Abstract
Early detection of gastrointestinal tumors improves patient survival. However, patients with these tumors are typically diagnosed at an advanced stage and have poor prognosis. The incidence and mortality of gastrointestinal cancers, including esophageal, gastric, liver, colorectal, and pancreatic cancers, are increasing worldwide. Novel diagnostic and therapeutic agents are required to improve patient survival and quality of life. The tumor microenvironment, which contains nontumor cells, signaling molecules such as growth factors and cytokines, and extracellular matrix proteins, plays a critical role in cancer cell proliferation, invasion, and metastasis. Transforming growth factor beta (TGF-β) signaling has dual roles in gastrointestinal tumor development and progression as both a tumor suppressor and tumor promoter. Here, we review the dynamic roles of TGF-β and its receptors in gastrointestinal tumors and provide evidence that targeting TGF-β signaling may be an effective therapeutic strategy.
Introduction
Transforming growth factor beta (TGF-β) is a cytokine that participates in both physiological and pathological processes including tumorigenesis [1]. During tumor progression, TGF-β signaling regulates immune/inflammatory response and tumor microenvironment. It also regulates tumor growth, epithelial-mesenchymal transition (EMT), and cancer cell stemness depending on tumor stage and cellular context [2], [3], [4]. Malignant gastrointestinal tumors such as esophageal, gastric, liver, colorectal, and pancreatic carcinomas are a major cause of cancer-related deaths worldwide [5]. Aberrant TGF-β signaling has been associated with gastrointestinal cancer progression [6]. Several TGF-β-based therapeutics have been developed for the treatment of gastrointestinal cancers and have displayed efficacy in clinical trials [7], [8]. Here, we review the roles of TGF-β and its receptors in gastrointestinal tumors and describe the evidence that targeting TGF-β signaling may be an effective therapeutic strategy.
TGF-βs and Their Receptors
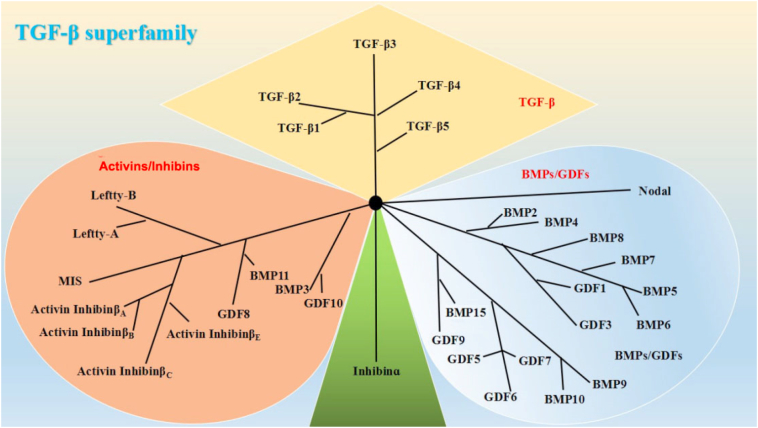
The TGF-β superfamily consists of at least 40 structurally and functionally related cytokines that are involved in various biological processes including embryonic development, extracellular matrix formation, immune regulation, inflammation, and cancer [1], [9], [10]. TGF-β family proteins are classified into several subtypes, including TGF-βs, activins/inhibins, and bone morphogenetic proteins (BMPs)/growth differentiation factors according to structural characteristics (Figure 1) [11].
Figure 1.
The TGF-β superfamily. Based on their structural features, the mammalian members of the TGF-β family are subdivided into (i) TGF-βs, (ii) activins/inhibins, and (iii) BMPs/growth and differentiation factors (GDFs).
Six TGF-β isoforms have been identified, which display variable sequence homology. TGF-β1, TGF-β2, and TGF-β3 are highly conserved and expressed in mammals [12]. TGF-β4 and TGF-β5 are predominantly expressed in birds and amphibians [13], while TGF-β6 is only expressed in fish [14]. TGF-β1 is the most abundant and ubiquitously expressed of the isoforms. TGF-β is synthesized in an inactive form (pre-proTGF-β), which contains a signal peptide, a pro region, and the mature coding region. Following removal of the signal peptide, the TGF-β dimer interacts with latency associated peptide, a protein derived from the N-terminal region of the proTGF-β, to form the small latent complex. This complex is secreted into the extracellular matrix upon binding to latent TGF-β-binding protein to form the large latent complex. TGF-β is then active until it is released from the large latent complex by proteases, integrins, pH, or reactive oxygen species. Active TGF-β consists of two identical peptide chains with a molecular weight of 25 kDa [15].
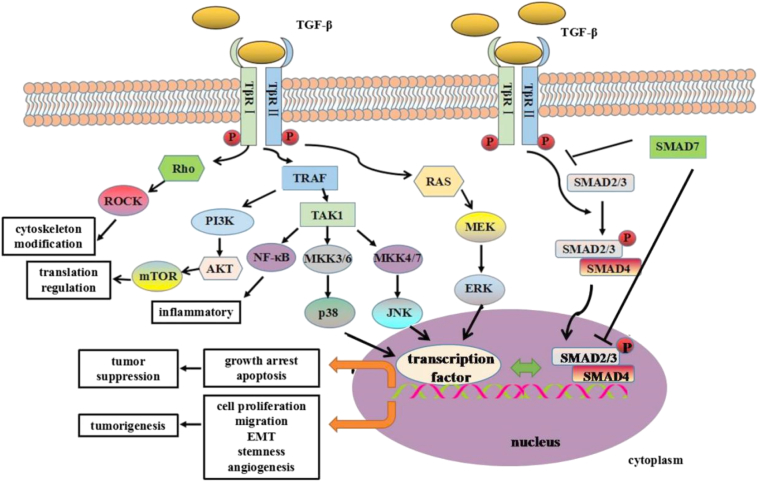
TGF-β signals through TGF-β receptors (TβRs) I and II to activate downstream signaling pathways [11], [16]. The TβRs are single-pass transmembrane proteins with serine/threonine kinase activity. Seven TβRIs and five different TβRIIs have been identified, which confer intracellular signaling specificity to all members of the TGF-β superfamily [1]. In the absence of ligand, TβRI and TβRII exist as monomers, homodimers, or heterodimers on the cell surface. Ligand binding promotes formation of a tetrameric complex between TβRII dimers and two TβRIs [17]. TβRIs and TβRIIs have N-terminal extracellular ligand binding domains, transmembrane segments, and C-terminal cytosolicserine/threonine kinase domains [11]. TGF-β binds specifically to the constitutively active TβRII, which activates TβRI by phosphorylating the glycine/serine-rich domain. Activated TβRI then phosphorylates downstream effectors to induce signal transduction (Figure 2) [17]. TβR activity is regulated by betaglycan, a type III TβR, and endoglin [16].
Figure 2.
Schematic representation of TGF-β signaling as well as the role of TGF-β signaling pathway in cancer onset and progression. TGF-β activates both Smad-dependent canonical and Smad-independent noncanonical signaling pathways. TGF-β binds to TβRII which then activates TβRI. TβRI-phosphorylated Smad2/3 form complexes with Smad4, entering nucleus and regulating the transcription of different targeted genes in both early and late stages of tumor development, contributing to tumor suppression and tumorigenesis, respectively. Smad7 antagonizes TGF-β signaling through blocking Smad2/3 activation and interfering with the formation of Smad2/3/4-DNA complex. In the noncanonical signaling pathways, TGF-β receptors initiate the signal through MAPKs, PI3K, and Rho family of small GTPases etc. Activated JNK/p38/ERK either interact with SMADs or induce their individual transcriptional programs directly to affect cancer cells. Rho-activated Rho-associated protein kinase is involved in cytoskeleton modification in the process of EMT. Through PI3K-AKT pathway, TGF-β can also activate mammalian target of rapamycin to regulate protein translation. In addition, TGF-β activation of the tumor necrosis factor receptor–associated factor proteins can also induce NF-κB signaling for inflammatory response.
Canonical TGF-β Signaling
Canonical TGF-β signaling is dependent upon Smad family proteins. Active TGFβI at the cell surface phosphorylates receptor-activated Smads (R-Smads). There are two sub-classes of R-Smads in which Smad2 and Smad3 mediate the TGFβ/activin pathway. Smad4 acts as a co-factor that binds to activated R-Smads to form a complex that translocates to the nucleus and regulates transcription (Figure 2) [1], [18], [19]. Interestingly, R-Smads have also been shown to interact with other proteins such as tripartite motif-containing 33 to regulate gene transcription [20].
TGF-β signaling is regulated through various mechanisms. Inhibitory Smad (Smad7) can inhibit TGF-β signaling by interacting with TβRI and R-Smads [21] (Figure 2). R-Smad stability is also modulated by phosphorylation [22]. Additionally, Smad ubiquitination regulatory factor (Smurf)–mediated TβRI degradation can dynamically regulate TGF-β signaling [23].
Noncanonical TGF-β Signaling
TβRs can also activate non–Smad-dependent signaling pathways, including the mitogen-activated protein kinase (MAPK) pathway mediated by p38, c-Jun amino terminal kinase (JNK), extracellular signal-regulated kinases (ERK), nuclear factor-κB (NF-κB), Rho, and phosphatidylinositol 3-kinase (PI3K)-Akt (Figure 2) [24], [25]. These non-Smad pathways might mediate signaling transduction alone or in cooperation with the canonical Smad pathway to modulate TGF-β signaling activity.
