Abstract
Therapeutic mAbs engage Fc γ receptor III (CD16) to elicit a protective cell-mediated response and destroy the target tissue. Newer drugs designed to bind CD16a with increased affinity surprisingly also elicit protective CD16b-mediated responses. However, it is unclear why IgG binds CD16a with more than 10-fold higher affinity than CD16b even though these receptors share more than 97% identity. Here we identified one residue, Gly-129, that contributes to the greater IgG binding affinity of CD16a. The CD16b variant D129G bound IgG1 Fc with 2-fold higher affinity than CD16a and with 90-fold higher affinity than the WT. Conversely, the binding affinity of CD16a-G129D was decreased 128-fold relative to WT CD16a and comparably to that of WT CD16b. The interaction of IgG1 Fc with CD16a, but not with CD16b, is known to be sensitive to the composition of the asparagine-linked carbohydrates (N-glycans) attached to the receptor. CD16a and CD16b-D129G displaying minimally processed oligomannose N-glycans bound to IgG1 Fc with about 5.2-fold increased affinity compared with variants with highly processed complex-type N-glycans. CD16b and the CD16a-G129D variant exhibited a smaller 1.9-fold affinity increase with oligomannose N-glycans. A model of glycosylated CD16b bound to IgG1 Fc determined to 2.2 Å resolution combined with a 250-ns all-atom molecular dynamics simulation showed that the larger Asp-129 residue deformed the Fc-binding surface. These results reveal how Asp-129 in CD16b affects its binding affinity for IgG1 Fc and suggest that antibodies engineered to engage CD16b with high affinity must accommodate the Asp-129 side chain.
Keywords: N-linked glycosylation, posttranslational modification (PTM), X-ray crystallography, protein–protein interaction, IgG, molecular dynamics, monoclonal antibody, fragment crystallizable region, immunity
Introduction
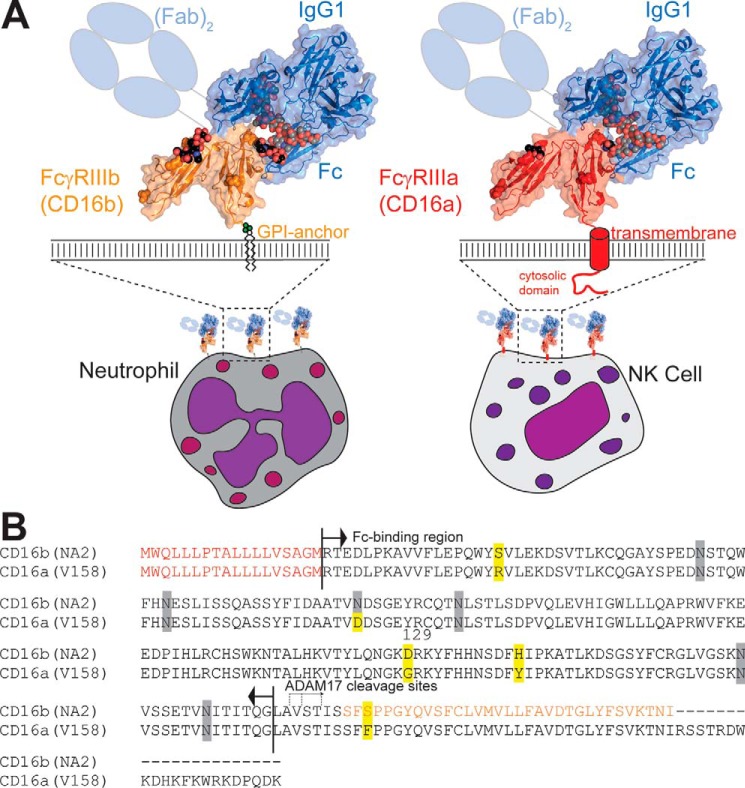
IgG opsonizes a previously encountered pathogen to target it for destruction by eliciting an immune response. IgG binds the target with high specificity through antigen binding fragments (Fabs) and interacts with innate immune system effector cells through the crystallizable fragment (Fc) by engaging Fc γ receptors (FcγRs), thus bridging the adaptive and innate immune systems (Fig. 1). There are three classes of cognate human FcγRs: FcγRI (CD64), FcγRII (CD32), and FcγRIII (CD16) (1). CD16 is of particular interest because it is the primary receptor responsible for antibody-dependent cell-mediated cytotoxicity and, thus, tumor clearance (2, 3). Two CD16 forms, CD16a and CD16b, bind to IgG1 with surprisingly different affinities considering the high degree of structural and sequence similarity (4, 5). In this manuscript, we probe the contribution of an amino acid residue to the different binding affinities of CD16a and CD16b.
Figure 1.
FcγR III/CD16 is expressed in two forms from two distinct genes. A and B, FcγRIIIb/CD16b is highly expressed on neutrophils, and FcγRIIIa/CD16a is the predominant FcγR expressed on NK cells. B, these proteins differ by only four amino acid residues (highlighted in yellow) in the mature proteins. The N-terminal signal peptide is indicated in red and is not found in the fully processed proteins. The C-terminal residues of CD16b shown in orange are cleaved prior to addition of the glycosylphosphatidylinositol anchor at the newly uncovered C-terminal serine. N-glycosylation sites are indicated with gray shading. Both proteins are cleaved by ADAM17 and released from the cell surface following cell activation. Resolved N-glycan residues are shown as spheres.
CD16a is an integral membrane protein predominantly expressed on NK2 cells and monocytes. CD16b is predominantly expressed with a glycosylphosphatidylinositol anchor on neutrophils, a subset of basophils and shows inducible expression on eosinophils (6–12) (Fig. 1). CD16a has two predominant alleles (V158 or F158) and CD16b has two predominant alleles, including human neutrophil antigen 1 (HNA-1 or NA1) and NA2 (13, 14). CD16a-V158 and CD16b-NA2 share >97% sequence identity and differ by only four amino acid residues in the extracellular antibody-binding domains (Fig. 1B). Despite the significant homology shared by CD16a and CD16b, CD16a binds antibodies more than 10-fold tighter, although it is unclear how differences in these four amino acids contribute to differences in binding affinity (5). Only one of these four differences, at position 129, forms part of the IgG1-binding interface.
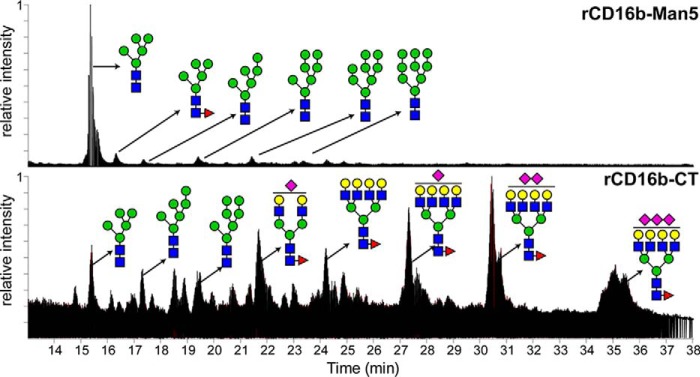
FcγRs are heavily modified with asparagine-linked carbohydrates (N-glycans; for a review of N-glycosylation, see Ref. 15). CD16a has as many as five N-glycans, and CD16b (NA2) has as many as six, although it is not clear whether all sites are fully occupied. The N-glycan composition of CD16 appears to be varied, depending on the type of expressing cell, and it has been shown that different cells bind IgG with different affinity through CD16 (16–19). Three recent studies reported that the binding affinity of CD16a is influenced by receptor N-glycan composition (19–21). In one example, CD16a with minimally processed Man5 N-glycans bound 12-fold tighter to IgG1 Fc (G0F) than CD16a, with highly processed and elaborated complex-type N-glycans, but glycan composition only impacted CD16b affinity by less than 2-fold (examples of different N-glycoforms are shown in Fig. 2) (19, 22). This result is surprising considering the significant homology shared by CD16a and CD16b (Fig. 1 and Fig. S1). The CD16a N-glycan at position 162 mediated this effect, which, likewise, may be influenced by the nearby Gly-129 residue (20).
