell from the host immune system.
The PCP domain has been implicated in immune evasion strategies of HEV. It has been suggested that HEV PCP acts as an antagonist to interferon (IFN)-stimulated gene 15 (ISG15) when expressed as a fusion protein containing both the Met and PCP domains (Karpe and Lole 2011). The PCP domain was shown be an IFN-β antagonist having a deubiquitinase activity that limits activation of the signaling proteins retinoic-acid-inducible gene-I (RIG-I) and TBK-1. Reduced levels of ubiquitin on RIG-I and TBK-1 reduce activation of the IFN-β promoter.
Hypervariable Region
Following the PCP domain is a region known as the HVR, described as such because of its considerable sequence diversity and length variation among HEV strains (Tsarev et al. 1992; Pudupakam et al. 2009; Smith et al. 2013). The HVR has also been referred to as the polyproline region (PPR) or PRR because of the existence of large numbers of proline residues within this region. The HVR is thought to potentially serve as a hinge between the X domain and the upstream Met, Y, and PCP regions because of inherent flexibility. Such flexibility could result from the large number of proline residues, which could lead to unstable tertiary structures (Koonin et al. 1992; Tsai et al. 2001; Dunker et al. 2008). Although not essential for viral replication or infectivity, the HVR appears to play a role in HEV replication efficiency in vivo (Pudupakam et al. 2009, 2011). Exchanging HVRs among different HEV genotypes results in genotype-specific differences in virus replication (Pudupakam et al. 2011). Within the disorder of the HVR, there appear to be conserved linear motifs among different genotypes including two protease cleavage sites, three ligand-binding sites, and two kinase phosphorylation sites (Purdy 2012; Purdy et al. 2012). Additional protein–protein interaction motifs were also predicted but will require further experimental verification.
The HVR sequences from gt1 and gt2 human HEVs are more conserved than those of gt3 and gt4, which infect both humans and a number of other animal species. The HVR might act as a modulator of infectivity in differing host environments, as evidenced by an intriguing variant strain of HEV isolated from a patient who was coinfected with HEV and human immunodeficiency virus (HIV) and who was chronically shedding HEV. This Kernow-C1 gt3 strain of HEV is thought to have recombined in the host to pick up a portion of the human ribosomal protein S17 sequence within the HVR (Shukla et al. 2011, 2012). The Kernow-C1 P6 strain containing the S17 insertion gained the ability to replicate in cell culture 500 times more efficiently than the parental strain, and it replicates in diverse cell lines derived from different animals, including those from mice, cats, dogs, and hamsters. In addition, the S17 HVR insertion bestowed a novel function to the ORF1 protein, allowing it to enter the nucleus and nucleoli (Kenney and Meng 2015a,b). Although this novel nuclear trafficking is not the sole determinant of enhanced viral replication, it serves to show that changes within the HVR can alter viral protein functions. It appears that the HVR has the ability to sample and test insertions of host protein sequences as, in addition to the aforementioned S17 sequence, viral quasispecies have also been identified that contained fragments of the sequences of ribosomal protein S19 and a small GTPase activating protein (Shukla et al. 2012). Understanding how captured host proteins aid in the virus life cycle may shed light on virus host interactions and specific needs of the virus to efficiently replicate within the host.
X Domain
The X domain sequence shows homology with cellular ADP-ribose-1′-monophosphatase (Appr-1′-pase) of the macrodomain protein family (Allen et al. 2003; Parvez 2015a). Appr-1′-pase has more than 300 orthologs in eukaryotes and can also be found in coronaviruses, alphaviruses, and rubella virus (Gorbalenya et al. 1991; Snijder et al. 2003; Karras et al. 2005; Ziebuhr 2005; Draker et al. 2006). Although the exact function the X domain plays in HEV replication is not fully understood, Appr-1′-pase is involved in the transfer RNA (tRNA) splicing pathway catalyzing the conversion of ADP-ribose-1′-monophosphate to ADP ribose. Mutation of amino acids Asn-809, His-812, Gly-816, and Gly-817 in the putative catalytic site of the X domain led to severe reductions in RNA replication (Parvez 2015a). The X domain also inhibits type I IFN induction in vitro similarly to the PCP domain, although at a different step in the IFN-β signaling pathway. The X domain prevented phosphorylation of IFN regulatory factor 3 (IRF-3), blocking induction of IFN-β activity (Nan et al. 2014).
Helicase
During the process of RNA capping, Hel is involved in phosphatase activity that catalyzes the initial cap formation. The Hel domain also possesses RNA duplex unwinding activities (Karpe and Lole 2010). Amino acid substitutions, Leu-1110 to Phe and Val-1120 to Ile, are frequently observed in gt1 HEV isolates derived from patients with fulminant hepatic failure (Devhare et al. 2014). These mutations were shown to alter ATPase activity but not RNA duplex unwinding via Hel (Devhare et al. 2014; Mhaindarkar et al. 2014).
RNA-Dependent RNA Polymerase
The largest functional domain within ORF1 is the RdRp. All positive-sense RNA viruses encode an RdRp, which are classified into three different supergroups based on sequence similarity. The HEV RdRp is considered to be in supergroup III together with alphavirus, rubivirus, and a number of plant virus RdRps (Koonin 1991), with the highest sequence similarity to the domains in rubella and beet necrotic yellow vein virus (Koonin and Gorbalenya 1992). When expressed as a fusion protein with green fluorescent protein (GFP), the RdRp localizes to the ER, a potential site for HEV replication (Rehman et al. 2008). The RdRp contains a magnesium-binding domain (GDD) amino acid motif that is critical for its activity. Additionally, the purified recombinant RdRp protein binds to the 3′ end of the HEV RNA through interactions with two stem-loop structures found at the 3′ end of genome and the 3′ poly(A) tail (Agrawal et al. 2001). The RdRp can sustain mutations resulting in escape from ribavirin-induced mutagenesis, as a Gly-1634 mutation has been found in association with the failure of ribavirin monotherapy in organ transplant patients infected with HEV (Debing et al. 2014; Lhomme et al. 2015; Todt et al. 2016). In addition to ribavirin resistance, RdRp mutations such as Cys-1483 to Trp and Asp-1530 to Thr appear to correlate with acute liver failure, being found in 25 of 25 patients afflicted with acute liver failure, whereas none of 30 patients with acute viral hepatitis without liver failure had these two mutations. These mutations also correlated with higher viral RNA loads, suggesting that mutations enhancing viral replication might lead to acute liver failure (Borkakoti et al. 2016).
