Abstract
Roots of Gmelina arborea (Gambhari) is a medicinally important raw drug traded in India. However, Gmelinaasiatica and Mallotus nudiflorus are also found in the raw drug markets as Gambhari. The current study aims to identify molecular markers based on the nuclear ribosomal DNA – ITS1 region to distinguish the authentic species from substitute/adulterants. The nuclear ribosomal internal transcribed spacer 1 (ITS1) was amplified to identify species-specific markers using universal primers. Based on the sequence of the ITS region, specific primers were designed for G. arborea, G. asiatica and M. nudiflorus which efficiently amplified 142 bp, 93 bp and 150 bp of the ITS1 region of the respective species. The notable feature of this molecular method is that it is technically accurate, practically convenient and suitable for analyzing large numbers of samples. This study demonstrates that the ITS1 region can be used for reliable authentication of medicinal plants and detection of adulterants and substitutes of Gambhari.
Keywords: Gmelina arborea, Gambhari, Gmelina asiatica, Mallotus nudiflorus, Internal transcribed spacer (ITS), DNA marker
1. Introduction
Gmelina arborea Roxb. Ex Sm. (Verbenacea) is a medicinally important plant distributed in the deciduous forests of South East Asia [1]. In India it is distributed in Western ghats, foot of north-western Himalaya to Chittagong and throughout Deccan peninsula (http://envis.frlht.org/indian-medicinal-plants-database.php). It is one of the vital ingredients of the Dasamoola herbal formulation with an annual trade demand of 1000 metric tons [2]. According to Ayurvedic Pharmacopoeia of India, the root of G. arborea has been used under the common name “Gambhari” with gmelinol as an active chemical marker [3]. G. arborea has been reported to have demulcent, stomachic, galactagogue, laxative, anti-helminthic, anti-inflammatory properties, and it has also been used in the treatment of anthrax, asthma, bronchitis, cholera, epilepsy, fever, hallucination, leprosy, rheumatism and snake poisoning [4], [5].
While demand for herbal medicinal products is surging, the distribution and availability of medicinal plants have gone down significantly. Because of these reasons, G. arborea is often adulterated or substituted with plants such as Gmelina asiatica and Mallotus nudiflorus (Euphorbiaceae) [2], [6]. Substitution and adulteration of Ayurvedic herbal ingredients reduce the therapeutic efficacy of the drug and may pose serious detrimental effects on consumers' health [7].
It is necessary to develop simple and accurate methods for distinguishing the authentic plants from its adulterants. Many recent studies have employed molecular techniques based on nucleotide sequencing in order to differentiate closely related plant species [8]. Since DNA markers are not affected by environmental conditions, organism age and physiological conditions, they are more reliable than morphological or chemical traits. The nuclear ribosomal internal transcribed spacer (ITS) region is one of the preferred genetic marker for molecular species identification, because it is highly repeated, contains variable regions flanked by more conserved DNA sequences and also universal primers are available for PCR amplification [9].
The objective of the present study was to identify different plant species traded as ‘Gambhari’ based on nuclear ribosomal DNA internal transcribed spacer (ITS1) sequences and to develop species-specific markers for distinguishing them.
2. Materials and methods
2.1. Plant material collection and authentication
Field and market samples of G. arborea, G. asiatica and M. nudiflorus were collected from different parts of south India and assigned with individual accession numbers (Supplementary Table 1). They were identified based on Flora of Tamil Nadu Carnatic [10], Flora of Orissa [11] and Flora of Bangalore District [12] by qualified field botanists at the Foundation for Revitalization of Local Health Traditions (FRLHT), Bangalore, India. The size, shape and arrangement of the leaves were considered as a key distinguishing character of the three species studied (details presented in Supplementary Table 2). The voucher samples were deposited in the Medicinal Plants Herbarium and Raw Drug Repository, FRLHT, Bangalore, India. The photographs of the flowering branches of the three species are presented in Supplementary Fig. 1. Market samples of Gambhari roots are shown in Supplementary Fig. 2.
