Abstract
Methyl-CpG–binding proteins (MBPs) are selective readers of DNA methylation that play an essential role in mediating cellular transcription processes in both normal and diseased cells. This physiological function of MBPs has generated significant interest in understanding the mechanisms by which these proteins read and interpret DNA methylation signals. Zinc finger and BTB domain–containing 38 (ZBTB38) represents one member of the zinc finger (ZF) family of MBPs. We recently demonstrated that the C-terminal ZFs of ZBTB38 exhibit methyl-selective DNA binding within the ((A/G)TmCG(G/A)(mC/T)(G/A)) context both in vitro and within cells. Here we report the crystal structure of the first four C-terminal ZBTB38 ZFs (ZFs 6–9) in complex with the previously identified methylated consensus sequence at 1.75 Å resolution. From the structure, methyl-selective binding is preferentially localized at the 5′ mCpG site of the bound DNA, which is facilitated through a series of base-specific interactions from residues within the α-helices of ZF7 and ZF8. ZF6 and ZF9 primarily stabilize ZF7 and ZF8 to facilitate the core base–specific interactions. Further structural and biochemical analyses, including solution NMR spectroscopy and electrophoretic mobility gel shift assays, revealed that the C-terminal ZFs of ZBTB38 utilize an alternative mode of mCpG recognition from the ZF MBPs structurally evaluated to date. Combined, these findings provide insight into the mechanism by which this ZF domain of ZBTB38 selectively recognizes methylated CpG sites and expands our understanding of how ZF-containing proteins can interpret this essential epigenetic mark.
Keywords: epigenetics, zinc finger, X-ray crystallography, DNA methylation, protein–DNA interaction, DNA reader, methyl-CpG binding protein, transcriptional regulation, zinc finger and BTB domain–containing protein
Introduction
Normal cellular function relies on exquisite control of transcriptional processes, which is in part mediated by the methylation status at CpG sites. In a number of disease conditions, patterns of DNA methylation become inappropriately distributed, leading to aberrant transcriptional outcomes that promote and maintain the disease state (1–7). Transcriptional regulation at many methylated gene loci is facilitated by methyl-CpG–binding proteins (MBPs),2 transcription factors that have evolved to selectively recognize methylated CpG (mCpG) sites (8–11). Upon binding, these epigenetic readers translate the mCpG signal into recruitment of protein assemblies that subsequently alter local chromatin architecture (12–18). Therefore, MBPs play a critical intermediary role in regulating gene activity in both normal and diseased cells, prompting significant interest in mechanistically understanding how these MBPs select and interpret DNA methylation signals.
In recent years, high-resolution structural investigations for several Cys2His2 zinc finger (ZF)–containing proteins in complex with their cognate methylated DNA sequences have been reported (19–26). Comparisons between these structural models have afforded insight into the similarities and differences in modes of mCpG recognition for this important class of MBPs (27–29). More recently, several high-throughput screening strategies have identified a number of additional ZF-containing proteins that appear to have selective recognition of methylated DNA (30–33) but remain to be biochemically and structurally characterized. It is therefore possible that additional modes of mCpG recognition by ZF MBPs exist.
Zinc finger and BTB domain–containing protein 38 (ZBTB38) belongs to the large broad complex, tramtrack, bric-á-brac, pox zinc finger (BTB/POZ) family of transcription factors and represents one of only three members of this extended family that has demonstrated selectivity for mCpG recognition (34). Methyl-specific DNA binding of ZBTB38 was originally characterized for the N-terminal ZFs 3–5 (Fig. 1A), which have high sequence commonality with the three ZFs of ZBTB33 (Kaiso), the founding member of the ZBTB MBPs (34–36). Recently, we identified a sequence-specific DNA binding site ((A/G)TmCG(G/A)(mC/T)(G/A)) for the C-terminal set of ZBTB38 ZFs and determined that motif recognition is also methyl-selective both in vitro and within cells (37). It was further demonstrated that, depending on the gene context, the C-terminal ZBTB38 ZFs participate in modulating transcriptional outcomes (37). These findings have significant implications for delineating the overall biological function of ZBTB38, given that its transcriptional activities have been linked to a number of core cellular processes such as cellular proliferation (38), apoptosis (35, 39), and replication/genomic stability (40).
Figure 1.
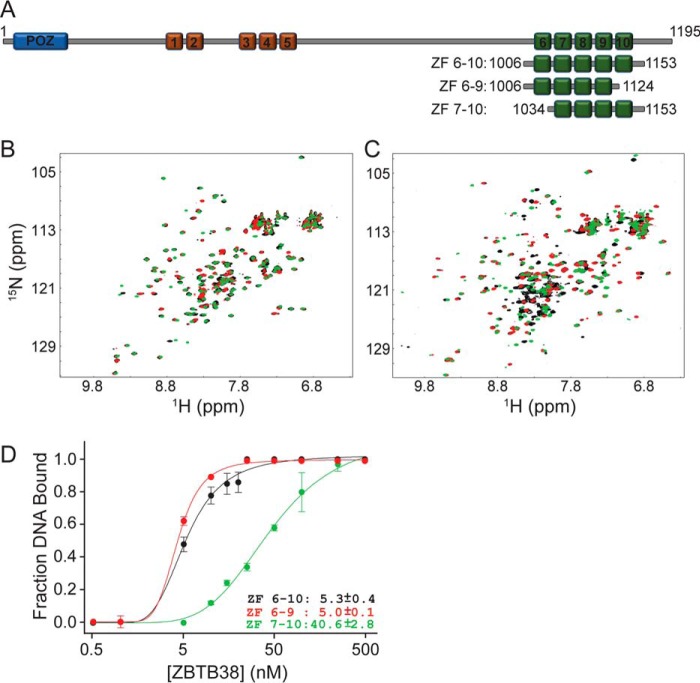
Optimization of the C-terminal ZBTB38 ZFs for use in structural studies with methylated DNA. A, domain organization of human ZBTB38 and construct design for the C-terminal ZF region used in this study (green boxes). Red boxes depict the N-terminal set of five ZFs, and the blue box designates the BTB/POZ protein:protein interaction domain. B, 1H-15N HSQC spectral overlay for free ZBTB38 ZFs 6–10 (black), ZFs 6–9 (red), and ZFs 7–10 (green). C, 1H-15N HSQC spectral overlay for ZBTB38 ZFs 6–10 (black), ZFs 6–9 (red), and ZFs 7–10 (green) each in complex with the mCZ38BS_21-mer. D, comparative binding isotherms for ZBTB38 ZFs 6–10, 6–9, and 7–10 in complex with mCZ38BS_27-mer. Each data point represents the average of triplicate data, with error bars depicting ± S.D.