ORF2
ORF2 encodes the viral capsid protein, which is the major structural component of the virion. In gt1 HEV, ORF2 is 1983 nucleotides in length, beginning 37 nucleotides downstream from the ORF1 stop codon and overlapping all but 14 nucleotides of ORF3 (Fig. 2). The stop codon for ORF2 occurs 65 nucleotides upstream of the poly(A) tail (Reyes et al. 1993). The ORF2 subgenomic mRNA (Fig. 2) encodes a protein composed of 660 amino acids and predicted to have a molecular mass of 72 kDa (Robinson et al. 1998). ORF2 protein contains an amino-terminal ER-targeting sequence for cotranslational targeting to the ER where it undergoes N-linked glycosylation (Ahmad et al. 2011). When expressed experimentally, the capsid protein produces an 88-kDa protein, which contains amino-terminally linked glycans with Asn-310 as the primary glycosylation site (Jameel 1999; Zafrullah et al. 1999). This protein can be further processed, form noncovalent homodimers, and transit to the cell membrane via an ER-targeting motif that is independent of glycosylation. Dimerization of the capsid protein appears to be dependent on Asp-562 (Xu et al. 2016). The carboxy-terminal 52 amino acids of the capsid are required to promote accurate encapsidation and stabilization of viral particles (Shiota et al. 2013).
The capsid protein is immunogenic with neutralizing antibodies targeting conformational epitopes within the P domain (Zhou et al. 2005). The commercial HEV 239 subunit vaccine produced in China contains a segment of the capsid protein (Zhu et al. 2010). The ORF2 protein product contributes significantly to the virus–host interactions, including targeting of the protein during assembly via ER localization signals (Jameel et al. 1996). ORF2 protein plays a modulatory role in the host cell altering the functions of elongation initiation factor 2A (eIF2a), activating transcription factor 4 (ATF-4), heat shock protein 72 (Hsp72), nuclear factor κB (NF-κB), and activation of the C/Ebp-homologous protein (CHOP) promoter (John et al. 2011; Surjit et al. 2012). In addition, the capsid protein interacts with the host factors 78-kDa glucose-regulated protein (GRP-78/BiP), α-tubulin, and heat shock protein 90 (HSP90) for viral attachment, uptake, and trafficking (Zheng et al. 2010). The ORF2 glycoprotein has been shown to inhibit NF-κB activity by blocking ubiquitin-mediated degradation of NF-κB inhibitor α (IκBα) in human hepatoma cells (Surjit et al. 2012).
Amino acid substitutions within the capsid protein sequence have been suggested as possible links to acute liver failure, as Pro-259 to Ser and Lys-350 to Glu amino acid substitutions were found in a cohort of Indian patients presenting with acute liver failure. Unfortunately, the contributions of these capsid gene mutations to enhanced pathology have yet to be studied (Borkakoti et al. 2014). The capsid protein also appears to act as a mediator in viral adaptation to cell culture as mutations within the 3′ end of ORF2 enhanced virus replication (Shukla et al. 2012).
ORF3
ORF3 begins 24 nucleotides downstream from the ORF1 stop codon and overlaps ORF2 by ∼300 nucleotides in a different reading frame (Fig. 2) (Graff et al. 2005b, 2006). ORF3 protein is translated from the bicistronic subgenomic RNA, and is multifunctional. Translation initiation occurs at the third AUG producing a protein with 113 amino acids and a molecular weight of 13 kDa, resulting in the ORF3 protein also being called VP13 (Graff et al. 2006; Huang et al. 2007). The ORF3 protein product appears to be fairly unique. There are two highly hydrophobic domains at its amino terminus, D1 (aa 7–23) and D2 (aa 28–53), and two proline-rich domains (P1 and P2) toward the carboxyl terminus. The D1 domain is rich in cysteine residues and is a binding site for microtubulin (Kannan et al. 2009), mitogen-activated protein kinase (MAPK) phosphatase (Kar-Roy et al. 2004), and the cytoskeleton (Zafrullah et al. 1997). The D2 hydrophobic domain acts as a binding site for hemopexin (Ratra et al. 2008). The P1 motif contains two possible overlapping phosphorylation kinase motifs at Ser-71, one a potential MAPK phosphorylation motif, and the other a cyclin-dependent kinase (CDK) phosphorylation motif (Korkaya et al. 2001). The P2 motif contains two PxxP motifs that are binding sites for Src homology 3 (SH3)-containing host structural and signal transduction molecules. Additionally, these motifs act as viral late domains, and interact with tumor suppressor gene 101 (TSG101) to facilitate virion release in association with a lipid quasi-envelope. Release of HEV as a quasi-enveloped virion appears to shield the virus from neutralizing antibody epitopes located on the capsid protein, as in hepatitis A virus (HAV) (Feng et al. 2013).
Initial analysis of the ORF3 sequence found no known homologous proteins in nature. However, the ORF3 protein product was recently found to have similarities to viroporins encoded by other enveloped viruses (Ding et al. 2017). ORF3 protein appears to form multimeric complexes within ER-derived membranes that act as ion channels and resemble class I viroporins from other viruses. ORF3 may facilitate ion fluxes across membranes, thereby disrupting the electrochemical gradient across the plasma membrane to stimulate viral budding. Replacing ORF3 with the influenza A virus M2 protein ion channel can facilitate particle release (Ding et al. 2017).
The ORF3 protein is a small phosphoprotein that has been shown to be involved in many facets of the viral life cycle. Although it is not essential for virus replication in tissue culture (Emerson et al. 2006), it is required for infection of macaques and pigs (Graff et al. 2006; Huang et al. 2007). The ORF3 protein interacts with a number of host cellular signaling proteins thought to enhance survival of the infected cell. It activates the external regulatory kinase (ERK) of the MAPK family independent of the traditional RAF/MEK pathway by inhibiting the phosphatase activity of dual specificity protein phosphatase 6 (PYST1) by binding to it, allowing for continuous operation of the MAPK pathway (Kar-Roy et al. 2004). Transient expression of ORF3 in hepatocytes also increases the phosphorylation of JNK1/2, which is favorable for host cell survival (Parvez and Al-Dosari 2015).
The subcellular localization of ORF3 protein to early and recycling endosomes delays activation of internalized epidermal growth factor receptor (EGFR) until the late endosome/lysosome, potentially prolonging the EGF-mediated intracellular signal. In addition, ORF3 protein delays the internalized degradation of the activated hepatocyte growth factor receptor (c-MET). This process appears to occur via a competitive binding of the ORF3 SH3 (PxxP) motif with the CIN85 SH3 domain competing with Cbl inhibiting lysosome degradation (Chandra et al. 2010). ORF3 also interacts with porcine liver-specific plasminogen and α2-antiplasmin. A potential novel role for ORF3 protein in the viral replication complex is emerging with the finding that ORF3 protein interacts with the HEV ORF1 X or macrodomain when it is in a complex with the viral methyltransferase (Anang et al. 2016). ORF3 has also been reported to suppress TLR-3-induced NF-κB signaling via TRADD and RIP1 (He et al. 2016). However, many of these ORF3 interaction studies have relied on overexpression of the ORF3 protein and may require further experimental verification in more relevant infectious virus-based assays.
