Abstract
North American Grapevine Yellows (NAGY) is a destructive disease of grapevines caused by phytoplasmas, wall-less bacteria that are insect-transmitted and found in plant phloem tissues. Although the disease was recognized in vineyards in the eastern United States since the 1980s, the identities of vectors remain unknown. The objectives of this study were to survey potential phytoplasma vector insects inhabiting Virginia vineyards that expressed NAGY symptoms and to evaluate their ability to transmit phytoplasmas associated with NAGY. Phytoplasmas were identified as ‘Candidatus Phytoplasma pruni’-related NAGYIIIβ strains and ‘Ca. Phytoplasma asteris’-related NAGYI-B strains. To determine the identities of the potential vectors, artificial feeding solution was used to evaluate the ability of leafhopper species to release phytoplasmas during feeding and phytoplasma strains were identified using molecular tools. Out of 49 insect species screened, Jikradia olitoria was the only insect that released phytoplasmas into the feeding solutions; all phytoplasmas, thus, detected were identified as NAGYIIIβ strains by nucleotide sequencing of three different genomic regions. No NAGYI-B strain was detected. To our knowledge, this is the first evidence of a potential insect vector of a specific phytoplasma associated with NAGY disease, and it is the first report of J. olitoria being a putative vector of a plant pathogenic phytoplasma.
Keywords: phytoplasma, vector, leafhopper, artificial diet, transmission analysis
North American Grapevine Yellows (NAGY) is a destructive disease of grapevines caused by phytoplasmas, wall-less, plant-pathogenic, insect-transmitted bacteria that are confined to plant phloem tissues. Phytoplasmas are taxonomically classified in the class Mollicutes and, unlike most other members of this class, have not been consistently cultured outside of their host plants and vector insects. Symptoms of NAGY are similar to other grapevine yellows diseases that occur globally, including Flavescence dorée (FD) (Boudon-Padieu 2015), bois noir (Daire et al. 1997, Sforza et al. 1998), and Australian Grapevine Yellows (Constable et al. 2003). Symptoms include leaf reddening in red-fruited cultivars (cvs.), leaf chlorosis in white-fruited cvs., downward rolling of leaf margins, dieback of shoot tips, abortion of fruit clusters, and nonuniform maturation of shoot stem periderm; affected vines often die within 2 or 3 years of symptom onset (Wolf 2015). Although there are common symptoms of all ‘grapevine yellows’ diseases, the specific pathogens, vectors, and alternative hosts often differ among these diseases, reflecting the different ecological conditions and geographical regions wherein the diseases occur. Grapevine yellows diseases require certain conditions, including susceptible hosts, one or more causal pathogens, effective vectors, and possibly alternative host plants to maintain a reservoir of the pathogens. Flavescence dorée, for example, is believed to have rapidly spread among French vineyards as a result of the importation of an effective insect vector, Scaphoideus titanus, from North America (Caudwell 1983, Tessitori et al. 2018). Although the disease was observed in Virginia since the 1980s, the ecology and epidemiology of NAGY remain poorly understood. Although considerable progress has been made in the characterization of the causal pathogens of NAGY (e.g., Davis et al. 2015, 2018) and the detection of alternative hosts (our unpublished data), the identification of the insect vector species is lacking.
The phytoplasmas causing NAGY began to be identified in the 1990s when restriction fragment length polymorphism (RFLP) analysis of polymerase chain reaction (PCR)-amplified 16S rDNA revealed the presence of two genetically different phytoplasmas in NAGY infected vines in Virginia (Prince et al. 1993, Davis et al. 1998). RFLP analysis of 16S rRNA sequences provided taxonomic means of classifying phytoplasmas into >30 16S ribosomal (16Sr) groups and >200 subgroups (Lee et al. 1998, Wei et al. 2007). One of the initially detected phytoplasmas was genetically similar to the 16SrIII phytoplasma responsible for X-disease in Prunus species, ‘Candidatus Phytoplasma pruni’. A second NAGY phytoplasma was described and differentiated from the group 16SrIII phytoplasma on the basis of 16S rDNA gene RFLP fragments (Davis et al. 1998). The second phytoplasma was closely related to strains classified in rRNA RFLP group 16SrI (‘Ca. Phytoplasma asteris’-related strains) or aster yellows group. Genotyping of the secY and ribosomal protein (rp) genes of the NAGY phytoplasma revealed that 16SrIII NAGY strains comprised two distinct sequevars, NAGYIIIα and NAGYIIIβ, both of them slightly different from strains of ‘Ca. Phytoplasma pruni’ (Davis et al. 2015). As such, the NAGYIII phytoplasma strains were termed ‘Ca. Phytoplasma pruni-related’ strains. The second NAGY phytoplasma, classified in the rRNA RFLP 16SrI aster yellows group, was recently assigned to 16SrI subgroup I-B and variant subgroup I-B* on the basis of genotypic analyses of the 16S rRNA and secY genes, complemented with 3D, in silico modeling of the SecY protein (Davis et al. 2018).
Insect transmission of NAGY is a logical assumption based on insect transmission of diverse phytoplasmas globally, the known role of leafhoppers in other grapevine yellows diseases (Weintraub and Beanland 2006), and the diversity of leafhoppers observed in infected vineyards (Beanland et al. 2006). Spatial analysis of symptomatic NAGY grapevines in two Virginia vineyards revealed a nonrandom, clustering incidence within the vineyard, with apparent ‘edge’ effects over eight years of observation (Beanland et al. 2006). Vector identification, however, is hampered by the extended latency between inoculation of grapevines and expression of disease symptoms.
Beanland et al. (2006) reported the seasonal abundance of a diverse range of leafhoppers and planthoppers, with three of them, Scaphoideus titanus, Osbornellus auronitens, and Jikradia olitorius (currently, J. olitoria), exhibiting seasonal movement into the vineyard that could explain the clustering of infected vines near the vineyard edge. In addition to the above-mentioned species, Graminella nigrifrons, Macrosteles quadrilineatus, and Deltocephalus flavicosta, which are recognized phytoplasma vectors in other yellows diseases (Weintraub and Beanland 2006), were also abundantly collected. Scaphoideus titanus was implicated in the original FD-like disease in New York, in part due to its abundance in and around affected vineyards, but also due to positive ELISA tests against antigens derived from FD-infected vines (Maixner et al. 1993). Although S. titanus was introduced to Europe, there is no evidence that the phytoplasmas specifically associated with NAGY were similarly introduced. In fact, there is no evidence to date of their occurrence outside of North America (Davis et al. 2015) and the only NAGY phytoplasma recently found in New York, a group III phytoplasma (Davis et al. 2015), is unrelated to the FD phytoplasma. Testing of leafhoppers found in and immediately outside NAGY-affected vineyards in Virginia occasionally produced positive PCR results for phytoplasmas detected in cultivated vines; however, actual transmission attempts were inconclusive (our unpublished data).
The objective of this study was to screen a wide range of Cicadellidae insects found in Virginia vineyards for their potential to transmit specific phytoplasmas responsible for NAGY.
Materials and Methods
Plant Material and Insects.
Cultivated grapevines
Mature leaves of grapevine (Vitis vinifera L.) cvs. Chardonnay, Cabernet Sauvignon, Riesling, and Tannat grown at three commercial vineyards in northern Virginia were used for identification of NAGY phytoplasmas and comparison to phytoplasmas recovered in artificial diet solutions. Leaves were sampled from May to September over the 2012–2016 period, they were kept at 4°C and processed within a few days.
