Naik et al. show that GATA3, Runx1, and E2A are essential for hierarchical assembly of a transcriptionally active RAG locus chromatin hub in CD4+CD8+ thymocytes. Signal-dependent down-regulation of RAG expression is associated with hub disassembly and depends on Ikaros.
Abstract
Expression of Rag1 and Rag2 is tightly regulated in developing T cells to mediate TCR gene assembly. Here we have investigated the molecular mechanisms governing the assembly and disassembly of a transcriptionally active RAG locus chromatin hub in CD4+CD8+ thymocytes. Rag1 and Rag2 gene expression in CD4+CD8+ thymocytes depends on Rag1 and Rag2 promoter activation by a distant antisilencer element (ASE). We identify GATA3 and E2A as critical regulators of the ASE, and Runx1 and E2A as critical regulators of the Rag1 promoter. We reveal hierarchical assembly of a transcriptionally active chromatin hub containing the ASE and RAG promoters, with Rag2 recruitment and expression dependent on assembly of a functional ASE–Rag1 framework. Finally, we show that signal-dependent down-regulation of RAG gene expression in CD4+CD8+ thymocytes depends on Ikaros and occurs with disassembly of the RAG locus chromatin hub. Our results provide important new insights into the molecular mechanisms that orchestrate RAG gene expression in developing T cells.
Introduction
The adaptive immune system generates highly diverse antigen receptors distributed on T and B cells by using a site-specific DNA recombination process called V(D)J recombination to assemble antigen receptor genes (Schatz and Swanson, 2011). The proteins encoded by recombination activating genes 1 and 2 (Rag1 and Rag2) form a heterotetrameric RAG recombinase complex that recognizes and cleaves DNA at recombination signal sequences flanking variable (V), diversity (D), and joining (J) gene segments of antigen receptor loci to initiate the V(D)J recombination reaction. Although RAG-mediated DNA breaks are essential for the creation of antigen receptor repertoires in developing lymphocytes, off-target RAG cleavage can have pathological consequences, including mutations and chromosomal translocations associated with lymphoid neoplasias (Gostissa et al., 2011). Therefore, the expression of Rag1 and Rag2 is tightly controlled in a cell- and developmental stage–specific manner (Kuo and Schlissel, 2009).
There are two waves of RAG gene expression during T and B lymphocyte development (Wilson et al., 1994). In developing thymocytes, RAG expression first occurs during the CD4−CD8− double-negative (DN) stage to catalyze Tcrb, Tcrg, and Tcrd gene rearrangement. RAG expression is permanently down-regulated in cells that successfully rearrange Tcrg and Tcrd to become γδ T cells and is transiently down-regulated in cells that successfully rearrange Tcrb and differentiate along the αβ pathway. The RAG genes are reexpressed as these thymocytes differentiate to the CD4+CD8+ double-positive (DP) stage, with RAG expression in DP thymocytes supporting Tcra gene recombination. Following successful Tcra recombination and assembly of an αβ TCR, RAG expression is permanently down-regulated in DP thymocytes receiving TCR signals associated with positive selection (Turka et al., 1991; Borgulya et al., 1992; Takahama and Singer, 1992). RAG genes are similarly expressed at two stages of B cell development, with expression in pro–B cells supporting Igh rearrangement and expression in pre–B cells supporting Igk and Igl rearrangement (Grawunder et al., 1995). Appropriate regulation of RAG gene expression is essential for the integrity of lymphocyte development. Absence of RAG expression results in blockade of lymphocyte development (Mombaerts et al., 1992; Shinkai et al., 1992). On the other hand, persistent Lck proximal promoter–driven RAG expression in the T lineage caused abnormal thymic development, changes in lymphoid tissue anatomy, and impaired cellular immune responses (Wayne et al., 1994a,b), and ubiquitous H-2K promoter–driven RAG expression in lymphoid and nonlymphoid cells caused abnormal B and T lymphopoiesis and reduced mouse lifespan (Barreto et al., 2001). The mechanisms that direct and fine-tune RAG gene and protein expression within developing lymphocytes are therefore of substantial interest.
Transcriptional regulation of RAG gene expression in B and T lymphocytes is complex (Kuo and Schlissel, 2009). RAG mRNAs have half-lives of only a few minutes, suggesting the need for continuous transcriptional bursting (Verkoczy et al., 2005). The Rag1 and Rag2 genes are convergently oriented, with their promoters separated by only ∼25 kb. A variety of studies have shown the Rag1 and Rag2 promoters to be coordinately controlled by additional cis-regulatory elements that differ between B and T cells. In B cells, the RAG promoters are controlled by proximal, distal, and Erag enhancers distributed upstream of Rag2 (Monroe et al., 1999; Hsu et al., 2003). Studies of the Erag enhancer have revealed FOXO1 as a positive regulator of RAG expression and Gfi1b, Ebf1, and c-Myb as negative regulators (Amin and Schlissel, 2008; Schulz et al., 2012; Timblin and Schlissel, 2013; Lee et al., 2017; Timblin et al., 2017). RAG expression in T cells is controlled by distinct cis-regulatory elements. In DN thymocytes, RAG expression is directed by sequences within 10 kb upstream of Rag2 (Yu et al., 1999). DP thymocytes express the RAG genes at levels severalfold higher than at any other stage of T or B lymphocyte development. In these cells, RAG gene expression is directed by a distal antisilencer element (ASE), located 73 kb upstream of Rag2 (Yannoutsos et al., 2004). The ASE was initially shown to function by counteracting the effect of an intergenic silencer, with ASE deletion causing a several hundred–fold reduction in RAG gene expression. Recently, we reported that the ASE acts as an enhancer that directly interacts with the Rag1 and Rag2 promoters (Hao et al., 2015). We also identified global chromatin organizer SATB1 as an ASE binding factor that promotes optimal RAG expression through effects on RAG locus organization. However, other factors required for ASE function and the organization and expression of the RAG genes in DP thymocytes are completely unknown.
The RAG promoters also appear to be distinctly regulated in different cell lineages (Kuo and Schlissel, 2009). A Rag1 promoter–driven reporter was expressed in both lymphoid and nonlymphoid cells, whereas a Rag2 promoter–driven reporter was active only in lymphoid cell lines (Brown et al., 1997; Lauring and Schlissel, 1999). In B cells, Pax5, c-Myb, lymphoid enhancer-binding factor 1 (LEF-1), Sp1, and Myc-associated zinc finger protein MAZ were all shown to bind to and regulate the Rag2 promoter (Lauring and Schlissel, 1999; Kishi et al., 2000; Jin et al., 2002; Miranda et al., 2002; Wu et al., 2004). In T cells, cMyb and GATA3 have been reported as positive regulators of Rag2 promoter activity (Kishi et al., 2000; Wang et al., 2000), whereas NFAT has been reported as a negative regulator (Patra et al., 2006). In contrast, the Rag1 promoter has been relatively less well studied. NF-Y was reported to bind the Rag1 promoter in B cells (Brown et al., 1997), and NFAT was shown to bind the Rag1 promoter in T cells (Patra et al., 2006). Zinc finger proteins 608 and 609 have been shown to regulate the Rag1 and Rag2 promoters in T cells, but it is not known if they do so by direct binding (Zhang et al., 2006; Reed et al., 2013).
In this study, by using the mouse DP thymocyte cell line VL3-3M2, we have characterized the mechanisms of ASE- and Rag1 promoter–mediated control of RAG gene expression. We identify GATA3 and E2A as critical regulators of the ASE, and Runx1 and E2A as critical regulators of the Rag1 promoter. These factors control RAG gene expression, at least in part, because they are necessary for the assembly of a transcriptionally active chromatin hub containing the ASE and both promoters. Notably, we reveal hierarchical assembly of this structure, with recruitment of the Rag2 promoter dependent on assembly of a functional ASE–Rag1 promoter framework. As such, Rag1 promoter mutations ablate Rag2 gene expression due to diminished contacts between the Rag2 promoter and both the ASE and Rag1. Finally, we show that signal-dependent down-regulation of RAG gene expression in DP thymocytes depends on Ikaros and occurs with disassembly of the RAG locus chromatin hub. Our results provide important new insights into the molecular mechanisms that orchestrate RAG gene expression in developing T cells.
Results
GATA3, E2A, Runx, and Ikaros are candidate regulators of ASE enhancer activity
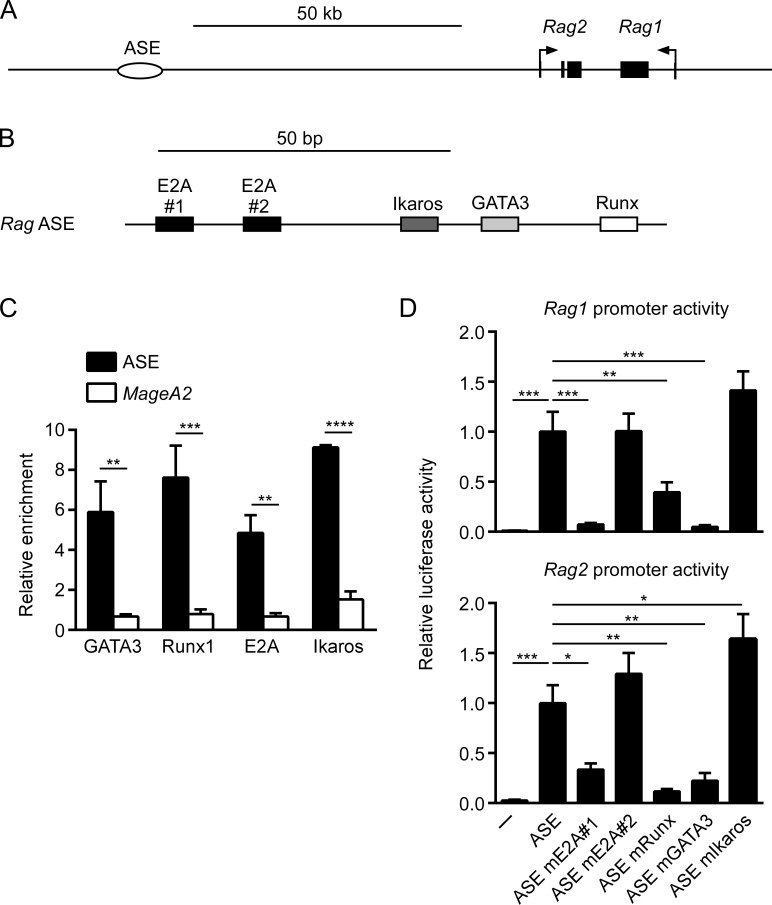
We previously identified a 140-bp core region of the mouse RAG ASE that was necessary for enhancer activity on the Rag1 and Rag2 promoters (Hao et al., 2015). For insight into enhancer function, we analyzed sequence conservation and found the ASE core to be highly conserved among eight vertebrate sequences available on the University of California, Santa Cruz (UCSC) genome browser (Fig. S1). Within this region we detected highly conserved binding sites for transcription factors E2A/HEB (hereafter E2A), Runx, GATA3, and Ikaros (Fig. S1 and Fig. 1, A and B). To evaluate whether these factors bound at the core ASE, we performed chromatin immunoprecipitation (ChIP) from extracts of sorted mouse DP thymocytes. The ASE core sequence was enriched in immunoprecipitates of all four transcription factors relative to the IgG control, whereas negative control MageA2 sequences were not (Fig. 1 C). Hence, E2A, GATA3, Runx, and Ikaros were all identified as potential regulators of ASE activity.
