During the phototropic response, plant cells reorient their cortical microtubule arrays by generating many new growing microtubules through severing. This study shows how new plus ends created by severing, which lack GTP-caps, are stabilized by CLASP to allow for microtubule amplification and array reorientation.
Abstract
Central to the building and reorganizing cytoskeletal arrays is creation of new polymers. Although nucleation has been the major focus of study for microtubule generation, severing has been proposed as an alternative mechanism to create new polymers, a mechanism recently shown to drive the reorientation of cortical arrays of higher plants in response to blue light perception. Severing produces new plus ends behind the stabilizing GTP-cap. An important and unanswered question is how these ends are stabilized in vivo to promote net microtubule generation. Here we identify the conserved protein CLASP as a potent stabilizer of new plus ends created by katanin severing in plant cells. Clasp mutants are defective in cortical array reorientation. In these mutants, both rescue of shrinking plus ends and the stabilization of plus ends immediately after severing are reduced. Computational modeling reveals that it is the specific stabilization of severed ends that best explains CLASP’s function in promoting microtubule amplification by severing and array reorientation.
Introduction
Many animal cells, and all higher plant cells, build interphase microtubule arrays with distinct architectures without benefit of a centrosome or other central organizer. In higher plants, interphase microtubules associated with the plasma membrane are organized into highly ordered arrays oriented transversely to the main axis of cell growth. These cortical arrays are crucial for determining the direction of cell expansion (Baskin, 2001; Wasteneys, 2002; Ehrhardt and Shaw, 2006), in part by guiding the deposition and thus orientation of cellulose as it is synthesized by protein complexes in the plasma membrane (Baskin, 2001; Paredez et al., 2006). New microtubules in these arrays are created by nucleation from γ-tubulin complexes distributed along the sides of existing microtubules (Wasteneys and Williamson, 1989; Chan et al., 2003; Murata et al., 2005; Nakamura et al., 2010) and by a second, recently discovered mechanism: the creation of new microtubule ends by katanin-mediated severing at locations where cortical microtubules intersect and cross over each other (Wightman and Turner, 2007; Lindeboom et al., 2013b; Zhang et al., 2013). This severing mechanism for microtubule generation plays an essential role in cortical array reorganization in response to light, a key environmental cue for plant development (Lindeboom et al., 2013b). When seedlings develop in the dark, cells in the embryonic axis, also known as the hypocotyl, undergo rapid axial elongation to push the seedling shoot into the light from the soil. Upon perception of blue light, principally through phototropin photoreceptors, transverse cortical arrays in hypocotyl epidermal cells undergo a remarkable reorientation to a transverse direction, a remodeling that takes place on the scale of ∼15 min. This reorganization occurs by up-regulation of katanin-mediated severing at crossovers, with a preference for severing of the newer of the two microtubules (Lindeboom et al., 2013b; Zhang et al., 2013). The repeated process of severing and growth of the new plus ends amplifies rapidly a new population of microtubules at 90° to the existing array. A functional consequence of this reorientation is redirection of the trajectories of cellulose synthase complexes as they build the cell wall (Paredez et al., 2006), thus remodeling cell wall structure.
Creation of new microtubules by severing and rescue has also been suggested in animal cells, particularly in neurons, epithelial cells, and meiocytes (Roll-Mecak, 2013). A critical question in all these systems is how the new plus ends created by severing are stabilized for severing to efficiently build a population of new microtubules. When new microtubules arrays are built by nucleation, the growing ends assembled at nucleation complexes are stabilized by the GTP state of newly added tubulin dimers (Mitchison and Kirschner, 1984) and by proteins that associate with growing ends (Akhmanova and Steinmetz, 2015), some of which are recruited by the GTP-cap (Akhmanova and Steinmetz, 2015). However, new plus ends generated by severing are likely created behind the GTP-cap. When created in vitro, these new plus ends shrink immediately (Walker et al., 1989; Tran et al., 1997). By contrast, when severing creates new plus ends in vivo, they frequently initiate new growth without observable shrinkage (Lindeboom et al., 2013b). The factors that act in vivo to stabilize new plus ends created by severing to permit generation of new microtubules are not known. Here we identify CLASP as a specific stabilization factor for new plus ends generated by katanin-mediated severing in the model plant Arabidopsis thaliana and demonstrate through quantitative imaging and computational modeling that plus end stabilization by CLASP after severing contributes to an effective reorganization of the microtubule array in response to environmental signals.
Results
Microtubule reorientation in +TIP mutants
Several proteins that accumulate at and track growing plus ends (+TIPs) have been identified in the model plant Arabidopsis (Bisgrove et al., 2004). We asked whether one or more of these +TIPs might act as a specific plus end rescue factor after severing to support creation of a new cortical array. Specifically, we assessed end binding protein 1 (EB1; Chan et al., 2003; Mathur et al., 2003), cytoplasmic linker–associated protein (CLASP; Ambrose et al., 2007; Kirik et al., 2007), and SPIRAL1 (SPR1; Nakajima et al., 2004; Sedbrook et al., 2004). EB1 and CLASP are both conserved across plants and animals. EB1 is encoded by three genes in Arabidopsis, EB1a–c; all three loci are disrupted in the triple mutant 3x-eb1 (Galva et al., 2014). CLASP, a protein with two TOG domains, is represented by a single gene in Arabidopsis, where loss of function (clasp) results in significant reduction of cell growth anisotropy (Ambrose et al., 2007; Kirik et al., 2007). SPR1 is specific to algae and plants (Furutani et al., 2000). In Arabidopsis, SPR1 is part of a six-gene family in which loss of SPR1 function itself (spr1) causes pronounced right-handed chiral growth of plant organs and loss of cell growth anisotropy.
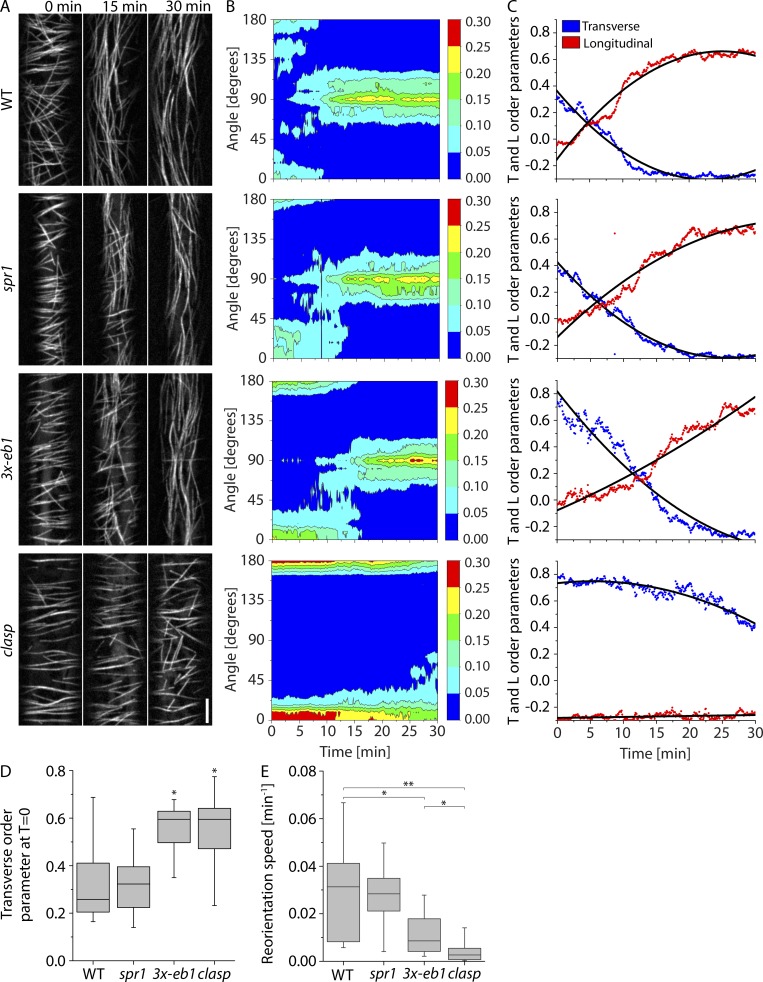
As a first step, we asked whether loss of function in any of these candidate proteins was impeded in blue light–stimulated cortical array reorganization in epidermal hypocotyl cells of dark-grown seedlings. Time-lapse imaging of microtubule organization was accomplished by introducing the tubulin marker 35S-YFP-TUA5 into the spr1, clasp, and 3x-eb1 mutants (Fig. 1 A and Video 1), and quantitation of local microtubule orientation over time was performed with the ImageJ (National Institutes of Health) plugin LOCO (Lindeboom et al., 2013b; Fig. 1, B and C). Two mutants, 3x-eb1 and clasp, showed altered cortical array organization before stimulation by blue light, with each displaying a higher degree of transverse order than WT (P < 0.05, Mann–Whitney U test compared with WT; Fig. 1 D). When the speed with which longitudinal order was built up over time was measured (Lindeboom et al., 2013b), both of these mutants were in addition significantly slower than WT (P < 0.05 and P < 0.002, 3x-eb1 and clasp, respectively, Mann–Whitney U; Fig. 1 E), with the median speed for clasp being approximately threefold slower than that of 3x-eb1 (P < 0.5, Mann–Whitney U test). By contrast, although spr1 mutants have pronounced morphogenetic phenotypes, no significant effect was measured in either initial array order or reorientation speed. Thus, both the 3x-eb1 and clasp mutants show significant impairment in light-stimulated cortical array reorientation, starting with similar and higher degrees of transverse order than WT, while at the same time showing a slower generation of longitudinal microtubule order, with clasp being the most defective.
Figure 1.
Microtubule reorientation in +TIP mutants. (A) Panels from confocal image time series of dark-grown hypocotyl cells expressing YFP-TUA5 in WT, spr1, 3- eb1, and clasp mutant backgrounds at 0, 15, and 30 min after induction of reorientation by blue light. See Video 1. Scale bar is 5 µm. (B) Representative contour plots of MT orientation over time corresponding to the videos shown in A. The color scale represents fraction of microtubules. (C) Longitudinal and transverse order parameters during MT reorientation for the videos shown in A. Black lines show quadratic fit. (D) Transverse order parameter at T = 0 for cortical microtubules imaged in WT, spr1, 3x-eb1, and clasp seedlings. n = 8, 9, 9, and 9 cells, respectively. A Kruskal–Wallis test showed significant differences among the genotypes (P < 0.01). Asterisks represent significant difference from WT by Mann–Whitney U test (P < 0.05). (E) Longitudinal reorientation speed (Lindeboom et al., 2013b) of imaged microtubule arrays in etiolated hypocotyl cells expressing YFP-TUA5 in WT, spr1, 3x-eb1, and clasp seedlings. A Kruskal–Wallis test showed significant differences for reorientation speed distributions among the genotypes (P < 0.01). n = 8, 9, 9, and 9 cells, respectively. Asterisks represent significant difference from WT in Mann–Whitney U test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Boxplots show the 25th and 75th percentile as box edges, the line in the box indicates median value, and the whiskers show the 2.5th and 97.5th percentile.