TGF-β–induced activation of ERK can phosphorylate transcription factors. Activated TGF-β receptors interact with tumor necrosis factor receptor–associated factor 6 and TGF-β-activated kinase 1 to activate multiple downstream kinases including JNK, p38 and IkB kinase. Activated JNK and p38 can phosphorylate the targeted transcription factors, while IkB kinase phosphorylates NF-κB. Activated Akt can control translation through mammalian target of rapamycin-1. These non–Smad-mediated translational and transcriptional responses cross talk with Smad-mediated transcriptional responses to contribute to either tumorigenesis or tumor suppression. Non-Smad proteins can also directly impact R-Smad activity. ERK can regulate the activity of R-Smads in a phosphorylation-dependent manner, while Akt regulates Smad3 activity by sequestering Smad3 in the cytoplasm. Furthermore, TGF-β also activates RhoA and Rho-associated protein kinase to induce actin polymerization involved in EMT process. Thus, cross talk between the canonical and noncanonical TGF-β signaling pathways contributes to tumor development (Figure 2) [25]. Cross talk has also been demonstrated between the TGF-β signaling pathway and the tumor necrosis factor-α and epidermal growth factor receptor pathways [24], [25], [26].
Role of TGF-β Signaling in Cancer
Altered TGF-β expression has been observed in several cancers [2], [7], [8]. Interestingly, TGF-β has dual roles in tumor progression, acting as both a tumor suppressor and tumor promoter in a stage- and context-dependent manner [4], [7], [8]. During tumor initiation, TGF-β signaling promotes cell cycle arrest and apoptosis, thereby acting as a tumor suppressor. In contrast, TGF-β has been shown to promote tumor cell proliferation, EMT, and stem-like behavior as well as fibrosis, inflammation, and angiogenesis during tumor progression (Figure 2) [4], [7], [8]. Upregulation of TGF-β expression was correlated with poor prognosis in patients with advanced-stage tumors. The accumulation of mutations in TGF-β signaling pathway components during tumor progression may contribute to the switch in TGF-β function from tumor-suppressive to tumor-promoting.
Role of TGF-β Signal in Gastrointestinal Cancers
TGF-β Signal and Esophageal Cancer
Esophageal cancer is the sixth leading cause of cancer-related death worldwide [27], [28]. Esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma are the most common histological subtypes of esophageal cancer [29]. Risk factors for esophageal cancer include smoking, consumption of hot liquids, poor oral health, and vitamin deficiency [28]. Recent studies have provided insight into the role of TGF-β signaling in esophageal cancer. Interestingly, higher serum TGF-β levels were observed in patients with esophageal cancer compared to healthy controls, and reduced levels were observed following radiotherapy [30]. Upregulation of TGF-β and concomitant overexpression of vascular endothelial growth factor (VEGF) were correlated with tumor size in ESCC [31]. Additionally, overexpression of TGF-β and reduced TβR expression were associated with depth of invasion and pathologic stage in ESCC [32]. Smad4, but not Smad2/3, expression was inversely correlated with invasion in ESCC [33]. Smurf2-induced Smad2 degradation may also contribute to tumor development and poor prognosis in ESCC [34]. Finally, TGF-β/Smad signaling has been shown to promote EMT in ESCC through PTEN/PI3K [35], [36]. Thus, TGF-β signaling plays a role in esophageal cancer progression.
MicroRNAs (miRNAs) are diagnostic and prognostic biomarkers in several cancers and have been shown to regulate TGF-β signaling in esophageal cancer [37], [38]. MiR-17/20a suppressed ESCC cell migration and invasion via the TGF-β/integrin β6 subunit pathway by targeting TβR2 and Smad anchor for receptor activation for degradation [39]. Additionally, TGF-β was shown to play a role in miR-455-3p–mediated ESCC progression [40]. MiR-655 suppressed ESCC progression by targeting zinc finger E-box binding homeobox 1and TβRII, which are required for TGF-β signaling, and suppressing EMT [41]. MiR-32 was recently found to promote ESCC metastasis through CXXC5-mediated inhibition of TGF-β signaling [42].
Cross talk between cytoskeleton-associated proteins, which are aberrantly expressed in esophageal cancer, and the TGF-β signaling pathway can promote ESCC progression. Reelin, which has a key role inneuronal migration, negatively regulates TGF-β–induced ESCC cell migration. Reelin expression was suppressed by the TGF-β pathway through the transcription factor Snail [43]. Fascin is an actin bundling protein that induces cell membrane protrusions. It regulates ESCC cell proliferation and invasion by modulating the levels of connective tissue growth factor and cysteine-rich protein 61 via the TGF-β pathway [44]. Overexpression of cysteine-rich protein 61 and connective tissue growth factor was associated with poor survival in ESCC [45]. Ezrin, a cytoskeletal cross-linking protein, also promotes ESCC cell proliferation and invasion through the TGF-β and MAPK signaling pathways [46]. Collectively, these data indicate that TGF-β signaling can promote esophageal cancer development and metastasis.
TGF-β Signal and Gastric Cancer
Gastric cancer (GC) is the second leading cause of cancer-related death worldwide. It has a particularly high mortality rate in Asia, which may be linked to both social and environmental factors [47], [48]. Despite early diagnosis and treatment with a combination of surgery, chemotherapy, and/or radiotherapy, the prognosis of GC patients is poor due to recurrence and distant metastasis [49]. Previous studies have demonstrated higher TGF-β levels in serum from GC patients compared to healthy controls. Elevated TGF-β levels were correlated with lymph node metastasis, worse overall survival, and poor prognosis in GC patients [50]. TGF-β was also increased in the gastric mucosa and in precancerous gastric cells [51]. Altered TGF-β signaling has been observed during GC progression. Additionally, mutations in TβRII have been implicated in gastric carcinogenesis [52]. Repression of TβRI transcription through CpG island methylation was also correlated with poor prognosis in GC patients [53]. Various mutations in the promoter regions of TGFB1 and TGFBR2 were associated with the riskofGC in a Chinese population [54]. In addition, an imbalance in Smad4/7 expression was associated with GC cell differentiation, metastasis, and apoptosis [55]. These data suggest TGF-β plays an important role in GC development.
EMT results in gastric epithelial cells acquiring mesenchymal characteristics and promotes stemness, invasion, and metastasis [56], [57]. EMT can be induced by pathogens, stress, and hypoxia [58]. EMT is modulated by the microenvironment in gastric and other cancers [58], [59]. TGF-β, Notch, and Wnt signaling can induce EMT in GC cells, thereby promoting tumor progression [58].
Several proteins regulate TGF-β-induced EMT in GC and have opposing effects. For example, ankyrin-repeat-containing, SH3-domain-containing, and proline-rich-region-containing protein 2 [60], grainyhead-like 2 [61] and metastasis suppressor protein 1 [62] suppress invasion and TGF-β–induced EMT in GC cells, whereas homeobox protein A13 [63], CC-chemokine receptor 7 [64], and homeoprotein Bapx1 [65] promote TGF-β–induced EMT. MiRNAs also regulate EMT in GC cells. MiR-381 inhibited TGF-β signaling and suppressed EMT in GC cells in part by targeting transmembrane member 16A [66]. In contrast, miR-21 enhanced TGF-β–induced EMT in GC cells by upregulating PTEN expression [67]. MiRNAs may contribute to GC progression by directly targeting components of the TGF-β signaling pathway. MiR-17-5p and miR-155 target TβRII in GC cells and are involved in proliferation and migration, while miR-424-5p, which is upregulated in GC cells, inhibits TGF-β signaling by targeting Smad3 and promotes GC cell proliferation [68], [69], [70]. Finally, miR-199a was found to target Smad4 in GC [71].
TGF-β Signal and Hepatocellular Carcinoma
Hepatocellular carcinoma (HCC) is the most common malignancy worldwide [72], [73]. The primary causes of HCC include toxins (e.g., alcohol and aflatoxin) and chronic hepatitis B virus or hepatitis C infection (account for approximately 80% of cases) [74]. Most patients are diagnosed at an advanced stage due to a lack of symptoms and rapid disease progression. The 5-year overall survival rate is less than 20% [75]. Surgical resection and liver transplantation are the best treatment options for HCC [76]. However, there is a high rate of recurrence and metastasis following curative hepatectomy [77]. A better understanding of the molecular mechanisms underlying HCC progression is required to identify more effective therapeutic targets.
Elevated TGF-β levels have been observed in patients with metastatic HCC compared to those with nonmetastatic disease [78], [79]. Immunostaining demonstrated expression of all three TGF-β isoforms in HCC cells and in the perineoplastic stroma, while little to no expression was detected in normal liver tissue [80]. Serum TGF-β levels were associated with disease progression and poor prognosis [81]. TGF-β may be a feasible marker for the detection of early-stage HCC because it has displayed higher sensitivity than traditional markers such as alpha-fetoprotein in past studies [82]. Loss of TβRII expression was correlated with tumor progression in HCC [83]. Additionally, mutations in Smad2 and Smad4 have been observed in a small minority of HCC patients [84]. TGF-β induces alterations in the cellular composition of the tumor microenvironment, which may also play an important role in HCC progression [85], [86]. A recent study stated that the combination of the expression level of c-Myc, TGF-β1, and embryonic liver fodrin (an adaptor protein of TGF-β signaling cascade) together can be used to accurately predict outcomes of patients with HCC [87].
