Abstract
Lysophosphatidic acid (LPA) activates cognate G protein-coupled receptors (GPCRs) to initiate biological signaling cascades. Lysophospholipid (LP) receptor binding properties remain incompletely assessed because of difficulties with ligand lipophilicity and lipid “stickiness.” These inherent attributes produce high levels of nonspecific binding within cell-membrane preparations used to assess GPCRs, as has been shown in classical binding assays using radiolabeled ligands, making accurate measurements of lipid binding kinetics difficult to achieve. Backscattering interferometry (BSI) is an optical technology that measures molecular binding interactions by reporting changes in the refractive index of a solution after binding events. Here, we report the use of BSI to assess LPA1 for its ability to bind to naturally occurring lipids and a synthetic LPA1 antagonist (ONO-9780307), under both primary- and competition-binding conditions. Assessment of 12 different lipids demonstrated that the known LP ligand, 1-oleoyl-LPA, as well as an endocannabinoid metabolite, anandamide phosphate, are specific ligands for LPA1, whereas other LPs tested were not. Newly determined dissociation constants (Kd values) for orthosteric lipid ligands approximated 10−9 M, substantially lower (i.e., with higher affinity) than measured Kd values in classical binding or cell-based assays. These results demonstrate that BSI may have particular utility in assessing binding interactions between lipid receptors and their lipid ligands and could provide new screening approaches for lipid receptor identification and drug discovery.
Keywords: lipid, phospholipids, endocannabinoid, optical measurement, molecular interaction, binding assay
Lysophospholipid (LP) signaling involving cognate G protein-coupled receptors (GPCRs) for lysophosphatidic acid (LPA), sphingosine-1-phosphate (S1P), and other lipids has revealed a vast biology affecting the development and function of most, if not all, organ systems and shown etiological or therapeutic involvement in diseases, including those of the nervous and immune systems, as well as in disease conditions like cancer and fibrosis (1–9). A key step in LP signaling is orthosteric GPCR engagement by binding of a cognate ligand to its receptor, followed by G-protein transduction (10, 11). LP receptor binding is thus a necessary step for both naturally occurring lipids and synthetic ligands that, combined with other members of the GPCR superfamily, account for ∼40% of currently marketed drugs (12).
LP GPCRs are members of the rhodopsin-like family of receptors (class A) from which ∼40 lipid GPCRs have now been identified (7). Crystal structures for seven lipid GPCRs have been solved including three LP receptors [LPA1 (13), LPA6 (14), and S1P1 (15)] (16).
Despite these receptor structural advances, classical pharmacological GPCR-ligand binding assays using radioligands have been, and remain, difficult, because the assays require nonbiological receptor overexpression to enable detectable specific binding, while still introducing high levels of nonspecific labeling of membranes which prevents saturation. Assays that involve competition- or displacement-binding protocols are important because they can provide quantitative information on the total number of GPCRs capable of binding ligand (Bmax), the equilibrium dissociation constants (Kd), and the inhibition constant (Ki) (17).
Backscattering interferometry (BSI) uses a label-free, optical technique measuring interactions in free solution (18–25). BSI is designed to acquire fast Fourier-transformed (FFT) light interference patterns generated by backscattered light from a microfluidics chip or glass capillary. It uses much smaller sample volumes (nanoliters) than other free-solution methods such as isothermal titration calorimetry and NMR (20). In addition, other techniques, such as surface plasmon resonance (SPR) (26, 27), require immobilization of one of the binding partners. BSI is broadly applicable for determining biomolecular interactions between proteins and a range of ligands that include small molecules (19), Ca2+ ions (21), carbohydrates (24), nucleotides (25), and proteins (22). However, its use has not been previously reported for measuring lipid-protein interactions. Here, we have tested BSI for determining binding parameters between a defined LP GPCR and endogenous lipid ligands, using direct and competition binding assays.
MATERIALS AND METHODS
Reagents
1-oleolyl-lysophosphatidic acid (1-oleoyl-LPA), 1-oleoyl-lysophosphatidylcholine, 1-oleoyl-lysophosphatidylethanolamine, 1-oleoyl-lysophosphatidylinositol, 1-oleoyl-lysophosphatidylserine, anandamide phosphate (AEAp), anandamide, 2-arachidonoylglycerol, 1-oleoylglycerol, S1P, and 1,2-dioleoyl-sn-glycero-3-phosphate (DOPA) were purchased from Avanti Polar Lipids. 2-oleoyl-LPA was enzymatically synthesized from DOPA using Rhizopus arrhizus lipase (Sigma) and stored in MeOH:DMSO:AcOH (89:10:1, v/v) at −80°C. LPA1 antagonist ONO-9780307 was generated as previously described (13).
