Abstract
Cannabis sativa is widely cultivated for medicinal, food, industrial, and recreational use, but much remains unknown regarding its genetics, including the molecular determinants of cannabinoid content. Here, we describe a combined physical and genetic map derived from a cross between the drug-type strain Purple Kush and the hemp variety “Finola.” The map reveals that cannabinoid biosynthesis genes are generally unlinked but that aromatic prenyltransferase (AP), which produces the substrate for THCA and CBDA synthases (THCAS and CBDAS), is tightly linked to a known marker for total cannabinoid content. We further identify the gene encoding CBCA synthase (CBCAS) and characterize its catalytic activity, providing insight into how cannabinoid diversity arises in cannabis. THCAS and CBDAS (which determine the drug vs. hemp chemotype) are contained within large (>250 kb) retrotransposon-rich regions that are highly nonhomologous between drug- and hemp-type alleles and are furthermore embedded within ∼40 Mb of minimally recombining repetitive DNA. The chromosome structures are similar to those in grains such as wheat, with recombination focused in gene-rich, repeat-depleted regions near chromosome ends. The physical and genetic map should facilitate further dissection of genetic and molecular mechanisms in this commercially and medically important plant.
Domesticated thousands of years ago (Li 1974), Cannabis sativa has been subjected to intensive breeding, resulting in extensive variation in morphology and chemical composition. It is perhaps best known for producing cannabinoids, a unique class of compounds that may function in chemical defense (Pate 1994) but also have pharmaceutical and psychoactive properties. Heat converts the cannabinoid acids (e.g., tetrahydrocannabinolic acid [THCA]) to neutral molecules (e.g., (–)-trans-Δ9–tetrahydrocannabinol [THC]) that bind to endocannabinoid receptors found in the vertebrate nervous system. This pharmacological activity leads to analgesic, antiemetic, and appetite-stimulating effects and may alleviate symptoms of neurological disorders, including epilepsy (Devinsky et al. 2014) and multiple sclerosis (van Amerongen et al. 2018). There are over 113 known cannabinoids (Elsohly and Slade 2005), but the two most abundant natural derivatives are THC and cannabidiol (CBD). THC is responsible for the well-known psychoactive effects of cannabis consumption, but CBD, while nonintoxicating, also has therapeutic properties and is specifically being investigated as a treatment for both schizophrenia (Osborne et al. 2017) and Alzheimer's disease (Watt and Karl 2017). Cannabis has traditionally been classified as having a drug (“marijuana”) or hemp chemotype based on the relative proportion of THC to CBD, but types grown for psychoactive use produce relatively large amounts of both. Cannabis containing high levels of CBD is increasingly grown for medical use.
THCA and CBDA are both synthesized from cannabigerolic acid (CBGA) by the related enzymes THCA synthase (THCAS) and CBDA synthase (CBDAS), respectively (Sirikantaramas et al. 2004; Taura et al. 2007). Expression of THCAS and CBDAS appear to be the major factor determining cannabinoid content, but the mechanisms that underlie the expression of these enzymes remain unresolved. Two competing theories are supported by existing data. In one, CBDAS and THCAS are mutually exclusive alleles (i.e., very different isoforms, as the protein sequences are only 84% identical). Genetic analysis supports this model, with approximately 1:2:1 segregation of chemotypes in a cross of drug type versus hemp (de Meijer et al. 2003). An alternative model is that THCAS and CBDAS are closely linked (i.e., adjacent on a chromosome), and one or the other is inactivated in drug-type or hemp strains. This model was motivated by the discovery of a THCAS-like gene in hemp plants (Kojoma et al. 2006) and is consistent with the possibility that these related genes are derived from an ancient tandem duplication. In addition, physical linkage of genes involved in specialized metabolic pathways has been repeatedly observed in plants, similar to operons in bacterial genomes (Nützmann and Osbourn 2014); such a cluster was recently described for benzylisoquinoline alkaloid biosynthesis genes in opium poppy (Guo et al. 2018). It is unknown whether genes involved in cannabinoid biosynthesis are clustered, although genetic analyses have previously indicated that at least one locus unlinked to THCAS/CBDAS contributes to cannabinoid content (Weiblen et al. 2015).
The draft genome and transcriptome of C. sativa described in 2011 (for a female plant of the drug-type strain Purple Kush [PK] and resequencing of a plant of the hemp variety “Finola” [FN]) (van Bakel et al. 2011) was unable to discriminate between these models due to high fragmentation. The C. sativa draft genome assembly, done largely with Illumina sequencing, was composed of 136,290 scaffolds, with an N50 of 16.2 kb. It was subsequently demonstrated that ∼70% of the C. sativa draft genome is composed of repetitive sequences (Pisupati et al. 2018). Measurement of single-nucleotide variants (SNVs) in four strains showed rates of heterozygosity ranging from 0.18%–0.26% and revealed that the drug-type and hemp-type strains were well separated by SNVs; the rate of occurrence of SNVs between these types was as high as 0.64% (van Bakel et al. 2011). Cytogenetic analysis has furthermore suggested a high degree of inter- and intracultivar karyotype polymorphisms (i.e., differences in homologous chromosomes that can be observed by microscopy), at least among hemp varieties (Razumova et al. 2016), which may further complicate genome assembly. To address these complications and to simultaneously leverage the high rate of SNVs between PK and FN, we coupled Pacific Biosciences (PacBio) long-read single-molecule real-time (SMRT) sequencing of PK and FN with Illumina resequencing of 99 F1 progeny between the two in order to generate a combined genetic and physical map. The combined map provides new insights into the arrangement of the chromosomes and the cannabinoid biosynthetic genes, including discovery of substantial rearrangement and gene duplications at the closely linked THC and CBD acid synthase gene loci.
Results
A combined genetic and physical map reveals that genes and recombination events are concentrated near chromosome ends
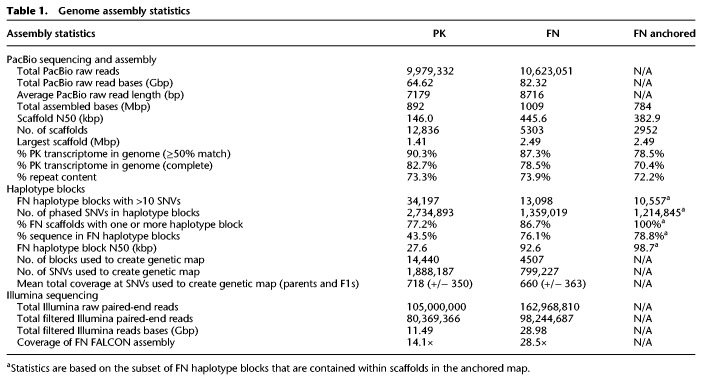
We performed PacBio SMRT sequencing of genomic DNA (gDNA) from the female parent PK and the male parent FN to a depth of ∼79× and ∼98×, respectively. We used these data to develop an initial set of scaffolds, using the FALCON assembler (Chin et al. 2016), with PK and FN analyzed separately (Table 1). The assemblies were further polished with Illumina data using Pilon (Walker et al. 2014) to correct indel errors associated with homopolymer repeats in the PacBio data. The FN assembly was more contiguous than the PK assembly (scaffold N50 of 445.6 vs. 146 kbp, respectively), likely reflecting the increased FN coverage and the use of a more recent sequencing chemistry, and each substantially improved on our original Illumina assembly (Supplemental Fig. S1; van Bakel et al. 2011). De novo repeat classification using RepeatModeler (http://www.repeatmasker.org/RepeatModeler/) confirmed that the sequence of both assemblies is highly repetitive (∼73%) (Supplemental Fig. S2), with hundreds of distinct families. The two sets of scaffolds largely mapped to each other one-to-one (Supplemental Fig. S3) but with differing breakpoints that mostly reflected differences in scaffold boundaries. The total size of the PK and FN assemblies was close to the haploid genome size estimated by flow cytometry (818 and 843 Mb for female and male, respectively) (Sakamoto et al. 1998). Overall, 90.3% of 30,074 previously described PK transcripts (van Bakel et al. 2011) mapped to the PK assembly (82.3% mapping completely within a single scaffold). Each assembly also contained >95% of eudicotyledon single-copy orthologs from OrthoDB, of which >97% were complete (Supplemental Fig. S4), indicating that both assemblies represented the vast majority of the cannabis gene space. An ortholog duplication rate of >14% and slightly larger than expected assembly sizes suggest that some regions of the diploid genomes were resolved into separate contigs, which can be an issue for polymorphic species (Shimizu et al. 2017).