Emerging evidence suggests TGF-β signaling has a dual or biphasic role in HCC development and progression [88], [89]. TGF-β acts as a tumor suppressor in early carcinogenesis but promotes tumor progression during later stages. Consistent with these observations, reduced TβRII correlated with increased tumor size and intrahepatic metastasis in HCC patients [83], and with the risk of HCC in a mouse model [90]. Additionally, reduced TβRII staining has been observed in approximately 25% of malignant compared to adjacent nonmalignant hepatocytes [91]. TGF-β was also found to suppress HCC proliferation through activating Hippo signaling [92]. NADPH oxidase 4 may mediate the antiproliferative and proapoptotic effects of TGF-β in HCC cells [93]. On the other hand, TGF-β1 contributes to HCC invasion and metastasis by promoting fibroblast growth factor receptor 4 expression via the noncanonical ERK pathway [94]. AGO1, the main component of RNA-induced silencing complexes, may promote HCC metastasis in a TGF-β–dependent manner [95]. Finally, miRNAs may regulate EMT in HCC cells by targeting TGF-β signaling. The miR-200 family of miRNAs was shown to suppress metastasis in human HCC cells [96], and miR-125b was found to suppress EMT by targeting Smad2 and Smad4 [97].
The mechanisms underlying the dual roles of TGF-β signaling during tumor progression have not been elucidated. Previous studies have suggested that HCC cells may become resistant to the antiproliferative effects of TGF-β but maintain sensitivity to the tumor-promoting effects [98]. Importantly, upregulation of growth factor–mediated survival signals, such as MAPK/ERKs, PI3K/Akt, and NF-κB, suppressed the antiproliferative effects of TGF-β in HCC cells [99]. TGF-β promotes the production of growth factors and cytokines such as PDGF and EGF, which contribute to tumor cell proliferation and invasion [99]. PDGF links TGF-β signaling to nuclear β-catenin accumulation during HCC progression [100]. EGF promotes HCC cell survival by protecting the cells from TGF-β–induced cell death [101]. A recent study demonstrated that abrogation of the posttranscriptional regulation of c-Myc in an epigenetic modification-dependent manner promoted HCC cell resistance to antiproliferative TGF-β signaling [102].
A temporal TGF-β-specific gene expression signature was established in mouse hepatocytes that could distinguish between HCC subtypes. Tumors expressing early TGF-β–responsive genes displayed the physiological growth inhibitory effects of TGF-β, whereas tumors expressing late TGF-β–responsive genes displayed aberrant upregulation of TGF-β signaling and the antiapoptotic and invasive effects of TGF-β. The TGF-β–specific gene expression signature could predict HCC invasion and liver metastasis [103].
TGF-β Signal and Colorectal Cancer
Colorectal cancer (CRC) is one of the three most common cancers worldwide [104], [105]. The incidence of CRC is higher in developed countries than in less developed countries, and nutrition is thought to be a major contributing factor [104]. The 5-year survival rate is less than 20%in patients with metastatic disease compared to 70%-90% in patients with nonmetastatic disease [104]. Approximately one third of all CRC cases are inherited [106]. Highly penetrant tumor susceptibility genes account for only 3%-6% of all CRC cases. The remaining fraction of the unexplained risk can likely be attributed to a combination of low to moderately penetrant genes and polygenic mechanisms [107].
Previous studies have demonstrated increased TGFB1 expression in CRC compared to benign adenoma and noncancerous tissue [108]. Elevated TGF-β levels were observed in primary tumor tissue and in plasma from CRC patients and were correlated with metastasis and poor prognosis [109], [110]. TGF-β was predominantly detected in the central regions of CRC liver metastases [111]. TGF-β induced growth arrest in moderately differentiated colon carcinomas but promoted proliferation in more aggressive tumors [112]. Reduced phosphorylation of the C-terminal region of Smad3 and activation of c-Jun NH(2)-terminal kinase–mediated phosphorylation of the linker region of Smad2/3 were observed in the human colorectal adenoma to carcinoma sequence, from which the majority of CRCs arise [113], [114]. Notably, inhibition of TGF-β can prevent CRC metastasis by unleashing a cytotoxic T-cell response against cancer cells, implying that TGF-β signaling suppresses cancer recognition by the immune system [115].
Mutational inactivation of the TGF-β signaling pathway is a key cause of CRC progression [107], [116]. Alterations in TGF-β signaling have been observed in nearly 50% of CRCs [107], [116]. For example, mutations in TGFBR2 that abolish TGF-β signaling have been frequently detected in CRCs [107], [117]. More than 80% of CRCs that display microsatellite instability harbor mutations in TGFBR2 (particularly frameshift mutations in exon 3) [116], [118]. The exact mechanisms by which TGFBR2 mutations lead to CRC have not been elucidated. However, a previous study demonstrated that inactivation of TβRII induced VEGF-A expression, which enhanced the metastatic potential of CRC cells [119]. TGF-β signaling was still active in some CRCs that displayed a high level of microsatellite instability despite frameshift mutations in TGFBR2 [120]. Mutations in TGFBR1 have also been detected in CRC [121]. For example, loss of three alanine residues within a stretch of nine alanine residues in the N-terminal region of TβRI (TGFBR1*6A) was associated with an increased risk of CRC [122], though these results have not been confirmed [123]. However, a recent study pointed out an oncogenic property of TGFBR1*6A which may promote the migration and invasion of colorectal cancer cells [124].
Mutations in downstream components of the TGF-β signaling pathway may also contribute to sporadic CRC development [107]. Smad4 is the most commonly disrupted Smad family protein in various cancers. It is mutated or lost in up to one third of all CRCs. Smad4 mutation or loss of expression has been frequently observed in late-stage tumors [125], [126], [127], [128]. Loss of Smad4 could alter BMP signaling to promote CRC metastasis through activation of Rho and Rho-associated protein kinase [129]. Mutations in Smad2 have been detected in approximately 3%-6% of CRC tumors [128], [130]. Mutations were more frequently observed in early-stage tumors. Both the Smad2 and Smad4 genes are located on chromosome 18q. This region is commonly deleted in CRC owing to a loss of the long arm of chromosome 18 (loss of heterozygosity) [131]. Smad7 is also located in this region [131].
Interestingly, a low-frequency coding variant, rs3764482 (c. 83C>T; p. S28F) in Smad7, was associated with the risk of CRC in a Chinese population [132]. A large-scale meta-analysis demonstrated that several single nucleotide polymorphisms in Smad7 were associated with CRC [133]. Mutations in Smad3 were also identified and had similar frequencies to those of the Smad2 mutations in sporadic CRCs [128]. In addition to the somatic mutations described above, germline mutations in Smads including Smad4 have been observed in several patients with juvenile polyposis syndrome [134], which can develop into CRC [135]. Thus, altered TGF-β signaling plays various stage-specific roles in CRC progression with mutations in TGFBR2 and Smad as the major contributor.
TGF-β Signal and Pancreatic Cancer
Pancreatic cancer is a leading cause of cancer-related death worldwide [136]. Risk factors for pancreatic cancer include tobacco use, obesity, and exposure to certain chemicals. In addition to genetic factors, age and gender are also correlated with risk of pancreatic cancer [136], [137]. Pancreatic ductal adenocarcinoma (PDAC) is the major histological subtype of pancreatic cancer (90% of cases). The median survival of PDAC patients is 6 months, and the 5-year survival rate is approximately 6% [138], [139].
Previous studies have demonstrated that increased TGF-β expression promotes disease progression in PDAC [140], [141]. Low circulating levels of TGF-β have been associated with prolonged survival in pancreatic cancer patients [142]. Increased TβR1 and TβR2 expression has been observed in the majority of pancreatic cancer subtypes [143]. Activation of TGF-β receptor signaling in PDAC cells resulted in increased Smad3 phosphorylation and nuclear translocation, leading to inhibition of cell growth. However, it also can result in activation of Smad7, which antagonizes Smad3, leading to activation of VEGF-A, vascularization, and metastasis [144].
TGF-β appears to have dual roles in PDAC progression [139]. Overexpression of TGF-β in early-stage PDACs was associated with reduced tumor cell proliferation and improved survival [145]. Mutations in TGF-β pathway proteins may explain the stage-dependent functions of TGF-β in PDACs. Mutations in Smad4 have been observed in approximately 50% of PDACs [146], [147]. Mutations in TβRII have been identified in 4%-7% of pancreatic cancers [146], [148].
