Abstract
Viruses hijack and modify host cell functions to maximize viral proliferation. Hepatitis C virus (HCV) reorganizes host cell metabolism to produce specialized membrane structures and to modify organelles such as double-membrane vesicles and enlarged lipid droplets (LDs), thereby enabling virus replication and assembly. However, the molecular bases of these host–HCV interactions are largely unknown. Here, using a chemical screen, we demonstrate that the benzamide derivative flutamide reduces the host capacity to produce infectious HCV. Flutamide disrupted the formation of enlarged LDs in HCV-infected cells, thereby abolishing HCV assembly. We also report that aryl hydrocarbon receptor (AhR), a known flutamide target, plays a key role in mediating LD accumulation and HCV production. This AhR function in lipid production was also observed in HCV-uninfected Huh-7 cells and primary human hepatocytes, suggesting that AhR signaling regulates lipid accumulation independently of HCV infection. We further observed that a downstream activity, that of cytochrome P450 1A1 (CYP1A1), was the primary regulator of AhR-mediated lipid production. Specifically, blockade of AhR-induced CYP1A1 up-regulation counteracted LD overproduction, and overproduction of CYP1A1, but not of CYP1B1, in AhR-inactivated cells restored lipid accumulation. Of note, HCV infection up-regulated the AhR–CYP1A1 pathway, resulting in the accumulation of enlarged LDs. In conclusion, we demonstrate that the AhR–CYP1A1 pathway has a significant role in lipid accumulation, a hallmark of HCV infection that maximizes progeny virus production. Our chemical–genetic analysis reveals a new strategy and lead compounds to control hepatic lipid accumulation as well as HCV infection.
Keywords: aryl hydrocarbon receptor (AhR) (AHR), cytochrome P450, Hepatitis C virus (HCV), lipid droplet, hepatocyte, virus assembly, fatty acid metabolism, triglyceride, CYP1A1, hepatitis
Introduction
Viruses have minimum structures and lack metabolic pathways. To ensure their own survival, viruses hijack host cellular machineries and reorganize cellular environments to achieve efficient production of progeny virus (1, 2). Hepatitis C virus (HCV)2 infection dynamically alters membrane structures or organelles in host hepatocytes, leading to the formation of unique membrane compartments. These novel structures include double membrane vesicles that serve as factories for viral genome replication, as well as lipid droplets (LDs) that allow efficient particle assembly (3–8). However, the molecular mechanism underlying this HCV-induced cellular reorganization remains poorly understood.
After replication of HCV, viral RNA accumulates inside the double membrane vesicles, which are isolated membrane structures derived from the endoplasmic reticulum/Golgi or related membranes. HCV RNA then combines with viral structural proteins to assemble into progeny particles in association with the LDs. We previously reported that the surface membranes of the enlarged LDs serve as a platform for particle assembly (3), suggesting that LD metabolism in hepatocytes is closely related to permissivity for HCV production. In addition, it has been reported that the overproduction of lipids is a risk factor for hepatocarcinogenesis; HCV core-transgenic mice exhibit hepatic steatosis and develop hepatocellular carcinoma (HCC), events that correlate with the hepatic lipid accumulation (9, 10). In clinical settings, HCV-infected patients with hepatic steatosis develop HCC at higher rates (11, 12). In nonalcoholic steatohepatitis (NASH), which is not related to viral hepatitis, hepatic lipid accumulation plays a pivotal role during HCC development (13, 14). Thus, clarification of the mechanisms underlying hepatic lipid accumulation is of significant importance for understanding liver pathogenesis as well as virus–host interaction. Here, we reveal a role for the aryl hydrocarbon receptor (AhR) in hepatic lipid accumulation.
AhR is a ligand-activated transcription factor classified as a member of the basic helix-loop-helix superfamily, members of which act as biological sensors by initiating gene expression programs in response to a variety of signals, including xenobiotics such as benzo[a]pyrene, tetrachlorodibenzo-p-dioxin (TCDD), and 3-methylcholanthrene (15). Upon ligand binding, AhR translocates into the nucleus and binds to its partner protein, AhR nuclear translocator; the resulting heterodimer is then recruited to the xenobiotic response element (XRE), a DNA-binding element located in the promoter regions of various genes, leading to the induction of a number of downstream genes (16). A list of the downstream genes includes those encoding members of the cytochrome P450 (CYP) family (e.g. CYP1A1, CYP1A2, and CYP1B1) (17) that are involved in the metabolism of xenobiotics. Notably, CYP1A1 is among the genes most strongly induced by AhR, and CYP1A1 protein directly hydroxylates or oxidizes the ligand xenobiotics that then can be excreted or themselves exert biological activities (18–20). Thus, the AhR–CYP pathway is implicated primarily in xenobiotic homeostasis. AhR also is involved in many other physiological processes, including immune regulation, cell development, and cell cycle regulation (21–24).
In the present study, we screened a chemical library using a HCV cell culture–based assay and identified flutamide based on the compound's ability to decrease the host capacity to support HCV assembly. Using flutamide as a chemical probe, we showed that the AhR–CYP1A1 pathway plays a significant role in the accumulation of LDs and thus the production of HCV. Furthermore, HCV infection activated this AhR pathway, a mechanism that likely maximizes viral assembly in infected hepatocytes. Thus, we identified a novel role for the AhR–CYP1A1 pathway in lipid metabolism and HCV production, which may serve as a drug target.
Results
Flutamide reduces the host cell capacity to produce infectious HCV
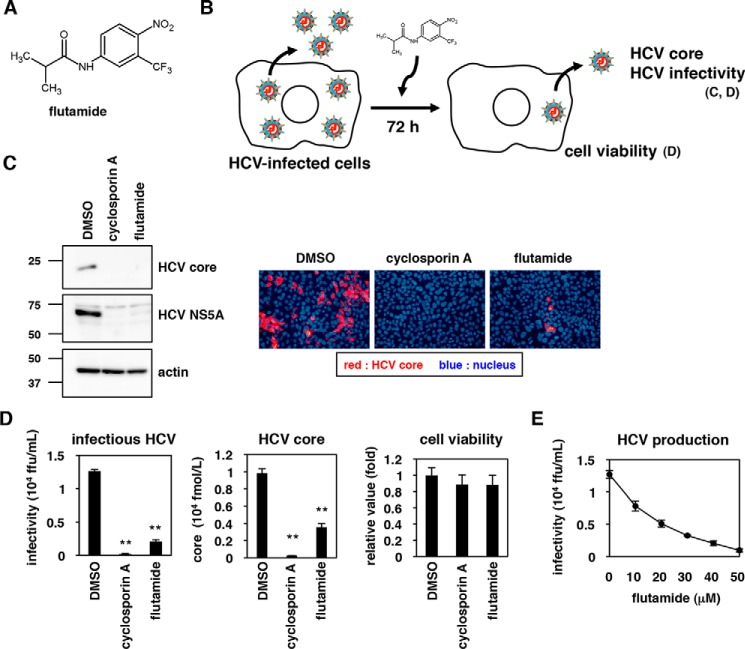
To identify pharmacological agents affecting HCV production, we screened a chemical library in HCV RNA-transfected Huh7-25 cells and measured changes in the production of infectious HCV following compound treatment (see “Experimental procedures”). This screen identified flutamide, a benzamide derivative (Fig. 1A), as a highly potent compound that reduced HCV production without apparent cytotoxicity. To confirm the anti-HCV effect of flutamide, HCV-infected Huh-7 cells were treated with flutamide, DMSO, or cyclosporin A (CsA; a known inhibitor of HCV replication (25)) for 72 h (Fig. 1B), and the supernatant was recovered. Infectivity of HCV in the supernatant was evaluated by inoculation to naive Huh-7.5.1 cells; HCV protein production then was assessed in the cells at 48 h post-inoculation by immunoblot and immunofluorescence (Fig. 1C). As shown in Fig. 1C, HCV in the culture supernatant of flutamide- as well as CsA-treated Huh-7 cells presented lower or even undetectable levels of HCV core and NS5A (nonstructural protein 5A) proteins (Fig. 1C). Quantitative analysis by the focus-formation assay using the culture supernatant showed that flutamide treatment yielded an ∼10-fold decrease in HCV infectivity, and an ∼3-fold decrease in HCV core protein secretion, without apparent cytotoxicity (Fig. 1D). This reduction in HCV infectivity depended on flutamide concentration (Fig. 1E). Thus, flutamide impairs the host capacity to support HCV production.
Figure 1.
Flutamide reduces the host cell capacity to produce infectious HCV. A, chemical structure of flutamide. B, schematic representation of the assay used to determine the effect of flutamide on HCV production. C, HCV-infected Huh-7 cells were cultured for 72 h in the presence or absence of the indicated compounds (0.1% DMSO, 3 μg/ml cyclosporin A, 40 μm flutamide) to allow HCV production. To evaluate the production of HCV, HCV produced in the culture supernatant was recovered and re-inoculated into naive Huh-7.5.1 cells to detect the resulting HCV core protein and HCV NS5A protein, as well as actin in the cells, at 48 h post-inoculation by immunoblot (left panels) and immunofluorescence (right panels). Red and blue signals in the right panels indicate HCV core protein and the nucleus, respectively. D, HCV-infected Huh-7 cells were cultured with or without the indicated compounds for 72 h as shown in A and C. The resultant culture supernatant was recovered to quantify the infectivity of HCV (left) and the amount of HCV core protein (center) by a focus-forming assay and CLEIA, respectively. Cell viability also was examined by an MTT assay of the cells (right). E, HCV production upon treatment with various concentrations (0, 10, 20, 30, 40, and 50 μm) of flutamide was measured as shown in A, C, and D. The data are presented as the means of three independent experiments, with error bars indicating S.D. Statistical significance was determined by Student's t test (*, p < 0.05; **, p < 0.01).