The reorientation defects we measured in the 3x-eb1 and clasp mutants could be due to specific effects on plus end stabilization after severing, changes in basic microtubule dynamics, or possibly severing itself. To assess these possibilities, we measured individual microtubule dynamics, severing rates at microtubule crossovers, and specific stabilization of plus ends after severing at the onset of reorientation. We then explored the consequences of measured changes in microtubule behaviors with modeling and computer simulation studies.
Microtubule dynamics in +TIP mutants
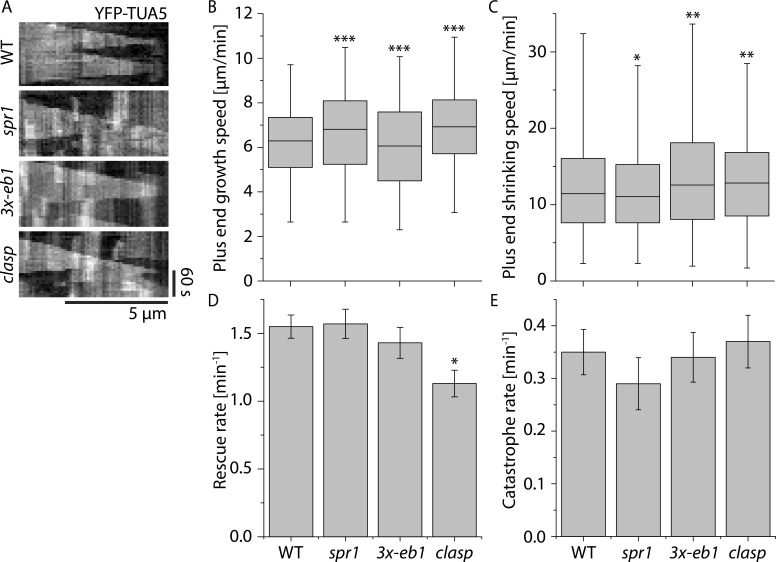
To measure basic microtubule dynamics, we generated kymographs from the reorientation videos (Fig. 2 A) and traced the trajectories of microtubule plus ends (see Materials and methods). In Arabidopsis cortical arrays, plus ends are easily distinguished from minus ends by their distinct dynamics, with plus ends showing extended episodes of growth whereas minus ends show primarily slow shrinkage and pause (Shaw et al., 2003). We observed quantitatively small but statistically significant differences in growth velocities between WT and all the tested +TIP mutants (P < 0.001, Mann–Whitney U test; Fig. 2 B and Table S1). spr1 and clasp showed higher growth velocities than WT, whereas plus ends in 3x-eb1 grew more slowly than WT. spr1 mutants tended to have slower shrinkage velocities than WT (P < 0.05, Mann–Whitney U test), whereas 3x-eb1 and clasp mutants both had higher shrinkage velocities than WT (Fig. 2 C; P < 0.05 and P < 0.01, respectively, Mann–Whitney U test). We observed that, when making transitions between shrinking to growth, the clasp mutants had a significant rescue defect compared with WT (Fig. 2 D; P < 0.05, rate ratio exact test). No significant differences were measured in catastrophe rates between WT and any of the other +TIP mutants (Fig. 2 E). In summary, we found small but significant differences in microtubule dynamics between WT and spr1, 3x- eb1, and clasp, the most notable being the rescue defect in clasp. The possible significance of these small but significant differences to the amplification of new microtubule arrays are explored below in computer modeling studies.
Figure 2.
Microtubule plus end dynamics in +TIP mutants. (A) Kymographs of microtubules during early stages of cortical array reorientation induced by blue light in WT, spr1, 3x-eb1, and clasp seedlings expressing YFP-TUA5. (B and C) Boxplot of microtubule plus end growth speeds (B) and plus end shrinkage speeds (C) in n = 2,148, 1,405, 1,837, and 1,516 segments measured in WT, spr1, 3x-eb1, and clasp seedlings, respectively. Boxplots show the 25th and 75th percentile as box edges, the line in the box indicates median value, and the whiskers show the 2.5th and 97.5th percentile. Asterisks indicate a significant difference from WT. (D) Microtubule rescue rates. n = 213, 135, 110, and 116 rescues in WT, spr1, 3x-eb1, and clasp mutant backgrounds, respectively. Error bars represent SEM. Asterisks indicate a significant difference by rate ratio test. (E) Microtubule catastrophe rates. n = 190, 119, 155, and 148 rescues in WT, spr1, 3x-eb1, and clasp mutant backgrounds, respectively. Error bars represent SEM. Asterisks indicate significant difference compared with WT by rate ratio test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data for all measurements are from six cells in six plants for each genotype.
Severing in +TIP mutants
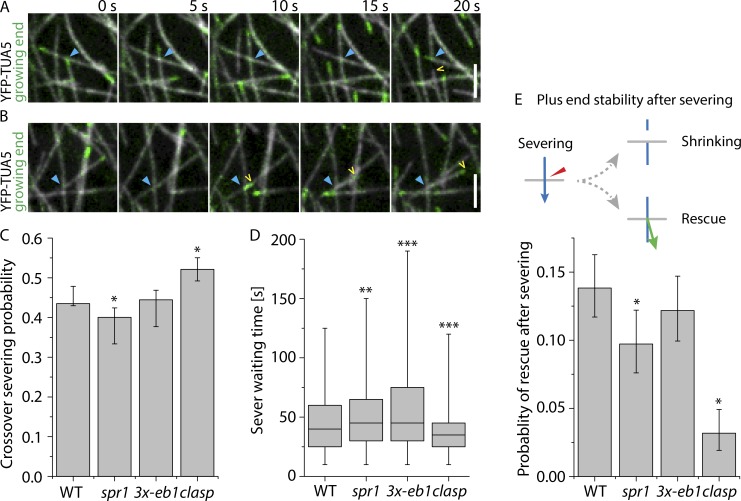
To assess the possible function of +TIPs in modulating severing and stabilizing microtubule ends after severing, we analyzed the outcomes of microtubule crossover formation in WT and spr1, 3x-eb1, and clasp mutant backgrounds expressing YFP-TUA5 as a microtubule marker. For each crossover, we marked the location, time of creation, time of resolution, whether the crossover was resolved by severing or by depolymerization of either crossover partner, and whether the old microtubule (the preexisting microtubule) or the new microtubule (the microtubule that crosses the preexisting one to form the crossover) at the crossover got severed (see Materials and methods). We also scored whether the newly created plus end was shrinking or growing as observed immediately from the crossover site (Fig. 3 A and Video 2; and Fig. 3 B and Video 3, respectively).
Figure 3.
Analysis of microtubule crossovers in +TIP mutants. (A and B) Examples of crossovers (blue arrowhead) where severing occurs, followed by depolymerization (A; see Video 2) or polymerization (B; see Video 3) of the new plus end (yellow arrowhead) in a WT cell expressing YFP-TUA5. Scale bars are 3 µm. (C) Severing probability per crossover in cells expressing YPF-TUA5 in WT, spr1, 3x-eb1, and clasp backgrounds (n = 2,027, 1,696, 1,718, and 1,147 crossovers in six cells, respectively). Error bars show 95% confidence internals. Asterisks indicate a significant difference from WT by Fisher’s exact test. (D) Waiting times from the observed moment of crossover generation until observed evidence for MT severing in WT, spr1, 3x-eb1, and clasp backgrounds (n = 2,027, 1,696, 1,718, and 1,147 crossovers in six cells, respectively). Boxplots show the 25th and 75th percentile as box edges, the line in the box indicates median value, and the whiskers show the 2.5th and 97.5th percentile. A Kruskal–Wallis test showed significant differences in sever waiting times among the genotypes (P < 0.001). Asterisks indicate significant difference from WT by Mann–Whitney U test. (E) Probability of new plus ends created by severing at crossovers being initially observed in a growing state (n = 882, 679, 764, and 598 crossover severing events in WT, spr1, 3x-eb1, and clasp backgrounds, respectively). Error bars show 95% confidence intervals. Asterisks indicate a significant difference from WT by Fisher’s exact test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We observed no significant difference (Fisher’s exact test) in the likelihood of observing severing after crossover formation between WT and 3x-eb1, with both showing a 44% probability of severing (Fig. 3 C). In the spr1 mutant, we measured a slight decrease in severing probability per crossover as compared with WT, at 42% (P < 0.05, Fisher’s exact test). By contrast, the clasp mutant showed a 54% probability of severing per crossover, a rate that is markedly higher than WT (P < 0.05, Fisher’s exact test; Fig. 3 C).
The severing likelihood depends both on the kinetics of severing itself (e.g., katanin recruitment, activation, and action) and on microtubule dynamics. Microtubule dynamics play an important role because crossovers get resolved both by severing and by microtubule ends depolymerizing beyond the crossover location (Nakamura et al., 2018). Once a crossover is resolved, it is no longer available to recruit katanin. To better assess severing activity itself, we measured the time from crossover formation to the time that optical evidence of severing was observed, a quantity we term the sever waiting time (Fig. 3 D and Fig. S1). When crossovers are resolved by microtubule dynamics, a sever waiting time is of course not calculated. When comparing the observed sever waiting time distributions, we found that both spr1 (P < 0.01, Mann–Whitney U test) and 3x-eb1 (P < 0.001, Mann–Whitney U test) have a significantly longer mean sever waiting time compared with WT. Strikingly, the clasp mutant shows a significant reduction in sever waiting time for microtubule severing compared with WT (P < 0.001, Mann–Whitney U test). In summary, microtubule severing activity is increased in the clasp mutant, suggesting that CLASP protein antagonizes severing activity. This was a surprise, because increased severing activity would be expected to aid, not inhibit, microtubule amplification (Lindeboom et al., 2013b).
Rescue after severing in +TIP mutants
During blue light–induced microtubule reorientation, most of the new growing microtubules are created by katanin-mediated severing (Lindeboom et al., 2013b). An outstanding question is how these new plus ends get stabilized immediately after severing, as they most likely lack the stabilizing GTP-cap. We therefore compared the fates of the new plus ends that are created by severing in WT and the +TIP mutants (Fig. 3 E). To score severing at crossovers, we asked whether there was evidence of either the formation of an optically resolved gap or a new growing end after a new crossover is made. In a previous study, we found that all gaps and new growing ends at crossovers were dependent on katanin-mediated severing (Lindeboom et al., 2013b). Gaps were always caused by shrinking of the new plus end. The frequency of stabilization and regrowth of new plus ends was determined by dividing the number of new growing ends emerging from crossovers by the total number of observed severing events (see Materials and methods for further details). In 3x-eb1 mutants, we observed that plus ends were in a growing state 12.2% of the time immediately after their creation by severing, a rate not significantly different from the 13.8% rate observed in WT (Fisher’s exact test). In the spr1 mutant, the probability of plus end regrowth from the crossover site after severing was significantly reduced to 9.7% (P < 0.05, Fisher’s exact test), a rate 70% of that observed in WT. Most strikingly, in the clasp mutant, we observed the new plus end created by severing in a growing state only 3.2% of the time (P < 0.05, Fisher’s exact test), a rate only 22% of that observed in WT. Thus, these tests of +TIP mutants reveal that SPR1 and CLASP both aid the stabilization of plus ends immediately after they are created by severing in epidermal cells during array reorientation, but CLASP is the dominant player, with stabilization being impaired nearly fivefold by loss of CLASP function alone.