Using a hybrid biophysical methodology and collective structural insight from available ZF–methylated DNA structures, we previously presented an atomistic model in which it was proposed that a key arginine common to ZF MBPs was replaced with a lysine (Lys-1055) in mCpG recognition (37). Here, using a combination of high-resolution structural and biochemical analyses, we characterize the interactions of the C-terminal ZBTB38 ZFs in complex with its methylated consensus motif, termed the methylated C-terminal ZBTB38 binding sequence (mCZ38BS). From this investigation, it was determined that Lys-1055 does function as an arginine surrogate in mCpG binding. This finding provides the first evidence that alternative modes of mCpG recognition by ZF MBPs may be utilized and expands our understanding of the mechanisms by which this essential epigenetic mark is read.
Results
Optimization of the C-terminal ZBTB ZF domain for structural investigations with methylated DNA
We determined previously that the C-terminal ZBTB38 ZFs 7–9 were necessary and sufficient for methyl-selective DNA recognition (37). However, it was also demonstrated that the highest-affinity binding was achieved in the presence of all five C-terminal ZFs (ZFs 6–10), suggesting that either or both ZF6 and ZF10 may contribute to the overall stability of the protein:DNA interaction. Thus, initial attempts to crystallize the C-terminal ZFs of ZBTB38 with the mCZ38BS focused on utilizing residues 1006–1153, encompassing ZFs 6–10 (Fig. 1A). Preliminary screening of crystallization conditions yielded several low-resolution diffracting crystals. Further optimization attempts, including varying the DNA length and introducing overhangs, failed to produce crystals with better than 7 Å diffraction despite obtaining a number of crystals with multiple morphologies.
Given these results, we postulated that removal of ZF6 or ZF10 might improve crystallization for this protein:DNA complex. Thus, we designed C-terminal constructs including ZFs 6–9 (residues 1006–1124) or ZFs 7–10 (residues 1034–1153) and assessed the consequence of removing either of these ZFs on mCZ38BS recognition (Fig. 1). Comparative 1H-15N heteronuclear single-quantum coherence (HSQC) analysis of free ZFs 6–9 and 7–10 showed that each protein adopted well-folded structural domains (Fig. 1B). Further, a large fraction of the resonances for ZFs 6–9 and 7–10 each superimposed with respective resonances from ZFs 6–10, indicating that the domain folds were preserved (Fig. 1B). In the presence of mCZ38BS, both ZFs 6–10 and 6–9 produced well-resolved spectra indicative of high-affinity binding interactions (Fig. 1C). Moreover, the majority of resonances outside of the random coil region observed for the ZF 6–10:mCZ38BS complex spectrum were largely recapitulated in the ZF 6–9:mCZ38BS complex spectrum, indicating similar structures for the protein: DNA complexes. In contrast, ZFs 7–10 in complex with the mCZ38BS resulted in significant signal loss for many resonances, suggestive of a much weaker binding interaction (Fig. 1C).
Consistent with the solution NMR observations, ZFs 6–10 and 6–9 had nearly identical low nanomolar binding affinities (Kd values of ∼5 nm) for mCZ38BS recognition, whereas ZFs 7–10 exhibited ∼8-fold weaker binding (Fig. 1D and Fig. S1A). This finding was also in agreement with our previously proposed atomistic model, where the α-helices of ZFs 6–9 could be readily positioned along the major groove of the DNA, whereas the exact positioning of ZF10 remained indeterminant (37). Although these observations do not fully preclude ZF10 from interacting nonspecifically with DNA, it is possible that another primary physiological role for this ZF domain exists. Indeed, it was shown that the region of ZBTB38 encompassing the C-terminal ZFs can also participate in protein:protein interactions (40). Combined, these findings suggest that ZBTB38 ZFs 6–9 constitute the minimal subset of ZFs necessary for high-affinity DNA recognition.
Overall structure of the C-terminal ZBTB38 ZF 6–9:mCZ38BS complex
ZBTB38 ZFs 6–9 (Fig. 2A) were co-crystallized with an 18-bp oligonucleotide sequence containing the previously determined high-affinity mCZ38BS motif (ATmCGGmCG) (37) (Fig. 2, B and C). Crystallization of the complexes succeeded with a shorter oligonucleotide duplex than what was used for the EMSA and initial solution NMR experiments (Table S1). No discernable differences in protein resonance chemical shifts were observed by solution NMR in complexation with either the 21-mer or 18-mer version of the mCZ38BS. The protein:DNA complex crystallized into space group C222(1), with one complex per asymmetric unit, and the structure was determined at a resolution of 1.75 Å (Table 1). Atomic positioning was afforded for all base atoms within the mCZ38BS and ZBTB38 residues 1009–1120, which excludes three and four flexible residues at the N and C termini, respectively.
Figure 2.
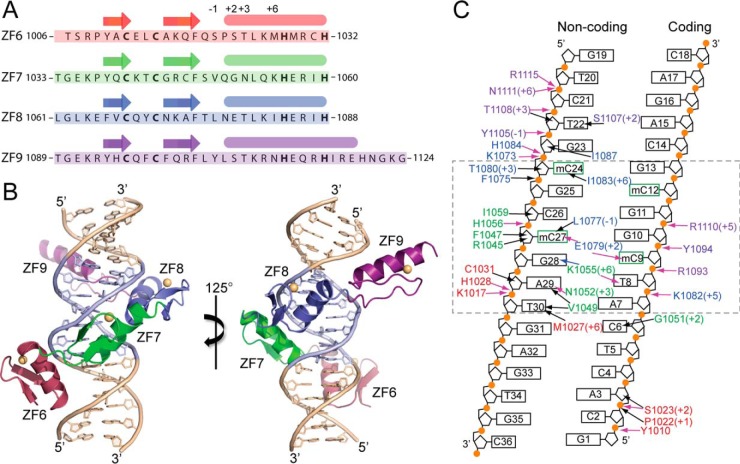
High-resolution structure of C-terminal ZBTB38 ZFs 6–9 in complex with its methylated DNA consensus motif. A, polypeptide sequence for ZBTB38 ZFs 6–9 (residues 1006–1124) utilized for structural analysis. Canonical amino acid positions along the ZF α-helices typically involved in DNA recognition are denoted above. B, X-ray crystal structure of ZBTB38 ZFs 6–9 in complex with the mCZ38BS_18-mer. The core ATmCGGmCG sequence is highlighted in blue, and zinc atoms for each ZF are depicted as yellow spheres. This figure was prepared using PyMOL. C, contact map summarizing the ZBTB38 ZF 6–9:mCZ38BS interactions. The dashed box denotes the location of the core mCZ38BS consensus motif. Residues are colored red, green, blue, and purple, corresponding to ZFs 6–9, respectively. Numbers in parentheses designate the residue position along the α-helix. Black arrows define van der Waals interactions, pink arrows define classical hydrogen bonds, and blue arrows define water-mediated hydrogen bonds. A complete list of contacts is available in Table S2.
Table 1.
