To better understand Rho GTPase signaling in neurite outgrowth, Bagonis et al. overcome persisting algorithmic limitations that have, thus far, stymied comprehensive studies of morphological variation among growth cones. They introduce Growth Cone Analyzer—a versatile computer vision tool for the quantification of filopodia-rich cell morphologies.
Abstract
Growth cones are complex, motile structures at the tip of an outgrowing neurite. They often exhibit a high density of filopodia (thin actin bundles), which complicates the unbiased quantification of their morphologies by software. Contemporary image processing methods require extensive tuning of segmentation parameters, require significant manual curation, and are often not sufficiently adaptable to capture morphology changes associated with switches in regulatory signals. To overcome these limitations, we developed Growth Cone Analyzer (GCA). GCA is designed to quantify growth cone morphodynamics from time-lapse sequences imaged both in vitro and in vivo, but is sufficiently generic that it may be applied to nonneuronal cellular structures. We demonstrate the adaptability of GCA through the analysis of growth cone morphological variation and its relation to motility in both an unperturbed system and in the context of modified Rho GTPase signaling. We find that perturbations inducing similar changes in neurite length exhibit underappreciated phenotypic nuance at the scale of the growth cone.
Introduction
Already in the mid-eighties, Bray and Chapman (1985) had postulated that a systematic assessment of morphology would be essential for unraveling the principles governing growth cone (GC) locomotion and navigation, events fundamental for neuronal development. A surprisingly limited number of efforts have picked up this thread. While recent work has tested correlations between neurite outgrowth rates and morphological/dynamic GC features (Hyland et al., 2014; Steketee et al., 2014; Ren and Suter, 2016), in these studies, quantification of GC morphology was performed by tedious visual inspection of hand-selected datasets, and/or considered only coarse descriptions of shape (Goodhill et al., 2015; Suo et al., 2015; Ren and Suter, 2016). Numerous studies have analyzed the effect of putative perturbations of GC regulation using endpoint metrics of neurite length observed at low resolution (e.g., Briançon-Marjollet et al., 2008; Chandran et al., 2016), without considering the effects on GC architecture. Such limited quantification is likewise pervasive in studies investigating the role of Rho GTPases, key regulators of cytoskeleton organization and dynamics (Hall and Lalli, 2010) in neuronal outgrowth. Hence, a proper validation of Bray’s paradigm that GC morphology and motility are coupled is still outstanding.
To enable acquisition of an unbiased inventory of GC morphologies and their motility output, we developed Growth Cone Analyzer (GCA) for fully automated analysis of high-resolution GC fluorescent movies. GCA addresses common limitations cited by current software for GC analysis (Misiak et al., 2014; Goodhill et al., 2015; Jacquemet et al., 2017; Urbančič et al., 2017), including poor detection of fine, low signal-to-noise structures known as filopodia, as well as difficulty in resolving crossed filopodia. Furthermore, while available GC analysis software offers generic schemes for the identification of veil protrusions (Tsygankov et al., 2014; Jacquemet et al., 2017; Urbančič et al., 2017), GCA includes a detection module specifically designed for complex, multi-scale, veil/stem morphologies and provides measurements of neurite outgrowth.
To showcase GCA, we provide a quantitative analysis of motile GCs in the presence/absence of Rho GTPase pathway perturbations previously identified to produce shifts in neurite outgrowth in an N1E-115 mouse neuroblastoma model system (Pertz et al., 2008; Fusco et al., 2016). The approach introduced here provides insight into how neurite outgrowth relates to processes at the scale of the GC and unveils novel layers of cytoskeletal regulation that illustrate the complexity of Rho GTPase signaling in neurite outgrowth.
Results
Morphological heterogeneity of GCs
N1E-115 mouse neuroblastoma cells, like many GC model systems (Kleitman and Johnson, 1989; Kozma et al., 1997; Mason and Wang, 1997; Sarner et al., 2000; Pertz et al., 2008; Dent et al., 2011; Özel et al., 2015; Fusco et al., 2016), adopt a wide variety of shape configurations (Fig. S1 A). This morphological heterogeneity is exacerbated by genetic manipulations targeting cytoskeleton regulation (Fig. S1 B). GC morphology, while diverse, is ultimately dictated by the integration of two stereotypically shaped actin structures: a veil-like lamellipodium interspersed with thin filopodia (Lowery and Van Vactor, 2009). Filopodia are curvilinear actin bundles, and hence can best be identified by common ridge detectors (Frangi et al., 1998; Jacob and Unser, 2004; Fig. S1 C, i). In contrast, lamellipodial veils can be quite amorphous and therefore require a different image processing method (Fig. S1 C, ii). Furthermore, veils are interjected by consolidated, ridge-like segments of the neurite, referred to here as the stem (Fig. S1 C, iii and iv). Therefore, the terminal end of the growing neurite can be viewed as a unique conglomerate of distinct geometric features requiring differential image processing for optimal detection of each component. Segmentation tools that rely on a single image processing approach such as global thresholding (Costantino et al., 2008; Tsygankov et al., 2014) fail on this type of data (Fig. S1 D).
GCA pipeline
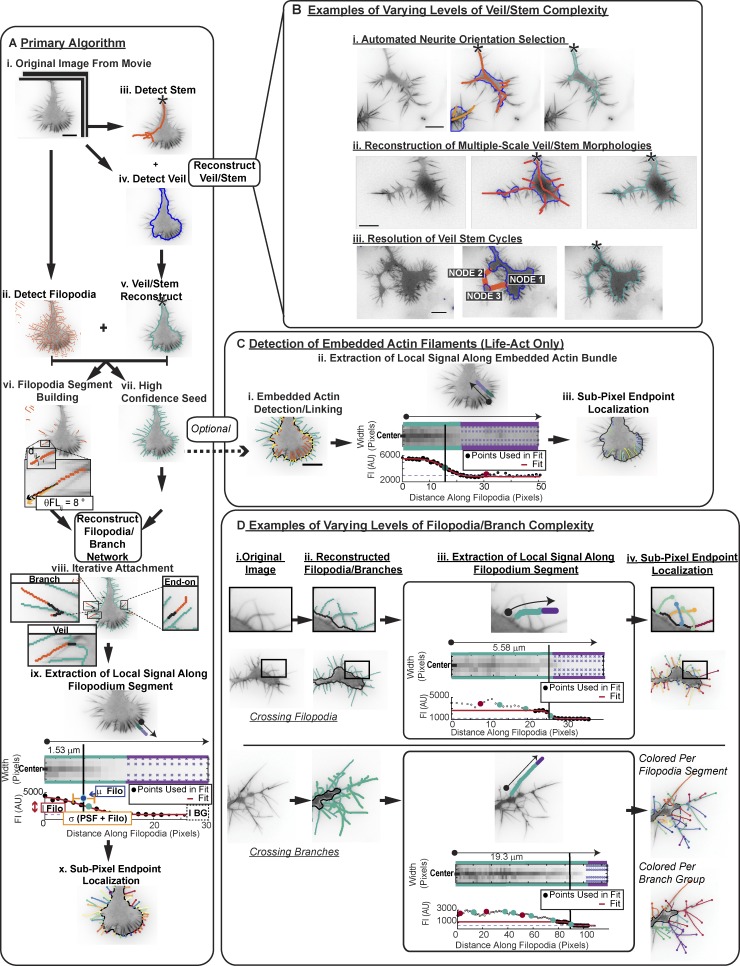
To address the challenges of quantifying GC morphology, we developed GCA (Fig. 1 and Fig. S2). GCA reconstructs the veil/stem and filopodia/branch system in two parallel detection steps optimized for each feature (Fig. 1 A, ii–v). It then recombines the information to create a high-fidelity segmentation (Fig. 1 A, vi–x; and Fig. S3). Fig. 1 outlines the general workflow of the algorithm for a canonical GC (Fig. 1, A and C; and Video 1), and highlights intersections where the algorithm automatically deviates from this workflow to address segmentation challenges posed by noncanonical GC morphologies (Fig. 1, B and D; Videos 2 and 3; and Fig. S3; see Materials and methods).
