Abstract
The attribution of a protein to an ultrastructural element by optical microscopy represents a major challenge in biology. Here, we report a method of near-native expansion microscopy (U-ExM), enabling the visualization of preserved ultrastructures of macromolecules by optical microscopy. Combined with super-resolution, U-ExM unveiled the centriolar chirality, only visualizable by electron microscopy. We demonstrate the general applicability of U-ExM by imaging different cellular structures including microtubules and mitochondria in cellulo.
Cells are constituted of organelles, large macromolecular assemblies that display specific structures that could for decades only be visualized by electron microscopy1. Although super-resolution fluorescence microscopy has evolved as a very powerful method for subdiffraction-resolution fluorescence imaging of cells, the visualization of ultrastructural details of macromolecular assemblies remains challenging2.
Recently an innovative method of expansion microscopy (ExM) emerged, which physically expands the immunolabeled sample enabling super-resolution imaging by standard fluorescence microscopy3,4 (Supplementary Fig. 1a, ExM). Alternative protocols of ExM such as Protein Retention ExM (ProExM)5 and Magnified Analysis of the Proteome (MAP)6 have been developed that cross-link proteins themselves in the polymer matrix and enable post-expansion immunostaining (Supplementary Fig. 1a, MAP). However, it remains unclear whether these methods preserve the molecular architecture of organelles.
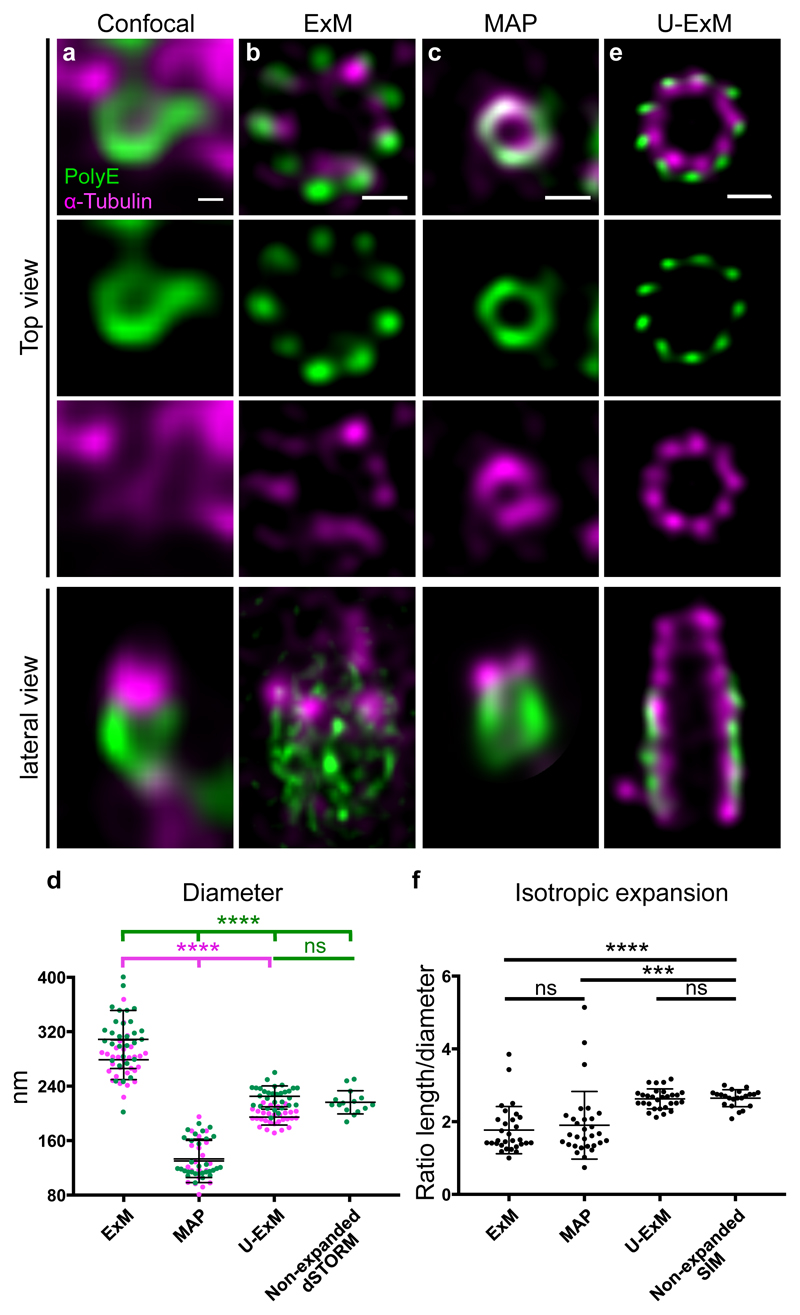
Here, we first set out to characterize the macromolecular expansion performance using established ExM and MAP protocols4,6. We used isolated Chlamydomonas centrioles as reference structure with a characteristic nine-fold microtubule triplet-based symmetry, forming a polarized cylinder ~ 500 nm long and ~ 250 nm wide 7 (Supplementary Fig. 1b). Isolated centrioles were immunolabeled for α-tubulin to visualize the centriolar microtubule wall and for polyglutamylated tubulin (PolyE) present only on the central region of the centriole7,8. While the cylindrical nature of the centriole is visible with the PolyE signal in confocal microscopy, it is impossible to reveal the canonical 9-fold symmetry of the microtubule triplets (Fig. 1a). Moreover, we noticed an antibody competition when co-staining for both α-tubulin and PolyE, with both antibodies recognizing epitopes on the C-terminal moiety of tubulin.
Figure 1. Centriole expansion using U-ExM.
(a-d) Non-expanded (a) and expanded (b-d) isolated centrioles stained with PolyE (green, Alexa488) and α-tubulin (magenta, Alexa568) imaged by confocal microscopy followed by HyVolution. Centrioles were expanded using ExM (b), MAP (c) or U-ExM (e). Scale bar in a: 100nm and b, c, e: 450nm. Representative images from 2 independent experiments (a) and 3 independent experiments (b, c, e). (d) Diameter of the centrioles in the different conditions. Green and magenta dots represent PolyE and α-tubulin diameters, respectively. Averages and standard deviation are as follows: PolyE: 308 nm ± 42 nm, 133 nm ± 27 nm, 225 nm ± 15 nm and 216 nm ± 17 for ExM, MAP, U-ExM and non-expanded dSTORM respectability. N= 30 centrioles for each condition (data from 3 independent experiments) except dSTORM where n= 15 non-expanded centrioles (1 experiment). α-tubulin: 279 nm ± 29, 130 nm ± 32 nm and 195 ± 12, for ExM (n= 29 centrioles), MAP (n= 20 centrioles) and U-ExM (n= 29 centrioles) respectability. Data from 3 independent experiments. Statistical significance was assessed by one-way ANOVA test: ****<0.0001, ns (non significant)=0.77. (f) Isotropic expansion measured as the ratio between the centriolar length and diameter. Average ratios and standard deviation are as follows: ExM=1.8 +/- 0.6 (n= 30 centrioles), MAP=1.9+/- 0.9 (n= 30 centrioles), U-ExM=2.6+/- 0.3 (n= 29 centrioles), Non-expanded SIM=2.6 +/- 0.2 (n= 22 centrioles). Data from 3 independent experiments except for SIM where it comes from a single experiment. Statistical significance was assessed by one-way ANOVA test: ****<0.0001, ***=0.0002, ns (non-significant)=0.84 for ExM vs MAP and 0.99 for U-ExM vs Non-Expanded SIM.
