Chakrabarty et al. propose a model in which slow axonal transport of actin can occur by a biased polymerization of actin filaments along the axon shaft without the involvement of microtubules (MTs) or MT-based motors. These dynamics are distinct from polymer sliding—the canonical mechanism thought to convey cytoskeletal cargoes in slow transport.
Abstract
Classic pulse-chase studies have shown that actin is conveyed in slow axonal transport, but the mechanistic basis for this movement is unknown. Recently, we reported that axonal actin was surprisingly dynamic, with focal assembly/disassembly events (“actin hotspots”) and elongating polymers along the axon shaft (“actin trails”). Using a combination of live imaging, superresolution microscopy, and modeling, in this study, we explore how these dynamic structures can lead to processive transport of actin. We found relatively more actin trails elongated anterogradely as well as an overall slow, anterogradely biased flow of actin in axon shafts. Starting with first principles of monomer/filament assembly and incorporating imaging data, we generated a quantitative model simulating axonal hotspots and trails. Our simulations predict that the axonal actin dynamics indeed lead to a slow anterogradely biased flow of the population. Collectively, the data point to a surprising scenario where local assembly and biased polymerization generate the slow axonal transport of actin without involvement of microtubules (MTs) or MT-based motors. Mechanistically distinct from polymer sliding, this might be a general strategy to convey highly dynamic cytoskeletal cargoes.
Introduction
Actin is a key cytoskeletal protein in neurons, with established roles in axon growth and synaptic homeostasis (Gomez and Letourneau, 2014; Coles and Bradke, 2015; Kevenaar and Hoogenraad, 2015; Papandréou and Leterrier, 2018). Although some actin can be synthesized locally in response to axonal guidance cues (Jung et al., 2014), the vast majority of actin, along with the other cytoskeletal proteins, tubulin and neurofilaments (NFs), are made in the neuronal soma and conveyed into axons via slow axonal transport as shown by classic in vivo pulse-chase radiolabeling studies (Black and Lasek, 1979; McQuarrie et al., 1986; Oblinger, 1988; Tashiro and Komiya, 1992; Galbraith and Gallant, 2000; Roy, 2014). Although these studies characterized overall actin transport, underlying mechanisms remained obscure as radiolabeling methods cannot visualize cargo movement.
More recently, live imaging revealed that GFP-tagged NF polymers move rapidly but intermittently in axons, and this infrequent movement is thought to result in a slow overall movement of the population—the “Stop and Go” model (Brown, 2000; Roy et al., 2000; Wang et al., 2000; for an alternate view, see Terada et al., 1996). Although short, motile structures resembling microtubules (MTs) are also seen in axons (Wang and Brown, 2002; He et al., 2005); other studies propose transport of unpolymerized tubulin (Terada et al., 2000; Maday et al., 2014). Conceptually, these studies advocate a model in which cytoskeletal polymers assemble in the neuronal soma and are translocated into axons by motor proteins. Unfortunately, such straightforward imaging strategies have not been useful for actin transport. First, actin is more dynamic than NFs or MTs. About half of the actin in squid axons is monomeric (Morris and Lasek, 1984), and GFP-tagged actin in cultured neurons only reveals a diffuse glow (Okabe and Hirokawa, 1990). Moreover, GFP-actin may not report formin-mediated actin behaviors—relevant in our setting (Chen et al., 2012).
Using probes that selectively bind to filamentous actin, we recently visualized actin dynamics in axons of cultured hippocampal neurons (Ganguly et al., 2015). We found that actin continuously polymerizes and depolymerizes at micrometer-sized “hotspots” along the axon spaced ∼3-4 µm apart, many of which were colocalized with stationary axonal endosomes. In addition, we saw rapidly elongating actin polymers extending along the shaft (we call these “actin trails”). Actin trails were formin (but not Arp 2/3) dependent, typically originated at hotspots, and helped enrich actin at presynaptic boutons. Based on these data, we proposed a model in which axonal actin nucleates on the surface of stationary endosomes, providing the nidus for polymers elongating along the axon shaft. More recently, actin hotspots and trails were also seen in Caenorhabditis elegans axons in vivo (Sood et al., 2018). Additionally, axons have a circumferential, periodic lattice of “actin rings” that wrap underneath the plasma membrane and are thought to be much more stable (Xu et al., 2013; Zhong et al., 2014; Ganguly et al., 2015; He et al., 2016). Though actin rings likely play important functions, hotspots/trails are the only known dynamic actin assemblies in mature axons, and it seems reasonable that the latter would somehow transport actin (Dubey et al., 2018). However, this is not straightforward to conceptualize as the nature of actin assembly/disassembly, occurring on the timescale of seconds, requires one to consider the biophysics of this exchange. Using a combination of live/superresolution imaging and quantitative modeling, we show in this study that a dynamic but polarized assembly of actin in axons can indeed lead to a slow anterograde bias of the population at rates consistent with slow transport.
Results and discussion
Dynamics of local actin assembly and polymer elongation in axon shafts
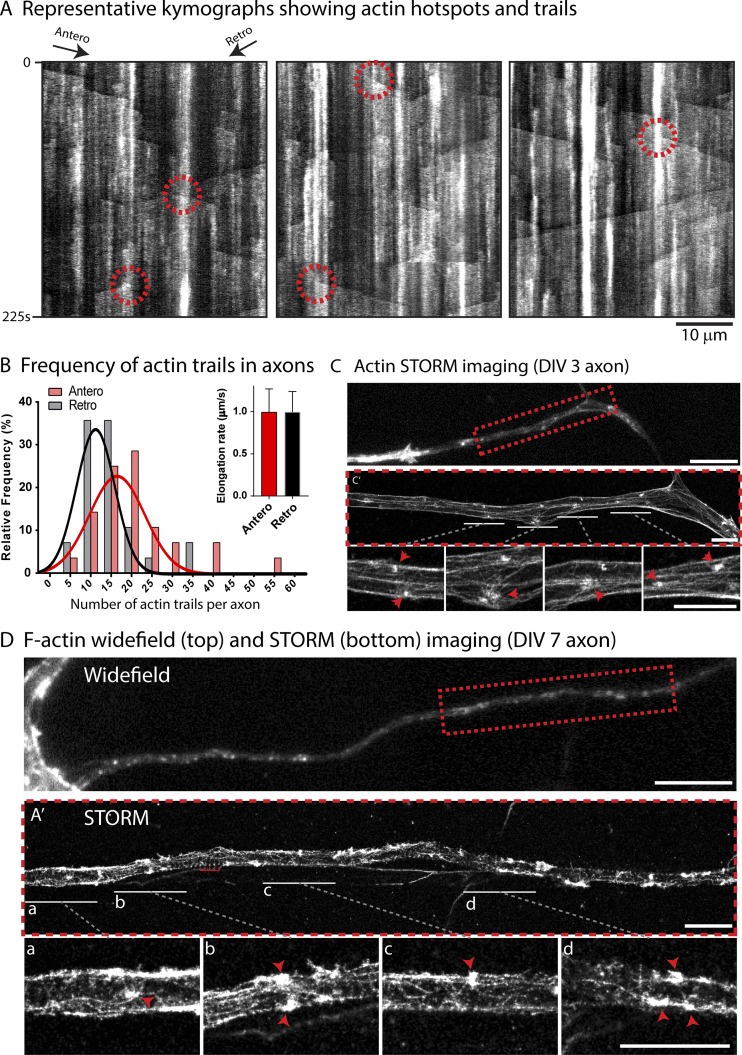
Kymographs in Fig. 1 A show examples of axons transfected with GFP:Utr-CH (GFP bound to the calponin homology domain of utrophin), a probe that selectively labels actin filaments (Burkel et al., 2007; see also Ladt et al., 2016). Note two key features: (1) repeated assembly/disassembly of actin (vertical interrupted lines; hotspots); and (2) bidirectionally elongating actin polymers appearing as diagonal plumes (actin trails). Also note that actin trails often originate from hotspots (Fig. 1 A; Ganguly et al., 2015). Interestingly, though the average elongation rate of actin trails was similar in both directions, the frequency of anterogradely elongating actin filaments was slightly higher (∼58% elongated anterogradely; Fig. 1 B).
Figure 1.
Dynamics and organization of actin in axon shafts. (A) Representative kymographs from axons transfected with GFP:Utr-CH to label filamentous actin. Note vertical “on/off” events representing dynamic local assembly (hotspots) and diagonal plumes of fluorescence representing bidirectionally elongating actin polymers (trails). Also note that actin trails often emerge from where the hotspots are situated (some marked by dashed circles; see also Ganguly et al., 2015). (B) Anterograde and retrograde frequency of actin trails (histogram) and polymer elongation rate (top right graph). Note the slightly higher frequency of anterograde trails (58/42%; mean elongation rates were 0.99 ± 0.001 µm/s [mean ± SEM] in both directions; data are from ∼1,000 total events; adapted from Ganguly et al., 2015). (C) Axon from a DIV3 neuron labeled with phalloidin (C′ shows STORM image of region within dashed red box). Note clusters of actin with radiating filaments (arrowheads in zoomed insets below) likely corresponding with actin hotspots seen by live imaging. (D) Axon from a DIV7 neuron labeled with phalloidin (A′ shows STORM image of region within dashed red box). Note clusters of actin and radiating filaments (red arrowheads in zoomed insets below; regions labeled a–e; actin rings marked by small comb in A′). Bars: 10 µm (C and D); 2 µm (A′, C′, and magnifications).
An emerging concept from these experiments is that axon shafts have submicrometer zones where actin filaments are nucleated. Using stochastic optical reconstruction microscopy (STORM) in younger axons that have relatively thicker profiles, we saw discrete clusters of actin with aster-like radiating actin filaments (Fig. 1 C) that may represent the hotspots seen by live imaging. Similar actin-asters are seen in more mature axons as well (Figs. 1 D and S1, A and B). Quantitative STORM data indicate that the average diameter of these actin-asters is ∼190 nm (Fig. S1 C). Interestingly, many of these actin-asters appear close to the axonal plasma membrane, a feature that cannot be appreciated by diffraction-limited microscopy, suggesting that there may be anatomical and/or mechanistic links between actin rings and trails.