Figure 1.
Automated segmentation of heterogeneous morphologies by GCA. (A) GCA segmentation pipeline: Canonical N1E-115 GC expressing LifeAct GFP. See Video 1. (B) GCA segments GCs of varying veil/stem complexity. See Video 2. (C) Detection of veil-embedded actin bundles. (D) GCA segments GCs of varying filopodia/branch complexity. See Video 3 and Materials and methods. Bar, 10 µm.
Validation of GCA algorithm
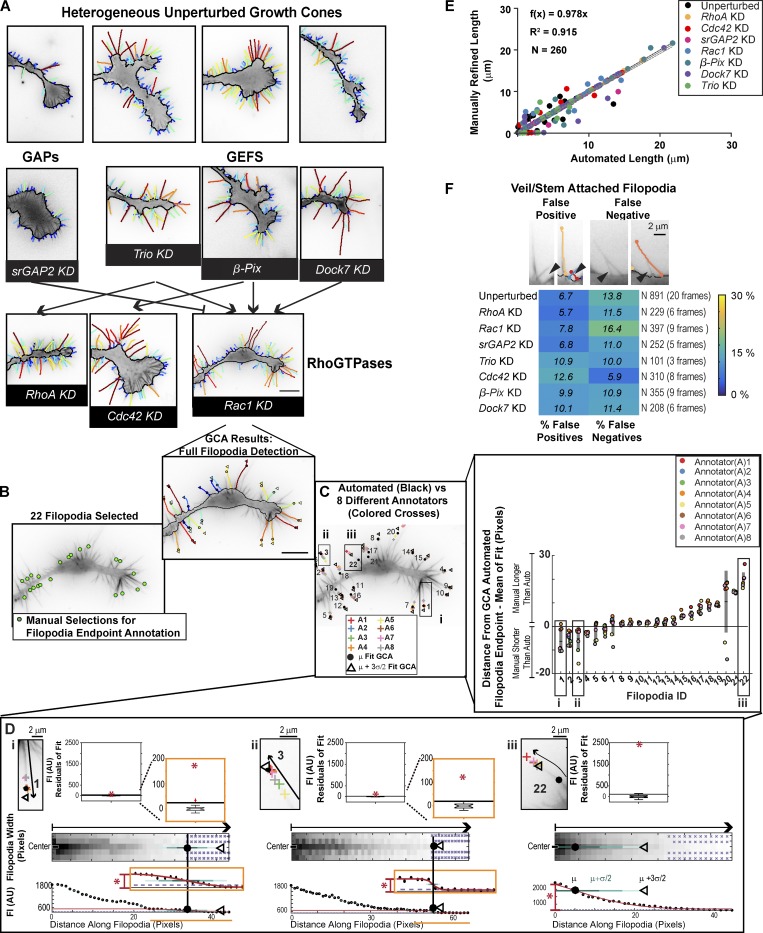
Fig. 2 (Videos 4, 5, and 6) shows the GCA segmentation for the set of images outlined in Fig. S1 A. Notably, the same filopodia/branch segmentation parameters (Table S1) were used for the entire N1E-115 GC dataset (n = 72 movies). While visual inspection is commonly considered the gold standard for validation of automated image analysis, given the complexity of GC architectures, and indeed most biological images, it is nontrivial to obtain an unbiased “ground truth” by manual annotation (Kozubek, 2016). For example, we have shown previously that manual annotation of whole-cell scale N1E-115 images differ, sometimes substantially, among annotators (Fusco et al., 2016). In the case of the N1E-115 GC scale images, we found even documentation of filopodia endpoint coordinates an impossible task for a group of manual annotators to perform systematically; ultimately, GCA provided a more standardized definition of filopodia length (Fig. 2, B–D; see Materials and methods). Given the above challenges, we devised a semi-automated approach to quantify the accuracy of the filopodia/actin bundle lengths (Fig. 2 E, Fig. S4, and Fig. S5 D) and detection numbers (Fig. 2 F and Fig. S5, A–C) extracted by GCA (see Materials and methods).
Figure 2.
Validation of GCA for segmentation of N1E-115 GCs. (A) GCA segmentation of N1E-115 GCs expressing GFP LifeAct on laminin. Embedded actin bundle detections not shown for clarity. (See Fig. S1, A and B, for raw images). GCs of KDs, as in Fig. S1 B, shown in context of their putative network interactions (see GC scale morphodynamic phenotypes section for references). See Videos 4, 5, and 6. Bar, 10 µm. (B–D) Variability in manual filopodia endpoint localization among annotators (see Materials and methods). (D) GCA filopodia linescan/fitting for example filopodia (i–iii). Boxplot insets (top): Distribution of the residuals for each fit compared with the fit amplitude (red star). Given the multiple scales of intensity decay in i and ii, fit regions, and the boxplots of the fit residuals, are magnified as indicated by orange insets. (E) Automated GCA versus manually refined filopodia lengths. See Fig. S4 A. Black line: Perfect correlation. Gray solid line: Linear fit to data. Gray dotted lines: CI of the fit. (F) GCA detection error for veil/stem attached filopodia. Top: Example overlays. Black arrowheads: Detection error. Bottom: Heat map of error rates.
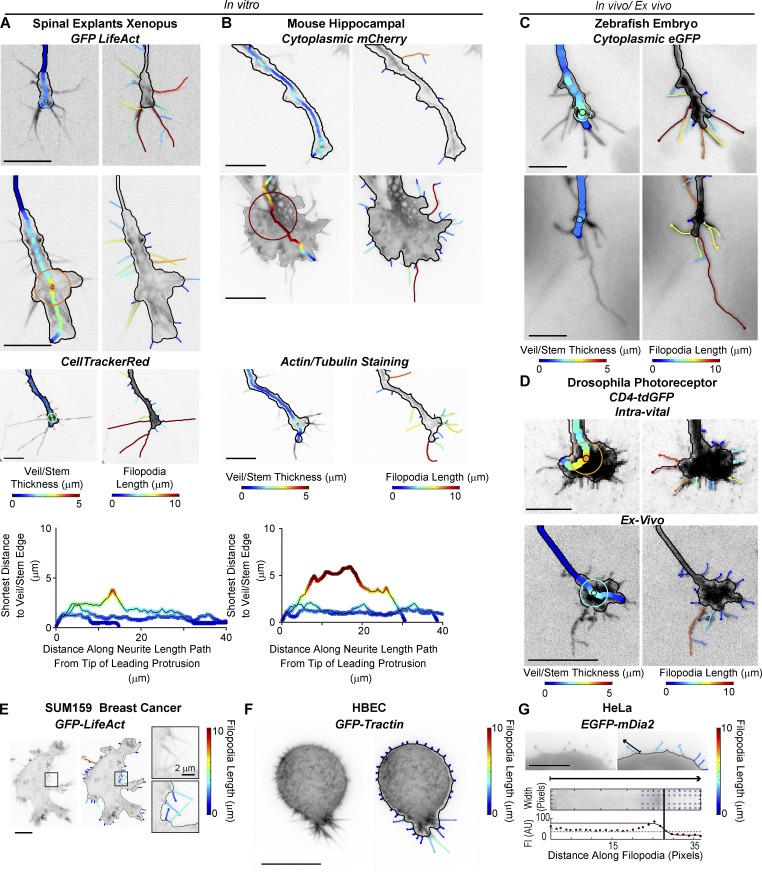
We found GCA to perform well, notably with minimal segmentation parameter changes, on GC images of different sizes, types, and fluorescent labels, imaged both in vitro and in vivo/ex vivo (Fig. 3, Fig. S6, and Videos 7 and 8), and GCA generalized well to non-GC images with filopodia (Fig. 3, E–G). These results underlie the robustness of the pipeline and indicate that while some heuristics were necessary to design the algorithm, the pipeline can be applied to vastly diverse imaging datasets with little user input.