Plus end stabilization and rescue are sensitive to local CLASP level
In higher plants, CLASP protein localizes to growing plus ends (Kirik et al., 2007) as described in animal cells (Akhmanova et al., 2001), but also accumulates along the lattice of cortical microtubules (Ambrose et al., 2007; Kirik et al., 2007), showing a heterogeneous distribution that includes brighter punctae (Kirik et al., 2007). We speculated that such distributed prepositioning along the lattice might be useful for both plus end rescue and plus end stabilization after severing. In support of this possibility, in in vitro studies of Schizosaccharomyces pombe, CLASP also showed heterogeneous localization of CLASP along microtubules and that local CLASP density was correlated with the probability of rescue (Al-Bassam et al., 2010).
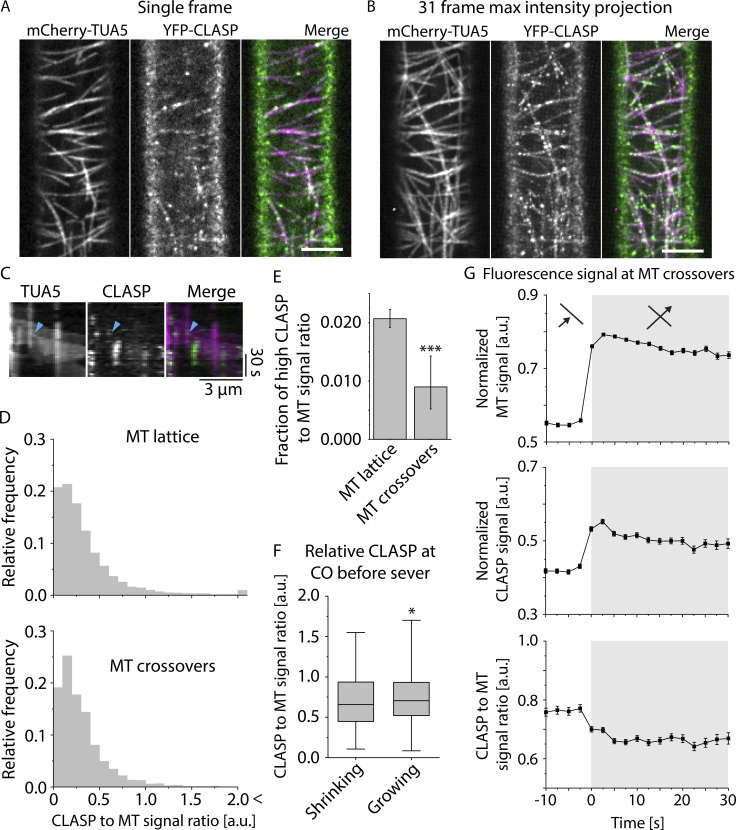
To investigate the relationship of CLASP protein localization to both rescue and stabilization after severing events, we imaged seedlings expressing YFP-CLASP expressed from the native CLASP promoter together with mCherry-TUA5 to label microtubules. We confirmed the previously described heterogeneous localization patterns of CLASP along the lattice in the etiolated hypocotyl cells used in this study (Fig. 4, A and B; and Video 4). In addition, we noted that YFP-CLASP accumulated in areas of high microtubule curvature (Fig. 5 C; and see especially dynamic examples in Video 5).
Figure 4.
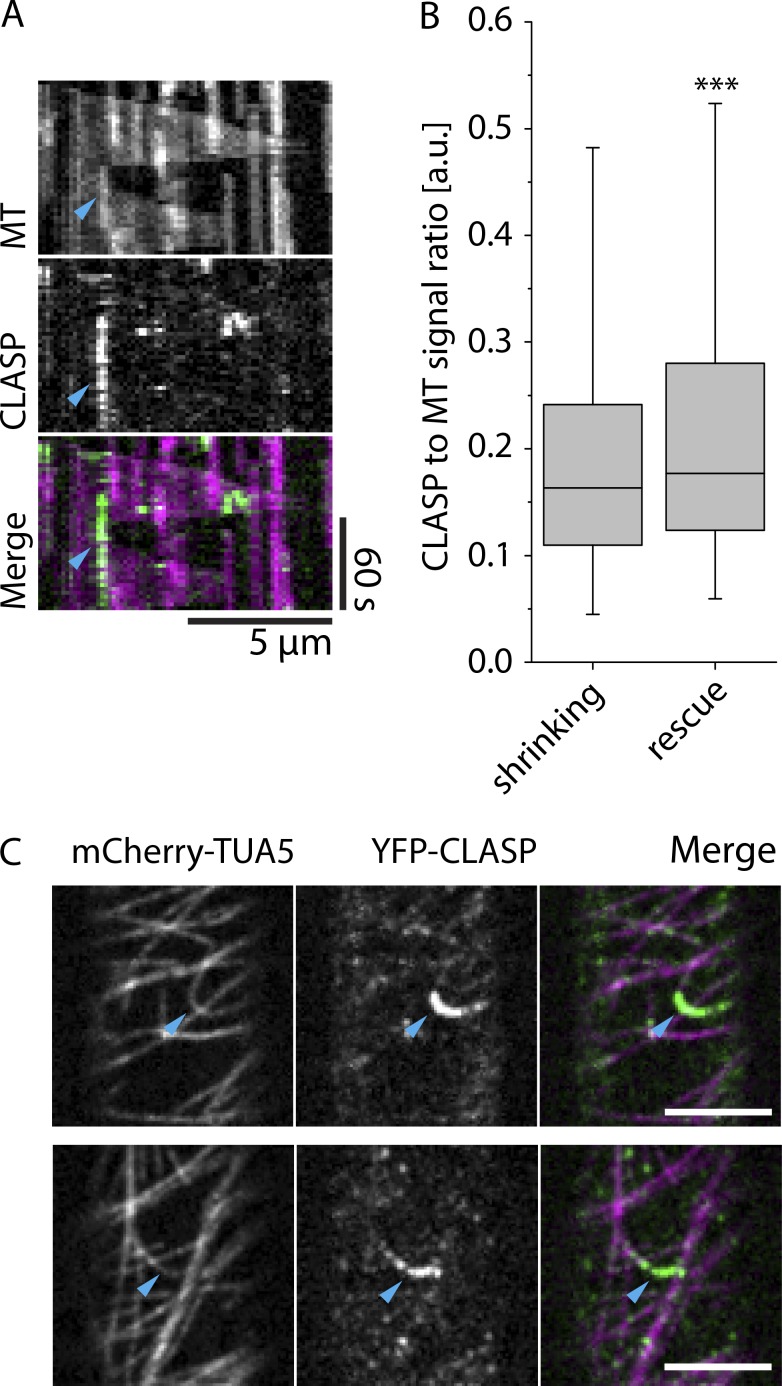
CLASP signal intensities at crossover sites. (A and B) Example of MT and CLASP signal distribution in a 3-d-old dark-grown hypocotyl epidermal cell of a clasp mutant that is complemented by a YFP-CLASP construct and in addition expresses mCherry-TUA5 (see Video 4). A single frame is shown in A, and B shows a maximum-intensity projection of the first 31 frames at a 5-s time interval. Scale bar is 5 µm. (C) Kymograph of MT plus end stabilization and regrowth after severing at a microtubule crossover, showing both MT and CLASP signal (see Video 6). Cyan arrowhead indicates the point of stabilization and regrowth of the new plus end as observed in the MT channel. (D) Histograms of the distribution of CLASP-to-MT signal ratio along the microtubules, excluding crossover sites. The top panel shows the distribution for free MT lattice, and the bottom panel shows the distribution exclusively for MT crossovers (n = 33,742 and n = 1,893 pixels for MT lattice and MT crossovers, respectively, with n = 396 crossovers in six cells). The distribution of CLASP-to-MT signal ratio for the crossovers is significantly different from that on the free MT lattice (P < 0.05, Mann–Whitney U test, one-tailed), with the crossover distribution depleted in the high-ratio values measured along the free lattice. (E) Bar graph depicting the frequency of high CLASP-to-MT signal ratio (>1.5) at MT lattice versus MT crossovers (n = 33,742 and n = 1,893 pixels for MT lattice and MT crossovers, respectively). The frequency of high CLASP-to-MT signal ratio is significantly lower at MT crossovers compared with MT lattice (***, P < 0.001, Fisher’s exact test, one-tailed; error bars show 95% confidence interval). (F) Boxplot of CLASP-to-MT signal intensity ratios at microtubule crossovers in the frame just before the lagging plus end was observed either growing (n = 100) or shrinking (n = 551). MTs observed growing immediately after severing showed modest but significantly higher CLASP-to-MT signal ratios than those observed shrinking after severing (*, P < 0.05, Mann–Whitney U test, one-tailed). Boxplots show the 25th and 75th percentile as box edges, the line in the box indicates median value, and the whiskers show the 2.5th and 97.5th percentile. (G) MT and CLASP signal intensities during crossover formation. The top panel shows the relative MT signal intensity, the middle panel the relative CLASP signal intensity, and the bottom panel shows the ratio of CLASP-to-MT signal intensity (n = 1,532 crossovers in six cells in six plants). The white background indicates the time before the crossovers are formed, and the gray background indicates the time at which the crossover is formed.
Figure 5.
CLASP signal and microtubule rescue. (A) Kymograph of a MT rescue event coinciding with high CLASP signal intensity. Cyan arrowhead indicates the point of MT rescue. (B) Boxplot of CLASP-to-MT signal intensity ratios for microtubules that continue shrinking and microtubules that get rescued. Boxplots show the 25th and 75th percentile as box edges, the line in the box indicates median value, and the whiskers show the 2.5th and 97.5th percentile. Microtubules were observed shrinking in 2,716 frames, and we observed rescue 301 times. CLASP-to-MT signal intensity ratios were shown to be significantly higher in locations where the rescues occurred by Mann–Whitney U test; ***, P < 0.001. (C) Examples of high CLASP signal intensity on highly curved microtubule in 3-d-old dark-grown hypocotyl epidermal cell expressing YFP-CLASP and mCherry-TUA5 (see Video 5). Scale bars are 5 µm.