Cell lines, flow cytometry, preparation of microsomal fractions, and Western blotting
The entire open reading frame of human Lpar1 with an additional sequence of the hemagglutinin (HA) epitope at the 5′ end was subcloned into the SacI and KpnI sites of the mammalian expression vector pCXN2.1. To establish stable cell lines, B103 cells (400,000 cells) were plated in 6-well dishes and transfected with a receptor-expressing or empty plasmids using Lipofectamine 2000 (Thermo Fisher Scientific), then stable expressing cells were selected by adding 1 mg/ml G418 (Gibco) for 2 weeks. The resultant drug-resistant cells were then stained with an anti-HA primary Ab (Clone 3F10, Roche), followed by a phycoerythrin-labeled anti-rat IgG secondary Ab, and HA-LPA1high populations were sorted and collected using a FACSAria II (BD Biosciences) cell sorter. Cells were maintained in DMEM high glucose (Gibco) containing 10% FBS (Gemini Bio Products) and penicillin-streptomycin (Gibco).
To prepare microsomal fractions, cells were starved for 16 h in DMEM high glucose containing 0.5% BSA (Gemini Bio Products), washed with ice-cold PBS, collected by scraping, and suspended in PBS containing cOmpleteTM protease inhibitor mixture (Roche). The collected cells were then sonicated for 1 min and centrifuged at 10,000 g for 10 min at 4°C, and the supernatant was recentrifuged at 100,000 g for 1 h. The pellet was homogenized in PBS cOmpleteTM protease inhibitor mixture (Roche) and stored at −80°C until use. The microsomal fraction was processed with a probe-type sonicator to generate nanovesicles for BSI. Protein concentration was determined by Bradford assay (Bio-Rad) using BSA as a standard.
To show that HA-tagged proteins were produced, 5 μg of protein from the microsomal fraction containing 5% 2-mercaptoethanol was run on an SDS-PAGE gel (Thermo Fisher Scientific) without heat denaturation and transferred to a PVDF membrane (Millipore). The membrane was blocked with a solution of 5% skimmed milk and probed with an anti-HA Ab (Clone 3F10, Roche). Following incubation with a HRP-conjugated anti-rat IgG Ab, the bands were visualized with a SuperSignalTM West Femto Maximum Sensitivity Substrate (Thermo Scientific) and a Bio-Rad ChemiDoc XRS+ imager.
GPCR binding assays using BSI
BSI has been described in detail previously (19–25). Briefly, a red helium-neon (HeNe) laser (λ) (632.8 nm) coupled to a collimating lens through a single-mode fiber produces a 450 µm diameter beam that illuminates the microfluidic channel. The laser beam reflects from the channel surface, resulting in a set of high-contrast interference fringes that are recorded on a linear array charge-coupled device (CCD). All data were collected in real time utilizing an in-house program written in LabView (National Instruments).
The nanovesicles obtained from control and receptor-expressing cells (20 µg/ml) were mixed with lipid ligands or an LPA1 antagonist, ONO-9780307, for 1 h at room temperature, followed by injection into a channel of the microfluidic chip. The channel was sequentially rinsed with methanol, water, and buffer between sample measurements. The specific binding signal was calculated by subtracting the signals of the control nanovesicle from that of the receptor nanovesicle, followed by normalization to the signals without ligands. The obtained signals displayed as milliradius (mrad) were fitted by nonlinear regression using the Michaelis-Menten equation using Prism software (GraphPad).
We found that it is critical to incorporate particular methodologies in order to obtain reliable results with BSI: 1) confirmation of nanovesicle quality by dynamic light scattering; 2) in order to avoid clogging and sample contamination, cleaning protocols were established to wash out residual sample in the microfluidic channel between experiments; and 3) precise preparation of matching references and samples, because a small fraction of high refractive-index chemicals (such as DMSO) can produce huge phase changes by themselves. An accurate BSI signal is highly dependent on controlling for varying refractive indexes of the solutions (buffers, solvents, etc.) used in each experiment. To this end, our methods included carefully matching the vehicles used in the references (containing LPA1-nanovesicles alone) and the samples (containing LPA1-nanovesicles mixed with ligands).