KRAS is mutated in nearly all PDACs and is a major driver of carcinogenesis [149]. However, mutation of KRAS alone is not sufficient for malignant transformation [150], [151]. One recent study demonstrated that mutant KRAS dosage along with other oncogenic gains like Myc drives the early progression of PDAC [152]. Additional mutations in tumor suppressors such as Smad4 and CDKN2A are required for PDAC initiation [151]. Smad4 loss of function may promote KRAS-driven malignant transformation of pancreatic ductal cells [150]. In addition, inactivation of retinoblastoma 1 converts TGF-β from a tumor suppressor to a protumorigenic factor that enhances PDAC cell proliferation [153]. Rac1 may also induce a switch in TGF-β signaling by antagonizing Smad2 and Smad3 activation in PDAC cells [154]. TGF-β also regulates the interaction between tumor cells and the surrounding stroma, which can promote PDAC initiation and metastasis [2], [155].
TGF-β–Based Therapies for Gastrointestinal Cancers
Elevated TGF-β expression in serum and tumor tissue was correlated with tumor stage and prognosis in gastrointestinal cancers [6]. Among different gastrointestinal cancers, dysfunction of TβR(s) is derived from multiple levels including transcription, translation, and mutation. It should be noted that TGFBR(s) mutation is frequently observed in pancreatic cancers, while alternation of TβR(s) level often appears in other types of gastrointestinal cancers. Additionally, downstream components of the TGF-β signaling pathway are also dysregulated during gastrointestinal tumorigenesis. Thus, alternation from these three levels (ligand, receptor, and signal transducer) together contributes to the development of gastrointestinal cancers, and therapeutics that target the TGF-β signaling pathway may be effective for the treatment of gastrointestinal cancers. TGF-β has dual roles in gastrointestinal cancer initiation and progression. Because it functions as a tumor suppressor during early-stage carcinogenesis and as a tumor promoter during later stages, the development of TGF-β–based therapeutics is challenging. Therapeutics could target TGF-β, TβRI or TβRII, or the Smads [156]. Indeed, small molecule therapeutics, antibodies, and inhibitors against these targets are currently in clinical trials [7], [156], such as antisense oligonucleotides against TGF-β or TβR to lower their synthesis, TGF-β–neutralizing monoclonal antibodies and anti-TβR monoclonal antibodies to interrupt ligand-receptor interaction, and small molecule inhibitor of TβR to prevent signaling transduction. Galunisertib, a TβRI kinase inhibitor, displayed considerable tumor suppression effects in PDAC and HCC and has entered the clinical phase II trial. Trabedersen, a TGF-β2 antisense oligodeoxynucleotide, showed certain effects in clinical trials in PDAC and CRC patients [7]. Although few drugs targeting TGF-β display efficacy in clinical trials, the future of TGF-β pathway-based strategies against gastrointestinal cancers is promising from these encouraging clinical trials [6].
Conclusions
Existing research allows for the conclusion that dysregulation of TGF-β signaling pathway is tightly associated with the development and progression of various gastrointestinal cancers. However, there is still much to learn about the biology of TGF-β signaling under normal conditions and how its dysfunction contributes to the different types of gastrointestinal cancers. TGF-β has critical roles at different stages during gastrointestinal cancers’ initiation and metastasis. Definitely, context is important to define the biological meaning of dynamic regulation of TGF-β signaling in the progression of these cancers. A better understanding of the molecular mechanisms underlying the roles of TGF-β, TβRs, and their downstream signaling pathways as well as the complicated cross talk among them during gastrointestinal cancer progression is important in order to develop more effective therapeutics for these diseases. We are still at the early stage in understanding what role TGF-β signaling plays in the development of gastrointestinal cancers. This stage is set to explore the therapeutics that target TGF-β signaling in gastrointestinal cancers, possibly in concert with other effective agents to maximize the therapeutic benefits and improve the outcome of these lethal cancers.
Consent for Publication
We have obtained consents to publish this paper from all the participants of this study.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13(10):616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Massague J. TGFbeta in cancer. Cell. 2008;134(2):215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabregat I, Fernando J, Mainez J, Sancho P. TGF-beta signaling in cancer treatment. Curr Pharm Des. 2014;20(17):2934–2947. doi: 10.2174/13816128113199990591. [DOI] [PubMed] [Google Scholar]
- 4.David CJ, Massague J. Contextual determinants of TGFbeta action in development, immunity and cancer. Nat Rev Mol Cell Biol. 2018;19(7):419–435. doi: 10.1038/s41580-018-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toomey PG, Vohra NA, Ghansah T, Sarnaik AA, Pilon-Thomas SA. Immunotherapy for gastrointestinal malignancies. Cancer Control. 2013;20(1):32–42. doi: 10.1177/107327481302000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katz LH, Likhter M, Jogunoori W, Belkin M, Ohshiro K, Mishra L. TGF-beta signaling in liver and gastrointestinal cancers. Cancer Lett. 2016;379(2):166–172. doi: 10.1016/j.canlet.2016.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neuzillet C, Tijeras-Raballand A, Cohen R, Cros J, Faivre S, Raymond E, de Gramont A. Targeting the TGFbeta pathway for cancer therapy. Pharmacol Ther. 2015;147:22–31. doi: 10.1016/j.pharmthera.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Colak S, Ten Dijke P. Targeting TGF-beta signaling in cancer. Trends Cancer. 2017;3(1):56–71. doi: 10.1016/j.trecan.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Frolik CA, Dart LL, Meyers CA, Smith DM, Sporn MB. Purification and initial characterization of a type beta transforming growth factor from human placenta. Proc Natl Acad Sci U S A. 1983;80(12):3676–3680. doi: 10.1073/pnas.80.12.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Massague J, Chen YG. Controlling TGF-beta signaling. Genes Dev. 2000;14(6):627–644. [PubMed] [Google Scholar]
- 11.Hinck AP. Structural studies of the TGF-betas and their receptors — insights into evolution of the TGF-beta superfamily. FEBS Lett. 2012;586(14):1860–1870. doi: 10.1016/j.febslet.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 12.Santibanez JF, Quintanilla M, Bernabeu C. TGF-beta/TGF-beta receptor system and its role in physiological and pathological conditions. Clin Sci. 2011;121(6):233–251. doi: 10.1042/CS20110086. [DOI] [PubMed] [Google Scholar]
- 13.Kajdaniuk D, Marek B, Borgiel-Marek H, Kos-Kudla B. Transforming growth factor beta1 (TGFbeta1) in physiology and pathology. Endokrynol Pol. 2013;64(5):384–396. doi: 10.5603/EP.2013.0022. [DOI] [PubMed] [Google Scholar]
- 14.Funkenstein B, Olekh E, Jakowlew SB. Identification of a novel transforming growth factor-beta (TGF-beta6) gene in fish: regulation in skeletal muscle by nutritional state. BMC Mol Biol. 2010;11:37. doi: 10.1186/1471-2199-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ten Dijke P, Arthur HM. Extracellular control of TGFbeta signalling in vascular development and disease. Nat Rev Mol Cell Biol. 2007;8(11):857–869. doi: 10.1038/nrm2262. [DOI] [PubMed] [Google Scholar]
- 16.de Caestecker M. The transforming growth factor-beta superfamily of receptors. Cytokine Growth Factor Rev. 2004;15(1):1–11. doi: 10.1016/j.cytogfr.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 17.Xu P, Liu J, Derynck R. Post-translational regulation of TGF-beta receptor and Smad signaling. FEBS Lett. 2012;586(14):1871–1884. doi: 10.1016/j.febslet.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 19.Schmierer B, Hill CS. TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol. 2007;8(12):970–982. doi: 10.1038/nrm2297. [DOI] [PubMed] [Google Scholar]
