Abstract
Phosphatidic acid (PA) phosphatase is an evolutionarily conserved enzyme that plays a major role in lipid homeostasis by controlling the cellular levels of its substrate, PA, and its product, diacylglycerol. These lipids are essential intermediates for the synthesis of triacylglycerol and membrane phospholipids; they also function in lipid signaling, vesicular trafficking, lipid droplet formation, and phospholipid synthesis gene expression. The importance of PA phosphatase to lipid homeostasis and cell physiology is exemplified in yeast, mice, and humans by a host of cellular defects and lipid-based diseases associated with loss or overexpression of the enzyme activity. In this review, we focus on the mode of action and regulation of PA phosphatase in the yeast Saccharomyces cerevisiae. The enzyme Pah1 translocates from the cytosol to the nuclear/endoplasmic reticulum membrane through phosphorylation and dephosphorylation. Pah1 phosphorylation is mediated in the cytosol by multiple protein kinases, whereas dephosphorylation is catalyzed on the membrane surface by an integral membrane protein phosphatase. Posttranslational modifications of Pah1 also affect its catalytic activity and susceptibility to degradation by the proteasome. Additional mechanistic understanding of Pah1 regulation should be instrumental for the identification of small-molecule inhibitors or activators that can fine-tune PA phosphatase function and thereby restore lipid homeostasis.
Keywords: diacylglycerol, triacylglycerol, Nem1-Spo7 protein phosphatase complex, obesity, lipodystrophy
Graphical Abstract
ROLES OF PA PHOSPHATASE IN LIPID METABOLISM AND ITS IMPORTANCE TO CELL PHYSIOLOGY
Phosphatidic acid (PA) phosphatase,2 the enzyme that catalyzes the Mg2+-dependent dephosphorylation of PA to produce diacylglycerol (DAG) (Fig. 1), has emerged as a vital regulator of lipid homeostasis in eukaryotic organisms (1). The PA phosphatase reaction was first characterized in 1957 from chicken liver extracts by Smith et al. (2). Yet, the existence of the enzyme had been implicated two years earlier by Kates (3) from a study on the hydrolysis of phosphatidylcholine with spinach chloroplasts. Among many attempts to purify PA phosphatase from diverse organisms, Lin and Carman (4) in 1989 could prepare the enzyme to near homogeneity from the yeast Saccharomyces cerevisiae. The enzyme-encoding gene PAH1 was identified from S. cerevisiae in 2006 by Han et al. (5), revealing that it is evolutionarily conserved in higher eukaryotes including human.
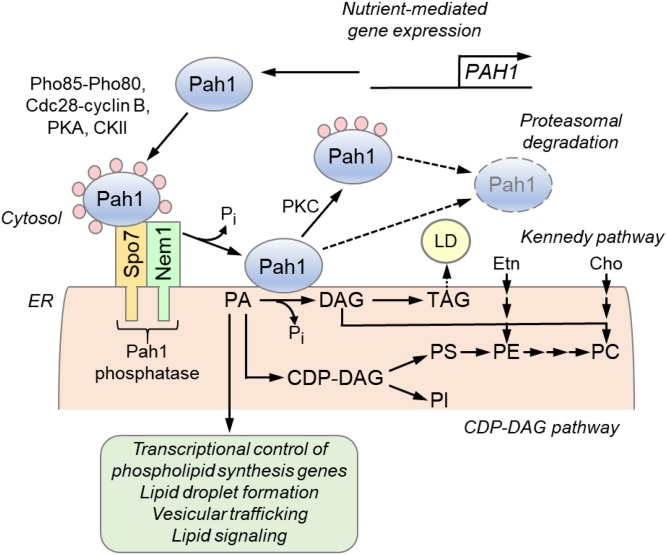
Fig. 1.