Figure 1.
Dissection of ASE enhancer activity using extrachromosomal reporter assays. (A and B) Diagrams show relative positioning of the ASE with respect to mouse Rag1 and Rag2 promoters (A) and conserved binding sites for transcription factors within the ASE core region (B). (C) ChIP assays of transcription factor binding to the ASE core region and the inactive MageA2 promoter in sorted DP thymocytes. The data represent mean ± SEM of three independent experiments, with enrichment expressed relative to control IgG ChIP. (D) Activity of wild-type and mutant ASEs tested in luciferase reporters containing the Rag1 promoter (top) or Rag2 promoter (bottom). Test ASE fragments of 1.2 kb were cloned downstream of Rag1 or Rag2 promoter–driven luciferase gene, and plasmids were assayed for luciferase activity following transient transfection into VL3-3M2 cells. The data represent mean ± SEM of four independent experiments, with values for mutant ASEs normalized to wild-type in each experiment and the average value for the wild-type ASE set as 1. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by two-way ANOVA with Holm-Sidak’s multiple comparisons test (C) or one-way ANOVA with Holm-Sidak’s multiple comparisons test (D).
To investigate the functional significance of transcription factor binding, we individually mutated the binding sites for these factors in luciferase reporter plasmids containing the 1.2-kb ASE region paired with either the Rag1 or Rag2 promoter (Table S1). We previously showed this ASE fragment to display potent enhancer activity on both promoters. Constructs were tested for transcriptional activity following transfection into the RAG-expressing, mouse DP thymoma cell line VL3-3M2. As previously reported, the Rag1 and Rag2 promoters were potently activated by the wild-type ASE (Fig. 1 D). However, when tested with either promoter, ASE activity was impaired by mutation of one of the two E2A sites, the Runx site, or the GATA3 site. In contrast, when tested on the Rag2 promoter, ASE activity was increased by mutation of the binding site for Ikaros, and ASE activity trended similarly when tested on the Rag1 promoter. Hence, GATA3, E2A, and Runx family transcription factors were candidate positive regulators of ASE activity, whereas Ikaros family transcription factors were implicated as potential negative regulators of ASE activity.
Intact E2A and GATA3 sites are essential for ASE activity at the endogenous RAG locus
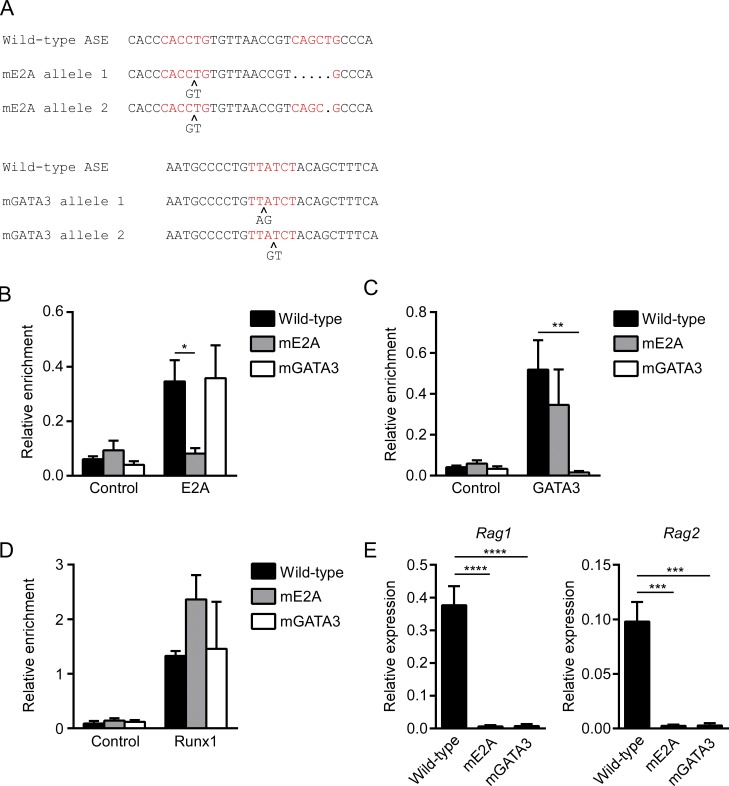
Although the luciferase experiments provided important insights into ASE function, these experiments test ASE activity in an extrachromosomal reporter plasmid that does not assemble native chromatin and does not reproduce the long-distance interactions that are important for ASE function in vivo (Hao et al., 2015). Therefore, we used clustered regularly interspaced short palindromic repeats (CRISPR)–Cas9 technology to disrupt the ASE GATA3 and E2A binding sites at the endogenous RAG locus in VL3-3M2 cells. We generated a clone of VL3-3M2 in which the GATA3 binding site was disrupted on both alleles and a clone in which both E2A binding sites were disrupted on both alleles (Fig. 2 A and Fig. S2). ChIP showed that the ASE core region with disrupted E2A binding sites displayed a dramatic loss of E2A binding (Fig. 2 B), whereas that with disrupted GATA3 binding sites displayed a dramatic loss of GATA3 binding (Fig. 2 C). Notably, loss of E2A binding did not affect GATA3 occupancy and vice versa, suggesting that the two factors bind independently to the ASE. Runx1 binding was also independent of E2A and GATA3 (Fig. 2 D).
Figure 2.
ASE function evaluated by CRISPR-Cas9 targeting of the endogenous VL3-3M2 ASE. (A) Nucleotide sequences showing wild-type ASE E2A and GATA3 binding sites and mutants generated by CRISPR-Cas9 gene targeting of the two alleles of individual VL3-3M2 clones. Consensus binding sites are highlighted in red. (B–D) ChIP compares E2A (B), GATA3 (C), and Runx1 (D) binding to the ASE core in wild-type and mutant VL3-3M2 clones. The data represent mean ± SEM of three independent experiments, with enrichment of ASE sequences in specific antibody and nonspecific IgG (control) immunoprecipitates expressed relative to the abundance of Tcra enhancer sequences (set to 1) in specific antibody immunoprecipitates in each cell line. (E) Rag1 and Rag2 transcript abundance assessed by RT-PCR in VL3-3M2 cell clones with wild-type and mutant ASEs. The data represent mean ± SEM of four independent experiments, with values for Rag1 and Rag2 normalized to those for Actb. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by two-way ANOVA with Holm-Sidak’s multiple comparisons test (B–D) or one-way ANOVA with Holm-Sidak’s multiple comparisons test (E).
Importantly, the ASE GATA3 and E2A binding site mutations caused dramatic reductions in the expression of the endogenous Rag1 and Rag2 genes (Fig. 2 E). In contrast, mutation of only one of the two E2A binding sites (E2A#1) caused only a modest decrease in RAG gene expression (data not shown). The results indicate that E2A and GATA3 binding are both critical for ASE function at the endogenous RAG locus.
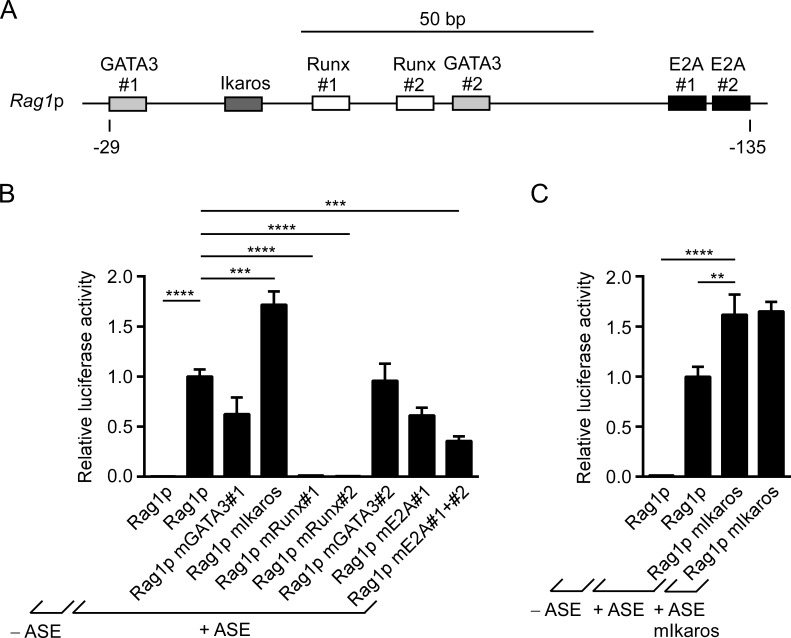
E2A, Runx, and Ikaros are candidate regulators of Rag1 promoter activity
To better understand ASE-dependent regulation of the RAG genes, we assessed the importance of transcription factor binding to the relatively understudied Rag1 promoter. Based on sequence alignments including eight vertebrate species, we identified highly conserved mouse Rag1 promoter binding sites for GATA3, Ikaros, Runx, and E2A (Fig. S2 and Fig. 3 A). Binding site mutants were then tested for effects on gene expression in ASE-containing luciferase reporters in VL3-3M2 cells (Fig. 3 B). The results demonstrated a complete loss of Rag1 promoter activity when either of two Runx binding sites was destroyed. Simultaneous disruption of a pair of E2A binding sites caused a modest reduction in promoter activity, whereas GATA3 site mutations had no effect. Notably, as was the case for the ASE, disruption of an Ikaros binding site caused a significant increase in Rag1 promoter activity. The results suggest that Runx and E2A family members may be positive regulators of the Rag1 promoter, whereas Ikaros family members may play a negative regulatory role. Notably, combined mutation of Ikaros binding sites in the ASE and Rag1 promoter yielded no further up-regulation than either mutation alone (Fig. 3 C). This suggests that negative regulation by Ikaros requires binding to both elements.
