SUMMARY
Virus-mediated expression of CRISPR/Cas9 is commonly used for genome editing in animal brains to model or treat neurological diseases, but the potential neurotoxicity of overexpressing bacterial Cas9 in the mammalian brain remains unknown. Through RNA sequencing (RNA-seq) analysis, we find that virus-mediated expression of Cas9 influences the expression of genes involved in neuronal functions. Reducing the half-life of Cas9 by tagging with geminin, whose expression is regulated by the cell cycle, maintains the genome editing capacity of Cas9 but significantly alleviates neurotoxicity. Thus, modification of Cas9 by shortening its half-life can help develop CRISPR/Cas9-based therapeutic approaches for treating neurological disorders.
Graphical Abstract
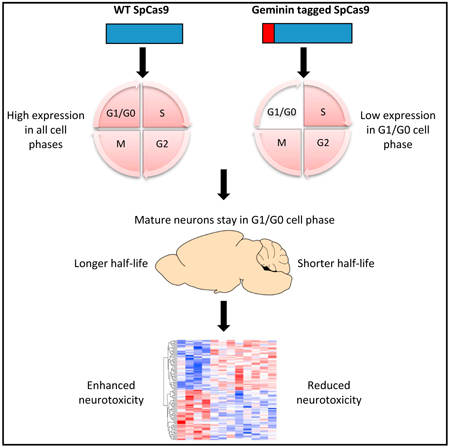
In Brief
Yang et al. use Geminin-tagged Cas9 to show that reducing the half-life of Cas9 diminishes neurotoxicity.
INTRODUCTION
CRISPR/Cas9, originally discovered as a prokaryotic defense system (Ishino et al., 1987), represents a new generation of RNA-directed genome editing technology (Doudna and Charpentier, 2014). CRISPR/Cas9 is easy to design and generate and has been successfully applied to a broad array of organisms (Hsu et al., 2014). Emerging studies indicate that CRISPR/Cas9-based genome editing approaches are able to ameliorate disease phenotypes in animal models of genetic disorders, including Duchenne muscular dystrophy (DMD) (Long et al., 2016; Nelson et al., 2016) and Huntington’s disease (HD) (Yang et al., 2017a), which spur the interests of utilizing this technology to treat genetic disorders in clinical settings. Nonetheless, all of the Cas9 proteins currently discovered, including the most commonly used type from Streptococcus pyogenes (SpCas9), consist of more than 1,000 amino acids. There are concerns that virus-mediated ectopic expression of bacterial Cas9 could elicit host responses or influence cellular functions in mammalian tissues (Chew et al., 2016; Wang et al., 2015). This is especially relevant to the CNS, as transgene expression in neurons is often achieved via viral transduction, such as adeno-associated virus (AAV), and terminally differentiated neurons may be sensitive to the overexpression of foreign proteins. Given that CRISPR/Cas9 can be used to eliminate mutant genes in the CNS for treating neurological disorders, it is imperative to clarify the influence of Cas9 on neurons and to alleviate its potentially detrimental effects.
As CRISPR/Cas9 permanently changes genomic sequence, we want to explore whether shortening its existence in the neurons may maintain its genome editing efficacy while minimizing the negative effects caused by its prolonged expression. Previous studies have generated modified versions of Cas9 with fast turnover in vitro (Gutschner et al., 2016; Tu et al., 2017) or chemically controlled split-Cas9 (Zetsche et al., 2015). For example, fusing Cas9 with Geminin, a cell-cycle regulator whose expression is absent during G0 and G1 phase (McGarry and Kirschner, 1998; Xouri et al., 2004), reduces its expression in cultured non-neuronal cells (Gutschner et al., 2016). Nonetheless, the turnover of these modified Cas9s and their genome editing capacity in the brain are unknown.
Although modifications of Cas9 can alter its half-life, two critical issues remain to be addressed. First, it is unclear whether a short-lived Cas9 still maintains its gene editing efficiency in the brain. Second, it remains to be determined whether shortening the half-life of Cas9 can alleviate its potential neurotoxicity in the brain. Based on the fact that neurons in the adult CNS stay at the quiescent phase (G0) of a cell cycle, we tested the effect of Cas9 that is fused with Geminin in the mouse brain. We found that shortening Cas9’s half-life does not affect its genome editing capacity in the mouse brain. Moreover, through RNA sequencing (RNA-seq) analysis, we found that AAV-mediated expression of Cas9 caused systemic changes in gene transcription related to neuronal functions, and such effects were attenuated by reducing its half-life.
