Abstract
Oncogenic FLT3 kinase is an important therapeutic target in acute myeloid leukemia (AML), however clinical responses to small molecule kinase inhibitors are short-lived due to rapid emergence of resistance as a consequence of point mutations or compensatory increases in FLT3 expression. We sought to develop a complementary pharmacological approach whereby proteasome-mediated FLT3 degradation could be promoted by identifying inhibitors of the deubiquitinating enzyme(s) (DUBs) responsible for cleaving ubiquitin from FLT3. As the relevant DUBs for FLT3 are not known, we assembled a focused library of most reported small molecule DUB inhibitors and performed a cellular phenotypic screen to identify compounds that could induce degradation of oncogenic FLT3. Subsequent target deconvolution efforts allowed us to identify USP10 as the critical DUB required to stabilize FLT3. Targeting USP10 showed efficacy in FLT3-ITD positive pre-clinical models of AML, including cell lines, primary patient specimens and mouse models of oncogenic FLT3-driven leukemia.
Introduction
The ubiquitin system plays a critical role in controlling protein homeostasis, a process necessary for cell health. Ubiquitination is a reversible post-translational modification whose most well-known and best characterized function is tagging proteins for proteolytic degradation[1]. However, its importance in protein activation/inactivation, localization, and lysosomal and autophagic degradation among other cellular processes is becoming increasingly appreciated[2]. Ubiquitin is a 76-amino acid protein attached to substrate proteins via iso-peptide bond formation between ubiquitin’s C-terminal glycine and a substrate lysine sidechain; linear and branched polyubiquitin chains are assembled via attachment of a new ubiquitin molecule to one of seven lysines or the N-terminal methionine of ubiquitin[3]. Ubiquitination is coordinated by the action of ubiquitin activating (E1), conjugating (E2), ligating (E3) and deubiquitinating (DUB) enzymes. DUBs have garnered significant interest as drug targets in recent years due to their role in stabilization of disease-causing proteins and oncology targets in particular[4].
At present, there are approximately 115 recognized human DUBs belonging to 6 distinct families[5, 6]. The substrates of DUBs, and contexts in which they are regulated, remain poorly understood[7]. Most studies aimed at identification of the DUB responsible for stabilization of a substrate of interest utilize a genetic-based screen measuring protein levels or a mass spectrometry-based approach to identify DUBs that interact with the target.[7, 8] Development of chemical probes to permit pharmacological interrogation of DUBs identified from such screens has followed with more than 40 DUB inhibitors now reported[9].
Screening of annotated enzyme family-specific small molecule libraries has been utilized successfully, in the kinase family for example[10, 11], as a complementary approach to discover disease targets. This middle of the road approach between a completely target unbiased small molecule phenotypic screen, in which target deconvolution can be extraordinarily difficult, and focusing inhibitor development on a single putative target that may not be ideal for pharmacological inhibition, can be a powerful approach for discovering novel and druggable dependencies of disease. This approach, to the best of our knowledge, has not been applied to DUBs, likely in large part due to a lack of well-characterized, commercially available DUB-targeting small molecule libraries.
Acute myeloid leukemia (AML) is the most common type of acute leukemia in adults. Approximately 30% of AML patients harbor activating mutations in FMS-like tyrosine kinase 3 (FLT3), a gene whose normal function is in controlling hematopoiesis. The most common type of FLT3 mutation results in internal tandem duplications (ITD) within the juxtamembrane domain, observed in 20–25% of AML patients and associated with markedly decreased survival[12]. An additional 7% of patients have point mutations within the “activation loop” of FLT3[12].
Mutant FLT3 is a clinically validated target. A number of FLT3 kinase domain inhibitors have been shown to induce partial, and usually brief, remissions in clinical trials of relapsed AML patients when administered as single agents[13]. In a large trial (RATIFY (CALGB 10603)) in newly diagnosed patients, however, midostaurin (PKC412) was shown to increase survival when combined with standard chemotherapy[14]. This study in particular supports the notion that inhibition of FLT3 is important, at least in patients with mutations in the FLT3 gene. Since drug resistance develops in some patients with newly diagnosed AML and virtually all patients with advanced disease, additional strategies to target FLT3 would be of value.
As is true for other receptor tyrosine kinases, there is ongoing synthesis and degradation of FLT3, thought to be accelerated by ligand binding. FLT3 turnover has been shown to be regulated via ubiquitin-mediated proteosomal and lysosomal degradation, and the E3 ubiquitin ligase c-Cbl targets FLT3 for ubiquitination and degradation[15]. In addition, inactivating point mutations in c-Cbl have been found in myeloid malignancies[16], which underscores the importance of tight choreography of FLT3 turnover in disease progression.
Here, we report the use of a focused DUB inhibitor library screen to identify USP10 as the DUB that stabilizes the FLT3-ITD oncoprotein via removal of a degradative ubiquitin tag. Furthermore, we show that pharmacological inhibition of USP10 promotes degradation of FLT3-ITD but not wild-type (wt) FLT3, leads to selective killing of oncogenic FLT3-expressing AML cells in vitro and in vivo, and overrides resistance to FLT3 kinase inhibitors caused by tyrosine kinase domain (TKD) mutations and other mechanisms. Our studies hence validate USP10 as a therapeutic target for FLT3 mutant AML. To the best of our knowledge, this is the first identification of a novel DUB substrate using a DUB targeting small molecule library screen and the first demonstration of stabilization of a mutant driver oncoprotein in AML by a DUB enzyme.
Results
HBX19818 and P22077 induce degradation of mutant FLT3
In order to identify novel targets and compounds that regulate protein homeostasis of oncogenic FLT3, we employed a whole cell phenotypic screen of 29 reported small molecule DUB inhibitors (Supplementary Results, Supplementary Table 1), which represents the majority of reported DUB inhibitors, annotated for inhibitory activity across a broad panel of DUBS ([17] and unpublished data), using oncogene-dependent and control cell lines followed by hit validation and target deconvolution and translational studies (Supplementary Results, Supplementary Figure 1a). The compounds were evaluated for ability to selectively kill growth factor-independent Ba/F3 cells expressing FLT3-ITD and Ba/F3 cells expressing FLT3-D835Y over IL-3-dependent parental Ba/F3 cells. Two chemically distinct hits from the screen, HBX19818 (1) and P22077 (2) (Figure 1a), both previously reported as USP7 inhibitors[18, 19], were confirmed to inhibit proliferation of mutant FLT3-positive Ba/F3 cells with EC50s in the single digit micromolar range (Figure 1b-c, Supplementary Results, Supplementary Figure 1b). The effects were partially IL-3 rescue-able indicating the growth suppression results from impaired FLT3 function. The anti-proliferative activity of HBX19818 and P22077 correlated with loss of FLT3 protein in Ba/F3-FLT3-ITD cells at the same concentrations and with a more modest loss of FLT3 protein in Ba/F3-D835Y cells (Figure 1d-e, Supplementary Results, Supplementary Figure 1c). Consistent with this, flow cytometry revealed loss of cell surface expression of FLT3-ITD with HBX19818 treatment (Supplementary Results, Supplementary Figure 1d). In contrast, FLT3 protein levels were unchanged in inhibitor-treated wt-FLT3-Ba/F3 cells (Figure 1f, Supplementary Results, Supplementary Figure 1e). Owing to a lack of FLT3-D835Y-positive cell lines, our subsequent studies focused on the FLT3-ITD mutation.
Figure 1. Targeted effects of HBX19818 and P22077 on mutant and wt FLT3-expressing Ba/F3 cells.
(a) Chemical structures of HBX19818 (1) and P22077 (2). (b) Effects of HBX19818 on Ba/F3-FLT3-ITD cells cultured in the absence or presence of 20% WEHI-conditioned media (used as a source of IL-3) following 72 hr of treatment. (n=2). (c) Effects of P22077 on Ba/F3-FLT3-ITD cells cultured in the absence or presence of 20% WEHI-conditioned media (used as a source of IL-3) following 22 hr of treatment. (n=2). (d) Effects of HBX19818 on FLT3 protein expression in Ba/F3-FLT3-ITD cells following 22 hr of treatment. (e) Effect of P22077 on FLT3 protein levels in Ba/F3-FLT3-ITD cells following 22 hr of treatment. (f) Effect of HBX19818 on FLT3 protein levels in Ba/F3-wtFLT3 cells following 24 hr of treatment. Immunoblots shown are representative of 1–2 additional studies for which similar results were observed.