Figure 3.
Dissection of Rag1 promoter activity using extrachromosomal reporter assays. (A) Diagram shows conserved binding sites for transcription factors within the Rag1 promoter, with numbering relative to the transcription start site. (B and C) Activity of wild-type and mutant Rag1 promoters tested in luciferase reporters containing a wild-type or mutated 1.2-kb ASE downstream of the luciferase gene. Plasmids were assayed for luciferase activity following transient transfection into VL3-3M2 cells. The data represent mean ± SEM of four independent experiments, with values normalized to those for wild-type Rag1p + ASE in each experiment and the average value for Rag1p + ASE set as 1. **, P < 0.01 ***, P < 0.001; ****, P < 0.0001 by one-way ANOVA with Holm-Sidak’s multiple comparisons test.
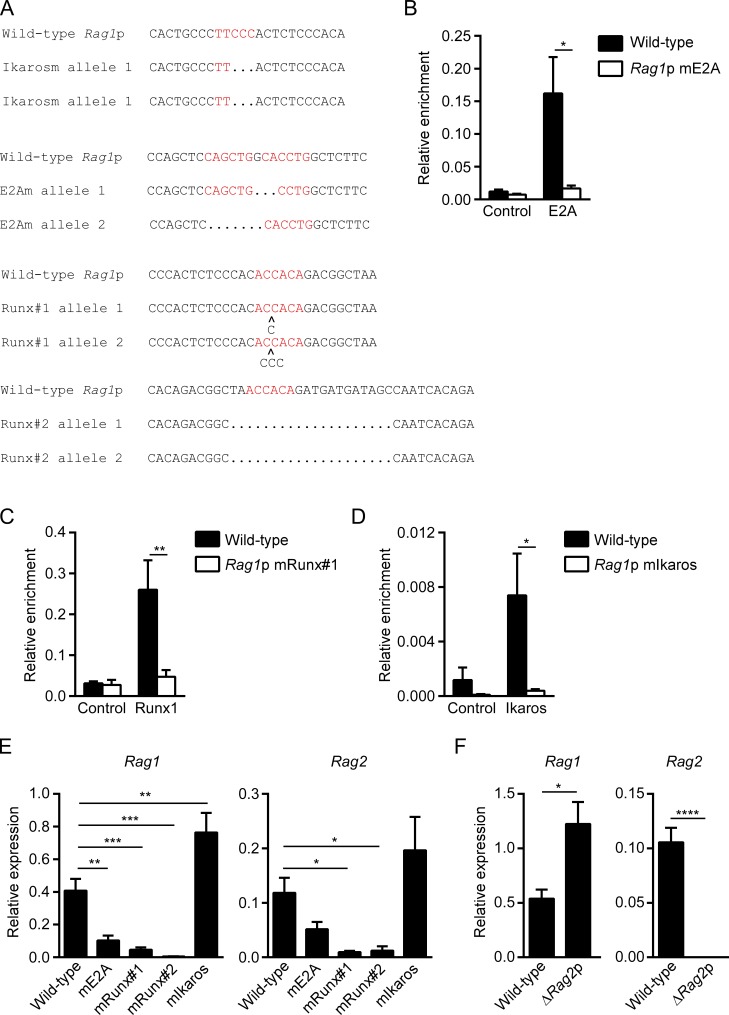
Intact Rag1 promoter E2A and Runx sites are required for maximal endogenous Rag1 and Rag2 expression
To assess the roles of E2A, Runx, and Ikaros transcription factors in endogenous Rag1 promoter function, we again used CRISPR-Cas9 to disrupt the relevant binding sites in the Rag1 promoter in VL3-3M2 cells (Table S2). We generated VL3-3M2 clones with mutations in Runx site #1 on both alleles or in Runx site #2 on both alleles, although the latter was a deletion that was substantially larger than the Runx site itself (Fig. 4 A). We generated a VL3-3M2 clone with disruption of one of two adjacent E2A sites on one allele and the second of these E2A sites on the other allele. We also generated a clone in which the Ikaros binding site was disrupted on both alleles. Although only one of two adjacent E2A motifs was disrupted on each allele in the mE2A clone, ChIP revealed nearly complete loss of E2A binding to the Rag1 promoter in these cells (Fig. 4 B). Runx1 binding to the Rag1 promoter was reduced to near background levels in cells with selective mutation of Runx site #1, suggesting that Runx1 may bind cooperatively to the two adjacent sites (Fig. 4 C). We also detected nearly complete loss of Ikaros occupancy in cells with an Ikaros binding site mutation (Fig. 4 D). Examination of Rag1 gene expression revealed that E2A and Runx site disruptions were associated with substantial reductions in promoter activity, whereas Ikaros site disruption caused increases in promoter activity (Fig. 4 E, left).
Figure 4.
Rag1 and Rag2 promoter function evaluated by CRISPR-Cas9 targeting of the endogenous VL3-3M2 Rag1 promoter. (A) Nucleotide sequences showing wild-type Rag1 promoter Ikaros, E2A, and Runx binding sites and mutants generated by CRISPR-Cas9 gene targeting of the two alleles of individual VL3-3M2 clones. Consensus binding sites are highlighted in red. (B–D) ChIP compares E2A (B), Runx1 (C), and Ikaros (D) binding to the Rag1 promoter in wild-type and mutant VL3-3M2 clones. The data represent mean ± SEM of three independent experiments, with enrichment of Rag1 sequences in specific antibody and nonspecific IgG (control) immunoprecipitates expressed relative to the abundance of Tcra enhancer sequences (set to 1) in anti-E2A (B) and anti-Runx1 (C) immunoprecipitates and relative to the abundance of IL2 receptor sequences (set to 1) in anti-Ikaros immunoprecipitates (D). (E) Rag1 and Rag2 transcript abundance assessed by RT-PCR in wild-type and Rag1 promoter mutant VL3-3M2 cells. The data represent mean ± SEM of six to seven independent experiments, with values for Rag1 and Rag2 normalized to those for Actb. (F) Rag1 and Rag2 transcript abundance measured in wild-type and Rag2 promoter–deleted (−119 to +53) VL3-3M2 cells. The data represent the mean ± SEM of five to six independent experiments using three independent ΔRag2p clones, with values expressed as in E. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by two-way ANOVA with Holm-Sidak’s multiple comparisons test (B–D), one-way ANOVA with Holm-Sidak’s multiple comparisons test (E), or unpaired Student’s t test (F).
Strikingly, VL3-3M2 clones with Runx site mutations in the Rag1 promoter displayed substantial reductions in Rag2 gene expression; those with E2A site mutation trended similarly (Fig. 4 E, right). Thus, Runx1 and likely E2A binding to the Rag1 promoter directly induce Rag1 gene expression and indirectly induce Rag2 gene expression. To further examine functional interdependency of the RAG promoters, we used CRISPR-Cas9 to delete 172 bp from the Rag2 promoter (−119 to +53) on both alleles of VL3-3M2 cells. This region includes the transcription start site as well as the known transcription factor binding sites (Lauring and Schlissel, 1999). This mutation inactivated Rag2 expression but caused a significant increase in Rag1 expression (Fig. 4 F). Thus, our data suggest that Rag2 promoter activity requires an intact and functional Rag1 promoter, whereas Rag1 promoter activity does not similarly require the Rag2 promoter; rather, an intact Rag2 promoter appears to diminish Rag1 promoter activity, suggesting the possibility of competition between the two promoters.