Figure 2.
Total ion current of procainamide-derived CD16b N-glycans expressed in two distinct glycoforms and analyzed by HILIC-MS. Top, CD16b was expressed to display Man5 N-glycans. Bottom, CD16b with CT N-glycans.
The N-glycan composition of the IgG1 Fc is a well-known variable in CD16 binding affinity (3, 23–26). This property directly impacts the development of therapeutic mAbs because increasing affinity for CD16 increased therapeutic efficacy (13, 25, 27–29). A major advance for mAb development occurred with the observation that IgG lacking a prevalent antibody modification, l-fucose attached to the core (1)GlcNAc residue of the N-glycan, increased binding affinity and antibody-dependent cell-mediated cytotoxicity (30). This observation inspired the development of many new glycoengineered mAb therapies (31–34). Notably, CD16b demonstrated a larger enhancement in binding nonfucosylated IgG1 Fc in vitro compared with CD16a in vitro (5). The current generation of glycoengineered antibodies is more effective at binding to CD16b on neutrophils and eliciting effector responses, showing greater therapeutic potential (35, 36). These studies support the development of mAbs that bind CD16b with higher affinity to mobilize an effective neutrophil response. However, the development of future mAbs is limited by a lack of information regarding the detailed mechanism and identification of which residues contribute to the reduced affinity of CD16b for IgG1 Fc compared with CD16a.
A comparison of the amino acid sequences for CD16b and CD16a reveals that only one of the four differences in the antibody-binding domains, at position 129, directly contributes the interface formed with IgG1 Fc (Fig. 1B). If Asp-129 of CD16b disrupts this interface, then CD16b D129G is expected to bind as tightly as CD16a, and CD16a G129D will bind with less affinity. Furthermore, if CD16a Gly-129 contributes to the sensitivity of affinity to receptor N-glycan composition, then CD16a G129D should no longer be sensitive to N-glycan composition. In this manuscript, we evaluate how Gly-129 and Asp-129 impact the affinity of CD16 for IgG1 Fc. We also determined how these residues modulate the differential sensitivity of CD16a and CD16b to N-glycan composition.
Results
N-glycomics of the recombinant proteins
To evaluate the impact of N-glycan composition and Asp-129 versus Gly-129 on CD16 structure and IgG1 Fc binding, we expressed four CD16 variants, including CD16a, CD16a G129D, CD16b, and CD16b D129G, using two cells lines that resulted in a panel of eight receptor variants. One cell line, HEK293F, contains a large repertoire of glycan-modifying enzymes and expressed CD16 with a heterogeneous mixture of highly branched complex-type N-glycans (Fig. 2 and Fig. S2). The second cell line, HEK293S, contains a defect in the GNT1 gene (37, 38) and expressed CD16 with predominantly Man5 oligomannose-type N-glycans, although small amounts of Man6, Man7, Man8, and Man9 were also present (Fig. 2).
rCD16b D129G-CT binds IgG1 Fc with an affinity comparable with rCD16a-CT
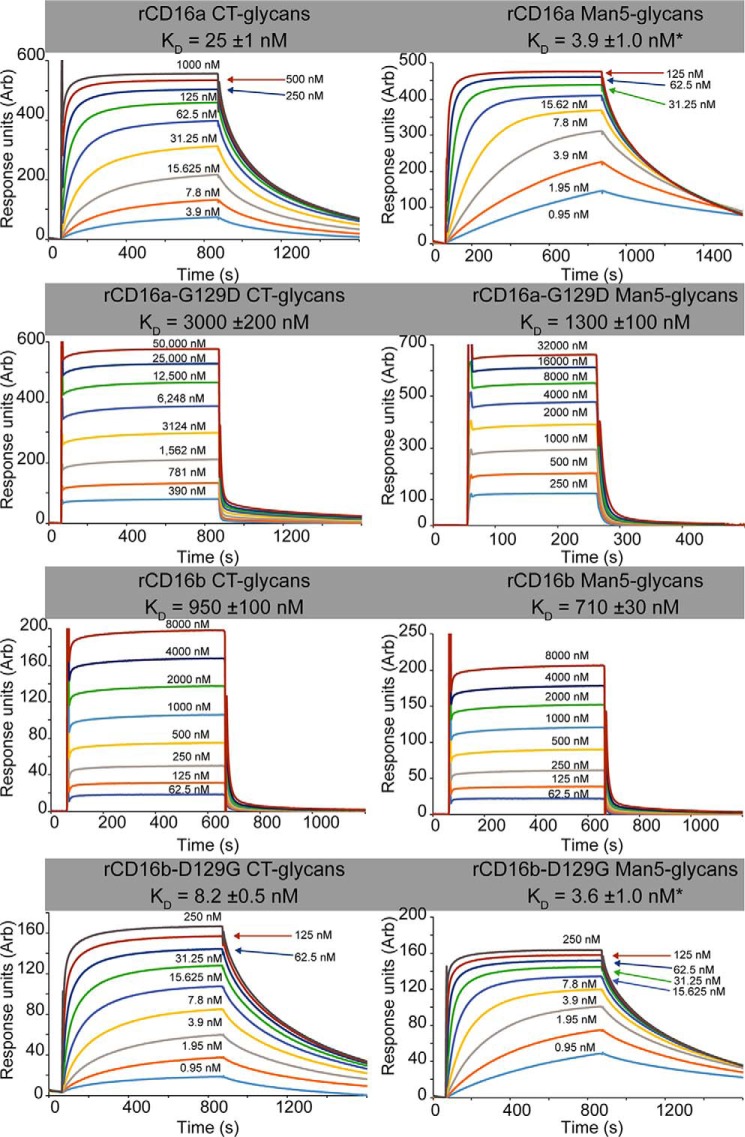
WT recombinant (r)CD16a-CT bound IgG1 Fc glycoforms with affinities between 38- and 60-fold tighter than rCD16b (Fig. 3 and Table 1). These values were comparable with previous results from our laboratory; however, the binding of rCD16a-CT is roughly 1.5 to 4-fold tighter (5, 20). As predicted, rCD16a-CT bound IgG1 Fc with more than 100-fold greater affinity than rCD16a G129D-CT. The rCD16a G129D-CT variant bound with an affinity comparable with rCD16b-CT (22 mm versus 6.4 mm, respectively; Fig. 4). In the complementary experiment, rCD16b-D129G-CT bound at least 64-fold tighter to three IgG1 Fc N-glycoforms than rCD16b-CT. Furthermore, the affinities of rCD16a-CT and rCD16b D129G-CT were less than 3.1-fold different, confirming the importance of a Gly at position 129 in high-affinity IgG1 Fc binding. It is possible that subtle differences in N-glycan composition resulting from expression conditions, in particular the N-glycan at Asn-162, impact the measured affinity. This effect is noted for CD16b D129G-CT, which contains a larger amount of oligomannose N-glycans compared with CD16a (Fig. S2).
Figure 3.
The binding affinity and binding kinetics of the CD16b D129G protein are comparable with the high-affinity CD16a protein. Shown are representative binding sensorgrams for the CD16 variants binding to IgG1 Fc (G0 glycoform). Dissociation constants were determined using equilibrium intensity values, with the exception of KD values marked with an asterisk, which were calculated by fitting kinetic data. Errors indicate uncertainties from the curve fitting procedures.