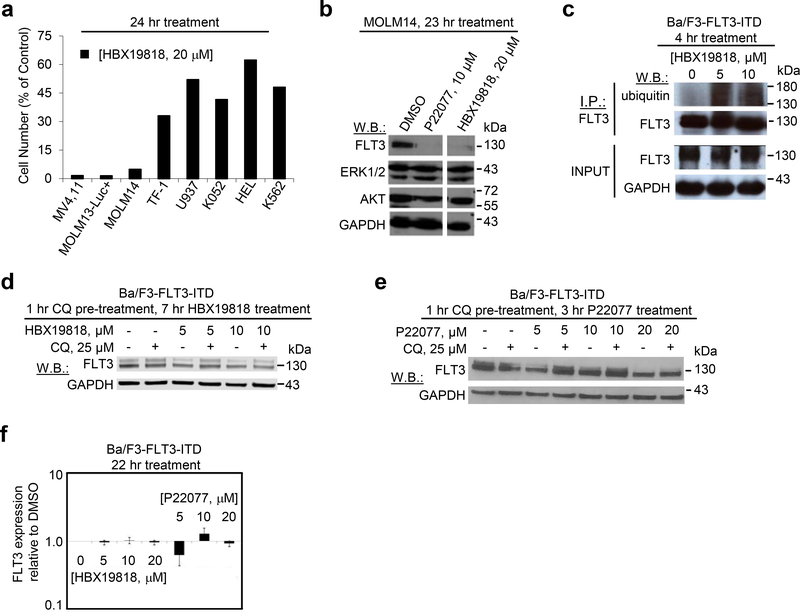
The effects of HBX19818 and P22077 on mutant FLT3-expressing cells were confirmed not to be unique to the Ba/F3 system. Both compounds suppressed the growth of the FLT3-ITD-positive AML cell lines, MOLM13-luc+, MOLM14, and MV4,11, in a dose-dependent manner with selectivity toward mutant FLT3-expressing cells versus wt or null FLT3-expressing cells (Figure 2a, Supplementary Results, Supplementary Figure 2a, Supplementary Table 2). It should be noted, however, that several human hematopoietic cell lines not driven by oncogenic FLT3 displayed relative sensitivity to P22077, which can likely be attributed to the multi-targeted nature of this agent. HBX19818 and P22077 treatment led to increased priming of mutant FLT3-expressing cells for apoptosis that significantly correlated with induction of apoptosis and that was stronger for FLT3 mutant cells than wt or null FLT3 cells (Supplementary Results, Supplementary Figure 2b-g). Consistent with data in the Ba/F3 system, HBX19818 and P22077 strongly induced FLT3 degradation in the FLT3-ITD positive lines MOLM13-luc+ and MOLM14 at 20 μM (Figure 2b, Supplementary Results, Supplementary Figure 3a-b) while having little to no impact on FLT3 protein in wt FLT3-expressing leukemia cell lines (Supplementary Results, Supplementary Figure 3c-d). As anticipated, inhibition of total cellular tyrosine phosphorylation was demonstrated in HBX19818-treated mutant FLT3-positive cells, which is consistent with drug-induced degradation of mutant FLT3 (Supplementary Results, Supplementary Figure 3e). Importantly, HBX19818 and P22077 treatment did not lead to degradation of signaling molecules downstream of FLT3, including AKT and ERK1/ERK2, suggesting that induction of mutant FLT3 degradation by HBX19818 and P22077 was selective (Figure 2b).
Figure 2. Targeted and selective effects of HBX19818 and P22077 on mutant FLT3-expressing human AML cells and investigation of mechanism of ubiquitin-mediated degradation.
(a) Analysis of proliferation of HBX19818-treated mutant FLT3-positive MV4,11, MOLM13-luc+ and MOLM14 cells as compared to null FLT3 or wt FLT3-expressing leukemia cells at a concentration of 20 μM following 24 hours of treatment. (n=2). (b) Analysis of FLT3, ERK1/ERK2, and AKT expression in MOLM14 cells treated with P22077 or HBX19818 for approximately 23 hours. The full immunoblot is shown in Supplementary Figure 4l. This immunoblot is representative of three independent experiments that yielded similar results. (c) Effect of HBX19818 on mutant FLT3 ubiquitination. This immunoblot is representative of four independent experiments for which similar results were observed. (d-e) Rescue of FLT3 degradation in HBX19818- and P22077 -treated Ba/F3-FLT3-ITD cells with the lysosome inhibitor, chloroquine (CQ). Results shown are representative of studies performed in duplicate for which similar studies were observed. (f) Effect of HBX19818 and P22077 on FLT3 transcription in Ba/F3-FLT3-ITD cells following 22 hours of treatment. Shown is FLT3 expression relative to GAPDH expression. Data shown are a composite of three independent experiments (n-=3).
Compounds induce ubiquitin-mediated degradation of FLT3
Consistent with the loss of FLT3 protein resulting from ubiquitin-dependent degradation, increased FLT3 ubiquitylation was observed for FLT3-ITD 4–8 hours after HBX19818 treatment (Figure 2c). Ubiquitin tags can encode proteosomal or lysosomal degradation. FLT3 has been reported to undergo degradation by both pathways. We established that both HBX19818- and P22077-induced FLT3-ITD degradation is partially rescued by inhibition of the lysosome (Figure 2d-e), and qPCR analysis confirmed the reduction in FLT3 levels occurred at the protein level only (Figure 2f).
Structure-activity relationship studies
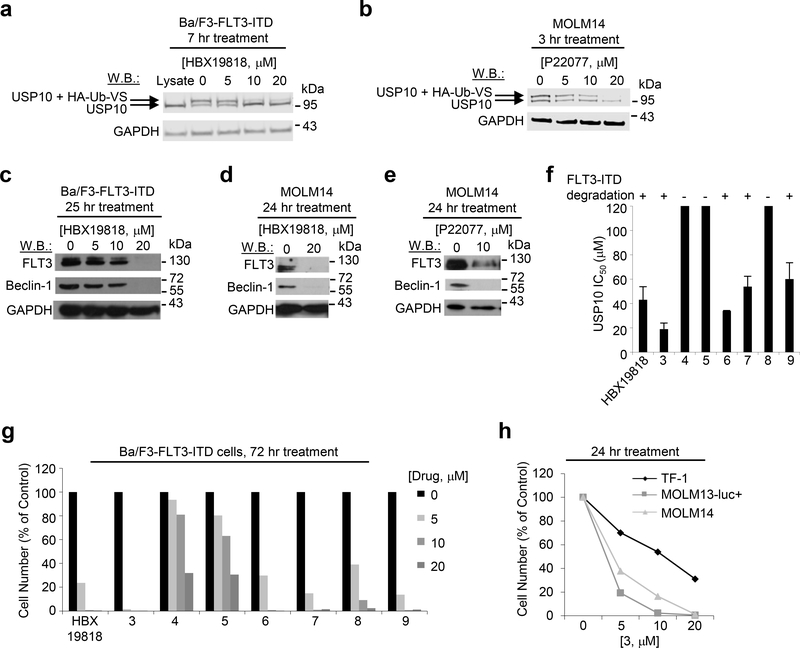
HBX19818 and P22077 were both reported as irreversible inhibitors of ubiquitin-specific protease 7 (USP7), a DUB best known for its role in stabilization of MDM2[18, 19]. Our profiling of the compounds in vitro against a panel of 33 recombinant DUB enzymes at a concentration of 10 μM using diubiquitin as substrate[17] identified USP10 as the most potently inhibited DUB of each compound (Supplementary Results, Supplementary Figure 4a-b). HBX19818 and P22077 inhibited USP10 with IC50s of 14 and 6 μM, respectively, and USP7 with IC50s of 57 and 10 μM, respectively, when tested in dose response in the same assay (Supplementary Results, Supplementary Figure 4a-b). Both compounds were confirmed to bind and inhibit USP10 in mutant FLT3-expressing cells using competitive activity-based protein profiling[20] and by evaluating protein levels of a known USP10 substrate, Beclin-1[21]. USP10 in lysates from live cells treated with either inhibitor was blocked from labeling with an HA-tagged ubiquitin probe modified to covalently label the active site cysteine of DUBs (HA-Ub-VS) with inhibitor concentrations in the low micromolar range (Figure 3a-b). In addition, protein levels of Beclin-1 and FLT3 were strongly decreased in 20 μM HBX19818-treated Ba/F3-FLT3-ITD and MOLM14 cells (Figure 3c-d). Beclin-1, like FLT3, was also strongly decreased in 10 μM P22077-treated MOLM14 cells (Figure 3e) and partially degraded in Ba/F3-FLT3-ITD cells (Supplementary Results, Supplementary Figure 4c). It should be noted that although only validated as USP10 and USP7 inhibitors, both compounds exhibit at least some degree of inhibitory activity against additional DUBs, and potentially non-DUB targets, which likely contributes to anti-proliferative effects observed at concentrations below where USP10 is well inhibited by compound.
Figure 3. Investigation of DUB targets of HBX19818 and P22077.
(a-b) Target engagement studies: Ba/F3-FLT3-ITD and MOLM14 cells were treated with the indicated concentration of compound, lysed, and incubated with 0.25 ug HA-Ub-VS for 30min at RT. The ability of compound to block USP10 labeling by HA-Ub-Vs indicates binding of the enzyme by inhibitor. These studies are representative of at least two independent studies for which similar results were observed. (c-e) Analysis of FLT3 and Beclin-1 levels in HBX19818- and P22077- treated Ba/F3-FLT3-ITD and MOLM14 cells. (f) USP10 biochemical IC50s of HBX19818 and HBX19818 analogs using Ub-AMC as substrate. The ability of each compound to promote loss of FLT3 is indicated by + or −. This study is representative of two independent studies for which similar results were observed (n=2). (g) Effects of HBX19818 and structural analogs of HBX19818 on proliferation of Ba/F3-FLT3-ITD cells following approximately 72 hours of treatment. (n=2). (h) Analysis of proliferation of 3-treated FLT3 null TF-1 cells versus FLT3-ITD-expressing MOLM13-luc+ and MOLM14 cells at 0, 5, 10, and 20 uM concentrations following 24 hr of treatment. (n=2).
