Abstract
Human papillomavirus (HPV) is a necessary but insufficient cause of a subset of oral squamous cell carcinomas (OSCCs) that is increasing markedly in frequency. To identify contributory, secondary genetic alterations in these cancers, we used comprehensive genomics methods to compare 149 HPV-positive and 335 HPV-negative OSCC tumor/normal pairs. Different behavioral risk factors underlying the two OSCC types were reflected in distinctive genomic mutational signatures. In HPV-positive OSCCs, the signatures of APOBEC cytosine deaminase editing, associated with anti-viral immunity, were strongly linked to overall mutational burden. In contrast, in HPV-negative OSCCs, T>C substitutions in the sequence context 5′-ATN-3′ correlated with tobacco exposure. Universal expression of HPV E6*1 and E7 oncogenes was a sine qua non of HPV-positive OSCCs. Significant enrichment of somatic mutations was confirmed or newly identified in PIK3CA, KMT2D, FGFR3, FBXW7, DDX3X, PTEN, TRAF3, RB1, CYLD, RIPK4, ZNF750, EP300, CASZ1, TAF5, RBL1, IFNGR1, and NFKBIA. Of these, many affect host pathways already targeted by HPV oncoproteins, including the p53 and pRB pathways, or disrupt host defenses against viral infections, including interferon (IFN) and nuclear factor kappa B signaling. Frequent copy number changes were associated with concordant changes in gene expression. Chr 11q (including CCND1) and 14q (including DICER1 and AKT1) were recurrently lost in HPV-positive OSCCs, in contrast to their gains in HPV-negative OSCCs. High-ranking variant allele fractions implicated ZNF750, PIK3CA, and EP300 mutations as candidate driver events in HPV-positive cancers. We conclude that virus-host interactions cooperatively shape the unique genetic features of these cancers, distinguishing them from their HPV-negative counterparts.
Human papillomavirus (HPV) is the cause of a distinct subset of oral squamous cell carcinomas (OSCCs) that is rising in incidence in the United States (Gillison et al. 2000; Chaturvedi et al. 2011). These HPV-positive head and neck cancers are different from their HPV-negative counterparts in many ways. Their predisposing risk factors are distinctive (sexual behavior vs. tobacco and alcohol use, respectively) (Gillison et al. 2008), and the prognosis of HPV-positive OSCCs is more favorable (Ang et al. 2010).
We originally proposed that HPV-positive OSCCs also comprise a distinct molecular entity when compared to HPV-negative OSCCs, based in part upon an inverse association between HPV presence and mutations in TP53 (Gillison et al. 2000). Subsequent analyses confirmed TP53 mutations in only 3% of HPV-positive versus 82% of HPV-negative head and neck cancers (Agrawal et al. 2011; Stransky et al. 2011; Pickering et al. 2013; The Cancer Genome Atlas Network 2015). Similarly, the pRB pathway is disrupted in HPV-positive cancers via E7 targeting for degradation by the proteasome, in contrast to mutation or methylation of CDKN2A (cyclin dependent kinase inhibitor 2A, encoding p16 protein10) and amplification of CCND1 (encoding cyclin D1) in HPV-negative cancers. A recent proteomic analysis of HPV-host protein interactions revealed novel HPV protein targets in additional pathways enriched for inactivating mutations in HPV-negative head and neck cancers (Eckhardt et al. 2018). For example, a novel interaction between HPV31 E1 and the NFE2L2 (also known as NRF2) transcriptional pathway was identified. Additional distinctions between HPV-positive and HPV-negative cancers have been identified by targeted analysis of somatic mutations and gene expression profiles (Slebos et al. 2006; Seiwert et al. 2015). Collectively, these data support the existence of common pathways, critical for the development of HPV-positive and HPV-negative head and neck cancers.
We note that prior genomic analyses focused predominantly on HPV-negative head and neck cancers arising in diverse anatomical sites and included limited numbers of HPV-positive OSCCs (Agrawal et al. 2011; Stransky et al. 2011; The Cancer Genome Atlas Network 2015). Here, we report a comprehensive analysis of genetic alterations in OSCCs (inclusive of oropharyngeal and oral cavity), including the largest collection of HPV-positive OSCCs studied to date by use of next generation DNA and RNA sequencing.
Results
Sequencing analysis of HPV-positive and HPV-negative OSCCs
We focused our analysis on the two head and neck cancer anatomic sites with subgroups increasing in incidence in the United States: HPV-positive (predominantly oropharyngeal) and HPV-negative (predominantly oral cavity) OSCCs. We analyzed sequencing data from 149 HPV-positive and 335 HPV-negative OSCC tumor-normal (T/N) pairs, from an Ohio cohort (n = 112) and TCGA (n = 372). Clinical and demographic information, along with details about sequencing data and analysis, are provided in Figure 1, Supplemental Figure S1, and Supplemental Table S1.
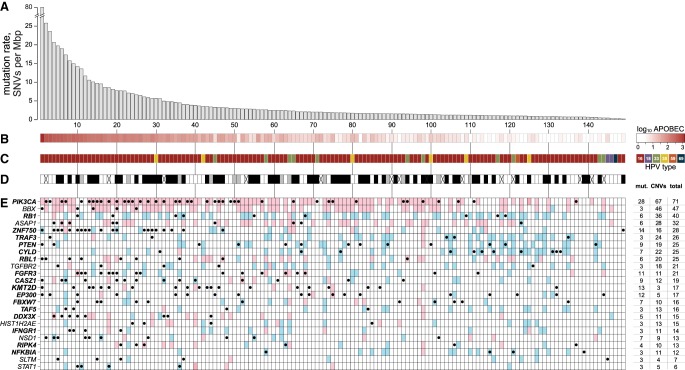
Figure 1.
Somatic mutations in HPV-positive OSCCs. (A) Somatic mutation rates in exons of 149 HPV-positive OSCC samples were determined from WGS and WES data. Mutation rates were calculated as the number of single nucleotide variants (SNVs) per megabase pair (Mbp) sequenced. Individual tumor samples were ranked by these rates (vertical alignments, left to right). (B) Heat map of log10-transformed counts of APOBEC-associated mutations in 5′-TCW-3′ sequence context in each cancer. (C) HPV viral types in cancers: key, right: dark red, HPV-16; lavender, 18; green, 33; yellow, 35; orange, 59; dark blue, 69. (D) Cigarette smoking history for each patient: black, ≥10 pack-years; gray, <10 pack-years; white, never smoker; X, no data. (E) Heat map of variants in 24 significantly mutated genes (left) identified by MutSig (adjusted P-value < 0.2) (Lawrence et al. 2013) in samples ordered as in A (x-axis). Key: red, copy number gain involving gene; blue, copy number loss; black dots, somatic variants including missense, nonsense, and splicing site SNVs, and frameshift or in-frame insertions or deletions (indels). Bold font (gene names): adjusted P-value < 0.1. Table at right: percentages of samples that have single nucleotide mutations and short indels; copy number variants (CNVs) (gains or losses); and total (combined). Individual genes were ranked by these combined frequencies (top to bottom). See also Supplemental Figure S1 and Supplemental Table S1.
We classified 149 cancers as HPV-positive based upon detection of viral transcripts (out of 482 OSCCs with available RNA-seq data) and DNA sequence reads that aligned to HPV reference genomes (out of 153 cases with available WGS data). Results were highly concordant. Additional confirmatory assays for tumor HPV status (e.g., HPV in situ hybridization and p16 protein immunohistochemical staining) were performed for the 112 cancers in the Ohio cohort. Again, results were highly concordant (Supplemental Fig. S1B–G; Supplemental Table S1A–H).
Of 149 HPV-positive cancers, viral typing showed that 86% were positive for HPV16, 7.4% for type 33, 4.0% for 35, 1.3% for 18, and 0.7% each for 59 and 69. Within the subset of 103 HPV-positive cancers studied by WGS, the median virus copy number was 11.4 per cell (range 0.1–247). Although many cancers lacked large segments of the viral genome, the full-length HPV E6 and E7 DNA coding regions were consistently present. Discordant DNA sequencing reads containing both HPV and host genome sequences, indicating viral integration into the host genome, were detected in 74% (Fig. 1C; Supplemental Fig. S1E; Supplemental Table S1I–K; Akagi et al. 2014).
Of the high-risk HPV types, HPV16 transcripts have been characterized most thoroughly; they undergo extensive splicing. Therefore, we assessed the expression of virus-specific transcripts using RNA-seq data from 126 HPV16-positive cancers. The predominant polycistronic transcript, highly expressed (median 96.1 FPKM [fragments per kilobase million]) in 100% of cancers, had coding potential for E6*1, E7, E1^E4, and E5. Transcripts with full-length HPV E6 coding potential also were expressed in 91% of HPV16-positive OSCC, albeit at substantially lower levels than E6*1 in most cases (Supplemental Fig. S1B–D; Supplemental Table S1L). Canonical transcripts for E5 were expressed in 71%.
A total of 23,291 somatic variants were identified in coding regions across 149 HPV-positive cancers. These included 14,290 missense single nucleotide variants (SNVs), 6044 synonymous SNVs, 1391 nonsense (stop-gain) SNVs, 227 splice donor and acceptor site variants, 263 frameshift and 75 in-frame insertion or deletion (indel) variants. Exonic mutation rates ranged broadly from 0.2 to 78 somatic variants (defined here as both SNVs and small indels) per megabase pair (Mbp) per tumor (Fig. 1A). The median rate of somatic variants in exons was 1.86 per Mbp, and 9.4% of cancers had ≥10 somatic variants per Mbp. These rates corresponded to counts ranging from 10 to 3959 exonic variants per tumor. Analysis of 103 HPV-positive cancers with WGS data identified a total of 1,342,816 somatic variants genome-wide. The median rate of genome-wide somatic variants was 2.04 per Mbp. Somatic mutation rates across genomes or exomes of HPV-positive cancers did not differ significantly from HPV-negative cancers. Details regarding variant rates in all OSCCs studied using various sequencing technologies (i.e., WGS and WES) and platforms are provided in Supplemental Materials and in Figure 1, Supplemental Figure S1A, H−M, and Supplemental Table S1M–P.
Overall, 81.1% of the somatic variants identified in expressed coding regions were confirmed using RNA-seq data, regardless of source of samples, sequencing platforms, and downstream SNV-calling bioinformatics pipelines. Excluding variants identified in HPV-positive OSCCs from TCGA WES data, 83.5% were confirmed by RNA-seq. These rates are similar to the 86% confirmation rate observed in TCGA (The Cancer Genome Atlas Network 2015). Moreover, 92% of somatic variants in 24 HPV-positive and HPV-negative OSCCs in our Ohio cohort were confirmed independently by targeted resequencing (Supplemental Table S1Q,R).
We assessed somatic mutation rates among 407 cases with available tobacco exposure data (including 135 HPV-positive and 272 HPV-negative OSCCs). Mutation rates were not significantly different in comparing heavy smokers (≥10 pack-years) to never or light smokers (<10 pack-years) in patients with HPV-positive OSCCs. In contrast, among HPV-negative OSCCs, mutation rates were significantly higher in heavy smokers. This result was consistent across sequencing platforms (Fig. 1D; Supplemental Fig. S1N–Q).
An HPV-positive tumor was an extreme outlier in overall mutation rate (Fig. 1). In this cancer, the DNA polymerase POLE was disrupted both by a missense mutation in a context previously associated with APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like) family member-mediated activity, as well as by a stop-gain mutation, resulting in its ultramutator phenotype (Rayner et al. 2016). As expected, mutation rates were significantly higher among HPV-positive cancers with deleterious mutations in DNA repair and mismatch repair genes (Supplemental Fig. S1R,S; Supplemental Table S1S–V).
Somatic variants and mutational signatures reflect distinct risk factors in OSCCs
Analysis of the six possible nucleotide substitutions in HPV-positive vs. HPV-negative OSCCs identified significant differences. Both C>T and C>G substitutions were significantly more frequent in HPV-positive than in HPV-negative cancers (mean frequencies 49.0% vs. 36.4%, adj. P = 1.8 × 10−15; and 18.6% vs. 14.8%, adj. P = 4.6 × 10−3, respectively). In contrast, the other four somatic variants each were significantly more frequent in HPV-negative OSCCs (Fig. 2; Supplemental Fig. S2; Supplemental Table S2).