Table 1.
Binding affinity measurements with two receptor glycoforms and three IgG1 Fc glycoforms determined by surface plasmon resonance
| Receptor | IgG1 Fc G0F |
IgG1 Fc G2F |
IgG1 Fc G0 |
|||
|---|---|---|---|---|---|---|
| KD (nm) | ± error | KD (nm) | ± error | KD (nm) | ± error | |
| rCD16a-CT | 10 | 70 | 2 | 25 | 1 | |
| rCD16a G129D-CT | 22,000 | 1,000 | 12,500 | 600 | 3,000 | 200 |
| rCD16b-CT | 6,400 | 400 | 4,200 | 300 | 950 | 100 |
| rCD16b D129G-CT | 100 | 10 | 48 | 3 | 8 | 1 |
| rCD16a-Man5 | 48 | 2 | 22 | 1 | 3.9a | 1.0 |
| rCD16a G129D-Man5 | 9,500 | 300 | 4,000 | 200 | 1,100 | 100 |
| rCD16b-Man5 | 5,200 | 300 | 2,900 | 200 | 560 | 30 |
| rCD16b D129G-Man5 | 13 | 1 | 6.0 | 0.3 | 3.6a | 1.0 |
| rCD16b | ||||||
| N38Q/N64Q/N74Q/N169Q-Man5 | 6,400 | 600 | 4,000 | 400 | 650 | 50 |
a KD values were calculated from kinetic data.
Figure 4.
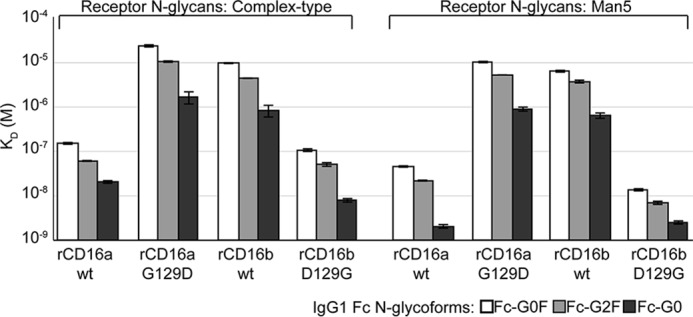
A glycine at position 129 of CD16 is essential for high-affinity IgG1 Fc binding. Dissociation constants indicate the impact of the IgG1 Fc N-glycan, CD16 in N-glycan, and the residue at position 129 on binding affinity. Error bars indicate the error of fit for the interactions.
Tighter-binding CD16 variants are sensitive to N-glycan composition
Comparing the binding affinities of the CD16 variants with different N-glycan composition provided insight into the role of Gly-129 in the higher-affinity interactions. All receptor variants bound tighter with oligomannose N-glycans; however, receptors with a Gly residue at position 129 showed an average affinity increase of 5.2 ± 2.6-fold compared with 1.9 ± 0.8-fold for receptors with an Asp residue at position 129 (Table 1).
The CD16 129 residue impacts association and dissociation of IgG1 Fc
The receptor variants that bound with lower affinity revealed very fast apparent association and dissociation kinetics using the relatively high concentrations of analytes required to reach saturation (CD16b and CD16a G129D). These rates were too rapid to accurately estimate from the surface plasmon resonance sensorgrams because binding was limited by the rate of analyte flow over the chip surface (Fig. 3). Conversely, variants that bound with higher affinity showed slower observed association and dissociation rates (CD16a and CD16b D129G). These observed rates appeared to be slow because of the small amounts of receptor used for the binding measurements and are not comparable with the association rates measured for the low-affinity variants that were tested at much higher analyte concentrations and likely bind with equal or slower on rates. As a result, the associations for the high-affinity variants failed to reach equilibrium after 800 s, and only an analysis of binding kinetics provided appropriate estimates of KD (Fig. 3 and Table 2). Reactions that permitted KD measurement using both equilibrium and kinetic data revealed comparable results and validated the direct comparison of KD values measured from both types of data (Tables 1 and 2).
Table 2.
Association and dissociation rate constants measured from kinetic fits of the surface plasmon resonance sensorgrams
| Receptor | IgG1 Fc G0F |
IgG1 Fc G0 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| kON (mm−1s−1) | ± error | kOFF (s−1) | ± error | KD (nm) | kON (mm−1s−1) | ± error | kOFF (s−1) | ± error | KD (nm) | |
| rCD16a-CT | 39 | 11% | 0.00506 | 14% | 130 | 98 | 13% | 0.00420 | 10% | 43 |
| rCD16b D129G-CT | 306 | 18% | 0.01853 | 16% | 61 | 317 | 4% | 0.00323 | 10% | 100 |
| rCD16a-Man5 | 437 | 19% | 0.01729 | 16% | 40 | 625 | 1% | 0.00242 | 29% | 3.9 |
| rCD16b D129G-Man5 | 422 | 2% | 0.00440 | 18% | 10 | 685 | 4% | 0.00246 | 20% | 3.6 |
Among the high-affinity variants, it is notable that rCD16a-CT bound to IgG1 Fc G0F and G0 with the slowest of the measured on rates (39 and 98 mm−1 s−1, respectively, compared with 306–685 mm−1 s−1; Table 2). Surprisingly, rCD16b D129G bound with a 3- to 8-fold faster kON compared with rCD16a but more similar kOFF values (0.8 to 3.7-fold difference). These differences contribute to KD values that were no more than 2.1 to 2.3-fold different. This result indicates that substitution at residue 129 does not transfer the complete kinetic properties between CD16a and CD16b.
The structure of glycosylated CD16b in complex with IgG1 Fc
Three models of IgG1 Fc bound to nonglycosylated CD16b revealed the structural features of low-affinity FcγRs bound to antibody (39, 40). However, the availability of moderate resolution diffraction (3.0–3.5 Å), moderate Rfree values (0.28–0.36), and high temperature factors for the Asp-129 backbone atoms (78–104 Å2) limit the ability to formulate hypotheses regarding the position of individual atoms at the Fc–receptor interface and compare with three higher quality structures of the CD16a:IgG1 Fc complex (2.2 Å with Rfree values of 0.24–0.26 (20, 41, 42)). One feature of the higher-quality CD16a complexes is the presence of the Asn-45 and Asn-162 glycans. Multiple reports highlight the importance of these two N-glycans and the dispensability of the remaining three N-glycans to IgG1 Fc binding affinity (20, 43, 44). To improve the crystal quality, we prepared glycosylated rCD16b-Man5 with four of the six N-glycosylation sites removed by mutating the modified Asn residue to a Gln (N38Q/N64Q/N74Q/N169Q). Indeed, this rCD16b-Man5 variant bound IgG1 Fc with 0.7- to 0.9-fold of the affinity observed using rCD16b-Man5 and indicated suitability for crystallization trials (Table 1).
Two crystal forms of the rCD16b-Man5–IgG1 Fc complex diffracted to high resolution. One form crystallized in the P212121 space group and diffracted to 2.5 Å (data not shown). A second crystal in the C2 space group diffracted to 2.22 Å with high completeness (Table S1). The CD16a–IgG1 Fc complex provided initial phases (20), and the iterative refinement of the higher-resolution data resulted in an Rfree of 0.2477 as well as a model with no Ramachandran outliers and only 1.2% of side chain rotamer outliers. As expected, the overall structure is highly similar to other CD16a and CD16b models (Fig. 5). This structural model has a root mean square deviation of 0.499 Å compared with the rCD16a-Man5–IgG1 Fc complex (PDB code 5VU0 (20)) and 2.054 Å compared with a nonglycosylated rCD16b–IgG1 Fc complex (PDB code 1T89 (40)).