- 20.Xi Q, Wang Z, Zaromytidou AI, Zhang XH, Chow-Tsang LF, Liu JX, Kim H, Barlas A, Manova-Todorova K, Kaartinen V. A poised chromatin platform for TGF-beta access to master regulators. Cell. 2011;147(7):1511–1524. doi: 10.1016/j.cell.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyazawa K, Miyazono K. Regulation of TGF-beta Family Signaling by Inhibitory Smads. Cold Spring Harb Perspect Biol. 2017;9(3) doi: 10.1101/cshperspect.a022095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wrighton KH, Lin X, Feng XH. Phospho-control of TGF-beta superfamily signaling. Cell Res. 2009;19(1):8–20. doi: 10.1038/cr.2008.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276(16):12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19(1):128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang YE. Non-Smad signaling pathways of the TGF-beta family. Cold Spring Harb Perspect Biol. 2017;9(2) doi: 10.1101/cshperspect.a022129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo X, Wang XF. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009;19(1):71–88. doi: 10.1038/cr.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Napier KJ, Scheerer M, Misra S. Esophageal cancer: a review of epidemiology, pathogenesis, staging workup and treatment modalities. World J Gastrointest Oncol. 2014;6(5):112–120. doi: 10.4251/wjgo.v6.i5.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19(34):5598–5606. doi: 10.3748/wjg.v19.i34.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin EW, Karakasheva TA, Hicks PD, Bass AJ, Rustgi AK. The tumor microenvironment in esophageal cancer. Oncogene. 2016;35(41):5337–5349. doi: 10.1038/onc.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun SP, Jin YN, Yang HP, Wei Y, Dong Z. Serum transforming growth factor-beta1 level reflects disease status in patients with esophageal carcinoma after radiotherapy. World J Gastroenterol. 2007;13(39):5267–5272. doi: 10.3748/wjg.v13.i39.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gholamin M, Moaven O, Memar B, Farshchian M, Naseh H, Malekzadeh R, Sotoudeh M, Rajabi-Mashhadi MT, Forghani MN, Farrokhi F. Overexpression and interactions of interleukin-10, transforming growth factor beta, and vascular endothelial growth factor in esophageal squamous cell carcinoma. World J Surg. 2009;33(7):1439–1445. doi: 10.1007/s00268-009-0070-y. [DOI] [PubMed] [Google Scholar]
- 32.Fukai Y, Fukuchi M, Masuda N, Osawa H, Kato H, Nakajima T, Kuwano H. Reduced expression of transforming growth factor-beta receptors is an unfavorable prognostic factor in human esophageal squamous cell carcinoma. Int J Cancer. 2003;104(2):161–166. doi: 10.1002/ijc.10929. [DOI] [PubMed] [Google Scholar]
- 33.Fukuchi M, Masuda N, Miyazaki T, Nakajima M, Osawa H, Kato H, Kuwano H. Decreased Smad4 expression in the transforming growth factor-beta signaling pathway during progression of esophageal squamous cell carcinoma. Cancer. 2002;95(4):737–743. doi: 10.1002/cncr.10727. [DOI] [PubMed] [Google Scholar]
- 34.Fukuchi M, Fukai Y, Masuda N, Miyazaki T, Nakajima M, Sohda M, Manda R, Tsukada K, Kato H, Kuwano H. High-level expression of the Smad ubiquitin ligase Smurf2 correlates with poor prognosis in patients with esophageal squamous cell carcinoma. Cancer Res. 2002;62(24):7162–7165. [PubMed] [Google Scholar]
- 35.Pang L, Li Q, Wei C, Zou H, Li S, Cao W, He J, Zhou Y, Ju X, Lan J. TGF-beta1/Smad signaling pathway regulates epithelial-to-mesenchymal transition in esophageal squamous cell carcinoma: in vitro and clinical analyses of cell lines and nomadic Kazakh patients from northwest Xinjiang, China. PLoS One. 2014;9(12) doi: 10.1371/journal.pone.0112300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang HY, Wang ZQ, Li YY, Wang F, Zeng QR, Gao Y, Xuan XY, Li SS. Transforming growth factor-beta1-induced epithelial-mesenchymal transition in human esophageal squamous cell carcinoma via the PTEN/PI3K signaling pathway. Oncol Rep. 2014;32(5):2134–2142. doi: 10.3892/or.2014.3453. [DOI] [PubMed] [Google Scholar]
- 37.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Esquela-Kerscher A, Slack FJ. Oncomirs — microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 39.Jing C, Ma G, Li X, Wu X, Huang F, Liu K, Liu Z. MicroRNA-17/20a impedes migration and invasion via TGF-beta/ITGB6 pathway in esophageal squamous cell carcinoma. Am J Cancer Res. 2016;6(7):1549–1562. [PMC free article] [PubMed] [Google Scholar]
- 40.Liu A, Zhu J, Wu G, Cao L, Tan Z, Zhang S, Jiang L, Wu J, Li M, Song L. Antagonizing miR-455-3p inhibits chemoresistance and aggressiveness in esophageal squamous cell carcinoma. Mol Cancer. 2017;16(1):106. doi: 10.1186/s12943-017-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harazono Y, Muramatsu T, Endo H, Uzawa N, Kawano T, Harada K, Inazawa J, Kozaki K. miR-655 Is an EMT-suppressive microRNA targeting ZEB1 and TGFBR2. PLoS One. 2013;8(5) doi: 10.1371/journal.pone.0062757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu YT, Zong D, Jiang XS, Yin L, Wang LJ, Wang TT, Zhu J, He X. miR-32 promotes esophageal squamous cell carcinoma metastasis by targeting CXXC5. J Cell Biochem. 2018:1–14. doi: 10.1002/jcb.27912. [DOI] [PubMed] [Google Scholar]
- 43.Yuan Y, Chen H, Ma G, Cao X, Liu Z. Reelin is involved in transforming growth factor-beta1-induced cell migration in esophageal carcinoma cells. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0031802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie JJ, Xu LY, Wu JY, Shen ZY, Zhao Q, Du ZP, Lv Z, Gu W, Pan F, Xu XE. Involvement of CYR61 and CTGF in the fascin-mediated proliferation and invasiveness of esophageal squamous cell carcinomas cells. Am J Pathol. 2010;176(2):939–951. doi: 10.2353/ajpath.2010.090118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou ZQ, Cao WH, Xie JJ, Lin J, Shen ZY, Zhang QY, Shen JH, Xu LY, Li EM. Expression and prognostic significance of THBS1, Cyr61 and CTGF in esophageal squamous cell carcinoma. BMC Cancer. 2009;9:291. doi: 10.1186/1471-2407-9-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie JJ, Xu LY, Xie YM, Zhang HH, Cai WJ, Zhou F, Shen ZY, Li EM. Roles of ezrin in the growth and invasiveness of esophageal squamous carcinoma cells. Int J Cancer. 2009;124(11):2549–2558. doi: 10.1002/ijc.24216. [DOI] [PubMed] [Google Scholar]
- 47.Rahman R, Asombang AW, Ibdah JA. Characteristics of gastric cancer in Asia. World J Gastroenterol. 2014;20(16):4483–4490. doi: 10.3748/wjg.v20.i16.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23(5):700–713. doi: 10.1158/1055-9965.EPI-13-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carcas LP. Gastric cancer review. J Carcinog. 2014;13:14. doi: 10.4103/1477-3163.146506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu WQ, Wang LW, Yuan JP, Yan SG, Li JD, Zhao HL, Peng CW, Yang GF, Li Y. High expression of transform growth factor beta 1 in gastric cancer confers worse outcome: results of a cohort study on 184 patients. Hepato-Gastroenterology. 2014;61(129):245–250. [PubMed] [Google Scholar]
- 51.Ma GF, Miao Q, Zeng XQ, Luo TC, Ma LL, Liu YM, Lian JJ, Gao H, Chen SY. Transforming growth factor-beta1 and -beta2 in gastric precancer and cancer and roles in tumor-cell interactions with peripheral blood mononuclear cells in vitro. PLoS One. 2013;8(1) doi: 10.1371/journal.pone.0054249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohue M, Tomita N, Monden T, Miyoshi Y, Ohnishi T, Izawa H, Kawabata Y, Sasaki M, Sekimoto M, Nishisho I. Mutations of the transforming growth factor beta type II receptor gene and microsatellite instability in gastric cancer. Int J Cancer. 1996;68(2):203–206. doi: 10.1002/(SICI)1097-0215(19961009)68:2<203::AID-IJC11>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 53.Kang SH, Bang YJ, Im YH, Yang HK, Lee DA, Lee HY, Lee HS, Kim NK, Kim SJ. Transcriptional repression of the transforming growth factor-beta type I receptor gene by DNA methylation results in the development of TGF-beta resistance in human gastric cancer. Oncogene. 1999;18(51):7280–7286. doi: 10.1038/sj.onc.1203146. [DOI] [PubMed] [Google Scholar]
- 54.Jin G, Wang L, Chen W, Hu Z, Zhou Y, Tan Y, Wang J, Hua Z, Ding W, Shen J. Variant alleles of TGFB1 and TGFBR2 are associated with a decreased risk of gastric cancer in a Chinese population. Int J Cancer. 2007;120(6):1330–1335. doi: 10.1002/ijc.22443. [DOI] [PubMed] [Google Scholar]
- 55.Leng A, Liu T, He Y, Li Q, Zhang G. Smad4/Smad7 balance: a role of tumorigenesis in gastric cancer. Exp Mol Pathol. 2009;87(1):48–53. doi: 10.1016/j.yexmp.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 56.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 57.Ye X, Weinberg RA. Epithelial-mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol. 2015;25(11):675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang L, Wu RL, Xu AM. Epithelial-mesenchymal transition in gastric cancer. Am J Transl Res. 2015;7(11):2141–2158. [PMC free article] [PubMed] [Google Scholar]