HCV assembly is impaired in flutamide-treated cells
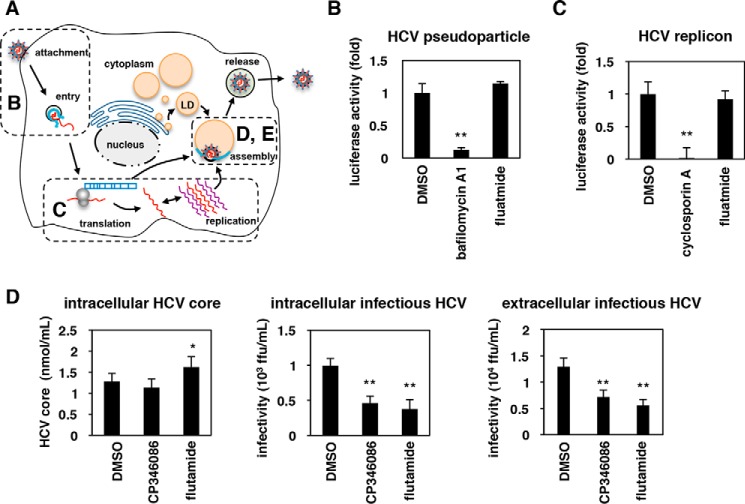
We investigated which process in the HCV life cycle was abolished in flutamide-treated cells (Fig. 2A). The earliest steps of the HCV life cycle, including attachment/entry, can be evaluated using HCV pseudoparticles (HCVpp), which consist of retrovirus-based pseudoviruses carrying HCV envelope proteins that enter into cells in an HCV envelope–dependent manner (see “Experimental procedures”). As shown in Fig. 2B, flutamide had little effect on HCVpp infection, in contrast to bafilomycin A1, a known inhibitor of HCV entry (26–28) (Fig. 2B). The middle phase of the life cycle, including translation and replication, is reproduced by a HCV subgenomic replicon system that encodes the HCV nonstructural proteins essential for RNA replication, but not the structural proteins required for viral assembly (see “Experimental procedures”). As shown in Fig. 2C, the replication activity was decreased by treatment with CsA, a known inhibitor of HCV replication, but not by treatment with flutamide (Fig. 2C). To investigate viral assembly, we extracted HCV that had accumulated inside cells to quantify the intracellular HCV infectivity in HCV RNA-transfected Huh7-25 cells (single-cycle virus production assay; see “Experimental procedures”) (Fig. 2A). Flutamide significantly decreased the amount of infectious HCV that accumulated inside the cells (Fig. 2D, center) without reducing the production of intracellular HCV core protein (Fig. 2D, left), as was the case with CP346086, a known inhibitor of HCV assembly (29) used as a positive control. Infectious HCV secreted outside of the cells was consistently reduced by flutamide treatment (Fig. 2D, right). Thus, flutamide-treated cells show a decreased ability to support the assembly of infectious HCV.
Figure 2.
The efficiency of HCV assembly is impaired in flutamide-treated cells. A, schematic representation of the HCV life cycle. HCVpp (B), the subgenomic replicon (C), and single-cycle virus production assay using the infection system (D and E) were used to evaluate the activity of individual steps (attachment/entry, translation/replication, and assembly, respectively). B, HCVpp assay for measuring the activity of HCV attachment/entry. Huh-7.5.1 cells pretreated with compounds (0.1% DMSO, 10 nm bafilomycin A1, and 40 μm flutamide) for 1 h were inoculated with HCVpp for 4 h in the presence of compounds. At 72 h post-inoculation, cells were lysed and assessed for luciferase activity generated by HCVpp infection. The relative luciferase activities are shown. C, HCV replicon assay monitoring translation/replication. Huh-7.5.1 cells transfected with an HCV subgenomic replicon RNA were treated with the compounds (0.1% DMSO, 3 μg/ml cyclosporin A, 40 μm flutamide) for 36 h. The luciferase activity driven from the replicon was determined, and the relative values are shown. D, single-cycle virus production assay to evaluate virus assembly. HCV RNA-transfected Huh7-25 cells (CD81(−)) were cultured for 72 h in the presence or absence of 0.1% DMSO, 30 μm flutamide, or 8 μm CP346086 to allow virus production. HCV particles accumulated inside the cells were extracted to quantify the intracellular HCV core protein level (left) and the infectivity of intracellular HCV (center). Infectivity of HCV secreted into the culture supernatant was also quantified (extracellular infectious HCV, right). The data are presented as the means of three independent experiments with error bars indicating S.D. Statistical significance was determined by Student's t test (*, p < 0.05; **, p < 0.01).
AhR supports the production of HCV
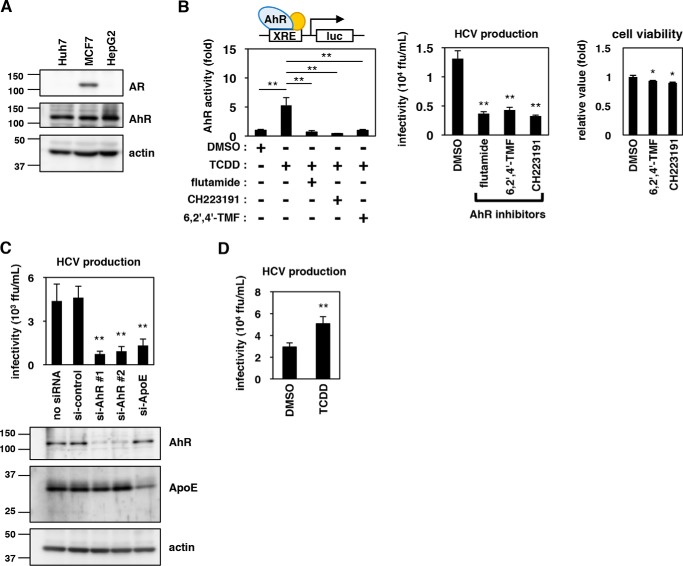
Flutamide is known to inhibit the transcriptional activity of androgen receptor (AR) and is used as a therapeutic agent against prostate cancer (30). However, AR was not detected by our immunoblot analysis of hepatocyte cell lines, including Huh-7 and HepG2 cells, in contrast to MCF7 cells, which are known to express AR and were used as a positive control (31) (Fig. 3A). Flutamide has also been reported to modulate the activity of another transcription factor, AhR (30). Notably, our initial chemical screening identified another AhR modulator, 3,3′-diindolylmethane, as a compound that impaired HCV production; we therefore examined AhR as a potential target of flutamide that might be responsible for the regulation of HCV assembly. We also tested other known AhR inhibitors, 6,2′,4′-trimethoxyflavone (6,2′,4′-TMF) and CH223191 (Fig. 3B, left), and found that all of these compounds clearly reduced the production of HCV (Fig. 3B, center) without showing remarkable cytotoxicity (Fig. 3B, right). In addition, knockdown of endogenous AhR expression significantly reduced Huh-7's capacity to produce infectious HCV (Fig. 3C). Moreover, Huh-7 cells treated with TCDD, a compound known to activate AhR (Fig. 3B, left), showed an elevated capacity to produce HCV (Fig. 3D). These results cumulatively suggested that AhR activity regulates the ability to support HCV production.
Figure 3.
AhR signaling mediates HCV production. A, endogenous protein expression of AR, AhR, and actin in Huh-7, HepG2, and MCF7 cells was determined by immunoblotting. B, left, AhR transcriptional activity was assessed using a reporter plasmid carrying a tandem repeat of the XRE upstream of the luciferase-encoding gene. Huh-7 cells transfected with this reporter plasmid were treated with the indicated compounds (DMSO, TCDD, flutamide, 6,2′,4′-TMF, and CH223191) for 48 h and were assessed for AhR transcriptional activity by a luciferase assay. Center, HCV production was quantified as shown in Fig. 1D following treatment with DMSO or AhR inhibitors (flutamide, 6,2′,4′-TMF, and CH223191) for 72 h. Right, cell viability was determined by MTT assay upon treatment with the indicated compounds. C, Huh-7 cells transfected with or without a randomized siRNA (si-control) or siRNAs targeting the genes encoding AhR (si-AhR #1 and si-AhR #2) or ApoE as a positive control (si-ApoE) were cultured for 72 h to evaluate HCV production as shown in Fig. 3B (right). Protein expression levels of AhR, ApoE, and actin in these cells were examined by immunoblot (left). D, HCV production was measured as shown in Fig. 3B following treatment with DMSO or TCDD, an AhR activator. The data are presented as the means of three independent experiments with error bars indicating S.D. Statistical significance was determined by Student's t test (*, p < 0.05; **, p < 0.01).