USP10 has been reported as a regulator of tumor suppressor p53 localization and stability.[22] Because a drug that degrades wt p53 could be undesirable, we sought to elucidate whether pharmacological USP10 inhibition impacted p53 levels in AML cell lines expressing the transcription factor. Treatment of MOLM13-luc+ and MOLM14 cells with HBX19818 or P22077 did not result in a decrease of p53 levels and in fact, if anything a modest increase in p53 levels was observed (Supplementary Results, Supplementary Figure 3a-b). USP10 hairpin knockdown did, as expected, result in decreased levels of p53. HBX19818 and P22077 were both originally reported as USP7 inhibitors.[18, 20] USP7 stabilizes MDM2 leading to increased ubiquitylation and degradation of p53. As expected, it has been shown that pharmacological USP7 inhibition decreases MDM2 levels and increases p53 levels.[19] The USP7 inhibitory activity of the inhibitors may counteract any potential effects on p53 degradation by USP10.
As a first assessment of a potential role for USP10 in FLT3 mutant AML, we evaluated a small series of HBX19818 analogs for USP10 inhibitory activity in a biochemical assay, impact on FLT3 protein levels and anti-proliferative effects against Ba/F3-FLT3-ITD (chemical structures shown in Supplementary Results, Supplementary Figure 4d). Good correlation among these parameters was observed, supporting USP10 as the relevant target of HBX19818 (Figure 3f-g and Supplementary Results, Supplementary Figure 4e-k). For example, 9, which inhibits USP10 comparably to HBX19818 (Figure 3f) but no longer inhibits USP7 (IC50 >> 100 μM, Supplementary Results, Supplementary Table 3), was observed to suppress cell growth and induce loss of FLT3 at similar concentrations (Figure 3g, Supplementary Results, Supplementary Figure 4k). The more potent USP10 inhibitor 3 (Figure 3f) has a lower anti-proliferation EC50 and induces FLT3 degradation at lower concentrations (Figure 3g, Supplementary Results, Supplementary Figure 4e), while 4 exhibited little inhibition of USP10 in a purified enzyme assay (IC50 >>100 μM) (Figure 3f), a significantly right-shifted anti-proliferation curve and no effect on FLT3 levels at the same concentrations that HBX19818 degraded FLT3 (Figure 3g, Supplementary Results, Supplementary Figure 4f). The more potent HBX19818 analog, 3, maintained specificity for FLT3 mutant MOLM13-luc+, MOLM14, and MV4,11 cell lines relative to the FLT3 null cell line, TF-1, and other leukemia lines not driven by FLT3 (Figure 3h and Supplementary Results, Supplementary Table 2), and led to a loss in cell surface FLT3 expression (Supplementary Results, Supplementary Figure 1d). Also similar to HBX19818, two analogs, 7 and 9, primed mutant FLT3-expressing cells more strongly than wt or null FLT3-expressing cells (Supplementary Results, Supplementary Figure 2h-i).
USP10 knockdown and overexpression studies
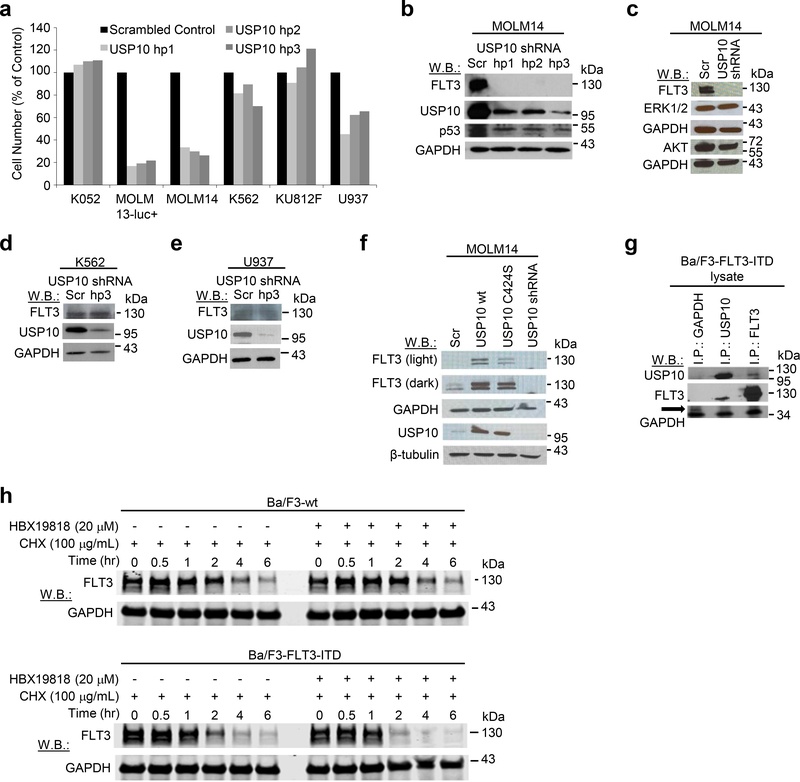
Having identified USP10 as the prime candidate for HBX19818- and P22077- induced degradation of mutant FLT3, we further investigated its role in FLT3 mutant AML by performing knockdown (KD) using three separate hairpins targeting each DUB. USP10 KD with each hairpin resulted in the robust degradation of FLT3-ITD, as well as substantial growth inhibition in FLT3-ITD-positive cells (MOLM13-luc+, MOLM14), as compared to the scrambled control hairpin (Figure 4a-c, Supplementary Results, Supplementary Figure 5a-b). As was observed for HBX19818 and P22077, USP10 KD had little to no impact on signaling molecules, including AKT and ERK1/2, downstream of FLT3 (Figure 4c). Effective USP10 KD by the same hairpins did not suppress growth of transformed human hematopoietic lines cell lines not driven by oncogenic FLT3 (K052, K562, KU812F, U937) (Figure 4a and Supplementary Results, Supplementary Figure 5c-f) and, similar to USP10-targeted small molecule inhibition, did not modulate wt FLT3 protein levels (Figure 4d-e and Supplementary Results, Supplementary Figure 5c-e). In addition, levels of USP10 were observed to be generally higher in most cell lines expressing higher levels of FLT3, including MOLM14 and MV4,11, consistent with a stabilizing role for USP10 in FLT3 protein regulation (Supplementary Results, Supplementary Figure 5g). In contrast to USP10 KD, there was little to no change in FLT3 or Beclin-1 levels observed in FLT3-ITD-expressing MOLM14 cells with USP7 KD (Supplementary Results, Supplementary Figure 6a), and transduction with the USP7 hairpins had little to no effect on cell viability as compared to the scrambled hairpin control (Supplementary Results, Supplementary Figure 6b). USP7 KD was demonstrated to be selective, as levels of USP10 decreased in USP10 KD cells but remained unchanged in USP7 KD cells (Supplementary Results, Supplementary Figure S6c-d). Furthermore, pharmacological inhibition of USP7 using the selective USP7 inhibitor, Compound 2[23], had less impact than HBX19818 on cell viability and did not lead to reduced FLT3 levels in Ba/F3-FLT3 ITD cells at concentrations up to 20 μM (Supplementary Results, Supplementary Figure 6e-g).
Figure 4. Investigation of USP10 as a mediator of FLT3-ITD and wt FLT3.
(a) Cell counts (Trypan Blue exclusion assay) determined approximately 1 week after puromycin selection of USP10 shRNA-infected cells. (n=2). (b) Effects of USP10 KD on FLT3 and p53 protein levels in MOLM14 cells. (c) Effects of USP10 KD on FLT3, AKT, and ERK1/2 protein levels in MOLM14 cells. (d-e) Effects of USP10 KD on FLT3 expression in wt FLT3-expressing K562 and U937 cells. (f) Analysis of FLT3 levels in MOLM14 cells overexpressing USP10 wt and catalytically inactive USP10, USP10C424S. Immunoblot shown is representative of 3 studies for which similar results were observed (Supplementary Figure 7A). (g) Association of endogenous USP10 with exogenously expressed FLT3-ITD in Ba/F3-FLT3-ITD cells. The GAPDH I.P. serves as a control. The arrow shown is pointing to GAPDH; the lower bands observed are likely the IgG light or L chain associated with the I.P. (h) HBX19818 shortens the half-life of FLT3-ITD to a greater extent than wt FLT3. This experiment is representative of three independent experiments for which similar results were observed (Supplementary Figure 8). CHX=cycloheximide; F/H – Flag/HA.
In the converse experiment, increased expression of USP10 was observed to correlate with higher stabilization of FLT3-ITD protein than wt FLT3 protein in stably transfected MOLM14 and transiently transfected HEK 293T cells (Figure 4f, Supplementary Results, Supplementary Figure 7a-b). It is important to note that, similar to oncogenic FLT3-driven AML cells, both HBX19818 and P22077 were able to induce degradation of FLT3 in HEK 293T cells, although approximately 2-fold higher concentrations were needed to replicate effects observed with both compounds in mutant FLT3-driven cells (Supplementary Results, Supplementary Figure 7c). Introduction of USP10 in which the catalytic cysteine has been replaced with serine, USP10C424S, into MOLM14 cells resulted in reduced stabilization of mutant FLT3 compared to wt confirming the importance of USP10 catalytic activity in regulating FLT3-ITD protein levels (Figure 4f). Taken together, the SAR, KD and overexpression studies are in strong support of USP10 being the critical regulator of FLT3-ITD stability but do not address whether the impact is direct or indirect. To answer this, we examined whether USP10 and FLT3 are in a complex in FLT3 mutant cells. We observed robust co-immunoprecipitation (co-I.P.) of USP10 with FLT3 in Ba/F3-FLT3-ITD cells; reverse co-I.P. studies confirmed the association of FLT3 with USP10 (Figure 4g). A similar interaction between USP10 and FLT3 was demonstrated in 293T cells engineered to exogenously express these proteins (Supplementary Results, Supplementary Figure 7d). Importantly, HBX19818 at 2, 4, and 6 hours and the chemokine, P22077, at 4 and 6 hours were observed to block the interaction between USP10 and FLT3-ITD (Supplementary Results, Supplementary Figure 7e).
