Mendiratta et al. review the interplay between the different regulatory layers that affect the transcription and dynamics of distinct histone H3 variants along the cell cycle.
Abstract
As the building blocks of chromatin, histones are central to establish and maintain particular chromatin states associated with given cell fates. Importantly, histones exist as distinct variants whose expression and incorporation into chromatin are tightly regulated during the cell cycle. During S phase, specialized replicative histone variants ensure the bulk of the chromatinization of the duplicating genome. Other non-replicative histone variants deposited throughout the cell cycle at specific loci use pathways uncoupled from DNA synthesis. Here, we review the particular dynamics of expression, cellular transit, assembly, and disassembly of replicative and non-replicative forms of the histone H3. Beyond the role of histone variants in chromatin dynamics, we review our current knowledge concerning their distinct regulation to control their expression at different levels including transcription, posttranscriptional processing, and protein stability. In light of this unique regulation, we highlight situations where perturbations in histone balance may lead to cellular dysfunction and pathologies.
Introduction
The eukaryotic genome is packaged and organized in chromatin. The nucleosome, the basic unit of chromatin, consists of an octamer with two copies each of the core histone H2A, H2B, H3, and H4 around which is wrapped ∼146 bp of DNA and a variable linker DNA associated with the linker histone H1. The core histones share a conserved histone-fold domain that mediates their head-to-tail heterodimerization. In this way, H2A and H2B form two dimers flanking a (H3-H4)2 tetramer. The less conserved linker histones, the H1 family, possess a central globular domain flanked by a short N-terminal tail and a long basic C terminus. All histone families exist as variants that are differentially expressed and can undergo several posttranslational modifications. A variety of nucleosomes thus use distinct histone variants, as well as posttranslational modifications with different properties often associated with specific chromatin states (Talbert and Henikoff, 2017; Reinberg and Vales, 2018). As the major protein component of chromatin, histones are critical for its dynamic organization, assembly, and disassembly during most DNA transactions. However, because of their basic nature, uncontrolled histone accumulation can lead to promiscuous interactions with any acidic component, forming aggregates that are ultimately cytotoxic. Nonnucleosomal histones are therefore constantly under check. From their synthesis to incorporation into chromatin, as well as during disassembly and disposal, histones are escorted by distinct histone chaperones (Gurard-Levin et al., 2014; Hammond et al., 2017). The first protein designated as a histone chaperone is nucleoplasmin, discovered as the major protein present in Xenopus laevis oocytes (Laskey et al., 1978). Mainly in charge of H2A-H2B, it functions along with the H3-H4 N1/N2 chaperones to provide a stockpile of maternal core histones. During early development, this storage of maternal core histones is required to assemble newly replicating DNA into chromatin, thereby sustaining the first rounds of rapid cell division (Woodland and Adamson, 1977; Earnshaw et al., 1980). Under this unusual situation, particular chaperones are thus needed to cope with massive amounts of soluble histones. In contrast, in cycling cells, the soluble reservoir is more limited and has long been ignored. Not only are histone chaperones in charge of the cytosolic reservoir of histones or histones in transit but, most importantly, they also promote the deposition, eviction, and recycling of specific histone variants during DNA replication, transcription, and repair. Physiological changes in the level of histone chaperones and histone variants occur at various times during development, such as the up-regulation of nucleoplasmin in Xenopus female germ cells to accommodate the pool of maternal histones (Laskey et al., 1978), the accumulation of H3.3 in rat postmitotic neurons when cells have exited from the cell cycle (Piña and Suau, 1987), and the up-regulation of the chaperone ASF1b in highly proliferating cells (Corpet et al., 2011). A major interest has thus arisen concerning the interplay between histone variants and their dedicated chaperones to maintain chromatin integrity during development, differentiation, and the entire lifespan of an individual (De Koning et al., 2007; Filipescu et al., 2013; Gurard-Levin et al., 2014; Hammond et al., 2017).
In this review, considering histone H3 variants and their expression during the cell cycle, we will describe how different mechanisms control the amounts of histones. H3 variants recently emerged as important actors in cancer biology, as they are altered or misregulated across different types of tumors (Vardabasso et al., 2014; Weinberg et al., 2017). Distinct variants are under tight regulation to meet various demands along the cell cycle and mark specific functional domains in the genome (Gurard-Levin et al., 2014; Sitbon et al., 2017). A notable example is CenH3CENP-A, a rapidly evolving variant that specifically marks the centromere. Other H3 variants show a high degree of sequence similarity and are associated with different domains. Based on their deposition pathway, H3 variants can be distinguished as replicative and non-replicative. Replicative forms are specialized variants whose expression peaks in S phase and whose incorporation is coupled to DNA synthesis. During S phase, a provision of replicative H3 variants (H3.1 and H3.2) supports the bulk assembly of chromatin onto newly synthesized DNA. In contrast, the constitutive expression of the non-replicative variant H3.3 sustains histone turnover throughout the cell cycle, representing the majority of histones in quiescent and terminally differentiated cells. The other non-replicative variant, CenH3CENP-A, is specifically deposited at centromeres and marks the site of kinetochore assembly. Its expression peaks in G2/M phase in mammalian cells, and its deposition occurs only late in mitosis/early G1. Interestingly, replicative histone genes show a particular organization in clusters not observed for replacement histones, providing a unique means for controlling their expression.
We will first briefly review our current knowledge on the dynamics and deposition of these histone variants onto DNA, considering both dedicated and general chaperones. Next, we will describe recent advances concerning the regulation of their expression and the impact on genome organization and function throughout the cell cycle. Finally, we will put forward a few examples of aberrant expression of histone genes and discuss the consequences of their imbalance.
Dedicated chaperones for dynamics and deposition of replicative and non-replicative histone variants
The deposition of H3 variants involves different histone chaperones and leads to a partitioning of the genome in distinct chromosomal domains (Fig. 1, A and B). In S phase, the doubling of genomic content requires a massive provision of histones to ensure the duplication of chromatin (Corpet and Almouzni, 2009; Alabert and Groth, 2012). Nucleosomes ahead of the replication fork are displaced, and histones are recycled onto newly replicated DNA along with de novo deposition of new histones to restore nucleosome density (Probst et al., 2009; Almouzni and Cedar, 2016). This leads to the mixing of new and parental histones along with their particular marks, thereby enabling the propagation of active and repressive states to subsequent cell generations (Ray-Gallet and Almouzni, 2010; Reinberg and Vales, 2018). Orchestration of histone incorporation and recycling during S phase involves mechanisms coupled to DNA synthesis and dedicated histone chaperone complexes. The chromatin assembly factor-1 (CAF-1) complex is the H3-H4 histone chaperone that promotes nucleosome assembly coupled to DNA synthesis during replication (Smith and Stillman, 1989) and repair (Gaillard et al., 1996). CAF-1 is recruited at replication forks through the interaction of its p150 subunit with the proliferating cell nuclear antigen (PCNA) (Moggs et al., 2000), a processivity factor for the DNA polymerase. The CAF-1 complex associates specifically with the replicative variant H3.1 and H3.2, coupling their deposition to DNA synthesis (Tagami et al., 2004; Latreille et al., 2014).
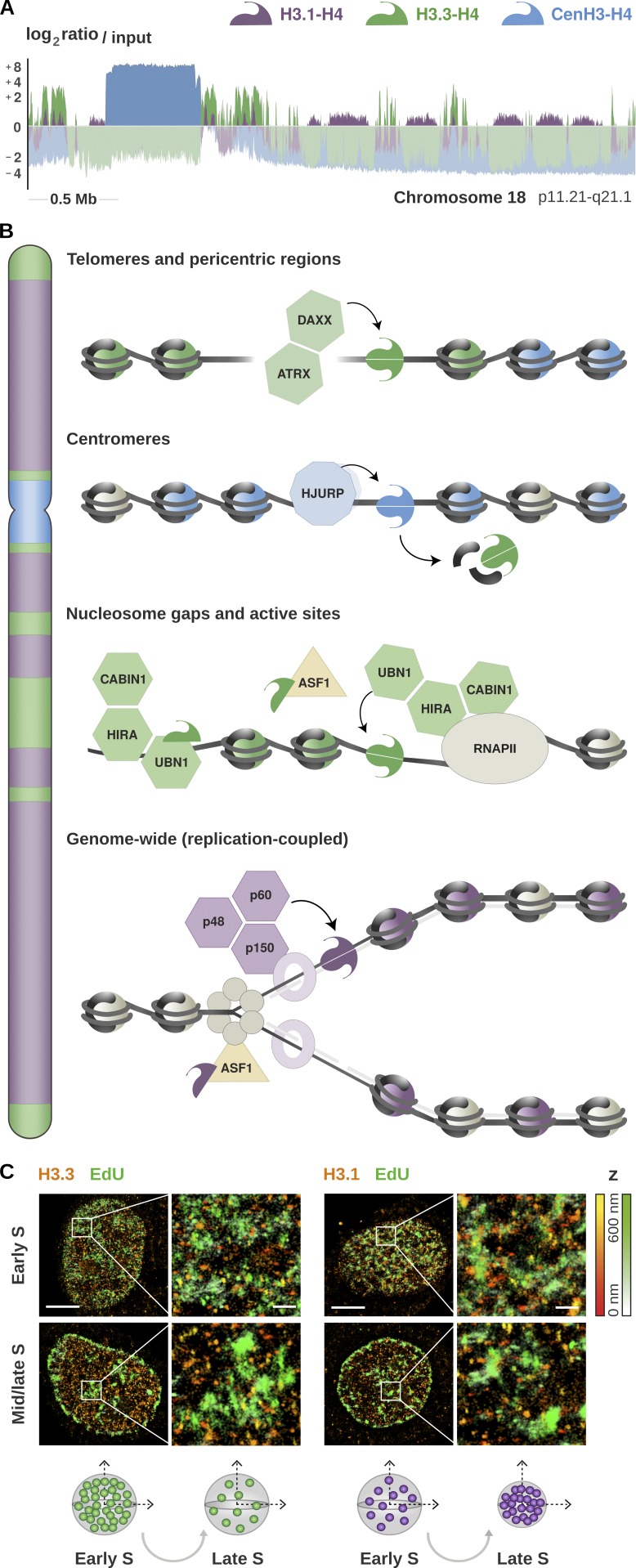
Figure 1.
Enrichment of histone H3 variants and their deposition by dedicated chaperones. (A) Genomic distribution of H3.1, H3.3, and CenH3 from published ChIP-Seq data in HeLa cells (Lacoste et al., 2014; Clément et al., 2018). The plot shows the enrichment relative to input for all variants at a representative region spanning the centromere and the proximal short and long arms of chromosome 18 (p11.21-q21.1). The enrichment is computed as the log2 ratio between the mean per-base number of reads from H3.1 (purple), H3.3 (green), and CenH3 (blue) and their respective input, at consecutive 10-kb bins (smoothed over five nonzero bins). Enriched regions are highlighted in darker colors, illustrating the partitioning of the genome into chromatin domains associated with specific variants. (B) Schematic representation of the histone chaperones involved in the deposition of H3 variants at distinct chromosomal locations. The DAXX/ATRX complex promotes the accumulation of the non-replicative variant H3.3 at telomeres and pericentric heterochromatin. H3.3 is also deposited at actively transcribed regions and regulatory sites by HIRA, a complex consisting of three subunits: CABIN1, HIRA, and UBN1. The non-replicative variant CenH3 is deposited specifically at centromeres by its dedicated chaperone HJURP, marking the site of kinetochore assembly. Both H3.1 and H3.3 are incorporated at centromeres during S phase, and H3.3 acts as a placeholder for CenH3 loading in late mitosis and early G1. H3.1 is deposited genome-wide by the CAF-1 complex during S phase, or at DNA repair sites throughout the cell cycle. The CAF-1 complex consists of three subunits: p48, p50, and p160. The p150 subunit interacts with PCNA and promotes CAF-1 recruitment at replication forks. This couples H3.1 deposition to DNA synthesis and ensures proper chromatin assembly during replication. ASF1 is a general chaperone that can handle both H3.1 and H3.3 and hands them over to their dedicated chaperones. (C) Illustration of high-resolution visualization by STORM of H3.1 and H3.3 along S phase (adapted from Clément et al., 2018). The STORM images show the nuclear distribution of H3.1 and H3.3 (HA staining, in red) at sites of DNA synthesis (EdU staining, in green) in early and mid/late S. Scale bars represent 5 µm. Insets represent enlarged images of selected areas where scale bars correspond to 600 nm. H3.3 clusters show stable volume, but there is a decrease in H3.3 density as S phase progresses; the late domains likely show a dilution of H3.3 during DNA replication. In contrast, H3.1 clusters change in both volume and density during S phase, with larger H3.1 clusters and low densities in early S and clusters with smaller volumes and higher density in mid/late S phase. See Clément et al. (2018) for further details.
Antisilencing function 1 (ASF1) is a H3-H4 chaperone viewed as an intermediary handing over distinct histone variants to their specific chaperones (Mello et al., 2002; Daganzo et al., 2003; Tang et al., 2006). ASF1 is rather promiscuous and can interact with all H3 variants, including H3.1, H3.3, and CenH3CENP-A. Initially implicated in the deposition of new histones (Tyler et al., 1999), ASF1 also regulates their supply during replication (Groth et al., 2005). Moreover, it plays a critical role in coupling histone dynamics with the progression of the replicative helicase during S phase (Groth et al., 2007). At replication forks, ASF1 forms a complex with the MCM2 subunit of the MCM helicase and binds H3-H4 dimers associated with the MCM2 histone-binding domain (Groth et al., 2007; Huang et al., 2015; Richet et al., 2015). MCM2 might act as a chaperone to unload parental H3-H4 tetramers ahead of the fork, while the association with two ASF1 could help dissociating the tetramers in two dimers of parental histones (Clément and Almouzni, 2015). ASF1 is thus ideally positioned to handle both parental and new histones. The distribution of global and parental H3.1 and H3.3 throughout S phase has been determined by superresolution microscopy (Fig. 1 C), which demonstrated that, in addition to causing a global loss of parental H3, ASF1 depletion leads to a relocalization of parental histones away from replication foci. This affects the distribution of both replicative and non-replicative H3 variants. Both exhibit a distinct nuclear distribution, and the effect of ASF1 loss differs between variants depending on replication timing (Clément et al., 2018).