Roles and regulation of PAH1-encoded PA phosphatase in lipid synthesis. The expression of the PAH1 gene that encodes the PA phosphatase protein Pah1 is regulated throughout growth by nutrient status. Pah1 in the cytosol is phosphorylated by multiple protein kinases during vegetative growth when the synthesis of phospholipids occurs at the expense of TAG. As cells progress into stasis, the phosphorylated Pah1 (pink circles) translocates to the ER membrane through its dephosphorylation by the Pah1 phosphatase, which is composed of Nem1 (catalytic subunit) and Spo7 (regulatory subunit). Dephosphorylated Pah1 that is associated with the ER membrane catalyzes the conversion of PA to DAG, which is then acylated to form TAG that is stored in lipid droplets (LD). Dephosphorylated Pah1 or PKC-phosphorylated Pah1 that is not phosphorylated at the seven target sites for Pho85-Pho80 protein kinase is degraded by the proteasome (dashed line arrows). PA is also utilized for the synthesis of membrane phospholipids via the CDP-DAG pathway, and it has signaling functions (green). When the CDP-DAG pathway for phospholipid synthesis is blocked, phosphatidylcholine (PC) or phosphatidylethanolamine (PE) may be synthesized from the DAG derived from the PA phosphatase reaction if cells are supplemented with choline (Cho) or ethanolamine (Etn) via the Kennedy pathway. PI, phosphatidylinositol; PS, phosphatidylserine.
DAG produced by PA phosphatase is acylated to produce the storage lipid triacylglycerol (TAG) at the ER membrane, which is then incorporated into lipid droplets (6) (Fig. 1). The DAG is also used to synthesize the membrane phospholipids phosphatidylcholine or phosphatidylethanolamine via the Kennedy pathway (7). The substrate PA is a precursor for membrane phospholipids that are synthesized via the liponucleotide intermediate CDP-DAG (7) (Fig. 1). In addition to their uses for lipid synthesis, PA/DAG play roles as regulatory molecules (e.g., lipid signaling, lipid droplet formation, vesicular trafficking, phospholipid synthesis gene expression) (8, 9). Thus, by the nature of its reaction, PA phosphatase controls the synthesis of TAG and membrane phospholipids, and the abundance of lipid signaling molecules.
The importance of PA phosphatase to lipid homeostasis and cell physiology is highlighted in yeast cells lacking the enzyme (e.g., the pah1Δ mutant) and in mice and humans containing the mutations of the enzyme genes (e.g., Lpin and LPIN, respectively). The pah1Δ mutant exhibits abnormal expansion of the nuclear/ER membrane (10), which is attributed to increases of the PA level and phospholipid synthesis that occur at the expense of TAG synthesis (5, 11). The increased phospholipid synthesis correlates with the derepression of phospholipid synthesis gene expression (10, 12), whereas the decreased TAG synthesis correlates with a decrease in lipid droplet formation (13–15). The pah1Δ mutant is susceptible to fatty acid-induced toxicity (13) and hypersensitive to oxidative stress (16) and exhibits a decrease in chronological life span (16). It also shows a defect in diverse cellular processes including vacuole fusion and acidification (17, 18), autophagy (19), cell wall integrity (20, 21), and growth on nonfermentable carbon sources (i.e., respiratory deficiency) (5, 22) and at elevated temperatures (5, 10, 22). Some of the pah1Δ mutant phenotypes (e.g., nuclear/ER membrane expansion, increased phospholipid synthesis gene expression and phospholipid content, and reduced lipid droplet formation) are suppressed by the loss of the DGK1 gene, which encodes CTP-dependent DAG kinase (1, 13, 14, 23), indicating that a proper balance of PA/ DAG is important to lipid metabolism and cell physiology. In mammals, lipin 1 influences lipid metabolism in multiple tissues. Lipin 1 deficiency in human and mice causes rhabdomyolysis (24, 25), and deficiency in mice is also characterized by hepatic steatosis during the neonatal period, lipodystrophy, insulin resistance, and peripheral neuropathy (26, 27). Lpin1 overexpression results in increased lipogenesis and obesity (28). Polymorphisms in the human LPIN1 gene are associated with insulin resistance and the metabolic syndrome (29). Human lipin 2 deficiency causes chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anemia (30, 31), whereas genetic variations in the human LPIN2 gene are associated with type 2 diabetes (32).