USP10 stabilizes FLT3-ITD to a greater extent than wt
The observed differential impact on FLT3 wt and mutant protein with USP10 pharmacological inhibition and KD as well as enzyme overexpression, is consistent with reports that activated FLT3 is more prone to ubiquitin-mediated degradation[15]. We analyzed the half-life of wt FLT3 and FLT3-ITD with and without over-expression of USP10 and in the absence and presence of HBX19818, to see if differences in protein stability might play a role in the differential responsiveness of the two proteins to DUB inhibitor treatment. In Ba/F3 cells, HBX19818 shortened the half-life of FLT3-ITD from 3–4 hr to around 2 hr, and was observed to shorten the half-life of FLT3-ITD to a greater extent than wt FLT3 (Figure 4h and Supplementary Results, Supplementary Figure 8). Data are suggestive that this differential responsiveness to HBX19818 between wt FLT3 and FLT3-ITD may be due to modest differences in the inherent overall stability/half-life of these proteins.
Degradation of mutant FLT3 overcomes resistance
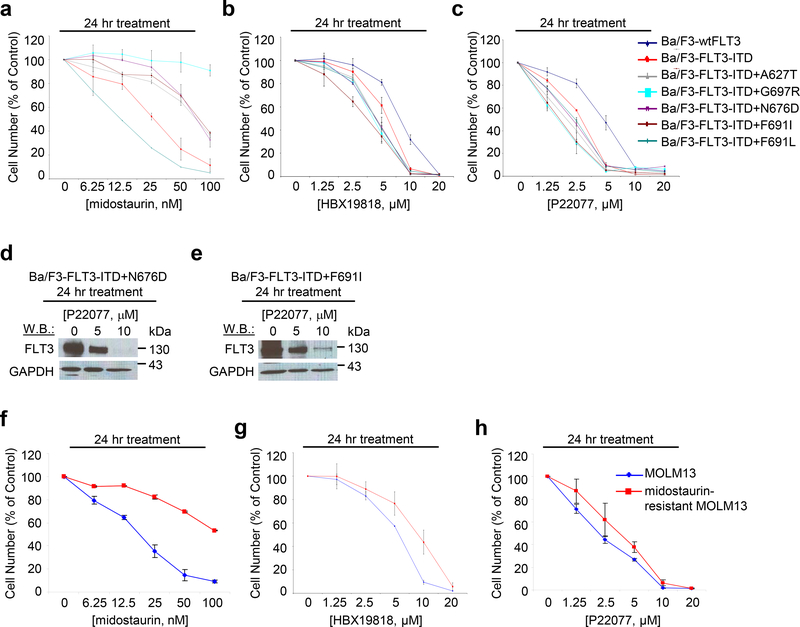
We sought to confirm that ubiquitin-mediated degradation could be advantageous compared to FLT3 kinase inhibition in terms of overriding drug resistance. Treatment of Ba/F3-FLT3-ITD cells expressing tyrosine kinase domain (TKD) point mutations with the FLT3 kinase inhibitors led to rightward shifts in the dose-response curves (Figure 5a, Supplementary Results, Supplementary Figure 9a-b), validating previously reported differential resistance to these inhibitors. In contrast, HBX19818 and P22077 treatments were equipotent against Ba/F3-FLT3-ITD cells versus Ba/F3-FLT3-ITD cells expressing the TKD point mutations, however less potent toward Ba/F3 cells engineered to over-express wt FLT3 (Figure 5b-c). Importantly, HBX19818 and P22077 induced degradation of FLT3 in the FLT3 kinase inhibitor-resistant cells at concentrations that were ineffective in promoting FLT3 degradation in Ba/F3-wt FLT3 cells (Figure 5d-e, Figure 1h, Supplementary Results, Supplementary Figure 1e, Supplementary Figure 9c-f). All TKD point mutants were confirmed to express constitutively activated FLT3 (Supplementary Results, Supplementary Figure 9g-h). In addition, HBX19818 and P22077 showed similar potency toward parental MOLM13 cells and MOLM13 cells rendered resistant to midostaurin following prolonged culture in the presence of the drug (Figure 5f-h). The midostaurin-resistant MOLM13 cells were characterized as highly over-expressing FLT3 protein, believed to contribute to their resistance.[24]
Figure 5. Targeted effects of HBX19818 and P22077 on cells resistant to FLT3 kinase inhibitors.
Approximately 24 hr treatment of Ba/F3-FLT3-ITD cells or Ba/F3-FLT3-ITD cells expressing TKD point mutants with midostaurin (a), HBX19818 (b), or P22077 (c). (n=2). (d-e) Effect of HBX19818 and P22077 on FLT3 expression in Ba/F3-FLT3-ITD cells expressing TKD point mutants following 24 hr of treatment. (f-h) Comparison of effects of midostaurin, HBX19818, and P22077 on proliferation of MOLM13 and midostaurin-resistant MOLM13 cells. (n=2).
HBX19818 synergizes with FLT3 kinase inhibitors
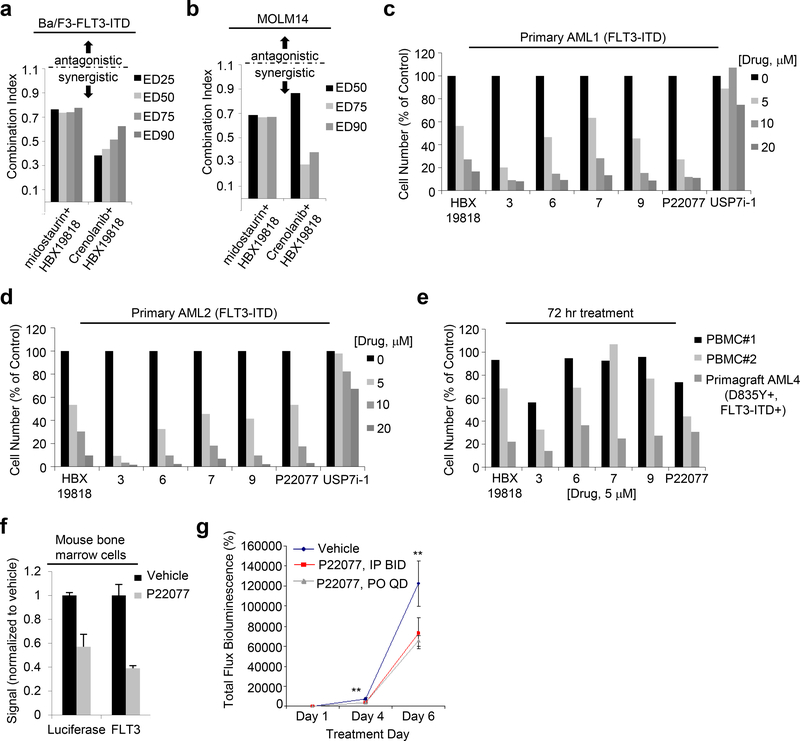
As further assessment of the therapeutic potential of USP10 inhibition, we investigated the ability of DUB inhibitors and FLT3 kinase inhibitors to interact synergistically. Specifically, a median-drug effect analysis was used, in which a combination index (CI) was calculated from growth inhibition curves using Calcusyn software (Biosoft, Cambridge, UK). Dual treatment of Ba/F3-FLT3-ITD, MOM13-luc+, and MOLM14 cells with HBX19818 and the FLT3 kinase inhibitors, midostaurin or crenolanib, at a fixed-ratio serial dilution resulted in decreased cell growth compared to single agent treatment (Supplementary Results, Supplementary Figure 9i-k). Combination index (CI) analysis indicated synergistic anti-proliferative effects (valued less than 1 indicate synergy) at 25%, 50%, 75% and 90% growth inhibition for MOLM13-luc+ cells and Ba/F3-FLT3-ITD cells and at 50%, 75%, and 90% growth inhibition for MOLM14 cells (Figure 6a,b, Supplementary Results, Supplementary Figure 9l) upon treatment with either kinase inhibitor and concomitant HBX19818 treatment.
Figure 6. Effects of combination of HBX19818 with FLT3 kinase inhibitors and targeted effects of USP10 inhibition on mutant FLT3-positive AML primary cells in vitro and in vivo.