At sites of severing at crossovers, we asked whether YFP-CLASP is present at crossovers, whether CLASP may be specifically recruited to crossovers, and whether CLASP quantity at crossovers influences the likelihood of plus end stabilization after severing. When we imaged YFP-CLASP in etiolated hypocotyl cells, we noticed that in addition to the previously described localization patterns, CLASP signal often appeared to be more prominent at crossovers than along the adjacent lattice (Fig. 4, A and B). Such increase in signal might indicate recruitment of CLASP to the crossover or a simple superimposition of CLASP signal associated with the lattice, just as the signal from labeled tubulin itself also increases at crossover sites. To distinguish between these possibilities, we measured YFP-CLASP and mCherry-TUA5 signal at crossover sites as crossovers are created and the ratio of these signals (Fig. 4 G). These measurements showed an increase and stable plateau of TUA5 signal as crossovers are formed, as expected, with a simultaneous increase and stable plateau of CLASP signal. However, the ratio of CLASP to TUA5 signals did not increase, but rather fell slightly (and significantly; Fig. 4 G). These results indicate that CLASP is associated with the two microtubule lattices at crossovers as they are formed, but the apparent increase in CLASP signal is due to signal superposition, not specific recruitment. The falloff in signal further suggests that although CLASP is present at crossovers, it may either interact less well with the lattice at crossovers or be competing with other proteins that are recruited to crossovers.
Prominent spots of intense CLASP signal are observed along the lattice of cortical microtubules. The above analysis examined CLASP signal at crossovers during and immediately after their formation. To ask whether prominent CLASP foci are recruited to crossovers specifically after their creation at any point, we measured the ratio of YFP-CLASP signal to mCherry-TUA5 signal at crossovers and along the lattice between crossovers in single image frames (Fig. 4 D). These measurements showed that the distribution of CLASP signal at crossover sites is similar to that on the rest of the lattice but is truncated for the higher-intensity values. To assess the significance of this truncation, we compared the frequency of CLASP-to-MT ratio intensity values above a threshold of 1.5 at both locations, values that represent the more distinct YFP-CLASP foci. Crossovers were significantly deficient in these higher CLASP signal intensities (Fig. 4 E), indicating that CLASP foci are underrepresented at crossovers, a result consistent with the dynamic crossover analysis.
Our data indicated that CLASP is present at crossovers and is required for efficient stabilization of new plus ends created by severing, but is stabilization sensitive to the level of CLASP at the crossover at the time of severing, as was observed in vitro for rescue with yeast CLASP (Al-Bassam et al., 2010)? We identified severing events in the tubulin channel for cells expressing YFP-CLASP and mCherry-TUA5, measured the CLASP signal at the crossover severing sites in the image frame just before severing was detected, and compared the results for events where plus ends that were stabilized and regrew to events where plus ends were not stabilized and shrunk immediately (Fig. 4 C; see Video 6). Stabilization events showed a significantly higher CLASP signal than did shrinking events (Fig. 4 F). Taken together, these data indicate that CLASP is present at crossovers and that stabilization after severing is positively correlated with the amount of CLASP at the crossover. At the same time, CLASP concentration at crossovers is slightly lower than along the lattice between crossovers and has less tendency to accumulate into prominent foci.
Al-Bassam et al. (2010) observed that rescue of shrinking plus ends was correlated with local CLASP concentration in vitro. To ask whether this may also be the case in living cells, we analyzed the ratio of CLASP to microtubule signal and related this to the behavior of shrinking microtubule ends (Fig. 5 A and see Materials and methods). We quantified the CLASP-to-MT signal ratio in 2,716 frames where the microtubule continued shrinking and 301 instances where the microtubule got rescued. When we compared the signal intensity ratio between shrinking and rescue frames, we measured a significantly higher CLASP-to-microtubule ratio at locations where rescue was observed (P < 0.001, Mann–Whitney U test, one-tailed; Fig. 5 B), indicating that locations of higher CLASP concentration on the microtubule lattice are preferential rescue sites for interphase microtubules in plants. Thus, high local CLASP concentration on the microtubule lattice aids rescue of shrinking plus ends, both in vitro and in vivo.
Rescue has also been reported recently to be associated with microtubule crossovers in animal cells (Aumeier et al., 2016). Although we did not observe crossovers to be hot spots for CLASP accumulation, we asked whether they nonetheless might be hotspots for rescue in Arabidopsis cells. We used kymograph analysis to compare the microtubule signal intensity measured at shrinking plus ends to that measured at locations where plus ends were rescued. Microtubule signal intensity is higher at crossovers because of the combined signal of microtubules at crossover sites. If indeed microtubule rescue is elevated at crossovers in plants, we would predict that microtubule signal should be higher on average at sites of rescue compared with locations where plus ends are shrinking. However, we found that the relative microtubule signal intensity was not significantly higher where plus ends were rescued (n = 301) compared with locations where they continued to shrink (n = 2,716) in Arabidopsis (P > 0.98, Mann–Whitney U test, one-tailed; Fig. S3). Thus, in etiolated hypocotyl cells, crossovers do not appear to be important sites of rescue.
Simulations for microtubule propagation efficiency by severing in +TIP mutants
Our measurements of microtubule array reorientation revealed that CLASP is a key player in cortical array remodeling in response to light stimulation. Measurements of microtubule behaviors revealed two differences that stood out when comparing clasp mutants to WT and the other +TIP mutants: a 28% reduction of the intrinsic rescue rate from 1.56 to 1.12 events/min, and a 4.7-fold decrease in the probability of plus end growth immediately after severing from 14% to 3%. Significant but quantitatively more modest changes in other parameters of polymer dynamics, such as growth and shrinkage rates, were also observed. To assess the contributions and significance of these changes in polymer behaviors and dynamics in amplifying the new population of microtubules that drives array reorientation, we designed and used a probabilistic model for generating longitudinal microtubules by severing and rescue (Nakamura et al., 2018). The simulations were parameterized with estimates from our in vivo measurements of microtubule dynamics and crossover outcomes in WT, spr1, 3x-eb1, and clasp backgrounds (Table S1), together with measurements of the spacing between transverse microtubules before reorientation (Fig. S2 and Materials and methods). In each simulation run, a single microtubule was initiated transversely to a grid of preexisting microtubules, and its fate and those of its progeny were assessed as a function of time. For each genotype or experimental test, n = 5 × 104 runs were performed to build up a probabilistic picture of microtubule fate. The results were analyzed in terms of two measures: the probability that a microtubule, and all of its progeny created at severing events, die out (extinction probability), and the expected number of surviving descendants of a microtubule at a fixed time point after initiation (Fig. 6, A and B).
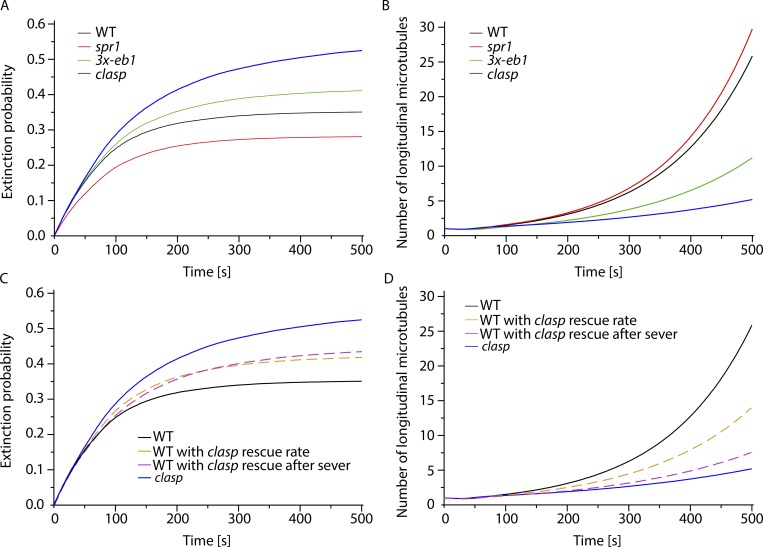
Figure 6.
Simulation of microtubule amplification driven by severing. (A) Time evolution of the fraction of extinctions in WT, spr1, 3x-eb1, and clasp mutants. Extinctions are more likely in less stable mutants, i.e., mutants that are less deep into the unbounded growth regimen. (B) Time evolution of the average number of microtubules during the amplification process shows that a lower probability of rescue after severing and a less deep unbounded growth regimen in 3x-eb1 and clasp mutants result in a slower amplification compared with spr1 and WT. (C and D) Time evolution of the fraction of extinctions in WT with clasp intrinsic rescue rate, and WT with clasp rescue after severing probability in silico mutants, shows that even if the two reduced rescue parameters have a similar effect on the extinction probability (C), the probability of rescue after severing Ps,+ has a greater impact on the amplification process (D). Indeed WT microtubules with clasp Ps,+ perform an amplification that almost completely reproduces the clasp results (dashed purple curve).
For all genotypes, simulations resulted in net microtubule amplification (Fig. 6 B) despite a finite probability of failure for each trial (Fig. 6 A). This outcome is consistent with experimental observations, where some degree of reorientation was observed for all genotypes, even in the most severe mutant, clasp. These results can be understood by examination of the values for the basic microtubule dynamics (growth and shrinkage speeds, transition frequencies), which for all genotypes are in the so-called unbounded growth regimen, a state where the measured polymer dynamics predict net polymer growth over time, rather than a state of equilibrium. The measure for this effect is the average growth velocity (Dogterom and Leibler, 1993). A positive average growth velocity itself leads to net amplification because individual microtubules are expected to survive on average in spite of being severed (Tindemans and Mulder, 2010).
Although all simulations showed a degree of amplification, the magnitudes of amplification and the probabilities of extinction varied markedly by genotype and mirrored the measured defects on reorientation speed, with clasp being the most defective in both amplification and having the highest likelihood of extinction, followed by 3x-eb1 and spr1 (Fig. 6, A and B). Thus, the measured microtubule dynamics and behaviors in clasp predict not only the lowest rate of net amplification, but also the highest likelihood of amplification failure.
To understand which of the two major changes in microtubule dynamics observed in the clasp mutant might more greatly affect the speed of amplification, the reduction in the intrinsic rescue rate (rr) or the quantitatively larger reduction in the probability of plus end stabilization after severing (Ps,+), we created two synthetic in silico mutants: WT with the clasp intrinsic rescue rate (WT/clasp-rr) and WT with the clasp probability of stabilization immediately after severing (WT/clasp-Ps,+). As shown in Fig. 6 C, both mutations have a similarly detrimental effect on extinction probability with respect to WT. However, amplification in WT/clasp-Ps,+ is slower than that in WT/clasp-rr, indicating that the change in Ps,+ dominates the amplification defect when CLASP function is lost (Fig. 6 D). Further, the reduction in Ps,+ by itself almost completely reproduces the clasp phenotype. Taken together, the simulation analyses quantify the importance of CLASP function in amplifying new longitudinal microtubules and support the hypothesis that the dominant function of CLASP in building a new array by severing-driven microtubule amplification is by its specific function in plus end stabilization immediately after severing.
