Salou et al. wondered what could differentiate MAIT and NKT cells, if not for TCR specificity. Once split according to RORγt and T-bet–expressing subsets, MAIT and NKT share almost identical transcriptional programs acquired in the thymus, which result in specific tissue residency patterns.
Abstract
Mucosal-associated invariant T (MAIT) cells are abundant T cells with unique specificity for microbial metabolites. MAIT conservation along evolution indicates important functions, but their low frequency in mice has hampered their detailed characterization. Here, we performed the first transcriptomic analysis of murine MAIT cells. MAIT1 (RORγtneg) and MAIT17 (RORγt+) subsets were markedly distinct from mainstream T cells, but quasi-identical to NKT1 and NKT17 subsets. The expression of similar programs was further supported by strong correlations of MAIT and NKT frequencies in various organs. In both mice and humans, MAIT subsets expressed gene signatures associated with tissue residency. Accordingly, parabiosis experiments demonstrated that MAIT and NKT cells are resident in the spleen, liver, and lungs, with LFA1/ICAM1 interactions controlling MAIT1 and NKT1 retention in spleen and liver. The transcriptional program associated with tissue residency was already expressed in thymus, as confirmed by adoptive transfer experiments. Altogether, shared thymic differentiation processes generate “preset” NKT and MAIT subsets with defined effector functions, associated with specific positioning into tissues.
Introduction
Conventional T cells develop in the thymus before populating the secondary lymphoid organs. The naive phenotype of conventional T cells is associated with their ability to constantly patrol the lymphoid organs by circulating through the lymph and blood. The large diversity of their TCR repertoire allows them to recognize a large variety of peptide antigens presented by classical MHC molecules. After encountering their cognate antigen in the secondary lymphoid organs, specific clones of naive T cells are activated, proliferate, exit the lymph nodes or the spleen, and reach inflamed tissues through the blood. Once in tissues, these clones mediate their effector activities and clear the pathogens. Some of them may remain in large numbers for a long period of time at the infection site and are even able to divide in situ following secondary infection (Schenkel and Masopust, 2014; Mackay et al., 2016; Milner et al., 2017). These so-called tissue-resident memory (TRM) T cells represent a set of T cells with various specificities and diverse effector activities that do not recirculate through the organism (Fan and Rudensky, 2016). Indeed, in parabiosis experiments, they do not exchange between the parabionts (Steinert et al., 2015). TRM T cells have been implicated in tissue homeostasis and defense against pathogens, mostly in the context of CD8+ T cell antiviral responses (Wakim et al., 2008; Schenkel and Masopust, 2014; Fan and Rudensky, 2016; Mackay et al., 2016; Milner et al., 2017). Differentiation of effector CD8+ T cells into TRM cells requires the expression of the transcription factors Runx3 (Milner et al., 2017), Hobit, and Blimp1 (Mackay et al., 2016). The resulting transcription program allows the definition of circulatory and tissue residency gene signatures, which encompass down- and up-regulated genes, respectively (Mackay et al., 2016; Milner et al., 2017). In particular, tissue residency is associated with the loss of CD62L and CCR7 expression and the up-regulation of CD69 and CD103 (ITGAE) expression. According to the organs, TRM may express different sets of integrins and chemokine receptors (Schenkel and Masopust, 2014).
In contrast to conventional T cells, mucosal-associated invariant T (MAIT) and natural killer T (NKT) cells display a TCR repertoire of limited diversity, recognizing glycolipids (in particular α-galactosyl-ceramide; αGC) or derivatives of a microbial vitamin B2 precursor (5-amino-ribityl-uracil; 5-A-RU) presented by the nonpolymorphic MHC class Ib molecules CD1d and MR1, respectively (Bendelac et al., 2007; Franciszkiewicz et al., 2016). NKT and MAIT cells exit the thymus with some memory characteristics in mice (Cui et al., 2015; Koay et al., 2016) and expression of specific surface markers such as CD161 and IL-18Rα in humans (Martin et al., 2009; Dusseaux et al., 2011) before locating both in lymphoid organs and tissues (Godfrey et al., 2015; Legoux et al., 2017; Ben Youssef et al., 2018). While a large body of knowledge is available on NKT cell characteristics and development in the thymus and in the periphery (Bendelac et al., 2007; Seiler et al., 2012; Lee et al., 2013; Godfrey et al., 2015; Engel et al., 2016; Gapin, 2016), much less is known about MAIT cells due to their rarity in the usual laboratory mice (Cui et al., 2015) and the lack of reagent allowing their easy identification in mice until recently. Availability of MR1 tetramers loaded with a MAIT ligand (5-OP-RU; Reantragoon et al., 2013) has been pivotal in understanding more about MAIT cell development in mice (Koay et al., 2016) and in humans (Koay et al., 2016; Ben Youssef et al., 2018). Although human MAIT cells exit the thymus as naive T cells, they already exhibit a particular differentiation program with expression of the transcription factor PLZF (ZBTB16), which is also expressed by mouse MAIT cells (Martin et al., 2009; Dusseaux et al., 2011; Walker et al., 2012; Koay et al., 2016; Ben Youssef et al., 2018) and has been linked to the tissue residency properties of murine NKT cells (Savage et al., 2008; Thomas et al., 2011). Indeed, parabiosis experiments demonstrated that liver NKT cells do not circulate between parabionts, despite being localized in the vasculature, as they are tightly bound to endothelial cells (Geissmann et al., 2005) in a process requiring LFA1/ICAM1 interactions (Thomas et al., 2011). Genetic ablation of PLZF abolishes tissue targeting of NKT cells, while PLZF overexpression in conventional T cells leads to tissue residency (Savage et al., 2008). Similarly, murine MAIT cells do not acquire a memory phenotype in the absence of PLZF (Koay et al., 2016).
Contrary to human MAIT cells that simultaneously express several transcription factors (RORγt, T-bet, EOMES, and HELIOS; Leeansyah et al., 2015; Dias et al., 2017), murine MAIT cells in the thymus can be divided into two mutually exclusive subsets according to expression of RORγt and T-bet (Koay et al., 2016). This contrasts with the three NKT subsets in mice: NKT1, NKT2, and NKT17, expressing T-bet, Gata3, or RORγt, respectively (Lee et al., 2013). In C57BL/6, NKT1 cells are far more numerous than any other NKT subset, while MAIT17 cells are more abundant than MAIT1 cells both in the thymus and the periphery (Koay et al., 2016; Mak et al., 2017). Still, the characteristics and properties of MAIT subsets in the different organs at steady state remain incomplete because of the scarcity of MAIT cells in laboratory mice. Besides sharing PLZF expression, the relationships between NKT and MAIT subsets are unclear. How similar or different the differentiation programs of NKT and MAIT subsets are has not been addressed.
Taking advantage of a congenic C57BL/6 mouse strain harboring increased MAIT cell numbers, we systematically analyzed MAIT cell distribution, location, and transcriptional features in various tissues, as compared with NKT cell subsets. We report that, once split into subsets according to the expression of RORγt, mouse MAIT and NKT subsets exhibit quasi-identical transcriptional programs enriched in tissue residency genes, a pattern also found in human MAIT cells. Parabiosis experiments in mice confirmed the residency properties of almost all MAIT and NKT subsets in various organs. We further demonstrate by transfer experiments that these cells are preprogrammed in the thymus to localize and become resident in nonlymphoid and lymphoid tissues.
Results
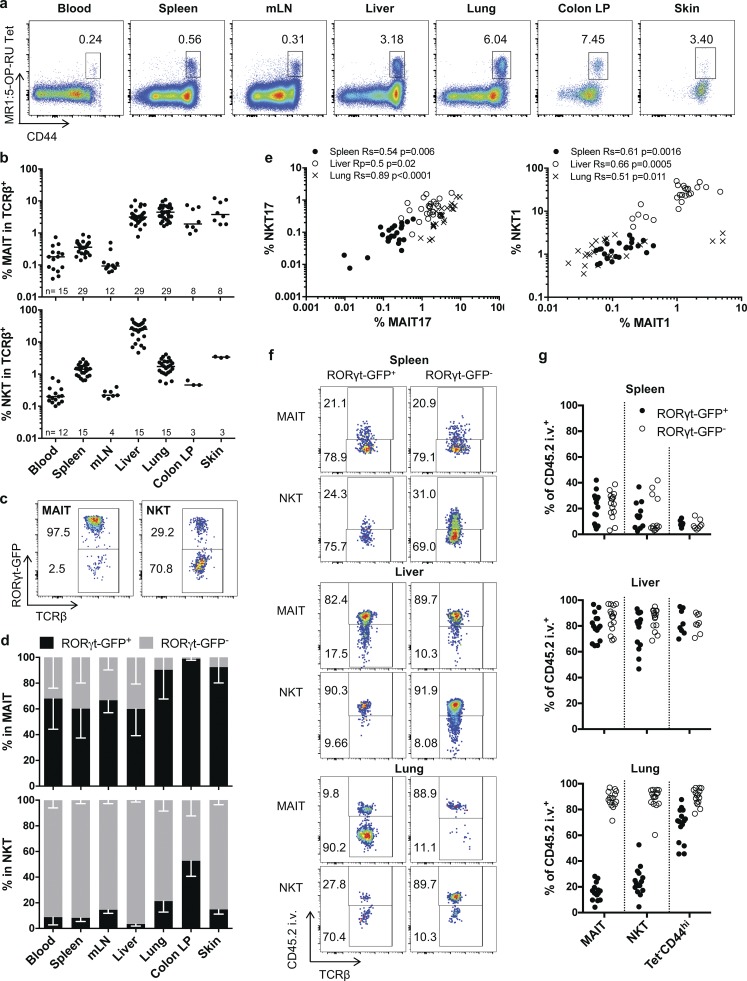
Specific but similar tissue locations of MAIT and NKT subsets
To better characterize the rules governing MAIT cell properties and tissue location, we analyzed the MAIThigh B6-MAITcast congenic strain using the recently available MR1:5-OP-RU tetramer (Reantragoon et al., 2013). In addition to having more MAIT cells than conventional laboratory mice (Cui et al., 2015), this strain expresses a RORγt-GFP reporter (Lochner et al., 2008). For comparison, we also studied NKT cells, the other subset expressing PLZF (Zbtb16). Both subsets were defined as Tet+CD44hi cells. Indeed, all the MR1:5-OP-RU tet+ cells expressed a high level of the memory marker CD44 (Koay et al., 2016) in various organs, with the exception of a very small subset of CD44lo Tet:MR1+ cells in the spleen and the mesenteric lymph nodes. Only the CD44hi subsets will be considered in the current study. As previously described (Cui et al., 2015; Koay et al., 2016), MAIT cells represent a large proportion of total T cells in the lungs, liver, colon lamina propria, and skin, but much less in the spleen and lymph nodes (Fig. 1, a and b). As compared with humans, we observed very few MAIT cells in the blood. In accordance with previous results in C57BL/6 mice (Koay et al., 2016; Mak et al., 2017), RORγt+ and RORγtneg MAIT and NKT subsets could be defined in the different organs (Fig. 1, c and d). For clarity, RORγt+ and RORγtneg subsets will be referred to as MAIT/NKT17 and MAIT/NKT1, respectively. Indeed, very few NKT2 and MAIT2 are present in the B6-MAITcast congenic strain, as shown by the very low number of PLZFhi RORγtlo cells (Fig. S1), a phenotype that defines the NKT2 subset (Lee et al., 2013). In addition, PLZFloRORγtlo NKT cells have been shown to express T-bet (Lee et al., 2013). While most MAIT cells express RORγt in mucosal organs, NKT cells are mainly NKT1, except in the colon, with a sizeable number of NKT17 cells. Most MAIT1 cells are found in the liver and the spleen.
Figure 1.
