Abstract
The potassium voltage-gated channel subfamily H member 2 (KCNH2) gene encodes the Kv11.1 potassium channel, which conducts the rapidly activating delayed rectifier current in the heart. KCNH2 pre-mRNA undergoes alternative polyadenylation and forms a functional, full-length Kv11.1a isoform if exon 15 is polyadenylated or a nonfunctional, C-terminally truncated Kv11.1a-USO isoform if intron 9 is polyadenylated. The molecular mechanisms that regulate Kv11.1 isoform expression are poorly understood. In this study, using HEK293 cells and reporter gene expression, pulldown assays, and RNase protection assays, we identified the RNA-binding proteins Hu antigen R (HuR) and Hu antigen D (HuD) as regulators of Kv11.1 isoform expression. We show that HuR and HuD inhibit activity at the intron 9 polyadenylation site. When co-expressed with the KCNH2 gene, HuR and HuD increased levels of the Kv11.1a isoform and decreased the Kv11.1a-USO isoform in the RNase protection assays and immunoblot analyses. In patch clamp experiments, HuR and HuD significantly increased the Kv11.1 current. siRNA-mediated knockdown of HuR protein decreased levels of the Kv11.1a isoform and increased those of the Kv11.1a-USO isoform. Our findings suggest that the relative expression levels of Kv11.1 C-terminal isoforms are regulated by the RNA-binding HuR and HuD proteins.
Keywords: potassium channel, ELAV-like protein 1 (HuR (human antigen R)), polyadenylation, RNA binding protein, RNA interference (RNAi), alternative polyadenylation, hERG, KCNH2, Kv11.1, Hu proteins
Introduction
KCNH2 or human ether-a-go-go-related gene 1 (hERG1) encodes the Kv11.1 voltage-gated potassium channel that conducts the rapidly activating delayed rectifier K+ current (IKr) in the heart (1–4). Kv11.1 channels contribute to the repolarization of cardiac action potential, and mutations in KCNH2 cause long QT syndrome type 2 (LQT2)2 (5). KCNH2 expresses several Kv11.1 isoforms, including Kv11.1a, Kv11.1b, Kv11.1a-USO, and Kv11.1b-USO (6). The Kv11.1a isoform represents the full-length Kv11.1 channel consisting of 1159 amino acids. Kv11.1b lacks the first 376 amino acids of Kv11.1a and has an alternate 36 amino acid N terminus. The C-terminal isoforms Kv11.1a-USO and Kv11.1b-USO contain the truncated USO C terminus, in which the last 359 amino acids of Kv11.1a/b are replaced by an alternate 88 residue C-terminal end. Functional studies have shown that Kv11.1a and Kv11.1b isoforms generate Kv11.1 currents with distinct gating properties (2–4, 7, 8), whereas Kv11.1a-USO and Kv11.1b-USO isoforms fail to form functional channels when expressed in mammalian cells (9–12). The relative expression of Kv11.1 isoforms is regulated in a tissue-specific manner (11). In the heart, two-thirds of KCNH2 pre-mRNA are processed to the nonfunctional Kv11.1a-USO isoform, whereas in the brain, the levels of Kv11.1a and Kv11.1a-USO are similar (9, 11). The importance of C-terminal Kv11.1 isoform expression is underscored by our recent finding that the LQT2-causing KCNH2 mutation IVS9–2delA leads to a switch in the expression of Kv11.1 isoforms from the functional Kv11.1a to the nonfunctional Kv11.1a-USO (13). Thus, the relative expression of Kv11.1a and Kv11.1a-USO isoforms plays an important role in the regulation of Kv11.1 channel function and the pathogenesis of LQT2.
The C-terminal Kv11.1 isoforms are generated by alternative polyadenylation of KCNH2 intron 9 (11). The full-length Kv11.1a isoform is produced by the splicing of intron 9 and use of a distal poly(A) site in exon 15, whereas the truncated Kv11.1a-USO isoform is generated by the activation of a proximal poly(A) site within intron 9. Alternative polyadenylation of KCNH2 pre-mRNA represents a novel posttranscriptional mechanism that regulates Kv11.1 isoform expression and channel function. Despite extensive studies of Kv11.1 channel function, regulation of Kv11.1 isoform expression by alternative polyadenylation is an unexplored area of Kv11.1 channel research.
Recent high-throughput sequencing studies reveal that 60–70% of human genes undergo alternative polyadenylation, leading to the generation of alternative mRNA transcripts with different coding sequences or variable lengths of 3′-untranslated regions (3′-UTRs) (14). We have previously shown that activity of the KCNH2 intron 9 poly(A) site plays an important role in relative expression of Kv11.1 isoforms. Elimination of the intron 9 poly(A) site results in predominant expression of Kv11.1a and an increase in channel current (11). Thus, factors that modulate polyadenylation activity may lead to the regulation of Kv11.1 isoform expression and channel function. Several RNA-binding proteins have been shown to enhance or inhibit polyadenylation (15–17). One example is Hu proteins, which are a group of RNA-binding proteins including the ubiquitously expressed protein HuR and the neuron-specific proteins HuB, HuC, and HuD. The primary function of Hu proteins is to regulate mRNA stability by binding to AU-rich elements (ARE) present in the 3′-UTR (18). Hu proteins have also been reported to block poly(A) sites that contain a U-rich sequence near cleavage sites (17). Recently, Hu proteins have been shown to modulate alternative polyadenylation by blocking a proximal poly(A) site of HuR mRNA and alter the relative expression of HuR mRNA transcripts with different lengths of 3′-UTRs (19, 20). Whether Hu proteins can regulate intronic polyadenylation and modulate relative expression of alternative mRNA transcripts with different coding sequences is unknown. In the present study, we tested the hypothesis that HuR and HuD can inhibit KCNH2 intron 9 poly(A) signal activity and up-regulate the functional Kv11.1a isoform. Our findings suggest that Hu proteins play an important role in the regulation of the relative expression of Kv11.1 isoforms.
