Abstract
Proprotein convertase subtilisin/kexin type 9 (PCSK9) targets the LDL receptor (LDLR) for degradation, increasing plasma LDL and, consequently, cardiovascular risk. Uptake of secreted PCSK9 is required for its effect on the LDLR, and LDL itself inhibits this uptake, though how it does so remains unclear. In this study, we investigated the relationship between LDL, the PCSK9:LDLR interaction, and PCSK9 uptake. We show that LDL inhibits binding of PCSK9 to the LDLR in vitro more impressively than it inhibits PCSK9 uptake in cells. Furthermore, cell-surface heparin-like molecules (HLMs) can partly explain this difference, consistent with heparan sulfate proteoglycans (HSPGs) acting as coreceptors for PCSK9. We also show that HLMs can interact with either PCSK9 or LDL to modulate the inhibitory activity of LDL on PCSK9 uptake, with such inhibition rescued by competition with the entire PCSK9 prodomain, but not its truncated variants. Additionally, we show that the gain-of-function PCSK9 variant, S127R, located in the prodomain near the HSPG binding site, exhibits increased affinity for HLMs, potentially explaining its phenotype. Overall, our findings suggest a model where LDL acts as a negative regulator of PCSK9 function by decreasing its uptake via direct interactions with either the LDLR or HLMs.
Keywords: atherosclerosis, cholesterol, lipoprotein metabolism, receptors/lipoprotein, dyslipidemias, low density lipoprotein receptor, single nucleotide polymorphism, familial hypercholesterolemia, coreceptor, mechanism, low density lipoprotein, proprotein convertase subtilisin/kexin type 9
Gain-of-function (GOF) mutations in the proprotein convertase subtilisin/kexin type 9 (PCSK9) gene, which encodes a self-cleaving protease, cause autosomal dominant familial hypercholesterolemia (1). Mechanistically, PCSK9 binds the epidermal growth factor precursor homology domain A (EGF-A) of the LDL receptor (LDLR) (2) and induces lysosomal-mediated degradation of the complex (3), thus limiting the availability of LDLR to recycle back to the cell surface and internalize LDL (4). The robust relationship between LDL and atherosclerosis (5) combined with a lack of ill effects in individuals devoid of functional PCSK9 (6, 7) has made PCSK9 an important therapeutic target against heart disease. Indeed, monoclonal antibodies inhibiting PCSK9 reduce serum LDL levels (8, 9), and also reduce cardiovascular events (10), even when added to current effective therapies, such as statins. Despite this impressive efficacy, these biologic therapies suffer from a lack of cost-effectiveness and a requirement for subcutaneous injections, leaving a need for additional therapeutic options (11, 12). As a result, several other strategies to inhibit PCSK9 function are under investigation (13). Because atherosclerosis is a chronic disease, a successful therapy may potentially be given for many years.
Endogenous regulation of PCSK9 occurs at both the transcriptional (14–17) and posttranscriptional levels (18), with secretion of PCSK9 itself an important regulatory step for its function (19). As a proprotein convertase, PCSK9 requires self-proteolysis between its prodomain and catalytic domain to permit exit from the ER (20–22). Protease-dead PCSK9 mutants do not undergo efficient secretion and have minimal effect on the LDLR (23). Despite the cleavage event, which serves as the overall rate-limiting step of PCSK9 secretion (24), the prodomain (residues 31-152) remains tightly but noncovalently bound to the catalytic domain for the remainder of PCSK9’s lifecycle (25–27). Yet despite the clear effects of PCSK9 on the LDLR and serum LDL, certain aspects of PCSK9 biology remain incompletely understood (18). Understanding this biology may uncover new pathways amenable to certain therapeutic strategies, or presage long-term effects of therapies, which we might not otherwise foresee. Why PCSK9 exists, for example, remains an unanswered question (28), as it seems rather odd for the predominant function of a gene to cause disease.
Despite the general requirement for self-cleavage and secretion for full PCSK9 function, the initial GOF PCSK9 mutant identified, S127R, has long been known to have paradoxically reduced self-processing, suggesting that additional mechanisms can overcome this functional restriction (21, 29). An unrelated series of experiments have shown that apoB100, the main protein component of LDL, binds PCSK9 both intra- and extracellularly, offering additional insights into the function of PCSK9 in lipid metabolism (30–32). This PCSK9:apoB100 interaction augments the secretion of apoB-containing lipids in an LDLR-independent manner (31) and inhibits the ability of serum PCSK9 to induce LDLR degradation in humans (32, 33). Therapeutically, the presence of LDL-bound PCSK9 in the serum permits PCSK9 removal during lipid apheresis treatments (34). The structural basis of the PCSK9:LDL association remains unknown, though the N terminus of the PCSK9 prodomain, which is dispensable for the PCSK9:LDLR interaction (35, 36), is required for PCSK9 to bind LDL (32). Recently, heparan sulfate proteoglycans (HSPGs) have been identified as coreceptors for PCSK9 on the hepatocyte cell surface to promote PCSK9 uptake and function, with a specific arginine-rich motif encompassing residues 93-139 in the PCSK9 prodomain as the likely binding site mediating this interaction (37).
Given the involvement of the PCSK9 prodomain in these interactions, and the potential for physically overlapping binding sites, we postulated that LDL and heparin-like molecules (HLMs), such as HSPGs, could each serve as reciprocal regulators with regard to PCSK9 function. In this study, we developed a luciferase-based assay to investigate conditions that modify PCSK9 uptake into hepatocytes. We found that while LDL is a potent inhibitor of PCSK9 binding to the LDLR in vitro, its ability to inhibit PCSK9 uptake into cells is comparatively modest. Consistent with an inhibitory effect on PCSK9:LDLR binding, we show that LDL-mediated inhibition of PCSK9 uptake is dependent on LDLR expression. Additionally, we show that both the PCSK9:HLM and the LDL:HLM interactions modulate LDL’s inhibitory effect on PCSK9 uptake. Moreover, we find that the entire PCSK9 prodomain, but not truncated variants, binds LDL in vitro and rescues PCSK9 uptake from LDL-mediated inhibition, suggesting that HLMs and LDL may share a common PCSK9 binding site. Lastly, we show that the S127R mutant of PCSK9, also in the prodomain, displays increased affinity for heparin, suggesting a potential mechanism to explain its GOF phenotype. Overall, our findings support a model wherein LDL negatively regulates PCSK9 uptake via both HLM-dependent and -independent mechanisms, providing a layer of feedback to modulate lipid homeostasis in the hepatocyte.
MATERIALS AND METHODS
Plasmid construction
All expression vectors were created by Gibson assembly (38) after PCR amplification of appropriate PCSK9 domains from previously described plasmids (22, 24), a plasmid encoding nanoluciferase (NLuc) (Promega, Madison, WI), and a plasmid encoding the Fc-Avi tag (generous gift from A. Martinko and J. Wells, University of California San Francisco). The PCSK9-NLuc plasmid and its variants were placed into the pcDNA5/FRT/TO backbone (Thermo Fisher Scientific, Waltham MA), and the ProPCSK9-Fc-Avi plasmid and its variants were placed into the pcDNA3.4 backbone (Thermo Fisher Scientific). Insertion of mutations (R93A, S127R, D374Y) and truncations was performed by site-directed mutagenesis (39) using custom synthesized oligonucleotide primers (Elim Biopharmaceuticals, Hayward CA). All constructs were extensively sequenced to ensure the absence of errors.
Cell culture
HEK 293T cells (ATCC, Manassas, VA) were maintained in high-glucose, pyruvate, and l-glutamine containing DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (Axenia BioLogix, Dixon, CA) at 37°C under 5% CO2 and dissociated for passage by 0.05% Trypsin-EDTA (Thermo Fisher Scientific). HepG2 cells (ATCC) were grown in low-glucose, pyruvate, and GlutaMAX containing DMEM (Thermo Fisher Scientific) supplemented with 10% FBS at 37°C under 5% CO2. To minimize cell clumping, HepG2 cells were dissociated by 0.25% Trypsin-EDTA (Thermo Fisher Scientific) and sent three times through a 21-gauge needle during each passage. For experiments requiring upregulation of cell-surface LDLRs, HepG2 cells were treated with sterol-depleting medium, which consisted of low-glucose, pyruvate, and GlutaMAX containing DMEM supplemented with 5% lipoprotein-deficient serum (Kalen BioMedical, Germantown, MD), 25 μM of mevastatin (Millipore Sigma), and 50 μM of mevalonolactone (Millipore Sigma). Flp-In T-Rex 293 cells (Thermo Fisher Scientific) were maintained in the same conditions as 293T cells, but additionally supplemented with 1 mg/ml Zeocin (InvivoGen, San Diego, CA) and 15 μg/ml blasticidin (InvivoGen) prior to selection, or 150 μg/ml Hygromycin B (InvivoGen) and 15 μg/ml blasticidin after selection of stable cell lines.
PCSK9-NLuc and ProPCSK9-Fc-Avi medium production
HEK 293T cells were seeded at 1 × 106 cells per T25 flask and incubated overnight. On the following day, the cells were transfected with Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer’s protocol using 3–6.9 μg of appropriate plasmid DNA per well. The medium was changed 6–12 h after transfections and harvested 24–72 h later. Relative amounts of PCSK9-NLuc were quantitated by luminescence assay (see below) and a human PCSK9 ELISA kit (R&D Systems, Minneapolis, MN) according to the manufacturer’s instructions. Relative amounts of ProPCSK9-Fc-Avi were quantitated using a Human Fc ELISA kit (Syd Labs, Natick, MA) according to the manufacturer’s instructions. For some experiments, a stable inducible PCSK9-NLuc cell line, derived from the Flp-In T-Rex 293 cell line (Thermo Fisher Scientific) according to the manufacturer’s instructions, was induced with 1 μg/ml doxycycline (Millipore Sigma, Burlington, MA) and conditioned medium was collected 24–48 h later.
Luminescence assays
Luciferase-containing samples were aliquoted into white 96-well or 384-well plates and mixed with equal volumes of a filtered 2× coelenterazine reagent for readout [300 mM sodium ascorbate (Millipore Sigma), 5 mM NaCl, 40 μM coelenterazine (Gold Biotechnology, St. Louis, MO), and either 0.1% BSA (Millipore Sigma) for medium-based nonlytic assays or 0.1% Triton X-100 (Millipore Sigma) for cell-based lytic assays]. The samples were incubated at room temperature in the absence of light with gentle shaking for 5 to 10 min followed by immediate readout of luminescence on a plate reader (Tecan Systems, San Jose CA).
PCSK9 in vitro binding assays
Inhibition of the PCSK9-LDLR interaction by LDL (Lee BioSolutions, Maryland Heights, MO) was evaluated using a CircuLex PCSK9-LDLR in vitro binding assay kit (MBL International, Woburn, MA) according to the manufacturer’s instructions with minor modifications. PCSK9 was incubated with LDL for 1 h prior to use in the assay. Relative PCSK9:LDLR(EGF-A) binding was defined as absorbance at 450 nm, with a value of 1 defined as the highest concentration of PCSK9 in the absence of LDL. To evaluate PCSK9 binding to heparin, conditioned medium containing PCSK9-NLuc or its variants was incubated in clear heparin-coated microplates (Bioworld, Dublin, OH) at room temperature with gentle shaking for 2 h. Input luminescence was measured as described above. Each well was then washed two times with 500 mM of NaCl, and luminescence assay was performed. The wash protocol was repeated with 5 mg/ml heparin (Millipore Sigma) and luminescence assay was repeated.
