Abstract
A DNA electrochemistry platform has been developed to probe proteins bound to DNA electrically. Here gold electrodes are modified with thiol-modified DNA, and DNA charge transport chemistry is used to probe DNA binding and enzymatic reaction both with redox-silent and redox-active proteins. For redox-active proteins, the electrochemistry permits the determination of redox potentials in the DNA-bound form, where comparisons to DNA-free potentials can be made using graphite electrodes without DNA modification. Importantly, electrochemistry on the DNA-modified electrodes facilitates reaction under aqueous, physiological conditions with a sensitive electrical measurement of binding and activity.
1. INTRODUCTION
The fundamental properties of DNA charge transport (DNA CT), particularly the picosecond timescale over which charge migrates (O’Neill, Becker, Wan, Barton, & Zewail, 2003) and the exquisite sensitivity of DNA CT to perturbations in the base pair π-stacking interactions (Arnold, Grodick, & Barton, 2016), facilitate the use of DNA electrochemistry in detecting the activity of many different DNA-binding proteins, as well as in sensing DNA damage (Fig. 1) (Boal et al., 2009, 2005; Boon et al., 2002; DeRosa et al., 2005; Gorodetsky, Ebrahim, et al., 2008; Grodick, Segal, Zwang, & Barton, 2014; Mui, Fuss, Ishida, Tainer, & Barton, 2011; Slinker et al., 2011). Here, we describe the characteristics, protocols, and platforms, we have used to detect and monitor these DNA-binding proteins electrically. This detection sensitively depends on an electrochemical signal readout from either a redox-active moiety in the DNA-binding protein (Boal et al., 2009, 2005; DeRosa et al., 2005; Grodick et al., 2014; Mui et al., 2011) or from a DNA-intercalating redox probe bound to the DNA electrode (Boon et al., 2002; Gorodetsky, Ebrahim, et al., 2008; Slinker et al., 2011). The DNA-mediated electrode platform can sense a DNA-binding protein because the protein kinks the DNA, interfering with DNA CT (Gorodetsky, Ebrahim, et al., 2008), or perhaps because of the protein binding and cutting DNA attached to the electrode (Boon et al., 2002; Slinker et al., 2011). The platform can even detect electrically the unwinding of a duplex substrate by a helicase enzyme (Grodick et al., 2014; Mui et al., 2011). DNA-modified electrodes thus serve as substrates and templates for a wide variety of DNA-binding proteins. Indeed, the limits of protein detection can be nanomolar concentrations (Boon et al., 2002; Gorodetsky, Ebrahim, et al., 2008; Slinker et al., 2011) and depend on protein binding affinity more so than any property of DNA CT.
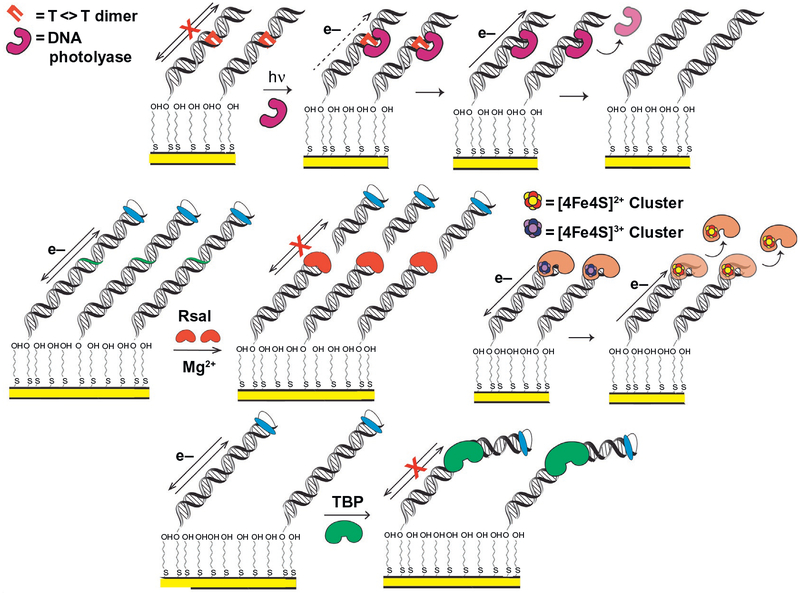
Fig. 1.
Electrochemical monitoring of DNA-binding protein activity on DNA-modified electrodes. (Top) DNA photolyase binds and repairs a thymine–thymine dimer on a DNA-modified electrode, restoring DNA CT and producing a signal from the flavin cofactor, through repaired DNA (DeRosa, Sancar, & Barton, 2005). (Center left) RsaI restriction enzyme cuts duplex DNA, removing covalently attached redox probe. Signal disappears after wash of surface, indicating that RsaI binding and cutting of DNA at recognition site occurs (Slinker, Muren, Renfrew, & Barton, 2011). (Center right) A bound [4Fe4S] enzyme is oxidized from the resting [4Fe4S]2+ state to the tightly bound [4Fe4S]3+ state through DNA CT; it can then be reduced from the tightly bound [4Fe4S]3+ state to the more weakly binding resting [4Fe4S]2+ state through DNA CT, promoting dissociation (Boal et al., 2009). (Bottom) TBP binding kinks duplex DNA, attenuating CT, and diminishing signal from a DNA-intercalating, covalently attached redox probe (Gorodetsky, Ebrahim, & Barton, 2008).
As an illustration, consider the transcription factor TATA-binding protein (TBP), which is responsible for activation of several different eukaryotic genes (Kornberg, 2007). TBP kinks duplex DNA approximately 90 degree when bound, significantly perturbing the π-stacking interactions of the DNA duplex (Boon et al., 2002; Gorodetsky, Ebrahim, et al., 2008)
(Fig. 1). This interaction perturbs DNA CT, and TBP binding is therefore detectable on DNA-modified electrodes (Gorodetsky, Ebrahim, et al., 2008). It was shown that when TBP binds and kinks duplex DNA containing the TATA box recognition sequence, CT attenuation occurs immediately. The DNA substrate in this assay contained the TBP recognition sequence, as well as a covalent, DNA-intercalating Nile Blue redox probe tethered at the distal end of the DNA duplex from the electrode surface. With nanomolar concentrations of TBP bound to the substrate, the DNA is kinked and the Nile Blue redox signal associated with DNA CT between the electrode surface and the redox probe is lost. The signal, moreover, could be easily regenerated upon washing the surface with KCl to remove TBP. This signal attenuation does not, importantly, occur when other proteins, which do not specifically bind the TATA box site or kink the substrate, are incubated on the electrode surface (Gorodetsky, Ebrahim, et al., 2008), nor when the 5’-TATA-3’ recognition sequence is not available.
DNA-intercalating redox probes can also be used to detect restriction enzyme activity upon binding specific recognition sequences on a DNA duplex (Fig. 1) (Boon et al., 2002; Slinker et al., 2011). Restriction enzymes RsaI and PvuII, for example, were each incubated on a DNA-modified electrode surface with a duplex substrate containing the respective restriction enzyme recognition sequence and a distal, covalently bound redox probe. These enzymes were given any necessary catalytic metal ions to perform their native function (Slinker et al., 2011), and they subsequently bound and cut the DNA substrate at the recognition site. This site was engineered in between the intercalated probe and the electrode surface, so the DNA no longer possessed a redox moiety once the restriction enzyme had cut the duplex at the appropriate site. This again did not occur when the substrate DNA lacked a recognition sequence, demonstrating that the observed effect depended on the reaction assayed. The restriction enzyme assay described is, additionally, adaptable to both the single electrode (Boon et al., 2002) and multiplexed chip (Slinker et al., 2011) setup.
In addition to detecting general protein binding and nuclease activity, these platforms have also facilitated the study of DNA-bound redox processes in biology. The repair of DNA by flavoenzyme DNA photolyase (Escherichia coli), for example, can be monitored in real time on DNA-modified electrodes (DeRosa et al., 2005). DNA photolyase is an enzyme that repairs cyclobutane thymine dimer (T<>T) lesions which attenuate DNA CT and are a result of photoinduced [2+2] cycloaddition between adjacent thymine bases. Photolyase repairs these lesions using a reductive catalytic cycle (Sancar, 2003), driven by photoexcitation of the flavin cofactor within the enzyme. The T<>T lesion is flipped out of the DNA helix in this reaction, and the flavin cofactor in photolyase initiates a redox reaction to reverse the damage. When the repaired thymine bases are then flipped back in to the DNA duplex, the substrate is able to perform DNA CT, generating a redox signal from the flavoenzyme (Fig. 1). Initially, no signal is observed on DNA-modified electrodes from photolyase in the presence of T<>T damaged duplex DNA. When the surface was irradiated with blue light, however, activating photolyase repair, a reversible redox signal at 40mV vs NHE appears on the electrode surface (DeRosa et al., 2005). This signal potential is within the expected range for photolyase, and it appears only after the T<>T lesion has been repaired. The redox signal is, moreover, attenuated when an abasic site is present in the duplex sequence between the gold electrode surface and the T<>T site, demonstrating that the electron transfer reaction is DNA mediated. This redox activity is observable using different electrochemical techniques, such as cyclic voltammetry (CV) and square wave voltammetry (SWV), and it is enhanced upon longer exposure times to blue light; more repaired lesions yield a larger electrochemical signal. These electrodes thus allow for observation of the redox activity involved in several different DNA-bound biochemical reactions.
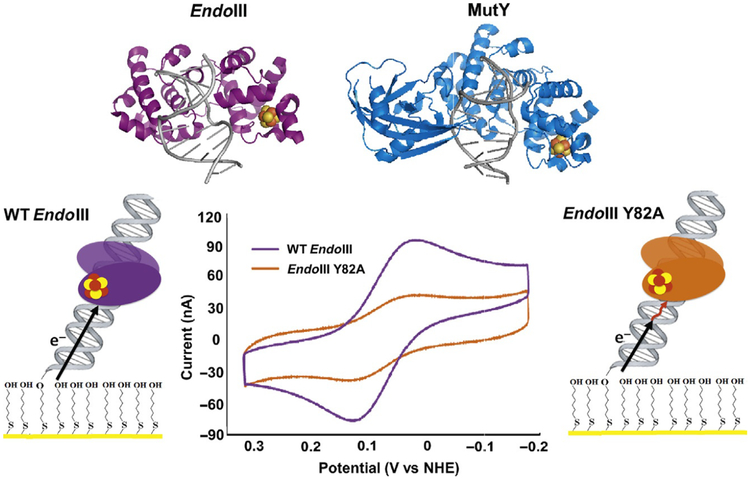
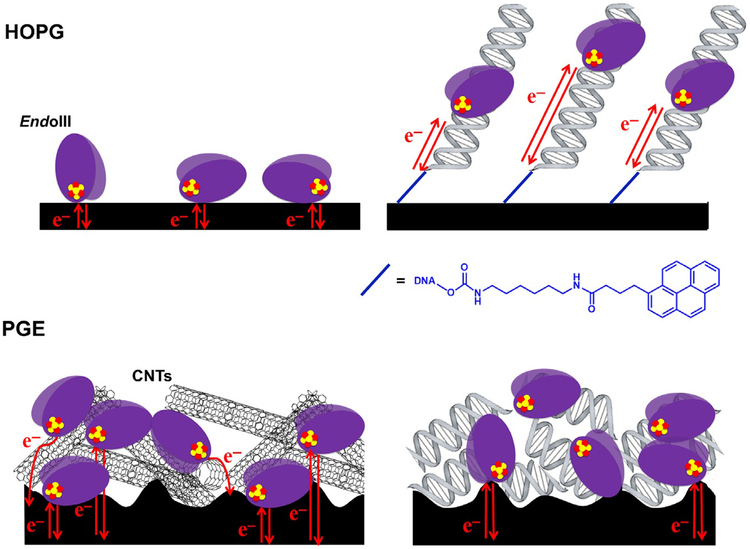
The surprising discovery of a [4Fe4S] cluster in the base excision repair (BER) glycosylase Endonuclease III (E. coli) (Cunningham et al., 1989) led to the investigation of several important questions about the role of these cofactors in DNA repair: Are [4Fe4S] clusters present in other DNA repair enzymes? Do they serve a structural or a biochemical purpose? The discovery of this cluster in Endonuclease III, for example, led to the prediction that it was also present in the homologous BER glycosylase MutY (Michaels, Pham, Nghiem, Cruz, & Miller, 1990), which, similar to Endonuclease III, catalyzes the removal of oxidative damage products from genomic DNA (Kim & Wilson, 2012). The [4Fe4S] cluster would eventually be shown to exist in several BER enzymes, including Endonuclease III, MutY (Guan et al., 1998), and uracil DNA glycosylase (UDG) in Anisocentropus fulgidus (Hinks et al., 2002). Several bioinformatics, structural, and spectroscopic studies contributed to these discoveries (Fu, O’Handley, Cunningham, & Johnson, 1992; Guan et al., 1998; Thayer, Ahern, Xing, Cunningham, & Tainer, 1995). The question of what role the [4Fe4S] clusters played, however, was less straightforward. These clusters are often associated with biological redox chemistry (Rees & Howard, 2003), yet early studies were unable to demonstrate a redox role for these clusters. The DNA-modified electrode platforms developed in our laboratory for protein detection proved instrumental in deciphering and demonstrating the redox chemistry performed by these clusters during DNA repair and represented a completely new tool in characterizing the redox chemistry of these DNA-binding proteins (Fig. 1) (Boal et al., 2009, 2005; Grodick et al., 2014; Mui et al., 2011).
2. DNA-MODIFIED ELECTRODES FOR ELECTROCHEMISTRY
2.1. Designs and Optimization of DNA Electrochemistry Substrates/Monolayers
The adaptability of the platform to various DNA substrates is in part why several different enzymatic reactions can be studied using DNA electro-chemistry. Optimal DNA substrates depend on the protein size and enzymatic function and must be suited for appending onto the DNA electrode surface. An alkanethiol moiety is generally tethered to one end of one strand comprising the final duplex oligonucleotide. This can be readily performed using standard phosphoramidite chemistry, or a thiol-modified oligonucleotide can be ordered from a company such as Integrated DNA Technologies (IDT). This moiety is instrumental in attaching the DNA to the electrode, as a covalent Au-thiol bond will form and give rise to a self-assembling DNA monolayer on the working electrode surface (Kelley et al., 1998) (Fig. 2). Pyrene linkers for DNA modification of graphite electrodes can be appended to the end of a DNA substrate in a similar manner (Gorodetsky, Boal, & Barton, 2006; Gorodetsky, Dietrich, et al., 2008; Gorodetsky, Ebrahim, et al., 2008). Special DNA modifications are commercially available as phosphoramidites from companies such as Glen Research and can be easily integrated into an oligonucleotide sequence on programmable devices such as the Applied Biosystems 3400 DNA Synthesizer (Boal et al., 2009, 2005; Grodick et al., 2014; Mui et al., 2011; Pheeney, Arnold, Grodick, & Barton, 2013; Slinker, Muren, Gorodetsky, & Barton, 2010; Slinker et al., 2011).
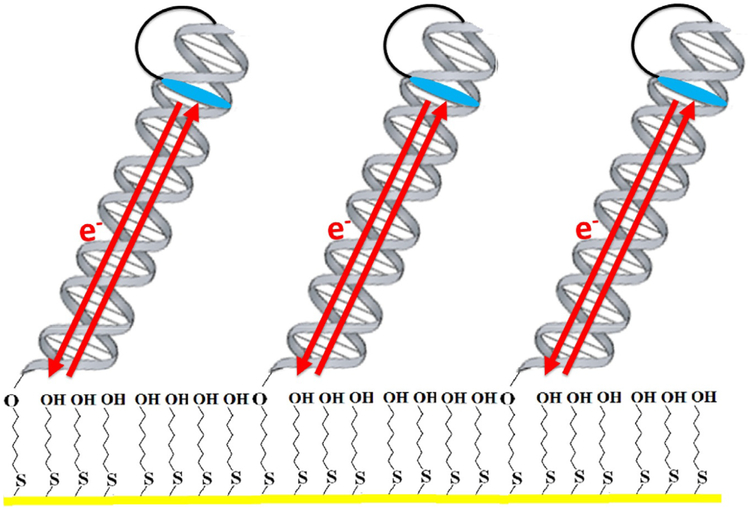
Fig. 2.
Electrochemical monitoring of DNA-mediated charge transport processes. In a typical setup, alkanethiol-modified DNA is annealed to its complement and allowed to form a self-assembled monolayer on a gold electrode. Gaps in the Au surface are filled in with 6-mercapto-1-hexanol, passivating the surface, and electrochemistry is carried out in a buffered solution. Redox-active probes, such as the intercalator Nile Blue, can be covalently tethered to one end of the DNA, or simply bound noncovalently. The DNA duplex then serves as a bridge for electron transfer between the probe and the gold electrode. Notably, charge transport through the DNA is very rapid, and electron transfer rates in this system are limited by tunneling through the alkanethiol linker.