- 59.Ma HY, Liu XZ, Liang CM. Inflammatory microenvironment contributes to epithelial-mesenchymal transition in gastric cancer. World J Gastroenterol. 2016;22(29):6619–6628. doi: 10.3748/wjg.v22.i29.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gen Y, Yasui K, Kitaichi T, Iwai N, Terasaki K, Dohi O, Hashimoto H, Fukui H, Inada Y, Fukui A. ASPP2 suppresses invasion and TGF-beta1-induced epithelial-mesenchymal transition by inhibiting Smad7 degradation mediated by E3 ubiquitin ligase ITCH in gastric cancer. Cancer Lett. 2017;398:52–61. doi: 10.1016/j.canlet.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 61.Xiang J, Fu X, Ran W, Wang Z. Grhl2 reduces invasion and migration through inhibition of TGFbeta-induced EMT in gastric cancer. Oncogenesis. 2017;6(1) doi: 10.1038/oncsis.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang H, Zhao Y, Cao L, Zhang J, Wang Y, Xu M. Metastasis suppressor protein 1 regulated by PTEN suppresses invasion, migration, and EMT of gastric carcinoma by inactivating PI3K/AKT signaling. J Cell Biochem. 2018:1–8. doi: 10.1002/jcb.27618. [DOI] [PubMed] [Google Scholar]
- 63.He YX, Song XH, Zhao ZY, Zhao H. HOXA13 upregulation in gastric cancer is associated with enhanced cancer cell invasion and epithelial-to-mesenchymal transition. Eur Rev Med Pharmacol Sci. 2017;21(2):258–265. [PubMed] [Google Scholar]
- 64.Ma H, Gao L, Li S, Qin J, Chen L, Liu X, Xu P, Wang F, Xiao H, Zhou S. CCR7 enhances TGF-beta1-induced epithelial-mesenchymal transition and is associated with lymph node metastasis and poor overall survival in gastric cancer. Oncotarget. 2015;6(27):24348–24360. doi: 10.18632/oncotarget.4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ouyang S, Zhu G, Ouyang L, Luo Y, Zhou R, Pan C, Bin J, Liao Y, Liao W. Bapx1 mediates transforming growth factor-beta- induced epithelial-mesenchymal transition and promotes a malignancy phenotype of gastric cancer cells. Biochem Biophys Res Commun. 2017;486(2):285–292. doi: 10.1016/j.bbrc.2017.03.029. [DOI] [PubMed] [Google Scholar]
- 66.Cao Q, Liu F, Ji K, Liu N, He Y, Zhang W, Wang L. MicroRNA-381 inhibits the metastasis of gastric cancer by targeting TMEM16A expression. J Exp Clin Cancer Res. 2017;36(1):29. doi: 10.1186/s13046-017-0499-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li C, Song L, Zhang Z, Bai XX, Cui MF, Ma LJ. MicroRNA-21 promotes TGF-beta1-induced epithelial-mesenchymal transition in gastric cancer through up-regulating PTEN expression. Oncotarget. 2016;7(41):66989–67003. doi: 10.18632/oncotarget.11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qu Y, Zhang H, Duan J, Liu R, Deng T, Bai M, Huang D, Li H, Ning T, Zhang L. MiR-17-5p regulates cell proliferation and migration by targeting transforming growth factor-beta receptor 2 in gastric cancer. Oncotarget. 2016;7(22):33286–33296. doi: 10.18632/oncotarget.8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei S, Li Q, Li Z, Wang L, Zhang L, Xu Z. miR-424-5p promotes proliferation of gastric cancer by targeting Smad3 through TGF-beta signaling pathway. Oncotarget. 2016;7(46):75185–75196. doi: 10.18632/oncotarget.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qu Y, Zhang H, Sun W, Han Y, Li S, Qu Y, Ying G, Ba Y. MicroRNA-155 promotes gastric cancer growth and invasion by negatively regulating transforming growth factor-beta receptor 2. Cancer Sci. 2018;109(3):618–628. doi: 10.1111/cas.13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Fan KJ, Sun Q, Chen AZ, Shen WL, Zhao ZH, Zheng XF, Yang X. Functional screening for miRNAs targeting Smad4 identified miR-199a as a negative regulator of TGF-beta signalling pathway. Nucleic Acids Res. 2012;40(18):9286–9297. doi: 10.1093/nar/gks667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McGlynn KA, London WT. The global epidemiology of hepatocellular carcinoma: present and future. Clin Liver Dis. 2011;15(2):223–243. doi: 10.1016/j.cld.2011.03.006. [vii-x] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wild CP, Hall AJ. Primary prevention of hepatocellular carcinoma in developing countries. Mutat Res. 2000;462(2-3):381–393. doi: 10.1016/s1383-5742(00)00027-2. [DOI] [PubMed] [Google Scholar]
- 74.Davis GL, Dempster J, Meler JD, Orr DW, Walberg MW, Brown B, Berger BD, O'Connor JK, Goldstein RM. Hepatocellular carcinoma: management of an increasingly common problem. Proceedings. 2008;21(3):266–280. doi: 10.1080/08998280.2008.11928410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tinkle CL, Haas-Kogan D. Hepatocellular carcinoma: natural history, current management, and emerging tools. Biologics. 2012;6:207–219. doi: 10.2147/BTT.S23907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin S, Hoffmann K, Schemmer P. Treatment of hepatocellular carcinoma: a systematic review. Liver Cancer. 2012;1(3-4):144–158. doi: 10.1159/000343828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang LY, Chang RM, Lau WY, Ou DP, Wu W, Zeng ZJ. Mesohepatectomy for centrally located large hepatocellular carcinoma: Indications, techniques, and outcomes. Surgery. 2014;156(5):1177–1187. doi: 10.1016/j.surg.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 78.Giannelli G, Fransvea E, Marinosci F, Bergamini C, Colucci S, Schiraldi O, Antonaci S. Transforming growth factor-beta1 triggers hepatocellular carcinoma invasiveness via alpha3beta1 integrin. Am J Pathol. 2002;161(1):183–193. doi: 10.1016/s0002-9440(10)64170-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.An Y, Gao S, Zhao WC, Qiu BA, Xia NX, Zhang PJ, Fan ZP. Transforming growth factor-beta and peripheral regulatory cells are negatively correlated with the overall survival of hepatocellular carcinoma. World J Gastroenterol. 2018;24(25):2733–2740. doi: 10.3748/wjg.v24.i25.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abou-Shady M, Baer HU, Friess H, Berberat P, Zimmermann A, Graber H, Gold LI, Korc M, Buchler MW. Transforming growth factor betas and their signaling receptors in human hepatocellular carcinoma. Am J Surg. 1999;177(3):209–215. doi: 10.1016/s0002-9610(99)00012-4. [DOI] [PubMed] [Google Scholar]
- 81.Lin TH, Shao YY, Chan SY, Huang CY, Hsu CH, Cheng AL. High serum transforming growth factor-beta1 levels predict outcome in hepatocellular carcinoma patients treated with sorafenib. Clin Cancer Res. 2015;21(16):3678–3684. doi: 10.1158/1078-0432.CCR-14-1954. [DOI] [PubMed] [Google Scholar]
- 82.Song BC, Chung YH, Kim JA, Choi WB, Suh DD, Pyo SI, Shin JW, Lee HC, Lee YS, Suh DJ. Transforming growth factor-beta1 as a useful serologic marker of small hepatocellular carcinoma. Cancer. 2002;94(1):175–180. doi: 10.1002/cncr.10170. [DOI] [PubMed] [Google Scholar]
- 83.Mamiya T, Yamazaki K, Masugi Y, Mori T, Effendi K, Du W, Hibi T, Tanabe M, Ueda M, Takayama T. Reduced transforming growth factor-beta receptor II expression in hepatocellular carcinoma correlates with intrahepatic metastasis. Lab Invest. 2010;90(9):1339–1345. doi: 10.1038/labinvest.2010.105. [DOI] [PubMed] [Google Scholar]
- 84.Yakicier MC, Irmak MB, Romano A, Kew M, Ozturk M. Smad2 and Smad4 gene mutations in hepatocellular carcinoma. Oncogene. 1999;18(34):4879–4883. doi: 10.1038/sj.onc.1202866. [DOI] [PubMed] [Google Scholar]
- 85.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rani B, Cao Y, Malfettone A, Tomuleasa C, Fabregat I, Giannelli G. Role of the tissue microenvironment as a therapeutic target in hepatocellular carcinoma. World J Gastroenterol. 2014;20(15):4128–4140. doi: 10.3748/wjg.v20.i15.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ji F, Zhang ZH, Zhang Y, Shen SL, Cao QH, Zhang LJ, Li SQ, Peng BG, Liang LJ, Hua YP. Low expression of c-Myc protein predicts poor outcomes in patients with hepatocellular carcinoma after resection. BMC Cancer. 2018;18(1):460. doi: 10.1186/s12885-018-4379-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meindl-Beinker NM, Matsuzaki K, Dooley S. TGF-beta signaling in onset and progression of hepatocellular carcinoma. Dig Dis. 2012;30(5):514–523. doi: 10.1159/000341704. [DOI] [PubMed] [Google Scholar]
- 89.Dooley S, ten Dijke P. TGF-beta in progression of liver disease. Cell Tissue Res. 2012;347(1):245–256. doi: 10.1007/s00441-011-1246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Im YH, Kim HT, Kim IY, Factor VM, Hahm KB, Anzano M, Jang JJ, Flanders K, Haines DC, Thorgeirsson SS. Heterozygous mice for the transforming growth factor-beta type II receptor gene have increased susceptibility to hepatocellular carcinogenesis. Cancer Res. 2001;61(18):6665–6668. [PubMed] [Google Scholar]