Figure 5.
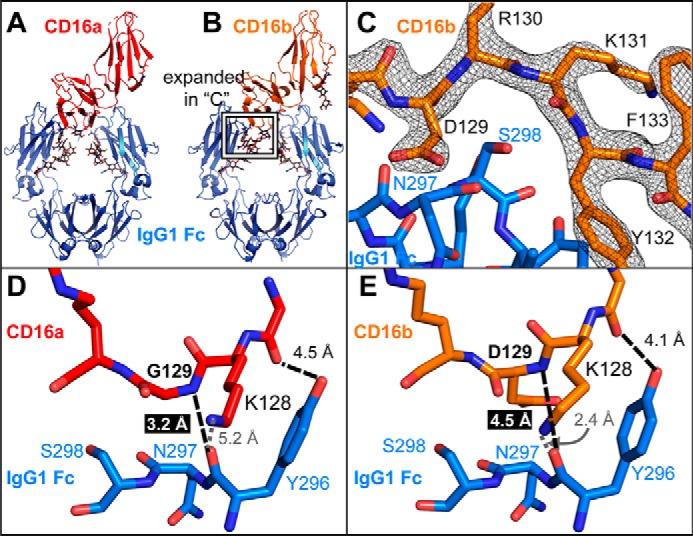
CD16a binds IgG1 Fc with greater affinity and makes a closer approach to IgG1 Fc through the loop containing residue 129. A and B, the overall complexes formed by CD16a (A) and CD16b (B) with IgG1 Fc are highly similar with a root mean square deviation of 0.499 Å. The protein backbone is shown as ribbons, and N-glycans are shown as sticks. C, the electron density of the CD16b region surrounding Asp-129 is well-resolved at 1.5 σ. D and E, the Gly-129 amide nitrogen atom of CD16a (D) is located 1.3 Å closer to the carbonyl carbon of the IgG1 Fc Asn-297 than the amide nitrogen of Asp-129 from CD16B (E).
The Asp-129 residue of CD16b distorts the Fc-binding interface
In addition to higher-resolution data and higher refinement scores compared with previous CD16b–IgG1 Fc complexes, the model calculated from the 2.22 Å dataset provided low temperature factors for the interface residues with values for the Asp-129 backbone atoms ranging from 42 to 43 Å2 (compared with 78–104 Å2 for previous CD16b models). These lower values allow comparison with CD16a models with similar quality (Fig. 5C). Despite the high degree of amino acid similarity, the interface formed between CD16b and IgG1 Fc is smaller and involves less residues than the interface formed by CD16a (Fig. S3). The buried surface area measured with this model is 860 Å2 and comparable with previous CD16b models (PDB code 1T89, 737 Å2; PDB code 1E4K, 816 Å2 (39, 40)). These areas are less than those measured with the CD16a–IgG1 Fc complex (PDB code 3SGJ, 1193 Å2; PDB code 3AY4, 924 Å2; PDB code 5VU0, 897 Å2 (20, 41, 42)).
Asp-129 represents the only amino acid difference between CD16b and CD16a that is found at the interface formed with IgG1 Fc. The close approach of the CD16a Gly-129 amide nitrogen to the IgG1 Fc Tyr-296 carbonyl (3.2 Å) is prevented by the CD16b Asp-129 residue, increasing this internuclear distance to 4.5 Å and distorting the CD16b backbone (Fig. 5, D and E). Surprisingly, it appears that this distortion is not present in the absence of IgG1 Fc; a model of unliganded CD16b reveals a backbone conformation identical to CD16a in the region of Asp-129 (Fig. 6A). The observed conformation in the unliganded CD16b model places the Asp-129 side chain within the van der Waals radii of the IgG1 Fc Asn-297 residue. Thus, CD16b must deform to accommodate IgG1 Fc, distorting the backbone. These changes at the Fc interface potentially explain why CD16b binds IgG1 Fc weaker than CD16a.
Figure 6.
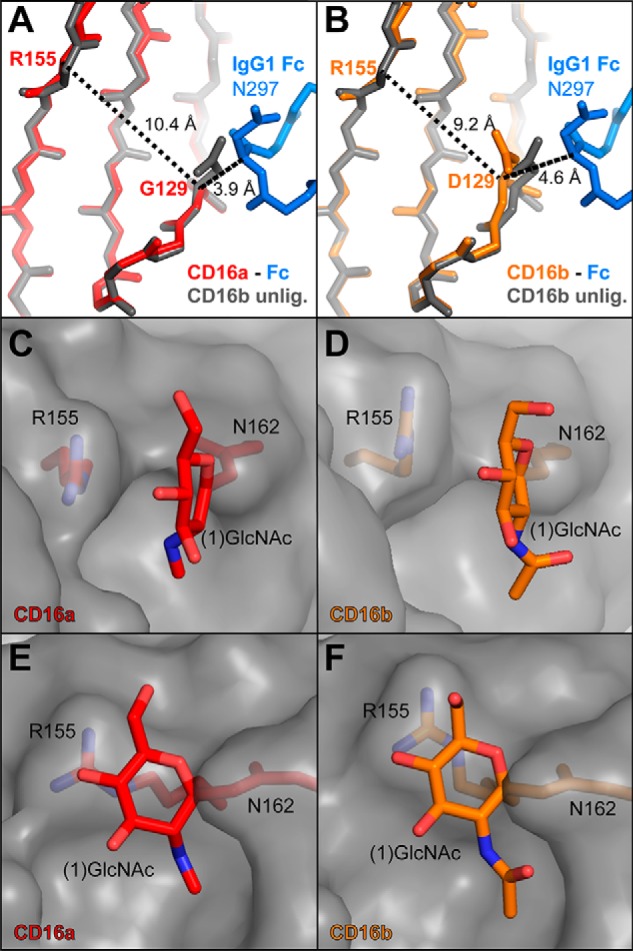
Asp-129 in CD16b decreases the distance between β strands and distorts the interface formed between the Asn-162-glycan and Arg-155 compared with Gly-129 and CD16a. A, the CD16a complex with glycosylated CD16a is shown in red (PDB code 5VU0 (20)) and superimposed on the unliganded (unlig.) CD16b (PDB code 1E4J (39)), shown in gray. Note that the position of the Asp-129 side chain clashes with the IgG1 Fc C'E loop. B, glycosylated CD16b in complex with IgG1 Fc is shown in orange and superimposed on the unliganded CD16b, shown in gray. IgG1 Fc binding distorts the conformation surrounding Asp-129, shortening the distance from Asp-129 to Arg-155. C and E, two views of CD16a, with the polypeptide surface shown in gray and relevant residues as red sticks. D and F, two poses of CD16b, with the polypeptide surface shown in gray and relevant residues as orange sticks. This Asp-129–mediated strand distortion perturbs the interface formed between the (1)GlcNAc residue of the CD16 Asn-162 glycan and Arg-155.
The backbone distortion because of Asp-129 impacts the local tertiary structure, deforming the β sheet and reducing the distance across the sheet by 1.2 Å compared with CD16a (as measured by the distance to Arg-155; Fig. 6, A and B). This change impacts the conformation of the Arg-155 side chain, which forms an interface with the Asn 162 glycan (1)GlcNAc residue in the CD16a complex. In the CD16b complex, the contacts between Arg-155 and the (1)GlcNAc residue are reduced as the Arg-155 side chain shifts away from the carbohydrate (Fig. 6, C–F). This intramolecular interface formed between the Asn-162 glycan (1)GlcNAc residue and the Arg-155 side chain may explain why the affinity of IgG1 for CD16a, but not CD16b, is sensitive to the receptor N-glycan composition (Fig. 4 and Table 1).