Live insect collections for transmission studies:
Leafhoppers were collected over the 2012–2017 period from vineyards with NAGY history located in Frederick, Fauquier and Loudoun Counties, Virginia, and therefore presumably with competent vectors present. Insects were collected using sweep nets from grapevines, vineyard floor, vineyard headlands, and mixed vegetation located immediately outside the vineyard, from May until October over the period 2012–2017. Net-collected leafhoppers and occasionally planthoppers were aspirated from nets into sample tubes using a mouth aspirator and transported in a cool container to the laboratory where they were identified and used in the transmission attempts explained below.
Geographically ‘extensive’ insect surveys:
To gain a sense of the diversity of Cicadellidae insects found in vineyards, leafhoppers and planthoppers were surveyed in 27 vineyards distributed from southeast Pennsylvania through Virginia and into the Yadkin Valley of North Carolina during the 2013 growing season. The vineyards typically included cv. Chardonnay and had exhibited some degree of NAGY incidence in recent years. Each vineyard was visited at least six times between late-May and early-September of 2013. Two methods were used to sample insects: Yellow sticky trap cards (10 × 30 cm) were placed on vineyard posts at two heights above ground level (0.6 and 1.5 m), near a vineyard’s edge located in close proximity of scrub or forest vegetation. For each vineyard, five such traps were placed at each height, for a total of 10 per vineyard per 7- to 14-d (generally) sampling period. Trap cards were transported to the laboratory after deployment and examined within one week under a dissecting microscope to identify and enumerate Cicadellidae specimens. Additionally, 100 sweeps of vine canopies and 100 sweeps of the vineyard floor were made at each vineyard with an insect sweep net (38-cm diameter) with insects of interest aspirated into storage tubes for identification and potential use in subsequent transmission studies.
Weekly ‘intensive’ insect surveys:
Insect surveys were also conducted weekly at two northern Virginia vineyards between 2013 and 2015 to estimate leafhopper diversity within and between seasons. The intensive sampling was conducted at ‘LV’ and ‘RdV’ vineyards, both located in Fauquier County, VA, and involved a 0.5-ha block of Chardonnay at LV, and two blocks of Cabernet Sauvignon at RdV vineyard, each approximately 0.8-ha in size. Both vineyards had a recent history of vine loss due to NAGY. Three yellow sticky trap cards were deployed in each block at the canopy height of the trellis (0.5 m at RdV and 1.2 m at LV) in three separate rows, and at least 20 m from any edge of the vineyard block. Cards were generally changed weekly, and occasionally biweekly, and the retrieved cards were examined under a dissecting microscope to identify and count leafhoppers and planthoppers. In addition, insects were also collected weekly at ground level using a sweep net (100 sweeps per vineyard block) along a 100-m length of row middle in each of the vineyard blocks. Surveyed periods of each year were 16 May to 17 October (2013), 23 May to 28 October (2014), and 27 April to 9 October (2015). Captured insects of interest were aspirated into plastic vials and cataloged in the laboratory.
For all insect surveys, adult leafhoppers and planthoppers were visually identified to genus and species where possible, using external morphology, coloration, and other external characteristics as referenced both by published taxonomic keys and by web-based, color photos. For the latter, detailed information and photos were utilized from the National Museum Wales natural history website (https://museum.wales/collections/natural-history/), the University of Georgia (https://www.forestryimages.org), and others. Identification of genera, and often species, was confirmed by Christopher H. Dietrich of the Illinois Natural Survey, Urbana, IL, using both external morphological characteristics and male genitalia structure.
Transmission Assays
Vineyard-collected live insects were either cooled on ice or briefly anesthetized with carbon dioxide in Petri dishes to slow their movement and allow identification and sorting for transmission studies. An artificial diet and feeding technique was adapted from Tanne et al. (2001) in an attempt to collect phytoplasmas secreted during the insect’s feeding activity. Leafhoppers were identified under a dissecting microscope and separated according to species. Individual insects were placed in clear, translucent, 1.5-ml microcentrifuge tubes whose caps were removed and replaced by yellow translucent caps (Tanne et al. 2001). The caps were first charged with 200 μl of 5% sucrose Tris-EDTA (TE) solution and sealed with tightly stretched Parafilm. The tube walls were pierced with small holes to permit air exchange and decrease condensation in the tubes. Tubes were held at room temperature and placed horizontally with the caps facing a source of light to encourage the insects to probe the sucrose solution through the Parafilm septum. Insects were either collected and directly transferred to artificial diet tubes (2012–2013) or were initially caged on NAGY-infected grapevine leaves in the field (2014–2015) for a 1-d acquisition access period (AAP). After AAP, insects were transferred to pots of barley (Hordeum vulgare L.) and white clover (Trifolium repens L.) plants for a 21-d latency period (LP), the length of time needed for most leafhoppers to become inoculative (Purcell 1982), before being transferred to the artificial diet feeding tubes. Insects were maintained in the feeding tubes until their death, which generally occurred within 72 h after being placed in the tubes. If not tested immediately, the transmission tubes containing insects and sucrose solution were stored at −20C. Ultimately, sucrose/saliva mixtures were tested for the presence of phytoplasmas using a seminested PCR aimed at amplifying the 16S region. Samples were used directly as the source of template DNA in PCRs. In total, 1,950 single-specimen artificial diet tests, used to feed 49 different leafhopper and planthopper species, were performed in 2012–2014. An additional 36 single-specimen transmission attempts were made in 2015. Given the results of such tests in 2012–2014, we focussed on J. olitoria captured in a mixed shrub/woodland setting within 100 m of a Chardonnay and Merlot research vineyard at the Alson H. Smith Jr. Agricultural Research and Extension Center (AHS Jr. AREC), located in Frederick County, Virginia, in 2016. This is the same setting from which J. olitoria specimens had been collected in August 2013. The research vineyard exhibited an increased incidence of NAGY-infected vines over the 2014–2016 period, with ~15 affected vines out of ~600 observed in 2016.
Finally, collection of J. olitoria was repeated at the LV vineyard between 28 July and 10 October 2017 and transmission trials with J. olitoria were also repeated in 2017 at the AHS Jr. AREC. The 2017 collections were prompted by our casual observation of an increased presence of J. olitoria specimens in these vineyards and our interest in repeating transmission trials with this species. More than 150 specimens of J. olitoria were sweep-net-collected from the vegetation outside of the vineyards between 3 August and 2 October 2017. Of these, 99 individual specimens were individually introduced to sucrose solution feeding tubes as described above. In addition to sweep netting, six yellow sticky cards were deployed and changed weekly at LV vineyard between 21 July and 10 October 2017. Three were located within the vineyard at the vine canopy height (1.2 m) on exposed areas of the trellis, and three were located at approximately the same height, facing out from the vineyard on a fence that separated the vineyard by ~20 m from mixed, deciduous woodland.
DNA Extraction
Grapevine leaves
Mature leaves from NAGY symptomatic field grapevines were collected and used for phytoplasma DNA extraction, PCR amplifications, and subsequent nucleotide sequence analysis. Excised leaves were initially washed in 1.2% sodium hypochlorite and rinsed five times in distilled water to reduce the bacterial population on the leaf surface. Midrib and major leaf veins were excised and frozen at −20°C until DNA extraction. Veins from two or three symptomatic leaves of the same infected shoot were pooled and ground using a FastPrep-24 Tissue and Cell Homogenizer (MP Biomedicals, Santa Ana, CA). DNA was extracted using a modified method described in Green et al. (1999). Briefly, 1 ml of extraction buffer composed of 100 mM Tris pH 8.0, 1.4 M NaCl, 50 mM EDTA pH 8.0, 2.5% (w/v) cetyltrimethylammonium bromide (CTAB), 1% (w/v) polyvinylpyrrolidone (PVP-40), and 0.2% (v/v) 2-mercaptoethanol (added just prior to use) was used per 1.5 g of plant tissue. Samples were then incubated for 30 min at 65°C in the presence of RNase A (Qiagen, Germantown, MD). The extraction then proceeded using a DNeasy Plant DNA Extract Kit (Qiagen), following the Qiagen protocol.