PCSK9-NLuc uptake assays
HepG2 cells were seeded into 12-well or 6-well plates at 5 × 105 to 1.5 × 106 cells per well with 0.5–1.5 ml of standard or sterol-depletion medium as appropriate and incubated overnight. Heparinase-treated cells were incubated with heparinase (Millipore Sigma) at 0.1–0.5 U/ml for 1 h and washed with PBS. All cells were provided fresh medium on the day of experiment, supplemented with 50 μM of chloroquine (Millipore Sigma) as appropriate to inhibit lysosome-mediated PCSK9 degradation, and then treated with appropriate conditioned PCSK9-NLuc medium and/or LDL or its variants as indicated. Relative amounts of conditioned PCSK9-NLuc medium were normalized by luminescence assay and anti-PCSK9 ELISA prior to coincubation to ensure uniform treatment for each experiment, and for some experiments, aliquots of the PCSK9-NLuc inputs were subjected to luminescence assay as an internal control. The blocking peptides, Pep2-8 (Ac-TVFTSWEEYLDWV-amide) and PepCtrl (Ac-TVATSAEEYLFWV-amide), were custom synthesized (Elim Biopharmaceuticals), reconstituted in PBS, and used at a working concentration of 100 μM. Typically, treated cells were incubated with treatments at 37°C (unless otherwise indicated) for 4 h, washed, and dissociated with 0.25% Trypsin-EDTA. The cells were centrifuged at 300 g for 5 min to remove supernatant, washed and respun, and lysed in lysis buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1× cOmplete protease inhibitor (Millipore Sigma), and 0.1% Nonidet P-40], and then clarified from insoluble fraction at 21,000 g for 15 min. Protein concentration was analyzed by a Micro BCA protein assay kit (Thermo Fisher Scientific), and equal amounts of lysate were then evaluated by luciferase assay.
Preparation of LDL variants
For experiments utilizing methylated LDL, LDL was reductively methylated as described in the literature (40, 41). Briefly, LDL (10–20 mg/ml) in 0.15 M NaCl was diluted to 1.5 total volume with 0.3 M sodium borate buffer (pH 9.0) and treated with 1 mg of sodium borohydride (Millipore Sigma) followed by six additions of 1 μl of 37% aqueous formaldehyde over 30 min at 4°C. The reaction was quenched by adding Tris-HCl (pH 7.4) to a final concentration of 100 μM, and the methylated LDL was dialyzed extensively against 150 mM NaCl at 4°C. Small dense and large LDL were separated from LDL using a one-layer system by ultracentrifugation using a TLA-100.3 rotor (Beckman Coulter, Brea, CA) at 350,000 g over an iodixanol density gradient medium (OptiPrep, CosmoBio, Tokyo, Japan) as per prior literature protocols (42).
LDL binding assays
Evaluation of PCSK9-NLuc and ProPCSK9-Fc-Avi fusion binding to LDL or its variants was performed as described in the literature with minor modifications (32). Briefly, conditioned medium containing equivalent amounts of the PCSK9-NLuc or ProPCSK9-Fc-Avi fusion (∼10 nM) was incubated with LDL or its variants (0.5 mg/ml) for 45 min at 37°C in either cell culture medium or PBS containing 1% BSA. Reaction mixtures were then separated using a three-layered iodixanol gradient and ultracentrifugation in a TLS55 rotor (Beckman Coulter) exactly as described (32). The LDL-free 500 μl upper fraction was removed by aspiration, and the 600 μl LDL-containing fraction, or its volumetric equivalent, was then collected and analyzed.
Western blots
Lysates and LDL-containing fractions from appropriate experiments were mixed 1:1 with 2× Laemmli sample buffer [125 mM Tris-HCl (pH 7.4), 20% glycerol, 4% SDS, 5% β-mercaptoethanol, 0.02% bromophenol blue] and heated to 95°C for 5 min. Equal amounts of lysate were resolved on 4–12% Bis-Tris NuPAGE gels (Thermo Fisher Scientific) in MOPS buffer (Thermo Fisher Scientific), transferred to nitrocellulose (Bio-Rad, Hercules, CA), and blocked in 5% BSA in TBS with 0.1% Tween-20. Membranes were probed overnight with antibodies against PCSK9 (1:1,000 dilution, D7U6L; Cell Signaling, Danvers, MA), β-actin (1:2,000 dilution, 8H10D10; Cell Signaling), apoB (1:338 dilution, MAB4124; R&D Systems), or Avi-Tag (1:500 dilution, A00674; GenScript, Piscataway, NJ), washed extensively, detected with secondary antibodies IRDye 800CW goat anti-rabbit IgG, IRDye 800CW goat anti-mouse IgG, and IRDye 680RD goat anti-mouse IgG (LI-COR Biotechnology, Lincoln, NE), and visualized on the ODYSSEY infrared imaging system (LI-COR Biotechnology).
LDL variant analysis
LDL subfractions were evaluated for total cholesterol content using a fluorometric assay kit (Cayman Chemical, Ann Arbor, MI) and triglyceride content using a colorimetric assay kit (Cayman Chemical) according to the manufacturer’s instructions.
Flow cytometry
HepG2 cells were prepared and treated in the PCSK9-NLuc uptake assay as described above, but were incubated with PCSK9-NLuc and its variants in the absence of the lysosomal inhibitor, chloroquine. Four hours after PCSK9-NLuc incubation, cells were washed with PBS, dissociated with 0.5 mM EDTA in PBS, transferred to 1.5 ml centrifuge tubes, and passaged through a 21-gauge needle thrice to minimize cell clumping. Cells were collected by centrifugation (300 g for 5 min), washed twice with solution A (PBS containing 0.5% BSA and 1 g/l glucose), and then labeled with 5 μl LDLR Alexa Fluor 488 conjugated antibody (FAB2148; R&D Systems) in 200 μl solution A for 30–40 min with gentle rotation at room temperature in the absence of light. Cells were washed again with solution A and then subjected to flow cytometric analysis using a FACS Calibur (Becton Dickinson, Franklin Lakes, NJ). Data analysis was performed using FlowJo v10 software (Ashland, OR), focusing on single viable cells as identified from forward-scatter and side-scatter gating.
Data analysis
Data and statistical analyses were performed using Prism 7.0 (GraphPad Software, La Jolla, CA). In general, raw luminescence or fluorescence outputs were normalized to positive and negative control samples, and experiments were performed in triplicate and repeated at least three times. P values indicate the results of unpaired t-tests with Welch’s correction. Error bars in all graphs other than flow cytometry represent SD. Error bars in graphs displaying data from flow cytometry represent 95% CIs of the mean.
RESULTS
A cell-based luciferase assay to measure PCSK9 uptake
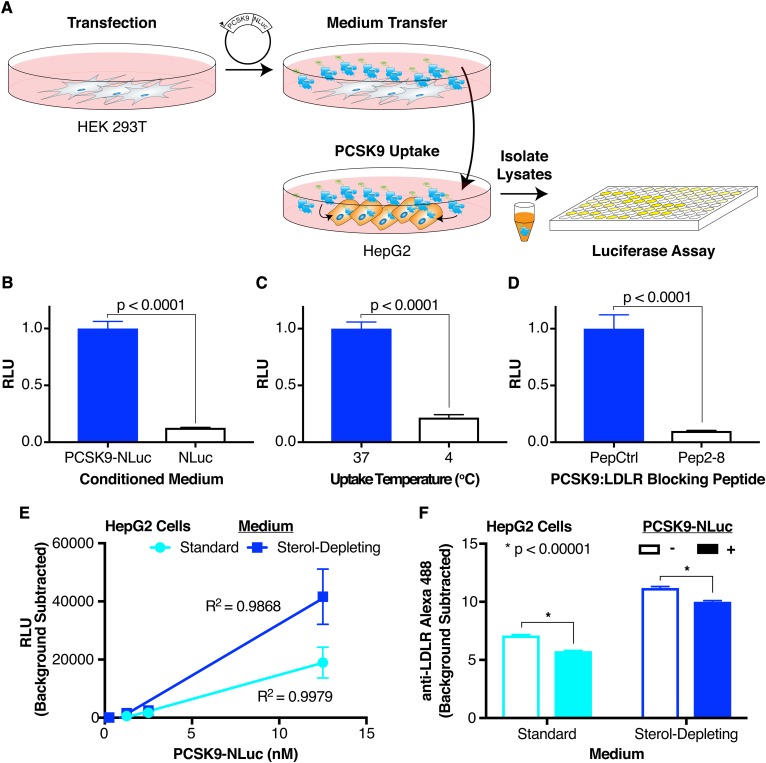
To investigate factors that affect PCSK9 internalization, we developed a system that would allow for a rapid luciferase-based readout as a proxy for PCSK9 uptake (Fig. 1A). Briefly, we overexpress a PCSK9-NLuc fusion in HEK 293T cells, then collect and transfer the conditioned medium to HepG2 cells in the presence of a lysosomal inhibitor to prevent degradation of the reporter (43). After coincubation, we dissociate and harvest the cells with trypsin, removing any uninternalized cell surface-bound PCSK9, and subject the cellular lysates to a luciferase assay. The luciferase reporter allows a quantitative evaluation of the PCSK9-NLuc concentration in the conditioned medium prior to transfer so as to ensure uniform inputs in any given experiment.
Fig. 1.
Luciferase-based assay for PCSK9 uptake. A: Overall schematic of assay. A PCSK9-NLuc fusion is heterologously expressed in HEK 293T cells, and the conditioned medium then transferred to HepG2 cells. Cell lysates are collected after the exogenous PCSK9-NLuc is taken up into cells, with the amount internalized quantified by luminescence. B: Relative luminescence of HepG2 cell lysates treated with PCSK9-NLuc (12.5 nM, blue bar) or an equivalent amount of free NLuc (open bar). Values are normalized to 1 for the PCSK9-NLuc treatment. C: Relative luminescence of HepG2 cell lysates treated with PCSK9-NLuc (12.5 nM) at an incubation temperature of 37°C (blue bar) or 4°C (open bar). Values are normalized to 1 for the 37°C condition. D: Relative luminescence of HepG2 cell lysates treated with PCSK9-NLuc (12.5 nM) and coincubated with an inactive peptide control (PepCtrl, blue bar) or a PCSK9:LDLR blocking peptide (Pep2-8, open bar). Values are normalized to 1 for the inactive peptide control. E: Luminescence, corrected for assay background, of equivalent amounts of HepG2 cell lysates treated with increasing concentrations of PCSK9-NLuc, stratified by the presence (blue) or absence (cyan) of sterol-starvation. Data include three separate experiments without normalization. Linear regression lines with the appropriate goodness-of-fit (R2) values are shown. F: Alexa Fluor 488 fluorescence, as a proxy for cell surface LDLR expression, and as determined by flow cytometric analysis, of HepG2 cells treated for 4 h with (filled bars) or without (open bars) PCSK9-NLuc and stratified by the presence (blue) or absence (cyan) of sterol-starvation.
The assay exhibited excellent specificity for the biologic process at hand. Experiments using conditioned medium containing equivalent concentrations of free NLuc as a control for nonspecific uptake reached only 13% of that containing the PCSK9-NLuc fusion [Fig. 1B, compare open bar, μRLU = 0.127 (RLU, relative luminescence unit), σRLU = 0.00348, to blue bar, μRLU = 1, σRLU = 0.0623]. Incubation of the recipient cells with the PCSK9-NLuc-conditioned medium at 4°C, so as to allow cell surface association of PCSK9 but prevent endocytosis (44), showed only 22% of the activity of the endocytosis-permissive 37°C incubation (Fig. 1C, compare open bar, μRLU = 0.217, σRLU = 0.0256, to blue bar, μRLU = 1, σRLU = 0.0584). Consistent with prior studies showing the dependence of PCSK9 function on PCSK9:LDLR binding (35), the assay required an intact PCSK9:LDLR interaction. Coincubation of the conditioned medium in the presence of Pep2-8, a PCSK9:LDLR(EGF-A) blocking peptide (45), resulted in a readout only 10% of the inactive peptide, PepCtrl (Fig. 1D, compare open bar, μRLU = 0.0998, σRLU = 0.00434, to blue bar, μRLU = 1, σRLU = 0.123).