Next, we expanded centrioles using both ExM and MAP protocols and imaged the samples by confocal microscopy followed by Hyvolution (Fig. 1b-c). The gels expanded ~4.2-fold (ExM) and ~3.5-fold (MAP), respectively. We noticed that the diameter of the centriole in ExM was markedly larger than expected from the determined expansion factor. Indeed, the PolyE signal depicts a 1.4x enlargement with an average diameter of 308 nm ± 42 nm after expansion compared to the diameter of 216 nm ± 17 nm determined from non-expanded centrioles imaged by dSTORM 9 (Fig. 1d, Supplementary Fig. 1c, Supplementary Fig. 2 and online methods), suggesting an anisotropic macromolecular expansion. Moreover, the tubulin signal appears inhomogeneous probably due to epitope masking of anti-PolyE antibodies (Fig. 1b, lateral view and Supplementary Fig. 3a-b). However, we noticed that the 9-fold symmetry of centrioles can be visualized, albeit not perfectly in ExM treated centrioles (Fig. 1b, Supplementary Fig. 3c-d). In contrast, we observed that the MAP treated centrioles appeared 1.6x smaller with an average diameter of 133 nm ± 27 nm (Fig. 1c and Fig. 1d), suggesting again inhomogeneous macromolecular expansion. As a consequence, the 9-fold symmetry of the PolyE is not apparent (Fig. 1c, Supplementary Fig. 3e). However, we observed a reduced antibody competition issue (Fig. 1c).
Based on these results, we set out to develop a new method of expansion microscopy preserving the overall ultrastructure of isolated organelles. Capitalizing on the MAP protocol6, we found that avoiding fixation and using a low formaldehyde (0.3-1% FA) and acrylamide (AA) concentration (0.15-1% AA) results in intact centriolar expansion with correct diameter (Supplementary Fig. 3f-h). Therefore, we termed it Ultrastructure Expansion Microscopy (U-ExM). U-ExM applied on isolated centrioles reveals unambiguously the nine-fold symmetry of the centriole as seen both for the α-tubulin and PolyE signal with correct diameters of 195 nm ± 12 nm and 225 nm ± 15 nm, respectively (Fig. 1d-e). U-ExM centrioles show a good overall preservation of the centriolar shape as compared to cryoEM images (Supplementary Fig. 4a-b) and a perfect isotropic expansion of centrioles compared to other methods (Fig. 1f). Moreover, we could alleviate the antibody competition as demonstrated by the central core decoration of the PolyE signal while retaining a complete tubulin decoration of the centriolar wall (Fig. 1e, lateral view). Finally, we found that the 9-fold symmetry is clearly visible and the centriolar roundness is best preserved in U-ExM compared to other expansion microscopy protocols (Supplementary Fig. 4c-d).
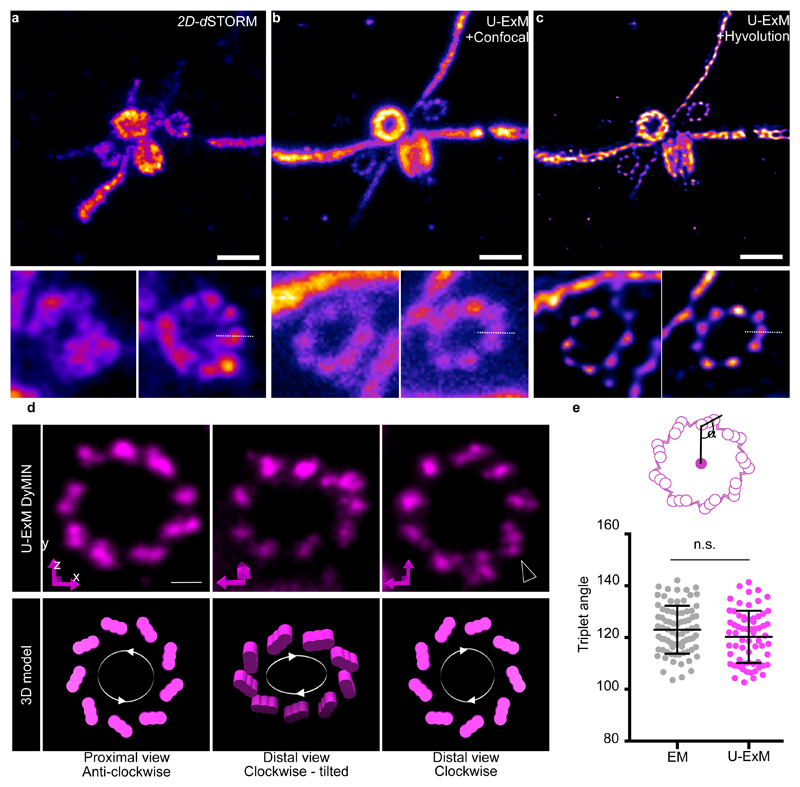
We then sought to test the potential of U-ExM by comparing confocal images of expanded pro-centrioles and non-expanded centrioles imaged by dSTORM, both stained for PolyE (Supplementary Fig. 5a-e and Fig. 2a-c). Here, the 9-fold symmetric microtubule blades cannot be visualized unambiguously by dSTORM unlike confocal images before and after deconvolution of U-ExM expanded samples (Fig. 2b-c, Supplementary Fig. 5f-h). Overall, U-ExM combined with confocal microscopy exhibits a higher labeling efficiency and apparent spatial resolution (Supplementary Fig. 5i-k) enabling the characterization of ultrastructural components of macromolecular assemblies.
Figure 2. U-ExM reaches the dSTORM precision limits.
(a) 2D dSTORM image of an isolated centriole. Scale bar: 250nm. (b) Confocal image of an expanded centriole using U-ExM (0.7%FA+1%AA). (c) Deconvoluted image in (b) using Hyvolution. Scale bar in (b, c): 1µm. Insets show the two procentrioles of the above image. The dotted white lines correspond to the plot line profile used to calculate the full width at half maximum (FWHM) shown in Supplementary Fig. 5i-k. Representative images from 1 experiment (a) and 3 independent experiments (b-c).
(d) Representative DyMIN images of procentrioles stained for α-tubulin (magenta, STAR RED) highlighting their anticlockwise or clockwise orientations. Below is the interpretation of such orientations in a 3D schematic model. Black arrowheads points to individual blades within a microtubule triplet (11 out of 90 procentrioles). Representative images from 2 independent experiments. Scale bar: 200nm. (e) Quantification of the angle between the center of the centriole and the microtubule triplet both from EM (123° ± 9°) (n= 77 triplets) and DyMIN (120° ± 10°) (n=65 triplets) images. Indicated values in brackets are averages with their associated standard errors. Unpaired two-tailed t test was as performed: P=0.0912.
We next set out to analyze precisely PolyE localization on the microtubule triplets using U-ExM (Supplementary Fig.6). U-ExM revealed that PolyE covers the outer surface of the tubulin signal with nine discrete puncta at both proximal and distal ends (Supplementary Fig. 6a, arrowhead and Supplementary Videos 1 and 2). To prevent any artifact due to the anisotropic resolution of confocal microscopy, we next performed an isotropic 3D reconstruction using a recent “reference-free” reconstruction approach 10 (see online methods) (Supplementary Fig. 6b-f). This result confirmed PolyE localization with clear nine discrete signals at both distal and proximal ends. By measuring the diameters of both PolyE and α-tubulin signals, we found that PolyE depicts a measured expanded diameter of 88 - 140 nm larger than that of tubulin (Supplementary Fig. 6g-i). Interestingly, by modeling several PolyE localizations on each microtubule triplet we found that PolyE localizes on the C microtubule establishing that U-ExM is able to distinguish a C-microtubule triplet localization for polyglutamylated tubulin in mature centrioles (Supplementary Fig. 7).
To further investigate the ability of U-ExM to reveal the molecular architecture of centrioles, we combined it with STED microscopy using either single (Fig. 2d) or dual color imaging (Supplementary Fig. 8a-b and Supplementary Video 3). As shown by electron microscopy (EM), centrioles are composed of nine microtubule triplets with a characteristic angle arranged in a clockwise manner as seen from the proximal side (Supplementary Fig. 8c). Strikingly, U-ExM-treated centriole pairs imaged using DyMIN11 allows a glimpse of the triplet structure of microtubules on the procentrioles (Supplementary Fig. 8d) as well as the visualization of the anticlockwise and clockwise orientation of microtubule triplets in procentrioles (Fig. 2d). In some cases, we could even identify three distinct fluorescent peaks for microtubule triplets that possibly correspond to the A-, B- and C-microtubule (Fig. 2d, arrowhead). Furthermore, we found a similar microtubule triplet angle of ~120° between EM and U-ExM (Fig. 2e) corroborating that U-ExM preserves the nanometric conformation of the sample.