Intracellular Ca2+ signaling
One day prior to performing the intracellular Ca2+ signaling assay, HA-LPA1-B103 and Vector-B103 cells were plated onto black clear-bottom 384-well plates (Greiner) at 20,000 cells/well. Cells were starved with FreeStyleTM 293 Expression Medium for at least 3 h, then loaded with Fluo-4 dye according to the manufacturer’s instructions (FLIPR Calcium 4 Assay Kit; Molecular Devices). Intracellular Ca2+ mobilization was monitored with a scanning fluorimeter (FDSS7000; Hamamatsu) at an excitation wavelength of 485 nm and an emission wavelength of 525 nm.
Statistical analyses
Results were expressed as means ± SEM. As appropriate, data were analyzed statistically by one-way ANOVA with Tukey’s or Bonferroni’s multiple comparisons test, using Prism software (GraphPad).
RESULTS
Nanovesicle preparation for BSI GPCR-lipid binding assays
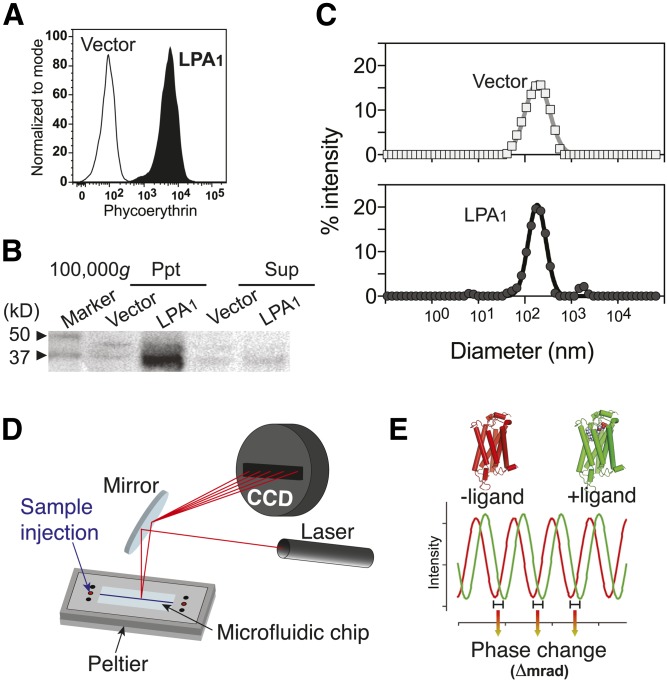
GPCR-containing nanovesicles prepared from receptor-expressing cells provided a useful and plentiful source of target GPCRs for BSI. Antibiotic selection and cell sorting were used to produce a polyclonal population of B103 rat neuroblastoma cells that stably expressed an N terminus HA epitope-tagged human LPA1 (HA-LPA1-B103 cells) (28). Cell-surface expression of HA-LPA1 was confirmed using flow cytometry (Fig. 1A) and in microsomal fractions by Western blotting (Fig. 1B). Microsomal fractions made from HA-hLPA1-B103 or control cells (vector-transfected cells) were sonicated to generate nanovesicles for BSI analyses. Dynamic light scattering was used to determine the average vesicle diameter, about 200 nm (Fig. 1C), an optimal size distribution for detecting BSI fringe patterns and phase shifts. Resulting nanovesicles mixed with nonlabeled ligands were simply injected into the BSI microfluidic channel (Fig. 1D), followed by FFT phase acquisition that identified phase changes occurring after GPCR-ligand binding (Fig. 1E).
Fig. 1.
LPA1 nanovesicle preparation (A–C), BSI setup (D), and a scheme of fringe shift (E). A: A histogram showing expression of HA-LPA1 on the cell surface of B103 cells. The surface expression level of the HA epitope on the LPA1 N terminus in HA-LPA1-B103 cells was confirmed by flow cytometry. Vector-B103 cells served as a negative control. B: A Western blot showing the expression of HA-LPA1 in the membrane fraction (100,000 g, ppt) prepared from HA-LPA1-B103 cells. Vector-B103 cells and supernatant fraction (100,000 g, sup) serve as negative controls. C: Size distributions of the resultant vector and LPA1-nanovesicles. Membrane fractions were processed with a probe sonicator to produce nanovesicles with an average diameter of 200 nm, as confirmed by dynamic light scattering. D: BSI setup. Proper irradiation by a HeNe laser of a semicircular cross-section flow path in the microfluidic chip generates optical interference, which is detected by a linear-array CCD camera. E: A schematic diagram of a typical fringe shift between protein with (green) or without (red) ligand. The phase under each condition is computed in real time using FFT, and then the phase change (Δ mrad) between the two different conditions is calculated.