Figure 3.
GCA applied to a variety of cellular images. (A–D) GCA veil thickness (left) and filopodia (right) overlays for GCs of different size, type, and fluorescent label imaged in vitro (A and B) and in vivo/ex vivo (C and D). Xenopus spinal explants (A) and mice hippocampal neurons (B): unpublished data courtesy of Lowery Laboratory, Boston College, and Gupton Laboratory, University of North Carolina at Chapel Hill, respectively. (A and B) Bottom: Veil/stem size profiles comparing the GCs shown in the first three rows of A (left) and B (right). (C) Rohon–Beard GCs in a zebrafish embryo: previously published raw images in St John and Key (2012). (D) Intra-vital (top) and ex vivo images (bottom) of Drosophila photoreceptor GCs: maximum intensity projections as in Langen et al. (2015) and Özel et al. (2015), respectively. See Video 7. (E and F) GCA segmentation of 2D maximum intensity projections of SUM159 triple negative human breast cancer cell (E) and transformed human bronchial epithelial cell (HBEC) in a collagen I matrix imaged using meSPIM (F; Welf et al., 2016). (G) GCA segmentation of a HeLa cell expressing a filopodium-tip localizing marker. Bottom: Automated line scan along sparsely labeled filopodium marked by arrow. Bar, 10 µm, unless noted.
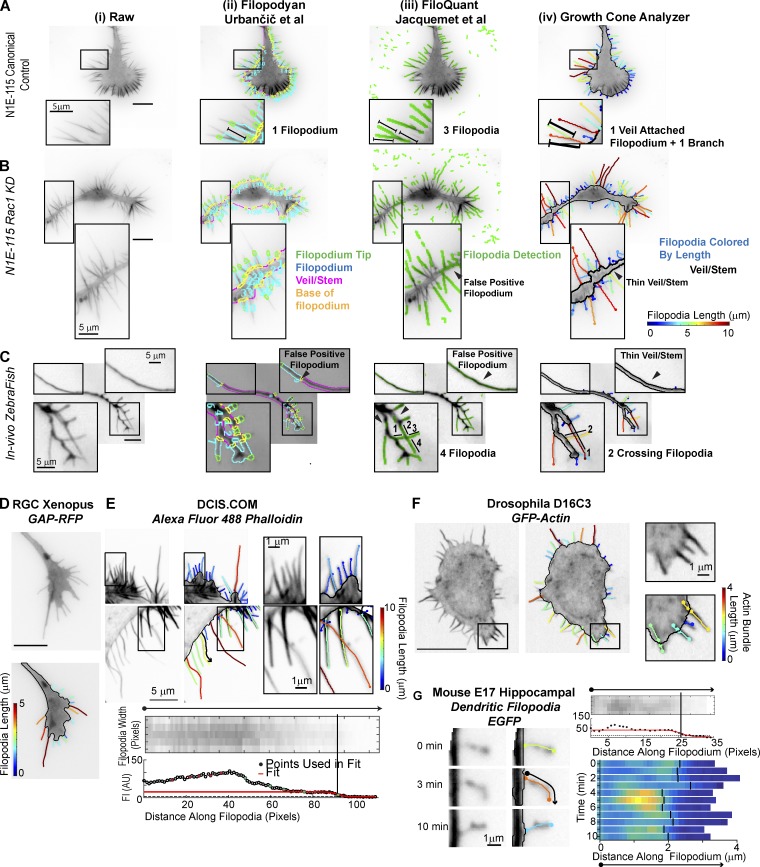
We also confirmed that contemporary software with similar goals to GCA (Jacquemet et al., 2017; Urbančič et al., 2017) is insufficient for the assessment of Rho GTPase perturbation of N1E-115 GCs and some in vivo morphologies (Fig. 4, A–C), even after extensive attempts to optimize control parameters. In contrast, GCA was able to segment test images included in these packages, with only minimal and intuitive segmentation parameter adjustments, and offered a number of substantial advances not available with the previous methods (Fig. 4, D–G; see Materials and methods).
Figure 4.
Comparison of GCA to contemporary filopodia detection software. (A–C) Segmentation overlays generated using Filopodyan (Urbančič et al., 2017; ii), FiloQuant (Jacquemet et al., 2017; iii), or GCA (iv). N1E-115 GCs expressing GFP-LifeAct (A and B) and a Rohon–Beard GC (C) in a zebrafish embryo (see Fig. 3 C). Visualization as in respective package. Zoomed regions highlight segmentation challenges. (D–G) GCA segmentation using previously published raw images available with contemporary software releases. (D and E) Filopodyan (Urbančič et al., 2017) and FiloQuant (Jacquemet et al., 2017). Bottom: Zoom and local intensity fit of filopodium marked by the black arrow in top image. (F and G) CellGeo (Tsygankov et al., 2014) and Dendritic Filopodia Motility Analyzer (Tárnok et al., 2015). Bar, 10 µm, unless noted.
Extraction of morphodynamic features
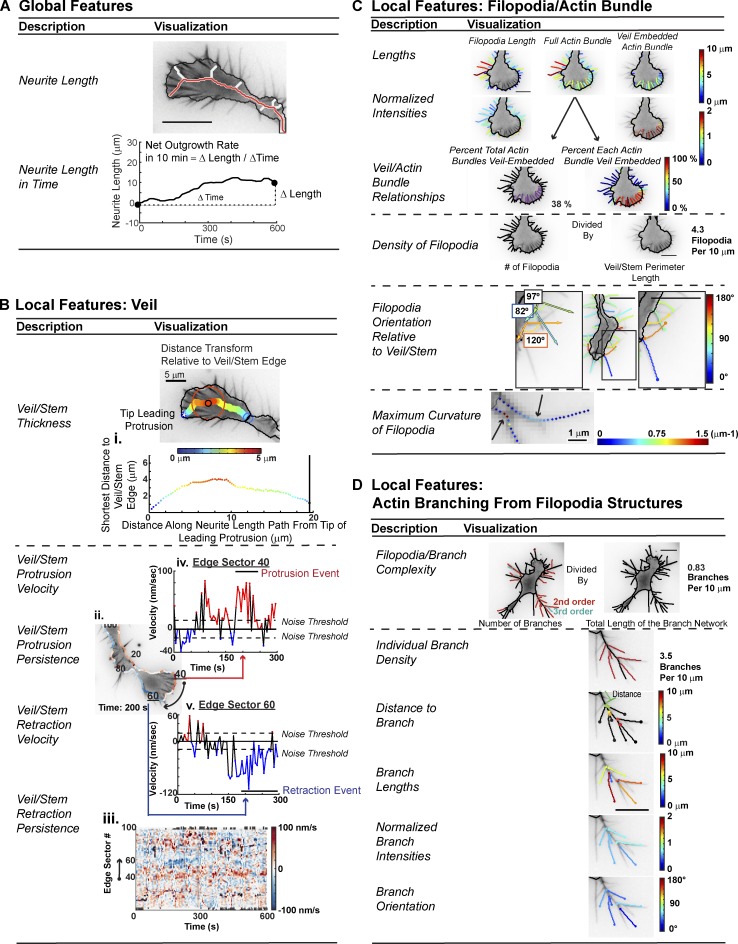
Segmentation via GCA enables quantification of many GC morphodynamic features (Fig. 5). These features include “global/functional” measurements, such as neurite length/outgrowth rate (Fig. 5 A), as well as more spatially localized measurements such as veil/stem thickness and dynamics (Fig. 5 B), the filopodia geometry and its integration with the veil (Fig. 5 C), and the branching pattern (Fig. 5 D; see Materials and methods). Notably, GCA extraction of filopodia lengths/densities (Mogilner and Rubinstein, 2005) and comparative analyses of local veil velocities (Sivadasan et al., 2016; Fig. S6 and Video 8) were consistent with manual quantification previously reported. Hence, GCA provides a robust framework for multi-variate feature extraction.
