This study provides mechanistic insight into how IL-17 receptor adopts EGFR to activate ERK5 axis in Lrig1+ stem cells for their proliferation and migration during wounding healing and tumorigenesis.
Abstract
Lrig1 marks a distinct population of stem cells restricted to the upper pilosebaceous unit in normal epidermis. Here we report that IL-17A–mediated activation of EGFR plays a critical role in the expansion and migration of Lrig1+ stem cells and their progenies in response to wounding, thereby promoting wound healing and skin tumorigenesis. Lrig1-specific deletion of the IL-17R adaptor Act1 or EGFR in mice impairs wound healing and reduces tumor formation. Mechanistically, IL-17R recruits EGFR for IL-17A–mediated signaling in Lrig1+ stem cells. While TRAF4, enriched in Lrig1+ stem cells, tethers IL-17RA and EGFR, Act1 recruits c-Src for IL-17A–induced EGFR transactivation and downstream activation of ERK5, which promotes the expansion and migration of Lrig1+ stem cells. This study demonstrates that IL-17A activates the IL-17R–EGFR axis in Lrig1+ stem cells linking wound healing to tumorigenesis.
Introduction
It is well documented that chronic tissue damage and inflammation can contribute to tumorigenesis. Although it has long been suggested that “tumor production is a possible overhealing” (Haddow, 1972; Dvorak, 1986), our understanding of how aberrant tissue repair leads to tumor formation continues to evolve. Recent efforts have been initiated to delineate the roles of tissue-specific stem cells in the tissue repair and tumorigenesis processes.
The epidermis, which is the epithelial component of skin, is composed of the interfollicular epidermis (IFE) and various adnexal structures, such as the pilosebaceous unit (PSU), with differing functions. Whereas the IFE provides the barrier that protect against the outside environment and fluid evaporation, the PSU is the site of hair follicle growth and sebum production. Distinct stem cell populations ensure the lifelong replenishment of units with these specific functions (Schepeler et al., 2014). Lrig1+ cells are stem cells restricted to the upper PSU in normal skin, which are responsible for either the maintenance of the upper part of the PSU, the infundibulum, and the sebaceous gland (SG). Fate mapping experiments have demonstrated that Lrig1+ stem cells are confined to the PSU in unchallenged skin, making no contribution to the IFE (Page et al., 2013). Upon wounding, Lrig1+ stem cell progenies acquire lineage plasticity and are rapidly recruited into the wounded region, subsequently making permanent contributions to the regenerated epidermis (Jensen et al., 2009; Page et al., 2013). Expression of the oncogenic K-Ras G12D in Lrig1-expressing cells drives SG and infundibula hyperplasia without affecting the IFE significantly. Interestingly, upon wounding, oncogene activation (K-Ras G12D) in Lrig1+ cells drives rapid tumor formation within days (Page et al., 2013), providing an attractive model to assess roles of new pathways for wound-induced tumorigenesis.
A growing body of evidence suggests that chronic inflammation is the instigating factor for the development of cancerous lesions following abnormal tissue repair. The proinflammatory cytokine IL-17A is emerging as an important cytokine in cancer initiation and progression, including skin cancer (Numasaki et al., 2005; Wang et al., 2009, 2014; He et al., 2012; Wu et al., 2015). While IL-17A has been shown to play an essential role in tissue repair in the skin (MacLeod et al., 2013), anti–IL-17A antibody (Cosentyx; Novartis) is highly efficacious in treating psoriasis (Langley et al., 2014; Blauvelt et al., 2017), an inflammatory skin disease due to excessive hyperproliferation of keratinocytes (Bata-Csörgö and Szell, 2012). The receptor for IL-17 (IL-17A) is a heterodimeric complex composed of two subunits, IL-17RA and IL-17RC (Toy et al., 2006; Gaffen, 2009; Zhang et al., 2014). Upon ligand binding, the adaptor, Act1 (also known as CIKS), is recruited to the receptor, where it mediates downstream signaling (Chang et al., 2006; Qian et al., 2007). TNF receptor-associated factor (TRAF) proteins are immediate binding partners of Act1 and required for downstream pathway activation (Hartupee et al., 2009; Bulek et al., 2011; Sun et al., 2011; Zepp et al., 2012). We recently identified a novel IL-17A signaling cascade via the specific interaction of Act1 with TRAF4 to mediate MEKK3-dependent ERK5 activation that is critically important for keratinocyte proliferation and tumor formation (Wu et al., 2015). This suggests that IL-17A is potentially the critical link between inflammation, tissue repair, and tumorigenesis.
In this study, we report that IL-17A via epidermal growth factor receptor (EGFR) is required for the expansion of Lrig1+ stem cells in PSU and the migration of Lrig1+ stem cell progenies into the IFE during wound healing and wound-induced tumorigenesis. Mechanistically, IL-17R recruits EGFR for IL-17A signaling in the Lrig1+ cells. The direct interaction between IL-17R and EGFR is mediated by TRAF4, whose expression is enriched in Lrig1+ stem cells. Lrig1-specific deletion of IL-17R–EGFR axis and TRAF4 deficiency impaired IL-17A–induced Lrig1+ cell expansion. Biochemically, we showed that the close proximity of IL-17R and EGFR allows the adaptor protein Act1 to recruit c-Src for IL-17A–induced EGFR transactivation and subsequent ERK5 activation, which plays a critical role in IL-17A–dependent expansion of Lrig1+ stem cells, epidermal hyperplasia, and skin tumorigenesis. Since Lrig1 is an inhibitory molecule for EGFR signaling, our results suggest that the skin has preserved Lrig1+ stem cells for tissue repair, which are called into action when IL-17A is expressed to cooperate with EGFR during wounding. This study is the first example of how a proinflammatory cytokine recruits a growth factor receptor to activate stem cells in response to wounding for tissue repair and tumorigenesis.
Results
IL-17A–induced signaling in Lrig1+ stem cells promotes the migration and expansion of Lrig1 progenies for wound healing
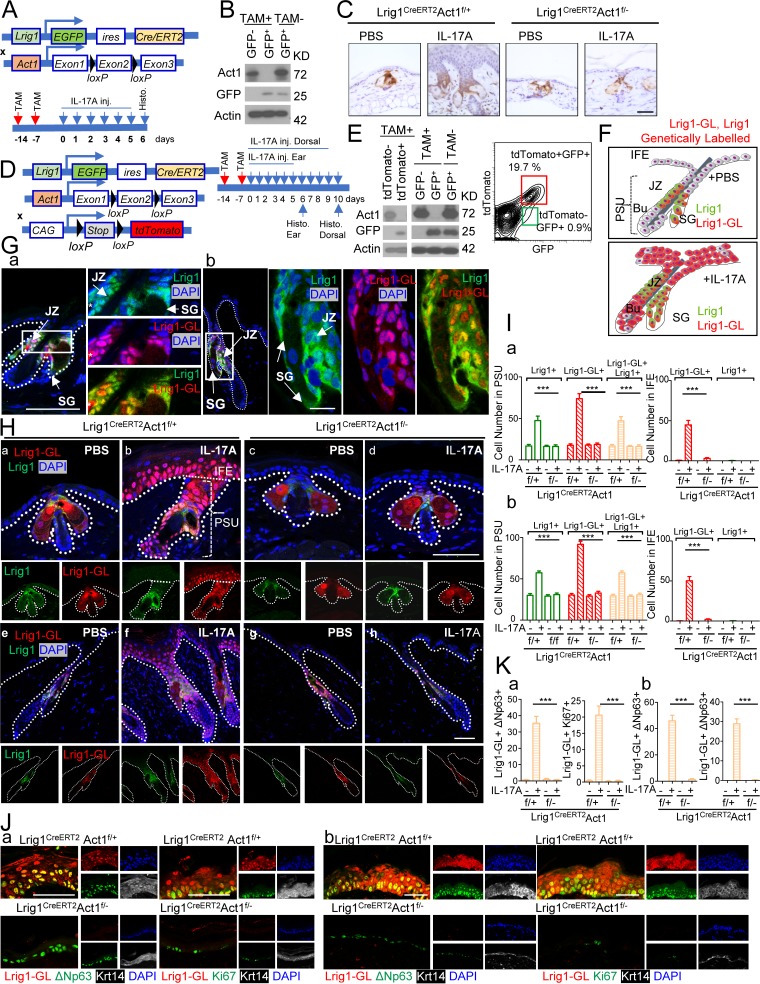
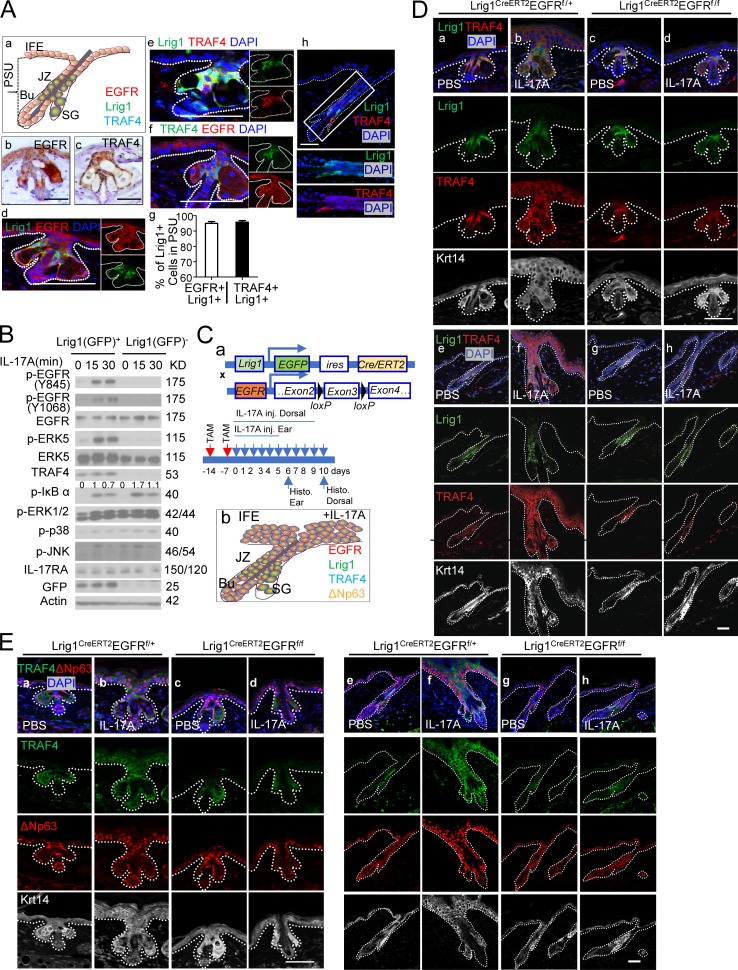
Lrig1 is a marker for stem cells that maintains the upper PSU, including the infundibulum and SG but not the IFE (Page et al., 2013). However, upon wounding, Lrig1+ stem cell progenies are rapidly recruited into the IFE for skin tissue repair, and the cells contribute to regenerating the epidermis (Page et al., 2013). Given the role of IL-17A signaling in skin wound healing (MacLeod et al., 2013), we hypothesize that IL-17A plays a role in the recruitment of Lrig1+ stem cell progenies into the wounded region for skin tissue repair. Upon stimulation with IL-17A, we indeed observed an expansion of Lrig1+ stem cells in the PSU (Fig. 1, A–C), suggesting that IL-17A may have a direct impact on the Lrig1+ stem cells. Deletion of Act1 in Lrig1+ cells (Fig. 1 A) abolished IL-17A–induced Lrig1+ stem cell expansion (Fig. 1, A–C) and reduced IL-17–mediated epidermal hyperplasia in the ear and dorsal skin (Fig. S1, A–C).
Figure 1.
IL-17A–induced signaling in Lrig1+ cells promotes the migration and expansion of Lrig1 progenies. (A) Schematic diagram for data shown in B and C. TAM, tamoxifen. (B) Epidermal cells from tamoxifen-treated Lrig1CreERT2Act1f/– mice before IL-17A injection were sorted and analyzed with Western blot. (C) Paraffin sections of the ear skin were stained for GFP. (D) Schematic diagram for E–K. (E) Epidermal cells from tamoxifen-treated Lrig1CreERT2Act1f/– ROSA26-lsl-tdTomato mouse were sorted for tdTomato or GFP followed by Western blot. Epidermal cells from ear skin of indicated mice were analyzed by flow cytometry for GFP and tdTomato. (F) Distribution of Lrig1+ cells and their progenies (Lrig1-GL, Lrig1 genetically labeled) cells in untreated or IL-17A–treated mouse skin. SG, sebaceous gland; JZ, junction zone; Bu, bulge. (G) Staining of Lrig1+ cells (green) and Lrig1 progenies (Lrig1-GL; red) in ear (a) and dorsal (b) PSU. The asterisk indicates autofluorescence. (H) Sections of PBS and IL-17A–injected ears (a–d) or dorsal skin (e–h) from littermate mice of the indicated genotypes. (I) Quantification of indicated cells in PBS- and IL-17A–injected ears (a) or dorsal skin (b). 10 PSUs and adjacent IFE were analyzed. n = 5 mice. (J and K) IL-17A–injected ear (a) and dorsal skin (b) from littermate mice were stained for ΔNp63, keratin 14, Ki67, and tdTomato. All bar graphs show means ± SEM. ***, P < 0.001, t test. Bars, 50 µm. All data are representative of three independent experiments.
