Abstract
Hydroxysteroid 17-β dehydrogenase 13 (HSD17B13) is a lipid droplet-associated protein; its gene-encoding variants affect the chronic liver diseases, including nonalcoholic fatty liver disease (NAFLD). To estimate the effect of rs72613567, a splice variant with an adenine insertion (A-INS), on NAFLD susceptibility and severity, we performed a case-control study with 609 individuals. We investigated the effect of carrying the A-INS allele in 356 patients with biopsy-proven disease and explored the relationship between rs72613567 genotypes and the hepatic transcriptome. The A-INS allele protected against NAFLD [odds ratio (OR) per adenine allele = 0.667; 95% CI, 0.486−0.916; P = 0.012]; this effect was nonsignificant when logistic regression analysis included BMI. The A-INS allele protected against nonalcoholic steatohepatitis (NASH) (OR = 0.612; 95% CI, 0.388−0.964; P = 0.033), ballooning degeneration (OR = 0.474; 95% CI, 0.267−0.842; P = 0.01), lobular inflammation (OR = 0.475; 95% CI, 0.275−0.821; P = 0.007), and fibrosis (OR = 0.590; 95% CI, 0.361−0.965; P = 0.035). In patients carrying A-INS, HSD17B13 levels decreased proportionally to allele dosage. Whole-transcriptome genotype profiling showed overrepresented immune response-related pathways. Thus, the rs72613567 A-INS allele reduces the risk of NASH and progressive liver damage and may become a therapeutic target.
Keywords: cirrhosis, genetics, mutations, fibrosis, heritability, hydroxysteroid 17-β dehydrogenase 13, protection, nonalcoholic steatohepatitis
Nonalcoholic fatty liver disease (NAFLD), which affects a high proportion of the global adult and child population, has emerged as the leading cause of end-stage liver disease (1, 2). A growing body of evidence indicates that NAFLD is a heritable (3) and polygenic complex trait (4). Variants located in genes that regulate lipid metabolism and lipid droplet (LD) biology, including patatin-like phospholipase domain containing 3 (PNPLA3)-rs738409 and transmembrane 6 superfamily member 2 transmembrane 6 superfamily member 2 (TM6SF2)-rs58542926, have been consistently replicated as major modifiers of the natural history of NAFLD (5–8), increasing the risk of developing severe histological forms of the disease, such as nonalcoholic steatohepatitis (NASH) and NASH-fibrosis. Of note, variants, more specifically SNPs, in the aforementioned genes have been discovered as a part of genome-wide (5) or exome-wide (7) association studies. Likewise, recent remarkable results from exome-sequence data coupled with electronic health records of 46,455 participants uncovered the association of the rs72613567 (−/adenine) deletion/insertion-variant in hydroxysteroid 17-β dehydrogenase 13 (HSD17B13) with a reduced risk of chronic liver disease, including NASH (9). Research findings further indicate that the rs72613567 variant is reproducibly associated with decreased alanine aminotransferase (ALT) and aspartate aminotransferase (AST) serum levels (9).
HSD17B13 encodes a protein that is involved in the regulation of lipid biosynthetic process (10); molecular analysis (11) and recent proteomic studies (10) have identified HSD17B13 as a novel liver lipid droplet-associated protein.
Findings yielded by recent functional studies demonstrate that the rs72613567, regarded as a splice region variant, is indeed a loss-of-function variant, which consists of an adenine insertion (A-INS) in the coding gene region (chr4:87310241, GRCh38.p7) adjacent to the donor splice site of intron 6 that results in a frame-shift and premature truncation of the HSD17B13 protein (9).
Of note, the protective effect of the rs72613567 A-INS allele against NASH was recently observed in a sample of morbidly obese patients recruited from a bariatric cohort (9). As a part of the same study, the original description of the protective effect of the rs72613567-insertion variant on chronic liver damage was examined in a population of European Americans among whom the A-INS frequency is ∼24% (9). Therefore, it remains to be established whether the observed influential effect of the variant on the course of NAFLD can be generalized to other populations. Hence, we explored the association between rs72613567 and NAFLD disease severity, including a putative protecting effect on histological features associated with progressive damage. Furthermore, we aimed to explore the impact of carrying the rs72613567 A-INS allele in abundance and cell localization of the HSD17B13 protein in the liver of the affected patients. To gain further biological insight into the role of rs72613567, we also explored differentially expressed genes of the whole transcriptome according to the variant genotypes.
METHODS
Study design and patient selection
We performed a case-control association study to estimate the effect of the rs72613567 variant on the susceptibility of NAFLD; this study involved a sample of 609 unrelated individuals, including 429 patients with NAFLD and 180 control subjects. Associations with NAFLD severity, including histological features known to be related to an aggressive course, were explored in a sample of 356 patients that showed the whole spectrum of the disease [nonalcoholic fatty liver (NAFL), NASH, and NASH-fibrosis], and who had histopathological confirmation of NAFLD.
Patients and controls were selected from hospital-based settings, including patients diagnosed with NAFLD and metabolic syndrome (MetS) (Hospital Abel Zubizarreta, Buenos Aires, Argentina) and morbidly obese patients that underwent bariatric surgery (Hospital de Alta Complejidad en Red “El Cruce”, Buenos Aires, Argentina).
Inclusion criteria were as follows: liver ultrasonographic examination was the initial screening criteria for selecting the cases and controls in the group of individuals included in the study of NAFLD and MetS, and it was performed in all the participants. Patients who presented fatty liver on liver ultrasonogram were indicated a liver biopsy if they showed either abnormal liver enzymes or severe insulin resistance [homeostatic model assessment of insulin resistance (HOMA-IR) value greater than 3] as explained earlier (6). A liver biopsy was performed in 268 patients who met the criteria; patients that showed ultrasonographic features of liver steatosis plus persistently normal liver enzymes during 12 months of follow-up were not included in the histological evaluation. All morbidly obese patients that were included in this study (n = 121) had histological confirmation of either NAFLD or normal liver by a liver biopsy that was performed during bariatric surgery.
Exclusion criteria were as follows: secondary causes of steatosis, including alcohol abuse (≥30 g for men and ≥20 g for women, of alcohol daily), total parenteral nutrition, hepatitis B and hepatitis C virus infection, and the use of drugs known to precipitate steatosis were excluded. In addition, patients with any of the following diseases were excluded: autoimmune liver disease, metabolic liver disease, Wilson’s disease, and α-1-antitrypsin deficiency.