Table 1.
Genome assembly statistics
We reasoned that a genetic map would provide an independent means to link scaffolds, in addition to being independently useful for genetic analysis. To generate a genetic linkage map, we employed the SOILoCo pipeline, created by Scaglione et al. (2016) to create a map of the artichoke genome. We applied the pipeline to F1 data from a cross between a PK female and FN male. SOILoCo requires phasing of the parental scaffolds into blocks in which parental haplotypes can be uniquely identified. It then uses SNVs in the offspring to determine which of the parental haplotypes is inherited for each F1 at each block. The inherited parental haplotypes are called using a hidden Markov model, which compensates for uncertainty in genotype calling caused by relatively low coverage typical of resequencing, by taking advantage of the multiple SNVs in each block. Because each of the four parental haplotypes is traced uniquely, recombination frequencies between blocks (and thus between scaffolds) can subsequently be calculated, and the recombination frequencies can be used to place blocks (and scaffolds) into linkage groups. Since the blocks of informative SNVs differ between the parental types, a separate genetic map is created for each parent (in this case, PK and FN). In our implementation, we identified phased haplotype blocks of physically linked unique SNVs in the FN assembly contained within PK or FN PacBio raw reads using HapCUT2 (Table 1; Edge et al. 2017), and scored them in 99 F1 progeny using Illumina sequencing (median coverage about 4×). We then ran the SOILoCo pipeline, followed by R/qtl (Broman et al. 2003) and MSTmap (Wu et al. 2008), to form linkage groups and order scaffolds within them.
The blocks formed 10 large linkage groups in both PK and FN, which we assume correspond to the established nine autosomes and X/Y (which contain a pseudoautosomal region and recombine) (Peil et al. 2003) and are hereafter referred to as chromosomes. The maps were largely consistent between PK and FN (Supplemental Fig. S5) and were therefore merged (MergeMap) (Wu et al. 2011). The merged genetic map is depleted for short scaffolds, repetitive sequence, and scaffolds containing a higher proportion of SNVs with segregation distortion (these SNVs are ignored by SOILoCo). The merged map contains 2952/5304 scaffolds, 784/1006 Mb (78%) of the initial sequence, 89% of eudicotyledon single-copy orthologs, and 21,168/30,074 of all PK transcripts (70.4%) (Table 1; Supplemental Fig. S4).
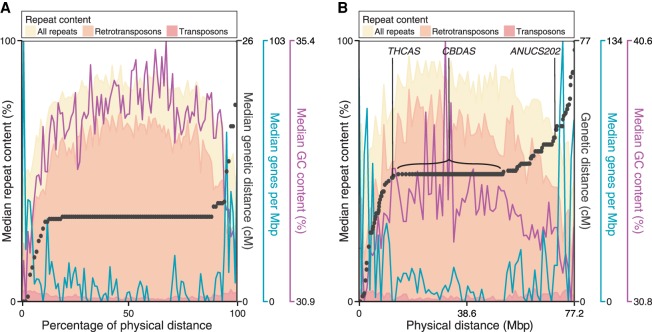
Figure 1A plots composite physical versus genetic distance across the chromosomes, with several major trends in the chromosomal sequences also illustrated (Supplemental Fig. S5 shows similar graphs and also plots of genetic vs. physical distance, as well as a comparison of recombination frequencies, for all individual chromosomes). First, there is a very strong tendency for recombination to occur near chromosome ends, while there are typically large blocks lacking recombination events across the middle of the chromosome. Second, genes are much more frequent near chromosome ends. Because promoters and enhancers are typified by open chromatin, which appears to promote crossovers in diverse species, including maize (Liu et al. 2009) and Arabidopsis thaliana (Choi et al. 2013), this arrangement may underlie the observed recombination frequencies. Third, the poorly recombining central parts of chromosomes not only are gene-poor but also have a higher repeat content, which may be methylated and could suppress recombination (Zamudio et al. 2015). Fourth, assuming that the centromere is located within the nonrecombining central segments of the chromosomes, then Chromosomes 5, 9, and 10 appear to be telocentric (i.e., behave as if they have a single long arm). These may represent the sex chromosome, one end of which is nonhomologous and thus nonrecombining, and Chromosomes 8 and 9 (as determined by cytogenetics) (Divashuk et al. 2014), which harbor 5S rDNA and 45S rDNA on one arm, respectively. The repetitive nature of these regions would be expected to impede both assembly and mapping. Indeed, four of five male-specific markers are found in the FN assembly, but none were placed on the genetic map, and the 45S and 5S rDNA are not in the assembly (Supplemental Table S1).
Figure 1.
Comparison of physical and genetic distance in Cannabis sativa and arrangement of sequence features on chromosomes. (A) Median values are indicated for all metacentric linkage groups (Chromosomes 5, 9, and 10 are excluded), scaled to the same physical length. Black points indicate the median increase in genetic distance every 1/100th of the physical distance. Shaded histograms superimposed show density of repeat sequences. Density of genes and GC content are also indicated by blue and purple lines. (B) Values for Chromosome 6, which contains the THCAS/CBDAS loci; here, black points are the representative of individual scaffolds.
Overall, the organization of C. sativa genes, repeats, and recombination frequency along chromosomes is similar to what is commonly observed in the grains (e.g., maize, barley, and wheat) (Gore et al. 2009; Liu et al. 2009; Mascher et al. 2017). To our knowledge, such an organization is unusual outside the grains: It has been observed in the walnut (Luo et al. 2015), but not thale cress (A. thaliana) (Meinke et al. 2009), apple (Di Pierro et al. 2016), strawberry (Davik et al. 2015), or mulberry (He et al. 2013), suggesting that this property is rare among Rosales.
Genomic organization of cannabinoid pathway genes
We next examined the positions of genes encoding known cannabinoid biosynthetic enzymes on the chromosomes. With the exception of the functional copies of CBDAS and THCAS, which are considered below, the cannabinoid-related genes are distributed in a mostly random fashion across the genome (indicated in Supplemental Fig. S5). The new map also finds that C. sativa encodes one copy of AAE1 (hexanoyl-CoA synthetase) and two tandem copies of tetraketide synthase (“olivetol synthase”). The genome sequences of both PK and FN also contain the THCAS-like gene described by Kojoma et al. (2006) which led to the two-locus THCAS/CBDAS hypothesis. This THCAS-like gene is 96% identical to THCAS at the nucleotide level and encodes a protein that is 93% identical to THCAS at the amino acid level. One copy of the THCAS-like gene is found in the PK assembly (scaffold 005500F: 2986–4620), and two are found in the FN assembly (scaffold 004887F, 13943–15577; 001793F, 69162–70796).