Our assay also showed a linear correlation (R2 ≈ 0.99) between input PCSK9 concentration and luminescence outputs over physiologic PCSK9 levels (46) from approximately 1 to 12.5 nM (Fig. 1D). Notably, at the concentrations of PCSK9 used, we were unable to detect internalized PCSK9-NLuc via Western blot with a PCSK9 antibody (data not shown), illustrating the sensitivity of this assay. Upregulation of cell surface LDLR on HepG2 cells by pretreatment in sterol-depleting medium (47) increased luciferase output [Fig. 1D, blue, for (PCSK9) = 12.5 nM: μRLU = 41627, σRLU = 9499] over treatment in standard medium [Fig. 1D, cyan, for (PCSK9) = 12.5 nM: μRLU = 18,955, σRLU = 5,338], further exhibiting the dependence of our assay on the PCSK9:LDLR interaction. To confirm that our system recapitulated the downstream physiologic effect on the LDLR, we evaluated cell-surface LDLR expression via flow cytometry. Our results show that treatment of HepG2 cells for 4 h with physiologic PCSK9-NLuc concentrations (12.5 nM) appropriately reduced cell-surface LDLR (Fig. 1E), both in cells cultured in sterol-depleting medium [Fig. 1E, blue, PCSK9 untreated (open bar): μRFU = 11.20, 95% CI = 11.06–11.34; PCSK9 treated (filled bar): μRFU = 10.00, 95% CI = 9.86–10.14] or standard medium [Fig. 1E cyan, PCSK9 untreated (open bar): μRFU = 7.11, 95% CI = 7.02–7.20; PCSK9 treated (filled bar): μRFU = 5.76, 95% CI = 5.68–5.84].
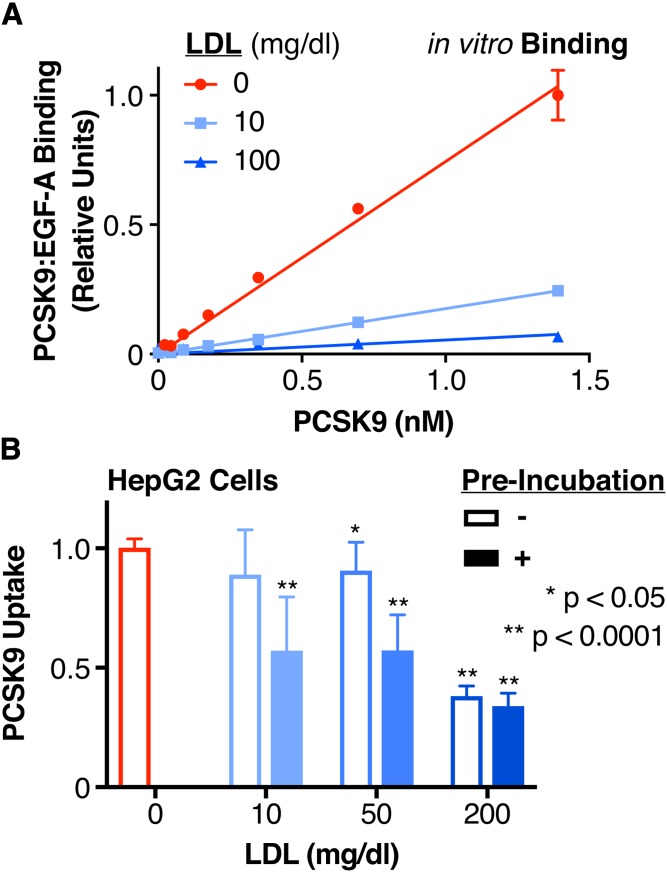
LDL inhibits the binding of PCSK9 to the LDLR in vitro more potently than PCSK9 uptake in tissue culture
We first sought to evaluate the inhibitory effect of LDL on complex formation between PCSK9 and the EGF-A domain of the LDLR. Prior FRET-based studies have shown that LDL can disrupt the binding of PCSK9 to the LDLR (33), though gel-shift assays have suggested that this mechanism does not involve a direct steric blockade between PCSK9 and the LDLR (32). Given that the latter study utilized a GOF D374Y variant with increased affinity for the EGF-A domain (35), we employed a commercially available ELISA using near physiologic levels of WT PCSK9 and saturating levels of EGF-A domain to probe the interaction (48). We found that both subphysiologic and physiologic levels of LDL potently inhibited PCSK9:LDLR(EGF-A) binding in a dose-dependent manner (Fig. 2A). Specifically, we found that at PCSK9 concentrations of 1.4 nM (100 ng/ml), 10 mg/dl of LDL inhibited PCSK9:LDLR(EGF-A) domain binding by 75% (Fig. 2A, cyan, μEGF-A Binding = 0.245, σEGF-A Binding = 0.0145), while 100 mg/dl of LDL inhibited binding by 93% (Fig. 2A, blue, μEGF-A Binding = 0.0667, σEGF-A Binding = 0.00369).
Fig. 2.
Effect of LDL on the PCSK9:LDLR interaction and PCSK9 uptake. A: Relative binding of increasing concentrations of PCSK9 to the EGF-A domain of LDLR in the absence (red) or presence of subphysiologic (10 mg/dl, cyan) or physiologic (100 mg/dl, blue) LDL, as measured by a commercially available PCSK9-LDLR ELISA assay. Raw absorbance data are normalized to 1 for the highest PCSK9 concentration in the absence of LDL and 0 in the absence of PCSK9. The linear regression lines are shown, with the extra-sum-of-squares F test indicating different slopes at P < 0.0001. B: Relative uptake, as measured by luminescence assay, of PCSK9-NLuc by HepG2 cells in the presence of increasing concentrations of LDL, shown by sequential addition (open bars) or preincubation (filled bars) of PCSK9 with LDL. Raw luminescence data are normalized to 1 for PCSK9-NLuc treatment in the absence of LDL and 0 in the absence of PCSK9-NLuc uptake. P values indicate comparison to the control (no LDL, red) arm.
As PCSK9 uptake is both dependent on the presence of the LDLR (49, 50) and increased by mutations (such as D374Y), which increase the binding affinity of PCSK9 for the LDLR (33), we sought to extend our in vitro findings to a cellular setting. We thus employed our luciferase-based system to evaluate the effects of LDL on PCSK9 uptake. We incubated conditioned PCSK9-NLuc medium with chloroquine-treated HepG2 cells in the presence of subphysiologic (10 mg/dl), low (50 mg/dl), and high (200 mg/dl) concentrations of LDL. In contrast to the results of the in vitro ELISA assay, LDL inhibited PCSK9 uptake modestly, with a marked effect only at the high concentration [Fig. 2B, open bars, for (LDL) = 200 mg/dl: μPCSK9 Uptake = 0.381, σPCSK9 Uptake = 0.0424]. We repeated our experiments by preincubating the conditioned PCSK9-NLuc medium with LDL prior to adding the mixture to HepG2 cells. This partially rescued the inhibitory activity of LDL, resulting in more impressive reductions of PCSK9 uptake [Fig. 2B, filled bars, for (LDL) = 10 mg/dl: μPCSK9 Uptake = 0.571, σPCSK9 Uptake = 0.225; for (LDL) = 50 mg/dl: μPCSK9 Uptake = 0.573, σPCSK9 Uptake = 0.148; for (LDL) = 200 mg/dl: μPCSK9 Uptake = 0.339, σPCSK9 Uptake = 0.0545]. The reductions in PCSK9 uptake were particularly notable for the 10 mg/dl and 50 mg/dl concentrations, though not quite to the levels of inhibition seen in the ELISA alone. We conclude from this data that a cell-based factor partially abrogates the ability of LDL to reduce PCSK9 uptake into cells. Furthermore, this cell-based factor appears most effective when PCSK9 and LDL are presented simultaneously to the cells.
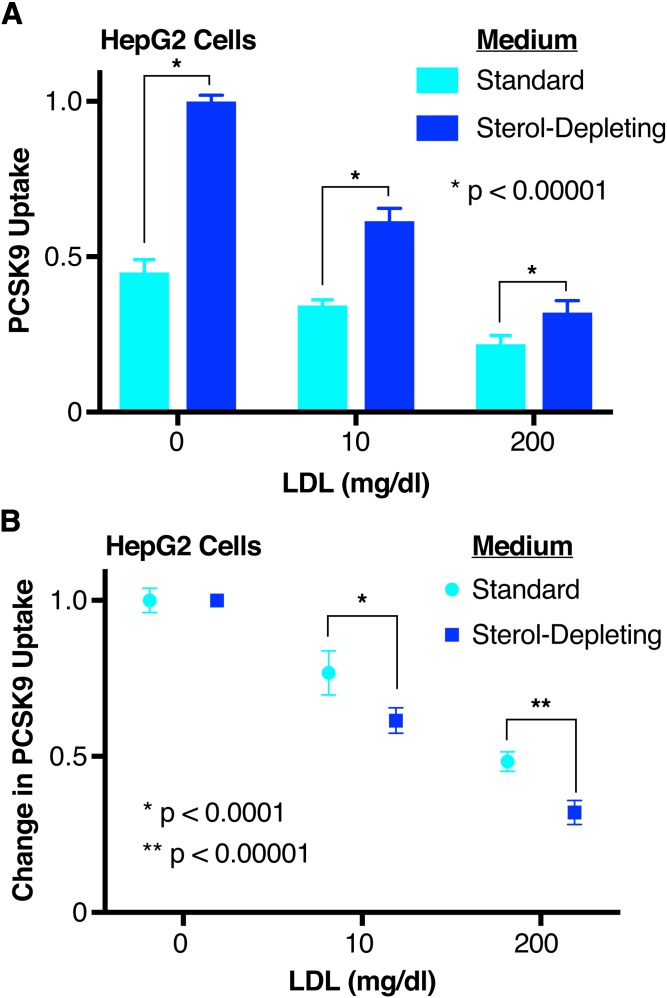
LDL-mediated inhibition of PCSK9 uptake is dependent upon LDLR expression
To investigate the mechanism by which LDL acts to decrease PCSK9 function, we asked whether PCSK9 uptake was dependent upon LDLR expression levels. Previous work has shown that LDL competes with PCSK9 for binding to LDLR (33), but PCSK9 uptake per se can occur in the absence of PCSK9:LDLR binding, even though the presence of LDLR itself is still required (32, 50). We thus treated HepG2 cells with standard or sterol-depleting medium to alter LDLR expression, and evaluated uptake of PCSK9 preincubated with subphysiologic (10 mg/dl) and high (200 mg/dl) LDL. LDL dose-dependently inhibited PCSK9 uptake for both cell treatments (Fig. 3A). Intriguingly, LDL served as a more effective inhibitor, both in absolute (Fig. 3A) and relative terms (Fig. 3B), in the cells with upregulated LDLR expression by sterol starvation. Specifically, 10 mg/dl LDL, relative to no LDL, decreased PCSK9 uptake by 23% in the cells cultured in standard medium (Fig. 3B, cyan, μPCSK9 Uptake = 0.768, σPCSK9 Uptake = 0.0707), but by 37% in the sterol-starved cells (Fig. 3B, blue, μPCSK9 Uptake = 0.615, σPCSK9 Uptake = 0.0411). Similarly, 200 mg/dl LDL, relative to no LDL, decreased PCSK9 uptake by 52% in the cells cultured in standard medium (Fig. 3B, cyan, μPCSK9 Uptake = 0.484, σPCSK9 Uptake = 0.0311), but by 68% in the sterol-starved cells (Fig. 3B, blue, μPCSK9 Uptake = 0.321, σPCSK9 Uptake = 0.0387). Overall, our results suggest that the inhibition of PCSK9 uptake by LDL is at least partially LDLR dependent, and are also consistent with a mechanism whereby LDL inhibits PCSK9 uptake by disrupting the PCSK9:LDLR interaction.