Critical roles for transcription factors GATA3, Runx1, E2A, and SATB1 in RAG gene expression
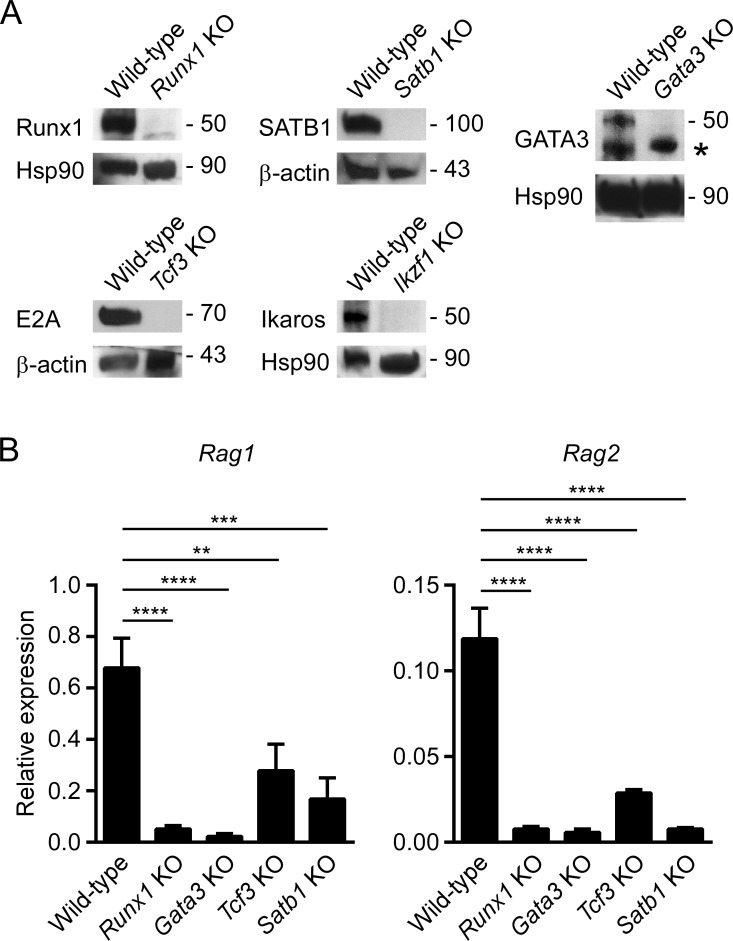
The above experiments implicated GATA3, Runx1, and E2A as positive regulators of RAG gene expression by binding to the ASE, the Rag1 promoter, or both. To independently assess the importance of these transcription factors in RAG gene expression, we used CRISPR-Cas9 to abrogate expression of these factors in VL3-3M2 cells. Loss of protein expression in each instance was confirmed by Western blotting (Fig. 5 A). To minimize concerns about off-target effects of CRISPR-Cas9, for each transcription factor we generated two independent mutant clones with disruptions in either exon 1 or exon 2 (Table S2). Because comparable results were obtained for exon 1 and exon 2 disruptions for each factor, the data from the two clones were combined (Fig. 5 B). Endogenous Rag1 and Rag2 gene expression was dramatically suppressed by the absence of Runx1 or GATA3 and was partially suppressed by the absence of E2A (encoded by Tcf3; Fig. 5 B). The same approach confirmed a role for SATB1 as a positive regulator of RAG gene expression in VL3-3M2 cells (Fig. 5, A and B), in accord with previous results in DP thymocytes in vivo (Hao et al., 2015). We also generated Ikzf1 mutants of VL3-3M2 to assess the role of Ikaros as a negative regulator of RAG gene expression (Fig. 5 A). These results will be described below.
Figure 5.
RAG gene expression analyzed in transcription factor KO VL3-3M2 cells. (A) Western blots of transcription factor protein expression in wild-type and KO VL3-3M2 cells. Bottom panels show loading controls. Approximate molecular weights are indicated in kilodaltons. Asterisk indicates a presumed nonspecific band detected by anti-GATA3. (B) Rag1 and Rag2 transcript abundance in wild-type and transcription factor KO VL3-3M2 cells assessed by RT-PCR. The data represent mean ± SEM of four to six independent experiments, with values for Rag1 and Rag2 normalized to those for Actb. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by one-way ANOVA with Holm-Sidak’s multiple comparisons test.
GATA3, Runx1, E2A, and SATB1 proteins regulate RAG locus conformation
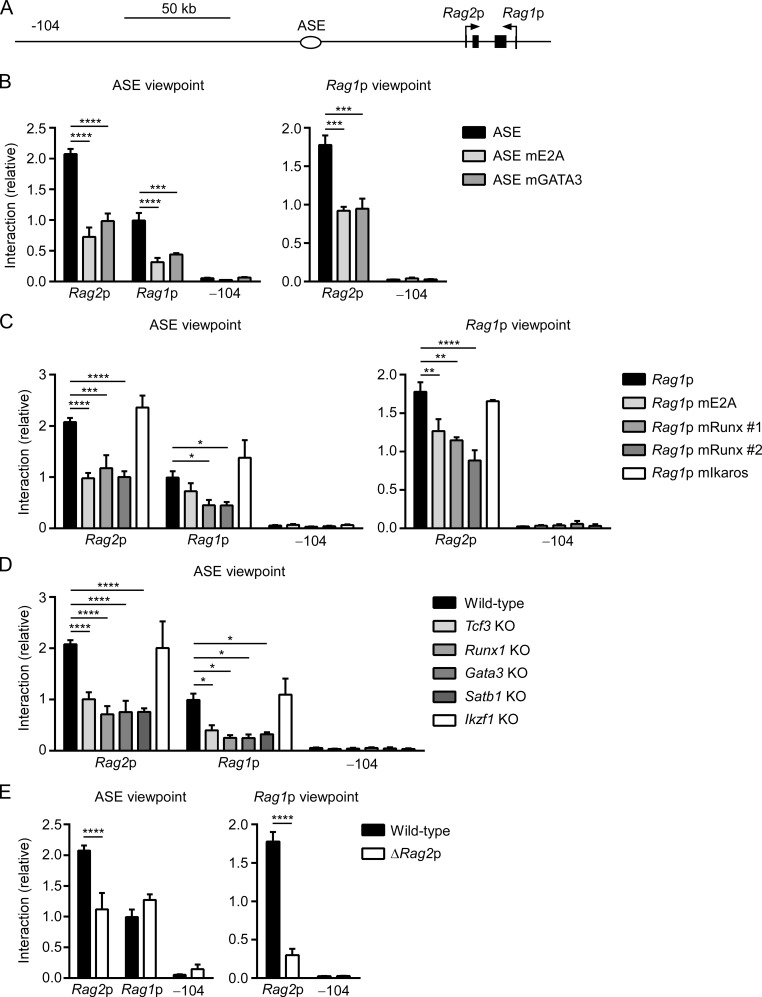
We previously showed that the ASE and Rag1 and Rag2 promoters interact in a developmental stage-specific fashion in DP thymocytes and that these interactions are mediated, in part, by chromatin organizer SATB1 (Hao et al., 2015). To better understand how GATA3, Runx1, and E2A regulate Rag1 and Rag2 gene expression, we measured the effects of binding site mutations and transcription factor KOs on RAG locus conformation by performing chromosome conformation capture (3C) in VL3-3M2 cell clones. Similar to mouse DP thymocytes, wild-type VL3-3M2 cells demonstrated frequent interactions between the ASE and the Rag1 and Rag2 promoters in assays using the ASE as a viewpoint (Fig. 6, A and B, left) and frequent interactions between the Rag1 and Rag2 promoters in assays using the Rag1 promoter as a viewpoint (Fig. 6 B, right). By comparison, interactions with a previously established negative control site located 104 kb distal to the ASE (Hao et al., 2015) were exceedingly low (−104; Fig. 6, A and B). ASE mutations in the E2A or GATA3 binding sites resulted in substantially reduced contacts between the ASE and the Rag1 and Rag2 promoters (Fig. 6 B, left) and also between the Rag1 and Rag2 promoters (Fig. 6 B, right). Thus, ASE binding sites for E2A and GATA3 are critical to physically organize a transcriptionally active RAG locus. Mutation of Rag1 promoter binding sites for E2A or Runx1 also resulted in reduced contact frequencies between the ASE and the Rag1 and Rag2 promoters (Fig. 6 C, left) and between the Rag1 and Rag2 promoters (Fig. 6 C, right). Thus, Rag1 promoter integrity is as important as ASE integrity for the detected pairwise interactions among the ASE and Rag1 and Rag2 promoters. In contrast, disruption of the Rag1 promoter Ikaros binding site had no effect on these interactions (Fig. 6 C).
Figure 6.
Regulation of RAG locus conformation by GATA3, E2A, Runx1, and SATB1. (A) RAG locus map identifying sites analyzed by 3C. (B–E) 3C analysis of interactions of BglII fragments with the (B) ASE (left) and Rag1 promoter (right) viewpoints in wild-type and ASE mutant VL3-3M2 cells, (C) ASE (left) and Rag1 promoter (right) viewpoints in wild-type and Rag1 promoter mutant VL3-3M2 cells, (D) ASE viewpoint in wild-type or transcription factor KO VL3-3M2 cells, and (E) ASE (left) and Rag1 promoter (right) viewpoints in wild-type and Rag2 promoter–deleted VL3-3M2 cells. In each case the −104 fragment served as a negative control. The data represent the mean ± SEM of three to four independent experiments, with interaction frequencies normalized to those of a nearest neighbor BglII fragment. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by two-way ANOVA with Holm-Sidak’s multiple comparisons test.
Consistent with the above observations, VL3-3M2 cells lacking expression of Runx1, GATA3, or E2A displayed reduced interactions between the ASE and Rag1 and Rag2 promoters, as did VL3-3M2 cells lacking SATB1 (Fig. 6 D). On the other hand, Ikaros KO VL3-3M2 cells demonstrated no change in ASE–promoter contacts (Fig. 6 D). Thus, by binding to the ASE or Rag1 promoter, GATA3, Runx1, E2A, and SATB1 are all essential for the formation of the transcriptionally active RAG locus chromatin interaction hub in VL3-3M2 cells.
The above experiments revealed that an intact Rag1 promoter is essential to recruit or maintain Rag2 promoter contacts with the Rag1 promoter and the ASE (Fig. 6 C). However, in cells with Rag2 promoter deletion, Rag2 lost contact with the ASE and Rag1 promoter, but Rag1 promoter–ASE interactions remained intact (Fig. 6 E). This result is fully consistent with the distinct effects of Rag1 and Rag2 promoter mutations on transcription from the reciprocal promoter (Fig. 4 C). We conclude that a transcriptionally active RAG locus is assembled in hierarchical fashion, with ASE–Rag1 promoter interaction creating a framework that allows stable Rag2 promoter recruitment to form a three-way complex. Nevertheless, the basis for apparent competition between the Rag1 and Rag2 promoters (Fig. 4 F) remains uncertain because Rag2 promoter deletion did not cause a significant increase in Rag1 promoter–ASE interactions (Fig. 6 E, left) or Rag1 promoter histone H3 acetylation (Fig. S3).