Phenotype and tissue distribution of MAIT subsets in RORγt-GfpTG B6-MAITCAST mice. (a) Representative CD44/MR1:5-OP-RU tetramer staining of TCRβ+ T cells in the indicated organs. (b) Quantification of MAIT (MR1:5-OP-RU Tet+CD44hi) or NKT (CD1d:PBS57 Tet+CD44hi) cells in TCRβ+ cells. Bar represents the median (n = 3–29 mice; at least three independent experiments, except for skin NKT cells). (c) Example of RORγt-GFP expression by NKT and MAIT cells in the lungs. (d) Proportion of MAIT (top) and NKT (bottom) cells expressing RORγt-GFP, in the same samples as in b. Mean with SD is represented. (e) Correlation between type 17 and type 1 MAIT/NKT subset frequencies in different organs (n = 24 mice in eight independent experiments). (f and g) Intra- or extravascular location of MAIT and NKT cell subsets: representative in vivo intravenous anti-CD45 staining (f) and quantification (g) on MAIT, NKT, and memory T cells (TCRβ+ MR1:5-OP-RU Tet− CD1d:PBS57 Tet− CD44hi) RORγt-GFP+ and RORγt-GFPneg subsets in the indicated organs (n = 8–15 mice in at least three independent experiments).
To consider representative patterns of NKT/MAIT subset distribution, we focused our analysis on the spleen, liver, and lungs. Contrary to NKT1, 2, and 17 subsets, for which differences in TCR Vβ repertoire have been reported (Lee et al., 2013), the proportion of Vβ6- or Vβ8-expressing MAIT cells was similar in all organs and in MAIT1 and MAIT17 subsets (Fig. S2). Interestingly, NKT1/17 and MAIT1/17 subset frequencies were highly correlated in the spleen, liver, and lungs, indicating common requirements for tissue seeding and/or the establishment of tissue populations, irrespective of antigen specificity (Fig. 1 e).
In humans, MAIT cells display a phenotype indicating tissue targeting (Dusseaux et al., 2011). To assess the intravascular or parenchymal location of the different subsets in mice, we performed a CD45 intravascular in vivo staining (Fig. 1, f and g). In accordance with the specific vascular structure of liver and spleen, as well as the known location of NKT cells in the marginal zone of the spleen and in liver sinusoids (Geissmann et al., 2005; Thomas et al., 2011; Barral et al., 2012), the vast majority of NKT and MAIT cells were labeled in the liver, whereas most of them were not in the spleen, irrespective of RORγt expression. In the lungs however, NKT/MAIT17 cells were unlabeled, contrary to most conventional memory T cells, thereby defining a parenchymal location. In contrast, both MAIT1 and NKT1 cells were intravascular (Fig. 1, f and g).
Altogether, these data indicate that MAIT and NKT cells can be found in the same peripheral organs, with inverse proportions of type 1 and 17 subsets. The RORγt versus T-bet status defines populations that are located in different lung compartments.
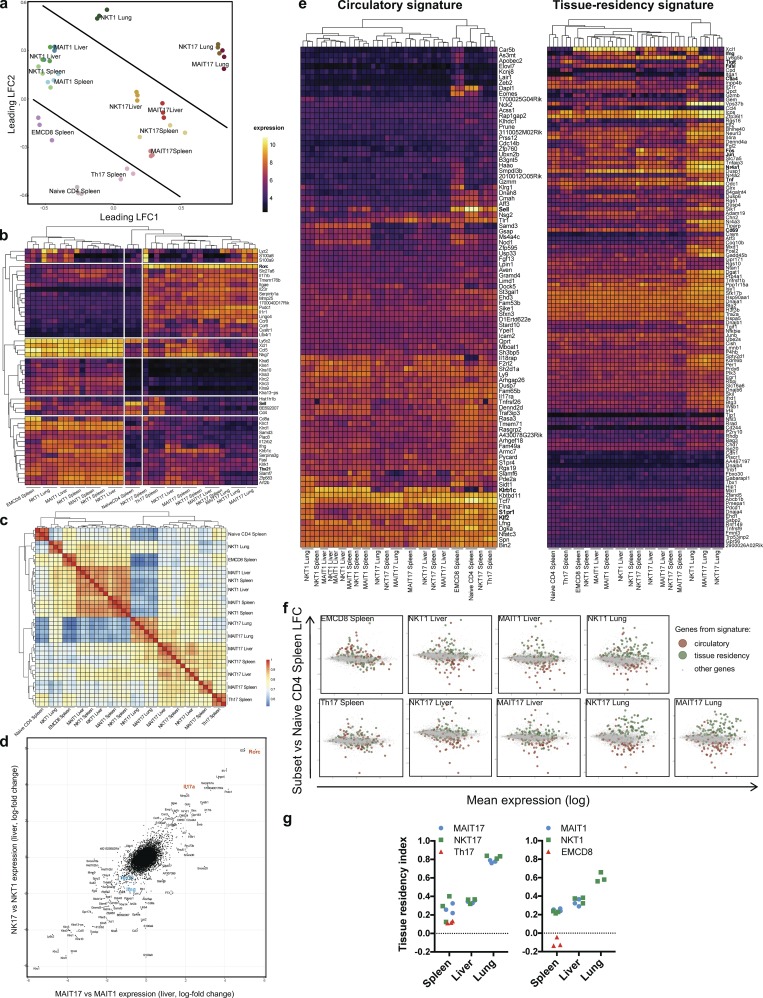
MAIT and NKT cell transcriptional programs are highly similar and further modulated by RORγt/T-bet expression and tissue location
The common features of MAIT and NKT cells, including memory phenotype, tissue location, presence of type 1 and 17 subsets, and their similar parenchymal or vascular location in various organs probably reflect a common differentiation program related to PLZF expression. To determine the nature of this program and to define the relationship between the different subsets, we analyzed the transcriptome of purified RORγt-GFPneg and RORγt-GFP+ MAIT and NKT subsets from the liver, the spleen, and the lungs, with the exception of the lung MAIT1 cells that were too few. As references, we studied Th17 (RORγt-GFP+CD4+), naive CD4+ (CD44loCD62LhiCD4+), and effector memory (EM) CD8+ (CD44hiCD62LloCD8+) T cells from the spleen.
To compare the transcriptional programs of these subsets, we used a multidimensional scaling (MDS) representation based on the euclidean distance between each pair of samples (Fig. 2 a). In this bidimensional representation, the distance is relative to the root-mean-square log-fold change (lfc) for the 10% most variable genes (1,668 genes) between each pair of samples. As expected, MAIT and NKT cells, irrespective of RORγt expression, cluster away from conventional T cell subsets (Fig. 2 a). This reflects different transcription programs between MAIT/NKT cells and conventional T cells, probably linked to PLZF expression. Moreover, type 1 or 17 subsets cluster together in all organs studied, independently of their MAIT or NKT nature, suggesting that the expression of RORγt/T-bet is one of the main determinants of the transcriptional profile of these cells. Finally, lung subsets cluster separately from the spleen/liver subsets, indicating that tissue location is associated with specific transcriptional program in the different subsets.
Figure 2.
NKT and MAIT subsets share a similar transcriptional program associated to tissue residency in peripheral organs. Gene expression of FACS-sorted subsets of MAIT, NKT, and conventional T cells was evaluated by microarray (Affymetrix) in triplicates (each replicate from pooled mice). (a) MDS scatter plot based on the 10% most variable transcripts, summarizing the euclidean distances between the cell subsets. (b) Heat map and unsupervised hierarchical clustering based on the expression of the top 50 most variable genes between samples. (c) Heat map representation and hierarchical clustering based on a correlation matrix (Pearson coefficient) from the gene expression values. (d) Scatter plot of gene expression ratios (lfc) of the type 17 and type 1 subsets in the liver illustrating the relationships between NKT and MAIT subsets. (e) Heat map and hierarchical clustering showing the expression level of genes associated to circulatory and tissue residency signatures (from Milner et al., 2017). Numerical values are depicted in Table S3. (f) Mean-difference scatter plots showing the expression ratio (lfc) of genes associated to the circulatory and tissue residency Runx3 signatures, as compared with naive CD4+ T cells. (g) Tissue residency index (see Materials and methods) evaluated in the different cell subsets.
When performing an unsupervised analysis and hierarchical clustering restricted to the 50 most variable genes for readability, the samples can be sorted in three main clusters (Fig. 2 b): (1) splenic naive CD4+ T cells; (2) RORγt-GFP+ samples, including Th17; and (3) RORγt-GFPneg NKT and MAIT subsets, alongside with the EM CD8+ T cells. The genes associated with each cluster are consistent with the sorting criteria. Indeed, naive CD4+ T cells express high level of Sell (L-selectin CD62L), an integrin involved in trafficking of lymphocytes from the blood to the lymph nodes. RORγt-GFP+ samples express high level of Rorc (RORγt), Il17rb, Il23r, Tmem176b, Ccr6, and Ccr8 genes, with a marked expression of Ccr8 in lung cells. In contrast, a potent Tbx21 (T-bet) expression is found in all RORγt-GFPneg subsets, further supporting the use of NKT1 and MAIT1 terms. These cells express Ifng and various NK cell lectin-like receptors: Klra1 (Ly49), Klrb1 (NKR-P1G), or Klrc2 (NKG2C), which may regulate cytotoxic properties, even if their precise role needs to be evaluated, especially in MAIT cells. A potent expression of the chemokines Ccl5 and Xcl1 was also found in type 1 NKT/MAIT cells, alongside with Zfp683 (Hobit), a transcription factor associated with tissue residency and known to be up-regulated by T-bet (Mackay et al., 2016). Finally, some calcium-binding proteins (S100a8 and S100a9), as well as Lyz2, show a tissue-specific expression pattern. Lyz2 has been previously reported to have a biased expression in the lungs (Yue et al., 2014). Zbtb16 (PLZF) was also very variably expressed across samples and restricted to NKT and MAIT subsets, but appears later in the list (57th most variable gene; see Table S1 a), probably because of the large proportion of NKT/MAIT samples in our dataset.
To assess the relationships between MAIT and NKT subsets in the different organs, we used a Pearson correlation method (Fig. 2 c). As expected, the expression profiles of MAIT/NKT1 cells correlated poorly with those of MAIT/NKT17 cells. However, the transcriptional patterns are highly correlated for cells from the same tissue and type 1 or 17 status. More specifically, cells from the lungs, or from the liver and the spleen show a strong correlation, suggesting a program specific to the lungs or to the liver/spleen.
To further estimate the proximity between NKT and MAIT subsets in peripheral tissues, irrespective of the program imparted by RORγt/T-bet expression, we used two approaches. First, we performed a graphical representation in which we compared the gene expression ratio of type 1 and 17 subsets for both MAIT and NKT cells. A more formal approach was also used, with a two-way ANOVA on factors “cell type” (MAIT/NKT) and “immune bias” (RORγt-GFP+ or RORγt-GFPneg). Both methods require a complete set of data among all the considered factors. In the absence of MAIT1 sample from the lungs, only liver samples were studied. A clear linear relationship of the RORγt/T-bet bias between liver MAIT and NKT cells was observed, with very few genes apart, strongly suggesting that MAIT and NKT are very similar cells (Fig. 2 d). MAIT/NKT17 had a high expression ratio of Rorc, Il17a, Pxdc1, and Mmp25. In contrast, MAIT/NKT1 expressed high levels of Tbx21, Ifng, Slamf7, and Mmp9. Among the 17,504 genes expressed by at least one sample, the two-way ANOVA identified only 36 genes differentially expressed according to the cell type factor (with a standard adjusted P value threshold of 0.05; Table S1, b and c), among which 28 did not vary according to the RORγt/T-bet factor. 19 genes were more expressed in MAIT cells (MAIT versus NKT factor, lfc > 1), including Cd8a, Ccl3, Ccl4, Itgae, Klrb1b, and Klra6, but also hepatocyte-specific genes such as Alb (albumine), BC021614 (gluthatione transferase), or Mup12 (major urinary protein) that may be related to higher hepatocytes contamination in the MAIT17 samples. Nine genes were more expressed in NKT cells (MAIT versus NKT factor; lfc < −1), including Il4 and Cd4, consistent with the known features of NKT cells as compared with MAIT cells. Interestingly, Xcl1 and Zfp683 (Hobit) were more expressed in NKT cells than in MAIT cells, as if NKT cells were more type 1–imprinted than MAIT cells. These results suggest that the major sources of variation between MAIT and NKT cells were mostly the consequence of technical artifacts. Consequently, once split according to type 1 or type 17 and taking into account the tissue location, MAIT and NKT cells are almost identical transcription-wise, if not for their TCR specificity.
