Abstract
Proteins involved in lipoprotein metabolism can modulate cardiovascular health. While often measured to assess adult metabolic diseases, little is known about the proteomes of lipoproteins and their relation to metabolic dysregulation and underlying inflammation in undernourished child populations. The objective of this population study was to globally characterize plasma proteins systemically associated with HDL, LDL, and triglycerides in 500 Nepalese children. Abnormal lipid profiles characterized by elevated plasma triglycerides and low HDL-cholesterol (HDL-C) concentrations were common, especially in children with subclinical inflammation. Among 982 proteins analyzed, the relative abundance of 11, 12, and 52 plasma proteins was correlated with LDL-cholesterol (r = −0.43∼0.70), triglycerides (r = −0.39∼0.53), and HDL-C (r = −0.49∼0.79) concentrations, respectively. These proteins included apolipoproteins and numerous unexpected intracellular and extracellular matrix binding proteins, likely originating in hepatic and peripheral tissues. Relative abundance of two-thirds of the HDL proteome varied with inflammation, with acute phase reactants higher by 4∼40%, and proteins involved in HDL biosynthesis, cholesterol efflux, vitamin transport, angiogenesis, and tissue repair lower by 3∼20%. Untargeted plasma proteomics detects comprehensive sets of both known and novel lipoprotein-associated proteins likely reflecting systemic regulation of lipoprotein metabolism and vascular homeostasis. Inflammation-altered distributions of the HDL proteome may be predisposing undernourished populations to early chronic disease.
Keywords: plasma proteomics, high density lipoprotein-cholesterol, low density lipoprotein-cholesterol, triglycerides
Lipoproteins, circulating complexes of lipid-bound proteins, play central roles in the transport and metabolism of lipids. They support energy metabolism, cholesterol and phospholipids supply to cell membranes, and fat-soluble vitamin transport. Apolipoproteins are integral constituents that determine the physical properties and the metabolic fate of lipoproteins, stabilizing their structure, shuttling cholesterol and triglycerides throughout the body, acting as ligands for cell surface receptors, and regulating enzymatic activities (1). For example, apoA-I provides major structural support for HDL, activates LCAT, and acts as a ligand for HDL receptors, facilitating HDL-cholesterol (HDL-C) efflux capacity (2). apoB, the major scaffold for LDL and a ligand for LDL receptors, plays an essential role in the delivery of cholesterol to peripheral tissues (3). apoCs and apoE, highly enriched in VLDL and chylomicrons, deliver fatty acids to tissues for energy metabolism and regulate the clearance of triglyceride-rich particles from the plasma (1). Except for apoB, other apolipoproteins are exchangeable between different lipoprotein complexes, while the core structural apolipoproteins confer the unique properties of different classes of lipoproteins (4).
Over the past decade, a more complete, if complex, picture of the roles of lipoproteins has been revealed in proteomics studies, expanding knowledge of the size, diversity, and heterogeneity of protein constituents associated with HDL (4–12), LDL (4, 5, 13, 14), and VLDL (4, 5, 15). The HDL proteome, in particular, includes proteins involved not only in lipid transport and metabolism but also in inflammation, the complement system, protease inhibitors, and wound healing, elucidating broader roles in anti-inflammatory, anti-oxidative, immune-regulatory, and anti-apoptotic capacities (6). Although newly discovered cotransported members of lipoprotein assemblies are less abundant and more transient than apolipoproteins (8), these minor proteins are important for lipoprotein metabolism and function, serving to modulate the atherogenic or cardioprotective properties of lipoproteins (16). Beyond proteins physically carried by lipoproteins, plasma proteomics can identify intracellular or extracellular matrix proteins involved either directly or indirectly in lipoprotein production, secretion, clearance, and a wide range of metabolic actions (17), as lipoproteins constantly interact with cells in hepatic and peripheral tissues, including the immune system.
As proteins are important cargoes of lipoproteins, changes in the composition of proteins can modify the metabolism and function of lipoproteins. For example, inflammation and infections induce multiple alterations in lipid and lipoprotein metabolism (18). Chronic inflammatory diseases and infections are associated with hypertriglyceridemia and low HDL-C, as well as atherosclerosis (19–22), likely affecting the innate immune response, which protects a host from pathogenesis. However, prolonged and unresolved inflammation can impair normal lipoprotein metabolism, partly due to changes in the protein components of lipoproteins, especially in protein-rich HDL particles (18). For example, chronic inflammation may induce marked redistribution of HDL protein subpopulations, including enzymes and lipid transfer proteins, compromising HDL functions (23). Systemic analysis of inflammation-induced changes in proteins metabolically linked to lipoproteins may offer insights into the mechanisms and causes of dysregulated lipid metabolism and dysfunctional lipoproteins that may compromise vascular health.
Compelling epidemiologic and clinical evidence suggests that the metabolic risk factors for cardiovascular disease can be detected and observed to progress throughout childhood (24). However, the family of plasma proteomes of lipoproteins [e.g., direct or indirect correlates of HDL-C and LDL-cholesterol (LDL-C)] in childhood, especially in low-resource societies where subclinical inflammation is common, has rarely been explored. In 2006–2008, we conducted a multipronged health and nutrition assessment of a large population cohort of 6–8-year-old children in the southern rural plains (Terai) of Nepal (25, 26). Typical of the region, the studied children were generally undernourished compared with the World Health Organization reference, but nearly 40% were dyslipidemic, characterized by high triglycerides (≥100 mg/dl) and low HDL-C (<40 mg/dl) (27). In a representative subset of these children, more than 25% showed elevated α-1-acid glycoprotein (AGP), a biomarker of chronic systemic inflammation (28). In the current study of 500 children, we globally characterized plasma proteins correlated with plasma HDL-C, LDL-C, and triglyceride concentrations and evaluated their association with inflammation applying a quantitative proteomics approach.
MATERIALS AND METHODS
Study population and field assessments
Children in this proteomics study are a subset of a large child follow-up cohort in the southeastern plains district of Sarlahi, Nepal. Briefly, a community-based cluster-randomized controlled trial of antenatal micronutrient supplementation was conducted between 1999 and 2001 (26). Among more than 4,000 liveborn infants during the trial, we revisited about 3,500 children when they were 6–8 years of age, from 2006 to 2008, to assess their health and nutritional status (27). Field workers collected data on child demographic and anthropometric characteristics, education, dietary and morbidity history in the past week, and household socioeconomic status during home visits. Phlebotomists visited households of children early in the morning and collected venous blood samples in 10 ml sodium heparin vacutainers to prevent blood clots. Collected blood samples were transported on ice to the project’s field laboratory in Sarlahi where plasma samples were separated by centrifugation (1,720 g for 15 min). Plasma samples were used for lipid assessments and remaining plasma was aliquoted into cryovials that were stored and shipped in liquid nitrogen tanks to the Center for Human Nutrition laboratory at Johns Hopkins University in Baltimore, MD where they were stored at −80°C until thawed for biochemical assessments and/or proteomics analysis (25). Among 2,130 children who met inclusion criteria of having available plasma samples and complete epidemiological data from the original maternal micronutrient supplementation trial and follow-up study, samples from 1,000 children, balanced across the five original maternal intervention groups (n = 200 in each) were randomly selected for micronutrient and inflammation status assessments (28). Of these, 50% of the samples (n = 100 from each maternal intervention group) were randomly selected for proteomics analysis (29). The original antenatal micronutrient supplementation trial was registered at ClinicalTrials.gov as NCT00115271. Oral informed consent was obtained from the parents of eligible children during the child follow-up due to high illiteracy in the study population. Ethical approval for the original maternal trial and child follow-up study was obtained from the institutional review board at the Johns Hopkins Bloomberg School of Public Health, Baltimore, MD and the Nepal Health Research Council in Kathmandu, Nepal. All methods were carried out in accordance with the principles of the Declaration of Helsinki.
Lipid and inflammation marker assessments
Assessments of lipid profiles and inflammation markers were previously described in detail (25, 28). Plasma concentrations of total cholesterol, HDL-C, and triglycerides were measured using a Cholestech LDX analyzer (Alere Inc.) by enzymatic colorimetric methods. This instrument has been certified for clinical application by the US Center for Disease Control’s Lipid Standardization Program (Atlanta, GA). The instrument is also robust and thus has been used for point-of-care diagnostic testing of lipid levels in remote areas of rural Nepal. The detectable ranges for total cholesterol, HDL-C, and triglycerides were 100–500, 15–100, and 45–650 mg/dl, respectively. Estimates of plasma LDL-C concentrations were calculated using the Friedewald formula (30). Quality control materials provided by the manufacturer were tested weekly and with the arrival of each new lot of cassette supplies for quality assurance. Plasma AGP was chosen as a biomarker of subclinical inflammation over C-reactive protein (CRP) because AGP is more likely to detect chronic inflammation. For example, in Nepal, AGP identified five times more children with systemic low-grade inflammation than CRP using conventional clinical thresholds (30% of children exhibited AGP >1.0 g/l and 6% of children showed CRP >5 mg/l) (31). AGP was measured using a radial immunodiffusion assay (Kent Laboratories; 0.90 ± 0.1 g/l, CV = 10.0%).
Proteomics analysis
The processes of immune-depletion of high abundant proteins and MS-based proteomics analysis have been previously described (29). Plasma specimens were depleted of six high abundance proteins (albumin, transferrin, immunoglobulin G, immunoglobulin A, anti-trypsin, and haptoglobin) using a Human-6 Multiple Affinity Removal System LC column (Agilent Technologies). Seven depleted samples randomly chosen from the plasma samples of 500 participants and one masterpool sample, which served as an internal standard (32), were digested at 37°C overnight with trypsin using a 1:10 enzyme to protein ratio. Samples were randomly labeled with eight different isobaric tags for relative and absolute quantification (iTRAQ) reagents, which have reporter ions to be detected for relative quantification, and were incubated at room temperature for 2 h. All labeled samples were combined and 90 μl of the combined peptide sample was dissolved in 4 ml of strong cation exchange loading buffer [25% v/v acetonitrile and 10 mK KH2PO4 (pH 2.8)]. The sample was fractionated into 24 fractions by strong cation exchange chromatography on an Agilent 1200 capillary HPLC system using a PolySulfoethyl A column. Peptides were loaded on to a reverse-phase nanobore column and eluted using a 2–50% acetonitrile and 0.1% formic acid gradient for 110 min at 300 nl/min. Eluting peptides were sprayed into an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific) and interfaced with a NanoAcquity ultra-HPLC (Waters). Precursors and the fragment ions were analyzed at resolutions of 30,000 and 15,000, respectively. Spectra from full MS scans and fragmented MS/MS scans were extracted with and without deconvolution using Thermo Scientific Xtract software and searched against the RefSeq 40 protein database. Peptides were identified from Mascot (Matrix Science v2.3) searches through the Proteome Discoverer software (v1.3; Thermo Scientific) with a confidence threshold of 5% false discovery rate (FDR). A total of 4,705 nonredundant proteins were detected at least one time among 72 iTRAQ experiments conducted to assess plasma samples of the 500 study children (supplemental Table S1). We included 982 proteins quantified in >10% of all 500 plasma proteins of children (n > 50) in the proteomics analysis.