RESULTS
Comparison of the Half-Lives of Different Cas9s In Vitro
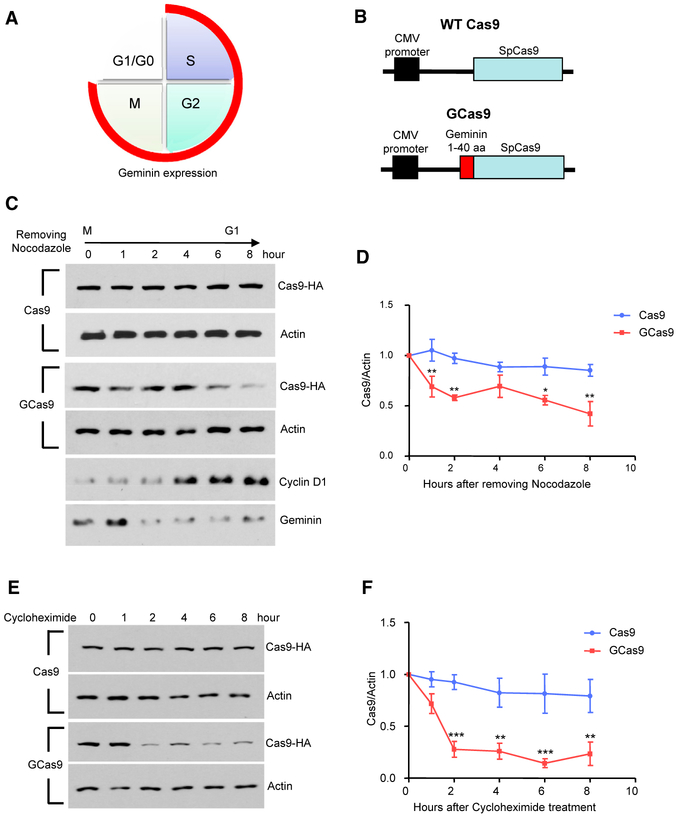
Due to the limited packaging capacity of AAV, we fused a short fragment, the first 40 amino acids of mouse Geminin, which harbors the destruction box (McGarry and Kirschner, 1998), to the N terminus of SpCas9 (GCas9) to restrict Cas9 expression in G0 and G1 phases (Figures 1A and 1B). We compared the expression of GCas9 with wild-type (WT) SpCas9 in the transfected N2a cells. The N2a cells were synchronized to the M phase by the treatment of 50 ng/mL of nocodazole overnight. On the next day, the old culture medium was replaced with fresh medium without nocodazole, and cell lysates were collected at different time points after the M phase following nocodazole removal. WT Cas9 remained stable as the cells transit from M phase to G1 phase. In contrast, the expression of GCas9 is significantly reduced in G1 phase (Figures 1C and 1D). We also measured the half-life of Cas9 after the transfected N2a cells reached full confluence via cycloheximide chase assay, in which we treated the cells with 50 μg/mL of cycloheximide, and collected cell lysates at different time points. The expression of WT Cas9 did not change 8 hr after cycloheximide treatment, whereas the expression of GCas9 dropped below 30% within 2 hr (Figures 1E and 1F). Therefore, the addition of N-terminal Geminin could facilitate Cas9 protein turnover based on the cell-cycle progression.
Figure 1. Validation of AAV-GCas9 In Vitro.
(A) Schematic representation of the expression of Geminin during cell-cycle progression. Geminin is expressed in the cycle phases indicated with red line and is degraded in the quiescent phase (G1/G0).
(B) Schematic representation of AAV-Cas9 and AAV-GCas9 constructs.
(C) The expression of Cas9 and GCas9 was examined in nocodazole-treated N2a cells at 0, 1, 2, 4, 6, and 8 hr after removing nocodazole. Actin was used as a loading control. Cyclin D1 and Geminin expression were used to indicate the progression of cell cycle from M to G1 phase.
(D) Quantitative analysis of western blotting results in (C) (n = 3; **p < 0.01; two-way ANOVA with Bonferroni post-tests). *p < 0.05.
(E) The expression of Cas9 and GCas9 was examined in N2a cells at 0, 1, 2, 4, 6, and 8 hr after cycloheximide treatment. Actin was used as a loading control.
(F) Quantitative analysis of western blotting results in (E) (n = 3; **p < 0.01; ***p < 0.001; two-way ANOVA with Bonferroni post-tests). Data are represented as mean ± SEM.
Comparison of the Genome Editing Capacities of Different Cas9s In Vivo
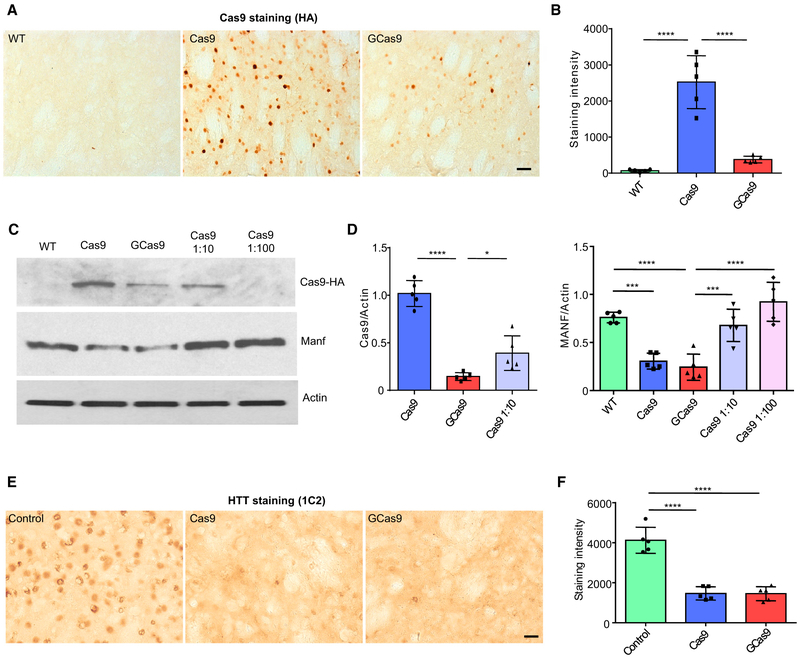
Next, we packaged WT Cas9 (AAV-Cas9) and GCas9 (AAV-GCas9) into AAV and delivered the AAVs (1 × 1013 vg) to the striatum of WT mice via stereotaxic surgery, in order to compare the expression and genome editing capacity of different versions of Cas9 in vivo. Despite the same amount of virus used, the expression of GCas9 was significantly lower than WT Cas9 two weeks after viral infection (Figures 2A and 2B), suggesting that GCas9 is less stable than WT Cas9 in the mouse brain. We then co-delivered AAV-Cas9 or AAV-GCas9 (1 × 1013 