Abstract
Bacterial microcompartments encapsulate enzymatic pathways that generate small, volatile, aldehyde intermediates. The Rhodococcus and Mycobacterium microcompartment (RMM) operon from Mycobacterium smegmatis encodes four enzymes, including (S)-1-amino-2-propanol dehydrogenase and a likely propionaldehyde dehydrogenase. We show here that a third enzyme (and its nonmicrocompartment-associated paralog) is a moderately specific (S)-1-amino-2-propanol kinase. We determined the structure of apo-aminopropanol kinase at 1.35 Å, revealing that it has structural similarity to hexosamine kinases, choline kinases, and aminoglycoside phosphotransferases. We modeled substrate binding, and tested our model by characterizing key enzyme variants. Bioinformatics analysis established that this enzyme is widespread in Actinobacteria, Proteobacteria, and Firmicutes, and is very commonly associated with a candidate phospholyase. In Rhizobia, aminopropanol kinase is generally associated with aromatic degradation pathways. In the RMM (and the parallel pathway that includes the second paralog), aminopropanol kinase likely degrades aminoacetone through a propanolamine-phosphate phospho-lyase–dependent pathway. These enzymatic activities were originally described in Pseudomonas, but the proteins responsible have not been previously identified. Bacterial microcompartments typically co-encapsulate enzymes which can regenerate required co-factors, but the RMM enzymes require four biochemically distinct co-factors with no overlap. This suggests that either the RMM shell can uniquely transport multiple co-factors in stoichiometric quantities, or that all enzymes except the phospho-lyase reside outside of the shell. In summary, aminopropanol kinase is a novel enzyme found in diverse bacteria and multiple metabolic pathways; its presence in the RMM implies that this microcompartment degrades aminoacetone, using a pathway that appears to violate some established precepts as to how microcompartments function.
Keywords: protein structure, enzyme kinetics, bioinformatics, biodegradation, microbiology, aminoacetone utilization microcompartment, aminopropanol kinase
Introduction
Bacterial microcompartments are specialized cellular inclusions built by diverse bacteria to mitigate the challenges presented by the toxic or volatile intermediates produced by certain metabolic pathways (1). Most microcompartments are catabolic, and contain a set of enzymes that produce, and then consume, aldehyde intermediates (2). Well-studied examples include the propanediol and ethanolamine utilization microcompartments, but more recently ethanol, choline, and rhamnose/fucose degrading microcompartments have also been proposed, and many more remain uncharacterized (3–5). The exceptions to this general rule are the environmentally important α- and β-carboxysomes, which encapsulate RubisCO and carbonic anhydrase and mediate anabolic carbon fixation in cyanobacteria and chemoautotrophs (6, 7). Microcompartments are defined by the presence of a thin icosahedral shell built from proteins from two distinct families (PF00936 and PF03319); this shell encapsulates key enzymes and is likely selectively permeable, allowing initial substrates and products to pass freely, whereas retaining enzymes, cofactors, and metabolic intermediates (8, 9).
Recent surveys of microbial genomes have uncovered a deep diversity in microcompartment families, many of which are poorly or wholly uncharacterized (2, 4). One such operon has been identified predominantly in species of the Actinobacteria phylum, and has been provisionally designated the RMM2 (Rhodococcus and Mycobacterium microcompartment) (2). Although formally uncharacterized, Urano and colleagues (10) previously showed that this operon is induced 100-fold by the addition of aminopropanol in the culture media. The RMM encoding operon contains a regulatory protein, an amino acid permease, and four structural proteins that are close homologs to those that form the only structurally characterized shell (9, 11), along with enzyme sequences with clear homology to alcohol dehydrogenases, aminoglycoside kinases, aminotransferases, and aldehyde dehydrogenases, respectively. Building on earlier work (10, 12), we recently published detailed structural and functional characterization of the alcohol dehydrogenase from Mycobacterium smegmatis MC2 155, identifying it as an NADP-dependent (S)-1-amino-2-propanol (aminopropanol) dehydrogenase (APDH) (13). However, the other enzymes in this pathway remain uncharacterized; specifically, understanding of the function of this microcompartment is hindered by the fact that no functions have been ascribed in the literature to any close homologs (i.e. with sequence identity >25%) of either the aminotransferase- or kinase-like enzymes.
The alcohol kinase homolog from the M. smegmatis MC2 155 microcompartment operon (msmeg_0270) is annotated as a member of the aminoglycoside phosphotransferase family (APH, PF01636), a group which is, in turn, part of the protein kinase-like domain superfamily (PK-like, SSF56112). Members of the PK-like family of enzymes have low sequence conservation but exhibit conserved tertiary structure including an N-terminal α/β-fold nucleotide triphosphate-binding domain and a mostly α-helical C-terminal substrate-binding domain (14). Enzymes of this family phosphorylate diverse substrates, ranging from proteins to small alcohols, using ATP (or, in some cases, GTP) as the donor. Kinases are not usually found in microcompartment pathways, likely because microcompartments generally lack the capacity to import stoichiometric quantities of cofactors, instead recycling a fixed cofactor pool through complementary enzymes (2, 15). Among microcompartments, the only identified phosphotransfer reaction is a phosphotransacylation reaction used to recycle CoA in the propanediol microcompartment, a reaction that occurs outside of the shell (16). Many organisms (mostly γ-Proteobacteria and Actinobacteria) also encode a homologous operon that has no shell proteins; interestingly, many mycobacterial strains encode both (RMM and non-RMM) variants of this operon. Early work by Turner and colleagues (17, 18) have shown that an enzyme with (S)-1-amino-2-propanol kinase (APK) activity can be purified from many of the same organisms that express APDH. Given the strict specificity of APDH, we therefore tested both the RMM-associated (msmeg_0270, henceforth APKMSM0270) and the non-RMM associated (msmeg_0780, henceforth APKMSM0780) APH homologs from M. smegmatis MC2 155, showing that both can act as aminopropanol kinase. We also report the structure, kinetics, and a model for substrate recognition for APKMSM0270.