2.2. Genomic DNA extraction
The root samples of G. arborea, G. asiatica and M. nudiflorus were cut into small pieces and dried in a dehydrator at 50 °C. Dried root samples were ground into powder with mortar and pestle in liquid nitrogen. Total genomic DNA was extracted from 100 mg of powdered sample following the protocol described by Milligan [13] with modifications. Extraction buffer containing (2% w/v CTAB, 1.4 M NaCl, 20 mM EDTA, 4 M LiCl, 100 mM Tris–HCl; pH 8, 1% PVP w/v and 0.2% v/v β mercaptoethanol; pH 7.5–8.0) was added to the powder and incubated at 60 °C in a water bath with occasional shaking. After incubation, extraction with equal volume of chloroform/isoamyl alcohol (24:1) was performed twice. After centrifugation at 10,000 rpm for 15 min, the supernatant was collected and precipitated with one volume of ice-cold 2-propanol and 1/10th volume of 3 M sodium acetate at 4 °C. The mixture was centrifuged for 15 min at 10,000 rpm. The collected DNA pellet was washed with 70% ethanol, air dried and dissolved in TE buffer (10 mM Tris–HCl, 1 mM EDTA; pH 8) after drying. The sample was treated with RNase A (20 μg/μl) for 30 min at 37 °C. Purity of DNA was checked using UV-VIS spectrophotometer (Shimadzu, Tokyo, Japan), by calculating the A260/280 ratio. The DNA stock concentration was maintained at 30–50 ng/ml in −20 °C.
2.3. Polymerase chain reaction (PCR) amplification of ITS sequence
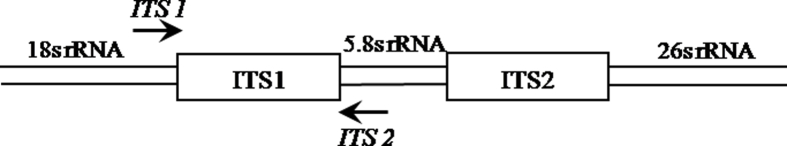
The nuclear DNA-ITS1 sequence was amplified from the extracted genomic DNA with the universal primers ITS 1 (forward primer; 5′-TCCGTAGGTGAACCTCGG-3′) and ITS 2 (reverse primer; 5′-GCTGCGTTCTTCATCGATGC-3′) [14] as summarized in Supplementary Table 3. The PCR amplicons were resolved on 2% agarose, 1× TAE buffer gels pre-stained with ethidium bromide (0.5 μg/ml). Simultaneously, 100 bp ladder (Bangalore Genei, Bangalore) was loaded to identify the size of amplicons. The gel was visualized under UV light in a gel documentation system (Bio-Rad, CA, USA) and image captured.
2.4. PCR product purification and sequencing
The PCR-amplified ITS1 regions of G. arborea, G. asiatica and M. nudiflorus were purified from agarose gel using QIAquick gel extraction kit (Qiagen, Maryland, USA), following manufacturer's instruction. Direct sequencing of purified PCR product was performed using primers ITS 1 and ITS 2 by means of the automatic ABI 3100 genetic sequencer (Applied Bio systems, CA, USA), in Bangalore Genei (Bangalore, India). Sequence quality checks were performed (details in supplementary data) and the nucleotide sequence of the ITS region for all the three species were submitted to GenBank (http://www.ncbi.nim.nih.gov/genbank/). Simultaneously, sequences were analyzed using ITSx software tool (version 1.0.11; http://microbiology.se/software/itsx/) to extract the ITS1 region from the whole sequence [15]. Primers were designed within the ITS1 region.
