Although centromeres usually associate with heterochromatic repetitive sequences, such repetitive sequences are not detected around neocentromeres. Nishimura et al. performed systematic 4C analysis of cells containing differently positioned neocentromeres and demonstrate that these neocentromeres commonly associate with distant heterochromatin-rich regions at the 3D level.
Abstract
The centromere is an important genomic locus for chromosomal segregation. Although the centromere is specified by sequence-independent epigenetic mechanisms in most organisms, it is usually composed of highly repetitive sequences, which associate with heterochromatin. We have previously generated various chicken DT40 cell lines containing differently positioned neocentromeres, which do not contain repetitive sequences and do not associate with heterochromatin. In this study, we performed systematic 4C analysis using three cell lines containing differently positioned neocentromeres to identify neocentromere-associated regions at the 3D level. This analysis reveals that these neocentromeres commonly associate with specific heterochromatin-rich regions, which were distantly located from neocentromeres. In addition, we demonstrate that centromeric chromatin adopts a compact structure, and centromere clustering also occurs in vertebrate interphase nuclei. Interestingly, the occurrence of centromere–heterochromatin associations depend on CENP-H, but not CENP-C. Our analyses provide an insight into understanding the 3D architecture of the genome, including the centromeres.
Graphical Abstract

Introduction
The centromere is the genomic locus where the kinetochore is formed, for ensuring faithful chromosomal segregation by interacting with the spindle microtubules. Various studies have revealed that the centromere is specified by sequence-independent epigenetic mechanisms involving the deposition of the centromere-specific histone H3 variant, CENP-A, into chromatin (Black and Cleveland, 2011; Perpelescu and Fukagawa, 2011; Allshire and Madhani, 2018). Studies on the neocentromere, which is newly formed on a noncentromeric locus after the inactivation of a native centromere and induces the formation of the kinetochore (du Sart et al., 1997; Marshall et al., 2008; Shang et al., 2013), largely support the notion that the centromeric position is epigenetically specified. The neocentromere was originally detected in human chromosomes that did not possess the α-satellite DNA sequence observed in native human centromeres (Voullaire et al., 1993; du Sart et al., 1997). After this initial discovery (Voullaire et al., 1993), neocentromeres were experimentally generated by inactivating the native centromeres in various model organisms such as Drosophila melanogaster (Maggert and Karpen, 2001), Schizosaccharomyces pombe (Ishii et al., 2008; Ogiyama et al., 2013), Candida albicans (Ketel et al., 2009; Thakur and Sanyal, 2013), and chicken DT40 cells (Shang et al., 2013). Because centromeres usually associate with highly repetitive sequences in most organisms, it is difficult to characterize their genomic features. However, as neocentromeres are formed in the nonrepetitive genomic regions in human and chicken cells, it is possible to characterize genomic features in the neocentromeric region (Alonso et al., 2010; Shang et al., 2013). For instance, by using specific antibodies against various histone modifications, centromere-specific histone modifications were identified based on chromatin immunoprecipitation (IP; ChIP) sequencing (ChIP-seq) analysis on nonrepetitive centromeres (Hori et al., 2014; Shang et al., 2016). Neocentromeres contain most of the centromeric proteins in quantities similar to those found in native centromeres, suggesting that the function of neocentromeres is equivalent to that of native centromeres (Saffery et al., 2000; Shang et al., 2013). It is therefore necessary to emphasize that nonrepetitive neocentromeres are powerful molecular entities for understanding the genomic features of centromeres.
Comparing the genomic features of the different neocentromeres from different species, it is observed that each neocentromere possesses distinct features. In S. pombe, the neocentromeres are preferentially formed near heterochromatinized regions, and the efficiency of the formation of neocentromeres was dramatically reduced when heterochromatin factors such as Swi6 (yeast homologue of HP1) were mutated (Ishii et al., 2008). In addition, synthetic heterochromatin, by tethering the H3K9 methyltransferase Clr4, induces centromere establishment de novo on fission yeast minichromosomes (Kagansky et al., 2009). However, the accumulation of heterochromatin proteins and histone modifications in the heterochromatin regions, such as the accumulation of histone H3 trimethylated at lysine 9 (H3K9me3), are not observed around the neocentromeres of human or chicken cells (Alonso et al., 2010; Shang et al., 2013; Hori et al., 2014). In addition to the association of centromeres with heterochromatin, centromeres and neocentromeres are clustered at particular positions in the nuclei of yeasts such as S. pombe or C. albicans (Funabiki et al., 1993; Thakur and Sanyal, 2012; Burrack et al., 2016). However, the formation of the centromere cluster is also not clear in vertebrate nuclei, owing to the appearance of multiple centromeric signals in the interphase nuclei.
Although some genomic features of each neocentromere appear to vary, the kinetochore is commonly formed on centromeres of all species, and therefore, there must be some similar genomic features in the centromeres of different species. Although heterochromatin regions are not detected near the neocentromeres of human or chicken cells, it might still be possible that the neocentromeres are physically associated with the heterochromatin regions of interphase nuclei.
Recently, the 3D genomic architecture of interphase nuclei has been extensively studied in various organisms (Dekker and Mirny, 2016). Microscopy-based approaches such as FISH revealed that certain loci in the interphase nuclei can physically interact even if the linear-genomic distances between these loci are large. In addition, using chromatin conformation capture (3C) technology, genome-wide long-range interactions between any pair of loci in the nuclei can be detected by cross-linking chromatin with formaldehyde (Dekker and Mirny, 2016). Although these interactions were originally identified by PCR using ligation fragments of digested cross-linked DNAs (3C-PCR), 3C technology is nowadays combined with the next-generation sequencing, including circular chromosome conformation capture (4C; Zhao et al., 2006), 5C (Dostie et al., 2006), Chromatin Interaction Analysis by Paired-End Tag Sequencing (ChIA-PET; Fullwood et al., 2009), and Hi-C (Lieberman-Aiden et al., 2009) analyses, which enables us to observe genome-wide interactions more efficiently.
The Hi-C technique was applied to yeast genomes, and the centromere cluster was found as a result of interchromosomal interactions (Mizuguchi et al., 2014; Varoquaux et al., 2015; Burrack et al., 2016) in yeasts. Observing the genome-wide interactions of centromeric regions in vertebrate cells using Hi-C is essential because centromeres in vertebrate cells usually contain repetitive sequences and full sequence information is absent in genomic databases. Although it is not technically easy to characterize the 3D structure of the vertebrate genome, including that of centromeres, understanding the organization of genomic regions, including centromeres in interphase nuclei, is vital.
To solve the issue, we used DT40 cells containing differently positioned neocentromeres (Shang et al., 2013). We removed the original native centromere on the Z chromosome by a genome engineering method and isolated those cell lines in which the neocentromeres were formed at various regions on the Z chromosome (Shang et al., 2013). Because the sequence of the neocentromeres remained unaltered both before and after the formation of the neocentromeres, it is possible to examine the sites of interactions of the neocentromeric regions by comparing the interaction sites both before and after neocentromere formation. In addition, by analyzing several neocentromeres, we could expect to find common interaction sites with the differently positioned neocentromeres. In this study, to identify centromere-specific interaction sites, we performed 4C-seq analyses (Zhao et al., 2006) using neocentromeric regions as viewpoints, which would reveal better high-resolution interaction profiles with neocentromeres than analyses with Hi-C. We compared the 4C profiles of three independent cell lines containing differently positioned neocentromeres with various viewpoints and found that these neocentromeres are commonly associated with specific heterochromatin-rich regions. Furthermore, our high-resolution 4C analysis revealed that frequent interactions occur within the 30–40-kb centromeric region to form more compact structures within the centromere. We also detected the centromere–centromere interactions of different chromosomes, which suggested that the centromeric chromatin contains common features. Interestingly, the long-range centromere–heterochromatin interactions depend on CENP-H but not CENP-C, suggesting that a class of centromere proteins contribute to the formation of the 3D architecture of the genome, including centromeres, in interphase nuclei.
Results
Establishment of the 4C-seq method for identifying regions interacting with the neocentromeres using chicken DT40 cells
To examine the 3D genomic architecture including neocentromeres, we performed 4C-seq analyses for cell lines containing differently positioned neocentromeres (Figs. 1 and S1). Cells were fixed with PFA, the genomic DNA was first digested with a six-base cutter restriction enzyme such as HindIII or EcoRI, and the cross-linked DNA fragments were subsequently ligated to construct the 3C library. For 4C-seq analyses, the 3C library was digested with a second four-base cutter restriction enzyme and re-ligated, and the sample was amplified by inverse PCR (4C-PCR; Fig. 1 A). We proceeded to prepare a primer set in a particular position (viewpoint) for analyzing the interactions. High-throughput sequencing of 4C-PCR products is used to identify all the fragments that interact with a viewpoint (Fig. 1 A; Splinter et al., 2012). As we amplified the 3C library by PCR for 4C-seq analysis, we used a statistical method (Splinter et al., 2012) for evaluating the frequency of interactions with the viewpoints, referred to as the site occupancy rate (SOR; Fig. 1 B). For this method, we treated multiple reads (≥1) from sequencing analysis as a single positive read to avoid possible PCR artifacts without considering the read numbers. The rate of the number of restriction sites with positive reads to the number of all the possible restriction sites in a region was calculated (Fig. 1 B). A typical example is represented in Fig. 1 B. If there were four or three restriction sites in a 150-kb region, and we mapped two positive sites in these regions, the SOR would be 0.5 (2/4) or 0.66 (2/3), respectively (Fig. 1 B). Within a 150-kb region of the chicken Z chromosome, the average number of HindIII sites was 55. As we can obtain a 4C profile at ∼3-kb resolution, it was decided that using a 150-kb window for the first restriction digestion with HindIII would be appropriate (Fig. 1 B). We used the SOR method for genome-wide 4C analysis, and one such example is represented in Fig. S1 (A and B). In Fig. S1 A, we chose a viewpoint within the neocentromeric region (3.8-Mb region on the Z chromosome) in #BM23 cells and demonstrated the SOR values along the Z chromosome. Detailed information for viewpoint positions in this study is shown in Table S1. Using different restriction enzymes—HindIII, BglII, or EcoRI—for each viewpoint in the same region, we obtained similar 4C profiles in #BM23 and #1320 cells (Fig. S1, A and B). Therefore, we concluded that our 4C analysis and SOR method are appropriate for analyzing the 3D architecture of the genome, including neocentromeres.