- 91.Yamazaki K, Masugi Y, Sakamoto M. Molecular pathogenesis of hepatocellular carcinoma: altering transforming growth factor-beta signaling in hepatocarcinogenesis. Dig Dis. 2011;29(3):284–288. doi: 10.1159/000327560. [DOI] [PubMed] [Google Scholar]
- 92.Zhang X, Fan Q, Li Y, Yang Z, Yang L, Zong Z, Wang B, Meng X, Li Q, Liu J. Transforming growth factor-beta1 suppresses hepatocellular carcinoma proliferation via activation of Hippo signaling. Oncotarget. 2017;8(18):29785–29794. doi: 10.18632/oncotarget.14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Carmona-Cuenca I, Roncero C, Sancho P, Caja L, Fausto N, Fernandez M, Fabregat I. Upregulation of the NADPH oxidase NOX4 by TGF-beta in hepatocytes is required for its pro-apoptotic activity. J Hepatol. 2008;49(6):965–976. doi: 10.1016/j.jhep.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 94.Huang J, Qiu M, Wan L, Wang G, Huang T, Chen Z, Jiang S, Li X, Xie L, Cai L. TGF-beta1 promotes hepatocellular carcinoma invasion and metastasis via ERK pathway-mediated FGFR4 expression. Cell Physiol Biochem. 2018;45(4):1690–1699. doi: 10.1159/000487737. [DOI] [PubMed] [Google Scholar]
- 95.Wang M, Zhang L, Liu Z, Zhou J, Pan Q, Fan J, Zang R, Wang L. AGO1 may influence the prognosis of hepatocellular carcinoma through TGF-beta pathway. Cell Death Dis. 2018;9(3):324. doi: 10.1038/s41419-018-0338-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yeh TS, Wang F, Chen TC, Yeh CN, Yu MC, Jan YY, Chen MF. Expression profile of microRNA-200 family in hepatocellular carcinoma with bile duct tumor thrombus. Ann Surg. 2014;259(2):346–354. doi: 10.1097/SLA.0000000000000223. [DOI] [PubMed] [Google Scholar]
- 97.Zhou JN, Zeng Q, Wang HY, Zhang B, Li ST, Nan X, Cao N, Fu CJ, Yan XL, Jia YL. MicroRNA-125b attenuates epithelial-mesenchymal transitions and targets stem-like liver cancer cells through small mothers against decapentaplegic 2 and 4. Hepatology. 2015;62(3):801–815. doi: 10.1002/hep.27887. [DOI] [PubMed] [Google Scholar]
- 98.Braun L, Gruppuso P, Mikumo R, Fausto N. Transforming growth factor beta 1 in liver carcinogenesis: messenger RNA expression and growth effects. Cell Growth Differ. 1990;1(3):103–111. [PubMed] [Google Scholar]
- 99.Fabregat I, Moreno-Caceres J, Sanchez A, Dooley S, Dewidar B, Giannelli G, Ten Dijke P, Consortium I-L. TGF-beta signalling and liver disease. FEBS J. 2016;283(12):2219–2232. doi: 10.1111/febs.13665. [DOI] [PubMed] [Google Scholar]
- 100.Fischer AN, Fuchs E, Mikula M, Huber H, Beug H, Mikulits W. PDGF essentially links TGF-beta signaling to nuclear beta-catenin accumulation in hepatocellular carcinoma progression. Oncogene. 2007;26(23):3395–3405. doi: 10.1038/sj.onc.1210121. [DOI] [PubMed] [Google Scholar]
- 101.Fabregat I, Herrera B, Fernandez M, Alvarez AM, Sanchez A, Roncero C, Ventura JJ, Valverde AM, Benito M. Epidermal growth factor impairs the cytochrome C/caspase-3 apoptotic pathway induced by transforming growth factor beta in rat fetal hepatocytes via a phosphoinositide 3-kinase–dependent pathway. Hepatology. 2000;32(3):528–535. doi: 10.1053/jhep.2000.9774. [DOI] [PubMed] [Google Scholar]
- 102.Sohn BH, Park IY, Lee JJ, Yang SJ, Jang YJ, Park KC, Kim DJ, Lee DC, Sohn HA, Kim TW. Functional switching of TGF-beta1 signaling in liver cancer via epigenetic modulation of a single CpG site in TTP promoter. Gastroenterology. 2010;138(5):1898–1908. doi: 10.1053/j.gastro.2009.12.044. [DOI] [PubMed] [Google Scholar]
- 103.Coulouarn C, Factor VM, Thorgeirsson SS. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology. 2008;47(6):2059–2067. doi: 10.1002/hep.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22(4):191–197. doi: 10.1055/s-0029-1242458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.De Rosa M, Pace U, Rega D, Costabile V, Duraturo F, Izzo P, Delrio P. Genetics, diagnosis and management of colorectal cancer (Review) Oncol Rep. 2015;34(3):1087–1096. doi: 10.3892/or.2015.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Burt R. Inheritance of colorectal cancer. Drug Discov Today Dis Mech. 2007;4(4):293–300. doi: 10.1016/j.ddmec.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu Y, Pasche B. TGF-beta signaling alterations and susceptibility to colorectal cancer. Hum Mol Genet. 2007;16(Spec 1):R14–R20. doi: 10.1093/hmg/ddl486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Buckhaults P, Rago C, Croix B St, Romans KE, Saha S, Zhang L, Vogelstein B, Kinzler KW. Secreted and cell surface genes expressed in benign and malignant colorectal tumors. Cancer Res. 2001;61(19):6996–7001. [PubMed] [Google Scholar]
- 109.Friedman E, Gold LI, Klimstra D, Zeng ZS, Winawer S, Cohen A. High levels of transforming growth factor beta 1 correlate with disease progression in human colon cancer. Cancer Epidemiol Biomarkers Prev. 1995;4(5):549–554. [PubMed] [Google Scholar]
- 110.Tsushima H, Kawata S, Tamura S, Ito N, Shirai Y, Kiso S, Imai Y, Shimomukai H, Nomura Y, Matsuda Y. High levels of transforming growth factor beta 1 in patients with colorectal cancer: association with disease progression. Gastroenterology. 1996;110(2):375–382. doi: 10.1053/gast.1996.v110.pm8566583. [DOI] [PubMed] [Google Scholar]
- 111.Turtoi A, Blomme A, Debois D, Somja J, Delvaux D, Patsos G, Di Valentin E, Peulen O, Mutijima EN, De Pauw E. Organized proteomic heterogeneity in colorectal cancer liver metastases and implications for therapies. Hepatology. 2014;59(3):924–934. doi: 10.1002/hep.26608. [DOI] [PubMed] [Google Scholar]
- 112.Schroy P, Rifkin J, Coffey RJ, Winawer S, Friedman E. Role of transforming growth factor beta 1 in induction of colon carcinoma differentiation by hexamethylene bisacetamide. Cancer Res. 1990;50(2):261–265. [PubMed] [Google Scholar]
- 113.Yamagata H, Matsuzaki K, Mori S, Yoshida K, Tahashi Y, Furukawa F, Sekimoto G, Watanabe T, Uemura Y, Sakaida N. Acceleration of Smad2 and Smad3 phosphorylation via c-Jun NH(2)-terminal kinase during human colorectal carcinogenesis. Cancer Res. 2005;65(1):157–165. [PubMed] [Google Scholar]
- 114.Armaghany T, Wilson JD, Chu Q, Mills G. Genetic alterations in colorectal cancer. Gastrointest Cancer Res. 2012;5(1):19–27. [PMC free article] [PubMed] [Google Scholar]
- 115.Tauriello DVF, Palomo-Ponce S, Stork D, Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, Sevillano M, Ibiza S, Canellas A, Hernando-Momblona X. TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554(7693):538–543. doi: 10.1038/nature25492. [DOI] [PubMed] [Google Scholar]
- 116.Jung B, Staudacher JJ, Beauchamp D. Transforming growth factor beta superfamily signaling in development of colorectal cancer. Gastroenterology. 2017;152(1):36–52. [Google Scholar]
- 117.Grady WM, Markowitz SD. Genetic and epigenetic alterations in colon cancer. Annu Rev Genomics Hum Genet. 2002;3:101–128. doi: 10.1146/annurev.genom.3.022502.103043. [DOI] [PubMed] [Google Scholar]
- 118.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science (New York, NY) 1995;268(5215):1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 119.Geng L, Chaudhuri A, Talmon G, Wisecarver JL, Wang J. TGF-Beta suppresses VEGFA-mediated angiogenesis in colon cancer metastasis. PLoS One. 2013;8(3) doi: 10.1371/journal.pone.0059918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.de Miranda NF, van Dinther M, van den Akker BE, van Wezel T, ten Dijke P, Morreau H. Transforming growth factor beta signaling in colorectal cancer cells with microsatellite instability despite biallelic mutations in TGFBR2. Gastroenterology. 2015;148(7):1427–1437.e8. doi: 10.1053/j.gastro.2015.02.052. [DOI] [PubMed] [Google Scholar]
- 121.Ku JL, Park SH, Yoon KA, Shin YK, Kim KH, Choi JS, Kang HC, Kim IJ, Han IO, Park JG. Genetic alterations of the TGF-beta signaling pathway in colorectal cancer cell lines: a novel mutation in Smad3 associated with the inactivation of TGF-beta-induced transcriptional activation. Cancer Lett. 2007;247(2):283–292. doi: 10.1016/j.canlet.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 122.Daley D, Morgan W, Lewis S, Willis J, Elston RC, Markowitz SD, Wiesner GL. Is TGFBR1*6A a susceptibility allele for nonsyndromic familial colorectal neoplasia? Cancer Epidemiol Biomarkers Prev. 2007;16(5):892–894. doi: 10.1158/1055-9965.EPI-06-0965. [DOI] [PubMed] [Google Scholar]