Discussion
In the cortical arrays of higher plants, growing microtubule plus ends are created both by microtubule nucleation (Wasteneys and Williamson, 1989; Nakamura et al., 2010) and by microtubule severing (Wightman and Turner, 2007; Lindeboom et al., 2013b; Zhang et al., 2013). During the reorientation of cortical arrays upon perception of blue light, creation by severing in fact dominates new microtubule generation, constituting more than 60% of the total initiation events. A recent in vitro study has shown that templated nucleation, whether from nucleation complexes or microtubule seeds, is an energetically unfavorable process that is made more efficient by microtubule associated proteins (Wieczorek et al., 2015). Likewise, the lack of a stabilizing GTP-cap presents an energetic challenge to the creation of new microtubules from severed ends. Our data identify CLASP as the major factor that acts to stabilize plus ends created by severing to promote the generation of new microtubules in plants.
The precise mechanisms by which CLASP might act to stabilize severed ends and promote regrowth have not been identified. CLASP action might lower the energetic barrier of templated polymer initiation from these ends, it might antagonize the transition to catastrophic disassembly in the absence of a GTP-cap, or perhaps both. Observation of free tubulin at sites of CLASP accumulation in vitro suggested that CLASP might act to concentrate tubulin locally to promote rescue of shrinking minus ends (Al-Bassam et al., 2010). This activity might also aid templated assembly from plus ends crated by severing. An alternative mechanism, not mutually exclusive, is that CLASP might antagonize catastrophe by providing increased stability as it interacts with the microtubule lattice. Proteins that influence microtubule stability often do so based on preferential binding to αβ-tubulin of specific curvature (Ayaz et al., 2012; Brouhard and Rice, 2014). We indeed observed examples of relatively higher CLASP fluorescence along highly curved microtubule regions (Fig. 5 C and Video 5). Therefore, CLASP might act to stabilize severed ends by halting the progression of curvature that occurs in microtubule protofilaments during depolymerization by binding to tubulin. A third, and intriguing, possibility is that CLASP might stabilize the lattice by facilitating repair of lattice defects. Crossover sites in animal cells and sites of lattice curvature have been shown to accumulate lattice defects that are self-repaired by incorporation of GTP-tubulin (Aumeier et al., 2016; de Forges et al., 2016), producing GTP islands that appear to stabilize the lattice as they are correlated with increased rescue frequency (Tropini et al., 2012; Aumeier et al., 2016; de Forges et al., 2016). In animal cells, lattice defects at crossovers were proposed to arise from friction as the microtubules collide. Although microtubules at the cortex of plants cells appear to be much more constrained by their lateral association with the cell membrane, this constraint may also produce local lattice strain and curvature at the crossover to create a hot spot for lattice defects and GTP island creation. Interestingly, the Roll-Mecak laboratory has recently shown that a previously unexpected result of katanin action is to also allow for the incorporation of GTP-tubulin dimers into the lattice as it removes GDP-tubulin dimers (Vemu et al., 2018). They propose that an important result of GTP-tubulin incorporation at sites of katanin action is to stabilize severed ends against catastrophe, allowing for more effective generation of new microtubules through severing. Thus, another possibly for CLASP action could be to facilitate the incorporation of GTP-tubulin into lattice defects, created either by local lattice curvature at crossovers or by the action of katanin, which is recruited to crossovers in plants (Lindeboom et al., 2013b) and itself binds preferentially to sites of lattice defects (Díaz-Valencia et al., 2011). Perhaps such a function could be related to the ability of CLASP to interact with free tubulin dimers and microtubule lattice at the same time as a local GTP-tubulin concentrating mechanism (Majumdar et al., 2018). GTP islands, including those at crossovers, have been also shown to recruit CLIP-170, a binding partner of CLASP in animal cells (de Forges et al., 2016). Although higher plants lack CLIP-170, it is possible that either an unidentified CLASP partner or CLASP itself could recognize sites of lattice defect and repair, recruiting CLASP. We found both that katanin-mediated severing at crossovers was repressed by CLASP, and that labeled CLASP protein become less abundant on the lattice as crossovers were made. These observations suggest that CLASP and katanin may compete at crossovers, consistent with the idea that these proteins recognize closely adjacent or even identical features on the lattice, including lattice defects and/or their repair. However, two observations that may argue against this defect repair hypothesis are that we did not observe CLASP to specifically accumulate at crossovers, nor did we observe crossovers to be favored sites of rescue. On the other hand, if CLASP is also competing with katanin and other proteins recruited to crossovers, such as the katanin subunits and SPR2 (Nakamura et al., 2018), then localization to sites appropriate to facilitate lattice repair may be masked by competitive crowding at less preferred sites. It will be interesting to dissect the role of competition of binding sites for the Katanin machinery and its severing function and CLASP and its stabilizing function in vitro.
Loss of CLASP function in Arabidopsis causes a range of altered phenotypes including defects in cell and tissue morphogenesis and cell division. Plant cell morphogenesis depends on interphase microtubule organization and function to establish directional, or anisotropic, extension of plant cell walls. Previous studies have proposed two mechanisms by which CLASP function might mediate interphase cortical array organization. First, loss of clasp function has been observed to reduce interaction of microtubules with the cell cortex in some cells (Ambrose et al., 2007). In these studies, a higher degree of parallel ordering was also observed. This defect was proposed to encourage nematic ordering by lateral interaction of the dissociated microtubules. Thus, normally, CLASP may function to promote normal patterns of organization by preventing such lateral interactions. Here, we measured an in increase in transverse ordering in clasp mutants before blue light stimulation, but we observed no evidence for pervasive membrane dissociation in the etiolated hypocotyl cells we studied, observing only two clear incidents of microtubule dissociation on ∼3,300 µm2 over 30 min in six cells of the CLASP and MT dual-labeled plants. Thus, higher baseline ordering is likely caused by a different mechanism in these cells. Second, CLASP has been observed to accumulate along high-curvature edges of newly divided cells and to be associated with the ability of cortical microtubules to grow around these edges, where they were otherwise observed to show a high likelihood of catastrophe (Ambrose et al., 2011). This activity was proposed to act as a permissive “gate” to bias the predicted self-ordering of plant cortical arrays (Deinum and Mulder, 2013) to direct array orientation in these cells. An entirely different mechanism of CLASP function that supports plant growth and development is the tethering of endosomal compartments near the plasma membrane (Ambrose et al., 2013), a function that is implicated in supporting the maintenance of the membrane proteins PIN2 (Ambrose et al., 2013), which transports the master plant hormone auxin, and BRI1 (Ruan et al., 2018), the receptor for the growth and cell division regulating protein BR that transports plant growth hormones auxin and brassinosteroid. Here we present evidence for a third mode of action of CLASP in cortical array organization and function, the stabilization of new plus ends created by severing to promoting microtubule generation and amplification. This mechanism is an essential engine in building a new array during stimulated array reorientation by blue light (Lindeboom et al., 2013b). Although our data indicate an important role for CLASP in building the new transverse array, they do not rule out a possibility that CLASP might also facilitate in some way the turnover of the original transverse array.
In addition to the clasp mutant, the 3x-eb1 mutant showed a significant reduction in reorientation speed as well. It remains to be determined whether this is because of EB1 function itself directly or perhaps through the direct interactions between EB1 and CLASP (Lawrence et al., 2018).
Severing is a fundamental tool used by cells across phyla to manipulate and regulate cytoskeletal arrays. Severing is proposed not only to aid in the disassembly of microtubules, but also to play an important role in building new arrays in important cell types including neurons, epithelial cells, and meiocytes (Roll-Mecak and Vale, 2006). Our observations and experiments in higher plant cells provided direct and quantitative evidence that severing can in fact act generatively to build new microtubule arrays. An important feature of this mechanism is how both new polymer ends created by severing are stabilized and subsequently regulated. We recently reported that microtubule minus ends in the treadmilling cortical arrays of higher plants are stabilized by a minus end tracking protein novel to higher plants, SPR2 (Nakamura et al., 2018). Without SPR2 function, minus ends show rapid depolymerization in vivo. This minus end stabilization activity was found to be essential for efficient microtubule amplification by severing and rapid cortical array reorientation. Our investigations here present a second important feature of this mechanism: the specific rescue by CLASP of the new plus ends created by severing. In animal cells, the proteins used to stabilize the new plus ends generated by severing have not been identified. As CLASP is conserved across animals and plants, our studies suggest that it may be a compelling candidate to investigate for this function to enable microtubule severing to rapidly build new cytoskeletal arrays.
Materials and methods
Plant material
Microtubules were marked by introducing the 35S-YFP-TUA5 construct (Shaw et al., 2003) into four genetic backgrounds of Arabidopsis—WT, spr1-6 (Sedbrook et al., 2004), clasp1-1 (Kirik et al., 2007), and 3x-eb1 (see below)—all ecotype Col-0. WT (Shaw et al., 2003) and spr1-6 (Sedbrook et al., 2004) transformants were described previously. For this study, 35S-YFP-TUA5 (Shaw et al., 2003) was introduced into clasp1-1 by Agrobacterium-mediated transformation. In this paper, spr1-6 and clasp1-1 are referred to as spr1 and clasp.
Introduction of 35S-YFP-TUA5 into a triple mutant eb1 background was accomplished as follows. The Ateb1a-2 T-DNA insertional mutant (WiscDsLox481-484J12; insertion in the seventh exon of AT3G47690) and the Ateb1b-2 T-DNA insertional mutant (WiscDsLox331A08; insertion in the sixth intron of At5g62500; see Galva et al. [2014] for additional details) were obtained from the Arabidopsis Knockout Facility (University of Wisconsin Biotechnology Center, Madison, WI; Woody et al., 2007). The Ateb1c-2 T-DNA insertional mutant (SALK_018475; insertion in the third exon of AT5G67270) was generated at the Salk Institute (San Diego, CA; Alonso et al., 2003) and obtained from the Arabidopsis Biological Resource Center. Ateb1a-2, Ateb1b-2, and Ateb1c-2 mutant plants were cross-pollenated then progeny self-pollenated to generate the Ateb1a-2 Ateb1b-2 Ateb1c-2 triple mutant. T-DNA insertions were tracked and homozygosity determined by performing PCR analysis (Klimyuk et al., 1993). Primers and primer pairs are listed in Table S2. We refer to this triple eb1 knockout mutant line as 3x-eb1 throughout the article. This 3x-eb1 line was then transformed with the 35S-YFP-TUA5 microtubule marker (Shaw et al., 2003).