Next we tested whether U-ExM can be applied in cellulo. Therefore, we first expanded unfixed cw15- C. reinhardtii cells. Although we observed a slight increase in centriole diameter in cellulo (236 nm +/- 18 for PolyE and 212 nm +/- 22 for tubulin), we could confirm correct isotropic expansion and 9-fold symmetry (Supplementary Fig. 9a-d). In addition, we found that U-ExM enables the visualization of the nine-fold symmetry of the axoneme with a tubulin diameter of 192 nm consistent with previous description12 (Supplementary Fig. 10 and supplementary Video 4). Moreover, we found that the nine microtubule doublets are highly polyglutamylated while the central pair is only weakly polyglutamylated (Supplementary Fig. 10a and Supplementary Fig. 11a-f). Interestingly, by modeling different polyglutamylation localizations on the axoneme, we found that polyglutamylation marks are deposited onto the surface of the B-tubule facing the flagellar lumen of Chlamydomonas axonemes as previously proposed both in Chlamydomonas and Tetrahymena13–16 (Supplementary Fig. 11g-q).
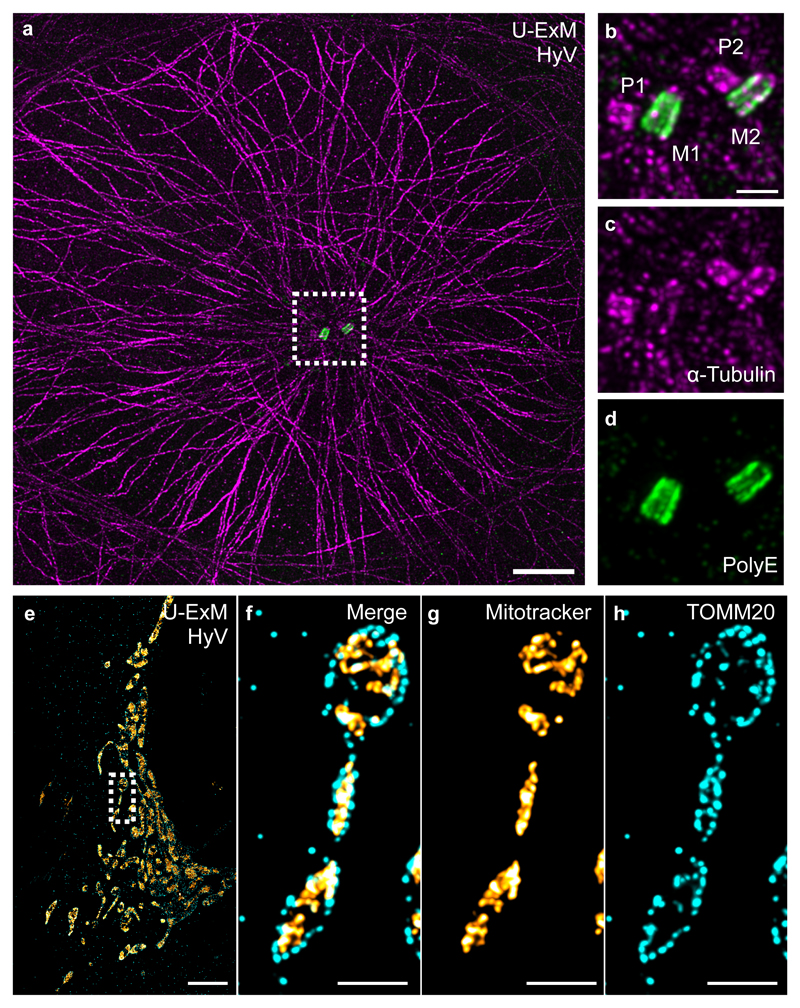
Second, we tested whether other dynamic cellular structures can be successfully expanded using U-ExM. Therefore, we first of all tested different fixation conditions combined with U-ExM on isolated centrioles for assessing structural preservation. We found that centrioles fixed with FA or methanol have an overall good structural preservation but a reduced centriole diameter while PFA/GA fixation does not allow full expansion of centrioles (Supplementary Fig. 12). Then we analyzed the effect of the different fixation conditions followed by incubation in acrylamide/formaldehyde solution (AA/FA) on human cells. We found that all fixation conditions tested preserve microtubules while PFA/GA fixation is best suited for structure preservation of mitochondria (Supplementary Fig. 13). Thus, we performed U-ExM with fixed mammalian cells and analyzed microtubules, mitochondria, and clathrin-coated pits as another membrane-bound structure. We found that U-ExM nicely expands methanol-fixed microtubules with a FWHM of 46 nm consistent with previous expansion microscopy methods4,5,17,18 as well as human centrioles with an average diameter of 190 nm ± 8 nm (Fig. 3 a-d, Supplementary Fig. 14 and Supplementary Fig. 15). Similarly, PFA+GA fixed mitochondria can be expanded using U-ExM with a good structural organization with the outer mitochondrial membrane translocase TOMM20 surrounding the overall mitotracker signal (Fig. 3e-h and Supplementary Fig. 16). For both microtubules and mitochondria, U-ExM shows uniform expansion with minimal distortions of 1.6% (microtubules) and 5% (mitochondria), similar to other expansion methods (Supplementary Fig. 17). Finally, we found that FA fixed clathrin-coated pits can also be visualized as hollow vesicles using U-ExM (Supplementary Fig. 18).
Figure 3. U-ExM applied on human cells.
(a) Representative Hyvolution confocal image of a U2OS cell fixed with methanol, expanded with U-ExM, and stained for α-tubulin (magenta) and PolyE (green). Scale bar: 10μm. (b-d) Insets (dotted square in a) show the centriolar pair, P: procentriole, M: mature centriole. Scale bar: 2μm. Representative images from 3 independent experiments. (e-h) Representative Hyvolution confocal image of a U2OS cell fixed with PFA/GA and stained for MitoTracker (orange) and the outer membrane mitochondrial translocase TOMM20 (cyan). Scale bar: 12μm. The dotted square shows a zoom of the highlighted region. Note that as expected TOMM20 signal is surrounding the MitoTracker signal. Scale bar: 3μm. Representative images from 1 experiment.
In summary, our results show that ExM protocols have to be carefully optimized to enable isotropic expansion of molecular assemblies. We demonstrated that U-ExM preserve ultrastructural details and can thus be used successfully to visualize the molecular architecture of multiprotein complexes of diverse nature. In comparison with a regular ExM protocol3,4,17,18, U-ExM can alleviate antibodies competition issues due to post-expansion labeling and prevent fluorophore loss. In addition, by avoiding chemical fixation of isolated protein complex, U-ExM improves the structural integrity as demonstrated with isolated centrioles. Importantly, in regular ExM approaches, the relative distance of the fluorophore to the epitope stays unchanged while our post-expansion labeling approach leads to a relative smaller antibody size compared to the expanded sample. Thereby, U-ExM couple to STED imaging unveils the chirality of the centriole, a structural feature that was only possible to reveal from the appendage proteins radiating 50 – 100 nm out of the centriole by super-resolution microscopy19. We are convinced that in the near future U-ExM will be successfully combined with single-molecule localization microscopy to enable fluorescence imaging of molecular details with unsurpassed spatial resolution.