Results
Enzyme kinetics
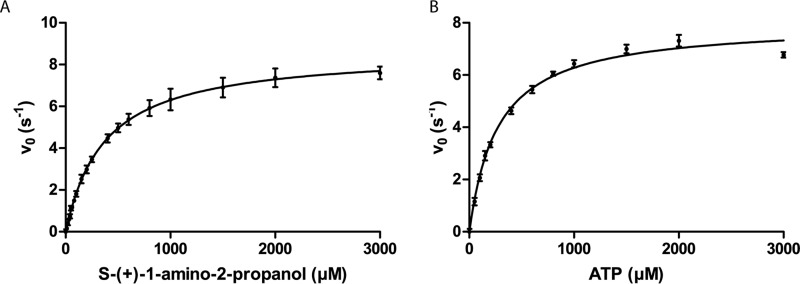
Kinetic parameters for APK were determined using a coupled assay. Briefly, ADP released by APK functioned as a limiting reagent for phosphoenolpyruvate conversion to pyruvate by pyruvate kinase; released pyruvate was reduced by lactate dehydrogenase at the expense of oxidizing NADH, and the resulting decrease in absorbance of NADH was monitored spectrophotometrically at 340 nm (Table 1). APKMSM0270 exhibits Michaelis-Menten kinetics against all substrates tested (Fig. 1). The best substrate proved to be (S)-aminopropanol, with a kcat of 8.7 s−1 and a Km of 0.373 mm. APKMSM0270 is enantioselective, but not absolutely so, with (R)-aminopropanol being phosphorylated with a 10-fold lower efficiency. (R,S)-1-Amino-2-butanol and ethanolamine were also both substrates, with kcat values similar to (S)-aminopropanol, although with Km roughly 3- and 600-fold higher, respectively. This suggests that APK may readily tolerate (slightly) larger substrates, but strongly discriminates against ethanolamine, which will commonly be present in cells for lipid biosynthesis, and whose diversion into degradation might be counterproductive.
Table 1.
Steady-state kinetic parameters for M. smegmatis MC2 155 APK variants
Reactions were assayed in 20 mm Tris, pH 8, and 150 mm NaCl at 25 °C. Errors for kinetic parameters are the standard deviation determined from nonlinear regression fitting of kinetic data as described in materials and methods. Nonlinear regression fittings had R2 values greater than 0.99.
| kcat | Km | kcat/Km | |
|---|---|---|---|
| s−1 | mm | m−1 s−1 | |
| APKMSM0270 WT | |||
| (S)-1-amino-2-propanol | 8.7 ± 0.05 | 0.373 ± 0.006 | 2.3 × 104 |
| (R)-1-amino-2-propanol | 3.2 ± 0.2 | 1.54 ± 0.15 | 2.1 × 103 |
| (R/S)-1-amino-2-butanol | 8.8 ± 0.2 | 1.11 ± 0.066 | 7.9 × 103 |
| Ethanolaminea | 8.1 ± 0.5 | 193 ± 13 | 4.2 × 101 |
| ATP | 8.0 ± 0.2 | 0.27 ± 0.02 | 3.0 × 104 |
| APKMSM0270 variantsb | |||
| E33Aa | 0.068 ± 0.003 | 26 ± 2 | 2.6 |
| R290Aa | 0.44 ± 0.07 | 72 ± 15 | 6.1 |
| D235A | NAc | NA | NA |
| R213A | 0.85 ± 0.01 | 2 ± 0.1 | 4.3 × 102 |
| APKMSM0780 Δ1–23 | |||
| S-(+)-1-Amino-2-propanol | 4.35 ± 0.03 | 0.069 ± 0.003 | 6.3 × 104 |
| ATP | 2.79 ± 0.02 | 0.074 ± 0.004 | 3.8 × 104 |
a Kinetic parameters are calculated by extrapolation from collected data due to inability to reach substrate saturation under assay conditions.
b Variant kinetic parameters are for S-(+)-1-amino-2-propanol with saturating concentrations of ATP.
c NA, not applicable.
Figure 1.
Enzyme kinetics. APKMSM0270 initial reaction rates were measured by changes in the NADH concentration due to ADP production coupled with pyruvate kinase/lactate dehydrogenase, and fit to the Michaelis-Menten equation by nonlinear regression. A, kinetics data for S-(+)-1-amino-2-propanol and B, kinetics data for ATP.
M. smegmatis also encodes a second APK homolog (APKMSM0780, YP_885185) from a nonmicrocompartment gene cluster, with sequence identity of 44% with APKMSM0270. The full sequence (as annotated by UniProt) proved insoluble, so the protein was recloned without the first 23 annotated amino acids, as alignment with APKMSM0270 suggested that the start methionine may be misannotated. APKMSM0780 was able to catalyze the phosphorylation of (S)-(+)-1-amino-2-propanol and is ∼3 times more efficient than its microcompartment-associated paralog due to a significantly lower Km (kcat of 4.35 s−1 and Km of 0.069 mm). This difference is intriguing, and possibly hints that the non-RMM pathway may preferentially operate when less substrate is available.
Structure of APKMSM0270
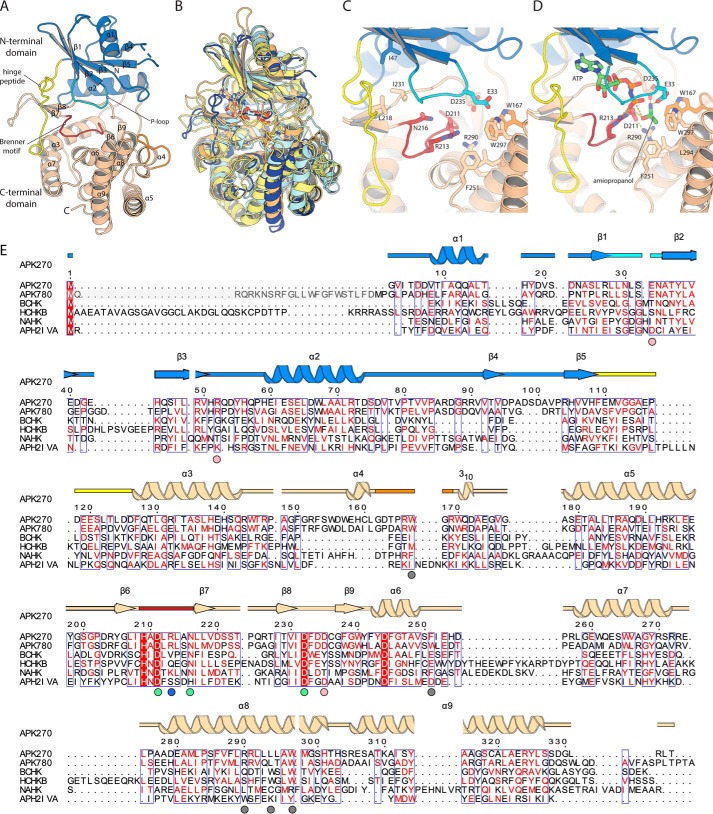
We determined the structure of APKMSM0270 using selenomethionine in space group C2, and then refined the structure against an isomorphous native dataset at 1.35 Å. A single protomer was found in the asymmetric unit, with most of the structure, excluding a short loop (97–101), being ordered. As predicted by the distant sequence homology, the fold of APK broadly resembles other atypical kinases of the aminoglycoside phosphotransferase family (APH; PF01636), with an N-terminal α/β-fold nucleotide-binding domain and a C-terminal, mostly α-helical substrate-binding domain (Fig. 2A). The N-terminal domain is built as a five-stranded anti-parallel β-sheet (topology 1,–2,3,–5,4) flanked by two α-helices. The C-terminal domain is built from seven helices (α3–α9), with four short β-strands (β6–β9) in two two-stranded antiparallel sheets, contributed by the long α5–α6 loop that packs into the cleft between the domains.