Control subjects that matched patients with NAFLD-MetS were selected from subjects attending our hospital for check-up purposes whose age and sex matched the NAFLD patients. In addition to the standard heath examination, all control individuals were subjected to a liver ultrasonographic examination. They were included in the study if they did not have evidence of fatty change or biochemical abnormalities. Furthermore, control subjects were confirmed not to have any of the features of MetS as defined by the National Cholesterol Education Program Adult Treatment Panel III and did not abuse alcohol.
In the population of morbidly obese patients, control subjects were obese patients who also underwent bariatric surgery and had no features of NAFLD demonstrated in the liver biopsy.
Liver biopsies from all patients as well as DNA samples from white blood cells from all the subjects included in this study were obtained with written informed consent in accordance with the Institutional Review Board-approved protocols. All the investigations performed in this study were conducted in accordance with the guidelines of the 1975 Declaration of Helsinki, as revised in 1993.
The case participants and control subjects were consecutively selected during the same study period from the same population of patients attending the participant institutions of Argentina, and all of them shared the same demographic characteristics (occupation, educational level, place of residence, and ethnicity).
Physical, anthropometric, and biochemical evaluation and histological assessment.
Health examinations included anthropometric measurements, a questionnaire on health-related behaviors, and biochemical determinations (see the supplemental information).
The disease severity was assessed by liver biopsy that was performed before any intervention with ultrasound guidance and a modified 1.4 mm-diameter Menghini needle (Hepafix, Braun, Germany) on an outpatient basis or during bariatric surgery. All liver biopsies were evaluated by the same pathologist. A portion of each liver biopsy specimen was routinely fixed in 40 g/l formaldehyde (pH 7.4), embedded in paraffin, and stained with hematoxylin and eosin, Masson’s trichrome, and silver impregnation for reticular fibers. All the biopsies were at least 3 cm in length and contained a minimum of eight portal tracts. The degree of steatosis was assessed according to the system developed by Kleiner et al. (12), based on the percentage of hepatocytes containing macrovesicular fat droplets. NASH and NAFLD activity score (NAS) (12, 13) were defined as reported previously, and a NAS threshold of 5 was used for further comparisons with variables of interest; NASH was defined as steatosis plus mixed inflammatory cell infiltration, hepatocyte ballooning and necrosis, Mallory’s hyaline, and any stage of fibrosis, including absent fibrosis (12, 13).
Intra-acinar (lobular) inflammation was defined as the presence of cellular components of inflammation (polymorphonuclear leukocytes, lymphocytes and other mononuclear cells, eosinophils, and microgranulomas) located in sinusoidal spaces, surrounding Mallory’s hyaline, or in hepatocellular necrosis (1, 13). It was graded 0–3 and was defined as 0 (absent) = no foci; 1 = <2 foci per 200× field; 2 = 2–4 foci per 200× field; and 3 = >4 foci per 200× field. Ballooning was scored as: 0 = none; 1 = rare or few; and 2 = many (12, 13). The severity of fibrosis was expressed on a four-point scale, as follows: 0 = none; 1 = perivenular and/or perisinusoidal fibrosis in zone 3; 2 = combined pericellular portal fibrosis; 3 = septal/bridging fibrosis; and 4 = cirrhosis (12, 13).
Genetic analysis
The genetic analyses were done on genomic DNA extracted from white blood cells (6). Genotyping of rs72613567 was performed using a custom TaqMan genotyping assay (see the supplemental information) (Applied Biosystems, Foster City, CA) according to manufacturer’s instructions, and confirmed by DNA Sanger sequencing (Macrogen, Inc., South Korea) (see the supplemental information and supplemental Fig. S1). Genotyping of the PNPLA3 rs738409 was performed as previously described (6).
To ensure genotyping quality, we included DNA samples as internal controls, hidden samples of known genotype, and negative controls (water). The overall genotype completion rate was 100%. To account for possible population stratification, we used a collection of 13 SNPs at different loci (located in chromosomes 4, 15, 17, 13, 1, and 3) and then analyzed the data with the Structure program version 2 (14) as we explained elsewhere (6, 15, 16). We found no evidence of stratification in our sample because the cases and the controls showed similar Q values and the Structure program assigned a similar distance to clusters with no further improvement in the fitting model by adding up to four clusters (the ln of likelihood was maximum for K = 1).
Using the CaTS power calculator for genetic association studies (17) and assuming a prevalence of NAFLD of 0.30, minor allele frequency A-INS 0.16 (the frequency found in our population, 0.84 for the risk allele), and an odds ratio (OR) of 1.8/0.56, our sample had 83% and 99%, for the additive and multiplicative genetic model, respectively.
Liver immunohistochemistry
Four micrometer sections were mounted onto silane-coated glass slides to ensure section adhesion through subsequent staining procedures. Briefly, sections were deparaffinized, rehydrated, washed in PBS, and treated with 3% H2O2 in PBS for 20 min at room temperature to block endogenous peroxidase. Following microwave heat-induced epitope retrieval in 0.1 M citrate buffer at pH 6.0 for 20 min, the slides were incubated with a dilution of 1:150 of rabbit polyclonal antibody for human anti-HSD17B13 [HSD17B13 antibody (OAAN01691) size 50 UL, Aviva Systems Biology, San Diego, CA]. Immunostaining was performed using the VECTASTAIN Elite ABC kit (Vector Laboratories, Burlingame, CA) detection system. Subsequently, slides were immersed in a 0.05% 3,3′-diaminobenzidine solution in 0.1 M Tris buffer, pH 7.2, containing 0.01% H2O2. After a brown color developed, slides were removed and the reaction was stopped by immersion in PBS. Negative controls were carried out with rabbit serum diluted to the same concentration as the primary antibody. HSD17B13 immunostaining was evaluated in a blinded fashion regarding any of the histological and clinical characteristics of the patients. The extent of staining was scored according to its amount and intensity by a four-point scoring system as follows: 0 = no staining, 1 = positive staining in less than 20% of cells, 2 = 21–50% of positive cells, and 3 = positive staining in more than 50% of cells. Histological specimens were assessed by a LEICA DM 2000 (Leica, Germany) trinocular microscope equipped with a high-definition camera (Leica MC190 HD); all images were recorded using the Leica Application Suite software.
Gene expression profiling of the liver transcriptome
Total RNA was prepared from fresh liver tissue using phenol extraction step method, with an additional DNase digestion. After extraction, RNA quantity was measured using the ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE), the ratio of absorbance 260 nm/280 nm being 1.9–2.0.
For NAFLD patients carrying the rs72613567 (−/−) homozygous genotype (n = 8), one target for each sample was prepared. For the reference group [rs72613567 (A/A) homozygous genotype], pooled samples from five patients were used. The same proportion of NASH and NAFL patients were included per rs72613567-genotype group; selection of cases was based on the availability of RNA of sufficient quality and quantity to perform the microarray experiment.