Statistical analyses
Standard cut-offs for low or high plasma lipid levels for children were derived from the National Cholesterol Education Program Expert Panel on Cholesterol Levels in Children (33). Estimation of relative abundance of proteins from reporter ion intensities across all obtained spectra have been previously described (32). Linear mixed effects (LME) models were employed to estimate linear associations between each log2 transformed-lipid or lipoprotein concentration as dependent variables and relative abundance of individual plasma proteins as a fixed effect, and each iTRAQ experiment as a random effect. Lipid values below the lower limit of detection were excluded from these analyses. We also adjusted for fasting status of children in all statistical models, as more than one-third of the children in the study were not fasted at the time of blood draw (25). We report summary statistics from the LME models, including number of child plasma samples in which a protein was detected, percent change (%) in lipid or lipoprotein concentration per 2-fold (100%) increase in relative abundance of a protein, P value calculated by using a two-sided test of a null hypothesis that there is no association between an individual protein and lipid or lipoprotein, q as FDR-adjusted P value to correct multiple comparisons (34), and r as a correlation coefficient between measured plasma lipid or lipoprotein concentration and its respective best linear unbiased prediction from the LME models (35). Proteins were considered significantly correlated with an outcome when passing a FDR threshold of 5% (q < 0.05). Because of their large number, we explored correlations between proteins associated with HDL-C concentration by calculating pairwise protein:protein correlation coefficients for proteins quantified in relative abundance within each iTRAQ experiment and averaged values across iTRAQ experiments.
Among proteins correlated with HDL-C, LDL-C, and triglyceride concentrations, we estimated and compared mean differences in relative protein abundance in plasma samples between children with and without inflammation (plasma AGP concentration >1.0 g/l or ≤1.0 g/l), adjusting for potential confounders, including child sex, age, ethnicity, fasting status, and stunting and underweight status (based on height-for-age and weight-for-age z-scores less than −2, respectively, derived from the World Health Organization growth reference for children) (36). P values were adjusted to control FDR using the Benjamini-Hochberg method (37). Gene symbols of protein GenInfo identifier (gi) numbers were derived from the Human Genome Organization (HUGO) gene annotation and used in the tables and figures (38). General descriptions of proteins were extracted from the NCBI protein database, UniProt, and in-depth review of literature (39, 40). Data visualization was performed using the Cytoscape (41). The datasets of lipid profiles and relative abundance of reported proteins in this study are available in supplemental Table S2. All analyses were performed using the R Environment for Statistical Computing (version 3.1.2; R Foundation for Statistical Computing, Vienna, Austria).
RESULTS
Lipid profiles and inflammation of children
In this cohort of 500 Nepalese children, median (interquartile range) values of plasma concentrations for total cholesterol, HDL-C, and LDL-C (n = 324) were 111 (100, 127), 27 (21, 33), and 71 (62, 83) mg/dl, respectively, and 89 (66, 118) mg/dl for triglycerides. These medians approximate plasma values less than fifth percentiles for total cholesterol and HDL-C, less than tenth percentile for LDL-C, and greater than ninetieth percentile for triglycerides of distributions among child populations between the ages of 6 and 9 in the United States (42, 43). The prevalence of high total cholesterol (≥200 mg/dl) and high LDL-C (≥130 mg/dl) was 0.4% and 0.6%, while the prevalence of high triglycerides (≥100 mg/dl) and low HDL-C (<40 mg/dl) was 40% and 90%, respectively. About 30% of children had inflammation indicated by elevated plasma AGP (>1.0 g/l). Child characteristics including lipid and lipoprotein profiles are compared by inflammation status in Table 1. Children with inflammation had lower plasma HDL-C (25 vs. 28 mg/dl; P = 0.005) and higher plasma triglyceride (97 vs. 87 mg/dl; P = 0.032) concentrations than children without inflammation, while there were no significant differences in plasma concentrations of total cholesterol and LDL-C. Stunting (48% vs. 36%; P = 0.015), underweight (57% vs. 45%; P = 0.017), and reported episodes of fever (16.8% vs. 4.6%; P < 0.001) and diarrhea (7.4% vs. 1.4%; P = 0.001) in the past week were more common in children with inflammation than without inflammation. No differences were observed in child demographic and educational characteristics, economics of household, and dietary history between the two groups.
TABLE 1.
Characteristics of 6- to 8-year-old children in rural Nepal by inflammation status (n = 500)
| Child Characteristics | Inflammationa (n = 149) | No Inflammationa (n = 351) | b |
| Demographic | |||
| Male, % | 52.3 | 48.7 | 0.519 |
| Age, years | 7.5 (0.4) | 7.4 (0.4) | 0.109 |
| Ethnicity | |||
| Pahadi (vs. Madheshi),c % | 29.5 | 32.8 | 0.545 |
| Anthropometryd | |||
| Height, cm | 113.6 (6.0) | 114.1 (5.4) | 0.372 |
| Weight, kg | 18.0 (2.3) | 18.4 (2.2) | 0.209 |
| BMI, kg/m2 | 13.9 (1.0) | 14.0 (1.0) | 0.315 |
| Stunting,e % | 47.7 | 35.5 | 0.015 |
| Underweight,e % | 57.0 | 44.9 | 0.017 |
| Low BMI,e % | 14.8 | 16.9 | 0.646 |
| Education, % | |||
| Ever sent to school | 67.8 | 66.4 | 0.841 |
| Literacy | 14.8 | 18.5 | 0.377 |
| Economics of household, % | |||
| Electricity | 51.0 | 51.0 | 1.000 |
| Land ownership | 77.2 | 76.9 | 1.000 |
| Diet in the past 7 days (≥3 times),f % | |||
| Bhat (rice) | 98.9 | 100.0 | 0.451 |
| Corn and wheat | 61.8 | 67.1 | 0.307 |
| Milk and curd | 60.4 | 53.3 | 0.173 |
| Fish, chicken, and other meat | 14.8 | 20.2 | 0.190 |
| Fruitg | 40.3 | 39.9 | 1.000 |
| Dark green leafy vegetables | 34.9 | 31.3 | 0.500 |
| Oil and ghyu (butter) | 100 | 99.7 | 1.000 |
| Morbidity in the past 7 days (≥1 time), % | |||
| Fever | 16.8 | 4.6 | <0.001 |
| Diarrhea | 7.4 | 1.4 | 0.001 |
| Productive cough | 4.0 | 3.7 | 1.000 |
| Rapid breathing and grunting | 3.4 | 2.6 | 0.846 |
| Lipid or lipoprotein concentrationsh | |||
| Total cholesterol, mg/dl | 107 (100, 124) | 112 (100, 128) | 0.165 |
| HDL-C, mg/dl | 25 (20, 31) | 28 (22, 34) | 0.005 |
| LDL-C, mg/dl | 71 (64, 81) | 72 (62, 83) | 0.835 |
| Triglycerides, mg/dl | 97 (69, 120) | 87 (65, 114) | 0.032 |
Inflammation was defined as plasma AGP concentration >1.0 g/l.
Values are mean (SD), percentages, or median (interquartile range).
P values were calculated using t-test for continuous variables with normal distributions, Mann-Whitney test for continuous variables with skewed distributions, and chi-square test for categorical variables.
Pahadi pertains to ethnic groups that originated in the Hills of Nepal; Madheshi pertains to groups whose origin is the southern plains (Tarai) of Nepal.
One outlier in weight (>45 kg) and two outliers in height (>140 cm) and BMI (>20 or <10 kg/m2) were excluded.
The z-scores were calculated based on the World Health Organization reference for 5–19 years (36). Stunting, height-for-age z-score less than −2; underweight, weight-for-age z-score less than −2; and low BMI, BMI-for-age z-score less than −2.
Data are missing for rice (n = 1), milk and curd (n = 2), and fruit (n = 23).
Fruit includes mango, papaya, jackfruit, guava, and orange.
The lowest detectable values were used for values of HDL-C (n = 36), triglycerides (n = 28), and total cholesterol (n = 146) below detection limits. Data are missing for LDL-C (n = 176).
Plasma proteins correlated with LDL-C concentrations
Eleven proteins were positively correlated with LDL-C (Table 2). apoB was most strongly correlated with plasma LDL-C (r = 0.69; q = 8.84 × 10−47). LDL-C concentrations increased by 121% per 100% increase in relative abundance of apoB. The rest of the proteins were mostly positive associates (r = 0.18∼0.58; q = 3.74 × 10−2 to 4.41 × 10−7), including kinesin family member 20B, platelet activating factor acetylhydrolase (also known as LDL-C-associated phospholipase 2), cholesteryl ester transfer protein (CETP), proteoglycan 4, and apoM, and four intracellular proteins observed in <100 children. Chondroadherin, which regulates chondrocyte growth, was the only protein negatively associated with LDL-C (r = −0.42; q = 2.77 × 10−2).
TABLE 2.