MD simulations of the CD16b–IgG1 Fc complex
Measurements of binding affinity pinpointed the residue at position 129 as a critical difference between CD16a and CD16b. X-ray crystallography highlighted differences in the interfaces formed by these receptors and IgG1 Fc. However, these data represent a single conformational snapshot, and it remained unclear whether X-ray crystallography only captured a representative state. The N-glycosylated CD16b–IgG1 Fc complex served as a starting point for a 250-ns all-atom molecular dynamics simulation to compare with a recent 250-ns simulation of the CD16a–IgG1 Fc complex to assess the stability of the observed interactions on this timescale (20). Although this simulation timescale is insufficient to probe all potential conformations accessed by these proteins, it may prove to be informative regarding differences in local conformation potentially governed by low energetic barriers that would be sampled in a relatively restricted simulation.
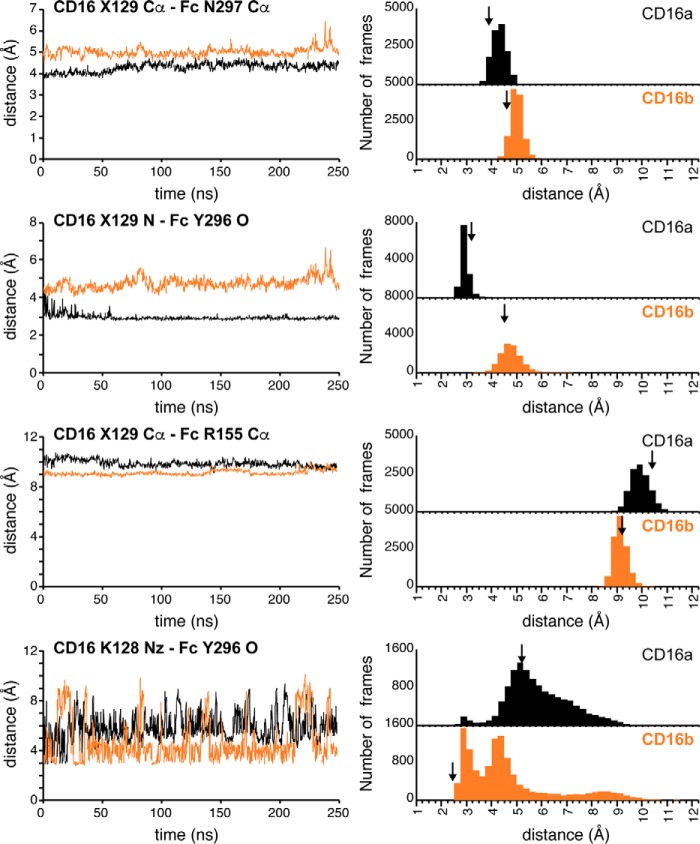
Distances separating the CD16 129 residue and the IgG1 Fc C'E loop showed values consistent with the models determined by X-ray crystallography: the CD16b Gly-129 Cα atom was 4.6 Å from the Fc Asn-297 Cα compared with 3.9 Å for the CD16a complex (Fig. 6). Although the average distances observed with MD are slightly larger (5.0 and 4.3 Å, respectively), the differences of 0.7 Å are identical (Fig. 7). These greater distances for CD16b separated the CD16a Gly-129 N and Fc Tyr-296 O atoms (average, 4.8 Å), which participated in an H-bond in the CD16a simulations (average, 3.0 Å). In the CD16b simulations and model determined by X-ray crystallography, this H-bond was replaced by an interaction with the CD16b Lys-128 Nz atom, which sampled distances within H-bond range in the CD16b simulation but not in the CD16a simulation (Fig. 7, bottom panels).
Figure 7.
Internuclear distances observed in crystallography are maintained during 250 ns all-atom molecular dynamics simulations of the CD16-IgG1 Fc complexes. Left panels, a 10-frame average of each distance over the course of the simulation. Right panels, histograms reporting the number of frames that correspond to each distance. Vertical arrows indicate the distance measured from structures determined by X-ray crystallography (CD16a-IgG1 Fc G0 (PDB code 5VU0 (20)); CD16b-IgG1 Fc G0, this work). Distances correspond to those measured in Figs. 5 and 6. X129 refers to residue Asp-129 of CD16b and Gly-129 of CD16a.
Simulation likewise preserved the observed deformation of the CD16b β sheet induced by Asp-129 that reduced the distance between the Arg-155 and 129 Cα atoms by 1.2 Å compared with CD16a (Fig. 6, A and B). Although the difference in averages is somewhat smaller in the simulations at 0.8 Å, the same trend is observed until the very end of the CD16b simulation (Fig. 7). Interestingly, CD16b drifts slightly away from IgG1 Fc in the final 30 ns of the simulation, as indicated by the increasing distances between CD16b Asp-129 and Fc residues Asn-297 and Tyr-296. At the same time, the β sheet deformation also disappears, and the distance between the CD16b Arg-155 and Gly-129 Cα atoms matches the CD16a simulation (9.6 Å). This observation supports the conclusion that complex formation and contact with the IgG1 Fc C'E loop impinges on the CD16b Asp-129 residue.
Discussion
The data presented here demonstrate that a single amino acid residue at position 129 accounts for the major differences in binding affinity, binding kinetics, and sensitivity to receptor N-glycan composition. The presence of a Gly at CD16 residue 129 supports high-affinity IgG1 Fc binding, but an Asp residue, as found in CD16b, reduces affinity. These data are supported by binding measurements and the structural model of the N-glycosylated CD16b–IgG1 Fc complex determined by X-ray crystallography. Extensive all-atom MD simulations further supported differences in the CD16–IgG1 Fc models.
The deformation of CD16b because of Asp-129 upon complex formation with IgG1 Fc likely contributes an energetic penalty that prevents CD16b from binding as tightly as CD16a. This is evident upon examining a model of unliganded CD16b that showed indistinguishable backbone geometry compared with CD16a in complex with IgG1 Fc (Fig. 6). Furthermore, this deformation reduced the buried surface area formed in the complex. With these data alone, it is not possible to quantify the impact of the deformation on IgG1 Fc binding affinity because it is not clear how Asp-129 impacts conformational sampling of unliganded CD16b. Robinson et al. (46) recently noted evidence for sampled conformation differences between CD16a and CD16b and developed a CD16a-selective affimer, AfG3, that binds between the two extracellular domains to a region with conserved amino acids in CD16a and CD16b. Although the authors believe that the selectivity of AfG3 is largely due to H-bonds formed by CD16a Arg-18 (CD16b NA2 S18), it is clear that amino acid differences distant from the IgG1-binding surface of CD16 can impact conformational sampling.
It is surprising that CD16a is more sensitive to N-glycan composition than CD16b and that this behavior is influenced by the residue at position 129. As stated, CD16a N-glycan composition affects affinity for binding IgG1 Fc (16, 19, 21). The data presented here further support the recent observation that CD16a is the only FcγR sensitive to receptor N-glycan composition (Table 1 and Ref. 22). Furthermore, analysis of the CD16 amino acid variants binding to IgG1 Fc highlights the central role of CD16a Gly-129 in this unexpected behavior. The structure of glycosylated CD16b in complex with IgG1 Fc provides some insight into the mechanism underlying this phenomenon: Asp-129 deforms the CD16b β sheet adjacent to the interface formed with IgG1 Fc, and this distortion perturbs contacts between the Asn-162 glycan and Arg-155 (Fig. 6). Although it is unclear why CD16a with Man5 N-glycans binds tighter to IgG1 Fc than CD16a with complex-type N-glycans, the ability to increase the sensitivity of CD16b to N-glycan composition by the D129G substitution indicates that residues surrounding Gly-129 are likely involved in this phenomenon.
