Abstract
Purpose
Retinal degenerative diseases can progress to severe reductions of vision. In general, the changes are permanent in higher vertebrates, including humans; however, retinal regeneration can occur in lower vertebrates, such as amphibians and teleost fish. Progranulin is a secreted growth factor that is involved in normal development and wound-healing processes. We have shown that progranulin promotes the proliferation of retinal precursor cells in mouse retinas. The purpose of this study was to investigate the role played by granulin 1 (grn1) in the retinal regeneration in zebrafish.
Methods
We injured the retina of zebrafish with needle puncturing, and the retinas were examined at different times after the injury. We also checked the proliferation and the expression of retinal regeneration–related genes after knockdown of grn1 by electroporation with morpholino oligonucleotides (MO) and intravitreal injection of recombinant grn1.
Results
Our results showed that the level of grn1 was highly increased after retinal injury, and it was expressed in various types of retinal cells. A knockdown of grn1 reduced the proliferation of Müller glial cells in zebrafish eyes undergoing retinal regeneration. The knockdown of grn1 also reduced the expression of achaete-scute homolog 1a (ascl1a), an important factor in retinal regeneration. An intravitreal injection of recombinant grn1 led to a proliferation of Müller glial cells and an increase in the expression of retinal regeneration–related genes, such as ascl1a and lin28.
Conclusions
These findings suggested that grn1 should be considered as a target for stimulating the dedifferentiation of Müller glial cells and retinal regeneration.
Keywords: grn1, granulin, Müller glia, retinal regeneration, zebrafish
Many retinal degenerative diseases, such as AMD and retinitis pigmentosa, are progressive,1 and the changes are permanent. Thus, developing therapies for these diseases could have a significant impact on the quality of life of the affected individuals. Recent clinical trials and basic laboratory studies have shown that transplantation of retinal or RPE cells can restore the vision in some of the eyes with degenerative diseases.2,3 The recent use of stem cell transplantation has also had some degree of success; however, the permanence of the degeneration of the retinal cells has made the task of restoring vision very difficult.
The retina of lower vertebrates, such as amphibians and teleost fish, can regenerate after an injury4–6; however, the retina of mammals, including humans, is not able to regenerate.7 In amphibians and zebrafish, Müller glia can respond to retinal injury, and then they acquire the function as stem cells that produce neural progenitors that become new neurons.8 In zebrafish, retinal injuries trigger signals inactivating glycogen synthase kinase (GSK) and 3β leading to the stabilization of β-catenin, an activating mitogen-activated protein kinase, extracellular signal-regulated kinase, and Janus kinase (JAK). These activations lead to a reprogramming of the Müller glial cells.9–12 The Achaete-scute homolog 1a (ascl1a), a member of the basic helix-loop-helix family of transcription factors, is induced by these signaling pathways and plays a key role in retinal regeneration in zebrafish.13,14 The level of ascl1a increases in the Müller cells soon after an injury of the retina, and it regulates genes such as Notch, lin28, and myc.15 Some growth factors and cytokines, such as fibroblast growth factor 2, heparin-binding epidermal (EGF)-like growth factor (HB-EGF), and tumor necrosis factor (TNF), are induced by retinal injury, and they, in turn, can induce the expression of ascl1a.10,16
Granulins are cleaved peptides of progranulin, which is one of the growth factor proteins. Progranulin is highly conserved in vertebrates.17 This peptide is expressed ubiquitously in neurons, epithelial cells, and immune cells.18 In the clinic, it has been recognized that loss-of-function mutations of the progranulin-encoding gene, GRN, cause familial frontotemporal dementia.19 Many studies have shown that progranulin exerts its anti-inflammatory effects through multiple pathways.20–22 The results of our laboratory showed that mammalian progranulin has neuroprotective effects on cells of the central nervous system during ischemia and on photoreceptors during exposure to phototoxic levels of illumination.23,24 Moreover, progranulin was found to promote the proliferation of retinal precursor cells and the differentiation of photoreceptors in the mouse retina.25 Progranulin was also found to be associated with retinal development in mice.26 These findings strongly suggested that progranulin could be involved in the retinal regeneration processes.
Zebrafish have four subtypes of progranulin: granulin (grn) a, grnb, grn1, and grn2. Grna and grnb have 9 to 10 repeats of the granulin/epithelin motif and are believed to be orthologues of human progranulin.27 Several studies have shown that grna is associated with liver morphogenesis,28 muscle growth and regeneration,29 retinal damage,30 and brain and retinal development.31 In zebrafish at 48 hours post fertilization, grna is exclusively expressed by microglia and/or microglial precursors within the brain and retina. The previous report also showed that grna governs neurogenesis by regulating cell cycle kinetics and the transition from proliferation to cell cycle exit and differentiation.31 However, the other subtypes of granulins, including grn1, have not been investigated in detail, and their functions remain undetermined.
Thus, the purpose of this study was to determine whether the granulins are involved in retinal regeneration. To accomplish this, we injured the retina of zebrafish and conducted time-course analysis of mRNA expressions of four types of granulins. As a result, we detected grn1 expression upregulated earlier than the other subtypes of granulins and it preceded upregulation of ascl1a. We then determined sites of grn1 expression in the retina. In addition, we examined whether an intravitreal injection of recombinant Grn1 increased the proliferation of Müller cells, and increased the level of expression of retinal regeneration–related genes including ascl1a.
Methods
Animals
Zebrafish from the AB line were raised at 28.5°C under a 14-hour light:10-hour dark cycle. All experimental protocols were approved by University of Michigan and Gifu Pharmaceutical University Committee on Use and Care of Animals and adhered to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Methods were carried out in accordance with the approved guidelines.