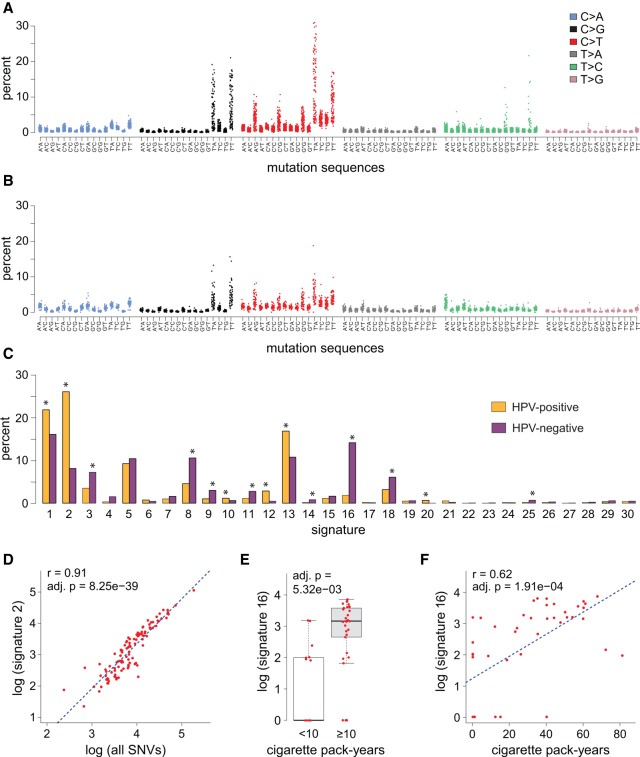
Figure 2.
Mutation signatures and molecular risk factors in OSCC. (A,B) Dot plots show (y-axis) fraction of (x-axis) SNVs in all 96 possible mutation 3-nt sequence contexts identified in each (A) HPV-positive (n = 149) and (B) HPV-negative (n = 50) OSCC studied by WGS. Key: Colors indicate the somatic SNVs located in (asterisk, x-axis labels, mutation sequences) the central mutation position in each 3-nt sequence. (C) Bar graph depicting the (y-axis) mean fraction of mutations in (x-axis) 30 mutation signatures across the collection of (yellow) HPV-positive and (purple) HPV-negative OSCCs with available WGS data. Similar results were obtained using WES and exonized WGS data. Signatures showing significant differences between HPV-positive and HPV-negative cancers included signature 2 and 16. (*) adjusted P < 0.01. (D) SNVs in signature 2, linked to APOBEC cytidine deaminase genome editing, were strongly associated with overall mutation burden in individual HPV-positive OSCCs (red dots; r = 0.91; adj. P = 8.25 × 10−39). A similar association was observed for signature 13, also linked to APOBEC activity (see Supplemental Fig. S2). (E) Box and whisker plot showing significant relative increase in SNVs in signature 16 in HPV-negative OSCC patients associated with heavy cigarette smoking (adj. P = 5.23 × 10−3). (F) Pack-years of cigarette smoking directly correlated with number of signature 16 mutations in HPV-negative OSCCs (red dots; r = 0.62; adj. P = 1.91 × 10−4). See also Supplemental Figure S2 and Supplemental Table S2.
Rates of SNVs occurring in all 96 possible three-nucleotide genomic sequence contexts were calculated for each of the 153 OSCCs with WGS data (Fig. 2A,B). Most pronounced among HPV-positive OSCCs were the fractions of C>T and C>G mutations observed in the context of 5′-T*CW-3′ genomic sequences associated with APOBEC cytidine deaminase editing. Thus, we compared the distribution of 30 mutation signatures as defined by COSMIC nomenclature among HPV-positive vs. negative OSCCs (Alexandrov et al. 2013), revealing several significant differences (Fig. 2C). Again, the contribution of signatures previously attributed to activity of the APOBEC family of cytidine deaminases (i.e., signatures 2 and 13) were significantly higher in HPV-positive than in negative OSCCs. Of all SNVs detected in HPV-positive cancers, 47.4% occurred in the context of mutation signatures 2 and 13, significantly more than just 21.7% of all SNVs in HPV-negative cancers (adj. P = 0). Similarly, on a per-tumor basis, the mean fractions of signature 2 and 13 mutations again were elevated in HPV-positive more than in HPV-negative OSCCs (35.0% vs. 18.3%, respectively; P = 5.12 × 10−10). Very strong correlations between numbers of signature 2 and 13 mutations and total SNVs indicated that APOBEC editing was the principal driver of the overall mutation burden in HPV-positive OSCCs (Figs. 1B, 2D; Supplemental Fig. S2A–M; Supplemental Table S2A–J).
Among HPV-negative OSCCs, mutations were distributed more broadly across the 30 mutational signatures when measured either as fractions or as total number of SNVs (Fig. 2). Notably, the contribution of signature 16, with a predominance of T>C substitutions occurring in a 5′-ATN-3′ sequence context, was significantly greater in HPV-negative than HPV-positive cancers, whether measured by mean fraction (14.2% vs. 1.8%, P = 5.05 × 10−7) or number (1457 vs. 106, P = 1.96 × 10−6) of variants (Alexandrov et al. 2016). A novel finding in our data was a statistically significant association between cigarette smoking exposure (≥10 vs. <10 pack-years) and higher fraction or number of signature 16 mutations in HPV-negative OSCCs (Fig. 2E). Moreover, pack-years of tobacco smoking positively correlated with number of signature 16 mutations (r = 0.39, P = 7.0 × 10−10) (Fig. 2F). We also observed weaker evidence supporting an association between intensity of current alcohol use and signature 16 mutations (Supplemental Fig. S2N). No such associations were observed in HPV-positive OSCCs, regardless of cut-point of tobacco exposure utilized in the analysis (e.g., never versus ever smoker). In HPV-negative OSCCs, no single signature accounted for a high fraction or number of overall SNVs. Several signatures (i.e., 8, 9, 10, 11, and 16) in addition to APOBEC signatures 2 and 13 correlated with total mutation burden. Inferences drawn from analyses utilizing WES data were similar (Supplemental Fig. S2C–N; Supplemental Table S2A–O).
In both HPV-positive and HPV-negative OSCCs, as the genome-wide mutation burden declined, the contribution of signature 1 mutations was significantly elevated. Therefore, tumors with low mutation burden were characterized by mutation signatures attributed to spontaneous deamination of 5-methylcytosines. No associations were observed with age (Supplemental Fig. S2; Supplemental Table S2).
Frequently mutated genes in HPV-positive OSCCs
In assessing somatic variants with predicted functional impacts in HPV-positive OSCCs, we identified statistically significant enrichment of mutations in 17 genes (q < 0.1) (Fig. 1E). Confirming previous reports, recurrent mutations in PIK3CA, KMT2D, FGFR3, FBXW7, and DDX3X were significantly increased among HPV-positive cancers (The Cancer Genome Atlas Network 2015; Seiwert et al. 2015). We found significantly increased rates of mutations in PTEN, TRAF3, RB1, CYLD, and RIPK4, confirming studies cataloging their presence in one or more cancers (Chung et al. 2015; Seiwert et al. 2015; Hajek et al. 2017). Novel to our analysis was significant enrichment of mutations in ZNF750, EP300, CASZ1, TAF5, RBL1, IFNGR1, and NFKBIA. When the false discovery rate was relaxed to q < 0.2, NSD1, ASAP1, BBX, SLTM, TGFBR2, HIST1H2AE, and STAT1 also were found to be recurrently mutated. Only ZNF750 and DDX3X mutations co-occurred in individual cancers at frequencies greater than expected by chance (Fig. 1E; Supplemental Table S3A,B).
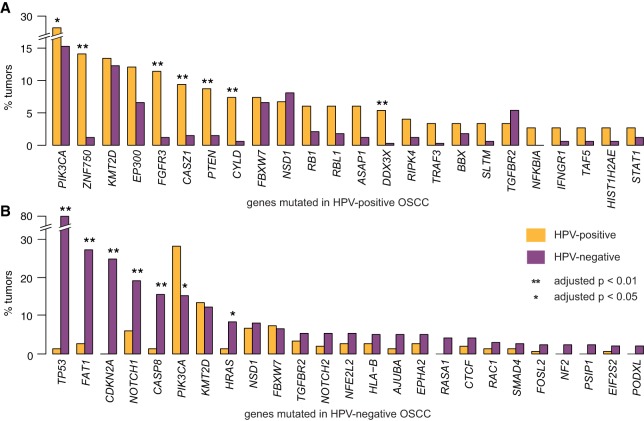
The most significantly mutated genes among HPV-positive OSCCs were markedly different from those in HPV-negative OSCCs (Fig. 3A,B; Supplemental Fig. S3; Supplemental Table S3). Mutations in PIK3CA, ZNF750, FGFR3, CASZ1, PTEN, CYLD, and DDX3X were significantly more common in HPV-positive than HPV-negative cancers (adjusted P < 0.05). In contrast, TP53, FAT1, CDKN2A, NOTCH1, CASP8, and HRAS were mutated significantly more frequently in HPV-negative OSCCs (adj. P < 0.05) (Agrawal et al. 2011). Common to both types of OSCCs were mutations in PIK3CA, KMT2D, FBXW7, NSD1, and TGFBR2 (Supplemental Table S3A–F).
Figure 3.
Comparison of mutation frequency in MutSig-identified significantly mutated genes (HPV-positive OSCCs vs. HPV-negative OSCCs). Bar graphs display fractions of all (yellow) HPV-positive and (purple) HPV-negative OSCCs bearing somatic variants (SNVs and small indels) (A) in 24 MutSig genes detected in HPV-positive OSCCs and (B) in 25 genes in HPV-negative OSCCs. (*) adjusted P-value < 0.05, (**) adjusted P-value < 0.01. See also Supplemental Figure S3 and Supplemental Table S3 for frequencies of somatic variants and CNVs in HPV-positive and HPV-negative OSCCs.
The natural history of HPV-positive OSCC patients with ≥10 pack-years smoking history more closely approximates that of HPV-negative OSCC patients (Ang et al. 2010). Therefore, we investigated whether or not the most highly mutated genes in HPV-negative OSCCs also were mutated frequently in this subset of HPV-positive cancers. While mutations commonly found in HPV-negative OSCCs also were detected in HPV-positive OSCCs, no associations with tobacco exposure (categorized as none, <10 pack-years, or ≥10 pack-years) were observed (Supplemental Table S3G).
To study mutations that may be common to HPV-associated cancers regardless of tissue type, we compared the frequency distribution of mutations enriched in HPV-positive OSCCs to those in cervical cancers, which are uniformly HPV-positive. Genes significantly enriched and occurring at similar mutational frequencies in both HPV-positive OSCCs and cervical cancers were PIK3CA, EP300, PTEN, FBXW7, and TGFBR2. In contrast, mutations in ZNF750, FGFR3, CASZ1, CYLD, and RIPK4 were significantly more frequent in HPV-positive OSCCs than in cervical cancer (Supplemental Fig. S3A; Supplemental Table S3H,I). A trend toward increased KRAS mutations was observed in cervical cancer. We conclude that among HPV-associated cancers, some genetic alterations may be shared across anatomic sites, whereas others may be specific to a particular anatomic site.
Predicted functional impacts of somatic variants in HPV-positive OSCCs
To explore the functional implications of somatic variants in the 24 most frequently mutated genes in HPV-positive OSCCs, we plotted their relative positions, counts, and types in the context of annotated protein domains (Fig. 4; Supplemental Fig. S4; Supplemental Table S4).
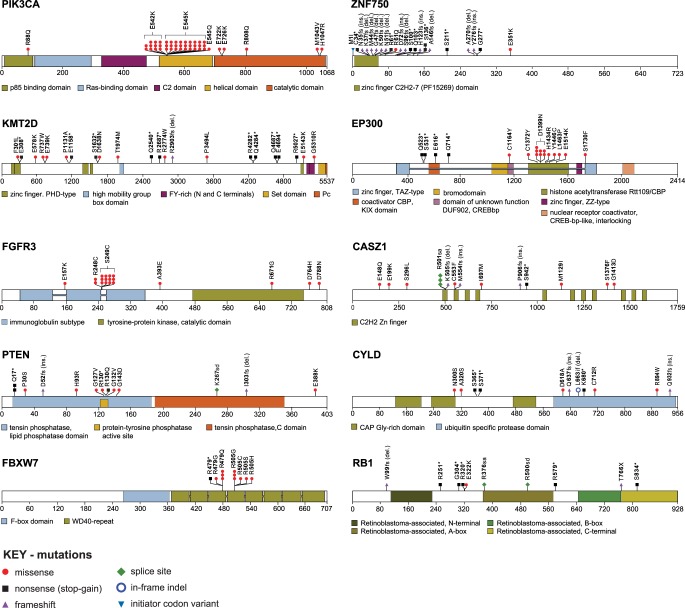
Figure 4.