CD16b has received much less attention than CD16a. Although engaging CD16a is essential for the therapeutic activity of most mAbs, CD16b has emerged as an important target (35). The recent realization that targeting CD16b may improve these drugs is partially due to the roughly 50-fold higher amount of CD16b in the human body compared with CD16a. We estimate that the average person contains more than 200 μg of CD16b from neutrophils in circulation compared with just 4 μg of CD16a from monocytes and NK cells (2, 19, 47). Perhaps because of this difference in abundance, mAbs that specifically target CD16b provide definite advantages. One glycoengineered anti-CD20 antibody, obinutuzumab, showed greater therapeutic efficacy than the standard anti-CD20 therapy rituximab because of higher affinity and an ability to activate neutrophils through CD16b but not CD32a (35). Another study also found that glycoengineered nonfucosylated rituximab similarly enhanced neutrophil activation through CD16b engagement (36). These findings demonstrate that increasing the CD16b binding affinity of mAbs provides better therapeutic responses and potentially lower doses in the clinic. Our finding that Asp-129 substantially decreases rCD16b affinity for IgG1 Fc indicates that designing antibodies to accommodate the Asp-129 side chain may improve CD16b binding affinity and contribute to improved therapeutic outcomes.
Experimental procedures
Materials
All materials were purchased from Sigma-Aldrich unless noted otherwise.
Protein expression and purification
CD16a (V158) and CD16b (NA2) were previously cloned into the Not1 and EcoR1 restriction sites of the pGen2 vector (5, 48, 49). CD16a and CD16b cloned into a pUC57 vector served as templates to generate genes encoding mutated receptors according to the QuikChange protocol (Agilent). Mutations were verified by DNA sequencing at the Iowa State University DNA facility. Verified mutant genes were subcloned into the pGen2 plasmid using the Not1 and EcoR1 restriction enzymes (49).
Expression and purification of CD16 with complex-type N-glycans was achieved by expression from HEK293F cells and purification as described previously (5). CD16 with oligomannose (Man5) N-glycans was expressed using the HEK293S cell line as described previously (19). Typical yields were ∼0.2 mg/ml of final culture volume. IgG1 Fc used for binding analyses and single IgG1 Fc glycoforms were prepared as described previously (5). Binding analyses for the previous report (5) and the results reported in this manuscript were collected within a few days of one another. IgG1 Fc lacking a fucose residue was used for crystallography and was prepared by expressing protein in the presence of 250 μm 2-fluoro-2-deoxy fucose as described previously (5). Glycomics analyses of PNGaseF-released N-glycans using HILIC-ESI-MS/MS were performed as described previously (19).
Crystallization of the CD16b–IgG1 Fc complex
CD16b-N38Q/N64Q/N74Q/N169Q with Man5 N-glycans was transferred to a buffer containing 50 mm Tris, 100 mm sodium chloride, and 0.5 mm EDTA (pH 8.0). A 1:50 molar ratio (receptor:tobacco etch virus–GFP) was added, and the samples were incubated overnight in the dark at room temperature. CD16 was separated from GFP by flowing over a nickel-nitrilotriacetic acid resin (Qiagen) and concentrating the flow-through fractions containing CD16. CD16 was exchanged into a buffer containing 20 mm MOPS and 100 mm sodium chloride (pH 7.2) with an Amicon Ultra centrifugal filter with a 10-kDa molecular weight cutoff (Merck Millipore). CD16 was then mixed with IgG1 Fc (-fucose) at a (1:1.5) molar ratio. Complex was separated from unbound IgG1 Fc by purification with an AKTA Pure chromatography system equipped with a Superdex 200 gel filtration column pre-equilibrated with a buffer containing 20 mm MOPS and 100 mm sodium chloride (pH 7.2) (GE Healthcare Bio-Sciences). Fractions containing the CD16–Fc complex were identified by SDS-PAGE and concentrated to 12 mg/ml. Crystals in the P212121 space group were obtained using the hanging drop method with a well solution of 50 mm MES, 10% PEG3350 (w/v), and 50 mm sodium chloride (pH 6.0). Crystals in the C2 space group were obtained using the hanging drop method with a well solution of 50 mm MES, 8% PEG3350 (w/v), and 60 mm sodium chloride. All crystals were cryoprotected with 15% ethylene glycol diluted with well solution and flash-frozen in liquid nitrogen. Diffraction data were collected at 1° increments for 200 frames with an exposure time of 0.5 s on beamline 23-ID-D at the Argonne National Laboratory Advanced Photon Source with a Pilatus3 6 M detector. The software HKL-3000 was used to index, scale, and merge datasets. Initial phases were determined with molecular replacements in Phenix-Phaser MR using PDB code 5VU0 (20). Final refinement was performed with Phenix using secondary structure restraints and noncrystallographic symmetry restraints. The structure was deposited in the PDB as 6EAQ. Measurements of interfaces and buried surface area were made using the PDBePISA utility (45). Molecular dynamics simulations of the glycosylated CD16b–Fc complex were performed using Amber16 as described previously (20).
Surface plasmon resonance binding experiments
Binding experiments were performed as described previously (5). For interactions that reached a clear equilibrium, the intensity values of the association phase were fitted with a binding isotherm to determine a dissociation constant for the interaction. For slower interactions where a clear equilibrium was not achieved, dissociation constants were calculated using the observed on and off rates as described previously (5). Values for Tables 1 and 2 include errors resulting from the least-squares fitting procedure.
Author contributions
J. T. R. and A. W. B. conceptualization; J. T. R. and A. W. B. data curation; J. T. R. and A. W. B. formal analysis; J. T. R. and A. W. B. validation; J. T. R. and A. W. B. investigation; J. T. R. and A. W. B. visualization; J. T. R. and A. W. B. methodology; J. T. R. and A. W. B. writing-original draft; J. T. R. and A. W. B. writing-review and editing; A. W. B. resources; A. W. B. supervision; A. W. B. funding acquisition; A. W. B. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Daniel J. Falconer for help with X-ray crystallography and molecular dynamics simulations and Dr. Aaron Marcella for help with X-ray crystallography.
This work was supported by NIGMS, National Institutes of Health under Award R01 GM115489, the Roy J. Carver Department of Biochemistry, Biophysics, and Molecular Biology at Iowa State University, and by the Department of Energy for synchrotron time at the Argonne National Laboratory Advanced Photon Source (GUP-48455). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the Department of Energy.
This article contains Figs. S1–S3 and Table S1.
The atomic coordinates and structure factors (code 6EAQ) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- NK
- natural killer
- MD
- molecular dynamics
- CT
- complex-type
- HILIC
- hydrophilic interaction chromatography
- ESI
- electrospray ionization.