ORF4
A novel ORF4 that is positioned entirely within the ORF1 coding sequence in an alternate reading frame has been identified only within gt1 HEV strains (Nair et al. 2016). Translation of ORF4 is dependent on an IRES-like element within nucleotides 2701–2787 and is activated in response to cellular ER stress. ORF4 protein interacts with multiple viral proteins forming a viral replication complex consisting of the viral RdRp, helicase, and X proteins. ORF4 protein appears to stimulate viral RdRp activity, thereby enhancing viral replication. The lack of ORF4 conservation among other HEV genotypes suggests a need for more information to elucidate its exact function(s).
REPLICATION STRATEGY
Attachment and Entry
The historical lack of efficient in vitro cell culture systems for HEV has prevented many aspects of the HEV replication cycle from being studied in depth. How the virus attaches to and enters cells are poorly understood. Structural and sequence analyses have suggested that a putative receptor binding motif within the capsid protein is conserved among mammalian HEV genotypes (Guu et al. 2009). Another study implies that amino acids 458–607 in the carboxy-terminal (M domain) region of the capsid form a receptor binding site (He et al. 2008). From studies using HEV VLPs that are composed of recombinant ORF2 protein, the capsid protein is thought to attach to cells via heparin sulfate proteoglycans (HSPGs) (Kalia et al. 2009). A virus overlay protein binding assay suggested that a protein with a molecular weight of 55 kDa could be the entry receptor; however, mass spectrometry analysis was inconclusive, suggesting that the entry receptor could be one of up to 31 different proteins (Zhang et al. 2011). Other candidates involved in HEV interactions with the cell surface include heat shock cognate protein 70 (HSC70) and Grp78. However, HSC70 and Grp78 are thought to be more essential in intracellular transport or in another essential role in HEV replication rather than as cellular receptors for the virus (Kalia et al. 2009; Yu et al. 2011; Cao and Meng 2012). Further biological confirmation of these proteins as true HEV receptors is necessary.
Once binding is accomplished on the cell surface, HEV particles are internalized through a clathrin and dynamin-2 based pathway that also requires cholesterol (Kapur et al. 2012; Holla et al. 2015). Recent reports suggest that there could be two different pathways for HEV entry, one for native unenveloped particles and one for quasi-enveloped virions. The quasi-enveloped virion pathway involves the small GTPases Rab5 and Rab7 and degradation of the quasi-envelope membrane within the lysosome (Yin et al. 2016). Studies using VLPs generated with a truncated capsid protein showed that an HSP90 inhibitor blocked intracellular transport without altering virus entry (Zheng et al. 2010). HEV capsid uncoating is even less well understood than attachment. The P domains of the capsid possess polysaccharide-binding sites that could be involved in capsid disassembly (Guu et al. 2009).
Translation and Transcription of Subgenomic RNA
Once the genomic RNA is uncoated and delivered to the cytosol, ORF1 is translated directly from it by cap-dependent translation. Consistent with this, eukaryotic translation initiation factors 4A, 4G, and 4E (found within the eIF4F complex) are required for HEV replication (Zhou et al. 2015). ORF2 and ORF3 are also translated in a cap-dependent fashion from the subgenomic RNA (Fig. 2) (Graff et al. 2006). Initial studies of experimentally infected macaque liver tissues suggested the presence of three RNA species, the 7.2-kb, full-length genomic RNA, and 3.7 kb and 2 kb RNA species, which were interpreted as subgenomic RNAs for ORF2 and ORF3, respectively (Tam et al. 1991). However, a more recent study in Huh7 liver cells transfected with replication-competent HEV RNA replicons identified only a single capped 2.2 kb subgenomic RNA that acts as a bicistronic mRNA for the translation of both ORF2 and ORF3 (Graff et al. 2006). The presence of only a single subgenomic, bicistronic mRNA was further confirmed by another in vitro study of gt3 HEV in PLC/PRF/5 hepatoma cells (Ichiyama et al. 2009). The ORF2 and ORF3 proteins are generated from this single subgenomic RNA by a process involving leaky ribosome scanning in which ribosomes bypassing the ORF3 initiator AUG scan downstream to initiate protein synthesis at the closely positioned ORF2 AUG initiator codon (Fig. 2) (Graff et al. 2006). Translation of the subgenomic RNA initiates at nucleotide position 5122 in gt1 HEV (Sar55 strain), which corresponds to the third methionine in ORF3 (Huang et al. 2007).
HEV genomic RNA contains two structured cis-active RNA elements (CREs) that are essential for virus replication (Fig. 2) (Cao et al. 2010; Parvez 2015b). Many RNA viruses contain CREs in both 5′ and 3′UTRs. However, HEV contains a short 26-nucleotide-long 5′UTR and no CRE was found within it, although an RNA element within the 5′ region of the ORF1 appears to be the RNA encapsidation sequence (Fig. 2) (Surjit et al. 2004). CRE 1 overlaps the 3′ end of ORF2 and continues into the 3′UTR where it forms two stem-loop structures—both of which interact with the RdRp and are essential for HEV replication (Agrawal et al. 2001; Emerson et al. 2001; Magden et al. 2001; Cao and Meng 2012). Viral replication does not appear to be hindered by swapping the 3′UTRs between different genotypes, but having the correct stem loop structure, sequence, and accessibility of the stem loops are critical, as a single nucleotide change at position 7106 in gt1 HEV significantly decreases virus replication (Emerson et al. 2001; Graff et al. 2005a). Additionally, short hairpin RNAs and ribozymes targeting the 3′UTR effectively inhibit viral replication (Sriram et al. 2003; Kumar et al. 2010). The second CRE is found within the intergenic region of the HEV genome and forms a stem-loop structure that is a possible promoter for the synthesis of the 2.2-kb subgenomic RNA (Cao et al. 2010).
Viral Genome Replication
Once the viral RdRp is synthesized, either as part of an unprocessed polyprotein or as a processed independent protein, a negative-sense RNA replicative intermediate is produced. This negative-sense RNA serves as the template for production of both positive-sense, progeny genomic RNA and the subgenomic bicistronic mRNA. The negative-sense intermediate has been detected in HEV-infected animal tissues (Nanda et al. 1994; Meng et al. 1998; Williams et al. 2001). The activity of the RdRp has been shown in vitro using the HEV replicon system (Agrawal et al. 2001; Graff et al. 2005a). An HEV replication complex appears to form on ER membranes or on vesicles composed of ER-derived membranes (Rehman et al. 2008). The composition of the replication complex is not well described. There is evidence that the gt1 replication complex contains the ORF4 protein, viral helicase, RdRp, and X domains of ORF1, as well as host eukaryotic elongation factor 1 isoform-1 (eEF1α1) and tubulin β (Nair et al. 2016). Replication of the viral RNA appears to be rather slow. In studies using an HEV replicon system in which GFP or luciferase was cloned into ORF2 in lieu of the capsid protein, peak reporter gene expression was shown to occur 8 or more days posttransfection of HEV RNA (Emerson et al. 2004).