Fig. 3.
Effect of LDLR upregulation on LDL-mediated PCSK9 uptake inhibition. A: Relative uptake, as measured by luminescence assay, of PCSK9-NLuc by HepG2 cells in the presence of increasing concentrations of LDL, and stratified by the presence (blue) or absence (cyan) of sterol-starvation. Note that, in this experiment, LDL was preincubated with PCSK9-NLuc prior to coincubation with the cells. Raw luminescence data are normalized to 1 for PCSK9-NLuc treatment in the absence of LDL and presence of sterol-starvation, and 0 in the absence of PCSK9-NLuc uptake. B: Same experiment (and source data) as in A, but each medium condition is normalized separately to 1 (in the absence of LDL) to highlight the relative impact of LDL under each.
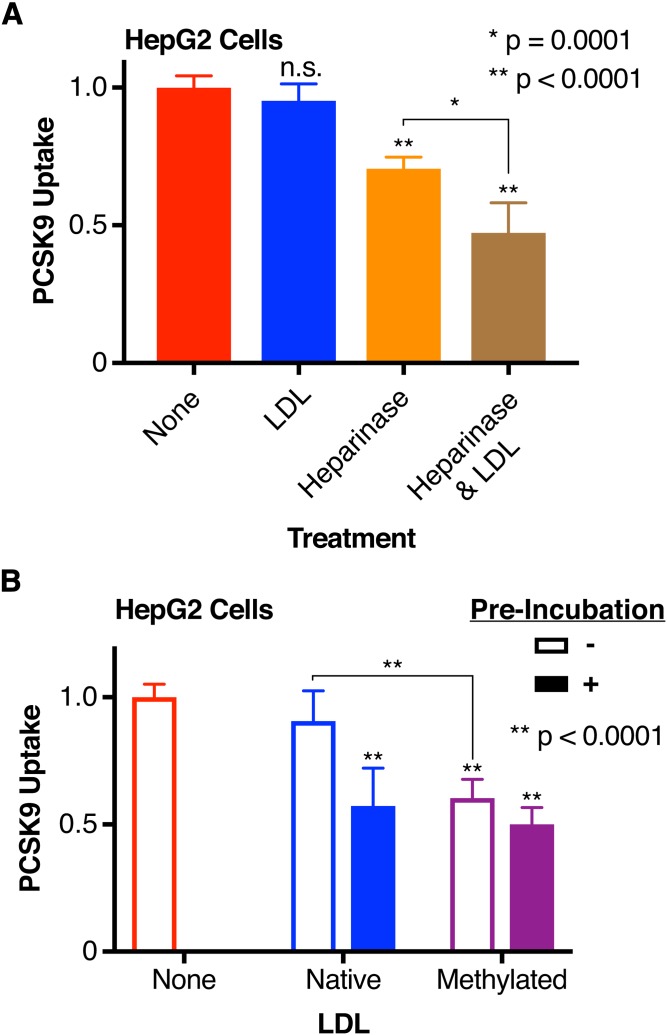
HLMs negatively regulate LDL as an inhibitor of PCSK9 uptake
Our initial results showing a preincubation requirement for LDL to exert its inhibitory effect suggested that a cell-based factor might be competing with LDL to bind PCSK9. Thus, we sought to identify this factor. Given that size and density of LDL have been identified as variables that alter the atherogenicity of the LDL particle (51–54), we investigated whether LDL density subfractions might differentially affect PCSK9 uptake. However, we found no difference in PCSK9:LDLR(EGF-A) binding in vitro nor in cell-based PCSK9 uptake when comparing small dense LDL to large LDL (supplemental Fig. S1). We next hypothesized that HSPGs on the cell surface might mediate this effect, given their recent description as coreceptors for PCSK9 and affinity for the PCSK9 prodomain (37). To test this, we evaluated PCSK9 uptake with reagents expected to modify HSPG activity. First, we treated HepG2 cells with heparinase to remove cell surface HLMs (including HSPGs) prior to evaluating the effect of LDL on our PCSK9-NLuc uptake assay. Unsurprisingly, and consistent with HSPGs acting as coreceptors for PCSK9 (37), heparinase treatment in the absence of LDL reduced PCSK9-NLuc uptake by about 30% (Fig. 4A, orange, μPCSK9 Uptake = 0.705, σPCSK9 Uptake = 0.0424), with its absolute effect similar to that of preincubation of PCSK9 with LDL. Physiologic levels of LDL (50 mg/dl) added to heparinase-treated cells, but not preincubated with PCSK9, reduced PCSK9 uptake further (Fig. 4A, brown, μPCSK9 Uptake = 0.4722, σPCSK9 Uptake = 0.110), to about 50% of the total for untreated cells (Fig. 4A, red, μPCSK9 Uptake = 1, σPCSK9 Uptake = 0.0431). This suggests that LDL can inhibit PCSK9 uptake even in the absence of HLMs, and remains consistent with our prior data showing that such inhibition is at least partially dependent on LDLR expression.
Fig. 4.
Effect of the LDL:HLM interaction on LDL-mediated PCSK9 uptake inhibition. A: Relative uptake, as measured by luminescence assay, of PCSK9-NLuc by HepG2 cells coincubated with LDL (blue), treated with heparinase (orange), or both (brown). Raw luminescence data are normalized to 1 for PCSK9-NLuc coincubation in the absence of LDL or heparinase treatment and 0 in the absence of PCSK9-NLuc. P values indicate comparison to the control arm (no treatment, red) unless otherwise indicated. B: Relative uptake, as measured by luminescence assay, of PCSK9-NLuc by HepG2 cells in the presence of either native (blue) or reductively methylated (purple) LDL, stratified by sequential addition (open bars) or preincubation (filled bars) of PCSK9 with LDL. Raw luminescence data are normalized to 1 for PCSK9-NLuc treatment in the absence of LDL and 0 in the absence of PCSK9-NLuc uptake. P values indicate comparison to the control arm (no LDL, red) unless otherwise indicated.
As native LDL binds negatively charged sulfated glycoproteins (such as HSPGs) (40, 55, 56), we also hypothesized that the direct interaction between LDL and HLMs was necessary to explain our differential effects on preincubation. To look specifically at this interaction, we subjected LDL to reductive methylation, which reduces the affinity of LDL to bind either HLMs or the LDLR itself (40, 41). Importantly, iodixanol gradient purifications of these LDL species confirmed that the methylated LDL retained the ability to bind PCSK9 (supplemental Fig. S2). Consistent with our hypothesis, methylated LDL, regardless of preincubation status, retained the ability to inhibit PCSK9 uptake [Fig. 4B, purple, no preincubation (open bar): μPCSK9 Uptake = 0.603, σPCSK9 Uptake = 0.0737; preincubation (filled bar): μPCSK9 Uptake = 0.501, σPCSK9 Uptake = 0.0653] to levels similar to preincubation with native LDL [Fig. 4B, blue, preincubation (filled bar): μPCSK9 Uptake = 0.573, σPCSK9 Uptake = 0.148]. Cells not treated with LDL, beyond that present in the medium, had higher uptake (Fig. 4B, red open bar: μPCSK9 Uptake = 1, σPCSK9 Uptake = 0.0513) than cells treated with the methylated LDL (Fig. 4B, purple bars), suggesting that our findings were specific to the disruption of the LDL:HLM interaction rather than disruption of the LDL:LDLR interaction. This is consistent with prior work showing that LDL can inhibit the binding of PCSK9 to the LDLR, even when the LDLR cannot itself bind LDL (32). Overall, when taken together with data from heparinase-treated cells, these data suggest that LDL-mediated inhibition of PCSK9 uptake is in part negatively regulated by the LDL:HLM interaction.
HLMs also interact with PCSK9 to negatively regulate LDL as an inhibitor of PCSK9 uptake
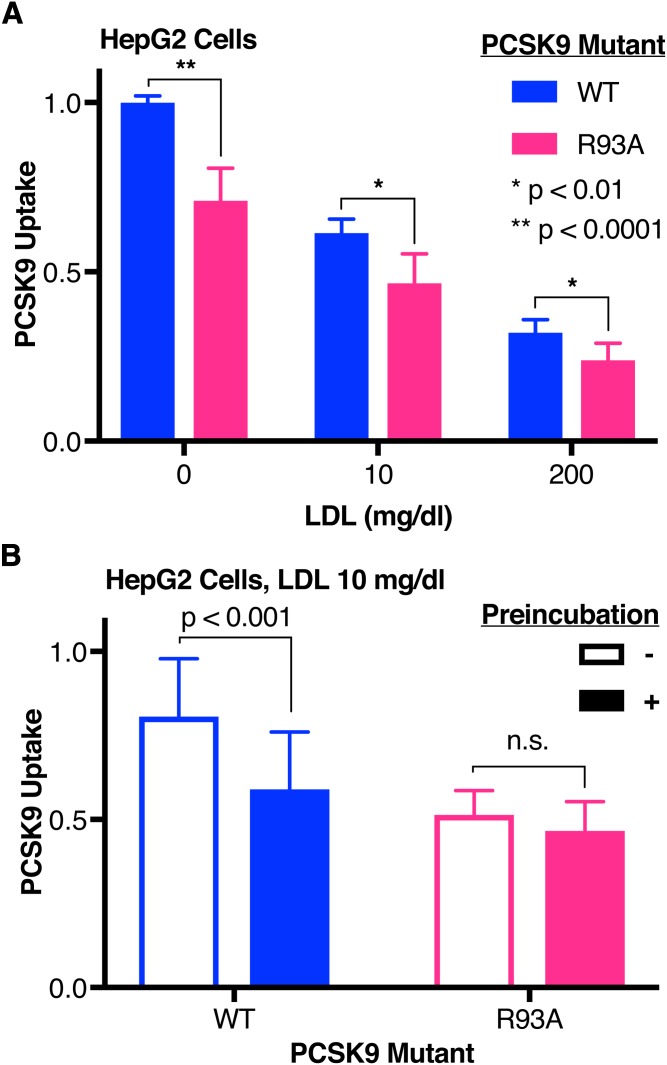
Having shown the importance of the LDL:HLM interaction, we next asked whether the interaction between PCSK9 and HLMs could also modulate the ability of LDL to inhibit PCSK9 uptake. Here, we used the R93A variant of PCSK9, which removes an arginine residue from the described HSPG binding motif in the prodomain and decreases the binding affinity of PCSK9 for heparin (37). As expected, in the absence of LDL, uptake of R93A PCSK9 is less robust (Fig. 5A, magenta, μPCSK9 Uptake = 0.710, σPCSK9 Uptake = 0.0962) than that of WT PCSK9 (Fig. 5A, blue, μPCSK9 Uptake = 1, σPCSK9 Uptake = 0.0202). LDL, when preincubated with PCSK9, inhibited uptake of both WT and R93A PCSK9 similarly (Fig. 5A). However, at subphysiologic levels of LDL (10 mg/dl), the requirement for preincubation to inhibit PCSK9 uptake was no longer present for the R93A variant [Fig. 5B, magenta, no preincubation (open bar): μPCSK9 Uptake = 0.514, σPCSK9 Uptake = 0.0723; preincubation (filled bar): μPCSK9 Uptake = 0.466, σPCSK9 Uptake = 0.0870]. These data suggest that the PCSK9:HLM interaction partly abrogates the effect of LDL as an inhibitor of PCSK9 uptake.