Figure 2.
Structure of APKMSM270. A, structure of the APKMSM0270 monomer. Secondary structure elements are labeled. B, superposition of APKMSM0270 with representative structural homologs. APKMSM0270 is shown in dark blue. N-Acetylhexosamine-1-phosphate kinase (PDB 4ocq) is shown in pale yellow. Choline kinase (PDB 4r78) in pale orange. Spectinomycin phosphotransferase (PDB 3i0o) is shown in pale cyan. C, details of the catalytic site of APKMSM0270. D, model of the ternary complex of APKMSM0270 with ATP and propanolamine. E, multiple sequence alignment of APK with structural homologs. The secondary structure is colored as described in A. APKMSM0270 is aligned with APKMSM0780 (greyed residues are present in UniProt but are likely a misannotation), S. pneumoniae bacterial choline kinase (BCHK, UniProt Q8DPI4), Homo sapiens choline kinase B (HCHKB, UniProt Q9Y259), B. longum N-acetylhexosamine 1-kinase (NAHK, UniProt E8MF12), and Enterococcus casseliflavus aminoglycoside phosphotransferase (2′)-Iva. The secondary structure and residue numbering of APKMSM0270 are aligned with the multiple sequence alignment. Colored circles below the multiple sequence alignment indicate residues of the hydrophobic methyl binding pocket (gray), substrate α-amino binding (pink), conserved Brenner motif (green), and the phosphate-product stabilizing residue (light blue).
Searching the entire PDB for structural homologs using DALI (19) retrieves 329 chains with Z-scores >10. The closest functionally characterized homologs are bacterial N-acetylhexosamine kinases (PDB code 4ocq), with Z = 22.2, an r.m.s. deviation of 3.4 Å, and 14% sequence identity (20) (Fig. 2B). High r.m.s. deviation values are typical for this enzyme family, and reflect inter-domain flexibility as well as considerable variability in the secondary structure complement of the C-terminal domain (14). Bacterial choline kinase (PDB code 4r78; Z = 20.1) and spectinomycin phosphotransferase (PDB code 3i0o, Z = 18.9) are similarly closely related. No retrieved homologs had r.m.s. deviation values <3.0 Å, or sequence identities >19%. APK is therefore a distant homolog of hexosamine kinases, choline kinases, and aminoglycoside phosphotransferase families, without being especially close to any individual enzyme group.
Active site modeling
APK's active site architecture resembles that of other APH family members, with three key conserved motifs that together form the ATP-binding site: the hinge loop (which connects the N- and C-terminal domains Phe109–Phe127, yellow in Fig. 2), the Brenner motif (H209XD211XXXXN216; dark red in Fig. 2), and the P-loop (Leu29–Asn34, cyan in Fig. 2) (21). ATP binding in APH enzymes is generally accompanied by major structural rearrangements including a 16° rotation of the N-terminal domain toward the C-terminal domain, and repositioning of the P-loop (as seen in comparing PDB code 4ock to PDB code 4ocp) (20). Comparisons to reported APH ATP analog costructures (including Bifidoacterium longum N-acetylhexosamine kinase (PDB code 4wh3), Streptococcus pneumonia choline kinase (PDB code 4r78), and Enterococcus faecalis 3′,5″-aminoglycoside phosphotransferase type IIIa (PDB code 1j7u)) indicates that in APK, the adenine ring likely binds with its nonpolar face stacked between Ile47, Leu218, Ile231, and with hydrogen bonds with the backbone groups contributed by the hinge loop. Conserved residues of the Brenner motif Asp211 and Asn216 along with Asp232 superpose closely with those used to coordinate the two magnesium ions that bridge the α-, β-, and γ-phosphates. Additional hydrogen bonds to ATP are generally contributed by backbone amide groups in the conserved phosphate-binding loop or P-loop. Arg213 in APK is less usual, and appears to be analogous to Lys210 in N-acetylhexosamine kinase lysine, which forms additional contacts with the γ-phosphate.
Although APK's substrate is very different from that of all well-studied APH family members, modeling of aminopropanol is simplified by the fact that this is a small, chemically simple molecule with few degrees of freedom. The position of the attacking hydroxyl group is also largely fixed by the position of the γ-phosphate, and the key catalytic residue Asp211 (from the Brenner motif), which activates this hydroxyl as a nucleophile (Fig. 2C). Arg290 is an additional charged residue conserved in the APK catalytic pocket. This residue is homologous to Arg284 in the active site of spectinomycin phosphotransferase (PDB code 3i0o), a residue that forms a hydrogen bond with the catalytic aspartate (Asp212) in the substrate complex, but whose role has not been specifically investigated by mutagenesis (22). In analogy with spectinomycin phosphotransferase, we suggest that this residue helps position the catalytic Asp211 in the reaction. Asp235 in APK is identically placed to Asp220 in aminoglycoside-2″-phosphotransferase-IVa (PDB code 3sg8 (23)), suggesting that the propanolamine amine forms a salt bridge to the carboxylate of this residue (as does Asp220 in PDB 3sg8 to tobramycin's 3′-N, a group analogously adjacent to the phosphorylation site 2′-OH); this residue may also help coordinate magnesium as seen in an unpublished aminoglycoside phosphotransferase structure (PDB code 4n57). An additional interaction with this amine group is possible with a second highly conserved acidic residue, Glu33, which is positioned close by in the apo structure (6 Å between the carboxylate groups), and is in a region of the structure that is typically rearranged upon substrate binding.
To test the roles of these possible catalytic residues, we generated and investigated the site-directed enzyme variations E33A, R213A, D235A, and R290A. All of these variants expressed well, were soluble, and showed cooperative unfolding in a fluorimetric thermal denaturation assay, confirming that they fold. E33A and D235A had a Tm very similar to the WT enzyme (53 °C), whereas the R213A and R290A had a Tm 7 and 10 °C lower, possibly indicating that these residues help stabilize the enzyme by neutralizing excess negative charge in the active site. All variants except D235A showed additional stabilization by 6–10 °C upon binding of Mg2+ and ATP; this suggests that Asp235 may be positioned to bridge the substrate amine and a magnesium ion. As suggested by our model, all of these variants resulted in significant losses in enzyme activity: the R213A variant has 2 orders of magnitude lower activity, the E33A and R290A variants have activity reduced by 4 orders of magnitude, and the D235A does not detectably turn over (Table 1).