vg) with AAV-guide RNA (gRNA) (0.25 × 1013 vg) targeting manf gene. Manf is a recently identified, noncanonical neurotrophic factor, which is neuroprotective in several disease conditions, including Parkinson’s disease, ischemic stroke, and spinocerebellar ataxia 17 (Lindahl et al., 2017). We previously utilized the same manf gRNA to study the function of Manf in mouse hypothalamus (Yang et al., 2017b). Both WT Cas9 and GCas9 reduced the expression of Manf to a similar level (Figures 2C and 2D). T7E1 assay and DNA sequencing confirmed the existence of mutations in the targeted genomic region caused by both versions of Cas9 (Figures S1A and S1B). Importantly, diluting the titer of AAV-Cas9 by 10 times significantly attenuated the knockdown effect, though the expression of WT Cas9 was still higher than GCas9 (Figures 2C and 2D). This result demonstrates that the low level of GCas9 in the CNS could efficiently carry out genome editing function, which cannot be replaced by simply reducing the amount of WT Cas9 delivered. It is possible that DNA cutting requires a sufficient level of initially expressed Cas9, but once DNA has been cut, the rapid degradation of Cas9 does not influence DNA mutations that have already been generated. To further confirm the efficiency of GCas9 in genome editing, we tested its effect on the expression of mutant huntingtin (HTT) in HD knockin (HD140Q) mice and compared with WT Cas9 that has been previously demonstrated to efficiently eliminate HTT aggregates (Yang et al., 2017a), Delivery of AAV-GCas9 (1 × 1013vg) with AAV-htt-gRNA (0.25 × 1013 vg; T1 and T3 gRNAs; same as used in our previous study) was able to produce a similar level of reduction of HTT accumulation or aggregates as AAV-Cas9 (1 × 1013 vg) with AAV-htt-gRNA (0.25 × 1013 vg) in the injected striatum (Figures2E and 2F). Through DNA sequencing, we identified genomic mutations that can terminate HTT expression (Figure S1C).
Figure 2. Validation of AAV-GCas9 In Vivo.
(A) Immunohistochemistry showing the expression of Cas9 protein using hemagglutinin (HA) antibody in the striatum of wild-type (WT) mice 2 weeks after injection of AAV-Cas9 or AAV-GCas9 (scale bar: 50 μm).
(B) Quantitative analysis of immunohistochemistry results in (A) (n = 5; ****p < 0.0001; one-way ANOVA with Tukey’s post-tests).
(C) Western blot examination of the expression of Cas9 and Manf in the striatum of 3-month-old WT mice injected with AAV-Cas9, AAV-GCas9, AAV-Cas9 diluted 10 times, or AAV-Cas9 diluted 100 times. Actin was used as a loading control.
(D) Quantitative analysis of western blotting results in (C) (n = 5; *p < 0.05; ***p < 0.001; ****p < 0.0001; one-way ANOVA with Tukey’s post-tests).
(E) Immunohistochemistry showed the removal of mutant Huntingtin in the striatum of heterozygous 3-month-old HD140Q KI mice injected with AAV-Cas9 or AAV-GCas9. Mutant huntingtin was detected by 1C2 antibody. Uninjected heterozygous HD140Q KI mice were used as a control (scale bar: 20 μm).
(F) Quantitative analysis of immunohistochemistry results in (E) (n = 5; ****p < 0.0001; one-way ANOVA with Tukey’s post-tests). Data are represented as mean ± SEM. See also Figure S1.
