Abstract
Human sialic acid–binding immunoglobulin-like lectin 14 (Siglec-14) is a glycan-recognition protein that is expressed on myeloid cells, recognizes bacterial pathogens, and elicits pro-inflammatory responses. Although Siglec-14 is a transmembrane protein, a soluble form of Siglec-14 is also present in human blood. However, the mechanism that generates soluble Siglec-14 and what role this protein form may play remain unknown. Here, investigating the generation and function of soluble Siglec-14, we found that soluble Siglec-14 is derived from an alternatively spliced mRNA that retains intron 5, containing a termination codon and thus preventing the translation of exon 6, which encodes Siglec-14's transmembrane domain. We also note that the translated segment in intron 5 encodes a unique C-terminal 7-amino acid extension, which allowed the specific antibody-mediated detection of this isoform in human blood. Moreover, soluble Siglec-14 dose-dependently suppressed pro-inflammatory responses of myeloid cells that expressed membrane-bound Siglec-14, likely by interfering with the interaction between membrane-bound Siglec-14 and Toll-like receptor 2 on the cell surface. We also found that intron 5 contains a G-rich segment that assumes an RNA tertiary structure called a G-quadruplex, which may regulate the efficiency of intron 5 splicing. Taken together, we propose that soluble Siglec-14 suppresses pro-inflammatory responses triggered by membrane-bound Siglec-14.
Keywords: biomarker, alternative splicing, G-quadruplex, inflammation, sialic acid, toll-like receptor (TLR), Siglec, soluble receptor, transmembrane signaling
Introduction
Integral membrane proteins are sometimes found in a soluble form. Many examples of soluble receptors have been reported that contribute to the fine-tuning of cellular responses (1). For example, soluble interleukin-6 (IL-6)3 receptor is generated by both alternative splicing and proteolysis, binds IL-6, and either agonizes or antagonizes IL-6 function (2). Soluble forms of the receptor of advanced glycation end-product are also produced by both alternative splicing and proteolysis and function as decoy receptors (3). These soluble receptors not only represent a mechanism to fine-tune biological responses but are, in some cases, also useful as biomarkers of clinical conditions (4–6).
Siglecs are a family of sialic acid-recognition proteins expressed primarily on leukocytes that have regulatory roles in the fine-tuning of leukocyte activities (7–10). The involvement of Siglecs in various diseases (including infectious diseases, cancer, and chronic diseases) has been reported, and therapeutic agents targeting Siglecs are being developed (11, 12). Although Siglecs are type 1 transmembrane proteins, soluble forms of Siglecs have also been reported (13). Recent studies have shown that the soluble form of Siglec-9 is produced by dental pulp stem cells and shows anti-inflammatory effects by regulating the polarization of macrophages (14–16). Thus, soluble Siglecs may also contribute to the fine regulation of immune cell activities.
Siglec-14 is expressed on myeloid cells, associates with signal adapter protein DNAX activating protein of 12 kDa (DAP12), and transduces immune cell-activating signals through the spleen tyrosine kinase (17). Genes encoding human Siglec-14 and its inhibitory counterpart Siglec-5 (SIGLEC14 and SIGLEC5, respectively) are located in tandem on chromosome 19. Recombination between SIGLEC14 and SIGLEC5 at the high-homology region (5′UTR through exon 3) yields an allele that encodes a SIGLEC14–SIGLEC5 fusion gene (encoding a protein identical to Siglec-5), which does not produce Siglec-14 protein (18). We previously demonstrated that this SIGLEC14-null allele is common (allele frequency >0.05) in all human populations tested, and its frequency is particularly high in East Asia (exceeding 0.5) (18). We have also reported that homozygous SIGLEC14-null patients with chronic obstructive pulmonary disease (COPD) are less prone to exacerbation (i.e. an acute worsening of disease symptoms, often caused by microbial airway infection) (19). In a previous study, we found that the soluble form of Siglec-14 is present in sera from COPD patients who have at least one functional SIGLEC14 allele (19). Furthermore, soluble Siglec-14 (sSiglec-14) was recently reported to be a potential early plasma biomarker of bronchopulmonary dysplasia (20), a condition affecting premature infants, and is accompanied by lung inflammation (21, 22). We hypothesized that sSiglec-14 may represent a negative feedback mechanism that regulates myeloid pro-inflammatory responses elicited by the engagement of membrane-bound Siglec-14 (mSiglec-14). For example, activation of myeloid cells is known to up-regulate several proteases that cleave membrane proteins (e.g. a disintegrin and metalloproteinase domain-containing protein (ADAM)10 and ADAM17 (23)) that “shed” these membrane proteins. If mSiglec-14 is proteolytically cleaved, this will uncouple the ligand recognition (extracellular) and signal transduction (intracellular) functions of the protein, possibly terminating the pro-inflammatory signal elicited by mSiglec-14. In addition, the shed extracellular domain of Siglec-14 may influence myeloid cell differentiation, in a similar manner as reported for soluble Siglec-9 (14–16).
To test these hypotheses, we investigated the generation mechanism of sSiglec-14, and we explored its potential functions. In this paper, we demonstrate that sSiglec-14 is a product of alternative splicing. The alternative splicing may be influenced by the presence of an RNA G-quadruplex structure in intron 5 of SIGLEC14 pre-mRNA. We also show that sSiglec-14 has anti-inflammatory properties through interference with the cis-interaction between mSiglec-14 and Toll-like receptor 2 (TLR2), a pattern recognition receptor recognizing bacterial lipoproteins. The possible biological implications of these findings will be discussed.
Results
Alternative mRNA splicing generates sSiglec-14
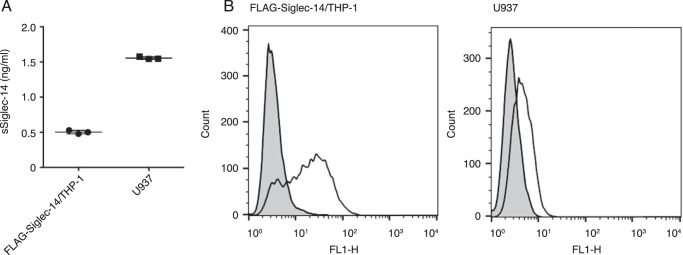
A soluble form of Siglec-14 may be generated by proteolysis of a membrane-bound form of Siglec-14 or by alternative mRNA splicing. To test whether proteolysis explains the generation of sSiglec-14, we first cultured Siglec-14/THP-1 cells that overexpress mSiglec-14 cDNA and tested whether sSiglec-14 is detected in the culture supernatant. By sandwich ELISA using a detection antibody that is specific to the third Ig-like domain of Siglec-14 (clone 40-1 (18, 19)), we detected a very low level of sSiglec-14 in the culture supernatant of Siglec-14/THP-1 (Fig. 1A). In contrast, the culture supernatant of U937 cells, which express lower levels of mSiglec-14 as compared with Siglec-14/THP-1 (Fig. 1B), contained higher levels of sSiglec-14 (Fig. 1A). These results prompted us to adopt an alternative hypothesis that sSiglec-14 is produced by alternative mRNA splicing, rather than by proteolysis of mSiglec-14.
Figure 1.
Production of soluble Siglec-14 is not proportional to the expression level of membrane-bound Siglec-14. A, ELISA of soluble Siglec-14 produced by FLAG-Siglec-14/THP-1 and U937 cell lines. Siglec-14–specific mouse mAb (clone 40-1) was used for detection. B, expression levels of membrane-bound Siglec-14 on FLAG-Siglec-14/THP-1 and U937 cell lines. Solid line, cells stained with anti-Siglec-14 antibody (clone 40-1); gray-shaded area, cells stained with control mouse antibody.
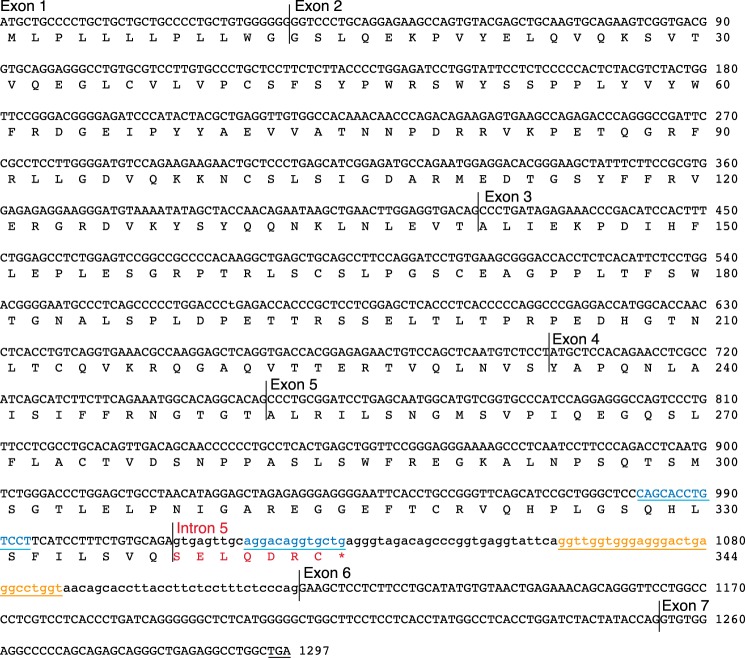
To test this hypothesis, we performed 3′-rapid amplification of cDNA ends (3′RACE) of SIGLEC14 using human bone marrow first-strand cDNA library (Fig. 2A). Sequencing of the obtained cDNA clones revealed that some clones represented a fully spliced form, whereas others retained intron 5 (or both introns 5 and 6). The splice variants containing intron 5 encode sSiglec-14, as intron 5 contains an in-frame stop codon that prevents the translation of exon 6 encoding the transmembrane domain (Fig. 3).
Figure 2.