Samples were further processed using the SurePrint G3 Unrestricted Gene Expression 8x60K microarray (product number G4140-90050; Agilent, Santa Clara, CA) according to instructions provided by the manufacturer. This array allows the testing of 26,083 Entrez Genes (https://www.ncbi.nlm.nih.gov/gene) and 30,606 long noncoding RNAs. The cDNA (cRNA) synthesis involved a single amplification for two color labeling; probes from rs72613567 (−/−) tissue and from the reference tissue [rs72613567 (A/A)] were differentially labeled by the incorporation of cyanine (Cy)3 and Cy5, respectively. Feature extraction, quantification of the intensity of fluorescent images, and scanned images were processed and analyzed by Agilent SureScan microarray scanner bundle, Agilent Scan control software, and Agilent GeneSpring 14.5-GX software, respectively. We used fold change values generated by the software from the array data as the ratio of Lowess normalized background subtracted Cy5/Cy3 signals if they passed the built-in quality control analysis of the software executed according to the manufacturer’s protocol for two-color experiments. Linearization of the data was undertaken by log2 transformation.
Gene enrichment analysis.
In addition to the liver whole-genome expression exploration, we conducted gene set enrichment analysis to investigate the pathways and biological processes potentially associated with the differentially expressed genes. FunRich v.3.1.3 (http://www.funrich.org) software was used; only pathways that reached significant threshold were considered.
Statistical analysis
Quantitative data are expressed as mean ± SD unless otherwise indicated. As a significant difference in SD was observed between the groups in most of the variables and the distribution was significantly skewed in most cases, we chose to be conservative and assessed the differences between the groups using nonparametric Mann-Whitney U or Kruskal-Wallis tests. The Cochran-Armitage test for trend was used in the categorical data analysis to assess the presence of association between the variant and the disease severity, as well as histological outcomes; chi-square (χ2), OR, and P value are informed. Ordinal variables were dichotomized as ballooning degeneration, lobular inflammation, and liver fibrosis: yes (1)/ no (0). In addition, we used generalized linear and nonlinear models (logistic regression and ANCOVA using binomial or ordinal multinomial distribution with logit and probit as link functions, respectively), adjusting for covariables, which were log-transformed. Then, adjustment for potential demographic, clinical, and genetic confounders (rs738409) was also performed. The CSS/Statistica program package version 6.0 (StatSoft, Tulsa, OK) was used in these analyses.
RESULTS
Clinical, biochemical, and histological features of patients with histological confirmation of NAFLD are shown in Table 1. While the studied population was primarily comprised of patients with NAFLD and features of MetS, the cohort also included 88 morbidly obese persons who represented 24.7% of the total sample. The control group characteristics are shown in supplemental Table S1.
TABLE 1.
Clinical and biochemical features of patients with NAFLD according to disease severity
| Variables | NAFL | NASH | P |
| Number of subjects (%) | 165 (46) | 191 (54) | |
| Female, % | 59.4 | 65 | NS |
| Age, years | 49.5 ± 12 | 50.5 ± 12 | NS |
| BMI, kg/m2 | 38.3 ± 13 | 36.5 ± 9.4 | NS |
| Arterial hypertension, % | 41.6% | 51.3% | NS |
| Glucose-related profile | |||
| Fasting plasma glucose, mg/dl | 100 ± 25 | 129 ± 110 | 1.0 × 10−8 |
| Fasting plasma insulin, μU/ml | 13.3 ± 8.8 | 19 ± 20 | 1.0 × 10−8 |
| HOMA-IR index | 3.34 ± 2.5 | 6.6 ± 16 | 1.0 × 10−8 |
| HbA1c | 6.3 ± 1.3 | 7.5 ± 4.4 | NS |
| Type 2 diabetes, % | 29 | 51.3 | 0.000038 |
| Lipid profile | |||
| Total cholesterol, mg/dl | 199 ± 50 | 204 ± 46 | NS |
| HDL-cholesterol, mg/dl | 49.5 ± 21 | 48 ± 14 | NS |
| LDL-cholesterol, mg/dl | 125 ± 44 | 125 ± 42 | NS |
| Triglycerides, mg/dl | 157 ± 88 | 187 ± 115 | 0.059 |
| Uric acid, mg/dl | 5.6 ± 7.5 | 5.0 ± 2.0 | NS |
| Liver phenotype | |||
| ALT, U/L | 49 ± 53 | 70 ± 55 | 1.0 × 10−8 |
| AST, U/L | 32 ± 17 | 49 ± 33 | 1.0 × 10−8 |
| GGT, U/L | 67 ± 57 | 88 ± 96 | 0.098 |
| AP, U/L | 183 ± 113 | 202 ± 115 | 0.098 |
| Histological features of NAFLD | |||
| Degree of steatosis (0–3) | 1.85 ± 0.81 | 2.4 ± 0.75 | 0.000068 |
| Lobular inflammation (0–3) | 0.52 ± 0.61 | 1.33 ± 0.68 | 1.0 × 10−8 |
| Hepatocellular ballooning (0–2) | 0.09 ± 0.28 | 0.87 ± 0.60 | 1.0 × 10−8 |
| Fibrosis stage | 0.06 ± 0.33 | 1.6 ± 0.70 | 1.0 × 10−8 |
| NAS | 2.4 ± 1.20 | 4.5 ± 1.2 | 1.0 × 10−8 |
Results are expressed as mean ± SD. P stands for statistical significance using Mann-Whitney U test, except for female/male proportion, arterial hypertension, and type 2 diabetes where P stands for statistical significance using the Chi-square test. HbA1c, glycated hemoglobin; GGT, γ-glutamyl-transferase; AP, alkaline phosphatase.
Genotype frequencies of rs72613567 (−/−: 436, −/A: 152, A/A: 21; P = NS) were in Hardy-Weinberg equilibrium; the frequency of the rs72613567 A-INS allele in our sample was 16%. Clinical, biochemical, and histological characteristics of the studied population according to rs72613567 genotypes are shown in Table 2. Significant differences in the BMI were observed between homozygous carriers (−/−) and those homozygous for the A-INS allele (A/A). Therefore, covariates that were significant in the univariate analysis were included in our logistic regression model. In further association analyses between disease-related binary outcomes (present/absent) and rs72613567 genotypes, BMI was systematically assessed as an independent variable.
TABLE 2.