X-ray diffraction data collection and refinement statistics
| Parameters |
||
|---|---|---|
| ZBTB38 ZF 6–9:mCZ38BS | ZBTB38 ZF 6–9_K1055R:mCZ38BS | |
| Data collection | ||
| Beamline | SSRL 12-2 | SSRL 12-2 |
| Wavelength (Å) | 0.9795 | 0.9795 |
| Space group | C222 (1) | C222 (1) |
| Unit cell parameters | ||
| a, b, c (Å) | 70.80, 77.27, 103.76 | 70.56, 77.61, 103.09 |
| Resolution range (Å) | 40.00–1.75 (1.81–1.75) | 38.80–1.59 (1.62–1.59) |
| Observed reflections | 193,907 | 487,692 |
| Unique reflections | 29,108 | 37,856 |
| Redundancy | 6.7 | 12.9 |
| Completeness (%) | 99.8 (99.8) | 99.3 (88.8) |
| Mosaicity (°) | 0.68 | 0.26 |
| <I>/σ(I) | 4.9 (0.8) | 9.4 (0.9) |
| CC½ | (0.572) | (0.610) |
| Refinement | ||
| Resolution range (Å) | 21.54–1.75 (1.81–1.75) | 35.40–1.60 (1.61–1.60) |
| No. of reflections | 29,043 | 37,492 |
| No. of reflections in Rfree set | 2,836 | 3,286 |
| Rwork | 0.1860 (0.2695) | 0.1939 (0.3029) |
| Rfree | 0.2112 (0.3055) | 0.2183 (0.3238) |
| RMSD bonds/angles (Å/°) | 0.007/1.800 | 0.007/1.041 |
| No. of atoms | ||
| Protein/DNA | 1679 | 1671 |
| Water | 152 | 202 |
| Others | 4 (Zn+2) | 4 (Zn+2) |
| B-factors (Å2) | ||
| Protein/DNA | 36.1 | 27.0 |
| Water | 38.5 | 32.3 |
| Others | 30.6 | 24.0 |
| Ramachandran favored (%) | 98.2 | 99.1 |
All four of the Cys2His2 ZFs adopt the canonical ββα zinc finger fold and wrap around the DNA with their α-helices oriented toward the major groove (Fig. 2, A and B). However, only ZF7 and ZF8 participate in base-specific DNA interactions, which are concentrated at the three core T8:A29, mC9:G28, and G10:mC27 base steps (Fig. 2C and Table S2). The α-helix of ZF6 spans the major groove and is primarily anchored through a series of electrostatic and direct hydrogen-bonding interactions with the phosphate backbone as well as by van der Waals contacts with the sugar rings on both DNA strands (Fig. 2C, Fig. S2A, and Table S2). These contacts from ZF6 are positioned toward the 5′ and 3′ ends of the coding and noncoding strands, respectively. ZF9 has an extended α-helix relative to ZFs 6–8, the C-terminal end of which orients away from the major groove of the DNA. This affords accumulation of a hydration layer within the corresponding major groove region of the DNA (Fig. S2B). Similar to ZF6, the N-terminal end of the ZF9 α-helix contributes analogous backbone interactions along both DNA strands toward the 3′ and 5′ ends of the coding and noncoding strands, respectively (Fig. 2C, Fig. S2C, and Table S2).
Both ZF6 and ZF9 also contribute interactions between residues along their respective α-helices (Met-1027 and Ser-1107) and the C5-methyl groups of thymine bases found along the noncoding strand (T30 and T22) (Fig. 2C, Fig. S2A, and Fig. S2C and Table S2). The side-chain interactions observed between the Cγ-methyl of Met-1027 (Fig. S1A) and the C5-methyl of T30 in conjunction with a contact between the Cγ2-methyl of Val-1049 (ZF7) and the C5-methyl of T30 may explain the slight observed preference for having an A:T step in the first position of the mCZ38BS (37) (corresponding to A7:T30 in Fig. 1C). Overall, it appears that ZFs 6 and 9 primarily function to stabilize ZF7 and ZF8 for making core base–specific interactions.
Molecular basis for recognition of the core TmCG consensus
Of the high-resolution structures currently available for ZF MBPs in complex with methylated DNA, the ZBTB38 ZF 6–9:mCZ38BS complex structure most resembles the ZBTB33:mDNA structures (19, 25) in that two ZF helices provide residues for mCpG recognition. Further, although the ZBTB33 methylated consensus motif contains two consecutive mCpG (mCpGmCpG) sites, structural and biochemical evidence has established that methyl sensitivity is primarily conferred by base-specific contacts at the 5′ mCpG, whereas methylation at the 3′ mCpG is less essential (19, 25). Similarly, the C-terminal ZBTB38 ZFs preferentially recognize the 5′ mCpG site within the mCZ38BS (7ATmCGGmCG13) (37). Although optimal binding between the ZBTB38 C-terminal ZFs and the mCZ38BS was observed when both CpG sites were methylated, biochemical analysis demonstrated that the methylation status of the 3′ mCpG contributed less to the overall binding interaction (37). Finally, the adjacent base preceding the 5′ mCpG in the mCZ38BS was identified from in vitro selection methodologies to be exclusively a thymine (37), which is consistent with the base-specific interactions from residues in ZF7 and ZF8 being centralized at the 5′ 8TmCG10 region.
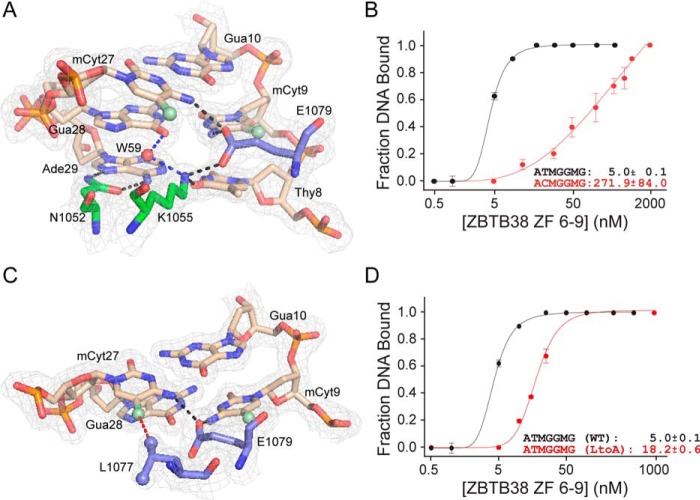
Direct binding interactions for the T8:A29 base pair are mediated by the side chains of Asn-1052 and Lys-1055. Specifically, the Nδ2 and Oδ1 of Asn-1052 each form hydrogen bonds with the N7 and N6 positions of A29, respectively (Fig. 3A). The Nζ of Lys-1055 makes a subsequent hydrogen bond with O4 of the pairing T8. Combined, these interactions afford selectivity in distinguishing a T:A from a G:C step. Indeed, mutation of T8:A29 to C8:G29 resulted in a significant ∼54-fold decrease in binding (Fig. 3B and Fig. S1B).
Figure 3.