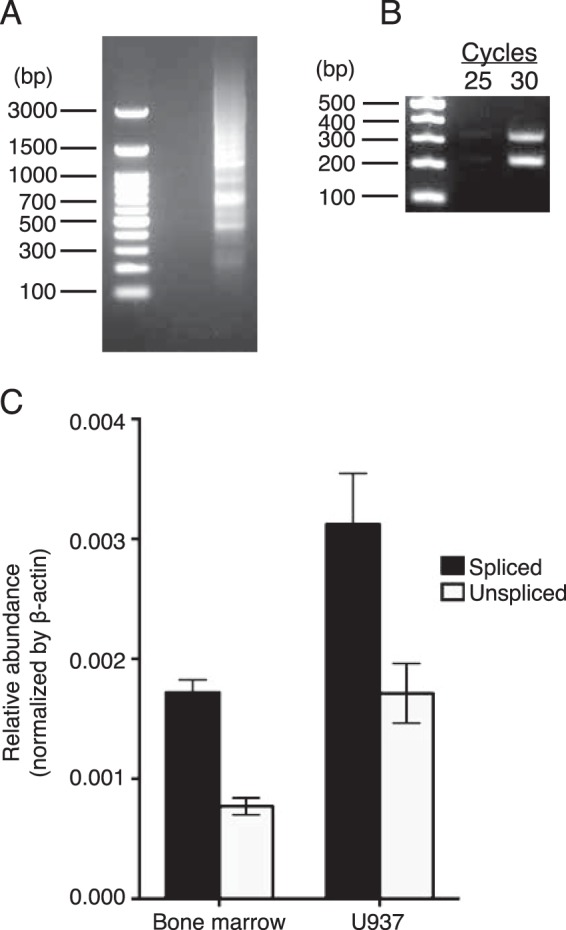
Human bone marrow cells produce alternatively spliced mRNA encoding soluble Siglec-14. Human bone marrow first-strand cDNA library (A–C) and first-strand cDNA prepared from U937 cells (C) were used as templates. A, agarose gel electrophoresis of 3′RACE PCR products obtained using a gene-specific primer (annealing to exon 5) and a universal primer. Products of nested PCR (second round) were separated by agarose gel electrophoresis. B, agarose gel electrophoresis of RT-PCR products with the primer pair flanking intron 5. C, relative abundances of membrane-bound (solid bars) and soluble (open bars) SIGLEC14 mRNA isoforms, normalized against ACTB mRNA.
Figure 3.
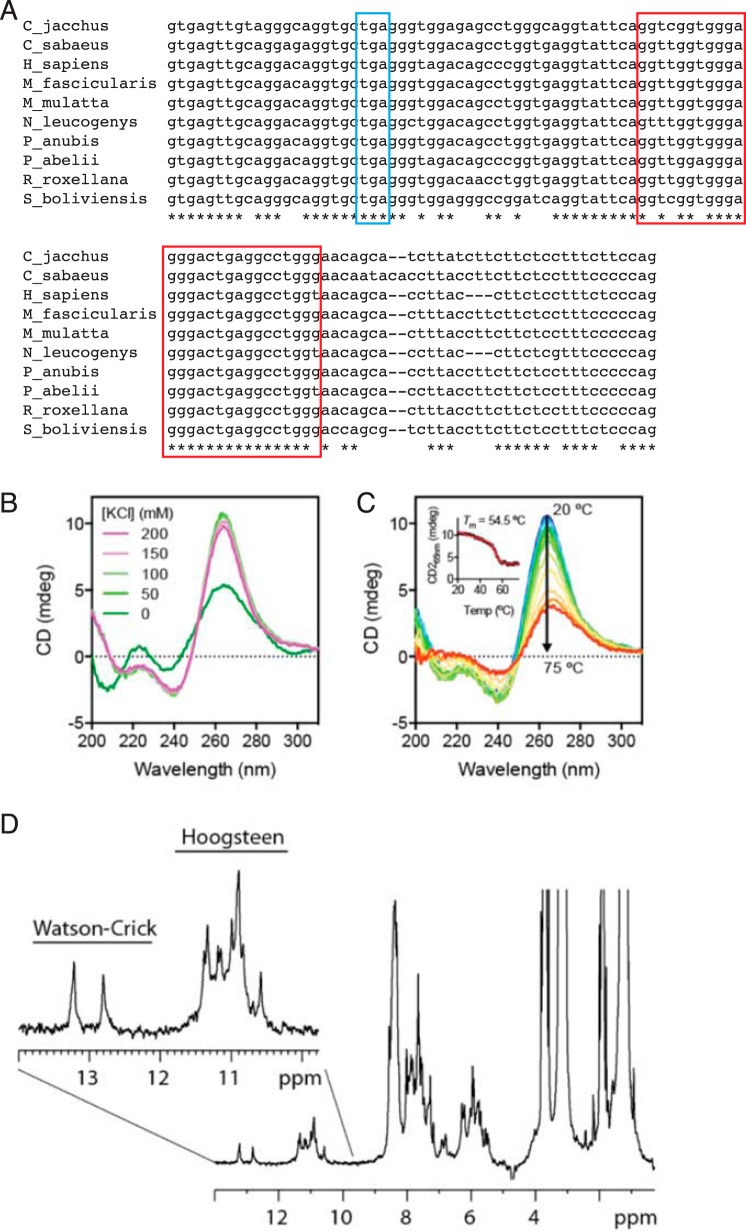
Sequence of soluble Siglec-14 cDNA and its translation. C-terminal heptapeptide unique to the soluble Siglec-14 synthesized from the alternatively spliced mRNA retaining intron 5 is shown in red. Stop codon in the intron 5-spliced form (encoding membrane-bound Siglec-14) is underlined. G-rich segment in intron 5 is shown in orange and underlined, and perfect inverted repeats flanking exon 5/intron 5 junction are shown in blue and underlined.
We analyzed the relative abundances of splice variants with or without intron 5 by RT-PCR using the same human bone marrow first-strand cDNA library as template and the primers that flank intron 5 and found that the relative abundances of mRNA that was spliced (mSiglec-14) versus unspliced (sSiglec-14) were similar, whereas the former appeared to be somewhat more abundant (Fig. 2B). Quantitative PCR using combinations of primers and probe that specifically identify the mRNA isoforms with or without intron 5 confirmed this observation (Fig. 2C).
sSiglec-14 has a unique C-terminal peptide segment
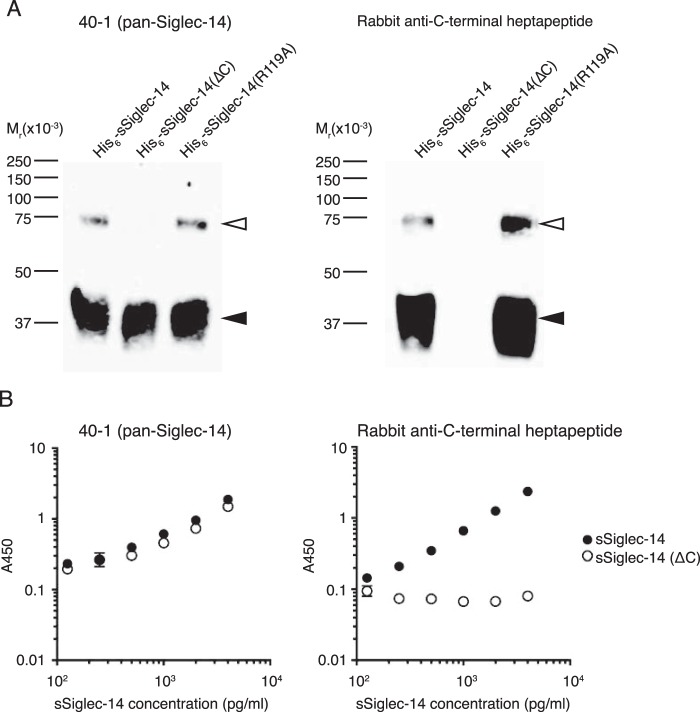
To demonstrate that the alternative splicing indeed accounts for sSiglec-14, we prepared a rabbit polyclonal antibody that recognizes a C-terminal heptapeptide that is unique to the sSiglec-14 generated by alternative splicing and used it in Western blotting (Fig. 4A). The data verified that the antibody is specific to sSiglec-14 with C-terminal heptapeptide. We used this antibody in sandwich ELISA (Fig. 4B) and found that sSiglec-14 with the C-terminal heptapeptide is indeed present in human serum. The concentration of sSiglec-14 with C-terminal heptapeptide in the human serum sample (a mixture of sera from multiple donors) was calculated to be 8.08 ± 0.35 ng/ml, which is lower than the concentration of total sSiglec-14 quantified by ELISA using the antibody (clone 40-1) that recognizes Siglec-14 regardless of the C-terminal peptide (29.1 ± 1.9 ng/ml). This result implies that sSiglec-14 undergoes gradual proteolysis from the C terminus and becomes undetectable by the polyclonal antibody, or the proteolytic cleavage of mSiglec-14 also contributes to the generation of sSiglec-14 in vivo. The results of the ELISA of U937 cell culture supernatant (sSiglec-14 with heptapeptide: 2.34 ± 0.06 ng/ml; total sSiglec-14: 2.48 ± 0.04 ng/ml) implies that the former mechanism likely contributes to the lower-than-expected concentration of sSiglec-14 with heptapeptide in human sera.
Figure 4.
Specificity of rabbit polyclonal antibody toward C-terminal heptapeptide of soluble Siglec-14. A, N-terminal His6-tagged recombinant soluble Siglec-14 proteins (full-length, C-terminally truncated (ΔC), or with R119A mutation) were subjected to nonreducing SDS-PAGE/Western blotting and stained with mouse mAb clone 40-1 (pan-Siglec-14 antibody; left panel) or with rabbit polyclonal antibody against soluble Siglec-14 C-terminal heptapeptide that is unique to the isoform generated by alternative mRNA splicing (right panel). Filled and open arrowheads indicate monomer and dimers of soluble Siglec-14, respectively. B, ELISA standard curves (capture: mouse mAb, clone 194128; detection: clone 40-1 (left panel) or rabbit polyclonal antibody against C-terminal heptapeptide (right panel)) for the standard proteins, N-terminal His6-tagged soluble Siglec-14 (solid circle), and N-terminal His6-tagged and C-terminal truncated soluble Siglec-14 (open circle). Error bars represent standard deviation of technical triplicates.
sSiglec-14 suppresses pro-inflammatory responses of cells that express mSiglec-14
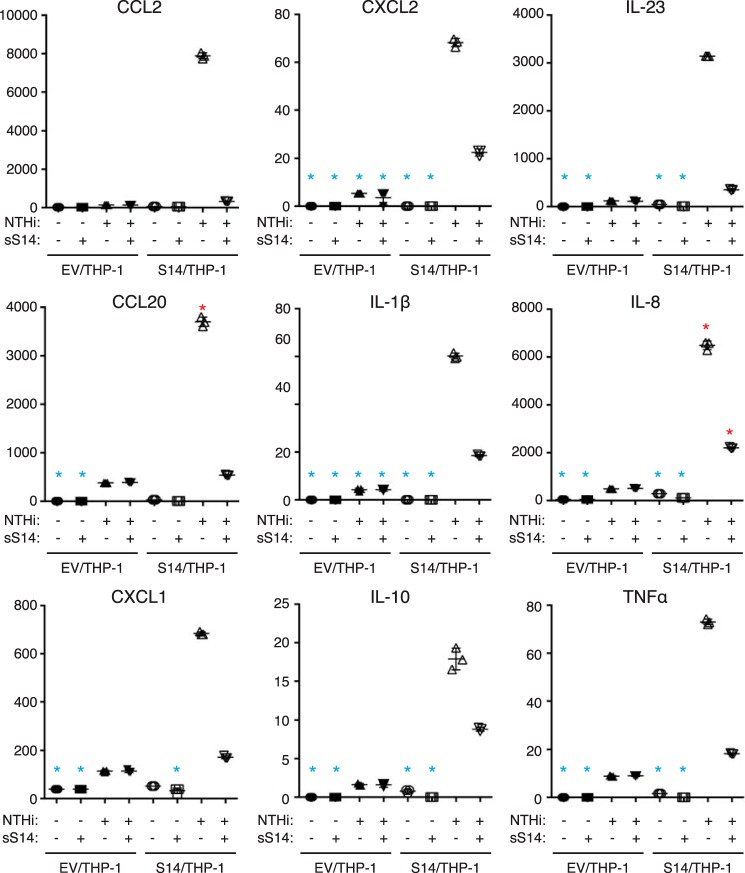
We have previously shown that the expression of Siglec-14 on THP-1 cells enhances pro-inflammatory responses elicited by nontypeable Haemophilus influenzae (NTHi) (19). To test whether sSiglec-14 has a role in the regulation of myeloid cell inflammatory responses, we prepared recombinant N-terminal His6-tagged sSiglec-14, and we stimulated Siglec-14/THP-1 cells with NTHi in the presence or absence of the recombinant sSiglec-14. As shown in Fig. 5A, recombinant sSiglec-14 suppressed IL-8 response in a dose-dependent manner. In addition, the production of several other cytokines and chemokines, chosen from genes that are specifically up-regulated in Siglec-14/THP-1 cells by NTHi stimulation (24), were also suppressed by recombinant sSiglec-14 (Fig. 6).