For the MT and CLASP dual-labeled line, we transformed clasp1-expressing ProAtCLASP-YFP-AtCLASP (Kirik et al., 2007) with a ProUBQ10-mCherry-TUA5 construct. This microtubule reporter was generated by amplifying a 634-bp genomic upstream regulatory fragment of UBQ10 from pUBN-GFP-DEST (Grefen et al., 2010). The CaMV 35S promoter was excised from the modified pMDC43 vector encoding mCherry (Kirik et al., 2007) and replaced by the UBQ10 fragment. LR recombinase (Invitrogen) was used to introduce the Arabidopsis TUA5 cDNA (Gutierrez et al., 2009) into the modified pMDC43 Gateway binary vector to generate a ProUBQ10-mCherry-TUA5 construct. The binary vector harboring the construct was then introduced into WT Arabidopsis plants (Col-0) by Agrobacterium-mediated transformation using the floral dip method (Clough and Bent, 1998). Transgenic T1 plants were selected for Hygromycin B selection. T2 progeny plants with normal growth and development (with the exception of mild twisting) were used for experiments.
Plant growth
Seeds were surface sterilized with a 70% ethanol solution, stratified for 3 d at 4°C, and sown on 1% agar containing Hoagland’s No. 2 salts at pH 5.7. After 1 h of light exposure on a clean bench, plates were wrapped in aluminum foil and set on edge at approximately a 10° angle to vertical, and seedlings were germinated and grown in darkness for 60–72 h at 22°C.
Specimen mounting
Foil-covered plates were unwrapped, and seedlings were mounted under red safelight conditions to prevent de-etiolation (Paredez et al., 2006). Seedlings were placed gently on a coverslip in sterile water and affixed with a 2-mm-thick 1% agarose pad. To reduce specimen drift, the mounted seedling was rested for 20–30 min before observation.
Microscopy
All observations were made in epidermal cells in the upper hypocotyl, a region where cell expansion is rapid in 3-d-old etiolated seedlings. The time-lapse imaging of the YFP-TUA5–labeled lines was performed at 5-s time intervals for a duration of 30 min on a CSU-X1 spinning-disk confocal head (Yokogawa) mounted on a DMI6000B microscope with Adaptive Focus Control (Leica) and a 100× Plan Apo 1.4-NA oil-immersion objective. Images were acquired with an Evolve 512 EMCCD camera (Photometrics). For each image, excitation was supplied by a 488-nm laser for 300 ms at 4.5 mW as measured at the end of optical fiber feeding the spinning disk unit.
We performed imaging of cells dual-labeled with YFP-CLASP and mCherry-TUA5 with a CSU-X1 spinning disk head (Yokogawa) mounted on an Eclipse Ti body (Nikon) with a 100× 1.4-NA Plan Apo oil-immersion objective and perfect focus system (Lindeboom et al., 2013a). Exposures were acquired with an Evolve 512 EMCCD camera (Photometrics) every 2.5 s, using a 491-nm laser at 8.2 mW to excite YFP-CLASP for 500 ms and a 591-nm laser at 8.2 mW to excite mCherry-TUA5 for 300 ms. All experiments were performed at ∼21°C.
Reorientation analysis
We used the ImageJ plugin LOCO (Lindeboom et al., 2013b) and a Matlab script to extract microtubule orientation output to analyze microtubule reorientation after blue light induction. We used a 21 × 3-pixel kernel that was rotated with increments of 9° over a range of 180°. Then, for each pixel, we assigned the angle for which the kernel contained the highest total fluorescence signal. To limit our measurements to pixels that contained microtubule signal, we performed a default threshold in ImageJ and applied this threshold to the angular data. For each angle, we then determined the fraction of the total pixels above the threshold level. From these angular fractions, we also extracted the longitudinal (L) and transverse (T) order parameters based on over- or underrepresentation in the longitudinal and transverse orientation, respectively (Lindeboom et al., 2013b). To calculate T, the fraction of angular values in a transverse bin 45° wide and centered at 90° was given a weighting of 1, and the fractions at all other orientations were given a weighting of –1/3. These products were summed to yield T. With these weightings, an isotropic distribution of angles yields a value T = 0, and a distribution angle entirely within the transverse bin yields a value T = 1. Similarly, L was constructed using a longitudinal bin 45° wide centered at 0°.
To ensure that there was sufficient transverse order at the start of stimulation to measure the transition to longitudinal orientation, datasets were selected for analysis where the initial transverse order parameter was lower than 0.1.
Analysis of microtubule dynamics
The growing and shrinking ends of individual labeled microtubules were visualized by time-phased subtraction of image time series acquired from seedlings expressing YFP-TUA5 as described previously (Lindeboom et al., 2013b). A time phase of 1 frame (5 s) was used to highlight plus end growth, and trajectories of segmented growing ends in selected regions of interest (ROIs) were created blindly using the PlusTipTracker software package (Applegate et al., 2011). These paths were then used to measure microtubule dynamic parameters using a custom pipeline in Matlab. First, the paths created by PlusTipTracker were used to generate kymographs. All kymographs generated were examined, and those showing clear evidence of plus end growth in at least five frames were then traced by hand using a custom graphical user interface in Matlab. Finally, polymerization and depolymerization velocities, as well as the frequency of rescue and severing rates, were calculated from the kymograph traces as reported earlier (Nakamura et al., 2018). This protocol allowed for less biased sampling and measurement of end dynamics than might result from selection by eye.
Analysis of severing and events at crossovers
To analyze the formation and resolution of microtubule crossovers in plants expressing YFP-TUA5 in the different genetic backgrounds, we time-phased and subtracted image series to highlight new microtubule growth (time phase of one frame; Lindeboom et al., 2013b). Using a custom-written environment in Matlab, we created defined ROIs in each image series in which were marked every new crossover, recording the x and y coordinates, the start of crossover formation, the time of crossover resolution, the angles of the old and new microtubules, and whether the old and/or new microtubule at the crossover got severed (Video 7). Severing was tallied either if an optically resolved gap in signal was formed at the crossover or if new end growth was observed to emerge from the site of the crossover. Our previous analysis showed that all observed gap and new growing end events are created by katanin-mediated severing (Lindeboom et al., 2013b). If a severing event took place at the crossover, we documented whether the new plus end created by severing was initially shrinking (gap formation) or growing (new growing end seen as a “comet” in the time-phased images).
This analysis of events at crossovers was also performed on the 2.5-s-interval image time series acquired for the analysis of CLASP and tubulin signal, with additional steps to register the two image channels and correct for photobleaching (Nakamura et al., 2018). In ImageJ (Rasband, 2012), we first registered the images in the microtubule channel using the plugin MultiStackReg. We then used the transformation matrix from this registration to register the CLASP channel. Each channel was subsequently corrected for photobleaching using an exponential fit to total signal intensity of the original image series. We then isolated a 7 × 7-pixel region centered around each of the crossover coordinates. The 1-pixel-wide outer border was used for determining the background signal intensity for both CLASP and MT signal. We discarded the highest 12-pixel values of these 28 pixels, as they typically represent the microtubule signal of the two microtubules forming the crossover. We took the median intensity value of the remaining pixels as the local background for the crossover for each individual frame of each crossover. To calculate the crossover signal intensity, we subtracted the background value calculated from the border from the mean intensity of the inner 5 × 5 pixels of the crossover ROI. We then normalized the signal intensity by dividing each individual crossover signal intensity value by the maximum value for each measurement series. The normalized crossover intensities were calculated for both the CLASP and MT signal, and the crossover event intensities were aligned with each other over time based on either the start or end frame of the crossover event. We calculated the CLASP-to-MT signal ratio by dividing the normalized CLASP intensity by the normalized MT intensity. The standard errors for this ratio estimate were calculated from the error propagation of the standard errors of the separate CLASP and MT signal intensities.
To quantify the CLASP-to-MT signal ratio at microtubule crossovers we used the method described above and extracted the CLASP-to-MT signal ratio in the frame before we observed evidence for severing at the crossover (T = −2.5 s) and grouped the signal intensity ratios based on whether the new plus end was observed growing or shrinking.
Quantification of microtubule and CLASP intensity at sites of rescue
For both the microtubule and CLASP channels, we first applied image registration and bleaching correction in ImageJ. For each frame, we normalized the intensity values per frame to the median intensity value after dividing the intensity values into 10 equal-sized bins and discarding the values in the highest and lowest bin to remove outliers. We then used these videos to extract kymographs using the method described in the Analysis of microtubule dynamics section. We traced the edge of the microtubule signal in the kymograph by hand and used a Matlab script to identify microtubule shrinkage and rescue events. Because CLASP in Arabidopsis cells has been reported to be a plus end tracking protein (Kirik et al., 2007), we wanted to exclude any bias of CLASP measurement that may occur because of rescue and subsequent recruitment of CLASP to the growing plus end. We therefore extracted the CLASP and microtubule intensity in a 3-pixel-wide ROI in the two frames immediately before that location is reached by the depolymerizing microtubule end. We assessed the averaged and normalized pixel intensity of these ROIs and sorted them into two groups for comparison: one group in which the shrinking microtubule continues to shrink in the following frame, and the other group in which the microtubule is rescued in the following frame.
Analysis of CLASP-to-MT signal intensity ratios along the microtubule lattice
To assess the distributions of CLASP signal intensity at microtubule crossovers and at the free microtubule lattice, we used spinning disk confocal images of plants expressing YFP-CLASP and mCherry-TUA5. To increase the proportion of crossovers, we selected a single frame 2.5 min after the start of the time-lapse video, manually traced all visible microtubules in the mCherry-TUA5 channel by hand, and recorded the line coordinates in Matlab. For these trace lines, we then determined where lines intersected and formed crossovers, so that we could determine which pixels in the images were associated with crossovers and which with microtubule lattice. Based on the point spread function of the microscope settings we used, we defined the crossover as the 5 pixels around the exact calculated intersection.
Starting with the raw images from the MT and CLASP channels, we performed a rolling ball background subtraction using ImageJ with a ball size of 30 pixels. After the background subtraction, we normalized the image intensities in both channels by the minimum and maximum to get a range of intensities between 0 and 1. From these normalized signal intensities, we calculated the CLASP-to-MT signal intensity ratios by dividing the normalized CLASP by the normalized MT signal intensity pixel by pixel. We used the Matlab function improfile to use the hand-traced coordinates and extract the signal intensities along those lines from the CLASP-to-MT ratio image (conserving the correct pixel length by providing the length of the line as an input). We then separated the CLASP-to-MT signal ratios into free lattice and microtubule crossovers based on the intersection points determined earlier. Every crossover consists of two optically resolved microtubules, and the crossover intensities are evaluated from the perspective of each microtubule separately. This means that the center of the crossover is evaluated twice, which we believe is appropriate considering that both microtubule at the crossover contribute to that local signal.
Microtubule bundle distances
We measured the distance between transversely oriented microtubules in WT, spr1, 3x-eb1, and clasp mutant backgrounds expressing YFP-TUA5 as described previously (Nakamura et al., 2018). Each time series was rotated so that the long axis of the cell was on the vertical axis of the image. After rolling ball subtraction (radius = 30 pixels), the intensity values were measured along the vertical midline of the cell image. Signal peaks were extracted using the Matlab function findpeaks. The normalized first derivative of peak prominence was used as a filter (values higher than 0.015) to identify microtubule signal peaks. The intermicrotubule distances were calculated from the positions of these peaks.