Clinical and biochemical features of subjects included in the study according to the rs72613567 genotypes
| Variables | rs72613567 (−/−) | rs72613567 (−/A) | rs72613567 (A/A) |
| Number of subjects (%) | 436 | 152 | 21 |
| Female, % | 63.5 | 65.3 | 66 |
| Age, years | 49 ± 13 | 49 ± 13 | 48 ± 12 |
| BMI, kg/m2 | 35 ± 11.5 | 34 ± 12 | 30 ± 10a |
| Glucose-related profile | |||
| Fasting plasma glucose, mg/dl | 106 ± 46 | 100 ± 33 | 95 ± 22 |
| Fasting plasma insulin, μU/ml | 14 ± 15 | 12 ± 10 | 11 ± 7.5 |
| HOMA-IR index | 4 ± 10 | 3 ± 3.4 | 2.7 ± 3 |
| HbA1c | 7 ± 3 | 6.3 ± 1.3 | 5 ± 1.4 |
| Lipid profile | |||
| Total cholesterol, mg/dl | 203 ± 48 | 194 ± 50 | 210 ± 41 |
| HDL-cholesterol, mg/dl | 49 ± 17 | 48 ± 17 | 48 ± 22 |
| LDL-cholesterol, mg/dl | 126 ± 43 | 120 ± 45 | 121 ± 50 |
| Triglycerides, mg/dl | 162 ± 101 | 145 ± 89 | 173 ± 90 |
| Liver phenotype | |||
| ALT, U/l | 53 ± 54 | 42 ± 38b | 38 ± 35 |
| AST, U/l | 37 ± 28 | 31 ± 19b | 32 ± 18 |
| GGT, U/l | 65 ± 72 | 68 ± 99 | 75 ± 119 |
| AP, U/l | 183 ± 111 | 171 ± 104 | 193 ± 127 |
Results are expressed as mean ± SD. P value is statistical significance using the Mann-Whitney U test, except for female/male proportion where p values (<0.01) are statistical significance using the Chi-square test. HbA1c, glycated hemoglobin; GGT, γ-glutamyl-transferase; AP, alkaline phosphatase.
Indicates rs72613567 (−/−) versus rs72613567 (A/A).
Indicates rs72613567 (−/−) vs. rs72613567 (−/A).
We found no interaction effect between the rs72613567 and PNPLA3-rs738409. Nevertheless, this result should be considered with caution because the sample size employed in our study did not allow sub-stratification of the population according to the different genotypes (nine combinations) with sufficient power to detect interactions.
Findings yielded by our additive model of inheritance indicate that the rs72613567 variant is associated with reduced odds of having NAFLD (OR per A-INS allele: 0.667; 95% CI, 0.486−0.916; P = 0.012). However, the protective effect of the rs72613567 variant on NAFLD was not significant (P = 0.076) when BMI was included in the logistic regression analysis.
The presence of the rs72613567 A-INS allele is associated with a protective effect against severe NAFLD histological features
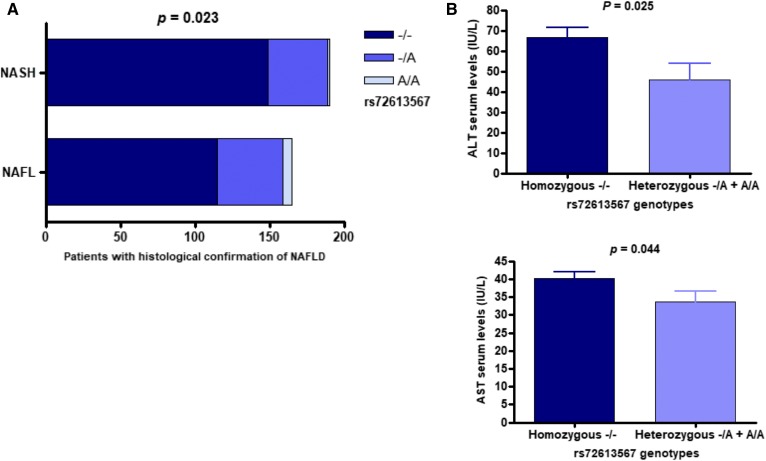
The results of genotypic Cochran-Armitage test for trend revealed a significant association of the rs72613567 and disease severity. The rs72613567 A-INS allele was associated with lower odds of having NASH (χ2 = 5.169; P = 0.023; cumulative OR: 0.568; 95% CI, 0.350−0.922; NASH versus NAFL) (Fig. 1A). Logistic regression analysis showed that the association of the rs72613567 variant and the histological disease severity was independent of BMI (P = 0.033; OR per A-INS allele: 0.612; 95% CI, 0.388−0.964).
Fig. 1.
Association of HSD17B13 rs72613567 with NAFLD disease severity and aminotransferase levels. A: Distribution of rs72613567 genotypes according to the histological disease severity (NASH vs. NAFL). P value denotes statistical significance in the Cochran-Armitage test for trend (additive model of inheritance). B: Analysis of the association between rs72613567 genotypes and serum levels of ALT and AST in the dominant model of inheritance (−/− vs. −/A + A/A); statistical differences between groups were evaluated using Mann-Whitney U test. Bars express mean ± SE.
In addition, presence of the rs72613567 A-INS allele was associated with a protective effect against severe histological outcomes, including ballooning degeneration (χ2 = 6.758; P = 0.009; cumulative OR: 0.461; 95% CI, 0.262−0.813) and lobular inflammation (χ2 = 11.444; P = 0.001; cumulative OR: 0.383; 95% CI, 0.217−0.674), according to the results yielded by the Cochran-Armitage test for trend. Protective associations of the rs72613567 variant and ballooning degeneration (P = 0.010; OR per A-INS allele: 0.474; 95% CI, 0.267−0.842) and lobular inflammation (P = 0.0074; OR per A-INS allele: 0.475; 95% CI, 0.275−0.821) were independent of BMI.
Likewise, carrying the rs72613567 A-INS allele reduced the likelihood of developing liver fibrosis (χ2 = 4.323; P = 0.038; cumulative OR: 0.567; 95% CI, 0.334−0.961). This protective effect against fibrosis was found to be independent of BMI (P = 0.035; OR per A-INS allele: 0.590; 95% CI, 0.361−0.965).
The rs72613567 A-INS allele was significantly associated with lower NASs (NAS <5 vs. NAS ≥5), χ2 = 9.249, P = 0.002, cumulative OR: 0.338, 95% CI, 0.171−0.668. This effect was also independent of BMI (P = 0.024; OR per A-INS allele: 0.369; 95% CI, 0.154−0.886). The degree of liver steatosis, as evaluated by liver biopsy and expressed as a percentage of hepatocytes infiltrated by fat was not associated with the rs72613567 variant.