Figure 5.
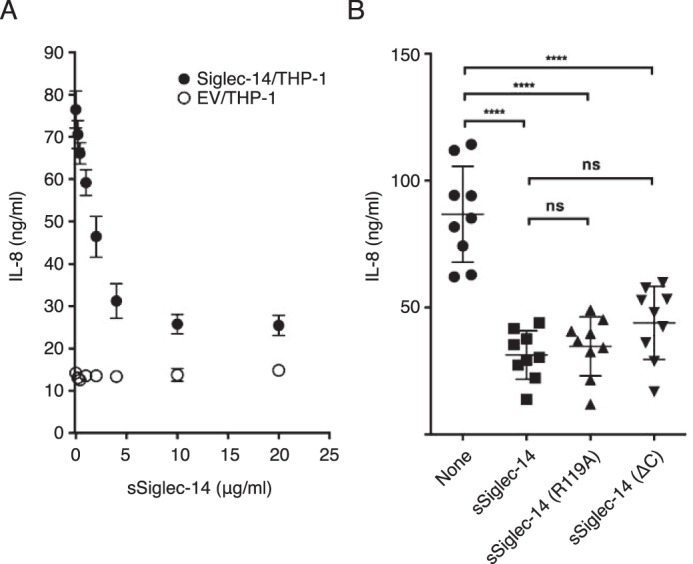
Soluble Siglec-14 inhibits production of IL-8 by myeloid cells expressing membrane-bound Siglec-14 upon NTHi stimulation. A, effect of soluble Siglec-14 on IL-8 production by Siglec-14/THP-1 (solid circle) stimulated with NTHi. EV/THP-1 (open circle) was used as a control. Error bars represent standard deviation of biological triplicates. B, roles of C-terminal heptapeptide and essential arginine residue (Arg-119) on the anti-inflammatory properties of soluble Siglec-14. Experiments were repeated nine times (biological nonuplicates), and each value is plotted. Error bars represent standard deviation. The dataset was analyzed by one-way ANOVA to test the differences among groups, and post hoc pairwise mean comparison was conducted using Bonferroni's multiple comparisons test. ****, adjusted p < 0.0001; ns, not significant (adjusted p > 0.05).
Figure 6.
Soluble Siglec-14 inhibits production of various cytokines and chemokines by myeloid cells expressing membrane-bound Siglec-14 upon NTHi stimulation. Analyte measured is indicated at the top of each panel. Blue asterisks represent the values below the range of calibration curve; red asterisks represent those above the range of calibration curve. Error bars represent standard deviation of technical triplicates.
We attempted to identify the structural element of sSiglec-14 that is required for its anti-inflammatory activity by introducing a mutation at the arginine residue that is required for sialic acid recognition (R119A) or by truncating the C-terminal heptapeptide (ΔC). Both variants suppressed IL-8 production to similar extents as the native form (Fig. 5B). This result implies that neither C-terminal heptapeptide nor sialic acid recognition is essential for the anti-inflammatory effect of sSiglec-14.
sSiglec-14 interferes with the interaction between mSiglec-14 and TLR2
As sSiglec-14 also suppressed the production of some cytokines/chemokines spontaneously produced at low levels in the absence of NTHi (Fig. 6), we hypothesized the following: 1) interaction between mSiglec-14 and a cis-ligand, likely a pattern-recognition receptor, triggers low-level production of cytokines/chemokines by Siglec-14/THP-1 in the absence of microbial stimulation, and 2) sSiglec-14, by interfering with the interaction between mSiglec-14 and the cis-ligand, cancels the enhanced production of cytokines/chemokines. To detect how sSiglec-14 suppresses production of pro-inflammatory cytokines/chemokines by Siglec-14/THP-1 cells, we sought to identify a protein that interacts with mSiglec-14 by a proximity labeling method (25). As shown in Fig. 7, biotin labels were introduced into many proteins in an mSiglec-14–dependent manner by proximity labeling, and biotinylated proteins were successfully enriched by streptavidin affinity purification. The proteins obtained by streptavidin affinity purification were analyzed by proteomics using LC-MS2. By applying a few screening criteria, we identified several proteins as cis-ligand candidates of mSiglec-14 (Table S1).
Figure 7.
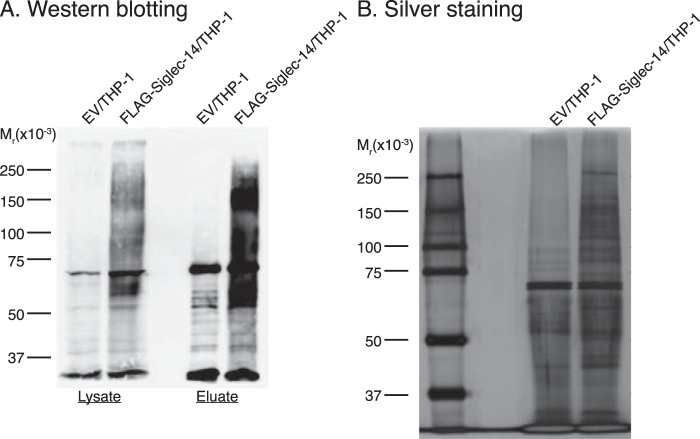
Proximity labeling of Siglec-14 cis-ligands on FLAG-Siglec-14/THP-1 cells. A, FLAG-Siglec-14/THP-1 cells were biotin-labeled with horseradish peroxidase-conjugated anti-FLAG antibody and biotin tyramide, and biotinylated proteins were purified from cell lysate with streptavidin beads. Cell lysates (Lysate) and eluates from streptavidin beads (Eluate) were subjected to SDS-PAGE/Western blotting and probed with streptavidin-HRP. B, streptavidin bead-purified proteins were subjected to SDS-PAGE/silver staining. EV/THP-1 was used as a negative control.
One of the cis-ligand candidates of mSiglec-14 was TLR2, a pattern recognition receptor that recognizes NTHi-associated lipoprotein and induces pro-inflammatory responses (26, 27). We therefore tested by co-immunoprecipitation whether TLR2 is interacting with mSiglec-14. As shown in Fig. 8A, TLR2 was indeed co-immunoprecipitated with mSiglec-14, verifying their interaction under steady state. We therefore tested whether the cis-interaction between mSiglec-14 and TLR2 explains the enhanced cytokine/chemokine production by Siglec-14/THP-1 cells. For this, we stimulated Siglec-14/THP-1 cells with N-palmitoyl-S-(2,3-bis(palmitoyloxy)propyl)cysteinyl-seryl-lysyl-lysyl-lysyl-lysine (Pam3CSK4), a specific agonist of the TLR1/2 heterodimer. Pam3CSK4 elicited IL-8 production to a similar extent as NTHi, which was suppressed by sSiglec-14 (Fig. 8B). We confirmed that a blocking antibody against TLR2 suppressed the Pam3CSK4-induced production of IL-8 and found that the addition of sSiglec-14 further suppressed IL-8 production to some extent (Fig. 8C; the anti-TLR2 antibody was used at nonsaturating concentration). Taken together, these results support our model that sSiglec-14 blocks production of pro-inflammatory cytokines/chemokines by interfering with the cis-interaction between mSiglec-14 and TLR2.
Figure 8.
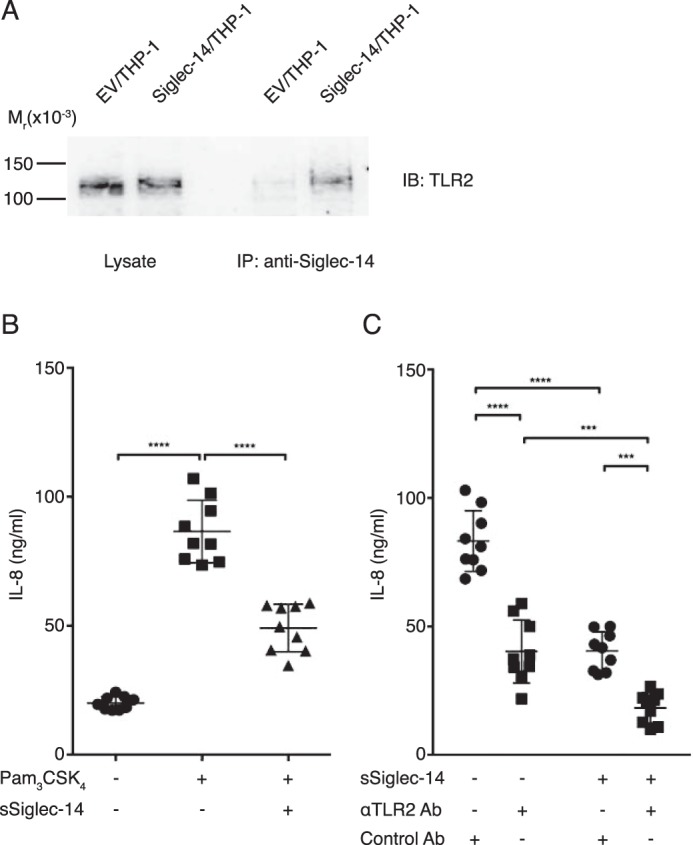
Soluble Siglec-14 blocks the cooperation between membrane-bound Siglec-14 and TLR2. A, co-immunoprecipitation (IP) of membrane-bound Siglec-14 and TLR2. B, effect of soluble Siglec-14 on IL-8 production by Siglec-14/THP-1 stimulated with Pam3CSK4, a TLR1/2 agonist. Experiments were repeated nine times (biological nonuplicates), and each value is plotted. Error bars represent standard deviation. The dataset was analyzed by one-way ANOVA to test the differences among groups, and post hoc pairwise mean comparison was conducted using Bonferroni's multiple comparisons test. ****, adjusted p < 0.0001. C, effect of soluble Siglec-14 and anti-TLR2 blocking antibody on IL-8 production by Siglec-14/THP-1 stimulated with Pam3CSK4. Experiments were repeated nine times (biological nonuplicates), and each value is plotted. Error bars represent standard deviation. The dataset was analyzed by two-way ANOVA to test the differences among groups, and post hoc pairwise mean comparison was conducted using Bonferroni's multiple comparisons test. ****, adjusted p < 0.0001; ***, 0.01 < Adjusted p < 0.001.