Figure 1.
Applying the 4C-seq analysis to DT40 cells. (A) An experimental scheme for 4C-seq analysis. The cells were fixed with 1% PFA, and the genome was initially digested with a six-base cutter restriction enzyme (RE; first RE digestion) and ligated (first ligation). The sample was decross-linked, and the genome was digested for a second time with a second four-base cutter restriction enzyme (second RE digestion) and ligated (second ligation). A primer set was prepared for a viewpoint (the region of interest where the interaction will be studied), and reverse PCR was performed (4C PCR). The PCR products were sequenced by a high-throughput sequencer and mapped to the reference genome. (B) Calculation of the SOR. The multiple reads mapped per fragment (≥1) were treated as a single positive read, and the rate of the number of positive restriction sites to the number of all the possible restriction sites in a region was calculated. For instance, if there were four or three restriction sites in a 150-kb region and two positive sites were in these regions, the SORs would be 0.5 (2/4) or 0.66 (2/3), respectively. (C) Representation of the cell lines used in this study. The native centromere (red) on the Z chromosome is located at the 42.6-Mb region in WT DT40 cells. The native centromere was removed, and the surviving cells were isolated and cloned. Each clone forms a neocentromere (blue) at a different position. The neocentromeres were formed at 3.8-, 35-, and 55-Mb regions in #BM23, #1320, and #1304 cell lines, respectively.
Neocentromeres are commonly associated with heterochromatin-rich regions
In this study, we focused on three cell lines containing differently positioned neocentromeres in which the neocentromeres were formed at 3.8-Mb (#BM23 cells), 35-Mb (#1320 cells), and 55-Mb (#1304 cells) regions on the Z chromosome, respectively, after removal of the original native centromere that was located at the 42.6-Mb region on the Z chromosome (Fig. 1 C; Shang et al., 2013). Although the growth rate of each neocentromere-containing cell line varies (Fig. S1 C), all cell lines grew well. Chromosome Z with neocentromere was stable in #1320 and #BM23 cells, but abnormal numbers of Z chromosomes were observed in half the population of #1304 cells (Fig. S1 D), suggesting that neocentromeres in #1320 and #BM23 cells are equivalent to normal centromeres, but neocentromeres in #1304 were not completely normal. Therefore, we mainly used #1320 and #BM23 cells for further 4C analysis, and data with #1304 cells were used as supplemental data, because the full activity of the neocentromere may be lost in #1304 cells. Viewpoints were set at the 3.8-, 35-, and 55-Mb regions on the Z chromosome for performing 4C analysis in all the three cell lines (Fig. 1 C), and the 4C profiles of the different cell lines at the same viewpoint were compared, which corresponded with either the centromeric or noncentromeric region, depending on the cell lines. For example, in #BM23 cell line, the 3.8-Mb region corresponds with the centromere; however, this does not hold true in the #1320 or #1304 cell lines (Fig. 1 C). By comparing the 4C profiles, it would be possible to identify the regions with which all the neocentromeres commonly associate.
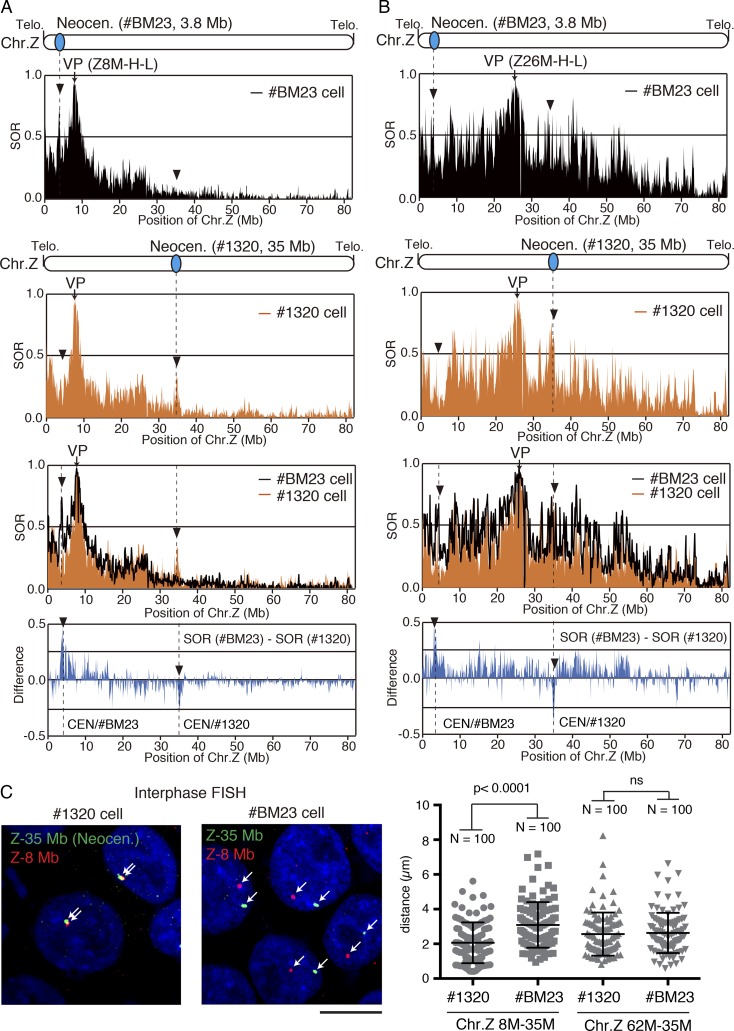
Then, we performed 4C analyses with a different combination of viewpoints with various cell lines. When we compared the 4C profiles for all the cell lines using the 3.8-Mb region as a viewpoint (Table S1), the 3.8-Mb region was found to specifically associate with two regions (the 8- and 26-Mb regions) in #BM23 cells; however, it did not associate with these two regions in the #1320 cell line (Fig. 2 A). We confirmed these results with a different set of primers in the same 3.8-Mb region (Fig. 2 B and Table S1). We also performed 4C analyses with the 35- and 55-Mb regions as viewpoints (Table S1) for #BM23, #1320, and #1304 cells (Fig. 2, C and D; and Fig. S2). The 8- and 26-Mb regions showed higher SOR values than when the 35-Mb (Fig. 2, C and D) and 55-Mb (Fig. S2) regions were used as viewpoints in #1320 cells and #1304 cells, respectively. It is important to note that the 35- and 55-Mb regions correspond with the centromeres in #1320 and #1304 cells, respectively (Fig. 1 C). The 4C profiles suggested that all the neocentromeres in the three cell lines (#BM23, #1320, and #1304) commonly associate with the 8- and 26-Mb regions (Figs. 2 and S2). In addition, we performed 4C analysis with WT DT40 cells using the native centromeric region on the Z chromosome as a viewpoint (42.6-Mb region; Table S1) and found that the native centromere also interacted with the 8- and 26-Mb regions (Fig. 3 A).
Figure 2.
4C profiles determined using different viewpoints in different cells containing various neocentromeres. (A) 4C profiles determined using #BM23 cells (second horizontal panel) and #1320 cells (third panel) with the 3.8-Mb region as a viewpoint (VP: Z3.8M-H-L). Top: ChIP-seq profile with CENP-A around the 3.8-Mb region in #BM23 cells. The two profiles were merged (fourth panel). Subtraction data: SOR values in #BM23 cells minus SOR values in #1320 cells are shown (bottom). Arrowheads indicate the 8- and 26-Mb regions. The position of the neocentromere in each cell line is indicated as broken lines. (B) 4C profiles determined using #BM23 cells (second horizontal panel) and #1320 cells (third panel) with the 3.8-Mb region as a viewpoint (VP: Z3.8M-H-R). Arrowheads indicate the 8- and 26-Mb regions. Although the same region was used for determining the 4C profiles in A, a different set of primers was used here (Table S1). (C) 4C profiles determined using #1320 cells (second horizontal panel) and #BM23 cells (third panel) with the 35-Mb region as a viewpoint (VP: Z35M-H-L). Top: ChIP-seq profile with CENP-A around the 35-Mb region in #1320 cells. The two profiles were merged (fourth panel). Subtraction data: SOR values in #1320 cells minus SOR values in BM23 cells are shown (bottom). Arrowheads indicate the 8- and 26-Mb regions. The position of the neocentromere in each cell line is indicated as broken lines. (D) 4C profiles determined using #1320 cells (second horizontal panel) and #BM23 cells (third panel) with the 35-Mb region as a viewpoint (VP: Z35M-H-R). Arrowheads indicate the 8- and 26-Mb regions. Although the same region was used for determining the 4C profiles in C, a different set of primers was used (Table S1).
Figure 3.
The native centromere on the Z chromosome associates with the 8- and 26-Mb regions. (A) 4C profiles determined using WT DT40 cells with the 42.6-Mb region (centromere region) as a viewpoint (top). Arrowheads indicate the 8- and 26-Mb regions. (B) ChIP-seq data on the Z chromosome with an antibody against H3K9me3 in DT40 cells. Arrowheads indicate the 8- and 26-Mb regions on the Z chromosome. Positions of the viewpoints (VPs) for Figs. 3 C, 4, and S3 experiments are shown. (C) 4C profiles determined using WT DT40 cells with the 8-Mb region (VP: Z8M-H-L) and 26-Mb region (VP: Z26M-H-L) as viewpoints. Arrowheads indicate the native centromere on the Z chromosome. (D) Analyses with two-color FISH in WT DT40 cells. Green-labeled BAC clone #279C6, comprising the 42.6-Mb centromere region, and red-labeled BAC clone #116B8, comprising the 26-Mb region, were used as probes (left). As a negative control, red-labeled BAC clone #279G5 comprising the 62-Mb region was used (right). Signals are shown by arrows. DNA was stained with DAPI (blue). Bar, 10 µm. The graph shows the distance between the two probes for each cell line.