Flutamide disrupts LD accumulation
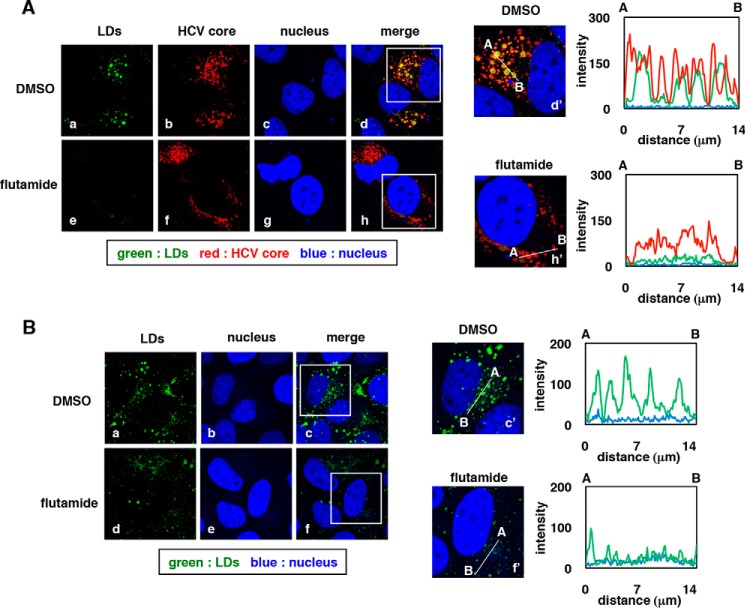
How might AhR modulators affect the host cell's capacity to support HCV assembly? We have previously reported that the viral assembly process occurs on the surfaces of the LDs, which apparently serve as platforms for the formation of infectious HCV (3). Notably, LDs (detected by BODIPY493/503 fluorescence) were markedly disrupted in HCV-infected cells following treatment with flutamide (Fig. 4A, a versus e). Colocalization of HCV core and LDs (Fig. 4A, d and d′ versus h and h′, shown by yellow) was also greatly reduced following flutamide treatment (fluorescence intensities are shown on the right graph in d′ and h′). As HCV core localization to the LDs is essential for HCV assembly (3, 4, 32), the observed loss of HCV core–LD colocalization is likely to be responsible for the reduced HCV assembly by flutamide. Following flutamide exposure, the HCV core protein level was reduced, although the change in the expression level was not as severe as that seen for the LDs themselves (Fig. 4A, b versus f). To examine whether the flutamide effect on LD disruption was direct or due to the elimination of HCV from infected cells, we examined the flutamide effect in uninfected Huh-7 cells. As shown in Fig. 4B, LDs in uninfected cells also were greatly reduced upon flutamide treatment (Fig. 4B, a versus d). As seen in Fig. 4A, the quantified intensity of LDs was decreased following flutamide exposure (Fig. 4B, c′ versus f′, right). Thus, these results suggested that flutamide disrupts LD accumulation independently of HCV infection.
Figure 4.
LDs are disrupted upon flutamide treatment. A, HCV-infected Huh-7 cells treated with DMSO (0.1%) or flutamide (40 μm) were fixed, and immunofluorescence was assessed following staining with anti-HCV core antibody (HCV core; red), BODIPY493/503 (LDs; green), and DAPI (nucleus; blue). d and h are merged images for a–c and e–g, respectively. d′ and h′ are large magnifications of the indicated insets in d and h, respectively. Right graphs indicate the intensity for green (LDs), red (HCV core), and blue (nucleus) signals on the line A–B inside the cell shown in d′ and h′. y and x axes indicate signal intensity and the distance from A (μm), respectively. B, uninfected Huh-7 cells treated with DMSO or flutamide were fixed and labeled with BODIPY493/503 (LDs; green) and DAPI (nucleus; blue). c and f, merged images for panels a and b and panels d and e, respectively. Presentation of data is the same as in A.
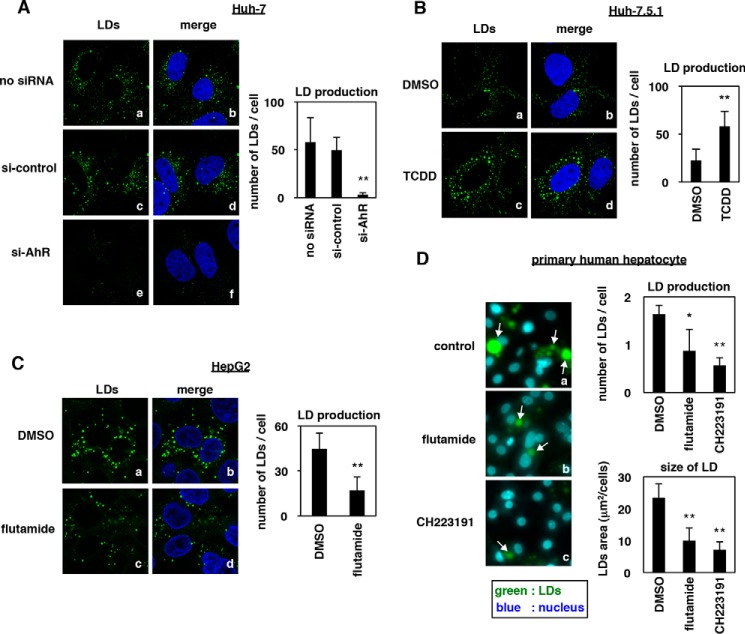
AhR activity regulates the accumulation of LDs
Consistent with the essential role of AhR in the effect of flutamide, LD production was reduced in Huh-7 cells transfected with an siRNA against AhR (Fig. 5A, a, c, and e) and was enhanced in Huh-7.5.1 cells treated with an AhR activator, TCDD (Fig. 5B, a and c). Consistent with these results, these phenomena were also seen in another hepatoma cell line, HepG2, and in primary human hepatocytes; LDs in HepG2 and primary human hepatocytes were reduced by flutamide or another AhR inhibitor, CH223191 (Fig. 5, C (a versus c) and D). These data suggested that AhR activity determines the LD level in hepatocytes.
Figure 5.
AhR activity regulates the accumulation of LDs. A, uninfected Huh-7 cells transfected with si-control or si-AhR or left untreated were fixed and stained with BODIPY493/503 (LDs; green) and DAPI (nucleus; blue). B–D, uninfected Huh-7.5.1 cells (B), HepG2 cells (C), or primary human hepatocytes (D) treated with DMSO, TCDD, flutamide, or CH223191 were stained to visualize LDs (green) and the nucleus (blue). The white arrows in D indicate LDs. Quantified number of LDs per cell (A–D) and the area of LDs per cell (D) are shown as the mean values. The data are presented as the means of three independent experiments with error bars indicating S.D. Statistical significance was determined by Student's t test (*, p < 0.05; **, p < 0.01).
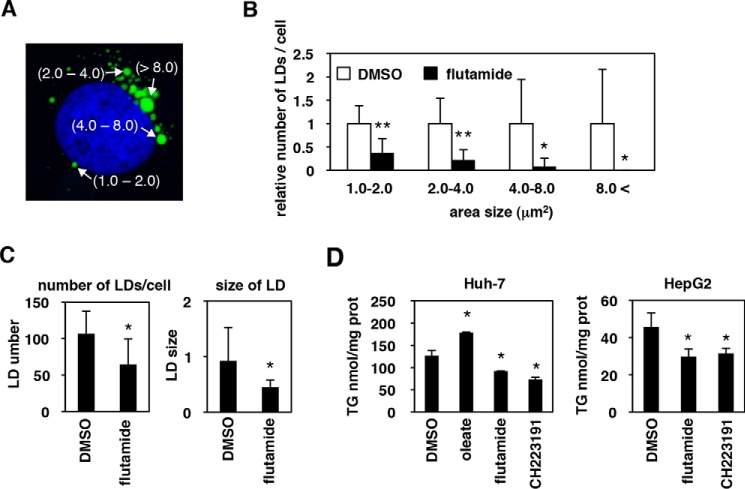
Triglyceride, as well as the size and number of LDs, are reduced in flutamide-treated cells
We further performed a fine quantification analysis for the size and the number of LDs in Huh-7 cells treated with or without flutamide. As shown in Fig. 6A, untreated cells showed a typical pattern upon staining with BODIPY493/503, with LDs visible at a range of different sizes within each cell (Fig. 6A). We classified each LD into four categories based on size (1–2, 2–4, 4–8, and >8 μm2) and counted the number of LDs for each category in cells treated with or without flutamide (see “Experimental procedures”). As seen in Fig. 6B, which shows the relative number of LDs/cell for each LD size category, flutamide-treated cells presented significantly reduced numbers of LDs of all sizes, with LDs of the largest sizes (4–8 and >8 μm2) showing notable decreases in number (Fig. 6B). The resulting mean number of LDs per cell and the mean LD size were accordingly reduced by treatment with flutamide (Fig. 6C). These data are in agreement with the possibility that a component of the LDs is depleted, rather than indicating that the LD budding/fusion process is attenuated upon flutamide exposure. Indeed, the total amount of triglyceride, one of the main component of LDs (33, 34), was significantly reduced in Huh-7 cells treated with flutamide or with CH223191 (Fig. 6D, left), although the sensitivity of the assay was not as high as immunofluorescence detection. Such a reduction in the amount of triglyceride by these inhibitors also was observed in HepG2 cells (Fig. 6D, right). Together, these results indicated that inactivation of AhR decreases the amounts of LDs and of triglyceride in hepatocytes.
Figure 6.