Proteins correlated with plasma LDL-C concentrations in children of rural Nepal aged 6–8 years (n = 324)
| Protein Name | na | r | R2 | P | q | Percent Changeb | Accessionc | Referenced |
| apoB | 324 | 0.69 | 0.48 | 6.39 × 10−50 | 8.84 × 10−47 | 120.8 | 105990532 | (5, 13, 14, 53, 76–78) |
| Kinesin family member 20B (KIF20B) | 208 | 0.47 | 0.22 | 6.38 × 10−10 | 4.41 × 10−7 | 37.2 | 46049114 | |
| Platelet-activating factor acetylhydrolase (PAF-AH) | 266 | 0.30 | 0.09 | 4.49 × 10−7 | 1.55 × 10−4 | 25.7 | 189095271 | (5, 13) |
| G2/mitotic-specific cyclin B3 (CCNB3) | 83 | 0.58 | 0.34 | 1.59 × 10−6 | 3.14 × 10−4 | 34.5 | 90669307 | |
| Zinc finger and BTB domain containing 1 (ZBTB1) | 74 | 0.58 | 0.33 | 4.29 × 10−6 | 6.70 × 10−4 | 50.8 | 182509178 | |
| Netrin receptor UNC5C (UNC5C) | 91 | 0.46 | 0.21 | 4.36 × 10−6 | 6.70 × 10−4 | 38.3 | 16933525 | |
| CETP, plasma | 319 | 0.21 | 0.05 | 1.24 × 10−4 | 1.01 × 10−2 | 19.7 | 169636439 | |
| Proteoglycan 4 (PRG4) | 260 | 0.23 | 0.05 | 2.50 × 10−4 | 1.79 × 10−2 | 27.0 | 189181724 | |
| Chondroadherin (CHAD) | 86 | −0.42 | 0.18 | 4.61 × 10−4 | 2.77 × 10−2 | −23.4 | 153251229 | |
| Poly (ADP-ribose) glycohydrolase (PARG) | 92 | 0.36 | 0.13 | 5.50 × 10−4 | 2.99 × 10−2 | 25.1 | 70610136 | |
| apoM | 324 | 0.18 | 0.03 | 8.38 × 10−4 | 3.74 × 10−2 | 24.7 | 22091452 | (5, 14, 53, 76, 77) |
Eleven proteins quantified by MS and estimated by LME modeling in >10% of the samples that were positively and negatively correlated with LDL-C adjusting for fasting status (q < 0.05), listed by the strength of association (in increasing order of q).
Number of child plasma samples in which each protein was detected and quantified by MS. Children with no LDL-C data due to under the detectable range of HDL-C (<15 mg/dl), total cholesterol (<100 mg/dl), or triglyceride (<45 mg/dl) were excluded (n = 176).
Percent change in LDL-C concentrations of children per 100% (two times) increase in relative abundance of a protein.
GenInfo sequence number as assigned to all protein sequences by the NCBI at the National Library of Medicine, National Institutes of Health.
Proteins have been previously reported to be physically associated with LDL particles (79).
Plasma proteins correlated with triglyceride concentrations
Three apoC proteins were most highly correlated with triglycerides (apoC-II/C-III/C-IV) (r = 0.51∼0.54; q = 8.24 × 10−13 to 7.22 × 10−17) (Table 3). Other positive correlates included cathelicidin antimicrobial peptide, proteoglycan 4, retinol-binding protein 4 (RBP4), and apoE. Five negatively associated proteins included extracellular matrix-related proteins, such as anthrax toxin receptor (ANTXR)2, neuropilin 1, and insulin-like growth factor binding protein 1, and lipid transfer proteins such as CETP and phospholipid transfer protein (PLTP).
TABLE 3.
Proteins correlated with plasma triglyceride concentrations in children of rural Nepal aged 6–8 years (n = 472)
| Protein Name | na | r | R2 | P | q | Percent Changeb | Accessionc | Referenced |
| apoC-II | 472 | 0.54 | 0.29 | 4.20 × 10−20 | 7.22 × 10−17 | 47.2 | 32130518 | (5, 53) |
| apoC-III | 472 | 0.53 | 0.29 | 3.31 × 10−19 | 2.84 × 10−16 | 48.8 | 4557323 | (5, 15, 53) |
| apoC-IV | 472 | 0.51 | 0.26 | 1.44 × 10−15 | 8.24 × 10−13 | 37.8 | 4502161 | (5, 15, 53) |
| Cathelicidin antimicrobial peptide (CAMP) | 394 | 0.44 | 0.19 | 1.18 × 10−6 | 5.07 × 10−4 | 25.1 | 39753970 | (53) |
| Proteoglycan 4 (PRG4) | 380 | 0.40 | 0.16 | 7.07 × 10−6 | 2.43 × 10−3 | 49.3 | 189181724 | |
| ANTXR2 | 346 | −0.40 | 0.16 | 1.85 × 10−5 | 5.28 × 10−3 | −28.3 | 50513243 | (53) |
| CETP, plasma | 465 | −0.40 | 0.16 | 2.33 × 10−5 | 5.71 × 10−3 | −23.9 | 169636439 | (53) |
| Neuropilin 1 (NRP1) | 432 | −0.39 | 0.15 | 5.14 × 10−5 | 1.10 × 10−2 | −25.4 | 66864913 | |
| RBP4 | 472 | 0.40 | 0.16 | 2.45 × 10−4 | 3.91 × 10−2 | 30.1 | 55743122 | |
| apoE | 472 | 0.40 | 0.16 | 2.45 × 10−4 | 3.91 × 10−2 | 30.9 | 4557325 | (53) |
| PLTP | 445 | −0.39 | 0.15 | 2.73 × 10−4 | 3.91 × 10−2 | −27.9 | 5453914 | (5, 15, 53) |
| Insulin-like growth factor binding protein 1 (IGFBP1) | 417 | −0.33 | 0.11 | 3.16 × 10−4 | 4.17 × 10−2 | −13.2 | 4504615 |
Twelve proteins quantified by MS and estimated by LME modeling in >10% of the samples that were positively and negatively associated with plasma triglycerides adjusting for fasting status (q < 0.05), listed by the strength of association (in increasing order of q).
Number of child plasma samples in which each protein was detected and quantified by MS. Children with triglyceride concentrations below a detection limit (<45 mg/dl) were excluded (n = 28).
Percent change in plasma triglycerides of children per 100% (two times) increase in relative abundance of a protein.
GenInfo sequence number as assigned to all protein sequences by the NCBI at the National Library of Medicine, National Institutes of Health.
Proteins have been previously reported to be physically associated with VLDL particles.
Plasma proteins correlated with HDL-C concentrations
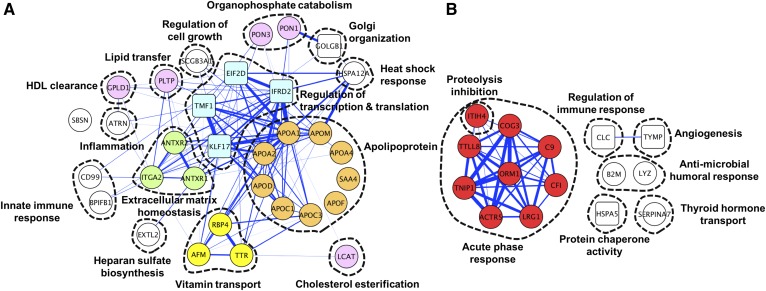
Thirty-six proteins were positively correlated with HDL-C (Table 4). apoA-I (r = 0.79; q = 1.06 × 10−91) and apoA-II (r = 0.58; q = 4.16 × 10−21) were two of the three most strongly associated proteins followed by seven other apolipoproteins [apoA-IV/apoC-I/apoC-III/apoD/apoM/apoF/serum amyloid A4 (SAA4)] (r = 0.45∼0.56; q = 4.23 × 10−3 to 2.72 × 10−16). Proteins with well-defined roles in HDL metabolism and function included PLTP and enzymes, such as LCAT and paraoxonase (PON)1 and PON3. Interestingly, intracellular proteins, such as interferon-related developmental regulator 2 (IFRD2), eukaryotic translation initiation factor 2D (EIF2D), Kruppel-like factor 17 (KLF17), and TATA element modulatory factor 1 (TMF1), were also strongly correlated with HDL-C. Proteins mediating cell-cell and cell-extracellular matrix interaction, such ANTXR1 and ANTXR2, were also among positive correlates. Associated fat-soluble vitamin transporters included retinol binding protein 4, transthyretin, and afamin. Sixteen proteins (Table 5), moderately negatively correlated with HDL-C (r = −0.33 ∼−0.49; q = 2.05 × 10−2 to 7.59 × 10−4), are primarily involved in the acute phase response (i.e., orosomucoid 1 or AGP and leucine-rich α-2-glycoprotein 1) and the complement system (i.e., complement component 9 and complement factor I). Protein-protein correlations as well as molecular network and functional clusters of the plasma HDL-C proteome are visualized in Fig. 1A and B. Prominent correlations among proteins positively associated with HDL-C (Fig. 1A) were found between apoA-I and apoA-II (r = 0.75, colored in orange) and apoA-I with other intracellular proteins, such as IFRD2, EIF2D, KLF17, and TMF1 (r = 0.53∼0.91, colored in blue). Among negative HDL-C correlates (Fig. 1B), proteins involved in the acute phase response proteolytic inhibition were strongly correlated with each other (r = 0.36∼0.90, colored in red). Correlates of HDL-C appear to be involved in a wide range of functions that extend beyond known functions of HDL, such as regulation of gene transcription and translation and extracellular matrix homeostasis.
TABLE 4.