Chromatin assembly is also required to sustain the turnover of histones that occurs independently of the cell cycle. This is mostly mediated by a replication- and cell cycle–independent pathway that involves the deposition of the histone variant H3.3 (Ahmad and Henikoff, 2002). The HIRA complex associates specifically with H3.3 (Tagami et al., 2004). The complex consists of three protein subunits: histone cell cycle regulator (HIRA), ubinuclein-1 (UBN1), and calcineurin binding protein 1 (CABIN1). H3.3 is incorporated into chromatin throughout the cell cycle and accumulates in postmitotic cells. It is mainly associated with transcribed regions and regulatory sites with high nucleosome turnover (Goldberg et al., 2010) but also, more broadly, at any given location where a gap occurs (Ray-Gallet et al., 2011). At active promoters, HIRA colocalizes with H3.3, UBN1, and ASF1a (Pchelintsev et al., 2013). HIRA coordinates with ASF1a to mediate the deposition of histone H3.3 in a replication-independent manner. If CAF-1 fails to assemble nucleosomes, the HIRA complex can provide a fallback strategy. The current model proposes that HIRA, owing to its ability to bind naked DNA in vitro, promotes H3.3 deposition in a gap-filling fashion at nucleosome-free regions (Ray-Gallet et al., 2011). The recently identified homotrimerization property of HIRA subunit is required for CABIN1 binding and is critical for its functional activity (Ray-Gallet et al., 2018). HIRA also promotes nucleosome reassembly after nonhomologous end joining in a replication-independent manner, and CAF-1 in a DNA synthesis-coupled manner (Li and Tyler, 2016). Thus, the two chaperones act in concert, balancing each other to ensure chromatin maintenance. This recently discovered homotrimerization property of the HIRA subunit resembles the trimerization of the yeast Ctf4, a component of the replication machinery present at replication forks (Simon et al., 2014), and is likely required for CABIN1 binding to ensure histone deposition at DNA damage sites (Ray-Gallet et al., 2018).
Besides transcribed regions, H3.3 is also deposited at specific chromosomal landmarks independent of HIRA. The death domain–associated protein DAXX and the chromatin remodeling factor α-thalassemia/mental retardation syndrome protein (ATRX) promote the enrichment of H3.3 at pericentric heterochromatin and telomeres (Drané et al., 2010; Goldberg et al., 2010). The DAXX/ATRX complex associates with H3.3 at telomeres in a replication-uncoupled manner. H3.3-H4 dimers directly interact with DAXX via its histone-binding domain. ATRX is dispensable for the interaction but contributes to target DAXX at specific chromatin domains, thereby promoting DAXX-dependent H3.3 accumulation (Lewis et al., 2010). In addition, the oncoprotein DEK has been involved in the differential distribution of H3.3 by HIRA and DAXX/ATRX in somatic and embryonic stem cells. DEK can help in loading ATRX, and hence H3.3, on telomeric regions, thereby maintaining telomere integrity (Ivanauskiene et al., 2014).
The dedicated chaperone Holliday junction recognition protein (HJURP) ensures CenH3CENP-A loading at centromeres (Dunleavy et al., 2009; Foltz et al., 2009). CenH3CENP-A specifically marks the centromere and is required for kinetochore assembly and proper chromosome segregation during cell division. During replication, CenH3CENP-A is diluted while H3.1 and H3.3 are deposited, and H3.3 is likely used as a placeholder for newly assembled CenH3CENP-A (Dunleavy et al., 2011), which is deposited later during late telophase/early G1 (Jansen et al., 2007; Bodor et al., 2013). Phosphorylation of HJURP is required to ensure proper timing of CenH3CENP-A incorporation, as nonphosphorylatable mutants localize to centromeres throughout the cell cycle (Müller et al., 2014b).
As outlined above, histone chaperones are generally in charge of dedicated histone cargos and promote the deposition of specific variants, whereas others, like ASF1, are more promiscuous. For further details on H3 variants and their chaperones, we direct the reader to recent reviews (Müller and Almouzni, 2017; Sitbon et al., 2017). Remarkably, under particular circumstances, some chaperones can substitute for each other when one is either missing or limiting. Thus, chaperone function can show some degree of overlap. This has been observed upon DAXX depletion when a fraction of the replacement variant H3.3 associates with the replicative assembly machinery (Drané et al., 2010), or when HIRA backs up CAF-1 to fill nucleosome gaps behind the fork (Ray-Gallet et al., 2011), or when DAXX handles excess CenH3CENP-A in place of HJURP upon CenH3CENP-A overexpression (Lacoste et al., 2014). Such cross-talk between histone chaperones and their choice of histone variant can thus provide robustness in the process of histone management and suggests a potential for chromatin plasticity.
Mechanisms for deposition and dynamics in and out of chromatin are thus critical. They involve DNA synthesis–coupled pathways for replicative variants and DNA synthesis–uncoupled pathways for non-replicative ones. Beyond handling histones, their regulation and production play an equally important role to meet different cellular demands. We will next discuss how the expression of replicative and non-replicative variants is regulated.
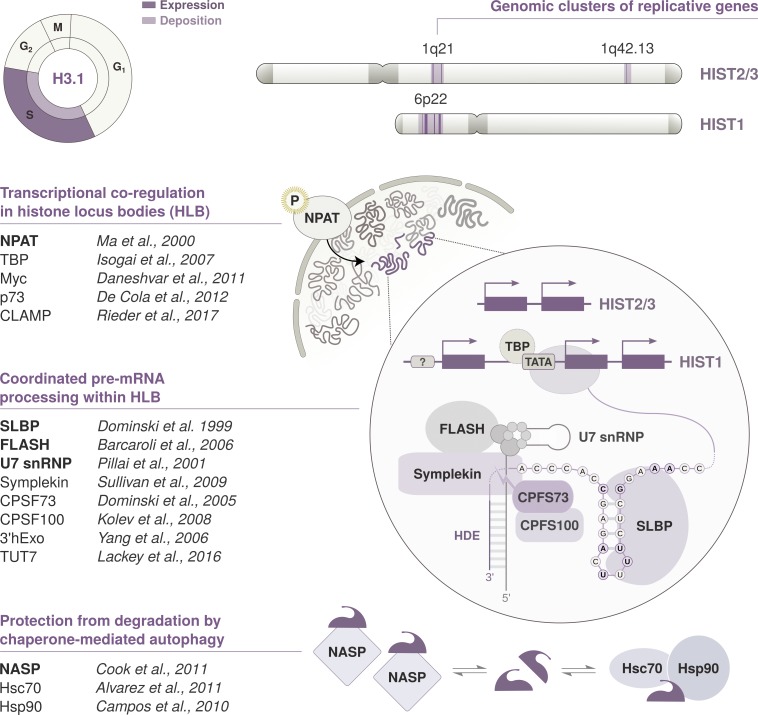
Replicative histone variants: A unique genomic organization and regulation at multiple levels
Chromatin assembly in S phase requires vast amounts of newly synthesized histones to ensure its restoration on the duplicated genome. Replicative histone variants share a unique transcriptional program that ensures higher expression levels for DNA synthesis–coupled deposition in S phase. In contrast to replacement variants, H3.1 and H3.2 genes show a peculiar organization in clusters that comprise multiple copies of all core histones and the H1 linker. This offers a potential means to optimize coregulation. Indeed, in the human and mouse genome, replicative histone genes cluster at three syntenic loci that remained physically linked through evolution (Marzluff et al., 2002). The human histone cluster 1 (HIST1) is located on chromosome 6 (6p22) and comprises more than 50 coding genes, while HIST2 and HIST3 are located on chromosome 1 (1q21 and q42) and contain, respectively, 10 and 3 coding genes. Besides their physical proximity, genes are compartmentalized and processed in nuclear bodies that concentrate the factors required for their transcription and processing. These bodies were initially thought to coincide with Cajal bodies, subnuclear compartments discovered at the beginning of the century (Cajal, 1903) and implicated in the biogenesis of ribonucleoproteins associated with histone pre-mRNAs (Frey and Matera, 1995; Calvi et al., 1998; Liu et al., 2000). The presence of separate histone locus bodies (HLBs) dedicated to the transcription of replicative histones was only recognized later (Liu et al., 2006). Similar to Cajal bodies, HLB foci are marked by coilin, but they form at distinct subnuclear locations and coilin is dispensable for their assembly (Liu et al., 2009). The nuclear compartmentalization of replicative histone genes is also reflected by their higher-order organization (Rao et al., 2014; Fritz et al., 2018). Higher-order structures can be revealed from self-interacting regions where contacts among genome sequences in physical proximity occur more frequently (topologically associating domains [TADs]). The three gene subsets in the human cluster 1 interact within separate TADs. Notably, these TADs further interact with each other over an ∼1.5-Mb distance, indicating that separate genes also establish long-range contacts and come in physical proximity within the nucleus (Fig. 2 A). The distinctive interplay among replicative histone genes is thus apparent at multiple dimensions, but it is still unclear how these layers of organization relate to each other. HLBs might represent a case of self-organization (Matera et al., 2009) or an example of phase-separated nuclear bodies that assemble via liquid demixing by macromolecular crowding (Zhu and Brangwynne, 2015; Duronio and Marzluff, 2017). This might resemble the phase separation recently implicated in the formation of heterochromatin protein 1 (HP1) foci (Larson et al., 2017; Strom et al., 2017) and repetitive RNA foci (Jain and Vale, 2017). It would be interesting to experimentally address the link between phase transitions and the assembly of HLBs. Transgenic assays in Drosophila melanogaster revealed that a sequence located between histone H3 and H4 genes is important to mediate HLB assembly. Neither the H3 and H4 coding region nor the 3′ signals are required for HLB formation (Salzler et al., 2013). Chromatin-linked adaptor for male-specific lethal (CLAMP) is a zinc finger protein that binds the GA repeat motif within bidirectional H3-H4 promoter and controls chromatin accessibility, thereby enhancing transcription and promoting HLB formation (Rieder et al., 2017). Much remains to be understood concerning how their transcriptional regulation exploits this particular 3D organization.
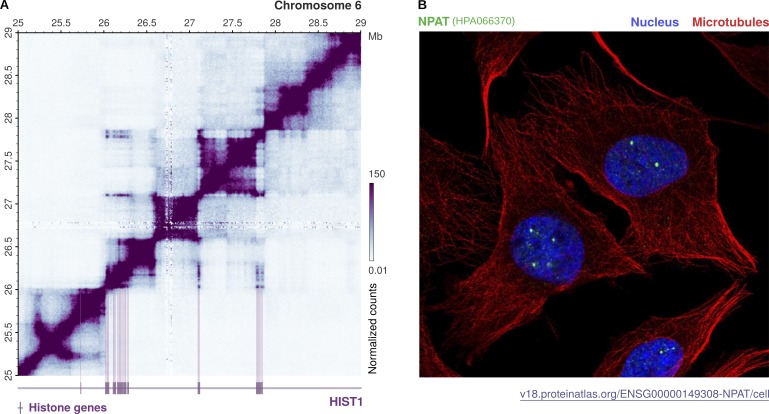
Figure 2.
Higher-order organization of replicative histone genes and compartmentalization of nuclear factors in HLBs. Replicative histones are redundantly encoded by multiple intronless genes that exhibit a conserved cluster organization across several lineages. (A) Location and spatial interactions among histone genes within the human histone cluster 1 (HIST1). The contact matrix was generated using iteratively corrected Hi-C data in GM12878 cells at 10-kb resolution (Rao et al., 2014) and plot with gcMapExplorer (Kumar et al., 2017). Genome bins where histone genes are located are marked in purple, illustrating the presence of three separate subsets within the HIST1 cluster. Neighboring genes in each subset interact within separate TADs, but all three subsets further engage in long-range interactions that bring distant genes together over an ∼1.5-Mb distance. This spatial organization might reflect the compartmentalization of replicative histone genes in the nucleus. Their pre-mRNAs are indeed transcribed and processed in dedicated nuclear bodies, called HLBs. (B) Nuclear distribution of NPAT, an essential factor driving HLB assembly and transcription initiation. The confocal image was retrieved from the Human Protein Atlas (v.18) and shows NPAT staining in U2-OS cells (in green). The nucleus and microtubules are stained in blue and red, respectively. NPAT concentrates at distinct subnuclear locations and marks the HLBs.