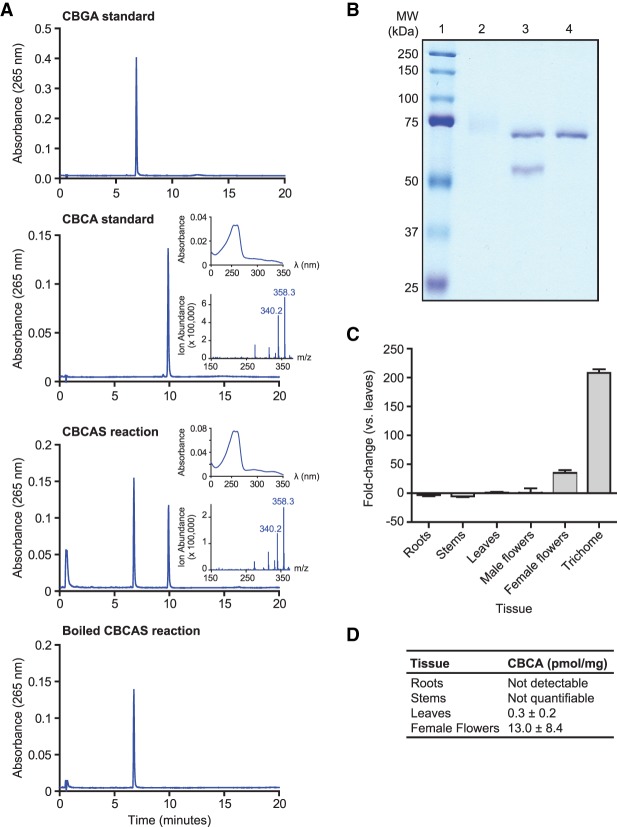
We examined the possibility that this THCAS-like gene encoded cannabichromenic acid (CBCA) synthase (CBCAS), which is found in both drug-type and hemp strains and resembles THCAS and CBDAS in its catalytic mechanism (Morimoto et al. 1997). We expressed the predicted open reading frame as a secreted protein in Pichia pastoris strain X-33. We then added CBGA substrate to clarified culture media to test for enzyme activity. The products of this reaction were analyzed by high-performance liquid chromatography (HPLC), which revealed a specific signal for CBCA (Fig. 2A). Purification of the Pichia secreted protein through a series of chromatographic steps yielded a 59-kDa product at the expected size of CBCAS without its secretory signal sequence (calculated to be 58.9 kDa) (Fig. 2B). We next determined the kinetic properties of CBCAS after optimizing reaction conditions using the purified protein (Supplemental Fig. S6). At the optimal temperature of 40°C and a pH of 5.5, the reaction followed Michaelis–Menten reaction kinetics with a Km of 9.3 ± 2.3 µM and a kcat of 0.02 sec−1. These values are similar to those reported for CBCAS purified from cannabis floral tissue (Km = 23 µM, kcat = 0.04 sec−1) (Morimoto et al. 1998). Finally, the accumulation of CBCA correlates well with the expression of CBCAS in various cannabis tissues, with the highest concentration observed in female floral tissue and minimal amounts in the leaf, stem, and root (Fig. 2C). Taken together, these data confirm that we identified the gene encoding CBCA synthase.
Figure 2.
Characterization of CBCAS activity and expression. (A) HPLC analysis of CBCAS activity detected in Pichia pastoris cell cultures. Chromatograms of the CBGA substrate and CBCA standards are shown together with chromatograms of the enzyme reaction in media sampled from Pichia expressing CBCA in the presence of CBGA substrate before and after boiling at 95°C for 10 min. Insets correspond to the UV-absorbance spectrum (top) and the mass spectrum derived from a single quadrapole mass spectrometer (bottom) of the compound that eluted at 10 min. (B) SDS-PAGE analysis of CBCAS expressed in P. pastoris and purified by protein chromatography. (Lane 1) Protein ladder. (2) Concentrated protein fraction exhibiting CBCAS activity. The high-molecular-weight smear is glycosylated CBCAS. (3) Same fraction as lane 2, treated with EndoHf (MW = 70 kDa). (4) EndoHf only. (C) qRT-PCR analysis of CBCAS expression in cannabis tissues. cDNA derived from cannabis tissues was used as a template for PCR reactions using CBCAS-specific primers and EF1α as a reference gene. Differential expression of CBCAS is depicted as fold-change between tissue types compared with leaves. Trichome tissue consisted of isolated trichome secretory cells. (D) Quantification of CBCA content of the developing seedlings by HPLC.
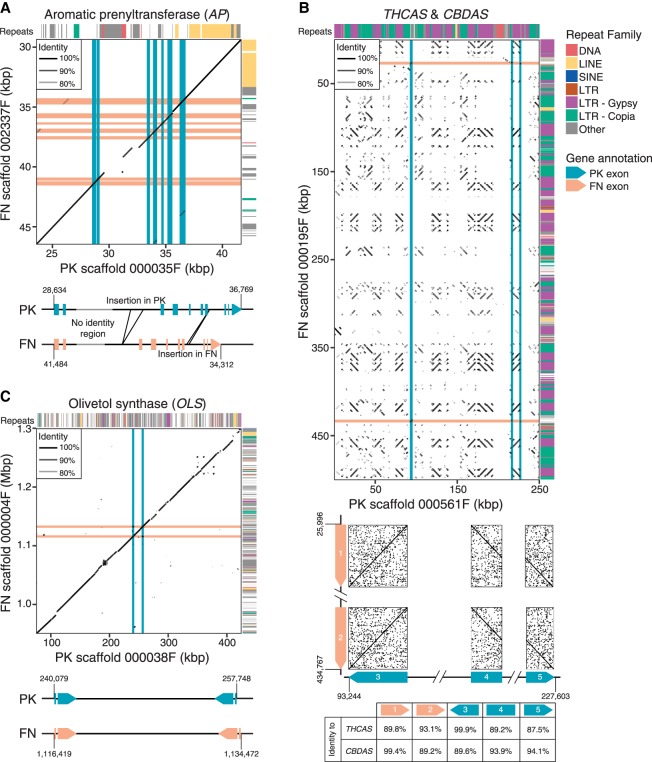
A previous study (Weiblen et al. 2015) used QTL analysis in C. sativa to associate 121 genetic markers with total cannabinoid content and THCA/CBDA ratio. Outside of THCAS/CBDAS, this study identified only one locus displaying a strong association with total cannabinoid content, at a distance of ∼1.2 cM between the trait and the marker. In our genetic map, this locus (marker ANUCS501) is linked to aromatic prenyltransferase (AP), which catalyzes the production of CBGA, the substrate of THCAS, CBDAS, and CBCAS, with a similar recombination frequency (2.1 cM in PK; 4 cM in FN). This observation suggests that either polymorphisms or differential regulation of AP contributes to cannabinoid production, presumably by controlling substrate concentration for THCAS and CBDAS. PK has greater than fivefold higher transcript levels of AP than FN (van Bakel et al. 2011), with no difference in copy number, suggesting that AP enzyme levels may be higher in drug-type plants partly due to differences in transcript levels. In addition to polymorphisms, there are multiple large (>100 bp) indels in and around the AP locus (including two within introns), which correspond mainly to LTRs, LINEs, and simple-repeat-like insertions, which could conceivably alter regulation of transcription or splicing (Fig. 3A).
Figure 3.