Sucrose solutions
We compared the PCR efficiency between nonextracted sucrose solution samples and samples whose DNA was extracted as described by Tanne et al. (2001) and we concluded that there was no reason to extract DNA from the artificial diets, as sucrose did not impair the PCR efficiency and the extraction method, in some cases, caused false negatives (our unpublished data). The extraction protocol was particularly unsuccessful on artificial diets used to feed individual insects, because it inevitably reduced the amount of phytoplasma DNA. Therefore, 2-μl samples of feeding media were used directly for PCR, without DNA extraction.
PCR Analysis
Phytoplasma detection for both grapevine leaves and the sucrose feeding media was carried out using nested or seminested PCR in a Bio-Rad C1000 thermocycler (Bio-Rad, Hercules, CA).
Briefly, a first amplification was diluted 1:30 and 2 μl were used in the second round of amplification. To amplify the 16S region, two primer combinations were used: P1 and P7 primers, followed by 16S-Sr and P1A; or P1 and 16S-Sr primers followed by P1A and 16S-Sr in a seminested PCR. Table 1 lists all the primers used in this work. The DNA (2 μl extract, approximately 20–60 ng) was amplified using AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen, Carlsbad, CA) in 25-μl reactions. Reaction mixtures contained 0.2 μM of each primer, 1 mM MgSO4, and 0.025 U/μl Taq polymerase. The following PCR conditions were used for both rounds: initial denaturation at 94°C for 2 min, followed by 38 cycles of 94°C for 1 min, 55°C for 2 min, and 72°C for 3 min, plus a final extension of 72°C for 7 min.
Table 1.
Primers used in PCR sequencing
| Primer name | Sequence | Reference |
|---|---|---|
| 16S-SR | 5′-GGT CTG TCA AAA CTG AAG ATG-3′ | Lee et al. (2004) |
| P1 | 5′-AAG AGT TTG ATC CTG GCT CAG GAT T-3′ | Deng and Hiruki (1991) |
| P1A | 5′-AAC GCT GGC GGC GCG CCT AAT AC-3′ | Lee et al. (2004) |
| P7 | 5′-CGT CCT TCA TCG GCT CTT-3′ | Schneider et al. (1995) |
| F5 | 5′-GGT TGT CGT CAG CTC GTG TCG-3′ | Lee, Pers. communication |
| P5 | 5′-CGG CAA TGG AGG AAA CT-3′ | Schneider et al. (1995) |
| R3 | 5′-GGC ACA TAG TTA GCC GGG GCT TAT-3′ | Lee, Pers. communication |
| R5 | 5′- CGA CAC GAG CTG ACG ACA ACC-3′ | Lee, pers. communication |
| L15F1A(III) | 5′-CTT CTG GTA AAG GAC ATA AAG G-3′ | Lee et al. (2010) |
| MapR1A(III) | 5′-GGT TCT TCG TGC AAT TGC AAA CC-3′ | Lee et al. (2010) |
| SecYF1(III) | 5′-CTA GAC CAG GTT TTG AAG G-3′ | Lee et al. (2010) |
| SecYR1(III) | 5′-GAC CTG CTT TTC TCA TTA TAG C-3′ | Lee et al. (2010) |
| SecYG-SF1 | 5′-CAG AAA ATG GTT TTT CCC AAT GGG T-3′ | Dally pers. communication |
| SecYG-SR1 | 5′-GTT TAG TAG CGT AAG TGA TTG G-3′ | Dally, Pers. communication |
| Tuf340a | 5′-GCT CCT GAA GAA ARA GAA CGT GG-3 | Makarova et al. (2012) |
| Tuf340b | 5′-ACT AAA GAA GAA AAA GAA CGT GG-3′ | Makarova et al. (2012) |
| Tuf400aM13F | 5′-GTA AAA CGA CGG CCA GTG AAA CAG AAA AAC GTC AYT ATG CTC A-3′ | Makarova et al. (2012) |
| Tuf400bM13F | 5′-GTA AAA CGA CGG CCA GTG AAA CTT CTA AAA GAC ATT ACG CTC A-3′ | Makarova et al. (2012) |
| Tuf400cM13F | 5′-GTA AAA CGA CGG CCA GTG AAA CAT CAA AAA GAC AYT ATG CTC A-3′ | Makarova et al. (2012) |
| Tuf400dM13F | 5′-GTA AAA CGA CGG CCA GTG AAA CAG AAA AAA GAC AYT ATG CTC A-3′ | Makarova et al. (2012) |
| Tuf400eM13F | 5′-GTA AAA CGA CGG CCA GTC AAA CAG CTA AAA GAC ATT ATY CTC A-3′ | Makarova et al. (2012) |
| Tuf835raT7 | 5′-TAA TAC GAC TCA CTA TAG GGA ACA TCT TCW ACH GGC ATT AAG AAA GG-3′ | Makarova et al. (2012) |
| Tuf835rbT7 | 5′-TAA TAC GAC TCA CTA TAG GGA ACA CCT TCA ATA GGC ATT AAA AAW GG-3′ | Makarova et al. (2012) |
| Tuf835rcT7 | 5′-TAA TAC GAC TCA CTA TAG GGA ACA TCT TCT ATA GGT AAT AAA AAA GG-3′ | Makarova et al. (2012) |
| Tuf890ra | 5′-ACT TGD CCT CTT TCK ACT CTA CCA GT-3′ | Makarova et al. (2012) |
| Tuf890rb | 5′-ATT TGT CCT CTT TCW ACA CGT CCT GT-3′ | Makarova et al. (2012) |
| Tuf890rc | 5′-ACC ATT CCT CTT TCA ACA CGT CCA GT-3′ | Makarova et al. (2012) |
For tuf amplification, Tuf 340/Tuf890 and Tuf400/Tuf835 primer cocktails were used in direct and nested PCR, respectively. Nested primers include the nucleotides of the general primers M13F and T7 (italics) used for sequencing.
SecY genomic region was amplified using the TaKaRa LA Taq DNA Polymerase (Takara Mirus Bio, Madison, WI) with primers L15F1A(III)/MapR1A(III), followed by the primer set SecYF1(III)/SecYR1(III) as described by Lee et al. (2010). PCR was carried out in mixtures containing 200 μM each dNTPs, 0.2 μM each primer, 2 mM MgCl2, and 0.05 U/μl Taq polymerase. For the first amplification round, the following conditions were used: 94°C for 1 min followed by 35 cycles of 94°C for 30 s each, 50°C for 1 min, and 68°C for 5 min, and a final extension step of 72°C for 10 min. The second amplification proceeded as follows: 94°C for 2 min followed by 38 cycles of 94°C for 1 min each, 50°C for 2 min and 72°C for 3 min, and a final extension step of 72°C for 7 min.
The PCR cocktail and thermal profile for NAGYI phytoplasmas were identical to those for NAGYIII group phytoplasmas except for primers. SecY group I primers were L15F1 and MapR1 for the first round (Lee et al. 2010) and AYSecYF1 and AYSecYR1 for the second round (Lee et al. 2006).