In addition to ensuring that a DNA substrate is modified for attachment to an electrode surface, the oligonucleotide sequence and design is important for monitoring the desired redox reaction. The most important component of a DNA substrate is the presence of a stable duplex segment, generally 15–40 base pairs in length, though DNA CT through up to 100 base pairs (34nm) has been observed electrochemically (Slinker et al., 2011). The duplex should contain at least 50% GC pairs, which is easily verified using the OligoAnalyzer tool on the IDT website. This GC content will prevent melting of a duplex on the electrode surface. A 5’- or 3’-ssDNA overhang, generally 3–15 nucleotides in length (Grodick et al., 2014; Mui et al., 2011), can also be engineered onto the end of the oligonucleotide extending into the electrolyte solution.
DNA monolayers on electrodes can additionally be formed with high or low duplex DNA substrate density, optimized for the size, and binding properties of the enzyme assayed (Pheeney et al., 2013). Some examples of the different densities of monolayers that can be formed on Au electrodes are shown in Fig. 3. Larger proteins, for example, may require low-density monolayers to access the substrate. They may also require a longer duplex sequence or single-stranded DNA (ssDNA) overhang segment to accommodate a larger DNA footprint. Finally, oligonucleotide sequences with mismatches, apurinic sites, or even oxidative lesions such as 8-oxo-guanine, the target lesion of MutY, can be engineered into a substrate duplex. This incorporation is achieved readily with phosphoramidite chemistry; many of these special sequences can alternatively be ordered from IDT.
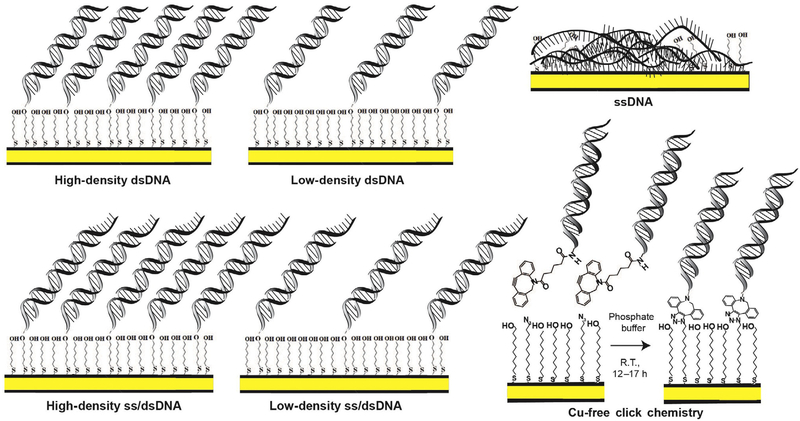
Fig. 3.
Different DNA monolayer morphologies formed on DNA-modified Au electrodes. When duplex DNA is incubated with Mg2+ on an Au surface (yellow), the substrate forms a high-density monolayer of duplex DNA (top left). When incubated on Au in the absence of Mg2+ a low-density duplex DNA monolayer results. DNA containing a single-stranded overhang segment at the interface of DNA monolayer and electrolyte can also be used to form high-density or low-density monolayers for assaying proteins with a preferred primed end substrate (bottom left). When single-stranded DNA is incubated on the Au electrode, the substrate adheres to the surface and passivates the Au, precluding observation of a redox signal (top right). Finally, Cu-free click chemistry can be used to form a DNA monolayer on an Au electrode surface (bottom right). Azide-terminated alkanethiol-modified Au electrode is incubated in 1:1 mix of mercaptoundecanol and 1-azidoundecane-11-thiol in ethanol for about 4h. 50 μM DBCO-modified dsDNA in DNA phosphate buffer is incubated with modified Au electrodes for 12–17h to let the cyclooctyne-based copper-free click reaction proceed. DBCO-modified DNA clicks only to the azide terminal groups, so that the binding density depends on the initial azide content. These monolayers all serve as useful conditions or controls when characterizing redox activity of a DNA-binding enzyme.
With respect to DNA-modified gold electrodes, we have worked with several platforms over time, each of which has its own particular uses. The overall strategy in forming DNA monolayers is the same in all cases, but each platform has distinct requirements in its preparation. The platform developed by our laboratory is unique because we modify our electrode surfaces with duplex DNA substrates, as opposed to ssDNA substrates. ssDNA adheres to and passivates the gold electrode, making the surface very heterogeneous, and precluding observation of a DNA CT-mediated redox signal (Pheeney et al., 2013). Later, we describe the procedures for DNA film preparation on three devices: the 16-electrode multiplexed chip, the standard gold rod electrode, and a gold on mica atomic force microscopy (AFM) surface adapted to fit a custom electrochemical cell (Fig. 4).
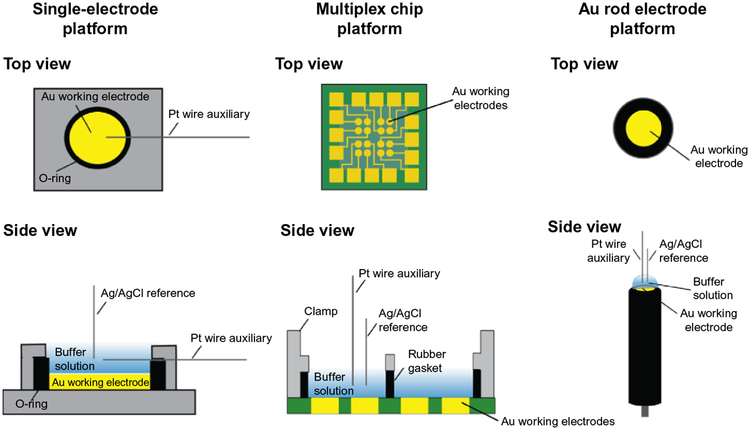
Fig. 4.
Different platforms for DNA electrochemistry. Single Au electrodes can be set up on either an Au on mica surface (left) or using a rod electrode (right). A multiplex platform (center) (Pheeney et al., 2013; Slinker et al., 2010) with 16-electrodes separated into four quadrants can also be used to assay multiple DNA substrates on a single surface, with replicates for each condition. Platforms are shown from the top (above) and from the side (below) with components of the setup.
2.2. Preparing a Self-Assembled Monolayer for DNA Electrochemistry
2.2.1. Sixteen-Electrode Chip Setup (Pheeney et al., 2013)
Notes:
This procedure takes ~2 days, with an overnight incubation step.
Incubate DNA monolayer 21–24h for best results.
Much of the material used for this setup is custom-made, but the mono-layer formation protocol is adaptable to different Au electrode surfaces, for example, the single Au on mica surface and rod electrode setup.
Thiol-modified ssDNA substrates should be re-reduced with dithiothreitol (DTT)/Cleland’s Reagent and repurified after 2–3 weeks of storage at −20°C in the reduced form.
The monolayer needs to be incubated in a moist environment; a pipette box with water in the bottom works well. The porous surface, raised from the water at the bottom, facilitates incubation of the electrode on a raised platform.
Solutions and Reagents:
Sixteen-electrode multiplex chip
Buna-N rubber gaskets, plastic clamps for setup
Isopropanol, Acetone, MQ water
1 M MgCl2
6-Mercapto-1-hexanol (stored under Argon, 100mM stock)
Purified, annealed thiol-modified dsDNA substrate/thiol-modified ssDNA for control
Instruments and Supplies:
Sonication bath (Branson Ultrasonic)
UV ozone cleaner
Small screwdriver for chip assembly
Chip Incubation Box
Argon Gun
Ag/AgCl Gel Tip Reference Electrode
Platinum Wire
Buffer Conditions:
Thiol-modified dsDNA: 5mM sodium phosphate, pH 7.0, 50mM NaCl 100 mM 6-Mercapto-1-hexanol: 5mM phosphate, pH 7.0, 50mM NaCl, 5% glycerol
Electrochemistry buffer (chip washing): 5mM sodium phosphate, pH7.0, 50mM NaCl, 5% glycerol
TBP buffer (chip washing): 5mM sodium phosphate, pH 7.0, 50mM NaCl, 5% glycerol, 4mM MgCl2, 4mM spermidine
Procedure:
- Retrieve annealed, thiol-modified dsDNA stock and make the desired stock for the monolayer (dilute 50% to 25μM with DNA storage buffer, 5mM sodium phosphate, pH 7.0, 50mM NaCl). Final volumes are approximately 20–25μL in each multiplex electrode quadrant. The prepared substrate stock should be prepared with 10%–20% more volume of DNA solution than will be used in electrochemistry experiment.
- High-density duplex: 25μM dsDNA, 0.1 M MgCl2. For MgCl2 addition, use the 1 M MgCl2 stock (American Bioanalytical) and add directly to the 25μM dsDNA stock to a final concentration of 0.1 M.
- Low-density duplex: 25μM dsDNA
- ssDNA control: Dilute thiol-modified ssDNA stock (approximately 150–800μM stock) fourfold into 5mM Pi, pH 7.0, 50mM NaCl. Add 1 M MgCl2 stock to a final concentration of 0.1 M.
Allow DNA stocks to thaw from storage at −20°C vortex, centrifuge, and prepare high-density or low-density duplex DNA.
Pour deionized water into a sonication bath (Branson Ultrasonic size and model is sufficient) before cleaning chip, clamp, gasket.
Place chip in a beaker alone with tweezers, and place the clamp and gasket into a separate beaker with tweezers.
- Wash chip in one beaker and clamp/gasket in a separate beaker in the sonication bath with the following four wash cycles:
- Chip: 3 washes of 10–20mL acetone, 1 wash of 10–20mL 100% isopropanol
- Clamp and gasket: 1 wash of 40–60mL 50% isopropanol in MQ water, 3 washes of 40–60mL MQ water
Dry the chip thoroughly with an argon gun and place in a UV ozone cleaner. Set the ozone cleaner to 10–20min cleaning time, depending on how long the chip has been stored in the hood under argon. Longer ozone cleaning times may be necessary for chips that have been stored outside a clean room environment for longer periods of time.
Dry the clamp and gasket thoroughly with the argon gun. Place them on clean surface, such as a clean room wipe or paper towel.
When the ozone cleaning cycle has finished, retrieve the chip with tweezers and set it on the center of the platform setup.
Align gasket first, then clamp on top of the chip. When the alignment is satisfactory, use the small screwdriver to fasten the setup in place. Tighten the screws thoroughly to avoid leakage of the DNA substrates between quadrants.
Deposit 20–25μL of each dsDNA substrate for monolayers from the prepared stock into the four quadrants. Avoid mixing the stock solutions or mixed monolayers will result.
When all monolayers are deposited, cover the top of the clamp with Parafilm and place the chip setup in the incubator box. Incubate the monolayers for 21–24h.
After the monolayers have incubated, wash the electrodes with 20–25μL volume per quadrant of DNA electrochemistry buffer (5mM sodium phosphate, pH 7.0, 50mM NaCl, 5% glycerol), five cycles through all four quadrants on the chip.
Passivate the electrode surface with 1mM 6-mercapto-1-hexanol, a 100-fold dilution of the 100mM stock in DNA electrochemistry buffer. Wash the electrode quadrants in the same manner as performed with DNA electrochemistry buffer, rinsing each quadrant three times with the passivation agent.
Incubate the backfilled surface in the humid box for 45min.
After 45min have passed, wash each quadrant ten more times with DNA electrochemistry buffer, in 20–25μL volumes per quadrant, to remove mercaptohexanol.
-
Optional: Wash all quadrants twice with TBP buffer (5mM sodium phosphate, pH 7.0, 5% glycerol, 4mM MgCl2, 4mM spermidine), in the same volumes as previous washes. This washing can aid in mono-layer formation and produce better CV scans. Add ~150–300μL of TBP buffer to the top of the solution. Assemble a circuit with a gel tip reference electrode, with a Pt wire fastened securely to the reference setup and proper alligator clip connections (white = reference, red = counter, green = ground) Scan on CHI software to ensure that a monolayer has formed.
CHI Software Parameters for Monolayer Scans:- CV technique
- 100mV/s scan rate
- High voltage = 0.1V, low voltage = −0.4V
- Sweep Segments: 6 =
- Sensitivity: 1 × 10−7 to 1 × 10−8 for initial buffer scans generally works well.
When the presence of a monolayer (observe capacitance of ~40nA on a 2mm2 electrode) has been verified, wash electrodes with protein storage buffer at least five times, through all four quadrants. Repeat the preparation and setup for scanning in TBP buffer, and scan a background of the protein buffer.
2.2.2. Single Au Rod Electrode Setup
Notes:
Total experiment time and monolayer incubation time of 21–24h for optimal results are the same for this platform as for the multiplex chip. Thiol-modified DNA which has been stored at −20°C for 2–3 weeks or longer should again be re-reduced and purified again before deposition onto this electrode platform.
A humid incubation environment is also necessary for this platform. The volume of DNA incubated on the rod electrodes, ~10–15μL of dsDNA substrate, is prone to evaporation and should be monitored during the incubation period to prevent evaporation of the DNA solution droplet. (The same incubation chamber appropriate for the chip can be used for this setup.)
Solutions and Reagents:
The same DNA substrates, 6-mercapto-1-hexanol passivating agent stock, buffers, and 1 M MgCl2 stock used for the chip setup can be used here.
Instruments and Supplies:
Au working rod electrode (1.6mm diameter model manufactured by Pine Research Instruments or Bioanalytical Systems is a typical example for this platform)
Buehler Diamond polish (0.05μm alumina)
Polish Pads
Ag/AgCl Gel Tip Reference Electrode
Platinum Wire
Buffer Conditions:
The same buffers (DNA storage buffer for substrate storage, DNA Electrochemistry Phosphate Buffer, and TBP buffer) as those used in the chip platform are used in this setup.
Procedure:
Thaw annealed, thiol-modified dsDNA stocks and prepare 25μM high-density or low-density DNA substrate solutions as described in the Procedure for the chip setup.
Deposit a small scoop of 0.05μM alumina polish onto the polishing pad. Mix with water to make a slurry of moderate thickness on the pad surface.
Wipe the Au rod electrode with a Kim Wipe and then press the Au surface of the electrode into the slurry on the polishing pad. To ensure thorough and even polishing of the surface, make figure-eight motions with the rod electrode surface, pressing into the slurry each time.
Rinse the polished electrode with deionized water until the slurry has been completely removed from the surface. Blot any excess water on the surface by touching a Kim Wipe to the edges of the electrode. Take care to avoid direct contact of the Kim Wipe with the polished surface, instead using the wipe to absorb the excess water.
Deposit the dry, polished electrode surface into a humid, secure position where the monolayer can incubate after dsDNA deposition.
Deposit high-density or low-density DNA onto the Au surface in a 10–15μL volume, so that a small droplet forms over the surface area of the Au working electrode. Take care to avoid touching the polished surface with a pipet tip, close the incubation chamber, and allow the monolayer to form for 21–24h.
Wash the surface of the rod electrode with the same reagents as described for the chip: five washes of DNA Electrochemistry Phosphate Buffer, three washes of 1mM 6-mercapto-1-hexanol in DNA Electrochemistry Phosphate Buffer, incubate 45min, wash 10 times with DNA Electrochemistry Phosphate Buffer. Optional washing with TBP buffer can also be performed here. All washes should be with a 10–15μL volume of buffer/passivating agent. When pipetting droplets on and off of the electrode surface, take care again to avoid touching the electrode surface, where the new monolayer of DNA has formed.
Check for capacitance on the electrode after the final wash. This is performed in the same manner as for the chip; the reference electrode must touch the droplet but not the surface of the DNA electrode.
2.2.3. Single Au Electrode Setup: Au on Mica Working Electrode (Boal et al., 2009, 2005; Mui et al., 2011).
Notes:
Total experiment time and monolayer incubation time of 21–24h for optimal results are the same for this platform as for the multiplex chip. Thiol-modified DNA which has been stored at −20°C for 2–3 weeks or longer should again be re-reduced and repurified before deposition onto this electrode platform.
A humid incubation environment is also necessary for this platform. The volume of DNA incubated on the rod electrodes, ~10–15μL of dsDNA substrate, is prone to evaporation and should be monitored during the incubation period to prevent evaporation of the DNA solution droplet. (The same incubation chamber appropriate for the chip can be used for this setup.)
Solutions and Reagents:
The same DNA substrates, 6-mercapto-1-hexanol passivating agent stock, buffers, and 1 M MgCl2 stock used for the chip setup can be used here.
Instruments and Supplies:
Au on mica surface (Molecular Imaging)
Platinum wire
Silver paint4. Rubber O-ring/Metal apparatus to fasten Au on mica surface
Ag/AgCl Gel Tip Reference Electrode
Buffer Conditions:
The same buffers (DNA storage buffer for substrate storage, DNA Electrochemistry Phosphate Buffer, and TBP buffer) as those used in the chip platform are used in this setup.
Procedure:
Attach Au on Mica onto the metal apparatus which will connect the Au surface in the three-electrode cell to the potentiostat for measurements. Typically, silver paint is an effective bonding agent to connect Au on mica to the apparatus.
Assemble the working electrode surface, fixing its area with the O-ring and top section of the apparatus to hold the electrode surface in a constant position.
Insert a platinum wire into the electrode solution area, using rubber to prevent leakage through the opening for the platinum counter electrode in the electrochemical cell.
Pipet approximately 40–50μL of 25μM thiol-modified dsDNA (high density or low density) onto the Au surface. Allow the monolayer to form, incubating in a humid environment for 21–24h.