Finally, the rs72613567 A-INS allele was associated with lower levels of serum ALT (P = 0.025) and AST (P = 0.044); however, this association was specifically found in the dominant model of inheritance (−/− vs. −/A + A/A) (Fig. 1B).
Logistic regression analysis showed that the rs72613567 variant remained significantly associated with the disease severity and liver damage-related histological outcomes (ballooning degeneration, fibrosis, and lobular inflammation) after adjusting for potential demographic, clinical, and genetic confounders in addition to BMI (Table 3).
TABLE 3.
Association analysis of rs72613567 after adjustment for potential confounding variables
| Trait | OR (95% CI) | P |
| Disease severity | 0.635 (0.404–0.9990 | 0.049 |
| Ballooning degeneration | 0.468 (0.278–0.817) | 0.007 |
| Lobular inflammation | 0.450 (0.263–0.770) | 0.003 |
| Fibrosis | 0.416 (0.174–0.997) | 0.048 |
Potential confounding variables: sex, age, BMI, HOMA-IR, and rs738409 (PNPLA3); P and OR stand for statistical significance and odds ratio, respectively, using analysis in a logistic regression model. BMI and HOMA-IR were log-transformed.
HSD17B13 protein expression is significantly decreased in the liver of patients with NAFLD carrying the rs72613567 A-INS allele
In a recent study, Abul-Husn et al. (9) used RNA sequencing to discover the functional effects of the rs72613567 splice variant. The authors reported that the rs72613567 A-INS allele was associated with a premature truncation of the protein.
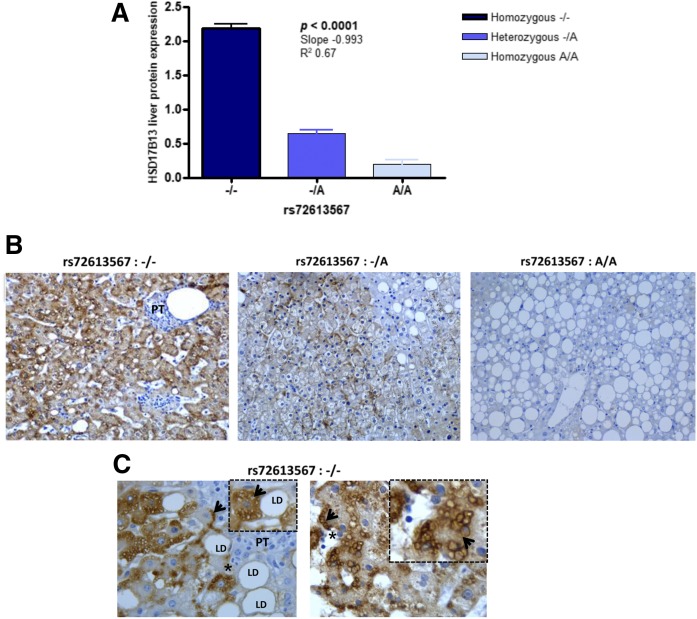
By using immunohistochemistry expression in the liver of patients with NAFLD, we explored the level of HSD17B13 according to the rs72613567 genotypes (Fig. 2A, B). We found significantly lower levels of HSD17B13 protein in the liver of patients carrying the A-INS allele compared with patients homozygous for the deletion (−/−) (Fig. 2A, B). HSD17B13 immunoreactivity product was significantly reduced in an allele dose-dependent manner (linear test for trend P < 0.0001, slope −0.993). Moreover, statistical analysis revealed significantly reduced HSD17B13 immunoreactivity in subjects carrying at least one copy of the rs72613567 A-INS allele compared with that noted in homozygous (−/−) patients (P = 0.0003, Mann-Whitney test). We observed selective HSD17B13 immunoreactivity within the hepatocytes, but not in other cell types of the liver; analysis of subcellular localization of HSD17B13 showed immunoreactivity primarily associated to cytoplasmic elements, including membrane-bound organelles and small LDs (Fig. 2C). We also observed robust immunostaining in the vicinity of the hepatocyte membrane, which is a potentially novel feature (Fig. 2C).
Fig. 2.
Expression and subcellular localization of HSD17B13 protein in the liver. A: The scores of HSD17B13 expression in patients with NAFLD evaluated by immunohistochemistry according to rs72613567 genotypes (−/−, n = 8; −/A, n = 7; A/A, n = 5). Values represented by vertical bars are mean ± SD; mean values of HSD17B13 immunoreactivity in the liver of rs72613567 homozygous A/A, heterozygous −/A, and homozygous −/− patients were compared by ANOVA and statistical differences between groups were evaluated using Newman-Keuls test. ANOVA posttest for linear trend was performed, and slope, R2, and P values are shown in the graph. B: Representative liver expression pattern of HSD17B13 in NAFLD patients carrying the rs72613567 (−/−), rs72613567 (−/A), and rs72613567 (A/A) genotypes. Original magnification: 200×. C: Patterns of subcellular localization of HSD17B13 in representative NAFLD patients carrying the rs72613567 −/− homozygous genotype. The asterisks indicate the inset (4× magnification of the original image) showing that the protein presents exclusively cytoplasmic immunoreactivity, which preferentially accumulates on the surface of small fat droplets (arrow) or close to the hepatocyte membrane (arrow). HSD17B13 immunoreactivity was examined using light microscopy of liver sections. Original magnification 400×. PT, portal tract.