Figure 5.
GCA morphodynamic feature extraction. (A) Neurite length and net outgrowth rate definitions. (B) Features of veil/stem. (C) Features of filopodia/actin bundles. (D) Features of filopodia branching. See Materials and methods. Bar, 10 µm, unless noted.
Relationship between neurite outgrowth and GC scale morphodynamics
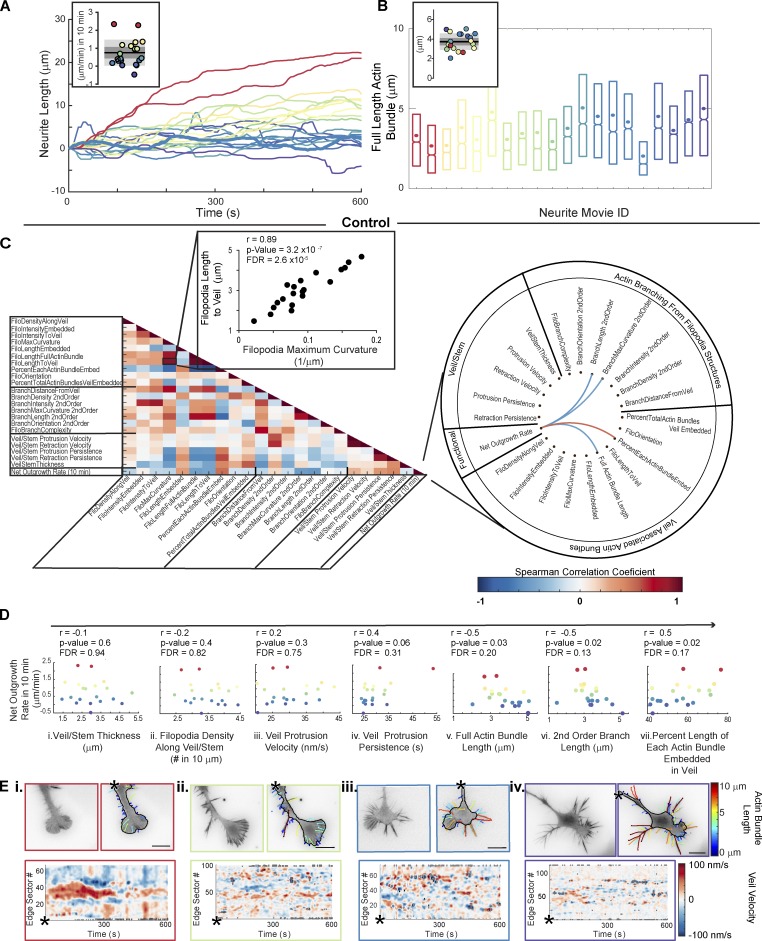
Equipped with this analytical pipeline, we returned to this question: Does GC motility relate to GC morphology? While molecular perturbations can indeed induce changes in GC morphology (Pertz et al., 2008; Fig. S1 B), they are also often accompanied by larger system adaptations that complicate interpretation of motility phenotypes. Therefore, we first sought to use the full distribution of GCA features to detect potential morphology–motility relations latent within the heterogeneous control population. We analyzed 20 movies (10-min) of unperturbed N1E-115 GCs and identified correlations between neurite-to-neurite variation in net outgrowth rate (Fig. 6 A) and temporally averaged GC morphology features (Fig. 6 B).
Figure 6.
Correlations between GC morphology and motility within an unperturbed, heterogeneous population. (A) Variation in neurite outgrowth rate across 20 control cells (inset, net outgrowth: 10 min. Bar, mean; dark gray regions, 95% CI of the mean; light gray regions, ±SD). (B) Distributions of actin bundle lengths for data in A, sorted by net outgrowth velocity (colors as in A; 3,047–8,213 actin bundles per movie). Boxplots: 25th, 50th, and 75th distribution quantiles. Circles: Mean per movie. Inset: Distribution of means. (C) Spearman correlation matrix of GCA features (left). Schema ball of strongest correlations between net outgrowth rate and GC features (P < 0.05 and false discovery rate (FDR) < 0.2; right). (D) Select morphology features versus net outgrowth rate (as in C). Each data point: single GC movie as in A. Spearman correlation coefficient, r. (E) GCA segmentation overlays (top) and veil-velocity maps (bottom) corresponding to key data points in the net neurite outgrowth rate distribution in A. See Video 4. Bar, 10 µm.
Testing for monotonic relationships between feature pairs (Fig. 6 C) revealed a positive correlation between filopodia length and curvature, and a negative correlation between filopodia length and veil protrusion persistence. However, we were particularly interested in identifying correlations between neurite outgrowth rate and GC morphology (Fig. 6, C and D). The strongest correlations included an inverse relationship between neurite outgrowth and filopodia actin bundle/branch length, as well as a positive relationship between outgrowth and the percentage each filopodium was embedded by veil (Fig. 6, C and D). Other actin bundle features, such as filopodia density, orientation, and branch complexity, were not significantly correlated to net neurite outgrowth in 10 min. Likewise, the veil stem thickness was a poor predictor, in line with Hyland et al. (2014). Visualization of select movies corresponding to key data points in the neurite outgrowth rate distribution (Fig. 6 A), namely the population extremes (Fig. 6 E, i and iv), and two movies within ±SD of the mean (Fig. 6 E, ii and iii) confirmed the structures of these GCs were consistent with the conclusions drawn from the correlation analysis (Fig. 6 E and Video 4).
GC scale morphodynamic phenotypes upon perturbation of Rho GTPase signaling
We next sought to test if the additional morphological variation introduced by siRNA-mediated perturbations of Rho GTPase signaling would support the correlative relations. We focused upon a subset of knockdowns (KDs) found to induce neurite length defects within an ∼20-h timescale (Fusco et al., 2016; Table S2). These included Rac1 and three structurally distinct, positive regulators of Rac1 activity, β-Pix (also known as ARHGEF7; ten Klooster et al., 2006), Dock7 (Watabe-Uchida et al., 2006, and Trio (Debant et al., 1996; Briançon-Marjollet et al., 2008). In addition, we examined perturbations resulting in enhanced neurite length (Fusco et al., 2016). This set included siRNA KD of (1) a negative regulator of Rac1 (srGAP2; Guerrier et al., 2009); (2) RhoA, whose downstream signaling via its effector ROCK has been implicated in neurite retraction in response to inhibitory stimuli (Kozma et al., 1997; Amano et al., 1998; Zhang et al., 2003); and (3) Cdc42, which, when reduced, is typically associated with shorter neurite lengths (Chandran et al., 2016), but exhibits robust up-regulation of neurite outgrowth in our system, similar to that observed upon RhoA depletion (Pertz et al., 2008; Fusco et al., 2016; Fig. 2 A and Videos 5 and 6).