BSI identifies specific lipid ligands for LPA1
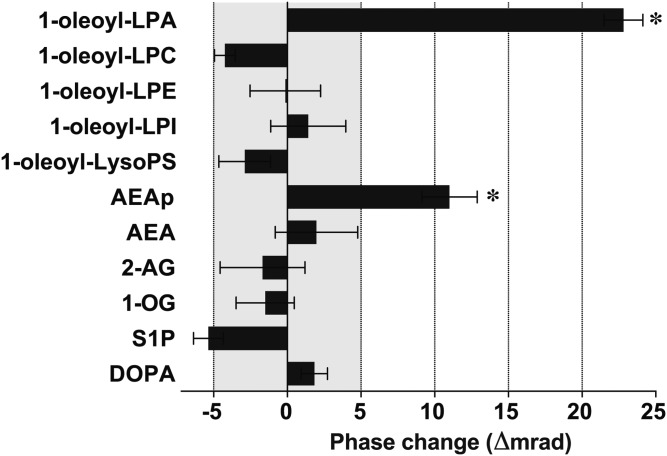
To evaluate whether BSI could be used to differentiate receptor interactions with known LPA1 lipid ligands from those with other lipids, a series of native lipids, including LPA1 ligands 1-oleoyl-LPA (29) and AEAp (13), were assayed for LPA1 binding using BSI (Fig. 2). The experiments were designed to measure phase changes between lipid-LPA1 mixture (sample) versus LPA1-nanovesicle alone (reference). The current BSI setup generates a ∼5 mrad range of machine noise, indicating that actual binding signals are likely higher than this range. Both 1-oleoyl-LPA and AEAp produced significant phase-shift signals that were statistically greater than the signals produced by nonspecific lipids. These data indicate that BSI can be used to quantify bona fide GPCR-lipid interactions and demonstrate its high sensitivity (true positive/false negative = 2/0) and specificity (true negative/false positive = 9/0), thus supporting its utility in lipid-receptor screening strategies.
Fig. 2.
BSI-based small-scale lipid screening identifies LPA and AEAp as endogenous ligands for LPA1. BSI signals were obtained as phase changes (Δmrad) between LPA1-nanovesicles, with indicated natural lipid ligands at 10 nM (sample), versus LPA1-nanovesicles alone (reference). Phase changes of less than 5 mrad were considered background noise level. Mean ± SEM. n = 5. * P < 0.05 by one-way ANOVA with Bonferroni’s multiple comparisons test as compared with other lipids. A result of a single experiment is shown. LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; LPI, lysophosphatidylinositol; LysoPS, lysophosphatidylserine; AEA, anandamide; 2-AG, 2-arachidonoyl glycerol; 1-OG, 1-oleoyl glycerol; DOPA, 1,2-dioleoyl-sn-glycero-3-phosphate.
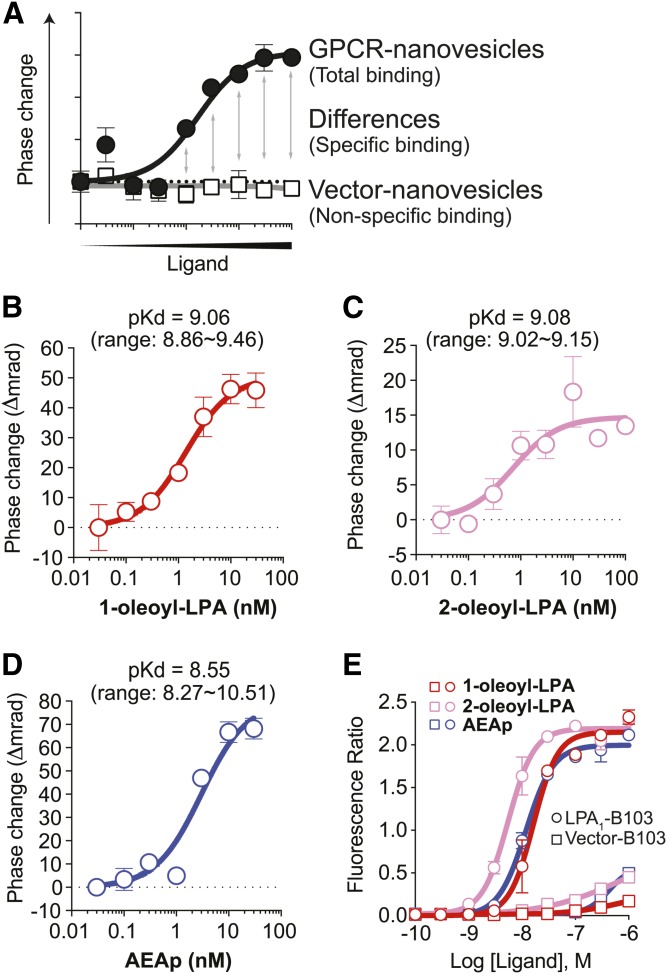
Kd values obtained from BSI may be more physiologically relevant than those obtained from traditional methods