Besides their distinctive genomic and subnuclear organization, replicative histones differ from replacement variants in terms of gene architecture and mRNA processing. Virtually all genes in the histone clusters lack introns, have relatively short UTRs, and produce transcripts that harbor a conserved 3′ stem-loop structure and do not undergo polyadenylation in many organisms (Marzluff et al., 2008; Duronio and Marzluff, 2017; Mei et al., 2017). The only processing needed to form a mature histone mRNA is the endonucleolytic cleavage of its precursor. This is mediated in cis by the 3′ stem-loop and a purine-rich sequence downstream of the cleavage site, the histone downstream element (HDE). These features are widely conserved in metazoans as well as unicellular eukaryotes (Dávila López and Samuelsson, 2008; Marzluff et al., 2008) but are not found in species, such as Saccharomyces cerevisiae, that lack DNA synthesis–coupled histone variants (Eriksson et al., 2012). In budding yeast, there is a single H3.3-like variant whose expression is induced in S phase together with other histone gene pairs (Osley et al., 1986). Histone repression outside of S phase is mediated by several factors, including the yeast orthologues of several known chaperones such as HIR1, HIR2, and HIR3 (Sherwood et al., 1993; Spector et al., 1997) as well as Asf1 (Sutton et al., 2001). In addition, histone transcription is also regulated by Spt10, a putative acetyltransferase, together with its partner Spt21 (Kurat et al., 2014). At the protein level, Rad53 participates as part of a surveillance mechanism that monitors the accumulation of excess histone proteins and triggers their degradation (Gunjan et al., 2005; Singh et al., 2010). This mode of regulation at the level of protein stability is not unique to budding yeast but is further exploited in other organisms. Indeed, the human histone chaperone nuclear autoantigenic sperm protein (NASP), similar to the Xenopus N1/N2, maintains a cytosolic soluble pool of H3-H4 dimers and protects them from degradation via chaperone-mediated autophagy (Cook et al., 2011).
In organisms with replicative histone variants, their expression throughout the cell cycle is controlled at multiple levels (Rattray and Müller, 2012). Replicative histone genes are transcribed and processed by several factors within the HLBs (Fig. 3). Transcription initiation is controlled by cyclin E/CDK2-dependent phosphorylation of the nuclear protein, ataxia-telangiectasia locus (NPAT) at the G1/S transition. Phosphorylated NPAT is required to initiate the assembly of HLBs and persists throughout S phase to activate the expression of replicative histone genes (Ma et al., 2000; Zhao et al., 2000; White et al., 2011). NPAT also interacts with FLICE-associated huge protein (FLASH), an essential cofactor involved in HLB assembly and 3′ processing of nascent transcripts (Barcaroli et al., 2006; Yang et al., 2009; White et al., 2011). Pre-mRNA maturation relies on the recognition of the cis-regulatory elements at the 3′ end by two factors that are specific to replicative histones: the stem-loop binding protein (SLBP) and the U7 snRNP (Mowry and Steitz, 1987; Dominski et al., 1999; Sullivan et al., 2001; Zhao et al., 2004). The U7 snRNP is a ribonucleoprotein complex composed of U7 snRNA, Sm proteins, and U7-specific Lsm10 and Lsm11 proteins (Pillai et al., 2001, 2003). The 5′ end of the complex binds histone pre-mRNAs via U7 snRNA hybridization to the 3′ HDE sequence (Cotten et al., 1988; Soldati and Schümperli, 1988). SLBP binds the stem-loop upstream and interacts with the U7 snRNP to stabilize its association. This stabilization is required for proper maturation and cleavage of histone pre-mRNAs (Pandey et al., 1994; Sullivan et al., 2001) but might be dispensable in vitro if the RNA duplex is sufficiently stable (Streit et al., 1993; Dominski et al., 1999).
Figure 3.
Cell cycle timing and regulation of replicative histone genes. The replicative variant H3.1 is deposited by CAF-1 in a DNA synthesis–coupled manner, mostly during replication in S phase. Expression peaks at the G1/S transition, and its transcription is regulated in concert with other replicative histone genes located within the histone clusters. Their pre-mRNAs are processed through a distinct pathway that involves the recognition of two unique cis-regulatory elements: a 3′ stem-loop structure and an HDE. Transcription and pre-mRNA processing are compartmentalized in the nucleus and coordinated in HLBs. HLB assembly and transcriptional activation is initiated by NPAT. NPAT is phosphorylated by the Cyclin E/CDK2 complex at the G1/S transition. The maturation of histone pre-mRNAs requires the endonucleolytic cleavage of its 3′ tail and is mediated by several factors recruited to HLBs. The U7 snRNP binds the HDE via hybridization of the U7 snRNA. It interacts with SLBP, a protein that specifically recognizes the 3′ stem-loop structure and stabilizes the U7 snRNP association. FLASH is another essential coactivator that promotes the recruitment of transcription factors and interacts with the Lsm11 subunit of the U7 snRNP to recruit the components of the histone cleavage complex. These transcription factors include Symplekin and the CPSF73/CPSF100 heterodimer that catalyzes the 3′ end endonucleolytic cleavage. Mature mRNAs are cleaved downstream of the 3′ stem-loop, which is required for mRNA degradation. SLBP is also degraded at the end of S phase after phosphorylation by the Cyclin A/CDK1 complex. H3 availability is further modulated at the protein level by NASP, a H3-H4 histone chaperone that protects soluble histones from degradation via chaperone-mediated autophagy counteracting Hsc70 and Hsp90.
Pre-mRNA cleavage is catalyzed by the CPSF73 endonuclease (Dominski et al., 2005; Mandel et al., 2006). The interaction between FLASH and the U7-specific protein Lsm11 is critical for CPSF73 recruitment at the cleavage site (Burch et al., 2011; Yang et al., 2013). SLBP, U7 snRNP, and FLASH contribute to recruit scaffolding factors and additional components of the histone cleavage complex. The core components include Symplekin (Kolev and Steitz, 2005; Tatomer et al., 2014) and CPSF100, which heterodimerizes with CPSF73 to catalyze the endonucleolytic cleavage of the 3′ end (Dominski et al., 2005; Kolev et al., 2008). Other factors (Duronio and Marzluff, 2017) might contribute to stabilize interactions in the cleavage complex that are specific to the processing of histone pre-mRNAs. mRNA levels of replicative histones decrease at the end of S phase, and SLBP is degraded upon phosphorylation by cyclin A/CDK1 (Koseoglu et al., 2008). The 3′ stem-loop structure is necessary and sufficient for the degradation of histone mRNAs (Graves et al., 1987; Pandey and Marzluff, 1987). Degradation is also regulated via 3′ end uridylation by the uridylyltransferase TUT7 (Mullen and Marzluff, 2008; Lackey et al., 2016), and the 3′hExo enzyme is needed in the initial steps (Yang et al., 2006). The mechanism underlying 3′ end recognition to allow 3′hExo to initiate degradation of the stem-loop is not well understood but might involve elements downstream of the stop codon (Graves et al., 1987).
Besides factors that are specific to the processing of replicative histones, several common targets and transcriptional regulators contribute to expression in the HLB (also summarized in Fig. 3). Human histone genes harbor TATA, CCAAT, and GC boxes in their promoter region, as well as putative binding sites for several transcription factors (Mariño-Ramírez et al., 2006). In human cells, FLASH participates in the recruitment of coactivators, such as p73, that contribute to the transcription of replicative histones (De Cola et al., 2012). In the tandemly arrayed linker and core histone clusters of Drosophila, the TATA-binding protein TBP regulates the transcription of core histones, while H1 genes have TATA-less promoters modulated by the TBP related factor, TRF2 (Isogai et al., 2007). The core and linker histones are indeed differentially expressed in flies, with H1 transcribed throughout the S phase and the core histones induced during a short pulse in early S phase (Guglielmi et al., 2013). Myc also colocalizes to the Drosophila HLB and contributes to the expression of replicative histone genes (Daneshvar et al., 2011). In mouse embryonic stem cells, several chromatin factors such as E2f1, Ctcf, Smad1, and Yy1 are likely to be involved in the regulation of both core and linker histones (Gokhman et al., 2013). The contribution of different factors to the transcription of specific genes is still unclear and might vary among lineages (Mariño-Ramírez et al., 2006) as well as in a tissue- and developmental-specific manner. The transcription elongation rate also affects the pre-mRNA folding at the 3′ end. Stem-loop formation is impaired in slow elongation conditions following UV irradiation or RNA polymerase II mutation, which leads to an accumulation of polyadenylated histone mRNAs (Saldi et al., 2018). The negative elongation factor (NELF) interacts with the nuclear cap binding complex (CBC) and plays a role in the 3′ processing. Their combined knockdown leads to increased expression of replicative histone genes, and CBC was shown to interact directly with SLBP. Both NELF and CBC physically associate with the histone gene body, and NELF accumulates in nuclear foci where histone cleavage factors localize (Narita et al., 2007). Genome-wide RNAi screens in Drosophila showed that depletion of H3.3 and H2Av disrupts the expression of replicative histones (Wagner et al., 2007). The U7 snRNP fails to accumulate at the HLB when H2Av is mutated. Although the effect may be indirect, this suggests that histone variants themselves might play a role in transcriptional processing.
The regulation of specific genes within the histone clusters has yet to be characterized systematically. Nonetheless, their unique cis-regulatory features and distinctive organization ensures a coordinated processing that enables a precise temporal control of their expression. This regulation is critical for cell cycle progression and has important implications for both genome and epigenome assembly. The cross-talk between 3D organization in the nucleus and transcription is surely a remarkable paradigm, and the link between HLBs and topological domains is a promising avenue for investigation. Such studies might also give important insights into the coregulation of genes induced synchronously by other cues.
Distinct transcriptional regulation and cell cycle expression of the centromeric variant CenH3
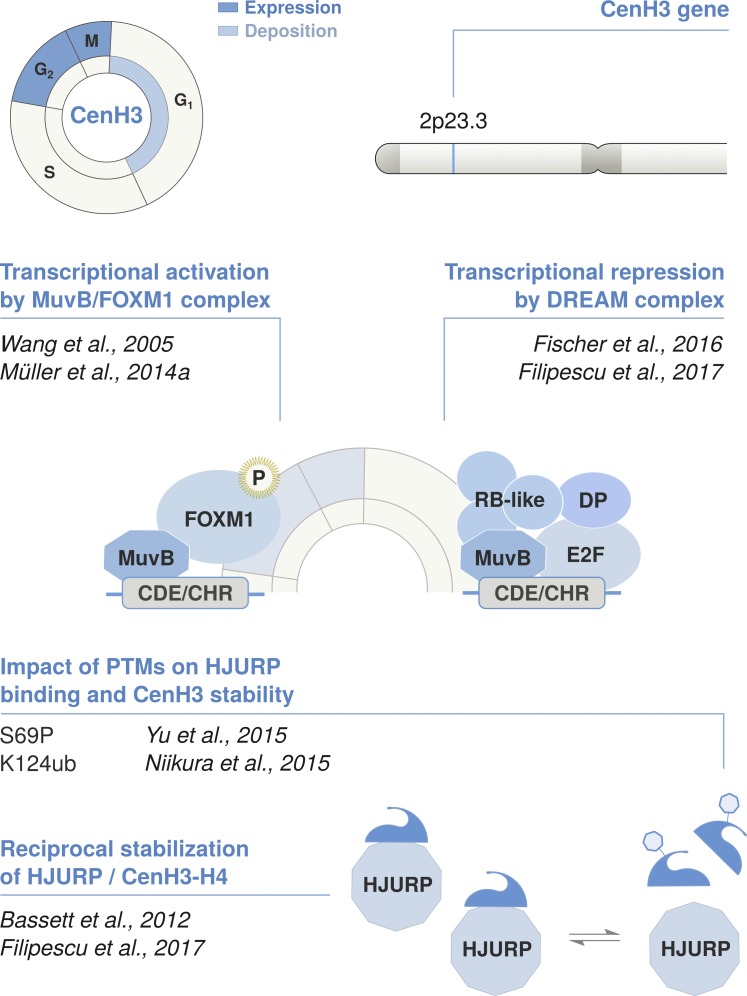
In contrast to replicative variants, CenH3CENP-A is encoded by a single multi-exon gene located outside of histone clusters (Sullivan et al., 1994; Régnier et al., 2003). CenH3CENP-A expression is regulated through a distinct, DNA synthesis–independent pathway (Fig. 4). Transcripts lack a 3′ stem-loop and undergo conventional processing through splicing and polyadenylation. The expression of CenH3CENP-A peaks in G2, and this temporal control is key for its centromeric targeting (Shelby et al., 1997). Induction of CenH3CENP-A during S phase leads to a loss of specific targeting and misassembly at chromosome arms. CenH3CENP-A deposition occurs in telophase/early G1 (Jansen et al., 2007). HJURP, its chaperone, is recruited at the centromere during this critical time window and is otherwise distributed throughout the nucleoplasm and at nucleoli (Dunleavy et al., 2009). A controlled expression of CenH3CENP-A is necessary to prevent promiscuous interactions with low-affinity chaperones and aberrant CenH3CENP-A loading (Lacoste et al., 2014; Shrestha et al., 2017). Thus, ensuring an exclusive handling by HJURP is likely critical for proper deposition of CenH3CENP-A. Its periodic regulation is indeed a widely conserved feature, and CenH3CENP-A mistargeting occurs in a variety of distant species whenever the gene is ectopically expressed at constitutively high levels (Van Hooser et al., 2001; Heun et al., 2006; Gascoigne et al., 2011; Mendiburo et al., 2011; Choi et al., 2012; Lacoste et al., 2014).
Figure 4.
Cell cycle timing and regulation of the centromeric variant CenH3. The non-replicative variant CenH3CENP-A is deposited at centromeres by its dedicated chaperone HJURP in telophase/early G1. Its expression is regulated in a cell cycle–dependent manner and peaks in G2/M. CenH3CENP-A is encoded by a single multi-exon gene located outside of the histone clusters that undergoes conventional pre-mRNA processing via splicing and polyadenylation. Transcription is coordinated in concert with other late cell cycle genes, including HJURP. This is regulated in cis by the CHR/CDE motif in their promoter region. The recruitment of the DREAM complex at the CHR is thought to promote transcriptional repression during G1. At the beginning of S phase, the MuvB core of the DREAM complex remains bound to the CHR/CDE while other components dissociate and are replaced by B-MYB. The B-MYB-MuvB (MMB) complex recruits FOXM1 in late S phase. B-MYB is degraded upon phosphorylation, whereas the progressive phosphorylation of FOXM1 leads to its activation in G2/M. Both CenH3 and HJURP are induced at the same time and mutually stabilize each other at the protein level. The reciprocal stabilization is affected by posttranslational modifications on the N-terminal tail of CenH3, which further regulate the timing of deposition. S69 phosphorylation prevents interaction with HJURP and premature loading, while K124 ubiquitylation favors HJURP binding and might contribute to their stabilization.