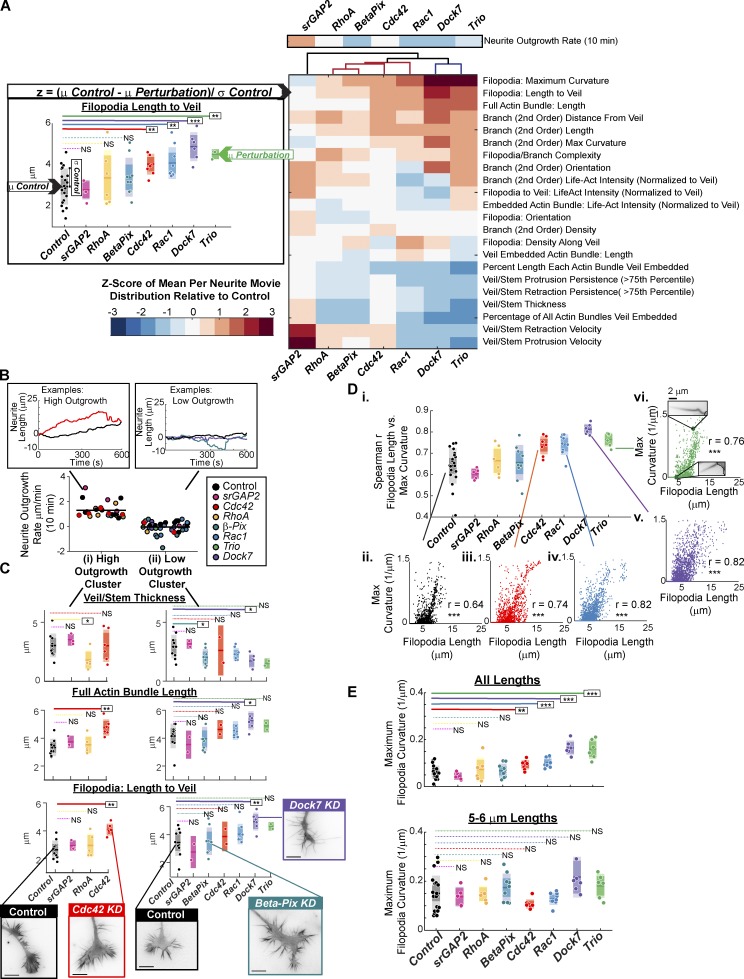
GC feature values were averaged per GC movie, and a z-score between control and perturbation calculated (Fig. 7 A, left). This z-score vector across all features defined a GC scale morphodynamic signature (MS) for each KD, similar to the whole-cell scale profiles extracted previously in this same system (Fusco et al., 2016). Notably, hierarchical clustering of the GC scale MSs generally grouped the perturbations according to their previously characterized whole-cell scale MSs (Fig. 7 A) and provided further insight as to how these neurite length phenotypes may arise. For instance, Rac1, Dock7, and Trio KD all showed reduced whole-cell scale neurite outgrowth dynamics, and in all cases, the corresponding GC scale MS was characterized by long filopodia/branches, low actin bundle veil embedment, and reduced veil dynamics (Videos 5 and 6). Analogous GC features were characteristic of stagnant neurites in the unperturbed population (Fig. 6, B–D). In contrast, β-Pix KD neurites were more dynamic at the whole-cell scale, despite their final short-neurite phenotype. Accordingly, GCs were characterized by a similar yet milder filopodia and veil dynamics phenotype (Fig. 7 A and Video 6)
Figure 7.
Morphodynamic analyses of select Rho GTPase pathway KDs. (See Fig. 2 A). (A) GC scale morphodynamic profiles extracted by GCA. Left: Example z-score calculation; scatter plots: mean filopodia length per 10-min GC movie (1,995–9,944 filopodia per movie). Midline bars: Mean of treatment distribution (3–20 GC movies per condition); dark colored regions: 95% CI of the mean; light colored regions: ±SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001: two-sided permutation t test of the means. Right: Hierarchical clustering of siRNA z-score vectors. Above clustering: z-scores for 10-min neurite outgrowth rate (a global/functional feature). (B) N1E-115 Rho GTPase perturbation dataset (n = 66 GC movies) separated into high (i) and low (ii) net neurite outgrowth clusters for further analysis in C. Insets: Prototypical neurite length trajectories for high (right) and low (left) neurite outgrowth clusters. (C) Robustly elongating Cdc42 KD GCs exhibit enhanced filopodia lengths with no change in veil/stem thickness. Select GC features for high (3–10 GC movies per condition; 2,743–10,216 actin bundles per movie; i) and low (2–10 GC movies per condition; 2,146–8,280 actin bundles per movie; ii) outgrowth neurites. Scatter plots as defined in A. *, P < 0.05; **, P < 0.01; NS, P > 0.05: two-sided permutation t test of the means. Insets: Example images, respective cluster. (D) Positive correlation between filopodia length and maximum filopodia curvature. (i) Scatter points: Spearman r between the two features per GC movie; 5–20 GC movies per treatment; 1,299–7,060 filopodia per movie. (ii–vi) Example correlation plots. Scatter points: single filopodium. N filopodia: (ii) 4,952; (iii) 3,739; (iv) 4,647; (v) 3,740; (vi) 2,328. ***, P < 0.001: Benjamini–Hochberg adjusted. (E) Enhancement of filopodia curvature observed upon Rho GTPase perturbation (top) is primarily a secondary effect of increased filopodia length (bottom). 5–20 GC movies per treatment; 1,299–7,060 filopodia per movie (top); 25–519 filopodia per movie (bottom). *, P < 0.05; **, P < 0.01; ***, P < 0.001: two-sided permutation t test of the means. See Videos 5 and 6.
Conversely, srGAP2 KD GCs, which likely display enhanced Rac1 activity (Guerrier et al., 2009), and exhibit elevated neurite elongation/branching (Pertz et al., 2008; Fusco et al., 2016) at the whole-cell scale, showed increased local veil protrusion velocities without major changes in filopodia morphology (Fig. 7 A and Video 6). The srGAP2 KD profile was also distinct from the other neurite length–enhancing perturbations tested. RhoA and Cdc42 KD GCs, while displaying enhanced neurite outgrowth at the whole-cell scale, failed to induce significant changes in the local veil velocity. Furthermore, Cdc42 KD resulted in a strong enhancement in filopodia length, while RhoA KD resulted in reduction of the veil/stem thickness (Fig. 7 A). Clustering the data by neurite outgrowth rate confirmed that robustly elongating Cdc42 KDs indeed exhibit longer actin bundles (Fig. 7 C, i), a feature typically associated with low outgrowth in the unperturbed population (Fig. 6 D, v). In contrast, with the exception of Dock7 KD, the initially striking actin bundle/filopodia length phenotypes exhibited by Rac1 and the Rac1 GEF KDs (Fig. 7 A) were diminished when normalizing for the effect of neurite outgrowth (Fig. 7 C, ii). These results indicate that GCs exhibiting features usually related to inefficient neurite outgrowth can display robust neurite elongation under certain signaling conditions, highlighting the complexity of the mechanisms coupling Rho GTPase activity, GC morphology, and the up-regulation of neurite outgrowth.