We proceeded to examine our previous data for ChIP-seq profiles using antibodies against various histone modifications on the Z chromosome (Hori et al., 2014) to identify the chromatin features of the 8- and 26-Mb regions on the Z chromosome. We found that the 8- and 26-Mb regions corresponded with regions highly enriched in H3K9me3, which is a marker of heterochromatin (Fig. 3 B). H3K9me3 is usually associated with the pericentromeric region (Fukagawa and Earnshaw, 2014), which is located around the centromere and consists of repetitive sequences. In fact, we found that H3K9me3 was enriched in the repetitive centromeres of chicken chromosomes, including chromosomes 1 and 2 (Hori et al., 2014). However, consistent with our previous study (Shang et al., 2013; Hori et al., 2014), H3K9me3 was not detected around the nonrepetitive centromeres on the Z chromosome, including around the different neocentromeres and the nonrepetitive centromere of chromosome 5. Consistent with our previous findings, the nonrepetitive human neocentromeres did not associate with H3K9me3 (Alonso et al., 2010). Nevertheless, our 4C data indicated that nonrepetitive centromeres, including neocentromeres, on the Z chromosome physically associate with heterochromatin-rich regions.
Verification of neocentromere–heterochromatin interactions with other methods
To verify the association of the neocentromeres with the 8- and 26-Mb regions, in which H3K9me3 was found to be enriched, we also performed 4C analyses using the 8- and 26-Mb regions as viewpoints (Table S1) in #BM23, #1320, and WT DT40 cells (Figs. 3 C, 4 [A and B], and S3 [A and B]). Using the 8-Mb region as a viewpoint, we found that the SOR values around the 3.8-Mb region, which corresponds with the centromeric region in #BM23 cells, were higher in #BM23 cells than in #1320 cells (Fig. 4 A). Additionally, the SOR values around the 35-Mb region, corresponding with the centromeric region in #1320 cells, were higher in #1320 cells than in #BM23 cells (Fig. 4 A). This observation was clearly reproducible using a different set of primers in the 8-Mb viewpoint region (Fig. S3 A and Table S1). We also used the 26-Mb region as a viewpoint (Table S1) for 4C analyses in #BM23 and #1320 cells (Figs. 4 B and S3 B). Similar to the observation of the 4C profiles using the 8-Mb region as a viewpoint, the 26-Mb region associated with the 3.8- and 35-Mb regions in #BM23 and #1320 cells, respectively (Figs. 4 B and S3 B). We also detected the native centromere using the 8- or 26-Mb region as a viewpoint in WT DT40 cells (Fig. 3 C). Based on 4C analyses using the heterochromatin regions as viewpoints, we confirmed that the neocentromere and native centromere regions on the Z chromosome are preferentially associated with the 8- and 26-Mb regions that were enriched with a heterochromatin marker in chicken DT40 cells.
Figure 4.
The neocentromeres are associated with heterochromatin-rich regions. (A) 4C profiles determined using #BM23 cells (top horizontal panel) and #1320 cells (second panel) with the 8-Mb region as a viewpoint (VP: Z8M-H-L). Detailed information pertaining to the VP is also represented in Fig. 3. The two profiles were merged (third panel). Subtraction data: SOR values of #BM23 cells minus SOR values of #1320 cells are shown (bottom). Arrows indicate the 3.8- and 35-Mb regions. The position of the neocentromere is indicated as broken lines. (B) 4C profiles determined using #BM23 cells (top horizontal panel) and #1320 cells (second panel) with the 26-Mb region as a viewpoint (VP: Z26M-H-L). Detailed information pertaining to the VP is also presented in Fig. 3. The two profiles were merged (third panel). Subtraction data: SOR values in BM23 cells minus SOR values in #1320 cells are shown (bottom). Arrows indicate the 3.8- and 35-Mb regions. The position of the neocentromere is indicated as broken lines. (C) Analyses with two-color FISH in #1320 and #BM23 cells. Green-labeled BAC clone #206E12, comprising the 35-Mb region (centromere in #1320 cells), and red-labeled BAC clone #261B8, comprising the 8-Mb region, were used as probes. Signals are shown by arrows. As a negative control, BAC clone #279G5 comprising the 62-Mb region was used. DNA was stained with DAPI (blue). Bar, 10 µm. The graph shows the distance between the two probes for each cell line.
In addition to 4C analyses using various viewpoints, we attempted to verify these interactions by analyses with FISH, using cell lines containing neocentromeres (Figs. 4 C and S3 C). We isolated BAC clones, #206E12, #261B8, and #116B8, which comprised the 35-, 8-, and 26-Mb regions of the Z chromosome, respectively, and these BAC clones were used as probes for performing FISH during interphase. We performed a two-color FISH analysis using green-labeled #206E12 as a probe for the 35-Mb region and red-labeled #261B8 as a probe for the 8-Mb region in #1320 and #BM23 cells. Using #1320 cells, in which the 35-Mb region corresponds with the centromeric region, the distance between the signals from #206E12 (the 35-Mb region) and #261B8 (the 8-Mb region) were obtained at closer distances in #1320 cells than in #BM23 cells. However, in comparison with that of #1320 cells, the distance between the signals was longer in #BM23 cells in which the 35-Mb region is not the centromere (Fig. 4 C), consistent with the 4C profiles obtained in Fig. 2. We also used the BAC clone (#279G5) comprising the 62-Mb region as a negative control probe, which is 27 Mb from the 35-Mb region and was not detected in association with the neocentromere region. Distance between the 35- and 62-Mb regions did not change in #1320 and #BM23 cells, suggesting that a close association of the 35-Mb region with the 62-Mb region was not observed in #1320 cells (Fig. 4 C). We also performed two-color FISH using the #206E12 green probe for the 35-Mb region and the #116B8 red probe for the 26-Mb region in #1320 and #BM23 cells. This combination also confirmed that the 35-Mb region corresponding with the centromeric region is closer to the 26-Mb region in #1320 cells than in #BM23 cells (Fig. S3 C). These measurements were performed using the Imaris software, which enabled us to measure the 3D distance in nuclei. We also measured nuclear volume in #1320 and #BM23 cells and confirmed that nuclear volume was constant (Fig. S3 C). Our FISH analysis also demonstrated a close association of the native centromere region (42.6 Mb) with the 26-Mb region (Fig. 3 D).
Neocentromeres and native centromere Z do not contain repetitive sequences, and 3D association of these centromeres with the heterochromatin-rich region might be specific for nonrepetitive centromeres. Because repetitive centromeres are closely located in the heterochromatin-rich region near the centromere such a long-distance association might not occur. Investigating the H3K9me3 profile, we found that a H3K9me3-rich region exists around 81.5 Mb on chromosome 1, which is 18.5 Mb from the repetitive centromere region on chromosome 1 (100-Mb region; Fig. S4 A). Then, we prepared probes for the centromere and 81.5-Mb regions and performed FISH analysis. We also prepared a probe for the 118.5-Mb region as a control. As shown in Fig. S4 B, we detected a significant association between the repetitive centromere on chromosome 1 and the 81.5-Mb heterochromatin-rich region, suggesting that a 3D association between repetitive centromeres and the heterochromatin-rich region also occurs.
Combining the results of FISH with 4C analyses in interphase nuclei, we conclude that neocentromeres, which do not contain repetitive sequences, and a repetitive centromere (on chromosome 1) are physically associated with heterochromatin-rich regions in the 3D genomic arrangement, although centromeres and heterochromatin-rich regions are physically distant at the 1D level.
Multiple interactions occur within a centromeric region
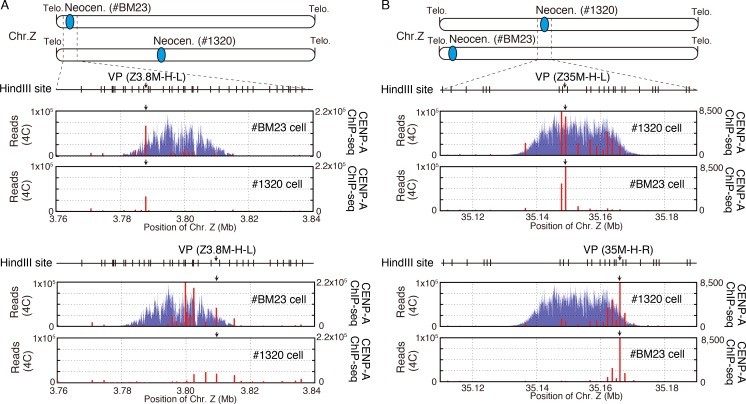
Owing to the sensitivity of the 4C analysis technique, it is possible to detect genomic interactions at the 3D level, at the resolution of several kilobases in a specific region of the genome. As previously demonstrated, neocentromeres are formed in regions spanning ∼30–40 kb (Shang et al., 2013; Hori et al., 2017), so we focused on 30–40-kb neocentromeric regions for detecting genomic interactions within the region both before and after the formation of neocentromeres. In this analysis, we evaluated interaction efficiency using the read numbers obtained from sequencing after normalization with total read numbers because this method is more appropriate for evaluating interactions within regions near the viewpoint (Splinter et al., 2012). We set up viewpoints near both edges of the 3.8-Mb CENP-A binding region in #BM23 cells and performed 4C analysis using these viewpoints in both #BM23 and #1320 cells. In #BM23 cells, these viewpoints detected multiple interaction sites in the 30-kb CENP-A binding region, but the number of interacting sites in this region was reduced in #1320 cells (Fig. 5 A). We also prepared two viewpoints in the CENP-A binding region in #1320 cells and performed 4C analysis using these viewpoints in #BM23 and #1320 cells (Fig. 5 B). In #1320 cells, these viewpoints detected multiple sites of interactions in the CENP-A binding region; however, the number of interaction sites in this region was less in #BM23 cells than in #1320 cells (Fig. 5 B). We also confirmed this observation by the 3C-PCR method (Fig. S4, C and D). In general, 3C-PCR analysis can be applied to detect interaction of two loci within a 1-Mb region, whereas 4C can detect interactions of two loci at a longer distance (Naumova et al., 2012). The results of 4C analyses within the two centromeric regions (3.8- and 35-Mb regions in #BM23 and #1320 cells, respectively) and 3C-PCR results indicated that multiple interactions frequently occur within the centromere region, suggesting that the centromeric chromatin forms a compact structure.