Online methods
Reagents
Formaldehyde (FA, 36.5-38%, F8775), sodium acrylate (SA, 97-99%, 408220), guanidine hydrochloride (8M, G7294), acrylamide (AA, 40%, A4058), N,N’-methylenbisacrylamide (BIS, 2%, M1533), PIPES (P6757) and poly-D-lysine (A-003-E) were purchased from Sigma-Aldrich. Proteinase K (>600 U/mL, EO0491), ammonium persulfate (APS, 17874), tetramethylethylendiamine (TEMED, 17919), DMEM supplemented with GlutaMAX (61965), fetal bovine serum (10270) and Penicillin-Streptomycin (15140) were obtained from ThermoFisher. DMEM/HAM′s F12 with L-glutamine (Sigma, D8062) supplemented with 10% FBS (Sigma, F7524) and penicillin (100 U/ml) and streptomycin (0.1 mg/ml) (Sigma, R8758). Paraformaldehyde (PFA, 15700) and glutaraldehyde (GA, 25%, 16200) were purchased from Electron Microscopy Sciences. Sodium dodecyl sulfate (SDS), Triton X-100 and Tween-20 were obtained from AppliChem and Tris from Biosolve. Nuclease-free water (AM9937) was purchased from Ambion™-ThermoFisher. TetraSpeck™ 0.1µm fluorescent beads (T7279) were obtained from ThermoFisher. BSA was purchased from Roche (reference 10 735 086 001). Cytoskeleton buffer: 10 mM MES (M8250, Sigma), 150 mM NaCl (Sigma), 5 mM EGTA (67-42-5, Sigma), 5 mM glucose (Sigma, G8270) and 5 mM MgCl2 (ApliChem, A4425); pH 6.1 (NaOH).
SA was diluted with nuclease-free water at a concentration of 38% (wt/wt) and stored at 4°C for 6 months. Monomer solutions (MS) of AA and SA were pre-mixed in different ratios and concentrations according to ExM, MAP or U-ExM protocols and kept as aliquots at -20°C. MS were thawed and cooled at 4°C before gel synthesis. Free radical initiator APS and polymerization catalyst TEMED were prepared as 10% (wt/wt) stock solutions in nuclease-free water and frozen at -20°C. For polymerization, APS and TEMED stocks were thawed and chilled on ice before adding to MS in desired concentrations. For the ExM protocol, Proteinase K was added to fresh prepared digestion buffer directly before use. For the MAP and U-ExM protocols, PFA/AA and FA/AA solutions were prepared fresh prior use.
Coverslips used for either sample loading (12mm) or image acquisition (24mm) were first washed with absolute ethanol and subsequently dried. Next, coverslips were coated with poly-D-lysine (0.1 mg/ml) and incubated for 1 h either at room temperature (RT) (12mm coverslips) or at 37°C (24mm coverslips), washed 3 times with ddH2O and stored at 4°C for 1 week.
For immunolabelling, the following primary and secondary antibodies were used in this study: rabbit polyclonal anti-polyglutamate chain (PolyE, IN105) (1:500, AG-25B-0030-C050, Adipogen), mouse monoclonal anti-α-tubulin (DM1α) (1:500, T6199, Sigma-Aldrich), rat monoclonal anti-α-tubulin (YL1/2) (1:500, ab6160, Abcam), rabbit monoclonal anti-TOMM20 (EPR15581-39) (1:200 for non-expanded cells or 1:100 for U-ExM, ab186734, Abcam), mouse anti-alpha-tubulin (B-5-1-2) (6.7 mg/ml, 1:500, T5168, Sigma), rabbit anti-clathrin heavy chain (1.0 mg/ml, 1:500, Abcam), MitoTracker™ Red CMXRos (100nM, M7512, Invitrogen, ThermoFisher), goat anti-rabbit Alexa Fluor 488 IgG (H+L) (A11008), goat anti-mouse Alexa Fluor 488 IgG (H+L) (A11029), goat anti-mouse Alexa Fluor 568 IgG (H+L) (A11004) (1:400, Invitrogen, ThermoFisher) and anti-rat Cy3 (1:400, Jackson ImmunoResearch). Alexa Fluor 488 F(ab′)2 of goat anti rabbit IgG (2mg/ml, 1:200, A11070, ThermoFisher), Se Tau-647-NHS (K9-4149, SETA BioMedicals) conjugated to F(ab’)2 of goat anti-Rabbit IgG (SA5-10225, ThermoFisher) (1.5 mg/ml, 1:200), DNA-dye Hoechst 3342 (10,0 mg/ml, 1:1000, C10340, Invitrogen). Secondary antibodies anti-rabbit STAR 580 and anti-mouse STAR RED (1:400, Abberior) were used for STED acquisition. Alexa647 F(ab′)2 of goat anti rabbit IgG (2mg/ml, 1:200, A-21246, ThermoFisher) were used for dSTORM imaging.
Chlamydomonas centrioles isolation and centrifugation on coverslips
Chlamydomonas centrioles were isolated from the cell wall-less Chlamydomonas strain CW15- and spun on coverslips as previously described8. Coverslips were next processed either for regular immunofluorescence or for expansion microscopy protocols.
Immunofluorescence of non-expanded isolated centrioles
Coverslips with isolated centrioles were fixed with 4% FA in PBS for 10 min at RT and washed in PBS. Coverslips, with centrioles facing down, were then placed on 75 μl of the primary antibody solution diluted in PBS/BSA 2% for 1 h at RT in a humid chamber. Coverslips were then washed in PBS 3 times for 5 min and subsequently incubated for 1 h at RT with 75 μl of the secondary antibody solution diluted in PBS/BSA 2% in a humid chamber, protected from light. Finally, coverslips were washed in PBS, 3 times for 5 min. Coverslips were then mounted on a glass slide using 6 μl DABCO containing mounting medium.
ExM protocol
Centrioles were processed as indicated above for immunofluorescence. After the last PBS wash, coverslips were incubated 10 min at RT in 0.25% GA in PBS in a 6-well plate, followed by washing in PBS, 3 times for 5 min and then processed for gelation. A small plastic box was covered with parafilm and put on ice to create a flat hydrophobic surface for gelation. A drop of 35 μl of ExM-MS (8.625% (wt/wt) SA, 20% (wt/wt) AA, 0.15% (wt/wt) BIS, 2M NaCl in 1X PBS) supplemented with 0.2% APS and 0.2% TEMED, with the initiator (APS) added last, was placed on the chilled parafilm and coverslips were carefully put on the drop with centrioles facing the gelling solution. Gelation proceeded for 1 min on ice and then shifted to 37°C in the dark for 1 h. Then, coverslips with attached gels were transferred into a 6-well plate for incubation in 2 ml digestion buffer (1X TAE buffer, 0.5% TritonX-100, 0.8M guanidine hydrochloride, pH ~8.3) supplemented with fresh Proteinase K at 8 units/ml for 45 min at 37°C. Finally, gels were removed with tweezers from the coverslips and placed in beakers filled with ddH2O for expansion. Water was exchanged at least twice every 30 min and then incubated in ddH2O overnight at RT. Gel expanded between 4X and 4.2X according to SA purity.
MAP protocol
Coverslips with isolated centrioles were incubated in a solution of 4% PFA with 30% AA in PBS for 4-5 h at 37°C, without a fixation step. Incubation time in PFA/AA was shortened compared to the original ‘cultured cell’ MAP protocol6, in order to be adapted to smaller specimen as isolated centrioles. Immediately after PFA/AA incubation, gelation was performed as described above in the ExM protocol. Coverslips with centrioles facing down were placed on 35 μl of MAP-MS (7% (wt/wt) SA, 20% (wt/wt) AA, 0.1% (wt/wt) BIS in 1X PBS) supplemented with 0.5% APS and 0.5% TEMED, with the initiator (APS) added last, on parafilm in a pre-cooled humid chamber. Gelation proceeded for 1 min on ice and then shifted to 37°C in the dark for 1 h. Coverslips with gels were then incubated in ~2 ml denaturation buffer (200 mM SDS, 200 mM NaCl and 50 mM Tris in nuclease-free water) in a 6-well plate for 15 min at RT. Gels were then removed from the coverslips with tweezers and moved in a 1.5 ml Eppendorf centrifuge tube filled with fresh denaturation buffer and incubated at 95°C for 30 min. After denaturation, gels were placed in beakers filled with ddH2O for an initial expansion. Water was exchanged at least twice every 30 min, at RT. Successively, gels were placed in PBS 2 times for 15 min to remove water excess before incubation with primary antibody solution. In this step, gels shrink back to ~50% of their expanded size. Next, gels were incubated with primary antibody diluted in PBS/BSA 2% overnight at RT, with gentle shaking. Gels were then washed in PBS + 0.1% Tween 20 (PBST), 3 times for 10 min with shaking, and incubated with secondary antibody solution diluted in PBS/BSA 2% for ~6 h at RT, with gentle shaking. Gels were then washed in PBST, 3 times for 10 min with shaking, and finally placed in beakers filled with ddH2O for final expansion. Water was exchanged at least twice every 30 min and then gels were incubated in ddH2O overnight. Gel expanded between 3.3X and 3.5X according to SA purity.