Liver gene expression profiling according to rs72613567 genotypes suggests differences in immune-mediated pathways
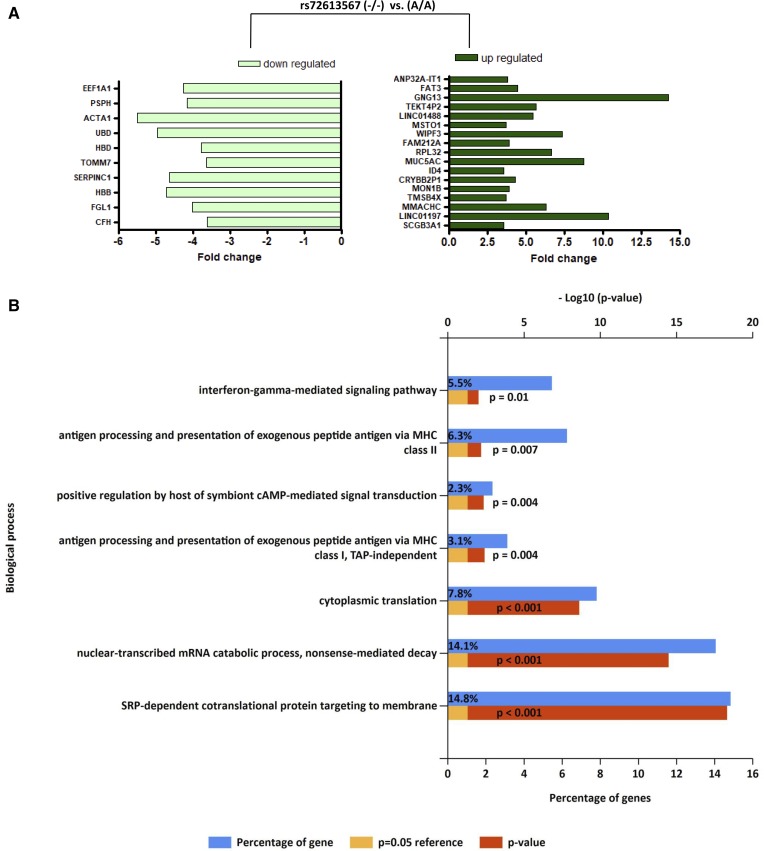
We hypothesized that rs72613567 could be associated with patterns of intrahepatic gene expression that explain the impact of the variant on the phenotype. Transcriptome analysis revealed differential expression of 161 genes. The most (≥3.5-fold) downregulated and upregulated genes in the liver of patients carrying the rs72613567 (−/−) genotype relative to the expression levels of patients homozygous for the A-INS allele are illustrated Fig. 3A, while the entire gene list, including fold changes and P values, is shown in supplemental Table S2. These results provide valuable insight into the putative global transcriptional response associated with the variant, which seems to be significantly enriched by biological pathways related to the immune system, including antigen presentation and interferon-related processes, cytokine signaling, and signal transduction (Fig. 3B). In addition, enrichment analysis findings indicated that the “signal recognition particle (SRP)-dependent cotranslational protein targeting to membrane” pathway was significantly overrepresented (Fig. 3B). This pathway involves a cytosolic SRP to form a complex, which in turn docks with an SRP receptor complex on the endoplasmic reticulum membrane (https://reactome.org/content/detail/R-HSA-1799339).
Fig. 3.
Liver whole gene expression profiling and pathway enrichment analysis in patients with NAFLD according to rs72613567 genotypes. A: Fold change of upregulated and downregulated genes in the liver of patients carrying the rs72613567 (−/−) genotype, considering as reference the expression of the whole transcriptome in the liver of patients carrying two copies of the rs72613567 A-INS allele. The most (≥3.5-fold) downregulated and upregulated genes that reached statistical significance after multiple testing correction are illustrated on the charts. The entire list of genes is shown in supplemental Table S2. CFH, complement factor H; FGL1, fibrinogen-like 1; HBB, hemoglobin β; SERPINC1, serpin peptidase inhibitor clade C (antithrombin) member 1; TOMM7, translocase of outer mitochondrial membrane 7 homolog (yeast); HBD, hemoglobin delta; UBD, ubiquitin D; ACTA1, actin α1 skeletal muscle; PSPH, phosphoserine phosphatase; EEF1A1, eukaryotic translation elongation factor 1 α1; SCGB3A1, secretoglobin family 3A member 1; LINC01197, long intergenic nonprotein coding RNA 1197; MMACHC, methylmalonic aciduria (cobalamin deficiency) cblC type with homocystinuria; TMSB4X, thymosin β4 X-linked; MON1B, MON1 secretory trafficking family member B; CRYBB2P1, crystallin, β B2 pseudogene 1; ID4, inhibitor of DNA binding 4 (dominant negative helix-loop-helix protein); MUC5AC, mucin 5AC (oligomeric mucus/gel-forming); RPL32, ribosomal protein L32; FAM212A, family with sequence similarity 212 member A; WIPF3, WAS/WASL interacting protein family member 3; MSTO1, misato 1 (mitochondrial distribution and morphology regulator); LINC01488, long intergenic nonprotein coding RNA 1488; TEKT4P2, tektin 4 pseudogene 2; GNG13, guanine nucleotide binding protein (G protein) γ 13; FAT3, FAT atypical cadherin 3; ANP32A-IT1, ANP32A intronic transcript 1 (non-protein coding). B: Gene enrichment analysis for biological processes (Gene Ontology database) using the FunRich (http://www.funrich.org) tool. Bonferroni and Benjamini-Hochberg and false discovery rate method were used to correct for multiple testing. Pathways were ranked according to the P value (red bar); a P value lower than 0.05 was considered significant. The blue bar indicates the percentage of altered genes in a whole pathway.
DISCUSSION
As a part of the present study, we found that the rs72613567 variant in the HSD17B13 gene confers protection against NAFLD severe histological disease. In line with the recent seminal findings reported by Abul-Husn et al. (9), we observed that the rs72613567 A-INS allele is associated with decreased risk of chronic liver damage in NAFLD patients, specifically with an ∼40% reduction in the odds of developing NASH. In addition, by exploring the association between the variant and histological outcomes of liver injury, we found a protective effect of the rs72613567 A-INS allele against ballooning degeneration, lobular inflammation, and liver fibrosis. In patients carrying the rs72613567 A-INS allele, the odds (per allele) of presenting ballooning degeneration and lobular inflammation are 50% lower relative to those pertaining to patients who do not carry the insertion allele. Furthermore, presence of the rs72613567 A-INS allele reduces the risk of liver fibrosis by ∼40% per allele. In addition, in agreement with recently published evidence (9), we observed that rs72613567 was associated with serum levels of transaminases in the dominant model of inheritance.
In the present study, we not only replicated the findings of Abul-Husn et al. (9), but also provided novel information regarding the protective effect of the variant against histological features associated with progressive liver disease. In our sample, we observed that all significant associations with histological traits remained independent of BMI. On the contrary, the variant does not seem to robustly influence the susceptibility of fatty liver, as the effect remained nonsignificant when BMI was included in the logistic regression model. Hence, these results suggest that the effects of rs72613567 on NAFLD pathogenesis are restricted to the cellular events associated with liver damage, rather than pertaining to those mediating fat accumulation in the hepatocyte. This observation is in agreement with the results of Abul-Husn et al. (9) that originally found the lack of association between the variant and NAFLD as disease trait.
However, a key aspect of the Abul-Husn et al. report (9) and the current study is that, in both studies, the effect of the variant is considerably large (ORs approaching 0.5). This is substantially important for a complex trait like liver fibrosis.