(a) Combination indices corresponding to co-treatment of Ba/F3-FLT3-ITD cells with midostaurin or crenolanib with HBX19818. (n=2) (b) Combination indices corresponding to co-treatment of MOLM14 cells with midostaurin or crenolanib with HBX19818. (n=2) (c-d) Effects of DUB inhibitors on FLT3-ITD-expressing primary AML patient cells following approximately 72 hrs of treatment. (n=2). (e) Effects of USP10 inhibitors on normal PBMCs versus mutant FLT3-expressing AML primagraft cells (D835Y+, FLT3-ITD+) following 72 hours treatment. (n=2). (f) Correlation between luciferase-positive leukemia burden as measured by Bright Glo assay and luminoskan (left panel) and percent FLT3 as measured by flow cytometry using a CD135-PE conjugated antibody (right panel) in bone marrow samples from vehicle- versus P22077 (50 mg/kg, IP BID)-treated mice (pilot study, 4 day treatments). (n=3). (g-h) Effect of P22077 treatment on Ba/F3-FLT3-ITD-luc+ cell growth in a non-invasive in vivo bioluminescence model of leukemia. (n = 7–8) Left panel: Total flux bioluminescence plotted as a graph. Error bars represent the standard error of the mean. Right panel: Bioluminescent images of representative mice with matched starting leukemia burden. Student t-test (two-sided): Vehicle vs IP BID: Day 4 (p=0.0069212), Day 6 (p=0.1033934). Vehicle vs PO QD: Day 4 (p=0.0034501), Day 6 (p=0.0425383).
Targeting FLT3 is efficacious in AML pre-clinical models
We next evaluated the therapeutic potential of our lead USP10 inhibitor series by testing their growth inhibitory effect on primary patient tumor samples and patient derived xenografts (PDXs) ex vivo. USP10 inhibitors HBX19818, selected HBX19818 analogs, and P22077 all caused a dose-dependent reduction in survival against two each of FLT3-ITD positive patient samples and primagrafts (Figure 6c-e and Supplementary Results, Supplementary Figure S10a-d). HBX19818 was less potent toward two donor peripheral blood mononucleated cell (PBMC) samples from healthy donors while P22077 was less potent toward one of two PBMCs samples tested against (Figure 6e, Supplementary Results, Supplementary Figure 10D, Supplementary Tables 4–7). The selective USP7 inhibitor Compound 2 had little to no effect on survival of these samples (Figure 6c-d and Supplementary Results, Supplementary Figure 10a, c). Enough cells were obtained from one primagraft to enable analysis of FLT3 levels via immunoblotting and indicated a strong reduction in FLT3 following 21 hours of treatment with either HBX19818 or P22077 at a concentration of 20 μM (Supplementary Results, Supplementary Figure S10e).
These results inspired testing of P22077 in vivo, using the same FLT3-ITD+, D835Y+ AML primagraft that P22077 induced FLT3 degradation with ex vivo. Following the methodology for in vivo administration of P22077 outlined in [25], we administered vehicle (n=3) and P22077 (15 mg/kg) (n= 3) to primagraft mice IP 1X daily once disease was observed by flow cytometry. Immunoblot analysis of protein lysates from mouse bone marrow cells, collected following 21 days of treatment and pooled respectively from each treatment group, showed a strong FLT3 signal in vehicle control-treated mice that was undetectable in P22077-treated mice (Supplementary Results, Supplementary Figure 10f), suggesting drug-induced FLT3 degradation in vivo. It is important to note that P22077 at 15mg/kg was generally well tolerated for the 21 day treatment period with little change in weight (approximately 2–3g on average loss for both vehicle-treated and P22077-treated; none of the mice were below 15% weight loss).
We also tested the ability of P22077 to suppress the growth of mutant FLT3-positive cells in a second model, a non-invasive in vivo bioluminescence model. Ba/F3-FLT3-ITD-luc+ cells were first confirmed to respond to midostaurin and P22077 similar to non-luciferase-expressing cells in terms of growth suppression and FLT3 degradation (Supplementary Results, Supplementary Figure 10g-j) and DUB inhibitor-induced loss of FLT3 surface expression (Supplementary Results, Supplementary Figure 10k). In a small pilot study Ba/F3-FLT3-ITD-luc+ harboring female NCR nude mice treated with 50 mg/kg P22077 twice daily via intraperitoneal injection (n=4) for 4 days had a lower percentage (approximately 2-fold) of FLT3 expression in extracted bone marrow as measured by flow cytometry using a CD35-PE conjugated antibody in comparison to bone marrow extracted from vehicle control mice (n = 4) (Figure 6f). Aliquots of the bone marrow samples showed a similar approximately 2-fold reduction in luciferase-positive signal in P22077-treated mouse bone marrow samples as compared to vehicles (Figure 6f). Taken together, these results suggest reduction in tumor burden via on-target effects. A larger 3-arm (n=8 per arm) study was then carried out, with administration of 50 mg/kg P22077 twice daily via intraperitoneal injection, P22077 once daily via oral gavage, or vehicle to Ba/F3-FLT3-ITD-luc+ harboring female NCR nude mice. P22077 treatment was observed to lead to killing of mutant FLT3-ITD-expressing cells in vivo as measured by in vivo bioluminescence measurements, with a statistically significant decrease in leukemia burden compared to vehicle control mice noted following 4–6 days of treatment (Figure 6g-h). Importantly, there was no significant difference in weight observed between vehicle- and drug-treated mice treated for up to 11 days (Supplementary Results, Supplementary Figure 10l-m). There was also generally no evidence of vital organ toxicity in the mouse studies.
Discussion
The five-year survival rate for AML patients is only 20% [26]. The prognosis is especially poor for patients harboring FLT3-ITD mutations as these are associated with aggressive and lethal disease [27]. Treatment with FLT3 kinase inhibitors unfortunately provides responses of only short duration due to emergence of drug resistance [28]. Additionally, patients treated with FLT3 kinase inhibitors experience side-effects such as myelosuppression as a result of inhibition of wt FLT3 [29]. These limitations warrant the development of novel, targeted agents. Therapeutic targeting of mutant FLT3 by promoting its degradation as opposed to inhibition of its kinase activity is a novel approach that is potentially beneficial for overcoming resistance to current FLT3 kinase inhibitors and may prove more efficacious than kinase inhibitors by simultaneously blocking both enzymatic and scaffolding functions of FLT3.
Here, we introduce the DUB USP10 as a critical effector enzyme of tumor growth and survival in FLT3-ITD-positive AML and present two chemical classes of USP10 inhibitor that promote FLT3 degradation and confer an anti-proliferative effect in vitro and in vivo. Most studies aimed at identification of the DUB responsible for stabilization of a substrate of interest start with a genetic-based screen, typically knockdown or over-expression of individual DUBs, measuring protein levels. We, however, took a novel approach, frequently utilized in the kinase field but to the best of our knowledge not yet reported in the DUB field, and performed a screen of small molecule DUB inhibitors for ability to selectively suppress growth of mutant FLT3-expressing cells over wt FLT3-expressing cells. This novel strategy was enabled by our assembly of a DUB inhibitor library and annotation of the library for inhibitory activity across a large panel of DUBs. The top hit from our screen, HBX19818, led to striking and selective anti-proliferative effects against mutant FLT3-positive cells, which we discovered results from inhibition of USP10, a previously unreported target of the compound. Our small molecule-centered approach enabled us to not only identify a novel mechanism for regulation of FLT3-ITD but also to rapidly interrogate the translational potential of pharmacological inhibition of USP10 in mutant FLT3-driven AML in preclinical models. The data generated across multiple models strongly supports the notion that USP10 inhibition may offer a novel strategy for targeting mutant FLT3 AML clinically and has the potential to overcome kinase inhibitor resistance mechanisms.
The observed selective degradation of mutant FLT3 may offer a significant clinical advantage over FLT3 kinase inhibitors that inhibit both wt and mutant enzyme by sparing wt FLT3 in normal hematopoietic cells. Phosphorylation of the FLT3 receptor has been shown to be necessary for FLT3 ubiquitination and degradation by the E3 ubiquitin ligase CBL [16, 30] and both we, and others, observe more significant phosphorylation of mutant FLT3 than wt FLT3 in the absence of FLT3 ligand, with the highest levels of autophosphorylation observed in cells expressing both FLT3-ITD and TKD mutations. Experimentally, we observed that the half-life of FLT3-ITD following HBX19818 treatment is shorter than wt FLT3. Our data are in support of other reports showing that autophosphorylated FLT3-ITD, as compared to wt FLT3, undergoes more rapid degradation via proteasome- and lysosome-mediated pathways, where degradation was facilitated by the E3 ubiquitin ligases c-Cbl and Cbl-b [15]. Together, these data suggest that mutant FLT3 exists in a state more prone to ubiquitination than wt FLT3, and this may account for our observed selective degradation of mutant FLT3 compared to wt FLT3 by USP10 inhibition.
It is early but exciting days for the DUB inhibitor field. Approximately 40 DUB inhibitors have been reported. While the majority exhibit modest potency (micromolar range) and are polypharmacological agents [9], recent reports of potent and selective inhibitors lend confidence to the notion that high quality probes and lead therapeutics targeting the gene family can be achieved in significant number [31, 32]. As an increasing number of papers implicate DUBs in disease, efforts are also increasing to identify novel inhibitors. We believe that with development of DUB-targeting libraries, stringent compound triaging following HTSs, and success in crystallography enabling structure-guided medicinal chemistry efforts, novel probes are on the horizon.
Overall, these studies indicate that therapeutic targeting of USP10 has potent suppressive effects on FLT3-ITD positive AML including kinase inhibitor resistant FLT3 mutants. This novel approach warrants further investigation as an alternative treatment strategy for this disease. We are currently applying the same screening approach to other mutant proteins that help drive AML in subsets of patients.