Notably, deletion of Act1 in Lrig1+ cells was sufficient to attenuate IL-17A–induced epidermal hyperplasia in the IFE (Fig. 1 C). Since Lrig1+ cells remained in the PSU upon IL-17A injection (Fig. 1 C), we hypothesized that the IL-17A induced the migration of Lrig1 progenies to the epidermis. To test this hypothesis, we crossed the Lrig1CreERT2Act1f/− and Lrig1CreERT2Act1f/+ mice onto a ROSA26-lsl-tdTomato background (Fig. 1 D). Tamoxifen treatment induced efficient recombination at the ROSA26 locus and robust deletion of Act1 (Fig. 1 E). The Lrig1+ stem cells (GFP+) remained in the PSU (Fig. 1, F and G), while Lrig1 progenies (tdTomato+ and Lrig1-GL) were expanded and migrated to the IFE in response to IL-17A injection (Fig. 1, F–I). Lrig1-specific Act1 deficiency abolished IL-17A–induced expansion of Lrig1+ stem cells in the PSU and expansion/migration of Lrig1 progenies (tdTomato+GFP–) into the IFE (Fig. 1, F–I).
We have previously reported that IL-17A induces the expansion of p63+ cells in the IFE (Wu et al., 2015). Interestingly, Lrig1-specific Act1 deficiency abolished IL-17A–induced expansion of p63+ cells (Fig. S1, D and E). The expanded p63+ cells were tdTomato+ and highly proliferative (Ki67+) in both ear and dorsal skin (Fig. 1, J and K), indicative of their origin as Lrig1+ stem cells. Taken together, the data suggest that IL-17A induces the migration of Lrig1 progenies to the epidermis, transitioning to p63+ and Ki67+ cells.
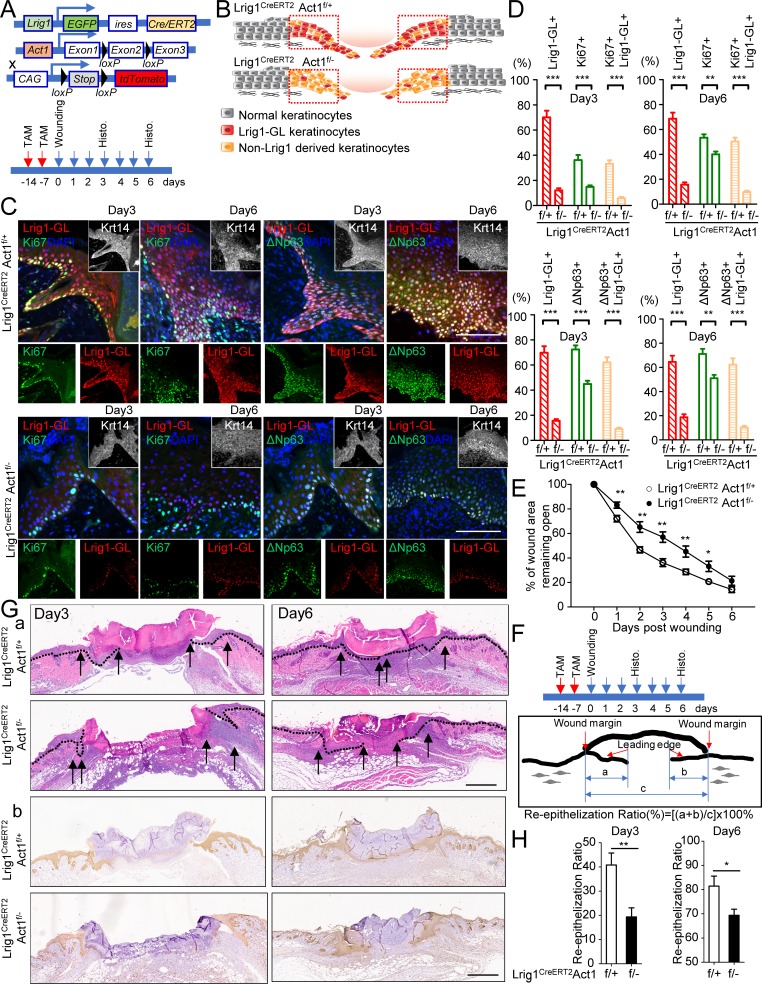
In response to tissue damage, the Lrig1 progenies rapidly migrate out of the PSU and contribute majorly to wound repair (Aragona et al., 2017). Tissue damage robustly induces a variety of inflammatory cytokines, including IL-17A (MacLeod et al., 2013). We found that IL-17RC–deficient mice exhibited impaired wound healing (Fig. S1, F and G), which prompted us to examine whether IL-17A–induced signaling in the Lrig1+ stem cells contributed to the wound-healing process. Lineage tracing confirmed that the Lrig1 progenies contributed majorly to the leading edge (Fig. 2, A–D). The migration and proliferation of Lrig1 progenies (Fig. 2, B–D) induced by wounding were greatly diminished in Lrig1CreERT2Act1f/− mice compared with the control mice. Consistently, deletion of Act1 in Lrig1+ cells led to delayed closure of the wound opening (Fig. 2 E). Although the gross sizes of the wound became comparable by day 6 (Fig. 2 E), the extent of the reepithelialization in the Lrig1CreERT2Act1f/− mice was significantly reduced compared with the control mice (Fig. 2, F–H). Additionally, histology analysis of the wound on day 3 (Fig. 2, F–H) showed both larger wound size and compromised reepithelization in Lrig1CreERT2Act1f/− mice compared with that in the control mice. Taken together, our data indicate that IL-17A–induced migration and expansion of Lrig1 progenies (Lrig1-GL) participate in reepithelization to promote wound healing.
Figure 2.
IL-17A–induced signaling in Lrig1+ cells is required for the migration and expansion of Lrig1 progenies for wound healing. (A) Schematic diagram for data in B–D. TAM, tamoxifen. (B) Distribution of Lrig1 progenies in a healing wound. (C) Paraffin sections of reepithelializing wound from littermate mice were stained for tdTomato (red) and Ki67 (green) or ΔNp63 (green). Bar, 50 µm. (D) Quantifications of the indicated cells as a percentage of Krt14+ cells in the leading edge of the wound area. n = 10. Data are representative of three independent experiments shown in C and D. (E) Wound-closure kinetics in littermate mice of indicated genotypes. Graph shows the mean percentage of surface area that remained open as a percentage of original size. n = 10 wounds. (F) Schematic diagram in reference to G and H. The lengths of leading edges and the initial wound length were defined as indicated. (G) H&E staining (a) and staining for Krt14 (b) in reepithelializing wound. Bar, 500 µm. (H) Measurement of reepithelization ratio in wound area of Lrig1CreERT2Act1f/+ and littermate Lrig1CreERT2Act1f/– mice. n = 10 wounds. Data are representative of three independent experiments shown in E–H. All bar graphs in Fig. 2 show means ± SEM. ***, P < 0.001; **, P < 0.01; *, P < 0.05, t test.
IL-17A–induced signaling in Lrig1+ stem cells drives Lrig1 progenies to participate in skin tumorigenesis
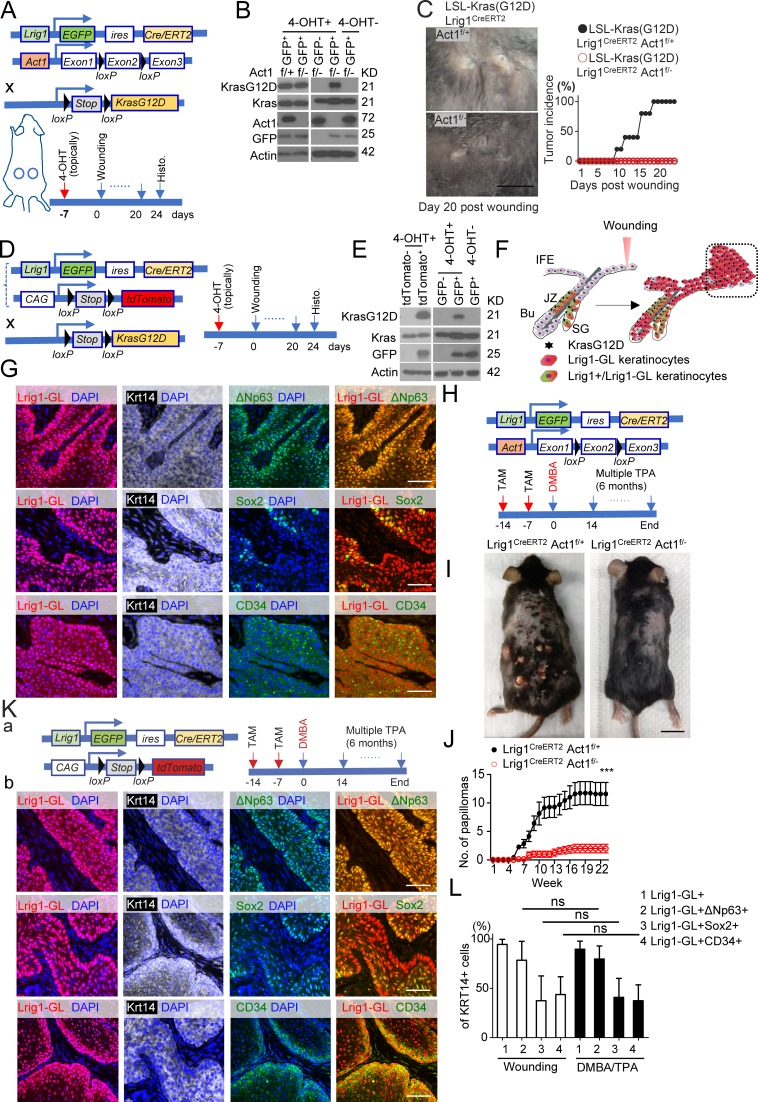
Oncogene activation (K-Ras G12D) in Lrig1+ cells drives wound-induced tumor formation within days (Page et al., 2013). Loss of Act1 impaired wound-induced tumorigenesis in the LSL-K-Ras (G12D)/Lrig1ERCreT2Act1f/– model (Fig. 3, A–C). Importantly, tamoxifen application induced efficient and specific KrasG12D expression and Act1 deletion (Fig. 3 B). In addition, KrasG12D was only activated in Lrig1 progenies tdTomato+ but not tdTomato− cells (Fig. 3 E). As a result, all the tumor cells in the wound-induced tumors were Lrig1 progenies (Fig. 3, D–G). Notably, these Lrig1 progenies in the tumor expressed stem cell markers, p63 (ΔNp63), Sox2, and CD34 (Boumahdi et al., 2014; Fig. 3, D–G). The data indicate that IL-17A–induced signaling in Lrig1+ cells is required to drive the oncogene-bearing Lrig1 progenies to form tumors in response to wounding (Fig. 3 F).
Figure 3.
IL-17A–induced signaling in Lrig1+ cells drives Lrig1 progenies to participate in skin tumorigenesis. (A) Schematic diagram showing Cre-mediated ablation of Act1 and activation of KrasG12D in Lrig1 lineage cells. Time frame refers to B and C. 4-OHT, 4-hydroxytamoxifen. (B) Epidermal cells sorted based on GFP from LSL-Kras(G12D)Lrig1CreERT2Act1f/– mice were analyzed by Western blot. (C) Representative macroscopic photograph (day 20; A) of wound-induced tumors in indicated littermate mice and quantification of tumor incidence as a function of time. 10 wounds were evaluated per group. Data are representative of two independent experiments. Bar, 1 cm. (D) Schematic diagram for data shown in E–G. (E) Western blot analysis of epidermal cells sorted based on tdTomato or GFP from mice before wounding. (F) Schematic diagram showing the migration of Lrig1 progenies to the wounding site for tumorigenesis. SG, sebaceous gland; JZ, junction zone; Bu, bulge. (G) Wound-induced tumors were stained for indicated markers. Bar, 50 µm. Data were verified in three independent experiments shown in E–G. (H) Schematic diagram for data shown in I and J. (I and J) Photographs (I) and average tumor numbers (J) of the DMBA/TPA. n = 7. Data are representative of three independent experiments. Bar, 2 cm. (K) Schematic diagram for lineage tracing in DMBA/TPA model (a). DMBA/TPA-induced tumors from Lrig1CreERT2 ROSA26-lsl-tdTomato mice were stained for ΔNp63+ (green), Sox2+ (green), or CD34+ (green; b). Bars, 50 µm. (L) Quantification of the percentages of indicated cells in the Krt14+ tumor cells. 56 tumors were analyzed for tdTomato and Krt14; 14 tumors were analyzed for Krt14 in combination with ΔNp63, Sox2, or CD34. All bar graphs in Fig. 3 show means ± SEM. ***, P < 0.001; ns, not significant (P > 0.05), t test.
We have previously shown that ablation of Act1 from the whole epidermis using ectoderm-Cre K5 protects mice from 7,12-dimenthylbenz[a]anthracene (DMBA)/12-O-tetradecaboylphorbol-13-acetate (TPA)–induced skin tumorigenesis (Wu et al., 2015). One important question is whether IL-17A–induced signaling in Lrig1+ stem cells might be required for skin tumorigenesis induced by DMBA/TPA. Lrig1-specific Act1 deficiency indeed protected mice from DMBA/TPA-induced carcinogenesis (Fig. 3, H–J). In line with this observation, Lrig1-specific IL-17RC deficiency also significantly reduced the DMBA/TPA-induced tumorigenesis (Fig. S2 A). Lineage tracing revealed that the tdTomato+ cells were specifically detected in the tumor cells but not in the stromal cells (Fig. 3, K and L). Furthermore, >90% of the constituent tumor cells were tdTomato+ (Fig. 3, K and L), indicating that the Lrig1 progenies derived from the Lrig1+ stem cells contributed greatly to the tumor mass (Fig. S2 B).