Somatic mutations alter protein structures and functions. Schematics of protein domains, displaying sites of mutations identified in the 12 most frequently mutated genes identified in HPV-positive OSCCs. Insets: color-coded, annotated protein domains (see also Supplemental Fig. S4; Supplemental Table S4).
Of the 280 somatic variants in the 24 genes identified as the most frequently mutated genes in HPV-positive OSCCs (Fig. 1), 121 (43%) lacked evidence for previous annotation in any human cancer (Supplemental Table S4A). Moreover, 197 (70%) of the somatic variants overall were not reported specifically in OSCCs and therefore are novel in these particular cancers. We found three previously unreported, recurrent somatic variants, each in at least two independent OSCC samples, disrupting a splice acceptor site in CASZ1, an initiator codon in DDX3X, and a stop-gain mutation in ZNF750 (Supplemental Table S4A). Of the 83 somatic variants that were reported previously, 16 (19%) were identified individually in single HPV-positive OSCC samples in our study as well, confirming their recurrent roles in these cancers.
The majority (87%, 39 of 45) of PIK3CA protein mutations in HPV-positive cancers were gain-of-function changes localized to two amino acids in the helical domain, i.e., 542E>K, 545E>K, and 545E>Q (Ng et al. 2018). Their nucleotide substitutions were consistent with APOBEC-mediated mutagenesis (Henderson et al. 2014). In contrast, the predominant mutations in PIK3CA (47%) in HPV-negative cancers were 1047H>R and 1047H>L in the catalytic domain, also reported as activating mutations (Ng et al. 2018).
We found that 7% of HPV-positive OSCCs harbored loss-of-function, missense, or nonsense mutations disrupting the ubiquitin ligase and tumor suppressor protein FBXW7. These included the recurrent mutations 479R>Q and 505R>G (Davis et al. 2014; The Cancer Genome Atlas Network 2015; Chung et al. 2015; Seiwert et al. 2015). Additional mutations in encoded proteins at recurrent amino acid positions included activating mutations in FGFR3 at 249S>C (Duperret et al. 2014; Ng et al. 2018); mutations in the histone acetyltransferase EP300 at 1399D>N; disruption of a splice acceptor site of the zinc finger transcription factor CASZ1 affecting amino acids downstream from 501R; and mutations in the phosphatase domain of PTEN at 130R (Ng et al. 2018). Also observed were mutations that cluster within specific protein domains, such as diverse frameshift or nonsense mutations in the zinc finger domain of ZNF750 and several distinct mutations in the active phosphatase site of PTEN. In contrast, the genes KMT2D, CYLD, and RB1 each harbored a range of diverse mutations, as did genes with mutation hotspots (Fig. 4; Supplemental Fig. S4; Supplemental Table S4A). Among such nonrecurrent, unclustered mutations, those in PIK3CA protein including 88R>Q and 726E>K have been found to be activating (Ng et al. 2018).
Ontology analysis of the 24 most frequently mutated genes in HPV-positive OSCCs revealed strong and statistically significant enrichment (≥10-fold, P < 0.01) of several biological processes, including apoptosis, regulation of sequence-specific DNA transcription factors, viral processes, and the innate immune response (Supplemental Table S4B–D).
Genomic copy number alterations in OSCCs
We note that analysis of somatic variants alone substantially underestimated the frequency at which the 24 most significantly mutated genes were disrupted in HPV-positive cancers (Fig. 5; Supplemental Fig. S5; Supplemental Table S5). While 28% of HPV-positive OSCCs had activating mutations in PIK3CA, gene amplification was also present in 67%. Moreover, transcript levels of amplified genes were increased significantly, as shown from RNA-seq data (e.g., PIK3CA, BBX, ASAP1, adjusted P < 0.01). Conversely, genes inactivated by somatic variants (e.g., RB1, PTEN, CYLD, EP300, FBXW7, TAF5, IFNGR1, NSD1, and STAT1) were deleted frequently and their expression significantly decreased. In some individual cancers, both somatic variants and CNV coincided, potentially resulting in biallelic disruption (e.g., PIK3CA and CYLD). After considering these impacts of CNV, genetic alterations in RB1, BBX, and TAF5 were more frequent in HPV-positive OSCCs and those in RAC1 were more frequent in HPV-negative OSCCs (in addition to significant differences in somatic variants noted above). Comparable, coincident disruptions of genes by SNVs and CNVs were observed in HPV-negative OSCCs (Fig. 1E; Supplemental Figs. S2C, S5A–F; Supplemental Table S5A–D).
Figure 5.
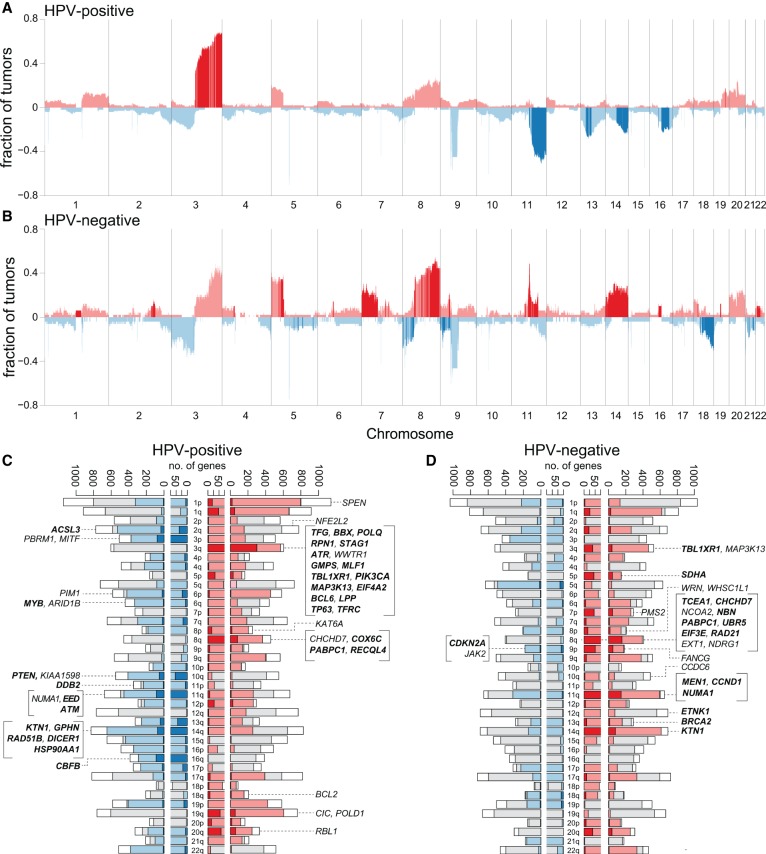
Recurrent chromosomal segment gains and losses in HPV-positive vs. HPV-negative OSCCs. Histograms show the cumulative fraction of samples with significant changes in genomic segment copy number gains (red) and losses (blue) for (A) 103 HPV-positive and (B) 50 HPV-negative OSCCs. Fractions of affected cancers (y-axis) were determined by summing up WGS samples with ploidy N < 1.5 (loss, blue, negative values) or N > 2.5 (gain, red, positive values), divided by the total number. x-axis, chromosomal coordinates Chr 1–22. Dark segments: genomic segments that were significantly enriched in either HPV-positive or in HPV-negative cancers, based on adjusted P < 0.01 using the proportion test to compare each 500-kb segment in the two types of samples. Sex chromosomes are not shown due to unreliable copy number calls. (C,D) Bar plots show the numbers of differentially expressed genes associated with copy number changes for each chromosome arm in (C) 101 HPV-positive OSCC and (D) 50 HPV-negative OSCC cancers. Insets: Listed are differentially expressed genes in CNV regions, already annotated as cancer-causing genes in Sanger Cancer Gene Census database and/or identified here as most highly mutated genes identified by MutSig. x-axis: numbers of genes; y-axis: chromosome arms. Each bar shows numbers of genes on each chromosome arm: (pink, red) in regions with copy number gains; (red) with increased expression that is significantly associated with local copy number gains, adjusted P-value < 0.05; (light blue, dark blue) in regions with copy number losses; (dark blue) with decreased expression that is significantly associated with local copy number losses, adjusted P-value < 0.05; (gray) lacking sufficient copy number changes; (white) with low variance of expression, so not tested. For each gene, the variance in gene expression was calculated for 101 HPV-positive and 50 HPV-negative cancers from batch-adjusted RNA-seq data. Differentially expressed, CNV genes were identified using the t-test to compare their expression in at least three samples with local copy number gain or loss vs. expression in samples with normal copy number at the gene (see also Supplemental Fig. S5; Supplemental Table S5).
A striking example of the complementary roles played by SNVs and CNVs was provided by the RB1 family of genes. Approximately 6% of cancers harbored deleterious SNVs in RB1, and deletions of the genetic locus were observed in another 34% (Fig. 1E). In addition, the retinoblastoma family member gene RBL1, encoding the protein p107, was disrupted by SNVs in 6% and by CNV in 20%. These observations were surprising, given the ubiquitous expression of the viral oncogene E7 in these cancers. The function of the E7 oncoprotein in degrading pRB family members by targeting them to the proteasome is well-established. The critical requirement for HPV E7 in cervical cancer development is demonstrated by its strictly conserved protein sequence in cancers, despite its widespread variation in infected controls (Mirabello et al. 2017). Thus, we investigated whether nonsynonymous mutations in E7 were associated with RB1 gene disruptions in HPV-positive OSCCs. No such association was observed (Supplemental Table S5E,F). The importance of complete genetic knockout of RB1 (encoding pRB) and RBL1 (p107) in the pathogenesis of OSCC, as suggested by these data, also has been corroborated by mouse models (Shin et al. 2012).
To investigate associations between CNV and alterations in gene expression, we analyzed WGS data to identify subchromosomal regions with recurrent gains and losses in 103 HPV-positive cancers. Regions with significant gains were detected in Chr 1q, 3q, 5p, 8q, 19q, 20p, and 20q, while significant losses were observed in Chr 2q, 3p, 4p, 7q, 8p, 9p, 9q, 10q, 11q, 13q, 14q, 15q, 16p, 16q, and 21p (Fig. 5A,B; Supplemental Fig. S5; Supplemental Table S5). Next, we identified alterations in expressed transcript levels by analyzing RNA-seq data and assessed their associations with the CNVs (Fig. 5C,D; Supplemental Table S5G–T). Only 9.0% of genes in regions of recurrent gain displayed concordant, statistically significant increases in their expression relative to cancers without regional CNV (adjusted P < 0.01). Ontology terms of these genes were enriched in cell cycle and DNA damage response. In the most frequently amplified region of Chr 3q, 570 genes were expressed, of which 301 (53%) displayed significantly increased expression. Among these were 20 cancer census genes, including PIK3CA, TP63, and SOX2. Similarly, only 8.0% of genes in regions of recurrent loss had significant decreases in their expression, and ontology terms again were enriched in cell cycle regulation. In the most frequently lost region of Chr 11q, 86 of 456 expressed genes showed significantly decreased expression, including four cancer census genes, NUMA1, EED, ATM, and FLI1 (Supplemental Table S5G–T). Corresponding analysis of HPV-negative cancers showed distinctive, recurrent gains and losses (Fig. 5A,B; Supplemental Fig. S5; Supplemental Table S5).
When we compared regions of recurrent copy number gains or losses in HPV-positive cancers to those in 50 HPV-negative OSCCs (Fig. 5A,B; Supplemental Table S5S,T), patterns were more distinctive than reported previously (Smeets et al. 2006). Significantly more frequent among HPV-positive cancers were gains in Chr 3q and losses of 11q, 13q, 14q, 16p, and 16q. Of these, the most frequently gained region, spanning ∼45 Mbp of Chr 3q25–29, was present in 66% of cancers, while a 56-Mbp region on 11q14-25 was lost in 43%. Particular subchromosomal regions that were significantly lost in HPV-positive OSCCs were, in contrast, gained in HPV-negative cancers. Genes were identified in these regions with concomitant decreases vs. increases in their expression. A region of Chr 11q13.3–q13.4 containing the gene CCND1 was lost in 17% of HPV-positive and inversely gained in 14% of HPV-negative OSCCs. Similarly, a region of Chr 14q24.3–q32.3 containing TRIP11, GOLGA5, DICER1, HSP90AA1, and AKT1 was deleted in 21.4% of HPV-positive but gained in 24% of HPV-negative OSCCs. We conclude that HPV-positive cancers are fundamentally different from HPV-negative cancers with regard to regions of chromosomal gains and losses and consequent associations with altered gene expression.