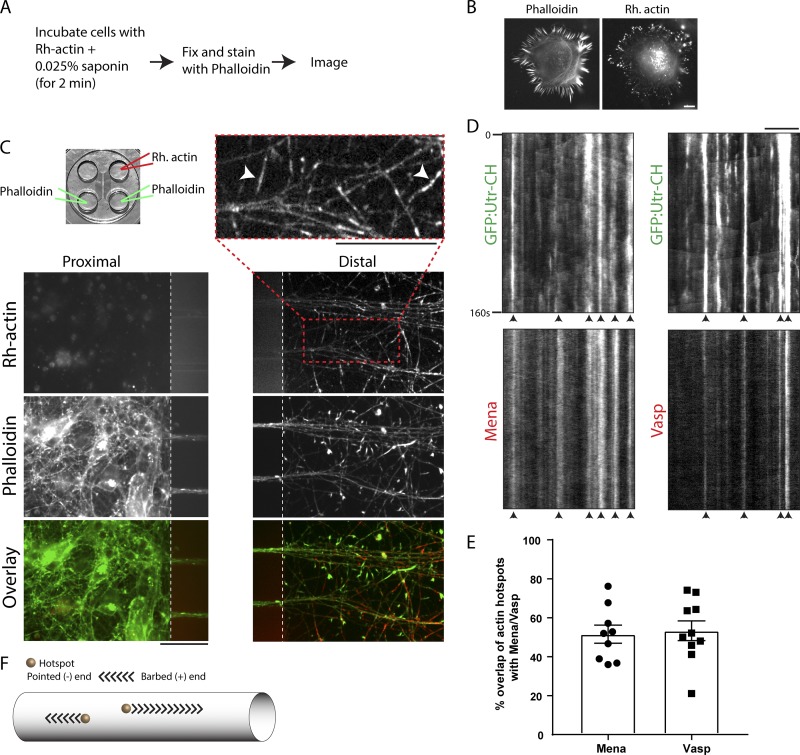
Next, we visualized actin nucleation in axons using an established method to label newly incorporated actin barbed ends (see Materials and methods and Fig. 2 A; Symons and Mitchison, 1991; Marsick et al., 2010; Marsick and Letourneau, 2011). Consistent with previous studies showing that barbed ends of actin filaments in filopodia are located at the tip of the filopodia (Mejillano et al., 2004), we saw preferential incorporation of labeled monomers along filopodial tips in CathA-differentiated (CAD) cells (Fig. 2 B). We adapted the technique to microfluidics where axon shafts can be physically and fluidically isolated from somatodendritic domains. Rhodamine-labeled monomers were only added to the axonal chamber (Fig. 2 C); thus, any labeling in axons is due to local monomer assembly (and not transport/diffusion from the soma/dendrites). Indeed, there was extensive rhodamine-actin labeling in axons (Fig. 2 C). Furthermore, barbed end–binding proteins Mena/Vasp (Breitsprecher et al., 2011) were localized to the hotspots (Fig. 2, D and E), suggesting that actin trails elongate bidirectionally with their barbed ends facing the hotspots (see schematic in Fig. 2 F).
Figure 2.
Labeling of newly incorporated barbed ends in axon shafts. (A) Protocol to label newly incorporated actin barbed ends. (B) Representative images of CAD cells after barbed end labeling. Note newly incorporated barbed ends at filopodial tips. (C) Top left: Microfluidic device to separate axons from somatodendritic compartments. Rhodamine (Rh)-actin was loaded only into the axonal chamber, and phalloidin staining was performed on both chambers as described in Materials and methods. Bottom: Rhodamine-actin and phalloidin labeling in somatodendritic (left) and axonal chambers (right). Note numerous small rhodamine-actin puncta in axons, indicating local incorporation of monomers (top right image). Also note some elongated structures (arrowheads in top right image) likely representing the rhodamine label incorporated into elongating actin filaments. (D and E) Kymographs from axons cotransfected with GFP:Utr-CH and mCherry:Mena or mCherry:Vasp and imaged near simultaneously. Note the colocalization of actin hotspots with Mena/Vasp (some marked with arrowheads). Colocalization is quantified in E. The plot depicts mean ± SEM. Bars: 10 µm (A and D); 50 µm (C). (F) Schematic showing actin trails elongating with their barbed ends facing the hotspots.
Biased anterograde flow of axonal actin
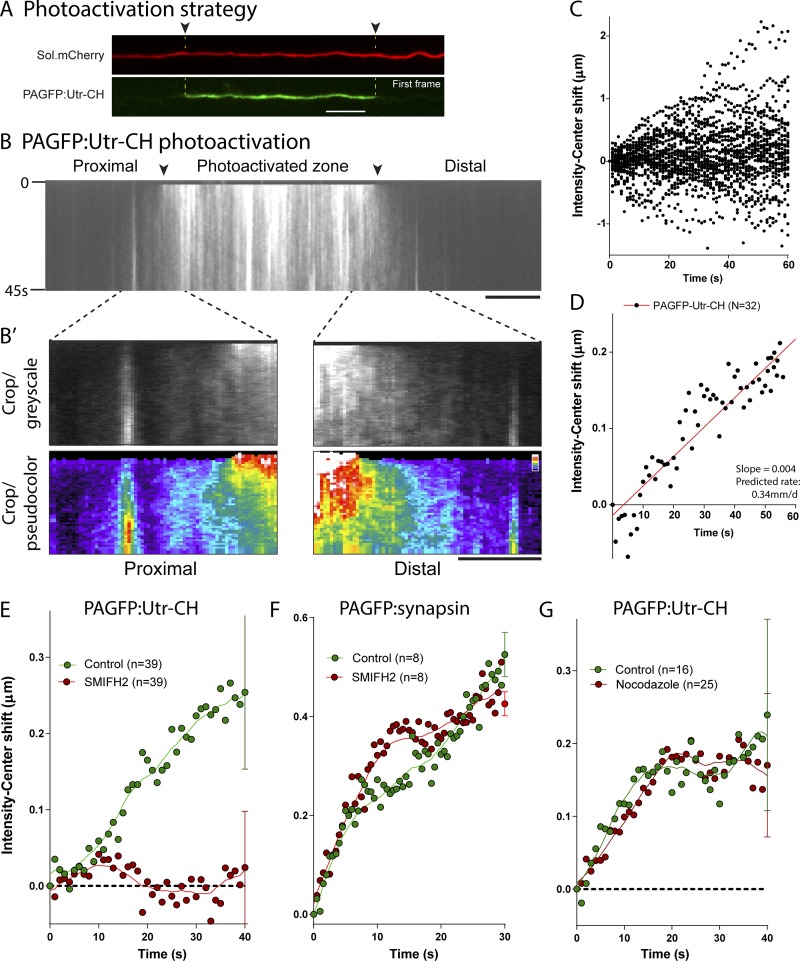
We and others have developed imaging assays to visualize slow axonal transport in cultured neurons and squid axons (Terasaki et al., 1995; Terada et al., 2000, 2010; Scott et al., 2011; Tang et al., 2013; Ganguly et al., 2017). In our experiments, cytosolic proteins were tagged to photoactivatable GFP (PAGFP) and transfected in cultured hippocampal neurons. Thereafter, a discrete pool was photoactivated in the axon, and the resultant fluorescent pool was tracked by live imaging. Dispersion of fluorescence is biased toward the axon tip, which can be quantified by analyzing the shift in the center of fluorescence over time (“intensity-center shift”). The biased movement is energy dependent and distinct from the rapid and unbiased diffusion of PAGFP alone (Scott et al., 2011). Using this assay with PAGFP:Utr-CH, we found a slow anterograde bias of the photoactivated actin population as shown in Fig. 3 (B, B′, and C). The overall rate of egress was ∼0.34 mm/d (Fig. 3 D), in line with previous pulse-chase radiolabeling research in central nervous system neurons (∼0.4 mm/d; see Oblinger, 1988). Interestingly, in squid axons, dispersion of injected rhodamine-phalloidin–labeled actin filaments was also anterogradely biased, although these observations were qualitative (Terasaki et al., 1995).
Figure 3.
Anterograde bias of actin in axons. (A) Axon cotransfected with PAGFP:Utr-CH and soluble mCherry. Note that a discrete ROI is photoactivated (green), and the mobility of the fluorescent pool is analyzed over time. (B and B′) Kymograph from a PAGFP:Utr-CH photoactivation experiment. The photoactivated zone is marked by arrowheads, and elapsed time in seconds is shown on left. (B′) Zoomed insets (grayscale and pseudocolor) highlight the anterogradely biased dispersion of axonal actin (note the higher signal in distal axon segment). (C and D) Raw (C) and mean (D) intensity center shifts (see Materials and methods) reveal a slow, anterograde bias of the actin population. (E and F) Intensity center shifts of PAGFP:Utr-CH and PAGFP:synapsin in axons treated with the formin inhibitor SMIFH2 (Rizvi et al., 2009). Note that while formin inhibition blocks the biased transit of actin, there is no effect on synapsin transport. (G) Anterograde bias of actin was unaffected by interruption of MT-based transport by nocodazole (10 µg/ml). The solid curves are smoothened fits of the data. Bars, 10 µm.