The sensitivity of GCA facilitated the interpretation of the visually striking kinked and buckling filopodia observed upon Dock7 or Trio KD (Fig. S1 B, Fig. 2 A, and Fig. 5 C). While, indeed, the curvature of filopodia in these perturbations was increased, further assessment of the length and curvature variation in the unperturbed GCs also indicated a strong correlation among these two measurements, both among the averaged neurite movie values (Fig. 6 C) and at the level of the individual filopodium (Fig. 7 D). Indeed, normalization for the effect of filopodia length on curvature (Fig. 7 E) suggests that the increases in filopodia curvature upon these perturbations is primarily associated with increased filopodia length. These results, combined with those highlighted in Fig. 7 (B and C), demonstrate that the extraction of multiple features is necessary for the biological interpretation of these GC phenotypes.
Identification of morphological transitions along a neurite outgrowth trajectory
Time-averaged analysis of unperturbed (Fig. 6) and Rac1 pathway perturbation data (Fig. 7) revealed consistent relationships between morphology and net neurite outgrowth on a 10-min timescale. However, the averaging may mask more nuanced relationships. We therefore sought an analytical framework to (1) confirm morphology/motility relationships at the level of individual neurite trajectories and (2) visualize potential subpopulations of GCs where these relationships may be altered. As fluctuations among morphological features may be nontrivially coupled, transitions between morphological states in a time-lapse sequence can likely be most robustly identified by combining multiple GCA features. Hence we asked if Hidden Markov/Bayesian model (HMM) selection of multi-variate GC feature trajectories could be employed to identify the timing of GC morphology changes, as recently proposed by Gordonov et al. (2016) for the detection of coarse-grain morphology transitions in migrating breast cancer cells.
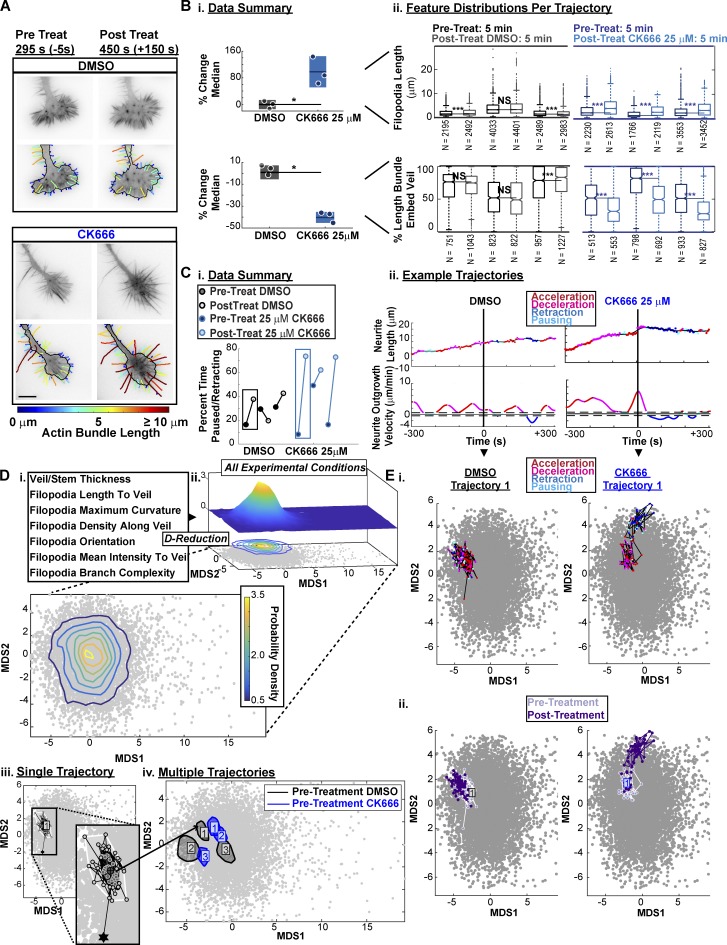
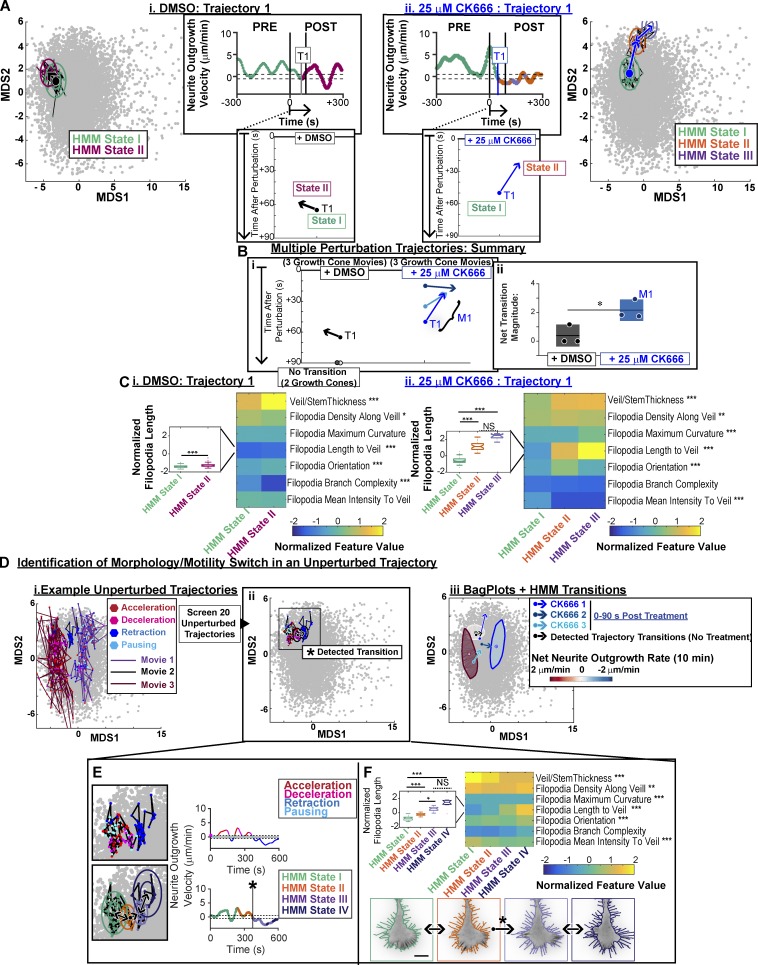
To validate the HMM, we used the small molecule CK666 (Nolen et al., 2009) to acutely inhibit the actin nucleator Arp2/3. This perturbation has been previously shown to induce defects in GC morphology (Yang et al., 2012) and neurite outgrowth (Fusco et al., 2016). Consistent with observations in Aplysia (Yang et al., 2012), CK666 treatment of N1E-115 GCs induced both the enhancement of filopodia length and decreases in the veil-embedment of individual actin bundles (Fig. 8, A and B). Furthermore, in line with predictions from our unperturbed correlation data (Fig. 6), neurites treated with CK666 consistently induced increases in the percent time the neurite spent paused or retracting (Fig. 8 C).
Figure 8.