Online Methods
Chemical compounds and biologic reagents
DUB inhibitors HBX19818 and P22077 were purchased from Medchem Express and dissolved in DMSO to obtain a 10 mM stock solution. HBX19818 analogs were purchased from ChemDiv and dissolved in DMSO to obtain a 10 mM stock solution. UPLC-MS analysis of all compounds was consistent with reported purity and molecular weight. Serial dilutions were then made, to obtain final dilutions for cellular assays with a final concentration of DMSO not exceeding 0.1%.
Antibodies
The following antibodies were purchased from Cell Signaling Technology (Danvers, MA): total AKT (rabbit, #9272) and total p44/42 MAPK (Erk1/2) (3A7) (mouse, #9107) were used at 1:1000. Anti-GAPDH (D16H-11) XP (R) (rabbit mAb, #5174) was used at 1:1000. Beclin-1 (rabbit, #3738) was used at 1:1000. USP10 (D7A5) (rabbit, #8501) was used at 1:1000. P53 (rabbit, #9282) was used at 1:1000. β-tubulin (rabbit, #2146s) was used 1:1000.
FLT3/Flk-2 (C-20) (sc-479) and Ub (P4D1) (mouse, sc-8017) were purchased from Santa Cruz Biotechnology, Inc., (Dallas, TX) and used at 1:1000 for immunoblotting. Anti-pTyr (mouse, clone 4G10) was purchased from Upstate Biotechnology (Lake Placid, NY) was used at 1:1000. Anti-HAUSP/USP7 antibody (rabbit, ab4080) and anti-ubiquitin antibody (rabbit, ab7780) were purchased from Abcam (Cambridge, MA) and used at 1:1000.
Cell lines and cell culture
FLT3-ITD- or FLT3-D835Y-containing MSCV retroviruses were transfected into the murine hematopoietic cell line Ba/F3 (IL-3-dependent) as previously described (Kelly et al., 2002). Nomo-1, P31-FUJ, and NB4 were obtained from Dr. Gary Gilliland. MV4,11 cells were obtained from Dr. Anthony Letai. Hel, K562, THP, U937, TF-1 and K052 cells were purchased from the American Type Culture Collection (ATCC) (Manassas, VA, USA). The human FLT3-ITD-postive AML line, MOLM14 (Matsuo et al., 1997), was obtained from Dr. Scott Armstrong, Dana Farber Cancer Institute (DFCI), Boston, MA. The human FLT3-ITD-positive AML line, MOLM-13 (DSMZ (German Resource Centre for Biological Material), was made to express luciferase fused to neomycin phosphotransferase (pMMP-LucNeo) as previously described (Armstrong et al.,2003).
All cell lines were cultured at a concentration of 2X105 to 5X105 in RPMI (Mediatech, Inc., Herndon, VA) with 10% fetal bovine serum (FBS) and supplemented with 2% L-glutamine and 1% penicillin/streptomycin. Exceptions include TF-1 and OCI-AML5 cells, which were cultured in RPMI media with 10% FBS and supplemented with 2% L-glutamine and 1% pen/strep and human GM-CSF (2 ng/mL). Parental Ba/F3 cells were cultured in RPMI with 10% FBS and supplemented with 2% L-glutamine and 1% penicillin/streptomycin and 15–20% WEHI (as a source of IL-3). All cell lines were passaged in 5% CO2 at 37oC.
Cell lines used in this study were submitted for cell line authentication within 6 months of manuscript preparation through cell line short tandem repeat (STR) profiling (DDC Medical, Fairfield, OH and Molecular Diagnostics Laboratory, Dana Farber Cancer Institute). All cell lines matched ≥80% with lines listed in the ATCC or DSMZ Cell Line Bank STR and were confirmed to be virus- and Mycoplasma-free.
PBMCs were generously provided by Dr. Steven Treon and Dr. Guang Yang.
Immunoblotting and immunoprecipitation
Preparation of protein lysates, immunoblotting, and immunoprecipitation were carried out as described previously (Weisberg et al., 2002).
Proliferation studies
The trypan blue exclusion assay was used for quantification of cells prior to seeding for CellTiter-Glo Luminescent Cell Viability assays (Promega, Madison, WI) and carried out as described previously (Weisberg et al., 2002). CellTiter-Glo Luminescent Cell Viability assays, carried out according to manufacturer instructions, were used for proliferation studies. Cell viability is reported as percentage of control (untreated) cells, and error bars represent the standard deviation for each data point.
AML patient cells
Mononuclear cells were isolated from mutant FLT3-positive AML patient samples. Cells were tested in liquid culture (DMEM, supplemented with 20% FBS) in the presence of drug. All blood and bone marrow samples from AML patients were obtained under approval of the Dana Farber Cancer Institute Institutional Review Board. Primary AML 1: Female; 59 years old; <5% bone marrow blasts; 2.6K WBC count; crit: 30; 1% peripheral blasts; previous therapy: 3+7 chemotherapy; cytogenetics: normal; mutations: IDH2 (5%), RUNX1 (15%), SRSF2 (16.8%), FLT3-ITD (24 aa). Primary AML2: Male; 69 years old; 90% bone marrow blasts; 23K WBC count; crit: 24; 5% peripheral blasts; previous therapy: azacytidine, cytarabline, high dose Ara-c; cytogenetics: normal; mutations: SRSF2 (54%), ASXL1 (46%), RUNX1 (39.4%), TET2 (ins) (46%), TET2 (point mutation) (2.8%), TET2 (del) (3.5%), FLT3-ITD (51 aa).
PEI transfection of 293T cells
Cell culture and transfection:
HEK 293T cells were cultured in DMEM containing 10% FBS, at 37°C, 5% CO2 incubator and transfected using Polyethylenimine (PEI) (Polysciences) according to the manufacturer’s instructions. Ba/F3-FLT3-ITD and MOLM14 cells were maintained in RPMI 1640 medium containing 10% FBS, at 37°C, 5% CO2 incubator.
For the endogenous ubiquitination assay, Ba/F3-FLT3-ITD or MOLM14 cells were treated with HBX19818 or P22077 or DMSO control for 4 or 24h at 0, 5, 10, 20uM, cells were collected and then lysed. Immunoprecipitation was carried out using an anti-FLT3 antibody. Immunoblots were analyzed using anti-ubiquitin or anti-FLT3 antibodies.
Drug combination studies
For synergy studies, cell viability was initially determined using the trypan blue exclusion assay to quantify cells for cell seeding. Following this, the CellTiter-Glo Luminescent Cell Viability assay (Promega, Madison, WI) was carried out to measure cell growth. Single agents were added simultaneously at fixed ratios to cells. Cell viability was expressed as the function of growth affected (FA) drug-treated versus control cells, and data were analyzed by Calcusyn software (Biosoft, Ferguson, MO and Cambridge, UK). The Calcusyn program was utilized for synergy measurement and based on isobologram generation and the method of Chou-Talalay (1984), which utilizes the median effect principle to quantify drug combination effects to determine whether the effects of agents administered together are greater than that expected from a simple addition of their individual effects. After determining the ED50 or IC50 of each drug, combinations were studied where the concentrations were multiples, or fractions, of the ED/IC50. Specifically, concentrations of DUB inhibitor and kinase inhibitor were tested alone and combined as follows: 0.25X IC50, 0.5X IC50, IC50, 2X IC50, and 4X IC50. Calcusyn program-generated combination index (CI) values allow for a quantitative measurement of synergism, where synergism is defined by a CI<1, an additive effect is defined by a CI=1, and antagonism is defined by a CI>1. Statistical analysis is automatically part of the computations.
Primagraft study
All animal studies were carried out according to protocols approved by the Dana-Farber Cancer Institute’s Institutional Animal Care and Use Committee.
Female NSG mice (6 weeks of age, Jackson Laboratories, Bar Harbor, ME) were administered either vehicle (10% DMSO, +90% D5W IP QD) (n=3) or P22077, 15 mg/kg IP QD (dissolved in 10% DMSO, + 90% D5W) (n=3) for a total of 21 days once leukemia burden reached the following levels as determined by percent double positive CD45+CD33+ cells in the peripheral blood: 2E#0 (vehicle) (3.07%), 2E#1 (vehicle) (0.34%), 2E#30 (vehicle) (1.63%), 2D#0 (P22077, 15mg/kg) (4.68%), 2D#1 (P22077, 15mg/kg) (1.5%), 2E#10 (P22077, 15mg/kg) (0.29%). Mice were sacrificed on day 21 of treatment. Bone marrow was flushed from mouse femurs, and spleens and livers were dissected and preserved first in formalin, followed 24 hours later by preservation in 70% ethanol.
All AML primagraft samples used in the studies in this manuscript were obtained through the Public Repository of Xenografts (proxe.org).
Flow cytometry
Flow cytometry was carried out as previously described, according to standard protocols (Weisberg et al., 2011). Briefly, a Fortessa flow cytometry machine equipped with FACSDiva analytical software was used for analyzing the percentage of FLT3-positive cells.
Non-invasive in vivo bioluminescence study
All animal studies were performed according to protocols approved by the Dana-Farber Cancer Institute’s Institutional Animal Care and Use Committee.