Like actin trails, the slow anterograde bias of actin in our photoactivation experiments was also formin dependent (Fig. 3 E; see also Fig. S2 A). Interestingly, the biased transport of the cytosolic cargo PAGFP:synapsin (Scott et al., 2011; Tang et al., 2013; Ganguly et al., 2017) was not formin dependent (Fig. 3 F), and there were also qualitative differences in kymographs from photoactivation of these two proteins in axons (Fig. S2 B). Finally, disruption of MTs did not have any effect on the anterograde bias of actin (Figs. 3 G and S2, C and D), in line with our previous observations that axonal actin hotspots and trails were independent of MTs (Ganguly et al., 2015). Since the anterogradely biased dispersion of synapsin is MT dependent (Scott et al., 2011; Tang et al., 2013), collectively, the data suggest two distinct mechanisms by which cytosolic proteins are conveyed in axons. Given the heterogeneity of cytosolic cargoes moving in slow transport, it has been long speculated that diverse mechanisms were at play, and our data support this idea.
Simulation of axonal actin hotspots and trails
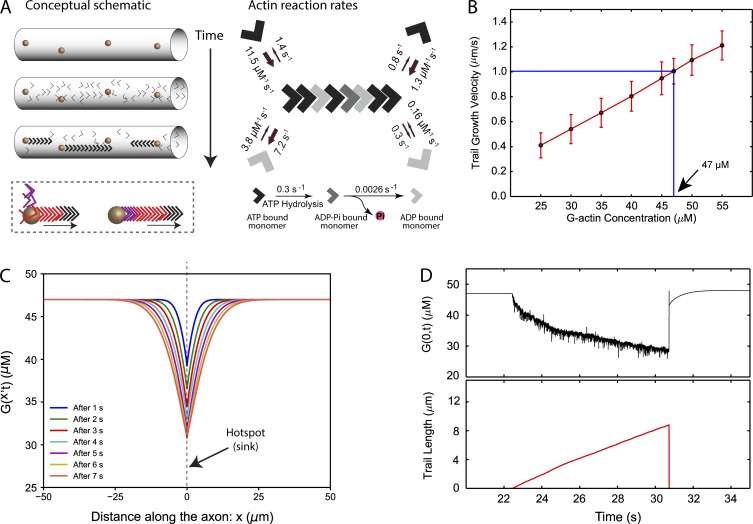
The model emerging from our preceding experiments is: (1) axons have discrete foci where actin monomers are nucleated (hotspots), (2) actin filaments elongate bidirectionally from the hotspots, with barbed ends facing the hotspots, and (3) actin has an overall slow anterograde bias in axons. Given that both hotspots/trails dynamics and slow actin transport share key characteristics (formin dependent and MT independent), we hypothesized that hotspots and trails drive the slow transport of actin along axons. Mechanistically, complex phases of actin monomer/polymer exchange would lead to periodic hotspot assembly and disassembly along with elongation/collapse of trails. To test this hypothesis, we designed a robust simulation of actin hotspots and trails in axons and then performed virtual photoactivation experiments. The simulations employed established biophysical principles of actin monomer/filament assembly, incorporating parameters from our imaging data whenever possible. Specifically, multiple virtual actin hotspots were allowed to originate linearly along a hypothetical axon cylinder (axon thickness and distance between the nucleating zones was based on imaging data from Ganguly et al. [2015]), with polymers extending from these hotspots. A schematic of the modeling is shown in Fig. 4 A (left). Note that monomers nucleate with their barbed ends facing the hotspots, with polymers extending in both anterograde and retrograde directions. Also note that in this scenario, addition of new monomers at the barbed ends of the elongating trails will lead to translocation of individual monomers toward the pointed end of the growing filament (Fig. 4 A, left, dashed inset at bottom; see also Video 1).
Figure 4.
Modeling of axonal actin hotspots and trails. (A) Left: Stationary hotspots (yellow spheres) are localized uniformly along the axon. Actin trails nucleate on the hotspots, with barbed ends facing the hotspots. Note that as new monomers (colored arrowheads) are incorporated at the barbed end, there is progressive translocation of individual monomers toward the pointed end (bottom box). Right: Known assembly, disassembly, and summary of reaction rates of actin. (B) Basal actin monomer concentration in the axon. Average elongation velocity of actin trails increases linearly (red line) with increase in basal monomer concentration. At a basal concentration of 47 µM, the average elongation velocity matches the experimentally observed average trail elongation velocity of ∼1 µm/s (blue lines). (C) Actin monomer concentration profiles along the axon during the growth of an actin trail. Multiple colored lines show declining concentration of actin monomers along the axon shaft at successive time points after trail nucleation. Note that the hotspot is located at x = 0, and the trail grows with its barbed end attached to this location. The trail collapses ∼9 s after nucleation, growing to a length of 8.8 µm. This wide concentration gradient drives in monomer from regions of higher concentration toward the hotspot. (D) Actin monomer depletion during trail elongation. An actin trail is randomly nucleated at t = 22.4 s, the trail grows (bottom, red trace), and monomer depletion from the barbed end (top, black trace) slows down the rate of trail elongation. Note that fluctuations of actin-monomer concentration reflect monomers randomly attaching/detaching from the barbed end. As the trail collapses at t = 30.7 s, actin-monomer concentration spikes back up.
The on/off kinetics of actin in our model is in accordance with known biophysical properties of monomer/polymer exchange. Briefly, actin monomers can bind to either ATP or ADP in its cleft, with distinct association/dissociation rates from the ends of actin polymers (Pollard, 1986). In the filaments, ATP-actin can hydrolyze irreversibly to form ADP-Pi-actin (Carlier et al., 1988), which undergoes Pi release to form ADP-actin (Carlier and Pantaloni, 1986). The known association, dissociation, and hydrolysis rate constants of ATP-actin and ADP-actin are summarized in Fig. 4 A (right; see Carlier and Pantaloni, 1986; Pollard, 1986; Blanchoin and Pollard, 2002). The monomer pool of G-actin is known to be predominantly ATP bound (Sun et al., 1995; thus, we assume that the monomer pool of G-actin is only ATP bound). There are two main reasons for this: (1) a high intracellular ATP/ADP ratio, which ensures that the basal ratio of ATP–G-actin/ADP–G-actin is nearly 20:1 (Atkinson et al., 2004; Tantama et al., 2013), and (2) actin-binding proteins such as profilin and thymosin-β4, which catalyze the exchange of ADP–G-actin to ATP–G-actin so that even a small fraction of ADP-bound monomers that can form by hydrolysis of ATP–G-actin are rapidly converted back (Goldschmidt-Clermont et al., 1992; Didry et al., 1998).
Once bound to the filament, subunits in our model randomly undergo hydrolysis at a rate of 0.3/s and release the phosphate group at a rate of 0.0026/s (Carlier and Pantaloni, 1986). It is known that actin filaments can linearly grow up to 17.7 µm before they start bending (called persistence length; see Gittes et al., 1993). Since the lengths of the axonal actin trails is much smaller (average 8.8 µm; see Ganguly et al., 2015), we modeled them as one-dimensional (1D) linear filaments elongating along the axon shaft (also note that they appear linear by live imaging; see Fig. 1 A for an example). Given that the typical diameter, d, of an axon visualized in our imaging experiment is ∼200 nm, the maximum angle of an elongating actin trail (of length Lt) with the long axis of the axon is only ~1° (Θ = tan−1(d/Lt) = 1.3° for a typical trail length of 8.8 µm), justifying a 1D mathematical model of axonal actin trails.
The nucleation of actin trails and their subsequent elongation by incorporating monomers from the axonal pool was modeled by Markov processes. Nucleation rates of anterograde and retrograde trails were calculated from imaging research of GFP:Utr-CH (Ganguly et al., 2015). Specifically, the average nucleation rate (rn) of anterograde and retrograde trails (for a given hotspot) was rn,a = 0.001885/s and rn,r = 0.001381/s, respectively (see Eq. 1 in Materials and methods). Probabilities of trail nucleation and of competing association and dissociation reactions were calculated at each time step using the nucleation rates rn,a and rn,r and established reaction rates. The effect of incorporating actin monomers from the axonal pool into the elongating trails and the subsequent release of actin back into the axonal pool was modeled using a 1D diffusion equation with sinks and sources. Based on previous research, the diffusion coefficient of actin monomers was assumed to be 6 µm2/s (McGrath et al., 1998). Using imaging data, we estimated that a monomer concentration of 47 µM would be needed to sustain the polymerization at this rate (see Fig. 4 B). Though actin concentration in cultured hippocampal axons is unknown, our estimate is within the range for other cell types (for example, concentration of monomeric actin in chick embryonic neurons is reported to be 30–37 µM; see Devineni et al., 1999). For more details of the simulation, see Materials and methods and Fig. S3 A.
In our imaging experiments, actin trails invariably collapsed after elongating for a few micrometers (Fig. 1 A; Ganguly et al., 2015). In our simulations, however, if unchecked, the trails would probably grow indefinitely, indicating that the collapse is not a simple consequence of monomer depletion but is mediated by yet-unknown mechanisms. To more closely mimic the actual experimental data, we forced the simulated elongating trails to collapse after reaching predetermined lengths (determined from the distribution range of polymer length data from Ganguly et al. [2015]; average 8.8 µm). The mean lifetime of trails is τmean = 8.8 s, and the mean rate of cessation of filaments is
Fig. 4 (C and D) highlights the kinetic monomer/polymer exchange in our model when an actin trail elongates and collapses. Note that as the filament grows, the monomer pool at the hotspot is depleted, supplying subunits to the elongating trail (Fig. 4 C). The dynamic actin subunit/polymer in our model is shown in Fig. 4 D. In this model, note that the actin monomer pool (black trace) is depleted as the filament grows over a few seconds (red trace). Actin depletion was also seen in denser actin networks such as lamellipodia, although the rate of depletion was severalfold higher (Boujemaa-Paterski et al., 2017). For molecular simulations of the monomer/polymer exchange and elongation/collapse of actin trails, see Videos 1 and 2.