Virus Assembly and Release
Neither the assembly nor release steps in the HEV life cycle are well characterized. It is thought that RNA binding to the amino-terminal domain of the capsid protein results in carboxy-terminal/carboxy-terminal dimers, which are critical for completing viral capsid assembly. The capsid protein packages the viral genome through binding to a putative RNA packaging signal comprised of a 76-nucleotide region near the 5′ end of the HEV genome, within the ORF1 coding sequence at about nucleotide position 50 (Surjit et al. 2004). Virion assembly seems to progress through a mechanism in which preformed capsid decamers become localized at each fivefold vertex, and hexameric rings are located at icosahedral threefold positions. The cellular location of capsid–capsid and capsid–RNA interaction remains unknown, as does the process by which genomic RNA is selected for encapsidation rather than transcription or translation. The unglycosylated form of ORF2 interacts with amino acid residues 57–81 of the ORF3 protein after the ORF3 protein is phosphorylated at Ser-80 (Tyagi et al. 2002). The ORF3 protein appears to be involved in viral egress via interaction with host late domain proteins (Yamada et al. 2009; Emerson et al. 2010). Enveloped HEV has been detected by electron microscopy within intracellular multivesicular bodies (MVBs) in association with trans-Golgi network protein 2 and CD63, suggesting capsids bud into intracellular vesicles to form MVBs followed by release of virus particles via an exosome-like release pathway. HEV particle release via this pathway is also sensitive to disruption of the host proteins VPS28a and Hrs (Nagashima et al. 2014). ORF3 also acts as a viroporin to facilitate ion permeability at the plasma membrane facilitating virus release (Ding et al. 2017). Cell polarity plays a role in directing ORF3 localization and hence virus release to the apical side of charged culture cells (Emerson et al. 2010).
CONCLUDING REMARKS
HEV is an understudied pathogen, and a lack of standardized commercial assays for detection has likely led to underreporting of the true number of individuals infected with the virus, especially in industrialized countries. Commercial meat products containing infectious HEV have been found in local supermarkets in many countries, thus posing a concern for zoonotic transmission through consumption of undercooked meats. Chronic HEV infection has been recognized to be a significant clinical problem in immunocompromised individuals, especially organ transplant recipients, in some regions. The complex pathogenicity of the virus and a historical lack of the means to study it in cell culture have hampered our ability to understand its interplay with the host, and as a consequence HEV-specific therapeutic drugs are almost nonexistent.
There are more questions than answers regarding the life cycle of the virus, largely owing to the lack of an efficient cell culture system to propagate HEV. Nonetheless, recent advances in cell culture techniques have shown that HEV possesses a number of steps within its replication cycle that are comparable to those of other positive-stranded RNA viruses. Thus, it may be possible to repurpose antiviral drugs developed previously against other viruses, or to develop new therapeutics based on conserved replication mechanics. Another aspect of the HEV life cycle shared with many other viruses is immune evasion. HEV uses multiple strategies to avoid the host immune system, including maintaining only a low level of virus replication, expressing viral proteins that interact with and disrupt host cellular signaling molecules involved in the immune response, and even cloaking of the viral capsid via envelopment of virus particles within the host. Many aspects of HEV replication and pathogenesis remain to be uncovered, such as factors regulating ORF1 polyprotein processing, how pregnancy vastly increases the chance for mortality during infection, how HEV infection leads to neurological sequelae, how HEV is able to traverse species barriers, and even more general aspects of how and where the virus traffics within host cells. Continued development of in vitro and in vivo models will help enhance our understanding of HEV replication, and aid our efforts to develop effective HEV-specific therapeutics.
ACKNOWLEDGMENTS
We thank members of the Kenney Laboratory, Leah Everitt and Zachary Fritts, for critically reviewing the manuscript. The authors’ work on HEV is funded by Grants from the National Institutes of Health (R01AI050611 and R01AI074667).
Footnotes
Editors: Stanley M. Lemon and Christopher Walker
Additional Perspectives on Enteric Hepatitis Viruses available at www.perspectivesinmedicine.org
REFERENCES
- Agrawal S, Gupta D, Panda SK. 2001. The 3′ end of hepatitis E virus (HEV) genome binds specifically to the viral RNA-dependent RNA polymerase (RdRp). Virology 282: 87–101. [DOI] [PubMed] [Google Scholar]
- Ahmad I, Holla RP, Jameel S. 2011. Molecular virology of hepatitis E virus. Virus Res 161: 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahola T, Karlin DG. 2015. Sequence analysis reveals a conserved extension in the capping enzyme of the alphavirus supergroup, and a homologous domain in nodaviruses. Biol Direct 10: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MD, Buckle AM, Cordell SC, Lowe J, Bycroft M. 2003. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J Mol Biol 330: 503–511. [DOI] [PubMed] [Google Scholar]
- Anang S, Subramani C, Nair VP, Kaul S, Kaushik N, Sharma C, Tiwari A, Ranjith-Kumar CT, Surjit M. 2016. Identification of critical residues in hepatitis E virus macro domain involved in its interaction with viral methyltransferase and ORF3 proteins. Sci Rep 6: 25133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari IH, Nanda SK, Durgapal H, Agrawal S, Mohanty SK, Gupta D, Jameel S, Panda SK. 2000. Cloning, sequencing, and expression of the hepatitis E virus (HEV) nonstructural open reading frame 1 (ORF1). J Med Virol 60: 275–283. [PubMed] [Google Scholar]
- Balayan MS, Andjaparidze AG, Savinskaya SS, Ketiladze ES, Braginsky DM, Savinov AP, Poleschuk VF. 1983. Evidence for a virus in non-A, non-B hepatitis transmitted via the fecal–oral route. Intervirology 20: 23–31. [DOI] [PubMed] [Google Scholar]
- Batts W, Yun S, Hedrick R, Winton J. 2011. A novel member of the family Hepeviridae from cutthroat trout (Oncorhynchus clarkii). Virus Res 158: 116–123. [DOI] [PubMed] [Google Scholar]
- Borkakoti J, Ahmed G, Hussain SA, Rai A, Kar P. 2014. Novel molecular alterations in the ORF 2 capsid gene of hepatitis E virus in patients with acute liver failure in North India. Arch Virol 159: 3391–3394. [DOI] [PubMed] [Google Scholar]
- Borkakoti J, Ahmed G, Kar P. 2016. Report of a novel C1483W mutation in the hepatitis E virus polymerase in patients with acute liver failure. Infect Genet Evol 44: 51–54. [DOI] [PubMed] [Google Scholar]