Wash the surface of the rod electrode with the same reagents as described for the chip: five washes of DNA Electrochemistry Phosphate Buffer, three washes of 1mM 6-mercapto-1-hexanol in DNA Electrochemistry Phosphate Buffer, incubate 45min, wash 10 times with DNA Electro-chemistry Phosphate Buffer. Optional washing with TBP buffer can also be performed here. All washes should be with a 40–50μL volume of buffer/passivating agent. When pipetting droplets on and off of the electrode surface, take care again to avoid touching the electrode surface, where the new monolayer of DNA has formed.
Check for capacitance on the electrode after the final wash. This is performed in the same manner as for the chip; the reference electrode must touch the droplet but not the surface of the DNA electrode.
2.3. DNA-Modified Au Electrodes Using Copper-Free Click Chemistry
Conventional DNA-modified surfaces are prepared through self-assembly of thiolated DNA duplexes on gold electrodes followed by backfilling with an alkanethiol to passivate any remaining exposed surface. By including or excluding 100mM MgCl2 during the incubation, one can form both high-density (30–50 pmol/cm2) (Kelley, Barton, Jackson, & Hill, 1997) and low-density (15–20 pmol/cm2) (Boon et al., 2002) monolayers on Au. While straightforward to fabricate, these films pose challenges for control over the spacing of the DNA molecules (Murphy, Cheng, Yu, & Bizzotto, 2009; Sam, Boon, Barton, Hill, & Spain, 2001). Close-packed DNA films limit the accessibility to individual helices during the event of the detection of very large proteins that target-specific sequences of DNA, or hybridization/dehybridization events (Peterson, Heaton, & Georgiadis, 2001). Although adjusting the ionic strength of the deposition solution with Mg2+, some control over the surface density is possible (~15–50 pmol/cm2), close packing still occurs among many helices (Furst, Hill, & Barton, 2013). In such films, the DNA helices cluster into exceedingly large domains of very high density within a sea of passivating thiol. The extensive clustering of helices can be somewhat problematic because it leads to variability across the electrode surface, with regions of close-packed helices in which access to specific base sequences may be inhibited.
The structural similarity of the components of a mixed monolayer-forming solution is a major determining factor for the degree of homogeneity within the resulting self-assembled monolayer (SAM) (Love, Estroff, Kriebel, Nuzzo, & Whitesides, 2005; Ulman, 1996). Thus an alternative approach to a low-density DNA film is to prepare a homogeneous mixed SAM without DNA, followed by DNA conjugation to the functionalized mixed monolayer (Fig. 3). Previous work showed the preliminary formation of a mixed alkanethiol monolayer on gold containing azide-terminated thiols, followed by copper-catalyzed click chemistry to tether single-stranded oligonucleotides to gold surfaces (Devaraj et al., 2005). While copper-catalyzed click chemistry shows high efficiency with mild reaction conditions, conventional copper (I) catalysts can damage DNA and are difficult to remove after the reaction has occurred.
We have developed a catalyst-free method of DNA conjugation to a mixed monolayer that capitalizes on ring strain to drive the [3+2] cycloaddition (Agard, Prescher, & Bertozzi, 2004; Baskin & Bertozzi, 2007). We first form a mixed azide-terminated monolayer, then add cyclooctyne-labeled DNA that spontaneously couples only to the azide via azide-alkyne cycloaddition. The resulting DNA-modified surfaces obtain a low density, more evenly spaced monolayer, while maintaining surface passivation against the redox reporter. Both electrochemical and imaging methods used to characterize these monolayers have been reported (Furst et al., 2013; Furst, Muren, Hill, & Barton, 2014; Muren & Barton, 2013). This approach offers several advantages over conventional preparations of DNA mono-layers: (i) it allows for precise control over the total amount of DNA by simply changing the fraction of thiol-azide present in the preliminary monolayer; (ii) the preliminary self-assembly step results in a passivated surface before the addition of DNA, minimizing undesirable direct interactions between the gold surface and DNA helices; and (iii) because the underlying azide conjugation sites are more evenly distributed in the preliminary mono-layer, DNA helices are less prone to cluster into large, high-density domains.
This platform facilitates DNA-mediated CT and is thus extremely sensitive to perturbations in the DNA, providing exquisite electrochemical discrimination between well-matched and -mismatched DNA duplexes. Additionally, this platform provides greater sensitivity to protein binding events than conventional high-density films due to the larger number of accessible surface-exposed binding sites. In particular, low-density films allow for the detection of as little as 4nM TBP and 5nM human methyltransferase DNMT1 (Furst et al., 2013, 2014; Muren & Barton, 2013). The enhanced detection with copper-free click chemistry adds another sensitive detection tool to the toolbox of electrochemical DNA detection strategies.
Here, we briefly describe the synthesis of azide-terminated alkanethiol linker, the preparation of cyclooctyne-modified DNA from a commercially available source, and the conditions for the copper-free click reaction for DNA-modified electrodes.
2.3.1. Dibenzo-Bicyclooctyne-Modified DNA
From the variety of cyclooctyne-based copper-free click reagents, we use a soluble dibenzo-bicyclooctyne (DBCO)-sulfo-NHS ester sodium salt for conjugation reactions with amino-modified oligonucleotides.
Solutions and Reagents:
DBCO-sulfo-NHS Ester (Glen Research)
Primary amine modified 5’ DNA samples (IDT)
GE Healthcare illustra NAP-5 column
Sodium bicarbonate conjugation buffer (pH 9)
Instruments and Supplies:
HPLC
UV–vis
Thermo cycler
Buffer Conditions:
DNA phosphate buffer (5mM sodium phosphate, 50mM NaCl, pH 7).
Procedure:
Dissolve DBCO-sulfo-NHS Ester at a concentration of 5.2mg per 60μL (~0.17 M solution) in water.
Use this stock solution to conjugate with amino-modified oligos in sodium bicarbonate conjugation buffer (pH 9).
For a 0.2μmol synthesis of a 5’ end amino-modified oligo: dissolve oligo in 500μL of conjugation buffer. Add 6μL of DBCO-sulfo-NHS Ester solution.
Vortex mixture and incubate at room temperature overnight.
Desalt conjugated oligo on a GE Nap 5 column to remove salts and organics. Nap 5 column protocol is followed from the supplier instruction.
Purify DBCO-modified DNA and its complementary strand using reverse-phase HPLC with a polymeric PLRP-S column (Agilent) and characterized by mass spectrometry.
To prepare duplexes, the DBCO-modified DNA and its complementary strand stocks were desalted, resuspended in DNA phosphate buffer, and quantified by UV–vis absorption at 260nm. Equimolar amounts (50μM) of complementary strands were combined and thermally annealed.
2.3.2. Copper-Free Click Reaction for DNA-Modified Electrodes
Proper cleaning of the gold surface is necessary to obtain high-quality thiol-gold-based SAM. For this purpose, rational methods for preparing highly reproducible gold surfaces, include the oxidative and reductive pretreatments (Campuzano, Pedrero, Montemayor, Fatas, & Pingarron, 2006; Kondo et al., 2007). Briefly, gold substrates could be oxidized to a positive charge state via conventional methods, such as ultraviolet/ozone, oxygen plasma, electrochemical oxidation, and piranha solution oxidation. The freshly prepared oxidized gold surfaces can be chemically reduced to zero state (metallic gold) after they were immersed in ethanol. The synthesis of azide-terminated thiol linker, 1-azidoundecane-11-thiol, is reported and adapted from a previously published procedure (Shon, Kelly, Halas, & Lee, 1999).
Solvents and Reagents:
Gold electrode (Au) for voltammetry 1.6mm diameter (Bioanalytical Systems)
0.05μm alumina polish powder (Buehler)
Piranha solution (1:3 H2O2/H2SO4)
SAM deposition solutions: dissolve the desired ratio of mercaptoundecanol (Sigma) and 1-azidoundecane-11-thiol in ethanol. The total thiol concentration is always 1mM.
DBCO-modified double-stranded DNA
Buffer Conditions:
DNA phosphate buffer (5mM sodium phosphate, 50mM NaCl, pH 7.0).
Procedure:
Au rod electrode was polished with alumina polish powder for 1min, rinse with deionized water.
The rod electrodes were immersed in piranha solution for 15min, rinse with deionized water.
Immerse in ethanol and sonicate for 10min, rinse with deionized water.
Cycle Au rod in 50mM H2SO4 between 1.4 and 0V vs Ag/AgCl to obtain an Au (111) single-crystal electrode. Rinse Au rod again with deionized water followed by ethanol (Kondo et al., 2007).
Immerse the cleaned gold substrates in the SAM deposition solution for 4h. After deposition, SAM is rinsed in ethanol and water in order to remove excess adsorbate and dried with N2 to remove residual solvent.
Rinse the Au rod with DNA phosphate buffer. Annealed DBCO-modified dsDNA are conjugated with 1-azidoundecane-11-thiol in phosphate buffer at room temperature for 12–17h.
Note:
For step 4, sharp anodic and cathodic peaks were observed at +1.30 and +0.91V, respectively in CV. The former peak can be assigned to the oxide formation and the latter to the reduction of oxide.
For the 16-electrode multiplex chip setup with the copper-free click chemistry, follow Section 2.2.1 procedures 1–9 for chip cleaning and preparation. For SAM preparation and click DNA coupling, follow Section 2.3.2 procedures 5–6.
Another copper-free click reaction using a cyclooctyne moiety (OCT) tethered 5’ DNA, a mixed monolayer of mercaptoethanol (MCE) as the passivating agent and 6-azido-1-hexanethiol was also reported (Furst et al., 2013). The azide-terminated SAM was formed by soaking the electrodes in an ethanol solution containing 1mM MCE and 0.25mM 6-azido-1-hexanethiol for 24h to form a monolayer composed of 20% azide, followed by an OCT-labeled duplexes, OCT–DNA, coupling to the film via azide-alkyne cycloaddition.
2.4. Characterization of DNA Self-Assembled Monolayers
As described earlier, DNA self-assembled monolayers (SAMs) can be formed on gold electrodes by spontaneous assembly of thiolated DNA or by clicking alkyne-modified DNA onto a preformed azide/thiol monolayer. Regardless of how the monolayer was formed, it is important to characterize the DNA surface coverage and monolayer morphology before proceeding with experiments, as both of these parameters can affect the ability of redox probes or proteins to undertake DNA-mediated processes. If the surface coverage is too sparse, probes and proteins may preferentially interact with the surface, while too much crowding provides steric hindrance that can block efficient protein binding.
We have developed several methods to assess these factors, including visualization of DNA-modified surfaces with AFM, quantification of 32P-labeled DNA, and quantification by [Ru(NH3)6]3+ groove binding (Furst et al., 2013; Sam et al., 2001). AFM is useful in providing a means of visualizing the overall arrangement of DNA on the electrode surface. By repetitive scanning at a high applied voltage, holes can be generated in the surface, permitting the absolute film height to be measured (Furst et al., 2013). Surface area and height can further be used to estimate surface coverage by DNA, although this estimate should be verified by either 32P-labeling or [Ru(NH3)6]3+ quantification. The primary limitations of AFM in surface characterization are that it does require access to an instrument and that such manipulations of the surface preclude further experiments with the particular film being examined. Nonetheless, AFM is an indispensable technique in characterizing the morphology of novel surfaces or monolayers.
2.4.1. AFM Imaging of DNA Films
Solvents and Reagents:
Gold metal (Kurt J. Lesker Industries)
Gold AFM surface (Novascan)
Ethanol (200 proof)
Hexanethiol
Instruments and Supplies:
Silicon AFM tips (Nanosensors Advanced TEC, force constant 0.2N)
Metal evaporator
Multimode Scanning Probe Microscope (Digital Instruments)
Buffer Conditions:
DNA phosphate buffer (5mM sodium phosphate, 50mM NaCl, pH 7.0).
Procedure:
Prepare DNA SAMs on gold electrodes or on Novascan AFM surfaces
Deposit 10nm gold onto the silicon AFM tips using a metal evaporator
Soak the AFM tips in 10mM hexanethiol in ethanol for 1h, and rinse thoroughly with ethanol prior to use
Mount surfaces containing DNA films on scanning probe microscope
Scan surface in contact mode
To measure monolayer height, apply 10V to the AFM tip and repetitively scan a 1μm square to remove the film in this region; after hole generation, measure the height profile by scanning in contact mode
2.4.2. 32P Labeling of DNA
32P labeling allows direct quantification of the DNA at the surface, and provides a 1:1 ratio of signal to DNA. 32P is easily appended to the 5’ end of DNA using commercially available T4 polynucleotide kinase and γ−32P ATP, and monolayers can be formed according to standard procedures. However, the safety concerns, limited half-life of the probe (14 days), and difficulty in measuring radioactivity on an electrode make this technique less appealing.
Solvents and Reagents:
T4 polynucleotide kinase (New England Biolabs)
T4 buffer (New England Biolabs)
10pmol ssDNA with free 5’ ends
γ−32P ATP (Perkin Elmer; 3000–6000 Ci/mmol)
MQ water
Ethylenediaminetetraacetic acid (EDTA)
Instruments and Supplies:
Benchtop incubators
MicroBioSpin6 columns (BioRad)
Tabletop centrifuge
1.5mL Eppendorf tubes
Procedure:
Prepare reactions mixes (50μL) in Eppendorf tubes by adding DNA, 5μL 10× concentrated T4 buffer, and MQ water; keep on ice
Thaw 32P-labeled ATP, and add 40μCi to each reaction tube (All steps involving radioactivity should be carried out behind a Lucite shield!)
Add 1.0μL T4 polynucleotide kinase (5units) to each tube, and start reactions by sealing the tube and incubating at 37°C for 30min
Stop reactions by adding EDTA to a final concentration of 10mM, and heat inactivate the kinase by incubation at 85°C for 10min
Isolate DNA by adding quenched reactions to a MicroBioSpin6 column and spinning for 4min at 1000 × g
2.4.3. DNA Quantification Using [Ru(NH3)6]3+
Due to the experimental ease relative to AFM and 32P-labeling, we generally favor the use of [Ru(NH3)6]3+ for DNA quantification. This method is quite simple, involving only the addition of [Ru(NH3)6]3+ to a surface and scanning (Furst et al., 2013). However, unlike 32P, the signal to DNA stoichiometry is not 1:1, as [Ru(NH3)6]3+ binds electrostatically to the DNA backbone in a ratio of 1 molecule per 3 DNA phosphates. Further drawbacks to this strategy are that it can be easy to underestimate the amount of DNA if saturation is not achieved, and surface accessibility can be an issue. To ensure accurate quantification with [Ru(NH3)6]3+, increasing concentrations should be added until signal saturation is achieved, with care being taken to use a total monovalent ionic strength of no greater than 5mM in the buffer to ensure access of [Ru(NH3)6]3+ to the DNA. The background signal can be determined by comparison with an alkane-thiol only SAM, and at this point, the amount of [Ru(NH3)6]3+ bound to DNA can be determined from either CV or chronocoulomtery. DNA surface coverage is then calculated from the following equation:
| (1) |
Γ is DNA surface coverage in mol/cm2, Q is total measured charge in coulombs from the Ru3+/2+ reduction, n is the number of electrons transferred per reduction (1 in the case of [Ru(NH3)6]3+), F is Faraday’s constant (96,485C/mol), A is electrode area in cm2, and #nt/Ru is the maximum number of [Ru(NH3)6]3+ molecules bound per nucleotide (Kissinger & Heineman, 1996). For the sake of comparison, values are typically reported in pmol/cm2 (Furst et al., 2013). Lastly, it should be noted that, due to the difficulty in washing such small molecules off of the surface, [Ru(NH3)6]3+ quantification should be the final step if further experiments with redox probes or proteins are planned.
Solvents and Reagents:
[Ru(NH3)6]Cl3 (Sigma-Aldrich)
Instruments and Supplies:
Potentiostat
Ag/AgCl reference electrode (Bioanalytical Systems)
Pt wire counter electrode (Kurt J. Lesker Industries)
DNA monolayers on gold electrode
Buffers Conditions:
DNA phosphate buffer (5mM sodium phosphate, pH 7.0).
Procedure:
Prepare DNA SAM in parallel with a monolayer without DNA
Add a small quantity (~1μM) [Ru(NH3)6]Cl3 in 5mM DNA phosphate buffer and scan at a low scan rate (20mV/s is ideal) by CV (or apply a negative potential and use chronocoulometry); the main reductive peak will be near 0mV vs NHE (~200mV vs Ag/AgCl)
Titrate increasingly high concentrations of [Ru(NH3)6]Cl3, scanning each time as in step 2
Repeat with the DNA-free surface
Quantify peak area and subtract DNA-free charge from that obtained with DNA to determine surface coverage
3. DETECTION OF REDOX-SILENT PROTEINS
Electrochemical assays that rely on the sensitivity of DNA-mediated charge transport (DNA CT) chemistry show particular promise for rapid biosensing. As DNA CT is mediated through the base pair π-stack formed by the double helix, this chemistry has unmatched structural sensitivity to perturbations of the π-stack. The nonredox-active DNA-binding proteins that we detect structurally distort the DNA. With a DNA-modified electrode, when a potential is applied to the electrode, DNA CT facilitates reduction of a redox probe, producing an electrochemical signal. DNA with a structural distortion to the π-stack shows an attenuated signal, relative to unperturbed DNA, thereby allowing for sensitive detection of the structural distortion. As most DNA-binding proteins bind specific DNA sequences, this property may be exploited to specifically detect a protein of interest. Electrodes can easily be modified with customized DNA-containing binding sites aimed at the specific detection of target proteins (Figs. 1 and 6). Thus DNA may be utilized in these electrochemical sensors of protein-DNA interactions as both the recognition element and the transducer.