Simulation of axonal actin photoactivation experiments
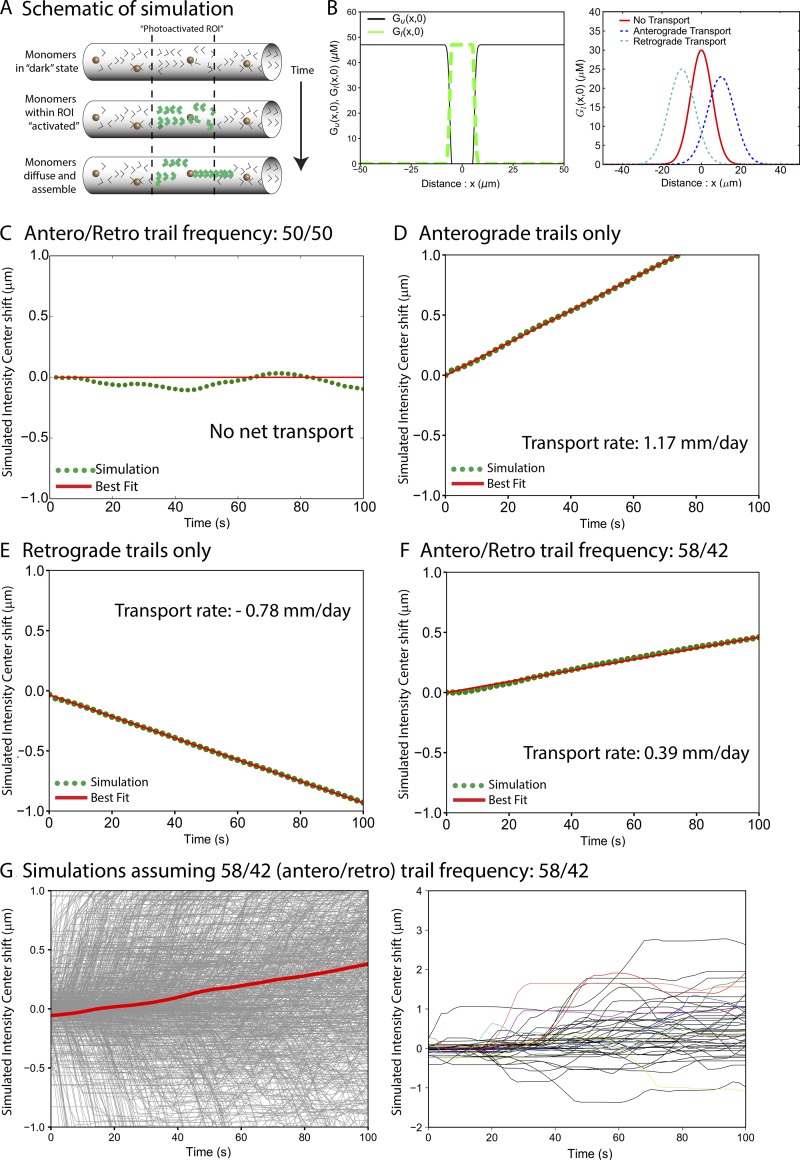
To determine whether the axonal actin dynamics (hotspots/trails) can generate an overall biased egress of the population, we performed virtual photoactivation experiments in axons with simulated hotspots and trails and asked whether there was a shift in the resultant virtual intensity center shift (as in our actual imaging experiments). In these simulations, all axonal actin monomers and filaments—nucleating and elongating according to the abovementioned model parameters—were initially considered to be in the “dark state.” Thereafter, actin in a 15-µm axon segment was converted to an activated state mimicking photoactivation (see schematic in Fig. 5 A). Distribution of the photoactivated actin monomer population for an axon is shown in Fig. 5 B (left). The virtual photoactivated actin population was subsequently tracked as it diffused along the axon, occasionally translocating when monomers got incorporated into the actin trails (see Materials and methods for more details). A schematic of the expected anterograde or retrograde intensity center shifts from simulated photoactivation experiments is depicted in Fig. 5 B (right).
Figure 5.
Virtual photoactivation paradigm to evaluate contribution of actin hotspots/trails to slow transport. (A) Simulation of photoactivation imaging paradigm. Initially, all actin is in monomeric state (top). After equilibration of the system, axonal actin within a 15-µm region is virtually “activated” (middle; green arrowheads represent photoactivated actin). Subsequently, the fluorescent actin either diffuses or translocates after incorporation into trails (bottom; see Materials and methods for more details). (B) Left: Labeled (green dotted line) and unlabeled (black solid line) actin concentration profiles along the axon shortly after photoactivation. Right: Expected outcomes of photoactivation simulations: For no net transport, the center of the labeled G-actin distribution is expected to be at its initial position (red line). For net anterograde or retrograde transport, the center is expected to move anterogradely (blue line) or retrogradely (cyan line), respectively. (C) Simulated intensity center shift for equal anterograde and retrograde trail nucleation rates. The center of fluorescent G-actin (green points) fluctuated about its initial position x = 0 as indicated by the red line. Results shown were averaged over 100 simulation runs. (D) Simulated intensity center shift when only anterograde trails were nucleated. The center of fluorescent G-actin (green points) moved anterogradely at a rate of 0.0135 µm/s (1.17 mm/d). Results were averaged over 1,000 simulation runs and fitted to a straight line (red line). (E) Simulated intensity center shift when only retrograde trails were nucleated. The center of fluorescent G-actin (green points) moved retrogradely at a rate of −0.009 µm/s (−0.78 mm/d). Results were averaged over 1,000 simulation runs and fitted to a straight line (red line). (F) Simulated intensity center shift for 58/42% bias of anterograde–retrograde trail frequencies. Center of fluorescent G-actin (green points) moved anterogradely at a rate of 0.0045 µm/s (0.39 mm/d). Results were averaged over 1,000 simulation runs and fitted to a straight line (red line). (G) Individual runs of simulated pulse-chase experiment (without averaging). Left: 1,000 individual simulation runs (gray lines) and the averaged (red line) intensity center shift. Right: 50 individual runs of the pulse-chase experiment.
Fig. 5 (C–F) shows cumulative average shifts from simulations, where we altered the number of elongating anterograde and retrograde trails (frequency) in axons. As expected, when the number of elongating anterograde and retrograde trails was equal (i.e., a 50/50 frequency), there was no net transport (Fig. 5 C; 100 simulations). If only anterograde trails were allowed, the entire population was transported anterogradely at a rate of 0.0135 µm/s or 1.17 mm/d (Fig. 5 D; 1,000 simulations). If only retrograde trails were allowed, the actin population had an overall retrograde bias at a rate of −0.009 µm/s or −0.78 mm/d (Fig. 5 E; 1,000 simulations). However, an anterograde/retrograde bias of 58/42% as seen in our imaging data led to an overall transport rate of 0.0046 µm/s or 0.39 mm/d (Fig. 5 F; 1,000 simulations). Note that the transport rates from these virtual experiments are reminiscent of the data from our real-imaging experiments (Fig. 3; rate = 0.004 µm/s or 0.34 mm/d) and are also comparable with rates determined by pulse-chase radiolabeling of central nervous system neurons in vivo (for example, mean actin transport rates in mouse corticospinal neurons was ∼0.4 mm/d; Oblinger, 1988). Raw intensity center shifts from all 1,000 simulations (the “58/42 ratio” group) are shown in Fig. 5 G (left). Fig. 5 G (right) shows 50 random intensity center shift patterns from the entire dataset. The stochastic pattern of intensity center shift seen in both real and virtual datasets (compare Fig. 3 C with Fig. 5 G, right) is likely due to arbitrary photoactivation of anterograde/retrograde trails that happened to be elongating in the photoactivated zone at the time of activation.
Finally, we also examined effects of actin-disrupting drugs in our virtual intensity center–shift simulations. For these studies, we first took the data from our previous experiments where we added latrunculin A and SMIFH2 to axons and then examined various actin trail dynamics (trail frequency, elongation length, and elongation rate) before and after adding these drugs (Ganguly et al., 2015). We then added these altered parameters to our virtual transport-simulation model, asking how the overall anterogradely biased flow of actin would change in response to these pharmacologic agents (running hundreds of simulations for each condition). As shown in Fig. S3 B, both latrunculin and SMIFH2 led to an attenuation of the slow anterograde bias of actin in these simulations, further underlining the critical importance of the hotspots and trails in generating the overall anterogradely biased transit of actin in axons.
Biased polymerization: An unusual mode of cytoskeletal transport
Though the form in which cytoskeletal proteins are transported—monomer or polymer—was heavily debated in the 1990s (Baas and Brown, 1997; Hirokawa et al., 1997), axonal transport of NF polymers has now been unequivocally demonstrated at least in cultured neurons (Yan and Brown, 2005). However, mechanisms underlying the axonal transport of actin have been mysterious for decades (Galbraith and Gallant, 2000). One striking difference between actin and other axonal cytoskeletal proteins is the highly dynamic nature of actin. For instance, NFs are extremely long-lasting polymers with half-lives of months to years (Barry et al., 2007), and neuronal MTs are also quite stable (Baas et al., 1991). In contrast, actin at the axonal growth cone is extremely dynamic (Gallo and Letourneau, 2004), and our data also indicate a remarkable dynamism in the axon shaft as well (Figs. 1 and 2; see also Ganguly et al., 2015).
Nevertheless, given the precedence of polymer transport in axons, we began these experiments expecting to see actin filaments translocate in axons. However, we were surprised to see unusual actin dynamics such as hotspots and trails along with elongating (not translocating) actin polymers, which eventually led us to this unusual model. Collectively, our imaging and modeling data advocate a new mode of cytoskeletal transport in which biased assembly and polymerization can lead to a slow anterograde movement of the population without the involvement of MTs or MT-based motors. Note that this biased polymerization model is fundamentally distinct from the Stop and Go model as there is no polymer sliding. In hindsight, most prevailing models of axonal transport do not take dynamic assembly/disassembly into account. Though this may have minimal consequences for stable cytoskeletal elements, monomer/polymer exchange is likely to have dramatic consequences for the transport for unstable cytoskeletal proteins such as actin. We propose that other dynamic cytoskeletal proteins in neurons (and perhaps also nonneuronal cells) might employ similar strategies for translocation. Polarized assembly of actin is also seen in other contexts, and the precise molecular mechanisms creating such bias are generally unclear (Allard and Mogilner, 2013). The underlying reason for the polarized elongation of actin trails in axons is also unclear, and an inventory of the molecules involved in triggering the hotspots/trails is likely necessary to solve this puzzle.