MAIT and NKT subsets exhibit a tissue residency transcriptomic signature in the different organs
The location of MAIT and NKT cells in tissues (Fig. 1, f and g), as well as their expression of PLZF (Fig. S1 a; Thomas et al., 2011) and the strong impact of the tissue on the transcriptional program (Fig. 2, a and c), suggest that they reside in tissues at steady state. Our transcriptome analysis enabled us to characterize the basis of tissue residency of type 1 and 17 MAIT/NKT subsets in the spleen, liver, and lungs by evaluating the expression of the core circulatory and Runx3-dependent tissue-resident signatures described in a recent work by Milner et al. (2017). Notably, these signatures are based on those generated by comparing circulating and tissue-resident antiviral effector CD8+ T cells (Wakim et al., 2012; Mackay et al., 2016; Milner et al., 2017). The latter subset does not express PLZF in mice and resides in tissues following an infection. In contrast, no evidence of MAIT and NKT cell priming has been described so far in the literature.
We first looked at the expression levels of the different genes from the two signatures in our different subsets (Fig. 2 e). Hierarchical clustering shows that the circulatory signature is less expressed in all NKT and MAIT subsets than in the conventional T cells from the spleen (naive CD4+, Th17, and EM CD8+). More specifically, MAIT and NKT subsets have a decreased expression of Klf2, Sell, S1pr1, Aff3, and Eomes (Fig. 2 e). In contrast, all NKT and MAIT cell subsets in the lungs express high levels of most of the tissue residency signature genes (98% are up-regulated), including markers of a direct TCR activation (Jun, Fos, Junb, Nfkbie, and Nr4a1). Some notable exceptions are found, with decreased expression in the NKT/MAIT17 subsets of several effector genes (Xcl1, Fasl, Ifng, and Ccl4), suggesting that RORγt expression modulates the residency program or that the residency core signature also includes type 1 effector molecules. More precisely, a gradient can be observed on our cell subsets with an increased tissue residency signature (including Cd69) and a decreased circulatory signature from splenic conventional T cells, to splenic/liver MAIT/NKT cells, and then lung MAIT/NKT cells, irrespective of RORγt expression. This is shown on mean-difference scatter plots (Fig. 2 f) and was quantified by a tissue residency index among samples (see Materials and methods; Fig. 2 g). Indeed, MAIT and NKT exhibit a higher residency index than conventional T cells, and although this index is similar between splenic and liver MAIT and NKT subsets, it strongly increases for the lung subsets, independently of the RORγt status. Altogether, these transcriptional data strongly suggest that, as previously described for liver NKT cells, MAIT cells are tissue-resident cells. This is even more pronounced in the lungs, suggesting a higher differentiation in this organ as compared with the spleen or the liver.
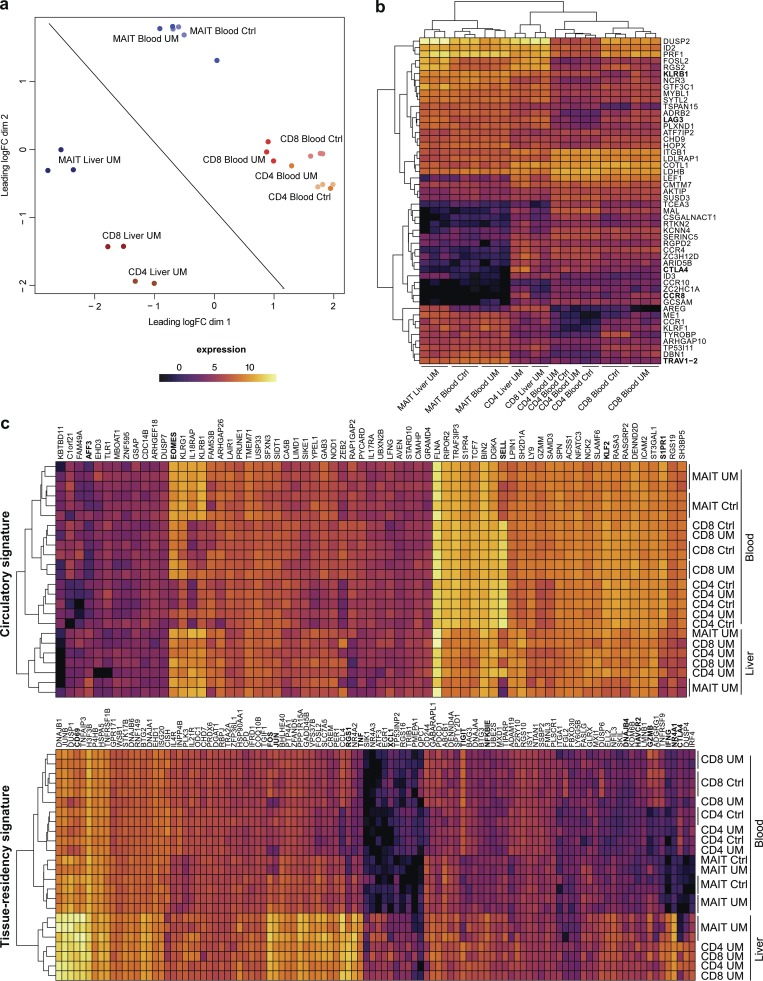
Human MAIT cells express a tissue-resident transcription program in the liver
At variance with what is observed in mice, no distinct MAIT1 or MAIT17 subsets exist in human blood, as all MAIT cells express a variety of transcription factors such as RORγt, T-bet, Eomes, and Helios (Leeansyah et al., 2015; Dias et al., 2017). However, they are also found in tissues in which they have specific effector functions (Gibbs et al., 2017; Salou et al., 2017). To determine whether a tissue residency program is also operating in humans, we analyzed the transcription pattern of MAIT cells as compared with conventional memory (CD45RA−CD27+) CD4+ and CD8+ T cells from human blood and liver. Since the paired samples of blood and liver cells were obtained from patients operated for metastatic uveal melanoma (UM; liver samples from a nonmetastatic liver fragment), we first checked whether subsets from the blood of patients were similar to those of healthy controls (Fig. 3 a). All cell subsets from both healthy and UM patients have very similar expression programs, as shown by their close proximity on the MDS plot. Consistent with previous reports (Turtle et al., 2011; Walker et al., 2012), the transcriptional program of blood MAIT cells was very different (distant) from that of conventional memory T cells. The great proximity in the MDS plot of memory CD4+ and CD8+ T cells, which are already known to have very different programs, further highlights the specific program of blood MAIT cells. As expected, blood MAIT cells expressed the transcription factors RORC (lfc = 1.82; blood MAIT vs. blood memory conventional cells), IKZF2 (Helios; lfc = 1.56), EOMES (lfc = 2.11), TBX21 (T-Bet; lfc = 1.73), and ZBTB16 (PLZF, lfc = 3.04) with the resulting expression of IL23R (lfc = 3.36), IL18RAP (lfc = 3.46), IFNGR1 (lfc = 1.51), IL12RB2 (lfc = 1.98), KLRF1 (lfc = 4.30), and KLRG1 (lfc = 1.86; Fig. 3 b and Table S2 a).
Figure 3.
Human MAIT cells express a tissue residency signature in the liver. (a) MDS scatter plot of gene expression by MAIT subsets as compared to conventional subsets in liver and blood. (b) Heat map and unsupervised hierarchical clustering based on the expression of the top 50 differentially expressed genes between blood MAIT and conventional T cells. (c) Heat map displaying expression of the human orthologs of the circulatory and tissue residency Runx3 core gene signature in the human samples.
Interestingly, the shift in the MDS plot for the liver samples as compared with blood samples was similar for all subsets, suggesting that the tissue location is associated with similar changes in gene expression pattern. As such, we observed in all cell subsets a potent induction of genes associated to T cell activation (NR4A1 all subsets in liver vs. all subsets in blood lfc = 9.17, NR4A2 lfc = 6.97, PDCD1 lfc = 2.23, and TNF lfc = 4.91) or direct TCR signaling (NFKBIA lfc = 4.20, JUN lfc = 4.43, and FOS lfc = 5.33). Most of these genes have orthologs in the tissue residency signature defined in mice (Fig. 3 c). Accordingly, in all subsets, RUNX3 (lfc = 2.01) and CD69 (lfc = 4.28) are more expressed in the liver than in the blood, supporting an increased tissue-resident program expression in the liver. In fact, on the MDS plot, the MAIT and CD8+ T cells are closer in the liver than in the blood, indicating converging transcription programs in the tissue. Surprisingly, ZBTB16 (PLZF) was expressed by liver CD4+ and CD8+ T cells, showing that induction of this transcription factor can occur outside the thymus in humans. Finally, CTLA4 and TIGIT, negative costimulatory molecules, were much less expressed in blood and liver MAIT cells than in conventional T cells, suggesting that MAIT cells may be less sensitive to inhibitory signals or that the negative feedback loops operating in MAIT cells are different from those of conventional T cells.
However, liver MAIT cells harbor specific tissue homing characteristics as compared with other liver T cells (Table S2 b). In particular, several chemokine receptors are strongly down-regulated (CCR4 lfc = −5.09, CCR8 lfc = −9.77, CXCR3 lfc = −3.74, CX3CR1 lfc = −4.60, CCR10 lfc = −4.02, and CXCR5 lfc = −2.20). Moreover, SELL is less expressed in MAIT cells (circulatory signature, CD62L lfc = −2.73) while CD69 is more expressed (tissue residency signature, lfc = 1.02). These results suggest that liver MAIT cells are less prone to recirculate than conventional memory T cells. Altogether, these data suggest that human MAIT from the liver have characteristics of tissue-resident T cells.