Results
HuR and HuD inhibit intron 9 poly(A) signal activity
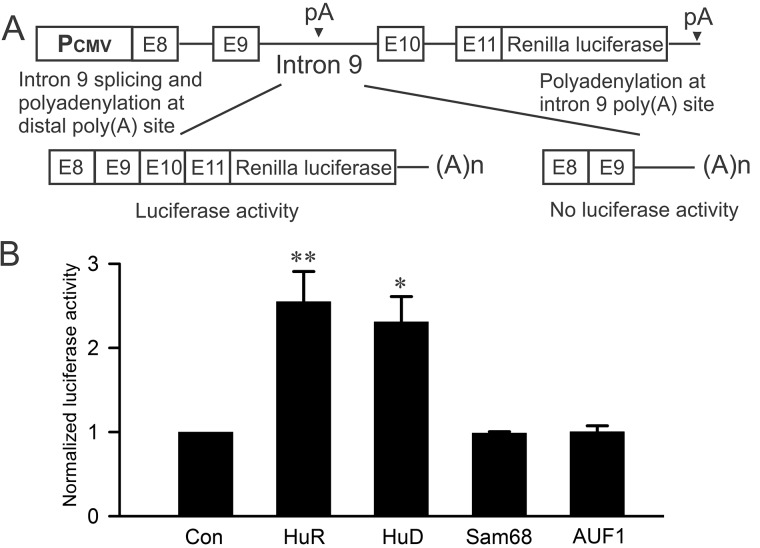
As a first step in demonstrating whether Hu proteins can regulate KCNH2 intron 9 alternative polyadenylation, we used a reporter construct containing the Renilla luciferase gene downstream of a splicing competent minigene composed of human KCNH2 genomic DNA from exon 8 to exon 11 (21, 22). In this KCNH2 minigene reporter construct, the splicing of intron 9 would generate active luciferase and polyadenylation of intron 9 would result in no luciferase activity (Fig. 1A). We co-transfected the reporter construct with HuR or HuD. As shown in Fig. 1B, both HuR and HuD significantly increased the luciferase activity. We also tested RNA-binding proteins Sam68 and AUF1. Sam68 is a KH-type RNA-binding protein that has been reported to modulate alternative polyadenylation of Aldh1a3 pre-mRNA and AUF1 is an ARE RNA-binding protein that generally promotes rapid decay of target mRNAs (16, 18). Sam68 and AUF1 had no effect on the luciferase activity. Expression of HuR, HuD, Sam68, and AUF1 in transfected cells was confirmed by immunoblot analysis (Fig. S1). These results suggest that HuR and HuD, but not Sam68 and AUF1, may inhibit intron 9 polyadenylation and promote intron 9 splicing, leading to an increase in the luciferase activity.
Figure 1.
Effect of HuR, HuD, Sam68, and AUF1 on KCNH2 intron 9 processing using a luciferase reporter construct. A, diagram of the KCNH2 minigene luciferase reporter construct. B, histogram showing the significant increase in luciferase activity following the co-transfection of HuR or HuD compared with vector-transfected control (**, p < 0.01; *, p < 0.05, n = 4, error bars, S.E.).
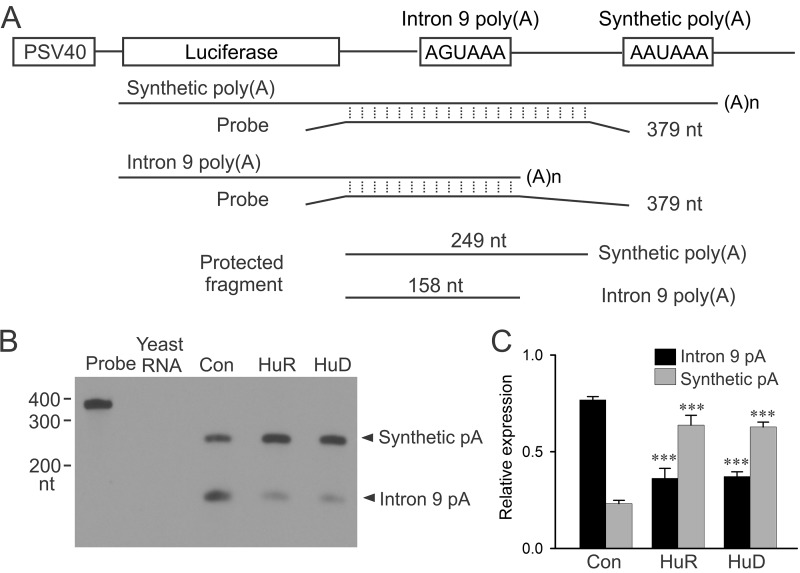
To demonstrate directly that HuR and HuD inhibit KCNH2 intron 9 polyadenylation, we performed a competition assay using a tandem poly(A) signal construct containing the KCNH2 intron 9 poly(A) signal AGUAAA and flanking sequences (−130/+172 nt) which are positioned upstream of a relatively strong synthetic poly(A) signal (Fig. 2A and Fig. S2). We co-transfected HuR or HuD with the KCNH2 tandem poly(A) signal construct and performed RPA analysis using a probe specific to 249 nt of KCNH2 intron 9 (11). This probe will generate a 158 nt fragment if the intron 9 poly(A) signal is used and a 249 nt fragment if the synthetic poly(A) signal is utilized (Fig. 2A). When the pcDNA3 vector was co-transfected with the tandem poly(A) signal construct, the transcription was predominantly terminated at the intron 9 poly(A) site (Fig. 2B). Co-transfection with HuR or HuD with the tandem poly(A) signal construct resulted in decreased usage of the intron 9 poly(A) site from 77 to 36% (HuR) or 37% (HuD), and concomitantly increased usage of the synthetic poly(A) site from 23 to 64% (HuR) or 63% (HuD) (p < 0.001, n = 3) (Fig. 2C). These results indicate that HuR and HuD are able to inhibit KCNH2 intron 9 poly(A) signal activity.
Figure 2.
Inhibition of the KCNH2 intron 9 poly(A) site by HuR and HuD. A, diagram of the tandem poly(A) signal construct and a schematic presentation of the RPA protocol. B, RPA analysis of relative usage of intron 9 poly(A) signal and synthetic poly(A) signal following the co-transfection of the tandem poly(A) signal construct with HuR, HuD, or vector-transfected control. Yeast RNA was used as a control for the complete digestion of the probes by RNase. C, histogram showing the usage of the intron 9 pA and synthetic pA site. RNA signals were quantified and plotted as the expression of intron 9 pA and synthetic pA relative to the total signal (intron 9 pA+synthetic pA). Co-transfection with HuR or HuD results in the significantly decreased polyadenylation at the intron 9 pA site and significantly increased polyadenylation at the synthetic pA site compared with vector-transfected control (***, p < 0.001, n = 4, error bars, S.E.).