Materials and methods
DNA constructs, cell cultures, and transfections
GFP:Utr-CH and PAGFP:Utr-CH were gifts from W. Bement (University of Wisconsin, Madison, WI). PAGFP, GFP:synapsin-Ia, synaptophysin:mRFP, mCherry:Mena, and mCherry:Vasp constructs were gifts from G. Patterson and J. Lippincott-Schwartz (National Institutes of Health [NIH], Bethesda, MD), G. Augustine (Nanyang Technological University, Seoul, South Korea), L. Laganado (Medical Research Council, Cambridge, UK), and Stephanie Gupton (University of North Carolina, Chapel Hill, NC), respectively. All mouse procedures were approved by the University of Wisconsin Committee on Animal Care and Use and were in line with NIH guidelines. Hippocampal cultures were obtained from brains of postnatal (P0–P1) CD-1 mice (either sex) and plated on 35-mm glass-bottomed dishes (MatTek) as previously described (Ganguly and Roy, 2014). In brief, the glass-bottomed dishes were coated with 100 µl of 1 mg/ml poly-d-lysine in 0.1 M borate buffer for 2 h at room temperature, washed thrice with double-distilled H2O, and air dried. Hippocampi from P0–P1 mice were dissected in ice-cold dissection buffer (HBSS buffer with 4.44 mM d-glucose and 6.98 mM Hepes, pH 7.3) and incubated in 0.25% Trypsin-EDTA for 15 min in a 37°C water bath. Hippocampi were transferred into a blocking medium (30% FBS and 1× PBS) followed by plating medium (neurobasal medium supplemented with 10% FBS, 2% B27, and 1% GlutaMAX). The hippocampi were dissociated in the plating medium and plated at a density of 25,000 neurons/cm2. Neurons were allowed to mature for 7–9 d in 2 ml neurobasal/B27 media (neurobasal media supplemented with 2% B27 and 1% GlutaMAX) in an incubator at 37°C and 5% CO2. Neurons were transfected with indicated constructs with Lipofectamine 2000/3000 (Thermo Fisher Scientific). For GFP-UtrCH construct, 0.3 µg DNA was used, while for all other constructs, 1.2 µg DNA was used. 12–16 h after transfection, neurons were transferred to an imaging buffer (Hibernate-E-Low fluorescence media [Brainbits] supplemented with 2% B27, 2 mM GlutaMAX, 0.4% d-glucose, and 37.5 mM NaCl) and imaged at 35.5–37°C (on a heated stage chamber; model STEV; World Precision Instrument, Inc.).
Imaging and image analyses
Live-imaging experiments were performed on an inverted epifluorescence microscope (Eclipse Ti-E; Nikon) equipped with CFI Plan Apochromat VC 100× 1.40 NA oil (Nikon) objectives. An electron-multiplying charge-coupled device camera (QuantEM:512SC; Photometrics) or a Scientific CMOS camera (Prime95B; Photometrics) and a LED illuminator (SPECTRA X; Lumencor) were used for most experiments. For imaging axonal actin, low-GFP:UtrCH–expressing neurons were selected using criteria specified by Ganguly et al. (2015) and Ladt et al. (2016). Axons were identified by morphology, and only neurons with unambiguously identified axons were selected for imaging (Roy et al., 2011; Ganguly and Roy, 2014). Axonal actin was imaged at 20% LED power (400 ms exposure) at a rate of one frame per second for a total duration of 10 min. For near-simultaneous dual-color imaging, exciting LED lights were rapidly switched (within microseconds) using the SPECTRA X LED illuminator. A dual-emission filter cube (Chroma Technology Corp) was used to collect GFP/RFP emission with subpixel registration. Photoactivation was done using a previously described setup (Roy et al., 2011) or using Mosaic3 device (Andor Technology) attached to an Eclipse Ti-E epifluorescence microscope. Briefly, a photoactivation region of interest (ROI) was selected along axons, the PAGFP:Utr-CH was photoactivated for 250 ms using 405-nm diode laser light, and the GFP fluorescence was imaged at one frame per second (with the 100× oil objective). For soluble PAGFP photoactivation, neurons were photoactivated for 30 ms, and GFP fluorescence was at imaged every 600 ms. For PAGFP:Synapsin, photoactivation was done for 1 s using a setup described by Roy et al. (2011), and postphotoactivation images were acquired every 500 ms. Kymographs were generated using the Kymograph function in the MetaMorph software (Molecular Devices). The intensity-center assay was performed using script written in Python by P. Dubey (based on the formula described by Roy et al. [2011] and Scott et al. [2011]). Briefly, after photoactivation, the videos were background corrected, and the photoactivated ROI was cropped. Kymographs were generated along the axon in the photoactivated ROI, and each kymograph was fed into the Python script to calculate center of mass for each time frame. Center of mass from first frame was subtracted from subsequent time frames to calculate shift in center of mass. As large puncta in images skew the intensity center, kymographs containing multiple large puncta were not taken into account for the analysis. For pharmacological inhibition experiments, neurons were treated with 30 µm SMIFH2 (Tocris Biosciences) for 30 min, 10 µg/ml nocodazole (Life Technologies) for 30 min, or 5 µm vincristine (Life Technologies) for 1 h, and the appropriate concentration of DMSO was used as a control.
Actin barbed end labeling in microfluidics
Neurons were grown in a microfluidics device (RD450; Xona Microfluidics) per the manufacturer’s instructions with a few modifications. New microfluidic devices were washed with 1% Alconox, followed by thorough washing with double-distilled H2O. Thereafter, devices were incubated with 70% ethanol for 30 min, washed with sterile distilled water, and air dried. The devices were then UV sterilized for 15 min. Meanwhile, 60-mm glass-bottomed MatTek dishes with 20-mm microwell diameter were coated with 1 µg/ml poly-d-Lysine (Sigma-Aldrich) and 10 µg/µl laminin (Sigma-Aldrich) for 2 h, washed, and air dried. The UV-sterilized microfluidic devices were kept in the microwell of the 60-mm glass-bottomed dish. Before neurons were plated on the device, both main chambers of the device were equilibrated with neurobasal/B27 media (supplemented with 2% B27 and 1% GlutaMAX) for 5 min. Subsequently, neurobasal medium was removed, and ∼400,000 hippocampal neuronal cells were plated on one side of the chamber (somatodendritic chamber). By day in vitro (DIV) 3–4, axons were found to enter the other chamber (axonal chamber).
The barbed end labeling protocol was adapted from Marsick and Letourneau (2011). First, 0.45 µm rhodamine-labeled actin (actin protein [rhodamine] human platelet; Cytoskeleton) solution was prepared in permeabilization buffer (138 mM KCl, 10 mM Pipes, 3 mM EGTA, 4 mM MgCl2, and 1% BSA, pH 6.9) containing 0.025% saponin and 0.2 mM ATP. For rhodamine-labeling experiments in microfluidics devices, 100 µl rhodamine-actin solution was loaded into the axonal chamber for 2 min. Immediately thereafter, both chambers were fixed with fixation solution (4% paraformaldehyde, 0.05% glutaraldehyde, and 10% sucrose made in PBS) for 5 min and washed thrice with PBS. The microfluidic device was then carefully removed without damaging the neuronal processes. Neurons were then permeabilized with 0.2% Triton X-100 in PBS, washed, and incubated 1 µM Alexa Fluor 488 phalloidin (Life Technologies) for 1 h. For rhodamine-actin–labeling experiments using CAD cells, the CAD cells were grown at a density of 30,000 cells/cm2 in DMEM/F12 (Thermo Fisher Scientific) with 8% FBS (HyClone) and 1% penicillin-streptomycin (Thermo Fisher Scientific). 1 d after plating, cells were incubated with 100 µl rhodamine-actin solution for 2 min, followed by fixation, permeabilization, and phalloidin staining as described for primary neurons above.
3D STORM imaging
Rat hippocampal neurons from E18 embryos were cultured on 18-mm coverslips at a density of 6,000/cm2 following guidelines established by the European Animal Care and Use Committee (86/609/CEE) and approval of the local ethics committee (agreement D13-055-8). After 3–8 DIV, neurons were fixed, processed, and imaged as previously described (Xu et al., 2013; Ganguly et al., 2015). After a brief extraction with 0.25% Triton X-100 and 0.3% glutaraldehyde in a cytoskeleton-preserving buffer, neurons were fixed with 2% glutaraldehyde in the same buffer for 15 min before quenching with 0.1% NaBH4 for 7 min. Then, the samples were blocked and stained with primary and secondary antibodies in immunocytochemistry buffer (0.22% gelatin and 0.1% Triton X-100 in phosphate buffer), followed by overnight incubation at 4°C with phalloidin–Alexa Fluor 647 (0.5 µM in phosphate buffer; Life Technologies). Coverslips were placed in STORM buffer (50 mM Tris, pH 8, 10 mM NaCl, 10% glucose, 100 mM ethanolamine, 3.5 U/ml pyranose oxidase, and 40 µg/ml catalase) and imaged on an N-STORM microscope (Nikon). Phalloidin (0.1–0.25 µM) was added in the STORM medium to mitigate progressive unbinding from actin filaments during imaging (phalloidin-PAINT). A series of 60,000 images (67 Hz frame rate) was acquired at full power of the 647-nm laser, with progressive reactivation with the 405-nm laser. Sequences of images were processed for localizations using the N-STORM software, and 2D projections of the 3D-STORM data were generated using the ThunderSTORM plugin for ImageJ (NIH; Ovesný et al., 2014). To measure the hotspot diameters, line profiles were drawn across the larger dimension of hotspots on high-magnification (4 nm/pixel) XY projections generated from the STORM data. The hotspot diameter was determined as the full width at half maximum of the resulting intensity profile using the ProFeatFit ImageJ script (Leterrier et al., 2015).