The identification of the PA phosphatase gene has been an exciting discovery with the anticipation that the enzyme could be a target for inhibition of TAG synthesis to ameliorate obesity. However, as indicated above, abolishing PA phosphatase activity may lead to other disease conditions, and thus the enzyme inhibition needs to be fine-tuned at various levels. The known disease conditions also indicate that a fine-tuning of enzyme activation is equally important. The roles and regulation of PA phosphatase are largely conserved from yeast to human, and because of its experimental advantages, yeast has been utilized to study the enzyme and its role in lipid metabolism (1). In this article, a progress in understanding the mode of action and regulation of yeast PA phosphatase is discussed with a perspective on research that might lead to discovering molecules that inhibit or stimulate activity to modulate lipid homeostasis.3
CONSERVED AND NON-CONSERVED REGIONS IN THE YEAST PA PHOSPHATASE PROTEIN Pah1
The yeast PA phosphatase protein Pah1 consists of the conserved and nonconserved regions (Fig. 2). The conserved N-LIP and C-LIP (HAD-like) domains (5, 26) are essential for catalytic activity and thus for its physiological function (33), and a conserved tryptophan residue within the sequence WRDPLVDID is also required for physiological function (34). The N-terminal sequence of Pah1 forming an amphipathic helix binds to the membrane (35), and its C-terminal acidic tail clustered with negatively charged amino acids is required to interact with the ER-associated Pah1 phosphatase (Nem1-Spo7 complex) (35–37). The nonconserved regions of Pah1, which are located between the conserved domains and after the C-LIP domain (Fig. 2), are unfolded and render the enzyme susceptible to proteasomal degradation (34). These regions contain most of the Pah1 phosphorylation sites and play a major role in the enzyme localization and in the control of its stability (35, 38–43).
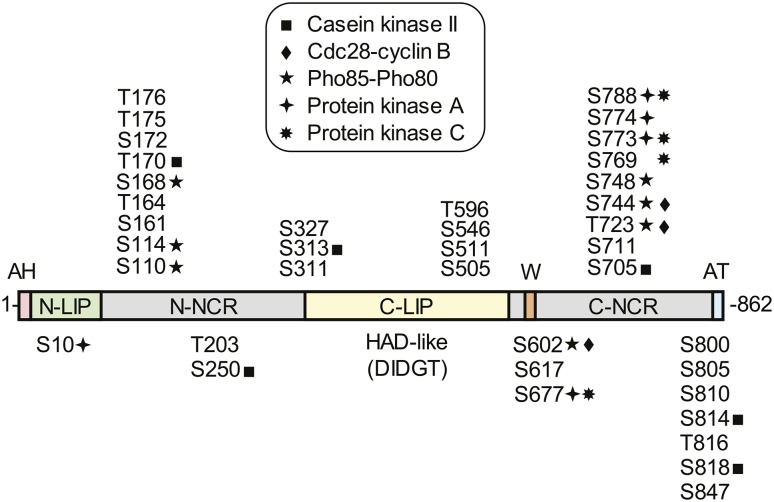
Fig. 2.
Domains/regions and phosphorylation sites in Pah1. The diagram shows the positions of the amphipathic helix (AH, pink) required for ER membrane interaction, the N-LIP (green) and C-LIP (contains HAD-like domain with DIDGT catalytic motif) (yellow) domains that are required for PA phosphatase activity, the acidic tail (AT, blue) required for interaction with Spo7 of the Pah1 phosphatase, N-terminal non-conserved region (N-NCR, grey), C-terminal non-conserved region (C-NCR, grey), the serine (S) and threonine (T) residues in their approximate regions that are known phosphorylation sites, and the sites that are phosphorylated by CKII, Cdc28-cyclin B, Pho85-Pho80, PKA, and PKC, and the tryptophan (W) residue within the C-terminal conserved sequence WRDPLVDID (orange) required for Pah1 function in vivo.