HuR interacts with the downstream region of the intron 9 poly(A) signal
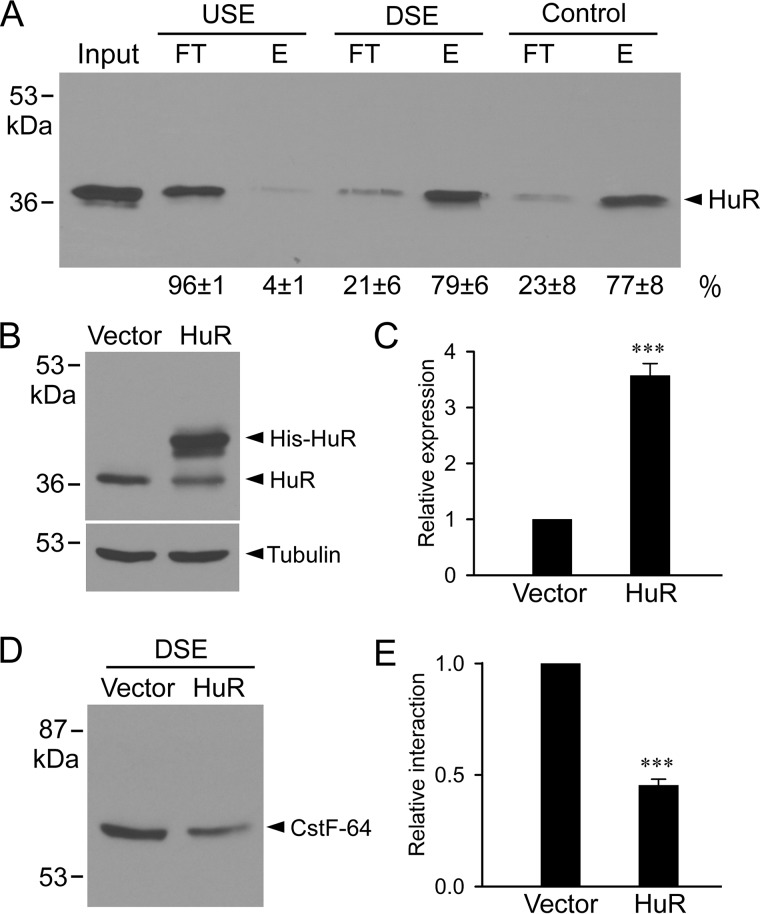
Hu proteins have been reported to have a strong binding affinity to U/GU-rich sequences (17, 19, 23). The downstream region of the KCNH2 intron 9 poly(A) signal contains U/GU-rich elements that are important for poly(A) signal activity (21). To test if HuR can interact with these downstream U/GU-rich sequence elements (DSE), we carried out pulldown assays using biotinylated RNA oligos and cell lysates from HEK293 cells (Fig. 3A and Fig. S2). The integrity of the biotinylated RNA oligos was confirmed using denaturing polyacrylamide gel (Fig. S3). HuR interaction with DSE was readily detected by the pulldown assay (Fig. 3A). Comparable interaction was observed when we used a positive control RNA oligo containing a known HuR-binding sequence in the 3′-UTR of androgen receptor mRNA (24). We also studied the interaction of AUF1 with DSE and found no association of AUF1 with DSE. AUF1 did show interaction with the 3′-UTR of androgen receptor mRNA as reported previously (Fig. S4) (24). In contrast to DSE, the HuR interaction with the upstream sequence elements (USE) was minimal. These results indicate that HuR is able to bind to the downstream region of the KCNH2 intron 9 poly(A) signal.
Figure 3.
Interaction of HuR with intron 9 poly(A) signal downstream sequence. A, cell lysates from HEK293 cells were incubated with biotinylated and streptavidin bead–bound RNA probes corresponding to the DSE, the USE, or the control HuR-binding sequence from the 3′-UTR of androgen receptor mRNA. The RNA-bound protein fractions (E) and unbound fractions (FT) were analyzed by immunoblotting with anti-HuR antibody. Signals were quantified and expressed as percentage of total (FT + E) protein (n = 4). B, immunoblot analysis of total nuclear extracts from vector or HuR transfected cells. C, histogram showing the significant increase in HuR expression following transfection compared with vector-transfected control (***, p < 0.001, n = 3, error bars, S.E.). Protein bands were quantified, normalized to the tubulin, and plotted as relative expression of the vector-transfected control. D, total nuclear extracts from HEK293 cells co-transfected with vector or HuR were incubated with biotinylated RNA oligo corresponding to DSE. Immunoblot analysis of RNA-bound protein fractions probed with anti-CstF-64 antibody. E, histogram showing the significant decrease in the interaction between CstF-64 and DSE following HuR transfection compared with vector-transfected control (***, p < 0.001, n = 3, error bars, S.E.). Protein bands were quantified and plotted as relative to the interaction in the vector-transfected control.
HuR inhibits the recruitment of the cleavage stimulation factor CstF-64 to DSE
Pre-mRNA polyadenylation normally requires binding of the CstF-64 subunit of the cleavage stimulation factor (CstF-64) to the U/GU-rich downstream elements (17, 19). We hypothesized that HuR binding to these elements may interfere with the CstF-64 recruitment to DSE, thereby inhibiting KCNH2 intron 9 polyadenylation. We overexpressed HuR by transfecting the HuR-pcDNA3.1/His plasmid into HEK293 cells. Immunoblot analysis showed that HuR was significantly overexpressed in the nuclear extract compared with vector-transfected control (increased 3.6-fold, p < 0.001, n = 3) (Fig. 3, B and C). We then analyzed the CstF-64 association with the DSE RNA oligo using the biotinylated RNA pulldown assay. The analysis of the streptavidin-retained fraction with a CstF-64–specific antibody showed that the CstF-64 interaction with DSE was significantly less efficient in the HuR overexpressed extract than in the vector-transfected extract (Fig. 3, D and E). As a control, we overexpressed AUF1 and showed that overexpressed AUF1 had no effect on the CstF-64 interaction with DSE (Fig. S4). Taken together, our results suggest that HuR interferes with CstF-64 recruitment to the DSE of the intronic polyadenylation signal, thereby inhibiting KCNH2 intron 9 polyadenylation.
Regulation of Kv11.1 isoform expression by HuR and HuD
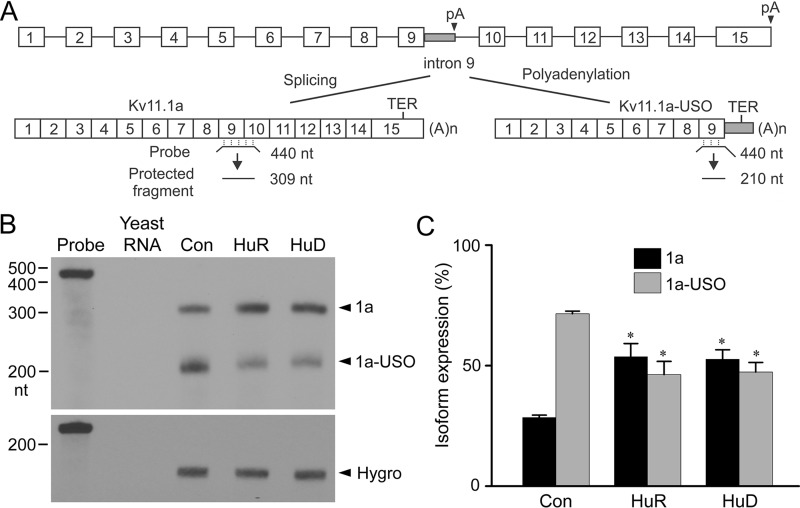
To test whether inhibition of intron 9 polyadenylation by HuR and HuD leads to modulation of the Kv11.1 isoform expression, we used a short KCNH2 gene construct (13). When expressed in HEK293 cells, the short KCNH2 gene construct undergoes alternative polyadenylation to generate the Kv11.1a and Kv11.1a-USO isoforms (Fig. 4A). The short KCNH2 gene construct allows us to study the regulation of Kv11.1 isoform expression at the mRNA, protein, and functional levels. We transiently transfected HuR or HuD into Flp-In HEK293 cells that stably express the short KCNH2 gene. RPA analysis showed that transfection of HuR or HuD resulted in an increase in the Kv11.1a transcript and a decrease in the Kv11.1a-USO transcript (Fig. 4, B and C). This result indicates that relative expression of Kv11.1 C-terminal isoforms can be regulated by HuR and HuD.