Comparison of scaffolds between PK and FN assemblies. Alignments of scaffolds from PK and FN FALCON assemblies containing key cannabinoid biosynthesis enzymes are shown. Locations of exons are indicated by pink and blue lines for FN and PK, respectively. Repeat classes given are from RepeatModeler. Individual repeat types indicated were identified by manual analysis. Features of genes are further described and compared beneath the alignments. (A) Aromatic prenyltransferase (AP). (B) THCAS and CBDAS. (C) Olivetol synthase (OLS, or tetraketide synthase).
Extensive rearrangement of the cannabinoid synthase locus underlies chemotype differences between PK and FN
Finally, we examined THCAS and CBDAS in the PK and FN genomes. The PK assembly contains only a single copy of THCAS and no exact copies of CBDAS: None have >95% identity to CBDAS at the nucleotide level. Similarly, the FN assembly contains only a single functional copy of CBDAS, while no THCAS gene is detected. These observations are confirmed by raw sequencing reads; no reads from FN map to THCAS, and no reads from PK map to CBDAS. Both genomes include the aforementioned CBCAS. This supports claims made using the draft genome and transcriptome (van Bakel et al. 2011). As expected from established segregation patterns, THCAS and CBDAS map to roughly the same region on Chromosome 6, near a known marker associated with THCA and CBDA content (ANUCS202) (Fig. 1B). However, the scaffolds that contain THCAS (in PK) and CBDAS (in FN) are dramatically different from each other, and neither has a clear counterpart in the other genome. The scaffold containing THCAS in PK does, however, contain a pseudogenic copy of CBDAS, with ∼94% identity to the known CBDAS sequence. The gene is likely nonfunctional as it has a gypsy element insertion at its center. Assuming these loci share common ancestry, there has clearly been extensive rearrangement since their divergence. The scaffold containing THCAS is ∼250 kb and that containing CBDAS is ∼750 kb, but the dotplot shown in Figure 3B illustrates almost complete lack of similarity over this span, with the exception of a large number of LTR-class retroelements. The extreme rearrangement clearly shows that these two genes do not have a simple isogenic relationship; Figure 3, A and C, illustrates more typical patterns of sequence similarity between PK and FN. The scaffold containing CBDAS is located within a much larger repeat-rich and gene-poor region of ∼39 Mb in the central section of Chromosome 6, encompassing 151 scaffolds with no recombination in either parent observed among the 99 F1s (Fig. 1B). The scaffold containing THCAS was separated from this region in a single recombination event among the 99 crosses, thus placing it at one end of this region and indicating that the THCAS and CBDAS scaffolds are at separate loci. We suggest that this repeat-rich segment of the chromosome may have hosted a series of tandem duplications and rearrangements amplifying an ancestral gene, leading to the present chromosomal organization; there is also a pseudogene with 89%–93% identity to each of THCAS, CBDAS, and CBCAS in this region. We note that this observation represents a modification of both previous models of CBDAS and THCAS arrangement: They are not isoforms at an otherwise equivalent locus, and no equivalent of THCAS (deactivated or not) is found in hemp.
Discussion
The combined sequence/genetic map presented here is consistent with the known C. sativa karyotype and genome size, contains the vast majority of known transcripts, and largely correlates between PK and FN. To completely finish the sequence, it will most likely be necessary to further improve the resolution of the genetic map and/or leverage hybrid scaffolding technologies, e.g., by incorporating single-molecule genomic maps (Pendleton et al. 2015) or Hi-C data that provides >1 Mb phasing information (Kronenberg et al. 2018). Another future goal will be to identify and fully assemble the X/Y Chromosomes. There are numerous scaffolds in both PK and FN with no obvious counterpart in the other genome, which could represent distinctive components of the sex chromosomes and which were not captured in our genetic map.
The identification of CBCAS allows for a number of potential applications. Cannabichromene (CBC) is a weaker agonist of the cannabinoid CB1 and CB2 receptors compared with THC and CBD. However, unlike THC, both CBD and CBC have been shown to decrease nociception both by blocking the activity of ankyrin-type transient receptor potential channels that play roles in the perception of pain-inducing signals and by inhibiting the reuptake of endocannabinoids such as anandamide (Maione et al. 2011). Furthermore, CBC operates as a gastrointestinal anti-inflammatory agent in mice and protects adult neuronal stem progenitor cells in vitro (Izzo et al. 2012; Shinjyo and Di Marzo 2013). It therefore may be useful to breed medical cannabis strains with higher quantities of CBCA to treat specific ailments such as inflammatory bowel disease and Crohn's disease. Finally, the high degree of sequence similarity between CBCAS, THCAS, and CBDAS and the presence of multiple pseudogenes suggest that gene duplication and divergence has been the key driver of cannabinoid end-product diversification in cannabis. Comparative sequence analysis of the enzymes will help ascertain which amino acids are important in catalysis, and may lead to the rational design of cannabinoid biosynthetic enzymes that produce novel cannabinoids not observed in nature.
Our identification of CBCAS also clarifies a puzzling finding of Kojoma et al. (2006), who used PCR to amplify a THCAS-like gene from “fiber-type” (hemp) cannabis that contained no THCA. Based on the sequence of the gene that we show has CBCAS activity, the THCAS-like gene amplified by Kojoma et al. (2006) is CBCAS. This result makes sense, since nondrug/hemp forms of cannabis also contain CBCA.
Cannabis and cannabinoids are increasingly employed in medicine and recently have been legalized for recreational use in many jurisdictions. The new map should facilitate vastly improved genetic analysis, including QTL mapping, which will accelerate crop improvement efforts. Drug prohibition has restricted access to cannabis by plant breeders and researchers, and as a result, it has received less attention than other crops. Cannabis suffers from insect pests and widespread fungal diseases and has a number of agronomic issues such as flowering time requirements that make it difficult to grow in some environments. In addition, breeding of cannabis types with specific cannabinoid and terpene profiles is desirable for the development of new varieties for medical and recreational use. The fact that a strong and interpretable result was obtained by re-examining a previously described marker correlating with total cannabinoid content (Weiblen et al. 2015) clearly shows the potential of this approach as it applies to cannabinoid metabolism. Due to the relatively high rate of polymorphism in cannabis, it should be possible to employ resequencing (e.g., low-coverage short-read Illumina protocols) either on crosses or at a population level to associate variants or variation with traits and genes, using the genetic map.
Methods
Plant cultivation and gDNA isolation
A female PK plant, produced through multiple vegetative propagation generations from the original source plant used to produce the draft C. sativa genome (van Bakel et al. 2011), was pollinated by a male FN plant in an indoor growth chamber. Seeds produced from this cross were germinated under standard conditions and grown to seedling stage. gDNA was isolated from young leaves using a GenElute Genomic Miniprep Kit (Sigma-Aldrich). The secure facilities used for plant growing were licensed by Health Canada.
PacBio SMRT sequencing of the PK and FN genomes
gDNA library preparation and sequencing were performed according to the manufacturer's instructions and reflect the P6-C4 sequencing enzyme and chemistry, respectively. PK and FN gDNA was first repurified using a 0.8× AMPure XP purification step (0.80× AMPure beads added, by volume, to each DNA sample dissolved in 200 µL EB, vortexed for 10 min at 2000 rpm, followed by two washes with 70% alcohol and finally diluted in EB), to remove small fragments and/or biological contaminant. The purified DNA sample was taken through DNA damage and end-repair steps. Briefly, the DNA fragments were repaired using DNA damage repair solution (1× DNA damage repeat buffer, 1× NAD+, 1 mM ATP high, 0.1 mM dNTP, and 1× DNA damage repeat mix) with a volume of 21.1 µL and incubated at 37°C for 20 min. DNA ends were repaired next by adding 1× end repair mix to the solution, which was incubated at 25°C for 5 min, followed by the second 0.45× Ampure XP purification step. Next, 0.75 µM of blunt adapter was added to the DNA, followed by 1× template prep buffer, 0.05 mM ATP low, and 0.75 U/µL T4 ligase to ligate (final volume of 47.5 µL) the SMRTbell adapters to the DNA fragments. This solution was incubated at 25°C overnight, followed by a 65°C 10-min ligase denaturation step. After ligation, the library was treated with an exonuclease cocktail to remove unligated DNA fragments using a solution of 1.81 U/µL Exo III 18 and 0.18 U/µL Exo VII and then incubated at 37°C for 1 h. Two additional 0.80× Ampure XP purifications steps were performed to remove <1000-bp molecular-weight DNA and organic contaminant.