A set of nonpolar residues (Trp167, Phe251, Leu294, and Trp297) contributed by the helical domain form a hydrophobic pocket is found immediately adjacent to this cluster of charged residues. Leu294 and Trp297 are found in successive turns of helix α8, whereas Phe251 is contributed by an extension of the α7–α8 loop. Trp167 is notable as this position is normally not positioned in the active site: in other APH enzymes, this residue forms part the long helix α4, whereas in APK this helix is broken into two short segments, with the center bulging out as a loop that positions Trp167 adjacent to Trp297 (orange in Fig. 2). Leu294 and Trp297 are the structural analogs of Trp251 and Trp254, which in the bacterial choline kinase LicA (PDB code 4r7b), interact with the methyl modifications of the betaine group of choline. We suggest that, in APK, this hydrophobic pocket is optimized to envelop the methyl group of the substrate. The importance of interactions with the methyl group is underscored by the very poor affinity (extrapolated Km of 190 mm) exhibited by ethanolamine.
Sequence and gene association analysis of APK homologs
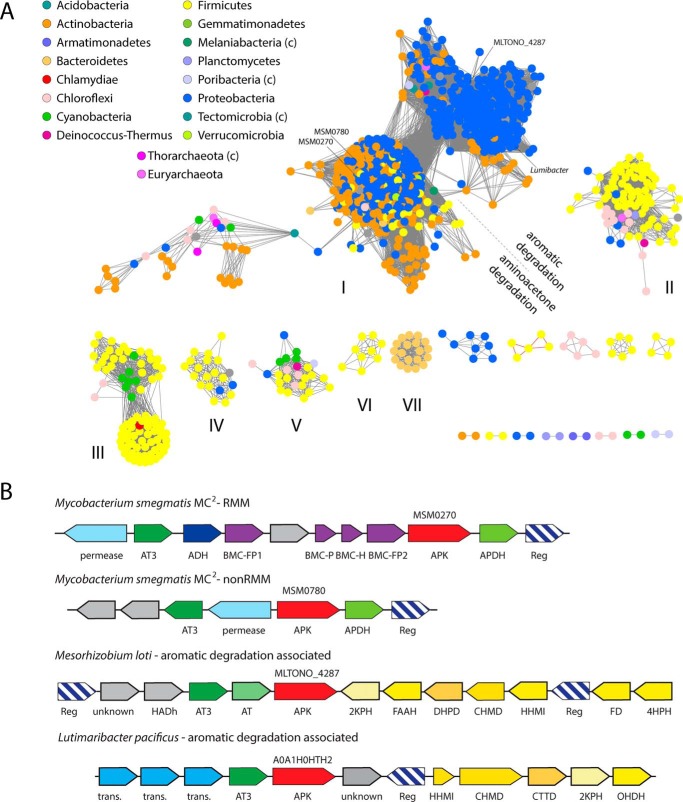
We used BLAST to search the UNIREF90 database (i.e. a subset of the full sequence databases with no pairs of sequences having >90% sequence identity) on the UniProt website with the APKMSM0270 sequence; sequences were accepted as being likely APK orthologs on the basis of conservation of amino acids Glu33, Trp167, Arg213, Phe251, Arg290, Leu294, and Trp297 (with three of these being experimentally verified as being functionally important residues). We found that a large group of proteins (>600) conserve all of these amino acids, in some cases with as little 34% overall sequence identity (e.g. UniProt number A0A1T5CUT6 from Sphingomonas; E value 2e-38, 34% sequence identity). Relaxing these criteria slightly, there is also a larger family of proteins that conserve most, but not all of these motifs (e.g. UniProt number A0A1H7ZUK6 from Gemmobacter aquatilis has an E value of 1e-20, and 31% sequence identity and conserves all of the identified residues except Phe251(Gln) and Leu294(Val). These proteins, whereas possibly not authentic APK orthologs, are likely to be significantly more similar to APK than any other functionally characterized enzyme. APK-like proteins were most abundant in Proteobacteria, Firmicutes, and Actinobacteria, with significant numbers also in Chloroflexi, Cyanobacteria, and Bacteroidetes, and a few scattered representatives also found in another dozen bacterial phyla, and two archaeal phyla. APK is thus a widespread and common enzyme, with high prevalence in three important bacterial phyla.
We clustered these sequences (selected using the more permissive 1e-20 cutoff) in EFI-EST using e-50 as an edge cutoff and displaying the results in cytoscape (Fig. 3A). This classification places most of the Actinobacterial and Proteobacterial sequences in a single large cluster, along with a subset of the Firmicute sequences and other sequences. This cluster is bilobed, and separates into two large groups (with some smaller clusters) at stringencies <e-60. We examined the association of genes within these clusters using EFI-GNT. In cluster I, a very tight associated cluster is found between APK and an aminotransferase III gene. This gene is found within 10 genes in 72% of 1271 organisms; inspection of individual organisms shows that both proteins are in a single operon, and are generally separated by no more than one ORF. Together this implies a tight functional association. Other proteins strongly co-occurring in this cluster include a short chain alcohol dehydrogenase (36%), an aldehyde dehydrogenase (27%), an amino acid permease (46%), and various proteins associated with the degradation of catechol-like molecules, such as isochorismatase (11%). Looking at how genes cluster in individual genomes reveals two distinct patterns underpinning the association between the APK and aminotransferase. In the left lobe of cluster I, with Actinobacterial strains dominant, APK, the aminotransferase, and the permease associate with an APDH homolog to form an operon, often with additional proteins. A subset (about 10% of cluster I) of these operons also encode BMC shell proteins and an aldehyde dehydrogenase to form an RMM (Fig. 3B). The microcompartment operon is found primarily in Actinobacteria (although we also noted examples here in Alphaproteobacteria and Bacilli, e.g. Bacillus azotoformans UniProt MEV2011). As argued below, we strongly suspect that both of these operons encode a pathway capable of incorporating aminoacetone into central metabolism as propionyl-CoA.
Figure 3.
Bioinformatics analysis of APK homologs. A, sequence similarity network of APK homologs. Sequences are connected by edges if E < e-50. Sequences are colored by phyla, as noted in the key. Phyla marked (c) are candidate phyla. APK homologs highlighted in B are indicated. B, gene organization of representative examples of APK homologs. Gene labels: BMC, bacterial microcompartment shell proteins; AT3, aminotransferase III; trans., transporter; 4HPH, 4-hydroxyphenylacetate 3-hydroxylase; FD, flavin reductase; HHMI, 5-carboxymethyl-2-hydroxymuconatedelta-isomerase; CHMD, 5-carboxy-2-hydroxymuconate semialdehyde dehydrogenase; DHPD, 3,4-dihydroxyphenylacetate 2,3-dioxygenase; FAAH, fumarylacetoacetate hydrolase; 2KPH, 2-keto-4-pentenoate hydratase; OHDH, 2-oxo-hept-3-ene-1,7-dioate hydratase; CTTD, catechol 2,3-dioxygenase.