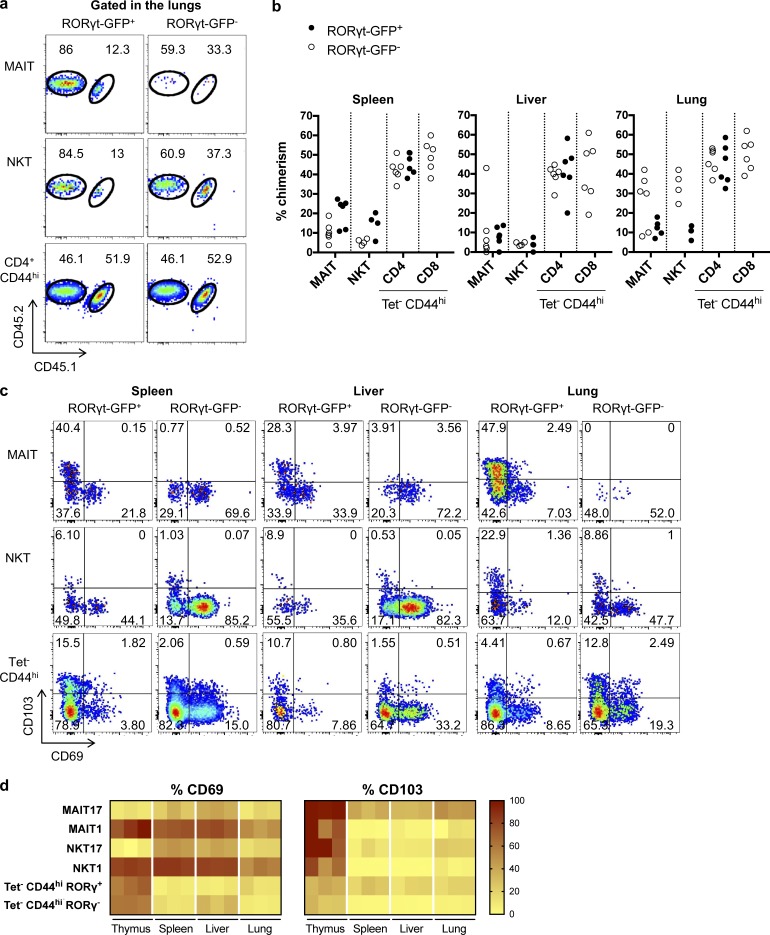
MAIT cells are tissue-resident memory cells that do not recirculate in parabiotic mice
To assess whether MAIT cells are truly tissue resident, as suggested by the transcriptional data, we generated parabiotic mice and studied the exchange of NKT, MAIT, and control CD44hi Tet− T cells in the spleen, the liver, and the lungs between the CD45.2 and CD45.1/2 parabionts. Both RORγt+ and RORγtneg conventional T cells reached equilibrium between the parabionts after 5 wk, while NKT and MAIT subsets from all the tissues did not exchange much, except for the NKT/MAIT1 subsets in the lungs (Fig. 4, a and b). Notably, despite their extravascular location, spleen NKT/MAIT17 cells exchanged a little bit more than the liver intravascular ones (Fig. 1, f and g). In all organs studied, two molecules associated with tissue residency, CD69 and CD103, were expressed by a large proportion of MAIT and NKT cells. Surprisingly, however, their expressions were mutually exclusive: CD103 was preferentially expressed by the NKT/MAIT17 subsets in accordance with the transcriptional data, whereas CD69 was more expressed by the NKT/MAIT1 subsets (Fig. 4, c and d). Interestingly, CD103 (encoded by Itgae) is among the few genes that are more expressed in MAIT17 as compared with NKT17 both at the RNA (Fig. 2 b) and protein level (Fig. 4 c). To date, the origin of this difference is unknown.
Figure 4.
MAIT cells are tissue-resident T cells. (a and b) CD45.1/2-congenically marked animals were linked as parabiotic pairs for 5 wk: example of staining in the lungs (a) and quantification in the indicated organs (b; n = 4–6 mice analyzed from three pairs). (c and d) CD69/CD103 expression by MAIT/NKT subsets as compared with conventional T cells: example of staining (c) and quantification (d; n = 3 mice; representative experiment).
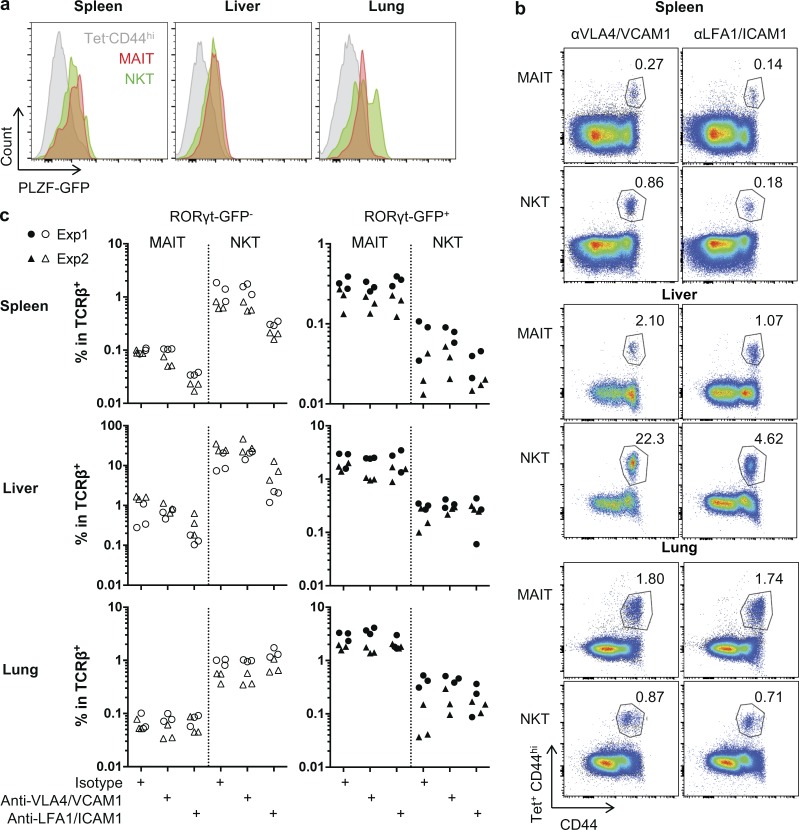
The mechanisms leading to tissue residency are unclear. Expression of PLZF has been associated with tissue residency for liver NKT cells (Geissmann et al., 2005; Savage et al., 2008; Thomas et al., 2011). Using a GFP-reporter mouse crossed onto the B6-MAITcast congenic strain, we showed that PLZF is expressed by all NKT/MAIT subsets (Fig. 5 a). Because LFA1/ICAM1 interactions are required for NKT attachment to the liver endothelium (Thomas et al., 2011), we studied NKT/MAIT subset frequencies in the different organs after blocking this interaction using anti-LFA1/ICAM1 antibodies. The frequency of NKT/MAIT1 subsets decreased by up to 10-fold in the liver and the spleen, but not in the lungs (Fig. 5, b and c). Of note, the anti-LFA1/ICAM1–induced decrease of NKT/MAIT1 cell frequencies in the spleen was observed, despite their extravascular location. The slight decrease observed in the spleen and the liver for NKT17 cells and in the lungs for NKT/MAIT17 cells was not constant from one experiment to another (Fig. 5, b and c). Thus, LFA1/ICAM1 interactions are important only for NKT/MAIT1 subset location in the spleen and liver, but the mechanisms of tissue retention of the other subsets are probably different.
Figure 5.
LFA1/ICAM1-dependent tissue retention of MAIT1 and NKT1 subsets. (a) PLZF expression by MAIT and NKT cells in the different organs using a GFP-PLZF reporter mouse crossed onto the B6-MAITCast background. (b and c) LFA1/ICAM1-blocking antibodies were injected in vivo 24 h before subset quantification: example of staining (b) and quantification (c; n = 6 mice; two independent experiments).
MAIT cells acquire their tissue-targeting properties in the thymus
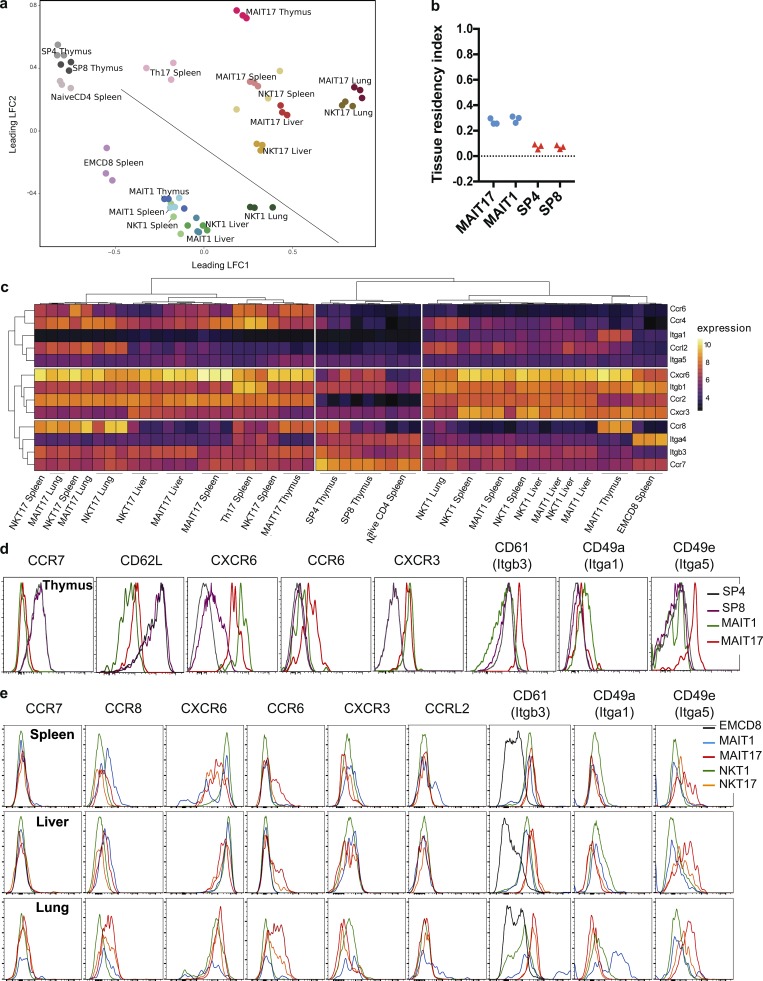
In mice, contrary to conventional T cells, MAIT and NKT cells acquire a memory phenotype (CD44hi) in the thymus in relation with PLZF expression (Savage et al., 2008; Koay et al., 2016). To assess whether MAIT subsets acquire tissue targeting properties in the thymus, we analyzed the transcriptome of purified MAIT17 (RORγt+) and MAIT1 (RORγtneg) from the thymus as compared with conventional mature (TCRβ+HSAlow) CD4+ and CD8+ T cells. Unsupervised analysis of the 10% most variable genes evidenced that MAIT1 and MAIT17 are very different from conventional CD4+ and CD8+ T cells, which cluster together, despite their known distinct functional characteristics. Moreover, MAIT1 and MAIT17 are also very different from one another (Fig. 6 a). The circulatory signature was significantly less expressed in MAIT1 or MAIT17 cells as compared with CD4+ or CD8+ conventional T cells, whereas the tissue residency signature was slightly up-regulated in the MAIT subsets (Fig. 6 b; Table S3). This was associated with an increase in the tissue residency index (Fig. 6 b), indicating an early induction of the tissue residency program in MAIT cells during their thymic differentiation. Accordingly, thymic MAIT cell subsets expressed low levels of CCR7 and CD62L, both at the transcriptional and the protein level (Fig. 6, c and d), suggesting an inability to efficiently reach lymph nodes. These data show that MAIT cells acquire their resident phenotype before leaving the thymus. To understand the basis of location in the different organs, we focused the analysis on integrins, selectins, and chemokine receptors differentially expressed between conventional T cells and among MAIT/NKT subsets.
Figure 6.
MAIT subsets acquire a tissue targeting program in the thymus. (a) MDS scatter plot summarizing the expression levels of the 10% most variable transcripts in the indicated samples. (b) Tissue residency index (see Materials and methods) evaluated in the different subsets. (c) Heat map displaying the expression of a selected list of chemokine receptors and integrins by thymic subsets. (d and e) Chemokine receptor and integrin protein expression by the different NKT and MAIT cell subsets in the thymus (d) and in the spleen, liver, and lungs (e; n = 3).
As compared with single-positive conventional T cells, thymic MAIT cells expressed high levels of Ccr2 (Fig. 6 c; recently implicated in diapedesis to inflamed tissues; Lee et al., 2018), Ccr8 (potentially targeting to skin and lung tissues), and Cxcr6 (gut-liver tropism), as well as Ccr6 (skin, gut, and brain tropism) for MAIT17 cells and Cxcr3 (homing to inflamed tissues) for MAIT1 cells (Fig. 6 c). These results are consistent with those obtained for thymic NKT subsets after single-cell analysis (Engel et al., 2016). Most of these results were confirmed at the protein level with the available antibodies (Fig. 6 d), except for CCR2, which we did not detect (data not shown), and CXCR3 expression, which was similar between thymic MAIT1 and MAIT17. These results suggest that MAIT cells leaving the thymus are targeted to reach various nonlymphoid tissues with a tropism that may vary according to type 1 versus 17 subsets.