The frequency of the rs72613567 A-INS allele in our sample was 16%, which is in line with that reported in the 1000 Genomes Project for the A-INS allele in the American population (1000 Genomes Project, phase 3; http://www.ensembl.org). However, this percentage is lower than that observed in European Americans (9). Regarding ethnicity, our sample of patients and controls might present both similarities and differences compared with the sample included in the study of Abul-Husn et al. (9). In fact, our sample is composed of subjects of self-reported European ancestry. Nevertheless, we cannot rule out the influence of other factors, including recombination events, different degrees of genetic diversity and linkage disequilibrium patterns, and environmental exposure, which together may suggest that our cohort could be regarded as a distinctive one compared with the sample of Abul-Husn et al. (9). Hence, one may a priori speculate that our replication study might support the generalizability of the seminal results (9).
Frequency diversity of rs72613567 across different populations might potentially explain differences in the prevalence and severity rates of NAFLD across ethnic groups. Nevertheless, this conclusion needs extensive validation because the role of the rs72613567 variant in the protection against NASH has not been extensively explored.
Earlier functional exploration of rs72613567 has robustly demonstrated that the splice variant associates with liver transcripts that encode a prematurely truncated HSD17B13 protein (9). Hence, it is postulated that the rs72613567 variant mediates its protective effect(s) against severe liver damage by disrupting HSD17B13 protein expression. Our results consistently showed that the rs72613567 A-INS allele is significantly associated with decreased levels (or absence) of HSD17B13 protein in the liver of heterozygous (−/A) and homozygous (A/A) patients with NAFLD, respectively, in an allele dose-dependent manner.
Whereas there is reproducible information regarding tissue-expression patterns of HSD17B13 protein (9, 10), including the findings reported by Horiguchi et al. (11) who demonstrated a predominant tissue distribution of HSD17B13 in the liver compared with the intestine, knowledge of the actual function and regulation of the HSD17B13 gene is relatively limited. The protein encoded by HSD17B13 belongs to the short-chain dehydrogenase/reductase family, which is involved in the metabolism of steroid hormones, prostaglandins, lipids, xenobiotics, and retinoids (18). Lipid-particle organization and regulation of lipid biosynthesis are principal biological processes associated with HSD17B13 protein activity (10, 11).
We confirmed the subcellular localization of HSD17B13 in the liver of patients with NAFLD, which was primarily observed in the cytoplasm of hepatocytes in close association with the surface of small fat droplets and the cell membrane facing adjacent hepatocytes (Fig. 2B, C). Furthermore, HSD17B13 expression was found to be negative in other cellular components of the liver tissue milieu, including inflammatory cells and vascular or bile ductular cells (Fig. 2B, C). One might speculate that the differential expression of the HSD17B13 protein on the surface of liver LDs (9–11, 19) may help explain associations with the phenotype. In fact, there is robust evidence indicating that liver LDs are multifunctional organelles, which are not only associated with other cellular structures, including endoplasmic reticulum, but also recruit cytosolic proteins and inflammatory mediators (20).
As shown by Zhang et al. (20) who evaluated the preferential distribution of proteins according to different subpopulations of LDs, small LDs are enriched of members of small molecular weight GTPases (known as small G proteins), which are involved in important cellular processes, including cell proliferation, cytoskeletal dynamics, and vesicle transport (21).
The interpretation of protein localization should be taken with caution because we cannot rule out the probability that HSD17B13 can localize to other organelles, including vesicles or lysosomes.
We have investigated the relationship between the rs72613567 genotypes and liver gene expression patterns. We found an overrepresentation of pathways associated with the immune response, including antigen processing and interferon γ-mediated signaling pathways, which resembles the inflammatory response of the autoimmune diseases.
It should be noted, however, that these results were derived from a comparison of a small sample of patients that presented severe NAFLD histological disease and the rs72613567 (−/−) genotype with patients with less severe NAFLD and the rs72613567 (A/A) genotype. Thus, it could be argued that the transcriptional landscape could reflect changes associated with the disease severity per se rather than the rs72613567 genotypes. Nonetheless, we carefully balanced the proportion of patients with severe histological disease that were included in the analysis per genotype.
On the other hand, results from animal studies, particularly Hsd17b13-KO mice, suggest a contrasting scenario (22) to that of observed in human studies. Hsd17b13-KO mice show increased hepatic de novo lipogenesis and are prone to develop severe NAFLD with portal inflammation (22). Hsd17b13-KO mice are neither obese nor insulin resistant, but present overexpression of proteins involved in fatty acid synthesis (22). It thus seems difficult to translate the experimental results noted above to humans, particularly because the genetic component of NAFLD is very much dependent on the interaction between genetic modifiers and environmental factors. Likewise, moderate interspecies homology in members of the 17β-hydroxysteroid dehydrogenase family (23) might explain difficulties in explaining results based on animal models.
Although our work provides insights on the effect of the rs72613567 variant in mitigating the chances of having NASH, it has some limitations. For example, we cannot draw broader conclusions about the role of the variant in protection against end-stage liver disease, including cirrhosis and hepatocellular carcinoma. In addition, our population did not include a representative sample of lean patients with NAFLD, in whom the role of the variant in protecting against severe histological outcomes requires specific validation.
In conclusion, the presence of the rs72613567 A-INS allele mitigates the prevalence of NASH, as well as the severity of histological lesions, in patients with NAFLD in our population. The phenotypic variance of NAFLD seems to be largely influenced by common (frequent) genetic variation in loci that regulate lipid metabolism and liver LD plasticity and dynamics in high interaction with the environment. It thus seems reasonable to presume that NAFLD pathogenesis is not limited to excessive fat deposition but also depends on how hepatocytes deal with excesses of lipids/fatty acids.
Findings regarding the protective effect of the rs72613567 A-INS allele on severe histological disease might predictably have impact on NAFLD diagnosis and management. For example, knowledge of a patient’s genotype might be useful for biomarker discovery and subject stratification in clinical trials, and may lead to more precise treatment decisions, especially when using novel drugs. Moreover, a comprehensive druggability assessment of HSD17B13 protein, including therapeutic gene inhibition by silencing strategies or use of small molecules for selective HSD17B13 enzyme inhibition, would be of remarkable impact in the translational field.