Hierarchical analysis of batch-corrected RNA-seq data from 147 of 149 HPV-positive OSCCs and all 335 HPV-negative OSCCs separated the cancers into three main groups (Supplemental Fig. S5I). The majority of HPV-positive cancers clustered in one group characterized by differential up-regulation of immune response genes (e.g., complement and immunoglobulin production) (Keck et al. 2015). In an exploratory univariate analysis, a subset of HPV-positive cancers that clustered by expression with most HPV-negative cancers had significantly worse survival (Supplemental Fig. S5L; Supplemental Tables S5O–Q). These exploratory findings require further validation.
Ranked variant allele fractions identify candidate cancer-causing driver genes
Previous genetic analysis of premalignant lesions led to the formulation of a genetic progression model for the formation of HPV-negative OSCCs (Califano et al. 1996). In contrast, a similar model for HPV-positive OSCCs has not been developed due to a lack of clinically identifiable premalignant lesions. Therefore, we sought to identify somatic variants whose variant allele fractions (VAFs) ranked highly across HPV-positive OSCCs, as they may represent early clonal events of potential pathophysiological significance.
First, to validate this approach, we calculated VAFs for all nonsynonymous, somatic variants in exons and ranked them for each of 329 HPV-negative OSCCs, thereby defining “ranked VAFs” (Supplemental Fig. S6). Next, we determined which of these ranked VAFs were significantly enriched among the top 5% VAFs across all of the cancers. Eleven of the 12 genes so identified were among the 25 genes that were mutated most frequently in HPV-negative OSCCs by MutSig analysis (Supplemental Fig. S6A,B; Supplemental Table S6). Notably, the ranked VAFs of mutations in CDKN2A and TP53 ranked most frequently within the top fifth percentile across all cancers. A heat map of ranked VAFs demonstrated that FAT1, CASP8, NOTCH1, and HRAS mutations also were significantly enriched within the top 5% among HPV-negative cancers (Supplemental Fig. S6C,D; Supplemental Table S6). Mutations in CDKN2A and TP53 have been reported as some of the earliest genetic events in a direct analysis of oral premalignant lesions, related to their genomic positions in regions of loss of heterozygosity (e.g., Chr 9p21 [CDKN2A] and Chr 17p13 [TP53]) (Califano et al. 1996). Moreover, these events are predictive of progression to cancer (Zhang et al. 2012). This analysis provided substantial support for our approach.
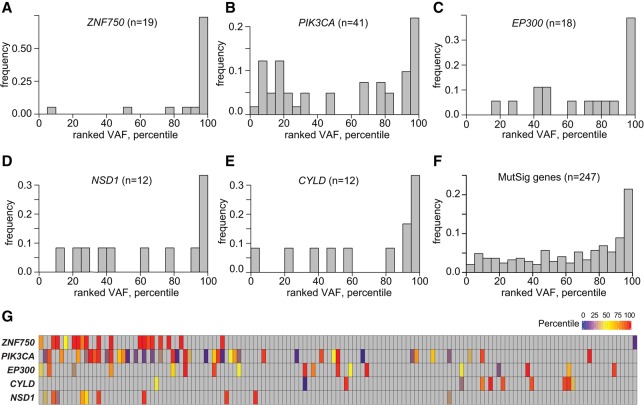
Extending from proof-of-concept analysis in HPV-negative cancers, we sought to identify candidate, early genetic progression events for HPV-positive cancers (Fig. 6). Biased distributions of ranked VAFs were observed in ZNF750, PIK3CA, and EP300 and the other highly mutated MutSig genes overall, as their SNVs were significantly enriched in the top 5% of all ranked VAFs in the HPV-positive OSCCs. Other genes showed no such bias (binomial test, q < 0.01) (Fig. 6A–C; Supplemental Fig. S6E,F; Supplemental Table S6). Additionally, while the distributions of ranked VAFs for SNVs in NSD1 and CYLD also appeared to be biased toward the top 5%, they did not reach statistical significance (Fig. 6D,E). The ranked VAFs of all highly mutated MutSig genes in HPV-positive OSCCs showed a similar enrichment of high percentiles (Fig. 6F; Supplemental Fig. S6E). We note that genes disrupted by SNVs with highly ranked VAFs also were associated frequently with copy number variations in both HPV-positive (e.g., PIK3CA) and HPV-negative cancers (e.g., CDKN2A) (Supplemental Fig. S6G). We conclude that inactivating mutations in ZNF750 and EP300 and activating mutations in PIK3CA may play an early role in the genetic progression of HPV-positive OSCCs (Fig. 6; Supplemental Fig. S6E–H).
Figure 6.
Ranked variant allele fractions in HPV-positive OSCCs. VAFs for somatic variants located inside exons were ranked in each tumor. Shown are genes with ranked VAFs that were significantly enriched in the top fifth percentile across all HPV-positive OSCCs. Distributions of ranked VAFs of somatic variants in (A) ZNF750 in 19 cases; (B) PIK3CA in 41 cases; (C) EP300 in 18 cases; (D) NSD1 in 12 cases; (E) CYLD in 12 cases; and (F) all highly mutated MutSig genes in 145 HPV-positive cases. Of these 247 somatic variants, 54 (22%) were identified within the top fifth percentile of VAFs (P = 2.2 × 10−23). (G) Heat map showing ranked VAF percentiles of mutations disrupting five highly mutated genes (left) in individual HPV-positive OSCCs, ordered by overall mutation frequency (vertical columns; see Fig. 1). Key, right: percentiles of ranked VAFs. See also Supplemental Figure S6 and Supplemental Table S6.
Discussion
Here, we have identified numerous genetic alterations among HPV-positive OSCCs that are distinct from HPV-negative OSCCs. We extended the unique characteristics of HPV-positive OSCCs to include mutational signatures, recurrent somatic mutations and candidate early driver genes, subchromosomal gains and losses affecting gene expression, and distinct gene expression profiles. These data improve the current understanding of the secondary genetic events that contribute to the development of HPV-positive OSCCs. Our data indicate that HPV oncoproteins and host genomic alterations cooperatively disrupt genomic stability, epithelial differentiation, apoptosis, transcriptional regulation and the anti-viral immune response, and induce cellular proliferation (Fig. 7). Together, these data support a hypothesis that viral-host interactions cooperatively shape the genomes of HPV-positive OSCCs (Mesri et al. 2014). We anticipate that these results will foster advances in diagnosis and therapy of this malignancy.
Figure 7.
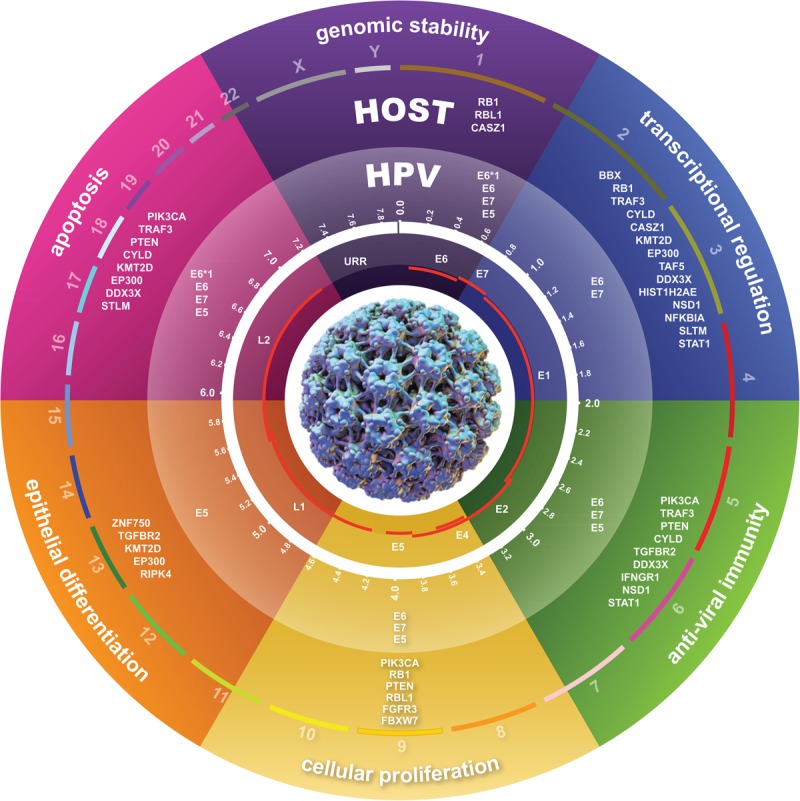
The genetic features of HPV-positive OSCCs. A schematic depicts host-virus genetic interactions and features of HPV-positive OSCCs. HPV infection, represented by the viral capsid (center), is a necessary initiating event but insufficient for the development of cancer. Secondary genetic alterations accumulate as a consequence of host genomic instability induced by HPV oncoproteins, APOBEC induced host genome editing, and other forms of DNA mutagenesis. HPV E6*1, E6, E7, and E5 oncoproteins and these recurrent somatic variants in host genes and pathways cooperatively disrupt host genomic stability, apoptosis, cellular proliferation, epithelial differentiation, the anti-viral immune response, and transcriptional regulation. These viral-host interactions coordinately shape the genomes of HPV-positive OSCCs to promote HPV infection persistence, carcinogenesis, and tumor immune evasion.
First and foremost, HPV-positive OSCCs were defined by the presence and expression of HPV E6*1/E6 and E7 oncogenes. Our observation that the truncated E6*1 isoform was expressed more highly and consistently than E6 is consistent with studies of cervical cancer, as its expression increased with severity of cervical dysplasia and cancer (Chen et al. 2014a). Although E6*1 protein may be less efficient at degrading TP53, it stabilized E6 and E7 proteins and facilitated DNA damage (Olmedo-Nieva et al. 2018). Also of potential pathological significance were full-length HPV E5 transcripts detected in 71% of HPV16-positive OSCCs. HPV E5 enhanced receptor tyrosine kinase signaling (e.g., by EGFR) and impaired expression of MHC class I and II proteins (Belleudi et al. 2015). Therefore, transcripts for all viral proteins with known oncogenic activity were present in a large majority of HPV-positive OSCCs.
Genomic instability caused by disruption of the p53 and pRB tumor suppressor pathways appears necessary for the pathogenesis of all OSCCs. The TP53 and RB1 tumor suppressors that control cell death and proliferation are targeted directly for proteolytic degradation by the HPV E6 and E7 oncoproteins, respectively (Hoppe-Seyler et al. 2018). We were surprised to find that many host pathways already targeted by HPV oncoproteins also were disrupted by host genome alterations. This included the p53 and pRB pathways. In addition to ubiquitin-mediated targeting to the proteosome, E6 also mediates TP53 degradation through binding to the histone acetyltransferase (HAT) E1A-binding protein p300 (EP300, encoded by EP300). In so doing, E6 inhibits EP300-mediated TP53 acetylation, thus promoting MDM2-mediated degradation of TP53 (Zimmermann et al. 1999). Despite universal expression of E6*1/E6 transcripts, we found EP300 was mutated in 12% of HPV-positive OSCCs. Moreover, the histone H3 lysine-4 methyltransferase KMT2D was also mutated in 13% (Stransky et al. 2011; The Cancer Genome Atlas Network 2015; Seiwert et al. 2015). This transcriptional enhancer recruits EP300 to TP53 target genes to promote transcription (Froimchuk et al. 2017). In another example, despite nearly universal expression of E7 transcripts, genetic mutation or deletion of RB1 was observed in 40% of HPV-positive cancers. RB1 and CCND1 deletion may compensate for high CDKN2A (p16 protein) expression, induced as a consequence of E7-mediated up-regulation of the histone lysine demethylase KDM6B (McLaughlin-Drubin et al. 2011). Based on these data, we speculate that genetic alterations in gene products targeted by viral oncoproteins may facilitate cancer progression more actively than viral-host protein-protein interactions alone.
We showed here that the risk-factor profiles that distinguish HPV-positive from HPV-negative OSCCs, i.e., HPV infection vs. tobacco and alcohol use, respectively (Gillison et al. 2008), were reflected in different mutational signatures in cancer genomes. In HPV-positive OSCCs, a strong correlation was observed between the contribution of APOBEC-mediated genome editing (i.e., signatures 2 and 13) and the number of somatic variants overall, thus implicating innate anti-viral immune responses as the principal driver of mutational burden. In contrast, in HPV-negative OSCCs, novel dose-response associations were observed between pack-years of tobacco use (and trends for current alcohol use) and mutational signature 16. Adding to these mutations in HPV-negative cancers were lower frequencies of APOBEC mutations, previously hypothesized as attributable to smoking-associated inflammation (Alexandrov et al. 2016). Tobacco and/or alcohol use also increase risk of esophageal and liver cancer, both of which are characterized by signature 16 mutations (Chang et al. 2017; Letouze et al. 2017). Thus, we have now directly linked epidemiological exposures to downstream genomic consequences in OSCC.