Transcriptome Profiling of the Effects of Different Cas9s on Neuronal Functions
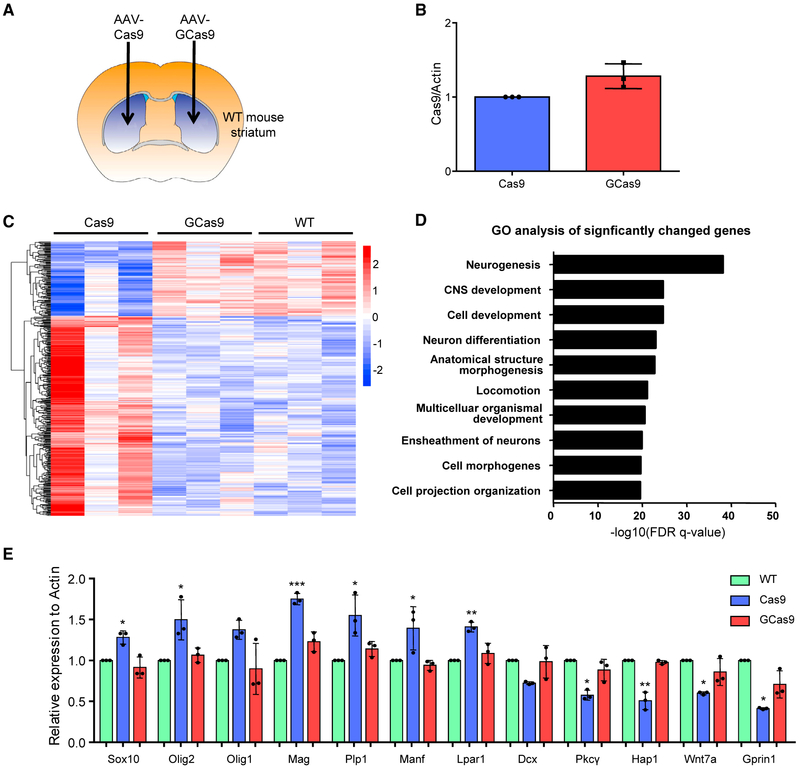
To further compare the effects of Cas9 expression on the CNS, we injected AAV-Cas9 to one side of the striatum and AAV-GCas9 to the other side (Figure 3A). Three weeks after the injection, RNA was extracted from the injected striatum. qRT-PCR showed similar levels of Cas9 and GCas9 mRNAs in the injected striatum (Figure 3B), indicating the lower level of GCas9 protein is caused by reduced protein stability. RNA-seq analysis identified 400 genes with significantly changed expression in AAV-Cas9-injected tissues, compared with AAV-GCas9-injected or uninjected WT tissues (Figure 3C). Gene Ontology (GO) enrichment analysis of the 400 genes revealed that the top 10 affected pathways are those closely linked to neuronal activities, including neurogenesis, neuron differentiation, and ensheathment of neurons (Figure 3D). In addition, a closer examination of the differentially expressed genes in protein interaction networks by the Web-based Gene Set Analysis Toolkit (WebGestalt; Wang et al., 2017) suggested that many of them are involved in chemical synaptic transmission and myelin sheath formation (Figure S2). To validate the RNA-seq analysis, we selected 12 genes for qRT-PCR, which showed consistent changes in gene expression as originally discovered in RNA-seq results (Figure 3E).
Figure 3. Effects of Cas9 Expression on Genes Involved in Neuronal Functions.
(A) Schematic representation of stereotaxic viral injection.
(B) The mRNA levels of AAV-Cas9 and AAV-GCas9 were compared three weeks after stereotaxic injection via qRT-PCR (n = 3; two-tailed Student’s t test).
(C) Heatmap view of 400 significantly changed genes in AAV-Cas9-injected striatal tissues compared with uninjected or AAV-GCas9-injected striatal tissues (n = 3; p < 0.05).
(D) Gene ontology (GO) analysis was performed on the 400 significantly changed genes. The top 10 pathways with the lowest false discovery rate (FDR) were shown.
(E) qRT-PCR was performed to compare the mRNA levels of various genes selected from the RNA-seq results (n = 3; *p < 0.05; **p < 0.01; ***p < 0.001; one-way ANOVA with Tukey’s post-tests). Data are represented as mean ± SEM. See also Figure S3.
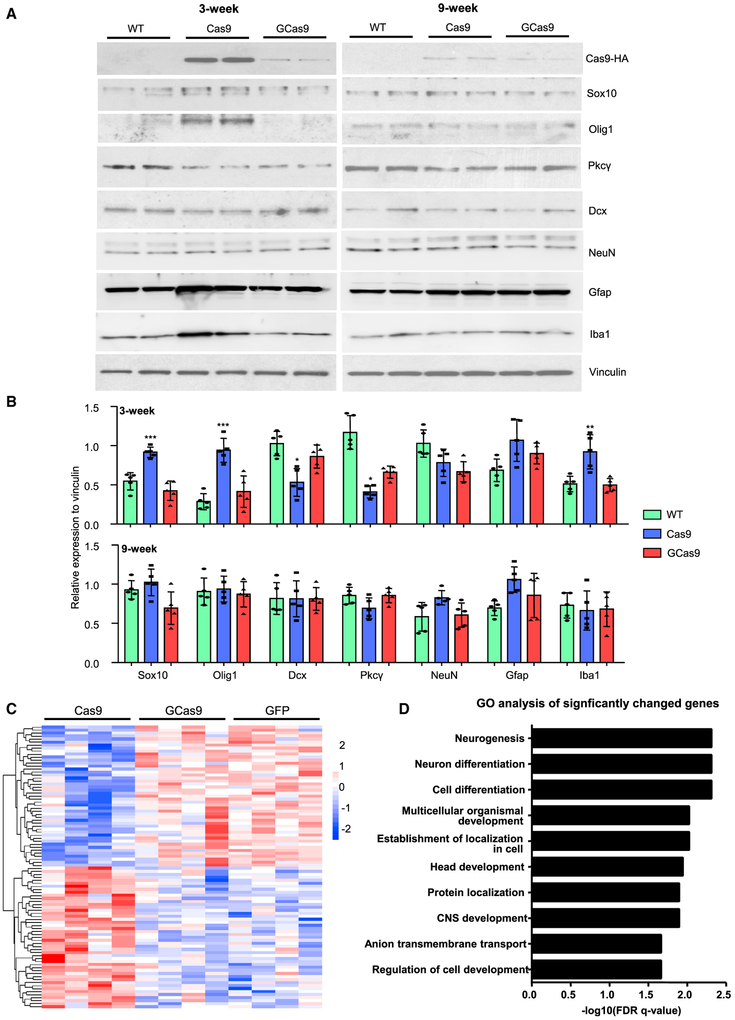
Moreover, we checked the protein level of several genes via western blotting. Three weeks after injection, the expression of Sox10 and Olig1, two transcription factors that are involved in myelination and upregulated during axon damage (Flung et al., 2015; Jessen and Mirsky, 2016; Xin et al., 2005), were significantly increased in AAV-Cas9-injected tissues, but not in AAV-GCas9-injected tissues. On the other hand, the expression of Dcx and Pkcγ, which mediate neurogenesis and neuronal survival, respectively (Couillard-Despres et al., 2005; Yang et al., 2014), were significantly reduced in AAV-Cas9-injected tissues, but not in AAV-GCas9-injected tissues (Figures 4A and 4B). We also examined the expression of NeuN, Gfap, and Iba1, which are markers for neurons, reactive astrocytes, and activated microglia, respectively. Interestingly, the expression of Iba1 was significantly increased in AAV-Cas9-injected tissues, but not in AAV-GCas9-injected tissues (Figures 4A and 4B). This result was further corroborated by Iba1 immunohistochemistry and qRT-PCR analysis of several cytokines released by activated microglia (Figures S3A–S3C). Therefore, consistent with previous studies (Chew et al., 2016; Wang et al., 2015), we found that virus-mediated expression of Cas9 could lead to immune responses in the brain, and this effect could be ameliorated by using GCas9 instead. We also tracked the effects of AAV-Cas9 expression by performing the same western blotting in striatal tissues 9 weeks after injection. At that time, Cas9 expression was much lower, and differences in protein expression between WT and Cas9- or GCas9-injected striatum were attenuated (Figures 4A and 4B).