Fig. 5.
Effect of the PCSK9:HLM interaction on LDL-mediated PCSK9 uptake inhibition. A: Relative uptake, as measured by luminescence assay, of WT (blue) or R93A (magenta) PCSK9-NLuc by HepG2 cells incubated with increasing concentrations of LDL. Raw luminescence data are normalized to 1 for WT PCSK9-NLuc treatment in the absence of LDL and 0 in the absence of PCSK9-NLuc. B: Relative uptake, as measured by luminescence assay, of WT (blue) or R93A (magenta) PCSK9-NLuc by HepG2 cells in the presence of 10 mg/dl LDL, shown by sequential addition (open bars) or preincubation (filled bars) of PCSK9 with LDL. Raw luminescence data are normalized to 1 for WT PCSK9-NLuc treatment in the absence of LDL and 0 in the absence of PCSK9-NLuc.
The full-length PCSK9 prodomain binds LDL and blocks the inhibitory effect of LDL on PCSK9 uptake
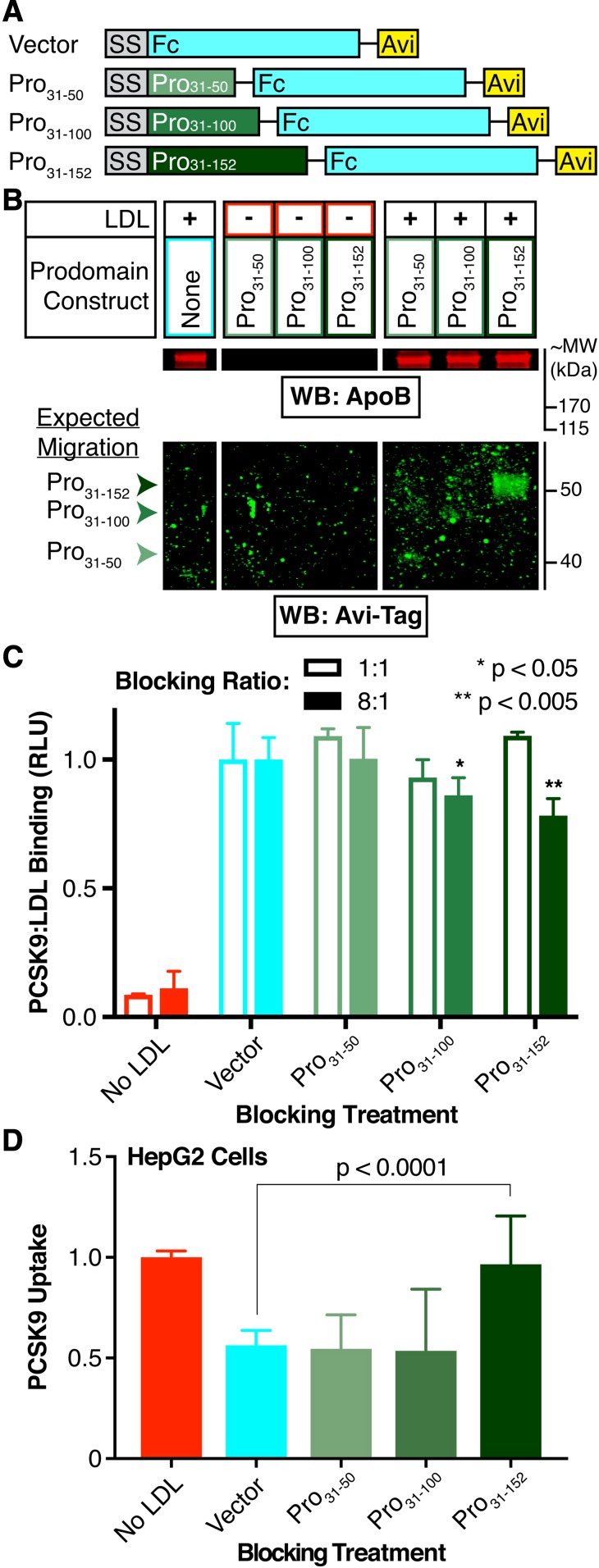
Given that both the LDL:HLM and PCSK9:HLM interactions dilute the inhibitory effect of LDL on PCSK9 uptake, we hypothesized that these interactions might directly compete with the PCSK9:LDL interaction. Prior work has shown the N terminus of the prodomain, specifically an unstructured region encompassing residues 31-52, as necessary for LDL binding (32). Deletion of these residues also improves PCSK9 binding to soluble LDLR (35) and increases the ability of PCSK9 to downregulate LDLR on cells (36). The binding site for HSPGs is also located on the PCSK9 prodomain, encompassing an arginine-rich motif spanning residues 93-139 (37). To ask whether the N-terminal prodomain residues were sufficient to mediate the inhibitory effect of LDL on PCSK9 uptake (and therefore nonoverlapping with the HSPG binding site), we created secreted PCSK9 prodomain variants to use in competition experiments. We designed our prodomain constructs as N-terminal fusions to secretable human Fc domains, a strategy similar to one previously used to permit secretion of the prodomain in the absence of the PCSK9 catalytic domain (57). Using the previously described binding sites as our guide, we generated three sequential C-terminal truncations of the prodomain encompassing residues 31-50, 31-100, and 31-152 (Fig. 6A). We expressed these prodomain-Fc fusions via transient transfections in 293T cells, quantified their production via a commercially available ELISA against the Fc tag, and tested each in various experiments.
Fig. 6.
Rescue of LDL-mediated inhibition of PCSK9 uptake by exogenous prodomain. A: Schematic of exogenous prodomain constructs. All constructs are driven by a cytomegalovirus promoter (not shown), and include the PCSK9 signal sequence (SS, gray), C-terminal Fc (cyan), and Avi (yellow) tags. The PCSK9 prodomain variants (shades of green) include residues (Pro)31-50, 31-100, or 31-152 (full-length). B: Detection of apoB and the Avi-Tag by Western blot, after SDS-PAGE analysis, of the LDL-containing fraction or its volumetric equivalent from gradient ultracentrifugation of in vitro binding assays between the prodomain constructs and LDL. Note that all lanes for a given antibody have equal exposures. C: Relative luminescence of the LDL-containing fraction or its volumetric equivalent from gradient ultracentrifugation of in vitro binding assays between PCSK9-NLuc and LDL in the absence (cyan) or presence (shades of green) of the indicated prodomain constructs. Open bars indicate a prodomain construct:PCSK9-NLuc ratio of 1:1, and closed bars indicate a prodomain construct:PCSK9-NLuc ratio of 8:1. Values are normalized to 1 for treatments in absence of blocking construct (cyan). D: Relative uptake, as measured by luminescence assay, of PCSK9-NLuc by HepG2 cells in the presence of LDL and the indicated exogenous prodomain Fc-Avi fusion. Note that, in these experiments, all LDL was preincubated with the PCSK9-NLuc and exogenous prodomain. Raw luminescence data are normalized to 1 for PCSK9-NLuc treatment in the absence of LDL and 0 in the absence of PCSK9-NLuc uptake.
First, we evaluated the ability for each prodomain variant to bind LDL in vitro. Here, we incubated the conditioned medium containing each fusion with LDL and then performed a three-layer iodixanol gradient ultracentrifugation to isolate the LDL-containing fractions, as per prior protocols (32). We then subjected the LDL-containing fractions to SDS-PAGE and Western blot using an antibody against the Avi tag. Our results show that the full-length prodomain comigrates with LDL along the density gradient, with no clear signal for the truncated prodomain variants (Fig. 6B). Importantly, the full-length prodomain fusion did not naturally migrate to the same layer as LDL in the iodixanol gradient, as shown by ultracentrifugation of the prodomains in the absence of LDL (Fig. 6B). To evaluate for competition of the exogenous prodomain for PCSK9:LDL binding, we coincubated the prodomain variants with PCSK9-NLuc in the LDL binding assay prior to LDL isolation. Luciferase assays of the LDL-containing fractions showed no differences in PCSK9-NLuc binding at an equimolar (i.e., 1:1) ratio of blocking construct to PCSK9-NLuc, but did show a modest decrease in PCSK9-NLuc binding at an 8-fold excess of blocking construct, which was most notable for the full-length prodomain (Fig. 6C, dark green filled bar, μRLU = 0.783, σRLU = 0.0655). To extend these findings to cells, and also evaluate the functional consequence of these prodomain variants, we then performed similar blocking experiments with these constructs in our PCSK9 uptake assays. Here, we incubated excess amounts of each construct with the tagged PCSK9-NLuc and LDL prior to evaluating the effects on PCSK9 uptake. Our results show that complete rescue of PCSK9 uptake required the full (i.e., nontruncated) prodomain (Fig. 6D, dark green, μPCSK9 Uptake = 0.966, σPCSK9 Uptake = 0.240). These data suggest a requirement for the entire prodomain, including the HSPG binding site, to permit the LDL particle to inhibit PCSK9 uptake. Moreover, these data are consistent with the prodomain:LDL binding interaction being sufficient for such an effect to occur.