Size-selection was confirmed using the Agilent bioanalyzer, and the mass was quantified using a Qubit assay before proceeding with primer annealing and DNA sequencing. For PK, 100 pM of SMRTbell libraries were mag bead loaded and sequenced with a combination of P5/C3 and P6/C4 chemistry on a PacBio RSII machine with 6-h movies. For FN, 3 pM of SMRTbell libraries were diffusion-loaded and sequenced on a Sequel machine with v2 chemistry and 10-h movies.
FALCON assembly and Illumina polishing
FALCON (Chin et al. 2016) was used to generate genome assemblies for PK (v0.4.0) and FN (v1.8.6). Briefly, raw subread data were filtered to remove the shortest reads to an approximate coverage of 70× for each genome, leaving 8,003,220 (80.2%) of subreads for PK and 6,646,226 (62.6%) of subreads for FN, or ∼58 Gbp for each. Preassembled reads (i.e., error-corrected reads) were then created with a length cutoff of ≥6000 bp for PK and ≥7000 bp for FN, resulting in 2,239,051 and 5,323,023 preassembled reads, respectively. The PK and FN genomes were then assembled using preassembled reads with a minimum length of 9 kbp or 7 kbp, respectively. Additional relevant assembly parameter settings for FN were as follows:
pa_HPCdaligner_option: -B128 -t16 -e0.8 -M24 -l1200 -k18 -h256 -w8 -s100 -T12
ovlp_HPCdaligner_option: -B128 -M24 -k24 -h600 -e.92 -l1800 -s100 -T12
falcon_sense_option: --output_multi --min_cov_aln 4 --min_idt 0.70 --min_cov 4
--max_n_read 200
falcon_sense_skip_contained: False
overlap_filtering_setting: --max_diff 120 --max_cov 120 --min_cov 4
Similar assembly parameters were used for PK, except that min_cov was set to 3.
Each FALCON assembly was corrected with paired-end Illumina reads using Pilon version 1.22 (Walker et al. 2014) after mapping available Illumina sequencing data (van Bakel et al. 2011) to the FALCON-assembled genomes using BWA-MEM (version 0.7.8) (Li 2013) with an average of 96× (PK) and 23× (FN) coverage. Correction was performed with the “diploid” flag and the “bases” flag set to correct only indels and SNPs. A total of 1,511,828 insertions and 228,876 deletions were corrected in the FN assembly, and 1,807,453 insertions and 283,918 deletions were corrected in the PK assembly.
Repeat content analysis
Repeats in the FN and PK genomes were predicted de novo and classified using RepeatModeler (v1.0.11; http://www.repeatmasker.org/RepeatModeler/). RepeatModeler was applied to each assembly with the “ncbi” engine (RMBlast v2.2.28) provided with RepeatModeler. Other prerequisite components installed with the RepeatModeler package included RECON v1.0.8 and RepeatScout v1.0.5 (Price et al. 2005), Tandem Repeat Finder v4.0.4 (Benson 1999), and Repbase-derived RepeatMasker libraries (http://www.girinst.org/server/RepBase/) from January 2017. The de novo repeat classification provided by RepeatModeler was filtered to remove families with a >1-kb BLAT (Kent 2002) alignment to PK transcripts. The final filtered RepeatModeler output was then used as input for RepeatMasker (Smit et al. 2013–2015) to produce a masked version of the assembly and obtain the genomic positions of annotated repeats.
Assessment of genome assembly completeness
The completeness of each genome assembly was assessed using BUSCO v3.0 (Simão et al. 2015) and the set of eudicotyledons single-copy orthologs from OrthoDB v10, with default arguments in the provided virtual machine instance.
Comparison of PK and FN scaffolds
PK and FN assemblies were aligned using LASTZ (Harris 2007) version 1.04.00 with the -ungapped and -notransitions options and a step of 20. Alignments with an identity of ≤95% and a length of ≤2000 bp were removed. To produce a dotplot, FN contigs were initially ordered by size along the y-axis. Next, PK contigs were ordered and orientated on the x-axis by the position of their best hit on the y-axis. FN contigs were then reordered on the y-axis according to their best hit to the newly ordered contigs on the x-axis. This process was repeated until the order of contigs on the x-axis, and the order of contigs on the y-axis converged.
Illumina sequencing of the FN and F1 individuals
Dual-indexed libraries were prepared using the Nextera DNA library preparation kit (Illumina), pooled equimolar, and sequenced on the HiSeq 2500 platform, yielding 529.9 Gbp total. FN was sequenced independently on the NextSeq 500 platform, yielding 49.9 Gbp.
Building the genetic map
Quality filtering
Barcode and adapter sequences were filtered from all FN and F1 Illumina PE reads. FN reads were further filtered using Sickle with the flags -q 20 -l 125 (https://github.com/najoshi/sickle) (version 1.33). PK Illumina 2×100 PE reads from the 2011 draft genome were also filtered using Sickle, with the flags -q 20 -l 90.
Variant calling
BWA-MEM (Li 2013) was used to map Illumina paired-end reads for FN, PK, and the F1s to the PK FALCON assembly, after which Picard (http://broadinstitute.github.io/picard/) was used for sorting, duplicate marking, and indexing the alignments. To call variants for the F1s, we used the mpileup function from bcftools (Li et al. 2009) over all of the F1 individuals and both parents to overcome spots of lower coverage in the F1s. Variants were also called individually for each parent using the GATK HaplotypeCaller (McKenna et al. 2010) to be used as input for haplotype phasing.
Phasing the parental haplotypes
Haplotypes for the parents were phased using HapCUT2 (Edge et al. 2017), using the –pacbio 1 argument to improve accuracy with PacBio reads and the –ea 1 argument to calculate switch quality scores. As input, parental SNPs called by HaplotypeCaller on the Illumina data were provided in conjunction with PacBio raw reads. This was done to both increase the length of the resulting haplotype blocks and boost confidence in the phasing by requiring agreement between the two sets of data. To further increase confidence, we only used SNPs that had a quality score greater than 25 and read coverage between six and 46 and that were more than five bases away from an indel. Haplotype blocks were then split if the switch quality score was less than 30. Finally, only blocks with more than 10 SNPs were retained to use as input for SOILoCo.
Genotyping the F1s
The SOILoCo method (Scaglione et al. 2016) was used to genotype the F1s at each haplotype block, using the output of HapCUT2 and the variants called by mpileup. Required values and divergence from the default parameters are as follows. For vcf2strings.pl, minor allele frequencies 1 and 2 (--MAF-1 and --MAF-2) were set to 0.25 and 0.75, respectively. This step allows the removal of any markers that may display segregation distortion (8.5% of markers show some degree of segregation distortion; scaffolds that do not get incorporated into the genetic map have an average of 20% of markers displaying significant distortion). When running gt-hmm.pl, the minimum number of variant calls in a haplotype block (--min-string) was set to six, the probability of a crossing-over event (--switch-prob) was set to 1 × 10−6, and the probability of having reads containing both alleles at a heterozygous site (--HCALL-prob) was set to 0.15. Lastly, population type (--pop) for calls2csvr.pl is set to cross pollinated (CP). This process is run separately for each parent, with the two respective sets of haplotype blocks.