Fig. 6.
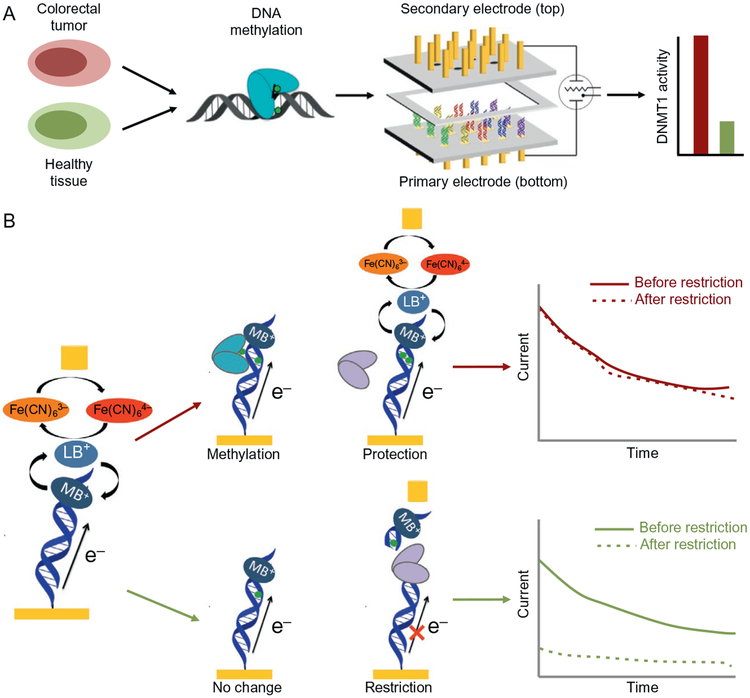
Overview of electrochemical DNMT1 analysis from tumors with two-electrode platform (top). Tumor and healthy tissues are lysed, and nuclear lysate is used to detect DNMT1 methyltransferase activity. The lysate is applied to a multiplexed, two working electrode platform that enables the conversion of methylation events into an electro-chemical signal. The electrochemical detection platform contains two electrode arrays, each with 15 electrodes (1mm diameter each) in a 5 × 3 array. Multiple DNAs are patterned covalently to the substrate electrode by an electrochemically activated click reaction initiated with the patterning electrode array. Once a DNA array is established on the substrate electrode platform, electrocatalytic detection is then performed from the top patterning/detection electrode. Generally, we find hyperactivity of DNMT1 in tumor samples as compared to the healthy adjacent tissue. Signal-on electrochemical assay for DNMT1 detection (bottom). Left: The bottom (primary) electrode modified with a dilute DNA monolayer is responsible for generating electrochemical signals through DNA-mediated (CT) amplified by electrocatalysis. Methylene blue (MB), a DNA-intercalating redox probe, is reduced by DNA CT and enters solution as leucomethylene blue (LB), where it can interact with an electron sink, ferricyanide. Upon interaction with LB, ferricyanide is reduced to ferrocyanide, reoxidizing the LB to MB in the process. Current is generated and detected at the secondary electrode from the reoxidation of ferrocyanide. The current generated is proportional to the amount of ferrocyanide oxidized. To detect DNMT1, crude lysate is added to the electrode. If DNMT1 (blue) is capable of methylating DNA (red arrow), the DNA on the electrode becomes fully methylated. If the protein is not active, the DNA remains hemimethylated or unmethylated (green arrow). A methylation-specific restriction enzyme BssHII (purple) is then added that cuts the unmethylated or hemimethylated DNA (green arrow), significantly attenuating the electrochemical signal, while leaving the fully methylated DNA (red arrow) untouched. Constant potential amperometry (right) is used to measure the percent change before and after restriction enzyme treatment. If the restriction enzyme does not affect the DNA (top), the signals overlay. If, however, the restriction enzyme cuts the DNA, the signal is significantly attenuated (bottom).
In order to measure the activities of nonredox-active DNA-binding proteins by DNA CT, a redox-active probe moiety is incorporated at or near the end of the DNA that is distal from the surface. For this purpose, noncovalent (Boon & Barton, 2003; Boon et al., 2003) and covalent (Buzzeo & Barton, 2008; Gorodetsky & Barton, 2007) redox probes have been employed as well as DNA-binding proteins that are redox active (Section 4). In the DNA-modified electrode, CT is mediated from the electrode surface to the redox probe via the intervening path of well-stacked DNA bases. Importantly, experiments with this platform are all performed in aqueous, buffered solution such that the DNA maintains a native, CT-active conformation.
With the electrochemical monitoring of the DNA-mediated CT, we are able to detect the activity of a sequence-specific restriction enzyme. The efficient cleavage by the restriction enzyme attenuates the DNA CT signals detected by a covalently attached Nile Blue redox probe at the 3’ end of the DNA probe (Section 3.1). The TBP severely kinks the DNA by 80 degree (Fig. 1). The attenuation in DNA CT caused by these structural perturbations can be detected by a covalently tethered methylene blue (MB) redox probe in buffer (Section 3.2). Proteins that bind but do not distort the DNA or proteins do not bind DNA, such as bovine serum albumin (BSA), do not cause this signal attenuation. Further work with the MB probe showed that its DNA-mediated signal may be amplified in an electrocatalytic cycle with ferricyanide (Kelley et al., 1997) and used to sensitively detect all base mismatches (Boon et al., 2002) and a variety of DNA lesions (Kelley, Boon, Barton, Jackson, & Hill, 1999) by an attenuation of DNA CT to the MB redox probe. We can therefore sensitively methyltransferase activity with the MB/ferricyanide electrocatalysis electrochemistry (Section 3.3). Lastly, incorporating the highly sensitivity of the electrocatalysis system, we have designed and fabricated a two-electrode electrochemical platform to detect methyltransferase activity from crude cell lysate (Section 3.4).
3.1. Detection of Restriction Enzyme AluI
We can demonstrate detection of DNA-binding proteins by measuring the sequence-specific activity of the AluI restriction endonuclease, which cleaves at the restriction site 5’-AGCT-3’, leaving blunt ends between the G and C bases. Covalent tethering of the redox probe Nile Blue on the DNA is the probe to monitor the restriction enzyme binding. Here we use the 16-electrode multiplex chip (Section 2.2.1, Fig. 4). The chip was prepared with 17-mer Nile Blue-modified DNA, where half of the electrodes were assembled with a sequence containing the AluI recognition site and the other half with a sequence lacking this site. The AluI restriction enzyme was titrated onto the chip, and the integrated CV peak areas were recorded at each concentration (Slinker et al., 2010). The threshold of AluI restriction activity for the sequence containing the restriction site was 400units/mL, corresponding to a concentration of approximately 10nM. As the total sample volume was 250μL, this corresponds to 2.5pmol of enzyme per chip, or 160fmol of enzyme per electrode. At concentrations greater than 1600units/mL, the charge at the electrodes lacking the restriction site decreases due to nonspecific restriction activity, also known as star activity. In this case, the DNA without the consensus restriction site contains a pseudo-site differing by only one base (5’-ATCT-3’). Thus, as expected at higher enzyme concentrations, restriction cleavage at this pseudo-site is apparent.
Several important implications arise from these observations. Cleavage by the AluI restriction endonuclease requires that the DNA on these chips is in its native conformation and accessible to the protein; one can therefore consider the DNA electrode surface equivalent to that in solution. Moreover, the observation of sequence-specific cleavage indicates that protein detection with DNA-mediated electrochemistry is highly selective. Also, by extension, incorporation of multiple DNA sequences with different protein binding characteristics on a single chip indicates that multiplex chips can serve as a robust platform to simultaneously monitor reactions on different oligonucleotides. Finally, this assay requires only microliter volumes of low protein concentrations, making it competitive with alternative detection methods.
Solutions and Reagents:
AluI restriction enzyme (New England Biolabs), stored at −20°C until use.
Nile Blue-modified DNA with the AluI restriction site (5’-AGCT-3’)
Nile Blue-modified DNA with the pseudo-site (5’-AGAT-3’)
Nile Blue perchlorate (laser grade, Acros)
Instruments and Equipment:
Slide-A-Lyzer mini dialysis kit (Pierce)
CH760B Electrochemical Analyzer and a 16-channel multiplexer module (CH Instruments)
Ag/AgCl reference electrode
Pt wire auxiliary electrode
Sixteen-electrode multiplex chip
Buffer Conditions:
DNA phosphate buffer (5mM sodium phosphate, 50mM NaCl, pH 7.0) Testing Phosphate buffer (DNA phosphate buffer supplemented with 4mM MgCl2, 4mM spermidine, 50μM EDTA and 10% glycerol, pH 7.0)
Tris buffer (50mM Tris–HCl, 10mM EDTA, and 10mM MgCl2, pH 7.8)
Procedure:
DNA-modified 16-electrode multiplex chip setup, see Section 2.2.1.
Prior to use, the AluI restriction enzyme aliquots were exchanged into Tris buffer using a Pierce Slide-A-Lyzer mini dialysis kit at 4°C with overnight stirring.
CV experiments were performed by a CH760B Electrochemical Analyzer and a 16-channel multiplexer module. Chips were tested with a common Pt auxiliary electrode and a common Ag/AgCl reference electrode. Electrochemistry was recorded at ambient temperature in either testing phosphate buffer or Tris buffer.
Dialyzed AluI in Tris buffer was titrated onto the chip with test concentrations ranging from 0 to 50nM (0–2000units/mL). The reaction was allowed to equilibrate at each point of the titration for approximately 30min before scanning the chip. The integrated CV peak areas were recorded at each concentration.
Note:
For the preparation of DNA-modified multiplex chips to measure restriction activity, MgCl2 was excluded from the DNA assembly solution in order to produce a lower density monolayer and grant greater access to the restriction enzyme.
For the electrochemical test, reference and counter electrodes can be patterned on the chip surface, though including other metals for a stable reference would increase the complexity of chip fabrication.
3.2. Detection of TBP Binding Activity
The transcriptional activator TBP has been easily detected on DNA-modified electrodes, given the large perturbation in DNA stacking associated with the binding of TBP. TBP binds to a TATA sequence in DNA and kinks the helix 80 degree at that location, leading to a significant DNA-mediated signal attenuation. In the presence of TBP, which binds to the specific TBP binding site (5’-TATAAAG-3’) and kinks the DNA, the charge accumulation is significantly attenuated (Furst et al., 2013). Protein binding, in kinking the DNA, acts essentially as a switch, turning off DNA CT. BSA, which does not bind to DNA, shows no signal change.
Solvents and Reagents:
MB-modified DNA with the TBP binding sites (5’-TATAAAG-3’)
Modified MB dye for coupling was synthesized as described previously (Pheeney & Barton, 2012)
TBP (ProteinOne), stored at −80°C until use
BSA (New England Biolabs), stored at −20°C until use
Mercaptohexanol (Sigma-Aldrich)
Instruments and Supplies:
Sixteen-electrode multiplex chip
CH760B Electrochemical Analyzer and a 16-channel multiplexer module (CH Instruments)
Ag/AgCl reference electrode
Pt wire counter electrode
Buffer Conditions:
Tris buffer (10mM Tris, 100mM KCl, 2.5mM MgCl2, 1mM CaCl2, pH 7.6)
DNA phosphate buffer (5mM sodium phosphate, 50mM NaCl, pH 7.0) TBP binding buffer (5mM sodium phosphate, 50mM NaCl, 4mM MgCl2, 4mM spermidine, 50μM EDTA, 10% glycerol, pH 7.0)
Procedure:
For the 16-electrode multiplex chip cleaning and preparation for the TBP binding test, see Section 2.2.1.
For all electrochemistry, CV scans were performed at a 100mV/s scan rate over the potential window of 0mV to −500mV. SWV was performed at 15Hz over the same potential range. Signal size was measured as the CV cathodic peak area or the SWV peak area.
For all protein binding experiments, after backfilling with mercaptohexanol, electrodes were backfilled with 3μM BSA in phosphate buffer for 45min at room temperature. After thorough rinsing by buffer exchange, background scans were performed in the TBP buffer TBP. After removing blank TBP buffer from the common well over the electrodes, a solution of the target protein in binding buffer was then added (200μL total volume).
Note:
In this electrochemical protein detection scheme, the protein binding buffer is also the electrochemical running buffer.
3.3. Methyltransferase Detection With Electrocatalysis
The redox-active intercalator MB binds to DNA and becomes electrochemically active on the DNA electrode as long as the individual duplexes that make up the film are completely Watson–Crick base paired. However, the presence of a single-base mismatch or other base-stacking perturbation between the electrode and the site of intercalation greatly attenuates the electrochemical response (Boon et al., 2002; Kelley, Boon, et al., 1999; Kelley, Jackson, Hill, & Barton, 1999). The sensitivity of DNA CT to perturbations in base pair stacking has been used as a platform for the development of electrochemical sensors for mutational analysis (Boon, Ceres, Drummond, Hill, & Barton, 2000), as well as protein/DNA interactions (Boon et al., 2002).
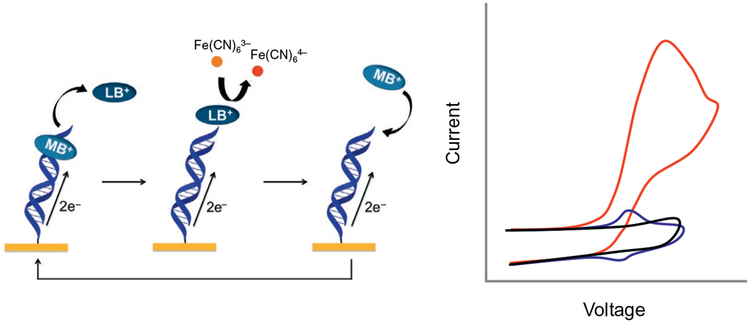
An electrochemical analysis strategy was developed that has improved sensitivity through the combination of electrocatalysis using MB and ferricyanide [Fe(CN)6]3− for signal amplification (Fig. 5). When a negative potential is applied to the DNA-modified electrode, the DNA-bound MB is reduced to leucomethylene blue (LB) via DNA CT and enters the solution. LB has a lower binding affinity to DNA than MB. In solution, ferricyanide [Fe(CN)6]3− is further reduced to ferrocyanide [Fe(CN)6]4− facilitated by the electrocatalytic reduction by MB. A key element is that electrostatic repulsion between the negatively charged DNA films prevents ferricyanide from penetrating and undergoing reduction without mediation by DNA. Instead the free-floating ferricyanide in solution receives electrons from MB through DNA CT. The auxiliary electrode inserted in solution measures the reduction signal of ferricyanide and shows a reading in current change. The positive oxidation potential reoxidizes the [Fe(CN)6]4− and LB is reoxidized to MB. The two-step electrocatalytically amplification has been used for methyltransferase DNMT1 activity detection (Muren & Barton, 2013). Once a DNA array is established on the Au electrodes platform, electrocatalytic detection is then performed with the three-electrode electrochemical cell. Importantly, whether the direct or catalytic reduction of MB is monitored, reduction of MB has been shown to take place via CT through the DNA base stack. DNA CT electrochemistry therefore provides an exquisitely sensitive means to monitor nucleic acid structure and stacking. Even small perturbations in base pair stacking, as is associated with some base lesions, diminish the efficiency of MB reduction (Boon et al., 2000; Kelley, Boon, et al. 1999).
Fig. 5.
Electrocatalytic cycle between free methylene blue (MB) and ferricyanide on a DNA-modified electrode. MB in its oxidized form is intercalated into the DNA base stack. Upon reduction of MB to leucomethylene blue (LB) via DNA-mediated CT, the affinity of the LB for DNA is lowered, and LB is no longer intercalated. The reduced LB is capable of reducing ferricyanide that is freely diffusing in solution. The LB is then reoxidized to MB and can reintercalate into the DNA. The ferricyanide acts as a diffusing electron sink in solution for the redox probe MB. Electrostatic repulsion prevents ferricyanide from penetrating the negatively charged DNA film. A cyclic voltammetry at a DNA-modified electrode of ferricyanide (black), MB (blue), and ferricyanide and MB (red).
DNA methylation is the most prominent form of epigenetic gene regulation and is a critical long-term gene silencing mechanism in mammals (Miranda & Jones, 2007). This covalent addition of a methyl group to the carbon-5 position of cytosine at predominantly 5’-CG-3’ sites is catalyzed by DNA methyltransferases, which use the cofactor S-adenosyl-l-methionine (SAM) as a methyl donor (Flynn & Reich, 1998). However, aberrant DNA methylation has been associated with multiple disease states (Baylin & Herman, 2000; Chen, Akbarian, Tudor, & Jaenisch, 2001; Esteller, 2002). DNMT1 transmits methylation patterns across cell divisions by completing methylation on newly replicated strands at 5’-CG-3’ sites that carry methylation on the template strand alone (Jeltsch, 2002). Thus DNMT1 is characterized as a maintenance methyltransferase and displays a significant preference for hemimethylated DNA substrates (Jeltsch, 2002). These inherently different activities contribute to the complex roles of methyltransferases that are now being elucidated in a growing number of cancers. We have developed an electrochemical platform that combines the ferricyanide/MB electrocatalysis signal-on detection of human DNMT1 activity (Muren & Barton, 2013). Due to the highly sensitivity of the redox probe, 4nM DNMT1 can be detected with the DNA-modified electrodes.