PHOSPHORYLATION/DEPHOSPHORYLATION IS A KEY MECHANISM FOR REGULATING PA PHOSPHATASE FUNCTION
Yeast PA phosphatase is regulated by genetic and biochemical mechanisms. On a transcriptional level, the PAH1 gene is regulated by cell growth and its expression is maximal in the stationary phase when nutrients are depleted (11, 18, 44). As expected, the level of PAH1 expression correlates with the extent of TAG synthesis (11, 18, 44). Biochemically, the enzyme activity of Pah1 is stimulated by negatively charged phospholipids (e.g., CDP-DAG and phosphatidylinositol) (45), but inhibited by positively charged sphingoid bases (e.g., phytosphingosine) (46) and by nucleotides (e.g., ATP and CTP) (47). In addition, Pah1 undergoes phosphorylation and dephosphorylation, and these posttranslational modifications are crucial to control its membrane localization, catalytic activity, and stability (1, 48) (Fig. 1). Pah1 is phosphorylated by several protein kinases, which include cyclin-dependent protein kinases [e.g., Pho85-Pho80 (40) and Cdc28-cyclin B (39)], protein kinase A (PKA) (41), protein kinase C (PKC) (42), and casein kinase II (CKII) (49) (Figs. 1, 2). The phosphorylation of Pah1 by Pho85- Pho80 (40), Cdc28-cyclin B (10, 39), and PKA (41) causes its localization in the cytosol apart from the substrate PA present in the membrane (Fig. 1). Moreover, Pah1 phosphorylated by Pho85-Pho80 (40) and PKA (41) has reduced catalytic activity. Its phosphorylation by CKII has little effect on catalytic activity but inhibits its subsequent phosphorylation by PKA (49). The PKC phosphorylation of Pah1 does not affect its localization or catalytic activity, but instead regulates its stability (42). This phosphorylation, which is favored without phosphorylation by Pho85-Pho80, promotes proteolysis by the 20S proteasome (42, 50) (Fig. 1).
Unlike its phosphorylation by multiple protein kinases, Pah1 is dephosphorylated by a single protein phosphatase (e.g., Pah1 phosphatase) localized in the nuclear/ER membrane that is composed of the Nem1 catalytic subunit and the Spo7 regulatory subunit (36) (Fig. 1). Both Nem1 and Spo7 contain two transmembrane-spanning regions that are responsible for their association with the nuclear/ER membrane (36). Nem1 binds to Spo7 through its conserved C-terminal region, and this association is responsible for the formation of the complex in the membrane bilayer (36). Like Pah1 (5, 29), Nem1 is a member of the HAD (haloacid dehalogenase) superfamily (51) and its phosphatase activity is dependent on the DXDX(TV) catalytic motif. The catalytic function of Nem1 on the Pah1 substrate requires its association with Spo7 (10, 36), and the specificity of the dephosphorylations is in the order of the sites phosphorylated by Pho85-Pho80 > PKA = CKII > Cdc28-cyclin B > PKC (43, 49). The dephosphorylation of Pah1 results in its translocation to the membrane (10, 35, 37–41, 52, 53) (Fig. 1). Moreover, dephosphorylated Pah1 is catalytically more active (38, 43). Given the requirement of the protein phosphatase complex on Pah1 function, cells lacking Nem1 and/or Spo7 exhibit phenotypes shown by cells lacking Pah1 (10, 11, 36). Overall, the modifications of Pah1 by phosphorylation and dephosphorylation ensure a precise control of its catalytic function on the target membrane.
Interestingly, while the Nem1-Spo7 complex functions to dephosphorylate Pah1, the complex itself has been shown to be phosphorylated (54, 55). However, the protein kinases involved and the specific effects of the phosphorylations on the complex function have yet to be elucidated.