Amplification of the tuf gene was carried out using the primer cocktails described in Makarova et al. (2012) and the AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen) in 25-μl reactions. Primer combination Tuf 340/890 was used for the first PCR round and Tuf 400/895 for the second. The following PCR conditions were used for both rounds of amplification: 94°C for 3 min followed by 38 cycles of 94°C for 45 s each, 54°C for 30 s and 72°C for 1 min, and a final extension step of 72°C for 7 min.
For 16S, secY, and tuf PCRs, expected molecular weight was determined by running 5 μl of each reaction on 1% agarose gel, and by visualization on an UV trans-illuminator, after staining the samples with SYBR Safe DNA Gel Stain (Invitrogen). Positive samples were purified using a MinElute PCR Purification Kit (Qiagen) according to the manufacturer’s instructions.
Nucleotide Sequencing
Nucleotide sequences were determined by sequencing the purified amplicons generated from the second rounds of PCR. DNA sequencing was performed at the Virginia Biocomplexity Institute (Virginia Tech, Blacksburg, VA). To sequence the 16S region, primers P1A, 16S-SR, P5, F5, R5, and R3 were used. SecY region was sequenced using primers SecYG-SF1, SecYG-SR1, SecYF1(III), and SecYR1(III). Primers M13F (5′-GTA AAA CGA CGG CCA GT-3′) and T7 (5′-TAA TAC GAC TCA CTA TAG GG-3′) were used for sequencing tuf gene. Sequences were aligned using the T-coffee method (Notredame et al. 2000) from the MacVector software (MacVector Inc., NC).
Results
Symptoms of NAGY observed on cultivated vines in this work were similar to those described in our previous NAGY reports (Beanland et al. 2006, Stoepler and Wolf 2013, Davis et al. 2015, Wolf 2015) and included discoloration, downward rolling, and early abscission of affected leaves, shoot die-back and failure of shoot stems to develop mature, brown periderm, and withering and abscission of fruit clusters (Supp Fig. 1 [online only]). Affected vines were marked by reduced vegetative capacity over multiple years, and eventual vine death.
Identification of Phytoplasma in Symptomatic, Cultivated Grapevines
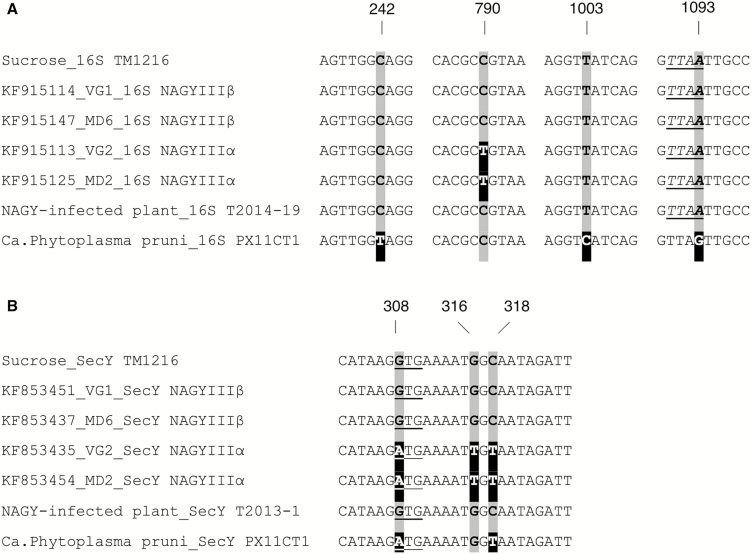
DNA from symptomatic grapevines was periodically collected and tested by PCR to confirm the presence of phytoplasmas. Nested PCR analysis using 16S, secY, and tuf gene primer combinations confirmed the presence of phytoplasmas in grapevine samples. Nucleotide sequencing of the PCR products confirmed that phytoplasmas associated with NAGY symptoms; in this, the course of this study was of the NAGYIIIβ sequevar (e.g., Fig. 1A and B).
Fig. 1.
Single-nucleotide polymorphisms (SNPs) in selected segments of genes encoding 16S rRNA (A) and SecY (B). Sequences derived from sucrose sample TM1216 were compared with sequences of NAGY phytoplasma sequevars NAGYIIIα and NAGYIIIβ, ‘Candidatus Phytoplasma pruni’ strain PX11CT1 and NAGY-infected plant samples (Riesling and Chardonnay in A and B, respectively). Nucleotides that differ from those of samples TM1216 are shaded black, whereas unchanged nucleotides are shaded gray. (A) Alignment of the 16S rDNA genomic region. Numbers on top indicate base positions of SNPs with reference to ‘Ca. Phytoplasma pruni’ strain PX11CT1, GenBank JQ044393 (16S rRNA; bases 242, 790, 1,003, and 1,093). MseI recognition site is underlined. (B) Alignment of the secY gene genomic region. Numbers on top indicate base positions of SNPs with reference to ‘Ca. Phytoplasma pruni’ strain PX11CT1, GenBank JQ268254 (secY bases 308, 316, and 318). Start codon is underlined.
Occurrence of J. olitoria in surveyed vineyards.
Both the extensive (N = 27 vineyards) survey of 2013 and the more intensive survey of LV and RdV vineyards over the 2013–2015 period illustrated and reinforced our previous knowledge about the diversity of leafhoppers found in, and immediately adjacent to, commercial Virginia vineyards. Approximately 40 genera of leafhoppers and planthoppers were found in ≥1 yr. The 20 most abundant leafhoppers found in the 2013 extensive survey are listed in Table 2 and include Graphocephala versuta, Empoasca fabae, and Agallia constricta. Jikradia olitoria was ranked 14 in relative abundance based on sticky trap card that captures within the vineyard and was found more frequently in vegetation adjacent to the vineyard than in the vineyard itself (Table 2). The more intensively sampled vineyards (RdV and LV) revealed a similar diversity of Cicadellidae with the eight most abundant leafhoppers of interest listed in Table 3. Noteworthy here was the absence of J. olitoria from either vineyard in 2013 and 2014, although a small number of J. olitoria were found at LV in September 2015. Vineyard surveys in each of the 3 yr (2013–2015) of insect monitoring revealed no apparent cases of NAGY in any of the three vineyard blocks at RdV or LV that were monitored for leafhoppers. This is noteworthy given that these vineyard blocks had exhibited multiple cases of NAGY destruction of vines in the 2011–2012 timeframe, which had prompted our inclusion of the blocks in our 2013–2015 surveys. The limited survey of 2017 revealed a surprising abundance of J. olitoria at both LV (Supp Fig. 2 [online only]) and the AHS Jr. AREC site, which provided an opportunity for additional phytoplasma transmission attempts with this species. Many of the specimens collected at the AHS Jr. AREC site were observed on wild grape (V. vulpina) and were subsequently swept or aspirated into sample tubes directly from this vegetation.
Table 2.