U-ExM protocol
In U-ExM, the sample was not or mildly fixed prior expansion. First, coverslips with unfixed isolated centrioles were incubated in a solution of 0.7% FA with 0.15% or 1% AA in PBS for 4-5 h at 37°C. Next, similarly to ExM and MAP protocols, the gelation was performed by incubating coverslips with centrioles facing down with 35μL of U-ExM-MS composed of 19% (wt/wt) SA, 10% (wt/wt) AA, 0.1% (wt/wt) BIS in 1X PBS supplemented with 0.5% APS and 0.5% TEMED, on parafilm in a pre-cooled humid chamber. Note that APS was added last. Importantly, the monomer solution was adapted specifically for U-ExM to achieve an expansion factor of ~4 fold. Briefly, to find the best expansion conditions, we increased SA and reduced AA concentrations in the monomer solution. The following combinations were tested to compare gel expansion: 20%AA/7%SA (original MAP MS), 10%AA/7%SA, 10%AA/19%SA (U-ExM MS), 5%AA/7%SA and 5%AA/19%SA (Supplementary Fig.19). Isolated centrioles were then embedded in gels made with monomer solutions having the different AA/SA combinations and expanded to check their quality. Only in gels made with 10%AA centrioles expanded isotropically and their shape looked preserved (Supplementary Fig.19). Two independent experiments were performed for each condition.
Gelation proceeded for 1 min on ice and then shifted to 37°C in the dark for 1 h. Coverslips with gels were then transferred into ~2 ml denaturation buffer (200 mM SDS, 200 mM NaCl and 50 mM Tris in ultrapure water) in a 6-well plate for 15 min at RT. Gels were then removed from the coverslips with flat tweezers and moved in a 1.5 ml Eppendorf centrifuge tube filled with fresh denaturation buffer and incubated at 95°C for 30 min. After denaturation, gels were placed in beakers filled with ddH2O for the first expansion. Water was exchanged at least twice every 30 min, at RT and then gels were incubated overnight in ddH2O. Next, to remove water excess before incubation with primary antibody solution, gels were placed in PBS 2 times for 15 min. Note that in this step, gels shrink back to ~50% of their expanded size. Incubation with primary antibody diluted in PBS/BSA 2% was at 37°C for ~3 h, with gentle shaking. Gels were then washed in PBS + 0.1% Tween 20 (PBST), 3 times for 10 min with shaking, and incubated with secondary antibody solution diluted in PBS/BSA 2% for ~3 h at 37°C, with gentle shaking. Gels were then washed in PBST, 3 times for 10 min with shaking, and finally placed in beakers filled with ddH2O for expansion. Water was exchanged at least twice every 30 min and then gels were incubated in ddH2O ON. Gel expanded between 4.0X and 4.5X according to SA purity.
Mounting and image acquisition
Before imaging, gel size was accurately measured with a caliper to calculate the fold expansion. The gel was then cut with a razor blade in pieces that fit in a 36 mm metallic chamber for imaging. Water excess was carefully removed from the piece of gel by placing it in between two laboratory wipes. The piece of gel was then mounted on 24 mm round #1.5 (high precision) poly-D-lysine coated coverslip, already inserted in the metallic chamber, and gently pressed with a brush to ensure adherence of the gel to the coverslip. This step is crucial to completely avoid gel drift during imaging. After a few seconds, a couple of drops of ddH2O were slowly added on top of the sample until the gel was completely covered with water, to avoid shrinking of the polymer. Confocal microscopy was performed on a Leica TCS SP8 using a 63x 1.4 NA oil objective, with the HyVolution mode20 to generate deconvolved images, with the following parameters. ‘HyVolution Grade’ at max Resolution, Huygens Essential as ‘Approach’, water as ‘Mounting Medium’ and Best Resolution as ‘Strategy’. 3D z-stacks at 0.12μm intervals were acquired with a pixel size of 35 nm.
For post-U-ExM imaging of clathrin, microutubles and DNA a RCM (Rescanning Confocal Microscope) was used which is based on the image scanning principle where pixel reassignment is purely achieved optomechanically 21. The RCM unit (Confocal.nl) is attached to a sideport of a Nikon TiE and equipped with a sCMOS Zyla 4.2P (Andor). The pinhole is fixed at 50µm pinhole size. As excitation source the laserunit Cobolt Skyra™ (Cobolt, Hübner Group) is fibercoupled and connected to the RCM unit. The laserunit has four laserlines (405nm, 488nm, 561nm, 640nm) with 50mW each (free beam). Various OD filters are introduced in the excitation path to attenuate the laserpower. The TiE is equipped with a motorized stage (Nikon) and a 60x water immersion objective (CFI Plan APO, 1.27 NA, Nikon). The setup is fully controlled with the NIS elements version 4.6 on windows 8.
STED imaging was performed on a commercial STED microscope (Expert Line, Abberior-Instruments, Germany) working at repetition rate of 40 Mhz22. Centrioles were immunostained using secondary antibodies conjugated to the dyes STAR RED and/or Star 580 (Abberior, Germany). STAR RED was imaged with excitation at a wavelength of 640 nm and time-gated fluorescence detection between 650-720 nm. STAR 580 was excited at 561 nm with time-gated detection between 580-630 nm. The STED laser had a wavelength of 775 nm and a pulse width of roughly 500 ps. The pinhole was set to 0.75 AU. For imaging a water-immersion objective lens was used (UPLSAPO 60XW, Olympus, Japan). To arrive at high fluorescence signals and STED resolutions for a clear structure representation, the recently published adaptive-illumination scan technique DyMIN was used 11. Non-expanded centrioles were imaged using a 100x 1.4 NA oil objective.
dSTORM imaging was conducted on an inverted microscope (Zeiss Axio Observer.Z1, Carl Zeiss Microscopy) equipped with a 100x oil-immersion objective (alpha Plan-Apochromat 100x/1.46 Oil DIC, Carl Zeiss Microscopy) and a 63x water objective lens (LD C-Apochromat 63x/1.15 W Corr M27, Carl Zeiss Microscopy). For illumination of the sample, a 640 nm diode laser (iBeam smart, Toptica Photonics) was used. The laser beam was adjusted to quasi-TIRF mode when using the oil objective and to epifluorescence configuration when using the water objective. Between twenty and fourty thousand frames were collected on an electron multiplying charge-couple device (EM-CCD) camera (Andor Ixon Ultra DU897U-CSO) at a frame rate of 50-80 Hz. An autofocus system (Zeiss Definite Focus) kept the focus stable during image acquisition. For 3D imaging a cylindrical lens (f=250 mm) was placed in the detection path of the microscopic setup. Samples were placed in freshly prepared photoswitching buffer consisting of 100 mM cysteamine hydrochloride (Sigma) in PBS (1x) (pH 7.5) supplemented with an oxygen scavenger system (2% Glucose (w/v), 2 U/ml Glucose-Oxidase (Sigma) and 200 U/ml catalase (Sigma)). 2D Super-resolution images were reconstructed using the ImageJ plugin ThunderSTORM 23 and for 3D images the open source software rapidSTORM 3.324 was used.