HOW DO WE DISCOVER MOLECULES TO CONTROL PA PHOSPHATASE FUNCTION WITHOUT DISTURBING LIPID HOMEOSTASIS?
Because of its role in the synthesis of TAG, PA phosphatase can be considered a drug target to ameliorate obesity and/or lipodystrophy. However, effector molecules specific for the enzyme have yet to be identified. The discovery of PA phosphatase regulators requires a systematic process that involves the library screening of natural products or synthetic compounds and/or the synthesis of substrate mimics serving as a specific inhibitor/activator. Rational drug design, which is commonly used by the pharmaceutical industry, requires the structural information of target proteins. For PA phosphatase, its structural determination has been a challenge because the enzyme is intrinsically unstable due to the unfolded regions. While the phosphorylation stabilizes the protein to some degree (50), it has been difficult to prepare it as a fully phosphorylated form that is suitable for crystallography. A genetically engineered protein that is functional in vivo but lacks the nonconserved regions (34) might be amenable to crystallization. However, it is the disordered regions that are so critical to enzyme regulation. Thus, at this point, rational drug design based on structure is dubious.
As discussed above, a potent inhibitory molecule that abolishes PA phosphatase activity would not be suitable as an obesity drug because it would disrupt lipid homeostasis and lead to other lipid-based diseases or even cancer. Accordingly, molecules that moderately affect PA phosphatase activity seem to be better in the control of TAG synthesis. The fine-tuning of PA phosphatase can also be achieved by the control of its cellular location as mediated by phosphorylation/dephosphorylation. Thus, understanding the phosphorylation and dephosphorylation as well as control of the posttranslational modifications could lead to the discovery of molecules to control PA phosphatase function. In addition, PA phosphatase activity can be indirectly controlled by molecules that affect the function of the Nem1-Spo7 complex through the control of its complex formation as well as the control of the phosphatase interaction with its substrate PA phosphatase. Additional studies are needed to gain a better understanding of these interactions and whether phosphorylation affects the protein-protein interactions. Yet another approach of regulating PA phosphatase function is to control its programmed degradation by the proteasome, which itself is influenced by phosphorylation/dephosphorylation. Clearly, more work is needed to fully understand the structure-function and control of PA phosphatase with the goal of using the enzyme as a target to control TAG-related diseases.
Footnotes
Abbreviations:
- CKII
- casein kinase II
- DAG
- diacylglycerol
- PA
- phosphatidic acid
- PKA
- protein kinase A
- PKC
- protein kinase C
- TAG
- triacylglycerol
This work was supported, in whole or in part, by National Institutes of Health United States Public Health Service Grants GM028140 and GM050679. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare that there are no conflicts of interest with the contents of this article.
REFERENCES
- 1.Pascual F., and Carman G. M.. 2013. Phosphatidate phosphatase, a key regulator of lipid homeostasis. Biochim. Biophys. Acta. 1831: 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith S. W., Weiss S. B., and Kennedy E. P.. 1957. The enzymatic dephosphorylation of phosphatidic acids. J. Biol. Chem. 228: 915–922. [PubMed] [Google Scholar]
- 3.Kates M. 1955. Hydrolysis of lecithin by plant plastid enzymes. Can. J. Biochem. 33: 575–589. [PubMed] [Google Scholar]
- 4.Lin Y-P., and Carman G. M.. 1989. Purification and characterization of phosphatidate phosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 264: 8641–8645. [PubMed] [Google Scholar]