RNA Isolation and PCR
All primers used are listed in the Table. Three fish were used for each experiment. Adult zebrafish were overdosed with tricaine, the eyes were enucleated, and the retinae were harvested. Total RNA was extracted from the isolated retinae with TRIzol (Thermo Fisher Scientific, Waltham, MA, USA), and cDNA synthesis and PCR were performed as described.13,14 Relative quantitative real-time PCR was performed in triplicate with SYBR Premix Ex TaqII (Takara Bio, Kusatsu, Japan) on the Thermal Cycler Dice (Takara Bio). In each sample, we prepared two retinas from a single fish. In each group, we prepared three fishes as a biological replicate. We conducted qPCR for each biological replicate run in triplicate. In each result, the error bars represent standard error among three biological replicates. Glyceraldehyde-3-phosphate dehydrogenase (gapdh) was used as an internal standard, and relative quantitative analysis was carried out by the ΔΔCt method. Semiquantitative analysis of RT-PCR was performed using the ImageJ software (http://imagej.nih.gov/ij/; provided in the public domain by the National Institutes of Health, Bethesda, MD, USA).
Table.
List of Primers
Retinal Lesions and Morpholino-Modified Antisense Oligonucleotide-Mediated Gene Knockdown
Retinas were injured and electroporated with morpholino oligonucleotides (MOs) as described in detail.13,14 Briefly, the zebrafish were anesthetized, and the right retina was stabbed once in each quadrant with a 30G needle inserted through the sclera to a depth of 1.5 mm. Lissamine-labeled MOs (Gene Tools, Philomath, OR, USA) were delivered at the time of injury using the same needle to inject the MOs. Approximately 0.5 μL MO (1-mM stock) was injected into the eye, and an uptake by the cells was facilitated by electroporation.13,14 Custom electrodes were placed across the head of the anesthetized fish with the cathode on the left eye and the anode on the right eye. An ECM 830 Electro Square Porator (BTX, San Diego, CA, USA) was used to deliver five consecutive 50-ms pulses at 70 V with a 950-ms interval between pulses. The following lissamine-labeled MOs were used:
control MO: 5′-CCTCTTACCTCAGTTACAATTTATA-3′
grn1 MO #1: 5′-CATCTTGCTGGCTGGGTTTCCTCCT-3′
grn1 MO #2: 5′-ACATTAACTCGAGATTCTGTTACCATTGAGTAAGGACA-3′
For bromodeoxyuridine (BrdU) incorporation, a solution of 20 μL of 20 mM BrdU was injected intraperitoneally into anesthetized fish at 4 days after injury.
Recombinant grn1 Injection and BrdU Incorporation
His-tagged recombinant Grn1 was synthesized with the TnT Quick Coupled Transcription/Translation System (Promega, Madison, WI, USA), and purified with the MagneHisProtein Purification System (Promega) and Zeba Spin Desalting Columns (Thermo Fisher Scientific) according to the protocols of the manufacturer.
The zebrafish were anesthetized, and the right eye was injected with 20 or 200 ng of the recombinant Grn1 that was dissolved in 1 μL of PBS. The left eye (control) was injected with 1 μL of vehicle used for the synthesis and purification of the mock plasmids. The solutions were injected into the vitreous through the cornea, being careful not to injure the retina. For BrdU incorporation, a solution of 20 μL of 20 mM BrdU was injected intraperitoneally into anesthetized fish at 4 days after the recombinant Grn1 was injected.
Quantification of BrdU-Positive Cell Number
At injured condition, the BrdU-positive cells were counted by using sum of 4 or 5 different serial transverse sections from the same fish surrounding an injury. At normal condition, the BrdU-positive cell number was counted by single transverse section including optic nerve.
Tissue Preparation, Immunohistochemistry, and In Situ Hybridization
Adult zebrafish were overdosed with tricaine, and the eyes were enucleated, the lens removed, and the eye cups were fixed in 4% paraformaldehyde. The fixed and frozen eye cups were sectioned at 12-μm thickness for immunofluorescence as described in detail.14,32,33 The antibodies used were diluted in PBS plus 0.1% Triton X-100 as described.14,32,33 For BrdU staining, sections were treated with 2 N HCl at 37°C for 20 minutes, rinsed in 0.1 M sodium borate solution (pH 8.5) for 10 minutes, and then processed using standard immunohistochemical procedures. In situ hybridization was performed as described in detail.34 Digoxigenin-labeled RNA probes were prepared using the DIG RNA labeling kit (Roche Diagnostics, Basel, Switzerland). Hybridization was performed by total immersion in a hybridization buffer that contained 50% formamide, 5× saline sodium citrate (SSC), 2% blocking reagent (Roche Diagnostics), 0.02% SDS, and approximately 100 ng/mL cRNA probe. Retinal sections on slides were pretreated by proteinase K for 5 minutes. Sections were hybridized at 65°C overnight with a 1-kb DIG-labeled zebrafish grn1 or ascl1a RNA probe. Washing steps included incubations in 2× SSC at 65°C. Sections were incubated at room temperature in 1% blocking reagent in maleic acid buffer, then in alkaline phosphatase–conjugated anti-DIG Fab fragments (1:5000 dilution; Roche Diagnostics), and developed 2 hours with 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium substrate (Kierkegaard and Perry Laboratories, Gaithersburg, MD, USA). Sections were rinsed several times in 100 mM Tris, 150 mM NaCl, 20 mM EDTA, pH 9.5, and coverslipped with glycerol gelatin (Sigma-Aldrich Corp., St. Louis, MO, USA). The source of the cDNA was a PCR product of adult zebrafish retina. grn1 cDNA was amplified from zebrafish retina RNA at 4 dpi using primer pairs grn1 F and grn1 ish. ascl1a probe was described previously.13
Statistical Analyses
Data are presented as the means ± SEM. The significance of the differences in the different groups was determined by Student's t-tests, Dunnett's tests, or Tukey's tests (GraphPad Prism; GraphPad, La Jolla, CA, USA). A P < 0.05 was taken to be statistically significant.