The number and the size of LDs, as well as triglyceride levels, are reduced upon AhR-inactivation. A, a typical image of the LDs (green) as well as the nucleus (blue) in untreated Huh-7 cells. LD sizes quantified with MetaMorph software are shown, classified into 1–2, 2–4, 4–8, and >8 μm2. B, the number of LDs in Huh-7 cells treated with (black) or without (white) flutamide was counted for each LD size category (1–2, 2–4, 4–8, and >8 μm2) and is shown as relative numbers of LDs in a given cell. C, mean numbers of total LDs in all categories in a given cell (left) and the mean size of LDs in all categories (right) were calculated and are shown in B. D, Huh-7 (left) and HepG2 cells (right) were cultured with the indicated compound for 72 h, and the total neutral lipid was extracted from these cells to quantify triglyceride as described under “Experimental procedures.” The data are presented as the means of three independent experiments with error bars indicating S.D. Statistical significance was determined by Student's t test (*, p < 0.05; **, p < 0.01).
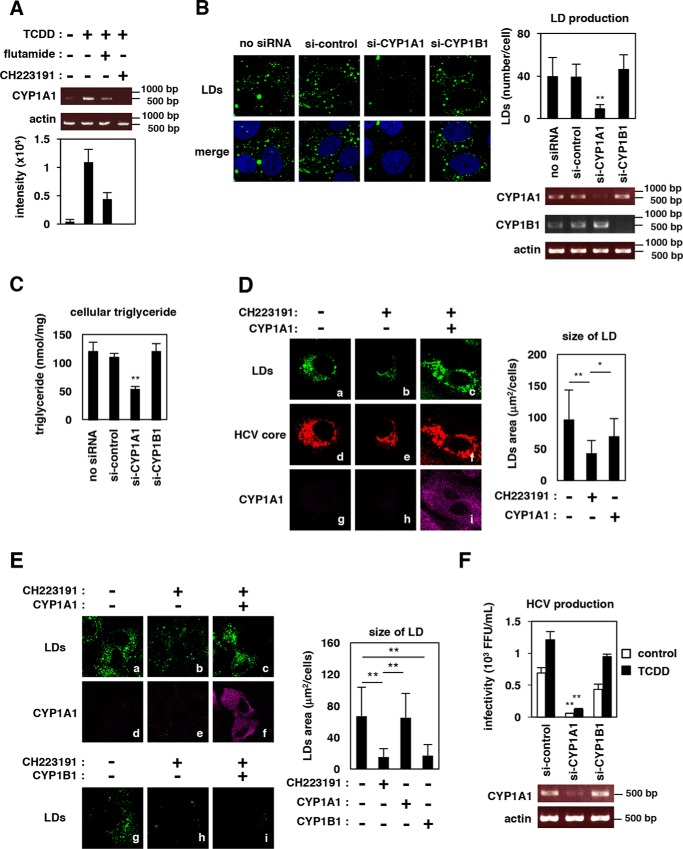
CYP1A1 plays a significant role in mediating lipid production
AhR is a ligand-activated transcription factor that regulates the expression of downstream genes involved in many physiological processes, particularly those employed in responding to environmental signals (15). Major downstream genes induced by AhR activation include members of the CYP family, such as CYP1A1, CYP1B1, and CYP1A2; the products of these genes mediate xenobiotic and hormone metabolism (15, 35). To investigate which AhR downstream gene is predominantly involved in the regulation of lipid metabolism, we first examined whether major components of the triglyceride synthetic pathway (Fig. S1A) were induced by AhR activation. Notably, however, expression of the examined genes (SREBP1 (sterol regulatory element–binding protein 1), ACC1 (acetyl-CoA carboxylase 1), FASN (fatty acid synthase), SCD1 (stearoyl-CoA desaturase 1), SLC13A5 (solute carrier family 13 member 5), CD36 (cluster of differentiation 36), DGAT1 (diacylglycerol o-acyltransferase 1), and DGAT2) was not affected by AhR activation (Fig. S1B). In contrast, the major AhR downstream gene, CYP1A1, was clearly up-regulated upon exposure of cells to TCDD, an effect that was reversed by co-treatment with CH223191 or flutamide (Fig. S1B and Fig. 7A). The role for CYP1-family proteins in hepatic lipid metabolism has not been well-understood. Interestingly, we observed that LDs in hepatocytes were reduced in cells knocked down for CYP1A1, but not in cells knocked down for CYP1B1 (Fig. 7B). The amount of cellular triglyceride was also significantly reduced by knockdown of CYP1A1, but not CYP1B1 (Fig. 7C). Significantly, sole overexpression of CYP1A1 in HCV-infected cells rescued the reduction in LD level caused by AhR inactivation (Fig. 7D, b versus c). This LD rescue by complementary expression of CYP1A1 was also observed in uninfected cells (Fig. 7E, b versus c); again, overexpression of CYP1B1 did not provide such a rescue (Fig. 7E, h versus i). These results suggested that CYP1A1 has a significant role in the AhR-mediated regulation of LD accumulation in hepatocytes. Consequently, Huh-7 cells depleted for CYP1A1, but not those depleted for CYP1B1, had a decreased capacity to support HCV production (Fig. 7F), further confirming that the AhR–CYP1A1 pathway is significant in the cell permissiveness for HCV production.
Figure 7.
CYP1A1 mediates lipid production. A, Huh-7 cells were treated with the indicated compounds for 24 h, and their total RNA was recovered and subjected to RT-PCR to detect mRNAs encoding CYP1A1 and actin. Intensity of the bands with three independent experiments was quantified by densitometry, and the average values are shown in the bottom graph. B, uninfected Huh-7 cells transfected with or without si-control or siRNA against CYP1A1 (si-CYP1A1) or CYP1B1 (si-CYP1B1) were stained to visualize the LDs (green) and the nucleus (blue). The mean number of LDs per cell is shown as in Fig. 5 (right). mRNA expressions for CYP1A1, CYP1B1, and actin are also shown at the bottom right. C, Huh-7 cells transfected with siRNAs as shown in B were recovered to quantify triglyceride in the cells as shown in Fig. 6D. D, HCV-infected Huh-7 cells in the presence or absence of an AhR inhibitor, CH223191, were transfected with an expression plasmid encoding Myc-tagged CYP1A1 or the corresponding empty vector. LDs (green), HCV core (red), and Myc-CYP1A1 (purple) in these cells were detected. Quantification for fluorescence showing the LD area is shown on the right. E, uninfected Huh-7 cells cultured in the presence or absence of CH223191 were transfected with an expression plasmid encoding Myc-tagged CYP1A1 or CYP1B1 or the corresponding empty vector and were observed as shown in D. Fluorescence was quantified as in D. F, Huh-7 cells transfected with si-control, si-CYP1A1, or si-CYP1B1 were infected with HCV at a multiplicity of infection of 0.2 for 4 h and then were treated with or without the AhR agonist TCDD for 48 h to allow HCV production. HCV production was quantified by the focus-formation assay as in Fig. 1D. The data are presented as the means of three independent experiments with error bars indicating S.D. Statistical significance was determined by Student's t test (*, p < 0.05; **, p < 0.01).
Effect of AhR inhibitors on the production of different HCV strains
The results were obtained using HCV JFH-1, a genotype 2a strain used for a standard HCV cell culture assays (36). We examined the effect of AhR inhibition on the production of different HCV strains, J6/JFH-1 chimeric HCV (named Jc1-n, genotype 2a) and a genotype 3a S310 strain (37). As shown in Fig. 8A, AhR inhibition by treatment with either flutamide or CH223191 significantly reduced the production of HCV J6/JFH-1 (Jc1-n) (Fig. 8A). In contrast, production of HCV S310 was not reduced by treatment with flutamide, CH223191, or another assembly inhibitor, CP346086 (29) (Fig. 8B). Thus, the dependence of HCV assembly on the AhR pathway is likely to be different among HCV strains or genotypes (see “Discussion”).
Figure 8.
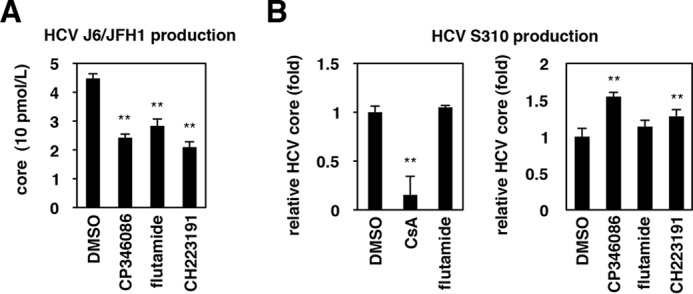
Effect of AhR inhibitors on the production of different HCV strains. A and B, Huh-7 cells transfected with HCV J6/JFH1 (Jc1-n) RNA (A) or infected with HCV S310 (B) were treated with or without the indicated compounds for 72 h. HCV core protein secreted into the culture supernatant was quantified as shown in Fig. 1D. Statistical significance was determined by Student's t test (*, p < 0.05; **, p < 0.01).
Activation of AhR in HCV-infected cells
AhR acts as a biological sensor and is modulated in response to environmental stresses such as xenobiotics, hypoxia, and circadian rhythm (38, 39). However, the relationship between virus infection and AhR activity remains poorly understood. As shown in Fig. 9A, the level of AhR-mediated transcription was increased in HCV JFH1-infected cells (Fig. 9A). Consistent with this observation, HCV-infected cells showed an accumulation of LDs (Fig. 9B), in confirmation of previous reports (3, 9, 40). When AhR activation in HCV-infected cells was counteracted by CH223191 treatment (Fig. 9A, lane 3), LD accumulation was attenuated (Fig. 9C, c). Together, these results suggested that HCV infection activates the AhR–CYP1A1 pathway to promote LD accumulation, thus enabling the efficient production of progeny virus (see “Discussion” and Fig. 10).
Figure 9.