Interestingly, Itga4 (CD49d) expression was lower in MAIT subsets as compared with conventional T cells in the thymus, whereas Itgb1 expression was higher (Fig. 6, c and d), suggesting that the latter chain associates with other integrin α chains such as Itga1 (resulting in VLA1, a laminin and collagen receptor) for MAIT1 cells. Thymic MAIT1 cells had lower Itgb3 (CD61) expression (both at the RNA and protein level) and higher Itgb1 expression as compared with thymic MAIT17 cells, further suggesting that, according to the MAIT subsets, the available α integrins pair with different β chains and may lead to differential tissue locations.
In the periphery (Fig. 6, c and e), CCR8 was more expressed in the lungs than in the spleen or liver, especially in the type 17 subsets. CXCR6 was less expressed in type 17 subsets than in type 1 subsets, whereas it was the opposite for CCR6. Interestingly, Itgb3 (CD61) was more expressed in all MAIT and NKT subsets as compared with EM CD8+ T cells with higher expression in type 17 subsets in the liver and lungs. The differential expression of Itga1 (CD49a) and Itga5 (CD49d) between type 17 and type 1 subsets in peripheral organs matched their expression in the thymus. Thus, the differential expression of chemokine receptors and integrins between conventional T cells and MAIT/NKT subsets, as well as between type 1 and type 17 MAIT/NKT subsets, which was acquired in the thymus, is selectively sustained in the different organs, probably leading to differential homing of the various subsets.
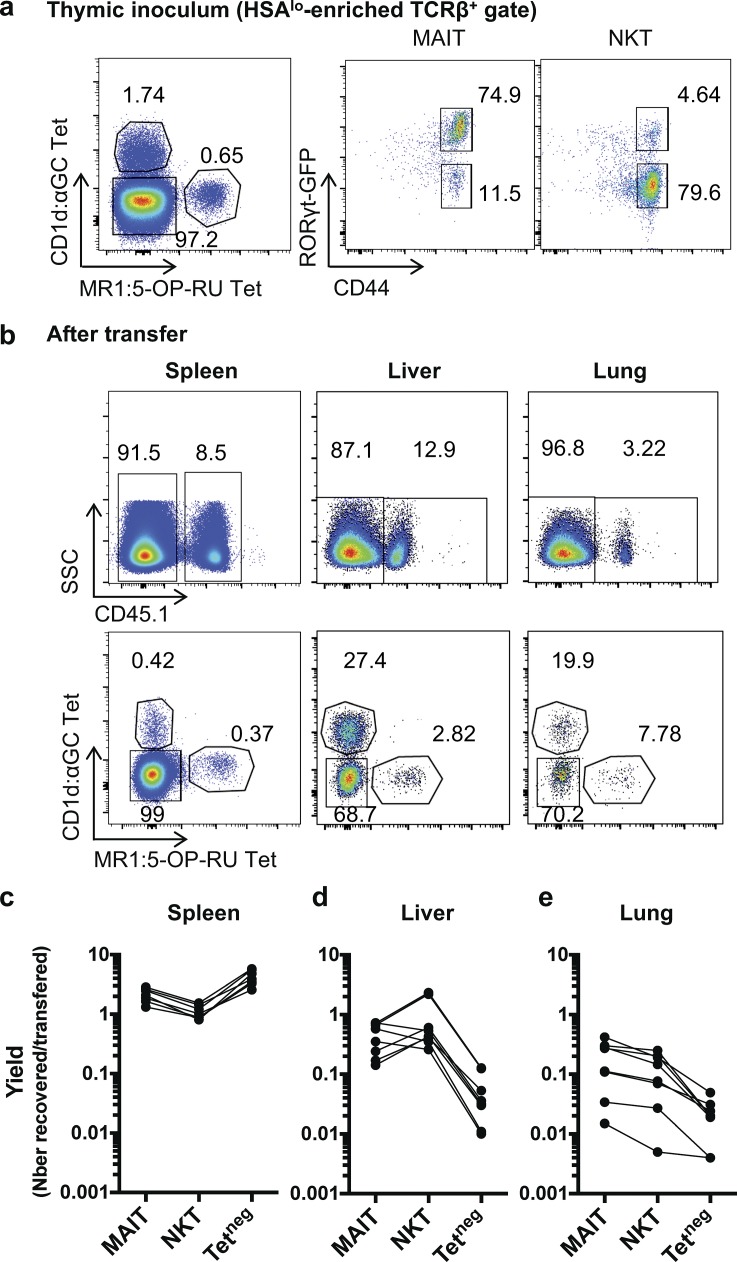
To further analyze whether thymic MAIT cells are already programmed to localize into tissues, we transferred mature thymocytes (containing MAIT and NKT) cells into congenically marked animals and 36 h later quantified transferred cells in the spleen, liver, and lungs. The results are expressed as a yield, i.e., the number of recovered cells in the observed organ over the number of transferred thymic cells, for MAIT, NKT, and conventional (Tetneg) T cells. Overall, most of the transferred cells are found in the spleen, for all subsets. Still, while the yield is more important for conventional T cells than MAIT and NKT cells in the spleen, the opposite is observed in the liver and the lungs in which MAIT and NKT migrate more than conventional T cells (increased yield by 10 to 100; Fig. 7). This is in accordance with the enrichment of the tissue residency program in MAIT thymic subsets and strongly illustrates the ability of MAIT and NKT cells to localize in tissues after thymus exit. Slight differences exist between MAIT and NKT cells, with a yield more important in the liver for NKT cells and in the lungs for MAIT cells. This is in accordance with the recovery observed for the type 1 and 17 subsets: both NKT1 and MAIT1 subsets migrate more to the liver than NKT17 and MAIT17 subsets. As most transferred NKT and MAIT cells were type 1 or type 17, respectively, this explains why the yield of total NKT in the liver is higher than the one of total MAIT and indicates specific tropism for the type 1 subset in the liver. Because of the low number of recovered cells in the lungs, no consistent trend regarding NKT/MAIT1 versus NKT/MAIT17 subset could be determined (data not shown). Altogether, these results confirm that NKT/MAIT subsets have already acquired in the thymus the properties directing them to specific peripheral tissues.
Figure 7.
NKT and MAIT cells from the thymus are more prone to locate to the liver and to the lungs than conventional T cells. Mature enriched (HSAlow) CD45.1+ thymocytes were transferred into CD45.2+ hosts, and the indicated organs were analyzed 36 h later for NKT (CD1d:PBS57Tet+), MAIT (MR1:5-OP-RU Tet+), and conventional T cell numbers. (a) example of staining of the inoculum (HSAhi-depleted thymocytes). (b) Examples of staining in the indicated organs after transfer. (c–e) Yield (no. recovered/no. injected cells) of the indicated subsets in the spleen (c), the liver (d), and the lungs (e; n = 6 transferred mice; three independent experiments).
Discussion
Herein, we show that MAIT and NKT subsets share common differentiation programs that are already expressed in the thymus. Two homologous subsets of NKT/MAIT cells can be defined according to RORγt and T-bet expression as early as the thymus. The proportion of type 1 and 17 subsets are the opposite for NKT and MAIT cells, and this correlates with different organ location: NKT1 cells are more numerous than NKT17 cells and mainly found in liver and spleen, whereas MAIT cells are more abundant in the lungs, with MAIT17 cells being far more numerous than MAIT1 cells. Still, both MAIT and NKT subsets share similar location in the lungs, the type 1 subsets being mostly located in the vasculature, while the type 17 subsets are in the parenchyma. Parabiosis experiments demonstrate for the first time that MAIT cells are tissue-resident cells, except for the type 1 subsets in the lungs. Tissue retention of the NKT/MAIT1 subsets in liver and spleen relies on LFA1/ICAM1 interactions, despite a different location, i.e., intravascular in the liver and parenchymal in the spleen. MAIT and NKT subsets are very similar transcription-wise, with progressive acquisition of a core tissue residency gene signature in the thymus, spleen/liver, and lungs. Altogether, these results support the use of preset T cells to designate MAIT and NKT cells, as previously proposed (Legoux et al., 2017).
In all the organs studied, MAIT1 and NKT1 as well as MAIT17 and NKT17 subsets share very similar transcription programs that are consistent with those already described (Fig. 2; Engel et al., 2016; Lee et al., 2016). The transcription pattern is very similar between the thymus and the spleen/liver for the preset-1 subsets, while it differs more for the preset-17 subsets (Fig. 6). In all cases, the lung samples displayed a more pronounced tissue residency signature, despite the fact that the lung preset-1 subsets are intravascular and recirculate between the parabionts. One hypothesis to explain this apparent discrepancy would be a short-term retention within the lungs, not indicated by our 5-wk parabiosis experiment. This could be related to the residual expression of Klf2 and CD62L (Sell) by the preset-1 subsets in the lungs, sufficient to allow rather rapid recirculation. Retention of preset-1 subsets in the liver vasculature relied on ICAM1/LFA1 interactions (Fig. 5), as already described for liver NKT cells (Thomas et al., 2011). Surprisingly, despite their location in the spleen parenchyma (Lee et al., 2015), preset-1 subsets also required ICAM1/LFA1 interactions to remain in this organ. In contrast, despite similar residency transcriptional program and intra-/extravascular location, liver and spleen preset-17 subsets are not released upon ICAM1/LFA1 interaction blockade. This could be related to different docking cell types for preset-1 and preset-17 subsets or to specific retention molecules induced by RORγt. Whether the same mechanisms are responsible for preset-17 retention in the lungs is unclear. Still, since the tissue residency transcription signature is stronger in this organ, the mechanisms may be different.
The basis of tissue residency could be related to the loss of a core gene circulatory signature, already apparent in the thymus, with additional expression of residency genes in the liver/spleen, further enhanced in the lung samples. This suggests that the residency signature is associated with peripheral tissue migration or localization, while the loss of circulating signature is already observed in the thymus. This loss of circulating signature is likely associated to PLZF expression, as PLZF regulates many genes involved in homing (such as Sell and Klf2), adhesion, and targeting (Mao et al., 2016). Interestingly, PLZF directly targets the promoter of Runx3, but down-regulates its expression as observed in PLZF gain- and loss-of-function models. Moreover, PZLF directly binds several genes present in the residency signature, as well as the consensus sequences for RUNX factors. Hence, the exact relationship between PLZF and the residency signature controlled by Runx3 remains unclear as both are tightly interweaved.
The rules governing targeting preset subsets to specific tissues remain to be determined. Since CCR7 and CD62L are not expressed by preset subsets exiting the thymus (our data; Cohen et al., 2013; Mingueneau et al., 2013), these cells cannot reach the lymph nodes through the blood. As shown from our thymocyte transfer experiments, preset-1 subsets are more prone to localize in the liver, while preset-17 subsets home to the lungs, as already observed for NKT subsets (Lee et al., 2015). Since NKT1/MAIT17 cells are more abundant in the thymus than NKT17/MAIT1 cells, this may explain why NKT cells are more abundant in the liver, while MAIT cells preferentially localize in the lungs. Tissue localization is consistent with the high expression of various chemokines receptors such as CCR6, CCR8, CXCR3, or CXCR6 in thymic preset T cell subsets. It is not clear whether specific organ targeting is determined in the thymus by distinct patterns of integrin and chemokine receptor expression or if migration to a given tissue is stochastic, only depending on subset-specific niches. Additional tissue-derived cues or differential in situ survival might further modulate the tissue population ratios. Selective genetic inhibition of chemokine receptor(s) or integrin(s) would be required to answer this question. One intriguing observation is the exclusive CD69 versus CD103 expression on preset T cells in all tissues, since the two markers are usually coexpressed on conventional TRM cells (Mackay et al., 2016; Milner et al., 2017; Beura et al., 2018). CD103 expression is important for brain TRM survival (Wakim et al., 2010), but we cannot exclude a different role in preset subset residency properties, especially as CD103 expression is regulated by PLZF (Mao et al., 2016). Since the proportion of CD103+ cells is higher in the preset-17 subsets, which are mainly in the lung parenchyma, it is possible that the expression of this integrin correlates to specific location inside the tissue as compared with CD69+ cells. The preferential expression of CD103 by MAIT17 cells is consistent with its known expression in mucosae and the abundance of MAIT17 cells in gut and lungs. Single-cell transcriptome analysis would enable to determine the basis of the exclusive CD69 versus CD103 expression.