The scaffold containing CBDAS and the scaffold from the PK FALCON assembly containing THCAS were genotyped separately. As both scaffolds do not have a counterpart in the other parental assembly, genotypes were extracted from variant loci that meet the following criteria: an allele frequency of 0.5 in the parent harboring the scaffold, no coverage in the opposing parent, an allele frequency of 0.5 in the F1s, and all F1s are homozygous. The scaffold containing THCAS is the only scaffold from the PK FALCON assembly that was placed in the genetic map.
Forming linkage groups
R/qtl (Broman et al. 2003) was employed to divide haplotype blocks for each parent separately across linkage groups using the formLinkageGroups function with maximum recombination frequency (max.rf) set to 0.05 and minimum LOD (min.LOD) set to 15. The resulting linkage groups were compared against one another to identify any pairs of linkage groups with a mean recombination frequency of greater than 0.8 between the haplotype blocks they contain, in which case the switchAlleles function was used to swap the alleles for all the haplotype blocks in the smaller linkage group, and formLinkageGroups was called again. Afterward, R/qtl functions checkAlleles, switchAlleles, and formLinkageGroups were run in succession two more times to further identify and fix haplotype blocks with swapped alleles. All linkage groups with more than 100 haplotype blocks were passed to the ordering step. For PK, there were 11 linkage groups with more than 100 haplotype blocks; however, two of them just missed the cutoff for being joined together and were therefore combined. Further support for combining these linkage groups came from a comparison with the FN map, in which the scaffolds held in these two PK linkage groups were held in a single FN linkage group.
Ordering scaffolds
Haplotype blocks were ordered within each linkage group using MSTmap (Wu et al. 2008) with the Kosambi distance function. Three rounds of ordering were done with a smoothing step in between carried out using the Perl implementation of the SMOOTH correction algorithm (van Os et al. 2005) that is provided with the SOILoCo pipeline using an error threshold of 0.85. Correspondence between the two parental sets of linkage groups was determined based on similarity in the sets of scaffolds belonging to each linkage group. To handle ambiguity in scaffold placement, if the haplotype blocks for any given scaffold were distributed over more than one linkage group within or between parental maps, a census was taken to determine the correct linkage group, and haplotype blocks that did not agree with the majority were removed. If fewer than half of the haplotype blocks were in agreement, all haplotype blocks for that scaffold were removed, and the scaffold was not placed in either parental map. Finally, for each scaffold within each map, a distribution of the genetic positions (in cM) for all haplotype blocks belonging to the scaffold was established, and any outlier blocks were removed. After removal of ambiguous haplotype blocks and scaffolds, a final round of ordering was carried out for each parental map.
Merging the genetic maps
To translate each parental map from haplotype blocks to scaffolds in order that they could be merged, scaffold placements were determined by averaging the locations of the haplotype blocks belonging to each scaffold. The genetic maps for PK and FN were then merged using MergeMap (Wu et al. 2011) with the weight of the FN genetic map set to two and the weight of the PK genetic map set to one because it was based off the FN FALCON assembly.
Gene cloning
CBDAS was amplified from DNA isolated from FN leaves using gene-specific primers (forward: 5′-CTGCAGGAATGAAGTACTCAACATTCTCCTTTTGG-3′; reverse: 5′-AAGCTTTCATGGTACCCCATGATGATGCCGTGGAAGAG-3′). PCR products were cloned into pCR8/GW/TOPO (Invitrogen), excised as PstI/KpnI fragments, and cloned into pPICz-alpa B (Invitrogen). The expression vectors were then transformed into P. pastoris strain X-33 (Invitrogen) by electroporation. Positive recombinants were selected for by plating transformed cells on YPD plates supplemented with 25 µg/mL phleomycin (Invivogen). To screen for activity, colonies were used to inoculate 5 mL BMG cultures, which were grown for 2 d at 37°C with shaking. The cells were then pelleted by centrifugation, resuspended in 5 mL BMM media, and grown for 4 d at 20°C with shaking with the addition of 1% methanol daily. Enzyme activity was tested by directly adding CBGA to clarified culture media, incubating overnight at 37°C, and then analyzing products by HPLC as previously described (Stout et al. 2012).
Quantitative PCR
RNA extraction, cDNA generation, and qRT-PCR conditions were identical to those previously reported (Stout et al. 2012). CBCAS primers (forward: 5′-CGGATGTACTGTTATGCTCCAA-3′; reverse: 5′-CATTCTCCATTAAAATAAGAAAGACAA-3′) were designed from alignments of THCAS-like genes identified in the cannabis genome to ensure their selectivity. Primers were tested using cloned THCAS, CBDAS, and CBCAS as templates. Any primer set that amplified a nontarget cDNA was discarded. Primer efficiencies were extrapolated from raw amplification data using LinRegPCR (Ruijter et al. 2009).
Recombinant CBCAS enzyme expression and purification
The culture with the highest CBCAS activity was selected for scaled up production. One milliliter of the initial culture was used to inoculate two 40 mL BMG cultures, which were grown for 2 d at 37°C. These cultures were then used to initiate two 400 mL modified BMM cultures that were buffered with 10 mM HEPES (pH 7) and were supplemented with riboflavin at 20 mg/L. These cultures were grown at 20°C with shaking at 100 RPM for 5 d, with methanol added to 1% by volume each day. The cultures were then clarified by centrifugation, and the resulting media were filtered and passed over two Bio-scale Mini CHT hydroxyapatite cartridges (Bio-Rad) at a flow rate of 1.5 mL/min at 4°C. The cartridges were then attached in series to an AKTA FPLC system (GE Healthcare) and eluted with a 75-mL linear gradient from 5 mM sodium phosphate (pH 7) to 500 mM sodium phosphate (pH 7). Active fractions were pooled, concentrated with a 30 kDa cutoff Centricon filter (Millipore), and buffer exchanged into 20 mM citrate (pH 4.7) using a PD10 column (GE Healthcare). The resulting fraction was then injected onto a MonoS 5/50 cation exchange column (GE Healthcare) and eluted with a 40-mL linear gradient of 20 mM citrate (pH 4.7) to 20 mM citrate (pH 4.7) + 500 mM NaCl. Active fractions were pooled, concentrated with a 30 kDa cutoff Centricon filter, and injected onto a Hiload 26/60 Superdex 200 size exclusion column (GE Healthcare). Proteins were eluted with a single column volume of 20 mM citrate (pH 5.0) + 150 mM NaCl. Throughout the purification, 1/10th volume of each fraction was retained for analysis to judge purity. Protein was isolated from each fraction using 15 µL of StrataClean resin (Stratagene) and analyzed by SDS PAGE.
Enzyme assays and HPLC quantification of reaction products
To test for CBCAS enzyme activity during the protein purification, 150 µL of protein fraction was mixed with 50 µL of 500 µM sodium citrate buffer (pH 5.0) and 20 µmol of CBGA and incubated overnight at 37°C. The reactions were then extracted twice with ethyl acetate, and the organic fractions were pooled and dried in a SpeedVac concentrator. The products were then resuspended in 16 µL 50% methanol, of which 10 µL were analyzed by HPLC as previously described (Stout et al. 2012). Reactions for enzyme kinetic analyses were composed of 1 µg of purified CBCAS, 100 mM sodium citrate (pH 5.0), and 100 mM NaCl. These reactions were performed under Michaelis–Menten conditions at 40°C for 1 h. Reaction product extraction and analyses were the same as above.