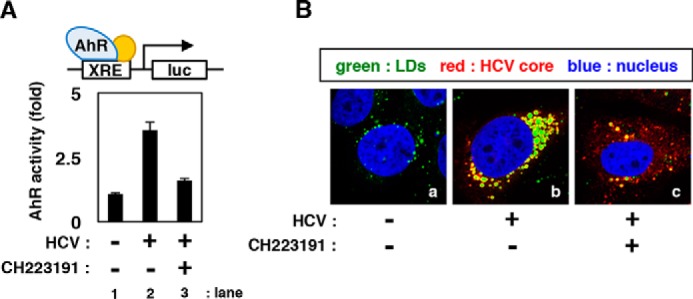
HCV triggers AhR activation and LD overproduction. A, HCV-infected Huh-7 cells transfected with an XRE-response luciferase reporter plasmid were treated with or without CH223191 for 72 h. AhR activity was quantified by measuring luciferase activity in the lysates of these cells; relative values are shown. B, HCV-infected or uninfected Huh-7 cells were treated with DMSO or CH223191 for 72 h. LDs (green), HCV core (red), and the nucleus (blue) were visualized. Error bars, S.D.
Figure 10.
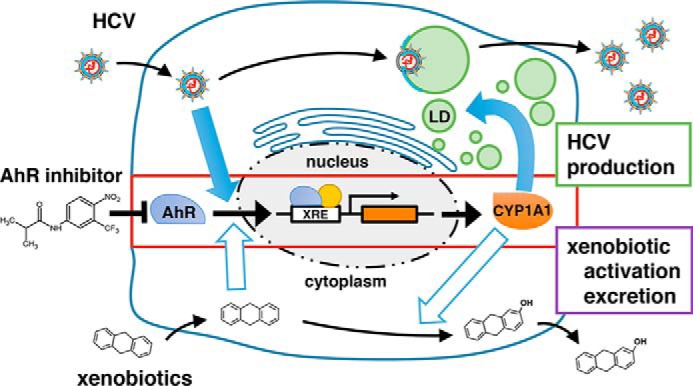
The AhR–CYP1A1 signaling pathway is critical for efficient HCV production. Shown is a schematic representation of the model for HCV to achieve efficient viral production. Top, HCV activates AhR-mediated transcription and up-regulates CYP1A1, the product of which induces LD overproduction. The accumulated LDs facilitate the efficient production of progeny particles. Bottom, xenobiotics such as TCDD (an AhR agonist) activate AhR and induce downstream genes, including CYP1A1. The products of these downstream genes hydrolyze the xenobiotics, yielding compounds that exhibit biological activities or are excreted. Flutamide or other AhR inhibitors block the dual role of the AhR–CYP1A1 pathway.
Discussion
In this study, a chemical screen identified flutamide as a compound that impaired the production of infectious HCV in hepatocytes. Using flutamide as a chemical probe, we showed that AhR regulated the production of triglyceride and the resultant LDs, thereby determining the host's capacity to produce HCV (Fig. 10). CYP1A1 was suggested to be a downstream gene that mediates AhR's regulation of lipid accumulation. On the other hand, HCV infection was shown to activate AhR with associated increases in the production of LDs. Based on the above evidence, we propose the model shown in Fig. 10 in which HCV activates the AhR–CYP1A1 pathway and reorganizes host cell functions to overproduce LDs, which serve as a platform for the production of progeny viruses (Fig. 10, top). This mechanism is in contrast to this pathway's original function, whereby incoming xenobiotics activate AhR and induce CYP1A1; the CYP1A1 protein then modifies xenobiotics, permitting the resulting compounds to exert biological activity or be excreted (Fig. 10, bottom). Flutamide and other AhR antagonists block both the xenobiotic response and the lipid accumulation that is critical for HCV production.
In this study, we mainly used HCV JFH-1, a genotype 2a strain used as a standard HCV in cell culture infection assays (36). Similar reduction in viral production upon flutamide treatment was observed using the Jc1-n (37) (Fig. 8A), another frequently used laboratory strain that harbors its core protein mainly on the endoplasmic reticulum but requires triglyceride synthesis for virus production (41). In contrast, production of HCV S310, a genotype 3 strain, was not decreased by either flutamide or CH223191 (Fig. 8B). Although the mechanism for assembly of genotype 3 HCV remains largely unknown, the dependence of viral assembly on triglyceride, and thus the AhR–CYP1A1 pathway, is likely to be diverse between HCV genotype 2 and 3 strains. It is interesting that the subcellular localization of core protein and LD content in S310-infected cells was different from those in JFH-1–infected cells (37). Further molecular analysis is needed to clarify the mechanisms underlying the assembly of multiple HCV genotypes, including the involvement of the AhR–CYP1A1 pathway.
AhR has been implicated in a variety of physiological events, including immune response, cell proliferation, and differentiation (21–24, 42). AhR, a well-characterized transcriptional regulator, was originally identified as a mediator of the xenobiotic response and homeostasis. Upon exposure of the cell to xenobiotics such as TCDD, AhR directly recognizes and binds to such xenobiotics; AhR then translocates to the nucleus, where the protein transactivates the expression of downstream genes. The activated genes include members of the CYP gene family (e.g. CYP1A1, CYP1A2, and CYP1B1) that encode proteins that directly hydrolyze the inducing ligands. There also have been reports (primarily using mouse models) suggesting that AhR functions in lipid metabolism, although these conclusions are diverse, depending on the experimental context. In one example, hepatic lipid was reported to accumulate in mouse liver following TCDD treatment and activation of AhR (43). In contrast, high-fat-diet–induced lipid accumulation was potentiated in the liver of AhR−/− mice compared with the effect in WT mice (44). Previous papers have also suggested a correlation between activation of the AhR signaling pathway and lipid metabolism–related genes, most notably with expression of the gene encoding the CD36, a free fatty acid transporter (45). Specifically, TCDD exposure yielded up-regulation of Cd36 in WT mice, whereas TCDD-induced lipid accumulation was attenuated in the liver of Cd36−/− mice. However, the majority of those reports used mouse models; there have been limited reports on the contribution of AhR and its downstream genes to lipid metabolism in human liver or human hepatocyte-derived cells. In the present paper, in contrast to the results in mice, TCDD-induced up-regulation of CD36 was not observed in human liver-derived cell lines. This result is in agreement with the observation that the XRE located in the mouse Cd36 promoter (at −1273 to −1263 bp with respect to the transcription start site), reported to be a key element for AhR regulation of Cd36 expression, is absent from the human CD36 promoter at the same position (45). In contrast, the human CYP1A1 promoter contains four putative XREs (46). These results suggest that distinct mechanisms of regulation are employed in different species for regulation of genes downstream of AhR. Whereas it has been reported that CYP1A1 is not necessarily abundant in expression in the liver (44), our data suggested that AhR's transcriptional activity was elevated in HCV-infected cells. This observation is consistent with a previous report showing that kynurenine, an endogenous AhR agonist, is produced to high levels in HCV-infected patients (47). Thus, HCV may achieve increased CYP1A1 expression in the liver of infected individuals even in the absence of stimulation with exogenous ligands (Fig. 10).
Besides serving as a platform for HCV production, lipid accumulation in the liver can be a basis for the development of HCC (10, 11). The AhR–CYP1A1 pathway may be closely associated with the lipid-induced pathogenesis that leads to HCC. Another cytochrome P450 protein, CYP2J2, has been reported to be associated with lipid accumulation and the related pathogenesis, including NASH; CYP2J2 transgenic mice showed a protective effect on the hepatic lipid accumulation induced by high-fat diet (33). CYP2J2-mediated regulation of bioactive epoxyeicosatrienoic acids, which show higher levels in both a mouse model and human biopsies of NASH, were shown to be a key to develop hepatic lipid accumulation and NASH (48). CYP1A1 is also reported to metabolize arachidonic acid and produce hydroxyeicosapentaenoic acids (49). Further studies are needed to clarify the mechanisms of how CYP1A1 mediates lipid production. In this study, it is significant that flutamide and other AhR antagonists identified in the present study are expected to be useful tools for clarifying the relevance of the AhR–CYP1A1 pathway in carcinogenesis while also serving as potential leads in drug development. Thus, the present study clarifies not only the mechanism of HCV-induced reorganization of host cells but also provides a potent novel target for developing a strategy against both virus infection and hepatic lipid accumulation.
Experimental procedures
Cell culture
Huh-7, Huh-7.5.1 (kindly provided by Dr. Francis Chisari at the Scripps Research Institute) (50), Huh7-25 (51), MCF7, and 293T cells were cultured in normal medium (Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Cell Culture Bioscience), 10 units/ml penicillin, 10 mg/ml streptomycin, 0.1 mm nonessential amino acids (Invitrogen), 1 mm sodium pyruvate, and 10 mm HEPES (pH 7.4)) at 37 °C in 5% CO2. For the experiment shown in Fig. 7, Huh-7 cells were cultured in lipid-depleted medium (DMEM-based normal medium with lipoprotein-deficient FBS (Sigma) in place of normal FBS). HepG2 cells were cultured with DMEM/F-12 + GlutaMax (Invitrogen) supplemented with 10 mm HEPES (Invitrogen), 200 units/ml penicillin, 200 μg/ml streptomycin, 10% FBS, and 5 μg/ml insulin at 37 °C in 5% CO2 (52). Primary human hepatocytes (Phoenixbio) were cultured as described previously (53).