The similar phenotype and transcriptome expressed by NKT and MAIT subsets indicate that the mechanisms of tissue targeting are common and not antigen specific. Accordingly, the frequencies of preset-1 or preset-17 subsets are correlated in the different organs (Fig. 1). This kind of correlation has also been observed between different T cell subsets expressing high levels of CD161 (indicating a common differentiation program) in human cord blood, as well as between blood NKT and MAIT cell populations from nonhomozygous twins (Ben Youssef et al., 2018). Altogether, these data suggest that common environmental cues determine the frequencies of the different preset subsets in different organs, irrespective of TCR specificity, and rules out a possible competition for peripheral niches between MAIT and NKT cells.
Antigen specificity, however, probably determines the proportion of preset-1 and -17 subsets generated in the thymus, which is very different for MAIT and NKT cells. For NKT cells, differences in Vβ repertoire between NKT1, NKT2, and NKT17, as well as differences in Nur77 expression during selection (Wei et al., 2006; Lee et al., 2013; Tuttle et al., 2018), suggest that the intensity of endogenous ligand/CD1d triggering is involved. We did not observe similar differences in Vβ6 or Vβ8 segment usage between MAIT subsets in any organs (Fig. S2). Still, a role for the fine TCR specificity during thymic selection cannot be excluded, as we studied the repertoire using antibodies against the most frequent MAIT Vβ chains. To strengthen this hypothesis, deep sequencing of the TCRβ CDR3 of MAIT1 and MAIT17 subsets with further TCRβ cloning and expression allowing avidity studies would be necessary, as MAIT TCRβ chains may lead to variable affinity to endogenous ligands (Eckle et al., 2014).
We checked whether human MAIT cells in the blood or in the liver would also display a tissue residency gene signature. This was indeed the case with a slightly down-regulated circulatory signature and slightly up-regulated tissue residency signature expressed by blood MAIT cells, as compared with memory CD8+ T cells. Notably, in human blood, expression of the transcription factor Cebpd is almost specific of MAIT cells (Table S2 a). Cebpd expression has been associated with migration to tissues through inflamed endothelium in a CCR2-dependent process (Lee et al., 2018). However, in mice, Cebpd was not found expressed in any of our transcriptome datasets, nor in publicly available NKT gene expression datasets. Because we could not study blood MAIT cells in mice because of their rarity, we cannot exclude that they also express Cebpd. Alternatively, the expression of chemokine receptors and integrin may be differentially regulated in mice and humans, as suggested by the simultaneous expression of many transcription factors in humans (Leeansyah et al., 2015; Dias et al., 2017), which is not the case in mice (our data; Koay et al., 2016). In the liver, human and mouse MAIT cells are very similar transcriptome-wise, with high expression of the core tissue residency gene signature. This highlights the importance of studying tissue MAIT cells when focusing on a tissue pathology. Notably, liver conventional CD4+ and CD8+ T cells expressed Zbtb16 (PLZF) similarly to blood MAIT cells, but still less than liver MAIT cells. Therefore, the significance of PLZF expression may be different in humans as compared with mice.
Altogether, our data indicate that MAIT and NKT subsets share most characteristics with regards to tissue location and probably also with regards to effector functions as indicated by transcriptomic analysis. Specific TCR triggering by endogenous or microbial antigens is possible in the different organs probably for all NKT/MAIT subsets (Mattner et al., 2005). However, the role of specific TCR-mediated interactions versus non-specific activation by lymphokines, which would be required in different pathophysiological contexts, remain to be determined for each subset (Vahl et al., 2013). If many functions are fulfilled independently of TCR triggering, this may lead to functional redundancy between MAIT and NKT subsets.
Materials and methods
Human samples and cell preparation
Paired blood and liver samples were obtained from three UM patients that underwent surgical intervention to remove liver metastasis in the context of a clinical trial (ClinicalTrials.gov accession no. NCT02849145) that had been approved by the Institut Curie Instituional Review Board as well as by the Comité de Protection des Personnes. All patients treated at Institut Curie are informed that samples obtained during diagnosis or therapeutic procedures can be used for research purpose. Liver samples correspond to fragments that were not invaded by the melanoma. To control for a potential impact of the melanoma, blood samples from three healthy volunteers were also studied. Blood mononuclear cells were isolated on a lymphoprep gradient using a Sepmate tube (StemCell Technologies) followed by red blood cell lysis (eBioscience). Liver biopsies were chopped in small pieces and digested in CO2-independent medium (Gibco) supplemented with 5% FCS (Gibco), 2 mg/ml collagenase I (Sigma), 25 µg/ml DNase (Roche), and 2 mg/ml hyaluronidase (Sigma) for 50 min at 37°C under constant agitation to obtain liver cell suspension. Cells were then stained using CD3 (clone UCHT1; BioLegend), CD4 (clone S3.5; Thermo Fisher), TCRγδ (clone B1.1; eBioscience), CD161 (clone 191B8; Miltenyi), Vα7.2 (clone 3C10; BioLegend), CD27 (clone 1A4CD27; Beckman), and CD45RA (clone HI100; eBiosciences) antibodies, sorted on an ARIA-III cell sorter (Becton Dickinson) and directly frozen in RNeasy lysis buffer (Qiagen) containing 5% beta mercaptoethanol (Qiagen). MAIT cells were defined as CD3+CD4−TCRγδ−CD161hiVα7.2+, EM CD4+ T cells as CD3+CD4+TCRγδ−CD27+CD45RA−, and EM CD8+ as CD3+CD4−TCRγδ− non-MAIT and CD27+CD45RA−.
RNA sequencing (RNAseq) sample preparation
RNA was extracted using the Single Cell RNA Purification kit (Norgen) according to the manufacturer’s instructions with an on-column DNA digestion (Qiagen). RNA quality was checked using Agilent pico chip (Agilent Technologies). Half of the sample was reverse transcribed and amplified using SMART-Seq v4 Ultra Low Input RNA kit for sequencing according to the manufacturer’s instructions (Takara Bio). Libraries were prepared from 0.6 ng of cDNA with the Nextera XT DNA Library Prep kit (Illumina) according to the manufacturer’s recommendations. 12 PCR cycles were set up to amplify and add barcodes to the libraries. Quality controls were performed with Qubit (Thermo Fisher) for quantification, and LabChip GX Touch (PerkinElmer) for fragments length determination. Molarity of the final pool was precisely quantified by qPCR with KAPA Library Quantification kit (Kapa Biosystems) and CFX96 Touch Real-Time PCR. High-throughput sequencing was performed on an Illumina HiSeq 2500 system in Rapid Run mode, using two flow cells with paired-end 100-read length.
RNAseq transcriptome analysis
The quality of paired reads were evaluated with Fastqc 0.11.5. Low quality reads were trimmed with cutadapt 1.14, with a quality threshold of Q ≥ 28. Duplicated PCR reads were identified and discarded with samtools 1.6. Transcript quantification was performed by alignment of high-quality reads (4.4–7.3 millions of reads per sample) with STAR 2.5.3a and by quasi-mapping with salmon 0.9.1 (with default k-mer length of 31; Patro et al., 2017). Both quantifications were performed on the reference genome GRCh38 release 90 (August 2017; built from ensembl sources). Quasi-mapping showed superior sensitivity on transcripts with highly conserved domains in our dataset (i.e., chemokine and chemokine receptors genes) and was therefore chosen as transcript quantification method, with corrections for random hexamer priming bias (–seqBias) and for fragment-level GC bias (–gcBias). Finally, estimated read counts were normalized and processed with edgeR 3.22.2, and downstream analysis was done with R version 3.5.0 (April 23, 2018) on a CentOS Linux 7 system (64-bit). Unsupervised clustering of the samples was based on a euclidean distance with the unweighted pair group method with arithmetic mean method. RNAseq expression values are represented as log(2) normalized expression in counts per million, with a prior read count of 1 to avoid taking log of 0. Datasets are available at the European Bioinformatics Institute under accession no. E-MTAB-7143.
Mouse strains, samples, and cell preparation
Unless stated otherwise, the B6-MAITcast congenic strain that harbors high MAIT cell numbers and a Rorc-GFP reporter gene were used (backcrossed for >10 generations; Lochner et al., 2008; Cui et al., 2015). For PLZF expression, a GFP reporter mouse (Constantinides et al., 2014) was crossed onto the B6-MAITCast background. Balb/c mice were from Charles River. All experiments were conducted in an accredited animal facility by the French Veterinarian Department following ethical guidelines and were approved by the relevant ethical committee (AP AF1S no. 6030-20 16070817147969 v2, authorization no. 118). To control for the variability in MAIT and NKT cell percentages in our mice, the experimental groups were designed to ensure even repartition of animals coming from the same breeding cages.
Spleen and thymus cells were obtained by mechanical dissociation trough a 40-µm cell strainer, followed by red blood cell lysis for spleen cells. Livers were mechanically dissociated on a 100-µm cell strainer. After perfusion and dissection, lungs were chopped in pieces and digested in CO2-independent medium containing 0.1 mg/ml DNase I (Roche) and 0.1 mg/ml liberase TL (Roche) using Gentlemacs operating system according to the manufacturer’s instructions (Miltenyi). Colons were chopped into pieces and incubated for 30 min at 37°C under continuous agitation in HBSS w/o (Life Technologies), 5 mM EDTA (Sigma), 5% FCS (Life Technologies), and 1 mM DTT. After rinsing for 20 min in HBSS w/o (Life Technologies), pieces were digested for 30 min at 37°C under continuous rotation in CO2-independent medium (Life Technologies) containing 0.17 U/ml liberase TM (Roche), 1 mg/ml collagenase D (Roche), and 125 g/ml DNase (Roche) and were further dissociated using Gentlemacs operating system according to the manufacturer’s instructions (Miltenyi). Skin from the back of the mouse was chopped into pieces and digested for 1 h at 37°C under continuous rotation in CO2-independent medium (Life Technologies) containing 2% FCS, 1% Hepes, 500 µg/ml hyaluronidase type IS (Sigma), 100 µg/ml Liberase TM (Roche), and 250 µg/ml DNase I (Roche). Further dissociation was obtained on the Gentlemacs operating system (Miltenyi) using the m_impTumor_01 program. Liver, lung, colon, and skin mononuclear cells were then isolated on a Percoll (40–80%) or lympholyte gradient. Blood cells were recovered by centrifugation after red blood cell lysis.
For CD45 intravascular staining, 3 µg CD45-biotinylated antibody was injected intravenously 3 min before mouse euthanasia.