ARP2/3 inhibition via CK666 treatment induces GC morphological changes and stagnation of neurite outgrowth. (A) GCA segmentation of N1E-115 GCs on laminin before (left) and after (right) treatment with DMSO or 25 µM CK666. Bar, 10 µm. (B) CK666 treatment increases filopodia length (top) and decreases the percent length each actin bundle is embedded in veil (bottom). (i) Percent change in the median of the actin bundle distribution (scatter points) before and after treatment. n = 3 GC movies per treatment type. Lines, mean of distribution per treatment condition; dark shaded regions, 95% CIs about the means. *, P = 0.1: two-tailed permutation t test of the means. (ii) Distributions of actin bundle measurements. Boxplot: 25th, 50th, and 75th distribution quantiles. Whiskers, 1.5 × (Q75% − Q25%). ***, P < 0.001: two-tailed permutation test of the medians. (C) ARP2/3 inhibition via CK666 treatment induces functional transitions from neurite elongation to neurite pausing/retraction. (i) Summary of the percent time individual neurites were paused or retracting before and after treatment. n = 3 GC movies per condition. (ii) Neurite outgrowth lengths/velocities colored by neurite outgrowth state. Dashed line: threshold for pausing. (D) Visualization of morphological heterogeneity among individual GC movies. (i) GCA features calculated per frame for D-reduction. (ii) MDS plot of morphology features (72 GC movies: 8,454 frames). Probability density isocontours for entire dataset visualized in 3D (top) and 2D (bottom). (iii) Example of full bagplot (i.e., 2D boxplot; Rousseeuw et al., 1999) for a single 5-min trajectory before acute perturbation. Dark shaded contour: bag; light shaded contour: fence; circles: individual data points. Black star: outlier data point. Cross: Tukey median. (iv) Bagplot “bags” for six GC movies before acute treatment with DMSO (black) or 25 µM CK666 (blue). (E) CK666 treatment induces a shift in morphology space, coupled to neurite outgrowth state. Two GC movies acutely treated with DMSO (left) or 25 µM CK666 (right). Per-frame coordinates in reduced morphology space colored by neurite outgrowth state (i) or timing relative to treatment (ii). Each movie: 10 min total, 5-s intervals.
Seven morphological GCA features (Fig. 5) were selected to generate a per-frame feature vector (Fig. 8 D, i) for each GC movie in the full N1E-115 dataset (n = 72). For visualization, these data were reduced to two dimensions via multi-dimensional scaling (MDS; see Materials and methods). We also used MDS coordinates to estimate the relative likelihood of a GC to maintain a particular combination of morphology features (Fig. 8 D, ii). Visualization of the pretreatment frames (Fig. 8 D, iii) indicated neurite-to-neurite heterogeneity in the GC shape before DMSO/drug application (Fig. 8 D, iv). However, upon CK666 treatment, GC trajectories diverged in morphology space (Fig. 8 E), and these morphological transitions appeared correlated with the switch from neurite elongation to pausing/retraction (Fig. 8, C [ii] and E [i]).
To identify potential morphology state transitions along each trajectory, HMM with Bayesian model selection (Posada and Buckley, 2004) was applied to the MDS coordinates for DMSO- and CK666-treated trajectories (Fig. 9 A). To confirm the reproducibility of the detected state transitions, we analyzed the HMM state transitions for all six movies within a 90-s window after application of treatment. CK666 treatment consistently resulted in morphology state transitions of similar direction, magnitude, and timing (Fig. 9, A [ii] and B; and Video 10). In contrast, after DMSO treatment, morphology transitions were either not observed (Fig. 9 B) or pointed in the opposing direction of the CK666-induced transitions (Fig. 9 A, i; and Video 9). Comparison of the feature value distributions corresponding to each of the HMM morphology states identified along the DMSO- (Fig. 9 C, i) and CK666-treated (Fig. 9 C, ii) GC movie trajectories revealed differences in the individual feature fluctuations, distinguishing the two transitions. This validates the HMM-based approach as an analytical method to isolate significant transitions in GC morphology in an unperturbed system or when the timing/effect of a system perturbation is not known a priori.
Figure 9.
Detection of significant transitions in GC morphology along a neurite outgrowth trajectory. (A) Time series modeling of a DMSO- (left and Video 9) and CK666 (right and Video 10)-treated GC movie in reduced morphology space. Colors: state ID inferred from the HMM/Bayesian model selection. Rings: 1× and 2× the SD of the state distribution centered on the mean. Bottom insets: Plot of detected transition within a 90-s window after respective treatment. (B) CK666 treatment induces reproducible GC morphology state transitions. (i) Plots of detected morphology transitions for six movies (as described in A, bottom inset) 90 s after treatment with DMSO (gray/black shaded vectors) or 25 µM CK666 (blue shaded vectors). (ii) Magnitude of the transition vectors in i. *, P < 0.05, two-tailed t test for equal means. (C) Median of feature distribution for each detected HMM morphology state detected in A (DMSO, left; CK666, right). *, P < 0.05; **, P < 0.01; ***, P <0.001: Kruskal–Wallis test. Insets: Boxplots of the distribution of filopodia lengths in each HMM state. *, P < 0.05; **, P < 0.01; ***, P < 0.001: Tukey–Kramer test. (D) Identification of morphology/motility switches in a heterogeneous population of unperturbed trajectories. (i) Three unperturbed GC movies overlaid on the morphological landscape, each frame colored by neurite outgrowth state as in Fig. 8 E, i. Identification of morphology/motility switch (starred black arrow) similar to that induced upon acute treatment with CK666 (A–C). Black rings: HMM states as defined in A. Colors as in D, i. (iii) “Bag” contours as defined in Fig. 8 D, iii, for GC movies in i. Arrows: Detected morphology state transitions from B and D, ii. Each movie: 10 min total, 5-s intervals. (E) Zoom (left) and neurite outgrowth velocity (right) of unperturbed GC movie identified in D, ii, frame colored by neurite outgrowth state (top) or HMM state ID as defined in A (bottom). (F) Feature/HMM state heat maps (as defined in C) for unperturbed movie identified in D, ii (top). Example GCA segmentations for each HMM morphology state (bottom).
We then used the MDS plots to visualize the dependence of neurite outgrowth on the morphological feature set. We examined the relative positioning of the two unperturbed GC movies exhibiting the highest and lowest net outgrowth (Fig. 6, A and E). This revealed a rough gradient, whereby higher MDS1 values correlated with poor neurite outgrowth within the unperturbed population (Fig. 9 D, i and iii). In addition, the direction of the CK666-induced transitions in GC morphology (blue arrows in Fig. 9, A [ii], B, and D [iii]) corresponded well with the direction of this outgrowth state gradient.
Based on these observations, we screened all unperturbed GC trajectories (Fig. 6) for morphological transitions. HMM automatically identified the timing of a spontaneous morphology transition in the unperturbed population (Fig. 9 D, ii), which was coupled to a switch between neurite elongation and retraction (Fig. 9 D, iii). Evaluation of the feature profiles of the HMM states along this trajectory (Fig. 9, E and F) confirmed that the transition was characterized by a significant enhancement of filopodia lengths, similar to the transition observed with CK666 treatment (Fig. 9 C, ii). These data show that similar relationships between GC morphology and motility are maintained in the larger population of unperturbed GCs for at least a subset of the trajectories.