The regulation of both CenH3CENP-A and HJURP is coordinated in concert with other late cell cycle genes involved in mitotic progression, such as CDC25B, AURKB, PLK1, and CENP-B (Wang et al., 2005). Expression of these genes peak in G2/M, and they harbor a conserved cell cycle–dependent element (CDE) and cell cycle genes homology region (CHR) in their promoter (Müller et al., 2014a). During G1 phase, CHR promotes their transcriptional repression via recruitment of the dimerization partner, RB-like, E2F, and MuvB (DREAM) complex (Sadasivam and DeCaprio, 2013). Binding of DREAM components to the CHR can be also facilitated by the upstream CDE sequence (Müller et al., 2012). When cells progress into S phase, DREAM components dissociate from the MuvB core complex and B-MYB binds to MuvB. The B-MYB-MuvB (MMB) complex recruits FOXM1 to the CHR site in late S phase. B-MYB is hence phosphorylated and undergoes proteasome degradation, whereas MuvB remains bound to FOXM1 (Down et al., 2012; Sadasivam et al., 2012). The progressive phosphorylation of FOXM1 by cell cycle–dependent kinases finally promotes its activation in G2/M (Fu et al., 2008; Laoukili et al., 2008; Chen et al., 2009), which leads to maximal induction of CHR-harboring genes bound by the MuvB-FOXM1 complex.
The CDE/CHR motif in CenH3CENP-A promoter was early recognized as a potential cis-regulatory element underlying its periodic expression (Shelby et al., 1997). CenH3CENP-A is a direct target of FOXM1 (Wang et al., 2005; Chen et al., 2013), together with HJURP and other mitotic genes. In human and mouse cells, FOXM1 depletion leads to reduced CenH3CENP-A expression and impaired mitotic progression (Wang et al., 2005). Both CenH3CENP-A and HJURP also proved to be potential targets of the DREAM repressor complex (Fischer et al., 2016). Notably, their CDE/CHR motif facilitates transcriptional repression upon p53-dependent recruitment of DREAM in mouse cells (Filipescu et al., 2017). Furthermore, p53 activation also leads to down-regulation of HJURP and CENP-A in human cells. Whether CenH3CENP-A repression during G1/S is likewise dependent on DREAM has yet to be confirmed. Nevertheless, the fine-tuning of CenH3CENP-A kinetics is likely constrained by the need to preserve its precise centromeric targeting.
Besides their transcriptional coregulation, CenH3CENP-A and HJURP are dedicated binding partners, and their interaction plays an important role in maintaining a homeostatic balance. CenH3CENP-A and HJURP coexist as a soluble complex in which each of them favors the reciprocal stabilization of the other (Bassett et al., 2012; Filipescu et al., 2017). The N-terminal portion of HJURP binds both CenH3 and H4, thereby protecting the dimers locally at the region of contact (Bassett et al., 2012). The specific interaction with HJURP is driven by the centromere targeting domain (CATD) of CenH3CENP-A and favors the stabilization of nonnucleosomal dimers. Exogenous overexpression of either CenH3CENP-A or HJURP leads to an increase in the endogenous levels of their binding partner (Filipescu et al., 2017). HJURP loss results in CenH3CENP-A depletion, and CenH3CENP-A knockdown leads to the proteasome-mediated degradation of HJURP. The interaction between the two could be further modulated by posttranslational modifications in CenH3CENP-A N-terminal tail. For instance, CDK1/cyclin B–dependent phosphorylation at serine 68 during G2/M hinders the interaction with HJURP, preventing premature loading of CenH3CENP-A at centromeres (Hu et al., 2011; Yu et al., 2015; Wang et al., 2017). Ubiquitylation at lysine 124 instead facilitates their interaction and might control the stability of the complex (Niikura et al., 2015). The CUL4A-RBX1-COPS8 complex mediates CenH3CENP-A lysine 124 ubiquitylation (Niikura et al., 2015, 2017), but the deubiquitylating enzyme has not yet been identified. Posttranslational modifications and their effect on CenH3 stability vary significantly across species, consistent with the low conservation of its N-terminal tail (Au et al., 2013; Bade et al., 2014). The underlying pathways might represent potential adaptations to consider in light of its rapid evolution (Malik and Henikoff, 2003; Kursel and Malik, 2017) or coevolution with their dedicated chaperone (Sanchez-Pulido et al., 2009; Rosin and Mellone, 2016).
Constitutive expression of the non-replicative H3.3 variant by independent paralogs
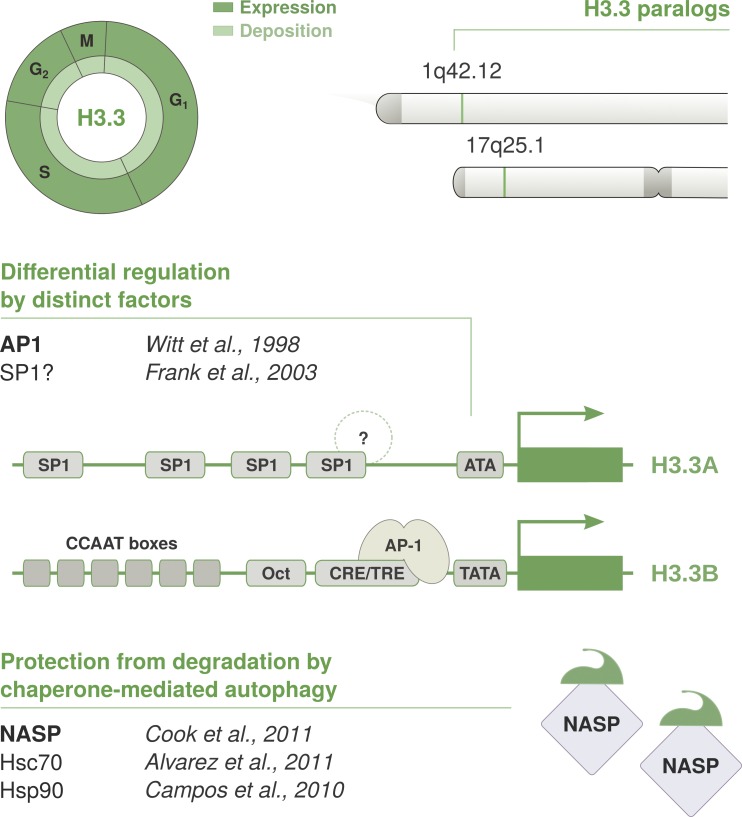
H3.3 is constitutively expressed in a cell cycle–independent manner, supporting histone turnover outside of S phase and in quiescent or postmitotic cells (Wu and Bonner, 1981; Wu et al., 1982, 1983) and histone replacement after fertilization in Drosophila (Loppin et al., 2005) and mice (Jang et al., 2015; Tang et al., 2015). H3.3 is redundantly encoded by two paralogous genes: H3.3A and H3.3B (Fig. 5). These are conserved in many organisms and, despite producing the same protein, they show different coding sequences, cis-regulatory targets, and intron-exon organization (Brush et al., 1985; Wells and Kedes, 1985; Chalmers and Wells, 1990; Akhmanova et al., 1995; Albig et al., 1995; Bramlage et al., 1997). Similar to CenH3CENP-A, H3.3A and H3.3B are solitary genes lacking 3′ stem-loop structure. Both give rise to polyadenylated transcripts, and H3.3B can generate up to three isoforms from alternative polyadenylation sites (Albig et al., 1995; Bramlage et al., 1997; Feng et al., 2005). Although both H3.3A and H3.3B can contribute to new H3.3 synthesis, the two genes show distinct expression profiles in the male and female germline, among different tissues, and during development (Krimer et al., 1993; Akhmanova et al., 1995; Bramlage et al., 1997; Couldrey et al., 1999; Jang et al., 2015; Maehara et al., 2015).
Figure 5.
Cell cycle timing and regulation of the replacement variant H3.3. The non-replicative variant H3.3 is deposited throughout the cell cycle by HIRA at regions of high turnover, such as regulatory sites and transcribed regions. H3.3 is expressed constitutively and redundantly encoded by two conserved paralogs: H3.3A and H3.3B. Both genes are located outside of histone clusters, contain introns, and give rise to polyadenylated mRNAs. H3.3A and H3.3B encode for the same protein, but their gene architecture is not conserved. They show distinct coding sequences, intron-exon organization, and cis-regulatory elements and are differentially expressed during development and among tissues. Putative binding sites have been identified in their respective promoter regions, and the CRE/TRE motif in H3.3B promoter was shown to mediate its activation via recruitment of AP-1 transcription factors. However, the basis of differences in expression is poorly understood. Similar to H3.1, the NASP histone chaperone can bind H3.3-H4 dimers and contributes to fine-tune protein levels via protection from chaperone-mediated autophagy.
The cis-regulatory elements in the human H3.3B promoter include an octamer (Oct) motif and a combined cAMP- and TPA-responsive element (CRE/TRE) flanked by a TATA box and six CCAAT boxes (Witt et al., 1997). Promoter deletion constructs show that the proximal promoter region, comprising Oct, CRE/TRE, and TATA elements, is sufficient to drive transcription in vitro (Witt et al., 1997). The CRE/TRE sequence in H3.3B promoter can recruit TPA-inducible AP-1 factors (Karin et al., 1997) and transcription factors of the CREB/ATF family that are typically activated via PKA-dependent phosphorylation in response to cAMP (Mayr and Montminy, 2001). However, cAMP treatment does not significantly affect H3.3B levels in vitro, whereas TPA induces a strong transcriptional response that is dependent on the presence of an intact CRE/TRE motif (Witt et al., 1998). This indicates a possible role for the recruitment of AP-1 factors through the TPA-inducible PKC pathway to induce H3.3B in vivo. However, additional players are likely involved in specific tissues or developmental stages and have not yet been characterized. Concerning H3.3A, it has a GC-rich promoter devoid of TATA and CCAAT boxes and instead harbors an ATA motif along with four SP1 binding sites. Stepwise deletion of the proximal promoter region, containing three SP1 motifs, reduces transcriptional activity in vitro in an additive manner (Frank et al., 2003); however, the binding partners and mechanistic basis are largely unexplored. Hence, factors promoting H3.3A induction or differential expression in various tissues remain largely unknown.
Consequences of H3 variant alterations and imbalances
As discussed, an incredible orchestration works to control the provision of histone proteins by multitasking at each possible level of regulation. Histones are always accompanied by chaperones, their guardians, throughout their cellular life (Gurard-Levin et al., 2014). The proper dosage of histone variants and chaperones plays an important role in defining the chromatin landscape during embryonic development, lineage commitment, and cell fate decisions (Filipescu et al., 2014). Xenopus oocytes are protected from the surplus of histones by nucleoplasmin, sustaining chromatin assembly through the rapid cell divisions that occur during early development. In mammalian cells, replicative aging is associated with impaired histone synthesis (O’Sullivan et al., 2010). This may be linked to chronic damage signals at telomeres that could affect histone levels (O’Sullivan et al., 2010). Histone supply also affects the length of S phase and cell cycle progression (Groth et al., 2005, 2007; Günesdogan et al., 2014). Interfering with SLBP, which reduces histone expression in human cells, also results in reduced cell growth and impaired S phase progression (Zhao et al., 2004). Intriguingly, the excess H3 during mitosis localizes to the centrosomes for proteasome-mediated degradation in worms, flies, and human cells (Wike et al., 2016). The cross-talk with the centrosome could hence play a role in preserving chromosome integrity by coordinating different signaling events. Genes in the HIST1 cluster are significantly up-regulated across different breast cancer cell lines and breast tumor specimens (Fritz et al., 2018). The chromatin organizer CTCF, mutated in several cancers, is found at the boundaries between TADs associated with distinct gene subclusters. Considering that the nuclear organization of the HLB is compromised in cancer cells (Ghule et al., 2009), this could contribute to the misregulation of histone genes in tumors.
CenH3CENP-A overexpression is common in many aggressive tumors (Tomonaga et al., 2003; Amato et al., 2009; Hu et al., 2010; Li et al., 2011; Wu et al., 2012b; Qiu et al., 2013; Filipescu et al., 2017). These CenH3CENP-A imbalances correlate with genomic instability and malignant progression as well as poor prognosis and response to treatment (Sun et al., 2016; Zhang et al., 2016). CenH3 overexpression also leads to ectopic recruitment of kinetochore components and mitotic defects in both Drosophila (Heun et al., 2006) and fission yeast (Choi et al., 2012; Gonzalez et al., 2014). In human cells, overexpression of CenH3CENP-A results in misassembly at chromosome arms (Lacoste et al., 2014) in a cell cycle–independent manner (Nechemia-Arbely et al., 2017). Ectopic deposition is mediated by ATRX/DAXX and confers higher tolerance to DNA damage, a potential mechanism for resistance. Excess CenH3CENP-A localizes at CTCF binding sites (Lacoste et al., 2014), DNase I hypersensitive sites, and transcription factor binding sites across the genome (Athwal et al., 2015). Subtelomeric regions prone to instability are also hotspots for CenH3CENP-A accumulation in overexpressing conditions (Athwal et al., 2015). Understanding how higher doses of CenH3CENP-A can confer a selective advantage in genomically unstable cells will be important. Overexpression is associated with better tolerance to damage, but higher CenH3CENP-A levels are also linked to chromosome instability and micronuclei formation in both cancer and stable diploid cells (Shrestha et al., 2017). Future studies will certainly be critical to understand how histone imbalances and chromatin misassembly may lead to severe pathological implications.