- 123.Carvajal-Carmona LG, Churchman M, Bonilla C, Walther A, Lefevre JH, Kerr D, Dunlop M, Houlston R, Bodmer WF, Tomlinson I. Comprehensive assessment of variation at the transforming growth factor beta type 1 receptor locus and colorectal cancer predisposition. Proc Natl Acad Sci U S A. 2010;107(17):7858–7862. doi: 10.1073/pnas.1002816107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou R, Huang Y, Cheng B, Wang Y, Xiong B. TGFBR1*6A is a potential modifier of migration and invasion in colorectal cancer cells. Oncol Lett. 2018;15(3):3971–3976. doi: 10.3892/ol.2018.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.De Bosscher K, Hill CS, Nicolas FJ. Molecular and functional consequences of Smad4 C-terminal missense mutations in colorectal tumour cells. Biochem J. 2004;379(Pt 1):209–216. doi: 10.1042/BJ20031886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Maitra A, Molberg K, Albores-Saavedra J, Lindberg G. Loss of Dpc4 expression in colonic adenocarcinomas correlates with the presence of metastatic disease. Am J Pathol. 2000;157(4):1105–1111. doi: 10.1016/S0002-9440(10)64625-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Takagi Y, Kohmura H, Futamura M, Kida H, Tanemura H, Shimokawa K, Saji S. Somatic alterations of the DPC4 gene in human colorectal cancers in vivo. Gastroenterology. 1996;111(5):1369–1372. doi: 10.1053/gast.1996.v111.pm8898652. [DOI] [PubMed] [Google Scholar]
- 128.Fleming NI, Jorissen RN, Mouradov D, Christie M, Sakthianandeswaren A, Palmieri M, Day F, Li S, Tsui C, Lipton L. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res. 2013;73(2):725–735. doi: 10.1158/0008-5472.CAN-12-2706. [DOI] [PubMed] [Google Scholar]
- 129.Voorneveld PW, Kodach LL, Jacobs RJ, Liv N, Zonnevylle AC, Hoogenboom JP, Biemond I, Verspaget HW, Hommes DW, de Rooij K. Loss of SMAD4 alters BMP signaling to promote colorectal cancer cell metastasis via activation of Rho and ROCK. Gastroenterology. 2014;147(1):196–208.e13. doi: 10.1053/j.gastro.2014.03.052. [DOI] [PubMed] [Google Scholar]
- 130.Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui LC, Bapat B, Gallinger S, Andrulis IL. MADR2 maps to 18q21 and encodes a TGFbeta-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996;86(4):543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 131.Grady WM, Pritchard CC. Molecular alterations and biomarkers in colorectal cancer. Toxicol Pathol. 2014;42(1):124–139. doi: 10.1177/0192623313505155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li J, Zou L, Zhou Y, Li L, Zhu Y, Yang Y, Gong Y, Lou J, Ke J, Zhang Y. A low-frequency variant in SMAD7 modulates TGF-beta signaling and confers risk for colorectal cancer in Chinese population. Mol Carcinog. 2017;56(7):1798–1807. doi: 10.1002/mc.22637. [DOI] [PubMed] [Google Scholar]
- 133.Huang Y, Wu W, Nie M, Li C, Wang L. SMAD7 polymorphisms and colorectal cancer risk: a meta-analysis of case-control studies. Oncotarget. 2016;7(46):75561–75570. doi: 10.18632/oncotarget.12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Houlston R, Bevan S, Williams A, Young J, Dunlop M, Rozen P, Eng C, Markie D, Woodford-Richens K, Rodriguez-Bigas MA. Mutations in DPC4 (SMAD4) cause juvenile polyposis syndrome, but only account for a minority of cases. Hum Mol Genet. 1998;7(12):1907–1912. doi: 10.1093/hmg/7.12.1907. [DOI] [PubMed] [Google Scholar]
- 135.Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138(6):2044–2058. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ilic M, Ilic I. Epidemiology of pancreatic cancer. World J Gastroenterol. 2016;22(44):9694–9705. doi: 10.3748/wjg.v22.i44.9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362(17):1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 138.Ying H, Dey P, Yao W, Kimmelman AC, Draetta GF, Maitra A, DePinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016;30(4):355–385. doi: 10.1101/gad.275776.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shen W, Tao GQ, Zhang Y, Cai B, Sun J, Tian ZQ. TGF-beta in pancreatic cancer initiation and progression: two sides of the same coin. Cell Biosci. 2017;7:39. doi: 10.1186/s13578-017-0168-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Javle M, Li Y, Tan D, Dong X, Chang P, Kar S, Li D. Biomarkers of TGF-beta signaling pathway and prognosis of pancreatic cancer. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0085942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Friess H, Yamanaka Y, Buchler M, Ebert M, Beger HG, Gold LI, Korc M. Enhanced expression of transforming growth factor beta isoforms in pancreatic cancer correlates with decreased survival. Gastroenterology. 1993;105(6):1846–1856. doi: 10.1016/0016-5085(93)91084-u. [DOI] [PubMed] [Google Scholar]
- 142.Zhao J, Liang Y, Yin Q, Liu S, Wang Q, Tang Y, Cao C. Clinical and prognostic significance of serum transforming growth factor-beta1 levels in patients with pancreatic ductal adenocarcinoma. Braz J Med Biol Res. 2016;49(8) doi: 10.1590/1414-431X20165485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lu Z, Friess H, Graber HU, Guo X, Schilling M, Zimmermann A, Korc M, Buchler MW. Presence of two signaling TGF-beta receptors in human pancreatic cancer correlates with advanced tumor stage. Dig Dis Sci. 1997;42(10):2054–2063. doi: 10.1023/a:1018814416903. [DOI] [PubMed] [Google Scholar]
- 144.Zhang H, Liu C, Kong Y, Huang H, Wang C, Zhang H. TGFbeta signaling in pancreatic ductal adenocarcinoma. Tumour Biol. 2015;36(3):1613–1618. doi: 10.1007/s13277-014-2757-4. [DOI] [PubMed] [Google Scholar]
- 145.Glazer ES, Welsh E, Pimiento JM, Teer JK, Malafa MP. TGFbeta1 overexpression is associated with improved survival and low tumor cell proliferation in patients with early-stage pancreatic ductal adenocarcinoma. Oncotarget. 2017;8(1):999–1006. doi: 10.18632/oncotarget.13533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lin X, Feng XH. Abrogation of transforming growth factor-beta signaling in pancreatic cancer. World J Surg. 2005;29(3):312–316. doi: 10.1007/s00268-004-7824-3. [DOI] [PubMed] [Google Scholar]
- 147.Yang G, Yang X. Smad4-mediated TGF-beta signaling in tumorigenesis. Int J Biol Sci. 2010;6(1):1–8. doi: 10.7150/ijbs.6.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Goggins M, Shekher M, Turnacioglu K, Yeo CJ, Hruban RH, Kern SE. Genetic alterations of the transforming growth factor beta receptor genes in pancreatic and biliary adenocarcinomas. Cancer Res. 1998;58(23):5329–5332. [PubMed] [Google Scholar]
- 149.Zeitouni D, Pylayeva-Gupta Y, Der CJ, Bryant KL. KRAS mutant pancreatic cancer: no lone path to an effective treatment. Cancers. 2016;8(4) doi: 10.3390/cancers8040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Leung L, Radulovich N, Zhu CQ, Wang D, To C, Ibrahimov E, Tsao MS. Loss of canonical Smad4 signaling promotes KRAS driven malignant transformation of human pancreatic duct epithelial cells and metastasis. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0084366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Eser S, Schnieke A, Schneider G, Saur D. Oncogenic KRAS signalling in pancreatic cancer. Br J Cancer. 2014;111(5):817–822. doi: 10.1038/bjc.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mueller S, Engleitner T, Maresch R, Zukowska M, Lange S, Kaltenbacher T, Konukiewitz B, Ollinger R, Zwiebel M, Strong A. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature. 2018;554(7690):62–68. doi: 10.1038/nature25459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gore AJ, Deitz SL, Palam LR, Craven KE, Korc M. Pancreatic cancer-associated retinoblastoma 1 dysfunction enables TGF-beta to promote proliferation. J Clin Invest. 2014;124(1):338–352. doi: 10.1172/JCI71526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ungefroren H, Groth S, Sebens S, Lehnert H, Gieseler F, Fandrich F. Differential roles of Smad2 and Smad3 in the regulation of TGF-beta1-mediated growth inhibition and cell migration in pancreatic ductal adenocarcinoma cells: control by Rac1. Mol Cancer. 2011;10:67. doi: 10.1186/1476-4598-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pickup M, Novitskiy S, Moses HL. The roles of TGFbeta in the tumour microenvironment. Nat Rev Cancer. 2013;13(11):788–799. doi: 10.1038/nrc3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Katz LH, Li Y, Chen JS, Munoz NM, Majumdar A, Chen J, Mishra L. Targeting TGF-beta signaling in cancer. Expert Opin Ther Targets. 2013;17(7):743–760. doi: 10.1517/14728222.2013.782287. [DOI] [PMC free article] [PubMed] [Google Scholar]