For APK orthologs in the right lobe of Cluster I (mostly Rhizobia and Actinobacteria), APK is associated with aminotransferase III in an operon that is adjacent to (although often divergently transcribed from) an operon encoding homologs of aromatic degradation pathway enzymes (24). These operons generally contain a 3,4-dihydroxyphenylacetate 2,3-dioxygenase and a 5-carboxy-2-hydroxymuconate semialdehyde dehydrogenase homolog; these two enzymes open the aromatic ring by oxygenation and then oxidize the resulting semialdehyde. Additional enzymes associated with preparatory steps (e.g. hydroxylating the ring) or further process of the resulting partially saturated hydrocarbon products into common metabolic intermediates (including isomerases, dehydratases, reductases, aldolases, and hydrolases) are also found associated. We propose that these pathways degrade a variety of phenyl derivatives with differing ring substituents; the aminotransferase and APK may be required to incorporate a 2-amino or 2-keto (these pathways commonly have an additional aminotransferase associated) product back into central metabolism.
Outside of the two main lobes of the largest cluster, gene associations are weaker, possibly implying a role in a variety of minor pathways. Acetyltransferases, amidohydrolases, glyoxalases, and HAD-like hydrolases are among the enzymes more commonly associated with APK homologs, but no individual enzyme seems associated in more than 10–20% of organisms. Hindering analysis is the fact that many of the associated proteins are poorly characterized. However, there are hints that APK plays a role in synthetic pathways. For example, APK is associated with an exopolysaccharide operon in Streptomyces sp. FBKL.4005, implying that aminopropanol may be incorporated into polysaccharides and other complex macromolecules.
Discussion
(S)-1-Amino-2-propanol O-kinase, as characterized here, represents a new group of atypical kinases within the APH group, related to hexosamine kinases, choline kinase, and aminoglycoside phosphotransferases. The conservation of key signature motifs within the substrate-binding site in a very diverse set of bacterial genomes argues that this enzyme family is widespread and important. The fairly weak substrate specificity APKMSM0270 displays, coupled with the occurrence of amino acid variants in the active site, argue that family members are likely involved in phosphorylating a variety of substrates, although most likely 1-amino-2-ols. In general, these enzymes appear to mostly occur in catabolic pathways, where after various preparatory steps, an amino alcohol is converted to an aldehyde by the sequential actions of a kinase and a phospho-lyase (see below). Another very common occurrence of this enzyme pair in Rhizobobacteria, where they co-occur with protocatechuate 2,3-like oxidizing catabolic pathways, is certainly worthy of further investigation. Some rarer homologs of APK are also associated with what appear to be biosynthetic pathways, for example, ones for lipid or polysaccharide biosynthesis. Of note is that aminopropanol is commonly found as a modification on cobalmin (although cobalmin is generally synthesized without APK involvement), and there is no reason to suspect that bacteria have not found other uses for 1-amino 2-propanol-O-phosphate.
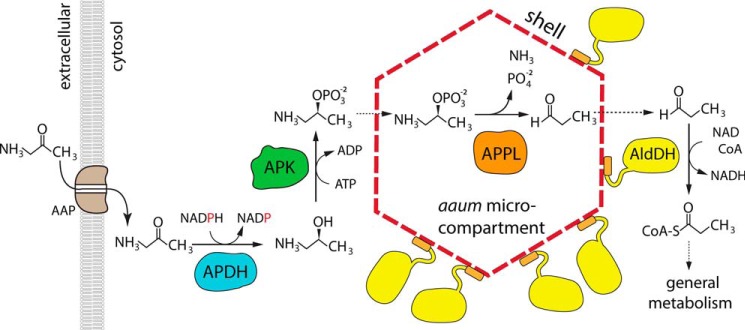
Our original interest in this enzyme arose from its association with the RMM microcompartment; this microcompartment is induced in Rhodococcus jostii by the addition of aminopropanol in the media (12), suggesting a possible role in aminoacetone (which commonly arises from an uncatalyzed reaction in threonine/glycine pathway) catabolism. Our previous work established that the short chain dehydrogenase is a highly specific (S)-1-amino-2-propanol dehydrogenase (13), whereas among the other three enzymes present in the operon, the aldehyde dehydrogenase has clear homology to CoA-dependent propionaldehyde dehydrogenase. The outstanding question remained how an APH-like enzyme and a class III aminotransferase could convert aminopropanol (or aminoacetone) to propionaldehyde. The demonstration that MSM0270 is an aminopropanol kinase allows the problem to be narrowed down to one of converting 1-amino-2-propanol phosphate to propionaldehyde, a reaction catalyzed by phospho-lyases. The only well characterized example is ethanolamine-phosphate phospho-lyase (EC 4.2.3.2.) (25, 26), a homolog of class III aminotransferases. Strongly supporting this idea, propanolamine phosphate phospho-lyase activity was found associated with APDH, APK, and propionaldehyde dehydrogenase activities in enriched Pseudomonas or Erwinia extracts by Turner and colleagues (17, 18) in the 70's. We therefore propose that the RMM microcompartment utilizes aminoacetone using the pathway shown in Fig. 4, and that this microcompartment should therefore be renamed the aminoacetone utilization microcompartment, or aaum. Of note, a second APK paralog (MSM0780) in M. smegmatis is found in an operon which parallels the enzymes of the aaum operon, but lacks the shell proteins; this version of the aminoacetone degradation pathway is phylogenetically more widespread than the aaum. This is, to our knowledge, the first instance where a catabolic pathway occurs in both a microcompartment-dependent and microcompartment-independent variant; in addition, the common co-occurrence of these two pathways in a single organism possibly suggests that these two pathway variants are likely optimized for distinct environmental conditions.
Figure 4.
Proposed organization of, and pathway mediated by, the aminoacetone utilization microcompartment. Aminoacetone is imported into the cell via a aminopropanol permease (AAP). It is then reduced to (S)-aminopropanol by APDH, which is in turn phosphorylated APK. Both of these reactions occur in the cytosol. Aminopropanol-phosphate then enters the microcompartment through the shell, and is transformed into propionaldehyde by (S)-1-amino-2-propanol-phosphate phospho-lyase (APPL). Propionaldehyde escapes through the shell, and is captured by propionaldehyde dehydrogenase (AldDH), which adheres to the exterior face of the microcompartment via its encapsulation peptide.