Figure 4.
Effect of HuR and HuD on Kv11.1 isoform expression. A, structure of the short KCNH2 gene construct and a schematic presentation of the RPA protocol for Kv11.1a and Kv11.1a-USO. B, RPA analysis of mRNA from Flp-In HEK293 cells stably expressing the short KCNH2 gene following transfection with vector control, HuR, or HuD. C, histogram showing modulation of Kv11.1a (1a) and Kv11.1a-USO (1a-USO) transcripts following HuR and HuD overexpression. RPA signals were quantified and shown as an isoform percentage of the total signal (1a + 1a-USO). The overexpression of HuR or HuD resulted in significantly increased expression of Kv11.1a transcripts and significantly decreased expression of Kv11.1a-USO transcripts compared with vector-transfected control (*, p < 0.05, n = 3, error bars, S.E.).
HuR and HuD up-regulate Kv11.1a channel protein on the plasma membrane
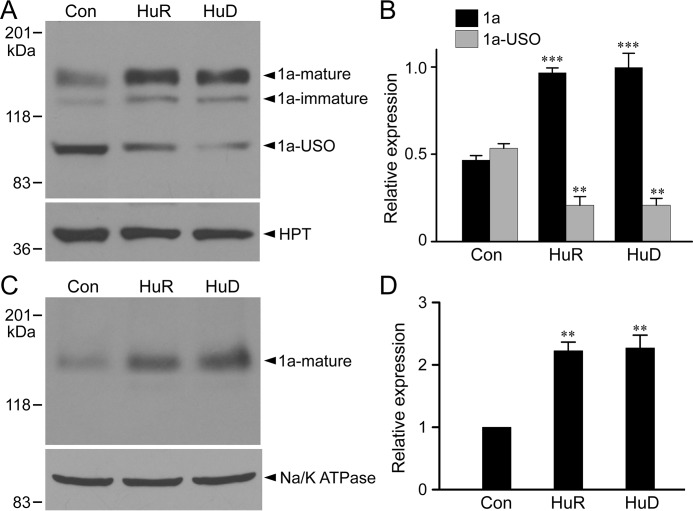
To determine whether HuR and HuD lead to isoform switch at the protein level, we analyzed Kv11.1 protein expression by immunoblotting. When expressed in HEK293 cells, the short KCNH2 gene construct produced three protein bands at 155 kDa, 135 kDa, and 100 kDa (Fig. 5A). The 155 kDa band represents the fully glycosylated mature form of Kv11.1a, the 135 kDa band represents the core-glycosylated immature form of Kv11.1a, and the 100 kDa band represents the core-glycosylated form of Kv11.1a-USO (13). Transient transfection of HuR or HuD into Flp-In HEK293 cells stably expressing the short KCNH2 gene significantly increased the level of Kv11.1a protein and decreased the Kv11.1a-USO protein level (Fig. 5, A and B). To determine whether up-regulation of Kv11.1a channel protein by HuR and HuD leads to an increase in the cell surface expression of the channel protein, we isolated cell surface proteins using biotinylation. As shown in Fig. 5, C and D, HuR and HuD significantly increased the 155 kDa, fully glycosylated, mature form of the Kv11.1a channel, suggesting that the cell surface density of Kv11.1a is up-regulated by HuR and HuD.
Figure 5.
Regulation of Kv11.1 isoform protein expression and cell surface expression by HuR and HuD. A, immunoblot analysis of Kv11.1 protein from Flp-In HEK293 cells stably expressing the short KCNH2 gene following transfection of vector control, HuR, or HuD. The expression level of HPT encoded by the hygromycin B resistance gene served as a loading control. Cell lysates were subjected to SDS-PAGE and probed with antibodies against the N terminus of Kv11.1 or HPT. B, histogram showing modulation of Kv11.1a (1a, including both 1a-mature and 1a-immature) and Kv11.1a-USO (1a-USO) isoforms following HuR and HuD overexpression. The protein bands were quantified, normalized to HPT, and plotted as relative expression of total Kv11.1 protein (1a + 1a-USO) in vector control. The overexpression of HuR or HuD resulted in significantly increased expression of the Kv11.1a isoform and significantly decreased expression of Kv11.1a-USO isoform compared with vector-transfected control (***, p < 0.001; **, p < 0.01, n = 3, error bars, S.E.). C, immunoblot showing the effect of HuR and HuD overexpression on the cell surface expression of Kv11.1 protein. Following transfection of vector control, HuR, or HuD, cell surface proteins were biotinylated, isolated, and analyzed with antibody against the N terminus of Kv11.1. The Na/K-ATPase served as a loading control. D, histogram showing the significant increase in cell surface expression of mature form of Kv11.1a following transfection of HuR or HuD compared with vector-transfected control (**, p < 0.01, n = 3, error bars, S.E.). Protein bands were quantified, normalized to Na/K-ATPase and plotted as relative expression of the vector control.
HuR and HuD increase Kv11.1 channel current
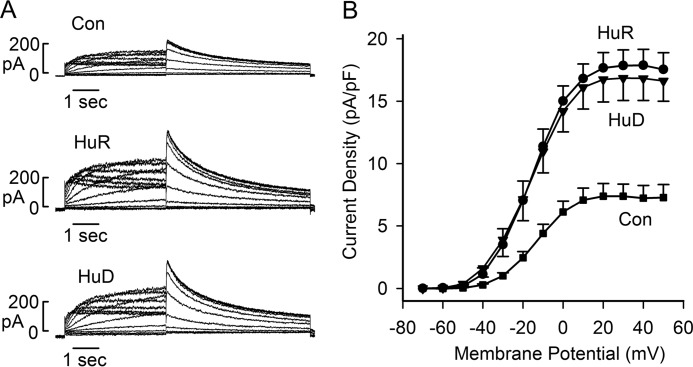
To study the functional effect of HuR- and HuD-induced isoform switch, we performed patch clamp recordings of Kv11.1 channel current. Cells stably expressing the short KCNH2 gene were transiently transfected with HuR+GFP or HuD+GFP plasmids. Transfection of HuR+GFP or HuD+GFP significantly increased Kv11.1 current compared with the GFP vector control (Fig. 6A). The maximum tail current densities in vector, HuR, and HuD were 7.2 ± 1.1 pA/pF (n = 7), 17.9 ± 1.3 pA/pF (p < 0.001, n = 8), and 16.9 ± 1.8 pA/pF (p < 0.001, n = 7) (Fig. 6B), respectively. These patch clamp experiments demonstrate that HuR and HuD increase Kv11.1 channel current.
Figure 6.
Effect of HuR and HuD on Kv11.1 channel current. A, representative currents from GFP-positive Flp-In HEK293 cells stably expressing the short KCNH2 gene following transfection of GFP vector (Con), HuR+GFP, or HuD+GFP. B, current-voltage plot of tail current densities measured at −50 mV following test voltages from −70 to +50 mV for vector-transfected control (square, n = 7), HuR (circle, n = 8) and HuD (triangle, n = 7), Error bars, S.E.