Statistical analysis
In several cases, we compare continuous variables among different genotypes, such as for reorientation speed, microtubule growth, shrinkage speeds, and sever waiting times. For all of these examples, measured values clearly did not have the appearance of a symmetrical distribution about a mean; therefore, we did not assume a normal distribution of these variables and chose to use nonparametric tests. We first performed a Kruskal–Wallis test to assess whether the compared samples are from the same distribution, thereby reducing the chance of a type I error. If the Kruskal–Wallis test indeed rejected the null hypothesis, we then performed a set of pairwise Mann–Whitney U tests between WT and each mutant. Normal distributions of values were also not assumed when comparing the CLASP-to-MT signal ratios between categories such as shrinking versus rescue and sever event versus nonsever event at crossovers; therefore, these comparisons were also made with Mann–Whitney U tests.
For comparing transition rates from growing to shrinking and vice versa, we assumed that the switching rates are the result of a single step Poisson process, for which the rate ratio Exact test is the appropriate statistical test. Although it is common in the field to assume that microtubule rescues and catastrophes are simple Poisson processes, we want to acknowledge that this assumption likely is an oversimplification.
For cases in which we compare binary outcomes between genotypes, such as severing probability and the probability of stabilization of the new plus end after severing, we used Fisher’s exact test. Fisher’s exact test and the χ2 test are both appropriate for assessing independence of binary outcomes and reach the same P values for higher sample sizes. However, when samples counts are relatively low, Fisher’s exact test is more precise. Because this is the case for stabilization of plus ends after severing in the clasp mutant, we opted to use the Fisher’s exact test consistently in our paper.
Stochastic model
To explore the importance of the stabilization of microtubule plus ends generated by severing, we built a stochastic model of longitudinal microtubules undergoing dynamic instability in a grid of stable transverse microtubules (Nakamura et al., 2018). In that article, the model addressed protein activities that affected both plus and minus end dynamics. Here, the activities we studied only affected plus ends by experimental measurement, and minus ends were stable by comparison. Thus, in the simulations presented here we kept plus ends dynamic while holding minus ends stable.
Dynamic parameters of the model are the growing speed v+, the shrinking speed v−, the catastrophe rate rc (the rate of transition from the growing to the shrinking state), and the rescue rates rr (the rate of transition from the shrinking to the growing state). The values of these parameters were measured in vivo in hypocotyl epidermal cells undergoing light-driven reorientation (Fig. 2 and Table S1). In addition, the model uses the probability of rescue after severing for the plus end, Ps,+. This parameter is the probability that the newly created plus end grows just after the severing event.
As the sole static model parameter was the distance d between neighboring transverse microtubules, this parameter was calculated according to the distribution:
where the length D is chosen such that the mean spacing between transverse microtubules davg is consistent with the distances experimentally measured between microtubules/bundles (Table S1 and Fig. S2).
When the plus end crosses a transverse microtubule, it creates a crossover. At every crossover, there is a competition between two different effects: either the crossover is erased by the shrinkage of the plus end caused by dynamic instability, or it leads to a severing event. Whether one effect wins against the other is determined by an intrinsic severing waiting time distribution Wk,θ(t), where Wk,θ(t) is the gamma probability density function (Papoulis, 1984):
with k and θ as scale and shape parameters of the distribution, and Γ(k) as the Euler gamma function (Abramowitz and Stegun, 1965):
Given the impossibility of measuring the intrinsic waiting time distribution for the severing resulting from the deletion of some crossovers because of dynamic instability, we have chosen k and θ such that the distribution of the severing waiting time conditional to the fact that the severing happened optimally fits the experimentally measured one (see Fig. S4). Given our assumption that the effect of katanin is different for different mutants because of difference in microtubule dynamics and severing properties, the parameters of the distribution are different for WT, 3x-eb1, spr1, and clasp mutants. Because we are interested in the behavior of longitudinal microtubules in only the first 500 s of the process, we limit severing to only the longitudinal microtubules.
Every simulation consists of one longitudinal microtubule nucleated in the grid of transverse microtubules, and it lasts for 500 simulated seconds. After that time, the output of the simulation can be either extinction, i.e., the initial microtubule and all its progeny have completely depolymerized because of dynamic instability, or amplification, i.e., there are one or more longitudinal microtubules still alive. We averaged our simulation results over 5 × 104 trials.
The main results of these simulation studies are reported in the results of the article. To quantify better the effect of Ps,+ and the effect of rr on the speed of amplification, we performed a sensitivity analysis. In the first case, having WT parameters as background we tuned Ps,+ from 0 to 0.25, and we counted the number of longitudinal microtubules in the simulation after 500 s. In the second case, we repeated the same procedure, keeping the WT value for Ps,+ = 0.15 and tuning the intrinsic rescue rate rr from 0.76 to 2.09 events/min. The reason for the choice of these two values for the intrinsic rescue rates is the following: given our interest in the behavior of microtubules in the unbounded growth regimen, the rate rr = 0.76 event/min is the smallest rescue rate required to be in the unbounded growth regimen, i.e., it is the value that allows the average growth speed (Dogterom and Leibler, 1993),
to be nonnegative, in particular to be J = 0. Taking WT values as our reference values, we can see that [Ps,+ = 0.15]:[Ps,+ = 0.25] = [rr = (1.56 − 0.76) events/min]:[rr = (2.09 − 0.76) events/min], motivating our choice to take 2.09 events/min as the intrinsic rescue rate. Fig. S5 shows that the effect on the amplification if we change Ps,+ is stronger than if we change rr, supporting our hypothesis that the key parameter for the speed of amplification is the fraction of rescue after severing.
Online supplemental material
Fig. S1 shows the observed sever waiting time distributions at crossovers in WT and + TIP mutant cells. Fig. S2 shows the measured intermicrotubule distances of initial transverse arrays in WT and + TIP mutant cells (these data used to parameterize models). Fig. S3 shows analysis of microtubule rescue at microtubule intersections. Fig. S4 shows the calculated intrinsic sever waiting distributions and comparison of the calculated conditional and observed sever waiting time distributions for WT and + TIP mutant cells. Fig. S5 shows sensitivity analysis of microtubule amplification to tuning of the intrinsic plus end rescue rate versus the rate of plus end stabilization and regrowth after severing. Table S1 lists the parameter values used for modeling of microtubule amplification. Table S2 lists the sequences of primers used in cloning. Video 1 displays the dynamic reorientation of cortical arrays upon stimulation by blue light in WT and + TIP mutant cells. Videos 2 and 3 show examples of severing events followed by plus end stabilization or lack of plus end stabilization. Videos 4, 5, and 6 show dynamic localization of CLASP to straight and curved microtubules, and relative to severing at crossovers. Video 7 shows an example of crossover identification during microtubule array reorientation as used in our analyses of experimental data.
Supplementary Material
Acknowledgments
We thank Zdeněk Lánský for advice on image analysis, David Quint and Luke Rice for excellent discussions, and anonymous reviewers for helpful suggestions.
This work was supported by the Carnegie Institution for Science (D.W. Ehrhardt), National Science Foundation grant 1158372 (D.W. Ehrhardt), and the Human Frontier Science Program (M. Nakamura). The work of M. Saltini is supported by the European Research Council Synergy grant MODELCELL. The work of B.M. Mulder is part of the research program of the Netherlands Organisation for Scientific Research.
The authors declare no competing financial interests.
Author contributions: D.W. Ehrhardt, J.J. Lindeboom, M. Nakamura, V. Kirik, T. Ketelaar, and A.M.C. Emons designed experimental strategy. J.J. Lindeboom, M. Nakamura, and A. Hibbel carried out experiments. J.J. Lindeboom, M. Nakamura, and D.W. Ehrhardt analyzed data. B.M. Mulder and M. Saltini designed and performed the simulations. J.J. Lindeboom, D.W. Ehrhardt, M. Nakamura, B.M. Mulder, and M. Saltini wrote the manuscript. A. Walia and J.C. Sedbrook provided biological materials. All authors contributed to editing the manuscript.