HPV-positive OSCCs arise from deep within the tonsillar crypt epithelium, thus limiting their early clinical detection and precluding development of a genetic progression model. We found biased, high-ranking somatic VAFs across many HPV-positive cancers, identifying candidate early driver mutations in ZNF750, PIK3CA, and EP300, and possibly also in NSD1 and CYLD. These genes contrasted markedly with early driver mutations that we confirmed in CDKN2A, TP53, FAT1, CASP8, NOTCH1, and HRAS among HPV-negative OSCCs. The latter finding is consistent with early loss of heterozygosity (LOH) on Chr 11q and 17q in premalignant lesions of the oral cavity (Califano et al. 1996). A recent observation that mutations in PIK3CA, EP300, NF1, or RB1 co-occurred with high-risk HPV infections in benign tonsil specimens suggested that they could serve as potential biomarkers for risk of progression (Ilmarinen et al. 2017). We anticipate that these data will inform potential secondary prevention strategies for HPV-positive OSCCs among individuals with oral high-risk HPV infections, via genomic profiling of oral rinse or circulating cell-free DNA specimens.
Nearly ubiquitous among HPV-positive OSCCs were genetic alterations that activate the PI3K-AKT-mTOR1 signaling pathway. Activation of this pathway by HPV E6, E7, and E5 is essential for several critical steps in the viral life cycle, including viral host cell entry (Surviladze et al. 2013), inhibition of autophagy to establish infection (Surviladze et al. 2013), viral replication (Spangle and Munger 2010), and resistance to T cell-mediated apoptosis (Noh et al. 2009). Considering the combined effects of somatic variants and CNVs, 80% of HPV-positive cancers harbored alterations in PIK3CA or PTEN. Moreover, activating mutations in FGFR3 (Eswarakumar et al. 2005) and inactivating mutations in the lysine 63 deubiquitinase CYLD (Yang et al. 2013) may activate this pathway. Our ranked VAF analysis also supported early onset of PI3K-AKT-mTOR1 pathway activation, consistent with in vitro raft cultures (Henken et al. 2011) and in vivo mouse models (Callejas-Valera et al. 2016), indicating that activation of this pathway is necessary for HPV-induced transformation.
Our data support disruption of epithelial differentiation as important for the pathogenesis of HPV-positive OSCCs (Fig. 7). The transcription factor ZNF750 was disrupted in 28% of HPV-positive OSCCs and was implicated by our ranked VAF analysis as an early driver (Figs. 1, 3, 6). ZNF750 is a ΔNp63-inducible gene essential for switching from self-renewal of basal keratinocytes to terminal differentiation (Cohen et al. 2012; Hazawa et al. 2017). Decreased ZNF750 expression promotes cellular proliferation while at the same time inhibiting terminal differentiation (Hazawa et al. 2017). ZNF750 is frequently mutated in squamous cell carcinomas such as esophageal and cervical cancers (Zhang et al. 2015; Campbell et al. 2018). Additional genes with known roles in regulation of epithelial differentiation that were recurrently mutated in HPV-positive OSCCs included lysine methyltransferase KMT2D (Lin-Shiao et al. 2018), the receptor-interacting protein kinase RIPK4 (Holland et al. 2002), the TGF receptor subunit TGFBR2 (Meyers et al. 2018), and EP300 (Wong et al. 2010). Overall, at least 37% of HPV-positive OSCCs had genetic mutations in regulators of epithelial differentiation, and 58% had mutations and/or CNV disrupting these genes. Productive HPV infection is dependent upon terminal differentiation of keratinocytes. The natural history of an HPV infection may diverge away from benign, productive infection toward tumorigenesis as a consequence of mutations that disrupt epithelial differentiation. Moreover, these mutations may explain the poorly differentiated or “basaloid” phenotype characteristic of HPV-positive OSCCs (Gillison et al. 2000).
We identified significant disruption of 17 genes by somatic variation among HPV-positive OSCCs, many of which encode potentially targetable proteins (e.g., FGFR3, PIK3CA, EP300) (Hayes et al. 2015). Most were highly distinct from those in HPV-negative OSCCs. When the affected host genes and pathways were considered collectively, we noted that cellular pathways that defend against viral infections were frequently disrupted by somatic mutations, including the Toll-like receptor (TLR), interferon gamma (IFNG), nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kappaB), and transforming growth factor beta (TGFbeta)-signaling pathways (Liang et al. 2015). Mutations in these genes and pathways could promote tumorigenesis by facilitating immune evasion by the virus and secondarily by the tumor.
Our CNV analysis confirmed that a focus on somatic variants alone would underestimate the overall frequency at which genes are disrupted in HPV-positive OSCCs. Coincident SNVs and CNVs at specific genetic loci suggested biallelic disruption (e.g., loss of heterozygosity) with enhanced functional impact. Our unbiased analysis of high-resolution WGS and RNA-seq data identified hundreds of genes whose expression was significantly altered by CNVs, precluding efforts to assign pathophysiological significance to any particular gene (The Cancer Genome Atlas Network 2015). Amplification of a region on Chr 3q25–29 common to all squamous cell carcinomas (Campbell et al. 2018) was more frequent among HPV-positive OSCCs (Fig. 5). Of particular interest were subchromosomal segments on Chr 11q, 14q, and 16q that were uniquely lost in HPV-positive and concomitantly gained in HPV-negative cancers, with commensurate but opposite changes in gene expression (e.g., inverse effects on CCND1, DICER1, and AKT1). Loss vs. gain in expression of the microRNA processing endoribonuclease DICER1 could have dramatic implications for gene expression profiles in HPV-positive vs. negative OSCCs. We are currently investigating these associations.
The anti-viral innate immune response leads to increased expression of type I IFN and IFN-stimulated genes, including the cytidine deaminase APOBEC3 family of viral restriction factors (Vieira and Soares 2013). A large majority of activating mutations in PIK3CA (Fig. 4) were products of APOBEC-mediated mutagenesis, corroborating a previous report (Henderson et al. 2014). More broadly, 47% of all SNVs detected in HPV-positive OSCCs occurred in an APOBEC signature context. The association between mutations in DNA repair genes and high APOBEC signature mutation burden supports a potential interaction between virus-induced APOBEC activity and inability to repair the consequent host genome editing. Although APOBEC3 family members inhibit HPV infectivity and promote HPV episomal loss (Ahasan et al. 2015), they paradoxically facilitate host genome mutagenesis (Warren et al. 2017). HPV E6 and E7 also induce transcription of and stabilize APOBEC3 family members (Vieira et al. 2014; Mori et al. 2017; Westrich et al. 2018), further contributing to accumulation of APOBEC signature mutations. These HPV-APOBEC interactions provide a striking example of virus-host interactions that cooperatively shape genomic alterations in these cancers.
Activation of the host anti-viral IFN signaling pathway is countered by HPV E6, E7, and E5 proteins to promote infection persistence (Westrich et al. 2017). For example, these viral proteins down-regulate expression of the dsDNA pattern recognition receptor TLR9 (Hasan et al. 2013) and repress transcription mediated by IFN regulatory factors 1, 3, and 9 (IRF1, IRF3, and IRF9) (Hong and Laimins 2017). Adding to these impacts of the virus were host genetic alterations in several genes in the TLR and IFN signaling pathways, including TRAF3, DDX3X, CYLD, and EP300. Consistent with prior reports (The Cancer Genome Atlas Network 2015; Chen et al. 2017; Hajek et al. 2017), 26% of HPV-positive OSCCs harbored mutation or deletion of a key mediator of TLR signaling, tumor necrosis factor receptor associated factor 3 (TRAF3). Also mutated in 5% was the DEAD-box RNA helicase 3 DDX3X, whose gene product directly interacts with TRAF3 to activate IRFs (Gu et al. 2017). DDX3X is frequently inactivated by viruses to suppress production of type I IFNs (Ariumi 2014), but to our knowledge no interactions with HPV oncoproteins have been reported. Moreover, the tumor suppressor cylindromatosis lysine 63 deubiquitinase (CYLD) (Bignell et al. 2000) was disrupted in 25% of HPV-positive OSCCs. Experimental knockout of CYLD reduced IFN-mediated signaling and expression of IFN-inducible genes (Zhang et al. 2011). Knockdown promoted proliferation, migration, and invasion, while preventing apoptosis of cervical cancer cells in vitro (Sanches et al. 2018). Triggering of the antiviral immune response is also dependent upon a complex between EP300 and IRF3, which together induce transcription of interferon and interferon-stimulated genes (Yoneyama et al. 1998).
Further highlighting the significance of an impaired anti-viral IFN signal transduction pathway in HPV-positive OSCCs were mutations detected in IFNGR1, which encodes a subunit of the IFNG receptor complex. A key transcription factor mediating IFN signaling, STAT1, also was recurrently mutated. Overall, 28% of HPV-positive OSCCs had genetic mutations in the IFN signaling pathway, and 52% had mutations and/or CNVs disrupting these genes. Collectively, virally mediated and host genetic alterations in TLR and IFN signaling, in the context of inactivated TP53, would prevent IFN-mediated growth arrest and facilitate persistence of HPV-infected cells (Hebner et al. 2007). Infected cells would also be resistant to IFNG-induced expression of MHC class I and II genes, thereby promoting immune evasion. Our findings thus have potential clinical implications for response to immunotherapy, given recent associations between loss of IFNG pathway genes and resistance to immune checkpoint blockade (Gao et al. 2016). We currently are investigating associations between these genetic alterations and the immune microenvironment of HPV-positive OSCCs.
Many of the IFN signaling pathway genes described above also are critical components of the NF-kappaB signaling pathway. Activation of this pathway is a common mechanism by which DNA tumor viruses, including HPV, promote cellular proliferation and inhibition of apoptosis (James et al. 2006; Xu et al. 2010; Sun and Cesarman 2011). Although NF-kappaB pathway activation may promote HPV clearance early in infection, in later stages it is associated with cervical cancer progression (Tilborghs et al. 2017). Among HPV-positive OSCCs, we observed mutually exclusive inactivating mutations in several negative regulators of the NF-kappaB signaling pathway, including TRAF3 (Guven-Maiorov et al. 2016), CYLD (Sun 2010; Lork et al. 2017), DDX3X (Xiang et al. 2016), and NFKBIA, the gene encoding NFKB inhibitor alpha (Natoli and Chiocca 2008). Although we confirmed NF-kappaB pathway-activating mutations in TRAF3 and CYLD (Hajek et al. 2017) and extended these to include an additional 17% of patients with DDX3X and NFKBIA mutations, no associations with these mutations and survival outcomes were observed (Supplemental Fig. S2O). Mutations in CYLD, TRAF3, and NFKBIA were enriched in Epstein-Barr virus-positive nasopharyngeal carcinoma (Li et al. 2017), implicating activation of the NF-kappaB pathway in promotion of both virus-caused cancers of the head and neck.
TGFB1 encodes a potent inhibitor of keratinocyte proliferation that is indirectly targeted by HPV16 E6, E7, and E5, resulting in promotion of cellular growth and immortalization (Nees et al. 2001). We identified mutations and deletions that disrupt a subunit of the TGFbeta receptor, TGFBR2, in ∼21% of HPV-positive OSCCs. These mutations may prevent TGFbeta-induced up-regulation of anti-viral IFNG expression in keratinocytes (Woodby et al. 2018) and therefore would be tumor-promoting. Moreover, they would disrupt growth inhibition induced by TGFbeta, whose expression otherwise is immunosuppressive in the tumor microenvironment (Travis and Sheppard 2014).