Intron 5 of mRNA splice variant encoding sSiglec-14 contains a G-quadruplex structure
To understand why the splice variant retaining intron 5 is relatively abundant, we sought a conserved element in intron 5 of primate SIGLEC14. As shown in Fig. 9A, intron 5 of SIGLEC14 contains a well-conserved guanosine (G)-rich segment, which could form a higher-order structure known as a G-quadruplex (G4) (28). On the one hand, DNA G4-forming sequences are preferentially enriched in telomeric and promoter regions, which are involved in telomere elongation and transcription regulations (29, 30). On the other hand, RNA G4–forming sequences have been demonstrated to modulate protein translation (31, 32) and alternative splicing (33, 34). Although the G-rich segment in SIGLEC14 intron 5 does not conform to a canonical G-quadruplex–forming sequence (GGGX2–4GGGX2–4GGGX2–4GGG) (35, 36), it still has the potential to form G4 structures because of the high density of guanosines. To test our hypothesis, we first performed far-UV CD analysis of the 27-nucleotide RNA corresponding to the conserved region in intron 5. As shown in Fig. 9B, the addition of 50 mm KCl induced significant changes in the far-UV CD spectrum of the 27-nucleotide RNA with strong positive and negative absorbance peaks at 264 and 240 nm, respectively, which is characteristic of all-parallel propeller-type G4 topology (36, 37). The spectral features closely resembled a well-characterized RNA G4 motif within the 5′-UTR of NRAS (31). Further titration of KCl up to 200 mm did not generate additional spectral changes, indicating efficient K+ uptake for G-tetrad coordination with 50 mm KCl. The 27-nucleotide RNA G4 motif is thermally stable, with an apparent melting temperature of 54.5 °C (Fig. 9C).
Figure 9.
Intron 5 of SIGLEC14 pre-mRNA contains an RNA G-quadruplex structure. A, alignment of primate SIGLEC14 intron 5 sequences. Stop codon is marked with a blue box, and the conserved G-rich segment is marked with a red box. Abbreviations in the panel represent the following species: Callithrix jacchus, common marmoset (C_jacchus); Chlorocebus sabaeus, green monkey (C_sabaeus); Homo sapiens, human (H_sapiens); Macaca fascicularis, crab-eating macaque (M_fascicularis); Macaca mulatta, rhesus macaque (M_mulatta); Nomascus leucogenys, Northern white-cheeked gibbon (N_leucogenys); Papio anubis, olive baboon (P_anubis); Pongo abelii, orangutan (P_abelii); Rhinopithecus roxellana, golden snub-nosed monkey (R_roxellana); Saimiri boliviensis, black-capped squirrel monkey (S_boliviensis). B, far-UV CD spectra of the 27-nucleotide RNA corresponding to the conserved segment in intron 5 of SIGLEC14, in the presence of various concentrations of KCl as indicated. C, thermal unfolding of the 27-nucleotide RNA monitored by far-UV CD spectroscopy. Full CD spectra are color-ramped from blue to red corresponding to sample temperatures from 20 to 75 °C. Inset, CD signal at 265 nm as a function of temperature, which is fitted to a two-state unfolding model with the melting temperature indicated. D, 1H NMR spectrum of the 27-nucleotide RNA in 100 mm KCl. The spectrum was collected at 303 K using a 600 MHz NMR spectrometer. The imino proton region is expanded with signals corresponding to Watson-Crick (12–14 ppm) and Hoogsteen (10–12 ppm) pairings indicated above. The presence of multiple well-resolved imino proton resonances in the Hoogsteen pairing region are indicative of the presence of a G-quadruplex structure.
To further characterize the nature of nucleotide pairing within the 27-nucleotide RNA, we employed one-dimensional proton NMR spectroscopy (Fig. 9D). Well-resolved imino proton resonances were observed at 10–14 ppm, corresponding to hydrogen-bonded imino protons of guanosine (G) and uridine (U) residues. The imino protons engaged in Watson-Crick pairing tend to exhibit chemical shifts in the range of 12–15 ppm. Only two resonances were observed in the Watson-Crick pairing region. Those involved in Hoogsteen pairing (typically seen in G4s) tend to be up-field–shifted to the range of 10–12 ppm (36, 38). More than 10 resolved resonances were observed in this region, in line with the expectation that the 27-nucleotide RNA sequence forms an ordered G4 structure.
Collectively, our spectroscopic analyses supported the ability of the 27-nucleotide RNA to form a G4 structure that is likely to be in an all-parallel topology, which is thermally stable under physiological conditions.
G-rich segment in intron 5 of SIGLEC14 pre-mRNA suppresses splicing of the intron
To test whether the G-rich segment in intron 5 of SIGLEC14 pre-mRNA negatively regulates the splicing of the intron, we prepared a “mini-gene” construct that contains intron 5 (consisting of exons 1–5 + intron 5 + exons 6–7 of SIGLEC14; designated Sig14+I5/pcDNA) and its variant that lacks the G-rich 27-nucleotide segment (Sig14+I5Δ27/pcDNA), and we evaluated the splicing efficiencies of intron 5 in transiently transfected 293T cells by quantitative PCR. The deletion of the G-rich 27-nucleotide segment in intron 5 indeed enhanced the efficiency of the intron 5 excision from pre-mRNA (ΔCT for the spliced versus unspliced isoforms in the following: Sig14+I5/pcDNA-transfected HEK293T cells, 4.33 ± 0.29; Sig14+I5Δ27/pcDNA-transfected HEK293T cells, 3.92 ± 0.43; seven biological replications; Fig. 10). Although the difference in the intron 5 splicing efficiencies between these two constructs was statistically significant (p = 0.0016, paired t test), the difference in ΔCT values for the two constructs was small (0.41 cycles on average, which translates to 20.41, ∼1.3 times difference in splicing efficiency). These results imply that the G-rich segment in intron 5 suppresses the splicing of intron 5, although the effect of this element alone may be relatively small.
Figure 10.
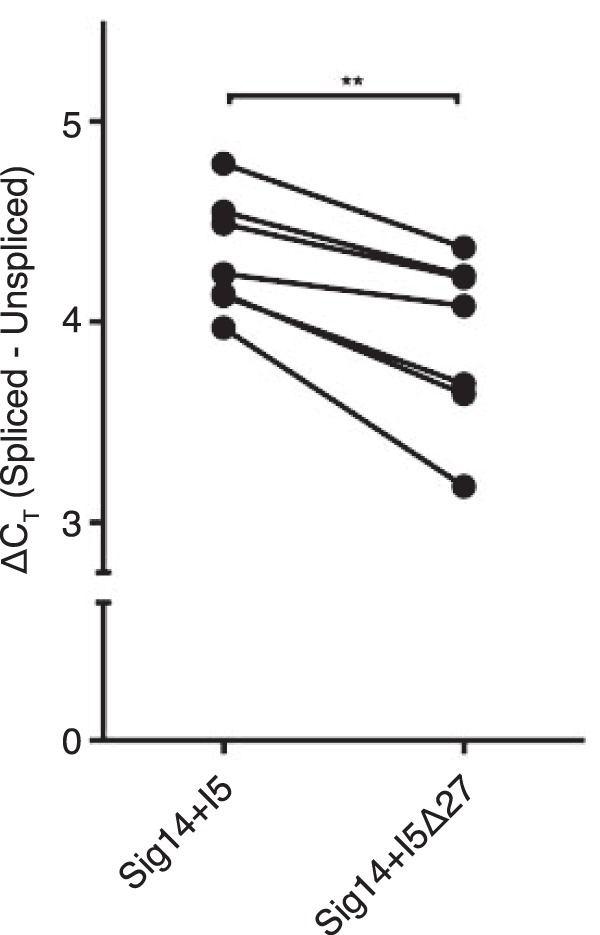
G-rich segment in intron 5 of SIGLEC14 pre-mRNA suppresses splicing of the intron. Differentials in the threshold cycle (ΔCT) between spliced (membrane-bound) versus unspliced (soluble) SIGLEC14 mRNA isoforms in 293T cells transfected with Sig14+I5/pcDNA (mini-gene construct with intron 5) or with Sig14+I5Δ27/pcDNA (mini-gene construct with intron 5 but lacking 27-nucleotide G-rich segment) were analyzed by quantitative RT-PCR. Each data point represents a mean of two to three technical replicates (i.e. two to three wells each of 293T cells were transfected with either Sig14+I5/pcDNA or Sig14+I5Δ27/pcDNA in parallel;, ΔCT for mRNA isoforms were analyzed by qRT-PCR for each well, and mean ΔCT was calculated for each construct). The experiments were repeated seven times. The dataset was analyzed by paired t test. **, p < 0.01.
Discussion
In this study, we demonstrated that alternative splicing generates sSiglec-14, and a sSiglec-14 isoform with a unique C-terminal heptapeptide is detectable in human sera. Several previous studies have reported the presence of alternatively spliced SIGLEC mRNA that encodes soluble forms (39–41), but the presence of corresponding protein has not been verified. Conversely, although the presence of soluble forms of Siglecs in human blood or cell culture supernatant has been reported (13, 14), these studies did not reveal the mechanism that generates these forms. Our study, to the best of our knowledge, is the first to demonstrate the connection between alternative splicing and soluble Siglec and to reveal the nature of any endogenous soluble Siglec present in human blood.
We demonstrated that sSiglec-14 suppresses pro-inflammatory cytokine production by interfering with the interaction between mSiglec-14 and TLR2. Our finding implies that sSiglec-14 is a mechanism employed by the innate immune system to fine-tune the inflammatory responses involving mSiglec-14, in addition to the antagonism by the membrane-bound Siglec-5, the inhibitory counterpart of mSiglec-14 (17–19, 42, 43). It was reported that Siglec-5 is down-regulated, whereas Siglec-14 is up-regulated, by lipopolysaccharide stimulation of neutrophils (43), implying the presence of a feed-forward loop that enhances myeloid inflammatory responses. Thus, it is tempting to speculate that the switching of Siglec-14 isoform from membrane-bound to soluble form through alternative splicing may function as a negative feedback mechanism to quell the inflammatory responses that could damage host tissues if uncontrolled. However, thus far we have not been able to identify a stimulus that robustly tips the balance between intron 5 spliced (membrane-bound) versus unspliced (soluble) forms of SIGLEC14 mRNA. Thus, the biological regulation afforded by sSiglec-14 is not yet fully understood.