Modeling: Assumptions
To model the dynamics of actin trails growing in the axon, we incorporate the available experimental data of their nucleation rates, elongation velocities, and length statistics in a 1D mathematical model. We discretize the axon into segments of length Δx(∼0.027 µm) and the time into intervals of 10−5 s and solved the diffusion equation by using a finite-difference method (Crank, 1979) using custom-made algorithms compiled using the commercially licensed Intel Fortran Compiler.
We summarize the simplifying assumptions used in our model below:
(A) The model assumes that hotspots are located linearly along the axon, uniformly spaced at a distance of 3.6 µm. We tested small variations in the hotspot spacing (up to experimentally observed values) and found no significant differences in our simulation results.
(B) The model assumes that the G-actin concentration is uniform along the radial direction of the axon. The justification for this assumption is that the time taken to equilibrate any concentration change in this direction is much smaller than the lifetime of a trail (see detailed discussion in the following sections).
(C) The model assumes that the monomer pool of G-actin is entirely ATP bound (see detailed discussion in the Results and discussion section).
(D) For the simplicity of computation, the model assumes the collapse of the trails to be an instantaneous process. When a trail reaches a predetermined length drawn from the length distribution of trails observed in the experiments, it collapses and deposits its monomer content into the axon.
(E) The photoactivated and nonphotoactivated subsets of the actin monomers are assumed to have no differences in their diffusion coefficients and binding or unbinding affinities to and from actin trails.
(F) We assume periodic boundary conditions for the fluorescently inactive G-actin to ensure flux balance in the axon. This signifies that actin lost at the proximal end of the axon is recovered back at the distal end. Since we only track the fluorescently active population until it is lost from the ends of the axon, we do not need this assumption for the photoactivated population.
The parameters used for our model are summarized in Table S1, and the detailed methods to obtain these are described in the following sections.
Simulating actin trail nucleation from stationary hotspots
In our model, hotspots are uniformly spaced and located linearly along the axon (Fig. 4 A). We assume further that actin trails grow either in proximal and distal directions with the same kinetics. The trail growth process starts with the nucleation at a hotspot. We determined the nucleation rates of the trails from the experimentally observed total number of actin trails nucleated in time intervals of 600 s in imaging windows of lengths Lw ranging between 50 µm and 80 µm (based on imaging data; see Ganguly et al., 2015).
The nucleation rates of actin trails are derived in terms of the number of trails nucleated from each hotspot per unit time. First, the total number of actin trails observed within an imaging window of length Lw in anterograde direction Na and retrograde direction Nr are counted within a fixed imaging time interval Ti. Denoting the average distance between hotspots by dh = 3.6 µm (see Ganguly et al., 2015), the expected number of hotspots within the imaging window is given by Lw/dh, and the nucleation rates of trails in antero- and retrograde directions are given by
| (1) |
In all imaging experiments, we found in the average Na = 20.4 anterograde trails and Nr = 14.9 retrograde trails for an imaging time of Ti = 600 s. With an average imaging window of 65 µm in length, we arrived at nucleation rates of anterograde and retrograde trails of rn,a = 0.001885/s and rn,r = 0.001381/s, respectively, for each hotspot.
Simulating actin trail elongation and subsequent G-actin dynamics
After the formation of a stable actin nucleus during the nucleation phase, the trail grows by assembling G-actin monomers from the available pool in the axon. We modeled actin trails as linear filaments growing parallel to the longitudinal axis of the axon. The nucleation of actin trails and their subsequent elongation by incorporating monomers from the available axonal pool was modeled by Markov processes. Probabilities of trail nucleation and of competing association and dissociation reactions were calculated at each time step using the nucleation rates rn,a and rn,r along with the reaction rates summarized in Fig. 4 A (right). The processes to occur at each time step were chosen stochastically using these probabilities.
The rate of elongation at the barbed or pointed end of the actin filament was directly proportional to the local concentration of G-actin at the respective ends (Pollard, 1986). As a monomer was added to or released from each end of the filament, the local concentration of G-actin was depleted or augmented, which affected the elongation rate of the filament.
We modeled the spatiotemporal G-actin concentration by the function , where denotes the position and t denotes the time. For an actin trail with its barbed end at and its pointed end at , the spatiotemporal change of the G-actin concentration () was governed by
| (2) |
The first term on the right-hand side of Eq. 2 describes the concentration change due to diffusion. The second term describes the instantaneous concentration change when a trail took up or released an actin monomer from its barbed end located at time t1. The time evolution of the prefactor kb(t) was determined stochastically based on which reaction had occurred at each time step (see discussion above). If a monomer was released from or added to the barbed end of the filament at time t1, then kb(t1) takes the value +1.0 or −1.0, respectively. Similarly, the third term describes the concentration change due to a single monomer being added to or released from the pointed end of the filament at time t = t2. A deterministic equivalent of Eq. 2 has been used by Novak et al. (2008) to describe G-actin dynamics in motile cells.
The length of the axons imaged in the experiments typically ranged from 150–200 µm, whereas their diameters were much smaller in comparison and ranged from 140–200 nm. As a consequence, the concentration profile of G-actin along the radial direction of the axon during trail growth varied at any instance of time only by a few percent, and we used a 1D representation of the actin concentrations along the length of the axon. For an actin-diffusion constant of 6 µm2/s and an axon diameter of 150 nm, we estimated the diffusion time of monomers across the radial dimension of the axon as . Hence, the time of equilibration of the G-actin concentration across the radial axonal section was much smaller than the lifetime of the trail (∼10 s), and we can assume that the radial concentration profile was approximately constant during trail growth. The resulting 1D diffusion equation (with appropriately renormalized parameters) for the distribution of G-actin along the axon read
| (3) |
To enforce a flux balance in the axon, i.e., the flux of G-actin monomers at the proximal end matches the flux at the distal end of the axon, we used a periodic boundary condition, i.e., G(−L,t) = G(L,t), to solve Eq. 3, where the axon extends from −L to L.
As the experiments with the Mena/Vasp proteins indicate (Fig. 2 D), actin trails grow with their barbed ends attached to the stationary hotspots. Therefore, the position of the barbed end of each trail, xb(t), was taken to be constant, while xp(t) increased or decreased as the trail grows. A consequence of the barbed-end attachment of the filament at the hotspots was that all the monomers attached to a growing trail were progressively moved toward the direction of trail growth (pointed end) when a new monomer was added to the barbed end of the filament (Fig. 4 A, bottom). The monomers attached to the filament were not moved when a new monomer attached at the pointed end of the filament. Since a much higher fraction (∼90%) of monomers attached to the barbed end of the filament, the monomers attached to a growing trail are predominantly pushed toward the direction of trail growth, giving rise to a directed transport of actin monomers.
Simulating the collapse of an actin trail
It is important to note that this model cannot explain the underlying causes behind the collapse of an actin trail. Therefore, to incorporate the collapse of actin trails in our model, we incorporated the length distribution of the trails observed in the imaging experiments (see Ganguly et al., 2015). Thus, each actin trail had a predetermined length at which it collapsed drawn from the observed frequency distribution of their lengths. We modeled the trail-collapse mechanism as an instantaneous process in which once the trail reached its collapse length, it disintegrated and deposited its monomer content back into the axon. If an actin trail growing with its barbed end at xb crashed when it reached length Lt, then monomers were added to the axoplasm, where Lm is the effective actin monomer length of ∼2.7 nm (Squire, 1981). These monomers were added to the sections of the axon that were previously occupied by the trail. If Δx is the length of each axonal section, then monomers were added to each section. Therefore, the concentration of each axonal section of length Δx and radius r, previously covered by the trail, increased by ΔG where
| (4) |
Simulating the photoactivation-based imaging paradigm in an axon
In our model of axonal actin trails growing in the axoplasm, actin monomers diffuse passively in the axon and occasionally are incorporated in trails. Due to the progressive motion of actin monomers when attached to a growing trail (Fig. 4 A, bottom) and the imbalance of anterograde and retrograde trails, a net anterograde transport of axonal actin emerges from our model describing a molecular hitchhiking process.
To quantify the rate of this transport, we computationally simulate a fluorescent pulse-chase imaging experiment. In this experiment, all actin (globular and filamentous) was photoactivated within a window of 15 µm in length. The fluorescently active actin population was subsequently tracked as it diffused in the axon and occasionally translocated when incorporated in trails.
We simulated actin trails in an axon of length L = 1,000 µm and a diameter of d = 170 nm. At the start of the simulation (t = − 50 s), actin was present only in monomeric form (Fig. 5 A, top). At t = 0, after a steady state had been reached in the axon, both G-actin and F-actin was photoactivated in a central zone of the axon 15 µm in length (Fig. 5 A, middle). The length of the axon allowed the fluorescently activated population to be tracked for ∼100 s before it was lost at the ends of the axon. Therefore, at t = 0, immediately after activation, the distribution of fluorescently active actin was given by
| (5) |
The boundaries of the photoactivated zone were smoothed using a Gaussian function to avoid any discontinuity in the first derivative of Gl(x,0) (Fig. 5 B, left, green trace) and to reflect experimental constraints. The resultant length of the photoactivation window was Lph ≈ 15 µm. The subsequent spatiotemporal dynamics of both the fluorescently active and inactive G-actin can be modeled using Eq. 3 for each species in conjunction with the stochastic elongation dynamics of the trails. However, we used different boundary conditions for each population. For the fluorescently inactive G-actin (Gu(x,t)), to maintain flux balance, we used periodic boundary conditions, i.e., fluorescently inactive actin leaving at the right end of the axon at x = L, enters the left end of the axon at x = −L, and vice versa. The fluorescently activated actin did not reach the boundaries x = ±L in the duration of our simulation and hence vanished there. Therefore,
| (6) |
As the fluorescently active G-actin monomers diffused through the axon, the distribution of monomers Gl(x,t) broadened over time. If there was no net active transport present (either no trails or equal anterograde and retrograde trails), the center of the fluorescently active distribution remained at its initial position (Fig. 5 B, right, red trace) since diffusion was not direction specific. If there were higher anterograde or retrograde trails nucleated, the center of the fluorescently active monomer population moved correspondingly in the anterograde (Fig. 5 B, right, blue trace) or retrograde direction (Fig. 5 B, right, cyan trace), respectively.