The 20 most abundant leafhopper species out of 85 morphospecies collected in geographically extensive survey of 27 vineyards across Virginia, North Carolina ,and Pennsylvania, 30 May to 24 Sep 2013
| Leafhopper | Within vineyard | In vegetation adjacent to vineyard | |||
|---|---|---|---|---|---|
| Cardsa | Ground sweepsb | Canopy sweepsb | Cardsa | Canopy sweepsb | |
| Graphocephala versuta | 13.08 | 0.29 | 0.49 | 10.99 | 0.83 |
| Empoasca fabae | 3.18 | 0.21 | 0.44 | 1.20 | 0.38 |
| Agallia constricta | 1.24 | 21.73 | 0.30 | 0.15 | 0.03 |
| Paraphlepsius irroratus | 0.39 | 0.25 | 0.00 | 0.41 | 0.03 |
| Oncometopia orbona | 0.39 | 0.00 | 0.00 | 0.23 | 0.04 |
| Xestocephalus desertorum | 0.28 | 0.07 | 0.00 | 0.28 | 0.00 |
| Erasmoneura vulnerata | 0.21 | 0.01 | 0.04 | 1.11 | 0.36 |
| Scaphytopius nigrifrons | 0.18 | 0.00 | 0.00 | 0.55 | 0.06 |
| Forcipata loca | 0.16 | 1.23 | 0.01 | 0.07 | 0.01 |
| Agalliopsis novella | 0.09 | 0.01 | 0.00 | 0.41 | 0.02 |
| Erythroneura curvata | 0.08 | 0.00 | 0.00 | 0.10 | 0.01 |
| Erythroneura sp. | 0.08 | 0.01 | 0.00 | 0.51 | 0.09 |
| Graphocephala coccinea | 0.06 | 0.00 | 0.01 | 0.48 | 0.04 |
| Jikradia olitoria | 0.05 | 0.00 | 0.00 | 0.48 | 0.09 |
| Graminella nigrifrons | 0.05 | 3.41 | 0.01 | 0.01 | 0.02 |
| Ossiannilssonola sp. | 0.04 | 0.00 | 0.00 | 0.01 | 0.00 |
| Scaphytopius acutus | 0.04 | 0.01 | 0.01 | 0.55 | 0.01 |
| Scaphoideus sp. (probably S. minor) | 0.04 | 0.00 | 0.00 | 0.20 | 0.02 |
| Erythroneura comes | 0.03 | 0.01 | 0.03 | 0.24 | 0.08 |
| Erythroneura tricincta | 0.02 | 0.00 | 0.01 | 0.24 | 0.18 |
Jikradia olitoria is in bold. Vineyard cultivars for ‘Within vineyard’ samples include V. vinifera cvs. Chardonnay, Cabernet Sauvignon, Merlot, and Riesling, and Vitis interspecific hybrid cv. Traminette. Adjacent vineyard vegetation included a mix of herbaceous and woody plants, the latter primarily wild grape (V. vulpina L.), and black cherry (Prunus serotina Ehrh.).
aRank order is based on within vineyard sticky trap card samples. Numbers for cards are the number of specimens collected per sampling method, averaged over the entire sampling period and across vineyards. Specifically, N = 71 cards in vegetation adjacent to vineyard and N = 496 cards in the vineyard canopy.
bNumbers for sweeps are the number of specimens collected per 100 sweeps, averaged across samples and vineyards. Specifically, N =157 within vineyard ground sweeps; N = 152 within vineyard canopy sweeps, and N = 159 adjacent vegetation sweeps.
Table 3.
Seasonal summation of weekly catches of selected leafhoppers at RdV and LV vineyards on yellow sticky cards or in vineyard floor vegetation sweeps over the 2013–2015 seasons
| 2013 | 2014 | 2015 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RdV | LV | RdV | LV | RdV | LV | |||||||
| Leafhopper | Cardsa | Sweepsb | Cardsa | Sweepsb | Cardsa | Sweepsb | Cardsa | Sweepsb | Cardsa | Sweepsb | Cardsa | Sweepsb |
| Agallia constricta | 34 | 645 | 1 | 131 | 16 | 131 | 9 | 71 | 16 | 134 | 20 | 370 |
| Graphocephala versuta | 194 | 26 | 205 | 8 | 46 | 2 | 50 | 2 | 18 | 1 | 18 | 3 |
| Paraphlepsius irroratus | 15 | 21 | 1 | 2 | 32 | 8 | 1 | 0 | 9 | 16 | 1 | 1 |
| Endria inimica | 2 | 61 | 0 | 5 | 1 | 48 | 0 | 5 | 0 | 14 | 0 | 3 |
| Exitanus exitiosus | 2 | 38 | 0 | 2 | 0 | 2 | 0 | 0 | 1 | 1 | 0 | 0 |
| Amblycellus curtisii | 0 | 3 | 0 | 2 | 1 | 12 | 1 | 19 | 0 | 0 | 1 | 10 |
| Liburniella ornata | 0 | 0 | 0 | 14 | 0 | 1 | 0 | 2 | 0 | 2 | 0 | 1 |
| Scaphytopius magdalensis | 1 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 2 | 1 | 0 |
| Jikradia olitoria | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 c | 0 |
See text for details of card locations and number of ground sweeps.
a Cards: Numbers are the mean number of specimens trapped on 3 cards and summed for all sampling periods of that season.
b Sweeps: Number of specimens collected per 100 ground sweeps and summed for all sampling periods of that season.
cMean of 2.33 J. olitoria per 3 cards counted on 8 Sep 2015, and mean of 0.33 per 3 cards counted on 25 Sep. 2015.
Transmission of Phytoplasma by J. olitoria on Artificial Diets
Out of 139 feeding solutions tested in 2012, none resulted in positive NAGY phytoplasma detection (Table 4). In 2013, 1,840 artificial diet transmission tests were conducted, of which four (0.2%) produced positive 16S PCR results (TM1216, TM1219, TM1221, TM1222; Supp Fig. 3A [online only]). The four phytoplasma-positive samples were from solutions that had been fed upon by four individuals of J. olitoria, captured in August 2013 at the AHS Jr. AREC (Table 4). PCR performed on the 36 artificial diet transmission tests conducted in 2014 with insects initially caged on NAGY-symptomatic grapevine leaves produced no positive results (Table 4). Similarly, none of the 37 artificial diet transmission trials resulted in positive results in 2015. Jikradia olitoria was not included in the AAP/LP, assays because we failed to capture this species during the 2014–2015 seasons. In 2016, 1 out of 29 single J. olitoria specimen transmission attempts resulted in positive transmission, and 3 out of 99 J. olitoria specimens in 2017 resulted in a positive transmission (Table 4). All of the positive transmissions occurred with specimens collected at the AHS Jr. AREC.
Table 4.
Leafhoppers and planthoppers evaluated using sucrose solution assay to gauge their competency in transmitting NAGY phytoplasmas
| Species | Sucrose media tested | |||
|---|---|---|---|---|
| 2012 | 2013 | 2014 | 2015 | |
| Acanaloniid planthopper | — | 1 | — | — |
| Agallia constricta | 5 | 585 | 17 | 1 |
| Agallia quadripunctata | — | 57 | — | — |
| Agalliopsis novella | 2 | 22 | — | — |
| Amblycellus curtisii | 5 | 25 | — | 1 |
| Arthaldeus pascuellus | — | — | — | 1 |
| Balclutha abdominalis | 1 | 10 | — | — |
| Chlorotettix galbanatus | 1 | 16 | — | — |
| Delphacodes sp. | — | 68 | — | — |
| Deltocephalus flavicosta | 12 | 132 | — | 3 |
| Dikraneura angustata | — | 15 | — | — |
| Draeculacephala sp. | 5 | 70 | — | — |
| Empoasca fabae | — | 60 | — | — |
| Empoasca vitis | — | 5 | — | — |
| Endria inimica | 14 | 61 | 6 | 10 |
| Erythroneura vulnerata | — | 38 | — | — |
| Erythroneura sp. | — | 23 | — | — |
| Erythroneura comes | — | 13 | — | — |
| Erythroneura diva | — | 9 | — | — |
| Erythroneura rubra | — | 6 | — | — |
| Erythroneura tricincta | — | 20 | — | — |
| Exitianus exitiosus | 49 | 102 | 3 | 9 |
| Forcipata loca | — | 54 | — | — |
| Graminella nigrifrons | 2 | 56 | — | 5 |
| Graphocephala coccinea | — | 4 | — | — |
| Graphocephala versuta | 40 | 91 | —— | — |
| Gyponana sp. | — | 2 | — | — |
| Hymetta balteata | — | 4 | — | —— |
| Jikradia olitoria a | — | 4/24 | — | — |
| Latalus sayi | — | 9 | — | 1 |
| Liburniella ornata | — | 16 | — | — |
| Macrosteles quadrilineatus | — | 5 | — | 1 |
| Leafhopper nymph (uncertain ID) | — | 40 | 3 | 1 |
| Oncometopia orbona | — | 6 | — | — |
| Orientus ishidae | — | 1 | — | — |
| Planthopper (unknown ID) | 1 | 8 | — | — |
| Platymetopius irroratus | — | 25 | — | — |
| Paraulacizes irrorata | — | 2 | — | — |
| Philaneus spumarius | — | — | 2 | — |
| Polyamia weedi | — | 18 | 2 | 2 |
| Scaphoideus spp.b | — | 33 | — | — |
| Scaphytopius acutus | — | 1 | — | — |
| Scaphytopius frontalis | — | 1 | — | — |
| Scaphytopius magdalensis | 2 | 1 | —— | —— |
| Scaphytopius nigrifrons | ——— | 3 | —— | — |
| Spangbergiella sp. | — | — | 2 | — |
| Spittle bug | — | 72 | — | — |
| Stirellus bicolor | — | 2 | — | 1 |
| Tylozygus bifidus | — | 2 | 1 | — |
| Xestocephalus desertorum | — | 1 | — | — |
Values are the numbers of individual nested PCR assays by insect species and year. Ratios (J. olitoria, in bold text) are the number of PCR-positive transmissions into sucrose solutions over total transmission attempts for that species, by year. See footnotes for years 2016 and 2017 in which only J. olitoria was tested.