Measurements of centriole diameter
Solely nearly perfect top view centrioles were selected for measurement of centriole diameter (see Supplementary Fig. 2). Briefly, we included in the analysis only centrioles where the most-distal and most-proximal regions were aligned. The line scan and plot profile tools of Fiji were used to determine the diameter. For each centriole, a line was drawn through the 9-fold symmetrical PolyE signal clearly seen at the end of the central core and the distance between the two peaks of intensity was measured. The diameter was obtained from the average of two measurements. Related to Figure 1, for each condition, 3 independent experiments were analysed. PolyE: n=30 centrioles for ExM, MAP and U-ExM, n=15 for non-expanded dSTORM; Tubulin: n=29 for ExM and U-ExM, n=20 for MAP. Related to Supplementary Fig. 12, FA: n=11 centrioles, 1 experiment; Methanol: n=16, 2 independent experiments; PFA/GA: n=13, 1 experiment. Related to Supplementary Fig. 14, the diameter of human centrioles in methanol fixed U2OS cells was calculated from 13 centrioles from 1 experiment.
Isotropic expansion of centrioles
Isotropic expansion was determined by comparing the ratio length/diameter of expanded centrioles to non-expanded centrioles. The line scan and plot profile tools of Fiji were used to measure length and diameter on α-tubulin staining of nearly perfect lateral view of centrioles. To determine the length, a line scan of a size able to cover the whole width of the centriole was drawn in a maximum projection image and the distance between the first peak and the last peak of intensity was measured. Measurements of the diameter were performed as described above. For each condition, 3 independent experiments were analyzed except for SIM experiment, which was performed once. N=30 centrioles for ExM and MAP, n=29 for U-ExM and n=22 for non-expanded SIM.
Analysis of the centriolar 9-fold symmetry
To graphically quantify the 9-fold symmetry of the PolyE signal, the Polar Transformer plugin (https://imagej.nih.gov/ij/plugins/polar-transformer.html), the line scan and plot profile tools of Fiji were used. For each centriole, a single Z plane (the same Z plane used to quantify the centriole diameter) was transformed to polar coordinates with the Polar Transformer plugin, generating “unwrapped” images (Supplementary Fig. 3c). Then, a straight line, wide enough to cover the whole signal, was drawn to obtain the plot profile. Curves from 30 centrioles for each condition from 3 independent experiments were merged together to create a unique averaged curve as follows. A sinusoidal model was generated to represent a theoretical plot profile of a polar transformed centriole. This model is described by the following formula:
where period can be calculated by dividing the length of an experimental plot profile by 9. To compare an experimental centriole plot profile to this model, the experimental values were rescaled between 0 and 1. Then, the plot profile is progressively moved along its x axis. For each shift, the cross-correlation between the experimental data and the model was calculated. The shift value giving the best cross-correlation is conserved and the plot profile was moved according to it. Once all plot profiles have been aligned on the sinusoidal model, an average profile was generated. The model and the cross-correlation search were done using the language R.
Analysis of centriolar shape
The shape descriptor tool of Fiji was used to analyze centriole shape quality. The single Z plane already used to measure centriole diameter was also used to analyze centriole shape. Using PolyE staining, a polygon with 9 vertexes was drawn around the circumference, joining the 9 microtubule (MT) triplets when visible. The shape descriptor tool gives several parameters, among them the roundness value, defined as minor axis/major axis of the figure. A value of 1 represents a perfect round shape. For each condition, 30 centrioles from 3 independent experiments were analysed.
Comparison between dSTORM and U-ExM
Isolated centrioles, either non-expanded or expanded with the U-ExM protocol, were stained for PolyE and imaged with dSTORM and confocal microscopes respectively. A straight line that bisected the microtubule triplets of procentrioles was drawn using the line tool in Fiji. Then, the line scan and plot profile tools were used to measure the fluorescence profile along this line and obtain the Full Width at Half Maxima (FWHM) of the curve for dSTORM (4 non-expanded procentrioles, total of 24 triplets, one experiment), U-ExM+Confocal (9 expanded procentrioles, total of 76 triplets, 3 independent experiments) and U-ExM+HyVolution (9 expanded procentrioles, total of 81 triplets, 3 independent experiments). Analysis was performed on maximum intensity projection of 3D stack acquisitions. To monitor the 9-fold symmetry of the PolyE signal the Polar Transformer plugin in Fiji was used as described above.
Isotropic 3D averaging
The particle averaging results for Chlamydomonas reinhardtii isolated centrioles were obtained with the method described in 10. The data is pre-processed to correct drifts between slices of the image stacks using the StackReg plugin25. We averaged 14 centrioles from one experiment that were selected using the software ImageJ. To model the point spread function (PSF), we acquired images of 0.1µm fluorescent beads embedded in an U-ExM gel. The final PSF volume is obtained by registering and averaging 15 images of beads. The reconstruction is realized in two steps.
The first step takes as input the PSF model and a restricted number L of particles representing the main orientations. It creates a coarse initial reconstruction without reference. Due to the cyclical symmetry of the centriole, most of the information can be captured with top and side views, which corresponds to L=2. The input volumes are down-sampled by a factor of 2 to accelerate computations. We used the bi-level and block-coordinate optimization approach described in 10. To further speed up computations, we replaced the stochastic optimization approach for the estimation of pose parameters by a deterministic search with coarse discretization. The second step refines the result of the first step by considering all the available data and a more accurate model. The volume and particle poses are alternately updated until convergence. In both steps, we apply a C9 symmetry constraint to the reconstructed volumes. The custom Matlab source code is available at: https://github.com/dfortun2/U-ExM. The reconstruction code with known angles uses the inverse problem library GlobalBioIm26. The parameters used for the reconstruction of Supplementary Fig. 6 are the default parameters accessible in the code.
Sub-triplet localization analysis
Isolated centrioles were expanded with the U-ExM protocol, co-stained for PolyE and α-tubulin and imaged with a confocal microscope. Before analysis, deconvolution was applied on images. A straight line that bisected the microtubule triplets starting from inside toward outside was drawn using the line tool in Fiji27. Then, the line scan and plot profile tools were used to measure the fluorescence profile of PolyE and α-tubulin along the same line and then normalized on the highest value, for both staining (Supplementary Fig. 7a-d). Both curves were aligned on the peak of PolyE as a reference point. N=8 centrioles from 3 independent experiments, for a total of 61 microtubule triplets.
To model the position of the fluorescence signal into the centriole, a 2D image extracted from cryoEM data was used and scaled to obtain a centriole with the expanded diameter after expansion. The resulting image gives a centriole with a diameter of 1125nm (Supplementary Fig. 7e) (Centriole diameter x expansion factor = 250 nm x 4.5 = 1,125 nm). The same operation was repeated for A-, B- and C-microtubules (Supplementary Fig. 7f-h). These images were then filtered at the resolution of HyVolution (140 nm) using a bandpass filter in ImageJ (Supplementary Fig. 7i-l). The final images (Supplementary Fig. 7m-o) were generated by merging each specific microtubule signal (Supplementary Fig. 7j-l) on the initial microtubule triplet image (Supplementary Fig. 7i). Sub-triplet localization analysis was done as described above.
Cryo-electron microscopy of Chlamydomonas centriole
Isolated Chlamydomonas centrioles8 were applied onto lacey carbon film grids (300Mesh, EMS), vitrified in liquid ethane. Grids were transferred into a JEM 2200FS cryo-electron microscope (JEOL) operating at 200 keV and equipped with a field emission gun. Images were collected using a 2048 × 2048 CCD camera (Gatan).
Quantification of MT-triplets angle
The angle tool of Fiji27 was used to measure the angle between the center of the centriole and the microtubule triplets. N= 77 microtubule triplets for EM images and n= 65 for U-ExM. Data from one experiment.
Chlamydomonas culture and expansion
Chlamydomonas cells were culture in Tris Acetate Phosphate (TAP) medium for 3 days at 23°C8. Cells were allowed to adhere onto 12mm poly-D-lysine coated coverslips for 15 min. This procedure, instead of spinning cells onto the coverslip, was used to increase the chance to find flagella positioned perpendicularly to the focal plane and to prevent deflagellation due to the centrifugation. Coverslips were next processed with the U-ExM protocol. 3D rendering was performed using the ImageJ28 plugin ClearVolume29.