Similar to the wound-induced tumors, the DMBA/TPA-induced tumors also expressed stem cell markers, p63, Sox2, and CD34. Interestingly, while Lrig1 was not induced in other subsets (tdTomato− cells) in the DMBA and TPA challenged dorsal skin before tumor formation (Fig. S2 C), we detected occasional foci of Lrig1+ cells in ∼20% of the DMBA/TPA-induced carcinomas but not in the other challenged conditions (Fig. S2 D). Importantly, the Lrig1+ cells in the DMBA/TPA-induced tumors were also positive for tdTomato, indicating that they had been derived from the Lrig1+ stem cells (Fig. 3, K and L; and Fig. S2 E). Since Lrig1 progenies lose the Lrig1 expression upon exiting the hair follicle (Fig. 1, F–I; Page et al., 2013), the Lrig1+ cells occasionally detected in the tumor most likely had reacquired the Lrig1 expression during the tumor progression.
IL-17A–induced EGFR activation in Lrig1+ stem cells is required for IL-17A–dependent wound healing and tumorigenesis
We reported that IL-17A–induced Act1/TRAF4-mediated ERK5 cascade stimulates epidermal hyperplasia and tumor formation. The ERK5 inhibitor effectively blocked IL-17A–induced expansion of Lrig1+ stem cells in the PSU and migration and expansion of Lrig1 progenies (Lrig1-GL and tdTomato) to the IFE of the ear and dorsal skins (Fig. S3 A). These results indicate that IL-17A–ERK5 axis plays a critical role in Lrig1+ stem cells for their expansion and migration.
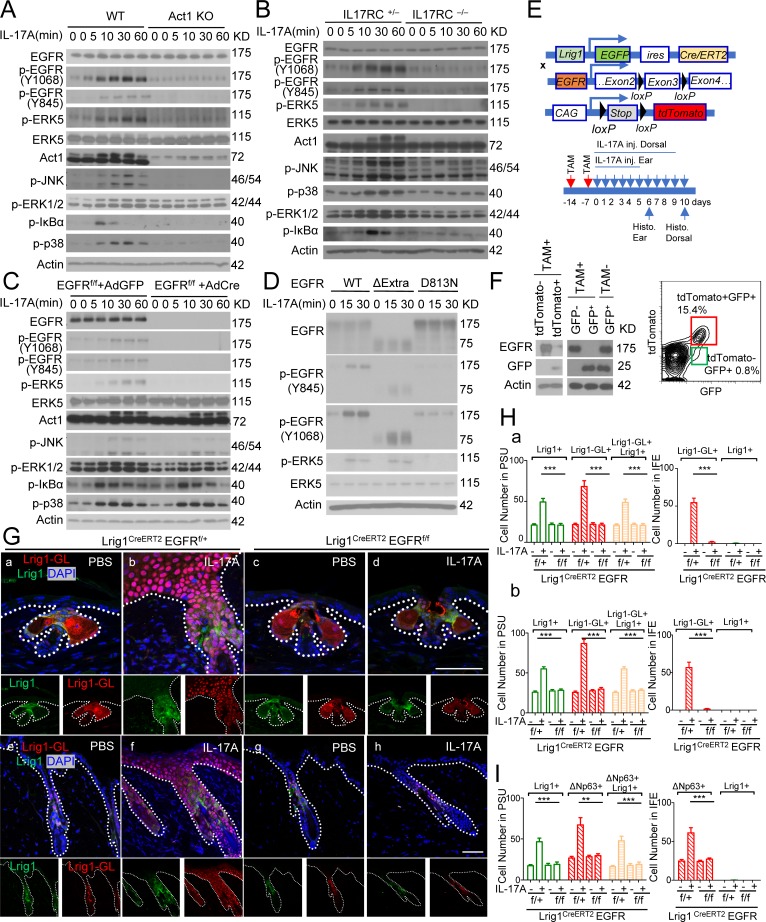
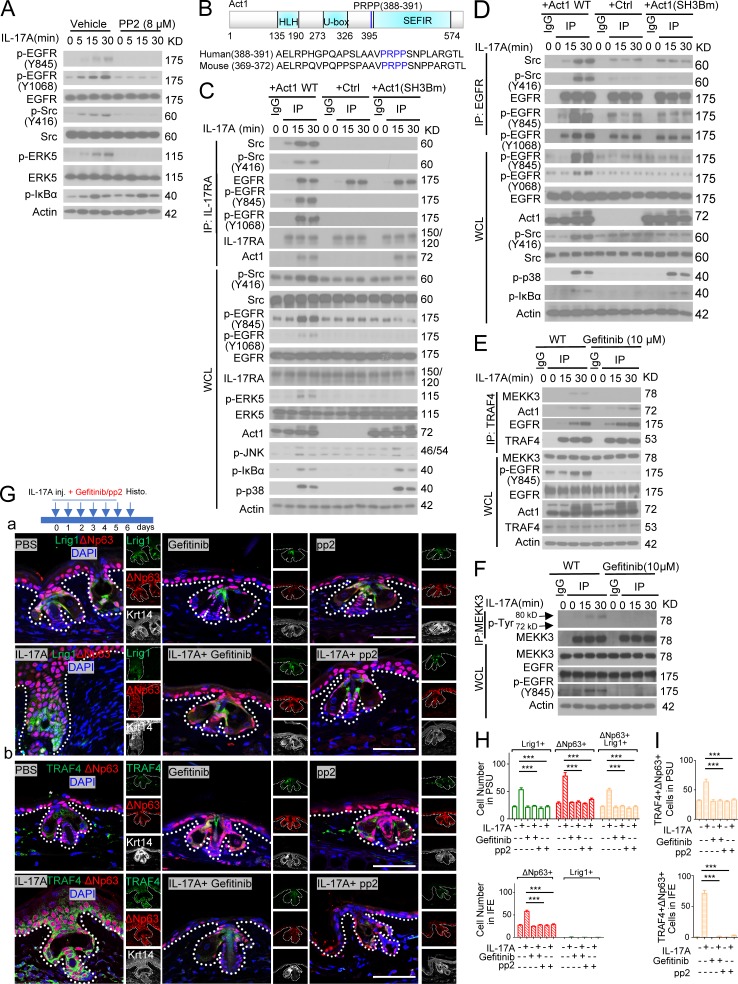
Tyrosine phosphorylation is implicated in the activation of MEKK3–ERK5 cascade (Chao et al., 1999; Su et al., 2001; Sun et al., 2001; Takeda et al., 2014). In searching for a tyrosine kinase, we found that IL-17 induced EGFR phosphorylation (Y-1068 and Y-845), which depended upon both IL-17RC and Act1 (Fig. 4, A and B; and Fig. S3 B). Importantly, IL-17A–induced p-ERK5, but not p-IκBα, p-p38, and p-JNK, was impaired in EGFR-deficient keratinocytes (Fig. 4 C). Consistently, IL-17A–induced ERK5 phosphorylation was also abolished in gefitinib-treated keratinocytes (Fig. S3 C). In EGFR-deficient HeLa cells, IL-17A–induced EGFR and ERK5 activation was restored by reconstituted WT EGFR and EGFR mutant without the extracellular domain but not with the kinase-inactive EGFR mutant (Fig. 4 D). Since EGFR deletion mutant without the extracellular domain was able to mediate IL-17A–induced ERK5 activation (Fig. 4 D), IL-17A–induced EGFR phosphorylation is unlikely to be due to enhanced shedding of EGFR ligands. Intradermal IL-17A injection resulted in activation of EGFR and ERK5, which were blocked by coinjection of EGFR inhibitor gefitinib (Fig. S3 D). IL-17A–induced epidermal hyperplasia was substantially reduced in the presence of EGFR inhibitor (Fig. S3 E), highlighting the role of EGFR kinase activity.
Figure 4.
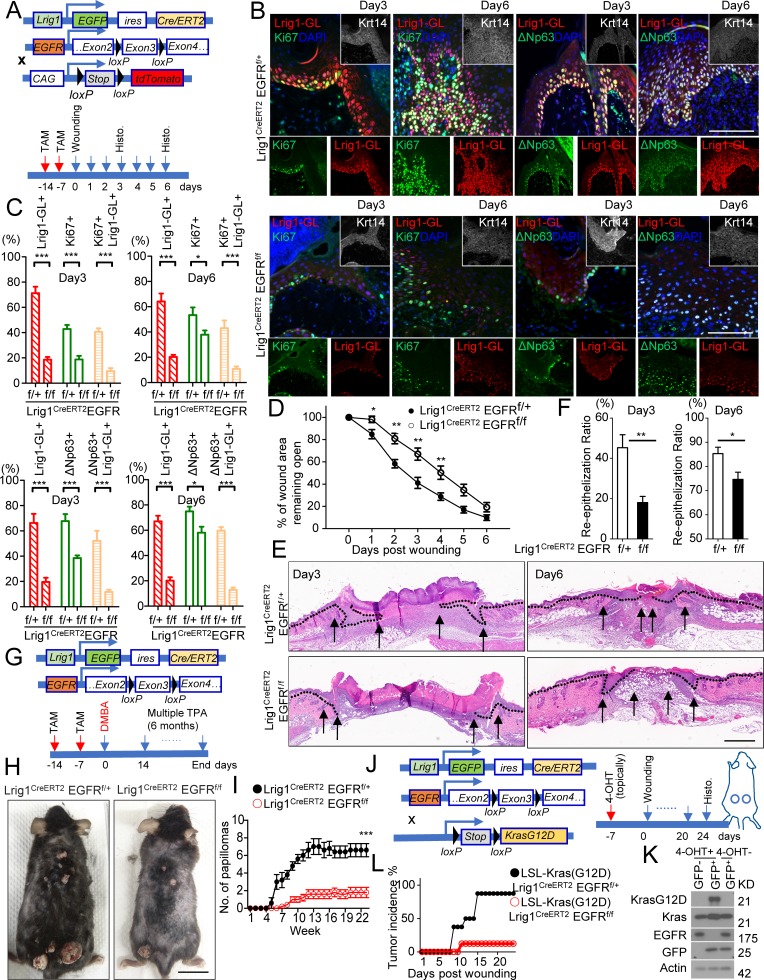
IL-17A–induced EGFR activation in Lrig1+ stem cells is required for the migration and expansion of Lrig1 progenies. (A and B) Keratinocytes isolated from indicated mice were stimulated with IL-17A followed by Western blot analysis. (C) EGFRf/f primary keratinocytes infected with adenovirus expressing Cre-recombinase or GFP were then stimulated with IL-17A, followed by Western blot analysis. (D) EGFR-deficient HeLa cells reconstituted with WT EGFR, extracellular domain deletion (ΔExtra), and kinase dead mutant (D813N) were stimulated with IL-17A followed by Western blot analysis. Data shown in A–D are representative of three experiments. (E) Schematic diagram for data in F–H. TAM, tamoxifen. (F) Recombination efficiency in epidermal cells isolated from tamoxifen-injected Lrig1CreERT2EGFRf/f ROSA26-lsl-tdTomato mice. (G and H) Sections of PBS and IL-17A–injected ears (G, a–d; and H, a) and dorsal skin (G, e–h; and H, b) of the indicated littermate mice were analyzed for Lrig1 (green) and tdTomato (red; G). Representative PSU and adjacent IFE are shown. Quantification of indicated cells is presented in H. 10 PSUs and adjacent IFE were analyzed. n = 5 mice. Bar, 50 µm. Data are representative of three independent experiments in G (a–d) and two independent experiments in G (e–h). (I) Quantification of indicated cells for Fig. S3 H. 10 PSUs and adjacent IFE were analyzed. n = 5 mice. All bar graphs show means ± SEM. ***, P < 0.001; **, P < 0.01, t test. Bars, 50 µm. Data are representative of three independent experiments.
We next examined the impact of Lrig1-specific EGFR deficiency on the expansion and migration of Lrig1 progenies in response to IL-17A stimulation. Using the same strategy (Fig. 1 D), we deleted EGFR specifically in Lrig1+ stem cells before IL-17A injection (Fig. 4, E and F). Deletion of EGFR in Lrig1+ cells abolished IL-17A–induced Lrig1+ cell expansion in the PSU and ablated the migration of Lrig1 progenies to the epidermis of both ear and dorsal skin (Lrig1-GL; Fig. 4, G and H). Lrig1-specific EGFR deficiency also diminished IL-17A–induced p63+ cell expansion in the epidermis (Fig. 4 I and Fig. S3, F–H) and attenuated epidermal hyperplasia (Fig. S3 I). Taken together, these results suggest that IL-17A–induced EGFR signaling in Lrig1+ stem cells promotes expansion of Lrig1+ stem cells and the migration of Lrig1 progenies.
Consistently, the migration and proliferation of Lrig1+ progenies induced by wounding was greatly impaired in Lrig1CreERT2 EGFRf/f mice compared with their respective control mice (Fig. 5, A–C). Deletion of EGFR in Lrig1+ stem cells led to delayed closure of the wound opening, accompanied with compromised reepithelization in Lrig1CreERT2EGFRf/f mice (Fig. 5, D–F). Moreover, Lrig1-specific EGFR deficiency protected mice from DMBA/TPA-induced carcinogenesis (Fig. 5, G–I) and also attenuated wounding-induced tumorigenesis on the back (Fig. 5, J–L). These results demonstrate that the IL-17A–EGFR axis plays a critical role in promoting the expansion of Lrig1+ stem cells and the migration of Lrig1 progenies to the IFE linking wound healing to skin tumorigenesis.
Figure 5.