Proteins positively correlated with plasma HDL-C concentrations in children of rural Nepal aged 6–8 years (n = 464)
| Protein Name | na | r | R2 | P | q | Percent Changeb | Accessionc | Referenced |
| apoA-I | 464 | 0.79 | 0.62 | 7.68 × 10−95 | 1.06 × 10−91 | 183.8 | 4557321 | (7, 10–12, 60, 80–91) |
| IFRD2 | 450 | 0.64 | 0.41 | 3.59 × 10−33 | 2.47 × 10−30 | 63.9 | 197333755 | |
| apoA-II | 464 | 0.58 | 0.34 | 9.04 × 10−24 | 4.16 × 10−21 | 76.9 | 4502149 | (7, 10–12, 60, 80–85, 87–91) |
| apoD | 464 | 0.56 | 0.31 | 8.22 × 10−19 | 2.72 × 10−16 | 53.2 | 4502163 | (7, 12, 60, 80–85, 87–91) |
| EIF2D | 238 | 0.57 | 0.33 | 9.85 × 10−19 | 2.72 × 10−16 | 61.0 | 56699485 | |
| apoC-I | 464 | 0.51 | 0.26 | 1.44 × 10−13 | 2.83 × 10−11 | 18.6 | 4502157 | (7, 10, 11, 60, 80–85, 87–91) |
| apoM | 464 | 0.51 | 0.26 | 7.09 × 10−13 | 1.22 × 10−10 | 54.8 | 22091452 | (7, 10–12, 60, 80–82, 84, 85, 87–91) |
| PLTP | 438 | 0.51 | 0.26 | 1.40 × 10−10 | 2.15 × 10−8 | 50.3 | 5453914 | (7, 60, 80, 81, 87, 88, 91) |
| ANTXR2 | 337 | 0.51 | 0.26 | 1.71 × 10−10 | 2.36 × 10−8 | 42.8 | 50513243 | (60) |
| TMF1 | 228 | 0.54 | 0.29 | 4.74 × 10−9 | 5.95 × 10−7 | 33.0 | 110347443 | |
| KLF17 | 258 | 0.50 | 0.25 | 6.99 × 10−9 | 8.04 × 10−7 | 26.4 | 104294874 | |
| PON1 | 464 | 0.49 | 0.24 | 8.81 × 10−9 | 9.35 × 10−7 | 27.3 | 19923106 | (7, 11, 12, 60, 80–85, 87–91) |
| ANTXR1 | 315 | 0.45 | 0.21 | 2.39 × 10−7 | 2.36 × 10−5 | 30.0 | 14149904 | (60) |
| SAA4 | 459 | 0.47 | 0.22 | 7.79 × 10−7 | 7.17 × 10−5 | 23.9 | 10835095 | (7, 10, 60, 80, 81, 83–85, 87–91) |
| PON3 | 457 | 0.46 | 0.21 | 1.23 × 10−6 | 1.06 × 10−4 | 28.1 | 29788996 | (7, 60, 80, 81, 88, 91) |
| BPI fold containing family B member 1 (BPIFB1) | 189 | 0.52 | 0.27 | 1.91 × 10−6 | 1.55 × 10−4 | 26.6 | 40807482 | (60) |
| apoF | 464 | 0.47 | 0.22 | 3.96 × 10−6 | 3.03 × 10−4 | 24.9 | 4502165 | (7, 60, 80–83, 85, 88–91) |
| Glycosylphosphatidylinositol specific phospholipase D1 (GPLD1) | 464 | 0.46 | 0.21 | 9.82 × 10−6 | 6.73 × 10−4 | 31.6 | 29171717 | (60, 80, 82) |
| Secretoglobin, family 3A member 1 (SCGB3A1) | 279 | 0.54 | 0.29 | 1.02 × 10−5 | 6.73 × 10−4 | 14.7 | 50363226 | (91) |
| LCAT | 464 | 0.45 | 0.20 | 2.94 × 10−5 | 1.69 × 10−3 | 52.3 | 4557892 | (7, 60, 80, 87, 88, 91) |
| apoC-III | 464 | 0.45 | 0.20 | 3.16 × 10−5 | 1.74 × 10−3 | 15.5 | 4557323 | (7, 10–12, 60, 80–91) |
| RBP4 | 464 | 0.45 | 0.21 | 4.47 × 10−5 | 2.37 × 10−3 | 23.4 | 55743122 | (7, 80, 82, 84, 85, 87, 88, 90) |
| CD99 molecule (CD99) | 444 | 0.41 | 0.17 | 4.78 × 10−5 | 2.39 × 10−3 | 17.1 | 171543879 | |
| DNA repair and recombination protein RAD54B (RAD54B) | 98 | 0.54 | 0.29 | 7.95 × 10−5 | 3.66 × 10−3 | 41.2 | 6912622 | |
| apoA-IV | 464 | 0.45 | 0.20 | 9.49 × 10−5 | 4.23 × 10−3 | 18.4 | 71773110 | (7, 10–12, 60, 80–91) |
| Transthyretin (TTR) | 464 | 0.45 | 0.20 | 1.02 × 10−4 | 4.42 × 10−3 | 32.3 | 4507725 | (7, 10–12, 80–82, 84–86, 88, 90, 91) |
| Integrin α2 (ITGA2) | 175 | 0.50 | 0.25 | 1.32 × 10−4 | 5.51 × 10−3 | 31.1 | 116295258 | (60) |
| Exostosin-like glycosyltransferase 2 (EXTL2) | 301 | 0.45 | 0.21 | 2.10 × 10−4 | 7.82 × 10−3 | 21.0 | 14149609 | |
| Golgin B1 (GOLGB1) | 322 | 0.44 | 0.20 | 2.28 × 10−4 | 8.28 × 10−3 | 19.5 | 148596984 | |
| Attractin (ATRN) | 464 | 0.45 | 0.20 | 2.94 × 10−4 | 1.04 × 10−2 | 34.7 | 21450863 | |
| Heat shock 70 kDa protein 12A (HSPA12A) | 107 | 0.47 | 0.22 | 6.29 × 10−4 | 2.01 × 10−2 | 43.2 | 119874213 | |
| Afamin (AFM) | 464 | 0.44 | 0.19 | 7.23 × 10−4 | 2.17 × 10−2 | 21.2 | 4501987 | (80, 82, 87, 90) |
| Sushi, von Willebrand factor type A, EGF and pentraxin domain-containing protein 1 (SVEP1) | 69 | 0.61 | 0.37 | 7.86 × 10−4 | 2.31 × 10−2 | 46.4 | 148886654 | |
| Aldehyde dehydrogenase 9 family member A1 (ALDH9A1) | 85 | 0.53 | 0.28 | 1.11 × 10−3 | 2.99 × 10−2 | 19.9 | 115387104 | |
| Suprabasin (SBSN) | 121 | 0.53 | 0.28 | 1.88 × 10−3 | 4.42 × 10−2 | 39.5 | 38348366 | |
| Plexin domain-containing protein 1 (PLXDC1) | 94 | 0.59 | 0.35 | 2.26 × 10−3 | 4.91 × 10−2 | 29.4 | 5011862 |
Thirty-six proteins quantified by MS and estimated by LME modeling in >10% of the samples that were positively associated with HDL-C adjusting for fasting status (q < 0.05), listed by the strength of association (in increasing order of q).
Number of child plasma samples in which each protein was detected and quantified by MS. Children with plasma HDL-C concentrations below a detection limit (<15 mg/dl) were excluded (n = 36).
Percent change in plasma HDL-C of children per 100% (two times) increase in relative abundance of a protein.
GenInfo sequence number as assigned to all protein sequences by the NCBI at the National Library of Medicine, National Institutes of Health.
Proteins have been previously reported to be physically associated with HDL particles (79).
TABLE 5.
Proteins negatively correlated with plasma HDL-C concentrations in children of rural Nepal aged 6–8 years (n = 464)
| Protein Name | na | r | R2 | P | q | Percent Changeb | Accessionc | Referenced |
| Complement component 9 (C9) | 464 | −0.45 | 0.20 | 1.28 × 10−5 | 7.59 × 10−4 | −20.7 | 4502511 | (7, 60, 82, 87, 90) |
| Orosomucoid 1 (ORM1) | 464 | −0.45 | 0.20 | 6.89 × 10−6 | 4.50 × 10−4 | −15.7 | 167857790 | (80, 85, 87–91) |
| Galectin 10 (CLC) | 377 | −0.48 | 0.23 | 3.85 × 10−5 | 2.09 × 10−3 | −7.0 | 20357559 | |
| Complement factor I (CFI) | 464 | −0.44 | 0.20 | 1.92 × 10−4 | 6.96 × 10−3 | −25.3 | 119392081 | (82, 87) |
| Leucine-rich α-2-glycoprotein 1 (LRG1) | 464 | −0.44 | 0.19 | 8.98 × 10−5 | 4.04 × 10−3 | −15.5 | 16418467 | (80, 87) |
| Beta-2-microglobulin (B2M) | 457 | −0.43 | 0.18 | 7.81 × 10−4 | 2.22 × 10−2 | −13.1 | 4757826 | (80, 87, 88) |
| Fatty acid binding protein 12 (FABP12) | 77 | −0.37 | 0.14 | 1.18 × 10−3 | 3.14 × 10−2 | −9.2 | 157427691 | |
| Thymidine phosphorylase (TYMP) | 315 | −0.44 | 0.19 | 1.81 × 10−3 | 4.03 × 10−2 | −13.0 | 4503445 | |
| Tubulin tyrosine ligase-like family member 8 (TTLL8) | 272 | −0.33 | 0.11 | 2.15 × 10−4 | 7.40 × 10−3 | −19.3 | 122937293 | |
| TNFAIP3 interacting protein 1 (TNIP1) | 356 | −0.45 | 0.20 | 8.82 × 10−4 | 2.45 × 10−2 | −13.6 | 116256481 | |
| Lysozyme (LYZ) | 457 | −0.44 | 0.19 | 1.07 × 10−3 | 2.92 × 10−2 | −15.0 | 4557894 | (84, 87) |
| Inter-α-trypsin inhibitor heavy chain family member 4 (ITIH4) | 464 | −0.45 | 0.20 | 1.25 × 10−3 | 3.19 × 10−2 | −27.2 | 31542984 | (7, 11, 60, 80, 82, 86, 87) |
| Conserved oligomeric Golgi complex subunit 3 (COG3) | 202 | −0.49 | 0.24 | 1.69 × 10−3 | 4.02 × 10−2 | −12.4 | 13899251 | |
| Heat shock 70 kDa protein 5 (glucose-regulated protein, 78 kDa) (HSPA5) | 458 | −0.42 | 0.17 | 2.01 × 10−3 | 4.30 × 10−2 | −19.1 | 16507237 | |
| Thyroxine-binding globulin (SERPINA7) | 464 | −0.44 | 0.19 | 1.70 × 10−3 | 4.02 × 10−2 | −19.5 | 205277441 | (87) |
| Actin-related protein 5 (ACTR5) | 233 | −0.42 | 0.17 | 7.08 × 10−4 | 2.05 × 10−2 | −13.7 | 151301041 |
aSixteen proteins quantified by MS and estimated by LME modeling in >10% of the samples that were negatively associated with HDL-C adjusting for fasting status (q < 0.05), listed by the strength of association (in increasing order of q).