aIn addition to years shown, 1/29 and 3/99 J. olitoria samples produced positive transmissions in 2016 and 2017, respectively.
bSpecimens included both S. minor and S. titanus as identified by C. Dietrich.
Molecular Identification of Phytoplasma Recovered From Artificial Diets
DNA sequences from 16S and secY were generated for four 2013 samples, one 2016 sample, and three 2017 samples. Sequences were not generated for tuf, but the PCR amplifications from the four 2013 samples were of the expected molecular weight. 16S consensus sequences from all eight samples shared 100% identity with each other; therefore, we used one sample, TM1216, for all further analysis. This sample was analyzed in iPhyClassifier (Zhao et al. 2009), which revealed a 99.8% similarity to the ‘Candidatus Phytoplasma pruni’ rrnA reference strain (GenBank JQ044393). The phytoplasma under study is, therefore, considered as a ‘Candidatus Phytoplasma pruni’ rrnA-related strain (Davis et al. 2015).
Because the phytoplasma-positive insects used for the transmission trials had not been purposefully fed on known NAGY-affected host plants, it was important to further characterize the sequences in order to confirm that the insects were indeed carrying NAGY phytoplasmas, rather than a similar strain. We aligned the TM1216 16S sequence with known NAGY sequences reported in Davis et al. (2015) belonging to sequevars NAGYIIIα (KF915113_VG2_16S, KF915125_MD2_16S) and NAGYIIIβ (KF915114_VG1_16S, KF915147_ MD6_16S), and with ‘Ca. Phytoplasma pruni’ PX11CT1 (GenBank JQ044393). TM1216 shared 100% similarity (1,492 bp) with the NAGYIIIβ sequevar (Fig. 1A and Supp Fig. 4 [online only]). TM1216 and NAGYIIIβ are different from ‘Ca. Phytoplasma pruni’ PX11CT1 (GenBank JQ044393) by four bp (1,488/1,492); the differences occur at sites 242, 790, 1003, and 1093 of JQ044393. Moreover, our TM1216 16S sequence was 99.9% identical (1,491/1,492) to sequence generated from a NAGY-infected Riesling grapevine (T2014-19) collected from a Virginia vineyard with high NAGY incidence.
Sequence alignment of the secY gene region also highlighted important differences between the phytoplasma found in the TM1216 sample and ‘Ca. Phytoplasma pruni’ (Fig. 1B). In particular, the triplet GTG corresponds to the start codon in the secY gene of our sample TM1216, which differentiates it from ‘Ca. Phytoplasma pruni’ and from the NAGYIIIα sequevar, whose translation initiation codon is ATG (Davis et al. 2015). Furthermore, our TM1216 sample presented a high percentage of similarity with the secY genomic region of a Chardonnay NAGY-infected plant from a Virginia vineyard (sample T2013-1 SecY; Supp Fig. 5 [online only]). The 2016 and 2017 positive samples differed from the secY sequence of TM1216 by one nucleotide (Supp Fig. 5 [online only]). Finally, tuf gene was used as an alternative reference; DNA sequencing of the tuf gene revealed 100% identity between our TM1216 sample and a NAGY-infected Chardonnay grapevine sampled at a Loudoun County, Virginia vineyard (sample T2014-16Tuf; Supp Fig. 6 [online only]).
Discussion
Working on the assumption that the vector(s) of NAGY is a leafhopper (Cicadellidae), we used a sucrose feeding solution to evaluate vector competency of a diverse range of leafhoppers. To competently transmit phytoplasmas, an insect must acquire the phytoplasma through feeding or probing the host plant. The phytoplasmas must then pass through the insect gut into the salivary glands and replicate, be incorporated into saliva, and then be injected into a new host or feeding solution during the insect’s foraging (Purcell 1982). For this reason, the detection of phytoplasma DNA in insects is not a sufficient indication of their competency to transmit phytoplasmas and was our rationale for not analyzing insects themselves for phytoplasmas. The use of sucrose feeding solutions to screen potential vectors is an established method that offers advantages over the conventional plant transmission trials: it reduces the time of the investigation by avoiding the need to wait for symptoms, reduces costs, and allows for individual assessment of insects as potential vectors (Tanne et al. 2001, Bressan et al. 2006, Lu et al. 2016). Of 49 field-collected insect species screened, J. olitoria was the only insect that secreted a NAGY phytoplasma into the sucrose feeding solution. The phytoplasmas recovered in the artificial diet were identified as NAGYIIIβ, a common phytoplasma sequevar found in NAGY-affected, cultivated grapevines in the eastern United States (Davis et al. 2015). This is the most compelling evidence yet of a competent NAGY vector, even though it does not rule out the fact that other leafhoppers could be potential vectors of this sequevar, the closely related NAGYIIIα sequevar, or the less commonly found 16SrI (aster yellows group) phytoplasma (Davis et al. 1998, Davis et al. 2018).
Jikradia olitoria is a member of the Membracoidea superfamily, Cicadellidae family, and Coelidiinae subfamily. Since its first description by Thomas Say in 1830 as Jassus olitorius, this species has undergone several taxonomic reclassifications. While still frequently referred to as Coelidia olitoria, the currently accepted name is Jikradia olitoria (C. Dietrich, personal communication). Jikradia sp. has been putatively associated with virus transmission in strawberry pallidosis (Frazier 1975). We found no reports of J. olitoria vectoring phytoplasmas. Relative to other leafhoppers that feed on the phloem of woody hosts, J. olitoria does not appear to be an abundant insect in commercial vineyards of the Mid-Atlantic United States; however, it is more commonly found in mixed, woody vegetation outside the vineyard. While we acknowledge that other survey methods, including nocturnal trapping, might have yielded more specimens, our findings are comparable with our earlier surveys. In a 3-yr period (2002–2004), Beanland et al. (2006) found 13–14 J. olitorius (syn. J. olitoria) specimens per season (May through October) in woodland settings bordering three survey vineyards, whereas only one to eight specimens per season were collected in sweep nets from the corresponding vineyard floors. The total number of phloem-feeding leafhoppers and planthoppers collected in that 3-yr period exceeded 10,000 and J. olitoria was ranked 15 in frequency. Greater specimen numbers were collected on yellow sticky cards (10–25 specimens per vineyard per year) used in the 2003 and 2004 seasons; however, the number of J. olitoria captured in woodland environments again outnumbered those captured on sticky trap cards in the vineyards by an average of 15:1 (Beanland et al. 2006). Beanland et al. (2006) also reported that the earliest seasonal capture of J. olitoria in malaise traps occurred in mid-July. Given the relatively low number of J. olitoria previously collected in this environment, the numbers found at LV vineyard (Supp Fig. 2 [online only]) and swept at the AHS Jr. AREC site in 2017 were remarkable. Anecdotally, most of the specimens collected at the AREC site were captured either by sweep-netting or by direct aspiration into sample tubes from wild grapevines outside the vineyard.