References
- 1. Hayes J. M., Cosgrave E. F., Struwe W. B., Wormald M., Davey G. P., Jefferis R., and Rudd P. M. (2014) Glycosylation and Fc receptors. Curr. Top. Microbiol. Immunol. 382, 165–199 [DOI] [PubMed] [Google Scholar]
- 2. Battella S., Cox M. C., Santoni A., and Palmieri G. (2016) Natural killer (NK) cells and anti-tumor therapeutic mAb: unexplored interactions. J. Leukoc. Biol. 99, 87–96 10.1189/jlb.5VMR0415-141R [DOI] [PubMed] [Google Scholar]
- 3. Guillerey C., Huntington N. D., and Smyth M. J. (2016) Targeting natural killer cells in cancer immunotherapy. Nat. Immunol. 17, 1025–1036 10.1038/ni.3518 [DOI] [PubMed] [Google Scholar]
- 4. Dekkers G., Treffers L., Plomp R., Bentlage A. E. H., de Boer M., Koeleman C. A. M., Lissenberg-Thunnissen S. N., Visser R., Brouwer M., Mok J. Y., Matlung H., van den Berg T. K., van Esch W. J. E., Kuijpers T. W., Wouters D., et al. (2017) Decoding the human immunoglobulin G-glycan repertoire reveals a spectrum of Fc receptor- and complement-mediated effector activities. Front. Immunol. 8, 877 10.3389/fimmu.2017.00877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Subedi G. P., and Barb A. W. (2016) The immunoglobulin G1 N-glycan composition affects binding to each low affinity Fc γ receptor. mAbs 8, 1512–1524 10.1080/19420862.2016.1218586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clarkson S. B., and Ory P. A. (1988) CD16. Developmentally regulated IgG Fc receptors on cultured human monocytes. J. Exp. Med. 167, 408–420 10.1084/jem.167.2.408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Haas M., Koene H. R., Kleijer M., de Vries E., Simsek S., van Tol M. J., Roos D., and von dem Borne A. E. (1996) A triallelic Fc γ receptor type IIIA polymorphism influences the binding of human IgG by NK cell Fc γ RIIIa. J. Immunol. 156, 2948–2955 [PubMed] [Google Scholar]
- 8. Edberg J. C., Barinsky M., Redecha P. B., Salmon J. E., and Kimberly R. P. (1990) Fc γ RIII expressed on cultured monocytes is a N-glycosylated transmembrane protein distinct from Fc γ RIII expressed on natural killer cells. J. Immunol. 144, 4729–4734 [PubMed] [Google Scholar]
- 9. Nagler A., Lanier L. L., Cwirla S., and Phillips J. H. (1989) Comparative studies of human FcRIII-positive and negative natural killer cells. J. Immunol. 143, 3183–3191 [PubMed] [Google Scholar]
- 10. Davoine F., Lavigne S., Chakir J., Ferland C., Boulay M. E., and Laviolette M. (2002) Expression of FcγRIII (CD16) on human peripheral blood eosinophils increases in allergic conditions. J. Allergy Clin. Immunol. 109, 463–469 10.1067/mai.2002.121952 [DOI] [PubMed] [Google Scholar]
- 11. Meknache N., Jönsson F., Laurent J., Guinnepain M.-T., and Daëron M. (2009) Human basophils express the glycosylphosphatidylinositol-anchored low-affinity IgG receptor FcγRIIIB (CD16B). J. Immunol. 182, 2542–2550 10.4049/jimmunol.0801665 [DOI] [PubMed] [Google Scholar]
- 12. Ravetch J. V., and Perussia B. (1989) Alternative membrane forms of Fc γ RIII(CD16) on human natural killer cells and neutrophils: cell type-specific expression of two genes that differ in single nucleotide substitutions. J. Exp. Med. 170, 481–497 10.1084/jem.170.2.481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koene H. R., Kleijer M., Algra J., Roos D., von dem Borne A. E., and de Haas M. (1997) FcγRIIIa-158V/F polymorphism influences the binding of IgG by natural killer cell FcγRIIIa, independently of the FcγRIIIa-48L/R/H phenotype. Blood 90, 1109–1114 [PubMed] [Google Scholar]
- 14. Salmon J. E., Edberg J. C., Brogle N. L., and Kimberly R. P. (1992) Allelic polymorphisms of human Fc γ receptor IIA and Fc γ receptor IIIB: independent mechanisms for differences in human phagocyte function. J. Clin. Invest. 89, 1274–1281 10.1172/JCI115712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moremen K. W., Tiemeyer M., and Nairn A. V. (2012) Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 13, 448–462 10.1038/nrm3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Edberg J. C., and Kimberly R. P. (1997) Cell type-specific glycoforms of Fc γ RIIIa (CD16): differential ligand binding. J. Immunol. 159, 3849–3857 [PubMed] [Google Scholar]
- 17. Kimberly R. P., Tappe N. J., Merriam L. T., Redecha P. B., Edberg J. C., Schwartzman S., and Valinsky J. E. (1989) Carbohydrates on human Fc γ receptors: interdependence of the classical IgG and nonclassical lectin-binding sites on human Fc γ RIII expressed on neutrophils. J. Immunol. 142, 3923–3930 [PubMed] [Google Scholar]
- 18. Yagi H., Takakura D., Roumenina L. T., Fridman W. H., Sautès-Fridman C., Kawasaki N., and Kato K. (2018) Site-specific N-glycosylation analysis of soluble Fcγ receptor IIIb in human serum. Sci. Rep. 8, 2719 10.1038/s41598-018-21145-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patel K. R., Roberts J. T., Subedi G. P., and Barb A. W. (2018) Restricted processing of CD16a/Fc gamma receptor IIIa N-glycans from primary human NK cells impacts structure and function. J. Biol. Chem. 293, 3477–3489 10.1074/jbc.RA117.001207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Falconer D. J., Subedi G. P., Marcella A. M., and Barb A. W. (2018) Antibody fucosylation lowers the FcγRIIIa/CD16a affinity by limiting the conformations sampled by the N162-glycan. ACS Chem. Biol. 13, 2179–2189 10.1021/acschembio.8b00342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hayes J. M., Frostell A., Karlsson R., Müller S., Martín S. M., Pauers M., Reuss F., Cosgrave E. F., Anneren C., Davey G. P., and Rudd P. M. (2017) Identification of Fc γ receptor glycoforms that produce differential binding kinetics for rituximab. Mol. Cell. Proteomics 16, 1770–1788 10.1074/mcp.M117.066944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Subedi G. P., and Barb A. W. (2018) CD16a with oligomannose-type N-glycans is the only “low affinity” Fc γ receptor that binds the IgG crystallizable fragment with high affinity in vitro. J. Biol. Chem. 293, 16842–16850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reusch D., and Tejada M. L. (2015) Fc glycans of therapeutic antibodies as critical quality attributes. Glycobiology 25, 1325–1334 10.1093/glycob/cwv065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matsumiya S., Yamaguchi Y., Saito J., Nagano M., Sasakawa H., Otaki S., Satoh M., Shitara K., and Kato K. (2007) Structural comparison of fucosylated and nonfucosylated Fc fragments of human immunoglobulin G1. J. Mol. Biol. 368, 767–779 10.1016/j.jmb.2007.02.034 [DOI] [PubMed] [Google Scholar]
- 25. Thomann M., Schlothauer T., Dashivets T., Malik S., Avenal C., Bulau P., Rüger P., and Reusch D. (2015) In vitro glycoengineering of IgG1 and its effect on Fc receptor binding and ADCC activity. PLoS ONE 10, e0134949 10.1371/journal.pone.0134949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chan A. C., and Carter P. J. (2010) Therapeutic antibodies for autoimmunity and inflammation. Nat. Rev. Immunol. 10, 301–316 10.1038/nri2761 [DOI] [PubMed] [Google Scholar]