Solvents and Reagents:
Methylene blue (Sigma-Aldrich)
Potassium ferricyanide K3Fe(CN)6 (Sigma-Aldrich)
Human DNMT1 (BPS Bioscience)
BSA (New England Biolabs, used as received)
SAM (New England Biolabs, used as received)
Restriction endonucleases BssHII (New England Biolabs, used as received)
Protease from Streptomyces griseus dry powder (Sigma-Aldrich), stored as a 250μM solution in protease buffer at −20°C
Instruments and Supplies:
Sixteen-electrode multiplex chip
CH760B Electrochemical Analyzer and a 16-channel multiplexer module (CH Instruments)
Ag/AgCl reference electrode
Pt wire auxiliary electrode
DNA-modified multiplex chip
Size exclusion spin column (10kDa cutoff, Amicon)
Incubator
Buffer Conditions:
DNA phosphate buffer (5mM sodium phosphate, 50mM NaCl, pH 7) Scanning buffer (5mM sodium phosphate, 50mM NaCl, 4mM MgCl2, 4mM spermidine, 50μM EDTA, 10% glycerol, pH 7)
DNMT1 activity buffer (50mM Tris–HCl, 1mM EDTA, 5% glycerol, pH 7.8)
Protease buffer (5mM sodium phosphate, 40% glycerol, pH 7) Methylation/restriction (M/R) buffer (10mM Tris–HCl, 50mM NaCl, 10mM MgCl2, pH 7.9)
Procedure:
For the 16-electrode multiplex chip cleaning and preparation test, see Section 2.2.1.
Rinse the chip with phosphate buffer. Scan DNA phosphate buffer first to ensure there is no extra signal/contamination anywhere. CV scan is from 0.4 to −0.4V at 0.1V/s scan rate.
Before the protein treatment, check the DNA-modified multiplex chip with MB and ferricyanide in the scanning buffer for surface passivation and the electrocatalysis signal from DNA monolayer. Replace solution three to five times with MB in scan buffer or ferricyanide in scan buffer at desired concentration.
DNMT1 with 100μg/mL of BSA and 160μM SAM were applied to individual chip quadrants, and chips were incubated at 37°C for 2h in a humidified container. Then chips were rinsed thoroughly with DNMT1 activity buffer and then protease buffer.
Chips were then treated with 1μM protease in DNA phosphate buffer for 1h at 37°C. Then chips were rinsed thoroughly with protease buffer and then M/R buffer.
Chips were treated with 1500units/mL of BssHII in M/R buffer at 37°C for 1h. Then chips were rinsed thoroughly with scanning buffer and scanned with 200μL of the MB and ferricyanide mixture in scanning buffer in a common well.
Note:
DNMT1 shows strong preferential activity at hemimethylated 5’-mCG-3’ sites, DNA substrates with a hemimethylated BssHII restriction site (5’-GmCGCGC-3’) were utilized.
BssHII requires full methylation of either 5’-CG-3’ site within its recognition sequence to prevent DNA restriction.
Buffer exchange of DNMT1 and BssHII prior to electrochemistry experiments is necessary to remove DTT, which disrupts DNA-modified electrodes upon heating. Buffer exchange by size exclusion spin column was performed on DNMT1 and BssHII. The exchange was performed according to manufacturer instructions at 4°C. DNMT1 was exchanged into DNMT1 activity buffer and BssHII was exchanged into M/R buffer.
A protease treatment step was introduced to remove bound DNMT1 following DNMT1 treatment, prior to BssHII treatment.
Including the methyltransferase and restriction enzyme incubations, the total assay time for DNMT1 is about 5h.
Signal size was measured as the CV cathodic peak area. The reported variation in the data represents the standard deviation across all electrodes measured for a given condition.
3.4. Direct Detection of Methyltransferase From Colorectal Cancer Cell Lysate With Two-Electrode Platform
Integrating the high sensitivity of the electrocatalysis system, we have developed a two-electrode platform with the click coupling of low-density DNA monolayers (Section 2.3) for direct detection in crude cancer cell lysates. As opposed to conventional electrochemical readout from the primary DNA-modified electrode, a secondary electrode coupled with ferricyanide/MB electrocatalytic signal amplification, enables more sensitive detection with spatial resolution on the DNA array electrode surface (Fig. 6). Using this two-electrode platform, arrays have been formed that facilitate differentiation between well-matched and -mismatched sequences, detection of transcription factors, and sequence-selective DNA hybridization, all with the incorporation of internal controls (Furst et al., 2013, 2014). For effective clinical detection, the two-electrode platform was multiplexed to contain two complementary arrays, each with 15 electrodes. With the sensitivity and selectivity obtained from the multiplexed, two working electrode array, an electrochemical signal-on assay for activity of the DNMT1 was incorporated.
A two-electrode detection system enables the determination of more specific spatial information on a single substrate electrode surface and leads to high sensitivity since the ferricyanide is only reduced at the secondary electrode, optimizing charge transport through the DNA. Our arrays are formed through selective electrochemical patterning of multiple DNA sequences onto a single-electrode surface containing a preformed mixed monolayer. Electrochemical readout is then accomplished via amperometric detection at a spatially isolated probe electrode controlled by a bipotentiostat. Because multiple DNA sequences are patterned onto a single substrate, different sequences can be examined under identical experimental conditions. With our assay, we now have the ability to incorporate both redundancy and internal controls onto the same electrode surface.
Extending from the signal-on DNMT1 assay, we incorporate the two-electrode electrochemical platform enabling label-free measurements from crude cultured colorectal cancer cell lysates (HCT116) and biopsied tumor tissues (Furst et al., 2014). The multiplexed detection system involving patterning and detection from a secondary electrode array combines low-density DNA monolayer patterning and electrocatalytically amplified DNA CT chemistry to measure selectively and sensitively DNMT1 activity within these complex and congested cellular samples. Based on differences in DNMT1 activity measured with this assay, we distinguish colorectal tumor tissue from healthy adjacent tissue. No difficult or time-consuming purification steps are necessary. For each electrode, only ~4000 cultured cells or ~500μg tissue sample are required. Importantly, because of the multiplexed nature of this platform, we are able to assay for substrate specificity while simultaneously measuring normal tissue and tumor tissue lysates. Therefore, with our platform, healthy tissue is easily distinguished from tumor tissue using very small amounts of sample. More generally, this work represents an important step in new electrochemical biosensing technologies.
Solution and Reagents:
12-Azidododecane-1-thiol (Sigma-Aldrich)
11-Mercaptoundecylphosphoric acid (Sigma-Aldrich)
HCT116 cells, either parent or DNMT1 −/− (received from Vogelstein Lab)
McCoy’s 5A media, with 10% FBS, 100units/mL penicillin, and 100μg/mL streptomycin
Trypsin
Instruments and Supplies:
Au rod electrodes (1mm in diameter)
0.05μm polish
1.5mm deep Teflon spacer
Tissue culture flasks (Corning Costar)
Cell culture incubator, 37°C under a humidified atmosphere containing 5% CO2
Centrifuge
−80°C freezer
Bipotentiostat (Bioanalytical Systems)
Ag/AgCl reference electrode
Pt wire auxiliary electrode
DNA-modified multiplex chip
Size exclusion spin column (10kDa cutoff, Amicon)
Incubator
Buffer Conditions:
1×Phosphate buffer for cell culture
DNA phosphate buffer (5mM sodium phosphate, 50mM NaCl, pH 7) Tris buffer (10mM Tris, 100mM KCl, 2.5mM MgCl2, 1mM CaCl2, pH 7.6)
DNMT1 activity buffer (50mM Tris–HCl, 1mM EDTA, 5% glycerol, pH 7.8)
Nuclear protein extraction kit (Pierce from Thermo Scientific) Bicinchoninic assay (BCA, Pierce)
Procedure:
The multiplexed setup consisted of two complementary arrays containing 15 × 1-mm-diameter gold rod electrodes embedded in Teflon. Gold surfaces were polished with 0.05-μm polish before monolayer assembly.
Thiol SAMs were formed on one of the plates by incubating with 1 M 12-azidododecane-1-thiol and 1 M 11-mercaptoundecylphosphoric acid in ethanol for 18–24h, followed by rinsing with ethanol and phosphate buffer.
The water-soluble [Cu(phendione)2]2+ (phendione =1,10-phenanthroline-5,6-dione) was synthesized by mixing two equivalents of phendione with copper sulfate in water. Covalent attachment of DNA to mixed monolayers containing 50% azide head group and 50% phosphate head group through electrochemically activated click chemistry was accomplished by applying a sufficiently negative potential to the secondary electrode. Specifically, a constant potential of −350mV was applied to a secondary electrode for 25min, allowing for precise attachment of the appropriate DNA to a primary electrode. 40μL of 100μM catalyst and 80μL of 50μM DNA in Tris buffer were added to the platform for covalent attachment.
All electrochemistry was performed as constant potential amperometry for 90s with an applied potential of 320mV to the patterning/detecting electrode array and −400mV to the substrate electrode array All scans were performed in Tris buffer with 4μM MB and 300μM ferricyanide.
To incubate electrodes with desired proteins or cell lysate, a 1.5-mm deep Teflon spacer was clipped to the primary electrode surface. Each electrode is isolated in an individual well that holds 4μL of solution.
HCT116 cells were grown in McCoy’s 5A media in tissue culture flasks in a cell culture incubator.
Approximately 6 million cells were harvested from adherent cell culture by trypsinization, followed by washing with cold PBS and pelleting by centrifugation at 500 × g for 5min.
A nuclear protein extraction kit was used for cell lysis, with buffer then exchanged by size exclusion spin column into DNMT1 activity buffer.
Cell lysate was immediately aliquoted and stored at −80°C until use.
BCA was used to quantify the total amount of protein in the lysate. The total protein concentration at which the lysate was frozen was 35,000–50,000μg/mL.
Cell lysate was combined with SAM to a final SAM concentration of 160μM; or the lysate was diluted in DNMT1 activity buffer to the desired total protein concentration and then combined with SAM to a final SAM concentration of 160μM. Incubate the electrodes at 37°C for 2h in a humidified chamber.
Then the electrodes were treated with protease and restriction enzyme BssHII as described in last section.
Note:
The two-electrode array contains two sets of fifteen gold electrodes, each embedded in a Teflon plate. Each electrode has a 1mm diameter (Fig. 6). The two complementary Teflon arrays are assembled with a 150-μm spacer between them, which were previously determined to be the optimal distance such that signals are not diffusion-limited. The electrodes of the primary (bottom) array are modified with DNA of the desired sequences such that DNA-mediated charge transport is detectable. The electrodes of the secondary (top) array are bare for electrochemical detection.
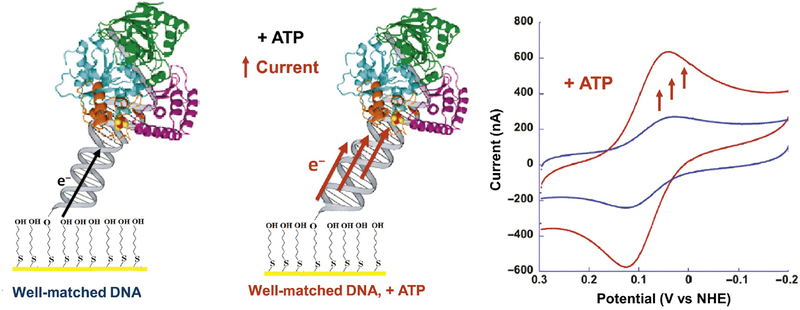
4. REDOX-ACTIVE ENZYMES IN DNA REPAIR MONITORING A REDOX-ACTIVE PROTEIN
Studies of DNA repair enzymes containing [4Fe4S] clusters repeatedly demonstrated that the proteins were isolated in an EPR-silent, [4Fe4S]2+ oxidation state (Boal et al., 2005; Cunningham et al., 1989; Hinks et al., 2002) and resistant to a change in cluster redox state even upon addition of powerful chemical oxidants and reductants to the protein solution. This lack of redox activity was observed in several spectroscopic and biophysical studies, leading to the early conclusion that the [4Fe4S] cluster played a structural rather than functional role in Endonuclease III (Cunningham et al., 1989; Fu et al., 1992; Thayer et al., 1995). In the case of MutY, a BER glycosylase with significant homology to Endonuclease III (Michaels et al., 1990); however, the [4Fe4S] cluster was demonstrated to be nonessential for structural integrity of the protein (Markkanen, Dorn, & Hübscher, 2013; Porello, Cannon, & David, 1998). A substrate-sensing role was thus proposed for the cluster in light of this discovery, but a chemical role for the cofactor in these BER enzymes continued to elude observation.
More structural and biochemical studies of both DNA-dissociated and DNA-bound forms of these proteins continued to make progress towards demonstrating a role for the [4Fe4S] cluster. DNA-bound, high-resolution X-ray crystal structures of both Endonuclease III and MutY (Fromme, Banerjee, Huang, & Verdine, 2004; Fromme & Verdine, 2003), as well as DNA-free structures, were determined, and it was clear that the protein conformation in DNA-bound structures of these proteins was not radically different from conformation in the DNA-dissociated structures (Fromme et al., 2004; Fromme & Verdine, 2003; Guan et al., 1998; Thayer et al., 1995). The [4Fe4S] cluster was, additionally, relatively close to the bound DNA substrate (approximately 20–30Å from the DNA) in both the MutY and EndoIII structures. The short distance from [4Fe4S] cluster to DNA is especially striking when considering that labile Fe2+ ions from such a cofactor can react with hydroxyl radicals and other reactive oxygen species, which are a natural consequence of aerobic respiration in cells, to damage DNA bases (Imlay, 2013). The design in Nature of placing a potentially harmful metal cofactor in a position so close to bound DNA suggested that the [4Fe4S] cluster plays a more significant role in these enzymes.
To probe directly any redox chemistry associated with the cluster, DNA-mediated electrochemistry studies were carried out (Boal et al., 2005; Gorodetsky et al., 2006). This platform is unique in that it facilitates the study of DNA-bound electron transfer activity of proteins, such as the vast family of redox-active, DNA-binding enzymes associated with genomic repair (Boal et al., 2009, 2005; DeRosa et al., 2005; Grodick et al., 2014; Mui et al., 2011). When BER glycosylases MutY (E. coli), Endonuclease III (E. coli), and UDG (A. fulgidus) were initially assayed for DNA-bound redox activity, all three displayed reversible redox signals in the physiologically relevant potential range, corresponding to cycling between the [4Fe4S]2+ and [4Fe4S]3+ oxidation states (Boal et al., 2005). They additionally all displayed similar midpoint potentials, near ~85mV vs NHE. Thus these enzymes appeared to be activated for redox activity at physiological potentials when bound to the DNA polyanion. A subsequent study of Endonuclease III on highly oriented pyrolytic graphite (HOPG) electrodes, comparing redox potentials in the presence and absence of DNA (Gorodetsky et al., 2006), provided direct evidence supporting this observation. The HOPG electrode study showed that while the DNA-dissociated potential of the [4Fe4S] cluster in Endonuclease III is outside the physiologically relevant potential range at approximately ~280mV vs NHE, the DNA-bound potential is near 80mV vs NHE (Gorodetsky et al., 2006). This 200mV negative shift in potential corresponds thermodynamically with a 1000-fold increase in DNA-binding affinity for the oxidized, [4Fe4S]3+ Endonuclease III relative to the reduced [4Fe4S]2+ Endonuclease III. Thus binding of DNA shifts the potential of the [4Fe4S] cluster in these enzymes into the physiologically relevant range, promoting oxidation to the [4Fe4S]3+ state and reversible biological redox activity.
DNA electrochemistry allows for observation of redox activity under physiologically relevant conditions and is adaptable to important control experiments for characterization of the redox signal. Since DNA CT is sensitive to base-pair mismatches and apurinic sites (Arnold et al., 2016; Grodick, Muren, & Barton, 2015), a duplex substrate on the DNA-modified electrode containing a mismatch or apurinic site attenuates a DNA-mediated redox signal. This control allows for confirmation that the protein redox activity is mediated by DNA CT. Additionally, charge transfer-deficient protein mutants, such as Endonuclease III Y82A, are easily assayed on this platform (Boal et al., 2009). A perturbed CT pathway through the protein also attenuates the DNA-bound signal and helps to characterize these mutants before they are used in other in vitro experiments, such as activity assays or genetics studies. Finally, proteins that bind nucleotide tri-phosphates, such as ATP, in their active form can be tested in the presence and absence of their cofactors (Grodick et al., 2014; Mui et al., 2011). This assay allows for comparison of redox activity in the active and inactive forms of the enzyme.