RNAi knockdown endogenous HuR reduces relative expression of the Kv11.1a isoform
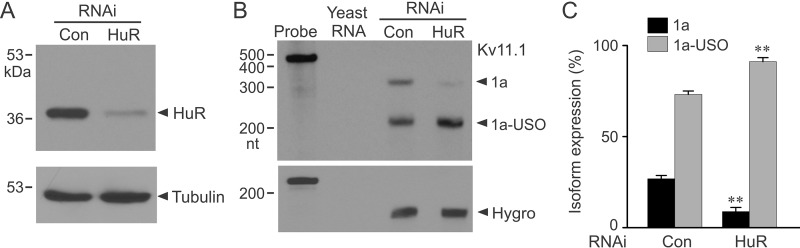
Although overexpression studies indicate a role of HuR in the regulation of Kv11.1 isoform expression (Fig. 4), these experiments did not prove that endogenous HuR contributes to such regulation. RNAi-mediated knockdown is an important complementary experiment to the overexpression studies. We co-transfected HuR siRNA with the short KCNH2 gene into HEK293 cells. The siRNA-mediated knockdown of HuR protein is shown in Fig. 7A. RPA analysis revealed that RNAi knockdown of HuR significantly decreased the level of the Kv11.1a mRNA and concomitantly increased the level of the Kv11.1a-USO mRNA (Fig. 7, B and C). These results indicate that endogenously expressed HuR plays a role in the regulation of Kv11.1 isoform expression.
Figure 7.
Effect of siRNA-mediated knockdown of HuR protein on Kv11.1 isoform expression. A, immunoblot showing that HuR siRNA reduced HuR protein expression. B, RPA analysis of mRNA from Flp-In HEK293 cells stably expressing the short KCNH2 gene following transfection of control siRNA (Con) or HuR siRNA. The same RNA probe as described in Fig. 4 was used. C, histogram showing modulation of Kv11.1a (1a) and Kv11.1a-USO (1a-USO) transcripts following HuR siRNA transfection. RPA signals were quantified and shown as an isoform percentage of the total signal (1a + 1a-USO). HuR siRNA treatment resulted in significantly decreased expression of Kv11.1a transcripts and significantly increased expression of Kv11.1a-USO transcripts compared with control siRNA (**, p < 0.01, n = 3, error bars, S.E.).
Discussion
Our present experiments reveal that HuR and HuD inhibit the poly(A) signal in KCNH2 intron 9 and modulate relative expression of Kv11.1 C-terminal isoforms. Co-expression of the short KCNH2 gene with HuR and HuD results in a shift from the nonfunctional Kv11.1a-USO isoform to the functional Kv11.1a isoform and an increase in Kv11.1 current. These findings suggest that RNA-binding proteins HuR and HuD play an important role in the regulation of Kv11.1 channel function.
Alternative polyadenylation is increasingly being recognized as an important mechanism of gene regulation (14). More than 60% of human genes contain two or more polyadenylation sites. Alternative poly(A) signals are commonly present in tandem within the region of the 3′-UTR, but are also frequently present in upstream intronic regions. Although the use of tandem alternative poly(A) signals leads to the generation of alternate mRNA transcripts with variable 3′-UTRs, the alternative polyadenylation at intronic sites results in the generation of alternate mRNA isoforms with different coding sequences. It has been reported previously that inhibition of the HuR upstream poly(A) signal by Hu proteins results in the increased utilization of a downstream polyadenylation site in the 3′-UTR of HuR pre-mRNA, leading to an up-regulation of the HuR mRNA isoform with a longer 3′-UTR (19, 20). Our findings show that HuR and HuD inhibit the intron 9 poly(A) signal, resulting in a switch from the truncated Kv11.1 isoform to the full-length Kv11.1 isoform. Thus, the present work is the first to demonstrate that Hu proteins can inhibit intronic polyadenylation and modulate the relative expression of the mRNA isoforms with different coding sequences.
We found that HuR binds to the downstream sequence but not the upstream sequence of the intron 9 poly(A) signal. We have previously shown that the downstream region of the intron 9 poly(A) signal contains two U/GU-rich elements important for KCNH2 intron 9 poly(A) signal activity. Mutations of these elements resulted in the predominant production of Kv11.1a and a marked increase in channel current. The binding of Hu proteins to U-rich regions of poly(A) sites has also been reported in SV40 late poly(A) site and calcitonin exon 4 poly(A) site (17). In addition, inhibition of polyadenylation of these poly(A) sites depends on binding of Hu proteins to the U-rich sequences. Similar to our finding, Hu proteins were shown to inhibit polyadenylation by interfering with the CstF-64 recruitment to these pre-mRNAs. Because the balance between splicing and polyadenylation of intron 9 is important for the relative expression of Kv11.1a and Kv11.1a-USO isoforms, inhibition of intron 9 polyadenylation by Hu proteins can shift the balance toward the splicing pathway, thereby leading to the predominant expression of the full-length Kv11.1a isoform.
The Hu family consists of four proteins, the neuron-specific proteins HuB, HuC, and HuD and the ubiquitously expressed protein HuR. We have previously demonstrated that the relative expression of Kv11.1 isoforms is regulated in a tissue-specific manner (11). The tissue-specific expression patterns of Hu proteins and Kv11.1 isoforms raise the possibility that Hu proteins may contribute to the tissue-specific expression of Kv11.1 isoforms. The expression level of HuR is often elevated in cancer cells, and reduced in senescent and quiescent cells including the brain and heart (25–27). The reduced expression of HuR may play a role in the relatively lower expression of full-length Kv11.1a isoform in the heart. In contrast, the expression neuron-specific Hu proteins HuB, HuC, and HuD may play a role in the relatively higher expression of the full-length Kv11.1a isoform in the brain.
It is well-documented that Hu proteins regulate mRNA stability by binding to ARE present in the 3′-UTR (18). To rule out the possibility that the effect of HuR and HuD on Kv11.1 isoform expression is caused by changes in mRNA stability of Kv11.1a and Kv11.1a-USO isoforms, we performed RNA stability assays. The half-lives of Kv11.1a and Kv11.1a-USO mRNAs are comparable and HuR had no effect on the stability of Kv11.1a and Kv11.1a-USO mRNAs (Fig. S5). This result is in line with the fact that no ARE is present in the 3′-UTR of the Kv11.1a or Kv11.1a-USO isoform.
The expression of HuR may undergo dramatic changes in specific physiological and pathological conditions. HuR is markedly reduced during heat shock as a result of proteasome-dependent degradation (28). In addition, HuR protein is predominantly in the nucleus but has been shown to relocalize to the cytoplasm during cellular stress (hypoxia and ischemia) and in response to alphavirus infection (25, 29–31). Because the regulation of alternative polyadenylation requires nuclear localization of Hu proteins, the decrease in HuR in the nucleus because of cytoplasmic relocalization may lead to down-regulation of the functional Kv11.1a isoform expression. Several disease conditions such as myocardial infarction and virus infection are frequently associated with arrhythmias (32, 33). Whether relocalization of HuR to the cytoplasm during these pathological conditions results in dysregulation of Kv11.1 isoform expression, leading to the development of arrhythmias, warrants future investigation.