- Cao D, Meng XJ. 2012. Molecular biology and replication of hepatitis E virus. Emerg Microbes Infect 1: e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Huang YW, Meng XJ. 2010. The nucleotides on the stem-loop RNA structure in the junction region of the hepatitis E virus genome are critical for virus replication. J Virol 84: 13040–13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V, Kalia M, Hajela K, Jameel S. 2010. The ORF3 protein of hepatitis E virus delays degradation of activated growth factor receptors by interacting with CIN85 and blocking formation of the Cbl-CIN85 complex. J Virol 84: 3857–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossaboom CM, Cordoba L, Dryman BA, Meng XJ. 2011. Hepatitis E virus in rabbits, Virginia, USA. Emerg Infect Dis 17: 2047–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossaboom CM, Heffron CL, Cao D, Yugo DM, Houk-Miles AE, Lindsay DS, Zajac AM, Bertke AS, Elvinger F, Meng XJ. 2016. Risk factors and sources of foodborne hepatitis E virus infection in the United States. J Med Virol 88: 1641–1645. [DOI] [PubMed] [Google Scholar]
- Debing Y, Gisa A, Dallmeier K, Pischke S, Bremer B, Manns M, Wedemeyer H, Suneetha PV, Neyts J. 2014. A mutation in the hepatitis E virus RNA polymerase promotes its replication and associates with ribavirin treatment failure in organ transplant recipients. Gastroenterology 147: 1008–1011.e1007; quiz e1015–1006. [DOI] [PubMed] [Google Scholar]
- Devhare P, Sharma K, Mhaindarkar V, Arankalle V, Lole K. 2014. Analysis of helicase domain mutations in the hepatitis E virus derived from patients with fulminant hepatic failure: Effects on enzymatic activities and virus replication. Virus Res 184: 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Heller B, Capuccino JM, Song B, Nimgaonkar I, Hrebikova G, Contreras JE, Ploss A. 2017. Hepatitis E virus ORF3 is a functional ion channel required for release of infectious particles. Proc Natl Acad Sci 114: 1147–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draker R, Roper RL, Petric M, Tellier R. 2006. The complete sequence of the bovine torovirus genome. Virus Res 115: 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Oldfield CJ, Meng J, Romero P, Yang JY, Chen JW, Vacic V, Obradovic Z, Uversky VN. 2008. The unfoldomics decade: An update on intrinsically disordered proteins. BMC Genomics 9: S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson SU, Zhang M, Meng XJ, Nguyen H, St Claire M, Govindarajan S, Huang YK, Purcell RH. 2001. Recombinant hepatitis E virus genomes infectious for primates: Importance of capping and discovery of a cis-reactive element. Proc Natl Acad Sci 98: 15270–15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson SU, Nguyen H, Graff J, Stephany DA, Brockington A, Purcell RH. 2004. In vitro replication of hepatitis E virus (HEV) genomes and of an HEV replicon expressing green fluorescent protein. J Virol 78: 4838–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson SU, Nguyen H, Torian U, Purcell RH. 2006. ORF3 protein of hepatitis E virus is not required for replication, virion assembly, or infection of hepatoma cells in vitro. J Virol 80: 10457–10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson SU, Nguyen HT, Torian U, Burke D, Engle R, Purcell RH. 2010. Release of genotype 1 hepatitis E virus from cultured hepatoma and polarized intestinal cells depends on open reading frame 3 protein and requires an intact PXXP motif. J Virol 84: 9059–9069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang SY, Han H. 2017. Hepatitis E viral infection in solid organ transplant patients. Curr Opin Organ Transplant 22: 351–355. [DOI] [PubMed] [Google Scholar]
- Feagins AR, Opriessnig T, Guenette DK, Halbur PG, Meng XJ. 2007. Detection and characterization of infectious hepatitis E virus from commercial pig livers sold in local grocery stores in the USA. J Gen Virol 88: 912–917. [DOI] [PubMed] [Google Scholar]
- Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496: 367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A, Aggarwal R. 2016. Advances in hepatitis E. II: Epidemiology, clinical manifestations, treatment and prevention. Exp Rev Gastroenterol Hepatol 10: 1065–1074. [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV, Lai MM. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, α- and coronaviruses. FEBS Lett 288: 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J, Nguyen H, Kasorndorkbua C, Halbur PG, St Claire M, Purcell RH, Emerson SU. 2005a. In vitro and in vivo mutational analysis of the 3′-terminal regions of hepatitis E virus genomes and replicons. J Virol 79: 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J, Nguyen H, Yu C, Elkins WR, St Claire M, Purcell RH, Emerson SU. 2005b. The open reading frame 3 gene of hepatitis E virus contains a cis-reactive element and encodes a protein required for infection of macaques. J Virol 79: 6680–6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J, Torian U, Nguyen H, Emerson SU. 2006. A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J Virol 80: 5919–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guu TS, Liu Z, Ye Q, Mata DA, Li K, Yin C, Zhang J, Tao YJ. 2009. Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc Natl Acad Sci 106: 12992–12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haqshenas G, Shivaprasad HL, Woolcock PR, Read DH, Meng XJ. 2001. Genetic identification and characterization of a novel virus related to human hepatitis E virus from chickens with hepatitis-splenomegaly syndrome in the United States. J Gen Virol 82: 2449–2462. [DOI] [PubMed] [Google Scholar]
- He S, Miao J, Zheng Z, Wu T, Xie M, Tang M, Zhang J, Ng MH, Xia N. 2008. Putative receptor-binding sites of hepatitis E virus. J Gen Virol 89: 245–249. [DOI] [PubMed] [Google Scholar]
- He M, Wang M, Huang Y, Peng W, Zheng Z, Xia N, Xu J, Tian D. 2016. The ORF3 protein of genotype 1 hepatitis E virus suppresses TLR3-induced NF-κB signaling via TRADD and RIP1. Sci Rep 6: 27597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holla P, Ahmad I, Ahmed Z, Jameel S. 2015. Hepatitis E virus enters liver cells through a dynamin-2, clathrin and membrane cholesterol-dependent pathway. Traffic 16: 398–416. [DOI] [PubMed] [Google Scholar]
- Huang FF, Sun ZF, Emerson SU, Purcell RH, Shivaprasad HL, Pierson FW, Toth TE, Meng XJ. 2004. Determination and analysis of the complete genomic sequence of avian hepatitis E virus (avian HEV) and attempts to infect rhesus monkeys with avian HEV. J Gen Virol 85: 1609–1618. [DOI] [PubMed] [Google Scholar]
- Huang YW, Opriessnig T, Halbur PG, Meng XJ. 2007. Initiation at the third in-frame AUG codon of open reading frame 3 of the hepatitis E virus is essential for viral infectivity in vivo. J Virol 81: 3018–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiyama K, Yamada K, Tanaka T, Nagashima S, Jirintai, Takahashi M, Okamoto H. 2009. Determination of the 5′-terminal sequence of subgenomic RNA of hepatitis E virus strains in cultured cells. Arch Virol 154: 1945–1951. [DOI] [PubMed] [Google Scholar]