Interactions between Siglecs and TLRs were previously reported (44). The authors of the previous study demonstrated that recombinant Siglec-5 and Siglec-9 can affinity-capture various TLRs from THP-1 cell lysate, whereas Siglec-14 was not included in the study. They demonstrated that NEU1, an endogenous sialidase, negatively regulates the interaction between mouse Siglec-E (ortholog of human Siglec-9) and TLR4. In contrast, our data (Fig. 5B) showed that sialic acid binding-deficient sSiglec-14 (R119A) still interferes with the interaction between mSiglec-14 and TLR2. Taken together, we tentatively conclude that the sialic acid dependence of the interaction between Siglec and its cis-ligand may differ, depending on the pair and the context.
Although the anti-inflammatory function of soluble Siglec-9 has been reported (14–16), these studies have not revealed the origin of soluble Siglec-9 (by alternative splicing or proteolysis) or its mechanism of action (by coordination/competition with membrane-bound Siglec-9 or by another mechanism). These studies primarily employed a recombinant soluble Siglec-9 fused with human IgG Fc as a surrogate of native soluble Siglec-9 in functional assays. Detailed analysis of the molecular form of natural soluble Siglec-9 may afford a deeper understanding of the mechanism of action. Likewise, a paper reporting the anti-inflammatory effect of “soluble Siglec-5” through engagement of P-selectin glycoprotein ligand-1 (PSGL-1) (45) also relied on recombinant proteins that are artificially made multivalent. Our previous study (19) demonstrated that soluble Siglec-5 detected by commercial sandwich ELISAs (13) may actually represent sSiglec-14, as neither Siglec-5 nor Siglec-14 was detected in the sera of SIGLEC14-null donors. The relevance of soluble Siglec-5 and Siglec-9 in homeostatic regulation of the immune system in vivo may deserve further investigation. Regardless, it is worth emphasizing that the studies by others indicated the possible utility of soluble Siglecs in translational research.
Regarding the suppression of pro-inflammatory cytokine production by sSiglec-14, we demonstrated one mechanism (inhibition of cis-interaction between mSiglec-14 and TLR2) that explains the observed phenomenon. Nevertheless, given that various proteins were biotin-labeled by proximity labeling pivoted on mSiglec-14 (Table S1), it is possible that the interactions between mSiglec-14 and other membrane proteins may also contribute to the suppression of inflammatory responses. In addition, sSiglec-14 may modulate myeloid cell functions other than inflammatory cytokine production. In addition to the experiments reported under “Results,” we also tested some hypotheses regarding the functions of sSiglec-14. For example, we investigated whether sSiglec-14 has bactericidal activity toward NTHi and observed neither direct killing nor enhancement of complement-mediated killing of NTHi by sSiglec-14 (Fig. 11). We also tested whether sSiglec-14 influences the polarization of macrophages (as reported for soluble Siglec-9), by adding recombinant sSiglec-14 in the in vitro differentiation system of human CD14+ monocytes to macrophages in the presence of macrophage colony-stimulatory factor. Analysis of the transcriptome by DNA microarray did not reveal any significant and consistent changes in the transcriptome that suggests the influence of sSiglec-14 on macrophage polarization (Table S2). Although it is beyond the scope of this study, further investigation may shed light on other aspects and the mechanisms by which sSiglec-14 modulates myeloid cell functions.
Figure 11.
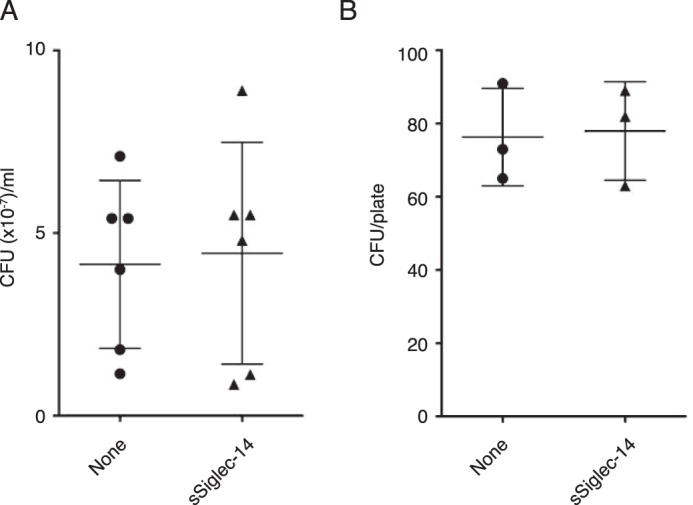
Soluble Siglec-14 lacks bactericidal activity against NTHi. A, direct bactericidal activity assay in the presence or absence of soluble Siglec-14 (6 biological replicates). B, complement-mediated bactericidal activity assay in the presence or absence of soluble Siglec-14 (three biological replicates). Experiments were performed as described under “Experimental procedures.” No statistically significant difference (by Student's t test) in bacterial counts between soluble Siglec-14–supplemented and control groups was observed.
The concentration of sSiglec-14 required for half-maximal inhibition of IL-8 production was estimated to be about 2 μg/ml (Fig. 5A), which is much higher than we observed in human serum (∼30 ng/ml in this study or ∼100 ng/ml in our previous study (19)); thus, we speculate that a local concentration of sSiglec-14 at the site of inflammation, in which myeloid cells accumulate, may attain biologically meaningful concentrations. Alternatively, the anti-inflammatory function of sSiglec-14 may require a special microenvironment, such as that found in granuloma tissue containing Mycobacterium tuberculosis. We noted that the concentrations of soluble Siglec-9 in the culture supernatant of dental pulp stem cells (∼600 pg/ml (14)) and in human sera (below the detection limit in three normal donor sera samples (13)) are much lower than those of sSiglec-14 present in U937 culture supernatant and in human serum.
The alternative splicing of SIGLEC14 mRNA is likely influenced by the presence of an RNA G4 structure in intron 5 of the pre-mRNA (Fig. 10). However, the effect of removal of the G4-forming segment from intron 5 appeared to be rather small (splicing efficiency increased by ∼1.3 times). In this regard, we noticed that there is a perfect inverted repeat (13 nucleotides) flanking the exon 5/intron 5 junction (marked with blue font and underlined in Fig. 3), which lies upstream of the G4-forming segment. Thus, although the effect of G4 alone may be small, the combination of G4 and the inverted repeat, along with RNA-binding proteins that regulate RNA splicing, may ultimately explain alternative splicing of SIGLEC14 pre-mRNA. Studies on the alternative splicing of myelin-associated glycoprotein (MAG), a Siglec expressed exclusively in the nervous system, demonstrated that the regulation of alternative splicing of MAG is complex, requiring heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1), a cytoplasmic isoform of quaking I that modifies the stability of hnRNP A1, and an RNA tertiary structure on MAG pre-mRNA to which hnRNP A1 binds (46–48). These studies also imply that 293T may not be the ideal system to study the effect of G4 because of the lack of an RNA-binding protein that regulates SIGLEC14 alternative splicing. Although it also falls outside of the focus of this study, identification of an RNA-binding protein that binds to the exon 5/intron 5 junction of SIGLEC14 pre-mRNA may ultimately reveal the mechanism of alternative splicing.
In summary, our study demonstrated that alternative splicing contributes to the generation of sSiglec-14, and sSiglec-14 manifests its anti-inflammatory properties by interfering with the cis-interaction between mSiglec-14 and TLR2. The alternative splicing of SIGLEC14 mRNA is likely influenced by the presence of an RNA G-quadruplex structure in intron 5 of the pre-mRNA. Whether soluble Siglec-14 has any translational value requires further investigation.
Experimental procedures
Cell lines and culture
U937 human histiocytic lymphoma cell line (CRL-1593.2; American Type Culture Collection), the THP-1 acute monocytic leukemia cell line (obtained from Human Science Research Resources Bank, Osaka, Japan; now part of the JCRB Cell Bank at the National Institutes of Biomedical Innovation, Health and Nutrition, Japan), and their derivatives were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (pen/strep). Siglec-14/THP-1 (a THP-1 sub-line expressing Siglec-14) and EV/THP-1 (a THP-1 sub-line transformed with an empty vector) were prepared as described (18). THP-1 cells expressing N-terminally FLAG-tagged membrane-bound Siglec-14 (FLAG-Siglec-14/THP-1) were prepared as described below. We obtained 293T cells from the American Type Culture Collection (CRL-3216) and maintained them in Dulbecco's modified Eagle's medium supplemented with 10% FBS and pen/strep.
Primers and DNA polymerase for PCR
Sequences of the primers used are listed in Table S3. Phusion High-Fidelity DNA polymerase (New England Biolabs) was used for PCRs unless otherwise stated.
3′-Rapid amplification of cDNA end (3′RACE) for SIGLEC14 mRNA
Human Bone Marrow Marathon-Ready cDNA (Clontech/Takara) was used as a template to amplify the 3′ end of SIGLEC14 mRNA, as instructed in the protocol provided by the manufacturer. Sequences of gene-specific primers (S14e5-2 and -3) used for the amplification are listed in Table S3. These gene-specific primers were designed based on the exon 5 sequence of SIGLEC14, as SIGLEC14 and SIGLEC5 mRNA show >99% sequence identity at exons 1–3, and the exon 4 of SIGLEC14 is short.
Determination of the relative abundance of mRNA variants by quantitative PCR
The abundance of mRNA splice variants encoding membrane-bound versus soluble Siglec-14 was analyzed by real-time quantitative PCR (RT-qPCR). In brief, total RNA was purified with a RNeasy Plus mini kit (Qiagen) in combination with RNase-Free DNase (Qiagen), and it was reverse-transcribed to cDNA with SuperScript III First-Strand Synthesis Super Mix (ThermoFisher Scientific). FastStart Universal Probe Master Mix (Roche Applied Science) was used for the amplification and detection of SIGLEC14 cDNA in a two-step quantitative PCR using a StepOnePlus Real-Time PCR System (ThermoFisher Scientific). Sequences of the primers and probes used are listed in Table S3.
Production of rabbit polyclonal antibody that recognizes C-terminal segment of sSiglec-14
Synthetic peptide corresponding to the C terminus of sSiglec-14 (sequence: C-SELQDRC; C at the N terminus was added for conjugation) was synthesized, conjugated to keyhole limpet hemocyanin, and used for the immunization of two rabbits (GenScript). The blood was collected from one of the rabbits after the third immunization, and the antibody fraction that recognized the peptide was purified by affinity purification using a column of agarose beads on which the antigen peptide was immobilized. Specificity of this antibody to sSiglec-14 with C-terminal 7-amino acid extension was verified by ELISA and Western blotting (Fig. 4).