RAG down-regulation is associated with loss of chromatin conformation and requires Ikaros
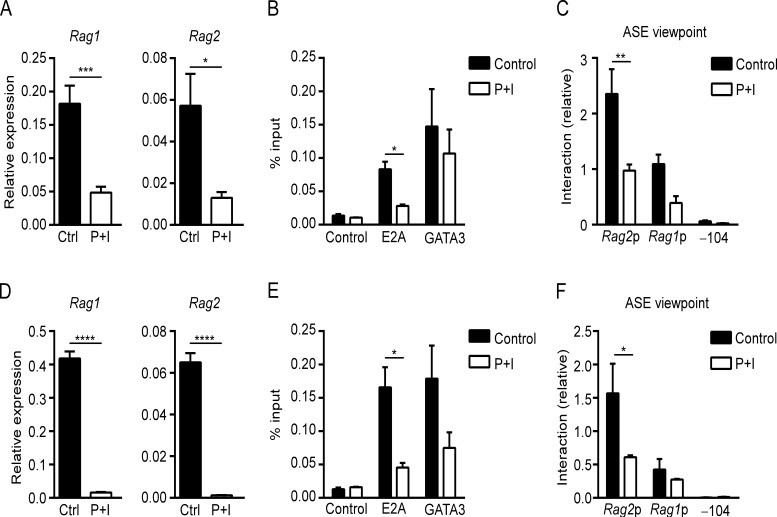
RAG expression is down-regulated upon positive selection in DP thymocytes, but the molecular basis for this down-regulation has not been established. In previous studies, the combination of PMA and ionomycin has proven effective in mimicking positive selection signals and down-regulating RAG gene expression in both DP thymocytes and VL3-3M2 cells (Turka et al., 1991; Brown et al., 1999). As expected, VL3-3M2 cells incubated with PMA and ionomycin demonstrated significant down-regulation of Rag1 and Rag2 transcripts as compared with control cells treated with the DMSO vehicle (Fig. 7 A). Transcriptional down-regulation occurred with loss of E2A from the ASE (Fig. 7 B). Moreover, 3C revealed that transcriptional down-regulation is associated with a disruption of RAG locus conformation, with a significant reduction in ASE–Rag2 promoter contacts and a reduction in ASE–Rag1 promoter contacts that fell just short of statistical significance (P = 0.06; Fig. 7 C). To substantiate these findings, we similarly treated primary DP thymocytes in culture. Consistent with previous studies (Turka et al., 1991), incubation with PMA and ionomycin resulted in a rapid and almost complete loss of Rag1 and Rag2 expression (Fig. 7 D). Moreover, as in VL3-3M2 cells, ChIP showed a significant reduction in E2A binding to the ASE (Fig. 7 E), and 3C showed a significant loss of ASE–Rag2 promoter interactions, although contacts between the ASE and the Rag1 promoter were not obviously perturbed (Fig. 7 F). Perturbation of ASE–Rag1 promoter interactions in VL3-3M2 but not in DP thymocytes could reflect the shorter time course of PMA and ionomycin stimulation in the latter (see Materials and methods). Preferential or early loss of Rag2 is reminiscent of the conformational state adopted in the absence of SATB1, supporting the notion that the Rag2 promoter is the most tenuously associated component of the RAG locus chromatin complex (Hao et al., 2015).
Figure 7.
RAG down-regulation is associated with loss of transcription factor binding and chromatin conformation. (A) Rag1 and Rag2 transcript abundance evaluated by RT-PCR in control and PMA plus ionomycin (P+I)–treated VL3-3M2 cells. The data represent mean ± SEM of six independent experiments, with values for Rag1 and Rag2 normalized to those for Actb. (B) ASE transcription factor occupancy evaluated by ChIP in control and PMA plus ionomycin–treated VL3-3M2 cells. The data represent mean ± SEM of three independent experiments. (C) 3C analysis of interactions of BglII fragments with the ASE viewpoint in control and PMA plus ionomycin–treated VL3-3M2 cells. The data represent the mean ± SEM of three independent experiments, with interaction frequencies normalized to those of a nearest neighbor BglII fragment. (D) Rag1 and Rag2 transcript abundance as in A using control and PMA plus ionomycin–treated sorted DP thymocytes. The data represent mean ± SEM of six independent experiments. (E) ASE transcription factor occupancy evaluated by ChIP in control and PMA plus ionomycin–treated sorted DP thymocytes. The data represent mean ± SEM of three independent experiments. (F) 3C analysis as in C using control and PMA plus ionomycin–treated sorted DP thymocytes. The data represent mean ± SEM of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired Student’s t test (A and D) or two-way ANOVA with Holm-Sidak’s multiple comparisons test (B, C, E, and F).
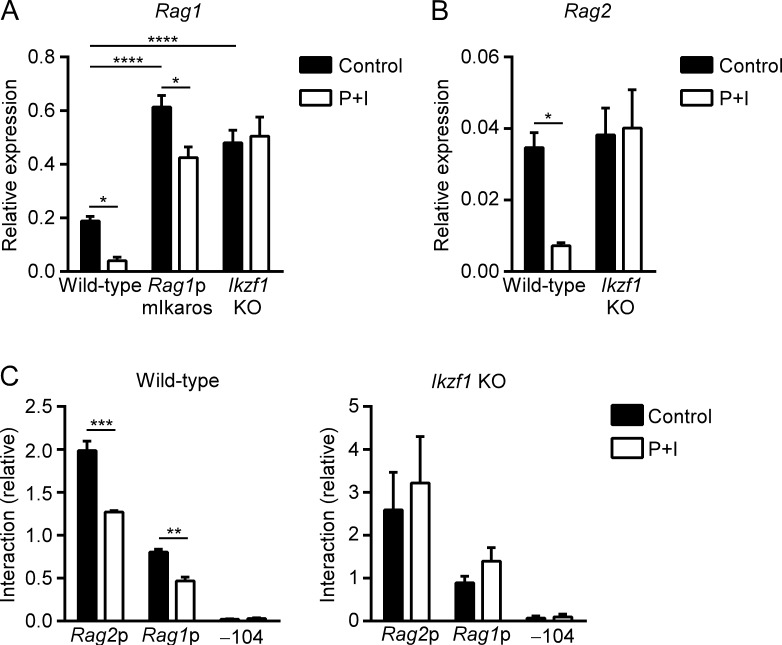
Previous work demonstrated that down-regulation of Dntt (encoding terminal deoxynucleotidyl transferase) in PMA- and ionomycin-stimulated DP thymocytes depends on Ikaros (Trinh et al., 2001). Since our experiments revealed Ikaros to be a negative regulator of Rag1 promoter function (Figs. 3 B and 4 E), we asked whether down-regulation of Rag1 in response to PMA and ionomycin requires Ikaros. As expected, mutation of the Rag1 promoter Ikaros binding site or KO of Ikzf1 caused a substantial up-regulation of Rag1 expression in untreated cells (Fig. 8 A). Notably, treatment with PMA and ionomycin caused an 80% reduction of Rag1 gene expression in wild-type VL3-3M2 but only a 30% reduction in cells with a mutated Rag1 promoter Ikaros binding site. Moreover, cells lacking Ikaros protein were incapable of down-regulating Rag1 gene expression under these conditions (Fig. 8 A). These results indicate that Ikaros binding to the Rag1 promoter is essential for Rag1 down-regulation during positive selection.
Figure 8.
RAG down-regulation and chromatin hub disassembly requires Ikaros. (A) Rag1 transcript abundance evaluated by RT-PCR in control and PMA plus ionomycin (P+I)–treated wild-type, Rag1p Ikaros site mutant, or Ikzf1 KO VL3-3M2 cells. The data represent mean ± SEM of five independent experiments. (B) Rag2 transcript abundance evaluated as in A in control and PMA plus ionomycin–treated wild-type and Ikzf1 KO VL3-3M2 cells. The data represent mean ± SEM of five independent experiments. (C) 3C analysis of interactions of BglII fragments with the ASE viewpoint in control and PMA plus ionomycin–treated wild-type (left) and Ikzf1 KO (right) VL3-3M2 cells. The data represent the mean ± SEM of three independent experiments, with interaction frequencies normalized to those of a nearest neighbor BglII fragment. *, P < 0.05; **, P < 0.01; ***, P < 0.001, ****, P < 0.0001 by two-way ANOVA with Holm-Sidak’s multiple comparisons test.
Ikzf1 KO did not cause up-regulation of Rag2 gene expression (Fig. 8 B). However, down-regulation of Rag2 gene expression in response to PMA and ionomycin was abrogated by Ikzf1 KO. Moreover, Ikzf1 KO prevented PMA and ionomycin-dependent loss of contacts between the ASE and the Rag1 and Rag2 promoters (Fig. 8 C). We conclude that Ikaros functions in nonredundant fashion to promote signal-induced down-regulation of Rag1 and Rag2 gene expression in VL3-3M2 DP thymocytes and that Ikaros functions in part by mediating disassembly of the RAG locus chromatin hub.
Discussion
Transcriptional regulation of the RAG genes is complex, with distinct cis-regulatory elements and transcription factors mediating RAG gene expression at different stages of B and T cell development (Kuo and Schlissel, 2009). Prior studies established the RAG ASE as an essential cis-regulator of RAG gene transcription in DP thymocytes, acting by engaging the Rag1 and Rag2 promoters in direct physical interactions to form an active chromatin hub (Yannoutsos et al., 2004; Hao et al., 2015). However, to date, chromatin organizer SATB1 is the only factor shown to function as a direct regulator of ASE-mediated RAG gene expression and locus conformation in DP thymocytes (Hao et al., 2015). Here we revealed E2A, GATA3, and Runx1 as additional transcriptional regulators that promote RAG gene expression and chromatin organization in DP thymocytes, GATA3 by binding to the ASE, Runx1 by binding to the Rag1 promoter, and E2A by binding to both elements. All three factors appear to play important roles in the assembly of the transcriptionally active conformation of the RAG locus.
We undertook several complementary approaches to assess the regulation of RAG gene expression in DP thymocyte cell line VL3-3M2: site-directed mutagenesis of extrachromosomal reporter plasmids, CRISPR-Cas9–mediated mutagenesis of the endogenous RAG locus, and CRISPR-Cas9–mediated KO of candidate transcriptional regulators. Individually, each of these approaches has limitations. Extrachromosomal reporter assays fail to replicate the constraints imposed by chromatin structure and distance at the endogenous locus. CRISPR-Cas9–mediated insertions and deletions disrupt transcription factor binding sites but also disrupt the spacing between otherwise unmanipulated binding sites that flank the sites of interest. Finally, transcription factor KOs can have pleiotropic effects and influence gene expression indirectly. Our conclusions gain power from concordant results obtained from the three complementary approaches, arguing persuasively that E2A, GATA3, and Runx1 function directly to positively regulate expression of the endogenous, chromatin-embedded RAG genes in DP thymocytes.