Figure 5.
The neocentromere forms a compact structure. (A) High-resolution 4C profile (red bars) within the centromeric region determined using #BM23 and #1320 cell lines. Two viewpoints (3.8M-H-L and 3.8M-H-R) were selected for the 3.8-Mb region. The CENP-A ChIP-seq profiles (blue) in #BM23 and #1320 cell lines around the 3.8-Mb region are also shown. The values on the vertical axis represent the number of sequence reads. HindIII sites around this region are also shown. Arrows show the viewpoint positions. (B) High-resolution 4C profile (red bars) within the centromere region determined using #BM23 and #1320 cell lines. Two viewpoints (35M-H-L and 35M-H-R) were used for the 35-Mb region. The CENP-A ChIP-seq profiles (blue) around the 35-Mb region in the #BM23 cells and #1320 cells are also shown. Arrows show the viewpoint positions. The values on the vertical axis represent the number of sequence reads. HindIII sites around this region are also shown.
Centromere clustering was observed from 4C analysis
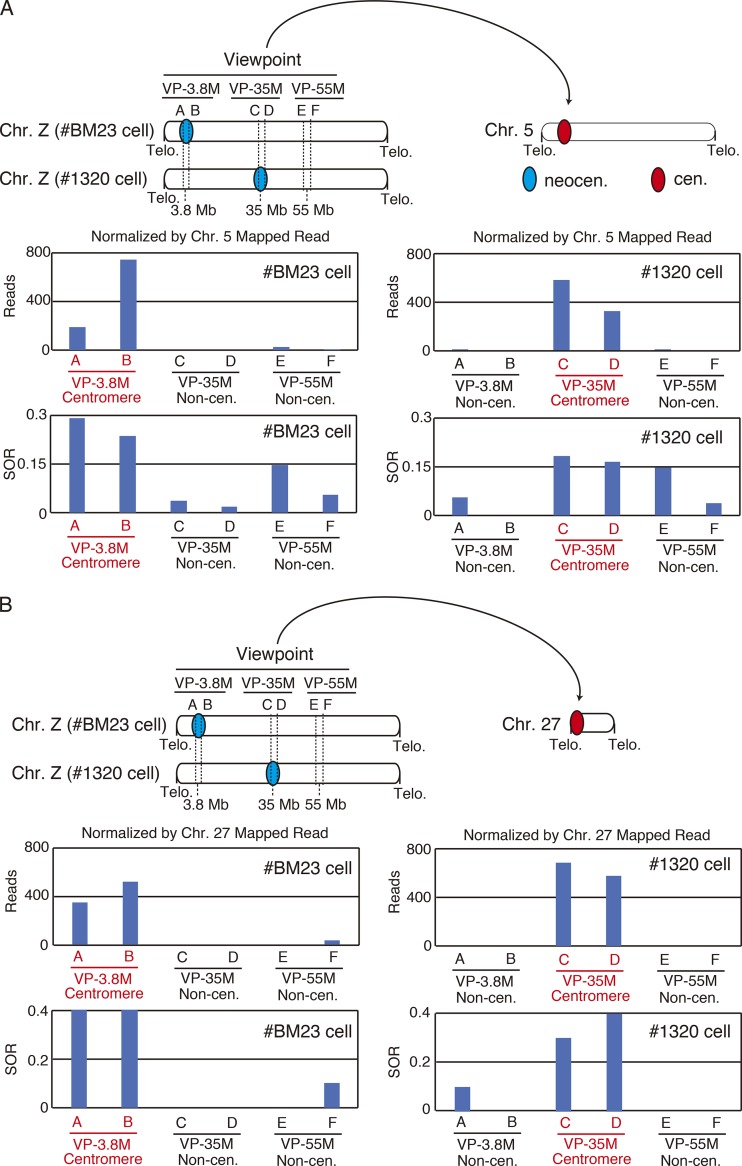
All the centromeres in yeasts are clustered, and this clustering is clearly visible as a single dot by microscopic observation (Funabiki et al., 1993). In addition, recent Hi-C analyses clearly demonstrated centromere clustering in yeast cells (Mizuguchi et al., 2014; Varoquaux et al., 2015; Burrack et al., 2016). However, it is not entirely certain whether the centromeres in vertebrate interphase nuclei are clustered because multiple punctate foci are observed during microscopic observation in interphase nuclei of vertebrate cells. By analyzing the 4C data, we intended to clarify whether centromere clustering occurs in DT40 cells (Fig. 6 and 7). We examined the number of sequence reads and SOR values, including that of the nonrepetitive centromere of chromosome 5 in #BM23 cells where the centromere is formed in the 3.8-Mb region, using 4C-seq data with six viewpoints (A–F of Fig. 6 A; and Table S1). We found that two viewpoints at the 3.8-Mb region (A and B in Fig. 6 A, left) associated with the sequence of the centromere on chromosome 5 in #BM23 cells. However, the other four viewpoints at the 35- and 55-Mb regions did not associate with the sequence of the centromere on chromosome 5 in #BM23 cells (Fig. 6 A, left). Because we calculated SOR values with the 150-kb window and the centromere region spans 30–40 kb, it is possible that the SOR may detect the noncentromere region. Although the two viewpoints at the 35-Mb centromeric region in #1320 cells (C and D in Fig. 6 A, right) were associated with the sequence of the centromere on chromosome 5, the other viewpoints were not associated with the centromere in #1320 cells (Fig. 6 A, right). We also examined the association of the neocentromeres with another nonrepetitive centromere on chromosome 27 (Fig. 6 B). As with the case for chromosome 5, the neocentromeric regions on chromosome 27 are associated with the centromere, but the noncentromeric viewpoints on the Z chromosome was not associated with the centromeres on chromosome 27 (Fig. 6 B).
Figure 6.
Neocentromeres associate with the centromeres of other chromosomes with nonrepetitive centromeres. (A) The number of sequence reads and SOR values containing the centromere sequence on chromosome 5 determined using six different viewpoints (A–F) in #BM23 and #1320 cell lines. The position of each viewpoint is shown. (B) The numbers of sequence reads and SOR values containing the centromere sequence on chromosome 27 determined using six different viewpoints (A–F) in #BM23 and #1320 cell lines. The position of each viewpoint is shown.
Figure 7.
Neocentromeres associate with the centromeres of other chromosomes with repetitive centromeres. (A) The number of sequence reads near repetitive centromere sequences on chromosome 1 determined using six different viewpoints (A–F) in #BM23 and #1320 cell lines. The position of each viewpoint is shown. (B) The number of sequence reads near repetitive centromere sequences on chromosome 2 determined using six different viewpoints (A–F) in #BM23 and #1320 cell lines. The position of each viewpoint is shown. (C) Measurements of numbers of centromere foci at various cell cycle stages using CENP-T as a centromere marker. Right image is a representative immune fluorescence image of DT40 cells with anti–CENP-T (red). Cell cycle stages are determined by EdU labeling (left; green). DNA was stained with DAPI (blue). Bar, 10 µm. The graph shows numbers of centromere foci at various cell cycle stages.
In addition to identifying the association of the neocentromeres on the Z chromosome with the nonrepetitive centromeres located on chromosomes 5 and 27, we examined whether the neocentromeres associate with repetitive centromeres. Most of the chicken chromosomes possess repetitive centromeres, but the sequences between the repetitive centromeres are fairly divergent (Shang et al., 2010). In spite of the intercentromeric sequence diversity, our analysis indicated that the sequences of both the centromeres of chromosomes 1 and 2 are associated with neocentromeres (Fig. 7, A and B). Considering these results, we concluded that neocentromeres associate with both repetitive and nonrepetitive centromeres and that centromere clustering does occur in the interphase nuclei of vertebrates. However, because punctate centromeric foci can be observed in the interphase nuclei of DT40 cells, this clustering should be of a transient nature. However, we scored the numbers of centromere foci in various cell cycle stages and found that the numbers of foci in interphase were reduced compared with those in mitosis (Fig. 7 C), which supports the centromere clustering observed by 4C analysis.
CENP-H but not CENP-C is involved in centromere–heterochromatin interactions
The results of the 4C and FISH analyses in this study demonstrated that centromeres are physically associated with heterochromatin-rich regions in the interphase nuclei even if they were distant at the sequence level. Next, it was important to address which molecules participate in establishing these interactions. H3K9 trimethylation is mediated by several H3 methyltransferases, and Suv39H is known as a major methyltransferase for H3K9me3 (Rea et al., 2000; Peters et al., 2001). Therefore, we tested the association of the 8- and 26-Mb regions with neocentromeres on the Z chromosome in Suv39H-deficient cells (Fig. S5, A–C). Chicken has two Suv39H genes (Suv39H1 and Suv39H2), and we disrupted both genes by CRIPSR/Cas9 using #1320 cells (Fig. S5 A). Consistent with a previous study (Peters et al., 2001), total H3K9me3 levels were reduced in these Suv39H-deficient cells based on immunoblot analysis with anti-H3K9me3 antibody (Hayashi-Takanaka et al., 2011), but H3K9me3 still existed (Fig. S5 B). We tested association of the 8-Mb region with the neocentromere (the 35-Mb region) in #1320 cells with or without Suv39H. As shown in Fig. S5 C, we found a tendency for this association to be reduced in Suv39H-deficient cells, but this dissociation was not statistically significant. There are some explanations for this observation. First, although total H3K9me3 levels were reduced in Suv39H-deficient cells, we could not evaluate by how much they had reduced in the 8-Mb region. The remaining H3K9me3 might be sufficient for the interaction. Second, Suv39H-deficient cells are viable, and H3K9me3 levels may have adapted to these cells. We may need conditional knockout cells for further analysis since it is possible that this interaction is not related to Suv39H. Nevertheless, with current data, we cannot conclude whether the association of the 8-Mb region with the neocentromere depends on H3K9me3.