2.5. Designing of species-specific primers and validation
Multiple sequence alignment of the ITS1 sequence of G. arborea, G. asiatica and M. nudiflorus was performed to observe the sequence variations for their discrimination. Based on the sequence variation, PCR primers capable of giving species specific amplification were designed using Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer) (Table 1). The primers were also checked using the in-silico PCR amplification tool (http://insilico.ehu.es/PCR/index.php) for their specificity and stringency. Oligonucleotides were custom synthesized by Bioserve biotechnologies (Hyderabad, India) and were used to amplify DNA extracted from all three ‘Gambhari’ samples. The conditions of PCR amplification with species-specific primers are summarized in Supplementary Table 3.
Table 1.
Details of the species-specific markers designed using ITS1 sequence.
| Plant species | Name of the marker | Primer sequence (5′→3′) | Expected amplicon size |
|---|---|---|---|
| G. arborea | GAR-F | GAGGAAGGATCAGGTCGAGA | 142 |
| GAR-R | GCGGAACGCTTCATTGAGAT | ||
| G. asiatica | GAS-F | GGTTAACGAACCCCGGC | 93 |
| GAS-R | GATCCCGCCCGATCACC | ||
| M. nudiflorus | TNF-F | GCTCTGCAGAACGACCC | 150 |
| TNF-R | CGCCGGGGTTGGTGTTA |
3. Results
The DNA extraction protocol described by Milligan [13] with some modifications yielded high molecular weight genomic DNA from the dried root samples of G. arborea, G. asiatica and M. nudiflorus. To the extraction buffer, 4 M LiCl was added in order to remove polyphenols and polysaccharides [16]. This DNA isolation protocol yielded high quality DNA. An absorbance (A 260/A280) ratio of 1.6–1.8 indicated insignificant levels of contaminating proteins and polysaccharides. The universal primers ITS 1 (forward) and ITS 2 (reverse) amplified the partial ITS1 region yielding an amplicon of approximately 300 bp with all accessions for the three species (Supplementary Fig. 3).
Direct sequencing of the gel purified amplicon yielded a 259 bases sequence of ITS1, partial 5.8s rRNA gene for Gmelina arborea (Genbank accession No. KJ704774). G. asiatica yielded a 328 bases sequence comprising of 18s ribosomal RNA gene partial, ITS1 complete and partial 5.8s rRNA gene (Genbank accession No. KJ704775). A 290 bases sequence of 18s ribosomal RNA gene, partial and ITS1, partial was obtained for M. nudiflorus (Genbank accession No. KJ704776).
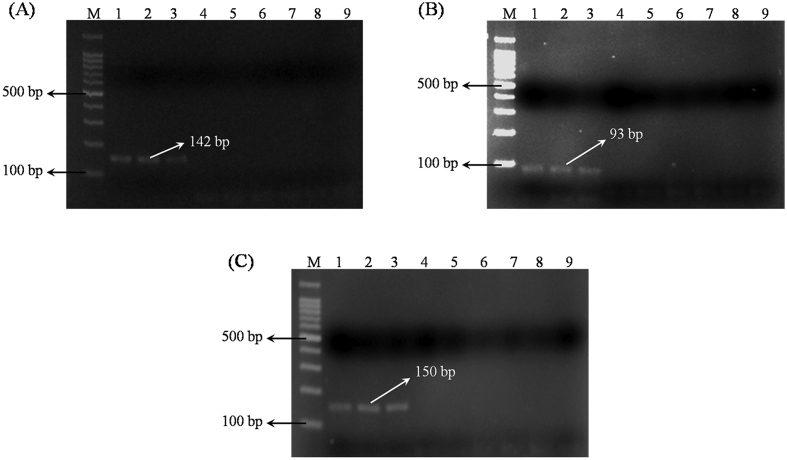
Species-specific primers GAR-F and GAR-R yielded 142 bp amplicon with G. arborea accessions (Fig. 1A) and did not produce any amplification with G. asiatica and M. nudiflorus. Similarly, primers GAS-F and GAS-R amplified the sequence size of 93 bp only with G. asiatica (Fig. 1B), while TNF-F and TNF-R produced amplification 150 bp of M. nudiflorus (Fig. 1C). Thus, the primers designed were found to be species specific.