Measurements of flagella diameter
Chlamydomonas cells, expanded with the U-ExM protocol, were co-stained for PolyE and α-tubulin and imaged with a confocal microscope. Before analysis, deconvolution was applied on images. Solely nearly perfect cilia cross section were selected for measurement of the diameter. The diameter was quantified with the use of the line scan and plot profile tools of Fiji 27. For each cilium, a line that bisected the center of the cilium was drawn to measure the fluorescence profile of both PolyE and α-tubulin along this line. A curve for each cilium was obtained from the average of two measurements and normalized on the highest value, for both staining. All curves were then aligned using as a reference point the centre between the two peaks of intensity. N=23 flagella from 3 independent experiments were analysed.
The modelling of PolyE on the B-microtubule (half or full) was done similarly to the sub-triplet localization on centrioles. Briefly, a cryoEM image of a cilium cross section was used and scaled to obtain a cilium with a diameter of 900nm (Centriole diameter x expansion factor = 200 nm x 4.5 = 900nm). These images were then filtered at the resolution of HyVolution (140 nm) using a bandpass filter in ImageJ.
Quantification of PolyE signal in Chlamydomonas flagella
Polyglutamylation of doublets and central pair MTs of Chlamydomonas flagella was analyzed on sprayed flagella (Supplementary Fig. 11a-f). For each flagellum, using the software Fiji27, a line scan of a few μm was drawn across the 9 microtubule doublets (MTD) and 2 central pair MTs to measure the plot profile of the PolyE signal. Next, the average of the 9 intensity peaks of the MTD and the average of the 2 intensity peaks of the central pair MTs were obtained. Finally the ratio between PolyE signal at the MTD and central pair MTs was calculated for each flagellum and the average between 4 sprayed flagella from 1 experiment was obtained.
Immunofluorescence of human cells
Human U2OS cells (gift from Erich Nigg) were seeded at a density of ~100,000 cells per well in a 6-well plate containing 12mm coverslips and incubated overnight at 37°C with 5% CO2 in DMEM supplemented with GlutaMAX, 10% fetal bovine serum and penicillin-streptomycin (100 µg/ml). To test the effect of fixation on microtubule preservation, four different conditions were tested. Cells were either 1) transferred into a solution of 0.7% FA and 1% AA in PBS (AA/FA U-ExM solution) without fixation, or 2) fixed for 7 min in -20°C methanol, or 3) fixed with 4% FA in CS buffer (10mM MES, 150mM NaCl, 5mM EGTA, 5mM glucose, 5mM MgCl2 pH 6.1) for 10 min at RT), or 4) rapidly pre-extracted in BRB80 solution with 0.5% Triton and fixed with 3% PFA+0.1%GA in PBS for 15 min at RT. After fixation, cells were quickly washed in PBS and then incubated for 5h in the AA/FA U-ExM solution at 37°C. Next, coverslips were incubated with primary antibody diluted in the antibody solution (PBS with 1% BSA and 0.05% Tween) for 1 h at RT in a humid chamber. For condition 1), cells were rapidly pre-extracted in BRB80 solution/0.5% Triton before incubation with the primary antibody solution. Coverslips were then washed in PBS 3 times for 5 min and subsequently incubated for 1 h at RT with secondary antibody diluted in the antibody solution in a humid and dark chamber. Finally, coverslips were washed in PBS, 3 times for 5 min, quickly dried and mounted on a glass slide using 3 μl DABCO containing mounting medium.
Similarly, the effect of different fixations on the preservation of mitochondria was tested proceeding with four different conditions. Cells were first incubated 15 min with MitoTracker (100nM, diluted in the culturing medium) and next either 1) transferred into the AA/FA U-ExM solution without fixation, or fixed 2) for 7 min in -20°C methanol, or 3) with 4% FA in PBS for 15 min at RT or 4) 3% PFA+0.1%GA in PBS for 15 min at RT. After fixation, cells were briefly washed in PBS followed by incubation with permeabilization/blocking buffer (PBS with 3% BSA; 0.3% Triton) for 30 min. Next, cells were rapidly washed in PBS and then transferred into the AA/FA U-ExM solution for 5 hours at 37°C. Finally, coverslips were briefly washed in PBS, quickly dried and mounted on a glass slide using 3 μl DABCO containing mounting medium.
Pre- and post-U-ExM on human cells for microtubule and mitochondria
Human U2OS cells were grown as described in the previous above. In this case, cells were seeded in a 6-well plate containing 24mm round #1.5 (high precision) coverslips. To compare microtubules in pre- and post-U-ExM, cells were fixed in -20°C methanol for 7 min followed by incubation in the AA/FA U-ExM solution for 5h at 37°C. Next, cells were stained with rat anti-α-tubulin (YL1/2) and secondary Cy3 as described in the previous paragraph. Then, cells were acquired to take pre-U-ExM images. Next, coverslips were processed with the U-ExM protocol with the following differences. Gelation was allowed to initiate on ice for 5 min before shifting to 37°C, 140μL of U-ExM-MS supplemented with 0.5% APS and 0.5% TEMED was used and denaturation proceeded for 1h 30 min (a longer time was chosen for cells compared to isolated centrioles to ensure maximal expansion in this complex specimen). For post-expansion staining, mouse anti-α-tubulin (DM1α) and Alexa Fluo 488 were used.
To compare mitochondria in pre- and post-U-ExM, cells were incubated 15 min with MitoTracker™ Red CMXRos (100nM, diluted in the culturing medium), fixed in 3% PFA+0.1%GA in PBS for 15 min at RT and then incubated in the AA/FA U-ExM solution for 5h at 37°C. Next, cells were acquired to take pre-U-ExM images. Following, U-ExM protocol was applied similarly to as described for microtubules, but denaturation proceeded for 1h (1h 30 min denaturation completely destroyed mitochondria, data not shown). For post-expansion staining, rabbit anti-TOMM20 and Alexa Fluo 488 were used. Note that the MitoTracker signal was retained after expansion. Thus, it was acquired and used to calculate the RMS-error between pre- and post-expansion images.
To recognize the region of the coverslip where cells were acquired, a mark was applied to the opposite side of the coverslip where cells were present. This allowed us to cut the piece of gel including only the cells acquired pre-expansion and facilitate their acquisition post-expansion.
Post-U-ExM of clathrin, microtubles and DNA
For Post-U-ExM labelling of clathrin coated pits, microtubules and DNA, COS-7 African Green monkey kidney cells (purchased from CLS Cell Line Servie GmbH) were cultured in DMEM/HAM′s F12 with L-glutamine supplemented with 10% FBS and penicillin (100 U/ml) and streptomycin (0.1 mg/ml) at 37°C and 5% CO2. 30000 cells per well were seeded on round 18 mm high precision coverglasses (No 1.5) in 12-well culture plates (TPP, 92012). Cells were grown for 24 hours at 37°C and 5% CO2 and subsequently fixed in 4% FA in cytoskeleton buffer at 37°C for 10 min. After fixation, cells were briefly washed with PBS and then transferred into AA/FA U-ExM solution for 5 h at 37°C. Gelation and denaturation of the sample was performed as described before for pre- and post- U-ExM on human cells but with 60µl of U-ExM-MS. For post-expansion staining, mouse anti-alpha-tubulin (B-5-1-2) and anti-clathrin heavy chain were diluted in PBS (1x) and incubated for three hours at 37°C simultaneously. Samples were washed in PBST three times for 20 min each time. Then secondary antibodies Al488- F(ab′)2 of goat anti rabbit IgG and Se Tau-647 conjugated to F(ab′)2 of goat anti mouse IgG were incubated simultaneously at 37°C for three hours in PBS (1x). The gels were washed two times in PBST for 20 min and once in PBS (1x) for 20 min. DNA-Hoechst dye in PBS (1x) was incubated for 20 min at room temperature. The gels were then fully expanded in ddH2O and immobilized on poly-D-lysine coated 24 mm coverslips as described before. The samples were imaged on a Rescanning Confocal Microscope.
Intensity profiles of manually chosen CCP′s that exemplary show typical central nulls were analyzed using Fiji. The Intensities were normalized to the maximum intensity value and double guassian fits were fitted to the Intensity profiles using the software Origin (OriginLab, Northampton, MA). To determine the diameter of the pits the distance of the centers of the single gaussian fits were calculated. The standard error of the diameter was calculated from the squareroot of the sum of the squared errors from the center values of the single gaussian fits.