- 5.Han G-S., Wu W-I., and Carman G. M.. 2006. The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281: 9210–9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorger D., and Daum G.. 2003. Triacylglycerol biosynthesis in yeast. Appl. Microbiol. Biotechnol. 61: 289–299. [DOI] [PubMed] [Google Scholar]
- 7.Henry S. A., Kohlwein S., and Carman G. M.. 2012. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 190: 317–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carman G. M., and Henry S. A.. 2007. Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 282: 37293–37297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrasco S., and Merida I.. 2007. Diacylglycerol, when simplicity becomes complex. Trends Biochem. Sci. 32: 27–36. [DOI] [PubMed] [Google Scholar]
- 10.Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., and Siniossoglou S.. 2005. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24: 1931–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pascual F., Soto-Cardalda A., and Carman G. M.. 2013. PAH1-encoded phosphatidate phosphatase plays a role in the growth phase- and inositol-mediated regulation of lipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 288: 35781–35792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han G-S., and Carman G. M.. 2017. Yeast PAH1-encoded phosphatidate phosphatase controls the expression of CHO1-encoded phosphatidylserine synthase for membrane phospholipid synthesis. J. Biol. Chem. 292: 13230–13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fakas S., Qiu Y., Dixon J. L., Han G-S., Ruggles K. V., Garbarino J., Sturley S. L., and Carman G. M.. 2011. Phosphatidate phosphatase activity plays a key role in protection against fatty acid-induced toxicity in yeast . J. Biol. Chem. 286: 29074–29085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adeyo O., Horn P. J., Lee S., Binns D. D., Chandrahas A., Chapman K. D., and Goodman J. M.. 2011. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han S., Bahmanyar S., Zhang P., Grishin N., Oegema K., Crooke R., Graham M., Reue K., Dixon J. E., and Goodman J. M.. 2012. Nuclear envelope phosphatase 1-regulatory subunit 1 (formerly TMEM188) Is the metazoan Spo7p ortholog and functions in the lipin activation pathway. J. Biol. Chem. 287: 3123–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park Y., Han G. S., Mileykovskaya E., Garrett T. A., and Carman G. M.. 2015. Altered lipid synthesis by lack of yeast Pah1 phosphatidate phosphatase reduces chronological life span. J. Biol. Chem. 290: 25382–25394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasser T., Qiu Q. S., Karunakaran S., Padolina M., Reyes A., Flood B., Smith S., Gonzales C., and Fratti R. A.. 2012. The yeast lipin 1 orthologue Pah1p regulates vacuole homeostasis and membrane fusion. J. Biol. Chem. 287: 2221–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherr G. L., LaMassa N., Li E., Phillips G., and Shen C. H.. 2017. Pah1p negatively regulates the expression of V-ATPase genes as well as vacuolar acidification. Biochem. Biophys. Res. Commun. 491: 693–700. [DOI] [PubMed] [Google Scholar]
- 19.Rahman M. A., Mostofa M. G., and Ushimaru T.. 2018. The Nem1/Spo7-Pah1/lipin axis is required for autophagy induction after TORC1 inactivation. FEBS J. 10.1111/febs.14448. [DOI] [PubMed] [Google Scholar]
- 20.Lussier M., White A. M., Sheraton J., di Paolo T., Treadwell J., Southard S. B., Horenstein C. I., Chen-Weiner J., Ram A. F., Capteyn J. C., et al. . 1997. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics. 147: 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz C., Cid V. J., Lussier M., Molina M., and Nombela C.. 1999. A large-scale sonication assay for cell wall mutant analysis in yeast. Yeast. 15: 1001–1008. [DOI] [PubMed] [Google Scholar]
- 22.Irie K., Takase M., Araki H., and Oshima Y.. 1993. A gene, SMP2, involved in plasmid maintenance and respiration in Saccharomyces cerevisiae encodes a highly charged protein. Mol. Gen. Genet. 236: 283–288. [DOI] [PubMed] [Google Scholar]