Bacterial microcompartments are thought to be organized around a signature enzyme, which generates an aldehyde, and an aldehyde dehydrogenase that consumes it. These are proposed to be always encapsulated together within a shell whose pores are likely evolved to minimize aldehyde escape (2). These pores generally do not passage large cofactors in stoichiometric quantities, so additional enzymes that regenerate cofactors, termed the enzyme core, need to be coencapsulated (2, 15). In short, every encapsulated enzyme that requires a cofactor must be paired with a second enzyme that regenerates that cofactor. In the aaum, there are three enzymes that together require four different coenzymes or cosubstrates: APDH requires NADPH (13), APK requires ATP, and the aldehyde dehydrogenase has all of the sequence hallmarks of a CoA and NAD+-dependent aldehyde dehydrogenase (including the Pro-His-Pro motif, which prevents NADP+ binding, and His387, which acts as a general base in the acylation reaction (27)). Note that in addition to the aldehyde dehydrogenase having cofactor requirements incompatible with APDH, aaum operons do not contain a phosphotransacylase gene, so the enzyme that in other microcompartments regenerates free CoA by transferring propionate to a phosphate group is missing (16). This strongly suggests that the aaum must either passage stoichiometric quantities of at least two cofactors (NAD+ and CoA) or localize all enzymes except the phospho-lyase (which regenerates its own cofactor) outside the shell. We propose that only the phospho-lyase is encapsulated, whereas the propionaldehyde dehydrogenase is cytosolic, but anchored on the exterior wall of the shell. The C terminus of this aldehyde dehydrogenase contains a conserved encapsulation peptide (28, 29) of the type known to bind the concave surface of BMC-H proteins (such as MSM0272) (30); however, the concave surface of a very closely homologous BMC-H was recently shown to localize to the cytoplasmic face of the shell, which presumably then targets the attached proteins to this face (9). This suggests an alternative model for microcompartment function, where only the “signature enzyme” that generates the aldehyde is encapsulated, whereas problematic cofactor recycling requirements are avoided by localizing the aldehyde dehydrogenase to the immediate exterior surface of the shell, where it can maximize capture of escaping aldehyde. This arrangement, and the existence of a parallel aminoacetone degradation pathway that does not depend on building a microcompartment, suggests a possible model for a “primitive,” less specialized microcompartment where encapsulation is nonessential but acts to more efficiently channel aldehydes to the aldehyde dehydrogenase by colocalizing these two key enzymes within a small volume in the cytosol.
Experimental procedures
Molecular biology
M. smegmatis MC2 155 was grown in 5 ml of LB (50 μg/ml of carbenicillin, 10 μg/ml of cycloheximide) over 5 days at 37 °C with shaking; genomic DNA was isolated by miniprep (Invitrogen PureLink). Genes encoding msmeg_0270 (UniProt number A0A0D6G1D9) and msmeg_0780 (UniProt number A0QQJ7) were amplified by PCR using PfuX7 polymerase (a generous gift from Dr. Dinesh Christendat, University of Toronto) for cloning into plasmids pET28a to include an N-terminal hexahistidine affinity tag (Table 2). Plasmids and amplified inserts were prepared by restriction digest, and ligated together after DNA purification. Ligated constructs were then transformed into DH5α cells for propagation. Plasmids suspected to contain the gene of interest were sequenced at the Advanced Analysis Center University of Guelph. Site-directed mutagenesis was performed using PCR with overlapping primers to generate the desired nucleotide substitutions and verified by sequencing (Table 2). BL21(DE3) or B834(DE3) (methionine-auxotrophic genotype for selenomethionine labeling) cells were transformed with plasmid containing the gene of interest for overexpression.
Table 2.
Cloning and mutagenesis primer sequences
| Cloning and primer sequences (5′-3′) | |
|---|---|
| Cloning primers | |
| APK270pet28aFWD | ATACAGCTCGAGTCAGGTCAGGCGAAGTCCGTCGGAGG |
| APK270pet28aREV | TTAGATCATATGGGCGTGATCACCGACGATGTCACCATC |
| APK780Δ1–23pet28aFWD | CGAAGCTAGCATGCCGGGACTTCCAGCTGACCACGAACTG |
| APK780Δ1–23pet28aREV | GCTAAAGCTTCTATGCTGTCGGGGTGAGGGGTGACGCAAA |
| Mutagenesis primers | |
| E33AFWD | AACCTGTCCGCGAACGCGACC |
| E33AREV | GGTCGCGTTCGCGGACAGGTT |
| D235AFWD | GATTTCGATGCCTGCGGATTC |
| D235AREV | GAATCCGCAGGCATCGAAATC |
| R213AFWD | GCCGACCTGGCGTTGGCCAAC |
| R213AREV | GTTGGCCAACGCCAGGTCGGC |
| R290AFWD | GTGTTCCTGGCCAGGCTCTTG |
| R290AREV | CAAGAGCCTGGCCAGGAACAC |
Protein expression and purification
BL21 cells were grown overnight in 5 ml of LB at 37 °C and used to inoculate 1 liter of 2YT media. Once an optical density of 0.8 at 600 nm was reached, cultures were induced for expression with 1 mm isopropyl β-d-1-thiogalactopyranoside and incubated for 18–20 h at 16 °C. Cells were pelleted by centrifugation at 4,400 × g and resuspended in 35 ml of lysis buffer (20 mm Tris, pH 8, 500 mm NaCl) prior to freezing at −20 °C for storage. Selenomethionine-labeled protein was expressed in B834 cells by an analogous protocol, except cells were pelleted once the culture reached an optical density of 0.6 at 600 nm and resuspended in selenomethionine media (prepared according to guidelines from the EMBL Heidelberg Protein Expression and Purification Core Facility https://www.embl.de/pepcore/pepcore_services/protein_expression/ecoli/seleno/)3 prior to induction with isopropyl β-d-1-thiogalactopyranoside.
Harvested cells were disrupted by sonication with a Sonicator XL2020 at a power level of 7 with 10-s on/20-s off pulses for 10 min of processing time. The insoluble fraction of the lysate was separated by centrifugation at 48,000 × g for 30 min at 4 °C. Lysate supernatant was loaded onto a 2 ml of Ni-IMAC column, and unbound proteins were eluted with 20 ml of lysis buffer. Partially purified protein was eluted with 10 ml of lysis buffer with added 500 mm imidazole. The buffer was exchanged by desalting the protein on a HiPrep 26/10 column equilibrated with 10 mm Tris, pH 8. Pure protein was isolated by anion exchange on a HiTrap Mono Q column with a gradient of 0 to 1 m NaCl (with 10 mm Tris, pH 8). Aliquots of pure target protein, as indicated by SDS-PAGE, were pooled and exchanged into 20 mm Tris, pH 8, 150 mm NaCl by desalting before being concentrated by centrifugal concentration and quantified spectrophotometrically by absorbance at 280 nm.
Protein crystallization
N-terminally hexahistidine-tagged protein was crystallized using serial seeding in a sitting drop configuration. One microliter of protein solution (30 mg/ml protein in 20 mm Tris, pH 8.0, 150 mm NaCl) was mixed with 1 μl of precipitating solution (1.1 m sodium malonate, pH 7, 0.1 m HEPES, pH 7, 1% (v/v) Jeffamine ED-2001 pH 7), and was then equilibrated against 50 μl of precipitating solution for 24 h. At this time, 0.2 μl of 1/1000 diluted crushed crystals were added in precipitating solution. Crystals grew as large, single monoclinic prisms and were cryoprotected using a 2 m sodium malonate soak followed by immersion in paratone N oil prior to freezing in liquid nitrogen.