Bioluminescence imaging was performed as described previously (Weisberg et al., 2005). Briefly, for administration to female NCR-nude mice (6–8 weeks of age; Taconic, NY), virus- and Mycoplasma-free Ba/F3-FLT3-ITD-luc+ cells were washed and resuspended in 1X PBS and administered via IV tail vein injection (0.5 × 10^6 cells/250 uL). A sample size of no less than 8 mice per treatment group was chosen to ensure statistical significance. Anesthesized mice were imaged two days following IV-injection of Ba/F3-FLT3-ITD-luc+ cells to generate a baseline used to establish treatment cohorts with matched tumor burden (mice were randomized and investigators were blinded to group allocation), and total body luminescence was measured as previously described (Armstrong et al., 2003). Drug treatment commenced two days after cell injection. Mice were treated with vehicle (10% DMSO in 90% [20%] HPBCD, IP BID) (n=8), P22077 (50 mg/kg, 10% DMSO in 90% [20%] HPBCD, IP BID) (n=8), P22077 (50 mg/kg, 10% NMP in 90% PEG300) for the indicated times. Note: One vehicle mouse that showed ≥ 10-fold lower leukemia burden than the other 7 vehicle mice in the vehicle treatment group across all time points was removed as an outlier from the final statistical analysis. One P22077 (PO, QD)- treated mouse died prematurely due to technical complications unrelated to treatment and consequently was not imaged with the other 7 mice from this treatment group.
For in vivo assessment of FLT3 protein levels in vehicle-treated and P22077-treated mice, 8 female NCR-nude mice (6–8 weeks of age; Taconic, NY), were administered Ba/F3-FLT3-ITD-luc+ cells via tail vein injection as described above. Mice were imaged and randomized 2 days later to generate a baseline used to establish treatment cohorts with matched tumor burden. At this point, mice were treated with vehicle (10% DMSO in 90% [20%] HPBCD, IP BID) (n=4) or P22077 (50 mg/kg, 10% DMSO in 90% [20%] HPBCD, IP BID) (n=4) for a total of 4 days. Bone marrow cell suspensions were then analyzed for FLT3 levels by flow cytometry using a CD135-PE conjugated antibody (Cat. # IM2234U, Beckman Coulter, Marseille, France). Flow cytometry was carried out as previously described, according to standard protocols (Weisberg et al., 2011). Briefly, a FACS Fortessa flow cytometry machine equipped with FACSDiva analytical software was used for analyzing the percentage of FLT3-positive cells.
The statistical significance in bioluminescence between two groups was determined by using the two-tailed Student’s t-test. A P < 0.05 was considered to be statistically significant. The data had similar variance, and met the assumptions of the tests.
Labeling with HA-ubiquitin-vinylmethylsulfone (HA-Ub-VS)
MOLM14 cells were treated for three hours with P22077 and Ba/F3-FLT3-ITD cells were treated for 7 hours with HBX-19818. Cells were harvested, washed with PBS, and lysed in 1% NP-40, 10% glycerol, 2% sodium orthovanadate, and HALT protease inhibitor cocktail (ThermoFisher). Lysate was diluted to 50 ug in 30 uL lysis buffer with 1 mM DTT and incubated on ice for 15 minutes. 0.25 ug HA-Ub-VS was added, and the sample was gently rocked at room temperature for 30 minutes, then denatured with LDS sample buffer. 12 ug lysate was separated by SDS-PAGE, transferred to a nitrocellulose membrane, blocked in milk, and treated with a USP10 antibody ((D7A5) (rabbit, #8501) (Cell Signaling, Danvers, MA). After washing, the membrane was treated with a 780-nm IRdye goat anti-rabbit IgG (Licor) and imaged using an Odyssey scanner (Licor).
Quantitative real-time polymerase chain reaction (qPCR)
Ba/F3 cells were treated with the indicated compounds for 23 hours, then harvested and washed with PBS. mRNA was extracted using the RNEasy Mini Kit (Qiagen) and converted to cDNA using SuperScript III reverse transcriptase (ThermoFisher) and a SimpliAmp thermal cycler (ThermoFisher). Real-time PCR was carried out in a 96-well plate using TaqMan probes and a 7500 FAST Real-Time PCR system (ThermoFisher). Relative gene expression was calculated by comparison to a GAPDH reference probe.
Chloroquine Rescue
Cells were plated in 24-well plates and 25 uM chloroquine was added. After 60 minutes, the indicated concentration of HBX-19818 or P22077 was added. After 3 or 7 hours for P22077 or HBX-19818, respectively, cells were harvested, washed with 1x PBS, and lysed. 30 ug lysate was separated by SDS-PAGE, transferred to a nitrocellulose membrane, blocked in milk, and treated with a FLT3 antibody (Santa Cruz). After washing, the membrane was treated with a horseradish peroxidase-conjugated goat anti-rabbit IgG, incubated with Peirce ECL Western Blotting Substrate (ThermoFisher) and imaged in a dark room.
Protein expression and purification
A construct of human USP10 covering residues 376–798 in the pET28a vector was over-expressed in E. coli BL21 (DE3) in TB medium in the presence of 50 mg/ml of kanamycin. Cells were grown at 37°C to an OD of 0.8, cooled to 17°C, induced with 500 μM isopropyl-1-thio-D-galactopyranoside, incubated overnight at 17°C, collected by centrifugation, and stored at −80°C. Cell pellets were sonicated in buffer A (50 mM HEPES (pH 7.5), 300 mM NaCl, 10% glycerol, 10 mM Imidazole, and 3 mM BME) and the resulting lysate was centrifuged at 30,000 xg for 30 min. Ni-NTA beads (Qiagen) were mixed with lysate supernatant for 30 min and washed with buffer A. Beads were transferred to an FPLC-compatible column and the bound protein was washed with 15% buffer B (50 mM HEPES (pH 7.5), 300 mM NaCl, 10% glycerol, 300 mM Imidazole, and 3 mM BME) and eluted with 100% buffer B. Thrombin was added to the eluted protein and incubated at 4°C overnight. The sample was then passed through a HiPrep 26/10 desalting column (GE Healthcare) pre-equilibrated with buffer A without imidazole, and the eluted protein was subjected to a second Ni-NTA step to remove His-tag and Thrombin. The eluent was concentrated and passed through a Superdex 200 10/300GL column (GE Healthcare) in a buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, and 1 mM DTT. Fractions were pooled, concentrated to 20 mg/ml, and frozen at −80°C.
Ubiquitin-AMC assay
USP10 activity assay. Recombinant USP10, residues 376–798, was tested for its activity in a Ubiquitin-AMC assay in presence or absence of inhibitors. For this assay, 10 nM USP10 were pre-incubated with different concentrations of inhibitors or DMSO as a control in 50 mM HEPES pH7.6, 0.5 mM EDTA, 11 uM ovalbumin, 5 mM DTT. The reaction was incubated for 6 hours at room temperature prior to the addition of 2 uM Ubiquitin-AMC (Boston Biochem) substrate. The initial rate of the reaction was measured by collecting fluorescence data at one minute interval over 30-minute period using a Clariostar fluorescence plate reader at excitation and emission wavelength of 345 and 445 nm respectively. The calculated initial rate values were plotted against inhibitor concentrations to determine IC50 values.
MALDI TOF DUB assays
31 human DUBs were freshly diluted in the reaction buffer (40mM Tris–HCl, pH 7.6, 5mM DTT, 0.005% BSA) at different concentrations (see supplementary table). Ubiquitin topoisomers (K63, K48, K11 and M1) were diluted to 0.2 μl/μg in dimer buffer (40mM Tris–HCl, pH 7.6, 0.005% BSA) and used as substrates at a fixed concentration (1.5 μM). The enzymes were pre-incubated with the compounds for 30 min at room temperature at 10 μM final concentration. 0.48 μl of di-ubiquitin topoisomers were added to the reaction mixture to initiate the reaction. The reaction was sealed and incubated for 30 min at room temperature and stopped by adding TFA to a final concentration of 2% (v/v). 1.050 μl of each reaction was copied in a fresh plate and spiked with 0.15 μl of 16 μM 15N-ubiquitin as internal standard and mixed 1:1 with 2.5 DHAP matrix freshly prepared (7.6 mg of 2,5 DHAP in 375 ml ethanol and 125 ml of an aqueous 12 mg/ml diammonium hydrogen citrate). Reaction and matrix were mixed and 200 nl of mixture was spotted in duplicate onto MTP AnchorChip 1,536 TF (600 mm anchor, Bruker Daltonics).
Mass spectrometry data was acquired on an UltrafleXtreme MALDI-TOF mass spectrometer (Bruker Daltonics) with Compass 1.3 control and processing software. The sample carrier was taught before each analysis to optimize and centre laser shooting. Internal calibration was performed before each analysis using the 15N-Ub peak [M+H]+ average = 8,569.3). Samples were analysed in automatic mode (AutoXecute, Bruker Daltonics). Ionization was achieved by a 2-kHz smartbeam-II solid state laser with a fixed initial laser power of 60% (laser attenuator offset 68%, range 30%) and detected by the FlashDetector at detector gain of x10. Reflector mode was used with optimized voltages for reflector-1 (26.45 kV) and reflector-2 (13.40 kV), ion sources (IonSource-1: 25.0 kV, IonSource-2: 22.87 kV) and pulsed ion extraction (320 ns). An amount of 3,500 shots were summed up in ‘random walk’ and with ‘large’ smartbeam laser focus. Spectra were automatically calibrated on the 15N-Ub m/z and processed using smoothing (Savitzky–Golay algorithm) and baseline subtraction (‘TopHat’) for reproducible peak annotation on non-resolved isotope distributions: one cycle, 0.2 m/z for the width. For area calculation, the complete isotopic distribution was taken into account. An in-house made script was used to report - 15N and mono-ubiquitin areas; plotting of graphs, calculation of standard deviation and coefficient of variation (%) were processed in Microsoft Excel.