Data access
The PacBio sequence read data generated for genome assembly, the Illumina sequencing data for the FN and F1 individuals, and the PK and FN genome assemblies from this study have been submitted to the NCBI BioProject database (http://www.ncbi.nlm.nih.gov/bioproject) under accession number PRJNA73819.
Competing interest statement
J.E.P. is the chief executive officer and shareholder of a for-profit cannabis science company, Anandia Laboratories, based in Vancouver, Canada. In this position he receives a salary. Anandia Laboratories performs analytical testing for licensed cannabis producers in Canada as well as works to develop new cannabis cultivars through breeding. L.H. was the chief research officer at CanniMed Therapeutics until May 2018. CanniMed is a for-profit company based in Saskatoon, Canada, that produces medical cannabis products for authorized patients. In this position he received compensation in the form of a salary and stock options. In June 2018 he moved to CB3 Life Sciences, where he is chief scientific officer. J.E.P. and J.M.S. have filed patent WO2015196275 on the nucleotide sequence encoding the enzyme CBCAS-based reagents, as well as methods for producing cannabinoids and/or altering cannabinoid production.
Supplementary Material
Acknowledgments
We thank the Donnelly Sequencing Centre (DSC) for assistance with Illumina sequencing. This work was supported by grants from the Canadian Institutes of Health Research (operating grant MOP-126070 to T.R.H., J.E.P., and H.v.B.; foundation grant FDN-148403 to T.R.H.). T.R.H. is a scholar of the Canadian Institute for Advanced Research (CIFAR) and holds the John W. Billes Chair of Medical Research at the University of Toronto. H.v.B. was supported in part by the NIH, National Institute of Allergy and Infectious Diseases, grant R01 AI119145. This work was supported in part through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.242594.118.
Freely available online through the Genome Research Open Access option.
References
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27: 573–580. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. 10.1093/bioinformatics/btg112 [DOI] [PubMed] [Google Scholar]
- Chin CS, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O'Malley R, Figueroa-Balderas R, Morales-Cruz A, et al. 2016. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods 13: 1050–1054. 10.1038/nmeth.4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Zhao X, Kelly KA, Venn O, Higgins JD, Yelina NE, Hardcastle TJ, Ziolkowski PA, Copenhaver GP, Franklin FC, et al. 2013. Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat Genet 45: 1327–1336. 10.1038/ng.2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davik J, Sargent DJ, Brurberg MB, Lien S, Kent M, Alsheikh M. 2015. A ddRAD based linkage map of the cultivated strawberry, Fragaria xananassa. PLoS One 10: e0137746 10.1371/journal.pone.0137746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Meijer EP, Bagatta M, Carboni A, Crucitti P, Moliterni VM, Ranalli P, Mandolino G. 2003. The inheritance of chemical phenotype in Cannabis sativa L. Genetics 163: 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devinsky O, Cilio MR, Cross H, Fernandez-Ruiz J, French J, Hill C, Katz R, Di Marzo V, Jutras-Aswad D, Notcutt WG, et al. 2014. Cannabidiol: pharmacology and potential therapeutic role in epilepsy and other neuropsychiatric disorders. Epilepsia 55: 791–802. 10.1111/epi.12631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pierro EA, Gianfranceschi L, Di Guardo M, Koehorst-van Putten HJ, Kruisselbrink JW, Longhi S, Troggio M, Bianco L, Muranty H, Pagliarani G, et al. 2016. A high-density, multi-parental SNP genetic map on apple validates a new mapping approach for outcrossing species. Horticult Res 3: 16057 10.1038/hortres.2016.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divashuk MG, Alexandrov OS, Razumova OV, Kirov IV, Karlov GI. 2014. Molecular cytogenetic characterization of the dioecious Cannabis sativa with an XY chromosome sex determination system. PLoS One 9: e85118 10.1371/journal.pone.0085118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edge P, Bafna V, Bansal V. 2017. HapCUT2: robust and accurate haplotype assembly for diverse sequencing technologies. Genome Res 27: 801–812. 10.1101/gr.213462.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsohly MA, Slade D. 2005. Chemical constituents of marijuana: the complex mixture of natural cannabinoids. Life Sci 78: 539–548. 10.1016/j.lfs.2005.09.011 [DOI] [PubMed] [Google Scholar]
- Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, Peiffer JA, McMullen MD, Grills GS, Ross-Ibarra J, et al. 2009. A first-generation haplotype map of maize. Science 326: 1115–1117. 10.1126/science.1177837 [DOI] [PubMed] [Google Scholar]
- Guo L, Winzer T, Yang X, Li Y, Ning Z, He Z, Teodor R, Lu Y, Bowser TA, Graham IA, et al. 2018. The opium poppy genome and morphinan production. Science 362: 343–347. 10.1126/science.aat4096 [DOI] [PubMed] [Google Scholar]
- Harris RS. 2007. “Improved pairwise alignment of genomic DNA.” PhD thesis, The Pennsylvania State University, University Park. [Google Scholar]
- He N, Zhang C, Qi X, Zhao S, Tao Y, Yang G, Lee TH, Wang X, Cai Q, Li D, et al. 2013. Draft genome sequence of the mulberry tree Morus notabilis. Nat Commun 4: 2445 10.1038/ncomms3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo AA, Capasso R, Aviello G, Borrelli F, Romano B, Piscitelli F, Gallo L, Capasso F, Orlando P, Di Marzo V. 2012. Inhibitory effect of cannabichromene, a major non-psychotropic cannabinoid extracted from Cannabis sativa, on inflammation-induced hypermotility in mice. Br J Pharmacol 166: 1444–1460. 10.1111/j.1476-5381.2012.01879.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT: the BLAST-like alignment tool. Genome Res 12: 656–664. 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojoma M, Seki H, Yoshida S, Muranaka T. 2006. DNA polymorphisms in the tetrahydrocannabinolic acid (THCA) synthase gene in “drug-type” and “fiber-type” Cannabis sativa L. Forensic Sci Int 159: 132–140. 10.1016/j.forsciint.2005.07.005 [DOI] [PubMed] [Google Scholar]
- Kronenberg ZN, Hall RJ, Hiendleder S, Smith TPL, Sullivan ST, Williams JL, Kingan SB. 2018. FALCON-Phase: integrating PacBio and Hi-C data for phased diploid genomes. bioRxiv 10.1101/327064 [DOI]
- Li H-L. 1974. An archaeological and historical account of cannabis in China. Econ Bot 28: 437–448. 10.1007/BF02862859 [DOI] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN].