IL-17A–induced EGFR activation in Lrig1+ stem cells is required for IL-17A–dependent wound healing and tumorigenesis. (A) Schematic diagram for data shown in B–D. TAM, tamoxifen. (B) Paraffin sections of reepithelializing wound from littermate mice were stained for tdTomato (red) and Ki67 (green) or ΔNp63 (green). Bar, 50 µm. (C) Quantifications of the indicated cells as a percentage of Krt14+ cells in the leading edge of the wound area. n = 10 wounds. Data are representative of three independent experiments shown in B and C. (D) Wound-closure kinetics in age-matched littermate mice of indicated genotypes. Graph shows the mean percentage of surface area that remained open as a percentage of original size of the wound. n = 10 mice per group. (E) H&E staining of reepithelializing wound. Bar, 500 µm. (F) Measurement of reepithelization ratio (leading edge ratio). Data are representative of three independent experiments shown in D–F. (G) Schematic diagram for data shown in H and I. (H and I) Photographs (H) and average tumor numbers (I) of representative mice after DMBA/TPA treatment. n = 5 mice per group. Data are representative of three independent experiments. Bar, 2 cm. (J) Schematic diagram for data shown in K and L. (K) Western blot analysis of recombinant efficiency at KrasG12D and EGFR loci in epidermal cells sorted based on GFP from LSL-Kras(G12D) Lrig1CreERT2EGFRf/f mice. (L) Tumor incidence of wound-induced tumorigenesis in littermate mice of indicated genotype. n = 8 wounds per group. Data are representative of three independent experiments. All bar graphs show ± SEM. ***, P < 0.001; **, P < 0.01; *, P < 0.05, t test.
TRAF4 is enriched in Lrig1+ stem cells and is required for IL-17A–induced IL-17RA–EGFR interaction
We then studied how IL-17A induces EGFR activation in Lrig1+ stem cells. Lrig1 has been implicated in inhibiting EGF binding to EGFR (Goldoni et al., 2007). As EGFR was colocalized with Lrig1 in the upper PSU (Fig. 6 A), the inhibition of EGF signaling by Lrig1 might be an important feature of Lrig1+ cells that is required for IL-17R to adopt EGFR for IL-17 signaling. Despite comparable IL-17R and EGFR expression, Lrig1+ cells were much more responsive to IL-17A–induced EGFR and ERK5 activation than that in Lrig1– cells (Fig. 6, A and B). On the other hand, TRAF4 protein levels were much higher in Lrig1+ cells than that in Lrig1− (Fig. 6 B), Lgr5+, and Lgr6+ cells (Fig. S4, B and C), which is consistent with the lack of IL-17A–induced ERK5 activation (Fig. 6 B and Fig. S4, B and C). Of note, IL-17RC deficiency in Lgr5 cells did not have a noticeable impact on IL-17A–induced skin hyperplasia or DMBA/TPA-induced skin tumorigenesis (Fig. S4, D and E). The data collectively imply a potential role for TRAF4 to mediate IL-17A–induced Lrig1+ cell expansion and the migration of Lrig1 progenies.
Figure 6.
TRAF4 is enriched in Lrig1+ stem cells. (A) Lrig1, EGFR, and TRAF4 expression in PSU and IFE in unchallenged skin. Paraffin sections of mouse ear were stained for EGFR, TRAF4, and Lrig1 (a–f). SG, sebaceous gland; JZ, junction zone; Bu, bulge. Quantification (g) of indicated cells over Lrig1+ cells in the PSU is presented. n = 15 hair follicles. Error bars represent ± SEM. Immunofluorescence staining for Lrig1 and TRAF4 was performed in dorsal skin (h). (B) Epidermal cells isolated from Lrig1CreERT2 mice sorted based on GFP (Lrig1) expression followed by Western blot analysis. Data are representative of three independent experiments. Normalized densitometric quantifications of p-IκBα are annotated above the blot. (C) Schematic diagram for data shown in D and E and Fig. S4 (F–I, a). Schematic summary of EGFR, Lrig1, TRAF4, and ΔNp63 expression pattern in IL-17A–injected skin (b). TAM, tamoxifen. (D and E) Ear and dorsal skin sections from indicated mice stained for Lrig1 (green) and TRAF4 (red; for ear [a–d] and dorsal skin [e–h]; D) or TRAF4 (green) and p63 (ΔNp63; red; for ear [a–d] and dorsal skin [e–h]; E). Representative PSU and adjacent IFE are shown. n = 5 mice per group. Data are representative of three independent experiments shown in D and E. Bars, 50 µm.
TRAF4 expression was indeed highly expressed in the infundibulum and SG (Fig. 6 A), which coincided with Lrig1 expression in the ear and dorsal skin (Fig. 6 A, e and h). Interestingly, IL-17A injection greatly induced the expansion of TRAF4+ cells in the PSU as well as in the IFE of both ear (Fig. 6 D, a–d) and dorsal skin (Fig. 6 D, e–h), colocalizing with p63+ cells (Fig. 6 E). Importantly, lineage tracing revealed that TRAF4 expression was exclusively induced in the Lrig1 and Lrig1 progenies in response to IL-17A injection and in wound- or DMBA/TPA-induced tumors (Fig. S4 A). Deletion of Act1 or EGFR in Lrig1+ cells abolished the expansion of TRAF4+ cells (Fig. 6, C–E; and Fig. S4, F–P). Importantly, IL-17A–induced Lrig1+ cell expansion was abolished in TRAF4-deficient mice (Fig. S4 Q). We detected EGFR–IL-17RA interaction in the PSU and IFE in IL-17A–treated epidermal tissue (Fig. S4 R), which was abolished by TRAF4 deficiency (Fig. S4 R). Based on these findings, we propose that TRAF4 may first facilitate the interaction of EGFR to IL-17RA in response to IL-17A stimulation, followed by bridging Act1 to MEKK3 for the activation of EGFR–ERK5 cascade (Wu et al., 2015), promoting Lrig1+ stem cell expansion and transition to p63+ epidermal progenitor cells.
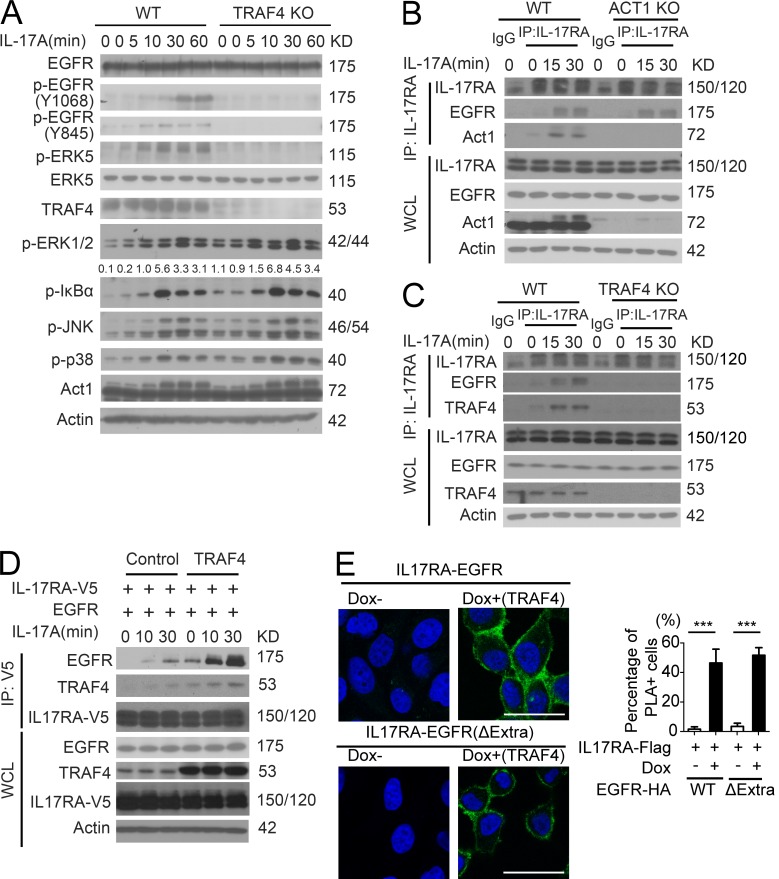
Notably, the cultured primary keratinocytes also expressed TRAF4 and were responsive to IL-17A treatment (Fig. 7 A). IL-17A–induced EGFR and ERK5 phosphorylation were abolished in TRAF4-deficient keratinocytes, suggesting that TRAF4 is required for IL-17A–induced EGFR phosphorylation and subsequent ERK5 activation (Fig. 7 A). IL-17A was still able to induce EGFR–IL-17RA interaction in Act1-deficient keratinocytes (Fig. 7 B), indicating that IL-17A–induced EGFR–IL-17RA complex formation was Act1 independent. In contrast, IL-17A failed to induce recruitment of EGFR to IL-17RA in TRAF4-deficient keratinocytes (Fig. 7 C). Furthermore, IL-17A–induced interaction between IL-17RA and EGFR was promoted by TRAF4 overexpression (Fig. 7 D). IL-17RA interacted with EGFR upon induction of TRAF4 by doxycycline (Fig. 7 E). Importantly, only TRAF4 but not TRAF3 or TRAF6 was able to promote the interaction between EGFR and IL-17RA, indicating that TRAF4 has the unique ability to bridge IL-17RA to EGFR (Fig. S4 S).
Figure 7.
TRAF4 is required for IL-17A–induced IL-17RA–EGFR interaction. (A) TRAF4 WT and KO primary keratinocytes were treated with IL-17A followed by Western blot analysis. Normalized densitometric quantifications of p-IκBα are annotated above the blot. (B and C) Primary keratinocytes from WT and Act1 KO mice (B) or TRAF4 KO mice (C) treated with 100 ng/ml IL-17A were subjected to coimmunoprecipitation with anti-mouse–IL-17RA followed by Western blot analysis. WCL, whole cell lysate; IP, immunoprecipitation. (D) HeLa cells were transfected with IL-17RA and a vector or TRAF4. Transfected cells were treated with IL-17A and subjected to coimmunoprecipitation with anti-V5 (IL-17RA) followed by Western blot analysis. Data are representative of three independent experiments shown in A–D. (E) Stable HeLa cells that expressed TRAF4 in response to doxycycline were transfected with HA-tagged EGFR or the extracellular domain deletion mutant (ΔExtra), together with IL-17RA-Flag. PLAs were performed using anti-FLAG and anti-HA after 24 h of induction for TRAF4. Data are representative of at least three experiments. Graph represents percentage of PLA-positive cells from 10 fields. Bar graphs show ± SEM. ***, P < 0.001, t test. Scale bars, 50 µm.
TRAF4 tethers IL-17RA and EGFR through the TRAF domain
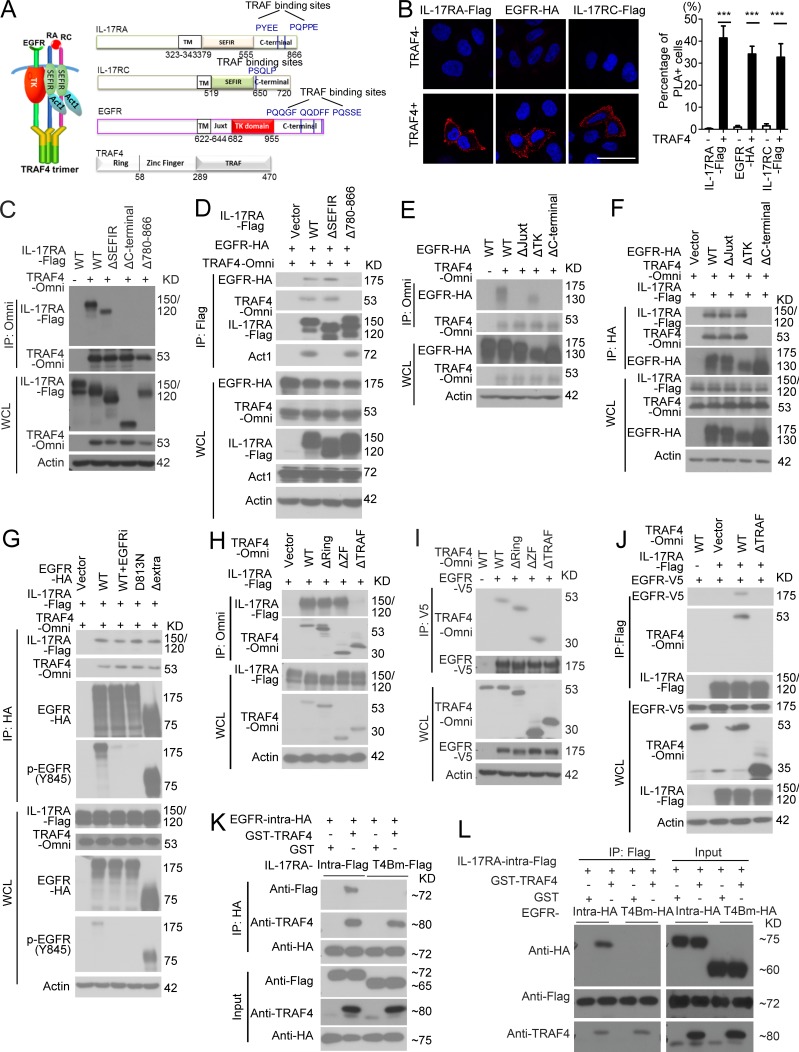
We then investigated how TRAF4 promotes EGFR–IL-17RA interaction. Notably, the intracellular domain of EGFR is sufficient for the TRAF4-promoted interaction between EGFR and IL-17RA (Figs. 8 A and 7 E). TRAF4 was able to interact with IL-17RA, IL-17RC, and EGFR (Fig. 8, A and B), suggesting that TRAF4 might bridge EGFR to IL-17RA-RC via its interaction with both receptors. While the deletion of SEFIR domain abolished the interaction of Act1 with the receptors (Qian et al., 2007; Liu et al., 2009), it had no impact on the interaction of TRAF4 with IL-17RA or IL-17RC (Fig. 8, C and D; and Fig. S5 A). Further deletion analysis showed that the C-terminal domain of IL-17RA-RC is required for their interaction with TRAF4 (Fig. 8, C and D; and Fig. S5 A). These results confirm that TRAF4, but not Act1, is required for EGFR’s recruitment to IL-17 receptor.