Number of child plasma samples in which each protein was detected and quantified by MS. Children with plasma HDL-C concentrations below a detection limit (<15 mg/dl) were excluded (n = 36).
Percent change in plasma HDL-C of children per 100% (two times) increase in relative abundance of a protein.
GenInfo sequence number as assigned to all protein sequences by the NCBI at the National Library of Medicine, National Institutes of Health.
Proteins have been previously reported to be physically associated with HDL particles (79).
Fig. 1.
Correlations among plasma proteins positively correlated (A) and negatively correlated (B) with plasma HDL-C concentrations (q < 0.05 and n > 100) in children of rural Nepal aged 6–8 years. The thickness of the lines reflects the strength of positive correlation coefficients between proteins (r ≥ 0.3 are presented). Circles and squares represent individual extracellular and intracellular proteins, respectively, indicated as their HUGO gene symbols. Dashed lines indicate molecular or functional clusters. Proteins with colors are mentioned in the text.
Changes in relative abundance of proteins correlated with LDL-C, triglyceride, and HDL-C concentrations by inflammation status
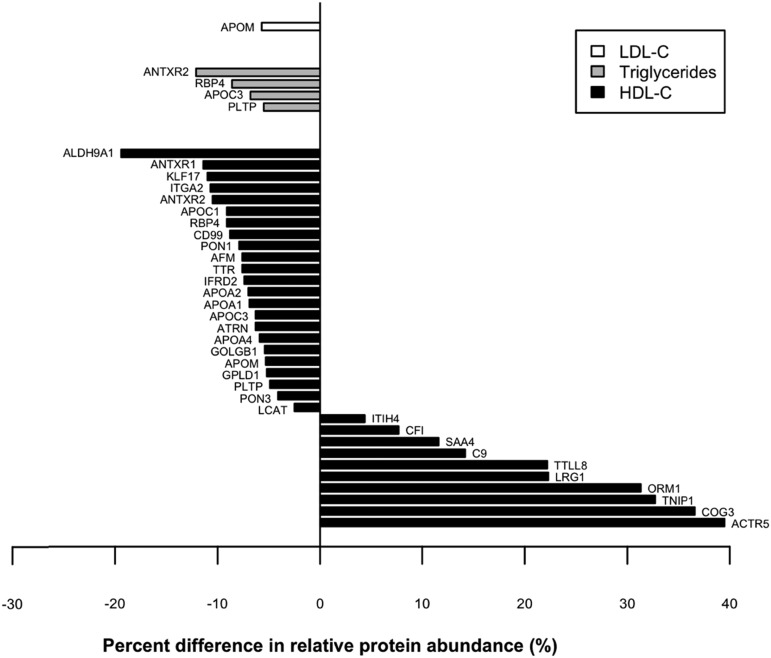
The relative abundance of proteins associated with the investigated lipids and lipoproteins was compared in children with inflammation (plasma AGP >1.0 g/l) to children without inflammation (AGP ≤1.0 g/l) (Fig. 2). Among proteins correlated with HDL-C, 10 were 4∼40% more abundant and 23 proteins were 3∼20% less abundant in children with inflammation, after adjusting for child sex, age, ethnicity, stunting, and underweight status and fasting status. Most proteins with higher and lower relative abundance in the presence of inflammation are negative and positive correlates of HDL-C, respectively. Thus, major structural proteins of HDL (apoA-I and apoA-II) and proteins involved in HDL biosynthesis (i.e., LCAT), vitamin transport (i.e., RBP4), protection against oxidative stress (i.e., PON1), and angiogenesis and tissue repair (i.e., ANTXR1 and ANTXR2), among others, were less abundant in children with inflammation than those without inflammation. An exception was SAA4, an apolipoprotein that was positively correlated with HDL-C (Table 4), but also a positive acute phase reactant, that was 12% higher in inflamed children. Among LDL and triglyceride proteomes, relative abundance of one (apoM) and four proteins (i.e., ANTXR2, RBP4, apoC-III, and PLTP), respectively, were reduced in children with inflammation compared with children without inflammation.
Fig. 2.
Percent difference in relative abundance of plasma proteins correlated with plasma LDL-C (white), triglyceride (gray), and HDL-C (black) concentrations in children with inflammation (plasma AGP concentration >1.0 g/l) compared with children without inflammation (AGP ≤1.0 g/l), adjusted for child sex, age, ethnicity, stunting, and underweight and fasting status (Benjamini-Hochberg corrected P value <0.05).
DISCUSSION
In this southern plains district of Nepal, reflecting undernourished living conditions for about 60% of the country’s population and that typify a wider rural region of Gangetic South Asia, abnormal lipid and lipoprotein profiles were common in young school-aged children, characterized by elevated plasma triglycerides, low HDL-C concentrations, and subclinical inflammation. Our untargeted plasma proteomics approach revealed distinct distributions of 11, 12, and 52 plasma proteins systemically associated with LDL-C, triglyceride, and HDL-C concentrations, respectively, whose functions extend beyond canonical lipid transport and metabolism. Inflammation was associated with remarkable differences in relative protein abundance, especially among biomarkers linked to HDL-C. These results collectively suggest that the family of plasma proteomes correlated with circulating lipoprotein concentrations in children comprise heterogeneous sets of proteins of known and novel biological function, and which are likely affected along a gradient of subclinical inflammation. The results of this large population study also suggest that many proteins not necessarily carried by lipoproteins reflect systemic regulation of lipoprotein metabolism.
The proteome of LDL-C in Nepalese children reveals a unique protein composition dominated by apoB. The strong positive correlation of apoB with LDL-C is consistent with evidence that apoB accounts for about 95% of the total protein mass in a LDL particle (44). Among the remaining proteins, kinesin family member 20B was the second strongest correlate of LDL-C, a motor protein known to transport intracellular vesicles along microtubules (45). A previous study showed elevated levels of serum cholesterol in kinesin family member 13B knockout mice, elucidating its novel role in the regulation of blood cholesterol by facilitating endocytosis of cell membrane receptor-binding LDL in hepatocytes (46). To our knowledge, this is the first human study to show that abundance of the same kinesin family member in plasma is positively correlated with plasma LDL-C concentrations, supporting a putative role of this cytoskeletal protein in regulating LDL clearance. Platelet-activating factor acetylhydrolase, which was previously reported to be physically carried by LDL particles (Table 2) (5, 13), was the third strongest correlate of LDL-C in this study. It modulates pro-inflammation and oxidative stress reactions (47), and has been identified as a risk factor for coronary heart disease (48, 49). Another positive correlate, CETP, participates in LDL remodeling from VLDL by transferring cholesteryl esters from HDL to VLDL particles in an exchange for triglyceride to HDL (50). LDL-C was also correlated with intracellular proteins involved in axon guidance (netrin receptor UNC5C), control of cell cycle (cyclin B3), and DNA damage response (zinc finger and BTB domain-containing protein 1), although their tissue origins and physical and functional relationships with LDL particles are unclear.
Proteins strongly correlated with triglycerides may represent proteins directly associated with triglyceride-rich lipoproteins. For example, apoC-II/C-III/C-IV and apoE are known to reside within VLDL and chylomicrons and play important roles in the metabolism of the triglyceride-rich lipoproteins by regulating activity of lipoprotein lipase and their uptake by peripheral tissues (1). Cathelicidin, an antimicrobial peptide, is a family member of antibacterial and lipopolysaccharide-binding proteins mostly found in neutrophil and macrophage granules (51). This protein has been shown to complex with LDL/VLDL particles (52, 53), suggesting that VLDL may serve as a plasma reservoir of this antimicrobial protein. Proteoglycan 4 correlated with both LDL-C and triglycerides in study children. Consistent with our results, Geyer et al. (17) also reported that proteoglycan 4 was the second most strongly correlated protein with both LDL-C and triglycerides, following apoB and apoC-III, respectively, and downregulated by weight loss in obese individuals. As proteoglycan 4 is a major adipocyte extracellular matrix protein (54), it may occur in plasma from tissue leakage.
The plasma HDL proteome comprised the highest number of proteins reflecting diverse biological functions of HDL particles (8). Among 36 positive correlates of HDL-C concentrations, nine were apolipoproteins, including apoA-I. Accounting for 70% of HDL protein, apoA-I has been shown to confer anti-inflammatory, anti-oxidative, and cardioprotective properties to the lipoprotein complex (2, 4). Less abundant apolipoproteins, such as apoA-II/A-IV/C-I/C-III/D/F/M and SAA4, are also known to coregulate metabolism, including interactions with other lipoproteins and cell-specific receptors (1). PLTP is the primary protein that transfers phospholipids from triglyceride-rich lipoproteins to HDL, representing the maturation of HDL particles (55). LCAT, the extracellular cholesterol-esterifying enzyme, promotes efflux of cholesterol depots in peripheral tissues to the liver to be catabolized and excreted (56). The PON1 enzyme prevents LDL oxidation and downstream events that can form atheroma (57). Correlations with RBP4, transthyretin, and afamin, which are all known to be physically bound to HDL particles, are consistent with their known roles in HDL of transporting fat-soluble vitamins A and E (29, 58).
Interestingly, our analysis has revealed several novel correlations between HDL-C and its constituent apoA-1 with intracellular proteins (Fig. 1A). IFRD2 shared the second strongest correlation with HDL-C, although its general functions remain unknown. EIF2D, KLF17, and TMF1 are known to coregulate transcriptional and translational processes. Although these proteins are not likely carried by lipoproteins and mechanisms giving rise to these strong correlations with HDL-C in plasma are unknown, we postulate that they may regulate expression of apoA-1 or other genes involved in HDL biogenesis and metabolism in hepatocytes (59). Lastly, positive correlations with ANXTR1 and ANXTR2, proteins found to be physically bound to HDL particles (60) and essential for homeostasis of extracellular matrix (61), could reflect HDL’s interactions with extracellular molecules and growth factors that promote resolution of inflammation and wound healing with epithelial proliferation (62).