- 23.Han G-S., O’Hara L., Carman G. M., and Siniossoglou S.. 2008. An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J. Biol. Chem. 283: 20433–20442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeharia A., Shaag A., Houtkooper R. H., Hindi T., de Lonlay P., Erez G., Hubert L., Saada A., de Keyzer Y., Eshel G., et al. . 2008. Mutations in LPIN1 cause recurrent acute myoglobinuria in childhood. Am. J. Hum. Genet. 83: 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang P., Verity M. A., and Reue K.. 2014. Lipin-1 regulates autophagy clearance and intersects with statin drug effects in skeletal muscle. Cell Metab. 20: 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Péterfy M., Phan J., Xu P., and Reue K.. 2001. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27: 121–124. [DOI] [PubMed] [Google Scholar]
- 27.Nadra K., De Preux Charles A-S., Medard J-J., Hendriks W. T., Han G-S., Grés S., Carman G. M., Saulnier-Blache J. S., Verheijen M. H., and Chrast R.. 2008. Phosphatidic acid mediates demyelination in Lpin1 mutant mice. Genes Dev. 22: 1647–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phan J., and Reue K.. 2005. Lipin, a lipodystrophy and obesity gene. Cell Metab. 1: 73–83. [DOI] [PubMed] [Google Scholar]
- 29.Wiedmann S., Fischer M., Koehler M., Neureuther K., Riegger G., Doering A., Schunkert H., Hengstenberg C., and Baessler A.. 2008. Genetic variants within the LPIN1 gene, encoding lipin, are influencing phenotypes of the metabolic syndrome in humans. Diabetes. 57: 209–217. [DOI] [PubMed] [Google Scholar]
- 30.Ferguson P. J., and El-Shanti H. I.. 2007. Autoinflammatory bone disorders. Curr. Opin. Rheumatol. 19: 492–498. [DOI] [PubMed] [Google Scholar]
- 31.Ferguson P. J., Chen S., Tayeh M. K., Ochoa L., Leal S. M., Pelet A., Munnich A., Lyonnet S., Majeed H. A., and El-Shanti H.. 2005. Homozygous mutations in LPIN2 are responsible for the syndrome of chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anaemia (Majeed syndrome). J. Med. Genet. 42: 551–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aulchenko Y. S., Pullen J., Kloosterman W. P., Yazdanpanah M., Hofman A., Vaessen N., Snijders P. J., Zubakov D., Mackay I., Olavesen M., et al. . 2007. LPIN2 is associated with type 2 diabetes, glucose metabolism and body composition. Diabetes. 56: 3020–3026. [DOI] [PubMed] [Google Scholar]
- 33.Han G-S., Siniossoglou S., and Carman G. M.. 2007. The cellular functions of the yeast lipin homolog Pah1p are dependent on its phosphatidate phosphatase activity. J. Biol. Chem. 282: 37026–37035. [DOI] [PubMed] [Google Scholar]
- 34.Park Y., Han G. S., and Carman G. M.. 2017. A conserved tryptophan within the WRDPLVDID domain of yeast Pah1 phosphatidate phosphatase is required for its in vivo function in lipid metabolism. J. Biol. Chem. 292: 19580–19589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karanasios E., Han G-S., Xu Z., Carman G. M., and Siniossoglou S.. 2010. A phosphorylation- regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc. Natl. Acad. Sci. USA. 107: 17539–17544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siniossoglou S., Santos-Rosa H., Rappsilber J., Mann M., and Hurt E.. 1998. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17: 6449–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karanasios E., Barbosa A. D., Sembongi H., Mari M., Han G-S., Carman G. M., and Siniossoglou S.. 2013. Regulation of lipid droplet and membrane biogenesis by the acidic tail of the phosphatidate phosphatase Pah1p. Mol. Biol. Cell. 24: 2124–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Hara L., Han G-S., Peak-Chew S., Grimsey N., Carman G. M., and Siniossoglou S.. 2006. Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J. Biol. Chem. 281: 34537–34548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi H-S., Su W-M., Morgan J. M., Han G-S., Xu Z., Karanasios E., Siniossoglou S., and Carman G. M.. 2011. Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of Ser602, Thr723, and Ser744 as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J. Biol. Chem. 286: 1486–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi H-S., Su W-M., Han G-S., Plote D., Xu Z., and Carman G. M.. 2012. Pho85p-Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism. J. Biol. Chem. 287: 11290–11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su W-M., Han G-S., Casciano J., and Carman G. M.. 2012. Protein kinase A-mediated phosphorylation of Pah1p phosphatidate phosphatase functions in conjunction with the Pho85p- Pho80p and Cdc28p-cyclin B kinases to regulate lipid synthesis in yeast. J. Biol. Chem. 287: 33364–33376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su W-M., Han G-S., and Carman G. M.. 2014. Cross-talk phosphorylations by protein kinase C and Pho85p-Pho80p protein kinase regulate Pah1p phosphatidate phosphatase abundance in Saccharomyces cerevisiae. J. Biol. Chem. 289: 18818–18830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su W-M., Han G-S., and Carman G. M.. 2014. Yeast Nem1-Spo7 protein phosphatase activity on Pah1 phosphatidate phosphatase is specific for the Pho85-Pho80 protein kinase phosphorylation sites. J. Biol. Chem. 289: 34699–34708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soto-Cardalda A., Fakas S., Pascual F., Choi H. S., and Carman G. M.. 2012. Phosphatidate phosphatase plays role in zinc-mediated regulation of phospholipid synthesis in yeast. J. Biol. Chem. 287: 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu W-I., and Carman G. M.. 1996. Regulation of phosphatidate phosphatase activity from the yeast Saccharomyces cerevisiae by phospholipids. Biochemistry. 35: 3790–3796. [DOI] [PubMed] [Google Scholar]
- 46.Wu W-I., Lin Y-P., Wang E., Merrill A. H. Jr., and Carman G. M.. 1993. Regulation of phosphatidate phosphatase activity from the yeast Saccharomyces cerevisiae by sphingoid bases. J. Biol. Chem. 268: 13830–13837. [PubMed] [Google Scholar]
- 47.Wu W-I., and Carman G. M.. 1994. Regulation of phosphatidate phosphatase activity from the yeast Saccharomyces cerevisiae by nucleotides. J. Biol. Chem. 269: 29495–29501. [PubMed] [Google Scholar]
- 48.Carman G. M., and Han G-S.. 2009. Phosphatidic acid phosphatase, a key enzyme in the regulation of lipid synthesis. J. Biol. Chem. 284: 2593–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsieh L-S., Su W-M., Han G-S., and Carman G. M.. 2016. Phosphorylation of yeast Pah1 phosphatidate phosphatase by casein kinase II regulates its function in lipid metabolism. J. Biol. Chem. 291: 9974–9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsieh L-S., Su W-M., Han G-S., and Carman G. M.. 2015. Phosphorylation regulates the ubiquitin- independent degradation of yeast Pah1 phosphatidate phosphatase by the 20S proteasome. J. Biol. Chem. 290: 11467–11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Madera M., Vogel C., Kummerfeld S. K., Chothia C., and Gough J.. 2004. The SUPERFAMILY database in 2004: additions and improvements. Nucleic Acids Res. 32: D235–D239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Z., Su W-M., and Carman G. M.. 2012. Fluorescence spectroscopy measures yeast PAH1- encoded phosphatidate phosphatase interaction with liposome membranes. J. Lipid Res. 53: 522–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barbosa A. D., Sembongi H., Su W-M., Abreu S., Reggiori F., Carman G. M., and Siniossoglou S.. 2015. Lipid partitioning at the nuclear envelope controls membrane biogenesis. Mol. Biol. Cell. 26: 3641–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swaney D. L., Beltrao P., Starita L., Guo A., Rush J., Fields S., Krogan N. J., and Villén J.. 2013. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods. 10: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dubots E., Cottier S., Peli-Gulli M. P., Jaquenoud M., Bontron S., Schneiter R., and De Virgilio C.. 2014. TORC1 regulates Pah1 phosphatidate phosphatase activity via the Nem1/Spo7 protein phosphatase complex. PLoS One. 9: e104194. [DOI] [PMC free article] [PubMed] [Google Scholar]