Reagents
Flutamide, cyclosporin A, TCDD, and DMSO were purchased from Sigma-Aldrich. CH223191 and 6,2′,4′-TMF were purchased from Cayman Chemical Co. Bafilomycin A1 was purchased from Wako.
Plasmid transfection
Transfection of plasmids was performed with Trans IT LT1 (Mirus) or Lipofectamine 2000 (Invitrogen) according to the respective manufacturer's protocol.
RNA synthesis and transfection
RNAs were synthesized in vitro using MEGAscript T7 (Ambion) and isolated using the RNeasy Mini kit (Qiagen). RNA was transfected to cells as described previously (54).
HCV cell culture assay
In this study, we mainly used HCV JFH-1 (36) (Figs. 1, 2, 3, 4, 7, and 9). HCV was recovered from the medium of Huh-7 cells transfected with HCV JFH-1 RNA as described (36). HCV was used to infect Huh-7 or Huh-7.5.1 cells at a multiplicity of infection of 0.1–0.2 for 4 h. After washing out of the HCV inoculum, the cells were cultured in the presence or absence of various compounds for 48 or 72 h. Production of HCV was quantified by measuring the infectivity of HCV and the amount of HCV core protein in the culture supernatant by the infectious focus–forming assay and chemiluminescent enzyme immunoassay (CLEIA) (Lumipulse, Fujirebio) (54). For the focus-forming assay, naive Huh-7.5.1 cells were inoculated with HCV (at different dilutions) for 4 h, and then HCV-positive cells were visualized at 48 h post-inoculation by detecting HCV core protein using immunofluorescence analysis to count the foci and calculating focus-forming units/ml of the HCV inoculum (54). HCV core protein level was measured by CLEIA according to the manufacturer's protocol (54).
For the single-cycle virus production assay (55), HCV RNA-transfected Huh7-25 cells, which are deficient for the CD81 HCV entry receptor and therefore do not support viral entry (51), were cultured in the presence or absence of compounds for 72 h. Infectivity of HCV that had accumulated in the cells (intracellular infectivity) was quantified by extracting intracellular HCV through four freeze–thaw cycles to quantify viral infectivity as described (55). For the measurement of intracellular HCV core protein, cells were lysed using passive lysis buffer (Promega) and sonication; HCV core protein in the lysate was quantified by CLEIA as described.
We used other HCV strains, J6/JFH-1 chimeric HCV (Jc1-n) (genotype 2a) and S310 (genotype 3a) (37) (Fig. 8). Jc1-n carries the 5′-UTR and the coding region from core to p7 that is derived from HCV J6 (37) with that from NS2 to NS5B and the 3′-UTR derived from JFH-1. Production of Jc1-n and S310 was evaluated by quantifying HCV core produced from the cells transfected with HCV RNA or infected with HCV.
Chemical screen
HCV RNA-transfected Huh7-25 cells were treated with compounds for 72 h, and infectious HCV in the culture supernatant was quantified. Compounds reducing the production of infectious HCV to less than 33% of that of the DMSO-treated control, but without showing serious cytotoxicity (retaining >80% of the value of the DMSO control in the MTT assay) were selected as “hits.”
MTT assay
The viability of cells was quantified using the Cell Proliferation Kit II (XTT) (Roche Diagnostics) as described previously (54).
Immunoblot analysis
Immunoblot analysis was performed as described previously (54). The primary antibodies used were anti-HCV core (2H9) (54), anti-HCV NS5A (TB0705#1) (56), anti-AhR (Santa Cruz Biotechnology, Inc.), anti-AR (Santa Cruz Biotechnology), anti-apolipoprotein E (ApoE) (Millipore), and anti-actin (Sigma-Aldrich) antibodies.
HCV replicon assay
Huh-7.5.1 cells transfected with an HCV subgenomic replicon RNA that encodes the firefly luciferase gene upstream of the HCV nonstructural region (SGR-JFH1/Luc) were incubated with or without compounds for 36 h and then were assessed for luciferase activity as described (54).
HCVpp assay
HCVpp is a retrovirus-based pseudovirus carrying the HCV E1 and E2 envelope proteins; the pseudoparticle enters cells in an E1E2-dependent manner (54). HCVpp was recovered from the culture supernatant of 293T cells transfected with the expression plasmids for HCV E1E2, murine leukemia virus Gag-Pol, and luciferase (kindly provided by Dr. Francois-Loic Cosset at University de Lyon) as described (57). HCVpp was used to inoculate Huh-7.5.1 cells by 4 h of exposure; after washing out of unbound HCVpp, cells were incubated for another 72 h, and luciferase activity in cell lysates was measured to assess HCV E1E2-dependent entry activity.
Immunofluorescence analysis
Indirect immunofluorescence analysis was performed by fixation of the cells in 4% paraformaldehyde followed by permeabilization with 0.05% saponin as described (3). HCV core, the LDs, and the nucleus were detected with an anti-HCV core antibody (2H9) (54), BODIPY493/503, and DAPI (respectively).
RNAi technique
Huh-7 cells were transfected with 30 nm siRNAs consisting of randomized siRNA (si-control) or siRNAs targeting AhR (si-AhR), APOE (si-ApoE), or CYP1A1 and CYP1B1 (ABI) using Lipofectamine RNAiMAX (Invitrogen); transfection was performed twice at a 2-day interval, according to the manufacturer's protocol.
Quantification of LDs
The sizes of each LD and the number of LDs were quantified with MetaMorph software (Molecular Devices) (37). LDs were categorized into four groups based on the sizes (areas) of the LDs (1–2, 2–4, 4–8, and >8 μm2), and the number of LDs for each category in a given cell was counted (Fig. 6B). The total number of LDs in a given cell also was counted (Fig. 6C). The mean size of total LDs was calculated (Fig. 6C). These data were determined by observing 5–15 randomly selected cells for each experiment.
Quantification of intracellular triglyceride
The accumulation level of triglyceride in Huh-7 cells or HepG2 cells was quantified using the Adipogenesis Assay Kit (Biovision). Colorimetric analysis of cellular triglyceride levels was carried out according to the manufacturer's protocol (58). Values were normalized to total protein levels in the cell lysate as determined by the BCA protein assay kit (Pierce); BSA was used as the standard.
AhR transcription assay
Huh-7 cells transfected with a reporter plasmid carrying a tandem repeat of the XRE upstream of the firefly luciferase gene (pXRE-luc) were stimulated with compounds for 24 h. Luciferase activities in the lysates of those cells were measured using a Luciferase Assay System (Promega) according to the manufacturer's protocol (54).
RT-PCR
RT-PCR analysis was performed as described previously (54), using RNA prepared from Huh-7 cells treated for 24 h with flutamide or CH223191 in the presence or absence of AhR agonist (TCDD). The primers used in this study were 5′-ACGGCAGCCCCTGTAACGACCACTGTGA-3′ and 5′-TGCCAAGATGGTTCCGCCACTCACCAGG-3′ for SREBP-1 transcripts, 5′-GCTGCTCGGATCACTAGTGAA-3′ and 5′-TTCTGCTATCAGTCTGTCCAG-3′ for ACC1 transcripts, 5′-CCCACCTACGTACTGGCCTA-3′ and 5′-CTTGGCCTTGGGTGTGTACT-3′ for FASN transcripts, 5′-GTACCGCTGGCACATCAACTT-3′ and 5′-TTGGAGACTTTCTTCCGGTCAT-3′ for SCD1 transcripts, 5′-CTTTGTGGCCACCCTGCTATTC-3′ and 5′-AGCAAATCCGCCCCCTAGTA-3′ for SLC13A5 transcripts, 5′-ACGCTGAGGACAACACAGTCTCT-3′ and 5′-GTAACAGGGTACGGAACCAAACTC-3′ for CD36 transcripts, 5′-GGCCTTCTTCCACGAGTACC-3′ and 5′-GGCCTCATAGTTGAGCACG-3′ for DGAT1 transcripts, 5′-AGTGGCAATGCTATCATCAT-3′ and 5′-GAGGCCTCGACCATGGAAGAT-3′ for DGAT2 transcripts, 5′-ACCACCAAGAACTGCTTAGCC-3′ and 5′-GAAGAGTGTCGGAAG-3′ for CYP1A1 transcripts, 5′-AAGGATTCCTATGTGGGC-3′ and 5′-CATCTCTTGCTCGAAGTC-3′ for actin transcripts, and 5′-GGTTCCTGTTGACGTCTTG-3′ and 5′-CTTCCAGTGCTCCGAGTAG-3′ for CYP1B1 transcripts.
Statistics
We basically performed three independent experiments with 3–5 replicates for each independent experiment to be subjected to statistics. Statistical significance was determined by Student's t test (*, p < 0.05; **, p < 0.01).
Author contributions
H. O. and K. W. conceptualization; H. O., S. N., S. Kim, R. S., M. F., and K. W. resources; H. O., S. N., and K. W. data curation; H. O., S. Kim, and K. W. software; H. O., S. N., S. Kim, R. S., H. A., S. Kamisuki, F. S., N. O., M. M., T. W., and K. W. formal analysis; H. O., K. N., and K. W. validation; H. O., K. N., S. N., M. F., and K. W. investigation; H. O., S. Kim, and K. W. visualization; H. O., K. N., S. N., M. F., and K. W. methodology; H. O. and K. W. writing-original draft; M. M. and K. W. writing-review and editing; K. W. supervision; H. O. and K. W. funding acquisition; K. W. project administration.