Figure 8.
TRAF4 tethers IL-17RA and EGFR through the TRAF domain. (A) A model for IL-17–induced IL-17R–EGFR complex formation. RA, IL-17RA; RC, IL-17RC; TK, tyrosine kinase domain; TM, transmembrane domain. (B) TRAF4-inducible HeLa clones were transfected with indicated plasmids, followed by induction of TRAF4 expression. Protein–protein interactions were detected by PLA (red dots). Graph represents mean percentage of PLA-positive cells from 10 fields. Error bars represent ± SEM. Data are representative of at least three experiments. ***, P < 0.001, t test. Bar, 50 µm. (C and D) IL-17RA-KO HeLa cells were transfected with indicated plasmids followed by coimmunoprecipitation. Deleted regions in mutant protein are shown in A. (E and F) EGFR-deficient HeLa cells were transfected with indicated plasmids and subjected to coimmunoprecipitation. Deleted regions in mutant protein are shown in A. (G) IL-17RA was cotransfected with EGFR (with/without pretreatment of gefitinib, EGFRi), kinase dead EGFR (D813N), or the extracellular domain deletion mutant (ΔExtra1-600) into EGFR-deficient HeLa cells with overexpression of TRAF4. Cell lysates were immunoprecipitated with anti-HA (EGFR), followed by Western blot analysis. (H) TRAF4-deficient HeLa cells were transfected with indicated TRAF4 plasmids (ΔRing, ΔZincFinger [ZF], and ΔTRAF) followed by coimmunoprecipitation. (I) TRAF4-deficient HeLa cells were transfected with the indicated plasmids followed by coimmunoprecipitation. (J) Structure–function analysis of TRAF4 for its ability to promote IL-17RA–EGFR interaction. All data are representative of three independent experiments shown in C–J. WCL, whole cell lysate; IP, immunoprecipitation. (K and L) Pull-down experiments with recombinant IL-17RA, TRAF4, and EGFR proteins. Data are representative of three independent pull-down experiments.
We then mapped the domain of EGFR required for its interaction with TRAF4. While deletion of kinase domain had no impact on the EGFR–TRAF4 interaction, removal of the juxtamembrane region or the C-terminal domain (Δ955–1186) of EGFR abolished its interaction with TRAF4 (Fig. 8 E). The C-terminal domain but not the juxtamembrane domain of EGFR was required for the recruitment of TRAF4 to EGFR and also EGFR’s interaction with IL-17RA (Fig. 8 F). Of note, EGFR inhibitor and mutation of the catalytic site of the EGFR kinase domain did not impact the recruitment of TRAF4 and EGFR to IL-17RA (Fig. 8 G). Importantly, the whole extracellular domain of EGFR is dispensable for the formation of IL-17RA–EGFR complex (Fig. 8 G and Fig. 7 E), confirming that the IL-17A–induced EGFR activation is not via recognition of EGF ligands.
The next question is which domains of TRAF4 are required for its interaction with IL-17RA and EGFR. Deletion of TRAF domain, but not Ring or Zinc finger domain, abolished the interaction of TRAF4 with IL-17RA and EGFR (Fig. 8, H and I). We found that deletion of TRAF domain of TRAF4 abolished the recruitment of EGFR to IL-17 receptor (Fig. 8 J). Notably, putative TRAF binding sites in the C-terminal regions of IL-17RA, IL-17RC, and EGFR were found to be important for the interaction with TRAF4 by our structure–function analysis (Fig. 8, A and C–F). In support of this, the recombinant WT but not TRAF-binding mutant of intracellular domains of IL-17RA and EGFR were able to directly interact with purified recombinant TRAF4 in vitro and interacted with each other in the presence of TRAF4 (Fig. 8, K and L; and Fig. S5 B). Taken together, these results suggest that TRAF4 directly mediates the interaction between IL-17RA and EGFR via their TRAF binding sites.
Tyrosine kinase c-Src is required for IL-17–induced EGFR transactivation
EGFR kinase is necessary for IL-17A–induced ERK5 activation (Fig. 4 D and Fig. S3 C). The IL-17R adaptor Act1 (via SEFIR–SEFIR interaction) is required for IL-17A–induced EGFR and ERK5 activation (Fig. 4 A), although Act1 is dispensable for IL-17–induced EGFR–IL-17RA complex formation (Figs. 7 B and 8, C and D). The question was then how Act1 mediates IL-17A–induced EGFR activation. Notably, Src kinase has been shown to play an important role in transactivation of EGFR (without EGF stimulation; Wu et al., 2002; Drube et al., 2006; Dewar et al., 2007; Hsieh et al., 2012). Interestingly, IL-17A–induced Src phosphorylation and Src inhibitor completely blocked IL-17A–induced EGFR and ERK5 activation (Fig. 9 A). IL-17A stimulation induced the interaction of Src to the IL-17R, which was abolished in Act1-deficient cells (Fig. 9 C), suggesting that Act1 is required for the recruitment of Src to the IL-17R–EGFR complex, resulting in EGFR activation. Src inhibitor blocked IL-17–induced EGFR activation without affecting the recruitment of EGFR to IL-17RA (Fig. S5 D), suggesting that EGFR’s interaction with IL-17R precedes EGFR activation.
Figure 9.
Tyrosine kinase c-Src is required for IL-17–induced EGFR transactivation. (A) Primary keratinocytes with and without pretreatment of Src inhibitor (PP2, 8 µM) were stimulated with IL-17A followed by Western blot analysis. (B) Putative SH3 binding motif in Act1. (C) Act1 KO keratinocytes were infected with retrovirus expressing Act1 WT, empty vector, or Act1 SH3 binding mutant (PRPP-ARAA, Act1SH3Bm) and treated with IL-17A. Treated cells were subjected to coimmunoprecipitation followed by Western blot. (D) Act1 KO keratinocytes were restored with WT Act1 and mutants as in C. Cells were then treated with IL-17A and subjected to coimmunoprecipitation. (E) Primary keratinocytes pretreated with 10 µM gefitinib were stimulated with IL-17A. Cell lysates were then immunoprecipitated with anti–TRAF4. (F) Primary keratinocytes with pretreatment of gefitinib were stimulated with IL-17A followed by coimmunoprecipitation Western blot. Data are representative of three independent experiments shown in A–F. WCL, whole cell lysate; IP, immunoprecipitation. (G) Lrig1CreERT2 mice were intradermally injected with PBS and IL-17A combined with gefitinib or pp2. Ear skin sections from these age-matched littermate mice were stained for Lrig1 (green) and ΔNp63 (red; a) or TRAF4 (green) and ΔNp63 (red; b). Bars, 50 µm. The asterisk indicates nonspecific staining. (H and I) Quantification of indicated cells for the data presented in Fig. 6 G. 10 PSUs and adjacent IFE were analyzed each group. Bar graphs show means ± SEM. ***, P < 0.001, t test. Data are representative of three independent experiments.
One important question is how Src is recruited to the IL-17R–EGFR complex. Src homology 3 (SH3) domain in Src protein is well known for protein–protein interactions necessary for Src-mediated signaling and function (Mayer and Baltimore, 1993; Feng et al., 1994; Holmes et al., 1996; Saksela and Permi, 2012). Interestingly, sequence analysis identified a putative SH3 binding motif in Act1 (Saksela and Permi, 2012; Fig. 9 B). We found that the SH3 binding motif in Act1 was indeed required for Src’s recruitment to IL-17R–EGFR complex in response to IL-17A stimulation (Fig. 9 C) and IL-17A–induced EGFR and Src activation (Fig. 9 D). In addition to Act1, IL-17A–induced Src’s recruitment to IL-17RA was also TRAF4 and EGFR dependent (Fig. S5, E and F). Furthermore, TRAF4 was also required for IL-17A–induced recruitment of Src to EGFR (Fig. S5 G). Our mutagenesis data suggest that Act1–Src interaction via the SH3 domain facilitates the recruitment of Src to the IL-17R/Act1–EGFR complex, providing a potential mechanism by which Src can be engaged in the IL-17RA–EGFR complex to mediate transactivation of EGFR in an IL-17A–dependent manner (Fig. S5 H).
Src kinase activity is required for IL-17A–induced EGFR/ERK5 phosphorylation (Fig. 9 A). Furthermore, Act1 SH3 binding mutant abolished Src-EGFR-ERK5 activation, without affecting IL-17A–induced IkBα phosphorylation (Fig. 9, C and D). Therefore, Act1-mediated Src recruitment is specifically required for IL-17A–induced EGFR/ERK5 activation. ERK5 is specifically activated by the MAPKK tyrosine–threonine kinase, MEK5, which is the substrate of the MEKK2/3 serine/threonine kinases (Chao et al., 1999; Sun et al., 2001; Takeda et al., 2014). In addition, we found that EGFR inhibitor blocked the IL-17A–induced MEKK3–TRAF4 complex without affecting the interaction between Act1 and TRAF4, suggesting that EGFR kinase activity is required for the recruitment of MEKK3 to TRAF4 (Fig. 9 E). In support of this, EGFR inhibitor was able to block IL-17A–induced MEKK3 tyrosine phosphorylation (Fig. 9 F).
Next, we examined whether Src-EGFR activation is required for IL-17A–induced Lrig1+ stem cell expansion and the migration of Lrig1 progenies to the IFE. Similar to ERK5 inhibitor (Fig. S3 A), we found Src and EGFR inhibitor effectively blocked IL-17A–induced expansion of the Lrig1+ stem cells and their progenies in the PSU and their transition to p63/TRAF4+ cells in the epidermis (Fig. 9, G–I). These results indicate that IL-17A–induced Src activation and Src-mediated transactivation of EGFR are required for Lrig1+ stem cell expansion and migration in response to IL-17A stimulation (Fig. S5 H).
Discussion
We have now identified a novel IL-17A–induced EGFR-mediated Act1–TRAF4–ERK5 signaling cascade that stimulates epidermal hyperplasia and tumor formation, providing a molecular mechanism tying together inflammation, tissue repair, and tumorigenesis. Interestingly, EGF intradermal injection or transgenic overexpression of EGFR ligands does not induce epidermal growth in adult mice (Cohen, 1962; Cohen and Elliott, 1963; Vassar and Fuchs, 1991). In contrast, we found that IL-17A induces EGFR activation and robust skin hyperplasia by acting on a stem cell population marked by an inhibitor for EGFR activation, Lrig1 (Goldoni et al., 2007). While Lrig1+ stem cells have been implicated in wound healing, our study is the first to report that IL-17A signaling is responsible for mobilizing these stem cells during wounding and tumorigenesis. Thus, our study identifies an unanticipated signaling and cellular mechanism through which IL-17A links wounding healing to tumorigenesis.
Notably, recent reports have shown that IL-17–mediated signaling also plays an important role in regulating stem cell population in other organs. IL-17A has been found to regulate the development of tuft cells and stem cell features in both mouse and human pancreatic cancer models and critically contribute to tumor growth and progression (Zhang et al., 2018). Similarly, IL-17 induces a unique progenitor cell population in the mouse colitis-associated tumorigenesis model, and IL-17 signaling increases the self-renewal of colon cancer stem cells (Lotti et al., 2013). Taken together, the evidence suggests that wound repair mechanism–associated cancer development may extend to other tissue organs.
Our data demonstrated Lrig1+ stem cells are the major cell type that are mobilized in response to IL-17A correlating with the high level of TRAF4 expression in Lrig1+ stem cells in unchallenged skin. It is important to note that TRAF4 expression was not detected in Lgr5+ and Lgr6+ stem cells sorted from the skin; neither of these cell populations exhibited EGFR and ERK5 activation after IL-17A stimulation. Lrig1 progenies contribute majorly to the tumor mass (>90% of the total tumor cells) induced by DMBA/TPA. Progenies of Lgr5+ and Lgr6+ stem cells in the DMBA/TPA model are barely detectable in induced skin tumors (van de Glind et al., 2016a,b). Topical application of TPA has been shown to potently induce IL-17A in the skin, which might explain the dominant role of IL-17A–mobilized Lrig1 lineage cells in the DMBA/TPA model. Of note, oncogenic β-catenin expression in Lgr5+, Lgr6+, and Lrig1+ cells results in tumors with different histological features (Kretzschmar et al., 2016), indicating that the different epidermal stem cells are compartmentalized and manifest distinct response during tumorigenesis. Future studies are required to determine whether IL-17A–induced signaling plays any roles in other epidermal stem cells for skin tumorigenesis driven by oncogene overexpression.