Supplementary Material
Footnotes
Abbreviations:
- A/A
- homozygous carriers for adenine insertion allele
- −/A
- heterozygous carriers for the adenine insertion allele
- A-INS
- adenine insertion
- ALT
- alanine aminotransferase
- AST
- aspartate aminotransferase
- Cy
- cyanine
- HOMA-IR
- homeostatic model assessment of insulin resistance
- HSD17B13
- hydroxysteroid 17-β dehydrogenase 13
- LD
- lipid droplet
- MetS
- metabolic syndrome
- NAFL
- nonalcoholic fatty liver
- NAFLD
- nonalcoholic fatty liver disease
- NAS
- NAFLD activity score
- NASH
- nonalcoholic steatohepatitis
- OR
- odds ratio
- PNPLA3
- patatin-like phospholipase domain containing 3
- SRP
- signal recognition particle
- TM6SF2
- transmembrane 6 superfamily member 2
- −/−
- homozygous carriers for the adenine deletion allele
This work was partially supported by Agencia Nacional de Promoción Científica y Tecnológica Grants PID-C2012-0061, PICT 2014-0432, PICT 2014-1816, and PICT 2015-0551. C.J.P., D.F., and S.S. are members of Consejo Nacional de Investigaciones Científicas (CONICET). The authors declare no conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Brunt E. M., Wong V. W., Nobili V., Day C. P., Sookoian S., Maher J. J., Bugianesi E., Sirlin C. B., Neuschwander-Tetri B. A., and Rinella M. E.. 2015. Nonalcoholic fatty liver disease. Nat. Rev. Dis. Primers. 1: 15080. [DOI] [PubMed] [Google Scholar]
- 2.Friedman S. L., Neuschwander-Tetri B. A., Rinella M., and Sanyal A. J.. 2018. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 24: 908–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Speliotes E. K., Yerges-Armstrong L. M., Wu J., Hernaez R., Kim L. J., Palmer C. D., Gudnason V., Eiriksdottir G., Garcia M. E., Launer L. J., et al. . 2011. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 7: e1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sookoian S., and Pirola C. J.. 2017. Genetic predisposition in nonalcoholic fatty liver disease. Clin. Mol. Hepatol. 23: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romeo S., Kozlitina J., Xing C., Pertsemlidis A., Cox D., Pennacchio L. A., Boerwinkle E., Cohen J. C., and Hobbs H. H.. 2008. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 40: 1461–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sookoian S., Castano G. O., Burgueno A. L., Gianotti T. F., Rosselli M. S., and Pirola C. J.. 2009. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J. Lipid Res. 50: 2111–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozlitina J., Smagris E., Stender S., Nordestgaard B. G., Zhou H. H., Tybjaerg-Hansen A., Vogt T. F., Hobbs H. H., and Cohen J. C.. 2014. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 46: 352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pirola C. J., and Sookoian S.. 2015. The dual and opposite role of the TM6SF2-rs58542926 variant in protecting against cardiovascular disease and conferring risk for nonalcoholic fatty liver: A meta-analysis. Hepatology. 62: 1742–1756. [DOI] [PubMed] [Google Scholar]
- 9.Abul-Husn N. S., Cheng X., Li A. H., Xin Y., Schurmann C., Stevis P., Liu Y., Kozlitina J., Stender S., Wood G. C., et al. . 2018. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N. Engl. J. Med. 378: 1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su W., Wang Y., Jia X., Wu W., Li L., Tian X., Li S., Wang C., Xu H., Cao J., et al. . 2014. Comparative proteomic study reveals 17beta-HSD13 as a pathogenic protein in nonalcoholic fatty liver disease. Proc. Natl. Acad. Sci. USA. 111: 11437–11442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horiguchi Y., Araki M., and Motojima K.. 2008. 17beta-Hydroxysteroid dehydrogenase type 13 is a liver-specific lipid droplet-associated protein. Biochem. Biophys. Res. Commun. 370: 235–238. [DOI] [PubMed] [Google Scholar]
- 12.Kleiner D. E., Brunt E. M., Van N. M., Behling C., Contos M. J., Cummings O. W., Ferrell L. D., Liu Y. C., Torbenson M. S., Unalp-Arida A., et al. . 2005. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 41: 1313–1321. [DOI] [PubMed] [Google Scholar]
- 13.Brunt E. M., Kleiner D. E., Wilson L. A., Belt P., and Neuschwander-Tetri B. A.. 2011. Nonalcoholic fatty liver disease (NAFLD) activity score and the histopathologic diagnosis in NAFLD: distinct clinicopathologic meanings. Hepatology. 53: 810–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian C., Gregersen P. K., and Seldin M. F.. 2008. Accounting for ancestry: population substructure and genome-wide association studies. Hum. Mol. Genet. 17: R143–R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sookoian S., Castano G. O., Scian R., Mallardi P., Fernandez T. G., Burgueno A. L., San Martino J., and Pirola C. J.. 2015. Genetic variation in transmembrane 6 superfamily member 2 and the risk of nonalcoholic fatty liver disease and histological disease severity. Hepatology. 61: 515–525. [DOI] [PubMed] [Google Scholar]
- 16.Sookoian S., and Pirola C. J.. 2011. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 53: 1883–1894. [DOI] [PubMed] [Google Scholar]
- 17.Skol A. D., Scott L. J., Abecasis G. R., and Boehnke M.. 2006. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 38: 209–213. [DOI] [PubMed] [Google Scholar]
- 18.Persson B., Kallberg Y., Bray J. E., Bruford E., Dellaporta S. L., Favia A. D., Duarte R. G., Jornvall H., Kavanagh K. L., Kedishvili N., et al. . 2009. The SDR (short-chain dehydrogenase/reductase and related enzymes) nomenclature initiative. Chem. Biol. Interact. 178: 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Y., Beliaeva O., Karki S., Brown P., Kedishvili N., and Rotman Y.. 2017. A novel retinol dehydrogenase, HSD17B13 contributes to the pathogenesis of non-alcoholic fatty liver disease (Abstract). Hepatology. 66: 1995. [Google Scholar]
- 20.Zhang S., Wang Y., Cui L., Deng Y., Xu S., Yu J., Cichello S., Serrero G., Ying Y., and Liu P.. 2016. Morphologically and functionally distinct lipid droplet subpopulations. Sci. Rep. 6: 29539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Csépányi-Kömi R., Lévay M., and Ligeti E.. 2012. Small G proteins and their regulators in cellular signalling. Mol. Cell. Endocrinol. 353: 10–20. [DOI] [PubMed] [Google Scholar]
- 22.Adam M., Heikela H., Sobolewski C., Portius D., Maki-Jouppila J., Mehmood A., Adhikari P., Esposito I., Elo L. L., Zhang F. P., et al. . 2018. Hydroxysteroid (17beta) dehydrogenase 13 deficiency triggers hepatic steatosis and inflammation in mice. FASEB J. 32: 3434–3447. [DOI] [PubMed] [Google Scholar]
- 23.Marchais-Oberwinkler S., Henn C., Moller G., Klein T., Negri M., Oster A., Spadaro A., Werth R., Wetzel M., Xu K., et al. . 2011. 17beta-Hydroxysteroid dehydrogenases (17beta-HSDs) as therapeutic targets: protein structures, functions, and recent progress in inhibitor development. J. Steroid Biochem. Mol. Biol. 125: 66–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.