Concerning H3.3, clear differences in expression between the two paralogs were observed in the fly and mouse male germlines (Akhmanova et al., 1995; Bramlage et al., 1997; Feng et al., 2005) and throughout preimplantation development in mammals (Couldrey et al., 1999; Kafer et al., 2010; Xue et al., 2013). In flies, H3.3A and H3.3B single knockouts are viable and fertile, whereas double-null mutants have low viability and are sterile (Hödl and Basler, 2009; Sakai et al., 2009). Hypomorphic H3.3B-null mice die postnatally, but single H3.3A knockouts are viable, with reduced male fertility (Couldrey et al., 1999; Tang et al., 2015). However, the effect of hypomorphic mutations depends on the genetic background (Jang et al., 2015), and it is unclear what drives this variability among the paralogs. Notably, differences in expression could have important implications in tumorigenesis, because mutations in H3.3A and H3.3B are linked to distinct types of cancers (Weinberg et al., 2017). Indeed, the impact and occurrence of somatic mutations differs between H3.3A and H3.3B (Behjati et al., 2013). K36 mutations, found in 95% of chondroblastomas and 7% of clear-cell chondrosarcomas, occur predominantly in the H3.3B gene. G34 and K27 mutations are instead nearly exclusive to H3.3A and linked to other types of tumors such as high-grade astrocytomas (Schwartzentruber et al., 2012; Sturm et al., 2012; Wu et al., 2012a; Aihara et al., 2014) and giant cell tumors of bone (Behjati et al., 2013; Sarungbam et al., 2016). The lysine 27 is encoded by an AAG codon in H3.3A and AAA in H3.3B; hence the chance of a specific substitution occurring in a given paralog can be explained in part by codon differences between H3.3A and H3.3B. K27 mutations are indeed also found in H3.1-coding genes, where, similar to H3.3A, the lysine is often encoded by a AAG codon (Kallappagoudar et al., 2015). However, differences in expression may affect the tissue specificity and outcome of specific amino acid substitutions, a possibility that has not been fully explored. Interestingly, duplication rates also vary between the two paralogs. Numerous H3.3 pseudogenes exist in both the human and mouse genome, and the majority are more closely related to H3.3A (Wells et al., 1987; Ederveen et al., 2011; Maehara et al., 2015). It is currently unknown whether genomic context or differential transcription has any impact on the outcome or propensity to genetic variation. Recent findings have provided insights into how alterations in multiple histone variants beyond H3 can cause changes in epigenome plasticity and genome stability, thereby driving cancer initiation and/or progression (Vardabasso et al., 2014; Park et al., 2016; Buschbeck and Hake, 2017). Understanding the regulation of distinct histone variants in normal conditions will be critical to gain insights into their role in cancer progression and open concrete avenues for future therapeutic strategies.
Concluding remarks
We have summarized the current knowledge concerning the transcriptional regulation of replicative and non-replicative histone variants. Their genomic organization and distinct features are a unique example of how an exquisite orchestration of events can contribute to handle the demands for histone variants under distinct physiological conditions. The coregulation of replicative variants, and the evolution of independent mechanisms that coordinate the production of non-replicative forms, may represent an interesting paradigm that could apply to other gene families. We have also learned a lot about the interplay among histone variants and their chaperones and how they shape the epigenome and sustain chromatin plasticity. In the future, it will be crucial to further investigate histone cross-talk in the variety of cell types that constitute an organism, under normal or pathological conditions. Multidisciplinary approaches and single-cell technologies will certainly play a pivotal role in the future to resolve the dynamics of chromatin architecture across different cells and over the lifetime of an individual.
Acknowledgments
We thank Dominique Ray-Gallet and Iva Simeonova for critical reading of the manuscript.
This work was supported by La Ligue Contre le Cancer (Equipe Labelisée 2016), Parisian Alliance of Cancer Research Institutes, Agence Nationale de la Recherche (11-LABX-0044 “DEEP,” 10-IDEX-0001-02 “PSL,” ANR-12-BSV5-0022-02 “CHAPINHIB,” ANR-14-CE16-0009 “Epicure,” ANR-14-CE10-0013 “CELLECTCHIP,” ANR-16-CE15-0018 “CHRODYT,” ANR-16-CE12-0024 “CHIFT,” and ANR-16-CE11-0028 “REPLICAF”), and European Research Council (ERC-2015-ADG project 694694 “ChromADICT” and ERC-2015-POC project 678563 “EPOCH28”). A. Gatto is supported by the Horizon 2020 Framework Programme for Research and Innovation (H2020 Marie Skłodowska-Curie Actions grant agreement 798106 “REPLICHROM4D”).
The authors declare no competing financial interests.
Author contributions: S. Mendiratta, A. Gatto, and G. Almouzni wrote the manuscript.
References
- Ahmad K., and Henikoff S.. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 9:1191–1200. 10.1016/S1097-2765(02)00542-7 [DOI] [PubMed] [Google Scholar]
- Aihara K., Mukasa A., Gotoh K., Saito K., Nagae G., Tsuji S., Tatsuno K., Yamamoto S., Takayanagi S., Narita Y., et al. 2014. H3F3A K27M mutations in thalamic gliomas from young adult patients. Neuro-oncol. 16:140–146. 10.1093/neuonc/not144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmanova A.S., Bindels P.C., Xu J., Miedema K., Kremer H., and Hennig W.. 1995. Structure and expression of histone H3.3 genes in Drosophila melanogaster and Drosophila hydei. Genome. 38:586–600. 10.1139/g95-075 [DOI] [PubMed] [Google Scholar]
- Alabert C., and Groth A.. 2012. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 13:153–167. 10.1038/nrm3288 [DOI] [PubMed] [Google Scholar]
- Albig W., Bramlage B., Gruber K., Klobeck H.-G., Kunz J., and Doenecke D.. 1995. The human replacement histone H3.3B gene (H3F3B). Genomics. 30:264–272. 10.1006/geno.1995.9878 [DOI] [PubMed] [Google Scholar]
- Almouzni G., and Cedar H.. 2016. Maintenance of Epigenetic Information. Cold Spring Harb. Perspect. Biol. 8:a019372 10.1101/cshperspect.a019372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez F., Muñoz F., Schilcher P., Imhof A., Almouzni G., and Loyola A.. 2011. Sequential establishment of marks on soluble histones H3 and H4. J. Biol. Chem. 286:17714–17721. 10.1074/jbc.M111.223453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato A., Schillaci T., Lentini L., and Di Leonardo A.. 2009. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol. Cancer. 8:119 10.1186/1476-4598-8-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athwal R.K., Walkiewicz M.P., Baek S., Fu S., Bui M., Camps J., Ried T., Sung M.-H., and Dalal Y.. 2015. CENP-A nucleosomes localize to transcription factor hotspots and subtelomeric sites in human cancer cells. Epigenetics Chromatin. 8:2 10.1186/1756-8935-8-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au W.C., Dawson A.R., Rawson D.W., Taylor S.B., Baker R.E., and Basrai M.A.. 2013. A novel role of the N terminus of budding yeast histone H3 variant Cse4 in ubiquitin-mediated proteolysis. Genetics. 194:513–518. 10.1534/genetics.113.149898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bade D., Pauleau A.-L., Wendler A., and Erhardt S.. 2014. The E3 ligase CUL3/RDX controls centromere maintenance by ubiquitylating and stabilizing CENP-A in a CAL1-dependent manner. Dev. Cell. 28:508–519. 10.1016/j.devcel.2014.01.031 [DOI] [PubMed] [Google Scholar]
- Barcaroli D., Bongiorno-Borbone L., Terrinoni A., Hofmann T.G., Rossi M., Knight R.A., Matera A.G., Melino G., and De Laurenzi V.. 2006. FLASH is required for histone transcription and S-phase progression. Proc. Natl. Acad. Sci. USA. 103:14808–14812. 10.1073/pnas.0604227103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett E.A., DeNizio J., Barnhart-Dailey M.C., Panchenko T., Sekulic N., Rogers D.J., Foltz D.R., and Black B.E.. 2012. HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev. Cell. 22:749–762. 10.1016/j.devcel.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behjati S., Tarpey P.S., Presneau N., Scheipl S., Pillay N., Van Loo P., Wedge D.C., Cooke S.L., Gundem G., Davies H., et al. 2013. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat. Genet. 45:1479–1482. 10.1038/ng.2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor D.L., Valente L.P., Mata J.F., Black B.E., and Jansen L.E.T.. 2013. Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol. Biol. Cell. 24:923–932. 10.1091/mbc.e13-01-0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramlage B., Kosciessa U., and Doenecke D.. 1997. Differential expression of the murine histone genes H3.3A and H3.3B. Differentiation. 62:13–20. 10.1046/j.1432-0436.1997.6210013.x [DOI] [PubMed] [Google Scholar]
- Brush D., Dodgson J.B., Choi O.R., Stevens P.W., and Engel J.D.. 1985. Replacement variant histone genes contain intervening sequences. Mol. Cell. Biol. 5:1307–1317. 10.1128/MCB.5.6.1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch B.D., Godfrey A.C., Gasdaska P.Y., Salzler H.R., Duronio R.J., Marzluff W.F., and Dominski Z.. 2011. Interaction between FLASH and Lsm11 is essential for histone pre-mRNA processing in vivo in Drosophila. RNA. 17:1132–1147. 10.1261/rna.2566811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschbeck M., and Hake S.B.. 2017. Variants of core histones and their roles in cell fate decisions, development and cancer. Nat. Rev. Mol. Cell Biol. 18:299–314. 10.1038/nrm.2016.166 [DOI] [PubMed] [Google Scholar]
- Cajal S.R. 1903. Un sencillo metodo de coloracion seletiva del reticulo protoplasmatico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab. Lab. Invest. Biol. (Madrid). 2:129–221. [Google Scholar]
- Calvi B.R., Lilly M.A., and Spradling A.C.. 1998. Cell cycle control of chorion gene amplification. Genes Dev. 12:734–744. 10.1101/gad.12.5.734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos E.I., Fillingham J., Li G., Zheng H., Voigt P., Kuo W.H., Seepany H., Gao Z., Day L.A., Greenblatt J.F., and Reinberg D.. 2010. The program for processing newly synthesized histones H3.1 and H4. Nat. Struct. Mol. Biol. 17:1343–1351. 10.1038/nsmb.1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers M., and Wells D.. 1990. Extreme sequence conservation characterizes the rabbit H3.3A histone cDNA. Nucleic Acids Res. 18:3075 10.1093/nar/18.10.3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Müller G.A., Quaas M., Fischer M., Han N., Stutchbury B., Sharrocks A.D., and Engeland K.. 2013. The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol. Cell. Biol. 33:227–236. 10.1128/MCB.00881-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-J., Dominguez-Brauer C., Wang Z., Asara J.M., Costa R.H., Tyner A.L., Lau L.F., and Raychaudhuri P.. 2009. A conserved phosphorylation site within the forkhead domain of FoxM1B is required for its activation by cyclin-CDK1. J. Biol. Chem. 284:30695–30707. 10.1074/jbc.M109.007997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E.S., Strålfors A., Catania S., Castillo A.G., Svensson J.P., Pidoux A.L., Ekwall K., and Allshire R.C.. 2012. Factors that promote H3 chromatin integrity during transcription prevent promiscuous deposition of CENP-A(Cnp1) in fission yeast. PLoS Genet. 8:e1002985 10.1371/journal.pgen.1002985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément C., and Almouzni G.. 2015. MCM2 binding to histones H3-H4 and ASF1 supports a tetramer-to-dimer model for histone inheritance at the replication fork. Nat. Struct. Mol. Biol. 22:587–589. 10.1038/nsmb.3067 [DOI] [PubMed] [Google Scholar]
- Clément C., Orsi G.A., Gatto A., Boyarchuk E., Forest A., Hajj B., Miné-Hattab J., Garnier M., Gurard-Levin Z.A., Quivy J.-P., and Almouzni G.. 2018. High-resolution visualization of H3 variants during replication reveals their controlled recycling. Nat. Commun. 9:3181 10.1038/s41467-018-05697-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A.J.L., Gurard-Levin Z.A., Vassias I., and Almouzni G.. 2011. A specific function for the histone chaperone NASP to fine-tune a reservoir of soluble H3-H4 in the histone supply chain. Mol. Cell. 44:918–927. 10.1016/j.molcel.2011.11.021 [DOI] [PubMed] [Google Scholar]
- Corpet A., and Almouzni G.. 2009. Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends Cell Biol. 19:29–41. 10.1016/j.tcb.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Corpet A., De Koning L., Toedling J., Savignoni A., Berger F., Lemaître C., O’Sullivan R.J., Karlseder J., Barillot E., Asselain B., et al. 2011. Asf1b, the necessary Asf1 isoform for proliferation, is predictive of outcome in breast cancer. EMBO J. 30:480–493. 10.1038/emboj.2010.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M., Gick O., Vasserot A., Schaffner G., and Birnstiel M.L.. 1988. Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the in vitro 3′ RNA processing reaction. EMBO J. 7:801–808. 10.1002/j.1460-2075.1988.tb02878.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couldrey C., Carlton M.B., Nolan P.M., Colledge W.H., and Evans M.J.. 1999. A retroviral gene trap insertion into the histone 3.3A gene causes partial neonatal lethality, stunted growth, neuromuscular deficits and male sub-fertility in transgenic mice. Hum. Mol. Genet. 8:2489–2495. 10.1093/hmg/8.13.2489 [DOI] [PubMed] [Google Scholar]
- Daganzo S.M., Erzberger J.P., Lam W.M., Skordalakes E., Zhang R., Franco A.A., Brill S.J., Adams P.D., Berger J.M., and Kaufman P.D.. 2003. Structure and function of the conserved core of histone deposition protein Asf1. Curr. Biol. 13:2148–2158. 10.1016/j.cub.2003.11.027 [DOI] [PubMed] [Google Scholar]