Supplementary Material
Acknowledgments
Huh-7 and Huh-7.5.1 cells were kindly provided by Dr. Francis Chisari at the Scripps Research Institute. Plasmids for preparing HCVpp were generous gifts from Dr. Francois-Loic Cosset at the University de Lyon.
This study was supported by Japan Society for the Promotion of Science KAKENHI Grants JP17H04085, JP18J14277, and JP66KT0111; the JST CREST program; Japan Agency for Medical Research and Development (AMED) Grants JP18fk0310114j0002, JP18fk0310101j1002, JP18fk0310103j0202, JP18fm0208019j0002, and JP18fk0210036j0001; the Suzuken Memorial Foundation; the Fuji Foundation for Protein Research; and the Japan Food Chemical Research Foundation. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Fig. S1.
- HCV
- hepatitis C virus
- AhR
- aryl hydrocarbon receptor
- LD
- lipid droplet
- HCC
- hepatocellular carcinoma
- NASH
- non-alcoholic steatohepatitis
- TCDD
- tetrachlorodibenzo-p-dioxin
- DAPI
- 4′,6-diamidino-2-phenylindole
- CLEIA
- chemiluminescent enzyme immunoassay
- XRE
- xenobiotic response element
- HCVpp
- HCV pseudoparticle
- AR
- androgen receptor
- 6,2′,4′-TMF
- 6,2′,4′-trimethoxyflavone
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- CsA
- cyclosporin A
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
References
- 1. Miller S., and Krijnse-Locker J. (2008) Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6, 363–374 10.1038/nrmicro1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hsu N. Y., Ilnytska O., Belov G., Santiana M., Chen Y. H., Takvorian P. M., Pau C., van der Schaar H., Kaushik-Basu N., Balla T., Cameron C. E., Ehrenfeld E., van Kuppeveld F. J., and Altan-Bonnet N. (2010) Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141, 799–811 10.1016/j.cell.2010.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., and Shimotohno K. (2007) The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9, 1089–1097 10.1038/ncb1631 [DOI] [PubMed] [Google Scholar]
- 4. Shavinskaya A., Boulant S., Penin F., McLauchlan J., and Bartenschlager R. (2007) The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 282, 37158–37169 10.1074/jbc.M707329200 [DOI] [PubMed] [Google Scholar]
- 5. Syed G. H., Amako Y., and Siddiqui A. (2010) Hepatitis C virus hijacks host lipid metabolism. Trends Endocrinol. Metab. 21, 33–40 10.1016/j.tem.2009.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Amako Y., Syed G. H., and Siddiqui A. (2011) Protein kinase D negatively regulates hepatitis C virus secretion through phosphorylation of oxysterol-binding protein and ceramide transfer protein. J. Biol. Chem. 286, 11265–11274 10.1074/jbc.M110.182097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moradpour D., Penin F., and Rice C. M. (2007) Replication of hepatitis C virus. Nat. Rev. Microbiol. 5, 453–463 10.1038/nrmicro1645 [DOI] [PubMed] [Google Scholar]
- 8. Egger D., Wölk B., Gosert R., Bianchi L., Blum H. E., Moradpour D., and Bienz K. (2002) Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76, 5974–5984 10.1128/JVI.76.12.5974-5984.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moriya K., Yotsuyanagi H., Shintani Y., Fujie H., Ishibashi K., Matsuura Y., Miyamura T., and Koike K. (1997) Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J. Gen. Virol. 78, 1527–1531 10.1099/0022-1317-78-7-1527 [DOI] [PubMed] [Google Scholar]
- 10. Moriya K., Fujie H., Shintani Y., Yotsuyanagi H., Tsutsumi T., Ishibashi K., Matsuura Y., Kimura S., Miyamura T., and Koike K. (1998) The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med. 4, 1065–1067 10.1038/2053 [DOI] [PubMed] [Google Scholar]
- 11. Ohata K., Hamasaki K., Toriyama K., Matsumoto K., Saeki A., Yanagi K., Abiru S., Nakagawa Y., Shigeno M., Miyazoe S., Ichikawa T., Ishikawa H., Nakao K., and Eguchi K. (2003) Hepatic steatosis is a risk factor for hepatocellular carcinoma in patients with chronic hepatitis C virus infection. Cancer 97, 3036–3043 10.1002/cncr.11427 [DOI] [PubMed] [Google Scholar]
- 12. Kurosaki M., Hosokawa T., Matsunaga K., Hirayama I., Tanaka T., Sato M., Yasui Y., Tamaki N., Ueda K., Tsuchiya K., Kuzuya T., Nakanishi H., Itakura J., Takahashi Y., Asahina Y., et al. (2010) Hepatic steatosis in chronic hepatitis C is a significant risk factor for developing hepatocellular carcinoma independent of age, sex, obesity, fibrosis stage and response to interferon therapy. Hepatol. Res. 40, 870–877 10.1111/j.1872-034X.2010.00692.x [DOI] [PubMed] [Google Scholar]
- 13. Ludwig J., Viggiano T. R., McGill D. B., and Oh B. J. (1980) Nonalcoholic steatohepatitis: Mayo Clinic experiences with a hitherto unnamed disease. Mayo Clin Proc. 55, 434–438 [PubMed] [Google Scholar]
- 14. Vernon G., Baranova A., and Younossi Z. M. (2011) Systematic review: the epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults. Aliment. Pharmacol. Ther. 34, 274–285 10.1111/j.1365-2036.2011.04724.x [DOI] [PubMed] [Google Scholar]
- 15. Nebert D. W., Dalton T. P., Okey A. B., and Gonzalez F. J. (2004) Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J. Biol. Chem. 279, 23847–23850 10.1074/jbc.R400004200 [DOI] [PubMed] [Google Scholar]
- 16. Mimura J., and Fujii-Kuriyama Y. (2003) Functional role of AhR in the expression of toxic effects by TCDD. Biochim. Biophys. Acta 1619, 263–268 10.1016/S0304-4165(02)00485-3 [DOI] [PubMed] [Google Scholar]
- 17. Fujii-Kuriyama Y., and Mimura J. (2005) Molecular mechanisms of AhR functions in the regulation of cytochrome P450 genes. Biochem. Biophys. Res. Commun. 338, 311–317 10.1016/j.bbrc.2005.08.162 [DOI] [PubMed] [Google Scholar]
- 18. Nebert D. W., Roe A. L., Dieter M. Z., Solis W. A., Yang Y., and Dalton T. P. (2000) Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem. Pharmacol. 59, 65–85 10.1016/S0006-2952(99)00310-X [DOI] [PubMed] [Google Scholar]
- 19. Shimada T., Oda Y., Gillam E. M., Guengerich F. P., and Inoue K. (2001) Metabolic activation of polycyclic aromatic hydrocarbons and other procarcinogens by cytochromes P450 1A1 and P450 1B1 allelic variants and other human cytochromes P450 in Salmonella typhimurium NM2009. Drug Metab. Dispos. 29, 1176–1182 [PubMed] [Google Scholar]
- 20. Rendic S., and Guengerich F. P. (2012) Contributions of human enzymes in carcinogen metabolism. Chem. Res. Toxicol. 25, 1316–1383 10.1021/tx300132k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Quintana F. J., Basso A. S., Iglesias A. H., Korn T., Farez M. F., Bettelli E., Caccamo M., Oukka M., and Weiner H. L. (2008) Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature 453, 65–71 10.1038/nature06880 [DOI] [PubMed] [Google Scholar]
- 22. Stevens E. A., Mezrich J. D., and Bradfield C. A. (2009) The aryl hydrocarbon receptor: a perspective on potential roles in the immune system. Immunology 127, 299–311 10.1111/j.1365-2567.2009.03054.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marlowe J. L., and Puga A. (2005) Aryl hydrocarbon receptor, cell cycle regulation, toxicity, and tumorigenesis. J. Cell. Biochem. 96, 1174–1184 10.1002/jcb.20656 [DOI] [PubMed] [Google Scholar]
- 24. Patel R. D., Kim D. J., Peters J. M., and Perdew G. H. (2006) The aryl hydrocarbon receptor directly regulates expression of the potent mitogen epiregulin. Toxicol. Sci. 89, 75–82 10.1093/toxsci/kfi344 [DOI] [PubMed] [Google Scholar]
- 25. Watashi K., Hijikata M., Hosaka M., Yamaji M., and Shimotohno K. (2003) Cyclosporin A suppresses replication of hepatitis C virus genome in cultured hepatocytes. Hepatology 38, 1282–1288 10.1053/jhep.2003.50449 [DOI] [PubMed] [Google Scholar]
- 26. Tscherne D. M., Jones C. T., Evans M. J., Lindenbach B. D., McKeating J. A., and Rice C. M. (2006) Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80, 1734–1741 10.1128/JVI.80.4.1734-1741.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blanchard E., Belouzard S., Goueslain L., Wakita T., Dubuisson J., Wychowski C., and Rouillé Y. (2006) Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 80, 6964–6972 10.1128/JVI.00024-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meertens L., Bertaux C., and Dragic T. (2006) Hepatitis C virus entry requires a critical postinternalization step and delivery to early endosomes via clathrin-coated vesicles. J. Virol. 80, 11571–11578 10.1128/JVI.01717-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jiang J., and Luo G. (2009) Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J. Virol. 83, 12680–12691 10.1128/JVI.01476-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jin U. H., Lee S. O., and Safe S. (2012) Aryl hydrocarbon receptor (AHR)-active pharmaceuticals are selective AHR modulators in MDA-MB-468 and BT474 breast cancer cells. J. Pharmacol. Exp. Ther. 343, 333–341 10.1124/jpet.112.195339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Macedo L. F., Guo Z., Tilghman S. L., Sabnis G. J., Qiu Y., and Brodie A. (2006) Role of androgens on MCF-7 breast cancer cell growth and on the inhibitory effect of letrozole. Cancer Res. 66, 7775–7782 10.1158/0008-5472.CAN-05-3984 [DOI] [PubMed] [Google Scholar]