Results
Induction of grn1 in Injured Retina
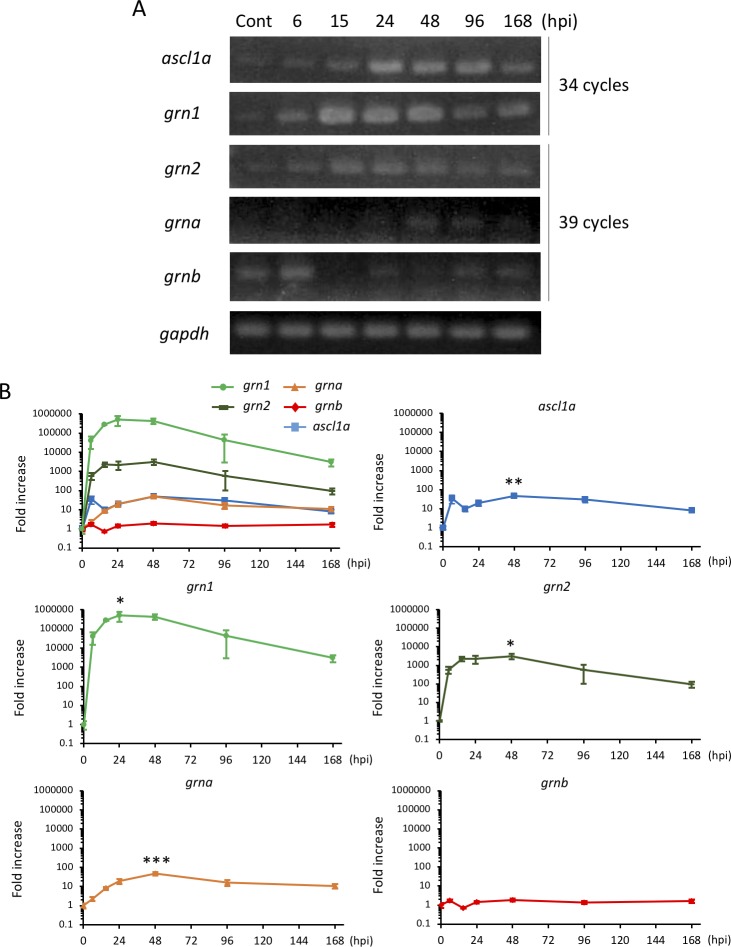
RT-PCR (Fig. 1A) and quantitative real-time PCR (qPCR; Fig. 1B) showed that grn1 and grn2 were induced soon after the retina was injured with the level of expression of grn1 higher than that of grn2 (Figs. 1A, 1B). On the other hand, grna, which is known to be induced by phototoxicity,30 was induced after grn1 and grn2 were induced, and grnb was highly expressed at control and unaltered after injury (Figs. 1A, 1B). The peak expression of grn1 was at 24 hours after the retinal injury, whereas that of grna and ascl1a were 48 hours (2 days) after the injury (Fig. 1B). Consequently, we found that ascl1a induction was preceded by grn1 upregulation. Therefore, after this experiment, we focused on grn1.
Figure 1.
Expression of grn induced by retinal injury in zebrafish. (A) RT-PCR showing the temporal expression pattern of genes encoding the grn family and ascl1a in the uninjured and injured retinas. (B) Quantitative PCR of grn1, grn2, grna, grnb, and ascl1a. The expression pattern of ascl1a is the same as our previous report.14 Data are the means ± SEMs (n = 3). ****P < 0.0001, *P < 0.05 versus ascl1a. Cropped blots are used in this figure and the full-length blots are presented in Supplementary Figure S5.
Grn1 Changes Expression Location in Retinal Regeneration
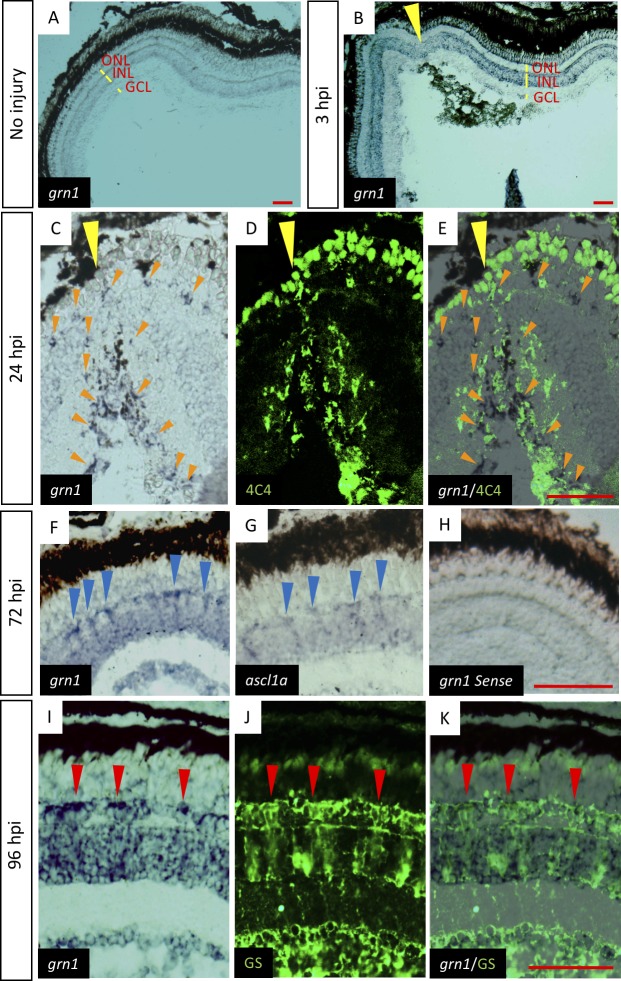
In situ hybridization was used to determine the sites of expression of the mRNA of grn1. In the uninjured control retina, grn1 was weakly expressed in the inner nuclear layer (INL) (Fig. 2A). grn1 was expressed in cells near the injury site at 3 hours post injury (hpi), especially in the retinal ganglion cell layer (GCL), photoreceptor cells, and INL (Fig. 2B). In contrast, grn1 was expressed in different retinal layers at 24 hpi (Fig. 2C).
Figure 2.