Core C-terminal ZBTB38 ZF residues involved in mCpG recognition. A, interactions between Asn-1052, Lys-1055, and Glu-1079 with the core T8:A29, mC9:G28 and G10:mC27 base pairs. Black dotted lines represent classical hydrogen bond interactions, and blue dotted lines represent water-mediated hydrogen bonds. Electron densities (2Fo-Fc) contoured at 1σ are shown. B, binding isotherms demonstrating the impact of mutating the T8:A29 base pair on the ability of ZBTB38 ZFs 6–9 to recognize its DNA target. Each data point represents the average of triplicate data, with error bars depicting ± S.D. C, interaction of Leu-1077 with 27mC. Glu-1079 is shown to provide a context for the Leu-1077:mC27 interaction. The red dotted line represents a van der Waals interaction, and the black dotted lines represent classical hydrogen bond interactions. Electron densities (2Fo-Fc) contoured at 1σ are shown. A and C were prepared using PyMOL. D, binding isotherms demonstrating a reduction in binding capability with the L1077A variant relative to the WT. Note that the binding curve for ZBTB38 ZFs 6–9 with the mCZ38BS from Fig. 1D has been replotted in B and D for comparison purposes. M denotes methylated cytosine. Each data point represents the average of triplicate data, with error bars depicting ± S.D.
A common mode of mCpG recognition among ZF MBPs involves hydrogen bond interactions between an arginine and the 3′ G and the formation of weaker van der Waals and/or CH···O-type hydrogen bonds between a glutamate and the mC methyl groups (19–21, 23–26). Although still considered a relatively weak nonbonding interaction, the importance of CH···O-type hydrogen bonds in stabilizing protein structure (41, 42) and facilitating protein binding at consensus DNA sites (43) is becoming increasingly recognized. Further structural insight from several ZF MBPs suggests that CH···O type hydrogen bonds are essential for conferring methyl-selective DNA recognition (19–21, 23–26). For ZBTB33 and Zfp57, which have much higher selectivity for mCpG over CpG relative to the other structurally characterized ZF MBPs, the core glutamate side-chain atoms can also participate in direct hydrogen-bonding interactions with the N4 amino group of one (Zfp57) or both (ZBTB33) mCs within the mCpG palindrome (19, 24, 25).
Similar to ZBTB33 and Zfp57, the C-terminal ZFs of ZBTB38 also exhibit high selectivity for mCpG over CpG sites. Consistent with this observation, Glu-1079 recognizes the 5′ palindromic mCpG site by forming cross-strand hydrogen bonds between its Oϵ2 atom and the N4 amino groups of mC9 and mC27. The positioning of Glu-1079 for making these DNA interactions is maintained through a hydrogen bond with Lys-1055 (Fig. 3A). In this orientation, the Oϵ1 and Oϵ2 atoms of Glu-1079 reside within the 3.7 Å van der Waals distance cutoff (41, 44) and adopt the H···O-C angle (110° ± 29° (43)), consistent with formation of CH···O-type hydrogen bonds with the C5-methyls of both T8 and mC9, respectively. Overall, given the number of core base contacts facilitated by Glu-1079, loss of this residue would be predicted to have a significant impact on the ability of the C-terminal ZFs of ZBTB38 to recognize DNA. Indeed, removal of Glu-1079 was assessed previously and found to result in an ∼10-fold decrease in DNA binding (37).
An additional contribution to mC binding specificity comes from the Cδ1-methyl of Leu-1077, which packs against the C5-methyl group of mC27 (Fig. 3C). An analogous Leu within ZBTB33 (Leu-533) was observed to participate in a similar interaction with its methylated DNA target (19). Substitution of Leu-1077 with an alanine did not affect the structural integrity of the protein (Fig. S3), as evidenced by the expected observation of chemical shift perturbations for only a few resonances relative to the WT but did result in an ∼4-fold decrease in DNA binding (Fig. 3D and Fig. S1C).
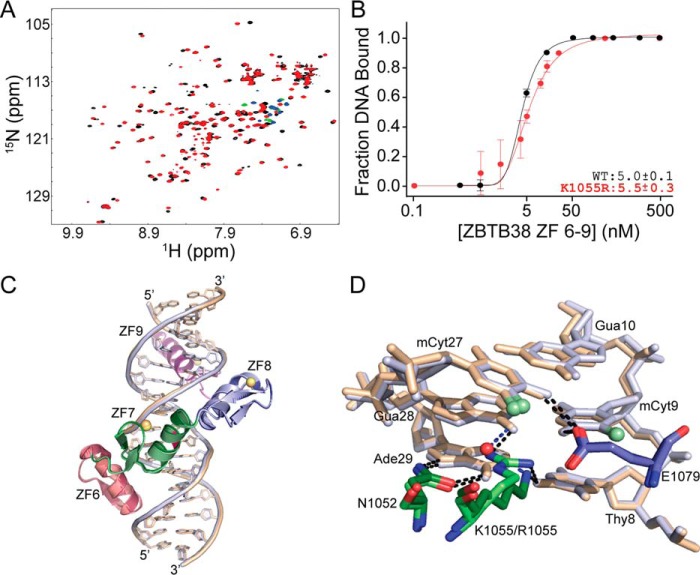
Notably, unlike the other ZF MBPs, ZF7 and ZF8 do not have an analogous arginine residue capable of hydrogen bonding with the mCpG guanines. However, Lys-1055 occupies a spatially comparable position in which its terminal Nζ group forms a water-mediated hydrogen bond with O6 of G28 (Fig. 3A). Overall, it is clear from the structure that Lys-1055 plays a key role in directly and indirectly mediating core recognition of the TmCG motif (Fig. 3A). As further evidence, removal of Lys-1055 was found previously to result in an ∼14-fold decrease in DNA binding (37). Nevertheless, to determine whether Lys-1055 functions as an arginine surrogate in mCpG recognition, a K1055R point variant was introduced into ZFs 6–9 and evaluated for mCZ38BS binding capability after confirming that any observed chemical shift perturbations from the WT were consistent with what would be expected for a conservative point mutation, indicating that protein structural integrity was maintained (Fig. S3).
Comparative 1H-15N HSQC analysis of WT ZFs 6–9 and ZFs 6–9_K1055R in complex with the mCZ38BS demonstrated that the K1055R variant also produced a well-resolved spectrum indicative of a high-affinity binding interaction, although there were chemical shift perturbation differences between the two complexes (Fig. 4A). Nevertheless, both proteins exhibit nearly identical low nanomolar binding affinities for recognition of the mCZ38BS (Fig. 4B and Fig. S1C). The ZBTB38 ZFs 6–9_K1055R construct was also capable of co-crystallizing with the mCZ38BS in the same space group as the WT complex, affording structural determination to a resolution of 1.60 Å (Table 1). The WT:mCZ38BS and K1055R:mCZ38BS structures are highly superimposable (protein backbone root mean square deviation between the crystal structures of 0.18 Å) and definitively demonstrate that Arg-1055 is able to accommodate the same direct hydrogen-bonding interactions at the 5′-8TmCG10-3′ region (Fig. 4, C and D, Fig. S2D, and Table S2). In addition, the ordered water molecule present in the WT:mCZ38BS complex (W54) is displaced by the Arg-1055 Nη2 atom, which can make a direct hydrogen bond with O6 of G28 (Fig. 4D and Table S2). This substitution of direct base interactions by Arg-1055 relative to the indirect water-mediated base contacts by Lys-1055 coincides with the observed chemical shift variances between these two complexes by solution NMR (Fig. 4A). Combined, the presented results indicate, for the first time, how a lysine residue, through the use of an ordered water molecule, can function as a surrogate for arginine in selective mCpG recognition. These findings expand the mechanisms by which ZF scaffolds can read and interpret this essential epigenetic mark.