Given the low frequency of new NAGY cases in Virginia vineyards (e.g., Beanland et al. 2006, typically <10% of original plantation), it is reasonable to propose that vectors of NAGY phytoplasmas either occur at a low frequency, or that the transmission efficiency of phytoplasmas and subsequent expression of NAGY symptoms is low, or both. The scarcity of J. olitoria at LV and RdV vineyards, coupled with the absence of NAGY cases in the surveyed blocks between 2013 and 2015, is notable, as is the occurrence of J. olitoria and the concomitant expression of NAGY affected vines, at the AHS Jr. AREC in 2015–2016. Admittedly, there might also be other leafhoppers that were not collected or tested for transmission that can competently transmit NAGY phytoplasmas. Our finding of ~35 genera of Cicadellids within the vineyard ecosystem approaches the 48 Cicadellidae genera found by Louis Stearns in a much more extensive ecological survey of Virginia in the early twentieth century (Stearns 1927).
Jikradia olitoria demonstrated an ability to transmit a specific NAGY phytoplasma into sucrose feeding solutions. The sequencing of PCR amplification products of 16S, secY, and tuf loci from phytoplasmas released in the artificial diets revealed the presence of NAGYIIIβ sequevar, a ‘Ca. Phytoplasma pruni’ – related strain, characterized and taxonomically classified by Davis et al. (2015). This specific phytoplasma was also detected in NAGY-infected grapevines cultivated in Virginia, both in the current study and previously (Davis et al. 2015). This is an important finding in this study, because it correlates the phytoplasmas found in the sucrose assays with those detected in vineyards with high incidence of NAGY-infections.
Using more than one molecular locus to identify phytoplasmas is a powerful strategy since it can help distinguish closely related strains, especially when the highly conserved 16S region provides insufficient resolution (Duduk et al. 2011, Al-Subhi et al. 2017). SecY is a single copy gene coding for a translocase subunit and it is one of the most variable markers used in phytoplasma classification (Lee et al. 2010). The presence of a GTG start codon, instead of ATG, in the secY gene is a clear confirmation of the presence of NAGYIIIβ in our positive samples. Other SNPs determined the differences with ‘Ca. Phytoplasma pruni’ and other related strains.
Makarova et al. (2012) proposed an elegant barcoding system based on the elongation factor Tu (tuf), as opposed to the conventional 16S gene. Their findings demonstrated how the short tuf gene could be used for phytoplasma identification, providing good resolution at both group and subgroup levels. In our study, we found 100% similarity with the tuf gene from infected grapevines. This work reports for the first time the use of the tuf gene as an alternative locus to identify NAGY phytoplasmas, and provides the first example of NAGY characterization collectively using 16S, secY, and tuf gene sequences.
While the molecular characterization of PCR amplicons in our artificial feeding solutions provides novel evidence of J. olitoria’s ability to vector a causal phytoplasma of NAGY, it does not rule out other potential vectors. Additional work is required to demonstrate the ability of J. olitoria to transmit phytoplasmas directly to grapevine or to other indicator plants. While not described in this report, we have, thus, far been unsuccessful with definitive insect-to-plant transmission of NAGY phytoplasmas; however, none of the plant transmission attempts done before 2017 used J. olitoria. Future transmission studies should include nymphal stages of J. olitoria and extended AAPs on NAGY symptomatic plants and LPs before transfer to indicator plants. Finally, only one phytoplasma, NAGYIIIβ, was confidently identified in the artificial feeding solutions, and the presence of at least two other phytoplasmas, one in the 16SrI, aster yellows group, and the NAGYIIIα sequevar, have previously been identified in Virginia vineyards (Davis et al. 1998, 2015), although neither were found in the course of the current study.
In summary, field-collected J. olitoria was the only species of 49 tested that proved capable of transmitting a specific NAGY phytoplasma into sucrose solution in artificial transmission attempts. The specific phytoplasma, NAGYIIIβ, was shown to be identical to a phytoplasma sequevar that causes NAGY in cultivated grapevine by analyzing three independent loci. The seasonal and interannual occurrence of J. olitoria is generally consistent with the occurrence of NAGY in sampled vineyards, although this requires long-term monitoring in additional vineyards to establish a more meaningful association.
Supplementary Material
Acknowledgments
We thank the Virginia Wine Board for financial support and the many grape growers who graciously allowed access to their vineyards for the purposes of this work. We thank Dr. LeAnn Beanland for her contributions to our understanding of NAGY in Virginia, Ellen Dally (formerly, USDA/ARS) for her guidance with molecular protocols, and Dana Melby (Virginia Tech) for her assistance with field work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author contributions
P.L., T.M.S., and D.J.M. conducted the wet lab experiments. P.L. and T.K.W. drafted the manuscript. T.K.W. conceived the project and supervised the laboratory. RED provided insights and guidance. All authors edited and approved the final version of the manuscript.