Distortion analysis
To estimate the sample deformation after expansion, the RMS error (Root Mean Square error) was calculated between two images of the same structure before and after expansion. This was done following the protocol described in Chozinski et al.4. This protocol also provides the scale factor between the images, thus giving the expansion factor of the experiment. For both microtubules and mitochondria, the data from 3 independent experiments were used.
Supplementary Material
Acknowledgments
We thank N. Klena for critical reading of the manuscript. We thank the Martinou lab and especially S. Zaganelli for helpful discussions and sharing mitochondrial reagents. D. Gambarotto and M. Schmidt-Cernohorska are supported by the European Reaserch Council ERC StG 715289 (ACCENT) and P. Guichard, V. Hamel and M. Le Guennec by the Swiss National Science Foundation (SNSF) PP00P3_157517. F. U. Zwettler and M. Sauer acknowledge support by the Deutsche Forschungsgemeinschaft (DFG) within the Collaborative Research Center 166 ReceptorLight (projects A04 and B04). M.Unser is supported by the ERC (GA No 692726 GlobalBioIm).
Footnotes
Statistics and reproducibility.
All experiments were carried out 3 times independently, except indicated otherwise in the figure legends. All data are expressed as the average (mean) +/- standard deviation (SD). The values n, which represents the number of centrioles or the number of triplets analyzed, is stated in figure legends and in online methods. Statistical one-way ANOVA test and unpaired two-tailed t tests were done and are indicated in figure legends.
Life sciences reporting summary.
Detailed information on experimental design and reagents can be found in the Life Sciences reporting summary.
Data availability statement. The data that support the findings of this study are available from the corresponding authors upon request.
Code availability statement. The custom Matlab source code is available at: https://github.com/dfortun2/U-ExM
Author Contributions
D.G., F.U.Z., M.S., V.H. and P.G. conceived and designed the project. M.S., V.H. and P.G. supervised the project. D.G. and F.U.Z. performed all ExM experiments. D.G. performed all U-ExM experiments with the help of S.B. and the data analysis. F.U.Z. performed the dSTORM imaging and the clathrin-coated pits experiment and analysis. J.H., G. S. and M.R. performed and analyzed the STED imaging. D.F. and M.U. performed the 3D averaging. M. S-C. initiated the U-ExM project. M.L.G performed the plot profile of the polar transform showing the 9-fold symmetry as well as the RMS calculation. E.B. helped in setting up ExM. All authors wrote and revised the final manuscript.
Competing Financial Interests
Authors declare no competing interests.
References
- 1.Koster AJ, Klumperman J. Electron microscopy in cell biology: integrating structure and function. Nat Rev Mol Cell Biol. 2003;Suppl:SS6–10. [PubMed] [Google Scholar]
- 2.Sahl SJ, Hell SW, Jakobs S. Fluorescence nanoscopy in cell biology. Nat Rev Mol Cell Biol. 2017;18:685–701. doi: 10.1038/nrm.2017.71. [DOI] [PubMed] [Google Scholar]
- 3.Chen F, Tillberg PW, Boyden ES. Optical imaging. Expansion microscopy. Science. 2015;347:543–8. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chozinski TJ, et al. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat Methods. 2016;13:485–488. doi: 10.1038/nmeth.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tillberg PW, et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat Biotechnol. 2016;34:987–992. doi: 10.1038/nbt.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ku T, et al. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat Biotechnol. 2016;34:973–981. doi: 10.1038/nbt.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamel V, et al. Identification of Chlamydomonas Central Core Centriolar Proteins Reveals a Role for Human WDR90 in Ciliogenesis. Curr Biol. 2017;27:2486–2498.e6. doi: 10.1016/j.cub.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klena N, et al. Isolation and Fluorescence Imaging for Single-particle Reconstruction of Chlamydomonas Centrioles. JoVE. 2018:e58109. doi: 10.3791/58109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heilemann M, et al. Subdiffraction-Resolution Fluorescence Imaging with Conventional Fluorescent Probes **. 2008:6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 10.Fortun D, et al. Reconstruction from Multiple Particles for 3D Isotropic Resolution in Fluorescence Microscopy. IEEE Trans Med Imaging. 2018:1–1. doi: 10.1109/TMI.2018.2795464. [DOI] [PubMed] [Google Scholar]
- 11.Heine J, et al. Adaptive-illumination STED nanoscopy. Proc Natl Acad Sci. 2017 doi: 10.1073/pnas.1708304114. 201708304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pigino G, et al. Cryoelectron tomography of radial spokes in cilia and flagella. J Cell Biol. 2011;195:673–87. doi: 10.1083/jcb.201106125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lechtreck KF, Geimer S. Distribution of polyglutamylated tubulin in the flagellar apparatus of green flagellates. Cell Motil Cytoskeleton. 2000;47:219–235. doi: 10.1002/1097-0169(200011)47:3<219::AID-CM5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 14.Kubo T, Yanagisawa H aki, Yagi T, Hirono M, Kamiya R. Tubulin Polyglutamylation Regulates Axonemal Motility by Modulating Activities of Inner-Arm Dyneins. Curr Biol. 2010;20:441–445. doi: 10.1016/j.cub.2009.12.058. [DOI] [PubMed] [Google Scholar]
- 15.Kubo T, Oda T. Electrostatic interaction between polyglutamylated tubulin and the nexin–dynein regulatory complex regulates flagellar motility. Mol Biol Cell. 2017;28:2260–2266. doi: 10.1091/mbc.E17-05-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suryavanshi S, et al. Tubulin Glutamylation Regulates Ciliary Motility by Altering Inner Dynein Arm Activity. Curr Biol. 2010;20:435–440. doi: 10.1016/j.cub.2009.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao M, et al. Expansion Stimulated Emission Depletion Microscopy (ExSTED) ACS Nano. 2018;12:4178–4185. doi: 10.1021/acsnano.8b00776. [DOI] [PubMed] [Google Scholar]
- 18.Halpern AR, Alas GCM, Chozinski TJ, Paredez AR, Vaughan JC. Hybrid Structured Illumination Expansion Microscopy Reveals Microbial Cytoskeleton Organization. ACS Nano. 2017;11:12677–12686. doi: 10.1021/acsnano.7b07200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang TT, et al. Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat Commun. 2018;9:1–11. doi: 10.1038/s41467-018-04469-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borlinghaus RT, Kappel C. HyVolution—the smart path to confocal super-resolution. Nat Methods. 2016;13:i–iii. [Google Scholar]
- 21.de Luca GMR, et al. Configurations of the Re-scan Confocal Microscope (RCM) for biomedical applications. J Microsc. 2017;266:166–177. doi: 10.1111/jmi.12526. [DOI] [PubMed] [Google Scholar]
- 22.Göttfert F, et al. Strong signal increase in STED fluorescence microscopy by imaging regions of subdiffraction extent. Proc Natl Acad Sci. 2017;114:2125–2130. doi: 10.1073/pnas.1621495114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ovesný M, Křížek P, Borkovec J, Švindrych Z, Hagen GM. ThunderSTORM: A comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolter S, et al. rapidSTORM : accurate, fast open-source software for localization microscopy orcae : online resource for community annotation of eukaryotes. Nat Methods. 2012;9:1040–1. doi: 10.1038/nmeth.2224. [DOI] [PubMed] [Google Scholar]
- 25.Thevenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 26.Unser M, Soubies E, Soulez F, McCann M, Donati L. GlobalBioIm: A Unifying Computational Framework for Solving Inverse Problems. Imaging Appl Opt 2017 (3D, AIO, COSI, IS, MATH, pcAOP) 2017;2017 CTu1B.1. [Google Scholar]
- 27.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider Ca, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Royer LA, et al. ClearVolume: Open-source live 3D visualization for light-sheet microscopy. Nat Methods. 2015;12:480–481. doi: 10.1038/nmeth.3372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.