- Daneshvar K., Khan A., and Goodliffe J.M.. 2011. Myc localizes to histone locus bodies during replication in Drosophila. PLoS One. 6:e23928 10.1371/journal.pone.0023928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dávila López M., and Samuelsson T.. 2008. Early evolution of histone mRNA 3′ end processing. RNA. 14:1–10. 10.1261/rna.782308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cola A., Bongiorno-Borbone L., Bianchi E., Barcaroli D., Carletti E., Knight R.A., Di Ilio C., Melino G., Sette C., and De Laurenzi V.. 2012. FLASH is essential during early embryogenesis and cooperates with p73 to regulate histone gene transcription. Oncogene. 31:573–582. 10.1038/onc.2011.274 [DOI] [PubMed] [Google Scholar]
- De Koning L., Corpet A., Haber J.E., and Almouzni G.. 2007. Histone chaperones: an escort network regulating histone traffic. Nat. Struct. Mol. Biol. 14:997–1007. 10.1038/nsmb1318 [DOI] [PubMed] [Google Scholar]
- Dominski Z., Zheng L.X., Sanchez R., and Marzluff W.F.. 1999. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol. 19:3561–3570. 10.1128/MCB.19.5.3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Yang X.C., and Marzluff W.F.. 2005. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 123:37–48. 10.1016/j.cell.2005.08.002 [DOI] [PubMed] [Google Scholar]
- Down C.F., Millour J., Lam E.W.-F., and Watson R.J.. 2012. Binding of FoxM1 to G2/M gene promoters is dependent upon B-Myb. Biochim. Biophys. Acta. 1819:855–862. 10.1016/j.bbagrm.2012.03.008 [DOI] [PubMed] [Google Scholar]
- Drané P., Ouararhni K., Depaux A., Shuaib M., and Hamiche A.. 2010. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24:1253–1265. 10.1101/gad.566910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy E.M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., and Almouzni-Pettinotti G.. 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 137:485–497. 10.1016/j.cell.2009.02.040 [DOI] [PubMed] [Google Scholar]
- Dunleavy E.M., Almouzni G., and Karpen G.H.. 2011. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G1 phase. Nucleus. 2:146–157. 10.4161/nucl.2.2.15211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio R.J., and Marzluff W.F.. 2017. Coordinating cell cycle-regulated histone gene expression through assembly and function of the Histone Locus Body. RNA Biol. 14:726–738. 10.1080/15476286.2016.1265198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W.C., Honda B.M., Laskey R.A., and Thomas J.O.. 1980. Assembly of nucleosomes: the reaction involving X. laevis nucleoplasmin. Cell. 21:373–383. 10.1016/0092-8674(80)90474-2 [DOI] [PubMed] [Google Scholar]
- Ederveen T.H.A., Mandemaker I.K., and Logie C.. 2011. The human histone H3 complement anno 2011. Biochim. Biophys. Acta. 1809:577–586. 10.1016/j.bbagrm.2011.07.002 [DOI] [PubMed] [Google Scholar]
- Eriksson P.R., Ganguli D., Nagarajavel V., and Clark D.J.. 2012. Regulation of histone gene expression in budding yeast. Genetics. 191:7–20. 10.1534/genetics.112.140145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng R., Tang X., Becker A., Berger A., Ye J., Akhmanova A., and Hennig W.. 2005. Regulation of the expression of histone H3.3 by differential polyadenylation. Genome. 48:503–510. 10.1139/g05-009 [DOI] [PubMed] [Google Scholar]
- Filipescu D., Szenker E., and Almouzni G.. 2013. Developmental roles of histone H3 variants and their chaperones. Trends Genet. 29:630–640. 10.1016/j.tig.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Filipescu D., Müller S., and Almouzni G.. 2014. Histone H3 variants and their chaperones during development and disease: contributing to epigenetic control. Annu. Rev. Cell Dev. Biol. 30:615–646. 10.1146/annurev-cellbio-100913-013311 [DOI] [PubMed] [Google Scholar]
- Filipescu D., Naughtin M., Podsypanina K., Lejour V., Wilson L., Gurard-Levin Z.A., Orsi G.A., Simeonova I., Toufektchan E., Attardi L.D., et al. 2017. Essential role for centromeric factors following p53 loss and oncogenic transformation. Genes Dev. 31:463–480. 10.1101/gad.290924.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M., Grossmann P., Padi M., and DeCaprio J.A.. 2016. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res. 44:6070–6086. 10.1093/nar/gkw523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E.T., Bailey A.O., Yates J.R. III, Bassett E.A., Wood S., Black B.E., and Cleveland D.W.. 2009. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 137:472–484. 10.1016/j.cell.2009.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D., Doenecke D., and Albig W.. 2003. Differential expression of human replacement and cell cycle dependent H3 histone genes. Gene. 312:135–143. 10.1016/S0378-1119(03)00609-7 [DOI] [PubMed] [Google Scholar]
- Frey M.R., and Matera A.G.. 1995. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc. Natl. Acad. Sci. USA. 92:5915–5919. 10.1073/pnas.92.13.5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz A.J., Ghule P.N., Boyd J.R., Tye C.E., Page N.A., Hong D., Shirley D.J., Weinheimer A.S., Barutcu A.R., Gerrard D.L., et al. 2018. Intranuclear and higher-order chromatin organization of the major histone gene cluster in breast cancer. J. Cell. Physiol. 233:1278–1290. 10.1002/jcp.25996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z., Malureanu L., Huang J., Wang W., Li H., van Deursen J.M., Tindall D.J., and Chen J.. 2008. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat. Cell Biol. 10:1076–1082. 10.1038/ncb1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard P.H., Martini E.M., Kaufman P.D., Stillman B., Moustacchi E., and Almouzni G.. 1996. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 86:887–896. 10.1016/S0092-8674(00)80164-6 [DOI] [PubMed] [Google Scholar]
- Gascoigne K.E., Takeuchi K., Suzuki A., Hori T., Fukagawa T., and Cheeseman I.M.. 2011. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 145:410–422. 10.1016/j.cell.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule P.N., Dominski Z., Lian J.B., Stein J.L., van Wijnen A.J., and Stein G.S.. 2009. The subnuclear organization of histone gene regulatory proteins and 3′ end processing factors of normal somatic and embryonic stem cells is compromised in selected human cancer cell types. J. Cell. Physiol. 220:129–135. 10.1002/jcp.21740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhman D., Livyatan I., Sailaja B.S., Melcer S., and Meshorer E.. 2013. Multilayered chromatin analysis reveals E2f, Smad and Zfx as transcriptional regulators of histones. Nat. Struct. Mol. Biol. 20:119–126. 10.1038/nsmb.2448 [DOI] [PubMed] [Google Scholar]
- Goldberg A.D., Banaszynski L.A., Noh K.-M., Lewis P.W., Elsaesser S.J., Stadler S., Dewell S., Law M., Guo X., Li X., et al. 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 140:678–691. 10.1016/j.cell.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M., He H., Dong Q., Sun S., and Li F.. 2014. Ectopic centromere nucleation by CENP—a in fission yeast. Genetics. 198:1433–1446. 10.1534/genetics.114.171173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves R.A., Pandey N.B., Chodchoy N., and Marzluff W.F.. 1987. Translation is required for regulation of histone mRNA degradation. Cell. 48:615–626. 10.1016/0092-8674(87)90240-6 [DOI] [PubMed] [Google Scholar]
- Groth A., Ray-Gallet D., Quivy J.-P., Lukas J., Bartek J., and Almouzni G.. 2005. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol. Cell. 17:301–311. 10.1016/j.molcel.2004.12.018 [DOI] [PubMed] [Google Scholar]
- Groth A., Corpet A., Cook A.J.L., Roche D., Bartek J., Lukas J., and Almouzni G.. 2007. Regulation of replication fork progression through histone supply and demand. Science. 318:1928–1931. 10.1126/science.1148992 [DOI] [PubMed] [Google Scholar]
- Guglielmi B., La Rochelle N., and Tjian R.. 2013. Gene-specific transcriptional mechanisms at the histone gene cluster revealed by single-cell imaging. Mol. Cell. 51:480–492. 10.1016/j.molcel.2013.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günesdogan U., Jäckle H., and Herzig A.. 2014. Histone supply regulates S phase timing and cell cycle progression. eLife. 3:e02443 10.7554/eLife.02443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunjan A., Paik J., and Verreault A.. 2005. Regulation of histone synthesis and nucleosome assembly. Biochimie. 87:625–635. 10.1016/j.biochi.2005.02.008 [DOI] [PubMed] [Google Scholar]
- Gurard-Levin Z.A., Quivy J.-P., and Almouzni G.. 2014. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu. Rev. Biochem. 83:487–517. 10.1146/annurev-biochem-060713-035536 [DOI] [PubMed] [Google Scholar]
- Hammond C.M., Strømme C.B., Huang H., Patel D.J., and Groth A.. 2017. Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 18:141–158. 10.1038/nrm.2016.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P., Erhardt S., Blower M.D., Weiss S., Skora A.D., and Karpen G.H.. 2006. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell. 10:303–315. 10.1016/j.devcel.2006.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hödl M., and Basler K.. 2009. Transcription in the absence of histone H3.3. Curr. Biol. 19:1221–1226. 10.1016/j.cub.2009.05.048 [DOI] [PubMed] [Google Scholar]
- Hu H., Liu Y., Wang M., Fang J., Huang H., Yang N., Li Y., Wang J., Yao X., Shi Y., et al. 2011. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 25:901–906. 10.1101/gad.2045111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Huang G., Sadanandam A., Gu S., Lenburg M.E., Pai M., Bayani N., Blakely E.A., Gray J.W., and Mao J.-H.. 2010. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res. 12:R18 10.1186/bcr2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Strømme C.B., Saredi G., Hödl M., Strandsby A., González-Aguilera C., Chen S., Groth A., and Patel D.J.. 2015. A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nat. Struct. Mol. Biol. 22:618–626. 10.1038/nsmb.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Y., Keles S., Prestel M., Hochheimer A., and Tjian R.. 2007. Transcription of histone gene cluster by differential core-promoter factors. Genes Dev. 21:2936–2949. 10.1101/gad.1608807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanauskiene K., Delbarre E., McGhie J.D., Küntziger T., Wong L.H., and Collas P.. 2014. The PML-associated protein DEK regulates the balance of H3.3 loading on chromatin and is important for telomere integrity. Genome Res. 24:1584–1594. 10.1101/gr.173831.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., and Vale R.D.. 2017. RNA phase transitions in repeat expansion disorders. Nature. 546:243–247. 10.1038/nature22386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C.-W., Shibata Y., Starmer J., Yee D., and Magnuson T.. 2015. Histone H3.3 maintains genome integrity during mammalian development. Genes Dev. 29:1377–1392. 10.1101/gad.264150.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen L.E.T., Black B.E., Foltz D.R., and Cleveland D.W.. 2007. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176:795–805. 10.1083/jcb.200701066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafer G.R., Lehnert S.A., Pantaleon M., Kaye P.L., and Moser R.J.. 2010. Expression of genes coding for histone variants and histone-associated proteins in pluripotent stem cells and mouse preimplantation embryos. Gene Expr. Patterns. 10:299–305. 10.1016/j.gep.2010.06.003 [DOI] [PubMed] [Google Scholar]
- Kallappagoudar S., Yadav R.K., Lowe B.R., and Partridge J.F.. 2015. Histone H3 mutations—a special role for H3.3 in tumorigenesis? Chromosoma. 124:177–189. 10.1007/s00412-015-0510-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M., Liu Z., and Zandi E.. 1997. AP-1 function and regulation. Curr. Opin. Cell Biol. 9:240–246. 10.1016/S0955-0674(97)80068-3 [DOI] [PubMed] [Google Scholar]
- Kolev N.G., and Steitz J.A.. 2005. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 19:2583–2592. 10.1101/gad.1371105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev N.G., Yario T.A., Benson E., and Steitz J.A.. 2008. Conserved motifs in both CPSF73 and CPSF100 are required to assemble the active endonuclease for histone mRNA 3′-end maturation. EMBO Rep. 9:1013–1018. 10.1038/embor.2008.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koseoglu M.M., Graves L.M., and Marzluff W.F.. 2008. Phosphorylation of threonine 61 by cyclin a/Cdk1 triggers degradation of stem-loop binding protein at the end of S phase. Mol. Cell. Biol. 28:4469–4479. 10.1128/MCB.01416-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimer D.B., Cheng G., and Skoultchi A.I.. 1993. Induction of H3.3 replacement histone mRNAs during the precommitment period of murine erythroleukemia cell differentiation. Nucleic Acids Res. 21:2873–2879. 10.1093/nar/21.12.2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Sobhy H., Stenberg P., and Lizana L.. 2017. Genome contact map explorer: a platform for the comparison, interactive visualization and analysis of genome contact maps. Nucleic Acids Res. 45:e152 10.1093/nar/gkx644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurat C.F., Lambert J.-P., Petschnigg J., Friesen H., Pawson T., Rosebrock A., Gingras A.-C., Fillingham J., and Andrews B.. 2014. Cell cycle-regulated oscillator coordinates core histone gene transcription through histone acetylation. Proc. Natl. Acad. Sci. USA. 111:14124–14129. 10.1073/pnas.1414024111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursel L.E., and Malik H.S.. 2017. Recurrent Gene Duplication Leads to Diverse Repertoires of Centromeric Histones in Drosophila Species. Mol. Biol. Evol. 34:1445–1462. 10.1093/molbev/msx091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey P.E., Welch J.D., and Marzluff W.F.. 2016. TUT7 catalyzes the uridylation of the 3′ end for rapid degradation of histone mRNA. RNA. 22:1673–1688. 10.1261/rna.058107.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste N., Woolfe A., Tachiwana H., Garea A.V., Barth T., Cantaloube S., Kurumizaka H., Imhof A., and Almouzni G.. 2014. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell. 53:631–644. 10.1016/j.molcel.2014.01.018 [DOI] [PubMed] [Google Scholar]