Alterations in expression of grn1 in retinal cells during retinal regeneration. Time-course in situ hybridization analysis shows that grn1 is expressed in different retinal layers at each time point. In retina, melanin mainly accumulates at the RPE and in situ labeled grn1 mainly localized inner retinal layer. In situ labeling also shows weaker signal than melanin. (A) In situ hybridization of grn1 at uninjured control retina. grn1 is weakly expressed in the INL. (B) In situ hybridization of grn1 at 3 hpi. Yellow arrowheads indicate the injury site. grn1 is expressed in the GCL, photoreceptor cells, and INL. The areas that are very close to the injury site express more grn1, whereas the injury site has lower expression of grn1. (C–E) Retina were stained at 24 hpi. Orange arrowheads indicate expression site of grn1. Yellow arrowheads indicate the injury site. (C) In situ hybridization of grn1. (D) Anti-4C4 antibody staining. (E) Merged image. (F–H) Retina were stained at 72 hpi. Blue arrowheads indicate the strong expression site of grn1 or ascl1a. (F) In situ hybridization of grn1. (G) In situ hybridization of ascl1a. (H) In situ hybridization using grn1 sense probe. (I–K) retina were stained at 96 hpi. Red arrowheads indicate the colocalization of grn1 and glutamine synthase (GS). (I) In situ hybridization of grn1. (J) Anti-GS antibody staining. (K) Merged image. Scale bars: 50 μm.
Microglia are known to migrate to the injury site, and sections were stained with anti-4C4 antibody,35 a microglial marker, to determine whether this occurred in the zebrafish (Fig. 2D). To check the migrating cells and grn1 colocalization, we chose 24 hpi based on the grn1 expression site. In earlier time points, like 3 hpi, grn1 expressed in the GCL and INL (Fig. 2B). At a later time point, like 72 hpi, grn1 expressed in GCL and INL (Fig. 2F) and the migration process is completed. The expression of grn1 did not colocalize with the 4C4-positive cells (Fig. 2E). Cells double stained by plastin, a marker for microglia and neutrophils, and 4C4 were observed peripheral to the injured site (Supplementary Fig. S1). The pattern of expression of grn1 was similar to that of ascl1a at 3 days post injury (dpi) (Fig. 2F, 2G), and it was also expressed in the GCL and INL, although the migrated cells were not observed at this time. A grn1 sense-strand probe gave no signal above background (Fig. 2H). Many of the grn1-positive cells were colocalized with glutamine synthase-positive cells at 4 dpi indicating the grn1 was expressed in a subset of Müller cells (Figs. 2I–K).
Grn1 Modulates Proliferation of Müller Glia Cells and Expression of ascl1a in Regenerating Retina
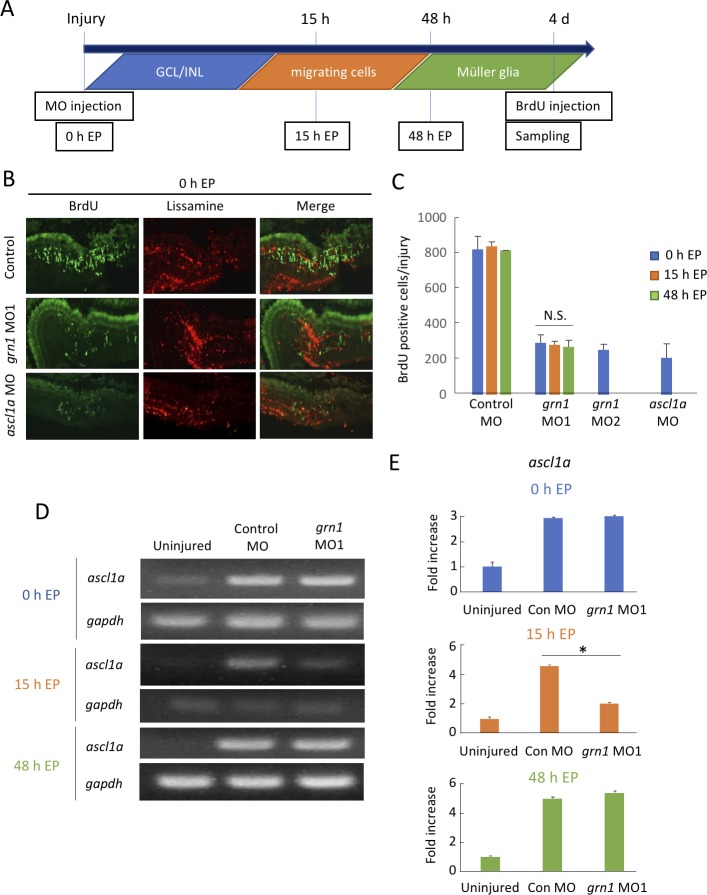
The sites of the expression of grn1 changed with time during the retinal regeneration processes. Therefore, next, we conducted time-dependent grn1 knockdown experiments using grn1 MO. To determine the time-specific influence of grn1, we conducted and compared electroporation (EP) at three different time points in grn1 or negative control MO-treated groups (Fig. 3A). In all groups, intravitreal MO injection was conducted at the same time as the injury. We considered that (1) 0 hour EP targeted grn1, which expressed in GCL and INL (Fig. 2B), (2) 15-hour EP targeted grn1, which expressed in migrating cells like neutrophils (Figs. 2C–E), and (3) 48-hour EP targeted grn1, which expressed in Müller glia. The MOs targeting ascl1a were also injected as positive controls as reported.13 The fish then received an intraperitoneal injection of BrdU 3 hours before they were killed on 4 dpi to quantify proliferating Müller glia and other retinal progenitor cells. Treatment with the MO of grn1 #1 and grn1 #2 decreased the number of BrdU-positive cells significantly compared with the negative control MO electroporated at 0 hpi (Figs. 3B, 3C). Injection of grn1 #1 MO by the electroporation at 15 hpi and 48 hpi also reduced the BrdU-positive cells; however, the reductions were not significantly different from that with the grn1 #1 MO (Fig. 3C). The expression of the mRNA of ascl1a showed that the level of ascl1a was significantly reduced only in the group that had been electroporated 15 hours after the grn1 MO injection (Fig. 3D, E). We also confirmed that lissamine-tagged control MO, which was electroporated at 15 hpi, merged with migrating cells that were stained by anti-4C4 antibody (Supplementary Fig. S2).
Figure 3.