BSI can be used to show specific ligand binding by subtracting nonspecific from total binding (Fig. 3A), enabling Kd determination by nonlinear regression and the Michaelis-Menten equation. Prior estimates of specific binding with the radioactive ligand [3H]1-oleoyl-LPA and overexpressed LPA1 resulted in a pKd = 7.16 (28). We used BSI to assess LPA1 binding to nonlabeled, natural lipid ligands, revealing the specific binding between LPA1 versus 1-oleoyl-LPA [pKd = 9.06 (0.87 ± 0.37 nM); Fig. 3B], 2-oleoyl-LPA [pKd = 9.08 (0.83 ± 0.09 nM); Fig. 3C], and AEAp [pKd = 8.55 (2.80 ± 0.89 nM); Fig. 3D]. Notably, the calculated Kd of 1-oleoyl-LPA to LPA1 is 77-fold less than the value obtained using radioactive binding (28). Importantly, lipid ligands mixed with vector nanovesicles did not produce positive BSI signals (supplemental Fig. S1), indicating that BSI estimates a more accurate Kd value in the elimination of the nonspecific binding signals.
Fig. 3.
BSI determines binding affinities between endogenous LPA1 ligands versus LPA1. A: An example of binding curves of nonspecific and total binding. Specific binding is calculated by subtracting nonspecific binding from total binding. B–D: Specific-binding curves between LPA1 versus 1-oleoyl-LPA (B), 2-oleoyl-LPA (C), and AEAp (D). Mean ± SEM. n = 5. pKd values were shown as mean with a range from two (B, Kd = 0.87 ± 0.37 nM; and C, Kd = 0.83 ± 0.09 nM) and three (D, Kd = 2.80 ± 0.89 nM) independent experiments. E: Ca2+ mobilization in LPA1-B103 and Vector-B103 cells. Cells were loaded with Fluo-4, and the intracellular Ca2+ mobilization response to the indicated ligands was monitored with a Hamamatsu FDSS7000. Mean ± SEM. n = 2. A result from two independent experiments with similar results is shown.
All of the endogenous lipid ligands described above are potent intracellular Ca2+-mobilizing agonists; therefore, we examined their potency in Ca2+ assays using HA-hLPA1-B103 cells and found a consistent order of ligand potency with that identified using BSI (2-oleoyl-LPA > 1-oleoyl-LPA = AEAp) (Fig. 3E).
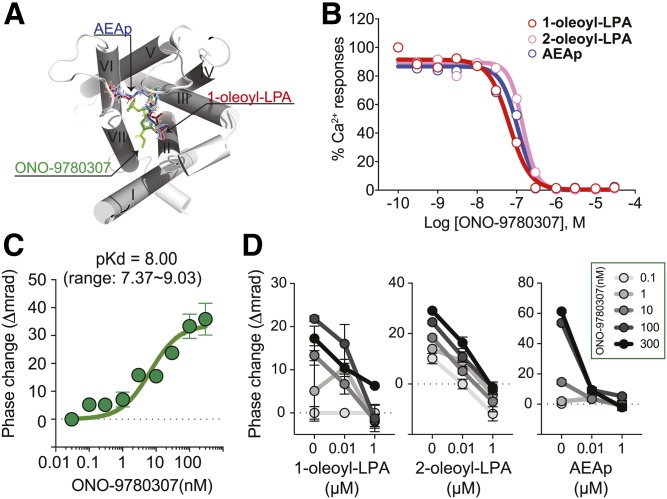
Kd determination of a synthetic LPA1 antagonist and its use in competition binding assays
ONO-9780307, an LPA1 antagonist that was used to solve the LPA1 crystal structure (13), binds to an orthosteric site in LPA1 (Fig. 4A), resulting in inhibition of Ca2+ mobilization that is normally produced by endogenous ligands with single-digit nanomolar IC50 values (Fig. 4B). BSI was used to determine the specific binding of ONO-9780307 to LPA1, yielding pKd = 8.00 (9.90 ± 4.55 nM) (Fig. 4C).
Fig. 4.