It is important to note that these Lrig1 progenies (Lrig1− cells) are phenotypically and functionally distinct from the Lrig1− cells in the IFE in unchallenged skin. Since IL-17A induced expansion of p63+ cells in the IFE (Wu et al., 2015), it was assumed that p63+ cells in the IFE were responsive to IL-17A. We now found that the expanded p63+ cells in the IFE induced by IL-17A challenge were actually Lrig1 progenies, which could be ablated by deletion of Act1 (IL-17 signaling) in Lrig1+ stem cells. Moreover, Lrig1-p63+ cells in the IFE in unchallenged skin do not express TRAF4, making them unable to activate IL-17A–induced EGFR and ERK5 for cell expansion. On the other hand, TRAF4 was highly induced exclusively in the Lrig1 progenies in response to challenge. We speculate that IL-17A may continue to activate EGFR-ERK5 in the Lrig1 progenies, given the high level of TRAF4 expression. A recent study has demonstrated that inflammatory memory in epidermal stem cells hastens barrier restoration in response to further tissue damage, predisposing the affected skin to hyperproliferative disorders such as cancer (Naik et al., 2017). It is possible that the IL-17A–induced Lrig1 progenies may survive and reside in the IFE and constitute part of the cell populations bearing the inflammatory memory with elevated TRAF4 expression. In addition, other inflammatory cytokines may very well enlist different epidermal stem cells to participate in these processes. Abrogating IL-17A signaling in the Lrig1+ cells did not completely abolish the wound-healing process, suggesting that additional cell types and cytokines also contribute to the reepithelization. Indeed, we detected occasional TRAF4+ cells that were tdTomato− in the leading edge. Moreover, it is important to note that Lrig1 marks a heterogeneous population of cells (Jensen et al., 2009; Page et al., 2013; Donati et al., 2017). Future studies are required to unravel the intricate and dynamic functions of these heterogeneous Lrig1+ progenies in wound healing and tumorigenesis.
It should be noted that the expression of stem cell markers, such Lrig1, Lgr5, and Lgr6, has been documented in transformed tumor cells. While the Lrig1 progenies lost Lrig1 expression as they migrated out of the PSU, ∼20% of DMBA/TPA-induced tumors with carcinoma features contained foci of Lrig1+ cells. Subsequent analysis showed that these Lrig1+ tumor cells were TdTomato+ tumor cells (Lrig1 progenies) derived from Lrig1+ stem cells in unchallenged skin, suggesting the Lrig1 progenies might have reacquired Lrig1 during tumor progression. Lrig1 is detected in human sebaceous carcinoma, and its expression is associated with poor differentiation and worse disease prognosis (Pünchera et al., 2016). It will be important to assess the activation of IL-17R–EGFR–ERK5 axis in the Lrig1+ cutaneous squamous cell carcinoma and other types of cancers. Several studies using primary human cancer models have shown that IL-17 signaling is required for the growth of a variety of carcinoma, including pancreatic and colon cancer (Lotti et al., 2013; Zhang et al., 2018), in addition to its role in tumor initiation. The key question is whether IL-17 is also required for the maintenance of cancer growth in squamous cell carcinoma. Future studies will be conducted to specifically delete IL-17R–Act1 in Lrig1+ and Lgr6+ tumor cells to determine the potential impact of this pathway on tumor progression.
Materials and methods
Animals
All experiments were conducted in accordance with Institutional Animal Care and Use Committee guidelines at the Cleveland Clinic Lerner Research Institute. The Lrig1-EGFP-ires-CreERT2 (labeled as Lrig1CreERT2) mice were kindly provided by Dr. Kim B. Jensen (Biotech Research & Innovation Centre, University of Copenhagen, Copenhagen, Denmark; Page et al., 2013). EGFR floxed mice were kindly provided by Dr. David W. Threadgill (Department of Veterinary Pathobiology, Texas A&M University, College Station, TX; Lee and Threadgill, 2009). WT C57BL/6 mice, LSL-K-Ras (G12D; Jackson et al., 2001), Rosa26-lsl-tdTomato (Madisen et al., 2010), and Lgr5-EGFP-ires-CreERT2 (labeled as Lgr5CreERT2; Barker et al., 2007) mice were purchased from The Jackson Laboratory. IL-17RC−/− and TRAF4−/− mice have been described previously (Shiels et al., 2000; Zheng et al., 2008). Lrig1CreERT2 mice were crossed back onto a C57BL/6 background for at least six generations. Act1 floxed mice and Act1−/− have been described previously (Qian et al., 2004). IL-17RC floxed mice were generated by Cyagen Biosciences Inc. Lrig1CreERT2/Rosa26-lsl-tdTomato (homozygous) mice were obtained after eight generations of breeding, which were then bred onto LSL-K-Ras (G12D) mice. Lrig1CreERT2/IL-17RCf/f were obtained after at least eight generations of breeding. Gender- and age-matched littermates were used for all experiments.
Cell line culture and virus infection
HeLa cells were maintained in DMEM supplemented with 10% (volume/volume) FBS (GIBCO BRL; Thermo Fisher Scientific), penicillin G (100 µg/ml), and streptomycin (100 µg/ml). Cells were starved overnight by incubating cells in DMEM (without serum) followed by IL-17A stimulation (100 ng/ml) for further experiments. Cells were pretreated with inhibitors for 30 min before IL-17A stimulation where applicable.
To generate EGFR, IL-17RA, and TRAF4 KO cell lines, two guide RNA (gRNA) sequences derived from the GeCKO (v2) library for each gene were used (Shalem et al., 2014), including 5′-TGAGCTTGTTACTCGTGCCT-3′ and 5′-GAGTAACAAGCTCACGCAGT-3′ for EGFR, 5′-CCAATGAACGTTTGTGCGTC-3′ and 5′-CCTGACGCACAAACGTTCAT-3′ for IL-17RA, and 5′-TGGGCCACTACGTCATCTAC-3′ and 5′-AGCCACAAAACTCGCACTTG-3′ for TRAF4. These CRISPR/Cas9 guide RNA sequences were cloned into pLenticrispr v2 (Sanjana et al., 2014). For each gene, two constructs were used as a pool for maximum KO efficiency in HeLa cells. HEK293FT cells were transfected with 3.0 µg lenti-CRISPR-v2 plasmids (gRNA inserted) or control plasmids, 2.0 µg psPAX2, and 1.0 µg VSV-G plasmids using PolyFect Transfection Reagent (Qiagen) according to the manufacturer’s instructions. Medium with lentivirus was harvested and filtered with 0.45 µM Millex-HP filter 36 h after transfection. HeLa cells were then infected with lentivirus (fresh medium:filtered medium containing virus = 1:1) for 24 h. Cells were then cultured in fresh medium for 72 h before puromycin selection (1 μg/ml for 1 wk). Single cell–derived clones were then picked up, expanded, and further identified by Western blot analysis (Fig. S5 C). While data are representative of at least three experiments with the same clone, experiments were also performed using at least two KO clones for each gene.
The Myc-tagged human TRAF4 sequence was cloned into pTRE3G vector to generate doxycycline-inducible TRAF4-expressing HeLa cells. HeLa cells were transfected with 6 µg pCMV-Tet3G plasmids (in 100-mm dish). 2 d after transfection, cells were selected with G418 (400 µg/ml) to generate a stable Tet-On 3G cell line constitutively expressing Tet-On 3G transactivator. Clones were then expanded and screened for optimal inducibility. Tet-On 3G HeLa cells (the clone with best inducibility) were then transfected with pTRE3G-TRAF4 plasmids together with a linear selection marker (puromycin-resistant structure from linearized empty pMXs-IRES-puro by EcoRI; molar ratio 1:1), followed by selection with puromycin (1 µg/ml). The puromycin-resistant clones were validated by Western blot analysis of TRAF4 expression in response to doxycycline (Dox, 1 µg/ml for 24 h); experiments were also performed using at least two clones.
Keratinocytes culture, virus infection, and sorting
Primary keratinocytes for signaling study has been described previously (Lichti et al., 2008; Wu et al., 2015). Generally, the dermis side of newborn (within 1–2 d) mouse skin was floated on cold trypsin solution (0.25% without EDTA, ∼5 ml trypsin in each well of a 6-well plate) and was incubated at 4°C overnight. The epidermis was then removed from the dermis after trypsin treatment. Collected epidermises were minced in PBS until the pieces were small enough to enter the tip of a 10-ml pipette. The suspension was triturated by pipetting up and down >10 times, then transferred to a 50-ml tube through a 100-µm nylon mesh strainer, while leaving most of the stratum corneum sheets behind. Cell suspension was then centrifuged at 350 g for 5 min at 4°C. The isolated keratinocytes were then cultured (as day 0) in keratinocyte-serum free medium (K-SFM), with a supplement (Life Technologies) of penicillin G (100 µg/ml) and streptomycin (100 µg/ml). Medium was changed every 2 d (cells could be cultured up to 1 wk). Keratinocytes were starved overnight by incubating cells in K-SFM without supplement, before cytokine stimulation for the indicated times.
For adenovirus infection, primary adenovirus expressing GFP or GFP/Cre recombinase was purchased from Vector Biolabs. Cultured keratinocytes on day 1 were then infected with a 1:10,000 dilution of the viruses for another 2 d. On day 3, cells were washed with PBS and cultured in fresh medium with supplement. Keratinocytes from two newborn (within 1–2 d) mice yield one nearly confluent 150-mm culture dish of keratinocytes by 5 d after plating.
For retrovirus infection, Act1-WT or Act1-SH3 binding mutant were cloned into pMXs-IRES-puro and transfected into Phoenix cells for viral packaging (6 µg for each 100-mm dish, 10 ml medium, RPMI-1640 with 10% FBS). Medium containing retrovirus was harvested and filtered with 0.45 µM Millex-HP filter 36 h after transfection. Keratinocytes on day 1 were then infected with retrovirus (K-SFM with supplement: freshly filtered viral medium = 1:1) for 24 h. Medium was then replaced with fresh K-SFM with supplement. On day 5, keratinocytes at ∼95% confluency in 150-mm dishes were starved overnight by incubating cells in K-SFM without supplement. Immunoprecipitation was then performed.
For Lrig1(GFP+) and Lgr5(GFP+) cell sorting (Fig. 6 B and Fig. S4 B), mouse dorsal skin was shaved (fur was removed by depilatory cream) and harvested. After the underlying fat was removed, the dermis side was floated on cold trypsin solution (0.25% without EDTA, ∼5 ml trypsin in each well of 6-well plate) and was incubated at 4°C overnight. The epidermis was mechanically separated from the dermis by scrapping with a scalpel to obtain single-cell suspensions, which was filtered with 100-µm cell strainers. Cells were resuspended with PBS (with 0.5% BSA) and immediately sorted by FACS for GFP+ and GFP− cells.
For other flow analyses or cell sorting, ear skin (split in to ventral halves) or dorsal skin (fur and underlying fat were removed) were floated on thermolysin solution (0.25 mg/ml in PBS without calcium) for 60 min at 37°C (Jensen et al., 2009; Agudo et al., 2018). The epidermis was separated, and single-cell suspensions were prepared (thermolysin was inactivated by addition of 5% FBS). Cells were then filtered with 100-µm cell strainers. After centrifugation (500 g, 3 min), cells were resuspended for flow analysis or immediately sorted by FACS.
Sorted cells for signaling study were recovered in K-SFM (without supplement) for 4 h at 37°C in 5% CO2, and further stimulated with 100 ng/ml mouse IL-17A at the indicated times.
Constructs
pcDNA3.1 was used to generate the expression constructs of IL-17RA, EGFR, TRAF4, and the corresponding mutants for protein interaction studies. pLenticrispr v2 was used for the expression constructs of IL-17RA, EGFR, and TRAF4 gRNA. pMXs-IRES-puro was used to generate the expression constructs of Act1 WT and Act1 SH3 binding mutant. IL-17RA and EGFR intracellular domain and their mutants (in TRAF4 binding sites shown in Fig. 8 A, including IL-17RA-intra-Flag, containing residues 341–866; IL-17RA-T4Bm-Flag, deletion of 780–866 from residues 341–866; EGFR-intra-HA, containing residues 644–1186; and EGFR-intra-T4BmHA, deletion of 977–1023 and 1118–1186 from residues 644–1186) were cloned into pET28a vector. Full length of TRAF4 was cloned into pGEXKG.
Tamoxifen induced Cre activity in vivo
To induce transient Cre activity in all the mice on LSL-K-Ras(G12D) background, mice were shaved and topically treated with 100 µg 4-hydroxytamoxifen (in 200 µl acetone)/cm2 7 d before wounding. To induce transient Cre activity in all other experiments, mice were intraperitoneally injected with two doses of tamoxifen (∼5 mg/25 g weight) 14 and 7 d before further procedures.
IL-17–induced skin hyperplasia
Mice in approximate telogen stage were used. For ear skin, IL-17A (500 ng/20 µl PBS) or IL-17F (1 µg/20 µl PBS; Carrier-free; R&D Systems) was injected intradermally for 6 d. Gefitinib (20 µl of 10 µM), XMD8-92 (20 µl of 5 µM), or pp2 (20 µl of 8 µM) was coinjected with IL-17A or PBS where applicable. On day 7, skin tissues were processed into paraffin tissue blocks or optimal cutting temperature (OCT) compound–embedded blocks for further analysis. For dorsal skin, IL-17A (1.25 µg/50 µl PBS each spot) was injected intradermally for 10 d (six spots each time), and skin tissues were then processed into paraffin tissue blocks or OCT-embedded blocks for further analysis. Mice on Lrig1CreERT2 or Lgr5CreERT2 background were intraperitoneally injected with two doses of tamoxifen (∼5 mg/25 g weight) 14 and 7 d before intradermal IL-17 injection.
DMBA/TPA skin cancer model
6–8-wk-old mice were intraperitoneally injected with two doses of tamoxifen (∼5 mg/25 g weight) 14 and 7 d before topical application of 200 µl 100 µM DMBA (dissolved in acetone) onto the shaved dorsal skin of each mouse. 2 wk after DMBA initiation, each mouse was topically treated with 30 µg of TPA in 200 µl of acetone twice a week up to 23 wk. Tumor incidence and numbers were monitored weekly. Tumors >1 mm were counted and recorded.