Figure 4.
Lys-1055 plays a surrogate role for arginine in mCpG recognition. A, comparative 1H-15N HSQC spectral overlay for WT ZBTB38 ZF 6–9 (black/green) and ZBTB38 ZF 6–9_K1055R (red/blue) in complex with mCZ38BS_18-mer. Green and blue cross-peaks indicate aliased arginine guanidinium side chain resonances that only appear upon DNA binding at neutral pH. B, binding isotherms comparing the binding affinity for WT ZBTB38 ZFs 6–9 (replotted from Fig. 1D) and ZBTB38 ZFs 6–9_K1055R with mCZ38BS_27-mer. Each data point represents the average of triplicate data, with error bars depicting ± S.D. C, overlay of the ZBTB38 ZF 6–9:mCZ38BS (dark colors, beige DNA, orange zinc atoms) and ZBTB38 ZF 6–9_K1055R:mCZ38BS (light colors, blue DNA, yellow zinc atoms) structures superimposed on their protein backbones with a root mean square deviation of 0.18 Å. The K1055R site is highlighted in magenta. D, comparison of interactions between Asn-1052, Lys-1055/Arg-1055, and Glu-1079 with the core T8:A29, mC9:G28, and G10:mC27 base pairs. The WT ZBTB38 ZF 6–9:mCZ38BS complex is depicted in dark colors and beige DNA, whereas the ZBTB38 ZF 6–9_K1055R:mCZ38BS complex is depicted in light colors and blue DNA. Black dotted lines represent classical hydrogen bond interactions, and blue dotted lines represent water-mediated hydrogen bonds.
Interactions for the 3′ mCpG site are limited to side chain contacts with the C5-methyl of mC24 as well as the presence of a water layer surrounding the C5-methyls of both mC24 and mC12 (Fig. S2B). Recognition of the C5-methyl of mC24 is mediated through the Cγ2-methyl of Ile-1083 and the ring edge of Tyr-1105 (Fig. S2C). The C5-methyl group of mC12 is devoid of direct or indirect interactions with the protein but has a couple of proximally localized ordered water molecules (Fig. S2B). Intriguingly, the majority of ZF MBP–methylated DNA complex structures solved to date have one mC within the mCpG palindrome that has a hydration shell around its methyl group (20, 23, 24, 26). This hydration of the mC methyl has been determined to improve protein binding by a factor of 2 (24). Although these side chain interactions and the methyl hydration observed in the ZF 6–9:mCZ38BS complex structure are individually relatively weak, they may collectively contribute to the moderate methyl selectivity observed at the 3′ mCpG site.
Discussion
The presented structure of ZBTB38 ZFs 6–9 in complex with the mCZ38BS illuminates how this ZF-containing domain elicits mCpG recognition. In particular, the structure affirms the prediction (37) that methyl-selective binding is preferentially localized at the 5′ mCpG site. In a manner comparable with ZBTB33 (19, 25), recognition of mC27pG28 is accommodated through a series of base-specific and van der Waals interactions contributed from the side chains of Lys-1055, Leu-1077, and Glu-1079 (Fig. 3 and Fig. S4). Additional canonical and CH···O-type hydrogen bonds from Glu-1079 facilitate recognition of the corresponding mC9 base within the 5′ mCpG palindrome.
In contrast, the 3′ mCpG is recognized through a series of side chain contacts between Ile-1083 and Tyr-1105 with the C5-methyl of mC24, which is consistent with this site contributing less to the overall DNA binding. Combined, it is evident that the majority of protein-mediated contacts for mCZ38BS recognition are preferentially localized to the noncoding strand (Fig. 2C and Table S2). This asymmetric DNA strand recognition is emerging as a common feature in ZF MBP binding (19–21, 23–26), although the physiological consequence of this capability is not yet fully understood.
Although it is evident that the C-terminal ZBTB38 ZFs display selectivity for recognition of mC over C at the 5′ site, contributions from interactions at the T8:A29 step in overall recognition of the mCZ38BS cannot be discounted. Indeed, loss of the combined hydrogen-bonding contributions from Asn-1052 and Lys-1055 as well as the CH···O-type hydrogen bond between Glu-1079 and the C5-methyl of T8 have a significant impact on DNA binding (Fig. 3B).
Notably, the presented structures definitively demonstrate that Lys-1055 is capable of functioning analogously to the spatially conserved arginine residue required for mCpG recognition in all other ZF MBPs investigated to date (19–26). Nevertheless, the functional equivalence of lysine and arginine residues for mCpG recognition by the C-terminal ZFs of ZBTB38 is intriguing for two main reasons. First, substitution of the corresponding mCpG recognizing Arg-178 with a lysine in Zfp57 severally impacted DNA binding and resulted in loss of methyl sensitivity (24), suggesting that not all ZF protein scaffolds can accommodate this interconversion. Secondly, structures from both the ZF and methyl-CpG–binding domain (MBD) families of MBPs in complex with methylated DNA demonstrate that an mC-Arg-G triad is utilized (28, 29) where, in addition to making a direct hydrogen bond with the mCpG guanine base, van der Waals interactions between the arginine side chain and mC 5C-methyl group are important for stabilizing the interaction and conferring some of the methyl sensitivity (28, 45). In the ZBTB38 ZF 6–9:mCZ38BS complex structure, these van der Waals contacts are missing.
This is seemingly due to the requirement of Lys-1055 to hydrogen-bond with the O6 of T8, which pulls the lysine side chain away from the C5-methyl of mC27 so that the distance is too great to accommodate these van der Waals interactions (Fig. 4D and Table S2). However, substitution of Lys-1055 with arginine results in a slight shift of the Arg-1055 side chain to a position more proximal to the C5-methyl of mC27 that re-establishes the van der Waals contacts (Fig. 4D, Fig. S2D, and Table S2). Given that the binding affinities for mCZ38BS are similar between the WT and K1055R variant proteins, it would seem that, in the context of the C-terminal ZFs of ZBTB38, loss of these van der Waal interactions can be compensated. This may be in part through interactions between the ordered water molecule (W59) and the C5-methyl of mC27 (Fig. 4D).