The negative correlates of the HDL proteome are widely involved in acute phase reactions and the complement system, with more than half previously reported to be associated with inflammation (Fig. 1B, colored in red) (31). These results are consistent with evidence that HDL hosts a large number of acute phase reactants and complement factors, providing a platform for assembly of immunomodulatory complexes (7). Although these negative correlations were relatively less strong than positive correlations with HDL-C, they affirm a recent notion that HDL is an integral component of the innate host defense system (6, 8).
In this thin pediatric population, elevated plasma triglycerides and low concentrations of HDL-C were common. Similar abnormal lipid profiles have also been observed in undernourished children in Brazil, Iran, and India (63–66). Predominantly cereal-based diets with low intakes of animal source foods in this population (67) may promote hypertriglyceridemia and low HDL-C and LDL-C in these children. Subclinical inflammation can also contribute to dyslipidemia, as high triglycerides and low HDL-C concentrations were prominent in these Nepalese children with inflammation, as consistently observed in patients with diverse inflammatory disorders and infectious diseases (22) and children with cystic fibrosis (68, 69) and bacterial infections (70). Substantial inflammation-associated changes in the relative abundance of HDL-correlated proteins appear to reflect both altered lipoprotein metabolism and dysfunction. For example, increased abundance of SAA4 (+12% among inflamed children) is a hallmark of a pro-inflammatory HDL phenotype, involving a displacement of other apolipoproteins and accelerated clearance of HDL (22, 71). Reduced abundance of PLTP (−5%) and LCAT (−3%) and their potent activators (apoA-1, apoA-IV, and apoC-I) can disturb the formation and maturation of HDL (72). Reduced PON1 (−8%) can promote oxidation of LDL and lead to increased accumulation of cholesterol in macrophages (73). It has been suggested that increased abundance of acute phase proteins residing in HDL particles are part of the innate response that provides rapid protection to a host (74). However, an unresolved chronic presence of pro-inflammatory agents with loss of anti-inflammatory and anti-oxidative proteins may compromise the atheroprotective capacity of HDL (74). Changes in the abundance of proteins correlated with LDL-C (n = 1) and triglycerides (n = 4) were also observed, but to a lesser extent than the HDL proteome. As triglyceride-rich lipoproteins carry less protein cargo than HDL, elevated concentrations of plasma triglyceride in these children are more likely to be affected by other factors, such as increased de novo hepatic synthesis and secretion of VLDL and low lipoprotein lipase activity during inflammation (22).
This is the first study, to our knowledge, that has profiled plasma proteomes of lipoproteins and triglyceride in a population of undernourished children, often considered to be at low risk of dyslipidemia. Extensive data on nutritional status and subclinical inflammation enabled us to estimate the potential effects of inflammation on a family of plasma proteomes correlated with lipid and lipoproteins. An unbiased quantitative plasma proteomics approach allowed us to discover a broadened scope of proteomes by discovering novel proteins originating probably in the liver, peripheral tissues, vascular endothelium, and immune cells that may be interpreted to reflect population homeostasis of lipoproteins and their likely interactions with multiple tissues. A major limitation is that we were unable to differentiate detected proteins by whether they were physically bound to lipoprotein particles because we did not isolate and purify lipoprotein particles by ultracentrifugation (75). Another limitation of this study is that high minimum detectable limits of lipid measurement resulted in many missing values, especially in LDL-C concentrations. Lastly, due to the relative scale of protein abundance quantified by iTRAQ MS, absolute abundance of individual proteins could not be compared. As some identified proteins serve as indicators of both lipid status and likely cardioprotective or atherosclerotic function, these proteins may be good candidates for targeted quantification and further evaluation.
In this generally undernourished population of school-aged children living in the rural southern plains of Nepal, global plasma proteomic characterization revealed a family of lipoproteomes with distinct proteins correlated with concentrations of LDL-C and triglyceride, and a large highly diverse HDL proteome that displayed substantial variation by inflammation status. As dyslipidemia and chronic inflammation, both major risk factors of cardiovascular disease, may also coexist among children in similar impoverished settings, further studies are warranted to evaluate clinical and public health implications of identified proteins as early childhood biomarkers of vascular health and disease in later life.
Supplementary Material
Acknowledgments
The authors thank the Johns Hopkins Nutriproteomics Research Team and the Nepal field team (Margia Arguello, Raghothama Chaerkady, Hongie Cui, Lauren R. DeVine, Jaime Johnson, Robert O’Meally, Steven C. LeClerq, Ashika Nanayakkara-Bind, Hee-Sool Rho, Sudeep Shrestha, and Fredrick Van Dyke). The authors also thank Ingo Ruczinski for his proteomics modeling and analytic guidance and C. Conover Talbot, Jr. for assistance with the HUGO gene annotation.
Footnotes
Abbreviations:
- AGP
- α-1-acid glycoprotein
- ANTXR
- anthrax toxin receptor
- CETP
- cholesteryl ester transfer protein
- CRP
- C-reactive protein
- EIF2D
- eukaryotic translation initiation factor 2D
- FDR
- false discovery rate
- HDL-C
- HDL-cholesterol
- HUGO
- Human Genome Organization
- IFRD2
- interferon-related developmental regulator 2
- iTRAQ
- isobaric tag for relative and absolute quantification
- KLF17
- Kruppel-like factor 17
- LDL-C
- LDL-cholesterol
- LME
- linear mixed effects
- PLTP
- phospholipid transfer protein
- PON
- paraoxonase
- RBP4
- retinol-binding protein 4
- SAA4
- serum amyloid A4
- TMF1
- TATA element modulatory factor 1
This study was supported by Bill and Melinda Gates Foundation Grants OPP 5241 (Plasma Nutriproteomics Study through the Assessment of Micronutrient Status by Nutriproteomics) and GH 614 (Global Control of Micronutrient Deficiency for the cohort study in Nepal from which plasma samples were obtained). The original field trial in Nepal from 1999 to 2001 in which mothers of studied children were enrolled was supported by United States Agency for International Development Grant HRN-A-00-97-00015-00 (Micronutrients for Health Cooperative Agreement between the Office of Health, Infectious Diseases, and Nutrition and the Center for Human Nutrition, Johns Hopkins Bloomberg School of Public Health). The Sight and Life Global Nutrition Research Institute, Baltimore, MD, and DSM Ltd, Kaiseraugst, Switzerland, provided additional assistance.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Dominiczak M. H., and Caslake M. J.. 2011. Apolipoproteins: metabolic role and clinical biochemistry applications. Ann. Clin. Biochem. 48: 498–515. [DOI] [PubMed] [Google Scholar]
- 2.Davidson W. S., and Thompson T. B.. 2007. The structure of apolipoprotein A-I in high density lipoproteins. J. Biol. Chem. 282: 22249–22253. [DOI] [PubMed] [Google Scholar]
- 3.Brown M. S., and Goldstein J. L.. 1985. The receptor model for transport of cholesterol in plasma. Ann. N. Y. Acad. Sci. 454: 178–182. [DOI] [PubMed] [Google Scholar]
- 4.Hoofnagle A. N., and Heinecke J. W.. 2009. Lipoproteomics: using mass spectrometry-based proteomics to explore the assembly, structure, and function of lipoproteins. J. Lipid Res. 50: 1967–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Zychlinski A., Williams M., McCormick S., and Kleffmann T.. 2014. Absolute quantification of apolipoproteins and associated proteins on human plasma lipoproteins. J. Proteomics. 106: 181–190. [DOI] [PubMed] [Google Scholar]
- 6.Gordon S. M., Hofmann S., Askew D. S., and Davidson W. S.. 2011. High density lipoprotein: it’s not just about lipid transport anymore. Trends Endocrinol. Metab. 22: 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaisar T., Pennathur S., Green P. S., Gharib S. A., Hoofnagle A. N., Cheung M. C., Byun J., Vuletic S., Kassim S., Singh P., et al. 2007. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Invest. 117: 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah A. S., Tan L., Long J. L., and Davidson W. S.. 2013. Proteomic diversity of high density lipoproteins: our emerging understanding of its importance in lipid transport and beyond. J. Lipid Res. 54: 2575–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidsson P., Hulthe J., Fagerberg B., and Camejo G.. 2010. Proteomics of apolipoproteins and associated proteins from plasma high-density lipoproteins. Arterioscler. Thromb. Vasc. Biol. 30: 156–163. [DOI] [PubMed] [Google Scholar]
- 10.Karlsson H., Leanderson P., Tagesson C., and Lindahl M.. 2005. Lipoproteomics II: mapping of proteins in high-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 5: 1431–1445. [DOI] [PubMed] [Google Scholar]
- 11.Gordon S. M., Deng J., Lu L. J., and Davidson W. S.. 2010. Proteomic characterization of human plasma high density lipoprotein fractionated by gel filtration chromatography. J. Proteome Res. 9: 5239–5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rezaee F., Casetta B., Levels J. H., Speijer D., and Meijers J. C.. 2006. Proteomic analysis of high-density lipoprotein. Proteomics. 6: 721–730. [DOI] [PubMed] [Google Scholar]