Wound-induced tumorigenesis
Mice on LSL-K-Ras(G12D) background were used for the wound-induced tumorigenesis assay. Mouse dorsal skin was shaved and topically treated with 100 µg 4-hydroxytamoxifen (in 200 µl acetone)/cm2 7 d before wounding. Two full-thickness wounds were created on the shaved dorsal skin of each of the 6–8-wk-old age- and gender-matched mice using 5-mm skin biopsy punches. Tumorigenesis (>0.5 mm) was monitored daily. Representative photographs were captured at day 20.
Wound-healing assay
Two full-thickness wounds were created on the shaved dorsal skin of each of the 6–8-wk-old female mice using 5-mm skin biopsy punches. Each wound diameter was determined as the average of longitudinal and lateral diameter. All measurements and quantifications for wound healing were performed by a blinded assessor. Wound closure was monitored, and skin sections were harvested at the indicated days. The wound areas remaining open (percentage of wound area at indicate days relative to the original wound) were calculated as follows: wound areas remaining open (%) = (open area on the indicated day/original wound area) × 100%. Reepithelization ratios (leading edge ratios) were measured and calculated by [(a + b)/c] × 100% (shown in Fig. 2F, where a and b are the length of the axes for the leading edges, and c indicates the axis of initial wound lengths; Shirakata et al., 2005; Safferling et al., 2013; Noguchi et al., 2014). Shown is ± SEM from three sections of each wound, for a total of 10 wounds; **, P < 0.01; *, P < 0.05 by t test.
Western blot and immunoprecipitation
Cells were washed with ice-cold PBS three times and lysed in lysis buffer (1% Triton X-100, 50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 12.5 mM β-glycerophosphate, 1.5 mM MgCl2, 10 mM NaF, 2 mM dithiothreitol, 2 mM sodium orthovanadate, 2 mM EGTA, and Protease Inhibitor Cocktail; Roche). Cell extracts were centrifuged at 12,000 rpm for 10 min at 4°C. Protein concentration was quantified (Bio-Rad Protein Assay Kit 5000006) before Western blot or immunoprecipitation. For immunoprecipitation, cell lysates were incubated overnight at 4°C with appropriate antibodies plus protein A/G Sepharose beads. Protein A/G Sepharose beads were preblocked by 5% BSA and washed five times with lysis buffer before incubation. After incubation, the beads were washed five times with lysis buffer, resolved by SDS-PAGE, and analyzed by Western blot. All of the data shown for the immunoprecipitation and Western blot are representative of at least three experiments.
Immunohisotochemistry/fluorescence staining and proximity ligation assay (PLA)
For paraffin sections, tissues were fixed with 10% formalin overnight and then kept in 70% ethanol at 4°C until processed into paraffin tissue blocks by AML Laboratories or Imaging Core at Cleveland Clinic Lerner Research Institute. Paraffin sections were subjected to heat-induced epitope retrieval recommended by the antibody manufacturer. For frozen sections, fresh tissues were embedded in OCT and sectioned (12 µm), which was further fixed in 4% PFA and washed with PBST (PBS+ 0.025% Tween 20) three times. Samples were incubated in blocking buffer for 2 h (PBS containing 2% donkey serum, 0.5% BSA, 0.5% fish skin gelatin, 0.05% Tween 20, and 0.1% Triton X-10, pH 7.2) before overnight incubation in primary antibody as listed. (1) For immunohistochemistry, blocked samples were treated with 0.3% H2O2, followed by incubation of biotinylated second antibodies and peroxidase streptavidin (Vector Laboratories). The 3′,3-diaminobenzidine substrate kit (BD PharMingen) was then used. (2) For fluorescence staining, blocked samples were incubated in the appropriate Alexa fluorophore–conjugated secondary donkey antibodies (Invitrogen). Monovalent Fab fragment of donkey anti-mouse IgG was used to blocking endogenous IgG before mouse-derived antibody incubation. (3) For the PLA (Duolink) assay, primary antibodies were listed as described, followed with the Duolink in situ Red/Green kit (Sigma-Aldrich; Gajadhar and Guha, 2010; Ritter et al., 2011). Immunohistochemical staining was captured with a DP71 digital camera (Olympus) attached to an Olympus BX41 microscope or with Keyence BZ-X700 microscope. Lineage tracing for wound healing was captured with a Keyence BZ-X700 microscope. All other fluorescent images were captured with a Leica Confocal Microscope provided by the Lerner Research Institute Imaging Core.
Protein purification and in vitro binding assay
IL-17RA and EGFR intracellular domain and their mutants (in TRAF4 binding sites) were cloned into the pET28a vector. Recombinant proteins were then expressed in Escherichia coli Rosetta and purified with Ni-NTA agarose (Thermo Fisher Scientific). Full length of TRAF4 was cloned into pGEX-KG. Recombinant GST-TRAF4 was expressed in E. coli Rosetta and purified with glutathione-agarose (Sigma-Aldrich). Proteins were further desalted and concentrated with an Ultra Centrifugal Filter (Millipore). For the in vitro binding assay, recombinant proteins (1 µg each) were incubated in 200 µl CelLytic M buffer (C2978; Sigma-Aldrich) at 4°C for 2 h. The protein–protein interactions of the purified recombinant proteins were examined by immunoprecipitation followed by Western blot analyses with the antibodies for the relevant proteins. Briefly, for the data presented in Fig. 8 K, the indicated recombinant proteins were incubated at 4°C for 2 h, followed by immunoprecipitation with mouse anti-HA and Western blot analyses with the indicated antibodies. For the data presented in Fig. 8 L, the indicated recombinant proteins were incubated at 4°C for 2 h, followed by immunoprecipitation with mouse anti-flag and Western blot analyses with the indicated antibodies: EGFR-intra-HA, containing residues 644–1186; EGFR-T4Bm-HA, deletion of 977–1023 and 1118–1186 from residues 644–1186; IL-17RA-intra-Flag, containing residues 341–866; IL-17RA-T4Bm-Flag, deletion of 780–866 from residues 341–866; and GST-TRAF4, containing full-length TRAF4.
Key antibodies and reagents
Key antibodies and reagents are listed in Table S1. Rabbit anti-mouse-IL-17RA antibody for immunoprecipitation was raised against the peptide Ac-KPIPDGDPNHKSKI-amide locating at extracellular domain of mouse IL-17RA and further subjected to affinity purification (services provided by Covance). Rabbit anti-TRAF4 antibody for immunoprecipitation was raised against the TRAF4 motif Ac-DYAKIYPDPELEVQ-amide and further subjected to affinity purification (services provided by Covance).
Quantification and statistical analysis
Statistical significance was determined using Student’s t test, as described for each experiment. All the data are representative of independent experiments as described separately. For cell quantification within the IFE and PSU, cross sections of 10 PSUs and adjacent IFE within 50 µm of both sides (or within 100 µm to single side, where applicable) of the hair follicle were analyzed. Five mice were analyzed for each genotype. Unless otherwise indicated, all error bars show SEM.
Online supplemental material
Fig. S1, related to Figs. 1 and 2, presents IL-17A–induced signaling in epidermal hyperplasia and wound healing. Fig. S2, related to Fig. 3, presents IL-17A–induced signaling in Lrig1+ cells that drives Lrig1 progenies to participate in skin tumorigenesis. Fig. S3, related to Fig. 4, presents IL-17A–induced EGFR activation in Lrig1+ stem cells, which is required for the migration and expansion of Lrig1 progenies. Fig. S4, related to Figs. 5, 6, and 7, presents TRAF4, which is enriched in Lrig1+ stem cells and required for IL-17A–induced IL-17RA–EGFR interaction. Fig. S5, related to Figs. 8 and 9, presents IL-17R–EGFR transactivation. Table S1 lists key antibodies and reagents used in this study.
Supplementary Material
Acknowledgments
We thank Dr. David W. Threadgill for kindly providing the EGFR floxed mice, J. Ma and W. Qian for technical support, and AML Laboratories for processing tissue samples for histology.
This work was supported by the National Institutes of Health grants 5P01CA062220 and 5P01 HL103453 to X. Li and the National Institutes of Health Shared Instrumentation Grant grant S10-OD019972 to the imaging core at the Lerner Institute at Cleveland Clinic.
The authors declare no competing financial interests.
Author contributions: X. Li, X. Chen, and G. Cai designed and analyzed all the experiments. X. Chen, G. Cai, C. Liu, J. Zhao, C. Gu, L. Wu, and C. Zhang were involved in performing and analyzing multiple experiments. K.B. Jensen provided the Lrig1-EGFP-ires-CreERT2 mice and consultation for experimental design. L. Zhu, J. Qin, J. Ko, A. Vidimos, S. Koyfman, and B.R. Gastman gave consultation for experiment design. T.A. Hamilton helped to revise the manuscript. X. Li and X. Chen wrote the paper with input from all coauthors. X. Li supervised all aspects of the study.
References
- Agudo J., Park E.S., Rose S.A., Alibo E., Sweeney R., Dhainaut M., Kobayashi K.S., Sachidanandam R., Baccarini A., Merad M., and Brown B.D.. 2018. Quiescent Tissue Stem Cells Evade Immune Surveillance. Immunity. 48:271–285.e5. 10.1016/j.immuni.2018.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona M., Dekoninck S., Rulands S., Lenglez S., Mascré G., Simons B.D., and Blanpain C.. 2017. Defining stem cell dynamics and migration during wound healing in mouse skin epidermis. Nat. Commun. 8:14684 10.1038/ncomms14684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., and Clevers H.. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 449:1003–1007. 10.1038/nature06196 [DOI] [PubMed] [Google Scholar]
- Bata-Csörgö Z., and Szell M.. 2012. The psoriatic keratinocytes. Expert. Rev. Dermatol. 7:473–481. 10.1586/edm.12.48 [DOI] [Google Scholar]
- Blauvelt A., Reich K., Tsai T.-F., Tyring S., Vanaclocha F., Kingo K., Ziv M., Pinter A., Vender R., Hugot S., et al. 2017. Secukinumab is superior to ustekinumab in clearing skin of subjects with moderate-to-severe plaque psoriasis up to 1 year: Results from the CLEAR study. J. Am. Acad. Dermatol. 76:60–69.e9. 10.1016/j.jaad.2016.08.008 [DOI] [PubMed] [Google Scholar]
- Boumahdi S., Driessens G., Lapouge G., Rorive S., Nassar D., Le Mercier M., Delatte B., Caauwe A., Lenglez S., Nkusi E., et al. 2014. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 511:246–250. 10.1038/nature13305 [DOI] [PubMed] [Google Scholar]
- Bulek K., Liu C., Swaidani S., Wang L., Page R.C., Gulen M.F., Herjan T., Abbadi A., Qian W., Sun D., et al. 2011. The inducible kinase IKKi is required for IL-17-dependent signaling associated with neutrophilia and pulmonary inflammation. Nat. Immunol. 12:844–852. 10.1038/ni.2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S.H., Park H., and Dong C.. 2006. Act1 adaptor protein is an immediate and essential signaling component of interleukin-17 receptor. J. Biol. Chem. 281:35603–35607. 10.1074/jbc.C600256200 [DOI] [PubMed] [Google Scholar]
- Chao T.H., Hayashi M., Tapping R.I., Kato Y., and Lee J.D.. 1999. MEKK3 directly regulates MEK5 activity as part of the big mitogen-activated protein kinase 1 (BMK1) signaling pathway. J. Biol. Chem. 274:36035–36038. 10.1074/jbc.274.51.36035 [DOI] [PubMed] [Google Scholar]
- Cohen S. 1962. Isolation of a mouse submaxillary gland protein accelerating incisor eruption and eyelid opening in the new-born animal. J. Biol. Chem. 237:1555–1562. [PubMed] [Google Scholar]
- Cohen S., and Elliott G.A.. 1963. The stimulation of epidermal keratinization by a protein isolated from the submaxillary gland of the mouse. J. Invest. Dermatol. 40:1–5. 10.1038/jid.1963.1 [DOI] [PubMed] [Google Scholar]
- Dewar B.J., Gardner O.S., Chen C.S., Earp H.S., Samet J.M., and Graves L.M.. 2007. Capacitative calcium entry contributes to the differential transactivation of the epidermal growth factor receptor in response to thiazolidinediones. Mol. Pharmacol. 72:1146–1156. 10.1124/mol.107.037549 [DOI] [PubMed] [Google Scholar]
- Donati G., Rognoni E., Hiratsuka T., Liakath-Ali K., Hoste E., Kar G., Kayikci M., Russell R., Kretzschmar K., Mulder K.W., et al. 2017. Wounding induces dedifferentiation of epidermal Gata6+ cells and acquisition of stem cell properties. Nat. Cell Biol. 19:603–613. 10.1038/ncb3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drube S., Stirnweiss J., Valkova C., and Liebmann C.. 2006. Ligand-independent and EGF receptor-supported transactivation: lessons from beta2-adrenergic receptor signalling. Cell. Signal. 18:1633–1646. 10.1016/j.cellsig.2006.01.003 [DOI] [PubMed] [Google Scholar]
- Dvorak H.F. 1986. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N. Engl. J. Med. 315:1650–1659. 10.1056/NEJM198612253152606 [DOI] [PubMed] [Google Scholar]
- Feng S., Chen J.K., Yu H., Simon J.A., and Schreiber S.L.. 1994. Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science. 266:1241–1247. 10.1126/science.7526465 [DOI] [PubMed] [Google Scholar]