In addition to the heterochromatin side, centromere proteins are suitable candidates that can influence the formation of these interactions, and we examined the contribution of the centromere proteins to the interaction. Once centromeres are epigenetically specified by the deposition of the histone H3 variant, CENP-A, into chromatin, a 16-subunit protein complex known as the constitutive centromere–associated network (CCAN) is assembled on the chromatin containing CENP-A in interphase nuclei. We therefore examined whether CCAN is involved during the formation of the centromere–heterochromatin interactions. We have previously shown that CCAN is divided into subgroups that have different roles during centromeric assembly (Fukagawa et al., 2001; Okada et al., 2006; Kwon et al., 2007; Hori et al., 2008; Nagpal et al., 2015; Nagpal and Fukagawa, 2016). Among them, the complex containing CENP-H is distinct from CENP-C in chicken interphase cells (Fukagawa et al., 2001; Kwon et al., 2007; Hori et al., 2008; Nagpal and Fukagawa, 2016). We thus focused on CENP-C and CENP-H and generated CENP-C– or CENP-H–conditional knockout cell lines using #1320 cells based on a method combining auxin-inducible degron (aid) tag with CRISPR/Cas9 (Nishimura and Fukagawa, 2017). In our method, the endogenous coding alleles were mutated by the CRISPR/Cas9 strategy, and an aid-tagged protein was stably expressed in the presence of the F-box protein TIR. Once auxin is added to the cells, the aid-tagged protein is rapidly degraded within 2 h (Nishimura et al., 2009; Nishimura and Fukagawa, 2017). It is therefore possible to create conditional knockouts of a target protein by the addition of auxin.
We confirmed protein degradation after mutating the endogenous proteins in #1320 cells expressing CENP-C–aid or CENP-H–aid after the addition of auxin. As shown in Fig. S5 D, endogenous CENP-C or CENP-H proteins were not detected in the individual knockout lines, and CENP-C–aid or CENP-H–aid were not detectable by Western blotting analysis within 2 h after the addition of auxin in #1320-based CENP-C– or CENP-H–conditional knockout lines, respectively. After the addition of auxin, we performed analyses with two-color FISH using the green-labeled BAC clone #206E12 containing the 35-Mb centromeric region and the red-labeled BAC clone #261B8 containing the 8-Mb heterochromatin region as probes for each knockout line at 0, 2, and 4 h after the addition of auxin (Fig. 8 A). Consistent with the analyses of FISH in #1320 cells (Fig. 4 C), the 8- and 35-Mb regions are associated in both lines at 0 h. In contrast, the distance between the 8- and 35-Mb regions increased in the CENP-H–knockout line after the addition of auxin; however, the distance was unaltered in CENP-C–knockout line even after the addition of auxin, suggesting that CENP-H but not CENP-C contributes to the formation of the centromere–heterochromatin interactions in #1320 cells.
Figure 8.
CENP-H but not CENP-C contributes to neocentromere–heterochromatin interactions. (A) Analyses with FISH using CENP-H– or CENP-C–conditional knockout #1320 cell lines. CENP-H and CENP-C are degraded upon adding auxin. Auxin was added at time 0, and green-labeled BAC clone #206E12 comprising the 35-Mb region (centromere in #1320 cells) as well as the red-labeled BAC clone #261B8 comprising of the 8-Mb region were used as probes. Signals are shown by arrows. DNA was stained with DAPI (blue). Bar, 10 µm. The graph on the right shows the distance between the two probes in each cell line at the indicated times after the addition of auxin. (B) 4C profiles determined with CENP-C–conditional knockout #1320 cell lines with the 35-Mb region as a viewpoint in the absence (top: CENP-C ON) or presence (middle: 2 h after auxin; CENP-C OFF) of auxin. The two profiles were merged (bottom). Arrowheads indicate the 8- and 26-Mb regions. The neocentromere was located at the 35-Mb region in these cells. (C) 4C profiles determined using #1320-based CENP-H–conditional knockout cell lines with the 35-Mb region as a viewpoint in the absence (top: CENP-H ON) or presence (middle: 2 h after auxin, CENP-H OFF) of auxin. CENP-H ON cells were treated with nocodazole for 2 h. The two profiles were merged (bottom). Arrows indicate the 8- and 26-Mb regions. The neocentromere was located at the 35-Mb region in these cells.
Finally, we performed 4C analyses using the 35-Mb region as a viewpoint in #1320-based CENP-C– or CENP-H–conditional knockout lines 0 or 2 h after the addition of auxin (Fig. 8, B and C). As CENP-C and CENP-H are centromere proteins and defects in these proteins can result in mitotic delays (Fukagawa et al., 2001; Kwon et al., 2007), high mitotic accumulation might affect the 4C profile. Therefore, we examined the cell-cycle profile of aid-based CENP-C– or CENP-H–conditional knockout cells after the addition of auxin by flow cytometry (Fig. S5 E). 2 h after the addition of auxin, mitotic accumulation was not observed in CENP-C–conditional knockout cells (Fig. S5 E), and we compared the 4C profiles of CENP-C–conditional knockout cells at 0 and 2 h after the addition of auxin. In contrast, the proportion of G2/M cells increased from 24.3% at 0 h to 31.4% at 2 h in CENP-H–conditional knockout cells after the addition of auxin (Fig. S5 E). This G2/M accumulation profile is similar to that of cells treated with the spindle poison nocodazole (32.8% at 2 h after the addition of nocodazole; Fig. S5 E). We thus compared the 4C profile of CENP-H–conditional knockout cells 2 h after the addition of auxin with that of the same cells treated with nocodazole for 2 h in the absence of auxin. As observed in the 4C analysis of #1320 cells (Fig. 2, C and D), the 35-Mb centromere region was observed to clearly interact with the 8- or 26-Mb heterochromatin regions in both CENP-C– and CENP-H–conditional knockout cells in the absence of auxin (Fig. 8, B and C). The interaction profiles between the centromere and heterochromatin regions remained unaltered in CENP-C–conditional knockouts 2 h after the addition of auxin (Fig. 8 B). However, the peaks at the 8- and 26-Mb regions noticeably decreased in the CENP-H–conditional knockout cells 2 h after the addition of auxin, in comparison with the cells in the control (Fig. 8 C). Moreover, the 4C profile near the centromere region was not changed in CENP-C– or CENP-H–knockout cells (Fig. S5 F).
Global 4C data for CENP-C– and CENP-H–knockout cells were consistent with the results obtained with FISH. Considering these data, we conclude that CENP-H but not CENP-C is the chief contributor to the formation of long-range centromere–heterochromatin interactions in interphase nuclei (Fig. 9).
Figure 9.
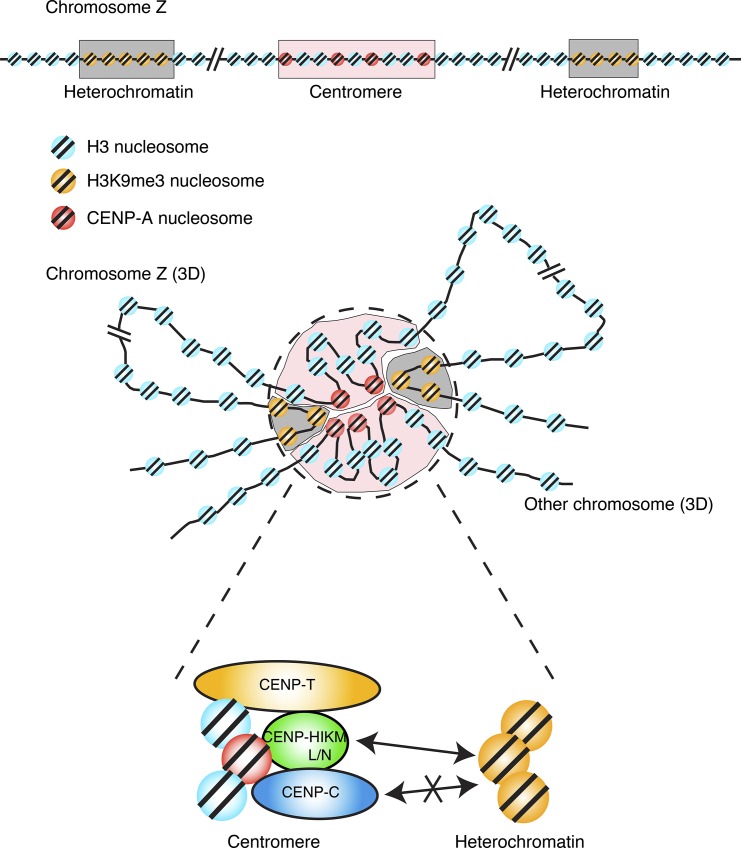
A proposed model of 3D architecture of the genome including the centromere in DT40 cells. The centromere is located at the 42.6-Mb region, and the H3K9me3 rich regions are located at 8- and 26-Mb regions on the chicken Z chromosome at the 1D level. Although the centromeric region is estimated to lie within 30–40 kb (∼200 nucleosomes) according to analyses with ChIP-seq using anti–CENP-A, there exist ∼30 CENP-A–containing nucleosomes, indicating that the CENP-A–containing nucleosomes are scattered in the centromeric region. Although distances between the centromere and heterochromatin regions are long at the 1D level, the centromere physically associates with regions of heterochromatin in the interphase nuclei. The nucleosomes containing CENP-A might indulge in self-interactions to form a compact structure. The formation of the 3D genomic architecture, including the centromere, depends on the CENP-H–associated complex but not on CENP-C.