Figure 4. Validation of RNA-Seq Results.
(A) Western blotting analysis was performed to compare the expression of selected proteins in the striatum 3 or 9 weeks after AAV-Cas9 or AAV-GCas9 injection. Vinculin was used as a loading control.
(B) Quantitative analysis of western blotting results in (A) (n = 5; *p < 0.05; **p < 0.01; ***p < 0.001; one-way ANOVA with Tukey post-tests).
(C) Heatmap view of 94 significantly changed genes in AAV-Cas9-injected striatal tissues compared with AAV-GFP- or AAV-GCas9-injected striatal tissues (n = 4; p < 0.05).
(D) GO analysis was performed on the 94 significantly changed genes. The top 10 pathways with the lowest FDR were shown. Data are represented as mean ± SEM. See also Figure S3.
Although comparison of AAV-Cas9 and AAV-GCas9 allowed us to identify altered gene expression specific to Cas9 rather than AAV infection, we included another RNA-seq experiment by using AAV-GFP-injected WT tissues as controls. Again, 94 differentially expressed genes, which fall into GO pathways, such as neurogenesis, neuron differentiation, and CNS development, were selectively altered in the Cas9-injected striatum as compared with the GFP- or GCas9-injected striatum (Figures 4C and 4D). To examine the potential off-target effects, we extracted genomic DNA from striatal tissues injected with either AAV-Cas9/AAV-manf-gRNA or AAV-GCas9/AAV-manf-gRNA for whole-genome sequencing. We found that the mutation rates in on-target and 16 most likely off-target loci were comparable between the two groups (Figure S4), suggesting that off targets are primarily determined by gRNA’s targeting specificity, and that both WT Cas9 and GCas9 have similar gene editing efficiency despite their different half-lives.
DISCUSSION
The safety issues related to Cas9 expression in mammalian cells require extra scrutiny. This issue is especially important for treating neurological diseases, as AAV injection, which enables robust transgene expression, is currently the major delivery method for expressing CRISPR/Cas9 in the brain and virus-mediated Cas9 expression has been reported to elicit host responses in liver and muscle (Chew et al., 2016; Wang et al., 2015). In this study, we systematically characterized the influence of AAV-Cas9 in the CNS and found that neuron-specific pathways were affected, indicating that indeed the ectopic overexpression of Cas9 could cause neurotoxicity.
Because neurons are postmitotic cells that are unable to proliferate and regenerate, toxic proteins are likely to accumulate in neurons to cause neuronal damages. Evidence to support this idea includes the fact that misfolded or mutant proteins preferentially accumulate in neuronal cells and affect their viability in age-dependent neurodegenerative diseases (Lim and Yue, 2015; Saxena and Caroni, 2011). Cas9 is a large bacterial protein, which may also readily accumulate in the neuronal cells and elicit detrimental effects. Indeed, our RNA-seq analysis demonstrated the adverse effects of Cas9 on the expression of neuronal genes. More importantly, the cell-cycle-dependent version of Cas9 (GCas9), although preserving the essential functions of Cas9, has a significantly shortened lifespan in the CNS, thereby alleviating the neurotoxicity. To minimize the potential toxicity of Cas9, several studies used self-destructing versions of Cas9, which remove Cas9 expression via a Cas9-specific gRNA or homologous recombination (Merienne et al., 2017; Wang et al., 2016). However, these approaches require multiple genetic modifications and additional transgene expression in a single neuron, which create another layer of risk for potential neurotoxicity. Our approach relies on post-translational modification of Cas9, which should work efficiently in all the infected neurons and perhaps other types of postmitotic cells, such as muscle cells, especially when these cells need to be transduced by Cas9 viruses.
We also found that shortening the half-life of Cas9 does not affect its gene editing efficiency. This finding suggests that gene editing is largely determined by the initial action of Cas9 rather than its prolonged expression. In support of this idea, off-target analysis showed no significant differences between unmodified Cas9 and GCas9 when the same gRNAs were used. Thus, our findings suggest that the short-lived Cas9 would be safer than the unmodified Cas9 for the future treatment of neurological diseases.