Flow cytometry analysis
Single cell suspensions were stained in PBS 0.5% BSA 2 mM EDTA complemented with rat anti-mouse CD16/CD32 antibody (clone 2.4G2 produced in-house, to block non-specific binding to Fcγ receptor) and combinations of monoclonal antibodies. Tetramer staining for both MAIT and NKT cells was performed in the same buffer at room temperature following the National Institutes of Health (NIH) Tetramer Core Facility instructions (both tetramers from the NIH Tetramer Core Facility; Emory University, Oxford, GA). The FoxP3 staining kit (eBioscience) was used for intranuclear staining according to the manufacturer’s instructions. DAPI or live/dead staining kit (according to the manufacturer’s instructions; BD Biosciences) were used to exclude dead cells. After staining, the samples were run on the cytometry platform of the Curie Institute, either on a Cytoflex (Beckman), a Fortessa (Becton Dickinson), or an LSRII (BD Biosciences) cytometer. Absolute numbers were calculated on the cytoflex cytometer, based on the volume acquired. Analysis was done using FlowJo (v10; Tree Star). In brief, T cells were identified after gating on lymphocytes using the forward scatter/side scatter plots and exclusion of doublets, dead cells, and B cells. Peripheral MAIT and NKT are defined as tet+CD44hi cells. In the thymus, mature MAIT and NKT cells were identified as CD44hiHSAlo. The following monoclonal antibodies were used: anti-TCRβ (clone H57-597), -CD44 (IM7), -CD19 (ID3), -B220 (RA3-6B2), -CD103 (M290), -CD69 (H1.2F3), -CD45.1 (A20), -CD45.2 (104), -CD45 (30-F11), -CD8 (53-6.7), -CD4 (RM4-5), -Vβ6 (RR4-7), -Vβ8 (F23.1), -HSA (M1/69), -CD49a (HMα1), -CD49e (5H10-27[MFRS]), -CD61 (2C9.G2), -CCRL2 (BZ2E3), -CXCR3 (CXCR3-173), -CXCR6 (SA051D1), -CCR6 (29-2L17), -CCR7 (4B12), -CCR8 (SA214G2), -RORγt (AFKJS-9), and -PLZF (9E12), purchased from BioLegend, BD Biosciences, eBioscience, or Thermo Fisher, depending on the fluorochrome needed.
Affymetrix sample preparation
Thymus, spleen, liver, and lung cell suspensions were sorted on an ARIA-III cell sorter. MAIT and NKT were defined as Tet+CD44hi cells. Control populations from the spleen were defined as follows, after excluding dead cells and doublets and gating on T cells: Th17 as RORγt-GFP+CD4+, naive CD4 as CD44loCD62LhiCD4+, and EM CD8 as CD44hiCD62LloCD8+. Cells were directly sorted in RNeasy lysis buffer. RNA was extracted using Qiagen MicroKit with an on column DNase incubation. Total RNA concentration and RNA integrity was monitored by electrophoresis (Agilent Bioanalyzer; RNA 6000 Pico Assay). Gene expression analysis was conducted using Mouse Clariom D chip (Thermo Fisher). In brief, 500 pg (Table S2) of total RNA were processed in parallel with an external Mouse Universal Reference RNA to control robustness and reproducibility of enzymatic steps. RNA samples were amplified with Ovation Pico WTA System v2 (Nugen) and labeled with Encore biotin module (Nugen). Array were hybridized with 5 µg of labeled DNA and assayed on a GeneChip Scanner 3000 7G (Affymetrix). Raw data were generated and controlled with Expression console (Affymetrix) at the Institut Curie Genomic facility. Datasets are available at European Bioinformatics Institute under accession no. E-MTAB-7133.
Microarray analysis
Probesets were annotated with the R oligo package 1.42.0 (Carvalho and Irizarry, 2010), with expression summarized at the transcript level. Between-array normalization was performed by robust multiarray average. Consistency of the results between hybridization batches was evaluated with external Mouse Universal Reference as internal controls. Ribosomal, mitochondrial, microRNAs, and predicted gene transcripts were filtered by pattern-matching with regular expressions based on the standard nomenclature defined by the International Committee on Standardized Genetic Nomenclature for Mice and excluded from analyses. TCR chain transcripts were analyzed separately after appropriate filtering. Identification of the most variable genes is based on the interquartile range of the dispersion in the expression intensities among all samples (calculated by the difference between the 25th and the 75th percentiles). Unsupervised clustering of the samples is based on a euclidean distance with the unweighted pair group method with arithmetic mean method. Post-processing and differential expression testing by linear models among the experimental groups were performed with limma 3.34.5 and edgeR 3.22.2 packages (McCarthy et al., 2012; Ritchie et al., 2015). All downstream analyses were performed with R version 3.5.0 on a CentOS Linux 7 system (64-bit). All microarray expression values are represented as log(2) normalized intensities.
Tissue residency index
Tissue residency and circulatory signatures defined by Milner et al. (2017) were used to estimate the residency phenotype of the different cell subsets as compared with splenic naive CD4+ conventional T cells. In each sample, the fraction of gene either up-regulated (lfc > 1) or down-regulated (lfc < −1) was computed for both signatures. The genes up-regulated in the tissue residency signature (TRMup) and down-regulated in the circulatory signature (Cirdown) were summed up as a bonus score. The genes down-regulated in the tissue residency signature (TRMdown) and up-regulated in the circulatory signature (Cirup) were summed up as a malus score. The final tissue residency index (TRI) is computed by the following formula:
Parabiotic mice
CD45.2 and CD45.1/2 B6-MAITCast mice were surgically joined as parabiotic pairs according to the technique described in Kamran et al. (2013). Spleen, liver, and lungs were collected 5 wk later.
LFA1/ICAM1 blockade
LFA1/ICAM1 blockade was performed as previously described (Thomas et al., 2011). In brief, blocking antibodies against LFA1 (100 µg/mouse; clone M17/4) and ICAM1 (200 µg/mouse; clone YNI.7.4), VLA4 (100 µg/mouse; clone PS/2), and VCAM1 (200 µg/mouse; clone M/K-2.7) or one isotype control (200 µg/mouse; clone 2A4) were injected intravenously 24 h before mouse euthanasia. MAIT and NKT cell numbers were evaluated by flow cytometry in spleen, liver, and lungs.
Adoptive T cells transfer
Thymic cells from B6-MAITcast mice were stained with an anti–CD24-FITC antibody and depleted of the CD24+ (HSA+) fraction using anti-FITC microbeads (Miltenyi) and LS columns (Miltenyi) according to the manufacturer’s instructions. Three thymuses were pooled for injection into one recipient, and the cell concentration was adjusted to transfer ∼40,000 MAIT cells among total enriched thymic cells. Cells were transferred intravenously into B6-MAITcast recipients, and the spleen, the liver, and the lungs were collected 36 h later for flow cytometry analysis.
Statistical method
A type I error threshold of P < 0.05 was used in all statistical tests of the present study. For the supervised analysis of the microarray and RNAseq datasets, inflation of the type I error among multiple comparisons was controlled with a false discovery rate fixed at level 0.05, adjusted according to the Benjamini–Hochberg procedure. For the microarray analysis, differential expression testing was conducted by ANOVA after linear model fitting of expression intensities with fixed terms “cell type” MAIT/NKT and “immune bias” Rorγt+/Rorγtneg (two-factor ANOVA) with the limma R package. For the RNAseq analysis, a single fixed term with levels matching the different cell types was considered. A contrast-based analysis was used to compare cell subsets to a common reference defined by (blood CD4 control + blood CD8 control)/2 and to compare the liver MAIT UM to the blood MAIT UM. Differential expression testing was conducted by likelihood-ratio test after generalized-linear model fitting of normalized read counts with the edgeR R package.
Online supplemental material
Fig. S1 shows the absence of MAIT2 in both Balb/c and B6-MAITcast strain. Fig. S2 shows the absence of Vβ6 and Vβ8 TCR bias in MAIT1 and MAIT17 subsets. Table S1 displays gene expression profile of murine cell subsets evaluated by microarray. Table S2 displays gene expression profile of human cell subsets evaluated by RNAseq. Table S3 displays expression levels (log2 normalized) of genes associated to the Runx3-related tissue residency signature and to the circulating signature in the thymus and the peripheral organs.
Supplementary Material
Acknowledgments
We thank Virginie Dangles-Marie, Isabelle Grandjean, Mickael Garcia, and the zootechnicians of the mouse facility platform of the Institut Curie, as well as the flow cytometry core (Zofia Maciorowski, Sophie Grondin, and Annick Viguier). For the Affymetrix data, we thank Aude Vieillefon, Audrey Rapinat, and David Gentien from the Genomics Platform of the Translational Research Department at Institut Curie. High throughput sequencing was performed by the ICGex NGS platform of the Institut Curie (Sylvain Baulande, Mylène Bohec, and Virginie Raynal) supported by the grants from the Agence Nationale de la Recherche (ANR; “Investissements d’Avenir” program; grant nos. ANR10EQPX03 [Equipex] and ANR10INBS0908 [France Génomique Consortium]), by the Canceropôle Ile de France, and by the SiRIC Curie program. We thank the NIH Tetramer Core Facility (Emory University) for providing CD1d:GC and MR1:5-OP-RU tetramers. The MR1/5-OP-RU tetramer technology was developed jointly by J. McCluskey, J. Rossjohn, and D. Fairlie, and the material was produced by the NIH Tetramer Core Facility as permitted to be distributed by the University of Melbourne.
This work was supported by the Institut National de la Santé et de la Recherche Médicale, Institut Curie, and ANR (Labex DCBIOL [ANR-10-IDEX-0001-02 PSL and ANR-11-LABX-0043] and Non-thematic call; neoMAIT, MAIT, and diabMAIT). O. Lantz’s group was supported by the “Equipe labellisée de la Ligue Contre le Cancer” program. F. Legoux was supported by a Marie-Skłodowska Curie individual fellowship (grant 706353) from the European Commission.
The authors declare no competing financial interests.
Author contributions: M. Salou, F. Legoux, and O. Lantz designed the research. M. Salou, F. Legoux, A. Darbois, A. du Halgouet, R. Alonso, A.-G. Goubet, L. Menger, E. Procopio, and V. Premel performed experiments. J. Gilet performed the bioinformatic analysis and W. Richer carried out the initial microarray analysis. C. Daviaud and F. Legoux performed the parabiotic experiments. M. Salou, F. Legoux, J. Gilet, and O. Lantz wrote the paper. O. Lantz supervised the whole project.