- 27. Mellor J. D., Brown M. P., Irving H. R., Zalcberg J. R., and Dobrovic A. (2013) A critical review of the role of Fc γ receptor polymorphisms in the response to monoclonal antibodies in cancer. J. Hematol. Oncol. 6, 1 10.1186/1756-8722-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Congy-Jolivet N., Bolzec A., Ternant D., Ohresser M., Watier H., and Thibault G. (2008) Fc γ RIIIa expression is not increased on natural killer cells expressing the Fc γ RIIIa-158V allotype. Cancer Res. 68, 976–980 10.1158/0008-5472.CAN-07-6523 [DOI] [PubMed] [Google Scholar]
- 29. Oboshi W., Watanabe T., Matsuyama Y., Kobara A., Yukimasa N., Ueno I., Aki K., Tada T., and Hosoi E. (2016) The influence of NK cell-mediated ADCC: structure and expression of the CD16 molecule differ among FcγRIIIa-V158F genotypes in healthy Japanese subjects. Hum. Immunol. 77, 165–171 10.1016/j.humimm.2015.11.001 [DOI] [PubMed] [Google Scholar]
- 30. Shields R. L., Lai J., Keck R., O'Connell L. Y., Hong K., Meng Y. G., Weikert S. H., and Presta L. G. (2002) Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human FcγRIII and antibody-dependent cellular toxicity. J. Biol. Chem. 277, 26733–26740 10.1074/jbc.M202069200 [DOI] [PubMed] [Google Scholar]
- 31. Natsume A., Wakitani M., Yamane-Ohnuki N., Shoji-Hosaka E., Niwa R., Uchida K., Satoh M., and Shitara K. (2006) Fucose removal from complex-type oligosaccharide enhances the antibody-dependent cellular cytotoxicity of single-gene-encoded bispecific antibody comprising of two single-chain antibodies linked to the antibody constant region. J. Biochem. 140, 359–368 10.1093/jb/mvj157 [DOI] [PubMed] [Google Scholar]
- 32. Niwa R., Sakurada M., Kobayashi Y., Uehara A., Matsushima K., Ueda R., Nakamura K., and Shitara K. (2005) Enhanced natural killer cell binding and activation by low-fucose IgG1 antibody results in potent antibody-dependent cellular cytotoxicity induction at lower antigen density. Clin. Cancer Res. 11, 2327–2336 10.1158/1078-0432.CCR-04-2263 [DOI] [PubMed] [Google Scholar]
- 33. Okazaki A., Shoji-Hosaka E., Nakamura K., Wakitani M., Uchida K., Kakita S., Tsumoto K., Kumagai I., and Shitara K. (2004) Fucose depletion from human IgG1 oligosaccharide enhances binding enthalpy and association rate between IgG1 and FcγRIIIa. J. Mol. Biol. 336, 1239–1249 10.1016/j.jmb.2004.01.007 [DOI] [PubMed] [Google Scholar]
- 34. Shinkawa T., Nakamura K., Yamane N., Shoji-Hosaka E., Kanda Y., Sakurada M., Uchida K., Anazawa H., Satoh M., Yamasaki M., Hanai N., and Shitara K. (2003) The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J. Biol. Chem. 278, 3466–3473 10.1074/jbc.M210665200 [DOI] [PubMed] [Google Scholar]
- 35. Golay J., Da Roit F., Bologna L., Ferrara C., Leusen J. H., Rambaldi A., Klein C., and Introna M. (2013) Glycoengineered CD20 antibody obinutuzumab activates neutrophils and mediates phagocytosis through CD16B more efficiently than rituximab. Blood 122, 3482–3491 10.1182/blood-2013-05-504043 [DOI] [PubMed] [Google Scholar]
- 36. Shibata-Koyama M., Iida S., Misaka H., Mori K., Yano K., Shitara K., and Satoh M. (2009) Nonfucosylated rituximab potentiates human neutrophil phagocytosis through its high binding for FcγRIIIb and MHC class II expression on the phagocytotic neutrophils. Exp. Hematol. 37, 309–321 10.1016/j.exphem.2008.11.006 [DOI] [PubMed] [Google Scholar]
- 37. Stanley P., Narasimhan S., Siminovitch L., and Schachter H. (1975) Chinese hamster ovary cells selected for resistance to the cytotoxicity of phytohemagglutinin are deficient in a UDP-N-acetylglucosamine–glycoprotein N-acetylglucosaminyltransferase activity. Proc. Natl. Acad. Sci. U.S.A. 72, 3323–3327 10.1073/pnas.72.9.3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reeves P. J., Callewaert N., Contreras R., and Khorana H. G. (2002) Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. U.S.A. 99, 13419–13424 10.1073/pnas.212519299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sondermann P., Huber R., Oosthuizen V., and Jacob U. (2000) The 3.2-A crystal structure of the human IgG1 Fc fragment–FcγRIII complex. Nature 406, 267–273 10.1038/35018508 [DOI] [PubMed] [Google Scholar]
- 40. Radaev S., Motyka S., Fridman W.-H., Sautes-Fridman C., and Sun P. D. (2001) The structure of a human type III Fcγ receptor in complex with Fc. J. Biol. Chem. 276, 16469–16477 10.1074/jbc.M100350200 [DOI] [PubMed] [Google Scholar]
- 41. Mizushima T., Yagi H., Takemoto E., Shibata-Koyama M., Isoda Y., Iida S., Masuda K., Satoh M., and Kato K. (2011) Structural basis for improved efficacy of therapeutic antibodies on defucosylation of their Fc glycans. Genes Cells 16, 1071–1080 10.1111/j.1365-2443.2011.01552.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ferrara C., Grau S., Jäger C., Sondermann P., Brünker P., Waldhauer I., Hennig M., Ruf A., Rufer A. C., Stihle M., Umaña P., and Benz J. (2011) Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcγRIII and antibodies lacking core fucose. Proc. Natl. Acad. Sci. U.S.A. 108, 12669–12674 10.1073/pnas.1108455108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shibata-Koyama M., Iida S., Okazaki A., Mori K., Kitajima-Miyama K., Saitou S., Kakita S., Kanda Y., Shitara K., Kato K., and Satoh M. (2009) The N-linked oligosaccharide at FcγRIIIa Asn-45: an inhibitory element for high FcγRIIIa binding affinity to IgG glycoforms lacking core fucosylation. Glycobiology 19, 126–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ferrara C., Stuart F., Sondermann P., Brünker P., and Umaña P. (2006) The Carbohydrate at FcγRIIIa Asn-162: an element required for high affinity binding to non-fucosylated IgG glycoforms. J. Biol. Chem. 281, 5032–5036 10.1074/jbc.M510171200 [DOI] [PubMed] [Google Scholar]
- 45. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 46. Robinson J. I., Baxter E. W., Owen R. L., Thomsen M., Tomlinson D. C., Waterhouse M. P., Win S. J., Nettleship J. E., Tiede C., Foster R. J., Owens R. J., Fishwick C. W. G., Harris S. A., Goldman A., McPherson M. J., and Morgan A. W. (2018) Affimer proteins inhibit immune complex binding to FcγRIIIa with high specificity through competitive and allosteric modes of action. Proc. Natl. Acad. Sci. U.S.A. 115, E72–E81 10.1073/pnas.1707856115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Summers C., Rankin S. M., Condliffe A. M., Singh N., Peters A. M., and Chilvers E. R. (2010) Neutrophil kinetics in health and disease. Trends Immunol. 31, 318–324 10.1016/j.it.2010.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Subedi G. P., Hanson Q. M., and Barb A. W. (2014) Restricted motion of the conserved immunoglobulin G1 N-glycan is essential for efficient FcγRIIIa binding. Structure 22, 1478–1488 10.1016/j.str.2014.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Barb A. W., Meng L., Gao Z., Johnson R. W., Moremen K. W., and Prestegard J. H. (2012) NMR characterization of immunoglobulin G Fc glycan motion on enzymatic sialylation. Biochemistry 51, 4618–4626 10.1021/bi300319q [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.