With physiologically relevant assay conditions and adaptability to different DNA and small-molecule substrates, DNA-mediated electrochemistry can be used to monitor several different kinds of enzymatic activity. Helicase activity from the [4Fe4S] enzymes XPD (Sulfolobus acidocaldarius) in the nucleotide excision repair (NER) pathway (Mui et al., 2011), as well as DinG (E. coli) in the R-loop maturation pathway (Grodick et al., 2014), has been measured on DNA-modified electrodes. XPD is an ATP-dependent, 5’- to 3’-helicase that is part of the TFIIH machinery, which is important in transcription and repair (Fan et al., 2008; Liu et al., 2008). Mutations in the [4Fe4S] domain of XPD furthermore lead to genetic disorders such as trichothiodystrophy (TTD), Cockayne syndrome (CS), and xeroderma pigmentosum (XP) (Fan et al., 2008; Liu et al., 2008; Mui et al., 2011). When assayed on a DNA-modified Au electrode, XPD was shown to have a similar DNA-bound redox potential to the BER glycosylase enzymes, near 80mV vs NHE. The redox signal for XPD increased in current upon addition of ATP, but did not change in the presence of a nonhydrolyzable ATP analogue (Mui et al., 2011). The positive correlation between ATP hydrolysis by XPD and the DNA-bound redox signal suggests that the active form of XPD is better coupled to the DNA duplex for DNA-mediated CT activity (Mui et al., 2011). A similar midpoint potential and ATP dependence was observed in the DinG signal on these electrodes (Grodick et al., 2014). Electrochemical experiments measuring the DNA-bound redox activity of glycosylase and helicase enzymes thus shaped the foundation for further biophysical and genetics studies (Boal et al., 2009; Grodick et al., 2014; Sontz, Mui, Fuss, Tainer, & Barton, 2012), which together shaped the foundation for a model in which DNA CT mediates the first step in searching the genome for damage.
A multiplexed electrode setup (Slinker et al., 2010, 2011) was adapted for DNA-bound, redox-active protein electrochemistry (Pheeney et al., 2013). Previously, electrochemistry on DNA-processing enzymes was performed on single surfaces; an AFM surface served as the Au working electrode (Boal et al., 2009, 2005; DeRosa et al., 2005; Mui et al., 2011; Romano, Sontz, & Barton, 2011). A different electrode surface was prepared to obtain each replicate of a single experiment, or to compare of CT activity between different substrates, such as well-matched versus mismatched DNA. The multiplexed setup, however (Fig. 4), has 16-Au electrodes at the center of a silicon chip, which can be patterned and fabricated using standard photolithography and metal evaporation techniques (Pheeney et al., 2013; Slinker et al., 2010, 2011). These electrodes are uniform in area and physically divided into four quadrants, so redox activity on as many as four different DNA substrates can be compared in parallel on a single surface. Though the single electrode platform with an Au AFM surface is robust and straightforward, this multiplexed system facilitates more efficient and ultimately more thorough characterization of DNA-bound redox enzyme activity.
4.1. Conditions for Protein Electrochemistry
When running a protein electrochemistry experiment, it is important to ensure that the buffer conditions, potential range, and protein concentrations used are not harmful to the DNA substrates or the protein in solution. It is also important that they facilitate protein binding to DNA, so a signal can be observed. The buffer pH should be near physiological pH; the general range depends on specific storage conditions for a protein and falls between7.0 and 8.0. Low pH conditions are not recommended, as they can promote glycosidic bond cleavage and depurination of DNA bases on the electrode surface (An et al., 2014). Phosphate is a salt with good buffer capacity, and it should be used as the buffer salt when possible. Some enzymes, such as helicases (Grodick et al., 2014; Mui et al., 2011) and polymerases (O’Brien et al., 2017), however, have phosphate binding sites for native nucleotide triphosphate substrates. These enzymes may have compromised ability to bind DNA or their NTP substrates in electrochemical assays, and in this case a buffer such as Tris or HEPES can be substituted. Tris buffer has a relatively large temperature dependence on pH, however (New England BioLabs, Inc., 2017) and should be used in electrochemical experiments only in a temperature-controlled environment.
Protein concentration is important when performing DNA-mediated electrochemistry. The general range of concentrations, though optimal conditions depend on the specific protein assayed, is approximately 5–50μM. Larger proteins often produce better signals with lower protein concentrations on the electrode, as this allows unhindered individual molecules to diffuse to the DNA substrate, and bind the substrate in a redox-active manner (Grodick et al., 2014; Mui et al., 2011). Though optimal concentration for each protein is determined empirically, the important consistency in these experiments is measurement of concentration for molecules of protein loaded with the redox cofactor. Proteins with a [4Fe4S] cluster have an absorbance at 410nm in the UV–visible spectrum, for example (Cunningham et al., 1989), and the concentration of cluster-loaded protein is most important because those macromolecules, unlike the apoprotein, will be capable of producing a redox signal. For CV experiments, the redox potential scanning range is typically 0.1 to 0.4V vs Ag/AgCl (0.3 to 0.2V vs NHE) (Boal et al., 2009; Grodick et al., 2014; Mui et al., 2011; Pheeney et al., 2013). A range of 0.4 to −0.3V vs NHE is acceptable for a signal with a different potential or widely split reductive and oxidative peaks, however. Bulk electrolysis potentials are generally higher or lower than these limits by only 0.1–0.2V, to ensure full reduction or oxidation of a sample. These mild potential values are important to use on this platform, as they will not strip the DNA monolayer and preclude the measure of DNA-mediated redox activity.
4.2. EndoIII and MutY: [4Fe4S] Proteins in BER
We have made extensive use of electrochemistry on DNA-modified gold electrodes as well as HOPG and pyrolytic graphite edge (PGE) in the presence and absence of DNA to investigate the role of the [4Fe4S] cluster in BER proteins (Bartels et al., 2017; Boal et al., 2005; Gorodetsky et al., 2006). Among these proteins, the first to be well characterized were the E. coli base excision (BER) glycosylases endonuclease III (EndoIII) and MutY (Fig. 7) (Cunningham et al., 1989; Fromme & Verdine, 2003; Porello et al., 1998; Thayer et al., 1995). EndoIII targets oxidized pyrimidines, excising the base as well as nicking the phosphate backbone. MutY is much more specific in its actions, excising adenine mispaired with 8-oxo guanine; unlike EndoIII, MutY lacks AP lyase activity and cannot nick the phosphate backbone. Both of these enzymes were first studied on DNA-modified gold using a 15-mer duplex containing a 5’ thiol modification on one strand (Boal et al., 2005). Using CV, reversible signals occurred at a midpoint potential of 60mV vs NHE for EndoIII and 90mV vs NHE for MutY (Fig. 7). These potentials are at the lower range of HiPIP [4Fe4S] proteins, which utilize the [4Fe4S]3+/2+ couple, and EPR experiments in this and other studies independently confirmed that this was the couple observed electrochemically (Boal et al., 2005; Yavin et al., 2005).
Fig. 7.
Electrochemistry of EndoIII and MutY on DNA-modified gold electrodes. EndoIII and MutY are BER glycosylases that target sites of oxidative damage in DNA; EndoIII (top left) excises oxidized pyrimidines, while MutY (top right) removes adenine mispaired with 8oxoG. When incubated on a DNA-modified electrode, both proteins (EndoIII depicted) display reversible single-electron redox peaks by CV, a process that can be disrupted by mutating critical amino acid residues in the CT pathway as illustrated by EndoIII Y82A (bottom). Structures are adapted from PDB structures IP59 (EndoIII) and 1RRQ (MutY); both are from Geobacillus stearothermophilus, but each shows high homology to the E. coli proteins used in electrochemistry. The CV is adapted from Pheeney, C. G., Arnold, A. R., Grodick, M. A., & Barton, J. K. (2013). Multiplexed electrochemistry of DNA-bound metalloproteins. Journal of the American Chemical Society, 135, 11869–11878.
Importantly, the total peak charge was significantly attenuated on films consisting of DNA with an abasic site positioned near the monolayer surface, indicating that the signal was DNA-mediated (Boal et al., 2005). This platform may also be used to elucidate the CT pathway between the DNA and the cluster, which is usually positioned ~15Å from the DNA (Fromme et al., 2004; Fromme & Verdine, 2003) and thus requires the assistance of aromatic amino acid residues to shuttle charge. With this purpose, we have investigated a range of CT-deficient EndoIII mutants, many of which have disease-relevance in the human homologues (Romano et al., 2011). Among these, EndoIII Y82A exhibited a particularly striking signal attenuation relative to WT, implicating this residue in the CT pathway (Boal et al., 2009).
Work carried out primarily with EndoIII has further revealed the importance of monolayer morphology to protein signals (Pheeney et al., 2013). On high-density films, close packing can render much of the DNA inaccessible, but the enhanced rigidity can make DNA-mediated signals easier to achieve. In contrast, low density films offer more accessible DNA, but the increased flexibility of the DNA can cause much of the signal to become surface-mediated, making the ability of a protein to signal through the DNA difficult to determine. Which monolayer is most appropriate depends largely on the protein itself. EndoIII shows smaller, but DNA-mediated, signals on high-density films, and larger, DNA-bound but surface-mediated signals on low density films; in contrast, bulkier proteins like MUTYH show the opposite effect (Bartels, et al., unpublished manuscript). For this reason, we find it is important to characterize any new protein on both types of DNA film, checking for mismatch or abasic site discrimination in each case as well.
Because EndoIII can be prepared at relatively high concentrations and in large quantities (Pheeney et al., 2013), it is possible to study this protein on graphite electrodes in the absence of DNA. We have used both HOPG and PGE for this purpose, with HOPG serving to make direct comparisons between DNA-free and DNA-bound proteins and PGE allowing us to determine the effects of nearby amino acids and non-DNA molecules on cluster potential. Using HOPG, SWV revealed a large, reversible DNA-mediated signal at 20mV vs NHE, while a smaller, irreversible signal was present around 250mV vs NHE, and the DNA-free [4Fe4S]2+/+ couple was also observed near −300mV vs NHE; these signals were also visible by CV, but the DNA-free signal was small with widely split peaks, impeding quantification (Gorodetsky et al., 2006). On a DNA-modified electrode, the protein is brought to the surface, but this does not occur without DNA modifications. Whether one is using HOPG or PGE, we have found stock concentrations of 50–75μM protein to be ideal; lower concentrations can be observed, but small and shallow peaks make the signal difficult to quantify.
To compare EndoIII and MutY in the absence of DNA, and to determine the effect of point mutations on cluster potential, we turned to thin film voltammetry on PGE with carbon nanotubes (CNTs), which provided large and readily quantifiable signals in the absence of DNA (Bartels et al., 2017). DNA tends to passivate the electrode surface in this system, however, so, while it is possible to see an effect of DNA-binding, the signals are far from ideal and noticeable only by SQWV. Limitations aside, using this system, we were able to show that DNA-binding has a large effect on the potential in both EndoIII and MutY, while poly-l-glutamate and several EndoIII point mutations (E200K, Y205H, and K208E) containing an altered charge distribution in the Fe–S domain do not appreciably alter the potential.
Overall, we have used DNA-modified gold, HOPG, and PGE electrodes to study BER, complemented by spectroscopic techniques, to study long-range electron transfer in [4Fe4S] BER glycosylases. Each of the electrochemical platforms described is useful for answering particular questions about these enzymes, and together they tell a more complete story than any single method alone. DNA-modified gold is well-characterized and easy to use, facilitating extensive characterization of DNA-bound proteins. This includes measurement of the DNA-bound potential, abasic site, or mismatch discrimination experiments to determine if the signal is DNA-mediated, and experiments with mutant proteins to identify the intraprotein CT pathway. The larger potential window provided by graphite electrodes allows both relevant redox couples of the [4Fe4S] proteins to be observed, and make direct comparisons of DNA-free and DNA-bound proteins possible. Among graphite electrodes, HOPG is ideal for comparing potentials on and off of DNA, while PGE is superior for examining differences between DNA-free proteins. In summary, the combination of these three platforms can provide a wealth of information, and their use in our lab has greatly improved the understanding of previously puzzling [4Fe4S] clusters in BER.
4.3. XPD: [4Fe4S] Proteins in NER
Some time after the discovery and characterization of DNA processing [4Fe4S] enzymes in BER, [4Fe4S] clusters were reported in proteins operating in several other repair pathways, including NER. In particular, our laboratory has worked extensively with the specialized helicases XPD (Fig. 8) and DinG, both superfamily 2 helicases (Grodick et al., 2014; Mui et al., 2011). XPD (Rad3 in yeast) is a component of the archaeal and eukaryotic transcription factor IIH (TFIIH) complex, responsible for unwinding DNA surrounding bulky lesions such as thymine dimers to facilitate their removal by endonucleases during NER (Fan et al., 2008). Although structurally homologous to XPD, DinG is a bacterial helicase specialized for unwinding RNA–DNA hybrids at sites of replication fork/transcription collisions (Ren, Duan, & Ding, 2009).
Fig. 8.
Electrochemistry of the NER helicase XPD on a substrate containing a 9-mer 5’ single-stranded overhang. On this substrate, electro-chemical experiments with XPD yielded a signal similar in potential and general form to those from BER proteins (left, blue CV at right). The addition of ATP, known to stimulate helicase activity in XPD, resulted in a substantial increase in current as a result of enhanced electronic coupling between the [4Fe4S] cluster and the DNA base stack (middle, red CV at right). All images in this figure were adapted from Mui, T. P., Fuss, J. O., Ishida, J. P., Tainer, J. A., & Barton, J. K. (2011) ATP-stimulated, DNA-mediated redox signaling by XPD, a DNA repair and transcription helicase. Journal of the American Chemical Society, 133, 16378–16381.
Electrochemistry with helicases on DNA-modified gold electrodes required some modifications from previous work. Unlike EndoIII and MutY, which can bind nonspecifically to dsDNA, XPD, as a 5’ → 3’ helicase, preferentially binds duplexes containing a 3’single-stranded overhang. To meet these criteria, a DNA substrate consisting of 20 base pairs of duplexed DNA and a 9-mer single-stranded overhang was prepared (Fig. 8). With respect to substrates of this type, we have had success with single-stranded overhangs ranging from 9–20 nucleotides in length (Mui et al., 2011); however, since ssDNA can be problematic on gold electrodes due to its tendency to adhere to the surface, overhangs substantially longer than 20 nucleotides are not recommended. Finally, as with EndoIII and MutY, the DNA monolayers were all formed in the absence of Mg2+ to improve substrate accessibility.
Electrochemistry with low density monolayers of the 20:29-mer substrate resulted in a substantial signal from XPD centered around 80mV vs NHE, similar in both appearance and potential to EndoIII and MutY (Mui et al., 2011) (Fig. 8). As long as the duplexed region of the DNA extends below the bound protein, mismatch discrimination experiments can be performed, and the incorporation of a CA mismatch into the base of this substrate did lead to substantial charge attenuation. Overall, these experiments revealed that, despite the single-stranded overhang, the surface-bound DNA was recognizable to a specialized helicase, and mismatch discrimination further indicated that this DNA was accessible in an upright conformation even to a protein as large as XPD (S. acidocaldarius XPD is ~64kDa).
In further contrast with EndoIII and MutY, XPD activity requires ATP hydrolysis to unwind DNA; this afforded an opportunity to observe enzymatic activity on an electrode. Addition of ATP, but not the poorly hydro-lyzed analogue ATP γ-S, led to a sharp increase in current (Fig. 8), indicative of improved coupling to the electrode upon helicase activity (Mui et al., 2011). It appears that XPD is able to signal its activity. Likely, this arises as a result of a conformational change associated with activity leading to more effective electronic coupling on the electrode. As mentioned previously, the electrochemical signal from a diffusing [4Fe4S] protein grows in overtime and eventually stabilizes before decreasing, which can complicate observation of signal enhancement by accessory factors such as ATP. To do so, ATP was added to the surface only after the signal had stabilized, and the change was recorded as the increase in current over time (current after ATP addition increased much more sharply than in either normal signal growth or following ATP γ-S addition).
In summary, DNA-modified gold electrodes can be prepared with diverse substrates, including those with single-stranded overhangs, and this platform can also be used to study the effects of cofactor binding and enzymatic activity on [4Fe4S] cluster coupling. Together, these features allow one to study a wide range of proteins under similarly diverse conditions, as exemplified by XPD.
4.4. Eukaryotic DNA Primase
The redox activity of human DNA primase on DNA illustrates the range of protein detection possible on the DNA-modified electrode platform (O’Brien et al., 2017). Eukaryotic primases are heterodimeric enzymes that contain a [4Fe4S] cluster cofactor in the large, regulatory subunit (Kuchta & Stengel, 2010). DNA primase, as well as the [4Fe4S] cluster domain of DNA primase, each have the intrinsic ability to bind with modest affinity a primed DNA substrate (Sauguet, Klinge, Perera, Maman, & Pellegrini, 2010; Vaithiyalingam, Warren, Eichman, & Chazin, 2010). It was initially observed on DNA-modified electrodes that the [4Fe4S] domain, as isolated in the [4Fe4S]2+ redox state, does not bind DNA tightly enough to couple the redox cofactor into the DNA duplex for CT. In light of the previously demonstrated disparity in DNA binding between the oxidized [4Fe4S]3+ state and the reduced, [4Fe4S]2+ state (Gorodetsky et al., 2006) of these enzymes, bulk electrolysis was performed in anaerobic conditions to monitor and compare the redox activity of an oxidized vs a reduced sample of the DNA primase [4Fe4S] domain. Upon performing bulk electrolysis to produce electrochemically reduced or oxidized samples, CV scans demonstrated that the reduced [4Fe4S]2+ protein, similar to the purified sample before electrolysis, was electrochemically inactive on DNA. The oxidized sample, however, displayed a large reductive peak that disappears after a single scan in CV to negative potentials but is regenerated upon iterative electrochemical oxidations. Electrochemical oxidation or reduction of a protein sample before CV scanning, as was previously demonstrated with EndoIII (Boal et al., 2005), facilitates detection of redox activity in identical enzymes under different cluster oxidation states. In the case of primase, the significant difference between the redox activity of the [4Fe4S]3+ and [4Fe4S]2+ states first observed on DNA-modified electrodes provided significant insight into the relationship between a change in [4Fe4S] cluster oxidation state and the primer synthesis/redox signaling activity of the enzyme.