Discussion
We previously generated DT40 cell lines containing differently positioned neocentromeres (Shang et al., 2013). By comparing the sequences of the different neocentromeres, we could not detect any obvious common features in sequences. However, despite the sequence divergence, all the proteins of the kinetochore generally assemble on the neocentromeres, suggesting the existence of common chromatin features among the different neocentromeres. Because our DT40 cell lines contain a variety of neocentromeres, they are excellent tools for identifying the common features among different neocentromeres. We attempted to identify common binding regions of the neocentromeres by analyses with 4C-seq. Although analyses with 4C-seq detected multiple binding regions for each neocentromere, we identified two major binding regions for the three neocentromeres. These two regions also associated with the native centromere containing nonrepetitive sequences on the Z chromosome. Interestingly, the heterochromatin marker H3K9me3 was found to be enriched in these two regions. Heterochromatin is usually formed at a region near the kinetochore known as the pericentromeric region, which flanks the centromeres and contains repetitive sequences. However, the enrichment of H3K9me3 has not been observed near all the neocentromeres of both chicken and human cells (Alonso et al., 2010; Shang et al., 2013). As heterochromatin contributes to the formation of the kinetochore (Folco et al., 2008; Sato et al., 2012; Allshire and Madhani, 2018), it was necessary to justify why no heterochromatin was detected near the centromeres containing nonrepetitive sequences. We demonstrated this by analyses with 4C-seq; centromeres with nonrepetitive sequences generally associate with heterochromatin regions in the 3D organization in spite of being distantly located from the neocentromeric regions at the sequence level. We believe that this is an interesting finding, explaining the role of heterochromatin. The heterochromatin located near the kinetochores is likely to have a preventive role, hindering the invasion of the centromeric region into regions of euchromatin. In our previous study (Hori et al., 2017), we observed that although the position of the centromere can alter, centromere drift is usually suppressed. It is likely that the pericentromeric heterochromatin functions as a barrier against centromere drift. However, it was difficult to understand why centromere drift is suppressed even in centromeres with nonrepetitive sequences because heterochromatin is not apparently detected near the nonrepetitive centromeres. Our finding that nonrepetitive centromeres associate with heterochromatin in the 3D arrangement explains why centromeric drift is suppressed even in nonrepetitive centromeres.
We also examined noncentromeric CENP-A levels in the 8- and 26-Mb regions because these regions might be potential sites for neocentromere formation. However, we did not observe significant accumulation of CENP-A in these regions, and we did not detect a clear correlation between centromere-binding sites, based on 4C, and neocentromere sites, suggesting that neocentromere formation is relatively stochastic.
Although we observed accumulation of H3K9me3 in the 8- and 26-Mb regions, H3K9me3-rich regions were not always neocentromere-binding sites. For example, one end of the Z chromosome is highly enriched with H3K9me3 (Fig. 3). This region consists of highly repetitive sequences and is heterochromatinized (Hori et al., 1996), but we did not detect significant interaction of this region with neocentromeres on the Z chromosome. Although we cannot conclude which heterochromatin regions preferentially associate with neocentromeres, more studies are needed to clarify the mechanisms of the interaction in the future.
Our 4C analysis also demonstrated that interactions within a centromeric region occurred frequently (Fig. 5). This suggested that the centromere forms a compact structure in the nuclei. ChIP-seq data with anti–CENP-A showed that centromeres with nonrepetitive sequences cover a 30–40-kb region in chicken chromosomes (Shang et al., 2010, 2013), but analyses of the copy number of centromere proteins suggest that there are ∼30 CENP-A nucleosomes per kinetochore (Johnston et al., 2010), indicating that CENP-A is scattered in the 30–40-kb centromeric region (∼200 total nucleosomes). Therefore, based on the 4C profiles for neocentromeres, we predicted that the scattered CENP-A nucleosomes associate with each other to form a compact structure (Fig. 9). Because some of the proteins of the kinetochore directly bind to the CENP-A nucleosome to form the structure of the kinetochore (Kato et al., 2013; Pentakota et al., 2017; Chittori et al., 2018; Tian et al., 2018), the assembled CENP-A cluster would be suitable for binding the downstream components of kinetochores. Although ChIP-seq profiles with CENP-A are obtained at base pair resolution, 4C detects HindIII fragments, which are ∼3 kb. On comparing 4C profiles with ChIP-seq profiles, we could not directly show a CENP-A–CENP-A interaction (Fig. 5). However, as CENP-A incorporation is an important feature for centromere formation, we prefer to use our proposed model.
In yeasts, such as S. pombe or C. albicans, centromeres including neocentromeres are clustered at a particular position in the nuclei (Funabiki et al., 1993; Thakur and Sanyal, 2012; Mizuguchi et al., 2014; Burrack et al., 2016). However, it is not obvious whether the centromere cluster is formed in vertebrate nuclei. Owing to the visibility of multiple centromeric signals in the interphase nuclei of vertebrates, it is clear that unlike in the nuclei of yeasts, all the centromeres in vertebrate nuclei are not clustered at one particular location. However, our 4C-seq analysis demonstrated that neocentromeres associate with other centromeres, including those containing repetitive and nonrepetitive sequences. Nevertheless, centromere–centromere interactions are perhaps not strong, because the sequence reads for centromeres on other chromosomes associated with the centromere of the Z chromosome were lower than with the interacting sequence on the Z chromosome. Based on 4C analysis, we conclude that centromere clustering does undoubtedly occur, but centromere–centromere interactions might be transient in the interphase nuclei of DT40 cells. Although transient, these interactions might have significance in the formation of the kinetochore and/or faithful chromosomal segregation. Interestingly, transient centromere clusters occur in prophase nuclei of early Drosophila embryos (Hiraoka et al., 1990); however, this might not be similar to our observation because our transient centromere–centromere association occurred at various stages during interphase. The next crucial challenge is to clarify the importance of centromere clustering in vertebrate nuclei.
Finally, we demonstrated that neocentromere–heterochromatin interactions in the 3D organization of the genome depend on CENP-H but not on CENP-C (Fig. 8). In our previous study, we showed that CENP-H and CENP-H–related proteins including CENP-T are distinct from CENP-C with respect to recruiting the outer kinetochore proteins (Hori et al., 2008, 2013; Fukagawa and Earnshaw, 2014). Although it is presently unknown as to how the long-range neocentromere–heterochromatin interactions occur, it is likely that heterochromatinized regions recognize a prekinetochore structure by CCAN rather than recognizing a particular protein. The localization of CENP-H–related proteins in the centromere of interphase nuclei is interdependent, but the localization of CENP-C in the interphase centromeres occurs downstream of the CENP-H–related proteins (Fukagawa et al., 2001; Kwon et al., 2007; Nagpal et al., 2015). This suggested that the CENP-H–related proteins form a prekinetochore structure before the localization of CENP-C to the CENP-A–containing centromeres in the interphase, which might be recognized by the heterochromatin regions. In this study, the inference that neocentromere–heterochromatin interactions depend on CENP-H but not CENP-C is consistent with this model. In addition, our preliminary data indicate that HP1 was detected in CENP-T IP but not in CENP-C IP.
Through 4C-seq analysis with cell lines containing variously positioned neocentromeres, we detected previously unidentified interaction sites with the neocentromeres. In addition to the interaction of heterochromatin regions with neocentromeres, we propose a model of the 3D architecture of the genome in cells containing neocentromeres (Fig. 9). Although the model has been developed by analyzing only three different DT40 cell lines containing neocentromeres, we believe that the 3D organization of the genome, including the neocentromere proposed in this study, might be applicable to other vertebrate cells including those of humans because heterochromatin is not clearly detectable near the neocentromeres of human cells (Alonso et al., 2010). We have developed additional cell lines containing neocentromeres located at various other positions. Combining other genome analysis techniques such as Hi-C or 5C with these cell lines or human cells containing neocentromeres would provide additional insights for understanding the role of centromeres in the formation of the 3D architecture of the genome. This study widens our understanding of the 3D architecture of genomes including centromeres.
Materials and methods
Cell culture
DT40 cells were cultured at 38.5°C in DMEM supplemented with 10% FBS (Sigma-Aldrich), 1% chicken serum (Gibco), and penicillin-streptomycin (Thermo Fisher Scientific; Hori et al., 2008). DT40 cell lines containing neocentromeres on the Z chromosome were created in previous research (Shang et al., 2013).
4C-seq
4C was performed as described previously (Splinter et al., 2012). Briefly, 2.0 × 107 cells were fixed in 1% PFA (Electron Microscopy Sciences) for 10 min and quenched in 125 mM glycine. Cells were lysed for 10 min in ice-cold lysis buffer (50 mM Tris-HCl [Sigma-Aldrich], pH 7.5, 150 mM NaCl [Nacalai Tesque], 5 mM EDTA [Nacalai Tesque], 0.5% Igepal CA-630 [Nacalai Tesque], 1% Triton X-100 [Nacalai Tesque], and 1× complete protease inhibitors [Roche]). Fixed and lysed cells were treated with 0.3% SDS (Nacalai Tesque) at 37°C with shaking at 900 RPM for 1 h with a Thermo-shaker MS-100 (Chiyoda Science) and quenched in 1% Triton X-100 at 37°C with shaking at 900 RPM for 1 h, and chromatin was digested with 900 units of HindIII (NEB) at 37°C with shaking at 900 RPM overnight. For EcoRI and BglII (NEB) digestion, 900 U of each enzyme were used for digestion. For inactivation of restriction enzyme digestion, digested chromatin was treated with 1% SDS at 65°C for 20 min. After dilution and quenching of SDS in 1% Triton X-100 at 37°C for 1 h, we performed DNA ligation reaction with addition of 50 U T4 ligase (Thermo Fisher Scientific) to samples, and samples were incubated at 16°C overnight. Ligated chromatin samples were decross-linked by addition of 300 µg proteinase K (Sigma-Aldrich) and incubation at 65°C overnight. DNA was isolated with phenol:chloroform (1:1; Wako) extraction and ethanol (Nacalai Tesque) precipitation, followed by RNase A (Nacalai Tesque) incubation for 45 min. Second digestion was performed with 100 U CviQI (NEB) at 25°C overnight. After heat inactivation of CviQI at 65°C for 30 min, DNA was re-ligated with 100 units of T4 ligase at 16°C overnight. After ethanol precipitation, DNA was purified by QIAquick PCR purification kit (28104; QIAGEN). 3.2 µg DNA was used for 4C-PCR as follows: 94°C for 2 min; 94°C for 10 s, 55°C for 1 min, and 68°C for 3 min, 29 repeats; 68°C for 5 min. After PCR amplification, PCR products were purified by AMpureXP (Beckman Coulter) to remove primer dimer. The 4C-library was sequenced with a Miseq sequencer (Illumina). Viewpoint positions and target oligonucleotide sequence for PCR are shown in Table S1. All sequence data were deposited in the DNA Data Bank of Japan (DDBJ) database, and accession numbers are shown in Table S2.