The GOF S127R PCSK9 variant has increased affinity for heparin and increased uptake sensitivity to heparinase
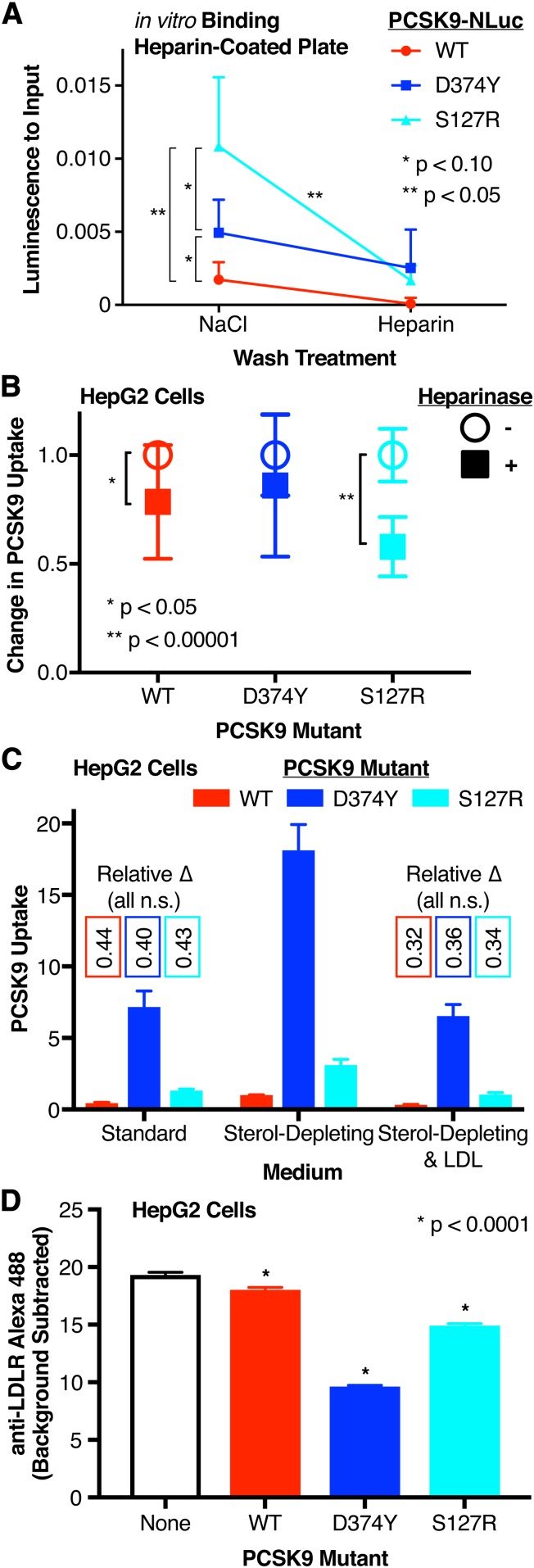
The initial discovery of PCSK9 as a Mendelian cause of familial hypercholesterolemia came from the PCSK9 S127R mutant (1). Biochemical characterization of this mutant has remained puzzling; both self-proteolysis and secretion are required for PCSK9 to exert its maximal effect on the LDLR, yet we and others have shown that the S127R variant is deficient in both proteolytic function and secretion (24, 58). Given the location of S127 within the prodomain and near the HSPG binding motif, we hypothesized that a positively charged arginine at this position could increase the affinity of PCSK9 for the negatively charged HSPGs, potentially explaining the GOF phenotype. To address this, we first evaluated the affinity of PCSK9 S127R for heparin in an in vitro assay. We generated WT, S127R, and D374Y PCSK9-NLuc constructs and produced the corresponding proteins via transient transfections in 293T cells. We then added the conditioned media to a heparin-coated plate and measured the relative luminescence of the PCSK9 inputs. We sequentially washed the plates with a high salt solution followed by a competitive free heparin solution, reading out luminescence after each step to monitor the remaining bound PCSK9-NLuc. Consistent with our hypothesis, after the salt wash, significantly more S127R PCSK9-NLuc remained bound (Fig. 7A, cyan, μRLU-Bound/RLU-Input = 0.0108, σRLU-Bound/RLU-Input = 0.00473), compared with WT (Fig. 7A, red, μRLU-Bound/RLU-Input = 0.00173, σRLU-Bound/RLU-Input = 0.00199) or the D374Y variant (Fig. 7A, blue, μRLU-Bound/RLU-Input = 0.00494, σRLU-Bound/RLU-Input = 0.00227). Furthermore, the S127R variant approached the basal level after the competitive wash with excess free heparin (Fig. 7A).
Fig. 7.
Effect of PCSK9 GOF variants on heparin binding and PCSK9 uptake. A: Relative in vitro binding affinity of WT (red), D374Y (blue), and S127R (cyan) PCSK9-NLuc to heparin after high-salt and heparin competition washes, as measured by luminescence assay. The Y axis shows relative luminescence compared with initial input on the heparin-coated plate. Only one side of each set of error bars is shown for clarity. B: Relative PCSK9 uptake, as measured by luminescence assay, by untreated (open circles) and heparinase-treated (filled squares) HepG2 cells incubated with WT (red), D374Y (blue), or S127R (cyan) PCSK9-NLuc-conditioned medium. Luminescence data are normalized to 1 for each PCSK9-NLuc treatment and 0 in the absence of PCSK9-NLuc treatment. C: Relative uptake, as measured by luminescence assay, of WT (red), D374Y (blue), or S127R (cyan) PCSK9-NLuc by HepG2 cells, stratified by sterol-starvation or LDL (200 mg/dl) coincubation. Luminescence data are normalized to 1 for WT PCSK9-NLuc treatment of sterol-starved cells and 0 in the absence of PCSK9-NLuc treatment. Numbers showing the relative change for each variant (compared with the uptake of that variant for sterol-starved cells) are listed. D: Alexa Fluor 488 fluorescence, as a proxy for cell surface LDLR expression, and as determined by flow cytometric analysis, of HepG2 cells treated in the absence (black) or presence of WT (red), D374Y (blue), or S127R (cyan) PCSK9-NLuc.
We then asked whether these findings would translate to cellular uptake of PCSK9. To do so, we repeated our cellular uptake assays using the WT, S127R, and D374Y PCSK9-NLuc variants with heparinase-treated and untreated HepG2 cells. Our results show that the relative decrease in uptake with the heparinase treatment was significantly greater for the S127R variant (Fig. 7B, cyan filled square, μPCSK9 Uptake = 0.579, σPCSK9 Uptake = 0.137) as compared with WT (Fig. 7B, red filled square, μPCSK9 Uptake = 0.785, σPCSK9 Uptake = 0.262) or D374Y PCSK9 (Fig. 7B, blue filled square, μPCSK9 Uptake = 0.861, σPCSK9 Uptake = 0.328). As expected, the degree of uptake of D374Y PCSK9 (Fig. 7C, blue, sterol-depleting medium, μPCSK9 Uptake = 18.1, σPCSK9 Uptake = 1.80) was about 18-fold greater than that of WT PCSK9 (Fig. 7C, red, sterol-depleting medium, μPCSK9 Uptake = 1, σPCSK9 Uptake = 0.0202), consistent with the increased affinity of the D374Y variant for the LDLR EGF-A domain (45). Intriguingly, S127R PCSK9 uptake (Fig. 7C, cyan, sterol-depleting medium, μPCSK9 Uptake = 3.11, σPCSK9 Uptake = 0.394) was about 3-fold greater than that of WT PCSK9. The potency of each species, with regard to effect on cell surface LDLR expression, also followed the relative differences in PCSK9 uptake, as evaluated by flow cytometry (Fig. 7D). Moreover, each species behaved similarly when tested in the absence of sterol-starvation or when preincubated with high levels of LDL (200 mg/dl) (Fig. 7C). Together, these data are consistent with the S127R mutant having higher affinity for cell surface HLMs (such as HSPGs), with this increased affinity driving increased uptake and ultimate function. However, our results suggest against a specific LDL interaction playing a key role in the pathogenesis of the S127R variant.
DISCUSSION
In this study, we examined the role of LDL as an inhibitor of PCSK9 uptake, developing a sensitive luciferase-based assay to specifically look at this step of the PCSK9 lifecycle. Consistent with prior studies, we found that LDL both inhibits PCSK9 uptake in cells and inhibits PCSK9:LDLR(EGF-A) binding in vitro (32, 33). We found that the in vitro binding results were more impressive than the effects in cells, prompting us to search for a cell-based factor that could modulate either PCSK9:LDLR binding or PCSK9 uptake directly. Though PCSK9 uptake is generally thought to be dependent upon the PCSK9:LDLR interaction (49), previous work has shown that such uptake can occur in the absence of direct PCSK9:LDLR binding, but still requires the presence of the LDLR (50). This suggests that an alternative PCSK9 internalization mechanism may exist. Here, we found that the inhibition of PCSK9 uptake by LDL was modulated by cell-surface HLMs in addition to LDLR expression itself. Additionally, the regulation by HLMs was dependent upon both the LDL:HLM and PCSK9:HLM interactions, as disruption of either augmented the ability of LDL to inhibit PCSK9 uptake. Furthermore, rescue of PCSK9 uptake from LDL-mediated inhibition occurred via competition from an exogenous full-length prodomain-Fc fusion encompassing a described HSPG binding site. When taken together, these data are consistent with the PCSK9:HLM and PCSK9:LDL interactions competing against each other, with both complexes potentially existing in a fluid equilibrium.
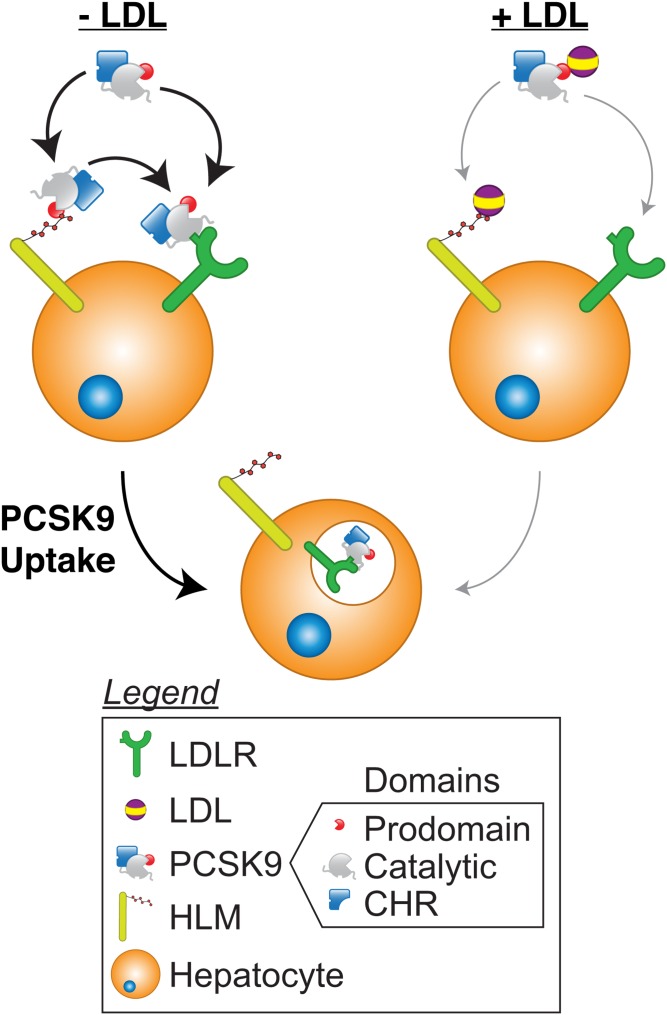
Though speculative, it is tempting to propose a model to incorporate these data into the known literature (Fig. 8). In the absence of LDL, PCSK9 may bind to the LDLR directly or be presented to the LDLR via binding to HLMs. Whether internalization can occur via the PCSK9:HLM interaction by itself remains to be determined. HLMs may also complex with available LDL to block the PCSK9:HLM interaction, thus reducing the ability of HLMs to present PCSK9 to the LDLR. In addition, the PCSK9:LDL complex likely has reduced affinity for free HLMs. As evidenced by the reduction in PCSK9 uptake by LDL in heparinase-treated cells, LDL can also mediate its inhibition through an HLM-independent mechanism, and likely does so by inhibiting the PCSK9:LDLR interaction allosterically. On the basis of our data alone, however, it remains unclear whether the HLM-dependent mechanism also requires PCSK9:LDLR binding or the presence of the LDLR itself. It is tantalizing to conjecture that HLMs may offer a way to shuttle PCSK9 into an endocytic vesicle without the need for the PCSK9:LDLR interaction, whereby PCSK9 can then direct itself into acidified endosomes (50, 59) and induce an interaction with the LDLR via its cysteine-histidine rich domain, which has known affinity for the LDLR at lower pH (60, 61). This could effectively bypass the need for the interaction between the PCSK9 catalytic and the LDLR EGF-A domains at the cell surface (50). However, it should be noted that this hypothesis remains untested and requires further study.
Fig. 8.