Regardless of the knowledge gained from the reported structures, the physiological rationale for why a lysine substitution was selected by the C-terminal ZFs of ZBTB38 for mCpG recognition remains unclear. Given that ZBTB38 appears to have two independent ZF domains that can each selectively recognize mCpG loci, perhaps the usage of Lys-1055 in the C-terminal ZFs provides an element for diversifying DNA target selection between the two ZF domains. Indeed, initial investigations within cells suggested that, depending on the gene context, the N- and C-terminal ZFs may work synergistically as well as mutually exclusively to modulate transcription (37). Nevertheless, the presented structural observations suggest that additional ZF-containing proteins that utilize this alternative mode of mCpG recognition may remain to be found. This is particularly relevant given that a number of uncharacterized ZFs have recently been identified as potential selective readers for methylated DNA (30–33).
Experimental procedures
Sample preparation
ZBTB38(1006–1153) encompassing ZFs 6–10 was cloned as described previously (37). The four ZF constructs depicted in Fig. 1A were PCR-amplified using ZBTB38(1006–1153) as a template. Protein variants were generated using standard site-directed mutagenesis methodologies with ZBTB38(1006–1124) (ZFs 6–9) as a template. All protein constructs were overexpressed in minimal medium enriched with 15N ammonium chloride using either autoinduction (ZFs 6–10) or isopropyl β-d-1-thiogalactopyranoside induction (ZFs 6–9 and 7–10), purified, and refolded as described previously (37).
All oligonucleotides utilized for NMR, X-ray crystallography, and EMSA were prepared synthetically (DNA/Peptide Facility, University of Utah), further desalted on Nap-5 or Nap-10 Sephadex G25 columns (GE Healthcare Life Sciences), and lyophilized. Duplex DNA formation was completed as described previously (46). DNA sequences utilized for solution NMR spectroscopy, X-ray crystallography, and EMSA are presented in Table S1.
Solution NMR spectroscopy
All C-terminal ZBTB38 ZF:DNA complexes were formed as described previously (46) in NMR buffer (10 mm Tris (pH 6.8), 1 mm tris(2-carboxyethyl)phosphine, 0.005% NaN3, and 10% D2O). All NMR spectra were recorded at 298 K on a Varian VNMRS 800 MHz spectrometer. Data were processed using NMRPipe (47) and analyzed with NMRView (48).
X-ray crystallography and structure refinement
All protein:DNA complexes were formed and validated by solution NMR spectroscopy as described above prior to crystallization. Robotic crystallization trials were performed for the various C-terminal ZBTB38 ZF:DNA complexes using an automated Crystal Gryphon robot (Art Robbins Instruments) within the PACT (pH, anion, cation crystallization trial) Premier HT-96 (Molecular Dimensions) and Natrix HT (Hampton Research) screens. Promising crystal conditions were further optimized using the sitting drop vapor diffusion method. Final crystals for the ZF 6–9:mCZ38BS complex were formed by mixing 1 μl of 8 mg/ml protein:DNA complex with 1 μl of crystallization buffer (0.25 m ammonium chloride, 0.1 m MES (pH 5.8), and 20% PEG 6000) and sealed in a well containing 1 ml of crystallization buffer. Final crystals for the ZF 6–9_K1055R:mCZ38BS complex were similarly formed by mixing 1 μl of 8 mg/ml protein:DNA complex with 1 μl of crystallization buffer (0.25 m ammonium chloride, 0.1 m MES (pH 6.0), and 19% PEG 6000) and sealed in a well containing 1 ml of crystallization buffer. Crystals for both complexes appeared in ∼6 days at 4 °C. Crystals were cryoprotected in crystallization buffer containing 30% (v/v) ethylene glycol. Diffraction data for both complexes were collected under cryogenic conditions from single crystals at the Stanford Synchrotron Radiation Lightsource, beamline 12-2 at a wavelength of 0.98 Å using a Dectris Pilatus3 S 6 M detector.
Data were processed using either HKL2000 (49) (ZF 6–9:mCZ38BS complex) or XDS (50) (ZF 6–9_K1055R:mCZ38BS complex). The ZF 6–9:mCZ38BS structure was determined by molecular replacement (MR) within PHENIX (51) using a subset of the Aart(residues 75–184):DNA(base steps 1–18) complex (PDB:2I13) (52), and refined to 1.75 Å. The ZF 6–9_K1055R:mCZ38BS structure was determined by MR within PHENIX using the ZF 6–9:mCZ38BS complex as a search model and refined to 1.60 Å. To avoid biasing during MR, DNA and ZF segments were systematically removed and then built back into the electron density using Coot (53, 54) prior to further refinement in PHENIX to validate each crystal structure. Crystallographic and refinement statistics for each complex are summarized in Table 1. Interactions at distances of <4.2 Å were identified using ENTANGLE (55) and are summarized in Fig. 2C and listed in Table S2.
EMSA
For Kd determination, duplex DNA (2 nm) was incubated with various concentrations of ZBTB38(1006–1153), ZBTB38(1006–1124) or mutant variants, or ZBTB38(1034–1153) in 20 μl of binding buffer (10 mm Tris (pH 7.0), 150 mm NaCl, 1 mm tris(2-carboxyethyl)phosphine, 0.005% NaN3, 100 μg/ml BSA and 10% (w/v) sucrose). Samples were incubated at room temperature for 30 min prior to electrophoretic separation in 10% 1× Tris borate (pH 8.3) nondenaturing polyacrylamide gels at 100 V for 1 h. Gels were soaked in 1× Tris borate supplemented with 1× SYBR Gold Nucleic Acid Gel Stain (Thermo Fisher Scientific) and imaged on an Amersham Biosciences Typhoon Biomolecular Imager (GE Healthcare Life Sciences) at 495 nm. Gel band intensities were quantified by ImageJ (56, 57). Kd values were obtained by first averaging the data from triplicate experiments and fitting the averaged fraction-bound DNA as a function of protein concentration to a Langmuir isotherm (SigmaPlot version 10.0 from Systat Software, Inc.), where the maximal fraction bound value was set to 1.
Author contributions
N. O. H. and B. A. B.-K. conceptualization; N. O. H., F. G. W., and B. A. B.-K. data curation; N. O. H., F. G. W., and B. A. B.-K. formal analysis; N. O. H. and F. G. W. investigation; N. O. H. and B. A. B.-K. visualization; N. O. H., F. G. W., and B. A. B.-K. methodology; N. O. H. and B. A. B.-K. writing-original draft; N. O. H., F. G. W., and B. A. B.-K. writing-review and editing; B. A. B.-K. supervision; B. A. B.-K. funding acquisition.
Supplementary Material
Acknowledgments
We thank Prof. C. P. Hill for providing access to X-ray crystallography equipment and for helpful discussions and Dr. P. F. Flynn for reading and suggestions for improving the manuscript. The NMR data included in this report were recorded in the David M. Grant NMR Center, a campus core facility of the University of Utah. Oligonucleotides were synthesized by the DNA/Peptide Facility, part of the Health Sciences Center Cores at the University of Utah. Sequencing was performed at the DNA sequencing Core Facility, University of Utah.