Experimental procedures
Plasmids, cell culture, and transfections
The minigene luciferase reporter construct was generated by subcloning the Renilla luciferase gene downstream of the splicing competent minigene composed of KCNH2 genomic DNA from exon 8 to exon 11 as described previously (21). Expression of the minigene luciferase reporter is driven by the CMV promoter. The vector also contains the firefly luciferase gene driven by the SV40 promoter, which was used as a control for transfection efficiency. HEK293 cells were transiently transfected with the minigene luciferase reporter construct using the Effectene method (Qiagen, Valencia, CA). After 48 h, cells were harvested and assayed for both firefly and Renilla luciferase activity using the Dual-Luciferase Assay kit (Promega, Madison, WI). Data were analyzed by normalizing Renilla luciferase activity to firefly luciferase activity.
The generation of the tandem poly(A) signal construct was described previously (11). The construct contained the SV40 promoter, the firefly luciferase gene, and 308 bp of KCNH2 intron 9 poly(A) signal and flanking sequences followed by a synthetic poly(A) signal. HEK293 cells were transiently transfected with the tandem poly(A) construct using the Effectene method.
The generation of a short KCNH2 gene construct in which the two longest introns, intron 2 (14.9 kb) and intron 5 (4.4 kb), are shortened to 600 bp was described previously (13). Stably transfected Flp-In HEK293 cells were generated by the co-transfection of the short KCNH2 gene construct (0.1 μg) with the Flp recombinase expression vector pOG44 (0.9 μg) using the Effectene method and selected with 100 μg/ml hygromycin. Flp-In HEK293 cells contain a single FRT genomic locus, allowing the integration of a single copy of the KCNH2 gene construct. Flp-In HEK293 cells were cultured in DMEM supplemented with 10% FBS.
HuR cDNA in pcDNA3.1/His-B vector was obtained from Dr. Luo (17) and HuD cDNA with Myc-tag in pcDNA3 vector was obtained from Dr. Perrone-Bizzozero (34). These plasmids were used in all HuR/HuD transfection experiments except patch clamp experiments where the plasmids expressing both GFP and HuR or HuD were used. For the HuR+GFP and HuD+GFP plasmids, the GFP coding sequence was subcloned into PGL-3 promoter vector at HindIII and XbaI sites, then the SV40 promoter–GFP fragment was exited at BglII and BamHI sites and subcloned into HuR-pcDNA3.1/His-B or HuD-pcDNA3 plasmid at BglII site. Sam68 cDNA was obtained from Mammalian Gene Collection and subcloned into pcDNA3.1/His-C at EcoRI and ApaI sites. AUF1 (HNRNPD) cDNA in the pFRT/TO/His/FLAG/HA-DEST vector was a gift from Dr. Markus Landthaler (Addgene plasmid no. 38066) (35). The His/FLAG/HA-tagged AUF1 cDNA was subcloned into pcDNA3 at HindIII and XhoI sites. The plasmids expressing HuR, HuD, HuR+GFP, or HuD+GFP were transiently transfected into the Flp-In HEK293 cells that stably express the short KCNH2 gene construct using PolyJet transfection reagent. HuR, HuD, Sam68, or AUF1 was transiently transfected together with the minigene luciferase reporter construct into HEK293 cells using the Effectene method. HuR or HuD was transiently transfected together with the tandem poly(A) signal construct into HEK293 cells using the Effectene method.
RNase protection assay
The RNase protection assay (RPA) was performed as described previously (11). Briefly, total RNA isolated from HEK293 cells were analyzed with the riboprobes using the RPAII and BrightStart BioDetect Kits (Ambion, Austin, TX). Briefly, antisense RNA riboprobes were transcribed in vitro in the presence of biotin-14-CTP. Yeast RNA was used as a control for the complete digestion of the probes by RNase. The relative intensity of each band was quantified using ImageJ software and adjusted for the number of biotin-labeled cytidines in each protected fragment. The expression level of the hygromycin B resistance gene from the short KCNH2 gene constructs was used to normalize relative expression of Kv11.1 isoforms.
Biotinylated RNA pulldown assays
The biotinylated RNA pulldown assay was performed using a Magnetic RNA-Protein Pull-Down Kit (Thermo Scientific). RNA oligos upstream and downstream of intron 9 poly(A) signal were custom synthesized by GenScript (Piscataway, NJ). The USE and DSE RNA oligos are 40 nt long (Fig. S2). A known HuR-binding sequence in the 3′-UTR of androgen receptor mRNA was used as a positive control RNA oligo (24). The RNA oligos were labeled with biotin using RNA 3′ End Desthiobiotinylation Kit (Thermo Scientific). The biotinylated RNAs were extracted with chloroform:isoamyl alcohol, precipitated with ethanol, rehydrated in nuclease-free water and bound to Streptavidin Magnetic Beads. The integrity of biotinylated RNA oligos was determined by electrophoresis with 15% denaturing polyacrylamide gel. After being transferred to nylon membrane, the bands were detected by alkaline phosphatase conjugated streptavidin and chemiluminescence. The cell lysates were prepared by Mammalian Protein Extraction Reagent (M-PER) (Thermo Scientific). The nuclear extracts were prepared using Nuclear and Cytoplasmic Extract Reagent (Thermo Scientific). Briefly, HuR/RNA complexes were allowed to form at 4 °C for 60 min in 50 μl mixtures containing 50 pmol biotinylated RNA probe, 50 μg of cell lysate in 1× binding buffer with 15% glycerol. To study the effect of HuR overexpression on CstF-64 binding to RNA oligos, the nuclear extract was used, as nuclear localized HuR is expected to modulate CstF-64 binding. CstF-64/RNA complexes were assembled at 30 °C for 30 min in 50 μl mixtures containing 50 pmol biotinylated RNA probe, 40% (v/v) nuclear extract diluted by 1× binding buffer to 2 mg/ml (i.e. 40 μg in total), 15% glycerol, and 1 mm ATP. The complexes were further stabilized by UV cross-linking at 254 nm, 1.0 J/cm2 with 0.01% Nonidet P-40, 2 mm DTT, and 20 mm phosphocreatine. Both HuR/RNA and the UV–cross-linked CstF-64/RNA complexes were washed with 50 μl of 1× wash buffer twice, and then bead-associated proteins were eluted with 50 μl of elution buffer for 30 min at 37 °C. For CstF-64/RNA complexes the elution buffer contains 1 mg/ml RNase A. The eluted samples were heated for 5 min at 95 °C in the presence of SDS-PAGE loading buffer and then analyzed by immunoblot.
RNAi knockdown of HuR
Small interfering RNA (siRNA) targeting HuR was obtained from Santa Cruz Biotechnology (Dallas, TX). The HuR siRNA (sc-35619) is a pool of two targeting-specific 19–25 nt siRNAs designed to knockdown expression of HuR. The Flp-In HEK293 cells stably expressing the short KCNH2 gene were transfected with control or HuR siRNAs using Lipofectamine 2000 (Invitrogen). After 48 h, cells were analyzed by RPA. The knockdown of the HuR protein was analyzed by immunoblotting.