- 32. Boulant S., Targett-Adams P., and McLauchlan J. (2007) Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J. Gen. Virol. 88, 2204–2213 10.1099/vir.0.82898-0 [DOI] [PubMed] [Google Scholar]
- 33. Walther T. C., and Farese R. V. Jr. (2012) Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81, 687–714 10.1146/annurev-biochem-061009-102430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Welte M. A. (2015) Expanding roles for lipid droplets. Curr. Biol. 25, R470–481 10.1016/j.cub.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee A. J., Cai M. X., Thomas P. E., Conney A. H., and Zhu B. T. (2003) Characterization of the oxidative metabolites of 17β-estradiol and estrone formed by 15 selectively expressed human cytochrome p450 isoforms. Endocrinology 144, 3382–3398 10.1210/en.2003-0192 [DOI] [PubMed] [Google Scholar]
- 36. Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z., Murthy K., Habermann A., Kräusslich H. G., Mizokami M., Bartenschlager R., and Liang T. J. (2005) Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11, 791–796 10.1038/nm1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim S., Date T., Yokokawa H., Kono T., Aizaki H., Maurel P., Gondeau C., and Wakita T. (2014) Development of hepatitis C virus genotype 3a cell culture system. Hepatology 60, 1838–1850 10.1002/hep.27197 [DOI] [PubMed] [Google Scholar]
- 38. Vorrink S. U., Severson P. L., Kulak M. V., Futscher B. W., and Domann F. E. (2014) Hypoxia perturbs aryl hydrocarbon receptor signaling and CYP1A1 expression induced by PCB 126 in human skin and liver-derived cell lines. Toxicol. Appl. Pharmacol. 274, 408–416 10.1016/j.taap.2013.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shimba S., and Watabe Y. (2009) Crosstalk between the AHR signaling pathway and circadian rhythm. Biochem. Pharmacol. 77, 560–565 10.1016/j.bcp.2008.09.040 [DOI] [PubMed] [Google Scholar]
- 40. Lerat H., Honda M., Beard M. R., Loesch K., Sun J., Yang Y., Okuda M., Gosert R., Xiao S. Y., Weinman S. A., and Lemon S. M. (2002) Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology 122, 352–365 10.1053/gast.2002.31001 [DOI] [PubMed] [Google Scholar]
- 41. Camus G., Herker E., Modi A. A., Haas J. T., Ramage H. R., Farese R. V. Jr, and Ott M. (2013) Diacylglycerol acyltransferase-1 localizes hepatitis C virus NS5A protein to lipid droplets and enhances NS5A interaction with the viral capsid core. J. Biol. Chem. 288, 9915–9923 10.1074/jbc.M112.434910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lahoti T. S., Hughes J. M., Kusnadi A., John K., Zhu B., Murray I. A., Gowda K., Peters J. M., Amin S. G., and Perdew G. H. (2014) Aryl hydrocarbon receptor antagonism attenuates growth factor expression, proliferation, and migration in fibroblast-like synoviocytes from patients with rheumatoid arthritis. J. Pharmacol. Exp. Ther. 348, 236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. He J., Hu B., Shi X., Weidert E. R., Lu P., Xu M., Huang M., Kelley E. E., and Xie W. (2013) Activation of the aryl hydrocarbon receptor sensitizes mice to nonalcoholic steatohepatitis by deactivating mitochondrial sirtuin deacetylase Sirt3. Mol. Cell. Biol. 33, 2047–2055 10.1128/MCB.01658-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wada T., Sunaga H., Miyata K., Shirasaki H., Uchiyama Y., and Shimba S. (2016) Aryl hydrocarbon receptor plays protective roles against high fat diet (HFD)-induced hepatic steatosis and the subsequent lipotoxicity via direct transcriptional regulation of Socs3 gene expression. J. Biol. Chem. 291, 7004–7016 10.1074/jbc.M115.693655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee J. H., Wada T., Febbraio M., He J., Matsubara T., Lee M. J., Gonzalez F. J., and Xie W. (2010) A novel role for the dioxin receptor in fatty acid metabolism and hepatic steatosis. Gastroenterology 139, 653–663 10.1053/j.gastro.2010.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kress S., Reichert J., and Schwarz M. (1998) Functional analysis of the human cytochrome P4501A1 (CYP1A1) gene enhancer. Eur. J. Biochem. 258, 803–812 10.1046/j.1432-1327.1998.2580803.x [DOI] [PubMed] [Google Scholar]
- 47. Larrea E., Riezu-Boj J. I., Gil-Guerrero L., Casares N., Aldabe R., Sarobe P., Civeira M. P., Heeney J. L., Rollier C., Verstrepen B., Wakita T., Borrás-Cuesta F., Lasarte J. J., and Prieto J. (2007) Upregulation of indoleamine 2,3-dioxygenase in hepatitis C virus infection. J. Virol. 81, 3662–3666 10.1128/JVI.02248-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wells M. A., Vendrov K. C., Edin M. L., Ferslew B. C., Zha W., Nguyen B. K., Church R. J., Lih F. B., DeGraff L. M., Brouwer K. L., Barritt A. S. 4th, Zeldin D. C., and Lee C. R. (2016) Characterization of the cytochrome P450 epoxyeicosanoid pathway in non-alcoholic steatohepatitis. Prostaglandins Other Lipid Mediat. 125, 19–29 10.1016/j.prostaglandins.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Arnold C., Konkel A., Fischer R., and Schunck W. H. (2010) Cytochrome P450-dependent metabolism of ω-6 and ω-3 long-chain polyunsaturated fatty acids. Pharmacol. Rep. 62, 536–547 10.1016/S1734-1140(10)70311-X [DOI] [PubMed] [Google Scholar]
- 50. Zhong J., Gastaminza P., Cheng G., Kapadia S., Kato T., Burton D. R., Wieland S. F., Uprichard S. L., Wakita T., and Chisari F. V. (2005) Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U.S.A. 102, 9294–9299 10.1073/pnas.0503596102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Akazawa D., Date T., Morikawa K., Murayama A., Miyamoto M., Kaga M., Barth H., Baumert T. F., Dubuisson J., and Wakita T. (2007) CD81 expression is important for the permissiveness of Huh7 cell clones for heterogeneous hepatitis C virus infection. J. Virol. 81, 5036–5045 10.1128/JVI.01573-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Iwamoto M., Watashi K., Tsukuda S., Aly H. H., Fukasawa M., Fujimoto A., Suzuki R., Aizaki H., Ito T., Koiwai O., Kusuhara H., and Wakita T. (2014) Evaluation and identification of hepatitis B virus entry inhibitors using HepG2 cells overexpressing a membrane transporter NTCP. Biochem. Biophys. Res. Commun. 443, 808–813 10.1016/j.bbrc.2013.12.052 [DOI] [PubMed] [Google Scholar]
- 53. Ishida Y., Yamasaki C., Yanagi A., Yoshizane Y., Fujikawa K., Watashi K., Abe H., Wakita T., Hayes C. N., Chayama K., and Tateno C. (2015) Novel robust in vitro hepatitis B virus infection model using fresh human hepatocytes isolated from humanized mice. Am. J. Pathol. 185, 1275–1285 10.1016/j.ajpath.2015.01.028 [DOI] [PubMed] [Google Scholar]
- 54. Nakajima S., Watashi K., Ohashi H., Kamisuki S., Izaguirre-Carbonell J., Kwon A. T., Suzuki H., Kataoka M., Tsukuda S., Okada M., Moi M. L., Takeuchi T., Arita M., Suzuki R., Aizaki H., et al. (2016) Fungus-derived neoechinulin B as a novel antagonist of liver X receptor, identified by chemical genetics using a hepatitis C virus cell culture system. J. Virol. 90, 9058–9074 10.1128/JVI.00856-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kato T., Choi Y., Elmowalid G., Sapp R. K., Barth H., Furusaka A., Mishiro S., Wakita T., Krawczynski K., and Liang T. J. (2008) Hepatitis C virus JFH-1 strain infection in chimpanzees is associated with low pathogenicity and emergence of an adaptive mutation. Hepatology 48, 732–740 10.1002/hep.22422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Masaki T., Matsunaga S., Takahashi H., Nakashima K., Kimura Y., Ito M., Matsuda M., Murayama A., Kato T., Hirano H., Endo Y., Lemon S. M., Wakita T., Sawasaki T., and Suzuki T. (2014) Involvement of hepatitis C virus NS5A hyperphosphorylation mediated by casein kinase I-α in infectious virus production. J. Virol. 88, 7541–7555 10.1128/JVI.03170-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bartosch B., Dubuisson J., and Cosset F. L. (2003) Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197, 633–642 10.1084/jem.20021756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McIntosh A. L., Huang H., Storey S. M., Landrock K. K., Landrock D., Petrescu A. D., Gupta S., Atshaves B. P., Kier A. B., and Schroeder F. (2014) Human FABP1 T94A variant impacts fatty acid metabolism and PPAR-alpha activation in cultured human female hepatocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 307, G164–G176 10.1152/ajpgi.00369.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.