- Jameel S. 1999. Molecular biology and pathogenesis of hepatitis E virus. Exp Rev Mol Med 1999: 1–16. [DOI] [PubMed] [Google Scholar]
- Jameel S, Zafrullah M, Ozdener MH, Panda SK. 1996. Expression in animal cells and characterization of the hepatitis E virus structural proteins. J Virol 70: 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John L, Thomas S, Herchenroder O, Putzer BM, Schaefer S. 2011. Hepatitis E virus ORF2 protein activates the pro-apoptotic gene CHOP and anti-apoptotic heat shock proteins. PLoS ONE 6: e25378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia M, Chandra V, Rahman SA, Sehgal D, Jameel S. 2009. Heparan sulfate proteoglycans are required for cellular binding of the hepatitis E virus ORF2 capsid protein and for viral infection. J Virol 83: 12714–12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamar N, Izopet J. 2014. Does chronic hepatitis E virus infection exist in immunocompetent patients? Hepatology 60: 427. [DOI] [PubMed] [Google Scholar]
- Kamar N, Garrouste C, Haagsma EB, Garrigue V, Pischke S, Chauvet C, Dumortier J, Cannesson A, Cassuto-Viguier E, Thervet E, et al. 2011. Factors associated with chronic hepatitis in patients with hepatitis E virus infection who have received solid organ transplants. Gastroenterology 140: 1481–1489. [DOI] [PubMed] [Google Scholar]
- Kamar N, Izopet J, Dalton HR. 2013. Chronic hepatitis E virus infection and treatment. J Clin Exp Hepatol 3: 134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan H, Fan S, Patel D, Bossis I, Zhang YJ. 2009. The hepatitis E virus open reading frame 3 product interacts with microtubules and interferes with their dynamics. J Virol 83: 6375–6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur N, Thakral D, Durgapal H, Panda SK. 2012. Hepatitis E virus enters liver cells through receptor-dependent clathrin-mediated endocytosis. J Viral Hepat 19: 436–448. [DOI] [PubMed] [Google Scholar]
- Karpe YA, Lole KS. 2010. RNA 5′-triphosphatase activity of the hepatitis E virus helicase domain. J Virol 84: 9637–9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpe YA, Lole KS. 2011. Deubiquitination activity associated with hepatitis E virus putative papain-like cysteine protease. J Gen Virol 92: 2088–2092. [DOI] [PubMed] [Google Scholar]
- Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG. 2005. The macro domain is an ADP-ribose binding module. EMBO J 24: 1911–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar-Roy A, Korkaya H, Oberoi R, Lal SK, Jameel S. 2004. The hepatitis E virus open reading frame 3 protein activates ERK through binding and inhibition of the MAPK phosphatase. J Biol Chem 279: 28345–28357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney SP, Meng XJ. 2015a. Identification and fine mapping of nuclear and nucleolar localization signals within the human ribosomal protein S17. PLoS ONE 10: e0124396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney SP, Meng XJ. 2015b. The lysine residues within the human ribosomal protein S17 sequence naturally inserted into the viral nonstructural protein of a unique strain of hepatitis E virus are important for enhanced virus replication. J Virol 89: 3793–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol 72: 2197–2206. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Gorbalenya AE. 1992. An insect picornavirus may have genome organization similar to that of caliciviruses. FEBS Lett 297: 81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Gorbalenya AE, Purdy MA, Rozanov MN, Reyes GR, Bradley DW. 1992. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: Delineation of an additional group of positive-strand RNA plant and animal viruses. Proc Natl Acad Sci 89: 8259–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkaya H, Jameel S, Gupta D, Tyagi S, Kumar R, Zafrullah M, Mazumdar M, Lal SK, Xiaofang L, Sehgal D, et al. 2001. The ORF3 protein of hepatitis E virus binds to Src homology 3 domains and activates MAPK. J Biol Chem 276: 42389–42400. [DOI] [PubMed] [Google Scholar]
- Kumar A, Panda SK, Durgapal H, Acharya SK, Rehman S, Kar UK. 2010. Inhibition of hepatitis E virus replication using short hairpin RNA (shRNA). Antiviral Res 85: 541–550. [DOI] [PubMed] [Google Scholar]
- Lee GH, Tan BH, Teo EC, Lim SG, Dan YY, Wee A, Aw PP, Zhu Y, Hibberd ML, Tan CK, et al. 2016. Chronic infection with camelid hepatitis E virus in a liver transplant recipient who regularly consumes camel meat and milk. Gastroenterology 150: 355–357.e353. [DOI] [PubMed] [Google Scholar]
- Lhomme S, Kamar N, Nicot F, Ducos J, Bismuth M, Garrigue V, Petitjean-Lecherbonnier J, Ollivier I, Alessandri-Gradt E, Goria O, et al. 2015. Mutation in the hepatitis E virus polymerase and outcome of ribavirin therapy. Antimicrob Agents Chemother 60: 1608–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Gillam S. 2000. Mutational analysis of the rubella virus nonstructural polyprotein and its cleavage products in virus replication and RNA synthesis. J Virol 74: 5133–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et al. 2012. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 380: 2095–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magden J, Takeda N, Li T, Auvinen P, Ahola T, Miyamura T, Merits A, Kaariainen L. 2001. Virus-specific mRNA capping enzyme encoded by hepatitis E virus. J Virol 75: 6249–6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr LD, Wang CY, Frey TK. 1994. Expression of the rubella virus nonstructural protein ORF and demonstration of proteolytic processing. Virology 198: 586–592. [DOI] [PubMed] [Google Scholar]
- Meng XJ, Halbur PG, Haynes JS, Tsareva TS, Bruna JD, Royer RL, Purcell RH, Emerson SU. 1998. Experimental infection of pigs with the newly identified swine hepatitis E virus (swine HEV), but not with human strains of HEV. Arch Virol 143: 1405–1415. [DOI] [PubMed] [Google Scholar]
- Mhaindarkar V, Sharma K, Lole KS. 2014. Mutagenesis of hepatitis E virus helicase motifs: Effects on enzyme activity. Virus Res 179: 26–33. [DOI] [PubMed] [Google Scholar]
- Nagashima S, Jirintai S, Takahashi M, Kobayashi T, Nishizawa T, Kouki T, Yashiro T, Okamoto H. 2014. Hepatitis E virus egress depends on the exosomal pathway, with secretory exosomes derived from multivesicular bodies. J Gen Virol 95: 2166–2175. [DOI] [PubMed] [Google Scholar]