See supplementary methods for enzyme dilution and DUB topoisomerase data.
Dynamic BH3 profiling (DBP)
To determine drug-induced changes in mitochondrial priming, we performed dynamic BH3 profiling as previously described (Montero et al., 2015; Pan et al., 2014). Briefly, 0.4×106 cells/well were exposed to drug treatment for 14 hours. At the end of incubation time, cells were washed in PBS, pelleted at 500xg for 5 min and resuspended in MEB buffer. 15μl of cell suspension was added to each well of 384 well plate containing 15μl of MEB buffer containing 20 μg/mL digitonin and BH3 peptides at twice their final concentration and incubated for 60 min at 26ºC to allow mitochondrial depolarization. Peptide exposure was then terminated by adding 10μl 4% formaldehyde in PBS for 15 min, followed by neutralization with N2 buffer (1.7M Tris, 1.25M glycine, pH 9.1) for 10 min. To determine cytochrome C levels, anti-cytochrome C clone 6H2.B4 conjugated to Alexafluor 647 (BD Bioscience) was diluted 1:50 in 10X staining buffer (10% BSA, 2% Tween-20 and 0.02% sodium azide in PBS) and 10μl of this antibody containing buffer was added to each well for a final dilution of 1:400. Cells were stained overnight at 4ºC in dark and data was acquired on BD LSR Fortessa analyzer (BD Biosciences). Priming change (Δ) is calculated by comparing cytochrome C abundance in treated cells to that of DMSO treated control cells.
KD of genes by shRNA
pLKO.1puro lentiviral shRNA vector particles against USP10 and USP7 were purchased from Sigma-Aldrich (St. Louis, MO). Cells were incubated with the viral particles in the presence of 8 μg/ml Polybrene for 24 hours, and the cells were selected with 1–2 μg/ml puromycin for 72 hours. Following selection, cells were used for the studies described.
Repeat USP10 knockdown studies in MOLM14 cells: Viral particles were produced co-transfecting pLKO.1 containing shRNA or scramble (purchased from Sigma-Aldrich) together with psPAX2 (addgene#12260) and pMD2.G (addgene#12259), concentrated using LENTI-X concentrator (Clontech). MOLM14 cells were then infected in presence of 5 ug/ml polybrene and selection was started 48h post infection using 1 ug/ml puromycin.
Overexpression of USP10 wild-type and mutant in MOLM14 cells
FLAG-HA-USP10 was a gift from Wade Harper lab [Addgene (#22543)] [7]. This construct was used to create the corresponding USP10 catalytic dead construct (USP10 C424S) using site directed mutagenesis according to the manufacturer’s instruction.Viral particles were produced co-transfecting USP10 WT, C424S or control vector together with GAG/POL and VSV-G containing vectors in 293T cells, and concentrated using LENTI-X concentrator (Clontech). MOLM14 cells were then infected in presence of 5 ug/ml polybrene and selection was started 48h post infection using 1 ug/ml puromycin. Expression of exogenous USP10 was confirmed by HA blot.
Supplementary Material
Acknowledgements
We would like to thank Darwin Ye and Dr. Sarah Walker for their assistance with assessment of luciferase expression indicative of leukemia cell burden in bone marrow of mice via the Bright-Glo Luciferase Assay System (Promega, Madison, WI). Work was funded by Dana-Farber Cancer Institute Accelerator Fund (S.J.B., E.L.W.), Leukemia and Lymphoma Society (S.J.B., E.L.W.), Chleck Family Foundation (N.J.S), National Science Fellowship Graduate Research Fellowship Program (L.D), and Claudia Adams Barr Award (S.J.B., E.L.W.).
Footnotes
Competing financial interests
The authors have no competing financial interests.
Data availability statement
All data generated during this study are included in this published article (and its supplementary files) or are available from the corresponding author upon request.
References
- 1.Hershko A, The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ, 2005. 12(9): p. 1191–7. [DOI] [PubMed] [Google Scholar]
- 2.Clague MJ, Coulson JM, and Urbe S, Cellular functions of the DUBs. J Cell Sci, 2012. 125(Pt 2): p. 277–86. [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM and Fushman D, Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol, 2004. 8(6): p. 610–6. [DOI] [PubMed] [Google Scholar]
- 4.Heideker J and Wertz IE, DUBs, the regulation of cell identity and disease. Biochem J, 2015. 465(1): p. 1–26. [DOI] [PubMed] [Google Scholar]
- 5.Clague MJ, et al. , Deubiquitylases from genes to organism. Physiol Rev, 2013. 93(3): p. 1289–315. [DOI] [PubMed] [Google Scholar]
- 6.Abdul Rehman SA, et al. , MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol Cell, 2016. 63(1): p. 146–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sowa ME, et al. , Defining the human deubiquitinating enzyme interaction landscape. Cell, 2009. 138(2): p. 389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams SA, et al. , USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell, 2011. 146(6): p. 918–30. [DOI] [PubMed] [Google Scholar]
- 9.Ndubaku C and Tsui V, Inhibiting the deubiquitinating enzymes (DUBs). J Med Chem, 2015. 58(4): p. 1581–95. [DOI] [PubMed] [Google Scholar]
- 10.Atwal RS, et al. , Kinase inhibitors modulate huntingtin cell localization and toxicity. Nat Chem Biol, 2011. 7(7): p. 453–60. [DOI] [PubMed] [Google Scholar]
- 11.Li Z and Rana TM, A kinase inhibitor screen identifies small-molecule enhancers of reprogramming and iPS cell generation. Nat Commun, 2012. 3: p. 1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levis M, FLT3 mutations in acute myeloid leukemia: what is the best approach in 2013? Hematology Am Soc Hematol Educ Program, 2013. 2013: p. 220–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weisberg E, et al. , FLT3 inhibition and mechanisms of drug resistance in mutant FLT3-positive AML. Drug Resist Updat, 2009. 12(3): p. 81–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stone RM, et al. , Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N Engl J Med, 2017. 377(5): p. 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oshikawa G, et al. , c-Cbl and Cbl-b ligases mediate 17-allylaminodemethoxygeldanamycin-induced degradation of autophosphorylated Flt3 kinase with internal tandem duplication through the ubiquitin proteasome pathway. J Biol Chem, 2011. 286(35): p. 30263–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sargin B, et al. , Flt3-dependent transformation by inactivating c-Cbl mutations in AML. Blood, 2007. 110(3): p. 1004–12. [DOI] [PubMed] [Google Scholar]
- 17.Ritorto MS, et al. , Screening of DUB activity and specificity by MALDI-TOF mass spectrometry. Nat Commun, 2014. 5: p. 4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reverdy C, et al. , Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem Biol, 2012. 19(4): p. 467–77. [DOI] [PubMed] [Google Scholar]
- 19.Chauhan D, et al. , A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell, 2012. 22(3): p. 345–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altun M, et al. , Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem Biol, 2011. 18(11): p. 1401–12. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, et al. , Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell, 2011. 147(1): p. 223–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan J, et al. , USP10 regulates p53 localization and stability by deubiquitinating p53. Cell, 2010. 140(3): p. 384–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kessler BM, Selective and reversible inhibitors of ubiquitin-specific protease 7: a patent evaluation (WO2013030218). Expert Opin Ther Pat, 2014. 24(5): p. 597–602. [DOI] [PubMed] [Google Scholar]
- 24.Weisberg E, et al. , Reversible resistance induced by FLT3 inhibition: a novel resistance mechanism in mutant FLT3-expressing cells. PLoS One, 2011. 6(9): p. e25351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan YH, et al. , USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death Dis, 2013. 4: p. e867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Kouchkovsky I and Abdul-Hay M, ‘Acute myeloid leukemia: a comprehensive review and 2016 update ’. Blood Cancer J, 2016. 6(7): p. e441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martelli MP, et al. , Mutational landscape of AML with normal cytogenetics: biological and clinical implications. Blood Rev, 2013. 27(1): p. 13–22. [DOI] [PubMed] [Google Scholar]
- 28.Weisberg E, et al. , Discovery and characterization of novel mutant FLT3 kinase inhibitors. Mol Cancer Ther, 2010. 9(9): p. 2468–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warkentin AA, et al. , Overcoming myelosuppression due to synthetic lethal toxicity for FLT3-targeted acute myeloid leukemia therapy. Elife, 2014. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavagna-Sevenier C, et al. , FLT3 signaling in hematopoietic cells involves CBL, SHC and an unknown P115 as prominent tyrosine-phosphorylated substrates. Leukemia, 1998. 12(3): p. 301–10. [DOI] [PubMed] [Google Scholar]
- 31.Dexheimer TS, et al. , Discovery of ML323 as a Novel Inhibitor of the USP1/UAF1 Deubiquitinase Complex, in Probe Reports from the NIH Molecular Libraries Program. 2010: Bethesda (MD). [Google Scholar]
- 32.Baez-Santos YM, St John SE, and Mesecar AD, The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res, 2015. 115: p. 21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.