A model of PCSK9 uptake. In the absence of LDL (left), PCSK9 can bind to the LDLR directly or bind to HLMs, such as HSPGs, to facilitate presentation to the LDLR. Both mechanisms can promote internalization. When LDL is present (right), it can complex with PCSK9 to reduce both PCSK9:LDLR and PCSK9:HLM binding, the former through an allosteric mechanism. LDL:HLM complex formation also downregulates PCSK9:HLM binding, and this reduction decreases the ability of HLMs to present PCSK9 to the LDLR for internalization. Whether HLMs can mediate PCSK9 internalization independent of PCSK9:LDLR binding remains to be seen. Though PCSK9 is depicted as bound to the LDLR in the endosome as part of a canonical pathway, PCSK9 internalization can occur in the absence of PCSK9:LDLR binding. CHR, cysteine-histidine rich domain.
Given the role of HLMs as promoters of PCSK9 uptake, we also evaluated their relationship to the S127R mutant, because the biochemical basis for its GOF phenotype remains unknown. Despite its reduction in processing efficiency, the S127R mutant still affects LDLR levels after being secreted (29). We found that uptake of the S127R mutant, as well as its ability to downregulate the LDLR, was enhanced over WT PCSK9, though less than the D374Y variant. While other studies have shown uptake of S127R PCSK9 to be similar to WT, these were performed in other cell types, where the relative contribution of HLMs may differ (33). Overall, our studies support the concept that increased binding affinity of the S127R variant to HLMs could promote cell surface association and uptake, providing a potential mechanism for its effects. This is consistent with other work showing that the GOF effect of the S127R and D374Y mutations are additive on PCSK9 function, arguing against directly overlapping mechanisms of action (62). Structurally, the side chain of S127 lies about 10 Å away from the guanidinium group of the R105 located within the HSPG binding motif of the prodomain. As such, the introduction of a positively charged residue could plausibly augment the binding interaction with negatively charged HLMs. Interestingly, a nearby mutant, D129G, shows a similar phenotype (58), and could follow a similar mechanism via the loss of a negatively charged aspartate. Importantly, LDL also inhibits uptake of S127R PCSK9, doing so in similar proportion to its effects on WT and D374Y PCSK9, suggesting that the mechanistic basis of the S127R mutant is not specific to an interaction with LDL alone.
Our results come with several important caveats. First, we have focused on PCSK9 uptake as a means to specifically investigate this stage of the PCSK9 lifecycle, as the sensitivity of our assay allowed us to probe perturbations that had a relatively modest downstream effect on the LDLR. But while uptake of PCSK9 is required for its ultimate function on the LDLR, such uptake does not necessarily imply that this function will occur (59).
Second, our study focused on in vitro and tissue culture models with physiologic concentrations of our parameters of interest. While our study remains consistent with prior work that LDL inhibits the ability of PCSK9 to induce LDLR degradation, this concept has only been tested in cell culture, and not directly proven in humans (32, 33). In this vein, it is notable that LDL may convey additional effects on PCSK9, including altering the equilibrium between the less active monomeric and more active oligomeric forms (63), as well as protecting serum PCSK9 from inactivation by furin-mediated cleavage (64). Thus, though outside the scope of our work, in vivo confirmation would be needed to understand whether the biochemical findings presented here have relevance to a clinical population with either normal or elevated serum cholesterol levels. To this end, it is notable that the relative effects seen by disrupting either the PCSK9:HLM or LDL:HLM interactions are only noticeable at low or subphysiologic LDL concentrations. Though circumstantial, this may provide insight into a time when evolutionary pressure existed to promote adequate serum LDL, giving a reason for PCSK9’s hypercholesterolemic function to exist.
Last, it must be noted that the results of our competition experiments with exogenous prodomain must be interpreted without confirmation of an organized structure, as despite their proper secretion, these prodomain-Fc fusions exist without cognate catalytic domains. Though the prodomain N terminus, known to be required for the prodomain:LDL interaction, is intrinsically unstructured, the extent to which the remainder of the prodomain can associate with either LDL or HLMs outside of the prodomain:catalytic domain complex remains a question. In this context, it is notable that the in vitro blocking experiments show only a modest decrease in PCSK9:LDL binding despite an 8-fold excess of prodomain. Whether this is due to a decreased affinity of “free” prodomain for LDL, as compared with the prodomain:catalytic domain complex present in mature PCSK9, remains a question. Moreover, because a structural basis for prodomain:HSPG binding has been proposed (37), it is conceivable that our free prodomain may simply not harbor the required structure to present the proper binding motif for HLMs. Thus, the free prodomain used in our blocking experiment may not interfere with PCSK9 binding to LDL and HLMs equally. Regardless, our data suggest that the prodomain N terminus is not, by itself, sufficient for binding LDL and mediating an inhibitory effect on PCSK9 uptake. Rather, our data suggest that PCSK9:LDL binding also requires a region of the prodomain that encompasses the HSPG binding motif, though structural information would be necessary to confirm this suspicion.
Overall, the interactions between LDL, PCSK9, and HLMs suggest a complex interplay to regulate ultimate PCSK9 function. Given the role of PCSK9 as a key driver of serum lipid levels, further understanding of these relationships and their regulatory mechanisms may allow the identification of potential targets that could ultimately upregulate the LDLR, lower serum LDL, and protect against atherosclerotic disease.
Acknowledgments
The authors thank Kevan Shokat (University of California San Francisco) for guidance, generous support, and critical reading of the manuscript. The authors also thank Sampath Parthasarathy (University of Central Florida) for suggesting the methylation of LDL and members of the Shokat laboratory for helpful discussion.
Footnotes
Abbreviations:
- EGF-A
- epidermal growth factor precursor homology domain A
- GOF
- gain-of-function
- HLM
- heparin-like molecule
- HSPG
- heparan sulfate proteoglycan
- LDLR
- LDL receptor
- NLuc
- nanoluciferase
- PCSK9
- proprotein convertase subtilisin/kexin type 9
- RLU
- relative luminescence unit
This work was supported by the Howard Hughes Medical Institute Medical Research Fellows Program (to A.M.G.), National Heart, Lung, and Blood Institute Grant K08 HL124068 and National Institutes of Health Grant LRP HMOT1243 (both to J.S.C.), the Hellman Foundation (to J.S.C.), and a Pfizer Foundation ASPIRE Cardiovascular Award (to J.S.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the other funding agencies.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Abifadel M., Varret M., Rabès J-P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D., et al. 2003. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34: 154–156. [DOI] [PubMed] [Google Scholar]
- 2.Zhang D-W., Lagace T. A., Garuti R., Zhao Z., McDonald M., Horton J. D., Cohen J. C., and Hobbs H. H.. 2007. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J. Biol. Chem. 282: 18602–18612. [DOI] [PubMed] [Google Scholar]
- 3.Maxwell K. N., Fisher E. A., and Breslow J. L.. 2005. Overexpression of PCSK9 accelerates the degradation of the LDLR in a post-endoplasmic reticulum compartment. Proc. Natl. Acad. Sci. USA. 102: 2069–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park S. W., Moon Y-A., and Horton J. D.. 2004. Post-transcriptional regulation of low density lipoprotein receptor protein by proprotein convertase subtilisin/kexin type 9a in mouse liver. J. Biol. Chem. 279: 50630–50638. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein J. L., and Brown M. S.. 2015. A century of cholesterol and coronaries: from plaques to genes to statins. Cell. 161: 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Z., Tuakli-Wosornu Y., Lagace T. A., Kinch L., Grishin N. V., Horton J. D., Cohen J. C., and Hobbs H. H.. 2006. Molecular characterization of loss-of-function mutations in PCSK9 and identification of a compound heterozygote. Am. J. Hum. Genet. 79: 514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hooper A. J., Marais A. D., Tanyanyiwa D. M., and Burnett J. R.. 2007. The C679X mutation in PCSK9 is present and lowers blood cholesterol in a Southern African population. Atherosclerosis. 193: 445–448. [DOI] [PubMed] [Google Scholar]
- 8.Stein E. A., Mellis S., Yancopoulos G. D., Stahl N., Logan D., Smith W. B., Lisbon E., Gutierrez M., Webb C., Wu R., et al. 2012. Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. N. Engl. J. Med. 366: 1108–1118. [DOI] [PubMed] [Google Scholar]
- 9.Sabatine M. S., Giugliano R. P., Wiviott S. D., Raal F. J., Blom D. J., Robinson J., Ballantyne C. M., Somaratne R., Legg J., Wasserman S. M., et al. 2015. Efficacy and safety of evolocumab in reducing lipids and cardiovascular events. N. Engl. J. Med. 372: 1500–1509. [DOI] [PubMed] [Google Scholar]
- 10.Sabatine M. S., Giugliano R. P., Keech A. C., Honarpour N., Wiviott S. D., Murphy S. A., Kuder J. F., Wang H., Liu T., Wasserman S. M., et al. ; FOURIER Steering Committee and Investigators. 2017. Evolocumab and clinical outcomes in patients with cardiovascular disease. N. Engl. J. Med. 376: 1713–1722. [DOI] [PubMed] [Google Scholar]
- 11.Fonarow G. C., Keech A. C., Pedersen T. R., Giugliano R. P., Sever P. S., Lindgren P., van Hout B., Villa G., Qian Y., Somaratne R., et al. 2017. Cost-effectiveness of evolocumab therapy for reducing cardiovascular events in patients with atherosclerotic cardiovascular disease. JAMA Cardiol. 2: 1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kazi D. S., Penko J., Coxson P. G., Moran A. E., Ollendorf D. A., Tice J. A., and Bibbins-Domingo K.. 2017. Updated cost-effectiveness analysis of PCSK9 inhibitors based on the results of the FOURIER trial. JAMA. 318: 748–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mullard A. 2017. Nine paths to PCSK9 inhibition. Nat. Rev. Drug Discov. 16: 299–301. [DOI] [PubMed] [Google Scholar]
- 14.Dubuc G., Chamberland A., Wassef H., Davignon J., Seidah N. G., Bernier L., and Prat A.. 2004. Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 24: 1454–1459. [DOI] [PubMed] [Google Scholar]
- 15.Jeong H. J., Lee H-S., Kim K-S., Kim Y-K., Yoon D., and Park S. W.. 2008. Sterol-dependent regulation of proprotein convertase subtilisin/kexin type 9 expression by sterol-regulatory element binding protein-2. J. Lipid Res. 49: 399–409. [DOI] [PubMed] [Google Scholar]
- 16.Shende V. R., Wu M., Singh A. B., Dong B., Kan C. F. K., and Liu J.. 2015. Reduction of circulating PCSK9 and LDL-C levels by liver-specific knockdown of HNF1α in normolipidemic mice. J. Lipid Res. 56: 801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tao R., Xiong X., DePinho R. A., Deng C-X., and Dong X. C.. 2013. FoxO3 transcription factor and Sirt6 deacetylase regulate low density lipoprotein (LDL)-cholesterol homeostasis via control of the proprotein convertase subtilisin/kexin type 9 (Pcsk9) gene expression. J. Biol. Chem. 288: 29252–29259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seidah N. G., Chrétien M., and Mbikay M.. 2018. The ever-expanding saga of the proprotein convertases and their roles in body homeostasis. Curr. Opin. Lipidol. 29: 144–150. [DOI] [PubMed] [Google Scholar]
