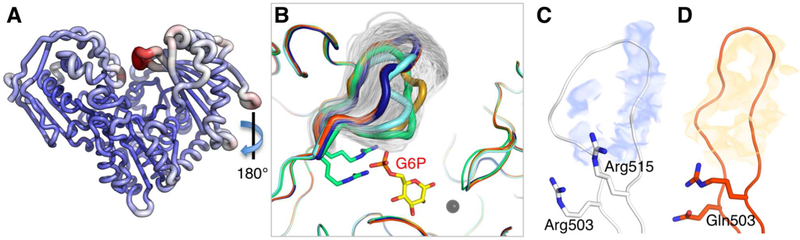
Figure 4. Analyses of D4 loop flexibility based on the MD simulations of human PGM1.
(A) RMSF values for WT enzyme mapped onto its structure. Increasing tube radius and a change from blue to red indicate higher RMSF values. (B) A sampling of the D4 loop conformers from the MD trajectory (one structure shown per 10 ps cycle) of WT PGM1. Sampled conformers (white) are shown only for the D4 loop; other regions of the polypeptide backbone are from the crystal structures of the missense variants and PGM1-G6P complex. Colors are as in Figure 2. View is 180° rotation relative to (A). Residence density analysis of the MD trajectories for the D4 loop (residues 505–513) of WT PGM1 (C) and the R503Q variant (D). Maps were calculated as described in Methods. Note the discontinuity of the density for WT D4 loop (blue) versus the continuous density for the R503Q loop (orange).