Competitive binding between endogenous LPA1 ligands versus LPA1 antagonist (ONO-9780307) as measured through BSI. A: The orthosteric binding site of LPA1 with an LPA1 antagonist, ONO-9780307 (green), 1-oleoyl-LPA (red), and AEAp (blue). B: Ca2+ mobilization in LPA1-B103 and Vector-B103 cells. Cells were loaded with Fluo-4, then the intracellular Ca2+ mobilization that was induced via the indicated ligands (300 nM), in the presence of ONO-9780307, was monitored with FDSS7000. Mean ± SEM. n = 2. A result from two independent experiments is shown. C: Specific binding curves between LPA1 versus ONO-9780307. A representative result from five independent experiments is shown. Mean ± SEM. n = 5. pKd values were shown as mean with a range from five independent experiments (Kd = 9.90 ± 4.55 nM). D: Competitive binding between ONO-9780307 versus endogenous ligands to LPA1. BSI signals were obtained from mixtures of LPA1-nanovesicles and ONO-9780307 (0.1, 1, 10, 100, and 300 nM) with endogenous natural lipid ligands (0, 0.01, and 1 µM). Mean ± SEM. n = 5. A result from two independent experiments is shown.
Competitive binding between ONO-9780307 and three endogenous LPA1 ligands was evaluated using BSI (Fig. 4D). ONO-9780307 binding produced dose-dependent BSI signals, and all three lipids abolished its binding signal in a dose-dependent manner, again within single-digit nanomolar ranges of detected IC50 values. These concordant results support the utility of BSI in competition assays in which antagonists and orthosteric ligands share the same binding pocket. The mechanism through which LPA1-ligand binding alters the BSI signal is unknown, but it may reflect the actions within the “baggy” LPA1 binding pocket (13, 16) whose occupancy with agonist or antagonist induces LPA1 conformational changes differently. Thus, BSI is also useful for assessing competitive binding, at least to an orthosteric binding site. Recent use of a commercial analysis of allosteric binding of nonlipid synthetic compounds to LPA1 support BSI’s use in GPCR binding studies (30).
DISCUSSION
Many biophysical and biochemical technologies for determining molecular interactions and conformational changes have been developed, such as SPR (26, 27) and fluorescence resonance energy transfer (31), yet all have limitations for lipid GPCR analyses. For example, fluorescent labeling of lipids fundamentally alters their chemical form. In addition, it is still challenging to maintain the native biological conformation of GPCRs upon their immobilization on an SPR sensor chip. Because lipids are extremely diverse in the length of their acyl chains, their degree of saturation, and their stereochemistry, a label-free technology for determining molecular interactions is preferred. Because BSI is an optical measurement technology not requiring labeling or immobilization of any binding partners, it provides such a solution for GPCR-lipid analyses, and potentially other applications, allowing for identification of binding partners with the potential for scalability using screening (Fig. 2).
This novel use of the BSI technology enables the characterization of the binding affinities between lipid GPCRs and their ligands (Fig. 3), and it is also useful for competitive binding analysis (Fig. 4). In this study, we demonstrated lower Kd values through the use of more physiologically relevant (naturally occurring, unlabeled) ligands than were observed in previous studies that utilized radioligands (28). We believe the higher Kd values obtained from radioligand-based experiments are caused by high background signals due to integration of radioactive “sticky” lipids into the lipid bilayer, which likely underestimates binding affinities. The BSI system largely eliminated background signals from nonspecific binding (supplemental Fig. S1), because integration of lipid ligands into the lipid bilayer has little effect on BSI signals; we believe this reduction in “noise” enabled more accurate measurements of Kd values, as compared with those obtained using radioactive binding assays. In addition, because isotopic substitutions can alter the rate of a reaction (known as the “isotope effect”) (32), BSI enabled us to avoid this artifact altogether.
Given the varied GPCR conformations among nonbinding, agonist-binding, and antagonist-binding states (10), we believe BSI most likely determined a refractive index for each GPCR conformational state. Interestingly, in the competitive binding assay (Fig. 4), the BSI signals of antagonist-bound LPA1 was canceled out by agonist-bound LPA1, although both conformational states separately produced positive signals. This complex, nonlinear behavior of BSI signals for GPCR binding is an interesting aspect that must be addressed in future studies. BSI continues to have several methodological limitations at present. For example, the current BSI assay is too low-throughput to use for a drug discovery and screening platform. It was also impossible to determine the receptor density within the nanovesicles, whereas it can be defined as Bmax in radioligand binding assays. Although BSI does provide more accurate measurements of binding affinities at present, its improvement regarding these issues will make BSI even more valuable.
The present study supports new uses for BSI in the discovery and screening of lipid GPCRs and demonstrated the potential for BSI to more accurately characterize other binding interactions with potential scalability. Moreover, the greater sensitivity in BSI over traditional binding assays may enable the study of GPCR pharmacology at a physiological, in vivo expression level in combination with gene editing in animal tissues or cells.