Our work implicates E2A and GATA3 as critical regulators of ASE function. Consistent with this, prior work had shown E2A and GATA3 to occupy the ASE enhancer core region and to do so as early as the DN2/DN3 stage of T cell development (Miyazaki et al., 2011, 2017; Zhang et al., 2012). Moreover, E2A- and HEB-deficient mice were reported to display substantially reduced RAG gene expression in DP thymocytes (D’Cruz et al., 2010), although it was not known whether these proteins regulated the RAG locus directly and, if so, whether through the ASE, the RAG promoters, or both. It is unclear what triggers ASE activity at the DN–DP transition. ASE occupancy by GATA3 has been examined in both DN and DP thymocytes and is substantially increased in the latter (Zhang et al., 2012). However, occupancy by E2A and SATB1 has not been examined in both compartments. Because Tcf3 and Gata3 expression is down-regulated between the DN2/3 and DP stages (Hernández-Hoyos et al., 2003), whereas Satb1 gene expression is up-regulated by two orders of magnitude across the same transition (Hao et al., 2015), SATB1 may represent the more likely candidate to trigger ASE activation and RAG gene expression in DP thymocytes. Yet SATB1 may not be the sole trigger, because RAG gene expression is reduced by only 75–80% in SATB1-deficient DP thymocytes (Hao et al., 2015).
Our work implicates Runx1 and E2A as critical regulators of the Rag1 promoter. Remarkably, disrupted binding of these factors to the Rag1 promoter caused reductions in Rag2 expression that were comparable to the reductions in Rag1 gene expression. Loss of Rag2 gene expression in Rag1 promoter mutants is likely secondary to the disruption of locus organization since in these mutants, the Rag2 promoter lost contact with both the Rag1 promoter and the ASE. Prior analysis indicated that Rag2 promoter contacts with both the ASE and the Rag1 promoter were disrupted in the absence of SATB1, even though ASE–Rag1 promoter interactions were maintained (Hao et al., 2015). This result, coupled with our current data, suggests that the ASE–Rag1 promoter interaction may represent the fundamental building block of locus organization and may serve as a platform for Rag2 promoter recruitment. Consistent with the notion of hierarchical assembly of a transcriptionally active RAG gene complex, deletion of the Rag2 promoter did not reciprocally impair Rag1 gene expression. In fact, Rag1 expression was elevated in Rag2 promoter deleted VL3-3M2 cells, suggesting that recruitment of Rag2 into the ASE–Rag1 promoter complex may result in competition between the two promoters for the binding of certain transcriptional regulators. Prior studies have shown that promoter–promoter interactions are common genome-wide and that promoters involved in such interactions tend to be coregulated and to interact with shared distal enhancers. Moreover, the activities of these promoters are often interdependent, at least in part because the promoters themselves often display enhancer activity (Li et al., 2012; Dao et al., 2017). Consistent with this possibility, the array of transcription factors recruited to the Rag1 promoter is similar to that recruited to the ASE and many other T cell–specific enhancers. The above considerations suggest that the RAG locus shares organizational and regulatory features characteristic of many complex loci across the genome.
RAG gene down-regulation in response to positive selection signals is essential to curtail ongoing Tcra gene rearrangement and thereby fix the TCR repertoire. We found that Ikaros plays a critical role in RAG gene down-regulation. In accord with our results, prior ChIP-sequencing analysis demonstrated Ikaros binding to the Rag1 promoter and the ASE in DP thymocytes; binding was detected at the Rag2 promoter as well (Gene Expression Omnibus accession no. GSE61148). Numerous studies have documented a role for Ikaros in transcriptional repression (Koipally et al., 1999; Sabbattini et al., 2001; Trinh et al., 2001; Su et al., 2004; Liang et al., 2017). Our results are reminiscent of a comparable role for Ikaros in Dntt gene down-regulation during signaling for positive selection (Trinh et al., 2001). Stimulation of VL3-3M2 and thymocytes with PMA and ionomycin caused dephosphorylation of Ikaros, resulting in increased DNA binding and suppressive function (Gurel et al., 2008). Ikaros can directly displace activating transcription factors (Trinh et al., 2001) or cause repression by recruiting chromatin remodeling complexes like NuRD and PRC2 (Liang et al., 2017; Heizmann et al., 2018). Ikaros may also function by relocating the RAG locus to pericentric heterochromatin (Brown et al., 1999). Further work will be required to assess the roles of SATB1 and Ikaros as potential triggers for RAG gene activation and inactivation, respectively, in DP thymocytes and to document epigenetic changes that may be required for permanent silencing of the RAG locus in mature thymocytes and peripheral T cells.
Materials and methods
Cells and cell culture
VL3-3M2 cells (Groves et al., 1995) were kindly provided by Dr. S. Sarafova (Davidson College, Davidson, NC) and were cultured at 37°C in RPMI 1640 supplemented with 10% (vol/vol) fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, 55 µM 2-mercaptoethanol, and 2 mM L-glutamine in an atmosphere of 5% CO2. DP thymocytes were obtained from C57BL/6 mice by cell sorting as described previously (Hao et al., 2015) and were cultured as above. PMA and ionomycin were obtained from Sigma and were added to the medium from a concentrated stock dissolved in DMSO to final concentrations of 20 ng/ml PMA and 250 ng/ml ionomycin. Control cells received an equivalent volume of DMSO. Treatment of VL3-3M2 cells was for 16 h, whereas treatment of DP thymocytes was limited to 2 h. All mice were used in accordance with protocols approved by the Duke University Animal Care and Use Committee.
CRISPR design and targeting
Guide RNAs were designed using a publicly available CRISPR design tool (Ran et al., 2013). Single guides targeting transcription factor binding sites were selected based on two considerations: proximity of PAM sequences to the consensus sequence and minimizing off target sites. For transcription factor KOs, guide RNA pairs were designed to target either exon 1 or exon 2. Guides were inserted into the pX458 vector (Addgene) containing coding sequences for Cas9 and GFP, as described earlier (Ran et al., 2013). VL3-3M2 cells were transfected with pX458 containing target guides by nucleofection using Amaxa Cell Line nucleofector kit V (Lonza) according to manufacturer’s instructions. Cells were grown for 48 h at 37°C in RPMI 1640 medium containing 10% (vol/vol) fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin, in an atmosphere of 5% CO2. The brightest GFP-positive cells were single-cell sorted into 96-well plates containing culture medium, and clones were grown for 7–10 d until visible colonies were obtained. Following preparation of genomic DNA, mutations were detected by analysis of Tm curves in real-time PCR using a Roche Lightcycler and were confirmed by sequencing. As many as six to eight sequences were analyzed for each clone to determine whether mutations were homozygous. VL3-3M2 cells with biallelic mutation at ASE E2A site #1 were retargeted to create a biallelic mutation in the second E2A binding site. Similarly, VL3-3M2 cells with monoallelic mutation in Rag1 promoter Runx site #1 were retargeted to create a mutation on the second allele.
ChIP
ChIP was performed using previously described methods (Chen et al., 2015). Briefly, 107 thymocytes or VL3-3M2 cells were cross-linked using 1% (vol/vol) formaldehyde for 10 min at 23°C, and cross-linking was terminated by addition of glycine to 0.125 M. Cells were washed and lysed in 1 ml of 5 mM Pipes, pH 8, 85 mM KCl, 0.5% (vol/vol) NP-40, 0.1 mM PMSF, and 0.1 mM benzamidine for 10 min on ice. Nuclei were collected by centrifugation and were lysed in 500 µl of 50 mM Tris-HCl, pH 8, 10 mM EDTA, 1% (wt/vol) SDS, 0.1 M benzamidine, and 0.1 M PMSF for 10 min at 23°C. Chromatin was sonicated using a Sonicator 3000 (Misonix) with cycles of 20 s on and 20 s off to generate 200- to 500-bp fragments. Sonicated samples were diluted into 2 ml of 0.01% (wt/vol) SDS, 1.1% (vol/vol) Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8, 167 mM NaCl and were precleared by incubation with 70 µl of 50% protein A agarose slurry containing salmon sperm DNA (Millipore). One-third of each sample was then incubated with specific antibodies or control IgG. Reagents included anti-GATA3 (Santa Cruz; HG 3-31) and anti-E2A (Santa Cruz; Yae) monoclonal antibodies and anti-Runx1 (Millipore; PC284), anti-E2A (Santa Cruz; N-649), and anti-Ikaros (Santa Cruz; H-100) polyclonal antibodies. Antibody-bound chromatin was pulled down using protein A agarose beads, which were washed twice with 1 ml of 0.01% (wt/vol) SDS, 1.1% (vol/vol) Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8, 167 mM NaCl, twice with 0.1% (wt/vol) SDS, 1% (vol/vol) Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8, 500 mM NaCl, twice with 100 mM Tris-HCl, pH 8, 500 mM LiCl, 1% NP-40, 1% deoxycholic acid, and twice with 10 mM Tris-HCl, pH 8, 1 mM EDTA. Bound chromatin was then eluted into 0.5 ml of 50 mM NaHCO3, 1% SDS, and cross-linking was reversed by adding 20 µl of 5M NaCl and incubation at 65°C for 16 h. DNA was then purified by phenol:chloroform extraction and ethanol precipitation. ChIP samples were analyzed using SYBR Green quantitative PCR as described earlier (Hao et al., 2015). ASE and MageA2 ChIP signals in DP thymocytes were expressed as enrichment relative to nonspecific IgG control immunoprecipitates. ASE and Rag1 promoter ChIP signals and IgG controls in wild-type and transcription factor binding site mutant VL3-3M2 cells were expressed by normalizing to positive control Tcra enhancer signals in E2A, GATA3, and Runx1 immunoprecipitates or to positive control IL2 promoter signals in Ikaros immunoprecipitates. For ChIP using antibodies specific for acetylated histone H3 (Millipore; 06-599) or control rabbit IgG (R&D Systems; ab-105-c), samples were prepared without cross-linking as described earlier (Hao and Krangel, 2011). Primers used were ASE ChIP F, 5′-CCACCTGTGTTAACCGTCAG-3′; ASE ChIP R, 5′-CTATCTTTGCAGCCCACCAA-3′; Rag1p F, 5′-GCTGTCTACTCTCTCCTTGCTC-3′; Rag1p R, 5′-TGTTTCTGCACTCAGGTCCC-3′; MageA2c-F, 5′-AACGTTTTGTGAACGTCCTGAG-3′; MageA2c-R, 5′-GACGCTCCAGAACAAAATGGC-3′; Eα ChIP F, 5′-CCCTGAAATGGGTAAGCTGG-3′; Eα ChIP R, 5′-TGTTCAGACCCAAACACCTG-3′; IL2p ChIP F, 5′-TAAGTGTGGGCTAACCCGA-3′; IL2p ChIP R, 5′-CAAGGAGCACAAGTGTCAATGTGA-3′; B2m F, 5′-CTGCTACTCGGCGCTTCAGT-3′; and B2m R, 5′-GAGAGGGGAAAGAGGCACTCA-3′.