Grn1 attenuates proliferation of Müller cells and expression of ascl1a. (A) Protocol for antisense MO injection into the zebrafish retina. MOs were injected intravitreally, causing retinal injury. EP was performed at the indicated time. (B) Images of grn1 MO-, ascl1a MO-, and control MO-treated retinas. All MOs were electroporated at 0 hpi. The representative images of BrdU by 15-hour EP or 48-hour EP groups are presented in Supplementary Figure S3. (C) Quantitative analysis of BrdU-positive cells after MO injections. Serial transverse sections were prepared, and the number of BrdU-positive cells per injury were counted. grn1 MO #2 and ascl1a MO were electroporated only at 0 hpi, whereas grn1 MO #1 was electroporated at 0, 15, and 48 hpi. (D) RT-PCR showing the alteration of ascl1a expression by grn1 knockdown at different times. (E) Semiquantitative analysis of RT-PCR. Data are the means ± SEMs; n = 3. P < 0.05 versus control MO-treated group. The cropped blots are used in this figure and the full-length blots are presented in Supplementary Figure S6.
Recombinant Zebrafish grn1 Protein Induces Proliferation of Retinal Cells and Retinal Regeneration-Associated Genes
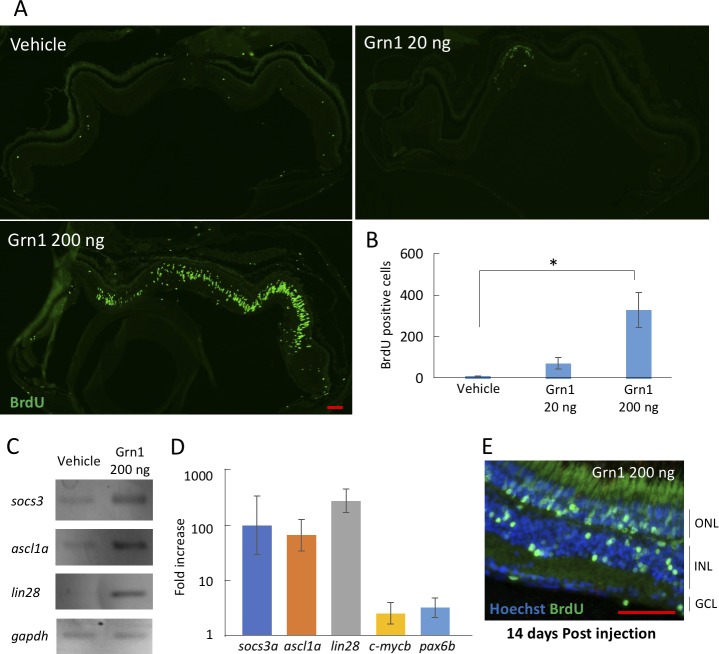
We investigated whether grn1 will induce retinal regeneration by injecting recombinant zebrafish grn1 protein intravitreally and vehicle in the control eyes (Fig. 4A). BrdU-positive cells were increased at 4 days after the injection of the recombinant grn1 protein in a dose-dependent manner (Figs. 4A, 4B).
Figure 4.
Recombinant grn1 induces retinal regeneration by the induction of retinal regeneration–related genes. (A) Intravitreal injection of recombinant grn1 induced BrdU-positive cells without retinal injury. BrdU was injected at 4 dpi. (B) Quantitative analysis of BrdU-positive cells on recombinant grn1 injection. The number of BrdU-positive cells was counted in a single section, including the optic nerve. (C) RT-PCR showing the induction of retinal regeneration–related genes by recombinant grn1 injection (200 ng). The RNA samples were obtained at 1 dpi of recombinant grn1. (D) Quantitative PCR shows recombinant grn1 highly induced socs3, ascl1a, and lin28, and weakly induced pax6 and c-mycb. (E) BrdU-positive cells have migrated to various retinal cell layers at 14 dpi of recombinant grn1 (200 ng). Data are the means ± SEMs; n = 3. P < 0.05 versus vehicle-treated group. N.S., not significant. Scale bar: 200 μm. The cropped blots are used in this figure and the full-length blots are presented in Supplementary Figure S7.
The levels of expression of the mRNAs of the genes associated with retinal regeneration were investigated at 1 day after the injection of the protein of recombinant grn1. ascl1a, lin-28, and socs3 were significantly increased after the recombinant grn1 injection, and in contrast, c-mycb and paired box 6b (pax6b) were only slightly increased (Figs. 4C, 4D).
We used a BrdU lineage-tracing strategy to test if recombinant grn1 injection stimulated proliferating cells, including Müller glia-derived progenitor cells, to migrate to all layers in the uninjured retina. For this experiment, grn1 was injected into the vitreous, and 3 days later the zebrafish received an intraperitoneal injection of BrdU and were then killed 10 days later. BrdU-positive cells were observed in all layers of the retina, especially in the outer nuclear layer (ONL) and INL (Fig. 4E).