- 13.Bancells C., Canals F., Benitez S., Colome N., Julve J., Ordonez-Llanos J., and Sanchez-Quesada J. L.. 2010. Proteomic analysis of electronegative low-density lipoprotein. J. Lipid Res. 51: 3508–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlsson H., Leanderson P., Tagesson C., and Lindahl M.. 2005. Lipoproteomics I: mapping of proteins in low-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 5: 551–565. [DOI] [PubMed] [Google Scholar]
- 15.Mancone C., Amicone L., Fimia G. M., Bravo E., Piacentini M., Tripodi M., and Alonzi T.. 2007. Proteomic analysis of human very low-density lipoprotein by two-dimensional gel electrophoresis and MALDI-TOF/TOF. Proteomics. 7: 143–154. [DOI] [PubMed] [Google Scholar]
- 16.Chait A., Han C. Y., Oram J. F., and Heinecke J. W.. 2005. Lipoprotein-associated inflammatory proteins: markers or mediators of cardiovascular disease? J. Lipid Res. 46: 389–403. [DOI] [PubMed] [Google Scholar]
- 17.Geyer P. E., Wewer Albrechtsen N. J., Tyanova S., Grassl N., Iepsen E. W., Lundgren J., Madsbad S., Holst J. J., Torekov S. S., and Mann M.. 2016. Proteomics reveals the effects of sustained weight loss on the human plasma proteome. Mol. Syst. Biol. 12: 901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khovidhunkit W., Kim M. S., Memon R. A., Shigenaga J. K., Moser A. H., Feingold K. R., and Grunfeld C.. 2004. Effects of infection and inflammation on lipid and lipoprotein metabolism: mechanisms and consequences to the host. J. Lipid Res. 45: 1169–1196. [DOI] [PubMed] [Google Scholar]
- 19.Haque S., Mirjafari H., and Bruce I. N.. 2008. Atherosclerosis in rheumatoid arthritis and systemic lupus erythematosus. Curr. Opin. Lipidol. 19: 338–343. [DOI] [PubMed] [Google Scholar]
- 20.Samarasekera E. J., Neilson J. M., Warren R. B., Parnham J., and Smith C. H.. 2013. Incidence of cardiovascular disease in individuals with psoriasis: a systematic review and meta-analysis. J. Invest. Dermatol. 133: 2340–2346. [DOI] [PubMed] [Google Scholar]
- 21.Becker A. E., de Boer O. J., and van Der Wal A. C.. 2001. The role of inflammation and infection in coronary artery disease. Annu. Rev. Med. 52: 289–297. [DOI] [PubMed] [Google Scholar]
- 22.Feingold K. R., and Grunfeld C.. 2015. The Effect of Inflammation and Infection on Lipids and Lipoproteins. Accessed September 7, 2018, at https://www.ncbi.nlm.nih.gov/books/NBK326741/. [Google Scholar]
- 23.Vaisar T., Tang C., Babenko I., Hutchins P., Wimberger J., Suffredini A. F., and Heinecke J. W.. 2015. Inflammatory remodeling of the HDL proteome impairs cholesterol efflux capacity. J. Lipid Res. 56: 1519–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman W. P. 3rd, Freedman D. S., Voors A. W., Gard P. D., Srinivasan S. R., Cresanta J. L., Williamson G. D., Webber L. S., and Berenson G. S.. 1986. Relation of serum lipoprotein levels and systolic blood pressure to early atherosclerosis. The Bogalusa Heart Study. N. Engl. J. Med. 314: 138–144. [DOI] [PubMed] [Google Scholar]
- 25.Stewart C. P., Christian P., Schulze K. J., Leclerq S. C., West K. P. Jr., and Khatry S. K.. 2009. Antenatal micronutrient supplementation reduces metabolic syndrome in 6- to 8-year-old children in rural Nepal. J. Nutr. 139: 1575–1581. [DOI] [PubMed] [Google Scholar]
- 26.Christian P., Khatry S. K., Katz J., Pradhan E. K., LeClerq S. C., Shrestha S. R., Adhikari R. K., Sommer A., and West K. P. Jr.. 2003. Effects of alternative maternal micronutrient supplements on low birth weight in rural Nepal: double blind randomised community trial. BMJ. 326: 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart C. P., Christian P., LeClerq S. C., West K. P. Jr., and Khatry S. K.. 2009. Antenatal supplementation with folic acid + iron + zinc improves linear growth and reduces peripheral adiposity in school-age children in rural Nepal. Am. J. Clin. Nutr. 90: 132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulze K. J., Christian P., Wu L. S., Arguello M., Cui H., Nanayakkara-Bind A., Stewart C. P., Khatry S. K., LeClerq S., and West K. P. Jr.. 2014. Micronutrient deficiencies are common in 6- to 8-year-old children of rural Nepal, with prevalence estimates modestly affected by inflammation. J. Nutr. 144: 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cole R. N., Ruczinski I., Schulze K., Christian P., Herbrich S., Wu L., Devine L. R., O’Meally R. N., Shrestha S., Boronina T. N., et al. 2013. The plasma proteome identifies expected and novel proteins correlated with micronutrient status in undernourished Nepalese children. J. Nutr. 143: 1540–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bachorik P. S., and Ross J. W.. 1995. National Cholesterol Education Program recommendations for measurement of low-density lipoprotein cholesterol: executive summary. The National Cholesterol Education Program Working Group on Lipoprotein Measurement. Clin. Chem. 41: 1414–1420. [PubMed] [Google Scholar]
- 31.Lee S. E., West K. P. Jr., Cole R. N., Schulze K. J., Christian P., Wu L. S., Yager J. D., Groopman J., and Ruczinski I.. 2015. Plasma proteome biomarkers of inflammation in school aged children in Nepal. PLoS One. 10: e0144279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herbrich S. M., Cole R. N., West K. P. Jr., Schulze K., Yager J. D., Groopman J. D., Christian P., Wu L., O’Meally R. N., May D. H., et al. 2013. Statistical inference from multiple iTRAQ experiments without using common reference standards. J. Proteome Res. 12: 594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Expert Panel on Integrated Guidelines for Cardiovascular Health and Risk Reduction in Children and Adolescents; National Heart, Lung, and Blood Institute. 2011. Expert panel on integrated guidelines for cardiovascular health and risk reduction in children and adolescents: summary report. Pediatrics. 128 (Suppl 5): S213–S256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Storey J. D. 2002. A direct approach to false discovery rates. J. R. Statist. Soc. B. 64: 479–498. [Google Scholar]
- 35.Robinson G. K. 1991. That BLUP is a good thing: the estimation of random effects. Stat. Sci. 6: 15–32. [Google Scholar]
- 36.de Onis M., Onyango A. W., Borghi E., Siyam A., Nishida C., and Siekmann J.. 2007. Development of a WHO growth reference for school-aged children and adolescents. Bull. World Health Organ. 85: 660–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benjamini Y., and Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 57: 289–300. [Google Scholar]
- 38.Gray K. A., Yates B., Seal R. L., Wright M. W., and Bruford E. A.. 2015. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 43: D1079–D1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.UniProt Consortium T. 2018. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 46: 2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.NCBI Resource Coordinators. 2017. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 45: D12–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T.. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamir I., Heiss G., Glueck C. J., Christensen B., Kwiterovich P., and Rifkind B. M.. 1981. Lipid and lipoprotein distributions in white children ages 6-19 yr. The Lipid Research Clinics Program Prevalence Study. J. Chronic Dis. 34: 27–39. [DOI] [PubMed] [Google Scholar]
- 43.Hickman T. B., Briefel R. R., Carroll M. D., Rifkind B. M., Cleeman J. I., Maurer K. R., and Johnson C. L.. 1998. Distributions and trends of serum lipid levels among United States children and adolescents ages 4–19 years: data from the Third National Health and Nutrition Examination Survey. Prev. Med. 27: 879–890. [DOI] [PubMed] [Google Scholar]
- 44.Segrest J. P., Jones M. K., De Loof H., and Dashti N.. 2001. Structure of apolipoprotein B-100 in low density lipoproteins. J. Lipid Res. 42: 1346–1367. [PubMed] [Google Scholar]
- 45.Vale R. D., Reese T. S., and Sheetz M. P.. 1985. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 42: 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanai Y., Wang D., and Hirokawa N.. 2014. KIF13B enhances the endocytosis of LRP1 by recruiting LRP1 to caveolae. J. Cell Biol. 204: 395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stafforini D. M., McIntyre T. M., Zimmerman G. A., and Prescott S. M.. 1997. Platelet-activating factor acetylhydrolases. J. Biol. Chem. 272: 17895–17898. [DOI] [PubMed] [Google Scholar]
- 48.Packard C. J., O’Reilly D. S., Caslake M. J., McMahon A. D., Ford I., Cooney J., Macphee C. H., Suckling K. E., Krishna M., Wilkinson F. E., et al. 2000. Lipoprotein-associated phospholipase A2 as an independent predictor of coronary heart disease. West of Scotland Coronary Prevention Study Group. N. Engl. J. Med. 343: 1148–1155. [DOI] [PubMed] [Google Scholar]
- 49.Lp-PLA(2) Studies Collaboration, Thompson A., Gao P., Orfei L., Watson S., Di Angelantonio E., Kaptoge S., Ballantyne C., Cannon C. P., Criqui M., et al. 2010. Lipoprotein-associated phospholipase A(2) and risk of coronary disease, stroke, and mortality: collaborative analysis of 32 prospective studies. Lancet. 375: 1536–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tall A. R. 1993. Plasma cholesteryl ester transfer protein. J. Lipid Res. 34: 1255–1274. [PubMed] [Google Scholar]
- 51.Kościuczuk E. M., Lisowski P., Jarczak J., Strzałkowska N., Jóźwik A., Horbańczuk J., Krzyźewski J., Zwierzchowski L., and Bagnicka E.. 2012. Cathelicidins: family of antimicrobial peptides. A review. Mol. Biol. Rep. 39: 10957–10970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sørensen O., Bratt T., Johnsen A. H., Madsen M. T., and Borregaard N.. 1999. The human antibacterial cathelicidin, hCAP-18, is bound to lipoproteins in plasma. J. Biol. Chem. 274: 22445–22451. [DOI] [PubMed] [Google Scholar]
- 53.Dashty M., Motazacker M. M., Levels J., de Vries M., Mahmoudi M., Peppelenbosch M. P., and Rezaee F.. 2014. Proteome of human plasma very low-density lipoprotein and low-density lipoprotein exhibits a link with coagulation and lipid metabolism. Thromb. Haemost. 111: 518–530. [DOI] [PubMed] [Google Scholar]