References
- Barral P., Sánchez-Niño M.D., van Rooijen N., Cerundolo V., and Batista F.D.. 2012. The location of splenic NKT cells favours their rapid activation by blood-borne antigen. EMBO J. 31:2378–2390. 10.1038/emboj.2012.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A., Savage P.B., and Teyton L.. 2007. The biology of NKT cells. Annu. Rev. Immunol. 25:297–336. 10.1146/annurev.immunol.25.022106.141711 [DOI] [PubMed] [Google Scholar]
- Ben Youssef G., Tourret M., Salou M., Ghazarian L., Houdouin V., Mondot S., Mburu Y., Lambert M., Azarnoush S., Diana J.S., et al. 2018. Ontogeny of human mucosal-associated invariant T cells and related T cell subsets. J. Exp. Med. 215:459–479. 10.1084/jem.20171739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K., Mitchell J.S., Thompson E.A., Schenkel J.M., Mohammed J., Wijeyesinghe S., Fonseca R., Burbach B.J., Hickman H.D., Vezys V., et al. 2018. Intravital mucosal imaging of CD8+ resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat. Immunol. 19:173–182. 10.1038/s41590-017-0029-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho B.S., and Irizarry R.A.. 2010. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 26:2363–2367. 10.1093/bioinformatics/btq431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen N.R., Brennan P.J., Shay T., Watts G.F., Brigl M., Kang J., and Brenner M.B.. ImmGen Project Consortium . 2013. Shared and distinct transcriptional programs underlie the hybrid nature of iNKT cells. Nat. Immunol. 14:90–99. 10.1038/ni.2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides M.G., McDonald B.D., Verhoef P.A., and Bendelac A.. 2014. A committed precursor to innate lymphoid cells. Nature. 508:397–401. 10.1038/nature13047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Franciszkiewicz K., Mburu Y.K., Mondot S., Le Bourhis L., Premel V., Martin E., Kachaner A., Duban L., Ingersoll M.A., et al. 2015. Mucosal-associated invariant T cell-rich congenic mouse strain allows functional evaluation. J. Clin. Invest. 125:4171–4185. 10.1172/JCI82424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias J., Sandberg J.K., and Leeansyah E.. 2017. Extensive Phenotypic Analysis, Transcription Factor Profiling, and Effector Cytokine Production of Human MAIT Cells by Flow Cytometry. Methods Mol. Biol. 1514:241–256. 10.1007/978-1-4939-6548-9_17 [DOI] [PubMed] [Google Scholar]
- Dusseaux M., Martin E., Serriari N., Péguillet I., Premel V., Louis D., Milder M., Le Bourhis L., Soudais C., Treiner E., and Lantz O.. 2011. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 117:1250–1259. 10.1182/blood-2010-08-303339 [DOI] [PubMed] [Google Scholar]
- Eckle S.B., Birkinshaw R.W., Kostenko L., Corbett A.J., McWilliam H.E., Reantragoon R., Chen Z., Gherardin N.A., Beddoe T., Liu L., et al. 2014. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J. Exp. Med. 211:1585–1600. 10.1084/jem.20140484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel I., Seumois G., Chavez L., Samaniego-Castruita D., White B., Chawla A., Mock D., Vijayanand P., and Kronenberg M.. 2016. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat. Immunol. 17:728–739. 10.1038/ni.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., and Rudensky A.Y.. 2016. Hallmarks of Tissue-Resident Lymphocytes. Cell. 164:1198–1211. 10.1016/j.cell.2016.02.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franciszkiewicz K., Salou M., Legoux F., Zhou Q., Cui Y., Bessoles S., and Lantz O.. 2016. MHC class I-related molecule, MR1, and mucosal-associated invariant T cells. Immunol. Rev. 272:120–138. 10.1111/imr.12423 [DOI] [PubMed] [Google Scholar]
- Gapin L. 2016. Development of invariant natural killer T cells. Curr. Opin. Immunol. 39:68–74. 10.1016/j.coi.2016.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann F., Cameron T.O., Sidobre S., Manlongat N., Kronenberg M., Briskin M.J., Dustin M.L., and Littman D.R.. 2005. Intravascular immune surveillance by CXCR6+ NKT cells patrolling liver sinusoids. PLoS Biol. 3:e113 10.1371/journal.pbio.0030113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs A., Leeansyah E., Introini A., Paquin-Proulx D., Hasselrot K., Andersson E., Broliden K., Sandberg J.K., and Tjernlund A.. 2017. MAIT cells reside in the female genital mucosa and are biased towards IL-17 and IL-22 production in response to bacterial stimulation. Mucosal Immunol. 10:35–45. 10.1038/mi.2016.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey D.I., Uldrich A.P., McCluskey J., Rossjohn J., and Moody D.B.. 2015. The burgeoning family of unconventional T cells. Nat. Immunol. 16:1114–1123. 10.1038/ni.3298 [DOI] [PubMed] [Google Scholar]
- Kamran P., Sereti K.I., Zhao P., Ali S.R., Weissman I.L., and Ardehali R.. 2013. Parabiosis in mice: a detailed protocol. J. Vis. Exp. (80):e50556. 10.3791/50556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koay H.F., Gherardin N.A., Enders A., Loh L., Mackay L.K., Almeida C.F., Russ B.E., Nold-Petry C.A., Nold M.F., Bedoui S., et al. 2016. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat. Immunol. 17:1300–1311. 10.1038/ni.3565 [DOI] [PubMed] [Google Scholar]
- Lee C.H., Zhang H.H., Singh S.P., Koo L., Kabat J., Tsang H., Singh T.P., and Farber J.M.. 2018. C/EBPδ drives interactions between human MAIT cells and endothelial cells that are important for extravasation. eLife. 7:e32532 10.7554/eLife.32532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Holzapfel K.L., Zhu J., Jameson S.C., and Hogquist K.A.. 2013. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol. 14:1146–1154. 10.1038/ni.2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Wang H., Starrett G.J., Phuong V., Jameson S.C., and Hogquist K.A.. 2015. Tissue-Specific Distribution of iNKT Cells Impacts Their Cytokine Response. Immunity. 43:566–578. 10.1016/j.immuni.2015.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Starrett G.J., Lee S.T., Yang R., Henzler C.M., Jameson S.C., and Hogquist K.A.. 2016. Lineage-Specific Effector Signatures of Invariant NKT Cells Are Shared amongst γδ T, Innate Lymphoid, and Th Cells. J. Immunol. 197:1460–1470. 10.4049/jimmunol.1600643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeansyah E., Svärd J., Dias J., Buggert M., Nyström J., Quigley M.F., Moll M., Sönnerborg A., Nowak P., and Sandberg J.K.. 2015. Arming of MAIT Cell Cytolytic Antimicrobial Activity Is Induced by IL-7 and Defective in HIV-1 Infection. PLoS Pathog. 11:e1005072 10.1371/journal.ppat.1005072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legoux F., Salou M., and Lantz O.. 2017. Unconventional or Preset αβ T Cells: Evolutionarily Conserved Tissue-Resident T Cells Recognizing Nonpeptidic Ligands. Annu. Rev. Cell Dev. Biol. 33:511–535. 10.1146/annurev-cellbio-100616-060725 [DOI] [PubMed] [Google Scholar]
- Lochner M., Peduto L., Cherrier M., Sawa S., Langa F., Varona R., Riethmacher D., Si-Tahar M., Di Santo J.P., and Eberl G.. 2008. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. J. Exp. Med. 205:1381–1393. 10.1084/jem.20080034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay L.K., Minnich M., Kragten N.A., Liao Y., Nota B., Seillet C., Zaid A., Man K., Preston S., Freestone D., et al. 2016. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 352:459–463. 10.1126/science.aad2035 [DOI] [PubMed] [Google Scholar]
- Mak J.Y., Xu W., Reid R.C., Corbett A.J., Meehan B.S., Wang H., Chen Z., Rossjohn J., McCluskey J., Liu L., and Fairlie D.P.. 2017. Stabilizing short-lived Schiff base derivatives of 5-aminouracils that activate mucosal-associated invariant T cells. Nat. Commun. 8:14599 10.1038/ncomms14599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao A.P., Constantinides M.G., Mathew R., Zuo Z., Chen X., Weirauch M.T., and Bendelac A.. 2016. Multiple layers of transcriptional regulation by PLZF in NKT-cell development. Proc. Natl. Acad. Sci. USA. 113:7602–7607. 10.1073/pnas.1601504113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E., Treiner E., Duban L., Guerri L., Laude H., Toly C., Premel V., Devys A., Moura I.C., Tilloy F., et al. 2009. Stepwise development of MAIT cells in mouse and human. PLoS Biol. 7:e54 10.1371/journal.pbio.1000054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattner J., Debord K.L., Ismail N., Goff R.D., Cantu C. III, Zhou D., Saint-Mezard P., Wang V., Gao Y., Yin N., et al. 2005. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 434:525–529. 10.1038/nature03408 [DOI] [PubMed] [Google Scholar]
- McCarthy D.J., Chen Y., and Smyth G.K.. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40:4288–4297. 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner J.J., Toma C., Yu B., Zhang K., Omilusik K., Phan A.T., Wang D., Getzler A.J., Nguyen T., Crotty S., et al. 2017. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature. 552:253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingueneau M., Kreslavsky T., Gray D., Heng T., Cruse R., Ericson J., Bendall S., Spitzer M.H., Nolan G.P., Kobayashi K., et al. Immunological Genome Consortium . 2013. The transcriptional landscape of αβ T cell differentiation. Nat. Immunol. 14:619–632. 10.1038/ni.2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R., Duggal G., Love M.I., Irizarry R.A., and Kingsford C.. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 14:417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reantragoon R., Corbett A.J., Sakala I.G., Gherardin N.A., Furness J.B., Chen Z., Eckle S.B., Uldrich A.P., Birkinshaw R.W., Patel O., et al. 2013. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J. Exp. Med. 210:2305–2320. 10.1084/jem.20130958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., and Smyth G.K.. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salou M., Franciszkiewicz K., and Lantz O.. 2017. MAIT cells in infectious diseases. Curr. Opin. Immunol. 48:7–14. 10.1016/j.coi.2017.07.009 [DOI] [PubMed] [Google Scholar]
- Savage A.K., Constantinides M.G., Han J., Picard D., Martin E., Li B., Lantz O., and Bendelac A.. 2008. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 29:391–403. 10.1016/j.immuni.2008.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel J.M., and Masopust D.. 2014. Tissue-resident memory T cells. Immunity. 41:886–897. 10.1016/j.immuni.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler M.P., Mathew R., Liszewski M.K., Spooner C.J., Barr K., Meng F., Singh H., and Bendelac A.. 2012. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat. Immunol. 13:264–271. 10.1038/ni.2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert E.M., Schenkel J.M., Fraser K.A., Beura L.K., Manlove L.S., Igyártó B.Z., Southern P.J., and Masopust D.. 2015. Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell. 161:737–749. 10.1016/j.cell.2015.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S.Y., Scanlon S.T., Griewank K.G., Constantinides M.G., Savage A.K., Barr K.A., Meng F., Luster A.D., and Bendelac A.. 2011. PLZF induces an intravascular surveillance program mediated by long-lived LFA-1-ICAM-1 interactions. J. Exp. Med. 208:1179–1188. 10.1084/jem.20102630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turtle C.J., Delrow J., Joslyn R.C., Swanson H.M., Basom R., Tabellini L., Delaney C., Heimfeld S., Hansen J.A., and Riddell S.R.. 2011. Innate signals overcome acquired TCR signaling pathway regulation and govern the fate of human CD161(hi) CD8α+ semi-invariant T cells. Blood. 118:2752–2762. 10.1182/blood-2011-02-334698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle K.D., Krovi S.H., Zhang J., Bedel R., Harmacek L., Peterson L.K., Dragone L.L., Lefferts A., Halluszczak C., Riemondy K., et al. 2018. TCR signal strength controls thymic differentiation of iNKT cell subsets. Nat. Commun. 9:2650 10.1038/s41467-018-05026-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahl J.C., Heger K., Knies N., Hein M.Y., Boon L., Yagita H., Polic B., and Schmidt-Supprian M.. 2013. NKT cell-TCR expression activates conventional T cells in vivo, but is largely dispensable for mature NKT cell biology. PLoS Biol. 11:e1001589 10.1371/journal.pbio.1001589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakim L.M., Waithman J., van Rooijen N., Heath W.R., and Carbone F.R.. 2008. Dendritic cell-induced memory T cell activation in nonlymphoid tissues. Science. 319:198–202. 10.1126/science.1151869 [DOI] [PubMed] [Google Scholar]
- Wakim L.M., Woodward-Davis A., and Bevan M.J.. 2010. Memory T cells persisting within the brain after local infection show functional adaptations to their tissue of residence. Proc. Natl. Acad. Sci. USA. 107:17872–17879. 10.1073/pnas.1010201107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakim L.M., Woodward-Davis A., Liu R., Hu Y., Villadangos J., Smyth G., and Bevan M.J.. 2012. The molecular signature of tissue resident memory CD8 T cells isolated from the brain. J. Immunol. 189:3462–3471. 10.4049/jimmunol.1201305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.J., Kang Y.H., Smith M.O., Tharmalingham H., Ramamurthy N., Fleming V.M., Sahgal N., Leslie A., Oo Y., Geremia A., et al. 2012. Human MAIT and CD8αα cells develop from a pool of type-17 precommitted CD8+ T cells. Blood. 119:422–433. 10.1182/blood-2011-05-353789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D.G., Curran S.A., Savage P.B., Teyton L., and Bendelac A.. 2006. Mechanisms imposing the Vbeta bias of Valpha14 natural killer T cells and consequences for microbial glycolipid recognition. J. Exp. Med. 203:1197–1207. 10.1084/jem.20060418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F., Cheng Y., Breschi A., Vierstra J., Wu W., Ryba T., Sandstrom R., Ma Z., Davis C., Pope B.D., et al. Mouse ENCODE Consortium . 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 515:355–364. 10.1038/nature13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.