- Nair VP, Anang S, Subramani C, Madhvi A, Bakshi K, Srivastava A, Shalimar, Nayak B, Ranjith Kumar CT, Surjit M. 2016. Endoplasmic reticulum stress induced synthesis of a novel viral factor mediates efficient replication of genotype-1 hepatitis E virus. PLoS Pathog 12: e1005521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan Y, Yu Y, Ma Z, Khattar SK, Fredericksen B, Zhang YJ. 2014. Hepatitis E virus inhibits type I interferon induction by ORF1 products. J Virol 88: 11924–11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda SK, Panda SK, Durgapal H, Jameel S. 1994. Detection of the negative strand of hepatitis E virus RNA in the livers of experimentally infected rhesus monkeys: Evidence for viral replication. J Med Virol 42: 237–240. [DOI] [PubMed] [Google Scholar]
- Paliwal D, Panda SK, Kapur N, Varma SP, Durgapal H. 2014. Hepatitis E virus (HEV) protease: A chymotrypsin-like enzyme that processes both non-structural (pORF1) and capsid (pORF2) protein. J Gen Virol 95: 1689–1700. [DOI] [PubMed] [Google Scholar]
- Panda SK, Ansari IH, Durgapal H, Agrawal S, Jameel S. 2000. The in vitro-synthesized RNA from a cDNA clone of hepatitis E virus is infectious. J Virol 74: 2430–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez MK. 2013. Molecular characterization of hepatitis E virus ORF1 gene supports a papain-like cysteine protease (PCP)-domain activity. Virus Res 178: 553–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez MK. 2015a. The hepatitis E virus ORF1 “X-domain” residues form a putative macrodomain protein/Appr-1″-pase catalytic-site, critical for viral RNA replication. Gene 566: 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez MK. 2015b. The intergenic-junction variant (genotype 2 isolate) of hepatitis E virus restores the CREX “stem-loop” structural integrity, essential for viral life cycle. Gene 559: 149–154. [DOI] [PubMed] [Google Scholar]
- Parvez MK. 2017. Mutational analysis of hepatitis E virus ORF1 “Y-domain”: Effects on RNA replication and virion infectivity. World J Gastroenterol 23: 590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez MK, Al-Dosari MS. 2015. Evidence of MAPK-JNK1/2 activation by hepatitis E virus ORF3 protein in cultured hepatoma cells. Cytotechnology 67: 545–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perttila J, Spuul P, Ahola T. 2013. Early secretory pathway localization and lack of processing for hepatitis E virus replication protein pORF1. J Gen Virol 94: 807–816. [DOI] [PubMed] [Google Scholar]
- Pudupakam RS, Huang YW, Opriessnig T, Halbur PG, Pierson FW, Meng XJ. 2009. Deletions of the hypervariable region (HVR) in open reading frame 1 of hepatitis E virus do not abolish virus infectivity: Evidence for attenuation of HVR deletion mutants in vivo. J Virol 83: 384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudupakam RS, Kenney SP, Cordoba L, Huang YW, Dryman BA, Leroith T, Pierson FW, Meng XJ. 2011. Mutational analysis of the hypervariable region of hepatitis E virus reveals its involvement in the efficiency of viral RNA replication. J Virol 85: 10031–10040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy MA. 2012. Evolution of the hepatitis E virus polyproline region: Order from disorder. J Virol 86: 10186–10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy MA, Lara J, Khudyakov YE. 2012. The hepatitis E virus polyproline region is involved in viral adaptation. PLoS ONE 7: e35974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratra R, Kar-Roy A, Lal SK. 2008. The ORF3 protein of hepatitis E virus interacts with hemopexin by means of its 26 amino acid N-terminal hydrophobic domain II. Biochemistry 47: 1957–1969. [DOI] [PubMed] [Google Scholar]
- Rehman S, Kapur N, Durgapal H, Panda SK. 2008. Subcellular localization of hepatitis E virus (HEV) replicase. Virology 370: 77–92. [DOI] [PubMed] [Google Scholar]
- Rein DB, Stevens GA, Theaker J, Wittenborn JS, Wiersma ST. 2012. The global burden of hepatitis E virus genotypes 1 and 2 in 2005. Hepatology 55: 988–997. [DOI] [PubMed] [Google Scholar]
- Reuter G, Boros A, Matics R, Kapusinszky B, Delwart E, Pankovics P. 2016. A novel avian-like hepatitis E virus in wild aquatic bird, little egret (Egretta garzetta), in Hungary. Infect Genet Evol 46: 74–77. [DOI] [PubMed] [Google Scholar]
- Reyes GR, Huang CC, Tam AW, Purdy MA. 1993. Molecular organization and replication of hepatitis E virus (HEV). Arch Virol Supplementum 7: 15–25. [DOI] [PubMed] [Google Scholar]
- Robinson RA, Burgess WH, Emerson SU, Leibowitz RS, Sosnovtseva SA, Tsarev S, Purcell RH. 1998. Structural characterization of recombinant hepatitis E virus ORF2 proteins in baculovirus-infected insect cells. Protein Expr Purif 12: 75–84. [DOI] [PubMed] [Google Scholar]
- Ropp SL, Tam AW, Beames B, Purdy M, Frey TK. 2000. Expression of the hepatitis E virus ORF1. Arch Virol 145: 1321–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG, Johnson JE. 1989. Icosahedral RNA virus structure. Annu Rev Biochem 58: 533–569. [DOI] [PubMed] [Google Scholar]
- Rozanov MN, Koonin EV, Gorbalenya AE. 1992. Conservation of the putative methyltransferase domain: A hallmark of the “Sindbis-like” supergroup of positive-strand RNA viruses. J Gen Virol 73: 2129–2134. [DOI] [PubMed] [Google Scholar]
- Sehgal D, Thomas S, Chakraborty M, Jameel S. 2006. Expression and processing of the hepatitis E virus ORF1 nonstructural polyprotein. Virol J 3: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota T, Li TC, Yoshizaki S, Kato T, Wakita T, Ishii K. 2013. The hepatitis E virus capsid C-terminal region is essential for the viral life cycle: Implication for viral genome encapsidation and particle stabilization. J Virol 87: 6031–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla P, Nguyen HT, Torian U, Engle RE, Faulk K, Dalton HR, Bendall RP, Keane FE, Purcell RH, Emerson SU. 2011. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc Natl Acad Sci 108: 2438–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla P, Nguyen HT, Faulk K, Mather K, Torian U, Engle RE, Emerson SU. 2012. Adaptation of a genotype 3 hepatitis E virus to efficient growth in cell culture depends on an inserted human gene segment acquired by recombination. J Virol 86: 5697–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Purdy MA, Simmonds P. 2013. Genetic variability and the classification of hepatitis E virus. J Virol 87: 4161–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Simmonds P; International Committee on Taxonomy of Viruses Hepeviridae Study Group; Jameel S, Emerson SU, Harrison TJ, Meng XJ, Okamoto H, Van der Poel WH, Purdy MA. 2014. Consensus proposals for classification of the family Hepeviridae. J Gen Virol 95: 2223–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Simmonds P; Members of the International Committee on the Taxonomy of Viruses Hepeviridae Study Group; Jameel S, Emerson SU, Harrison TJ, Meng XJ, Okamoto H, Van der Poel WH, Purdy MA. 2015. Consensus proposals for classification of the family Hepeviridae. J Gen Virol 96: 1191–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Simmonds P, Izopet J, Oliveira-Filho EF, Ulrich RG, Johne R, Koenig M, Jameel S, Harrison TJ, Meng XJ, et al. 2016. Proposed reference sequences for hepatitis E virus subtypes. J Gen Virol 97: 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, Guan Y, Rozanov M, Spaan WJ, Gorbalenya AE. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol 331: 991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram B, Thakral D, Panda SK. 2003. Targeted cleavage of hepatitis E virus 3′ end RNA mediated by hammerhead ribozymes inhibits viral RNA replication. Virology 312: 350–358. [DOI] [PubMed] [Google Scholar]