References Cited
- Al-Subhi A., S. A. Hogenhout R. A. Al-Yahyai, and Al-Sadi A. M.. 2017. Classification of a new phytoplasmas subgroup 16SrII-W associated with Crotalaria witches’ broom diseases in Oman based on multigene sequence analysis. BMC Microbiol. 17: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beanland L., Noble R., and Wolf T. K.. 2006. Spatial and temporal distribution of North American Grapevine Yellows disease and of potential vectors of the causal phytoplasmas in Virginia. Environ. Entomol. 35: 332–344. [Google Scholar]
- Boudon-Padieu E. 2015. Flavescence dorée, Pp. 109–110. In Wilcox W. F., Gubler W. D. and J. K. Uyemoto (eds.), Compendium of grape diseases, disorders, and pests, 2nd ed American Phytopathological Society Press, St. Paul Minnesota. [Google Scholar]
- Bressan A., D. Clair O. Sémétey, and Boudon-Padieu E.. 2006. Insect injection and artificial feeding bioassays to test the vector specificity of flavescence dorée phytoplasma. Phytopathology. 96: 790–796. [DOI] [PubMed] [Google Scholar]
- Caudwell A. 1983. L’origine des jaunisses à mycoplasmes (MLO) des plantes et l’exemple des jaunisses de la vigne. Agronomie. 3: 103–111. [Google Scholar]
- Constable F. E., Gibb K. S., and Symons R. H.. 2003. Seasonal distribution of phytoplasmas in Australian grapevines. Plant Pathol. 52: 267–276. [Google Scholar]
- Daire X., Clair D., Reinert W., and Boudon-Padieu E.. 1997. Detection and differentiation of grapevine yellows phytoplasmas belonging to the elm yellows group and to the stolbur subgroup by PCR amplification of non-ribosomal DNA. Eur. J. Plant Pathol. 103: 507–514. [Google Scholar]
- Davis R. E., Jomantiene R., Dally E. L., and Wolf T. K.. 1998. Phytoplasmas associated with grapevine yellows in Virginia belong to group 16SrI, subgroup (tomato big bud phytoplasma subgroup), and group 16SrIII, new subgroup I. Vitis. 37: 131–137. [Google Scholar]
- Davis R. E., Dally E. L., Zhao Y., Lee I. -M., Wei W., Wolf T. K., Beanland L., LeDoux D. G., Johnson D. A., Fiola J. A.,. et al. 2015. Unraveling the etiology of North American Grapevine Yellows (NAGY): novel NAGY phytoplasma sequevars related to ‘Candidatus Phytoplasma pruni’. Plant Dis. 99: 1087–1097. [DOI] [PubMed] [Google Scholar]
- Davis R. E., Dally E. L., Zhao Y., and Wolf T. K.. 2018. Genotyping points to divergent evolution of ‘Candidatus Phytoplasma asteris’ strains causing North American grapevine yellows and strains causing aster yellows. Plant Dis. 102: 1696–1702. [DOI] [PubMed] [Google Scholar]
- Deng S., and Hiruki C.. 1991. Amplification of 16S rRNA genes from culturable and nonculturable Mollicutes. J. Microbiol. Methods. 14: 53–61. [Google Scholar]
- Duduk B., and Bertaccini A.. 2011. Phytoplasma classification: taxonomy based on 16S ribosomal gene, is it enough?Phytopathogenic Mollicutes. 1: 3–13. [Google Scholar]
- Frazier N. W. 1975. Possible transmission of strawberry pallidosis by the leafhopper Coelidia olitoria. Plant Dis. Rep. 59: 40–41. [Google Scholar]
- Green M. J., Thompson D. A., and MacKenzie D. J.. 1999. Easy and efficient DNA extraction from woody plants for the detection of phytoplasmas by polymerase chain reaction. Plant Dis. 83: 482–485. [DOI] [PubMed] [Google Scholar]
- Lee I. M., D. E. Gundersen-Rindal, and Bertaccini A.. 1998. Phytoplasma: ecology and genomic diversity. Phytopathology. 88: 1359–1366. [DOI] [PubMed] [Google Scholar]
- Lee I. M., M. Martini C. Marcone, and Zhu S. F.. 2004. Classification of phytoplasma strains in the elm yellows group (16SrV) and proposal of ‘Candidatus Phytoplasma ulmi’ for the phytoplasma associated with elm yellows. Int. J. Syst. Evol. Microbiol. 54: 337–347. [DOI] [PubMed] [Google Scholar]
- Lee I. M., Y. Zhao, and Bottner K. D.. 2006. SecY gene sequence analysis for finer differentiation of diverse strains in the aster yellows phytoplasma group. Mol. Cell. Probes. 20: 87–91. [DOI] [PubMed] [Google Scholar]
- Lee I. M., K. D. Bottner-Parker Y. Zhao R. E. Davis, and Harrison N. A.. 2010. Phylogenetic analysis and delineation of phytoplasmas based on secY gene sequences. Int. J. Syst. Evol. Microbiol. 60: 2887–2897. [DOI] [PubMed] [Google Scholar]
- Lu H., Wilson B. A. L., Ash G. J., Woruba S. B., Fletcher M. J., You M., Yang G., and Gurr G. M.. 2016. Determining putative vectors of the Bogia Coconut Syndrome phytoplasma using loop-mediated isothermal amplification of single-insect feeding media. Sci. Rep. 6: 35801 10.1038/srep35801(accessed 24 November 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maixner M., Pearson R. C., Boudon-Padieu E., and Caudwell A.. 1993. Scaphoideus titanus, a possible vector of grapevine yellows in New York. Plant Dis. 77: 408–413. [Google Scholar]
- Makarova O., N. Contaldo S. Paltrinieri G. Kawube A. Bertaccini, and Nicolaisen M.. 2012. DNA barcoding for identification of ‘Candidatus Phytoplasmas’ using a fragment of the elongation factor Tu gene. Plos One. 7: e52092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C., D. G. Higgins, and Heringa J.. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302: 205–217. [DOI] [PubMed] [Google Scholar]
- Prince J. P., Davis R. E., Wolf T. K., Lee I.-M., Mogen B. D., Dally E. L., Bertaccini A., Credi R. C., and Barba M.. 1993. Molecular detection of diverse mycoplasma-like organisms (MLOs) associated with grapevine yellows and their classification with aster yellows, X- disease and elm yellows MLOs. Phytopathol. 83: 1130–1137. [Google Scholar]
- Purcell A. H. 1982. Insect vector relationships with procaryotic plant pathogens. Ann. Rev. Phytopathol. 20: 397–417. [Google Scholar]
- Schneider B., Seemüller E., Smart C. D., and Kirkpatrick B. C.. 1995. Phylogenetic classification of plant pathogenic mycoplasma-like organisms or phytoplasmas, p. 369–380. InRazin S. and J.G. Tully (eds.), Molecular and diagnostic procedures in mycoplasmology, Vol. 1 Academic Press, San Diego, CA. [Google Scholar]
- Sforza R., Clair D., Daire X., Larrue J., and Boudon-Padieu E.. 1998. The role of Hyalesthes obsoletus (Hemiptera: Cixiidae) on the occurrence of bois noir of grapevines in France. J. Phytopathol. 146: 549–556. [Google Scholar]
- Stearns L. A. 1927. The Cicadellidae (Homoptera) of Virginia. Technical Bulletin 31. Virginia Polytechnic Institute, Virginia Agricultural Experiment Station, Blacksburg, VA. [Google Scholar]
- Stoepler T. M., and Wolf T. K.. 2013. North American Grapevine Yellows Disease: current knowledge and management recommendations for wine growers. Virginia Cooperative Extension Publication AREC-48P. Virginia Tech, Blacksburg, VA. [Google Scholar]
- Tanne E., E. Boudon-Padieu D. Clair M. Davidovich S. Melamed, and Klein M.. 2001. Detection of phytoplasma by polymerase chain reaction of insect feeding medium and its use in determining vectoring ability. Phytopathology. 91: 741–746. [DOI] [PubMed] [Google Scholar]
- Tessitori M., La Rosa R., and Marzachi C.. 2018. Flavescence dorée and bois noir diseases of grapevine are evolving pathosystems. Plant Health Progress. 19: 136–138. [Google Scholar]
- Wei W., R. E. Davis I. M. Lee, and Zhao Y.. 2007. Computer-simulated RFLP analysis of 16S rRNA genes: identification of ten new phytoplasma groups. Int. J. Syst. Evol. Microbiol. 57: 1855–1867. [DOI] [PubMed] [Google Scholar]
- Weintraub P. G. and Beanland L.. 2006. Insect vectors of phytoplasmas. Annu. Rev. Entomol. 51: 91–111. [DOI] [PubMed] [Google Scholar]
- Wolf T. K. 2015. North american grapevine yellows, pp. 111–113. InWilcox W. F., Gubler W. D., and J. K. Uyemoto (eds.), Compendium of grape diseases, disorders, and pests, 2nd ed American Phytopathological Society Press, St. Paul, MN. [Google Scholar]
- Zhao Y., W. Wei I. M. Lee J. Shao X. Suo, and Davis R. E.. 2009. Construction of an interactive online phytoplasma classification tool, iPhyClassifier, and its application in analysis of the peach X-disease phytoplasma group (16SrIII). Int. J. Syst. Evol. Microbiol. 59: 2582–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.