The HPV genome is chromatinized in both its episomal and integrated forms. Viral proteins alter epigenetic regulation of host gene expression to promote viral genome expression (Soto et al. 2017). Several chromatin modifiers and transcriptional regulators were recurrently mutated in HPV-positive OSCCs, including histone acetyltransferase EP300, lysine methyltransferases KMT2D and NSD1, the core histone HIST1H2AE, and transcription factors CASZ1, BBX, and TAF5. Such mutations can dramatically alter transcriptomes and cancer phenotypes, as demonstrated recently by NSD1 mutations. NSD1 mutations were linked to a subset of head and neck cancers characterized by global DNA hypomethylation, an “immune-cold” phenotype, and decreased expression of pro-inflammatory chemokines (Brennan et al. 2017; Papillon-Cavanagh et al. 2017). Novel to our analysis were missense and stop-gain mutations in the zinc-finger transcription factor CASZ1, potentially involved in regulation of transcription-coupled DNA repair (Lui et al. 2015). CASZ1 is a candidate tumor suppressor in neuroblastoma (Liu et al. 2011; Virden et al. 2012), and its biallelic loss via HPV integration and LOH has been reported in cervical cancer (Schmitz et al. 2012). We identified genetic alteration of another novel gene, BBX, in ∼47% of HPV-positive OSCCs. BBX is a member of the high-mobility group (HMG) box-containing family of transcription factor genes that includes SOX2 and HMGB1. Model systems support its role in regulation of the G1/S cell cycle transition (Chen et al. 2014b) and stem cell differentiation (Wang et al. 2016). BBX is located on the frequently amplified region on Chr 3q and is coordinately overexpressed. Further research is needed to clarify a role for BBX in the pathogenesis of HPV-positive OSCCs.
We acknowledge several limitations of our study. First, although we generated and studied a large data set, the statistical power afforded by 149 cancers nevertheless precluded reliable detection of recurrent mutations below ∼3% prevalence as well as accurate subclassification of tumors based on gene expression profiles. Second, our ranked VAFs in individual OSCCs were subject to several potential confounders (e.g., copy number variation, tumor purity, and subclonal heterogeneity of somatic variants). We acknowledge that only 40% of TP53 mutations ranked in the top 5% of all VAFs in HPV-negative OSCCs, lower than expected from an accepted genetic progression model (Califano et al. 1996). Ongoing analytical approaches designed to correct for local copy number changes and cancer sample purity are likely to help identify which highly ranked VAFs are the most reliable indicators of early mutagenesis. Third, differences in sequencing protocols and platforms used over time necessitated bioinformatics adjustments, including batch correction of RNA-seq data. We did not detect substantial changes in variant calls or in overall transcript levels when various alignment software versions or reference genome assembly releases GRCh37 (hg19) vs. GRCh38 (hg38) were compared (Supplemental Figs. S1K, S5K). Fourth, copy number estimates for sex chromosomes can be unreliable, but this impacted only DDX3X on Chr X. And finally, we did not profile epigenetic marks using methods such as chromatin immunoprecipitation sequencing (ChIP-seq) or methylation sequencing (Methyl-seq). These and other genomics and epigenomics data will add important new insights about epigenetic regulation and transcription in HPV-positive OSCCs beyond what we analyzed here.
We conclude that HPV-positive OSCCs are genetically distinct from HPV-negative OSCCs in numerous ways. HPV-positive OSCCs are characterized by secondary genetic alterations in genes and pathways frequently targeted by HPV oncoproteins and those defending against viral infections, including the interferon signaling pathway. These genetic alterations would promote HPV infection persistence, carcinogenesis, and tumor immune evasion. We conclude that viral-host interactions cooperatively shape the distinctive genomic features of HPV-positive OSCCs.
Methods
Additional detailed methods are provided in the Supplemental Material.
Study population
Patients newly diagnosed with oral cavity or oropharyngeal squamous cell carcinoma at The Ohio State University Comprehensive Cancer Center from 2011 to 2016 were eligible to participate in a study of the genomics of oral cancer. Patients provided written, informed consent for genomics studies including WGS of T/N pairs and RNA-seq, prospective collection of clinical data, and risk factor data by computer-assisted self-interview. The study was approved by Institutional Review Boards at Ohio State University and at the University of Texas MD Anderson Cancer Center.
The overall study population included 112 subjects from the Ohio cohort and 372 from The Cancer Genome Atlas project (TCGA) (Supplemental Table S1A). The 149 HPV-positive OSCC cases included 103 studied by whole genome sequencing (WGS) and 46 by whole exome sequencing (WES). Of the WGS T/N pairs, 86 were in the Ohio cohort and 17 were downloaded from the TCGA website at https://gdc.cancer.gov/. The 335 HPV-negative OSCC cases included 50 T/N pairs studied by WGS (26 from our Ohio cohort; 24 from TCGA) and 285 pairs studied by WES (TCGA). Sequencing, clinical, and demographic data were downloaded from TCGA.
HPV testing in the Ohio cohort
Tumor DNA samples (see Supplemental Methods for details) were tested for 37 HPV types using the Roche LINEAR ARRAY (Roche Molecular Diagnostics). Viral types were confirmed by identification of type-specific HPV E6/E7 genes using TaqMan quantitative PCR (qPCR) assay (Koshiol et al. 2011). Expression of type-specific HPV E6/E7 transcripts was quantified by reverse transcription followed by TaqMan qPCR assay (Jordan et al. 2012). Corresponding formalin-fixed and paraffin-embedded cancer samples were analyzed by p16 protein immunohistochemistry and HPV in situ hybridization (Jordan et al. 2012).
Genomic DNA sequencing
A total of 103 HPV-positive and 50 HPV-negative OSCC T/N pairs were studied by WGS, including 86 HPV-positive and 26 HPV-negative T/N pairs from the Ohio cohort. Details of genomic DNA sequencing and RNA-seq libraries and analysis are available in the Methods section of the Supplemental Materials.
HPV transcript analysis
RNA-seq reads were aligned against a custom, hybrid genome comprised of human reference hg19 with 13 appended HPV type genome sequences, i.e., types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 69 (Akagi et al. 2014) using GSNAP (version 2014-12-20) (Wu and Nacu 2010). To quantify HPV16 transcript isoforms in 126 HPV16-positive OSCCs, we used Cufflinks (Trapnell et al. 2012) with a hybrid template of RefSeq hg19 plus HPV16 transcript isoforms (Zheng and Baker 2006). Further details are available in the Methods section of the Supplemental Materials.
Based on clinical annotations for OSCC samples from TCGA, 482 oral cavity and oropharyngeal cancers with available RNA-seq data were selected for downloading and analysis. RNA-seq reads were aligned against reference genomes for the same 13 HPV types using BWA-MEM (version 0.5.9) (Li and Durbin 2010). Virus status was determined based on detection of >1 virus transcript fragment per kilobasepair per million aligned fragments (FPKM).
Somatic variant detection
For WGS samples sequenced at Complete Genomics (CGI), somatic variants (i.e., SNVs and short indels) were identified using the CGI Cancer Genome Pipeline v. 2. Variants with CGI somatic quality scores <0 were excluded from further analysis. Somatic variants in Illumina WGS tumors from the Ohio cohort or TCGA were detected using MuTect v1.1.4 (Cibulskis et al. 2013), LoFreq v2 (Wilm et al. 2012), and Strelka v.1.0.13 (Saunders et al. 2012) to detect SNVs. SomaticIndelDetector v.2.3-9 (part of GATK, Broad Institute) and Strelka v.1.0.13 (Saunders et al. 2012) were used to detect small indels (all at default settings). Somatic variants were filtered based on the default quality pass filter for each caller. Variants identified by only one somatic variant caller were excluded. Somatic variant calls from samples studied with WES were downloaded and counted without further modification from the TCGA portal (BI_IlluminaGA_DNASeq_automated, level 2). Counts of somatic variants were determined from “exonized” WGS data (both CGI and Illumina), based on the Agilent SureSelect Human All Exon v.5 panel. Somatic variant functional consequences were annotated using Variant Effect Predictor (VEP v. 79) (McLaren et al. 2016) using the GRCh37 (hg19) human genome assembly option. The details of somatic variant confirmation and annotation of somatic SNVs are available in the Methods section of the Supplemental Materials.
Mutation signatures
The 3-nt sequence context for each SNV (out of 96 possible genomic 3-nt sequences) was extracted and counted based on mapped chromosomal coordinates and the reference human genome hg19. The 3-nt mutation frequencies per tumor were used as inputs for a signature calling tool, deconstructSigs (Rosenthal et al. 2016) which de-convoluted them into 30 pre-defined COSMIC cancer mutation signatures (http://cancer.sanger.ac.uk/cosmic/signatures). Potential associations between signatures and candidate risk factors were assessed by t-test or Pearson's correlation using the t-test or corr.test functions in R, respectively (R Core Team 2018).
Identification of significantly mutated genes
To identify significantly mutated genes, MutSigCV 1.4 was run at default settings (Lawrence et al. 2013), using as inputs both variants identified from WES data sets, downloaded from TCGA without modification, and variants called from WGS data as annotated by VEP v.79. MutSig gene candidate outputs with q-value <0.2 were filtered further to impose minimum thresholds of expression and prevalence in OSCC patients, so RNA-seq expression values of each gene candidate were required to have median FPKM > 1 across the panel of OSCCs, and >2% of patients would have coding-change variants in each gene.
The details of Gene Ontology analyses, CNV detection, and frequency distribution of copy number gains or losses are available in the Methods section of the Supplemental Materials.
Association between CNVs and altered gene expression
Using WGS data, the longest genomic fragment with a single estimated copy number (i.e., “estimated ploidy”) overlapping with each gene was identified. This estimated ploidy was assigned to each gene as defined by GENCODE release 18 (based on GRCh37.p12) (https://www.gencodegenes.org/human/release_18.html). Genes were identified for which three or more samples harbored local chromosomal gains or losses, with gains defined by estimated ploidy N > 2.5 and losses by ploidy N < 1.5. Gene expression data were normalized as described for RNA-seq analysis (details are available in the Methods section of the Supplemental Materials). Autosomal protein-coding genes were included in further analysis, while sex chromosome genes were excluded. After normalization, genes showing expression variance <0.3 (due to very low expression levels across almost all samples) were not studied further. The t-test statistic was calculated to compare expression levels in samples with normal copy numbers vs. gains, or separately between normal vs. losses. Multiple testing correction with a false discovery rate (FDR) method was performed.
Ranked VAF analysis
For each called somatic variant, high quality reads mapping to the reference or alternative alleles were counted. For CGI WGS variants, read counts were extracted from masterVar output files produced by CGI Cancer Sequencing pipeline v2.2. For Illumina WGS variants, read counts were extracted from MuTect, LoFreq, or Strelka outputs for SNVs, or SomaticIndelDetector or Strelka outputs for small indels. For TCGA WES variants, aligned sequence reads were downloaded in BAM format from the TCGA portal (BI_IlluminaGA_DNASeq_automated, Level 2), and counted using SAMtools mpileup (Li et al. 2009). Somatic variants were included for further VAF analysis when localized within gene exons (Ensembl v. 79), with >20 mapped reads and VAF >5%. A total of 26,646 somatic variants in 145 HPV-positive samples (i.e., Ohio CGI, n = 34; Ohio Illumina WGS, n = 52; TCGA-WGS, n = 17; and TCGA-WES, n = 42; with four other TCGA-WES samples unavailable for this analysis) and 46,610 somatic variants in 329 HPV-negative samples (i.e., Ohio CGI, n = 25; Ohio Illumina, n = 1; TCGA-WGS, n = 24; and TCGA-WES, n = 279) met these criteria. Since VAF distributions differed dramatically between cancers, they were ranked relative to other somatic variants within individual OSCCs. The distribution of ranked VAFs was plotted for all HPV-positive or HPV-negative cancers. To assess the statistical significance of ranked VAF distributions, they were assigned to 20 bins of five-percentile increments. For genes with three or more somatic variants in different cancer samples, we used the binomial statistic to compare the observed vs. expected numbers of ranked VAFs in the top fifth percentile bin. Multiple testing correction using the FDR method was performed to calculate adjusted P-values.
Data access
WGS and RNA-seq data from this study have been submitted to the European Genome-phenome Archive (EGA; https://ega-archive.org/) hosted by the European Bioinformatics Institute (EBI), European Molecular Biology Laboratory (EMBL), under accession numbers EGAS00001002393, EGAS00001003228, and EGAS00001003237.
Supplementary Material
Acknowledgments
We thank the patients with oropharyngeal and oral cavity cancers at Ohio State University who enrolled in our research study. We thank Drs. Jeffrey N. Myers and Faye M. Johnson at MD Anderson Cancer Center and members of the Gillison and Symer laboratories for insightful comments at various stages of this study; TCGA for access to whole exome sequencing and RNA-seq data; Elisa Venturini, Karen Bunting, Benjamin Hubert, and Dayna M. Ochswald for expert help with project management at the New York Genome Center; the Genomics Shared Resource at Ohio State University Comprehensive Cancer Center for DNA and RNA quality assays (supported by National Cancer Institute grant P30CA016058); and Anthony S. Baker (OSU), and Jordan Pietz and Jennifer McGee (MDA), for help preparing graphical figures. This study was funded by the Oral Cancer Foundation (M.L.G.), Ohio State University Comprehensive Cancer Center (M.L.G., D.E.S.), University of Texas MD Anderson Cancer Center (M.L.G., D.E.S.), Ohio Supercomputer Center (PAS0425; D.E.S.), Ohio Cancer Research Associate grant (GRT00024299; K.A.), Cancer Prevention Research Institute of Texas (CPRIT; M.L.G.), and National Cancer Institute grant R50CA211533 (K.A.). M.L.G. is a CPRIT Scholar.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.241141.118.