Fig. 1.
PCR amplification of genomic DNA with species-specific markers. (A) Markers GAR-F and GAR-R gave a 142-base amplicon with Gmelina arborea samples (Lanes 1–3); Lanes 4 to 6 – G. asiatica; Lanes 7 to 9 – M. nudiflores. (B) GAS-F and GAS-R markers gave a 93-base amplicon with only G. asiatica samples (Lanes 1–3); Lanes 4 to 6 – G.arborea; Lanes 7 to 9 – M. nudiflores. (C) Markers TNF-F and TNF-R gave 150-base amplicon with only M. nudiflorus samples (Lanes 1–3); Lanes 4 to 6 – G. arborea; Lanes 7 to 9 – G. asiatica. Lane M, 100 bp DNA ladder.
4. Discussion
An accurate and straightforward discrimination of authentic plant species is very important in order to ensure safety and quality of herbal drugs [17]. Although morphological, microscopic and chromatography based identification methods are simple, the degree to which these methods accurately identify the correct species strongly depends on the skill and expertise of the identifiers [18]. Moreover, it is very likely that the related plant species and substitutes or adulterants may share similar characters, making the conventional approaches less accurate, particularly if the substances at hand are plant parts such as root and barks [19].
DNA-based methods have become important tool for species identification of plants [8]. The nuclear ribosomal ITS region has been reported to display a high degree of divergence between species but is often highly conserved within species; hence they are most preferred genetic markers for species level identification [20]. Because of the high copy number of the ITS region in genomic DNA, the chances of getting amplification from the processed or old herbal materials are good [8]. ITS1 has been successfully used in distinguishing medicinal herbs like Amomum villosum (Zingiberacea) [21] and also in differentiating the species Boerhavia diffusa (Punarnava) from Boerhavia erecta [22]. Rai et al. [23] have used ITS2 sequences to distinguish members of Asparagaceae and Asclepiadaceae.
5. Conclusion
The primers introduced in this study target the nuclear ribosomal ITS1 region, and we hope that they will be useful in assessing the botanical identity of raw drugs traded under the name of Gambhari.
Sources of funding
National Medicinal Plants Board (NMPB), Ministry of AYUSH and Ministry of Environment and Forests, Govt. of India under “Centre of Excellence” scheme.
Conflict of interest
None.
Acknowledgements
Thanks to Dr. K. Ravi Kumar and Mr. Murthy for providing plant accessions and authentication of medicinal plant species. Acknowledging Dr. Murugan, SASTRA University for the plant photographs and plant description. Ms. Krithika is thankfully acknowledged for technical inputs on sequence data analysis. Authors also wish to thank Dr. K.V. Krishnamurthy and Dr. K. Subrahmanya Kumar for their valuable inputs to this study.
Footnotes
Peer review under responsibility of Transdisciplinary University, Bangalore.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.jaim.2017.10.001.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
figs1.

figs2.

figs3.

figs4.
figs5.