Luciferase assay
Luciferase assays were performed as described previously (Hao et al., 2015).
Quantitative RT-PCR
Total RNA was isolated from thymocytes and VL3-3M2 cells using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. cDNA was generated using 1 μg of RNA and an iScript kit (Bio-Rad) according to the manufacturer’s instructions. Real-time quantitative PCR and primers for Actb, Rag1, and Rag2 were as described (Hao et al., 2015).
Western blot
Antibodies specific for Runx1 (Abcam; ab23980), GATA3 (Santa Cruz; HG 3-31), E2A (Santa Cruz; Yae), Ikaros (Santa Cruz; M-20), SATB1 (Cell Signaling; L-745), HSP90 (Santa Cruz; H-114), and Actin (Santa Cruz; I-19) were used to perform Western blotting using a Bio-Rad apparatus according to the manufacturer’s instructions. Molecular weights were determined using a multicolor broad-range protein ladder (Spectra; 26634).
3C
3C was performed as previously described (Hao et al., 2015) with the following modifications: 107 thymocytes or 5 × 106 VL3-3M2 cells were cross-linked using 1% formaldehyde for 10 min at 23°C. Cross-linking was terminated by addition of glycine to 0.125 M and incubation for 5 min at 23°C. Cells were then pelleted, washed once with Dulbecco’s PBS without Ca2+ and Mg2+, and subjected to lysis by incubation in 10 mM Tris-HCl, pH 8, 10 mM NaCl, 0.2% (vol/vol) NP-40, 0.1 M benzamidine, and 0.1 mM PMSF for 10 min on ice. Nuclei were pelleted, washed with PBS, and resuspended in 0.5 ml of New England Biolabs buffer 3.1 containing 0.3% SDS. After incubation for 1 h at 37°C, Triton X-100 was added to 2% final concentration, and incubation was continued for 1 h at 37°C. The chromatin was then digested by incubation with 200 U of BglII for 16 h at 37°C, followed by addition of 200 U BglII for an additional 6 h. Digestion was stopped by addition of SDS to 0.8% (wt/vol) and incubation for 20 min at 65°C. Chromatin was then diluted to 7 ml in 30 mM Tris HCl, pH 8, 10 mM MgCl2, and 1% (vol/vol) Triton X-100 and incubated at 37°C for 1 h. A 200-µl aliquot was collected to assess the efficiency of digestion, and the remainder was supplemented with dithiothreitol to 1 mM and ATP to 0.1 mM and was subjected to ligation by addition of 200 U of T4 DNA ligase for 16 h incubation at 16°C. Ligation was then continued by addition of 200 U of T4 DNA ligase for an additional 6 h. The ligated chromatin was then subjected to reverse cross-linking by addition of proteinase K to 200 µg/ml and incubation for 16 h at 65°C and was purified by phenol:chloroform extraction and isopropanol precipitation. Ligated products were quantified using BglII digested and ligated BACs 374F10 and 2141E7 (BACPAC, CHORI) and PCR primers and probes for Taqman-based real-time quantitative PCR as described earlier (Hao et al., 2015).
Online supplemental material
Fig. S1 shows sequence conservation of the ASE core. Fig. S2 shows sequence conservation of the Rag1 promoter. Fig. S3 shows ChIP analysis of Rag1 promoter histone acetylation in VL3-3M2 cells with intact or deleted Rag2 promoter. Table S1 shows transcription factor binding site mutations generated for luciferase assays. Table S2 shows the list of guide RNAs used in this study.
Supplementary Material
Acknowledgments
We thank D. Dauphars and S. Chen for their valuable comments on the manuscript.
This research was supported in part by National Institutes of Health grant GM41052 to M.S. Krangel.
The authors declare no competing financial interests.
Author contributions: A.K. Naik and M.S. Krangel designed the study; A.K. Naik, A.T. Byrd, and A.C.K. Lucander performed the experiments; A.K. Naik and M.S. Krangel analyzed the data; and A.K. Naik and M.S. Krangel wrote the manuscript.
References
- Amin R.H., and Schlissel M.S.. 2008. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat. Immunol. 9:613–622. 10.1038/ni.1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto V., Marques R., and Demengeot J.. 2001. Early death and severe lymphopenia caused by ubiquitous expression of the Rag1 and Rag2 genes in mice. Eur. J. Immunol. 31:3763–3772. [DOI] [PubMed] [Google Scholar]
- Borgulya P., Kishi H., Uematsu Y., and von Boehmer H.. 1992. Exclusion and inclusion of α and β T cell receptor alleles. Cell. 69:529–537. 10.1016/0092-8674(92)90453-J [DOI] [PubMed] [Google Scholar]
- Brown K.E., Baxter J., Graf D., Merkenschlager M., and Fisher A.G.. 1999. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell. 3:207–217. 10.1016/S1097-2765(00)80311-1 [DOI] [PubMed] [Google Scholar]
- Brown S.T., Miranda G.A., Galic Z., Hartman I.Z., Lyon C.J., and Aguilera R.J.. 1997. Regulation of the RAG-1 promoter by the NF-Y transcription factor. J. Immunol. 158:5071–5074. [PubMed] [Google Scholar]
- Chen L., Carico Z., Shih H.Y., and Krangel M.S.. 2015. A discrete chromatin loop in the mouse Tcra-Tcrd locus shapes the TCRδ and TCRα repertoires. Nat. Immunol. 16:1085–1093. 10.1038/ni.3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Cruz L.M., Knell J., Fujimoto J.K., and Goldrath A.W.. 2010. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat. Immunol. 11:240–249. 10.1038/ni.1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao L.T.M., Galindo-Albarrán A.O., Castro-Mondragon J.A., Andrieu-Soler C., Medina-Rivera A., Souaid C., Charbonnier G., Griffon A., Vanhille L., Stephen T., et al. 2017. Genome-wide characterization of mammalian promoters with distal enhancer functions. Nat. Genet. 49:1073–1081. 10.1038/ng.3884 [DOI] [PubMed] [Google Scholar]
- Gostissa M., Alt F.W., and Chiarle R.. 2011. Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annu. Rev. Immunol. 29:319–350. 10.1146/annurev-immunol-031210-101329 [DOI] [PubMed] [Google Scholar]
- Grawunder U., Leu T.M., Schatz D.G., Werner A., Rolink A.G., Melchers F., and Winkler T.H.. 1995. Down-regulation of RAG1 and RAG2 gene expression in preB cells after functional immunoglobulin heavy chain rearrangement. Immunity. 3:601–608. 10.1016/1074-7613(95)90131-0 [DOI] [PubMed] [Google Scholar]
- Groves T., Katis P., Madden Z., Manickam K., Ramsden D., Wu G., and Guidos C.J.. 1995. In vitro maturation of clonal CD4+CD8+ cell lines in response to TCR engagement. J. Immunol. 154:5011–5022. [PubMed] [Google Scholar]
- Gurel Z., Ronni T., Ho S., Kuchar J., Payne K.J., Turk C.W., and Dovat S.. 2008. Recruitment of ikaros to pericentromeric heterochromatin is regulated by phosphorylation. J. Biol. Chem. 283:8291–8300. 10.1074/jbc.M707906200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao B., and Krangel M.S.. 2011. Long-distance regulation of fetal Vδ gene segment TRDV4 by the Tcrd enhancer. J. Immunol. 187:2484–2491. 10.4049/jimmunol.1100468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao B., Naik A.K., Watanabe A., Tanaka H., Chen L., Richards H.W., Kondo M., Taniuchi I., Kohwi Y., Kohwi-Shigematsu T., and Krangel M.S.. 2015. An anti-silencer- and SATB1-dependent chromatin hub regulates Rag1 and Rag2 gene expression during thymocyte development. J. Exp. Med. 212:809–824. 10.1084/jem.20142207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heizmann B., Kastner P., and Chan S.. 2018. The Ikaros family in lymphocyte development. Curr. Opin. Immunol. 51:14–23. 10.1016/j.coi.2017.11.005 [DOI] [PubMed] [Google Scholar]
- Hernández-Hoyos G., Anderson M.K., Wang C., Rothenberg E.V., and Alberola-Ila J.. 2003. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 19:83–94. 10.1016/S1074-7613(03)00176-6 [DOI] [PubMed] [Google Scholar]
- Hsu L.Y., Lauring J., Liang H.E., Greenbaum S., Cado D., Zhuang Y., and Schlissel M.S.. 2003. A conserved transcriptional enhancer regulates RAG gene expression in developing B cells. Immunity. 19:105–117. 10.1016/S1074-7613(03)00181-X [DOI] [PubMed] [Google Scholar]
- Jin Z.X., Kishi H., Wei X.C., Matsuda T., Saito S., and Muraguchi A.. 2002. Lymphoid enhancer-binding factor-1 binds and activates the recombination-activating gene-2 promoter together with c-Myb and Pax-5 in immature B cells. J. Immunol. 169:3783–3792. 10.4049/jimmunol.169.7.3783 [DOI] [PubMed] [Google Scholar]
- Kishi H., Wei X.C., Jin Z.X., Fujishiro Y., Nagata T., Matsuda T., and Muraguchi A.. 2000. Lineage-specific regulation of the murine RAG-2 promoter: GATA-3 in T cells and Pax-5 in B cells. Blood. 95:3845–3852. [PubMed] [Google Scholar]