CDKN2A encodes a number of distinct protein isoforms. p16 (INK4) is structurally unrelated and functionally different from an alternate open reading frame (ARF) encoding a protein product that functions as a tumor suppressor (also known as p14).
References
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et al. 2011. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 333: 1154–1157. 10.1126/science.1206923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahasan MM, Wakae K, Wang Z, Kitamura K, Liu G, Koura M, Imayasu M, Sakamoto N, Hanaoka K, Nakamura M, et al. 2015. APOBEC3A and 3C decrease human papillomavirus 16 pseudovirion infectivity. Biochem Biophys Res Commun 457: 295–299. 10.1016/j.bbrc.2014.12.103 [DOI] [PubMed] [Google Scholar]
- Akagi K, Li J, Broutian TR, Padilla-Nash H, Xiao W, Jiang B, Rocco JW, Teknos TN, Kumar B, Wangsa D, et al. 2014. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res 24: 185–199. 10.1101/gr.164806.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. 2013. Signatures of mutational processes in human cancer. Nature 500: 415–421. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Ju YS, Haase K, Van Loo P, Martincorena I, Nik-Zainal S, Totoki Y, Fujimoto A, Nakagawa H, Shibata T, et al. 2016. Mutational signatures associated with tobacco smoking in human cancer. Science 354: 618–622. 10.1126/science.aag0299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, Westra WH, Chung CH, Jordan RC, Lu C, et al. 2010. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 363: 24–35. 10.1056/NEJMoa0912217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariumi Y. 2014. Multiple functions of DDX3 RNA helicase in gene regulation, tumorigenesis, and viral infection. Front Genet 5: 423 10.3389/fgene.2014.00423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belleudi F, Nanni M, Raffa S, Torrisi MR. 2015. HPV16 E5 deregulates the autophagic process in human keratinocytes. Oncotarget 6: 9370–9386. 10.18632/oncotarget.3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, Green H, Brown C, Biggs PJ, Lakhani SR, et al. 2000. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet 25: 160–165. 10.1038/76006 [DOI] [PubMed] [Google Scholar]
- Brennan K, Shin JH, Tay JK, Prunello M, Gentles AJ, Sunwoo JB, Gevaert O. 2017. NSD1 inactivation defines an immune cold, DNA hypomethylated subtype in squamous cell carcinoma. Sci Rep 7: 17064 10.1038/s41598-017-17298-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Califano J, van der Riet P, Westra W, Nawroz H, Clayman G, Piantadosi S, Corio R, Lee D, Greenberg B, Koch W, et al. 1996. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res 56: 2488–2492. [PubMed] [Google Scholar]
- Callejas-Valera JL, Iglesias-Bartolome R, Amornphimoltham P, Palacios-Garcia J, Martin D, Califano JA, Molinolo AA, Gutkind JS. 2016. mTOR inhibition prevents rapid-onset of carcinogen-induced malignancies in a novel inducible HPV-16 E6/E7 mouse model. Carcinogenesis 37: 1014–1025. 10.1093/carcin/bgw086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JD, Yau C, Bowlby R, Liu Y, Brennan K, Fan H, Taylor AM, Wang C, Walter V, Akbani R, et al. 2018. Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep 23: 194–212.e6. 10.1016/j.celrep.2018.03.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. 2015. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517: 576–582. 10.1038/nature14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Tan W, Ling Z, Xi R, Shao M, Chen M, Luo Y, Zhao Y, Liu Y, Huang X, et al. 2017. Genomic analysis of oesophageal squamous-cell carcinoma identifies alcohol drinking-related mutation signature and genomic alterations. Nat Commun 8: 15290 10.1038/ncomms15290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, et al. 2011. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol 29: 4294–4301. 10.1200/JCO.2011.36.4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Xue Y, Poidinger M, Lim T, Chew SH, Pang CL, Abastado JP, Thierry F. 2014a. Mapping of HPV transcripts in four human cervical lesions using RNAseq suggests quantitative rearrangements during carcinogenic progression. Virology 462-463: 14–24. 10.1016/j.virol.2014.05.026 [DOI] [PubMed] [Google Scholar]
- Chen T, Zhou L, Yuan Y, Fang Y, Guo Y, Huang H, Zhou Q, Lv X. 2014b. Characterization of Bbx, a member of a novel subfamily of the HMG-box superfamily together with Cic. Dev Genes Evol 224: 261–268. 10.1007/s00427-014-0476-x [DOI] [PubMed] [Google Scholar]
- Chen T, Zhang J, Chen Z, Van Waes C. 2017. Genetic alterations in TRAF3 and CYLD that regulate nuclear factor κB and interferon signaling define head and neck cancer subsets harboring human papillomavirus. Cancer 123: 1695–1698. 10.1002/cncr.30659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CH, Guthrie VB, Masica DL, Tokheim C, Kang H, Richmon J, Agrawal N, Fakhry C, Quon H, Subramaniam RM, et al. 2015. Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing. Ann Oncol 26: 1216–1223. 10.1093/annonc/mdv109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. 2013. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31: 213–219. 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen I, Birnbaum RY, Leibson K, Taube R, Sivan S, Birk OS. 2012. ZNF750 is expressed in differentiated keratinocytes and regulates epidermal late differentiation genes. PLoS One 7: e42628 10.1371/journal.pone.0042628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ, Welcker M, Clurman BE. 2014. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell 26: 455–464. 10.1016/j.ccell.2014.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duperret EK, Oh SJ, McNeal A, Prouty SM, Ridky TW. 2014. Activating FGFR3 mutations cause mild hyperplasia in human skin, but are insufficient to drive benign or malignant skin tumors. Cell Cycle 13: 1551–1559. 10.4161/cc.28492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt M, Zhang W, Gross AM, Von Dollen J, Johnson JR, Franks-Skiba KE, Swaney DL, Johnson TL, Jang GM, Shah PS, et al. 2018. Multiple routes to oncogenesis are promoted by the human papillomavirus–host protein network. Cancer Discov 8: 1474–1489. 10.1158/2159-8290.CD-17-1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. 2005. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev 16: 139–149. 10.1016/j.cytogfr.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Froimchuk E, Jang Y, Ge K. 2017. Histone H3 lysine 4 methyltransferase KMT2D. Gene 627: 337–342. 10.1016/j.gene.2017.06.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, Chen T, Roszik J, Bernatchez C, Woodman SE, et al. 2016. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 167: 397–404.e9. 10.1016/j.cell.2016.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE, et al. 2000. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst 92: 709–720. 10.1093/jnci/92.9.709 [DOI] [PubMed] [Google Scholar]
- Gillison ML, D'Souza G, Westra W, Sugar E, Xiao W, Begum S, Viscidi R. 2008. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J Natl Cancer Inst 100: 407–420. 10.1093/jnci/djn025 [DOI] [PubMed] [Google Scholar]
- Gu L, Fullam A, McCormack N, Hohn Y, Schroder M. 2017. DDX3 directly regulates TRAF3 ubiquitination and acts as a scaffold to co-ordinate assembly of signalling complexes downstream from MAVS. Biochem J 474: 571–587. 10.1042/BCJ20160956 [DOI] [PubMed] [Google Scholar]
- Guven-Maiorov E, Keskin O, Gursoy A, VanWaes C, Chen Z, Tsai CJ, Nussinov R. 2016. TRAF3 signaling: competitive binding and evolvability of adaptive viral molecular mimicry. Biochim Biophys Acta 1860: 2646–2655. 10.1016/j.bbagen.2016.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajek M, Sewell A, Kaech S, Burtness B, Yarbrough WG, Issaeva N. 2017. TRAF3/CYLD mutations identify a distinct subset of human papillomavirus-associated head and neck squamous cell carcinoma. Cancer 123: 1778–1790. 10.1002/cncr.30570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan UA, Zannetti C, Parroche P, Goutagny N, Malfroy M, Roblot G, Carreira C, Hussain I, Muller M, Taylor-Papadimitriou J, et al. 2013. The human papillomavirus type 16 E7 oncoprotein induces a transcriptional repressor complex on the Toll-like receptor 9 promoter. J Exp Med 210: 1369–1387. 10.1084/jem.20122394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes DN, Van Waes C, Seiwert TY. 2015. Genetic landscape of human papillomavirus–associated head and neck cancer and comparison to tobacco-related tumors. J Clin Oncol 33: 3227–3234. 10.1200/JCO.2015.62.1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazawa M, Lin DC, Handral H, Xu L, Chen Y, Jiang YY, Mayakonda A, Ding LW, Meng X, Sharma A, et al. 2017. ZNF750 is a lineage-specific tumour suppressor in squamous cell carcinoma. Oncogene 36: 2243–2254. 10.1038/onc.2016.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebner C, Beglin M, Laimins LA. 2007. Human papillomavirus E6 proteins mediate resistance to interferon-induced growth arrest through inhibition of p53 acetylation. J Virol 81: 12740–12747. 10.1128/JVI.00987-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S, Chakravarthy A, Su X, Boshoff C, Fenton TR. 2014. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep 7: 1833–1841. 10.1016/j.celrep.2014.05.012 [DOI] [PubMed] [Google Scholar]
- Henken FE, Banerjee NS, Snijders PJ, Meijer CJ, De-Castro Arce J, Rösl F, Broker TR, Chow LT, Steenbergen RD. 2011. PIK3CA-mediated PI3-kinase signalling is essential for HPV-induced transformation in vitro. Mol Cancer 10: 71 10.1186/1476-4598-10-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland P, Willis C, Kanaly S, Glaccum M, Warren A, Charrier K, Murison J, Derry J, Virca G, Bird T, et al. 2002. RIP4 is an ankyrin repeat-containing kinase essential for keratinocyte differentiation. Curr Biol 12: 1424–1428. 10.1016/S0960-9822(02)01075-8 [DOI] [PubMed] [Google Scholar]
- Hong S, Laimins LA. 2017. Manipulation of the innate immune response by human papillomaviruses. Virus Res 231: 34–40. 10.1016/j.virusres.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe-Seyler K, Bossler F, Braun JA, Herrmann AL, Hoppe-Seyler F. 2018. The HPV E6/E7 oncogenes: key factors for viral carcinogenesis and therapeutic targets. Trends Microbiol 26: 158–168. 10.1016/j.tim.2017.07.007 [DOI] [PubMed] [Google Scholar]
- Ilmarinen T, Munne P, Hagström J, Haglund C, Auvinen E, Virtanen EI, Haesevoets A, Speel EJM, Aaltonen LM. 2017. Prevalence of high-risk human papillomavirus infection and cancer gene mutations in nonmalignant tonsils. Oral Oncol 73: 77–82. 10.1016/j.oraloncology.2017.08.010 [DOI] [PubMed] [Google Scholar]
- James MA, Lee JH, Klingelhutz AJ. 2006. Human papillomavirus type 16 E6 activates NF-κB, induces cIAP-2 expression, and protects against apoptosis in a PDZ binding motif-dependent manner. J Virol 80: 5301–5307. 10.1128/JVI.01942-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan RC, Lingen MW, Perez-Ordonez B, He X, Pickard R, Koluder M, Jiang B, Wakely P, Xiao W, Gillison ML. 2012. Validation of methods for oropharyngeal cancer HPV status determination in US cooperative group trials. Am J Surg Pathol 36: 945–954. 10.1097/PAS.0b013e318253a2d1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck MK, Zuo Z, Khattri A, Stricker TP, Brown CD, Imanguli M, Rieke D, Endhardt K, Fang P, Bragelmann J, et al. 2015. Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin Cancer Res 21: 870–881. 10.1158/1078-0432.CCR-14-2481 [DOI] [PubMed] [Google Scholar]