Discussion
The results showed that an intravitreal injection of zebrafish grn1, a short form of progranulin, induced retinal regeneration. grna is believed to be the orthologue of human progranulin,27 and several studies have reported that grna is associated with liver morphogenesis,28 growth and regeneration of muscle tissues,29 and retinal regeneration.30 However, the function of grn1 is little known. Progranulin is cleaved into 6-kDa granulin peptides by many proteinases, including neutrophil elastase and matrix metalloproteinases.20 It is not clear which single 6-kDa granulin mediates its biological function. Grn1 has one-half of the granulin motif, thus grn1 may have functions that are similar to that of single 6-kDa granulin. In our study, we focused on the grn1 function against retinal regeneration because of the remarkable changes in expression levels of grn1 mRNA in all granulin subtypes after needle puncturing (Fig. 1). A previous report showed that zebrafish grn1, grn2, and grna are linked to a Hox gene cluster,27 but grnb is not linked. Hox gene cluster is important to development and also tissue regeneration in some organs, including eyes.36,37 Possibly, these features are reflected as the different expression pattern in our results. However, further studies are needed to uncover the underlying mechanism. Grn1 was expressed in various cell layers, and the amount varied at different times (Figs. 1, 2). The number of BrdU-positive cells was decreased by grn1 knockdown even though EP was done at 2 days after the injection (Figs. 3A, 3C, Supplementary Fig. S3). Thus, grn1 may be associated with the proliferation of Müller glia-derived progenitors after retinal injury.38,39 Moreover, the grn1 knockdown study using MO showed that grn1 strongly relates with the migration process of neutrophil and the blockage of grn1 at this time point only attenuated the expression of ascl1a (Figs. 3D, 3E). In Supplementary Figure S1, we conducted double staining using 24 hpi retina by 4C4 and plastin antibody. Plastin is expressed in both microglia and neutrophils, but 4C4 is expressed in microglia. Thus, we could detect neutrophils as plastin-positive and 4C4-negative cells (green in the merged image of Supplementary Fig. S1). The expression patterns and morphologies of these cells closely resembled the expression pattern of grn1 mRNA at 24 hpi (Figs. 2C, 2E). Therefore, in our view, grn1 possibly expressed in neutrophils. In our needle puncturing injury model, choroid is also punctured by needle. We considered the choroid vessels mainly as the source of the neutrophils. Actually, we found slight hemorrhage when sampling eyeballs at the early time points, such as 6 to 24 hpi. We also observed punctured cells that expressed plastin and mpeg at 3 hpi (Supplementary Fig. S4). Thus, amebocytes, such as neutrophils, may migrate into the injury site. Neutrophils secrete neutrophil elastase that can cleave progranulin protein. Hence, proteolytic modifications may be important for the functioning of grn1 on ascl1a expression. Several studies have reported that neutrophils are important for tissue regeneration.40,41 At present, no reports have shown the relationships between ascl1a and migrating cells in detail. In our experiments, we showed the importance of grn1 function at the specific time point (15 hpi) by grn1 knockdown using MO on ascl1a induction (Figs. 3D, 3E). This result indicated that grn1 acts in an autocrine fashion in migrating cells to regulate ascl1a induction; however, at present, the mechanism of this interaction is almost unclear. Therefore, more detailed studies on neutrophils and retinal regeneration will be necessary. Moreover, grn1 mRNA was also expressed in photoreceptor cells at 3 hpi. However, this expression was attenuated at later time points (Fig. 2). We cannot provide appropriate explanation about the roles and reasons of grn1 expression at photoreceptor cells. It also needs further investigation.
An intravitreal injection of the recombinant grn1 protein induced a proliferation of retinal cells including progenitor cells and an upregulation of the genes involved in retinal regeneration (Fig. 4). These findings suggest that grn1 is one of the early response genes that can trigger the dedifferentiation of Müller cells during retinal regeneration, such as HB-EGF.10 Moreover, various factors can induce retinal regeneration in zebrafish.5,12,42–44 Grn1 induces socs3a, which is downstream of Jak-Stat3 signaling, but grn1 had little effect of the induction of pax6b, which is downstream of GSK 3β-β-catenin signaling.8 Socs3a is also regulated by TNF receptor-associated factor 645 or insulin signaling.46 However, this signaling is also linked to Jak-Stat3 signaling pathway–associated retinal regeneration.16,47 Therefore, we considered grn1 also associated with Jak-Stat3 signaling like these factors and is less involved in the GSK3β signaling pathway. Rat progranulin and its single 6-kDa peptide, granulin A, interact with the EGF-like domain of cartilage oligomeric matrix protein, and they promote cell proliferation.48 Thus, grn1 may exert its regenerative effects by coordinating HB-EGF signaling. On the other hand, progranulin is associated with hepatocyte growth factor signaling,24,25,28 and it interacts with various proteins, such as the TNF receptor,49 which are associated with retinal regeneration.16 However, the effect of grn1 knockdown on the expression of ascl1a was limited. Thus, grn1 may have a supportive effect on the initiation factors of the retinal regeneration. The association of grn1 with other factors involved in the retinal regeneration are important areas of future investigations. The BrdU-positive cells induced by grn1 migrated to all retinal cell layers at 14 dpi (Fig. 4E), suggesting they may differentiate into various retinal cells. Further studies will determine whether grn1 can promote cell differentiation and the fate of these cells.
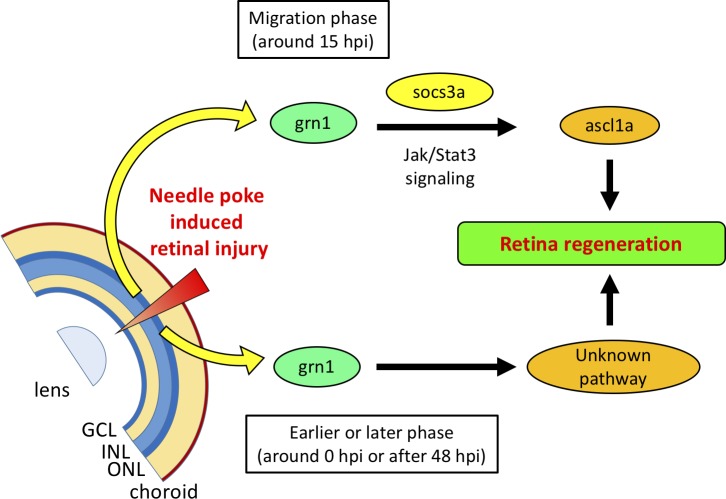
In conclusion, we have determined that grn1 is involved in both the dedifferentiation of Müller cells and proliferation of progenitor cells. Our results suggest that grn1 may play different roles depending on the cells types and the timing of its expression (Fig. 5). Determination of grn1 signaling will be necessary to understand its roles in retinal regeneration and its application to mammals.
Figure 5.
Schematic diagram of relationships between grn1 and retinal regeneration process after needle puncturing.
Supplementary Material
Acknowledgments
The authors thank members of the Goldman laboratory for comments and advice on these studies, and Hiromi Murase for technical support and fish care.
Supported by Takeda Science Foundation, the NOVARTIS Foundation (Japan), and NIH Grant from the National Eye Institute RO1 EY 018132. Some of this work took place while KT was a visiting scientist in the Molecular and Behavioral Neuroscience Institute at the University of Michigan.