- 19.Chen X-W., Wang H., Bajaj K., Zhang P., Meng Z-X., Ma D., Bai Y., Liu H-H., Adams E., Baines A., et al. 2013. SEC24A deficiency lowers plasma cholesterol through reduced PCSK9 secretion. eLife. 2: e00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seidah N. G., Benjannet S., Wickham L., Marcinkiewicz J., Jasmin S. B., Stifani S., Basak A., Prat A., and Chretien M.. 2003. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. USA. 100: 928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjannet S., Rhainds D., Essalmani R., Mayne J., Wickham L., Jin W., Asselin M-C., Hamelin J., Varret M., Allard D., et al. 2004. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J. Biol. Chem. 279: 48865–48875. [DOI] [PubMed] [Google Scholar]
- 22.Chorba J. S., and Shokat K. M.. 2014. The proprotein convertase subtilisin/kexin type 9 (PCSK9) active site and cleavage sequence differentially regulate protein secretion from proteolysis. J. Biol. Chem. 289: 29030–29043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maxwell K. N., and Breslow J. L.. 2004. Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc. Natl. Acad. Sci. USA. 101: 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chorba J. S., Galvan A. M., and Shokat K. M.. 2018. Stepwise processing analyses of the single-turnover PCSK9 protease reveal its substrate sequence specificity and link clinical genotype to lipid phenotype. J. Biol. Chem. 293: 1875–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cunningham D., Danley D. E., Geoghegan K. F., Griffor M. C., Hawkins J. L., Subashi T. A., Varghese A. H., Ammirati M. J., Culp J. S., Hoth L. R., et al. 2007. Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia. Nat. Struct. Mol. Biol. 14: 413–419. [DOI] [PubMed] [Google Scholar]
- 26.Hampton E. N., Knuth M. W., Li J., Harris J. L., Lesley S. A., and Spraggon G.. 2007. The self-inhibited structure of full-length PCSK9 at 1.9 A reveals structural homology with resistin within the C-terminal domain. Proc. Natl. Acad. Sci. USA. 104: 14604–14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piper D. E., Jackson S., Liu Q., Romanow W. G., Shetterly S., Thibault S. T., Shan B., and Walker N. P. C.. 2007. The crystal structure of PCSK9: a regulator of plasma LDL-cholesterol. Structure. 15: 545–552. [DOI] [PubMed] [Google Scholar]
- 28.Seidah N. G., Abifadel M., Prost S., Boileau C., and Prat A.. 2017. The proprotein convertases in hypercholesterolemia and cardiovascular diseases: emphasis on proprotein convertase subtilisin/kexin 9. Pharmacol. Rev. 69: 33–52. [DOI] [PubMed] [Google Scholar]
- 29.McNutt M. C., Kwon H. J., Chen C., Chen J. R., Horton J. D., and Lagace T. A.. 2009. Antagonism of secreted PCSK9 increases low density lipoprotein receptor expression in HepG2 cells. J. Biol. Chem. 284: 10561–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan D., Yancey P. G., Qiu S., Ding L., Weeber E. J., Linton M. F., and Fazio S.. 2008. Self-association of human PCSK9 correlates with its LDLR-degrading activity. Biochemistry. 47: 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun H., Samarghandi A., Zhang N., Yao Z., Xiong M., and Teng B-B.. 2012. Proprotein convertase subtilisin/kexin type 9 interacts with apolipoprotein B and prevents its intracellular degradation, irrespective of the low-density lipoprotein receptor. Arterioscler. Thromb. Vasc. Biol. 32: 1585–1595. [DOI] [PubMed] [Google Scholar]
- 32.Kosenko T., Golder M., Leblond G., Weng W., and Lagace T. A.. 2013. Low density lipoprotein binds to proprotein convertase subtilisin/kexin type-9 (PCSK9) in human plasma and inhibits PCSK9-mediated low density lipoprotein receptor degradation. J. Biol. Chem. 288: 8279–8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fisher T. S., Lo Surdo P., Pandit S., Mattu M., Santoro J. C., Wisniewski D., Cummings R. T., Calzetta A., Cubbon R. M., Fischer P. A., et al. 2007. Effects of pH and low density lipoprotein (LDL) on PCSK9-dependent LDL receptor regulation. J. Biol. Chem. 282: 20502–20512. [DOI] [PubMed] [Google Scholar]
- 34.Tavori H., Giunzioni I., Linton M. F., and Fazio S.. 2013. Loss of plasma proprotein convertase subtilisin/kexin 9 (PCSK9) after lipoprotein apheresis. Circ. Res. 113: 1290–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwon H. J., Lagace T. A., McNutt M. C., Horton J. D., and Deisenhofer J.. 2008. Molecular basis for LDL receptor recognition by PCSK9. Proc. Natl. Acad. Sci. USA. 105: 1820–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benjannet S., Saavedra Y. G. L., Hamelin J., Asselin M-C., Essalmani R., Pasquato A., Lemaire P., Duke G., Miao B., Duclos F., et al. 2010. Effects of the prosegment and pH on the activity of PCSK9: evidence for additional processing events. J. Biol. Chem. 285: 40965–40978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gustafsen C., Olsen D., Vilstrup J., Lund S., Reinhardt A., Wellner N., Larsen T., Andersen C. B. F., Weyer K., Li J., et al. 2017. Heparan sulfate proteoglycans present PCSK9 to the LDL receptor. Nat. Commun. 8: 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibson D. G., Young L., Chuang R-Y., Venter J. C., Hutchison C. A., and Smith H. O.. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 6: 343–345. [DOI] [PubMed] [Google Scholar]
- 39.Liu H., and Naismith J. H.. 2008. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weisgraber K. H., Innerarity T. L., and Mahley R. W.. 1978. Role of lysine residues of plasma lipoproteins in high affinity binding to cell surface receptors on human fibroblasts. J. Biol. Chem. 253: 9053–9062. [PubMed] [Google Scholar]
- 41.Mahley R. W., Weisgraber K. H., Melchior G. W., Innerarity T. L., and Holcombe K. S.. 1980. Inhibition of receptor-mediated clearance of lysine and arginine-modified lipoproteins from the plasma of rats and monkeys. Proc. Natl. Acad. Sci. USA. 77: 225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graham J. M., Griffin B. A., Davies I. G., and Higgins J. A.. 2001. Fractionation of lipoprotein subclasses in self-generated gradients of iodixanol. Methods Mol. Med. 52: 51–59. [DOI] [PubMed] [Google Scholar]
- 43.Steinman R. M., Mellman I. S., Muller W. A., and Cohn Z. A.. 1983. Endocytosis and the recycling of plasma membrane. J. Cell Biol. 96: 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeVay R. M., Yamamoto L., Shelton D. L., and Liang H.. 2015. Common proprotein convertase subtilisin/kexin type 9 (PCSK9) epitopes mediate multiple routes for internalization and function. PLoS One. 10: e0125127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y., Eigenbrot C., Zhou L., Shia S., Li W., Quan C., Tom J., Moran P., Di Lello P., Skelton N. J., et al. 2014. Identification of a small peptide that inhibits PCSK9 protein binding to the low density lipoprotein receptor. J. Biol. Chem. 289: 942–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lakoski S. G., Lagace T. A., Cohen J. C., Horton J. D., and Hobbs H. H.. 2009. Genetic and metabolic determinants of plasma PCSK9 levels. J. Clin. Endocrinol. Metab. 94: 2537–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheng Z., Otani H., Brown M. S., and Goldstein J. L.. 1995. Independent regulation of sterol regulatory element-binding proteins 1 and 2 in hamster liver. Proc. Natl. Acad. Sci. USA. 92: 935–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Min D. K., Lee H. S., Lee N., Lee C. J., Song H. J., Yang G. E., Yoon D., and Park S. W.. 2015. In silico screening of chemical libraries to develop inhibitors that hamper the interaction of PCSK9 with the LDL receptor. Yonsei Med. J. 56: 1251–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lagace T. A., Curtis D. E., Garuti R., McNutt M. C., Park S. W., Prather H. B., Anderson N. N., Ho Y. K., Hammer R. E., and Horton J. D.. 2006. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J. Clin. Invest. 116: 2995–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeVay R. M., Shelton D. L., and Liang H.. 2013. Characterization of proprotein convertase subtilisin/kexin type 9 (PCSK9) trafficking reveals a novel lysosomal targeting mechanism via amyloid precursor-like protein 2 (APLP2). J. Biol. Chem. 288: 10805–10818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berneis K. K., and Krauss R. M.. 2002. Metabolic origins and clinical significance of LDL heterogeneity. J. Lipid Res. 43: 1363–1379. [DOI] [PubMed] [Google Scholar]
- 52.Tabas I., Williams K. J., and Boren J.. 2007. Subendothelial lipoprotein retention as the initiating process in atherosclerosis: update and therapeutic implications. Circulation. 116: 1832–1844. [DOI] [PubMed] [Google Scholar]
- 53.Otvos J. D., Mora S., Shalaurova I., Greenland P., Mackey R. H., and Goff D. C.. 2011. Clinical implications of discordance between low-density lipoprotein cholesterol and particle number. J. Clin. Lipidol. 5: 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Austin M. A., Breslow J. L., Hennekens C. H., Buring J. E., Willett W. C., and Krauss R. M.. 1988. Low-density lipoprotein subclass patterns and risk of myocardial infarction. JAMA. 260: 1917–1921. [PubMed] [Google Scholar]
- 55.Goldstein J. L., Basu S. K., Brunschede G. Y., and Brown M. S.. 1976. Release of low density lipoprotein from its cell surface receptor by sulfated glycosaminoglycans. Cell. 7: 85–95. [DOI] [PubMed] [Google Scholar]
- 56.Vijayagopal P., Srinivasan S. R., Radhakrishnamurthy B., and Berenson G. S.. 1981. Interaction of serum lipoproteins and a proteoglycan from bovine aorta. J. Biol. Chem. 256: 8234–8241. [PubMed] [Google Scholar]
- 57.Saavedra Y. G., Zhang J., and Seidah N. G.. 2013. PCSK9 prosegment chimera as novel inhibitors of LDLR degradation. PLoS One. 8: e72113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Homer V. M., Marais A. D., Charlton F., Laurie A. D., Hurndell N., Scott R., Mangili F., Sullivan D. R., Barter P. J., Rye K-A., et al. 2008. Identification and characterization of two non-secreted PCSK9 mutants associated with familial hypercholesterolemia in cohorts from New Zealand and South Africa. Atherosclerosis. 196: 659–666. [DOI] [PubMed] [Google Scholar]
- 59.Nguyen M-A., Kosenko T., and Lagace T. A.. 2014. Internalized PCSK9 dissociates from recycling LDL receptors in PCSK9-resistant SV-589 fibroblasts. J. Lipid Res. 55: 266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamamoto T., Lu C., and Ryan R. O.. 2011. A two-step binding model of PCSK9 interaction with the low density lipoprotein receptor. J. Biol. Chem. 286: 5464–5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poirier S., Hamouda H. A., Villeneuve L., Demers A., and Mayer G.. 2016. Trafficking dynamics of PCSK9-induced LDLR degradation: Focus on human PCSK9 mutations and C-terminal domain. PLoS One. 11: e0157230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pandit S., Wisniewski D., Santoro J. C., Ha S., Ramakrishnan V., Cubbon R. M., Cummings R. T., Wright S. D., Sparrow C. P., Sitlani A., et al. 2008. Functional analysis of sites within PCSK9 responsible for hypercholesterolemia. J. Lipid Res. 49: 1333–1343. [DOI] [PubMed] [Google Scholar]
- 63.Tavori H., Fan D., Blakemore J. L., Yancey P. G., Ding L., Linton M. F., and Fazio S.. 2013. Serum proprotein convertase subtilisin/kexin type 9 and cell surface low-density lipoprotein receptor evidence for a reciprocal regulation. Circulation. 127: 2403–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fazio S., Minnier J., Shapiro M. D., Tsimikas S., Tarugi P., Averna M. R., Arca M., and Tavori H.. 2017. Threshold effects of circulating angiopoietin-like 3 levels on plasma lipoproteins. J. Clin. Endocrinol. Metab. 102: 3340–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]