Acknowledgments
The authors thank Mr. Michael N. Kammer (Vanderbilt University) for helping set up BSI; Ms. Yuka Takada (Ono Pharmaceutical Co., Ltd.,) for generating 2-acyl-LPA; Dr. Mike Hansen (GPCR Consortium) for providing Protein Data Bank files of LPA1 structures; Mr. Naoki Morita (Shimadzu Corp.) for generous support; Drs. Yasuyuki Fujii, Kyoko Noguchi, Julian Wong, Kazufumi Nagai, and Mr. Yuji Kawahara (SBP) for discussions; and Dr. Laura Wolszon and Ms. Danielle Jones for editorial assistance.
Footnotes
Abbreviations:
- 1-oleoyl-LPA
- 1-oleolyl-lysophosphatidic acid
- AEAp
- anandamide phosphate
- BSI
- backscattering interferometry
- FFT
- fast Fourier-transformed
- GPCR
- G protein-coupled receptor
- HA
- hemagglutinin
- LP
- lysophospholipid
- LPA
- lysophosphatidic acid
- mrad
- milliradius
- S1P
- sphingosine-1-phosphate
- SPR
- surface plasmon resonance
This work was supported by a grant from Ono Pharmaceutical Co., Ltd. and National Institute of Neurological Disorders and Stroke Grant R01 NS084398 (J.C.), and fellowships from the Uehara Memorial Foundation, the Kanae Foundation for the Promotion of Medical Science, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, and the Human Frontier Science Program, and National Institutes of Health Grant R01 NS103940 (Y.K.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Choi J. W., Herr D. R., Noguchi K., Yung Y. C., Lee C. W., Mutoh T., Lin M. E., Teo S. T., Park K. E., Mosley A. N., et al. 2010. LPA receptors: subtypes and biological actions. Annu. Rev. Pharmacol. Toxicol. 50: 157–186. [DOI] [PubMed] [Google Scholar]
- 2.Kihara Y., Maceyka M., Spiegel S., and Chun J.. 2014. Lysophospholipid receptor nomenclature review: IUPHAR Review 8. Br. J. Pharmacol. 171: 3575–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yung Y. C., Stoddard N. C., and Chun J.. 2014. LPA receptor signaling: pharmacology, physiology, and pathophysiology. J. Lipid Res. 55: 1192–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chun J., Hla T., Moolenaar W., and Spiegel S., editors. 2014. Lysophospholipid Receptors: Signaling and Biochemistry Wiley, Hoboken, NJ. [Google Scholar]
- 5.Yung Y. C., Stoddard N. C., Mirendil H., and Chun J.. 2015. Lysophosphatidic acid signaling in the nervous system. Neuron. 85: 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheng X., Yung Y. C., Chen A., and Chun J.. 2015. Lysophosphatidic acid signalling in development. Development. 142: 1390–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kihara Y., Mizuno H., and Chun J.. 2015. Lysophospholipid receptors in drug discovery. Exp. Cell Res. 333: 171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groves A., Kihara Y., and Chun J.. 2013. Fingolimod: direct CNS effects of sphingosine 1-phosphate (S1P) receptor modulation and implications in multiple sclerosis therapy. J. Neurol. Sci. 328: 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun J., Kihara Y., Jonnalagadda D., and Blaho V.. 2018. Fingolimod: lessons learned and new opportunities for treating multiple sclerosis and other disorders. Annu. Rev. Pharmacol. Toxicol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manglik A., and Kobilka B.. 2014. The role of protein dynamics in GPCR function: insights from the beta2AR and rhodopsin. Curr. Opin. Cell Biol. 27: 136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flock T., Hauser A. S., Lund N., Gloriam D. E., Balaji S., and Babu M. M.. 2017. Selectivity determinants of GPCR-G-protein binding. Nature. 545: 317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stevens R. C., Cherezov V., Katritch V., Abagyan R., Kuhn P., Rosen H., and Wuthrich K.. 2013. The GPCR Network: a large-scale collaboration to determine human GPCR structure and function. Nat. Rev. Drug Discov. 12: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chrencik J. E., Roth C. B., Terakado M., Kurata H., Omi R., Kihara Y., Warshaviak D., Nakade S., Asmar-Rovira G., Mileni M., et al. 2015. Crystal structure of antagonist bound human lysophosphatidic acid receptor 1. Cell. 161: 1633–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taniguchi R., Inoue A., Sayama M., Uwamizu A., Yamashita K., Hirata K., Yoshida M., Tanaka Y., Kato H. E., Nakada-Nakura Y., et al. 2017. Structural insights into ligand recognition by the lysophosphatidic acid receptor LPA6. Nature. 548: 356–360. [DOI] [PubMed] [Google Scholar]