- Suppiah S, Zhou Y, Frey TK. 2011. Lack of processing of the expressed ORF1 gene product of hepatitis E virus. Virol J 8: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M, Jameel S, Lal SK. 2004. The ORF2 protein of hepatitis E virus binds the 5′ region of viral RNA. J Virol 78: 320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M, Varshney B, Lal SK. 2012. The ORF2 glycoprotein of hepatitis E virus inhibits cellular NF-κB activity by blocking ubiquitination mediated proteasomal degradation of IκBα in human hepatoma cells. BMC Biochem 13: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam AW, Smith MM, Guerra ME, Huang CC, Bradley DW, Fry KE, Reyes GR. 1991. Hepatitis E virus (HEV): Molecular cloning and sequencing of the full-length viral genome. Virology 185: 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todt D, Gisa A, Radonic A, Nitsche A, Behrendt P, Suneetha PV, Pischke S, Bremer B, Brown RJ, Manns MP, et al. 2016. In vivo evidence for ribavirin-induced mutagenesis of the hepatitis E virus genome. Gut 65: 1733–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Ma B, Sham YY, Kumar S, Nussinov R. 2001. Structured disorder and conformational selection. Proteins 44: 418–427. [DOI] [PubMed] [Google Scholar]
- Tsarev SA, Emerson SU, Reyes GR, Tsareva TS, Legters LJ, Malik IA, Iqbal M, Purcell RH. 1992. Characterization of a prototype strain of hepatitis E virus. Proc Natl Acad Sci 89: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Korkaya H, Zafrullah M, Jameel S, Lal SK. 2002. The phosphorylated form of the ORF3 protein of hepatitis E virus interacts with its non-glycosylated form of the major capsid protein, ORF2. J Biol Chem 277: 22759–22767. [DOI] [PubMed] [Google Scholar]
- van der Poel WH, Verschoor F, van der Heide R, Herrera MI, Vivo A, Kooreman M, de Roda Husman AM. 2001. Hepatitis E virus sequences in swine related to sequences in humans, The Netherlands. Emerg Infect Dis 7: 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tong H, Hoan NX, Wang B, Wedemeyer H, Bock CT, Velavan TP. 2016. Hepatitis E virus mutations: Functional and clinical relevance. eBioMedicine 11: 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TP, Kasorndorkbua C, Halbur PG, Haqshenas G, Guenette DK, Toth TE, Meng XJ. 2001. Evidence of extrahepatic sites of replication of the hepatitis E virus in a swine model. J Clin Microbiol 39: 3040–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Kato K, Li T, Takeda N, Miyamura T, Hammar L, Cheng RH. 1999. Recombinant hepatitis E capsid protein self-assembles into a dual-domain T = 1 particle presenting native virus epitopes. Virology 265: 35–45. [DOI] [PubMed] [Google Scholar]
- Xing L, Li TC, Mayazaki N, Simon MN, Wall JS, Moore M, Wang CY, Takeda N, Wakita T, Miyamura T, et al. 2010. Structure of hepatitis E virion-sized particle reveals an RNA-dependent viral assembly pathway. J Biol Chem 285: 33175–33183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Behloul N, Wen J, Zhang J, Meng J. 2016. Role of asparagine at position 562 in dimerization and immunogenicity of the hepatitis E virus capsid protein. Infect Genet Evol 37: 99–107. [DOI] [PubMed] [Google Scholar]
- Yamada K, Takahashi M, Hoshino Y, Takahashi H, Ichiyama K, Nagashima S, Tanaka T, Okamoto H. 2009. ORF3 protein of hepatitis E virus is essential for virion release from infected cells. J Gen Virol 90: 1880–1891. [DOI] [PubMed] [Google Scholar]
- Yin X, Ambardekar C, Lu Y, Feng Z. 2016. Distinct entry mechanisms for nonenveloped and quasi-enveloped hepatitis E viruses. J Virol 90: 4232–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Li S, Yang C, Wei M, Song C, Zheng Z, Gu Y, Du H, Zhang J, Xia N. 2011. Homology model and potential virus-capsid binding site of a putative HEV receptor Grp78. J Mol Model 17: 987–995. [DOI] [PubMed] [Google Scholar]
- Yugo DM, Cossaboom CM, Meng XJ. 2014. Naturally occurring animal models of human hepatitis E virus infection. ILAR J 55: 187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafrullah M, Ozdener MH, Panda SK, Jameel S. 1997. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J Virol 71: 9045–9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafrullah M, Ozdener MH, Kumar R, Panda SK, Jameel S. 1999. Mutational analysis of glycosylation, membrane translocation, and cell surface expression of the hepatitis E virus ORF2 protein. J Virol 73: 4074–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L, Dai X, Meng J. 2006. Hepatitis E virus genotyping based on full-length genome and partial genomic regions. Virus Res 120: 57–69. [DOI] [PubMed] [Google Scholar]
- Zhang M, Purcell RH, Emerson SU. 2001. Identification of the 5′ terminal sequence of the SAR-55 and MEX-14 strains of hepatitis E virus and confirmation that the genome is capped. J Med Virol 65: 293–295. [DOI] [PubMed] [Google Scholar]
- Zhang W, Hua X, Shen Q, Yang S, Yin H, Cui L. 2011. Identification of genotype 4 hepatitis E virus binding proteins on swine liver cells. Virol J 8: 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bilic I, Troxler S, Hess M. 2017. Evidence of genotypes 1 and 3 of avian hepatitis E virus in wild birds. Virus Res 228: 75–78. [DOI] [PubMed] [Google Scholar]
- Zheng ZZ, Miao J, Zhao M, Tang M, Yeo AE, Yu H, Zhang J, Xia NS. 2010. Role of heat-shock protein 90 in hepatitis E virus capsid trafficking. J Gen Virol 91: 1728–1736. [DOI] [PubMed] [Google Scholar]
- Zhou YH, Purcell RH, Emerson SU. 2005. A truncated ORF2 protein contains the most immunogenic site on ORF2: Antibody responses to non-vaccine sequences following challenge of vaccinated and non-vaccinated macaques with hepatitis E virus. Vaccine 23: 3157–3165. [DOI] [PubMed] [Google Scholar]
- Zhou X, Xu L, Wang Y, Wang W, Sprengers D, Metselaar HJ, Peppelenbosch MP, Pan Q. 2015. Requirement of the eukaryotic translation initiation factor 4F complex in hepatitis E virus replication. Antiviral Res 124: 11–19. [DOI] [PubMed] [Google Scholar]
- Zhu FC, Zhang J, Zhang XF, Zhou C, Wang ZZ, Huang SJ, Wang H, Yang CL, Jiang HM, Cai JP, et al. 2010. Efficacy and safety of a recombinant hepatitis E vaccine in healthy adults: A large-scale, randomised, double-blind placebo-controlled, phase 3 trial. Lancet 376: 895–902. [DOI] [PubMed] [Google Scholar]
- Ziebuhr J. 2005. The coronavirus replicase. Curr Top Microbiol Immunol 287: 57–94. [DOI] [PMC free article] [PubMed] [Google Scholar]