This work was supported by MCB-1715370 from the National Science Foundation. The Stanford Synchrotron Radiation Lightsource Structural Molecular Biology Program is supported by the Department of Energy Office of Biological and Environmental Research and by NIGMS, National Institutes of Health (including P41GM103393). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4 and Tables S1 and S2.
The atomic coordinates and structure factors (codes 6E93 and 6E94) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- MBP
- methyl-CpG–binding protein
- mCpG
- methylated CpG
- ZF
- zinc finger
- BS
- binding sequence
- HSQC
- heteronuclear single-quantum coherence
- EMSA
- electrophoretic mobility shift assay
- MR
- molecular replacement.
References
- 1. Esteller M. (2005) Aberrant DNA methylation as a cancer-inducing mechanism. Annu. Rev. Pharmacol. Toxicol. 45, 629–656 10.1146/annurev.pharmtox.45.120403.095832 [DOI] [PubMed] [Google Scholar]
- 2. Jin Z., and Liu Y. (2018) DNA methylation in human diseases. Genes Dev. 5, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jones P. A., and Baylin S. B. (2007) The epigenomics of cancer. Cell 128, 683–692 10.1016/j.cell.2007.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lv J., Liu H., Su J., Wu X., Liu H., Li B., Xiao X., Wang F., Wu Q., and Zhang Y. (2012) DiseaseMeth: a human disease methylation database. Nucleic Acids Res. 40, D1030–D1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Robertson K. D. (2005) DNA methylation and human disease. Nat. Rev. Genet. 6, 587–610 10.1038/ni1200 [DOI] [PubMed] [Google Scholar]
- 6. Xiong Y., Wei Y., Gu Y., Zhang S., Lyu J., Zhang B., Chen C., Zhu J., Wang Y., Liu H., and Zhang Y. (2017) DiseaseMeth version 2.0 a major expansion and update of the human disease methylation database. Nucleic Acids Res. 45, D888–D895 10.1093/nar/gkw1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baylin S. B., and Jones P. A. (2011) A decade of exploring the cancer epigenome: biological and translational implications. Nat. Rev. Cancer 11, 726–734 10.1038/nrc3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klose R. J., and Bird A. P. (2006) Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31, 89–97 10.1016/j.tibs.2005.12.008 [DOI] [PubMed] [Google Scholar]
- 9. Defossez P.-A., and Stancheva I. (2011) Biological functions of methyl-CpG-binding proteins. Prog. Mol. Biol. Transl. Sci. 101, 377–398 10.1016/B978-0-12-387685-0.00012-3 [DOI] [PubMed] [Google Scholar]
- 10. Fournier A., Sasai N., Nakao M., and Defossez P.-A. (2012) The role of methyl-binding proteins in chromatin organization and epigenome maintenance. Brief. Funct. Genomics 11, 251–264 10.1093/bfgp/elr040 [DOI] [PubMed] [Google Scholar]
- 11. Sansom O. J., Maddison K., and Clarke A. R. (2007) Mechanism of disease: methyl-binding domain proteins as potential therapeutic targets in cancer. Nat. Clin. Pract. Oncol. 4, 305–315 10.1038/ncponc0812 [DOI] [PubMed] [Google Scholar]
- 12. Nan X., Ng H. H., Johnson C. A., Laherty C. D., Turner B. M., Eisenman R. N., and Bird A. (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393, 386–389 10.1038/30764 [DOI] [PubMed] [Google Scholar]
- 13. Jones P. L., Veenstra G. J., Wade P. A., Vermaak D., Kass S. U., Landsberger N., Strouboulis J., and Wolffe A. P. (1998) Methylated DNA and MeCP2 recruit histone deactylase to repress transcription. Nat. Genet. 19, 187–191 10.1038/561 [DOI] [PubMed] [Google Scholar]
- 14. Zhang Y., Ng H. H., Erdjument-Bromage H., Tempst P., Bird A., and Reinberg D. (1999) Analysis of the NuRD subunits reveals a histone deacetylases core complex and a connection with DNA methylation. Genes Dev. 13, 1924–1935 10.1101/gad.13.15.1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ng H. H., Zhang Y., Hendrich B., Johnson C. A., Turner B. M., Erdjument-Bromage H., Tempst P., Reinberg D., and Bird A. (1999) MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 23, 58–61 10.1038/12659 [DOI] [PubMed] [Google Scholar]
- 16. Sasai N., Matsuda E., Sarashina E., Ishida Y., and Kawaichi M. (2005) Identification of a novel BTB-zinc finger transcriptional repressor, CIBZ, that interacts with CtBP corepressor. Genes Cells 10, 871–885 10.1111/j.1365-2443.2005.00885.x [DOI] [PubMed] [Google Scholar]
- 17. Weber A., Marquardt J., Elzi D., Forster N., Starke S., Glaum A., Yamada D., Defossez P.-A., Delrow J., Eisenman R. N., Christiansen H., and Eilers M. (2008) Zbtb4 represses transcription of P21CIP1 and controls the cellular responses to p53 activation. EMBO J. 27, 1563–1574 10.1038/emboj.2008.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yoon H. G., Chan D. W., Reynolds A. B., Qin J., and Wong J. (2003) N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol. Cell 12, 723–734 10.1016/j.molcel.2003.08.008 [DOI] [PubMed] [Google Scholar]
- 19. Buck-Koehntop B. A., Stanfield R. L., Ekiert D. C., Martinez-Yamout M. A., Dyson H. J., Wilson I. A., and Wright P. E. (2012) Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso. Proc. Natl. Acad. Sci. U.S.A. 109, 15229–15234 10.1073/pnas.1213726109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hashimoto H., Olanrewaju Y. O., Zheng Y., Wilson G. G., Zhang X., and Cheng X. (2014) Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes Dev. 28, 2304–2313 10.1101/gad.250746.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hashimoto H., Wang D., Horton J. R., Zhang X., Corces V. G., and Cheng X. (2017) Structural basis for the versatile and methylation-dependent binding of CTCF to DNA. Mol. Cell. 66, 711–720.e3 10.1016/j.molcel.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hashimoto H., Zhang X., Zheng Y., Wilson G. G., and Cheng X. (2016) Denys-Drash syndrome associated WT1 glutamine 369 have altered sequence preferences and altered responses to epigenetic modifications. Nucleic Acids Res. 44, 10165–10176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu Y., Olanrewaju Y. O., Zheng Y., Hashimoto H., Blumenthal R. M., Zhang X., and Cheng X. (2014) Structural basis for Klf4 recognition of methylated DNA. Nucleic Acids Res. 42, 4859–4867 10.1093/nar/gku134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu Y., Toh H., Sasaki H., Zhang X., and Cheng X. (2012) An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev. 26, 2374–2379 10.1101/gad.202200.