Data collection and structure determination
Datasets were collected at the Canadian Light Source beamline 08ID at 100 K, and were processed using XDS and scaled using XSCALE (31). Data collected at the anomalous peak from a selenomethionine-derivatized crystal diffracted to 1.6 Å and was phased by single wavelength anomalous diffraction using AutoSol in Phenix (32). All four encoded methionine residues (after post-translational methionine trimming) were found as anomalous peaks, producing a phase solution with a 0.276 figure of merit. Subsequent automated model building by Phenix.autobuild was able to trace most of the structure. After manual rebuilding in Coot (33) and automated refinement by Phenix.refine, this model was used to phase an isomorphous 1.35 Å native dataset by molecular replacement using Phaser in Phenix (34), manual rebuilding in Coot, and automated refinement by Phenix.refine. In the final structure, residues 7–334 are traced in the map, except residues 99–101, which are too disordered to reliably trace. The structure contains additional density in the nucleotide-binding site that appears to be benzoate (source unknown) and catalytic site density that was tentatively identified as a terminal group of Jeffamine ED-2001. Data collection and structure refinement statistics are presented in Table 3. Structure figures were prepared in PyMOL version 2.0.
Table 3.
Data collection and refinement statistics
| APK270 Selmet | APK270 native | |
|---|---|---|
| RCSB ID | 6EF6 | |
| Data collectiona | ||
| Wavelength (Å) | 0.97921 | 0.97949 |
| Space group | C2 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 72.18, 70.19, 60.83 | 72.01, 70.05, 60.72 |
| α, β, γ (°) | 90.00, 95.36, 90.000 | 90.00, 95.12, 90.00 |
| Resolution (Å) | 50–1.60 (1.64–1.60 | 50–1.35 (1.39–1.35) |
| Rsym | 0.061 (0.55) | 0.058 (0.52) |
| I/σI | 16.7 (2.57) | 11.0 (2.04) |
| Completeness (%) | 99.9 (99.9) | 99.6 (98.4) |
| Redundancy | 3.8 (3.7) | 3.3 (2.8) |
| Refinement | ||
| Resolution (Å) | 50–1.35 (1.39–1.35) | |
| No. reflections | 65,635 | |
| Rwork/Rfree (%) | 14.9/16.8 | |
| No. atoms | ||
| Protein | 2638 | |
| Ligand/ion | 21 | |
| Water | 231 | |
| B-factors | ||
| Protein | 28.5 | |
| Ligand/ion | 42.9 | |
| Water | 34.5 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.011 | |
| Bond angles (°) | 1.16 | |
| Ramachandran plot | ||
| Favored (%) | 96.3 | |
| Allowed (%) | 99.7 | |
a The dataset was collected from a single crystal. Values in parentheses are for the highest-resolution shell.
Enzyme kinetics
Enzyme kinetic constants were determined by monitoring the ADP released by APK using a coupled pyruvate kinase/lactate dehydrogenase enzyme pair. Experiments were conducted on a MultiSkan FC plate reader at 340 nm at 25 °C. Data were collected for 8 replicates of 300-μl volumes containing 200 μm NADH, 2.5 mm phosphoenolpyruvate, 50 mm KCl, 10 mm MgCl2, 2 μl/ml of pyruvate kinase/lactate dehydrogenase solution (P0294 Sigma), and an APK concentration that was adjusted to optimize the signal (2–5 μg/ml of WT enzyme or 20–100 μg/ml of variant enzyme) with one substrate (alcohol substrate or ATP) held at 1 mm and the other varied over a range of concentrations. Kinetic constants were calculated by nonlinear regression in SigmaPlot fitting the data to the Michaelis-Menten equation.
Thermal denaturation
Structural stability of protein variants were tested by the thermofluor method using an Applied Biosystems StepOnePlus thermocycler real-time PCR system. 50-μl Reaction aliquots contained with 0.2 mg/ml of protein (or 0.2 mg/ml of protein plus 2 mm MgCl2 and 2 mm ATP) in 20 mm Tris, pH 8.0, 150 mm NaCl, and 1× SYPRO orange dye. Solution temperatures were increased in 1 °C increments and fluorescence was monitored at 570 nm. Each condition was evaluated in triplicate and melting temperatures are reported as the average temperature to denature half of the protein in the reaction.
Bioinformatics analysis
The sequence of APKMSM0270 was searched using EXPASY BLAST against the UniProt90 database. Sequence similarity networks were generated using EFI-EST, and a gene neighborhood network was generated using EFI-GNT (35). The resulting data relationships were visualized using cytoscape (36).
Author contributions
E. M. and M. S. K. formal analysis; E. M. investigation; E. M. methodology; E. M. writing-original draft; E. M. and M. S. K. writing-review and editing; M. S. K. conceptualization; M. S. K. supervision; M. S. K. funding acquisition; E. M. and M. S. K. visualization.
Acknowledgments
We thank P. Grochulski and S. Labiuk at Canadian Light Source for assistance in collecting diffraction data.
This work was supported by National Science and Engineering Council of Canada Grant 04045–2015 (to M. S. K.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 6EF6) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- RMM
- Rhodococcus and Mycobacterium microcompartment
- aaum
- aminoacetone utilization microcompartment
- APDH
- (S)-1-amino-2-propanol dehydrogenase
- APK
- (S)-1-amino-2-propanol kinase
- APKMSM0270
- the APK encoded by the RMM operon in M. smegmatis
- APKMSM0780
- the APK encoded by the non-RMM operon in M. smegmatis
- APH
- aminoglycoside phosphotransferase family
- PDB
- Protein Data Bank
- r.m.s.
- root mean square.