Production of N-terminal His6-tagged sSiglec-14 expression construct
Sequences of the primers used are listed in Table S3. The cDNA of sSiglec-14 was amplified by nested PCR using the first-strand cDNA from U937 as a template and the primers “Siglec-14 Forward” and “Siglec-14 Reverse” in the first round and “Siglec-14 expr Xba1” and “Siglec-14 Sol R Hind3” in the second round. The PCR product was digested with XbaI and HindIII and cloned to XbaI–HindIII sites of pcDNA3.1(−). This plasmid was used as a template to PCR-amplify cDNA segment lacking signal peptide-coding sequence and stop codon, using the primers “Sig14 dSP IBA42 Sac2” and “Sig14 dTerm IBC42 R XhoI,” and cloned to SacII–XhoI sites of pEXPR-IBA42 vector (IBA). The resulting construct encoded a recombinant protein consisting of BM40 signal peptide and His6 tag at the N terminus, followed by sSiglec-14 (without native signal peptide) and a C-terminal Strep tag. The construct devoid of the C-terminal Strep tag was prepared by PCR amplification of the necessary segment using “pEXPR-IBA Seq F” and “Siglec-14 Sol R Hind3,” followed by digestion with XbaI and HindIII and cloning to XbaI–HindIII sites of pcDNA3.1(−). The resulting construct encodes a protein consisting of N-terminal BM40 signal peptide and His6 tag, followed by sSiglec-14 (without native signal peptide). Expression constructs for mutant sSiglec-14 proteins were prepared using this construct as a template, appropriate primers designed using on-line software (see Table S3 for sequences), and Q5 site-directed mutagenesis kit (New England Biolabs). All plasmids were sequence-verified.
Production of recombinant N-terminal His6-tagged sSiglec-14 protein
The constructs prepared as above were transfected into Expi293F cells (ThermoFisher Scientific), as instructed by the manufacturer. The culture supernatant was collected at the 5th and 8th day post-transfection by centrifugation, and His6-tagged sSiglec-14 protein was purified using nickel-nitrilotriacetic acid–agarose resin (Agarose Bead Technologies), in accordance with the manufacturer's instructions. The purified proteins were analyzed for the presence of endotoxin using an E-TOXATE Kit (Sigma) and were consistently found to contain very low levels of endotoxin (<100 EU/mg protein).
NTHi culture
NTHi strain L-378 (BCRC 17029) was obtained from the Bioresource Collection and Research Center (Hsin-Chu, Taiwan). NTHi was cultured in HTM broth at 37 °C and harvested by centrifugation (2,000 × g for 15 min) when the culture reached A600 = 0.3. The bacteria were washed once with PBS and fixed in 1% formaldehyde/PBS, adjusted at 1 × 109 cells/ml, and kept at −20 °C until use.
Stimulation of THP-1 pro-inflammatory responses with NTHi or Pam3CSK4
THP-1 cells were suspended in Macrophage-SFM (ThermoFisher Scientific) at 1 × 106 cells/ml and preincubated for 1 h with recombinant N-terminal His6-tagged sSiglec-14 (up to 20 μg/ml), with or without anti-human TLR2-blocking antibody (maba2-htlr2, Invivogen, 2 μg/ml) or control antibody (maba2-ctrl, Invivogen, 2 μg/ml), as indicated. NTHi (at multiplicity of infection = 10) or Pam3CSK4 (tlrl-pms, Invivogen; at 300 ng/ml) was added to the cells, which were further incubated in a CO2 incubator for 24 h. Culture supernatant was collected by centrifugation (800 × g for 3 min), and the concentration of IL-8 was determined using BD OptEIA Human IL-8 ELISA Set (BD Biosciences) after appropriate dilution (100–1,000 times) with Assay Diluent (BD Biosciences).
Concentration of various chemokines and cytokines (including CC chemokine ligand 2 (CCL2), CCL20, CXC chemokine ligand 1 (CXCL1), CXCL2, IL-1β, IL-8, IL-10, IL-23, and tumor necrosis factor-α) in the culture supernatant were also measured in parallel using custom-made Magnetic Luminex Screening Assays (R&D Systems), in accordance with the protocol provided by the manufacturer. Quantification of these soluble factors was performed by using a Bio-Plex MAGPIX Multiplex Reader (Bio-Rad) and Luminex xPONENT4.2 for MAGPIX software (Bio-Rad).
Preparation of THP-1 cells expressing N-terminally FLAG-tagged Siglec-14 (FLAG-Siglec-14/THP-1)
Sequences of the primers used are listed in Table S3. The cDNA encoding mSiglec-14 was amplified by PCR using Siglec-14/pcDNA (17) as a template and “Hind3 hSig14-dSP Fwd” and “Not1 hSig14 Rev” as primers, digested with HindIII and NotI, and subcloned to HindIII-NotI sites of p3×FLAG-CMV9 (Sigma). This construct encodes a protein consisting of preprotrypsin signal peptide (MSALLILALVGAAVA) and a triple FLAG tag (DYKDHDGDYKDHDIDYKDDDK) followed by mSiglec-14 without native signal peptide (amino acids 17–396). The cDNA of N-terminally FLAG-tagged mSiglec-14 was amplified using this construct as a template and primers “Xho1 PPT Fwd” and “p3×FLAG Seq Rev” (annealing to the downstream of ORF), digested with XhoI and NotI, and cloned to XhoI-NotI sites of pMSCVpuro (Clontech/Takara). The plasmid was designated FLAG-Siglec-14/pMSCVpuro. All plasmids were sequence-verified.
Recombinant amphotropic retroviral particles were prepared by transient transfection of PLAT-A packaging cell line (49) with the FLAG-Siglec-14/pMSCVpuro transfer vector, and the supernatant containing recombinant virus particles was collected. THP-1 cells were infected with the recombinant virus particles in the presence of RetroNectin (Takara) and cultured with medium containing puromycin (1 μg/ml) for 7 days to select transduced cells. The cells were designated as FLAG-Siglec-14/THP-1.
Proximity biotin labeling and identification of Siglec-14-interacting proteins
Proximity biotin labeling and identification of proteins interacting with mSiglec-14 were performed by applying the method previously described (25). FLAG-Siglec-14/THP-1 cells (5 × 106 cells) were incubated with 5 μg of anti-FLAG M2–peroxidase antibody (A8592, Sigma) on ice for 30 min and washed twice with PBS. Cells were suspended in PBS containing biotin-tyramide (10 μm) and hydrogen peroxide (10 mm) and incubated for 5 min at room temperature. Cells were washed three times with PBS and lysed in 500 μl of cell lysis buffer containing protease inhibitor (Roche Applied Science). Biotinylated proteins were purified with 200 μl of Dynabeads MyOne Streptavidin C1 paramagnetic beads (ThermoFisher Scientific), eluted with 50 μl of SDS-PAGE sample buffer, and subjected to brief SDS-PAGE. The gel area containing proteins was excised, in-gel trypsin–digested, and subjected to LC-coupled tandem MS for protein identification using a TLQ-Orbitrap Velos mass spectrometer (ThermoFisher Scientific), as described previously (25).
Acquired data were analyzed using MaxQuant version 1.6.0.1 (50), and the proteins that were identified only as post-translationally modified forms, identified in the reversed database, or considered possible contaminants were excluded from the list. Subsequently, the proteins that fulfilled all the following criteria were selected (Table S1): 1) proteins whose abundance in the sample purified from labeled FLAG-Siglec-14/THP-1 was 10 times or more than from labeled EV/THP-1; 2) proteins identified with three or more unique peptides; and 3) proteins that are known or predicted to be membrane proteins. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (51) partner repository with the dataset identifier PXD009748.
Co-immunoprecipitation of mSiglec-14 with TLR2
Siglec-14/THP-1 cells (8 × 106 cells) were washed twice with 10 ml of PBS and cross-linked with 3,3′-dithiobis(sulfosuccinimidyl propionate) (ThermoFisher Scientific) at room temperature for 30 min. The reaction was quenched with 40 μl of 1 m Tris-HCl buffer (pH 7.5) at room temperature for 15 min. The cells were lysed in 1 ml of lysis buffer (150 mm NaCl, 1% Nonidet P-40, and 50 mm Tris-HCl (pH 8.0)). Cell lysate (0.75 mg) was subjected to immunoprecipitation with biotinylated anti-Siglec-5/14 antibody (6 μg; clone 194128; R&D Systems) and Dynabeads MyOne Streptavidin C1 beads (30 μl) at 4 °C overnight. Beads were washed twice with 500 μl of PBS containing 0.1% Tween 20 (PBST) and boiled in 40 μl of 1× SDS-PAGE Sample Buffer (Bio-Rad) supplemented with 100 mm DTT at 100 °C for 5 min. The supernatant after the removal of magnetic beads was subjected to SDS-PAGE using the Laemmli buffer system. Proteins were transferred to polyvinylfluoride membrane (PolyScreen PVDF Hybridization Transfer Membrane; PerkinElmer Life Sciences) by semi-dry blotting using Towbin buffer (15 V, 60 min). The membrane was blocked with 3% (w/v) BSA in 20 mm Tris-HCl (pH 7.5), 150 mm NaCl, and 0.1% Tween 20 (TBST), probed with goat anti-TLR2 polyclonal antibody (AF2616, R&D Systems) overnight at 4 °C, followed by peroxidase-conjugated donkey anti-rabbit antibody (705-035-147, Jackson ImmunoResearch) at room temperature for 1 h and further with Western Lightning ECL Pro (PerkinElmer Life Sciences). Protein bands were visualized with an ImageQuant LAS4000 Mini Biomolecular Imager (GE Healthcare).
Far-UV CD spectroscopy of synthetic 27-nucleotide RNA
Synthetic 27-nucleotide RNA (5′-GGUUGGUGGGAGGGACUGAGGCCUGGU-3′) corresponding to the conserved region in intron 5 of Siglec-14 was generated by solid-phase synthesis (GenScript; purity > 93% by HPLC). The 27-nucleotide RNA was dissolved at 10 μm in 10 mm Tris-HCl buffer (pH 7.4) and annealed by heating to 95 °C followed by overnight cooling. Native CD spectra in the presence of 0, 50, 100, 150, and 200 mm KCl were obtained at 20 °C using a temperature-controlled CD spectrometer (model J-815, Jasco, Japan) as described previously (52). The thermal stability of the RNA tertiary structure was assessed by collecting CD spectra between 20 and 75 °C. The sample was buffered in 10 mm Tris-HCl buffer (pH 7.4) with 100 mm KCl.