Immunoblot analysis
Immunoblot analysis was performed as described previously (11). Cell lysates were subjected to SDS-PAGE and transferred to nitrocellulose membranes electrophoretically. The Kv11.1 isoforms were detected using an anti-Kv11.1 antibody directed against the N terminus of Kv11.1a and Kv11.1a-USO (H-175) (Santa Cruz Biotechnology) at a 1:600 dilution and visualized with the ECL detection kit (Amersham Biosciences). The expression of hygromycin B phosphotransferase (HPT) encoded by the hygromycin B resistance gene was used to normalize the relative expression of Kv11.1 isoform proteins (11). Other antibodies used are anti-HuR (3A2, Santa Cruz Biotechnology), anti-CstF-64 (H-1, Santa Cruz Biotechnology), anti-AUF1 (Abcam, Cambridge, MA), anti-Na/K-ATPase (C464.6, Santa Cruz Biotechnology), anti-Xpress (Invitrogen), anti-c-Myc (Convance), and anti-FLAG M2 (Sigma-Aldrich).
Biotinylation and isolation of cell surface proteins
Biotinylation and isolation of cell surface proteins were performed using Pierce Cell Surface Protein Isolation Kit (Thermo Scientific). Cells cultured in 100 mm dishes were washed twice with 8 ml of ice-cold PBS, and then incubated with 8 ml of ice-cold PBS containing sulfo-NHS-SS-Biotin for 30 min at 4 °C. After two washes with ice-cold PBS, the cells were incubated in 8 ml of ice-cold PBS with 400 μl of quenching solution for 10 min at 4 °C and washed again with ice-cold PBS. Cells were scraped into ice-cold PBS and collected by centrifugation. Cell pellets were suspended in 200 μl of lysis buffer containing Protease Inhibitor Mixture (Thermo Scientific). The cells were disrupted by sonication on ice using five 1-s bursts and incubated on ice for 30 min. Cell lysates were collected after centrifugation at 10,000 × g for 2 min at 4 °C. The biotin-labeled cell surface proteins were isolated using NeutrAvidin Agarose columns (Thermo Scientific), eluted with SDS-PAGE sample buffer, and analyzed by immunoblotting.
Patch clamp recordings
Membrane currents were recorded in whole cell configuration as described previously (4). Cells were bathed in a solution containing 137 mm NaCl, 4 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, 10 mm glucose, and 10 mm HEPES (pH 7.4). The pipette solution contained 130 mm KCl, 1 mm MgCl2, 5 mm EGTA, 5 mm MgATP, and 10 mm HEPES (pH 7.2). Patch clamp experiments were performed using suction pipettes at 22 to 23 °C. Data were recorded using an Axopatch-200B amplifier and analyzed with pCLAMP10 software (Molecular Devices, Sunnyvale, CA).
Data analysis
Data are presented as mean ± S.E. Student's t test was used for comparison between two groups. Analysis of variance (ANOVA) was used for comparisons between more than two groups. p < 0.05 is considered statistically significant.
Author contributions
Q. G., M. R. S., and Z. Z. conceptualization; Q. G., M. R. S., and Z. Z. data curation; Q. G., M. R. S., and Z. Z. formal analysis; Q. G., M. R. S., and Z. Z. writing-original draft; Q. G., M. R. S., and Z. Z. writing-review and editing; Z. Z. funding acquisition.
Supplementary Material
Acknowledgments
We thank Drs. Hua Lou and Nora Perrone-Bizzozero for reagents.
This work was supported in part by the National Institutes of Health Grant R01 HL068854 and American Heart Association Grant 15GRNT23020018. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S5.
- LQT2
- long QT syndrome type 2
- ARE
- AU-rich elements
- nt
- nucleotide
- RPA
- RNase protection assay
- DSE
- downstream sequence elements
- USE
- upstream sequence elements
- pA/pF
- picoamperes/picofarads
- HPT
- hygromycin B phosphotransferase.
References
- 1. Warmke J. W., and Ganetzky B. (1994) A family of potassium channel genes related to eag in Drosophila and mammals. Proc. Natl. Acad. Sci. U.S.A. 91, 3438–3442 10.1073/pnas.91.8.3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sanguinetti M. C., Jiang C., Curran M. E., and Keating M. T. (1995) A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell 81, 299–307 10.1016/0092-8674(95)90340-2 [DOI] [PubMed] [Google Scholar]
- 3. Trudeau M. C., Warmke J. W., Ganetzky B., and Robertson G. A. (1995) HERG, a human inward rectifier in the voltage-gated potassium channel family. Science 269, 92–95 10.1126/science.7604285 [DOI] [PubMed] [Google Scholar]
- 4. Zhou Z., Gong Q., Ye B., Fan Z., Makielski J. C., Robertson G. A., and January C. T. (1998) Properties of HERG channels stably expressed in HEK 293 cells studied at physiological temperature. Biophys. J. 74, 230–241 10.1016/S0006-3495(98)77782-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Curran M. E., Splawski I., Timothy K. W., Vincent G. M., Green E. D., and Keating M. T. (1995) A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell 80, 795–803 10.1016/0092-8674(95)90358-5 [DOI] [PubMed] [Google Scholar]
- 6. Larsen A. P. (2010) Role of ERG1 isoforms in modulation of ERG1 channel trafficking and function. Pflugers Arch. 460, 803–812 10.1007/s00424-010-0855-8 [DOI] [PubMed] [Google Scholar]
- 7. Lees-Miller J. P., Kondo C., Wang L., and Duff H. J. (1997) Electrophysiological characterization of an alternatively processed ERG K+ channel in mouse and human hearts. Circ. Res. 81, 719–726 10.1161/01.RES.81.5.719 [DOI] [PubMed] [Google Scholar]
- 8. London B., Trudeau M. C., Newton K. P., Beyer A. K., Copeland N. G., Gilbert D. J., Jenkins N. A., Satler C. A., and Robertson G. A. (1997) Two isoforms of the mouse ether-a-go-go-related gene coassemble to form channels with properties similar to the rapidly activating component of the cardiac delayed rectifier K+ current. Circ. Res. 81, 870–878 10.1161/01.RES.81.5.870 [DOI] [PubMed] [Google Scholar]
- 9. Kupershmidt S., Snyders D. J., Raes A., and Roden D. M. (1998) A K+ channel splice variant common in human heart lacks a C-terminal domain required for expression of rapidly activating delayed rectifier current. J. Biol. Chem. 273, 27231–27235 10.1074/jbc.273.42.27231 [DOI] [PubMed] [Google Scholar]