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Yeh CT, Ji T, Ying K, Wu H, Tang HM, Fu Y, Nettleton D, Schnable PS. 2009. Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet 5: e1000733 10.1371/journal.pgen.1000733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MC, You FM, Li P, Wang JR, Zhu T, Dandekar AM, Leslie CA, Aradhya M, McGuire PE, Dvorak J. 2015. Synteny analysis in Rosids with a walnut physical map reveals slow genome evolution in long-lived woody perennials. BMC Genomics 16: 707 10.1186/s12864-015-1906-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maione S, Piscitelli F, Gatta L, Vita D, De Petrocellis L, Palazzo E, de Novellis V, Di Marzo V. 2011. Non-psychoactive cannabinoids modulate the descending pathway of antinociception in anaesthetized rats through several mechanisms of action. Br J Pharmacol 162: 584–596. 10.1111/j.1476-5381.2010.01063.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J, et al. 2017. A chromosome conformation capture ordered sequence of the barley genome. Nature 544: 427–433. 10.1038/nature22043 [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke D, Sweeney C, Muralla R. 2009. Integrating the genetic and physical maps of Arabidopsis thaliana: identification of mapped alleles of cloned essential (EMB) genes. PLoS One 4: e7386 10.1371/journal.pone.0007386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto S, Komatsu K, Taura F, Shoyama Y. 1997. Enzymological evidence for cannabichromenic acid biosynthesis. J Nat Prod 60: 854–857. 10.1021/np970210y [DOI] [Google Scholar]
- Morimoto S, Komatsu K, Taura F, Shoyama Y. 1998. Purification and characterization of cannabichromenic acid synthase from Cannabis sativa. Phytochemistry 49: 1525–1529. 10.1016/S0031-9422(98)00278-7 [DOI] [PubMed] [Google Scholar]
- Nützmann HW, Osbourn A. 2014. Gene clustering in plant specialized metabolism. Curr Opin Biotechnol 26: 91–99. 10.1016/j.copbio.2013.10.009 [DOI] [PubMed] [Google Scholar]
- Osborne AL, Solowij N, Babic I, Huang XF, Weston-Green K. 2017. Improved social interaction, recognition and working memory with cannabidiol treatment in a prenatal infection (poly I:C) rat model. Neuropsychopharmacology 42: 1447–1457. 10.1038/npp.2017.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pate D. 1994. Chemical ecology of Cannabis. J Int Hemp Assoc 2: 32–37. [Google Scholar]
- Peil A, Flachowsky H, Schumann E, Weber WE. 2003. Sex-linked AFLP markers indicate a pseudoautosomal region in hemp (Cannabis sativa L.). Theor Appl Genet 107: 102–109. 10.1007/s00122-003-1212-5 [DOI] [PubMed] [Google Scholar]
- Pendleton M, Sebra R, Pang AW, Ummat A, Franzen O, Rausch T, Stutz AM, Stedman W, Anantharaman T, Hastie A, et al. 2015. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat Methods 12: 780–786. 10.1038/nmeth.3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisupati R, Vergara D, Kane NC. 2018. Diversity and evolution of the repetitive genomic content in Cannabis sativa. BMC Genomics 19: 156 10.1186/s12864-018-4494-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA. 2005. De novo identification of repeat families in large genomes. Bioinformatics 21 Suppl 1: i351–i358. 10.1093/bioinformatics/bti1018 [DOI] [PubMed] [Google Scholar]
- Razumova OV, Alexandrov OS, Divashuk MG, Sukhorada TI, Karlov GI. 2016. Molecular cytogenetic analysis of monoecious hemp (Cannabis sativa L.) cultivars reveals its karyotype variations and sex chromosomes constitution. Protoplasma 253: 895–901. 10.1007/s00709-015-0851-0 [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45 10.1093/nar/gkp045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Akiyama Y, Fukui K, Kamada H, Satoh S. 1998. Characterization; genome sizes and morphology of sex chromosome in hemp (Cannabis sativa L.). Cytologia 63: 459–464. 10.1508/cytologia.63.459 [DOI] [Google Scholar]
- Scaglione D, Reyes-Chin-Wo S, Acquadro A, Froenicke L, Portis E, Beitel C, Tirone M, Mauro R, Lo Monaco A, Mauromicale G, et al. 2016. The genome sequence of the outbreeding globe artichoke constructed de novo incorporating a phase-aware low-pass sequencing strategy of F1 progeny. Sci Rep 6: 19427 10.1038/srep19427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Tanizawa Y, Mochizuki T, Nagasaki H, Yoshioka T, Toyoda A, Fujiyama A, Kaminuma E, Nakamura Y. 2017. Draft sequencing of the heterozygous diploid genome of satsuma (Citrus unshiu Marc.) using a hybrid assembly approach. Front Genet 8: 180 10.3389/fgene.2017.00180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinjyo N, Di Marzo V. 2013. The effect of cannabichromene on adult neural stem/progenitor cells. Neurochem Int 63: 432–437. 10.1016/j.neuint.2013.08.002 [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Sirikantaramas S, Morimoto S, Shoyama Y, Ishikawa Y, Wada Y, Shoyama Y, Taura F. 2004. The gene controlling marijuana psychoactivity: molecular cloning and heterologous expression of Δ1-tetrahydrocannabinolic acid synthase from Cannabis sativa L. J Biol Chem 279: 39767–39774. 10.1074/jbc.M403693200 [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. 2013–2015. RepeatMasker Open-4.0. http://www.repeatmasker.org.
- Stout JM, Boubakir Z, Ambrose SJ, Purves RW, Page JE. 2012. The hexanoyl-CoA precursor for cannabinoid biosynthesis is formed by an acyl-activating enzyme in Cannabis sativa trichomes. Plant J 71: 353–365. 10.1111/j.1365-313X.2012.04949.x [DOI] [PubMed] [Google Scholar]
- Taura F, Sirikantaramas S, Shoyama Y, Yoshikai K, Shoyama Y, Morimoto S. 2007. Cannabidiolic-acid synthase, the chemotype-determining enzyme in the fiber-type Cannabis sativa. FEBS Lett 581: 2929–2934. 10.1016/j.febslet.2007.05.043 [DOI] [PubMed] [Google Scholar]
- van Amerongen G, Kanhai K, Baakman AC, Heuberger J, Klaassen E, Beumer TL, Strijers RL, Killestein J, van Gerven J, Cohen A, et al. 2018. Effects on spasticity and neuropathic pain of an oral formulation of Δ9-tetrahydrocannabinol in patients with progressive multiple sclerosis. Clin Ther 40: 1467–1482. 10.1016/j.clinthera.2017.01.016 [DOI] [PubMed] [Google Scholar]
- van Bakel H, Stout JM, Cote AG, Tallon CM, Sharpe AG, Hughes TR, Page JE. 2011. The draft genome and transcriptome of Cannabis sativa. Genome Biol 12: R102 10.1186/gb-2011-12-10-r102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Os H, Stam P, Visser RG, van Eck HJ. 2005. SMOOTH: a statistical method for successful removal of genotyping errors from high-density genetic linkage data. Theor Appl Genet 112: 187–194. 10.1007/s00122-005-0124-y [DOI] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt G, Karl T. 2017. In vivo evidence for therapeutic properties of cannabidiol (CBD) for Alzheimer's disease. Front Pharmacol 8: 20 10.3389/fphar.2017.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiblen GD, Wenger JP, Craft KJ, ElSohly MA, Mehmedic Z, Treiber EL, Marks MD. 2015. Gene duplication and divergence affecting drug content in Cannabis sativa. New Phytol 208: 1241–1250. 10.1111/nph.13562 [DOI] [PubMed] [Google Scholar]
- Wu Y, Bhat PR, Close TJ, Lonardi S. 2008. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet 4: e1000212 10.1371/journal.pgen.1000212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Close TJ, Lonardi S. 2011. Accurate construction of consensus genetic maps via integer linear programming. IEEE/ACM Trans Comput Biol Bioinform 8: 381–394. 10.1109/TCBB.2010.35 [DOI] [PubMed] [Google Scholar]
- Zamudio N, Barau J, Teissandier A, Walter M, Borsos M, Servant N, Bourc'his D. 2015. DNA methylation restrains transposons from adopting a chromatin signature permissive for meiotic recombination. Genes Dev 29: 1256–1270. 10.1101/gad.257840.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.