NMR spectroscopy of synthetic 27-nucleotide RNA
The same synthetic 27-nucleotide RNA was dissolved at 137 μm in 10 mm Tris-HCl (pH 7.4) containing 100 mm KCl and 10% D2O. The one-dimensional 1H NMR spectra of the synthetic 27-nucleotide RNA were collected at 293, 298, and 303 K using an AVANCE III 600 NMR spectrometer (Bruker, Germany) equipped with a TCI cryogenic probe head. Water suppression was achieved using a WATERGATE pulse program as described previously (52, 53).
Preparation of mini-gene constructs for SIGLEC14 containing intron 5
Sequences of the primers used are listed in Table S3. A mini-gene construct consisting of SIGLEC14 exons 1–5, intron 5, and exons 6–7 was prepared by nested PCR using first-strand cDNA from U937 cells and nested primer sets (first PCR: Siglec-14 Forward + Siglec-14 1st R; second PCR: HsSig14 XhoI Nhe1-F + HsSig14 ER5h-R). The PCR product was digested with XhoI and EcoRV and cloned to XhoI–EcoRV sites of pcDNA3.1(−). The construct was designated “Sig14+I5/pcDNA.” Its variant that lacks G-rich 27-nucleotide was prepared using Q5 site-directed mutagenesis kit with primers S14 d27nt F and S14 d27nt R, and designated “Sig14+I5Δ27/pcDNA.” These constructs were transfected to 293T cells using Lipofectamine 2000 (ThermoFisher Scientific) according to the manufacturer's instructions. Total RNA was extracted 48 h post-transfection and subjected to isoform-specific RT-qPCR, as described above.
Bactericidal assays
Direct bactericidal assay
Freshly cultured NTHi (1 × 106 cells) was mixed with 1 μm (∼38 μg/ml) recombinant sSiglec-14 in 200 μl of HTM broth and incubated at 37 °C for 2 h. The mixture was serially diluted with HTM broth, spread onto Chocolate agar plates, and incubated overnight at 37 °C. Colonies were counted manually.
Complement-mediated bactericidal assay
Freshly cultured NTHi (200 cells) in 180 μl of Hanks' balanced salt solution (Ca2+ and Mg2+) was mixed with 20 μl of baby rabbit complement (Pel-Freez Biologicals). Recombinant sSiglec-14 protein (10 μg/ml) was introduced into the mixture and incubated at 37 °C for 30 min. The mixture was spread onto Chocolate agar plates and incubated overnight at 37 °C. Colonies were counted manually.
In vitro macrophage differentiation and transcriptome analysis
Human monocytes (5 × 106 cells, purchased from Zen-Bio) were differentiated in the presence of human macrophage colony stimulatory factor (100 ng/ml, R&D Systems) in RPMI 1640 medium containing 10% FBS and pen/strep for 7 days. Recombinant sSiglec-14 (1 μg/ml) was introduced at day 7, and the cells were further cultured for 24 h. Total RNA was extracted with RNeasy Plus mini kit (Qiagen), and subjected to gene expression analysis.
Affymetrix GeneChip assays were performed by the Affymetrix Gene Expression Service Lab supported by Academia Sinica. Total RNA (300 ng) was used for the serial syntheses of double-stranded cDNA and biotin-labeled antisense complementary RNA (cRNA) by in vitro transcription, followed by cRNA fragmentation, in accordance with the manufacturer's instructions (GeneChip Expression Analysis Technical Manual revision 5, Affymetrix). Labeled and fragmented cRNA (11 μg) was hybridized to GeneChip Human Genome U133 Plus 2.0 (Affymetrix) at 45 °C for 16.5 h. The chip was washed and stained with Fluidic Station-450 and was scanned with Affymetrix GeneChip Scanner 7G.
Author contributions
P.-C. J. H., P.-Y. L., I. W., and T. A. data curation; P.-C. J. H., I. W., S.-T. D. H., and T. A. formal analysis; P.-C. J. H., S.-T. D. H., and T. A. writing-original draft; P.-C. J. H., S.-T. D. H., and T. A. writing-review and editing; S.-T. D. H. and T. A. supervision; S.-T. D. H. and T. A. funding acquisition; T. A. conceptualization; T. A. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Meng-Ru Ho of the Biophysical Instrumentation Laboratory at the Institute of Biological Chemistry, Academia Sinica, Taiwan, for providing technical assistance and support with data from the far-UV CD experiments. The NMR spectra were obtained at the High-Field NMR Center of Academia Sinica, supported by the National Core Facility Program for Biotechnology, Taiwan. We also thank Dr. Shu-Yu Lin at Academia Sinica Common Mass Spectrometry Facilities, Academia Sinica, Taiwan, for instruction in LC-MS2 experiments and help in the analysis of proteomic data. Affymetrix GeneChip assays were performed by the Affymetrix Gene Expression Service Lab supported by Academia Sinica. We thank H. Nikki March, Ph.D., from Edanz Group for editing a draft of this manuscript.
This work was supported by Ministry of Science and Technology, Republic of China, Grants MOST 104-2321-B-001-070, 105-2321-B-001-051, and 106-2321-B-001-032 (to T. A.) and intramural funding from the Institute of Biological Chemistry, Academia Sinica (to S. T. D. H. and T. A.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1–S3.
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier accession no. PXD009748.
The GeneChip transcriptome analysis data have been deposited to Gene Expression Omnibus Database of National Center for Biotechnology Information with the accession number GSE121311.
- IL-6
- interleukin-6
- ADAM
- a disintegrin and metalloproteinase domain-containing protein
- COPD
- chronic obstructive pulmonary disease
- CXCL
- CXC chemokine ligand
- FBS
- fetal bovine serum
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- MAG
- myelin-associated glycoprotein
- mSiglec-14
- membrane-bound Siglec-14
- NTHi
- nontypeable Haemophilus influenzae
- Pam3CSK4
- N-palmitoyl-S-(2,3-bis(palmitoyloxy)propyl)cysteinyl-seryl-lysyl-lysyl-lysyl-lysine
- PSGL-1
- P-selectin glycoprotein ligand-1
- RACE
- rapid amplification of cDNA end
- RT-qPCR
- real-time quantitative PCR
- sSiglec-14
- soluble Siglec-14
- TLR2
- toll-like receptor 2
- TNF
- tumor necrosis factor
- pen/strep
- penicillin/streptomycin
- ANOVA
- analysis of variance
- EV
- empty vector.
References
- 1. Levine S. J. (2008) Molecular mechanisms of soluble cytokine receptor generation. J. Biol. Chem. 283, 14177–14181 10.1074/jbc.R700052200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jones S. A., Horiuchi S., Topley N., Yamamoto N., and Fuller G. M. (2001) The soluble interleukin 6 receptor: mechanisms of production and implications in disease. FASEB J. 15, 43–58 10.1096/fj.99-1003rev [DOI] [PubMed] [Google Scholar]
- 3. Ramasamy R., Yan S. F., and Schmidt A. M. (2009) RAGE: therapeutic target and biomarker of the inflammatory response–the evidence mounts. J. Leukoc. Biol. 86, 505–512 10.1189/jlb.0409230 [DOI] [PubMed] [Google Scholar]
- 4. Yonchuk J. G., Silverman E. K., Bowler R. P., Agustí A., Lomas D. A., Miller B. E., Tal-Singer R., and Mayer R. J. (2015) Circulating soluble receptor for advanced glycation end products (sRAGE) as a biomarker of emphysema and the RAGE axis in the lung. Am. J. Respir. Crit. Care Med. 192, 785–792 10.1164/rccm.201501-0137PP [DOI] [PubMed] [Google Scholar]
- 5. Ye W., Hu Y., Zhang R., and Ying K. (2014) Diagnostic value of the soluble triggering receptor expressed on myeloid cells-1 in lower respiratory tract infections: a meta-analysis. Respirology 19, 501–507 10.1111/resp.12270 [DOI] [PubMed] [Google Scholar]
- 6. Steis R. G., Marcon L., Clark J., Urba W., Longo D. L., Nelson D. L., and Maluish A. E. (1988) Serum soluble IL-2 receptor as a tumor marker in patients with hairy cell leukemia. Blood 71, 1304–1309 [PubMed] [Google Scholar]
- 7. Varki A., and Angata T. (2006) Siglecs-the major subfamily of I-type lectins. Glycobiology 16, 1R–27R 10.1093/glycob/cwj008 [DOI] [PubMed] [Google Scholar]
- 8. Crocker P. R., Paulson J. C., and Varki A. (2007) Siglecs and their roles in the immune system. Nat. Rev. Immunol. 7, 255–266 10.1038/nri2056 [DOI] [PubMed] [Google Scholar]
- 9. Pillai S., Netravali I. A., Cariappa A., and Mattoo H. (2012) Siglecs and immune regulation. Annu. Rev. Immunol. 30, 357–392 10.1146/annurev-immunol-020711-075018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Macauley M. S., Crocker P. R., and Paulson J. C. (2014) Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 14, 653–666 10.1038/nri3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Angata T. (2014) Associations of genetic polymorphisms of Siglecs with human diseases. Glycobiology 24, 785–793 10.1093/glycob/cwu043 [DOI] [PubMed] [Google Scholar]
- 12. Angata T., Nycholat C. M., and Macauley M. S. (2015) Therapeutic targeting of Siglecs using antibody- and glycan-based approaches. Trends Pharmacol. Sci. 36, 645–660 10.1016/j.tips.2015.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Biedermann B., Gil D., Bowen D. T., and Crocker P. R. (2007) Analysis of the CD33-related siglec family reveals that Siglec-9 is an endocytic receptor expressed on subsets of acute myeloid leukemia cells and absent from normal hematopoietic progenitors. Leuk. Res. 31, 211–220 10.1016/j.leukres.2006.05.026 [DOI] [PubMed] [Google Scholar]
- 14. Ishikawa J., Takahashi N., Matsumoto T., Yoshioka Y., Yamamoto N., Nishikawa M., Hibi H., Ishigro N., Ueda M., Furukawa K., and Yamamoto A. (2016) Factors secreted from dental pulp stem cells show multifaceted benefits for treating experimental rheumatoid arthritis. Bone 83, 210–219 10.1016/j.bone.2015.11.012 [DOI] [PubMed] [Google Scholar]
- 15. Matsubara K., Matsushita Y., Sakai K., Kano F., Kondo M., Noda M., Hashimoto N., Imagama S., Ishiguro N., Suzumura A., Ueda M., Furukawa K., and Yamamoto A. (2015) Secreted ectodomain of sialic acid-binding Ig-like lectin-9 and monocyte chemoattractant protein-1 promote recovery after rat spinal cord injury by altering macrophage polarity. J. Neurosci. 35, 2452–2464 10.1523/JNEUROSCI.4088-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shimojima C., Takeuchi H., Jin S., Parajuli B., Hattori H., Suzumura A., Hibi H., Ueda M., and Yamamoto A. (2016) Conditioned medium from the stem cells of human exfoliated deciduous teeth ameliorates experimental autoimmune encephalomyelitis. J. Immunol. 196, 4164–4171 10.4049/jimmunol.1501457 [DOI] [PubMed] [Google Scholar]