STAR ★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Xiao-Jiang Li (xli2@emory.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse lines and stereotaxic injection
Wild-type C57BL/6 (Jackson Laboratory) and HD140Q knock-in (provided by Dr. Michael Levine at UCLA) mice were housed and handled in accordance with policies of the Emory University Environmental Enrichment Program for Research Animals. The Animal Care Facility at Emory University meets the requirements of Federal Law (89-544, 91-579) and NIH regulations; the facility is accredited by the American Association for Accreditation of Laboratory Care (AAALC). The Department of Animal Resources provides veterinary care by laboratory animal veterinarians who are well trained in the laboratory use of these species. The mice (5 per cage maximum) were housed in a 12-hour light (7:00 AM - 7:00 PM) and 12-hour dark (7:00 PM - 7:00 AM) cycle at a controlled temperature (22°C), and fed ad libitum a regular chow diet.
For stereotaxic viral injection into the striatum, the mice were anesthetized with 1.5% isoflurane inhalation, and stablized in a stereotaxic instrument (David Kopf Instruments). The location of the injected site was determined according to the distance from bregma: anteriorposterior = + 0.55 mm, medial-lateral = ± 2.0 mm, dorsal-ventral = − 3.5 mm. Small holes were drilled on the skull, and a 30-gauge Hamilton microsyringe was used to deliver the virus at a speed of 100 nL per minute. Meloxicam (2 mg/kg) was given as analgesics, and after surgery the mice were placed on a heated blanket to recover from anesthetic.
Cell culture
Mycoplasma-free N2a cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 100 U ml−1 penicillin. Lipofectamine 2000 (ThermoFisher Scientific) was used for transfection. Cycloheximide and Nocodazole were purchased from Sigma.
METHOD DETAILS
Plasmid construction
To generate AAV-GCas9 plasmid, the DNA sequence corresponding to the first 40 amino acids of Geminin was cloned from mouse geminin cDNA, with primers (forward: 5′-TAACCGGTGCCACCGCCACCATGAATCCCAGTATGAAGCAGAAAC-3′; reverse: 5′-TAACCGGTTCTTCCAACAAGAGATCCAGATGC-3′). The amplified fragment was inserted into the modified PX551 plasmid containing the CMV promoter (Yang et al., 2017a) via Agel restriction digestion. The generation of AAV-manf-gRNA and AAV-htt-gRNA were described in our previous studies (Yang et al., 2017a; Yang et al., 2017b).
Virus generation
The AAV particles (AAV9 serotype, 1 × 1013 vg/ml) were packaged by the Viral Vector Core at Emory University. Briefly, AAV plasmids and AAV helper packaging plasmids were transfected into AAV293 cells. 72 hours after transfection, both the cells and culture medium were collected and lyzed, and loaded to iodixanol gradient for centrifugation. The viruses were dialyzed and concentrated using Amicon Ultra-15 centrifugal filter units. Virus titration was performed by using quantative RT-PCR.
Western blotting and immunohistochemistry
For western blotting, mouse brain tissues or harvested cells were lysed in ice-cold RIPA buffer containing Halt protease inhibitor cocktail and phosphatase inhibitors. The lysates were incubated on a rocker at 4°C for 30 min, sonicated, and centrifuged at top speed for 10 min. The supernatants were collected and subjected to SDS-PAGE. The proteins on the gel were transferred to a nitrocellulose membrane. The blot was blocked with 5% milk/PBS for 1 h at room temperature and incubated with a primary antibody in 3% BSA/PBS overnight at 4°C. After 3 washes in PBS, the blot was incubated with HRP-conjugated secondary antibodies in 5% milk/PBS for 1 h at room temperature. The blot was then washed 3 times in PBS, and developed using ECL Prime (GE Healthcare).
For immunohistochemistry, the mice were perfused with 0.9% saline and 4% paraformaldehyde sequentially, and the brains were further fixed in 4% paraformaldehyde overnight. The brains were transferred to 30% sucrose overnight, and then sectioned into 40 μm slices in cryostat. The brain sections were blocked with 3% bovine serum albumin in PBS supplemented with 0.2% Triton X-100 for 30 min at room temperature, and then incubated with primary antibodies at 4°C overnight. On the next day, the sections were washed with PBS 3 times, incubated in biotinylated secondary antibody (Vector Labs) for 2 hours, washed with PBS 3 times, incubated in VECTASTAIN ABC kit (Vector Labs) for 1 hour, washed with PBS 3 times, and developed with SIGMAFAST 3,3 Diaminobenzidine tablets (Sigma, D4293). Images were acquired with a Zeiss Axio Imager 2. ImageJ software was used to quantify the staining intensity. Briefly, the images were converted to 8-bit; the background was subtracted by using the Threshold function; and the integrated density was acquired using the Measure function. For each sample, the staining intensity was averaged from 5 randomly selected 20X images.
RNA-seq library preparation and analysis
RNA-seq analysis was conducted at the Yerkes Nonhuman Primate Genomics Core Laboratory (http://www.yerkes.emory.edu/nhp_genomics_core/). RNA was purified using QIAGEN Micro RNEasy columns, and RNA quality was assessed using Agilent Bioanalyzer. 10 ng of total RNA was used as input for mRNA amplification using 5′ template-switch PCR with the Clontech SMART-Seq v4 Ultra Low Input RNA kit, according to manufacturer’s instructions. Amplified mRNA was fragmented and appended with dual indexed bar codes using Illumina NexteraXT DNA Library Prep kits. Libraries were validated by capillary electrophoresis on an Agilent 4200 TapeStation, pooled, and sequenced on an Illumina HiSeq 3000 using (100 bp SR) at an average read depth of 20 M.