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nikolova E. N., Stanfield R. L., Dyson H. J., and Wright P. E. (2018) CH.O hydrogen bonds mediate highly specific recognition of methylated CpG sites by the zinc finger protein Kaiso. Biochemistry 57, 2109–2120 10.1021/acs.biochem.8b00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zandarashvili L., White M. A., Esadze A., and Iwahara J. (2015) Structural impact of complete CpG methylation within target DNA on specific complex formation of the inducible transcription factor Egr-1. FEBS Lett. 589, 1748–1753 10.1016/j.febslet.2015.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Buck-Koehntop B. A., and Defossez P.-A. (2013) On how mammalian transcription factors recognize methylated DNA. Epigenetics 8, 131–137 10.4161/epi.23632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Y., Zhang X., Blumenthal R. M., and Cheng X. (2013) A common mode of recognition for methylated CpG. Trends Biochem. Sci. 38, 177–183 10.1016/j.tibs.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ren R., Horton J. R., Zhang X., Blumenthal R. M., and Cheng X. (2018) Detecting and interpreting DNA methylation marks. Curr. Opin. Struct. Biol. 53, 88–99 10.1016/j.sbi.2018.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu S., Wan J., Su Y., Song Q., Zeng Y., Nguyen H. N., Shin J., Cox E., Rho H. S., Woodard C., Xia S., Liu S., Lyu H., Ming G.-L., Wade H., et al. (2013) DNA methylation presents distinct binding sites for human transcription factors. eLife 2, e00726 10.7554/eLife.00726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Spruijt C. G., Gnerlich F., Smits A. H., Pfaffeneder T., Jansen P. W., Bauer C., Münzel M., Wagner M., Müller M., Khan F., Eberl H. C., Mensinga A., Brinkman A. B., Lephikov K., Müller U., et al. (2013) Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159 10.1016/j.cell.2013.02.004 [DOI] [PubMed] [Google Scholar]
- 32. Yin Y., Morgunova E., Jolma A., Kaasinen E., Sahu B., Khund-Sayeed S., Das P. K., Kivioja T., Dave K., Zhong F., Nitta K. R., Taipale M., Popov A., Ginno P. A., Domcke S., et al. (2017) Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356, eaaj2239 10.1126/science.aaj2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu H., Wang G., and Qian J. (2016) Transcription factors as readers and effectors of DNA methylation. Nat. Rev. Genet. 17, 551–565 10.1038/nrg.2016.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Filion G. J., Zhenilo S., Salozhin S., Yamada D., Prokhortchouk E., and Defossez P.-A. (2006) A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol. Cell. Biol. 26, 169–181 10.1128/MCB.26.1.169-181.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oikawa Y., Matsuda E., Nishii T., Ishida Y., and Kawaichi M. (2008) Down-regulation of CIBZ, a novel substrate of caspase-3, induces apoptosis. J. Biol. Chem. 283, 14242–14247 10.1074/jbc.M802257200 [DOI] [PubMed] [Google Scholar]
- 36. Oikawa Y., Omori R., Nishii T., Ishida Y., Kawaichi M., and Matsuda E. (2011) The methyl-CpG-binding protein CIBZ suppresses myogenic differentiation by directly inhibiting myogenin expression. Cell Res. 21, 1578–1590 10.1038/cr.2011.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pozner A., Hudson N. O., Trewhella J., Terooatea T. W., Miller S. A., and Buck-Koehntop B. A. (2018) The C-terminal zinc fingers of ZBTB38 are novel selective readers of DNA methylation. J. Mol. Biol. 430, 258–271 10.1016/j.jmb.2017.12.014 [DOI] [PubMed] [Google Scholar]
- 38. Nishii T., Oikawa Y., Ishida Y., Kawaichi M., and Matsuda E. (2012) CtBP-interacting BTB zinc finger protein (CIBZ) promotes proliferation and G1/S transition in embryonic stem cells via Nanog. J. Biol. Chem. 287, 12417–12424 10.1074/jbc.M111.333856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cai Y., Li J., Yang S., Li P., Zhang X., and Liu H. (2012) CIBZ, a novel BTB domain-containing protein, is involved in mouse spinal cord injury via mitochondrial pathway independent of p53 gene. PLoS ONE 7, e33156 10.1371/journal.pone.0033156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miotto B., Chibi M., Xie P., Koundrioukoff S., Moolman-Smook H., Pugh D., Debatisse M., He F., Zhang L., and Defossez P.-A. (2014) The RBBP6/ZBTB38/MCM10 axis regulates DNA replication and common fragile site stability. Cell Rep. 7, 575–587 10.1016/j.celrep.2014.03.030 [DOI] [PubMed] [Google Scholar]
- 41. Horowitz S., and Trievel R. C. (2012) Carbon-oxygen hydrogen bonding in biological structure and function. J. Biol. Chem. 287, 41576–41582 10.1074/jbc.R112.418574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yesselman J. D., Horowitz S., Brooks C. L. 3rd, and Trievel R. C. (2015) Frequent side chain methyl carbon-oxygen hydrogen bonding in proteins revealed by computational and stereochemical analysis of neutron structures. Proteins 83, 403–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mandel-Gutfreund Y., Margalit H., Jernigan R. L., and Zhurkin V. B. (1998) A role for CH·O interactions in protein-DNA recognition. J. Mol. Biol. 277, 1129–1140 10.1006/jmbi.1998.1660 [DOI] [PubMed] [Google Scholar]
- 44. Derewenda Z. S., Lee L., and Derewenda U. (1995) The occurrence of C-H·O hydrogen bonds in proteins. J. Mol. Biol. 252, 248–262 10.1006/jmbi.1995.0492 [DOI] [PubMed] [Google Scholar]
- 45. Zou X., Ma W., Solov'yov I. A., Chipot C., and Schulten K. (2012) Recognition of methylated DNA through methyl-CpG binding domain proteins. Nucleic Acids Res. 40, 2747–2758 10.1093/nar/gkr1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Buck-Koehntop B. A., Martinez-Yamout M. A., Dyson H. J., and Wright P. E. (2012) Kaiso uses all three zinc fingers and adjacent sequence motifs for high affinity binding to sequence-specific and methyl-CpG DNA targets. FEBS Lett. 586, 734–739 10.1016/j.febslet.2012.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 48. Johnson B. A., and Blevins R. A. (1994) NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 10.1007/BF00404272 [DOI] [PubMed] [Google Scholar]
- 49. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 50. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Segal D. J., Crotty J. W., Bhakta M. S., Barbas C. F. 3rd, and Horton N. C. (2006) Structure of Aart, a designed six-finger zinc finger peptide, bound to DNA. J. Mol. Biol. 363, 405–421 10.1016/j.jmb.2006.08.016 [DOI] [PubMed] [Google Scholar]
- 53. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 54. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Allers J., and Shamoo Y. (2001) Structure-based analysis of protein-RNA interactions using the program ENTANGLE. J. Mol. Biol. 311, 75–86 10.1006/jmbi.2001.4857 [DOI] [PubMed] [Google Scholar]
- 56. Abramoff M. D., Magalhaes P. J., and Ram S. J. (2004) Image processing with ImageJ. Biophotonics Int. 11, 36–42 [Google Scholar]
- 57. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.