- 10. Guasti L., Crociani O., Redaelli E., Pillozzi S., Polvani S., Masselli M., Mello T., Galli A., Amedei A., Wymore R. S., Wanke E., and Arcangeli A. (2008) Identification of a posttranslational mechanism for the regulation of hERG1 K+ channel expression and hERG1 current density in tumor cells. Mol. Cell. Biol. 28, 5043–5060 10.1128/MCB.00304-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gong Q., Stump M. R., Dunn A. R., Deng V., and Zhou Z. (2010) Alternative splicing and polyadenylation contribute to the generation of hERG1 C-terminal isoforms. J. Biol. Chem. 285, 32233–32241 10.1074/jbc.M109.095695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stump M. R., Gong Q., and Zhou Z. (2012) Isoform-specific dominant-negative effects associated with hERG1 G628S mutation in long QT syndrome. PLoS One 7, e42552 10.1371/journal.pone.0042552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gong Q., Stump M. R., Deng V., Zhang L., and Zhou Z. (2014) Identification of Kv11.1 isoform switch as a novel pathogenic mechanism of long-QT syndrome. Circ. Cardiovasc. Genet. 7, 482–490 10.1161/CIRCGENETICS.114.000586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tian B., and Manley J. L. (2013) Alternative cleavage and polyadenylation: The long and short of it. Trends Biochem. Sci. 38, 312–320 10.1016/j.tibs.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hall-Pogar T., Liang S., Hague L. K., and Lutz C. S. (2007) Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3′-UTR. RNA 13, 1103–1115 10.1261/rna.577707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. La Rosa P., Bielli P., Compagnucci C., Cesari E., Volpe E., Farioli Vecchioli S., and Sette C. (2016) Sam68 promotes self-renewal and glycolytic metabolism in mouse neural progenitor cells by modulating Aldh1a3 pre-mRNA 3′-end processing. Elife, 5, e20750 10.7554/eLife.20750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu H., Zhou H. L., Hasman R. A., and Lou H. (2007) Hu proteins regulate polyadenylation by blocking sites containing U rich sequences. J. Biol. Chem. 282, 2203–2210 10.1074/jbc.M609349200 [DOI] [PubMed] [Google Scholar]
- 18. Brennan C. M., and Steitz J. A. (2001) HuR and mRNA stability. Cell. Mol. Life Sci. 58, 266–277 10.1007/PL00000854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dai W., Zhang G., and Makeyev E. V. (2012) RNA-binding protein HuR autoregulates its expression by promoting alternative polyadenylation site usage. Nucleic Acids Res. 40, 787–800 10.1093/nar/gkr783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mansfield K. D., and Keene J. D. (2012) Neuron-specific ELAV/Hu proteins suppress HuR mRNA during neuronal differentiation by alternative polyadenylation. Nucleic Acids Res. 40, 2734–2746 10.1093/nar/gkr1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gong Q., Stump M. R., and Zhou Z. (2014) Upregulation of functional Kv11.1 isoform expression by inhibition of intronic polyadenylation with antisense morpholino oligonucleotides. J. Mol. Cell. Cardiol. 76, 26–32 10.1016/j.yjmcc.2014.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gong Q., Stump M. R., and Zhou Z. (2018) Upregulation of functional Kv11.1a isoform expression by modified U1 small nuclear RNA. Gene 641, 220–225 10.1016/j.gene.2017.10.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ince-Dunn G., Okano H. J., Jensen K. B., Park W. Y., Zhong R., Ule J., Mele A., Fak J. J., Yang C., Zhang C., Yoo J., Herre M., Okano H., Noebels J. L., and Darnell R. B. (2012) Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron 75, 1067–1080 10.1016/j.neuron.2012.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barker A., Epis M. R., Porter C. J., Hopkins B. R., Wilce M. C., Wilce J. A., Giles K. M., and Leedman P. J. (2012) Sequence requirements for RNA binding by HuR and AUF1. J. Biochem. 151, 423–437 10.1093/jb/mvs010 [DOI] [PubMed] [Google Scholar]
- 25. Govindaraju S., and Lee B. S. (2013) Adaptive and maladaptive expression of the mRNA regulatory protein HuR. World J. Biol. Chem. 4, 111–118 10.4331/wjbc.v4.i4.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang W., Yang X., Cristofalo V. J., Holbrook N. J., and Gorospe M. (2001) Loss of HuR is linked to reduced expression of proliferative genes during replicative senescence. Mol. Cell. Biol. 21, 5889–5898 10.1128/MCB.21.17.5889-5898.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Masuda K., Marasa B., Martindale J. L., Halushka M. K., and Gorospe M. (2009) Tissue- and age-dependent expression of RNA-binding proteins that influence mRNA turnover and translation. Aging 1, 681–698 10.18632/aging.100073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abdelmohsen K., Srikantan S., Yang X., Lal A., Kim H. H., Kuwano Y., Galban S., Becker K. G., Kamara D., de Cabo R., and Gorospe M. (2009) Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 28, 1271–1282 10.1038/emboj.2009.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barnhart M. D., Moon S. L., Emch A. W., Wilusz C. J., and Wilusz J. (2013) Changes in cellular mRNA stability, splicing, and polyadenylation through HuR protein sequestration by a cytoplasmic RNA virus. Cell Rep. 5, 909–917 10.1016/j.celrep.2013.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dickson A. M., Anderson J. R., Barnhart M. D., Sokoloski K. J., Oko L., Opyrchal M., Galanis E., Wilusz C. J., Morrison T. E., and Wilusz J. (2012) Dephosphorylation of HuR protein during alphavirus infection is associated with HuR relocalization to the cytoplasm. J. Biol. Chem. 287, 36229–36238 10.1074/jbc.M112.371203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gu L., Wang H., Wang J., Guo Y., Tang Y., Mao Y., Chen L., Lou H., and Ji G. (2017) Reconstitution of HuR-inhibited CUGBP1 expression protects cardiomyocytes from acute myocardial infarction-induced injury. Antioxid. Redox Signal. 27, 1013–1026 10.1089/ars.2016.6880 [DOI] [PubMed] [Google Scholar]
- 32. Schwartz P. J., and Wolf S. (1978) QT interval prolongation as predictor of sudden death in patients with myocardial infarction. Circulation 57, 1074–1077 10.1161/01.CIR.57.6.1074 [DOI] [PubMed] [Google Scholar]
- 33. Alvarez M. F., Bolívar-Mejía A., Rodriguez-Morales A. J., and Ramirez-Vallejo E. (2017) Cardiovascular involvement and manifestations of systemic Chikungunya virus infection: A systematic review. Version 2. F1000Res. 6, 390 10.12688/f1000research.11078.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anderson K. D., Morin M. A., Beckel-Mitchener A., Mobarak C. D., Neve R. L., Furneaux H. M., Burry R., and Perrone-Bizzozero N. I. (2000) Overexpression of HuD, but not of its truncated form HuD I+II, promotes GAP-43 gene expression and neurite outgrowth in PC12 cells in the absence of nerve growth factor. J. Neurochem. 75, 1103–1114 [DOI] [PubMed] [Google Scholar]
- 35. Baltz A. G., Munschauer M., Schwanhäusser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M., Wyler E., Bonneau R., Selbach M., Dieterich C., and Landthaler M. (2012) The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 46, 674–690 10.1016/j.molcel.2012.05.021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.