Reads were aligned to an assembly of Mus musculus (GRCm38.p3) using STAR software (v2.5.2b) (http://code.google.com/p/rna-star/). Transcripts were annotated with GENCODE M3 release (https://www.gencodegenes.org/mouse/release_M3.html). Estimates of gene-wise and isoform-wise expression levels for individual genes were performed using DESeq2(Love et al., 2014). Genes differentially expressed (p ≤ 0.05) between Cas9 and WT (or GFP), GCas9 and WT (or GFP), and Cas9 and GCas9 samples were identified using negative binomial generalized linear models, uploaded to the MSigDB Molecular Signatures Database v6.1 “Investigate Gene Sets” tool (http://software.broadinstitute.org/gsea/login.jsp;jsessionid=2C9DB5577151F391255FE9FC8ABED5EA). Annotation enrichment analysis was performed using the C5 GO_BP gene sets. Heatmaps representing log2 transformed expression values were supervised on the abscissa (samples) and organized by unsupervised hierarchical clustering on the ordinate (transcripts). Expression values of each transcript (rows) were scaled to unity standard deviation. Unsupervised hierarchical clustering of transcripts (rows) was performed Euclidean distance and Complete linkage methods.
Quantitative RT-PCR
RNA was reverse transcribed into cDNA using High Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific, 4368814). Quantitative RT-PCR was performed with Power SYBR Green PCR Master Mix (ThermoFisher Scientific, 4367659), in RealPlex4 MasterCycler (Eppendorf).
Whole genome sequencing
Genomic DNA extracted from mouse striatum was sent to GENEWIZ for library preparation and whole genome sequencing. For data analysis, Bcl2fastq v. 2.17 converted .bcl files into fastq files and performed de-multiplexing according to barcodes for the samples. Data amount and quality were examined. Using HAS 2.1, sequence reads were first aligned to the Mus musculus (Ensembl GRCm38) reference genome with the Issac Aligner. The Issac Variant Caller was used to compare the DNA sequence in the samples with the reference genome. As a result, a list of germline SNPs and small InDels (up to 50bp) were detected in the samples. After alignment was completed, structural variations, including large InDels were detected using program Manta. All VCF files (excluding gVCFs) were annotated against the GRCm38.86 ENSEMBL annotated reference genome using SnpEff. Mutations within genes were identified and their effects on gene products were reported. VCF files were further filtered to retrieve variants located in the regions of interest. The relative sequencing depth was deducted using the method described in our previous study (Yang et al., 2017a).
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were generated from three or more experiments, and the results were expressed as mean ± SEM. Statistical significances were calculated based on either two-tailed Student’s t test, one-way or two-way ANOVA. A P value < 0.05 was considered significant.
DATA AND SOFTWARE AVAILABILITY
The accession numbers for the sequencing data reported in this paper are SRA: PRJNA498233 (whole-genome sequencing) and GEO: GSE122072 (RNA-seq).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Manf | LSBio | Cat# B2688; RRID: AB_1664451 |
| Actin | Sigma | Cat# A5060; RRID: AB_476738 |
| HA | Cell signaling | Cat# 3724; RRID: AB_1549585 |
| Vinculin | Sigma | Cat# V9131; RRID: AB_477629 |
| Cyclin D1 | Cell signaling | Cat# 2978; RRID: AB_2259616 |
| Geminin | Santa Cruz | Cat# 13015; RRID: AB_2263394 |
| HTT | Millipore | Cat# MAB1574; RRID: AB_94263 |
| Sox10 | Santa Cruz | Cat# 365692; RRID: AB_10844002 |
| Olig1 | R&D system | Cat# MAB2417; RRID: AB_2157534 |
| Dcx | Santa Cruz | Cat# 8066; RRID: AB_2088494 |
| Pkcγ | Santa Cruz | Cat# 211; RRID: AB_632234 |
| NeuN | Millipore | Cat# MAB377; RRID: AB_2298772 |
| Gfap | Cell signaling | Cat# 3670; RRID: AB_561049 |
| Iba1 | Wako | Cat# 019-19741; RRID: AB_839504 |
| Iba1 | Wako | Cat# 016-20001; RRID: N/A |
| Bacterial and Virus Strains | ||
| AAV9-CMV-Cas9 | This paper | N/A |
| AAV9-CMV-GCas9 | This paper | N/A |
| AAV9-manf-gRNA | This paper | N/A |
| AAV9-htt-gRNA | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cycloheximide | Sigma | C7698 |
| Nocodazole | Sigma | M1404 |
| Deposited Data | ||
| Whole genome sequencing data | This paper | SRA: PRJNA498233 |
| RNA-seq data | This paper | GEO: GSE122072 |
| Experimental Models: Cell Lines | ||
| N2a cell | ATCC | CCL-131 |
| Experimental Models: Organisms/Strains | ||
| Wild type C57BL/6 mouse | Jackson Laboratory | 000664 |
| HD140Q KI mouse | Dr. Michael Levine | N/A |
| Oligonucleotides | ||
| See Table S1 for oligonucleotide sequences | This paper | N/A |
Highlights.