- 15.Hanson M. A., Roth C. B., Jo E., Griffith M. T., Scott F. L., Reinhart G., Desale H., Clemons B., Cahalan S. M., Schuerer S. C., et al. 2012. Crystal structure of a lipid G protein-coupled receptor. Science. 335: 851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blaho V. A., and Chun J.. 2018. ‘Crystal’ clear? Lysophospholipid receptor structure insights and controversies. Trends Pharmacol. Sci. 39: 953–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flanagan C. A. 2016. GPCR-radioligand binding assays. Methods Cell Biol. 132: 191–215. [DOI] [PubMed] [Google Scholar]
- 18.Baksh M. M., and Finn M. G.. 2017. An experimental check of backscattering interferometry. Sens. Actuators B Chem. 243: 977–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baksh M. M., Kussrow A. K., Mileni M., Finn M. G., and Bornhop D. J.. 2011. Label-free quantification of membrane-ligand interactions using backscattering interferometry. Nat. Biotechnol. 29: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bornhop D. J., Kammer M. N., Kussrow A., Flowers R. A. 2nd, and Meiler J.. 2016. Origin and prediction of free-solution interaction studies performed label-free. Proc. Natl. Acad. Sci. USA. 113: E1595–E1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bornhop D. J., Latham J. C., Kussrow A., Markov D. A., Jones R. D., and Sorensen H. S.. 2007. Free-solution, label-free molecular interactions studied by back-scattering interferometry. Science. 317: 1732–1736. [DOI] [PubMed] [Google Scholar]
- 22.Kussrow A., Baksh M. M., Bornhop D. J., and Finn M. G.. 2011. Universal sensing by transduction of antibody binding with backscattering interferometry. ChemBioChem. 12: 367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kussrow A., Enders C. S., and Bornhop D. J.. 2012. Interferometric methods for label-free molecular interaction studies. Anal. Chem. 84: 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kussrow A., Kaltgrad E., Wolfenden M. L., Cloninger M. J., Finn M. G., and Bornhop D. J.. 2009. Measurement of monovalent and polyvalent carbohydrate-lectin binding by back-scattering interferometry. Anal. Chem. 81: 4889–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olmsted I. R., Xiao Y., Cho M., Csordas A. T., Sheehan J. H., Meiler J., Soh H. T., and Bornhop D. J.. 2011. Measurement of aptamer-protein interactions with back-scattering interferometry. Anal. Chem. 83: 8867–8870. [DOI] [PubMed] [Google Scholar]
- 26.Navratilova I., Besnard J., and Hopkins A. L.. 2011. Screening for GPCR ligands using surface plasmon resonance. ACS Med. Chem. Lett. 2: 549–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salamon Z., Tollin G., Alves I., and Hruby V.. 2009. Chapter 6. Plasmon resonance methods in membrane protein biology applications to GPCR signaling. Methods Enzymol. 461: 123–146. [DOI] [PubMed] [Google Scholar]
- 28.Yanagida K., Masago K., Nakanishi H., Kihara Y., Hamano F., Tajima Y., Taguchi R., Shimizu T., and Ishii S.. 2009. Identification and characterization of a novel lysophosphatidic acid receptor, p2y5/LPA6. J. Biol. Chem. 284: 17731–17741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukushima N., Kimura Y., and Chun J.. 1998. A single receptor encoded by vzg-1/lpA1/edg-2 couples to G proteins and mediates multiple cellular responses to lysophosphatidic acid. Proc. Natl. Acad. Sci. USA. 95: 6151–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellery J., Dickson L., Cheung T., Ciuclan L., Bunyard P., Mack S., Buffham W. J., Farnaby W., Mitchell P., Brown D., et al. 2018. Identification of compounds acting as negative allosteric modulators of the LPA1 receptor. Eur. J. Pharmacol. 833: 8–15. [DOI] [PubMed] [Google Scholar]
- 31.Kauk M., and Hoffmann C.. 2018. Intramolecular and intermolecular FRET sensors for GPCRs—monitoring conformational changes and beyond. Trends Pharmacol. Sci. 39: 123–135. [DOI] [PubMed] [Google Scholar]
- 32.Swiderek K., and Paneth P.. 2013. Binding isotope effects. Chem. Rev. 113: 7851–7879. [DOI] [PubMed] [Google Scholar]