- Koshiol J, Rotunno M, Gillison ML, Van Doorn LJ, Chaturvedi AK, Tarantini L, Song H, Quint WG, Struijk L, Goldstein AM, et al. 2011. Assessment of human papillomavirus in lung tumor tissue. J Natl Cancer Inst 103: 501–507. 10.1093/jnci/djr003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. 2013. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499: 214–218. 10.1038/nature12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letouze E, Shinde J, Renault V, Couchy G, Blanc JF, Tubacher E, Bayard Q, Bacq D, Meyer V, Semhoun J, et al. 2017. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat Commun 8: 1315 10.1038/s41467-017-01358-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Chung GT, Lui VW, To KF, Ma BB, Chow C, Woo JK, Yip KY, Seo J, Hui EP, et al. 2017. Exome and genome sequencing of nasopharynx cancer identifies NF-κB pathway activating mutations. Nat Commun 8: 14121 10.1038/ncomms14121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Oh BH, Jung JU. 2015. Novel functions of viral anti-apoptotic factors. Nat Rev Microbiol 13: 7–12. 10.1038/nrmicro3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Shiao E, Lan Y, Coradin M, Anderson A, Donahue G, Simpson CL, Sen P, Saffie R, Busino L, Garcia BA, et al. 2018. KMT2D regulates p63 target enhancers to coordinate epithelial homeostasis. Genes Dev 32: 181–193. 10.1101/gad.306241.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yang X, Li Z, McMahon C, Sizer C, Barenboim-Stapleton L, Bliskovsky V, Mock B, Ried T, London WB, et al. 2011. CASZ1, a candidate tumor-suppressor gene, suppresses neuroblastoma tumor growth through reprogramming gene expression. Cell Death Differ 18: 1174–1183. 10.1038/cdd.2010.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lork M, Verhelst K, Beyaert R. 2017. CYLD, A20 and OTULIN deubiquitinases in NF-κB signaling and cell death: so similar, yet so different. Cell Death Differ 24: 1172–1183. 10.1038/cdd.2017.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui GY, Kovacevic Z, Richardson V, Merlot AM, Kalinowski DS, Richardson DR. 2015. Targeting cancer by binding iron: dissecting cellular signaling pathways. Oncotarget 6: 18748–18779. 10.18632/oncotarget.4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F. 2016. The Ensembl Variant Effect Predictor. Genome Biol 17: 122 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Crum CP, Münger K. 2011. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci 108: 2130–2135. 10.1073/pnas.1009933108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesri EA, Feitelson MA, Munger K. 2014. Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe 15: 266–282. 10.1016/j.chom.2014.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers JM, Grace M, Uberoi A, Lambert PF, Munger K. 2018. Inhibition of TGF-β and NOTCH signaling by cutaneous papillomaviruses. Front Microbiol 9: 389 10.3389/fmicb.2018.00389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabello L, Yeager M, Yu K, Clifford GM, Xiao Y, Zhu B, Cullen M, Boland JF, Wentzensen N, Nelson CW, et al. 2017. HPV16 E7 genetic conservation is critical to carcinogenesis. Cell 170: 1164–1174.e6. 10.1016/j.cell.2017.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Takeuchi T, Ishii Y, Yugawa T, Kiyono T, Nishina H, Kukimoto I. 2017. Human papillomavirus 16 E6 upregulates APOBEC3B via the TEAD transcription factor. J Virol 91: e02413-16 10.1128/JVI.02413-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Chiocca S. 2008. Nuclear ubiquitin ligases, NF-κB degradation, and the control of inflammation. Sci Signal 1: pe1 10.1126/stke.11pe1 [DOI] [PubMed] [Google Scholar]
- Nees M, Geoghegan JM, Hyman T, Frank S, Miller L, Woodworth CD. 2001. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-κB-responsive genes in cervical keratinocytes. J Virol 75: 4283–4296. 10.1128/JVI.75.9.4283-4296.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PK, Li J, Jeong KJ, Shao S, Chen H, Tsang YH, Sengupta S, Wang Z, Bhavana VH, Tran R, et al. 2018. Systematic functional annotation of somatic mutations in cancer. Cancer Cell 33: 450–462.e10. 10.1016/j.ccell.2018.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh KH, Kang TH, Kim JH, Pai SI, Lin KY, Hung CF, Wu TC, Kim TW. 2009. Activation of Akt as a mechanism for tumor immune evasion. Mol Ther 17: 439–447. 10.1038/mt.2008.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo-Nieva L, Muñoz-Bello JO, Contreras-Paredes A, Lizano M. 2018. The role of E6 spliced isoforms (E6*) in human papillomavirus-induced carcinogenesis. Viruses 10: 45 10.3390/v10010045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papillon-Cavanagh S, Lu C, Gayden T, Mikael LG, Bechet D, Karamboulas C, Ailles L, Karamchandani J, Marchione DM, Garcia BA, et al. 2017. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat Genet 49: 180–185. 10.1038/ng.3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering CR, Zhang J, Yoo SY, Bengtsson L, Moorthy S, Neskey DM, Zhao M, Ortega Alves MV, Chang K, Drummond J, et al. 2013. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov 3: 770–781. 10.1158/2159-8290.CD-12-0537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- Rayner E, van Gool IC, Palles C, Kearsey SE, Bosse T, Tomlinson I, Church DN. 2016. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat Rev Cancer 16: 71–81. 10.1038/nrc.2015.12 [DOI] [PubMed] [Google Scholar]
- Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. 2016. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 17: 31 10.1186/s13059-016-0893-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanches JGP, Xu Y, Yabasin IB, Li M, Lu Y, Xiu X, Wang L, Mao L, Shen J, Wang B, et al. 2018. miR-501 is upregulated in cervical cancer and promotes cell proliferation, migration and invasion by targeting CYLD. Chem Biol Interact 285: 85–95. 10.1016/j.cbi.2018.02.024 [DOI] [PubMed] [Google Scholar]
- Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. 2012. Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics 28: 1811–1817. 10.1093/bioinformatics/bts271 [DOI] [PubMed] [Google Scholar]
- Schmitz M, Driesch C, Beer-Grondke K, Jansen L, Runnebaum IB, Dürst M. 2012. Loss of gene function as a consequence of human papillomavirus DNA integration. Int J Cancer 131: E593–E602. 10.1002/ijc.27433 [DOI] [PubMed] [Google Scholar]
- Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker T, Brown C, Pugh TJ, Stojanov P, Cho J, et al. 2015. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res 21: 632–641. 10.1158/1078-0432.CCR-13-3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin MK, Pitot HC, Lambert PF. 2012. Pocket proteins suppress head and neck cancer. Cancer Res 72: 1280–1289. 10.1158/0008-5472.CAN-11-2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slebos RJ, Yi Y, Ely K, Carter J, Evjen A, Zhang X, Shyr Y, Murphy BM, Cmelak AJ, Burkey BB, et al. 2006. Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res 12: 701–709. 10.1158/1078-0432.CCR-05-2017 [DOI] [PubMed] [Google Scholar]
- Smeets SJ, Braakhuis BJ, Abbas S, Snijders PJ, Ylstra B, van de Wiel MA, Meijer GA, Leemans CR, Brakenhoff RH. 2006. Genome-wide DNA copy number alterations in head and neck squamous cell carcinomas with or without oncogene-expressing human papillomavirus. Oncogene 25: 2558–2564. 10.1038/sj.onc.1209275 [DOI] [PubMed] [Google Scholar]
- Soto D, Song C, McLaughlin-Drubin ME. 2017. Epigenetic alterations in human papillomavirus-associated cancers. Viruses 9: 248 10.3390/v9090248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangle JM, Munger K. 2010. The human papillomavirus type 16 E6 oncoprotein activates mTORC1 signaling and increases protein synthesis. J Virol 84: 9398–9407. 10.1128/JVI.00974-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. 2011. The mutational landscape of head and neck squamous cell carcinoma. Science 333: 1157–1160. 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC. 2010. CYLD: a tumor suppressor deubiquitinase regulating NF-κB activation and diverse biological processes. Cell Death Differ 17: 25–34. 10.1038/cdd.2009.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC, Cesarman E. 2011. NF-κB as a target for oncogenic viruses. Curr Top Microbiol Immunol 349: 197–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surviladze Z, Sterk RT, DeHaro SA, Ozbun MA. 2013. Cellular entry of human papillomavirus type 16 involves activation of the phosphatidylinositol 3-kinase/Akt/mTOR pathway and inhibition of autophagy. J Virol 87: 2508–2517. 10.1128/JVI.02319-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilborghs S, Corthouts J, Verhoeven Y, Arias D, Rolfo C, Trinh XB, van Dam PA. 2017. The role of Nuclear Factor-κ B signaling in human cervical cancer. Crit Rev Oncol Hematol 120: 141–150. 10.1016/j.critrevonc.2017.11.001 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis MA, Sheppard D. 2014. TGF-β activation and function in immunity. Annu Rev Immunol 32: 51–82. 10.1146/annurev-immunol-032713-120257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira VC, Soares MA. 2013. The role of cytidine deaminases on innate immune responses against human viral infections. Biomed Res Int 2013: 683095 10.1155/2013/683095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira VC, Leonard B, White EA, Starrett GJ, Temiz NA, Lorenz LD, Lee D, Soares MA, Lambert PF, Howley PM, et al. 2014. Human papillomavirus E6 triggers upregulation of the antiviral and cancer genomic DNA deaminase APOBEC3B. MBio 5: e02234-14 10.1128/mBio.02234-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virden RA, Thiele CJ, Liu Z. 2012. Characterization of critical domains within the tumor suppressor CASZ1 required for transcriptional regulation and growth suppression. Mol Cell Biol 32: 1518–1528. 10.1128/MCB.06039-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Tang MC, Chang WC, Furushima K, Jang CW, Behringer RR, Chen CM. 2016. PiggyBac transposon-mediated mutagenesis in rats reveals a crucial role of Bbx in growth and male fertility. Biol Reprod 95: 51 10.1095/biolreprod.116.141739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CJ, Westrich JA, Doorslaer KV, Pyeon D. 2017. Roles of APOBEC3A and APOBEC3B in human papillomavirus infection and disease progression. Viruses 9: 233 10.3390/v9080233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westrich JA, Warren CJ, Pyeon D. 2017. Evasion of host immune defenses by human papillomavirus. Virus Res 231: 21–33. 10.1016/j.virusres.2016.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westrich JA, Warren CJ, Klausner MJ, Guo K, Liu C, Santiago ML, Pyeon D. 2018. Human papillomavirus 16 E7 stabilizes APOBEC3A protein by inhibiting Cullin 2-dependent protein degradation. J Virol 92: e01318-17 10.1128/JVI.01318-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N. 2012. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 40: 11189–11201. 10.1093/nar/gks918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong PP, Pickard A, McCance DJ. 2010. p300 alters keratinocyte cell growth and differentiation through regulation of p21Waf1/CIP1. PLoS One 5: e8369 10.1371/journal.pone.0008369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodby BL, Songock WK, Scott ML, Raikhy G, Bodily JM. 2018. Induction of interferon κ in human papillomavirus 16 infection by transforming growth factor β-induced promoter demethylation. J Virol 92: e01714-17 10.1128/JVI.01714-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26: 873–881. 10.1093/bioinformatics/btq057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang N, He M, Ishaq M, Gao Y, Song F, Guo L, Ma L, Sun G, Liu D, Guo D, et al. 2016. The DEAD-box RNA helicase DDX3 interacts with NF-κB subunit p65 and suppresses p65-mediated transcription. PLoS One 11: e0164471 10.1371/journal.pone.0164471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Katzenellenbogen RA, Grandori C, Galloway DA. 2010. NFX1 plays a role in human papillomavirus type 16 E6 activation of NFκB activity. J Virol 84: 11461–11469. 10.1128/JVI.00538-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WL, Jin G, Li CF, Jeong YS, Moten A, Xu D, Feng Z, Chen W, Cai Z, Darnay B, et al. 2013. Cycles of ubiquitination and deubiquitination critically regulate growth factor–mediated activation of Akt signaling. Sci Signal 6: ra3 10.1126/scisignal.2003197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J 17: 1087–1095. 10.1093/emboj/17.4.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Lee AJ, Wu X, Sun SC. 2011. Regulation of antiviral innate immunity by deubiquitinase CYLD. Cell Mol Immunol 8: 502–504. 10.1038/cmi.2011.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Poh CF, Williams M, Laronde DM, Berean K, Gardner PJ, Jiang H, Wu L, Lee JJ, Rosin MP. 2012. Loss of heterozygosity (LOH) profiles—validated risk predictors for progression to oral cancer. Cancer Prev Res (Phila) 5: 1081–1089. 10.1158/1940-6207.CAPR-12-0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhou Y, Cheng C, Cui H, Cheng L, Kong P, Wang J, Li Y, Chen W, Song B, et al. 2015. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet 96: 597–611. 10.1016/j.ajhg.2015.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng ZM, Baker CC. 2006. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci 11: 2286–2302. 10.2741/1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann H, Degenkolbe R, Bernard HU, O'Connor MJ. 1999. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J Virol 73: 6209–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.