To quantify the transport rate, we calculated the velocity of the fluorescence center. The center of fluorescence was measured by averaging over hundreds of independently seeded simulation runs using the equation
We also examined the effect of actin-disrupting drugs like latrunculin A and SMIFH2 on the actin transport rate. For these simulations, we used the data for the actin trail nucleation rates, trail length, and elongation rates before and after adding the drugs from Ganguly et al. (2015). The effect of latrunculin A was modeled by removing a fraction of the available monomers from the axon since latrunculin A is known to bind to G-actin with 1:1 stoichiometry and prevent them from polymerizing (Yarmola et al., 2000). The effect of SMIFH2 was modeled by attenuating the binding rates of the actin monomers (Goode and Eck, 2007). We then repeat our fluorescence pulse-chase experiments (see Fig. S3 B) over 250 simulation runs to measure the transport rate.
Statistical analysis
All graphs were plotted, and all statistical analyses were performed in Prism (GraphPad Software). Significance was calculated using a two-tailed t test in Prism. Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 shows superresolution images of hotspots and quantification of their diameters. Fig. S2 shows intensity center shift analysis of soluble PAGFP (A), comparison of PAGFP:synapsin and PAGFP:Utr-CH kymographs (B), effect of vincristine treatment on number of anterograde actin trails (C), and effect of vincristine treatment on transport of fast axonal transport cargo synaptophysin:DsRed. Fig. S3 shows the probability distribution of elongation velocity of actin trails obtained from the simulations (A) and simulation of transport rate of actin in the presence of latrunculin A or SMIFH2 (B). Table S1 lists the parameters used in the simulations. Video 1 shows the translocation of monomers attached to a trail due to monomer addition at the barbed end. Video 2 shows an overview of the transport mechanism.
Supplementary Material
Acknowledgments
The authors thank Stephanie Gupton (University of North Carolina, Chapel Hill, NC) for the Mena/Vasp constructs.
This work was supported by an National Institutes of Health grant to S. Roy (R01NS075233). C. Leterrier is supported by a Centre National de la Recherche Scientifique ATIP2016 grant.
The authors declare no competing financial interests.
Author contributions: The mathematical modeling of actin transport was designed and executed by N. Chakrabarty and P. Jung. Most of the experimental data were designed, obtained, and analyzed by P. Dubey. Y. Tang, A. Ganguly, and K. Ladt contributed with key experiments. All superresolution imaging was designed, performed, and analyzed by C. Leterrier. S. Roy conceived the project, helped design the experiments, managed collaborations, and wrote the manuscript.
References
- Allard J., and Mogilner A.. 2013. Traveling waves in actin dynamics and cell motility. Curr. Opin. Cell Biol. 25:107–115. 10.1016/j.ceb.2012.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson S.J., Hosford M.A., and Molitoris B.A.. 2004. Mechanism of actin polymerization in cellular ATP depletion. J. Biol. Chem. 279:5194–5199. 10.1074/jbc.M306973200 [DOI] [PubMed] [Google Scholar]
- Baas P.W., and Brown A.. 1997. Slow axonal transport: the polymer transport model. Trends Cell Biol. 7:380–384. 10.1016/S0962-8924(97)01148-3 [DOI] [PubMed] [Google Scholar]
- Baas P.W., Slaughter T., Brown A., and Black M.M.. 1991. Microtubule dynamics in axons and dendrites. J. Neurosci. Res. 30:134–153. 10.1002/jnr.490300115 [DOI] [PubMed] [Google Scholar]
- Barry D.M., Millecamps S., Julien J.-P., and Garcia M.L.. 2007. New movements in neurofilament transport, turnover and disease. Exp. Cell Res. 313:2110–2120. 10.1016/j.yexcr.2007.03.011 [DOI] [PubMed] [Google Scholar]
- Black M.M., and Lasek R.J.. 1979. Axonal transport of actin: slow component b is the principal source of actin for the axon. Brain Res. 171:401–413. 10.1016/0006-8993(79)91045-X [DOI] [PubMed] [Google Scholar]
- Blanchoin L., and Pollard T.D.. 2002. Hydrolysis of ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry. 41:597–602. 10.1021/bi011214b [DOI] [PubMed] [Google Scholar]
- Boujemaa-Paterski R., Suarez C., Klar T., Zhu J., Guérin C., Mogilner A., Théry M., and Blanchoin L.. 2017. Network heterogeneity regulates steering in actin-based motility. Nat. Commun. 8:655 10.1038/s41467-017-00455-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D., Kiesewetter A.K., Linkner J., Vinzenz M., Stradal T.E.B., Small J.V., Curth U., Dickinson R.B., and Faix J.. 2011. Molecular mechanism of Ena/VASP-mediated actin-filament elongation. EMBO J. 30:456–467. 10.1038/emboj.2010.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. 2000. Slow axonal transport: stop and go traffic in the axon. Nat. Rev. Mol. Cell Biol. 1:153–156. 10.1038/35040102 [DOI] [PubMed] [Google Scholar]
- Burkel B.M., von Dassow G., and Bement W.M.. 2007. Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motil. Cytoskeleton. 64:822–832. 10.1002/cm.20226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M.F., and Pantaloni D.. 1986. Direct evidence for ADP-Pi-F-actin as the major intermediate in ATP-actin polymerization. Rate of dissociation of Pi from actin filaments. Biochemistry. 25:7789–7792. 10.1021/bi00372a001 [DOI] [PubMed] [Google Scholar]
- Carlier M.-F., Pantaloni D., Evans J.A., Lambooy P.K., Korn E.D., and Webb M.R.. 1988. The hydrolysis of ATP that accompanies actin polymerization is essentially irreversible. FEBS Lett. 235:211–214. 10.1016/0014-5793(88)81264-X [DOI] [PubMed] [Google Scholar]
- Chen Q., Nag S., and Pollard T.D.. 2012. Formins filter modified actin subunits during processive elongation. J. Struct. Biol. 177:32–39. 10.1016/j.jsb.2011.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles C.H., and Bradke F.. 2015. Coordinating neuronal actin-microtubule dynamics. Curr. Biol. 25:R677–R691. 10.1016/j.cub.2015.06.020 [DOI] [PubMed] [Google Scholar]
- Crank J. 1979. The Mathematics of Diffusion. Oxford University Press. 414 pp. [Google Scholar]
- Devineni N., Minamide L.S., Niu M., Safer D., Verma R., Bamburg J.R., and Nachmias V.T.. 1999. A quantitative analysis of G-actin binding proteins and the G-actin pool in developing chick brain. Brain Res. 823:129–140. 10.1016/S0006-8993(99)01147-6 [DOI] [PubMed] [Google Scholar]
- Didry D., Carlier M.F., and Pantaloni D.. 1998. Synergy between actin depolymerizing factor/cofilin and profilin in increasing actin filament turnover. J. Biol. Chem. 273:25602–25611. 10.1074/jbc.273.40.25602 [DOI] [PubMed] [Google Scholar]
- Dubey P., Jorgenson K., and Roy S.. 2018. Actin Assemblies in the Axon Shaft - some Open Questions. Curr. Opin. Neurobiol. 51:163–167. 10.1016/j.conb.2018.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith J.A., and Gallant P.E.. 2000. Axonal transport of tubulin and actin. J. Neurocytol. 29:889–911. 10.1023/A:1010903710160 [DOI] [PubMed] [Google Scholar]
- Gallo G., and Letourneau P.C.. 2004. Regulation of growth cone actin filaments by guidance cues. J. Neurobiol. 58:92–102. 10.1002/neu.10282 [DOI] [PubMed] [Google Scholar]
- Ganguly A., and Roy S.. 2014. Using photoactivatable GFP to track axonal transport kinetics. Methods Mol. Biol. 1148:203–215. 10.1007/978-1-4939-0470-9_13 [DOI] [PubMed] [Google Scholar]
- Ganguly A., Tang Y., Wang L., Ladt K., Loi J., Dargent B., Leterrier C., and Roy S.. 2015. A dynamic formin-dependent deep F-actin network in axons. J. Cell Biol. 210:401–417. 10.1083/jcb.201506110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A., Han X., Das U., Wang L., Loi J., Sun J., Gitler D., Caillol G., Leterrier C., Yates J.R. III, and Roy S.. 2017. Hsc70 chaperone activity is required for the cytosolic slow axonal transport of synapsin. J. Cell Biol. 216:2059–2074. 10.1083/jcb.201604028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittes F., Mickey B., Nettleton J., and Howard J.. 1993. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J. Cell Biol. 120:923–934. 10.1083/jcb.120.4.923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont P.J., Furman M.I., Wachsstock D., Safer D., Nachmias V.T., and Pollard T.D.. 1992. The control of actin nucleotide exchange by thymosin beta 4 and profilin. A potential regulatory mechanism for actin polymerization in cells. Mol. Biol. Cell. 3:1015–1024. 10.1091/mbc.3.9.1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez T.M., and Letourneau P.C.. 2014. Actin dynamics in growth cone motility and navigation. J. Neurochem. 129:221–234. 10.1111/jnc.12506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode B.L., and Eck M.J.. 2007. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76:593–627. 10.1146/annurev.biochem.75.103004.142647 [DOI] [PubMed] [Google Scholar]