- 54.Mariman E. C., and Wang P.. 2010. Adipocyte extracellular matrix composition, dynamics and role in obesity. Cell. Mol. Life Sci. 67: 1277–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huuskonen J., Olkkonen V. M., Jauhiainen M., and Ehnholm C.. 2001. The impact of phospholipid transfer protein (PLTP) on HDL metabolism. Atherosclerosis. 155: 269–281. [DOI] [PubMed] [Google Scholar]
- 56.Santamarina-Fojo S., Lambert G., Hoeg J. M., and Brewer H. B. Jr.. 2000. Lecithin-cholesterol acyltransferase: role in lipoprotein metabolism, reverse cholesterol transport and atherosclerosis. Curr. Opin. Lipidol. 11: 267–275. [DOI] [PubMed] [Google Scholar]
- 57.Mackness M. I., and Durrington P. N.. 1995. HDL, its enzymes and its potential to influence lipid peroxidation. Atherosclerosis. 115: 243–253. [DOI] [PubMed] [Google Scholar]
- 58.West K. P. Jr., Cole R. N., Shrestha S., Schulze K. J., Lee S. E., Betz J., Nonyane B. A., Wu L. S., Yager J. D., Groopman J. D., et al. 2015. A plasma alpha-tocopherome can be identified from proteins associated with vitamin E status in school-aged children of Nepal. J. Nutr. 145: 2646–2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kardassis D., Gafencu A., Zannis V. I., and Davalos A.. 2015. Regulation of HDL genes: transcriptional, posttranscriptional, and posttranslational. Handb. Exp. Pharmacol. 224: 113–179. [DOI] [PubMed] [Google Scholar]
- 60.Riwanto M., Rohrer L., Roschitzki B., Besler C., Mocharla P., Mueller M., Perisa D., Heinrich K., Altwegg L., von Eckardstein A., et al. 2013. Altered activation of endothelial anti- and proapoptotic pathways by high-density lipoprotein from patients with coronary artery disease: role of high-density lipoprotein-proteome remodeling. Circulation. 127: 891–904. [DOI] [PubMed] [Google Scholar]
- 61.Besschetnova T. Y., Ichimura T., Katebi N., St. Croix B., Bonventre J. V., and Olsen B. R.. 2015. Regulatory mechanisms of anthrax toxin receptor 1-dependent vascular and connective tissue homeostasis. Matrix Biol. 42: 56–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gordts S. C., Singh N., Muthuramu I., and De Geest B.. 2014. Pleiotropic effects of HDL: towards new therapeutic areas for HDL-targeted interventions. Curr. Mol. Med. 14: 481–503. [DOI] [PubMed] [Google Scholar]
- 63.Carvajal I., Malave I., Correa C., Castillo C., Perez M., Hammar S., and Camejo G.. 1992. Changes in the serum lipid fractions of malnourished children with and without clinical infection. Paradoxical hypertriglyceridemia in malnutrition. Arch. Latinoam. Nutr. 42: 250–258. [PubMed] [Google Scholar]
- 64.Hovsepian S., Kelishadi R., Djalalinia S., Farzadfar F., Naderimagham S., and Qorbani M.. 2015. Prevalence of dyslipidemia in Iranian children and adolescents: a systematic review. J. Res. Med. Sci. 20: 503–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khalil A., Gupta S., Madan A., and Venkatesan M.. 1995. Lipid profile norms in Indian children. Indian Pediatr. 32: 1177–1180. [PubMed] [Google Scholar]
- 66.Veiga G. R., Ferreira H. S., Sawaya A. L., Calado J., and Florencio T. M.. 2010. Dyslipidaemia and undernutrition in children from impoverished areas of Maceio, state of Alagoas, Brazil. Int. J. Environ. Res. Public Health. 7: 4139–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Campbell R. K., Talegawkar S. A., Christian P., LeClerq S. C., Khatry S. K., Wu L. S., and West K. P. Jr.. 2014. Seasonal dietary intakes and socioeconomic status among women in the Terai of Nepal. J. Health Popul. Nutr. 32: 198–216. [PMC free article] [PubMed] [Google Scholar]
- 68.Levy E., Gurbindo C., Lacaille F., Paradis K., Thibault L., and Seidman E.. 1993. Circulating tumor necrosis factor-alpha levels and lipid abnormalities in patients with cystic fibrosis. Pediatr. Res. 34: 162–166. [DOI] [PubMed] [Google Scholar]
- 69.Figueroa V., Milla C., Parks E. J., Schwarzenberg S. J., and Moran A.. 2002. Abnormal lipid concentrations in cystic fibrosis. Am. J. Clin. Nutr. 75: 1005–1011. [DOI] [PubMed] [Google Scholar]
- 70.Henter J. I., Carlson L. A., Hansson M., Nilsson-Ehle P., and Ortqvist E.. 1993. Lipoprotein alterations in children with bacterial meningitis. Acta Paediatr. 82: 694–698. [DOI] [PubMed] [Google Scholar]
- 71.Cabana V. G., Siegel J. N., and Sabesin S. M.. 1989. Effects of the acute phase response on the concentration and density distribution of plasma lipids and apolipoproteins. J. Lipid Res. 30: 39–49. [PubMed] [Google Scholar]
- 72.Steinmetz A., Hocke G., Saile R., Puchois P., and Fruchart J. C.. 1989. Influence of serum amyloid A on cholesterol esterification in human plasma. Biochim. Biophys. Acta. 1006: 173–178. [DOI] [PubMed] [Google Scholar]
- 73.Steinberg D., Parthasarathy S., Carew T. E., Khoo J. C., and Witztum J. L.. 1989. Beyond cholesterol. Modifications of low-density lipoprotein that increase its atherogenicity. N. Engl. J. Med. 320: 915–924. [DOI] [PubMed] [Google Scholar]
- 74.Jahangiri A. 2010. High-density lipoprotein and the acute phase response. Curr. Opin. Endocrinol. Diabetes Obes. 17: 156–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Holzer M., Kern S., Birner-Grunberger R., Curcic S., Heinemann A., and Marsche G.. 2016. Refined purification strategy for reliable proteomic profiling of HDL2/3: impact on proteomic complexity. Sci. Rep. 6: 38533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Banfi C., Brioschi M., Barcella S., Wait R., Begum S., Galli S., Rizzi A., and Tremoli E.. 2009. Proteomic analysis of human low-density lipoprotein reveals the presence of prenylcysteine lyase, a hydrogen peroxide-generating enzyme. Proteomics. 9: 1344–1352. [DOI] [PubMed] [Google Scholar]
- 77.Karlsson H., Mortstedt H., Lindqvist H., Tagesson C., and Lindahl M.. 2009. Protein profiling of low-density lipoprotein from obese subjects. Proteomics Clin. Appl. 3: 663–671. [DOI] [PubMed] [Google Scholar]
- 78.Ståhlman M., Davidsson P., Kanmert I., Rosengren B., Borén J., Fagerberg B., and Camejo G.. 2008. Proteomics and lipids of lipoproteins isolated at low salt concentrations in D2O/sucrose or in KBr. J. Lipid Res. 49: 481–490. [DOI] [PubMed] [Google Scholar]
- 79.The Davidson/Shah Lab. 2015. LDL and HDL Proteome Watch. Accessed September 7, 2018, at http://homepages.uc.edu/~davidswm/Labpage.html.
- 80.Alwaili K., Bailey D., Awan Z., Bailey S. D., Ruel I., Hafiane A., Krimbou L., Laboissiere S., and Genest J.. 2012. The HDL proteome in acute coronary syndromes shifts to an inflammatory profile. Biochim. Biophys. Acta. 1821: 405–415. [DOI] [PubMed] [Google Scholar]
- 81.Davidson W. S., Silva R. A., Chantepie S., Lagor W. R., Chapman M. J., and Kontush A.. 2009. Proteomic analysis of defined HDL subpopulations reveals particle-specific protein clusters: relevance to antioxidative function. Arterioscler. Thromb. Vasc. Biol. 29: 870–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gordon S. M., Deng J., Tomann A. B., Shah A. S., Lu L. J., and Davidson W. S.. 2013. Multi-dimensional co-separation analysis reveals protein-protein interactions defining plasma lipoprotein subspecies. Mol. Cell. Proteomics. 12: 3123–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Heller M., Stalder D., Schlappritzi E., Hayn G., Matter U., and Haeberli A.. 2005. Mass spectrometry-based analytical tools for the molecular protein characterization of human plasma lipoproteins. Proteomics. 5: 2619–2630. [DOI] [PubMed] [Google Scholar]
- 84.Holzer M., Birner-Gruenberger R., Stojakovic T., El-Gamal D., Binder V., Wadsack C., Heinemann A., and Marsche G.. 2011. Uremia alters HDL composition and function. J. Am. Soc. Nephrol. 22: 1631–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Holzer M., Wolf P., Curcic S., Birner-Gruenberger R., Weger W., Inzinger M., El-Gamal D., Wadsack C., Heinemann A., and Marsche G.. 2012. Psoriasis alters HDL composition and cholesterol efflux capacity. J. Lipid Res. 53: 1618–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hortin G. L., Shen R. F., Martin B. M., and Remaley A. T.. 2006. Diverse range of small peptides associated with high-density lipoprotein. Biochem. Biophys. Res. Commun. 340: 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mangé A., Goux A., Badiou S., Patrier L., Canaud B., Maudelonde T., Cristol J. P., and Solassol J.. 2012. HDL proteome in hemodialysis patients: a quantitative nanoflow liquid chromatography-tandem mass spectrometry approach. PLoS One. 7: e34107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shao B., de Boer I., Tang C., Mayer P. S., Zelnick L., Afkarian M., Heinecke J. W., and Himmelfarb J.. 2015. A cluster of proteins implicated in kidney disease is increased in high-density lipoprotein isolated from hemodialysis subjects. J. Proteome Res. 14: 2792–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sreckovic I., Birner-Gruenberger R., Obrist B., Stojakovic T., Scharnagl H., Holzer M., Scholler M., Philipose S., Marsche G., Lang U., et al. 2013. Distinct composition of human fetal HDL attenuates its anti-oxidative capacity. Biochim. Biophys. Acta. 1831: 737–746. [DOI] [PubMed] [Google Scholar]
- 90.Watanabe J., Charles-Schoeman C., Miao Y., Elashoff D., Lee Y. Y., Katselis G., Lee T. D., and Reddy S. T.. 2012. Proteomic profiling following immunoaffinity capture of high-density lipoprotein: association of acute-phase proteins and complement factors with proinflammatory high-density lipoprotein in rheumatoid arthritis. Arthritis Rheum. 64: 1828–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weichhart T., Kopecky C., Kubicek M., Haidinger M., Doller D., Katholnig K., Suarna C., Eller P., Tolle M., Gerner C., et al. 2012. Serum amyloid A in uremic HDL promotes inflammation. J. Am. Soc. Nephrol. 23: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.