- Gaffen S.L. 2009. Structure and signalling in the IL-17 receptor family. Nat. Rev. Immunol. 9:556–567. 10.1038/nri2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajadhar A., and Guha A.. 2010. A proximity ligation assay using transiently transfected, epitope-tagged proteins: application for in situ detection of dimerized receptor tyrosine kinases. Biotechniques. 48:145–152. 10.2144/000113354 [DOI] [PubMed] [Google Scholar]
- Goldoni S., Iozzo R.A., Kay P., Campbell S., McQuillan A., Agnew C., Zhu J.X., Keene D.R., Reed C.C., and Iozzo R.V.. 2007. A soluble ectodomain of LRIG1 inhibits cancer cell growth by attenuating basal and ligand-dependent EGFR activity. Oncogene. 26:368–381. 10.1038/sj.onc.1209803 [DOI] [PubMed] [Google Scholar]
- Haddow A. 1972. Molecular repair, wound healing, and carcinogenesis: tumor production a possible overhealing? Adv. Cancer Res. 16:181–234. 10.1016/S0065-230X(08)60341-3 [DOI] [PubMed] [Google Scholar]
- Hartupee J., Liu C., Novotny M., Sun D., Li X., and Hamilton T.A.. 2009. IL-17 signaling for mRNA stabilization does not require TNF receptor-associated factor 6. J. Immunol. 182:1660–1666. 10.4049/jimmunol.182.3.1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D., Li H., Yusuf N., Elmets C.A., Athar M., Katiyar S.K., and Xu H.. 2012. IL-17 mediated inflammation promotes tumor growth and progression in the skin. PLoS One. 7:e32126 10.1371/journal.pone.0032126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes T.C., Fadool D.A., Ren R., and Levitan I.B.. 1996. Association of Src tyrosine kinase with a human potassium channel mediated by SH3 domain. Science. 274:2089–2091. 10.1126/science.274.5295.2089 [DOI] [PubMed] [Google Scholar]
- Hsieh H.L., Lin C.C., Chan H.J., Yang C.M., and Yang C.M.. 2012. c-Src-dependent EGF receptor transactivation contributes to ET-1-induced COX-2 expression in brain microvascular endothelial cells. J. Neuroinflammation. 9:152 10.1186/1742-2094-9-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson E.L., Willis N., Mercer K., Bronson R.T., Crowley D., Montoya R., Jacks T., and Tuveson D.A.. 2001. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15:3243–3248. 10.1101/gad.943001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K.B., Collins C.A., Nascimento E., Tan D.W., Frye M., Itami S., and Watt F.M.. 2009. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell. 4:427–439. 10.1016/j.stem.2009.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar K., Weber C., Driskell R.R., Calonje E., and Watt F.M.. 2016. Compartmentalized Epidermal Activation of β-Catenin Differentially Affects Lineage Reprogramming and Underlies Tumor Heterogeneity. Cell Reports. 14:269–281. 10.1016/j.celrep.2015.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley R.G., Elewski B.E., Lebwohl M., Reich K., Griffiths C.E., Papp K., Puig L., Nakagawa H., Spelman L., Sigurgeirsson B., et al. FIXTURE Study Group . 2014. Secukinumab in plaque psoriasis--results of two phase 3 trials. N. Engl. J. Med. 371:326–338. 10.1056/NEJMoa1314258 [DOI] [PubMed] [Google Scholar]
- Lee T.C., and Threadgill D.W.. 2009. Generation and validation of mice carrying a conditional allele of the epidermal growth factor receptor. Genesis. 47:85–92. 10.1002/dvg.20464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichti U., Anders J., and Yuspa S.H.. 2008. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat. Protoc. 3:799–810. 10.1038/nprot.2008.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Qian W., Qian Y., Giltiay N.V., Lu Y., Swaidani S., Misra S., Deng L., Chen Z.J., and Li X.. 2009. Act1, a U-box E3 ubiquitin ligase for IL-17 signaling. Sci. Signal. 2:ra63 10.1126/scisignal.2000382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotti F., Jarrar A.M., Pai R.K., Hitomi M., Lathia J., Mace A., Gantt G.A. Jr., Sukhdeo K., DeVecchio J., Vasanji A., et al. 2013. Chemotherapy activates cancer-associated fibroblasts to maintain colorectal cancer-initiating cells by IL-17A. J. Exp. Med. 210:2851–2872. 10.1084/jem.20131195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod A.S., Hemmers S., Garijo O., Chabod M., Mowen K., Witherden D.A., and Havran W.L.. 2013. Dendritic epidermal T cells regulate skin antimicrobial barrier function. J. Clin. Invest. 123:4364–4374. 10.1172/JCI70064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer B.J., and Baltimore D.. 1993. Signalling through SH2 and SH3 domains. Trends Cell Biol. 3:8–13. 10.1016/0962-8924(93)90194-6 [DOI] [PubMed] [Google Scholar]
- Naik S., Larsen S.B., Gomez N.C., Alaverdyan K., Sendoel A., Yuan S., Polak L., Kulukian A., Chai S., and Fuchs E.. 2017. Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature. 550:475–480. 10.1038/nature24271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi F., Nakajima T., Inui S., Reddy J.K., and Itami S.. 2014. Alteration of skin wound healing in keratinocyte-specific mediator complex subunit 1 null mice. PLoS One. 9:e102271 10.1371/journal.pone.0102271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numasaki M., Watanabe M., Suzuki T., Takahashi H., Nakamura A., McAllister F., Hishinuma T., Goto J., Lotze M.T., Kolls J.K., and Sasaki H.. 2005. IL-17 enhances the net angiogenic activity and in vivo growth of human non-small cell lung cancer in SCID mice through promoting CXCR-2-dependent angiogenesis. J. Immunol. 175:6177–6189. 10.4049/jimmunol.175.9.6177 [DOI] [PubMed] [Google Scholar]
- Page M.E., Lombard P., Ng F., Göttgens B., and Jensen K.B.. 2013. The epidermis comprises autonomous compartments maintained by distinct stem cell populations. Cell Stem Cell. 13:471–482. 10.1016/j.stem.2013.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pünchera J., Barnes L., and Kaya G.. 2016. Lrig1 expression in human sebaceous gland tumors. Dermatopathology (Basel). 3:44–54. 10.1159/000446427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y., Qin J., Cui G., Naramura M., Snow E.C., Ware C.F., Fairchild R.L., Omori S.A., Rickert R.C., Scott M., et al. 2004. Act1, a negative regulator in CD40- and BAFF-mediated B cell survival. Immunity. 21:575–587. 10.1016/j.immuni.2004.09.001 [DOI] [PubMed] [Google Scholar]
- Qian Y., Liu C., Hartupee J., Altuntas C.Z., Gulen M.F., Jane-Wit D., Xiao J., Lu Y., Giltiay N., Liu J., et al. 2007. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat. Immunol. 8:247–256. 10.1038/ni1439 [DOI] [PubMed] [Google Scholar]
- Ritter L.M., Khattree N., Tam B., Moritz O.L., Schmitz F., and Goldberg A.F.. 2011. In situ visualization of protein interactions in sensory neurons: glutamic acid-rich proteins (GARPs) play differential roles for photoreceptor outer segment scaffolding. J. Neurosci. 31:11231–11243. 10.1523/JNEUROSCI.2875-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safferling K., Sütterlin T., Westphal K., Ernst C., Breuhahn K., James M., Jäger D., Halama N., and Grabe N.. 2013. Wound healing revised: a novel reepithelialization mechanism revealed by in vitro and in silico models. J. Cell Biol. 203:691–709. 10.1083/jcb.201212020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksela K., and Permi P.. 2012. SH3 domain ligand binding: What’s the consensus and where’s the specificity? FEBS Lett. 586:2609–2614. 10.1016/j.febslet.2012.04.042 [DOI] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepeler T., Page M.E., and Jensen K.B.. 2014. Heterogeneity and plasticity of epidermal stem cells. Development. 141:2559–2567. 10.1242/dev.104588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., and Zhang F.. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 343:84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels H., Li X., Schumacker P.T., Maltepe E., Padrid P.A., Sperling A., Thompson C.B., and Lindsten T.. 2000. TRAF4 deficiency leads to tracheal malformation with resulting alterations in air flow to the lungs. Am. J. Pathol. 157:679–688. 10.1016/S0002-9440(10)64578-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakata Y., Kimura R., Nanba D., Iwamoto R., Tokumaru S., Morimoto C., Yokota K., Nakamura M., Sayama K., Mekada E., et al. 2005. Heparin-binding EGF-like growth factor accelerates keratinocyte migration and skin wound healing. J. Cell Sci. 118:2363–2370. 10.1242/jcs.02346 [DOI] [PubMed] [Google Scholar]
- Su B., Cheng J., Yang J., and Guo Z.. 2001. MEKK2 is required for T-cell receptor signals in JNK activation and interleukin-2 gene expression. J. Biol. Chem. 276:14784–14790. 10.1074/jbc.M010134200 [DOI] [PubMed] [Google Scholar]
- Sun D., Novotny M., Bulek K., Liu C., Li X., and Hamilton T.. 2011. Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing-regulatory factor SF2 (ASF). Nat. Immunol. 12:853–860. 10.1038/ni.2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Kesavan K., Schaefer B.C., Garrington T.P., Ware M., Johnson N.L., Gelfand E.W., and Johnson G.L.. 2001. MEKK2 associates with the adapter protein Lad/RIBP and regulates the MEK5-BMK1/ERK5 pathway. J. Biol. Chem. 276:5093–5100. 10.1074/jbc.M003719200 [DOI] [PubMed] [Google Scholar]
- Takeda A.N., Oberoi-Khanuja T.K., Glatz G., Schulenburg K., Scholz R.P., Carpy A., Macek B., Remenyi A., and Rajalingam K.. 2014. Ubiquitin-dependent regulation of MEKK2/3-MEK5-ERK5 signaling module by XIAP and cIAP1. EMBO J. 33:1784–1801. 10.15252/embj.201487808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toy D., Kugler D., Wolfson M., Vanden Bos T., Gurgel J., Derry J., Tocker J., and Peschon J.. 2006. Cutting edge: interleukin 17 signals through a heteromeric receptor complex. J. Immunol. 177:36–39. 10.4049/jimmunol.177.1.36 [DOI] [PubMed] [Google Scholar]
- van de Glind G.C., Out-Luiting J.J., Rebel H.G., Tensen C.P., and de Gruijl F.R.. 2016a Lgr5+ stem cells and their progeny in mouse epidermis under regimens of exogenous skin carcinogenesis, and their absence in ensuing skin tumors. Oncotarget. 7:52085–52094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Glind G.C., Rebel H.G., Out-Luiting J.J., Zoutman W., Tensen C.P., and de Gruijl F.R.. 2016b Lgr6+ stem cells and their progeny in mouse epidermis under regimens of exogenous skin carcinogenesis, and their absence in ensuing skin tumors. Oncotarget. 7:86740–86754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R., and Fuchs E.. 1991. Transgenic mice provide new insights into the role of TGF-alpha during epidermal development and differentiation. Genes Dev. 5:714–727. 10.1101/gad.5.5.714 [DOI] [PubMed] [Google Scholar]
- Wang K., Kim M.K., Di Caro G., Wong J., Shalapour S., Wan J., Zhang W., Zhong Z., Sanchez-Lopez E., Wu L.W., et al. 2014. Interleukin-17 receptor a signaling in transformed enterocytes promotes early colorectal tumorigenesis. Immunity. 41:1052–1063. 10.1016/j.immuni.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Yi T., Kortylewski M., Pardoll D.M., Zeng D., and Yu H.. 2009. IL-17 can promote tumor growth through an IL-6-Stat3 signaling pathway. J. Exp. Med. 206:1457–1464. 10.1084/jem.20090207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Chen X., Zhao J., Martin B., Zepp J.A., Ko J.S., Gu C., Cai G., Ouyang W., Sen G., et al. 2015. A novel IL-17 signaling pathway controlling keratinocyte proliferation and tumorigenesis via the TRAF4-ERK5 axis. J. Exp. Med. 212:1571–1587. 10.1084/jem.20150204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Graves L.M., Gill G.N., Parsons S.J., and Samet J.M.. 2002. Src-dependent phosphorylation of the epidermal growth factor receptor on tyrosine 845 is required for zinc-induced Ras activation. J. Biol. Chem. 277:24252–24257. 10.1074/jbc.M200437200 [DOI] [PubMed] [Google Scholar]
- Zepp J.A., Liu C., Qian W., Wu L., Gulen M.F., Kang Z., and Li X.. 2012. Cutting edge: TNF receptor-associated factor 4 restricts IL-17-mediated pathology and signaling processes. J. Immunol. 189:33–37. 10.4049/jimmunol.1200470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Liu C., Qian W., Han Y., Li X., and Deng J.. 2014. Structure of the unique SEFIR domain from human interleukin 17 receptor A reveals a composite ligand-binding site containing a conserved α-helix for Act1 binding and IL-17 signaling. Acta Crystallogr. D Biol. Crystallogr. 70:1476–1483. 10.1107/S1399004714005227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zoltan M., Riquelme E., Xu H., Sahin I., Castro-Pando S., Montiel M.F., Chang K., Jiang Z., Ling J., et al. 2018. Immune Cell Production of Interleukin 17 Induces Stem Cell Features of Pancreatic Intraepithelial Neoplasia Cells. Gastroenterology. 155:210–223.e3. 10.1053/j.gastro.2018.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Valdez P.A., Danilenko D.M., Hu Y., Sa S.M., Gong Q., Abbas A.R., Modrusan Z., Ghilardi N., de Sauvage F.J., and Ouyang W.. 2008. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14:282–289. 10.1038/nm1720 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.