- He J., Zhou R., Wu Z., Carrasco M.A., Kurshan P.T., Farley J.E., Simon D.J., Wang G., Han B., Hao J., et al. 2016. Prevalent presence of periodic actin-spectrin-based membrane skeleton in a broad range of neuronal cell types and animal species. Proc. Natl. Acad. Sci. USA. 113:6029–6034. 10.1073/pnas.1605707113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Francis F., Myers K.A., Yu W., Black M.M., and Baas P.W.. 2005. Role of cytoplasmic dynein in the axonal transport of microtubules and neurofilaments. J. Cell Biol. 168:697–703. 10.1083/jcb.200407191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N., Funakoshi S.T., and Takeda S.. 1997. Slow axonal transport: the subunit transport model. Trends Cell Biol. 7:384–388. 10.1016/S0962-8924(97)01133-1 [DOI] [PubMed] [Google Scholar]
- Jung H., Gkogkas C.G., Sonenberg N., and Holt C.E.. 2014. Remote control of gene function by local translation. Cell. 157:26–40. 10.1016/j.cell.2014.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevenaar J.T., and Hoogenraad C.C.. 2015. The axonal cytoskeleton: from organization to function. Front. Mol. Neurosci. 8:44 10.3389/fnmol.2015.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladt K., Ganguly A., and Roy S.. 2016. Axonal actin in action: Imaging actin dynamics in neurons. Methods Cell Biol. 131:91–106. 10.1016/bs.mcb.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leterrier C., Potier J., Caillol G., Debarnot C., Rueda Boroni F., and Dargent B.. 2015. Nanoscale Architecture of the Axon Initial Segment Reveals an Organized and Robust Scaffold. Cell Reports. 13:2781–2793. 10.1016/j.celrep.2015.11.051 [DOI] [PubMed] [Google Scholar]
- Maday S., Twelvetrees A.E., Moughamian A.J., and Holzbaur E.L.F.. 2014. Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron. 84:292–309. 10.1016/j.neuron.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsick B.M., and Letourneau P.C.. 2011. Labeling F-actin barbed ends with rhodamine-actin in permeabilized neuronal growth cones. J. Vis. Exp. 17:2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsick B.M., Flynn K.C., Santiago-Medina M., Bamburg J.R., and Letourneau P.C.. 2010. Activation of ADF/cofilin mediates attractive growth cone turning toward nerve growth factor and netrin-1. Dev. Neurobiol. 70:565–588. 10.1002/dneu.20800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J.L., Tardy Y., Dewey C.F. Jr., Meister J.J., and Hartwig J.H.. 1998. Simultaneous measurements of actin filament turnover, filament fraction, and monomer diffusion in endothelial cells. Biophys. J. 75:2070–2078. 10.1016/S0006-3495(98)77649-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuarrie I.G., Brady S.T., and Lasek R.J.. 1986. Diversity in the axonal transport of structural proteins: major differences between optic and spinal axons in the rat. J. Neurosci. 6:1593–1605. 10.1523/JNEUROSCI.06-06-01593.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejillano M.R., Kojima S., Applewhite D.A., Gertler F.B., Svitkina T.M., and Borisy G.G.. 2004. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell. 118:363–373. 10.1016/j.cell.2004.07.019 [DOI] [PubMed] [Google Scholar]
- Morris J.R., and Lasek R.J.. 1984. Monomer-polymer equilibria in the axon: direct measurement of tubulin and actin as polymer and monomer in axoplasm. J. Cell Biol. 98:2064–2076. 10.1083/jcb.98.6.2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak I.L., Slepchenko B.M., and Mogilner A.. 2008. Quantitative analysis of G-actin transport in motile cells. Biophys. J. 95:1627–1638. 10.1529/biophysj.108.130096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oblinger M.M. 1988. Biochemical composition and dynamics of the axonal cytoskeleton in the corticospinal system of the adult hamster. Metab. Brain Dis. 3:49–65. 10.1007/BF01001353 [DOI] [PubMed] [Google Scholar]
- Okabe S., and Hirokawa N.. 1990. Turnover of fluorescently labelled tubulin and actin in the axon. Nature. 343:479–482. 10.1038/343479a0 [DOI] [PubMed] [Google Scholar]
- Ovesný M., Křížek P., Borkovec J., Svindrych Z., and Hagen G.M.. 2014. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 30:2389–2390. 10.1093/bioinformatics/btu202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandréou M.-J., and Leterrier C.. 2018. The functional architecture of axonal actin. Mol. Cell. Neurosci. 91:151–159. 10.1016/j.mcn.2018.05.003 [DOI] [PubMed] [Google Scholar]
- Pollard T.D. 1986. Mechanism of actin filament self-assembly and regulation of the process by actin-binding proteins. Biophys. J. 49:149–151. 10.1016/S0006-3495(86)83630-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi S.A., Neidt E.M., Cui J., Feiger Z., Skau C.T., Gardel M.L., Kozmin S.A., and Kovar D.R.. 2009. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol. 16:1158–1168. 10.1016/j.chembiol.2009.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S. 2014. Seeing the unseen: the hidden world of slow axonal transport. Neuroscientist. 20:71–81. 10.1177/1073858413498306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Coffee P., Smith G., Liem R.K., Brady S.T., and Black M.M.. 2000. Neurofilaments are transported rapidly but intermittently in axons: implications for slow axonal transport. J. Neurosci. 20:6849–6861. 10.1523/JNEUROSCI.20-18-06849.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Yang G., Tang Y., and Scott D.A.. 2011. A simple photoactivation and image analysis module for visualizing and analyzing axonal transport with high temporal resolution. Nat. Protoc. 7:62–68. 10.1038/nprot.2011.428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D.A., Das U., Tang Y., and Roy S.. 2011. Mechanistic logic underlying the axonal transport of cytosolic proteins. Neuron. 70:441–454. 10.1016/j.neuron.2011.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood P., Murthy K., Kumar V., Nonet M.L., Menon G.I., and Koushika S.P.. 2018. Cargo crowding at actin-rich regions along axons causes local traffic jams. Traffic. 19:166–181. 10.1111/tra.12544 [DOI] [PubMed] [Google Scholar]
- Squire J. 1981. The Structural Basis of Muscular Contraction. Plenum Press, New York: 698 pp. 10.1007/978-1-4613-3183-4 [DOI] [Google Scholar]
- Sun H.-Q., Kwiatkowska K., and Yin H.L.. 1995. Actin monomer binding proteins. Curr. Opin. Cell Biol. 7:102–110. 10.1016/0955-0674(95)80051-4 [DOI] [PubMed] [Google Scholar]
- Symons M.H., and Mitchison T.J.. 1991. Control of actin polymerization in live and permeabilized fibroblasts. J. Cell Biol. 114:503–513. 10.1083/jcb.114.3.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Scott D., Das U., Gitler D., Ganguly A., and Roy S.. 2013. Fast vesicle transport is required for the slow axonal transport of synapsin. J. Neurosci. 33:15362–15375. 10.1523/JNEUROSCI.1148-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantama M., Martínez-François J.R., Mongeon R., and Yellen G.. 2013. Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio. Nat. Commun. 4:2550 10.1038/ncomms3550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro T., and Komiya Y.. 1992. Organization and slow axonal transport of cytoskeletal proteins under normal and regenerating conditions. Mol. Neurobiol. 6:301–311. 10.1007/BF02780559 [DOI] [PubMed] [Google Scholar]
- Terada S., Nakata T., Peterson A.C., and Hirokawa N.. 1996. Visualization of slow axonal transport in vivo. Science. 273:784–788. 10.1126/science.273.5276.784 [DOI] [PubMed] [Google Scholar]
- Terada S., Kinjo M., and Hirokawa N.. 2000. Oligomeric tubulin in large transporting complex is transported via kinesin in squid giant axons. Cell. 103:141–155. 10.1016/S0092-8674(00)00094-5 [DOI] [PubMed] [Google Scholar]
- Terada S., Kinjo M., Aihara M., Takei Y., and Hirokawa N.. 2010. Kinesin-1/Hsc70-dependent mechanism of slow axonal transport and its relation to fast axonal transport. EMBO J. 29:843–854. 10.1038/emboj.2009.389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M., Schmidek A., Galbraith J.A., Gallant P.E., and Reese T.S.. 1995. Transport of cytoskeletal elements in the squid giant axon. Proc. Natl. Acad. Sci. USA. 92:11500–11503. 10.1073/pnas.92.25.11500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., and Brown A.. 2002. Rapid movement of microtubules in axons. Curr. Biol. 12:1496–1501. 10.1016/S0960-9822(02)01078-3 [DOI] [PubMed] [Google Scholar]
- Wang L., Ho C.L., Sun D., Liem R.K., and Brown A.. 2000. Rapid movement of axonal neurofilaments interrupted by prolonged pauses. Nat. Cell Biol. 2:137–141. 10.1038/35004008 [DOI] [PubMed] [Google Scholar]
- Xu K., Zhong G., and Zhuang X.. 2013. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 339:452–456. 10.1126/science.1232251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., and Brown A.. 2005. Neurofilament polymer transport in axons. J. Neurosci. 25:7014–7021. 10.1523/JNEUROSCI.2001-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarmola E.G., Somasundaram T., Boring T.A., Spector I., and Bubb M.R.. 2000. Actin-latrunculin A structure and function. Differential modulation of actin-binding protein function by latrunculin A. J. Biol. Chem. 275:28120–28127. [DOI] [PubMed] [Google Scholar]
- Zhong G., He J., Zhou R., Lorenzo D., Babcock H.P., Bennett V., and Zhuang X.. 2014. Developmental mechanism of the periodic membrane skeleton in axons. eLife. 3:e04581 10.7554/eLife.04581 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.