Using bulk electrolysis to oxidize and reduce samples of DNA-binding [4Fe4S] proteins, before electrochemically monitoring the redox activity with CV or SWV scanning, thus provides a method of directly comparing DNA-mediated redox activity of the protein in the oxidized, [4Fe4S]3+ state, and the reduced, [4Fe4S]2+ state. This method of electrochemical oxidation is advantageous, as it produces a sample of protein in a specific redox state without the damaging effects of chemical oxidants or reductants. Although electrochemical oxidation and reduction avoids potentially damaging the cluster, which for example can degrade to the [3Fe4S]+ species upon oxidation by Co(phen)33+(phen = 1,10-phenanthroline) Boal et al., 2005), there are important limits to this method of cluster oxidation/reduction. The sample cannot be stoichiometrically oxidized/reduced on a feasible timescale for a protein experiment on a DNA electrode. Generally, yields of 60%–80% oxidized protein result upon bulk electrolysis for ~5–10min. The other important consideration in these electrochemical experiments is that the sample must be electrochemically converted on a DNA-modified electrode in an anaerobic atmosphere, with deoxygenated reagents, to avoid atmospheric oxidation of the cluster (Imlay, 2013) and to fully control the redox state of the sample assayed.
5. GRAPHITE ELECTRODES FOR DIRECT ELECTROCHEMISTRY IN THE PRESENCE AND ABSENCE OF DNA
DNA-modified gold is an invaluable tool in studying redox-active proteins as well as in detection, as described in subsequent sections, but the limited potential window available on gold monolayers is a disadvantage when attempting to compare potentials of redox-active [4Fe4S] proteins on and off DNA. On DNA-modified gold electrodes, the potentials of DNA-bound [4Fe4S] proteins all ranged between 65 and 95mV vs NHE, but little to no signal occurred on films lacking DNA (Boal et al., 2005) and in-solution DNA-free proteins were largely inert to electron transfer (Cunningham et al., 1989; Porello et al., 1998). Taken together, these observations implied that DNA-binding shifted the potential to some extent, but the degree of this change could not readily be investigated because the potential window on gold SAMs is limited by desorption of alkanethiols from gold at reducing potentials (≤ −0.4V vs NHE) (Imabayashi et al., 1997; Walczak et al., 1991) and gold oxidation at higher potentials. Although gold oxidation occurs at ~1.5 V for bare gold in concentrated acid, this potential decreases by hundreds of mV with increasing pH and is further lowered in the presence of alkanethiol mono-layers (Benck, Pinaud, Gorlin, & Jaramillo, 2014; Esplandiú, Hagenström, & Kolb, 2001). Finally, the window on gold was too narrow to observe the effect of DNA binding on the lower potential [4Fe4S]2+/+ couple (potentials for this couple typically range from −300 to −700 mV vs NHE).
Pyrolytic graphite electrodes, with an available potential window of 2V, offered a solution to these problems (Fig. 9) (Gorodetsky et al., 2006). Carbon electrodes are commonly used in the study of redox-active proteins, and several forms of graphite surface have been well characterized (Armstrong, Bond, Hill, Oliver, & Psalti, 1989; Banks & Compton, 2006; Blanford & Armstrong, 2006), including HOPG and PGE. Both of these electrodes are formed from stacks of conductive graphite sheets, but they differ in the nature of the exposed electroactive surface (Banks & Compton, 2006): HOPG exposes the largely flat basal plane of the uppermost sheet, and electron transfer is through the stacked sheets, while the PGE surface consists of the perpendicular edge plane with electron transfer occurring through individual sheets. These properties lend advantages and disadvantages to each surface, and the choice is largely dictated by the nature of the experimental questions at hand; indeed, we have successfully used both in our investigations of DNA-binding proteins containing [4Fe4S] clusters.
Fig. 9.
Graphite platforms for protein electrochemistry. Two general platforms are commonly used for protein electrochemistry: HOPG (top) and PGE (bottom). HOPG consists of a pristinely flat, strongly hydrophobic surface, while PGE is rough and often contains surface oxides that lower the hydrophobicity. Proteins can adsorb directly to HOPG, although this interaction is weak, but the ability of pyrene-modified DNA to form a noncovalent bond with the surface allows a direct comparison of DNA-free and DNA-bound proteins. In contrast, PGE provides ample surface area for binding DNA-free proteins, and signals can be further enhanced by the addition of carbon nanotubes (CNTs); DNA can be incorporated into a film on PGE, but in this environment, it tends to passivate the surface and the random orientation prevents the observation of DNA-mediated signals.
Redox-active proteins can be observed directly on HOPG, although the scarcity of suitable binding sites generally results in small, highly split redox peaks in electrochemistry (Armstrong et al., 1989; Gorodetsky et al., 2006). However, because the HOPG surface is composed of a layer of sp2-hybridized carbon, noncovalent stacking interactions between the surface- and pyrene-modified molecules are favored and it is possible to form monolayers with pyrene-modified molecules. In our laboratory, we have established a procedure for preparing pyrene-modified DNA; characterization by AFM, [Ru(NH3)6]3+, and 32P labeling indicates that these monolayers are similar to those formed on gold electrodes (Gorodetsky & Barton, 2006). Pyrene modification of DNA is relatively simple, involving a series of couplings that can be undertaken under ambient conditions, requiring only the exclusion of water. Once prepared, pyrenated DNA monolayers are relatively straightforward to form on HOPG, and the surface can be backfilled with octane; which blocks the exposed surface analogously to alkanethiols on gold.
5.1. Pyrene Modification of DNA
Solvent and Reagents:
Dry acetonitrile
Dry methanol
Dry dioxane
Dry dichloromethane
Diisopropylethylamine (DIEA)
1,1’-Carbonyldiimidazole (CDI)
Hexanediamine
1-Pyrenebutyric acid N-hydroxysuccinimide ester
Ammonium hydroxide
HPLC-grade acetonitrile
50mM ammonium acetate (filtered)
Ethanol
3 M NaCl
Instruments and Supplies:
Cylindrical glass cell with frit at base
Rubber septum
Parafilm
Shaker
3mL syringe
Needles
Aspirator
Tabletop centrifuge
Benchtop incubator
HPLC
PLRPS column for HPLC
UV–visible spectrophotometer
Buffer Conditions:
DNA phosphate buffer (5mM sodium phosphate, pH 7.0, 50mM NaCl).
Procedure:
Add freshly synthesized ssDNA bound to solid CPG beads to a glass cell connected to an aspirator; seal the exposed end with a septum secured by Parafilm. If the DNA was prepared directly on a DNA synthesizer, the terminal 5’-DMT group should be cleaved; alternatively, if the DNA is not attached to a solid support, one end must be blocked with a phosphate group to achieve selective functionalization.
Wash the beads successively with 3mL dry acetonitrile (4×) and 3mL dioxane (3×).
Add 1mL CDI in dioxane and shake at RT for 3h.
After 3h, wash 3× with 3mL dioxane.
Add 1mL hexanediamine in dioxane and shake for 30min at RT.
Wash successively with 3mL dioxane (03×), 3mL dichloromethane (3×), 3mL acetonitrile (3×), and 3mL methanol (3×).
Add 1mL 1-pyrenebutyric acid N-hydroxysuccinimide ester (dissolved in 90% dichloromethane/10% DIEA) and shake overnight at RT.
Wash successively with 3mL dichloromethane (3×), 3mL acetonitrile (3×), and 3mL methanol.
Cleave DNA from CPG beads with 800μL fresh NH4OH (8h, 60°C).
Isolate cleaved DNA by centrifugation in spin columns; discard the beads and dry the flow-through on a speed vacuum overnight.
Dissolve DNA in 600μL phosphate buffer (5mM sodium phosphate, pH 7.0, 50mM NaCl) and purify by HPLC (acetonitrile/50mM ammonium acetate in a gradient ranging from 95% to 85% ammonium acetate over 35min, followed by a return to 95% over 5min), making sure to collect the peak with absorbance at 260, 280, and 345nm.
Freeze the sample in liquid nitrogen and dry overnight by lyophilization.
Desalt DNA: add 100μL water, 50μL 3 M NaCl, and 1mL 100% ethanol; freeze in liquid nitrogen, spin down (12,000rpm, 25min), and dry overnight on a speed vacuum.
Quantify DNA by UV–vis and anneal with complement in a 1:1 ratio in phosphate buffer; annealing should be carried out by 5’-incubation at 95°C followed by slow cooling to RT over 2–3h.
5.2. DNA Monolayer Assembly on HOPG
Although very useful for DNA-mediated electrochemistry, the highly hydrophobic HOPG surface is not ideal for direct electrochemistry without DNA, and PGE is instead favored for this purpose. PGE has a rough surface that provides abundant, readily accessible electroactive sites which improve the coupling of redox-active species to the electrode. To minimize protein diffusion, it is common practice to immobilize redox-active enzymes in a thin film when working with PGE. Proteins immobilized in such films have been shown to be catalytically active even after several days, indicating that adsorption to the electrode does not alter their native conformations (Baffert et al., 2012). Although the PGE surface can be very effective in facilitating direct electrochemistry, the incorporation of conductive single-walled CNTs into the film can greatly enhance signal size by providing additional three-dimensional area for protein binding (Yin, Lu, Wu, & Cai, 2005). To anchor the CNTs in place and prevent film dispersal, the entire protein-CNT film can be secured by a capping layer such as Nafion (Yin et al., 2005). Indeed, we have achieved large signals from DNA-free [4Fe4S] proteins using several (3–6) CNT-protein layers capped with 5% aqueous Nafion (Bartels et al., 2017). It should be noted that, despite the advantages of PGE in direct protein electrochemistry, this surface is not ideal for studying DNA-bound proteins. In particular, homogenous DNA films cannot be prepared on this surface, necessitating the addition of DNA along with the protein. In this state, the orientation of the DNA prevents experiments with mismatch or abasic site discrimination, which rely on upright DNA, and it causes severe surface passivation; furthermore, CNTs were found to hinder DNA binding by EndoIII, and thus had to be excluded from these thin films (Bartels et al., 2017). Nonetheless, very small signals can still be obtained with DNA present, but they are of much lower quality than those obtained on gold or HOPG.
Solvents and Reagents:
Pyrene-modified DNA (preannealed)
Octane
Glycerol
Ethanol
MQ Water
Protein storage buffer
Instruments and Supplies:
HOPG electrode, either a commercial rod electrode (Pine Research Instrumentation provides a high-quality product) or surfaces (SPI Supplies sells these in several different grades)
5μm Silica polish (if using a rod electrode)
3 M Double-sided Scotch tape (if using surfaces)
Electrochemical cell (if desired; inverted drop cells can be purchased from Pine Research Instrumentation or custom made)
Sonicator (if using a rod electrode)
50mL Falcon tubes
Buffer Conditions:
DNA phosphate buffer (5mM sodium phosphate, pH 7.0, 50mM NaCl).
Procedure:
-
a. If using a commercially prepared rod electrode, clean the surface with 0.05μm silica polish. The polish can then be rinsed off by sonication for 0.5–1min first in ethanol and then in water.
b. Alternatively, if mounting a square of HOPG in a drop cell, clean HOPG can be exposed by pressing 3 M Scotch tape on the square and rapidly pulling back. This process will ideally remove a single layer, but it often takes several attempts before a truly pristine surface is obtained. Large defects are visible as pits on the surface and should be kept to a minimum; as with rod electrodes, a cursory buffer scan should be used to verify the absence of electroactive impurities.
To ensure the absence of surface defects and electroactive impurities, a CV scan of the surface should be taken in the protein storage buffer. Defects representing surface oxides on exposed edge plane give a broad, reversible electrochemical signal around 200mV vs NHE. These defects can enhance direct electrochemistry of adsorbed proteins, but too many of them will inhibit the attachment of pyrenated DNA.
Once the surface is sufficiently clean, add 25μM annealed DNA (pyrenated strand + complement) in phosphate buffer to the surface and incubate overnight; if high-density surfaces are desired, 100mM MgCl2 may be added along with the DNA in this step.
Rinse two to three times with phosphate buffer to remove unbound DNA, taking care not to scratch the surface. This step should be carried out very gently if pipetting to avoid removing excess DNA; if this is problematic, the electrode can instead be immersed in a Falcon tube containing 5–10mL phosphate buffer.
If DNA-free protein adsorption is a problem, the remaining bare electrode surface can be blocked by backfilling with octane. To backfill, add phosphate buffer containing 20% glycerol and 10% octane by volume and incubate 15–30min at RT; rinse in phosphate buffer containing 20% glycerol as in step 4. This process should be repeated two to three times.
After backfilling, the electrode should be rinsed and scanned in protein storage buffer. Once the background scans have been taken, protein solution can be added to the surface and scanned. High concentrations are ideal if possible; in the case of EndoIII, 50μM protein worked well, but even stronger signals were obtained using 200μM protein.
5.3. Protein Thin-Film Voltammetry With CNTs
Solutions and Reagents:
Ethanol
MQ water
Single-walled CNTs
Nafion (diluted to 5% in water, purchased as 10% solution from Sigma-Aldrich)
Instruments and Supplies:
PGE electrode (we use the relevant product from Pine Research Instrumentation, but others are available)
400 grit sandpaper or diamond polish
Sonicator
Electrochemical cell (if desired)
Potentiostat6. Reference electrode (usually Ag/AgCl)
Counter electrode (Pt wire)
Buffer Conditions:
Protein storage buffer
Procedures:
To roughen the surface for protein adsorption, the electrode should first be abraded with 400 grit sandpaper. This is done by applying water to a sheet of sandpaper and gently “polishing” the electrode. Sandpaper provides deep ridges for protein adsorption, and worked well for EndoIII and MutY. If sandpaper treatment is ineffective, diamond polish can be used as an alternative; diamond polish generates pitting across the surface, which may be more suitable for some proteins.
To remove electroactive impurities, sonicate the electrode in ethanol (30–60s) followed by water (30–60s). At this point, a drop of protein storage buffer can be applied and the background scanned to ensure the absence of impurities. It should be noted that the rough edge plane is oxide rich, and surface species will generate a broad peak around 200mV vs NHE; this should not interfere with protein voltammetry and can even assist in adsorption.
- To prepare thin films, first dry on a layer of CNTs, and then apply several layers of protein solution as follows:
- Suspend single-walled CNTs in water (0.25mg/mL) by vigorous sonication and add a droplet to the electrode surface, and dry in air or under an argon stream.
- Apply a droplet of protein solution (ideally 50–75μM) to the surface, and dry as with the CNTs. Repeat several times until the surface appears coated with protein.
- Secure the film with 5% Nafion in water; this should be applied to the electrode and dried as with the other layers.
Once the thin film is dried and secured, apply a 30–50μL droplet to the vertical electrode, drop reference and auxiliary electrodes into the bulk solution, and scan. Alternatively, the electrode can be inverted and placed in an electrochemical cell containing a bulk buffer solution.
In summary, graphite electrodes have the advantage of providing a wide accessible potential window that covers the entire range of potentials accessed by [4Fe4S] proteins, allowing the observation of redox events not possible on gold. Specifically, particular types of graphite electrode are better for addressing distinct aspects of protein electrochemistry. Because HOPG can facilitate both direct and DNA-mediated electrochemistry, this electrode is the best choice for making direct comparisons of DNA-free and DNA-bound potentials. However, the hydrophobicity of HOPG makes direct electrochemistry challenging, and if one is interested in studying redox-active proteins in the absence of DNA, thin-film voltammetry on PGE is the best option.
ACKNOWLEDGMENTS
We are grateful to the NIH for their support of this research and to our coworkers and collaborators for their insights, efforts, and hard work.