Calculation of SOR and occupancy comparison values
Adaptor sequences used for priming inverse PCR (4C-PCR) reactions were removed from the output file of each raw sequence. Sequences that had two or more first-restriction enzyme recognition sequences were divided at restriction sites and treated as different reads. Processed reads were mapped onto the chicken genome sequence using bowtie2. Uniquely hit reads and multiple-hit reads with mapped scores higher than those of the second best hits were extracted. Mapped positions that were located far from first-restriction enzyme sites were removed. The number of hits at each first-restriction enzyme site was calculated by counting the numbers of reads that mapped to each site.
Reference genomes were binned into 150-kb windows with a 100-kb moving step size. The ratio of enzyme sites with at least one read (≥1) to the total number of enzyme sites in a genomic window was termed the occupancy rate data. Comparison values were calculated by comparing the occupancy rate data from two samples within the same window as follows:
Comparison values were plotted as histograms and fitted to the exponential distribution (P(x) = λe(−λx)). Finally, the top 0.1% most-detected regions were found to be proximal to neocentromere regions.
Two-color FISH
Bacterial artificial chromosome (BAC) clones 261B8, 116B8, and 206E12 cover 8-, 26-, and 35-Mb (neocentromere region in #1320 cells) regions on the Z chromosome, respectively, and were used as FISH probes. Each BAC clone was labeled by a nick-translation method with DNase I (Roche), DNA polymerase I (Boehringer Mannheim), and Cy3-3-dUTP (NEL578; PerkinElmer) or FITC-12-dUTP (NEL412001EA; PerkinElmer). For double-color FISH analysis, DT40 cells were fixed with 3% PFA for 10 min after cytospinning to a slide glass (Matsunami). Fixed cells were treated with PBS:methanol:acetic acid (8:3:1) for 15 min and incubated in methanol:acetic acid (3:1) for 15 min. Samples were denatured with labeled probes at 73°C for 1 h. After overnight incubation at 39°C, cells were washed with 2× SSC twice, with formamide; 2× SSC (1:1) twice; and 2× SSC twice at 37°C by each wash for 5 min, respectively. After DAPI staining, cells were washed with PBS twice and mounted with Vectashield mounting reagent (Vector Laboratories). FISH images were captured by sCMOS camera (Zyla 4.2; Andor) mounted on ECLIPSE Ti microscope (Nikon) with an objective lens (Plan Apo lambda 100×/1.45 NA; Nikon) and CSU-W1 confocal scanner unit (Yokogawa) controlled by NIS elements (Nikon). The distance between two signals was measured by Imaris software (Bitplane).
Creation of auxin-based conditional knockout cell lines
Auxin-based CENP-C– or CENP-H–conditional knockout DT40 cell lines were created with #1320 cell line by an efficient method to make aid mutants (Nishimura and Fukagawa, 2017). CENP-H mutants were already created previously (Nishimura and Fukagawa, 2017). For creation of the CENP-C–knockout cell line, linearized pAID plasmid containing CENP-C cDNA and pX330-expressing single guide RNA (targeting for chicken CENP-C genome) were transfected into #1320 cells by Neon Transfection system (Thermo Fisher Scientific). After 1-wk selection in DMEM containing 30 µg/ml blasticidin S (Wako), drug-resistant colonies were picked up and examined for gene disruption by genomic PCR, sequencing, and Western blotting analysis.
Flow cytometry
Cells (1–5 × 106 cells in 10 ml) were labeled with 20 µM BrdU for 20 min and collected. The cells were washed with 10 ml ice-cold PBS and fixed in 70% ethanol. After washing with 1 ml 1% BSA in PBS, the samples were incubated in 1 ml of 4 N HCl with 0.5% Triton X-100 at room temperature for 30 min and washed with 1 ml of 1% BSA in PBS three times. The cells were treated with anti-BrdU (BD) and stained with FITC-labeled anti–mouse IgG (Jackson ImmunoResearch). DNA was stained with 1 ml of 10 µg/ml propidium iodide in 1% BSA in PBS at 4°C overnight. The stained cells were applied to Guava easyCyte (Merck). Obtained data were analyzed with InCyte software (Merck).
Quantification and statistical analysis
Data analyses for 4C-seq were described above. Results of FISH and immunofluorescence data in Figs. 3, 4, 8, S3, and S4 were made using Prism (GraphPad Software), and significance was evaluated by Student’s t test. A P value <0.0001 was defined as significant. Means and SD are indicated by a horizontal long bar and two short bars, respectively, in each graph. Sample numbers: n = 100 (WTDT40 cell; Chr.Z42.6M-Chr.Z8M) and 100 (WTDT40 cell; Chr.Z42.6M-Chr.Z62M) for Fig. 3; n = 100 (#1320 cell; Chr.Z35M-Chr.Z8M), 100 (#BM23 cell; Chr.Z35M-Chr.Z8M), 100 (#1320 cell; Chr.Z35M-Chr.Z62M), and 100 (#BM23 cell; Chr.Z35M-Chr.Z62M) for Fig. 4; n = 100 (0 h CENP-C–Caid), 100 (2 h CENP-C–Caid), 100 (4 h CENP-C–Caid), 100 (0 h CENP-H–Caid), 100 (2 h CENP-H–Caid), and 100 (4 h CENP-HC–Caid) for Fig. 8; n = 100 (#1320 cell; Chr.Z35M-26M), 88 (#BM23 cell; Chr.Z35M-26M), n = 100 (#1320 cell; nuclear size), and 88 (#BM23 cell; nuclear size) for Fig. S3; and n = 100 (WTDT40 cell; Chr.1-81.5M-Chr.1-100M) and 100 (WTDT40 cell; Chr.1-118.5M-Chr.1-100M) for Fig. S4.
Accession numbers
The DDBJ accession number for all sequence data in this study is DRA006803. Sample ID information about sequence data are summarized in Table S2.
Online supplemental material
Fig. S1 (A and B) shows 4C profiles of #BM23 and #1320 cells, and Fig. S1 (C and D) presents characterization of cell lines used in this study. Fig. S2 shows 4C profiles of the #1304 cell line using the neocentromere region as a viewpoint. Fig. S3 shows association of the 8- and 26-Mb regions with neocentromere in #1320 cells using 4C and FISH analyses. Fig. S4 (A and B) shows that repetitive centromere on chromosome 1 associates with a heterochromatin-rich region; Fig. S4 (C and D) presents 3C-PCR analysis in neocentromere regions. Fig. S5 (A–C) shows creation and characterization of Suv39H-deficient cells, and Fig. S5 (D–F) show creation and characterization of CENP-C– and CENP-H–deficient cells. Table S1 summarizes target oligonucleotide sequences in each viewpoint. Table S2 shows sample ID information for each sequence data of 4C analysis.
Supplementary Material
Acknowledgments
The authors are very grateful to Yuko Fukagawa, Kaori Ohshimo, and Reika Fukuoka for technical assistance.
This work was supported by Japan Society for the Promotion of Science KAKENHI grants 15H05972, 16H06279, and 17H06167 to T. Fukagawa; 15H05979 to T. Itoh; and 17K15041 to K. Nishimura.
The authors declare no competing financial interests.
Author contributions: K. Nishimura performed all experiments in this study. M Komiya and T. Itoh performed next-generation sequencing and analyzed these data. T. Hori made and characterized several cell lines and helped with FISH analysis. T. Fukagawa designed and supervised all experiments and wrote the manuscript, discussing with all authors.