Disclosure: K. Tsuruma, None; Y. Saito, None; H. Okuyoshi, None; A. Yamaguchi, None; M. Shimazawa, None; D. Goldman, None; H. Hara, None
References
- 1.Katherine JW, Jonathan HL, Stephen HT. General pathophysiology in retinal degeneration. Dev Ophthalmol. 2014;53:33–43. doi: 10.1159/000357294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mandai M, Fujii M, Hashiguchi T, et al. iPSC-derived retina transplants improve vision in rd1 end-stage retinal-degeneration mice. Stem Cell Reports. 2017;8:69–83. doi: 10.1016/j.stemcr.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mandai M, Watanabe A, Kurimoto Y, et al. Autologous induced stem-cell–derived retinal cells for macular degeneration. N Engl J Med. 2017;376:1038–1046. doi: 10.1056/NEJMoa1608368. [DOI] [PubMed] [Google Scholar]
- 4.Sherpa T, Fimbel SM, Mallory DE, et al. Ganglion cell regeneration following whole-retina destruction in zebrafish. Dev Neurobiol. 2008;68:166–181. doi: 10.1002/dneu.20568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mensinger AF, Powers MK. Visual function in regenerating teleost retina following cytotoxic lesioning. Vis Neurosci. 1999;16:241–251. doi: 10.1017/s0952523899162059. [DOI] [PubMed] [Google Scholar]
- 6.Lindsey AE, Powers MK. Visual behavior of adult goldfish with regenerating retina. Vis Neurosci. 2007;24:247–255. doi: 10.1017/S0952523806230207. [DOI] [PubMed] [Google Scholar]
- 7.Xia X, Ahmad I. Unlocking the neurogenic potential of mammalian muller glia. Int J Stem Cells. 2016;9:169–175. doi: 10.15283/ijsc16020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldman D. Müller glial cell reprogramming and retina regeneration. Nat Rev Neurosci. 2014;15:431–442. doi: 10.1038/nrn3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramachandran R, Zhao X-F, Goldman D. Ascl1a/Dkk/beta-catenin signaling pathway is necessary and glycogen synthase kinase-3 inhibition is sufficient for zebrafish retina regeneration. Proc Natl Acad Sci U S A. 2011;108:15858–15863. doi: 10.1073/pnas.1107220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan J, Ramachandran R, Goldman D. HB-EGF is necessary and sufficient for Müller glia dedifferentiation and retina regeneration. Dev Cell. 2012;22:334–347. doi: 10.1016/j.devcel.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson CM, Gorsuch RA, Bailey TJ, Ackerman KM, Kassen SC, Hyde DR. Stat3 defines three populations of muller glia and is required for initiating maximal muller glia proliferation in the regenerating zebrafish retina. J Comp Neurol. 2012;520:4294–4311. doi: 10.1002/cne.23213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyers JR, Hu L, Moses A, Kaboli K, Papandrea A, Raymond PA. β-catenin/Wnt signaling controls progenitor fate in the developing and regenerating zebrafish retina. Neural Dev. 2012;7:30. doi: 10.1186/1749-8104-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fausett BV, Gumerson JD, Goldman D. The proneural basic helix-loop-helix gene ascl1a is required for retina regeneration. J Neurosci. 2008;28:1109–1117. doi: 10.1523/JNEUROSCI.4853-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates Müller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat Cell Biol. 2010;12:1101–1107. doi: 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramachandran R, Zhao XF, Goldman D. Insm1a-mediated gene repression is essential for the formation and differentiation of Müller glia-derived progenitors in the injured retina. Nat Cell Biol. 2012;14:1013–1023. doi: 10.1038/ncb2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson CM, Ackerman KM, O'Hayer P, Bailey TJ, Gorsuch RA, Hyde DR. Tumor necrosis factor-alpha is produced by dying retinal neurons and is required for Muller glia proliferation during zebrafish retinal regeneration. J Neurosci. 2013;33:6524–6539. doi: 10.1523/JNEUROSCI.3838-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cenik B, Sephton CF, Cenik BK, Herz J, Yu G. Progranulin: a proteolytically processed protein at the crossroads of inflammation and neurodegeneration. J Biol Chem. 2012;287:32298–32306. doi: 10.1074/jbc.R112.399170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniel R, He Z, Carmichael KP, Halper J, Bateman A. Cellular localization of gene expression for progranulin. J Histochem Cytochem. 2000;48:999–1009. doi: 10.1177/002215540004800713. [DOI] [PubMed] [Google Scholar]
- 19.Petkau TL, Leavitt BR. Progranulin in neurodegenerative disease. Trends Neurosci. 2014;37:388–398. doi: 10.1016/j.tins.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Jian J, Konopka J, Liu C. Insights into the role of progranulin in immunity, infection, and inflammation. J Leukoc Biol. 2013;93:199–208. doi: 10.1189/jlb.0812429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Y, Xu X, Liu L, et al. Progranulin deficiency leads to severe inflammation, lung injury and cell death in a mouse model of endotoxic shock. J Cell Mol Med. 2016;20:506–517. doi: 10.1111/jcmm.12756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menzel L, Kleber L, Friedrich C, et al. Progranulin protects against exaggerated axonal injury and astrogliosis following traumatic brain injury. Glia. 2017;65:278–292. doi: 10.1002/glia.23091. [DOI] [PubMed] [Google Scholar]