- 17. Angata T., Hayakawa T., Yamanaka M., Varki A., and Nakamura M. (2006) Discovery of Siglec-14, a novel sialic acid receptor undergoing concerted evolution with Siglec-5 in primates. FASEB J. 20, 1964–1973 10.1096/fj.06-5800com [DOI] [PubMed] [Google Scholar]
- 18. Yamanaka M., Kato Y., Angata T., and Narimatsu H. (2009) Deletion polymorphism of SIGLEC14 and its functional implications. Glycobiology 19, 841–846 10.1093/glycob/cwp052 [DOI] [PubMed] [Google Scholar]
- 19. Angata T., Ishii T., Motegi T., Oka R., Taylor R. E., Soto P. C., Chang Y. C., Secundino I., Gao C. X., Ohtsubo K., Kitazume S., Nizet V., Varki A., Gemma A., Kida K., and Taniguchi N. (2013) Loss of Siglec-14 reduces the risk of chronic obstructive pulmonary disease exacerbation. Cell. Mol. Life Sci. 70, 3199–3210 10.1007/s00018-013-1311-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Förster K., Sass S., Ehrhardt H., Mous D. S., Rottier R. J., Oak P., Schulze A., Flemmer A. W., Gronbach J., Hübener C., Desai T., Eickelberg O., Theis F. J., and Hilgendorff A. (2018) Early identification of bronchopulmonary dysplasia using novel biomarkers by proteomic screening. Am. J. Respir. Crit. Care Med. 197, 1076–1080 10.1164/rccm.201706-1218LE [DOI] [PubMed] [Google Scholar]
- 21. Higgins R. D., Jobe A. H., Koso-Thomas M., Bancalari E., Viscardi R. M., Hartert T. V., Ryan R. M., Kallapur S. G., Steinhorn R. H., Konduri G. G., Davis S. D., Thebaud B., Clyman R. I., Collaco J. M., Martin C. R., et al. (2018) Bronchopulmonary dysplasia: executive summary of a workshop. J. Pediatr. 197, 300–308 10.1016/j.jpeds.2018.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ryan R. M., Ahmed Q., and Lakshminrusimha S. (2008) Inflammatory mediators in the immunobiology of bronchopulmonary dysplasia. Clin. Rev. Allergy Immunol. 34, 174–190 10.1007/s12016-007-8031-4 [DOI] [PubMed] [Google Scholar]
- 23. Edwards D. R., Handsley M. M., and Pennington C. J. (2008) The ADAM metalloproteinases. Mol. Aspects Med. 29, 258–289 10.1016/j.mam.2008.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Angata T., Ishii T., Gao C., Ohtsubo K., Kitazume S., Gemma A., Kida K., and Taniguchi N. (2014) Association of serum interleukin-27 with the exacerbation of chronic obstructive pulmonary disease. Physiol. Rep. 2, e12069 10.14814/phy2.12069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chang L., Chen Y. J., Fan C. Y., Tang C. J., Chen Y. H., Low P. Y., Ventura A., Lin C. C., Chen Y. J., and Angata T. (2017) Identification of Siglec ligands using a proximity labeling method. J. Proteome Res. 16, 3929–3941 10.1021/acs.jproteome.7b00625 [DOI] [PubMed] [Google Scholar]
- 26. Lugade A. A., Bogner P. N., Murphy T. F., and Thanavala Y. (2011) The role of TLR2 and bacterial lipoprotein in enhancing airway inflammation and immunity. Front. Immunol. 2, 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shuto T., Xu H., Wang B., Han J., Kai H., Gu X. X., Murphy T. F., Lim D. J., and Li J. D. (2001) Activation of NF-κB by nontypeable Hemophilus influenzae is mediated by toll-like receptor 2-TAK1-dependent NIK-IKK α/β-IκBα and MKK3/6-p38 MAP kinase signaling pathways in epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 98, 8774–8779 10.1073/pnas.151236098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hänsel-Hertsch R., Di Antonio M., and Balasubramanian S. (2017) DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 18, 279–284 10.1038/nrm.2017.3 [DOI] [PubMed] [Google Scholar]
- 29. Waller Z. A., Sewitz S. A., Hsu S. T., and Balasubramanian S. (2009) A small molecule that disrupts G-quadruplex DNA structure and enhances gene expression. J. Am. Chem. Soc. 131, 12628–12633 10.1021/ja901892u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dash J., Shirude P. S., Hsu S. T., and Balasubramanian S. (2008) Diarylethynyl amides that recognize the parallel conformation of genomic promoter DNA G-quadruplexes. J. Am. Chem. Soc. 130, 15950–15956 10.1021/ja8046552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kumari S., Bugaut A., Huppert J. L., and Balasubramanian S. (2007) An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 3, 218–221 10.1038/nchembio864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bugaut A., Rodriguez R., Kumari S., Hsu S. T., and Balasubramanian S. (2010) Small molecule-mediated inhibition of translation by targeting a native RNA G-quadruplex. Org. Biomol. Chem. 8, 2771–2776 10.1039/c002418j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weldon C., Behm-Ansmant I., Hurley L. H., Burley G. A., Branlant C., Eperon I. C., and Dominguez C. (2017) Identification of G-quadruplexes in long functional RNAs using 7-deazaguanine RNA. Nat. Chem. Biol. 13, 18–20 10.1038/nchembio.2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weldon C., Dacanay J. G., Gokhale V., Boddupally P. V. L., Behm-Ansmant I., Burley G. A., Branlant C., Hurley L. H., Dominguez C., and Eperon I. C. (2018) Specific G-quadruplex ligands modulate the alternative splicing of Bcl-X. Nucleic Acids Res. 46, 886–896 10.1093/nar/gkx1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huppert J. L., and Balasubramanian S. (2007) G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 35, 406–413 10.1093/nar/gkl1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patel D. J., Phan A. T., and Kuryavyi V. (2007) Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 35, 7429–7455 10.1093/nar/gkm711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hsu S. T., Varnai P., Bugaut A., Reszka A. P., Neidle S., and Balasubramanian S. (2009) A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J. Am. Chem. Soc. 131, 13399–13409 10.1021/ja904007p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fürtig B., Richter C., Wöhnert J., and Schwalbe H. (2003) NMR spectroscopy of RNA. Chembiochem 4, 936–962 10.1002/cbic.200300700 [DOI] [PubMed] [Google Scholar]
- 39. Takei Y., Sasaki S., Fujiwara T., Takahashi E., Muto T., and Nakamura Y. (1997) Molecular cloning of a novel gene similar to myeloid antigen CD33 and its specific expression in placenta. Cytogenet. Cell Genet. 78, 295–300 10.1159/000134676 [DOI] [PubMed] [Google Scholar]
- 40. Connolly N. P., Jones M., and Watt S. M. (2002) Human Siglec-5: tissue distribution, novel isoforms and domain specificities for sialic acid-dependent ligand interactions. Br. J. Haematol. 119, 221–238 10.1046/j.1365-2141.2002.03808.x [DOI] [PubMed] [Google Scholar]
- 41. Kitzig F., Martinez-Barriocanal A., López-Botet M., and Sayós J. (2002) Cloning of two new splice variants of Siglec-10 and mapping of the interaction between Siglec-10 and SHP-1. Biochem. Biophys. Res. Commun. 296, 355–362 10.1016/S0006-291X(02)00885-9 [DOI] [PubMed] [Google Scholar]
- 42. Yasui K., Angata T., Matsuyama N., Furuta R. A., Kimura T., Okazaki H., Tani Y., Nakano S., Narimatsu H., and Hirayama F. (2011) Detection of anti-Siglec-14 alloantibodies in blood components implicated in nonhaemolytic transfusion reactions. Br. J. Haematol. 153, 794–796 10.1111/j.1365-2141.2010.08488.x [DOI] [PubMed] [Google Scholar]
- 43. Ali S. R., Fong J. J., Carlin A. F., Busch T. D., Linden R., Angata T., Areschoug T., Parast M., Varki N., Murray J., Nizet V., and Varki A. (2014) Siglec-5 and Siglec-14 are polymorphic paired receptors that modulate neutrophil and amnion signaling responses to group B Streptococcus. J. Exp. Med. 211, 1231–1242 10.1084/jem.20131853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen G. Y., Brown N. K., Wu W., Khedri Z., Yu H., Chen X., van de Vlekkert D., D'Azzo A., Zheng P., and Liu Y. (2014) Broad and direct interaction between TLR and Siglec families of pattern recognition receptors and its regulation by Neu1. Elife 3, e04066 10.7554/eLife.04066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pepin M., Mezouar S., Pegon J., Muczynski V., Adam F., Bianchini E. P., Bazaa A., Proulle V., Rupin A., Paysant J., Panicot-Dubois L., Christophe O. D., Dubois C., Lenting P. J., and Denis C. V. (2016) Soluble Siglec-5 associates to PSGL-1 and displays anti-inflammatory activity. Sci. Rep. 6, 37953 10.1038/srep37953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao L., Mandler M. D., Yi H., and Feng Y. (2010) Quaking I controls a unique cytoplasmic pathway that regulates alternative splicing of myelin-associated glycoprotein. Proc. Natl. Acad. Sci. U.S.A. 107, 19061–19066 10.1073/pnas.1007487107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zearfoss N. R., Clingman C. C., Farley B. M., McCoig L. M., and Ryder S. P. (2011) Quaking regulates Hnrnpa1 expression through its 3′ UTR in oligodendrocyte precursor cells. PLoS Genet. 7, e1001269 10.1371/journal.pgen.1001269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zearfoss N. R., Johnson E. S., and Ryder S. P. (2013) hnRNP A1 and secondary structure coordinate alternative splicing of Mag. RNA 19, 948–957 10.1261/rna.036780.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., and Kumagai H. (2003) Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31, 1007–1014 10.1016/S0301-472X(03)00260-1 [DOI] [PubMed] [Google Scholar]
- 50. Tyanova S., Temu T., and Cox J. (2016) The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 10.1038/nprot.2016.136 [DOI] [PubMed] [Google Scholar]
- 51. Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., et al. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 10.1038/nbt.2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang Z. F., Li M. H., Hsu S. T., and Chang T. C. (2014) Structural basis of sodium-potassium exchange of a human telomeric DNA quadruplex without topological conversion. Nucleic Acids Res. 42, 4723–4733 10.1093/nar/gku083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang Z. F., Li M. H., Chen W. W., Hsu S. T., and Chang T. C. (2016) A novel transition pathway of ligand-induced topological conversion from hybrid forms to parallel forms of human telomeric G-quadruplexes. Nucleic Acids Res. 44, 3958–3968 10.1093/nar/gkw145 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.