REFERENCES
- Agard NJ, Prescher JA, & Bertozzi CR (2004). A strain-promoted [3+2] azidealkyne cycloaddition for covalent modification of blomolecules in living systems. Journal of the American Chemical Society, 126, 15046–15047. [DOI] [PubMed] [Google Scholar]
- An R, Jia Y, Wan B, Zhang Y, Dong P, Li J, et al. (2014). Non-enzymatic depurination of nucleic acids: Factors and mechanisms. PloS One, 9, e115950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong FA, Bond AM, Hill HAO, Oliver N, & Psalti ISM (1989). Electrochemistry of cytochrome c, plastocyanin, and ferredoxin at edge- and basal-plane graphite electrodes interpreted via a model based on electron transfer at electroactive sites of microscopic dimensions in size. Journal of the American Chemical Society, 111, 9185–9189. [Google Scholar]
- Arnold AR, Grodick MA, & Barton JK (2016). DNA charge transport: From chemical principles to the cell. Cell Chemical Biology, 23, 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baffert C, Sybirna K, Ezanno P, Lautier T, Hajj V, Meynial-Salles I, et al. (2012). Covalent attachment of FeFe hydrogenases to carbon electrodes for direct electron transfer. Analytical Chemistry, 84, 7999–8005. [DOI] [PubMed] [Google Scholar]
- Banks CE, & Compton RG (2006). New electrodes for old: From carbon nanotubes to edge plane pyrolytic graphite. Analyst, 131, 15–21. [DOI] [PubMed] [Google Scholar]
- Bartels PL; O’Brien E; Barton JK in collaboration with McDonnell K; Chemler JA; et al. unpublished manuscript.
- Bartels PL, Zhou A, Arnold AR, Nuñez NN, Crespilho FN, David SS, et al. (2017). Electrochemistry of the [4Fe4S] cluster in base excision repair proteins: Tuning the redox potential with DNA. Langmuir, 33, 2523–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin JM, & Bertozzi CR (2007). Bioorthogonal click chemistry: Covalent labeling in living systems. QSAR & Combinatorial Science, 26, 1211–1219. [Google Scholar]
- Baylin SB, & Herman JG (2000). DNA hypermethylation in tumorigenesis: Epigenetics joins genetics. Trends in Genetics, 16, 168–174. [DOI] [PubMed] [Google Scholar]
- Benck JD, Pinaud BA, Gorlin Y, & Jaramillo TF (2014). Substrate selection for fundamental studies of electrocatalysts and photoelectrodes: Inert potential windows in acidic, neutral, and basic electrolyte. PloS One, 9, e107942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanford CF, & Armstrong FA (2006). The pyrolytic graphite surface as an enzyme substrate: Microscopic and spectroscopic studies. Journal of Solid State Electrochemistry, 10, 826–832. [Google Scholar]
- Boal AK, Genereux JC, Sontz PA, Gralnick JA, Newman DK, & Barton JK (2009). Redox signaling between repair proteins for efficient lesion detection. Proceedings of the National Academy of Sciences of the United States of America, 106, 15237–15242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boal AK, Yavin E, Lukianova OA, O’Shea VL, David SS, & Barton JK (2005). DNA-bound redox activity of DNA repair glycosylases containing [4Fe-4S] clusters. Biochemistry, 44, 8397–8407. [DOI] [PubMed] [Google Scholar]
- Boon EM, Barton JK, Pradeepkumar PI, Isaksson J, Petit C, & Chattopadhyaya J (2002). An electrochemical probe of DNA stacking in an antisense oligonucleotide containing a C3’-endo-locked sugar. Angewandte Chemie, International Edition, 41, 3402–3405. [DOI] [PubMed] [Google Scholar]
- Boon EM, & Barton JK (2003). DNA electrochemistry as a probe of base pair stacking inA-, B-, and Z-form DNA. Bioconjugate Chemistry, 14, 1140–1147. [DOI] [PubMed] [Google Scholar]
- Boon EM, Ceres DM, Drummond TG, Hill MG, & Barton JK (2000). Mutation detection by electrocatalysis at DNA-modified electrodes. Nature Biotechnology, 18, 1096–1100. [DOI] [PubMed] [Google Scholar]
- Boon EM, Jackson NM, Wightman MD, Kelley SO, Hill MG, & Barton JK (2003). Intercalative stacking: A critical feature of DNA charge-transport chemistry. The Journal of Physical Chemistry B, 107, 11805–11812. [Google Scholar]
- Buzzeo MC, & Barton JK (2008). Redmond red as a redox probe for the DNA-mediated detection of abasic sites. Bioconjugate Chemistry, 19, 2110–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campuzano S, Pedrero M, Montemayor C, Fatas E, & Pingarron JM (2006). Characterization of alkanethiol-self-assembled monolayers-modified gold electrodes by electrochemical impedance spectroscopy. Journal of Electroanalytical Chemistry, 586, 112–121. [Google Scholar]
- Chen RZ, Akbarian S, Tudor M, & Jaenisch R (2001). Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nature Genetics, 27, 327–331. [DOI] [PubMed] [Google Scholar]
- Cunningham RP, Asahara H, Bank JF, Scholes CP, Salerno JC, Surerus K, et al. (1989). Endonuclease III is an iron-sulfur protein. Biochemistry, 28, 4450–4455. [DOI] [PubMed] [Google Scholar]
- DeRosa MC, Sancar A, & Barton JK (2005). Electrically monitoring DNA repair by photolyase. Proceedings of the National Academy of Sciences of the United States of America, 102, 10788–10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj NK, Miller GP, Ebina W, Kakaradov B, Collman JP, Kool ET, et al. (2005). Chemoselective covalent coupling of oligonucleotide probes to self-assembled monolayers. Journal of the American Chemical Society, 127, 8600–8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esplandiú MJ, Hagenström H, & Kolb DM (2001). Functionalized self-assembled Alkanethiol monolayers on au(111) electrodes: 1. Surface structure and electrochemistry. Langmuir, 17, 828–838. [Google Scholar]
- Esteller M (2002). CpG island hypermethylation and tumor suppressor genes: A booming present, a brighter future. Oncogene, 21, 5427–5440. [DOI] [PubMed] [Google Scholar]
- Fan L, Fuss JO, Chemg QJ, Arval AS, Hammel M, Roberts VA, et al. (2008). XPD helicase structures and activities: Insights into the cancer and aging phenotypes from XPD mutations. Cell, 133, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J, & Reich N (1998). Murine DNA (cytosine-5-)-methyltransferase: Steady-state and substrate trapping analyses of the kinetic mechanism. Biochemistry, 37, 15162–15169. [DOI] [PubMed] [Google Scholar]
- Fromme JC, Banerjee A, Huang SJ, & Verdine GL (2004). Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature, 427, 652–656. [DOI] [PubMed] [Google Scholar]
- Fromme JC, & Verdine GL (2003). Structure of a trapped endonuclease III-DNA covalent intermediate. The EMBO Journal, 22, 3461–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, O’Handley S, Cunningham RP, & Johnson MK (1992). The role of the iron-sulfur cluster in Escherichia coli endonuclease III: A resonance Raman study. The Journal of Biological Chemistry, 267, 16135–16137. [PubMed] [Google Scholar]
- Furst AL, Hill MG, & Barton JK (2013). DNA-modified electrodes fabricated using copper-free click chemistry for enhanced protein detection. Langmuir, 29, 16141–16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furst AL, Muren NB, Hill MG, & Barton JK (2014). Label-free electrochemical detection of human methyltransferase from tumors. Proceedings of the National Academy of Sciences of the United States of America, 111, 14985–14989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorodetsky AA, & Barton JK (2006). Electrochemistry using self-assembled DNA monolayers on highly oriented pyrolytic graphite. Langmuir, 22, 7917–7922. [DOI] [PubMed] [Google Scholar]
- Gorodetsky AA, & Barton JK (2007). DNA-mediated electrochemistry of disulfides on graphite. Journal of the American Chemical Society, 129, 6074–6075. [DOI] [PubMed] [Google Scholar]
- Gorodetsky AA, Boal AK, & Barton JK (2006). Direct electrochemistry of endonuclease III in the presence and absence of DNA. Journal of the American Chemical Society, 128, 12082–12083. [DOI] [PubMed] [Google Scholar]
- Gorodetsky AA, Dietrich LE, Lee PE, Demple B, Newman DK, & Barton JK (2008a). DNA binding shifts the redox potential of transcription factor SoxR. Proceedings of the National Academy of Sciences of the United States of America, 105, 3684–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorodetsky AA, Ebrahim A, & Barton JK (2008b). Electrical detection of TATA binding protein at DNA-modified microelectrodes. Journal of the American Chemical Society, 130, 2924–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodick MA, Muren NB, & Barton JK (2015). DNA charge transport within the cell.Biochemistry, 54, 962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodick MA, Segal HM, Zwang TJ, & Barton JK (2014). DNA-mediated signaling by proteins with 4Fe-4S clusters is necessary for genomic integrity. Journal of the American Chemical Society, 136, 16470–16478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Manuel RC, Arvai AS, Parikh SS, Mol CD, Miller JH, et al. (1998). MutY catalytic core, mutant, and bound adenine structures define specificity for DNA repair enzyme superfamily. Nature Structural Biology, 5, 1058–1064. [DOI] [PubMed] [Google Scholar]
- Hinks JA, Evans MCW, de Miguel Y, Sartori AA, Jiricny J, & Pearl LH (2002). An iron-sulfur cluster in the family 4 uracil-DNA glycosylases. The Journal of Biological Chemistry, 277, 16936–16940. [DOI] [PubMed] [Google Scholar]
- Imabayashi S, Iida M, Hobara D, Feng ZQ, Niki K, & Kakiuchi T (1997). Reductive desorption of carboxylic-acid terminated alkanethiol monolayers from Au(111) surfaces. Journal of Electroanalytical Chemistry, 428, 33–38. [Google Scholar]
- Imlay JA (2013). The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nature Reviews. Microbiology, 11, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A (2002). Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem, 3, 274–293. [DOI] [PubMed] [Google Scholar]
- Kelley SO, Barton JK, Jackson NM, & Hill MG (1997). Electrochemistry of methylene blue bound to a DNA-modified electrode. Bioconjugate Chemistry, 8, 31–37. [DOI] [PubMed] [Google Scholar]
- Kelley SO, Barton JK, Jackson NM, McPherson LD, Potter AB, Spain EM, et al. (1998). Orienting DNA helices on gold using applied electric fields. Langmuir, 14, 6781–6784. [Google Scholar]
- Kelley SO, Boon EM, Barton JK, Jackson NM, & Hill MG (1999a). Single-base mismatch detection based on charge transduction through DNA. Nucleic Acids Research, 27, 4830–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley SO, Jackson NM, Hill MG, & Barton JK (1999b). Long-range electron transfer through DNA films. Angewandte Chemie, International Edition, 38, 941–945. [DOI] [PubMed] [Google Scholar]
- Kim Y-J, & Wilson DM III. (2012). Overview of base excision repair biochemistry.Current Molecular Pharmacology, 5, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger PT, & Heineman WR (Eds.), (1996). Laboratory techniques in electroanalytical chemistry (2nd ed.). New York, NY: Marcel Dekker. [Google Scholar]
- Kondo T, Morita J, Hanaoka K, Takakusagi S, Tamura K, Takahasi M, et al. (2007). Structure of Au(111) and Au(100) single-crystal electrode surfaces at various potentials in sulfuric acid solution determined by in situ surface X-ray scattering. Journal of Physical Chemistry C, 111, 13197–13204. [Google Scholar]
- Kornberg RD (2007). The molecular basis of eukaryotic transcription. Proceedings of the National Academy of Sciences of the United States of America, 104, 12955–12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchta RD, & Stengel G (2010). Mechanism and evolution of DNA primases. Biochimica et Biophysica Acta, 1804, 1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Rudolf J, Johnson KA, McMahon SA, Oke M, Carter L, et al. (2008).Structure of the DNA repair helicase XPD. Cell, 133, 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love JC, Estroff LA, Kriebel JK, Nuzzo RG, & Whitesides GM (2005). Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chemical Reviews, 105, 1103–1169. [DOI] [PubMed] [Google Scholar]
- Markkanen E, Dorn J, & Hübscher U (2013). MUTYH DNA glycosylase: The rationale for removing undamaged bases from the DNA. Frontiers in Genetics, 4, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels ML, Pham L, Nghiem Y, Cruz C, & Miller JH (1990). MutY, an adenine glycosylase active on G-A mispairs, has homology to endonuclease III. Nucleic Acids Research, 18, 3841–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda TB, & Jones PA (2007). DNA methylation: The nuts and bolts of repression.Journal of Cellular Physiology, 213, 384–390. [DOI] [PubMed] [Google Scholar]
- Mui TP, Fuss JO, Ishida JP, Tainer JA, & Barton JK (2011). ATP-stimulated, DNA-mediated redox signaling by XPD, a DNA repair and transcription helicase. Journal of the American Chemical Society, 133, 16378–16381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muren NB, & Barton JK (2013). Electrochemical assay for the signal-on detection of human DNA methyltransferase activity. Journal of the American Chemical Society, 135, 16632–16640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JN, Cheng AKH, Yu HZ, & Bizzotto D (2009). On the nature of DNA self-assembled monolayers on au: Measuring surface heterogeneity with electrochemical in situ fluorescence microscopy. Journal of the American Chemical Society, 131, 4042–4050. [DOI] [PubMed] [Google Scholar]
- New England BioLabs, Inc. (2017). pH vs. temperature for Tris buffer. Retrieved from https://www.neb.com/tools-and-resources/usage-guidelines/ph-vs-temperature-for-tris-buffer.
- O’Brien E, Holt ME, Thompson MK, Salay LE, Ehlinger AC, Chazin WJ, et al. (2017). The [4Fe4S] cluster of human DNA primase functions as a redox switch using DNA charge transport. Science, 355, 813, eaag1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill MA, Becker H-C, Wan C, Barton JK, & Zewail AH (2003). Ultrafast dynamics in DNA-mediated electron transfer: Base gating and the role of temperature. Angewandte Chemie, International Edition, 42, 5896–5900. [DOI] [PubMed] [Google Scholar]
- Peterson AW, Heaton RJ, & Georgiadis RM (2001). The effect of surface probe density on DNA hybridization. Nucleic Acids Research, 29, 5163–5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pheeney CG, Arnold AR, Grodick MA, & Barton JK (2013). Multiplexed electrochemistry of DNA-bound metalloproteins. Journal of the American Chemical Society, 135, 11869–11878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pheeney CG, & Barton JK (2012). DNA electrochemistry with tethered methylene blue. Langmuir, 28, 7063–7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porello SL, Cannon MJ, & David SS (1998). A substrate recognition role for the [4Fe-4S]2+ cluster of the DNA repair glycosylase MutY. Biochemistry, 37, 6465–6475. [DOI] [PubMed] [Google Scholar]
- Rees DC, & Howard JB (2003). The interface between the biological and inorganic worlds: Iron-sulfur metalloclusters. Science, 300, 929–931. [DOI] [PubMed] [Google Scholar]
- Ren B, Duan X, & Ding H (2009). Redox control of the DNA damage-inducible protein DinG helicase activity via its iron-sulfur cluster. The Journal of Biological Chemistry, 284, 4829–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano CA, Sontz PA, & Barton JK (2011). Mutants of the base excision repair glycosylase, endonuclease III: DNA charge transport as a first step in lesion detection. Biochemistry, 50, 6133–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sam M, Boon EM, Barton JK, Hill MG, & Spain EM (2001). Morphology of 15-mer duplexes tethered to Au(111) probed using scanning probe microscopy. Langmuir, 17, 5727–5730. [Google Scholar]
- Sancar A (2003). Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chemical Reviews, 103, 2203–2237. [DOI] [PubMed] [Google Scholar]
- Sauguet L, Klinge S, Perera RL, Maman JD, & Pellegrini L (2010). Shared active site architecture between the large subunit of eukaryotic primase and DNA photolyase. PloS One, 5, e10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shon YS, Kelly KF, Halas NJ, & Lee TR (1999). Fullerene-terminated alkanethiolate SAMs on gold generated from unsymmetrical disulfides. Langmuir, 15, 5329–5332. [Google Scholar]
- Slinker JD, Muren NB, Gorodetsky AA, & Barton JK (2010). Multiplexed DNA-modified electrodes. Journal of the American Chemical Society, 132, 2769–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slinker JD, Muren NB, Renfrew SE, & Barton JK (2011). DNA charge transport over 34 nm. Nature Chemistry, 3, 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontz PA, Mui TP, Fuss JO, Tainer JA, & Barton JK (2012). DNA charge transport as a first step in coordinating the detection of lesions by repair proteins. Proceedings of the National Academy of Sciences of the United States of America, 109, 1856–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer MM, Ahern H, Xing D, Cunningham RP, & Tainer JA (1995). Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. The EMBO Journal, 14, 4108–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulman A (1996). Formation and structure of self-assembled monolayers. Chemical Reviews,96, 1533–1554. [DOI] [PubMed] [Google Scholar]
- Vaithiyalingam S, Warren EM, Eichman BF, & Chazin WJ (2010). Insights into eukaryotic priming from the structure and functional interactions of the 4Fe-4S cluster domain of human DNA primase. Proceedings of the National Academy of Sciences of the United States of America, 107, 13684–13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak M, Popenoe DD, Deinhammer RS, Lamp BD, Chung C, & Porterm MD (1991). Reductive desorption of alkanethiolate monolayers at gold: A measure of surface coverage. Langmuir, 7, 2687–2693. [Google Scholar]
- Yavin E, Boal AK, Stemp EDA, Boon EM, Livingston AL, O’Shea VL, et al. (2005). Protein-DNA charge transport: Redox activation of a DNA repair protein by a guanine radical. Proceedings of the National Academy of Sciences of the United States of America, 102, 3546–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Lu Y, Wu P, & Cai C (2005). Direct electrochemistry of redox proteins and enzymes promoted by carbon nanotubes. Sensors, 5, 220–234. [Google Scholar]