References
- Abramowitz M., and Stegun I.A.. 1965. Handbook of mathematical functions with formulas, graphs, and mathematical tables. Dover Publications, New York. 1 pp. [Google Scholar]
- Akhmanova A., and Steinmetz M.O.. 2015. Control of microtubule organization and dynamics: two ends in the limelight. Nat. Rev. Mol. Cell Biol. 16:711–726. 10.1038/nrm4084 [DOI] [PubMed] [Google Scholar]
- Akhmanova A., Hoogenraad C.C., Drabek K., Stepanova T., Dortland B., Verkerk T., Vermeulen W., Burgering B.M., De Zeeuw C.I., Grosveld F., and Galjart N.. 2001. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 104:923–935. 10.1016/S0092-8674(01)00288-4 [DOI] [PubMed] [Google Scholar]
- Al-Bassam J., Kim H., Brouhard G., van Oijen A., Harrison S.C., and Chang F.. 2010. CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev. Cell. 19:245–258. 10.1016/j.devcel.2010.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 301:653–657. 10.1126/science.1086391 [DOI] [PubMed] [Google Scholar]
- Ambrose C., Allard J.F., Cytrynbaum E.N., and Wasteneys G.O.. 2011. A CLASP-modulated cell edge barrier mechanism drives cell-wide cortical microtubule organization in Arabidopsis. Nat. Commun. 2:430 10.1038/ncomms1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C., Ruan Y., Gardiner J., Tamblyn L.M., Catching A., Kirik V., Marc J., Overall R., and Wasteneys G.O.. 2013. CLASP interacts with sorting nexin 1 to link microtubules and auxin transport via PIN2 recycling in Arabidopsis thaliana. Dev. Cell. 24:649–659. 10.1016/j.devcel.2013.02.007 [DOI] [PubMed] [Google Scholar]
- Ambrose J.C., Shoji T., Kotzer A.M., Pighin J.A., and Wasteneys G.O.. 2007. The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell. 19:2763–2775. 10.1105/tpc.107.053777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applegate K.T., Besson S., Matov A., Bagonis M.H., Jaqaman K., and Danuser G.. 2011. plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J. Struct. Biol. 176:168–184. 10.1016/j.jsb.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumeier C., Schaedel L., Gaillard J., John K., Blanchoin L., and Théry M.. 2016. Self-repair promotes microtubule rescue. Nat. Cell Biol. 18:1054–1064. 10.1038/ncb3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz P., Ye X., Huddleston P., Brautigam C.A., and Rice L.M.. 2012. A TOG:αβ-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science. 337:857–860. 10.1126/science.1221698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin T.I. 2001. On the alignment of cellulose microfibrils by cortical microtubules: a review and a model. Protoplasma. 215:150–171. 10.1007/BF01280311 [DOI] [PubMed] [Google Scholar]
- Bisgrove S.R., Hable W.E., and Kropf D.L.. 2004. +TIPs and microtubule regulation. The beginning of the plus end in plants. Plant Physiol. 136:3855–3863. 10.1104/pp.104.051037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard G.J., and Rice L.M.. 2014. The contribution of αβ-tubulin curvature to microtubule dynamics. J. Cell Biol. 207:323–334. 10.1083/jcb.201407095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J., Calder G.M., Doonan J.H., and Lloyd C.W.. 2003. EB1 reveals mobile microtubule nucleation sites in Arabidopsis. Nat. Cell Biol. 5:967–971. 10.1038/ncb1057 [DOI] [PubMed] [Google Scholar]
- Clough S.J., and Bent A.F.. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16:735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- de Forges H., Pilon A., Cantaloube I., Pallandre A., Haghiri-Gosnet A.M., Perez F., and Poüs C.. 2016. Localized mechanical stress promotes microtubule rescue. Curr. Biol. 26:3399–3406. 10.1016/j.cub.2016.10.048 [DOI] [PubMed] [Google Scholar]
- Deinum E.E., and Mulder B.M.. 2013. Modelling the role of microtubules in plant cell morphology. Curr. Opin. Plant Biol. 16:688–692. 10.1016/j.pbi.2013.10.001 [DOI] [PubMed] [Google Scholar]
- Díaz-Valencia J.D., Morelli M.M., Bailey M., Zhang D., Sharp D.J., and Ross J.L.. 2011. Drosophila katanin-60 depolymerizes and severs at microtubule defects. Biophys. J. 100:2440–2449. 10.1016/j.bpj.2011.03.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogterom M., and Leibler S.. 1993. Physical aspects of the growth and regulation of microtubule structures. Phys. Rev. Lett. 70:1347–1350. 10.1103/PhysRevLett.70.1347 [DOI] [PubMed] [Google Scholar]
- Ehrhardt D.W., and Shaw S.L.. 2006. Microtubule dynamics and organization in the plant cortical array. Annu. Rev. Plant Biol. 57:859–875. 10.1146/annurev.arplant.57.032905.105329 [DOI] [PubMed] [Google Scholar]
- Furutani I., Watanabe Y., Prieto R., Masukawa M., Suzuki K., Naoi K., Thitamadee S., Shikanai T., and Hashimoto T.. 2000. The SPIRAL genes are required for directional control of cell elongation in Aarabidopsis thaliana. Development. 127:4443–4453. [DOI] [PubMed] [Google Scholar]
- Galva C., Kirik V., Lindeboom J.J., Kaloriti D., Rancour D.M., Hussey P.J., Bednarek S.Y., Ehrhardt D.W., and Sedbrook J.C.. 2014. The microtubule plus-end tracking proteins SPR1 and EB1b interact to maintain polar cell elongation and directional organ growth in Arabidopsis. Plant Cell. 26:4409–4425. 10.1105/tpc.114.131482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., and Blatt M.R.. 2010. A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64:355–365. 10.1111/j.1365-313X.2010.04322.x [DOI] [PubMed] [Google Scholar]
- Gutierrez R., Lindeboom J.J., Paredez A.R., Emons A.M.C., and Ehrhardt D.W.. 2009. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11:797–806. 10.1038/ncb1886 [DOI] [PubMed] [Google Scholar]
- Kirik V., Herrmann U., Parupalli C., Sedbrook J.C., Ehrhardt D.W., and Hülskamp M.. 2007. CLASP localizes in two discrete patterns on cortical microtubules and is required for cell morphogenesis and cell division in Arabidopsis. J. Cell Sci. 120:4416–4425. 10.1242/jcs.024950 [DOI] [PubMed] [Google Scholar]
- Klimyuk V.I., Carroll B.J., Thomas C.M., and Jones J.D.. 1993. Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J. 3:493–494. 10.1111/j.1365-313X.1993.tb00169.x [DOI] [PubMed] [Google Scholar]
- Lawrence E.J., Arpag G., Norris S.R., and Zanic M.. 2018. Human CLASP2 specifically regulates microtubule catastrophe and rescue. Mol. Biol. Cell. 29:1168–1177. 10.1091/mbc.E18-01-0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom J.J., Lioutas A., Deinum E.E., Tindemans S.H., Ehrhardt D.W., Emons A.M.C., Vos J.W., and Mulder B.M.. 2013a Cortical microtubule arrays are initiated from a nonrandom prepattern driven by atypical microtubule initiation. Plant Physiol. 161:1189–1201. 10.1104/pp.112.204057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom J.J., Nakamura M., Hibbel A., Shundyak K., Gutierrez R., Ketelaar T., Emons A.M.C., Mulder B.M., Kirik V., and Ehrhardt D.W.. 2013b A mechanism for reorientation of cortical microtubule arrays driven by microtubule severing. Science. 342:1245533 10.1126/science.1245533 [DOI] [PubMed] [Google Scholar]
- Majumdar S., Kim T., Chen Z., Munyoki S., Tso S.-C., Brautigam C.A., and Rice L.M.. 2018. An isolated CLASP TOG domain suppresses microtubule catastrophe and promotes rescue. Mol. Biol. Cell. 29:1359–1375. 10.1091/mbc.E17-12-0748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J., Mathur N., Kernebeck B., Srinivas B.P., and Hülskamp M.. 2003. A novel localization pattern for an EB1-like protein links microtubule dynamics to endomembrane organization. Curr. Biol. 13:1991–1997. 10.1016/j.cub.2003.10.033 [DOI] [PubMed] [Google Scholar]
- Mitchison T., and Kirschner M.. 1984. Dynamic instability of microtubule growth. Nature. 312:237–242. 10.1038/312237a0 [DOI] [PubMed] [Google Scholar]
- Murata T., Sonobe S., Baskin T.I., Hyodo S., Hasezawa S., Nagata T., Horio T., and Hasebe M.. 2005. Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat. Cell Biol. 7:961–968. 10.1038/ncb1306 [DOI] [PubMed] [Google Scholar]
- Nakajima K., Furutani I., Tachimoto H., Matsubara H., and Hashimoto T.. 2004. SPIRAL1 encodes a plant-specific microtubule-localized protein required for directional control of rapidly expanding Arabidopsis cells. Plant Cell. 16:1178–1190. 10.1105/tpc.017830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Ehrhardt D.W., and Hashimoto T.. 2010. Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat. Cell Biol. 12:1064–1070. 10.1038/ncb2110 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Lindeboom J.J., Saltini M., Mulder B.M., and Ehrhardt D.W.. 2018. SPR2 protects minus ends to promote severing and reorientation of plant cortical microtubule arrays. J. Cell Biol. 217:915–927. 10.1083/jcb.201708130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoulis A. 1984. Probability, random variables, and stochastic processes. Second edition McGraw-Hill Companies, New York. 852 pp. [Google Scholar]
- Paredez A.R., Somerville C.R., and Ehrhardt D.W.. 2006. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 312:1491–1495. 10.1126/science.1126551 [DOI] [PubMed] [Google Scholar]
- Rasband W.S. 2012. ImageJ: Image processing and analysis in Java. Astrophysics Source Code Library. 1:06013. [Google Scholar]
- Roll-Mecak A. 2013. Botany. Shining light at microtubule crossroads. Science. 342:1180–1181. 10.1126/science.1248235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A., and Vale R.D.. 2006. Making more microtubules by severing: a common theme of noncentrosomal microtubule arrays? J. Cell Biol. 175:849–851. 10.1083/jcb.200611149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y., Halat L.S., Khan D., Jancowski S., Ambrose C., Belmonte M.F., and Wasteneys G.O.. 2018. The Microtubule-associated protein CLASP sustains cell proliferation through a brassinosteroid signaling negative feedback loop. Curr. Biol. 28:2718–2729.e5. 10.1016/j.cub.2018.06.048 [DOI] [PubMed] [Google Scholar]
- Sedbrook J.C., Ehrhardt D.W., Fisher S.E., Scheible W.-R., and Somerville C.R.. 2004. The Arabidopsis sku6/spiral1 gene encodes a plus end-localized microtubule-interacting protein involved in directional cell expansion. Plant Cell. 16:1506–1520. 10.1105/tpc.020644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw S.L., Kamyar R., and Ehrhardt D.W.. 2003. Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science. 300:1715–1718. 10.1126/science.1083529 [DOI] [PubMed] [Google Scholar]
- Tindemans S.H., and Mulder B.M.. 2010. Designing colloidal ground-state patterns using short-range isotropic interactions. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 82:021404 10.1103/PhysRevE.82.021404 [DOI] [PubMed] [Google Scholar]
- Tran P.T., Walker R.A., and Salmon E.D.. 1997. A metastable intermediate state of microtubule dynamic instability that differs significantly between plus and minus ends. J. Cell Biol. 138:105–117. 10.1083/jcb.138.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropini C., Roth E.A., Zanic M., Gardner M.K., and Howard J.. 2012. Islands containing slowly hydrolyzable GTP analogs promote microtubule rescues. PLoS One. 7:e30103 10.1371/journal.pone.0030103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemu A., Sczesna E., Zehr E.A., Spector J.O., Grigorieff N., Deaconescu A.M., and Roll-Mecak A.. 2018. Severing enzymes amplify microtubule arrays through lattice GTP-tubulin incorporation. Science. 361:eaau1504 10.1126/science.aau1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker R.A., Inoué S., and Salmon E.D.. 1989. Asymmetric behavior of severed microtubule ends after ultraviolet-microbeam irradiation of individual microtubules in vitro. J. Cell Biol. 108:931–937. 10.1083/jcb.108.3.931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys G.O. 2002. Microtubule organization in the green kingdom: chaos or self-order? J. Cell Sci. 115:1345–1354. [DOI] [PubMed] [Google Scholar]
- Wasteneys G.O., and Williamson R.E.. 1989. Reassembly of microtubules in Nitella tasmanica: assembly of cortical microtubules in branching clusters and its relevance to steady-state microtubule assembly. J. Cell Sci. 93:705–714. [Google Scholar]
- Wieczorek M., Bechstedt S., Chaaban S., and Brouhard G.J.. 2015. Microtubule-associated proteins control the kinetics of microtubule nucleation. Nat. Cell Biol. 17:907–916. 10.1038/ncb3188 [DOI] [PubMed] [Google Scholar]
- Wightman R., and Turner S.R.. 2007. Severing at sites of microtubule crossover contributes to microtubule alignment in cortical arrays. Plant J. 52:742–751. 10.1111/j.1365-313X.2007.03271.x [DOI] [PubMed] [Google Scholar]
- Woody S.T., Austin-Phillips S., Amasino R.M., and Krysan P.J.. 2007. The WiscDsLox T-DNA collection: an arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J. Plant Res. 120:157–165. 10.1007/s10265-006-0048-x [DOI] [PubMed] [Google Scholar]
- Zhang Q., Fishel E., Bertroche T., and Dixit R.. 2013. Microtubule severing at crossover sites by katanin generates ordered cortical microtubule arrays in Arabidopsis. Curr. Biol. 23:2191–2195. 10.1016/j.cub.2013.09.018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.