- Laoukili J., Alvarez M., Meijer L.A.T., Stahl M., Mohammed S., Kleij L., Heck A.J.R., and Medema R.H.. 2008. Activation of FoxM1 during G2 requires cyclin A/Cdk-dependent relief of autorepression by the FoxM1 N-terminal domain. Mol. Cell. Biol. 28:3076–3087. 10.1128/MCB.01710-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson A.G., Elnatan D., Keenen M.M., Trnka M.J., Johnston J.B., Burlingame A.L., Agard D.A., Redding S., and Narlikar G.J.. 2017. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 547:236–240. 10.1038/nature22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey R.A., Honda B.M., Mills A.D., and Finch J.T.. 1978. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 275:416–420. 10.1038/275416a0 [DOI] [PubMed] [Google Scholar]
- Latreille D., Bluy L., Benkirane M., and Kiernan R.E.. 2014. Identification of histone 3 variant 2 interacting factors. Nucleic Acids Res. 42:3542–3550. 10.1093/nar/gkt1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P.W., Elsaesser S.J., Noh K.-M., Stadler S.C., and Allis C.D.. 2010. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. USA. 107:14075–14080. 10.1073/pnas.1008850107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., and Tyler J.K.. 2016. Nucleosome disassembly during human non-homologous end joining followed by concerted HIRA- and CAF-1-dependent reassembly. eLife. 5:e15129 10.7554/eLife.15129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhu Z., Zhang S., Yu D., Yu H., Liu L., Cao X., Wang L., Gao H., and Zhu M.. 2011. ShRNA-targeted centromere protein A inhibits hepatocellular carcinoma growth. PLoS One. 6:e17794 10.1371/journal.pone.0017794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Hebert M.D., Ye Y., Templeton D.J., Kung H., and Matera A.G.. 2000. Cell cycle-dependent localization of the CDK2-cyclin E complex in Cajal (coiled) bodies. J. Cell Sci. 113:1543–1552. [DOI] [PubMed] [Google Scholar]
- Liu J.-L., Murphy C., Buszczak M., Clatterbuck S., Goodman R., and Gall J.G.. 2006. The Drosophila melanogaster Cajal body. J. Cell Biol. 172:875–884. 10.1083/jcb.200511038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.-L., Wu Z., Nizami Z., Deryusheva S., Rajendra T.K., Beumer K.J., Gao H., Matera A.G., Carroll D., and Gall J.G.. 2009. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol. Biol. Cell. 20:1661–1670. 10.1091/mbc.e08-05-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppin B., Bonnefoy E., Anselme C., Laurençon A., Karr T.L., and Couble P.. 2005. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 437:1386–1390. 10.1038/nature04059 [DOI] [PubMed] [Google Scholar]
- Ma T., Van Tine B.A., Wei Y., Garrett M.D., Nelson D., Adams P.D., Wang J., Qin J., Chow L.T., and Harper J.W.. 2000. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 14:2298–2313. 10.1101/gad.829500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehara K., Harada A., Sato Y., Matsumoto M., Nakayama K.I., Kimura H., and Ohkawa Y.. 2015. Tissue-specific expression of histone H3 variants diversified after species separation. Epigenetics Chromatin. 8:35 10.1186/s13072-015-0027-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik H.S., and Henikoff S.. 2003. Phylogenomics of the nucleosome. Nat. Struct. Biol. 10:882–891. 10.1038/nsb996 [DOI] [PubMed] [Google Scholar]
- Mandel C.R., Kaneko S., Zhang H., Gebauer D., Vethantham V., Manley J.L., and Tong L.. 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 444:953–956. 10.1038/nature05363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariño-Ramírez L., Jordan I.K., and Landsman D.. 2006. Multiple independent evolutionary solutions to core histone gene regulation. Genome Biol. 7:R122 10.1186/gb-2006-7-12-r122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W.F., Gongidi P., Woods K.R., Jin J., and Maltais L.J.. 2002. The human and mouse replication-dependent histone genes. Genomics. 80:487–498. 10.1006/geno.2002.6850 [DOI] [PubMed] [Google Scholar]
- Marzluff W.F., Wagner E.J., and Duronio R.J.. 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 9:843–854. 10.1038/nrg2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A.G., Izaguire-Sierra M., Praveen K., and Rajendra T.K.. 2009. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev. Cell. 17:639–647. 10.1016/j.devcel.2009.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr B., and Montminy M.. 2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2:599–609. 10.1038/35085068 [DOI] [PubMed] [Google Scholar]
- Mei Q., Huang J., Chen W., Tang J., Xu C., Yu Q., Cheng Y., Ma L., Yu X., and Li S.. 2017. Regulation of DNA replication-coupled histone gene expression. Oncotarget. 8:95005–95022. 10.18632/oncotarget.21887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello J.A., Silljé H.H.W., Roche D.M.J., Kirschner D.B., Nigg E.A., and Almouzni G.. 2002. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 3:329–334. 10.1093/embo-reports/kvf068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo M.J., Padeken J., Fülöp S., Schepers A., and Heun P.. 2011. Drosophila CENH3 is sufficient for centromere formation. Science. 334:686–690. 10.1126/science.1206880 [DOI] [PubMed] [Google Scholar]
- Moggs J.G., Grandi P., Quivy J.P., Jónsson Z.O., Hübscher U., Becker P.B., and Almouzni G.. 2000. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol. Cell. Biol. 20:1206–1218. 10.1128/MCB.20.4.1206-1218.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry K.L., and Steitz J.A.. 1987. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA’s. Science. 238:1682–1687. 10.1126/science.2825355 [DOI] [PubMed] [Google Scholar]
- Mullen T.E., and Marzluff W.F.. 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 22:50–65. 10.1101/gad.1622708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S., and Almouzni G.. 2017. Chromatin dynamics during the cell cycle at centromeres. Nat. Rev. Genet. 18:192–208. 10.1038/nrg.2016.157 [DOI] [PubMed] [Google Scholar]
- Müller G.A., Quaas M., Schümann M., Krause E., Padi M., Fischer M., Litovchick L., DeCaprio J.A., and Engeland K.. 2012. The CHR promoter element controls cell cycle-dependent gene transcription and binds the DREAM and MMB complexes. Nucleic Acids Res. 40:1561–1578. 10.1093/nar/gkr793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller G.A., Wintsche A., Stangner K., Prohaska S.J., Stadler P.F., and Engeland K.. 2014a The CHR site: definition and genome-wide identification of a cell cycle transcriptional element. Nucleic Acids Res. 42:10331–10350. 10.1093/nar/gku696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S., Montes de Oca R., Lacoste N., Dingli F., Loew D., and Almouzni G.. 2014b Phosphorylation and DNA binding of HJURP determine its centromeric recruitment and function in CenH3(CENP-A) loading. Cell Reports. 8:190–203. 10.1016/j.celrep.2014.06.002 [DOI] [PubMed] [Google Scholar]
- Narita T., Yung T.M.C., Yamamoto J., Tsuboi Y., Tanabe H., Tanaka K., Yamaguchi Y., and Handa H.. 2007. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell. 26:349–365. 10.1016/j.molcel.2007.04.011 [DOI] [PubMed] [Google Scholar]
- Nechemia-Arbely Y., Fachinetti D., Miga K.H., Sekulic N., Soni G.V., Kim D.H., Wong A.K., Lee A.Y., Nguyen K., Dekker C., et al. 2017. Human centromeric CENP-A chromatin is a homotypic, octameric nucleosome at all cell cycle points. J. Cell Biol. 216:607–621. 10.1083/jcb.201608083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura Y., Kitagawa R., Ogi H., Abdulle R., Pagala V., and Kitagawa K.. 2015. CENP-A K124 Ubiquitylation Is Required for CENP-A Deposition at the Centromere. Dev. Cell. 32:589–603. 10.1016/j.devcel.2015.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura Y., Kitagawa R., and Kitagawa K.. 2017. CENP-A Ubiquitylation Is Required for CENP-A Deposition at the Centromere. Dev. Cell. 40:7–8. 10.1016/j.devcel.2016.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan R.J., Kubicek S., Schreiber S.L., and Karlseder J.. 2010. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat. Struct. Mol. Biol. 17:1218–1225. 10.1038/nsmb.1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley M.A., Gould J., Kim S., Kane M.Y., and Hereford L.. 1986. Identification of sequences in a yeast histone promoter involved in periodic transcription. Cell. 45:537–544. 10.1016/0092-8674(86)90285-0 [DOI] [PubMed] [Google Scholar]
- Pandey N.B., and Marzluff W.F.. 1987. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol. Cell. Biol. 7:4557–4559. 10.1128/MCB.7.12.4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey N.B., Williams A.S., Sun J.H., Brown V.D., Bond U., and Marzluff W.F.. 1994. Point mutations in the stem-loop at the 3′ end of mouse histone mRNA reduce expression by reducing the efficiency of 3′ end formation. Mol. Cell. Biol. 14:1709–1720. 10.1128/MCB.14.3.1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-M., Choi E.-Y., Bae M., Kim S., Park J.B., Yoo H., Choi J.K., Kim Y.-J., Lee S.-H., and Kim I.-H.. 2016. Histone variant H3F3A promotes lung cancer cell migration through intronic regulation. Nat. Commun. 7:12914 10.1038/ncomms12914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pchelintsev N.A., McBryan T., Rai T.S., van Tuyn J., Ray-Gallet D., Almouzni G., and Adams P.D.. 2013. Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Reports. 3:1012–1019. 10.1016/j.celrep.2013.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R.S., Will C.L., Lührmann R., Schümperli D., and Müller B.. 2001. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 20:5470–5479. 10.1093/emboj/20.19.5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R.S., Grimmler M., Meister G., Will C.L., Lührmann R., Fischer U., and Schümperli D.. 2003. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 17:2321–2333. 10.1101/gad.274403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piña B., and Suau P.. 1987. Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Dev. Biol. 123:51–58. 10.1016/0012-1606(87)90426-X [DOI] [PubMed] [Google Scholar]
- Probst A.V., Dunleavy E., and Almouzni G.. 2009. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 10:192–206. 10.1038/nrm2640 [DOI] [PubMed] [Google Scholar]
- Qiu J.-J., Guo J.-J., Lv T.-J., Jin H.-Y., Ding J.-X., Feng W.-W., Zhang Y., and Hua K.-Q.. 2013. Prognostic value of centromere protein-A expression in patients with epithelial ovarian cancer. Tumour Biol. 34:2971–2975. 10.1007/s13277-013-0860-6 [DOI] [PubMed] [Google Scholar]
- Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., and Aiden E.L.. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 159:1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray A.M.J., and Müller B.. 2012. The control of histone gene expression. Biochem. Soc. Trans. 40:880–885. 10.1042/BST20120065 [DOI] [PubMed] [Google Scholar]
- Ray-Gallet D., and Almouzni G.. 2010. Molecular biology. Mixing or not mixing. Science. 328:56–57. 10.1126/science.1188653 [DOI] [PubMed] [Google Scholar]
- Ray-Gallet D., Woolfe A., Vassias I., Pellentz C., Lacoste N., Puri A., Schultz D.C., Pchelintsev N.A., Adams P.D., Jansen L.E.T., and Almouzni G.. 2011. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell. 44:928–941. 10.1016/j.molcel.2011.12.006 [DOI] [PubMed] [Google Scholar]
- Ray-Gallet D., Ricketts M.D., Sato Y., Gupta K., Boyarchuk E., Senda T., Marmorstein R., and Almouzni G.. 2018. Functional activity of the H3.3 histone chaperone complex HIRA requires trimerization of the HIRA subunit. Nat. Commun. 9:3103 10.1038/s41467-018-05581-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Régnier V., Novelli J., Fukagawa T., Vagnarelli P., and Brown W.. 2003. Characterization of chicken CENP-A and comparative sequence analysis of vertebrate centromere-specific histone H3-like proteins. Gene. 316:39–46. 10.1016/S0378-1119(03)00768-6 [DOI] [PubMed] [Google Scholar]
- Reinberg D., and Vales L.D.. 2018. Chromatin domains rich in inheritance. Science. 361:33–34. 10.1126/science.aat7871 [DOI] [PubMed] [Google Scholar]