Linking N-terminal Geminin promotes the degradation of Cas9 in G1/G0 cell phase
Geminin-tagged Cas9 has a shorter half-life in mouse brain than wild-type Cas9
AAV transduction of wild-type Cas9 affects genes involved in neuronal functions
Geminin-tagged Cas9 has less impact on genes involved in neuronal functions
ACKNOWLEDGMENTS
We thank James M. Billingsley (Yerkes Nonhuman Primate Genomics Core, Emory University) and Bing Yao (Department of Human Genetics, Emory University) for their help with RNA-seq analysis. This work was supported by NIH grants in the United States (NS101701 and NS106120 to X.-J.L. and NS0405016 to S.L.). This research project was supported in part by the Viral Vector Core of the Emory Neuroscience NINDS Core Facilities grant, P30NS055077.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and one table and can be found with this article online athttps://doi.org/10.1016/j.celrep.2018.11.019.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Chew WL, Tabebordbar M, Cheng JK, Mali P, Wu EY, Ng AH, Zhu K, Wagers AJ, and Church GM (2016). A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods 13, 868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillard-Despres S, Winner B, Schaubeck S, Aigner R, Vroemen M, Weidner N, Bogdahn U, Winkler J, Kuhn HG, and Aigner L (2005). Doublecortin expression levels in adult brain reflect neurogenesis. Eur. J. Neurosci 21, 1–14. [DOI] [PubMed] [Google Scholar]
- Doudna JA, and Charpentier E (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. [DOI] [PubMed] [Google Scholar]
- Gutschner T, Haemmerle M, Genovese G, Draetta GF, and Chin L (2016). Post-translational regulation of Cas9 during G1 enhances homology-directed repair. Cell Rep. 14, 1555–1566. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, and Zhang F (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung HA, Sun G, Keles S, and Svaren J (2015). Dynamic regulation of Schwann cell enhancers after peripheral nerve injury. J. Biol. Chem 290, 6937–6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y, Shinagawa H, Makino K, Amemura M, and Nakata A (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol 169, 5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen KR, and Mirsky R (2016). The repair Schwann cell and its function in regenerating nerves. J. Physiol 594, 3521–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, and Yue Z (2015). Neuronal aggregates: formation, clearance, and spreading. Dev. Cell 32, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M, Saarma M, and Lindholm P (2017). Unconventional neurotrophic factors CDNF and MANF: Structure, physiological functions and therapeutic potential. Neurobiol. Dis 97 (Pt B), 90–102. [DOI] [PubMed] [Google Scholar]
- Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, and Olson EN (2016). Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351, 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry TJ, and Kirschner MW (1998). Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93, 1043–1053. [DOI] [PubMed] [Google Scholar]
- Merienne N, Vachey G, de Longprez L, Meunier C, Zimmer V, Perriard G, Canales M, Mathias A, Herrgott L, Beltraminelli T, et al. (2017). The self-inactivating KamiCas9 system for the editing of CNS disease genes. Cell Rep. 20, 2980–2991. [DOI] [PubMed] [Google Scholar]
- Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, et al. (2016). In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351, 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S, and Caroni P (2011). Selective neuronal vulnerability in neurode-generative diseases: from stressor thresholds to degeneration. Neuron 71, 35–48. [DOI] [PubMed] [Google Scholar]
- Tu Z, Yang W, Yan S, Yin A, Gao J, Liu X, Zheng Y, Zheng J, Li Z, Yang S, et al. (2017). Promoting Cas9 degradation reduces mosaic mutations in non-human primate embryos. Sci. Rep 7, 42081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Mou H, Li S, Li Y, Hough S, Tran K, Li J, Yin H, Anderson DG, Sontheimer EJ, et al. (2015). Adenovirus-mediated somatic genome editing of Pten by CRISPR/Cas9 in mouse liver in spite of Cas9-specific immune responses. Hum. Gene Ther 26, 432–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wei D, Zhu X, Pan J, Zhang P, Huo L, and Zhu X (2016). A ‘suicide’ CRISPR-Cas9 system to promote gene deletion and restoration by electroporation in Cryptococcus neoformans. Sci. Rep 6, 31145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Vasaikar S, Shi Z, Greer M, and Zhang B (2017). WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45 (W1), W130–W137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Yue T, Ma Z, Wu FF, Gow A, and Lu QR (2005). Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J. Neurosci 25, 1354–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xouri G, Lygerou Z, Nishitani H, Pachnis V, Nurse P, and Taraviras S (2004). Cdt1 and geminin are down-regulated upon cell cycle exit and are over-expressed in cancer-derived cell lines. Eur. J. Biochem 271, 3368–3378. [DOI] [PubMed] [Google Scholar]
- Yang S, Huang S, Gaertig MA, Li XJ, and Li S (2014). Age-dependent decrease in chaperone activity impairs MANF expression, leading to Purkinje cell degeneration in inducible SCA17 mice. Neuron 81, 349–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Chang R, Yang H, Zhao T, Hong Y, Kong HE, Sun X, Qin Z, Jin P, Li S, and Li XJ (2017a). CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J. Clin. Invest 127, 2719–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Yang H, Chang R, Yin P, Yang Y, Yang W, Huang S, Gaertig MA, Li S, and Li XJ (2017b). MANF regulates hypothalamic control of food intake and body weight. Nat. Commun. 8, 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Volz SE, and Zhang F (2015). A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol 33, 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.