References
- Allshire R.C., and Madhani H.D.. 2018. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 19:229–244. 10.1038/nrm.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A., Hasson D., Cheung F., and Warburton P.E.. 2010. A paucity of heterochromatin at functional human neocentromeres. Epigenetics Chromatin. 3:6 10.1186/1756-8935-3-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.E., and Cleveland D.W.. 2011. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 144:471–479. 10.1016/j.cell.2011.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrack L.S., Hutton H.F., Matter K.J., Clancey S.A., Liachko I., Plemmons A.E., Saha A., Power E.A., Turman B., Thevandavakkam M.A., et al. 2016. Neocentromeres Provide Chromosome Segregation Accuracy and Centromere Clustering to Multiple Loci along a Candida albicans Chromosome. PLoS Genet. 12:e1006317 10.1371/journal.pgen.1006317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittori S., Hong J., Saunders H., Feng H., Ghirlando R., Kelly A.E., Bai Y., and Subramaniam S.. 2018. Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N. Science. 359:339–343. 10.1126/science.aar2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., and Mirny L.. 2016. The 3D Genome as Moderator of Chromosomal Communication. Cell. 164:1110–1121. 10.1016/j.cell.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J., Richmond T.A., Arnaout R.A., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C., et al. 2006. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 16:1299–1309. 10.1101/gr.5571506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Sart D., Cancilla M.R., Earle E., Mao J.I., Saffery R., Tainton K.M., Kalitsis P., Martyn J., Barry A.E., and Choo K.H.. 1997. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat. Genet. 16:144–153. 10.1038/ng0697-144 [DOI] [PubMed] [Google Scholar]
- Folco H.D., Pidoux A.L., Urano T., and Allshire R.C.. 2008. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 319:94–97. 10.1126/science.1150944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., and Earnshaw W.C.. 2014. The centromere: chromatin foundation for the kinetochore machinery. Dev. Cell. 30:496–508. 10.1016/j.devcel.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Mikami Y., Nishihashi A., Regnier V., Haraguchi T., Hiraoka Y., Sugata N., Todokoro K., Brown W., and Ikemura T.. 2001. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 20:4603–4617. 10.1093/emboj/20.16.4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood M.J., Liu M.H., Pan Y.F., Liu J., Xu H., Mohamed Y.B., Orlov Y.L., Velkov S., Ho A., Mei P.H., et al. 2009. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 462:58–64. 10.1038/nature08497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H., Hagan I., Uzawa S., and Yanagida M.. 1993. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 121:961–976. 10.1083/jcb.121.5.961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Takanaka Y., Yamagata K., Wakayama T., Stasevich T.J., Kainuma T., Tsurimoto T., Tachibana M., Shinkai Y., Kurumizaka H., Nozaki N., and Kimura H.. 2011. Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic Acids Res. 39:6475–6488. 10.1093/nar/gkr343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y., Agard D.A., and Sedat J.W.. 1990. Temporal and spatial coordination of chromosome movement, spindle formation, and nuclear envelope breakdown during prometaphase in Drosophila melanogaster embryos. J. Cell Biol. 111:2815–2828. 10.1083/jcb.111.6.2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Suzuki Y., Solovei I., Saitoh Y., Hutchison N., Ikeda J.E., Macgregor H., and Mizuno S.. 1996. Characterization of DNA sequences constituting the terminal heterochromatin of the chicken Z chromosome. Chromosome Res. 4:411–426. [DOI] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.H., Suzuki E., Okawa K., et al. 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 135:1039–1052. 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- Hori T., Shang W.H., Takeuchi K., and Fukagawa T.. 2013. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J. Cell Biol. 200:45–60. 10.1083/jcb.201210106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Shang W.H., Toyoda A., Misu S., Monma N., Ikeo K., Molina O., Vargiu G., Fujiyama A., Kimura H., et al. 2014. Histone H4 Lys 20 monomethylation of the CENP-A nucleosome is essential for kinetochore assembly. Dev. Cell. 29:740–749. 10.1016/j.devcel.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Kagawa N., Toyoda A., Fujiyama A., Misu S., Monma N., Makino F., Ikeo K., and Fukagawa T.. 2017. Constitutive centromere-associated network controls centromere drift in vertebrate cells. J. Cell Biol. 216:101–113. 10.1083/jcb.201605001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K., Ogiyama Y., Chikashige Y., Soejima S., Masuda F., Kakuma T., Hiraoka Y., and Takahashi K.. 2008. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 321:1088–1091. 10.1126/science.1158699 [DOI] [PubMed] [Google Scholar]
- Johnston K., Joglekar A., Hori T., Suzuki A., Fukagawa T., and Salmon E.D.. 2010. Vertebrate kinetochore protein architecture: protein copy number. J. Cell Biol. 189:937–943. 10.1083/jcb.200912022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagansky A., Folco H.D., Almeida R., Pidoux A.L., Boukaba A., Simmer F., Urano T., Hamilton G.L., and Allshire R.C.. 2009. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science. 324:1716–1719. 10.1126/science.1172026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Jiang J., Zhou B.R., Rozendaal M., Feng H., Ghirlando R., Xiao T.S., Straight A.F., and Bai Y.. 2013. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 340:1110–1113. 10.1126/science.1235532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketel C., Wang H.S., McClellan M., Bouchonville K., Selmecki A., Lahav T., Gerami-Nejad M., and Berman J.. 2009. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet. 5:e1000400 10.1371/journal.pgen.1000400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M.S., Hori T., Okada M., and Fukagawa T.. 2007. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol. Biol. Cell. 18:2155–2168. 10.1091/mbc.e07-01-0045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O., et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 326:289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggert K.A., and Karpen G.H.. 2001. The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics. 158:1615–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall O.J., Chueh A.C., Wong L.H., and Choo K.H.. 2008. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 82:261–282. 10.1016/j.ajhg.2007.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi T., Fudenberg G., Mehta S., Belton J.M., Taneja N., Folco H.D., FitzGerald P., Dekker J., Mirny L., Barrowman J., and Grewal S.I.S.. 2014. Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature. 516:432–435. 10.1038/nature13833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal H., and Fukagawa T.. 2016. Kinetochore assembly and function through the cell cycle. Chromosoma. 125:645–659. 10.1007/s00412-016-0608-3 [DOI] [PubMed] [Google Scholar]
- Nagpal H., Hori T., Furukawa A., Sugase K., Osakabe A., Kurumizaka H., and Fukagawa T.. 2015. Dynamic changes in CCAN organization through CENP-C during cell-cycle progression. Mol. Biol. Cell. 26:3768–3776. 10.1091/mbc.e15-07-0531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova N., Smith E.M., Zhan Y., and Dekker J.. 2012. Analysis of long-range chromatin interactions using Chromosome Conformation Capture. Methods. 58:192–203. 10.1016/j.ymeth.2012.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., and Fukagawa T.. 2017. An efficient method to generate conditional knockout cell lines for essential genes by combination of auxin-inducible degron tag and CRISPR/Cas9. Chromosome Res. 25:253–260. [DOI] [PubMed] [Google Scholar]
- Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., and Kanemaki M.. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods. 6:917–922. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Ogiyama Y., Ohno Y., Kubota Y., and Ishii K.. 2013. Epigenetically induced paucity of histone H2A.Z stabilizes fission-yeast ectopic centromeres. Nat. Struct. Mol. Biol. 20:1397–1406. 10.1038/nsmb.2697 [DOI] [PubMed] [Google Scholar]
- Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R. III, Desai A., and Fukagawa T.. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446–457. 10.1038/ncb1396 [DOI] [PubMed] [Google Scholar]
- Pentakota S., Zhou K., Smith C., Maffini S., Petrovic A., Morgan G.P., Weir J.R., Vetter I.R., Musacchio A., and Luger K.. 2017. Decoding the centromeric nucleosome through CENP-N. eLife. 6:e33442 10.7554/eLife.33442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perpelescu M., and Fukagawa T.. 2011. The ABCs of CENPs. Chromosoma. 120:425–446. 10.1007/s00412-011-0330-0 [DOI] [PubMed] [Google Scholar]
- Peters A.H., O’Carroll D., Scherthan H., Mechtler K., Sauer S., Schöfer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., et al. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 107:323–337. 10.1016/S0092-8674(01)00542-6 [DOI] [PubMed] [Google Scholar]
- Rea S., Eisenhaber F., O’Carroll D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., and Jenuwein T.. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 406:593–599. 10.1038/35020506 [DOI] [PubMed] [Google Scholar]
- Saffery R., Irvine D.V., Griffiths B., Kalitsis P., Wordeman L., and Choo K.H.. 2000. Human centromeres and neocentromeres show identical distribution patterns of >20 functionally important kinetochore-associated proteins. Hum. Mol. Genet. 9:175–185. 10.1093/hmg/9.2.175 [DOI] [PubMed] [Google Scholar]
- Sato H., Masuda F., Takayama Y., Takahashi K., and Saitoh S.. 2012. Epigenetic inactivation and subsequent heterochromatinization of a centromere stabilize dicentric chromosomes. Curr. Biol. 22:658–667. 10.1016/j.cub.2012.02.062 [DOI] [PubMed] [Google Scholar]
- Shang W.H., Hori T., Toyoda A., Kato J., Popendorf K., Sakakibara Y., Fujiyama A., and Fukagawa T.. 2010. Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 20:1219–1228. 10.1101/gr.106245.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang W.H., Hori T., Martins N.M., Toyoda A., Misu S., Monma N., Hiratani I., Maeshima K., Ikeo K., Fujiyama A., et al. 2013. Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev. Cell. 24:635–648. 10.1016/j.devcel.2013.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang W.H., Hori T., Westhorpe F.G., Godek K.M., Toyoda A., Misu S., Monma N., Ikeo K., Carroll C.W., Takami Y., et al. 2016. Acetylation of histone H4 lysine 5 and 12 is required for CENP-A deposition into centromeres. Nat. Commun. 7:13465 10.1038/ncomms13465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E., de Wit E., van de Werken H.J., Klous P., and de Laat W.. 2012. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: from fixation to computation. Methods. 58:221–230. 10.1016/j.ymeth.2012.04.009 [DOI] [PubMed] [Google Scholar]
- Thakur J., and Sanyal K.. 2012. A coordinated interdependent protein circuitry stabilizes the kinetochore ensemble to protect CENP-A in the human pathogenic yeast Candida albicans. PLoS Genet. 8:e1002661 10.1371/journal.pgen.1002661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J., and Sanyal K.. 2013. Efficient neocentromere formation is suppressed by gene conversion to maintain centromere function at native physical chromosomal loci in Candida albicans. Genome Res. 23:638–652. 10.1101/gr.141614.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T., Li X., Liu Y., Wang C., Liu X., Bi G., Zhang X., Yao X., Zhou Z.H., and Zang J.. 2018. Molecular basis for CENP-N recognition of CENP-A nucleosome on the human kinetochore. Cell Res. 28:374–378. 10.1038/cr.2018.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoquaux N., Liachko I., Ay F., Burton J.N., Shendure J., Dunham M.J., Vert J.P., and Noble W.S.. 2015. Accurate identification of centromere locations in yeast genomes using Hi-C. Nucleic Acids Res. 43:5331–5339. 10.1093/nar/gkv424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voullaire L.E., Slater H.R., Petrovic V., and Choo K.H.. 1993. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am. J. Hum. Genet. 52:1153–1163. [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Tavoosidana G., Sjölinder M., Göndör A., Mariano P., Wang S., Kanduri C., Lezcano M., Sandhu K.S., Singh U., et al. 2006. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 38:1341–1347. 10.1038/ng1891 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.