References
- 1.Dhakulkar S., Ganapathi T.R., Bhargava S., Bapat V.A. Induction of hairy roots in Gmelina arborea Roxb. and production of verbascoside in hairy roots. Plant Sci. 2005;169(5):812–818. [Google Scholar]
- 2.Ved D.K., Goraya G.S. Bishen Singh Mahendra Pal Singh; Dehra Dun: 2008. Demand and supply of medicinal plants in India. [Google Scholar]
- 3.Anonymous . vol. I. Government of India, Ministry of Health & Family Welfare, Department of AYUSH; New Delhi:: 2008. The ayurvedic Pharmacopoeia of India, Part I. [Google Scholar]
- 4.Kirtikar K.R., Basu B.D. 2nd ed. Taj Offset Press; Delhi: 1984. Indian medicinal plants. [Google Scholar]
- 5.Nadkarni A.K. 3rd ed. Popular Prakasan; Bombay: 1976. Indian materia medica. [Google Scholar]
- 6.Vaidya B. Chaukhambha Orientalia; Varanasi: 2005. Some controversial drugs in Indian medicine. [Google Scholar]
- 7.Newmaster S.G., Grguric M., Shanmughanandhan D., Ramalingam S., Ragupathy S. DNA barcoding detects contamination and substitution in North American herbal products. BMC Med. 2013;11:222–235. doi: 10.1186/1741-7015-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Balasubramani S.P., Murugan R., Ravikumar K., Venkatasubramanian P. Development of ITS sequence based molecular marker to distinguish, Tribulus terrestris L. (Zygophyllaceae) from its adulterants. Fitoter. 2010;81(6):503–508. doi: 10.1016/j.fitote.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Hillis D.M., Dixon M.T. Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol. 1991;66:411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- 10.Matthew K.M. 3 vols. The Rapinat Herbarium, St Joseph's College; , Tiruchirappalli, India: 1981–84. The Flora of Tamil Nadu Carnatic. [Google Scholar]
- 11.Saxena H.O., Brahman M. 4 vols. Regional Research Laboratory; Bhubaneswar, India: 1994–96. Flora of Orissa. [Google Scholar]
- 12.Ramaswamy S.V., Razi B.A. University of Mysore; India: 1973. Flora of Bangalore District. [Google Scholar]
- 13.Milligan B.G. Total DNA isolation. In: Hoelzel A.R., editor. Molecular genetic analysis of populations. IRL Press; Oxford: 1998. pp. 29–64. [Google Scholar]
- 14.White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., Thomas J., editors. PCR protocols: a guide to methods and applications. Academic Press; San Diego: 1990. [Google Scholar]
- 15.Bengtsson-Palme J., Ryberg M., Hartmann M., Branco S., Wang Z., Godhe A. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol. 2013;4(10):914–919. [Google Scholar]
- 16.Pirttila A.M., Hirsikorpi M., Kamarainen T., Jaakola L., Hohtola A. DNA isolation methods for medicinal and aromatic plants. Plant Mol Biol Rep. 2001;19(3):273. [Google Scholar]
- 17.WHO . World Health Organization; Geneva: 1998. Quality control methods for medicinal plant materials. [Google Scholar]
- 18.Liu Z., Zeng X., Yang D., Chu G., Yuan Z., Chen S. Applying DNA barcodes for identification of plant species in the family Araliaceae. Gene. 2012;499(1):76–80. doi: 10.1016/j.gene.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 19.Joshi K., Chavan P., Warude D., Patwardhan B. Molecular markers in herbal drug technology. Curr Sci. 2004;87:159–165. [Google Scholar]
- 20.Cheng T., Xu C., Lei L., Li C., Zhang Y., Zhou S. Barcoding the kingdom Plantae: new PCR primers for ITS regions of plants with improved universality and specificity. Mol Ecol Resour. 2016;16(1):138–149. doi: 10.1111/1755-0998.12438. [DOI] [PubMed] [Google Scholar]
- 21.Qiao C., Han Q., Zhao Z., Wang Z., Xu L., Xu H.X. Sequence analysis based on ITS1 region of nuclear ribosomal DNA of Amomum villosum and ten species of Alpinia. J Food Drug Anal. 2009;17(2):142–145. [Google Scholar]
- 22.Selvaraj D., Shanmughanandhan D., Sarma R.K., Joseph J.C., Srinivasan R.V., Ramalingam S. DNA barcode ITS effectively distinguishes the medicinal plant Boerhavia diffusa from its adulterants. Genom Proteom Bioinform. 2012;10(6):364–367. doi: 10.1016/j.gpb.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rai P.S., Bellampalli R., Dobriyal R.M., Agarwal A., Satyamoorthy K., Narayana D.A. DNA barcoding of authentic and substitute samples of herb of the family Asparagaceae and Asclepiadaceae based on the ITS2 region. J Ayurveda Integr Med. 2012;3(3):136–140. doi: 10.4103/0975-9476.100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.