- Koipally J., Renold A., Kim J., and Georgopoulos K.. 1999. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 18:3090–3100. 10.1093/emboj/18.11.3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T.C., and Schlissel M.S.. 2009. Mechanisms controlling expression of the RAG locus during lymphocyte development. Curr. Opin. Immunol. 21:173–178. 10.1016/j.coi.2009.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring J., and Schlissel M.S.. 1999. Distinct factors regulate the murine RAG-2 promoter in B- and T-cell lines. Mol. Cell. Biol. 19:2601–2612. 10.1128/MCB.19.4.2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.S., Lee B.K., Iyer V.R., Sleckman B.P., Shaffer A.L. III, Ippolito G.C., Tucker H.O., and Dekker J.D.. 2017. Corrected and Republished from: BCL11A is a critical component of a transcriptional network that activates RAG expression and V(D)J recombination. Mol. Cell. Biol. 38:e00362-17 10.1128/MCB.00362-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Ruan X., Auerbach R.K., Sandhu K.S., Zheng M., Wang P., Poh H.M., Goh Y., Lim J., Zhang J., et al. 2012. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 148:84–98. 10.1016/j.cell.2011.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z., Brown K.E., Carroll T., Taylor B., Vidal I.F., Hendrich B., Rueda D., Fisher A.G., and Merkenschlager M.. 2017. A high-resolution map of transcriptional repression. eLife. 6:e22767 10.7554/eLife.22767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda G.A., Villalvazo M., Galic Z., Alva J., Abrines R., Yates Y., Evans C.J., and Aguilera R.J.. 2002. Combinatorial regulation of the murine RAG-2 promoter by Sp1 and distinct lymphocyte-specific transcription factors. Mol. Immunol. 38:1151–1159. 10.1016/S0161-5890(02)00007-X [DOI] [PubMed] [Google Scholar]
- Miyazaki M., Rivera R.R., Miyazaki K., Lin Y.C., Agata Y., and Murre C.. 2011. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat. Immunol. 12:992–1001. 10.1038/ni.2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M., Miyazaki K., Chen K., Jin Y., Turner J., Moore A.J., Saito R., Yoshida K., Ogawa S., Rodewald H.R., et al. 2017. The E-Id protein axis specifies adaptive lymphoid cell identity and suppresses thymic innate lymphoid cell development. Immunity. 46:818–834.e4. 10.1016/j.immuni.2017.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P., Iacomini J., Johnson R.S., Herrup K., Tonegawa S., and Papaioannou V.E.. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 68:869–877. 10.1016/0092-8674(92)90030-G [DOI] [PubMed] [Google Scholar]
- Monroe R.J., Chen F., Ferrini R., Davidson L., and Alt F.W.. 1999. RAG2 is regulated differentially in B and T cells by elements 5′ of the promoter. Proc. Natl. Acad. Sci. USA. 96:12713–12718. 10.1073/pnas.96.22.12713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra A.K., Drewes T., Engelmann S., Chuvpilo S., Kishi H., Hünig T., Serfling E., and Bommhardt U.H.. 2006. PKB rescues calcineurin/NFAT-induced arrest of Rag expression and pre-T cell differentiation. J. Immunol. 177:4567–4576. 10.4049/jimmunol.177.7.4567 [DOI] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed N.P., Henderson M.A., Oltz E.M., and Aune T.M.. 2013. Reciprocal regulation of Rag expression in thymocytes by the zinc-finger proteins, Zfp608 and Zfp609. Genes Immun. 14:7–12. 10.1038/gene.2012.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbattini P., Lundgren M., Georgiou A., Chow C., Warnes G., and Dillon N.. 2001. Binding of Ikaros to the lambda5 promoter silences transcription through a mechanism that does not require heterochromatin formation. EMBO J. 20:2812–2822. 10.1093/emboj/20.11.2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz D.G., and Swanson P.C.. 2011. V(D)J recombination: mechanisms of initiation. Annu. Rev. Genet. 45:167–202. 10.1146/annurev-genet-110410-132552 [DOI] [PubMed] [Google Scholar]
- Schulz D., Vassen L., Chow K.T., McWhirter S.M., Amin R.H., Möröy T., and Schlissel M.S.. 2012. Gfi1b negatively regulates Rag expression directly and via the repression of FoxO1. J. Exp. Med. 209:187–199. 10.1084/jem.20110645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y., Rathbun G., Lam K.P., Oltz E.M., Stewart V., Mendelsohn M., Charron J., Datta M., Young F., Stall A.M., et al. 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 68:855–867. 10.1016/0092-8674(92)90029-C [DOI] [PubMed] [Google Scholar]
- Su R.C., Brown K.E., Saaber S., Fisher A.G., Merkenschlager M., and Smale S.T.. 2004. Dynamic assembly of silent chromatin during thymocyte maturation. Nat. Genet. 36:502–506. 10.1038/ng1351 [DOI] [PubMed] [Google Scholar]
- Takahama Y., and Singer A.. 1992. Post-transcriptional regulation of early T cell development by T cell receptor signals. Science. 258:1456–1462. 10.1126/science.1439838 [DOI] [PubMed] [Google Scholar]
- Timblin G.A., and Schlissel M.S.. 2013. Ebf1 and c-Myb repress Rag transcription downstream of Stat5 during early B cell development. J. Immunol. 191:4676–4687. 10.4049/jimmunol.1301675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timblin G.A., Xie L., Tjian R., and Schlissel M.S.. 2017. Dual mechanism of Rag gene repression by c-Myb during Pre-B cell proliferation. Mol. Cell. Biol. 37:e00437-16 10.1128/MCB.00437-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh L.A., Ferrini R., Cobb B.S., Weinmann A.S., Hahm K., Ernst P., Garraway I.P., Merkenschlager M., and Smale S.T.. 2001. Down-regulation of TDT transcription in CD4+CD8+ thymocytes by Ikaros proteins in direct competition with an Ets activator. Genes Dev. 15:1817–1832. 10.1101/gad.905601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turka L.A., Schatz D.G., Oettinger M.A., Chun J.J., Gorka C., Lee K., McCormack W.T., and Thompson C.B.. 1991. Thymocyte expression of RAG-1 and RAG-2: termination by T cell receptor cross-linking. Science. 253:778–781. 10.1126/science.1831564 [DOI] [PubMed] [Google Scholar]
- Verkoczy L., Aït-Azzouzene D., Skog P., Märtensson A., Lang J., Duong B., and Nemazee D.. 2005. A role for nuclear factor kappa B/rel transcription factors in the regulation of the recombinase activator genes. Immunity. 22:519–531. 10.1016/j.immuni.2005.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.F., Lauring J., and Schlissel M.S.. 2000. c-Myb binds to a sequence in the proximal region of the RAG-2 promoter and is essential for promoter activity in T-lineage cells. Mol. Cell. Biol. 20:9203–9211. 10.1128/MCB.20.24.9203-9211.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne J., Suh H., Misulovin Z., Sokol K.A., Inaba K., and Nussenzweig M.C.. 1994a A regulatory role for recombinase activating genes, RAG-1 and RAG-2, in T cell development. Immunity. 1:95–107. 10.1016/1074-7613(94)90103-1 [DOI] [PubMed] [Google Scholar]
- Wayne J., Suh H., Sokol K.A., Petrie H.T., Witmer-Pack M., Edelhoff S., Disteche C.M., and Nussenzweig M.C.. 1994b TCR selection and allelic exclusion in RAG transgenic mice that exhibit abnormal T cell localization in lymph nodes and lymphatics. J. Immunol. 153:5491–5502. [PubMed] [Google Scholar]
- Wilson A., Held W., and MacDonald H.R.. 1994. Two waves of recombinase gene expression in developing thymocytes. J. Exp. Med. 179:1355–1360. 10.1084/jem.179.4.1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.X., Zhao W.P., Kishi H., Dokan J., Jin Z.X., Wei X.C., Yokoyama K.K., and Muraguchi A.. 2004. Activation of mouse RAG-2 promoter by Myc-associated zinc finger protein. Biochem. Biophys. Res. Commun. 317:1096–1102. 10.1016/j.bbrc.2004.03.159 [DOI] [PubMed] [Google Scholar]
- Yannoutsos N., Barreto V., Misulovin Z., Gazumyan A., Yu W., Rajewsky N., Peixoto B.R., Eisenreich T., and Nussenzweig M.C.. 2004. A cis element in the recombination activating gene locus regulates gene expression by counteracting a distant silencer. Nat. Immunol. 5:443–450. 10.1038/ni1053 [DOI] [PubMed] [Google Scholar]
- Yu W., Misulovin Z., Suh H., Hardy R.R., Jankovic M., Yannoutsos N., and Nussenzweig M.C.. 1999. Coordinate regulation of RAG1 and RAG2 by cell type-specific DNA elements 5′ of RAG2. Science. 285:1080–1084. 10.1126/science.285.5430.1080 [DOI] [PubMed] [Google Scholar]
- Zhang F., Thomas L.R., Oltz E.M., and Aune T.M.. 2006. Control of thymocyte development and recombination-activating gene expression by the zinc finger protein Zfp608. Nat. Immunol. 7:1309–1316. 10.1038/ni1397 [DOI] [PubMed] [Google Scholar]
- Zhang J.A., Mortazavi A., Williams B.A., Wold B.J., and Rothenberg E.V.. 2012. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 149:467–482. 10.1016/j.cell.2012.01.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.