- Richet N., Liu D., Legrand P., Velours C., Corpet A., Gaubert A., Bakail M., Moal-Raisin G., Guerois R., Compper C., et al. 2015. Structural insight into how the human helicase subunit MCM2 may act as a histone chaperone together with ASF1 at the replication fork. Nucleic Acids Res. 43:1905–1917. 10.1093/nar/gkv021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder L.E., Koreski K.P., Boltz K.A., Kuzu G., Urban J.A., Bowman S.K., Zeidman A., Jordan W.T. III, Tolstorukov M.Y., Marzluff W.F., et al. 2017. Histone locus regulation by the Drosophila dosage compensation adaptor protein CLAMP. Genes Dev. 31:1494–1508. 10.1101/gad.300855.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin L., and Mellone B.G.. 2016. Co-evolving CENP-A and CAL1 Domains Mediate Centromeric CENP-A Deposition across Drosophila Species. Dev. Cell. 37:136–147. 10.1016/j.devcel.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadasivam S., and DeCaprio J.A.. 2013. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat. Rev. Cancer. 13:585–595. 10.1038/nrc3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadasivam S., Duan S., and DeCaprio J.A.. 2012. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 26:474–489. 10.1101/gad.181933.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A., Schwartz B.E., Goldstein S., and Ahmad K.. 2009. Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr. Biol. 19:1816–1820. 10.1016/j.cub.2009.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldi T., Fong N., and Bentley D.L.. 2018. Transcription elongation rate affects nascent histone pre-mRNA folding and 3′ end processing. Genes Dev. 32:297–308. 10.1101/gad.310896.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzler H.R., Tatomer D.C., Malek P.Y., McDaniel S.L., Orlando A.N., Marzluff W.F., and Duronio R.J.. 2013. A sequence in the Drosophila H3-H4 Promoter triggers histone locus body assembly and biosynthesis of replication-coupled histone mRNAs. Dev. Cell. 24:623–634. 10.1016/j.devcel.2013.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L., Pidoux A.L., Ponting C.P., and Allshire R.C.. 2009. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 137:1173–1174. 10.1016/j.cell.2009.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarungbam J., Agaram N., Hwang S., Lu C., Wang L., Healey J., and Hameed M.. 2016. Symplastic/pseudoanaplastic giant cell tumor of the bone. Skeletal Radiol. 45:929–935. 10.1007/s00256-016-2373-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J., Korshunov A., Liu X.-Y., Jones D.T.W., Pfaff E., Jacob K., Sturm D., Fontebasso A.M., Quang D.-A.K., Tönjes M., et al. 2012. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 482:226–231. 10.1038/nature10833 [DOI] [PubMed] [Google Scholar]
- Shelby R.D., Vafa O., and Sullivan K.F.. 1997. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 136:501–513. 10.1083/jcb.136.3.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood P.W., Tsang S.V., and Osley M.A.. 1993. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:28–38. 10.1128/MCB.13.1.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha R.L., Ahn G.S., Staples M.I., Sathyan K.M., Karpova T.S., Foltz D.R., and Basrai M.A.. 2017. Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells. Oncotarget. 8:46781–46800. 10.18632/oncotarget.18108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A.C., Zhou J.C., Perera R.L., van Deursen F., Evrin C., Ivanova M.E., Kilkenny M.L., Renault L., Kjaer S., Matak-Vinković D., et al. 2014. A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature. 510:293–297. 10.1038/nature13234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R.K., Liang D., Gajjalaiahvari U.R., Kabbaj M.-H.M., Paik J., and Gunjan A.. 2010. Excess histone levels mediate cytotoxicity via multiple mechanisms. Cell Cycle. 9:4236–4244. 10.4161/cc.9.20.13636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitbon D., Podsypanina K., Yadav T., and Almouzni G.. 2017. Shaping Chromatin in the Nucleus: The Bricks and the Architects. Cold Spring Harb. Symp. Quant. Biol. 82:1–14. 10.1101/sqb.2017.82.033753 [DOI] [PubMed] [Google Scholar]
- Smith S., and Stillman B.. 1989. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 58:15–25. 10.1016/0092-8674(89)90398-X [DOI] [PubMed] [Google Scholar]
- Soldati D., and Schümperli D.. 1988. Structural and functional characterization of mouse U7 small nuclear RNA active in 3′ processing of histone pre-mRNA. Mol. Cell. Biol. 8:1518–1524. 10.1128/MCB.8.4.1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector M.S., Raff A., DeSilva H., Lee K., and Osley M.A.. 1997. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol. Cell. Biol. 17:545–552. 10.1128/MCB.17.2.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit A., Koning T.W., Soldati D., Melin L., and Schümperli D.. 1993. Variable effects of the conserved RNA hairpin element upon 3′ end processing of histone pre-mRNA in vitro. Nucleic Acids Res. 21:1569–1575. 10.1093/nar/21.7.1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom A.R., Emelyanov A.V., Mir M., Fyodorov D.V., Darzacq X., and Karpen G.H.. 2017. Phase separation drives heterochromatin domain formation. Nature. 547:241–245. 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm D., Witt H., Hovestadt V., Khuong-Quang D.-A., Jones D.T.W., Konermann C., Pfaff E., Tönjes M., Sill M., Bender S., et al. 2012. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 22:425–437. 10.1016/j.ccr.2012.08.024 [DOI] [PubMed] [Google Scholar]
- Sullivan E., Santiago C., Parker E.D., Dominski Z., Yang X., Lanzotti D.J., Ingledue T.C., Marzluff W.F., and Duronio R.J.. 2001. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 15:173–187. 10.1101/gad.862801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K.F., Hechenberger M., and Masri K.. 1994. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 127:581–592. 10.1083/jcb.127.3.581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K.D., Steiniger M., and Marzluff W.F.. 2009. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol. Cell. 34:322–332. 10.1016/j.molcel.2009.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Clermont P.L., Jiao W., Helgason C.D., Gout P.W., Wang Y., and Qu S.. 2016. Elevated expression of the centromere protein-A(CENP-A)-encoding gene as a prognostic and predictive biomarker in human cancers. Int. J. Cancer. 139:899–907. 10.1002/ijc.30133 [DOI] [PubMed] [Google Scholar]
- Sutton A., Bucaria J., Osley M.A., and Sternglanz R.. 2001. Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription. Genetics. 158:587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H., Ray-Gallet D., Almouzni G., and Nakatani Y.. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 116:51–61. 10.1016/S0092-8674(03)01064-X [DOI] [PubMed] [Google Scholar]
- Talbert P.B., and Henikoff S.. 2017. Histone variants on the move: substrates for chromatin dynamics. Nat. Rev. Mol. Cell Biol. 18:115–126. 10.1038/nrm.2016.148 [DOI] [PubMed] [Google Scholar]
- Tang M.C.W., Jacobs S.A., Mattiske D.M., Soh Y.M., Graham A.N., Tran A., Lim S.L., Hudson D.F., Kalitsis P., O’Bryan M.K., et al. 2015. Contribution of the two genes encoding histone variant h3.3 to viability and fertility in mice. PLoS Genet. 11:e1004964 10.1371/journal.pgen.1004964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Poustovoitov M.V., Zhao K., Garfinkel M., Canutescu A., Dunbrack R., Adams P.D., and Marmorstein R.. 2006. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat. Struct. Mol. Biol. 13:921–929. 10.1038/nsmb1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatomer D.C., Rizzardi L.F., Curry K.P., Witkowski A.M., Marzluff W.F., and Duronio R.J.. 2014. Drosophila Symplekin localizes dynamically to the histone locus body and tricellular junctions. Nucleus. 5:613–625. 10.4161/19491034.2014.990860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T., Matsushita K., Yamaguchi S., Oohashi T., Shimada H., Ochiai T., Yoda K., and Nomura F.. 2003. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 63:3511–3516. [PubMed] [Google Scholar]
- Tyler J.K., Adams C.R., Chen S.R., Kobayashi R., Kamakaka R.T., and Kadonaga J.T.. 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 402:555–560. 10.1038/990147 [DOI] [PubMed] [Google Scholar]
- Van Hooser A.A., Ouspenski I.I., Gregson H.C., Starr D.A., Yen T.J., Goldberg M.L., Yokomori K., Earnshaw W.C., Sullivan K.F., and Brinkley B.R.. 2001. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114:3529–3542. [DOI] [PubMed] [Google Scholar]
- Vardabasso C., Hasson D., Ratnakumar K., Chung C.-Y., Duarte L.F., and Bernstein E.. 2014. Histone variants: emerging players in cancer biology. Cell. Mol. Life Sci. 71:379–404. 10.1007/s00018-013-1343-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E.J., Burch B.D., Godfrey A.C., Salzler H.R., Duronio R.J., and Marzluff W.F.. 2007. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol. Cell. 28:692–699. 10.1016/j.molcel.2007.10.009 [DOI] [PubMed] [Google Scholar]
- Wang I.-C., Chen Y.-J., Hughes D., Petrovic V., Major M.L., Park H.J., Tan Y., Ackerson T., and Costa R.H.. 2005. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol. Cell. Biol. 25:10875–10894. 10.1128/MCB.25.24.10875-10894.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Yu Z., Liu Y., and Li G.. 2017. Ser68 Phosphorylation Ensures Accurate Cell-Cycle-Dependent CENP-A Deposition at Centromeres. Dev. Cell. 40:5–6. 10.1016/j.devcel.2016.12.015 [DOI] [PubMed] [Google Scholar]
- Weinberg D.N., Allis C.D., and Lu C.. 2017. Oncogenic Mechanisms of Histone H3 Mutations. Cold Spring Harb. Perspect. Med. 7:a026443 10.1101/cshperspect.a026443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells D., and Kedes L.. 1985. Structure of a human histone cDNA: evidence that basally expressed histone genes have intervening sequences and encode polyadenylylated mRNAs. Proc. Natl. Acad. Sci. USA. 82:2834–2838. 10.1073/pnas.82.9.2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells D., Hoffman D., and Kedes L.. 1987. Unusual structure, evolutionary conservation of non-coding sequences and numerous pseudogenes characterize the human H3.3 histone multigene family. Nucleic Acids Res. 15:2871–2889. 10.1093/nar/15.7.2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A.E., Burch B.D., Yang X.-C., Gasdaska P.Y., Dominski Z., Marzluff W.F., and Duronio R.J.. 2011. Drosophila histone locus bodies form by hierarchical recruitment of components. J. Cell Biol. 193:677–694. 10.1083/jcb.201012077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wike C.L., Graves H.K., Wason A., Hawkins R., Gopalakrishnan J., Schumacher J., and Tyler J.K.. 2016. Excess free histone H3 localizes to centrosomes for proteasome-mediated degradation during mitosis in metazoans. Cell Cycle. 15:2216–2225. 10.1080/15384101.2016.1192728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt O., Albig W., and Doenecke D.. 1997. Transcriptional regulation of the human replacement histone gene H3.3B. FEBS Lett. 408:255–260. 10.1016/S0014-5793(97)00436-5 [DOI] [PubMed] [Google Scholar]
- Witt O., Albig W., and Doenecke D.. 1998. cAMP/phorbol ester response element is involved in transcriptional regulation of the human replacement histone gene H3.3B. Biochem. J. 329:609–613. 10.1042/bj3290609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodland H.R., and Adamson E.D.. 1977. The synthesis and storage of histones during the oogenesis of Xenopus laevis. Dev. Biol. 57:118–135. 10.1016/0012-1606(77)90359-1 [DOI] [PubMed] [Google Scholar]
- Wu R.S., and Bonner W.M.. 1981. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell. 27:321–330. 10.1016/0092-8674(81)90415-3 [DOI] [PubMed] [Google Scholar]
- Wu G., Broniscer A., McEachron T.A., Lu C., Paugh B.S., Becksfort J., Qu C., Ding L., Huether R., Parker M., et al. St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project . 2012a Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 44:251–253. 10.1038/ng.1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Qian Y.-M., Zhao X.-L., Wang S.-M., Feng X.-J., Chen X.-F., and Zhang S.-H.. 2012b Expression and prognostic significance of centromere protein A in human lung adenocarcinoma. Lung Cancer. 77:407–414. 10.1016/j.lungcan.2012.04.007 [DOI] [PubMed] [Google Scholar]
- Wu R.S., Tsai S., and Bonner W.M.. 1982. Patterns of histone variant synthesis can distinguish G0 from G1 cells. Cell. 31:367–374. 10.1016/0092-8674(82)90130-1 [DOI] [PubMed] [Google Scholar]
- Wu R.S., Tsai S., and Bonner W.M.. 1983. Changes in histone H3 composition and synthesis pattern during lymphocyte activation. Biochemistry. 22:3868–3873. 10.1021/bi00285a023 [DOI] [PubMed] [Google Scholar]
- Xue Z., Huang K., Cai C., Cai L., Jiang C.Y., Feng Y., Liu Z., Zeng Q., Cheng L., Sun Y.E., et al. 2013. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 500:593–597. 10.1038/nature12364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.C., Purdy M., Marzluff W.F., and Dominski Z.. 2006. Characterization of 3’hExo, a 3’ exonuclease specifically interacting with the 3′ end of histone mRNA. J. Biol. Chem. 281:30447–30454. 10.1074/jbc.M602947200 [DOI] [PubMed] [Google Scholar]
- Yang X.C., Burch B.D., Yan Y., Marzluff W.F., and Dominski Z.. 2009. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol. Cell. 36:267–278. 10.1016/j.molcel.2009.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.-C., Sabath I., Dębski J., Kaus-Drobek M., Dadlez M., Marzluff W.F., and Dominski Z.. 2013. A complex containing the CPSF73 endonuclease and other polyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3′-end processing. Mol. Cell. Biol. 33:28–37. 10.1128/MCB.00653-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Zhou X., Wang W., Deng W., Fang J., Hu H., Wang Z., Li S., Cui L., Shen J., et al. 2015. Dynamic phosphorylation of CENP-A at Ser68 orchestrates its cell-cycle-dependent deposition at centromeres. Dev. Cell. 32:68–81. 10.1016/j.devcel.2014.11.030 [DOI] [PubMed] [Google Scholar]
- Zhang W., Mao J.-H., Zhu W., Jain A.K., Liu K., Brown J.B., and Karpen G.H.. 2016. Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat. Commun. 7:12619 10.1038/ncomms12619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Kennedy B.K., Lawrence B.D., Barbie D.A., Matera A.G., Fletcher J.A., and Harlow E.. 2000. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 14:2283–2297. 10.1101/gad.827700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., McKillop-Smith S., and Müller B.. 2004. The human histone gene expression regulator HBP/SLBP is required for histone and DNA synthesis, cell cycle progression and cell proliferation in mitotic cells. J. Cell Sci. 117:6043–6051. 10.1242/jcs.01523 [DOI] [PubMed] [Google Scholar]
- Zhu L., and Brangwynne C.P.. 2015. Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr. Opin. Cell Biol. 34:23–30. 10.1016/j.ceb.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]