References
- 1. Chowdhury C., Sinha S., Chun S., Yeates T. O., and Bobik T. A. (2014) Diverse bacterial microcompartment organelles. Microbiol. Mol. Biol. Rev. 78, 438–468 10.1128/MMBR.00009-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Axen S. D., Erbilgin O., and Kerfeld C. A. (2014) A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comp. Biol. 10, e1003898 10.1371/journal.pcbi.1003898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jorda J., Lopez D., Wheatley N. M., and Yeates T. O. (2013) Using comparative genomics to uncover new kinds of protein-based metabolic organelles in bacteria. Protein Sci. 22, 179–195 10.1002/pro.2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zarzycki J., Erbilgin O., and Kerfeld C. A. (2015) Bioinformatic characterization of glycyl radical enzyme-associated bacterial microcompartments. Appl. Environ. Microbiol. 81, 8315–8329 10.1128/AEM.02587-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Craciun S., and Balskus E. P. (2012) Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl. Acad. Sci. U.S.A. 109, 21307–21312 10.1073/pnas.1215689109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Espie G. S., and Kimber M. S. (2011) Carboxysomes: cyanobacterial RubisCO comes in small packages. Photosyn. Res. 109, 7–20 10.1007/s11120-011-9656-y [DOI] [PubMed] [Google Scholar]
- 7. Kerfeld C. A., and Melnicki M. R. (2016) Assembly, function and evolution of cyanobacterial carboxysomes. Curr. Opin. Plant Biol. 31, 66–75 10.1016/j.pbi.2016.03.009 [DOI] [PubMed] [Google Scholar]
- 8. Yeates T. O., Jorda J., and Bobik T. A. (2013) The shells of BMC-type microcompartment organelles in bacteria. J. Mol. Microbiol. Biotechnol. 23, 290–299 10.1159/000351347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sutter M., Greber B., Aussignargues C., and Kerfeld C. A. (2017) Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell. Science 356, 1293–1297 10.1126/science.aan3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Urano N., Kataoka M., Ishige T., Kita S., Sakamoto K., and Shimizu S. (2011) Genetic analysis around aminoalcohol dehydrogenase gene of Rhodococcus erythropolis MAK154: a putative GntR transcription factor in transcriptional regulation. Appl. Microbiol. Biotechnol. 89, 739–746 [DOI] [PubMed] [Google Scholar]
- 11. Mallette E., and Kimber M. S. (2017) A complete structural inventory of the mycobacterial microcompartment shell proteins constrains models of global architecture and transport. J. Biol. Chem. 292, 1197–1210 10.1074/jbc.M116.754093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kataoka M., Nakamura Y., Urano N., Ishige T., Shi G., Kita S., Sakamoto K., and Shimizu S. (2006) A novel NADP+-dependent l-1-amino-2-propanol dehydrogenase from Rhodococcus erythropolis MAK154: a promising enzyme for the production of double chiral aminoalcohols. Lett. Appl. Microbiol. 43, 430–435 10.1111/j.1472-765X.2006.01970.x [DOI] [PubMed] [Google Scholar]
- 13. Mallette E., and Kimber M. S. (2018) Structure and kinetics of the S-(+)-1-amino-2-propanol dehydrogenase from the RMM microcompartment of Mycobacterium smegmatis. Biochemistry 57, 3780–3789 10.1021/acs.biochem.8b00464 [DOI] [PubMed] [Google Scholar]
- 14. Scheeff E. D., and Bourne P. E. (2005) Structural evolution of the protein kinase–like superfamily. PLoS Comp. Biol. 1, e49–e23 10.1371/journal.pcbi.0010049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huseby D. L., and Roth J. R. (2013) Evidence that a metabolic microcompartment contains and recycles private cofactor pools. J. Bacteriol. 195, 2864–2879 10.1128/JB.02179-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Erbilgin O., Sutter M., and Kerfeld C. A. (2016) The structural basis of coenzyme A recycling in a bacterial organelle. PLos Biol. 14, e1002399 10.1371/journal.pbio.1002399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Faulkner A., and Turner J. M. (1974) Microbial metabolism of amino alcohols: aminoacetone metabolism via 1-aminopropan-2-ol in Pseudomonas sp. N.C.I.B. 8858. Biochem. J. 138, 263–276 10.1042/bj1380263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jones A., Faulkner A., and Turner J. M. (1973) Microbial metabolism of amino alcohols: metabolism of ethanolamine and 1-aminopropan-2-ol in species of Erwinia and the roles of amino alcohol kinase and amino alcohol o-phosphate phospho-lyase in aldehyde formation. Biochem. J. 134, 959–968 10.1042/bj1340959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang K. C., Lyu S. Y., Liu Y. C., Chang C. Y., Wu C. J., and Li T. L. (2014) Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes. Acta Crystallogr. D Biol. Crystallogr. 70, 1401–1410 10.1107/S1399004714004209 [DOI] [PubMed] [Google Scholar]
- 21. Brenner S. (1987) Phosphotransferase sequence homology. Nature 329, 21–21 10.1038/329021a0 [DOI] [PubMed] [Google Scholar]
- 22. Fong D. H., Lemke C. T., Hwang J., Xiong B., and Berghuis A. M. (2010) Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila. J. Biol. Chem. 285, 9545–9555 10.1074/jbc.M109.038364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shi K., Houston D. R., and Berghuis A. M. (2011) Crystal structures of antibiotic-bound complexes of aminoglycoside 2″-phosphotransferase IVa highlight the diversity in substrate binding modes among aminoglycoside kinases. Biochemistry 50, 6237–6244 10.1021/bi200747f [DOI] [PubMed] [Google Scholar]
- 24. Fuchs G., Boll M., and Heider J. (2011) Microbial degradation of aromatic compounds: from one strategy to four. Nat. Rev. Microbiol. 9, 803–816 10.1038/nrmicro2652 [DOI] [PubMed] [Google Scholar]
- 25. Cuetos A., Steffen-Munsberg F., Mangas Sanchez J., Frese A., Bornscheuer U. T., Höhne M., and Grogan G. (2016) Structural basis for phospholyase activity of a class III transaminase homologue. ChemBioChem. 17, 2308–2311 10.1002/cbic.201600482 [DOI] [PubMed] [Google Scholar]
- 26. Schiroli D., Ronda L., and Peracchi A. (2015) Kinetic characterization of the human O-phosphoethanolamine phospho-lyase reveals unconventional features of this specialized pyridoxal phosphate-dependent lyase. FEBS J. 282, 183–199 [DOI] [PubMed] [Google Scholar]
- 27. Tuck L. R., Altenbach K., Ang T. F., Crawshaw A. D., Campopiano D. J., Clarke D. J., and Marles-Wright J. (2016) Insight into coenzyme A cofactor binding and the mechanism of acyl-transfer in an acylating aldehyde dehydrogenase from Clostridium phytofermentans. Sci. Rep. 6, 22108 10.1038/srep22108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fan C., Cheng S., Liu Y., Escobar C. M., Crowley C. S., Jefferson R. E., Yeates T. O., and Bobik T. A. (2010) Short N-terminal sequences package proteins into bacterial microcompartments. Proc. Natl. Acad. Sci. U.S.A. 107, 7509–7514 10.1073/pnas.0913199107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fan C., and Bobik T. A. (2011) The N-terminal region of the medium subunit (PduD) packages adenosylcobalamin-dependent diol dehydratase (PduCDE) into the Pdu microcompartment. J. Bacteriol. 193, 5623–5628 10.1128/JB.05661-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fan C., Cheng S., Sinha S., and Bobik T. A. (2012) Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments. Proc. Natl. Acad. Sci. U.S.A. 109, 14995–15000 10.1073/pnas.1207516109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 12 10.1107/S09074449090473375–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adams P. D., Grosse-Kunstleve R. W., Hung L.-W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., and Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 10.1107/S0907444902016657 [DOI] [PubMed] [Google Scholar]
- 33. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 34. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gerlt J. A., Bouvier J. T., Davidson D. B., Imker H. J., Sadkhin B., Slater D. R., and Whalen K. L. (2015) Enzyme function initiative-enzyme similarity tool (EFI-EST): a web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 1854, 1019–1037 10.1016/j.bbapap.2015.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]