- 23.Egashira Y, Suzuki Y, Azuma Y, et al. The growth factor progranulin attenuates neuronal injury induced by cerebral ischemia-reperfusion through the suppression of neutrophil recruitment. J Neuroinflammation. 2013;10:105. doi: 10.1186/1742-2094-10-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsuruma K, Yamauchi M, Sugitani S, et al. Progranulin, a major secreted protein of mouse adipose-derived stem cells, inhibits light-induced retinal degeneration. Stem Cells Transl Med. 2014;3:42–53. doi: 10.5966/sctm.2013-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuse Y, Tsuruma K, Sugitani S, et al. Progranulin promotes the retinal precursor cell proliferation and the photoreceptor differentiation in the mouse retina. Sci Rep. 2016;6:23811. doi: 10.1038/srep23811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuse Y, Tsuruma K, Mizoguchi T, Shimazawa M, Hara H. Progranulin deficiency causes the retinal ganglion cell loss during development. Sci Rep. 2017;7:1679. doi: 10.1038/s41598-017-01933-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cadieux B, Chitramuthu BP, Baranowski D, Bennett HPJ. The zebrafish progranulin gene family and antisense transcripts. BioMed Cent Genomics. 2005;6:156. doi: 10.1186/1471-2164-6-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li YH, Chen MHC, Gong HY, et al. Progranulin A-mediated MET signaling is essential for liver morphogenesis in zebrafish. J Biol Chem. 2010;285:41001–41009. doi: 10.1074/jbc.M110.138743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li YH, Chen HY, Li YW, et al. Progranulin regulates zebrafish muscle growth and regeneration through maintaining the pool of myogenic progenitor cells. Sci Rep. 2013;3:1176. doi: 10.1038/srep01176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Craig SEL, Calinescu A-A, Hitchcock PF. Identification of the molecular signatures integral to regenerating photoreceptors in the retina of the zebra fish. J Ocul Biol Dis Infor. 2008;1:73–84. doi: 10.1007/s12177-008-9011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh CE, Hitchcock PF. Progranulin regulates neurogenesis in the developing vertebrate retina. Dev Neurobiol. 2017;77:1114–1129. doi: 10.1002/dneu.22499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fausett BV, Goldman D. A role for 1 tubulin-expressing Muller glia in regeneration of the injured zebrafish retina. J Neurosci. 2006;26:6303–6313. doi: 10.1523/JNEUROSCI.0332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramachandran R, Reifler A, Parent JM, Goldman D. Conditional gene expression and lineage tracing of tuba1a expressing cells during zebrafish development and retina regeneration. J Comp Neurol. 2010;518:4196–4212. doi: 10.1002/cne.22448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barthel LK, Raymond PA. In situ hybridization studies of retinal neurons. Methods Enzymol. 2000;316:579–590. doi: 10.1016/s0076-6879(00)16751-5. [DOI] [PubMed] [Google Scholar]
- 35.Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial Muller glia that function as retinal stem cells. J Neurosci. 2007;27:7028–7040. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Del Rio-Tsonis K, Washabaugh CH, Tsonis PA. Expression of pax-6 during urodele eye development and lens regeneration. Proc Natl Acad Sci U S A. 1995;92:5092–5096. doi: 10.1073/pnas.92.11.5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben Khadra Y, Said K, Thorndyke M, Martinez P. Homeobox genes expressed during echinoderm arm regeneration. Biochem Genet. 2014;52:166–180. doi: 10.1007/s10528-013-9637-2. [DOI] [PubMed] [Google Scholar]
- 38.Powell C, Grant AR, Cornblath E, Goldman D. Analysis of DNA methylation reveals a partial reprogramming of the Muller glia genome during retina regeneration. Proc Natl Acad Sci U S A. 2013;110:19814–19819. doi: 10.1073/pnas.1312009110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagashima M, Barthel LK, Raymond PA. A self-renewing division of zebrafish Müller glial cells generates neuronal progenitors that require N-cadherin to regenerate retinal neurons. Development. 2013;140:4510–4521. doi: 10.1242/dev.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keightley MC, Wang CH, Pazhakh V, Lieschke GJ. Delineating the roles of neutrophils and macrophages in zebrafish regeneration models. Int J Biochem Cell Biol. 2014;56:92–106. doi: 10.1016/j.biocel.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 41.Neirinckx V, Coste C, Franzen R, Gothot A, Rogister B, Wislet S. Neutrophil contribution to spinal cord injury and repair. J Neuroinflammation. 2014;11:150. doi: 10.1186/s12974-014-0150-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin Wan, Xiao-Feng Zhao, Anne Vojtek DG. Retinal injury, growth factors and cytokines converge on β-catenin and pStat3 signaling to stimulate retina regeneration. Cell Rep. 2014;29:997–1003. doi: 10.1016/j.celrep.2014.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorsuch RA, Lahne M, Yarka CE, Petravick ME, Li J, Hyde DR. Sox2 regulates Müller glia reprogramming and proliferation in the regenerating zebrafish retina via Lin28 and Ascl1a. Exp Eye Res. 2017;161:174–192. doi: 10.1016/j.exer.2017.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao X-F, Wan J, Powell C, Ramachandran R, Myers MG, Jr, Goldman D. Leptin and IL-6 family cytokines synergize to stimulate müller glia reprogramming and retina regeneration. Cell Rep. 2014;9:272–284. doi: 10.1016/j.celrep.2014.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frobøse H, Groth Rønn S, Heding PE, et al. Suppressor of cytokine signaling-3 inhibits interleukin-1 signaling by targeting the TRAF-6/TAK1 complex. Mol Endocrinol. 2006;20:1587–1596. doi: 10.1210/me.2005-0301. [DOI] [PubMed] [Google Scholar]
- 46.Rui L, Yuan M, Frantz D, Shoelson S, White MF. SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. J Biol Chem. 2002;277:42394–42398. doi: 10.1074/jbc.C200444200. [DOI] [PubMed] [Google Scholar]
- 47.Fischer AJ, McGuire CR, Dierks BD, Reh TA. Insulin and fibroblast growth factor 2 activate a neurogenic program in Müller glia of the chicken retina. J Neurosci. 2002;22:9387–9398. doi: 10.1523/JNEUROSCI.22-21-09387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu K, Zhang Y, Ilalov K, et al. Cartilage oligomeric matrix protein associates with Granulin-Epithelin Precursor (GEP) and potentiates GEP-stimulated chondrocyte proliferation. J Biol Chem. 2007;282:11347–11355. doi: 10.1074/jbc.M608744200. [DOI] [PubMed] [Google Scholar]
- 49.Tang W, Lu Y, Tian Q-Y, et al. The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science. 2011;332:478–484. doi: 10.1126/science.1199214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.