Rho GTPase pathway perturbed GCs in the context of the morphology landscape
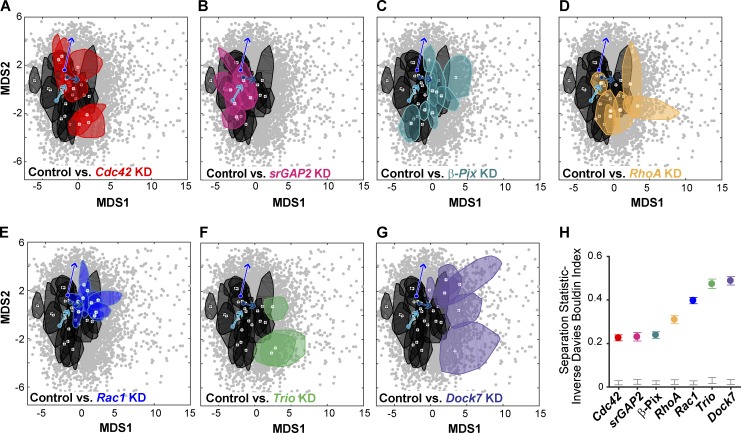
Visualization of the siRNA perturbations within the reduced morphology space (Fig. 10, A–G) reinforced the morphology/motility patterns found in the unperturbed population (Fig. 9 D). For example, Rac1, Dock7, and Trio KD, perturbations which at whole-cell scale produce the most profound dampening of neurite outgrowth (Fusco et al., 2016), maintained high MDS1 values (Fig. 10, E–G). In contrast, a majority of the β-Pix KD movies (Fig. 10 C), as well as outgrowth-enhancing perturbations, showed more overlap with the control (Fig. 10, A, B, D, and H). Finally, the acute inhibition of ARP2/3 activity via CK666 consistently shifted GC morphologies toward the state of GCs likely to have reduced Rac1 activity (Fig. 10, C, and E–G), though the endpoint of a CK666-induced transition depended upon the GC state at the time of drug application. In summary, these results demonstrate that features extracted from the GCA software can be combined with time series modeling (Gordonov et al., 2016) to identify the timing of significant transitions in GC shape and to study the relationship of such morphological transitions with the neurite’s outgrowth behavior.
Figure 10.
Rho GTPase pathway perturbed GCs in the context of the morphology landscape. (A–G) “Bag” contours as defined in Fig. 8 D, iii, for control (black) and siRNA (colored) movies. 3–20 GC movies per condition. (H) Separation statistics: control versus KD. Colored bars: 95th percentile bootstrapped CIs. Gray bars: CI between 5th and 95th quartile of the randomized, null distribution. Gray scatter points: same as in Fig. 8, D and E, and Fig. 9. Arrows: Morphology transitions of three example GC trajectories within 90 s after CK666 treatment (25 µM) as identified in Fig. 9 B.
Discussion
Technological advances for the automated analysis of GC morphology
Unbiased quantification of the relations between GC structure and neurite outgrowth is crucial for understanding GC function. While several toolkits for GC analysis (Costantino et al., 2008; Misiak et al., 2014; Goodhill et al., 2015; Jacquemet et al., 2017; Urbančič et al., 2017) or generic filopodia detection (Nilufar et al., 2013; Tsygankov et al., 2014; Barry et al., 2015; Saha et al., 2016) have been developed, they maintain too many technical limitations to allow the study of morphology/motility relationships in diverse cellular datasets (Fig. 4, A–C). We developed GCA to remove the vast majority of these technological barriers (Fig. 1 and Fig. 4). GCA provides a versatile segmentation framework by introducing (1) multi-scale ridge filter analysis to detect stem and filopodia; (2) filopodia-endpoint detection based on an adaptive, self-configuring model of the fluorescence decay along these structures; and (3) assembly by graph-matching of detected filopodia fragments into a complete representation of filopodia networks containing branches and apparent crossings present in 2D. Combined, these algorithms generate a segmentation and parameterization of GC morphology that is robust across a wide spectrum of GC types (Figs. 2, 3, and 4). Hence, we were able to capture the natural as well as experimentally induced variation in GC structure and apply various post-processing tools to model putative relations between GC morphology and motility.
Mining heterogeneity of unperturbed GCs
The robust quantification afforded by GCA allowed us to mine latent information in neurite-to-neurite heterogeneity observed within an unperturbed population of N1E-115 GCs (Fig. 6). In doing so, we found that net neurite outgrowth within a 10-min window is most strongly correlated with GC structures characterized by filopodia length and branching, as well as increased actin bundle veil-embedment (Fig. 6 D). The dominant relationships between GC morphology and outgrowth discovered in our system corroborate two classical, more qualitative studies that tested correlations between neurite outgrowth rate and subjectively defined GC morphological classifications (Kleitman and Johnson, 1989; Mason and Wang, 1997). However, if and how these morphology/motility relationships are dependent on GC subtype and extracellular environment remains an open question. The adaptability of GCA (Fig. 3, A–D; Fig. S6; and Videos 7 and 8) should enable rapid and unbiased screening of such relationships in other GC systems.
Perturbation of Rac1 signaling corroborates GC morphology/motility relations discovered in the unperturbed population
Multi-scale, morphodynamic quantification of perturbations converging on the Rac1 pathway (Fig. 2 A, Fig. 7, and Fig. 8) further confirm the relationship between GC morphology and motility unveiled by the unperturbed population (Fig. 6, Fig. 8, Fig. 9 D, and Fig. 10). Specifically, KD of Rac1, or two Rac1 guanine nucleotide exchange factors (GEFs; Trio and Dock7), reduced neurite outgrowth (Fig. 7 A; Fusco et al., 2016) and induced GCs with veils of diminished dynamics and long, curved filopodia (Fig. 7 A and Videos 5 and 6). These changes are consistent with Rac1’s well-established role as an activator of branched actin network expansion (Miki et al., 1998; Takenawa and Suetsugu, 2007; Tahirovic et al., 2010). Importantly, the filopodial morphology of these KDs (Fig. 2 and Fig. 7) is reminiscent of those poorly elongating neurites observed at the morphological boundaries of the unperturbed population (Fig. 6, A and E; and Fig. 9 D, iii), though in the case of Dock7 KD, these features were often exaggerated (Fig. 10, F and G).
Interestingly, the GCs of the β-Pix KD displayed a less conspicuous phenotype. β-Pix KD induced decreases in the veil-stem thickness and veil protrusion persistence time, while veil protrusion velocities and filopodia actin bundle length were unaffected (Fig. 7 A). A majority of β-Pix KD GCs shared features with pausing GCs in the unperturbed population (Video 6). This more subtle GC phenotype observed for β-Pix GCs is consistent with previous whole-cell scale results (Fusco et al., 2016), which found that unlike Rac1, Dock7, and Trio KD, β-Pix KD did not disrupt the frequency of neurite elongation events, but still induced an overall neurite length deficit. Given our combined GC/whole-cell scale results, we speculate that β-Pix maintains the spatial veil coordination necessary for sustained neurite elongation, while Dock7/Trio potentially facilitates the initiation of veil protrusions.
Conversely, KD of srGAP2, a Rac1-specific GTPase activating protein (GAP; Guerrier et al., 2009), led to increased neurite outgrowth and branching at the whole-cell scale (Fusco et al., 2016), and a pronounced increase in GC veil dynamics without any profound effect on GC morphology (Fig. 7 A), phenocopying unperturbed GCs that exhibit robust outgrowth rates (Fig. 9 D, iii; and Fig. 10, B and H). The opposing veil phenotypes of srGAP2 versus Rac1, Trio, or Dock7 KD suggest increased versus decreased Rac1 activity at the veil (Fig. 7 A), inducing specific morphologies that reside at opposing ends of the GC morphological landscape (Fig. 10). This spectrum of phenotypes associated with Rac1 pathway perturbations forms a similar gradient in both morphology and neurite outgrowth to that observed with acute perturbation of ARP2/3 (Fig. 10, B, C, and E–G). The agreement between GC phenotypes associated with diminished Rac1 activity and those associated with direct perturbation of Arp2/3 supports Arp2/3’s role as a WAVE complex effector (Takenawa and Miki, 2001; Takenawa and Suetsugu, 2007; Tahirovic et al., 2010), localized to the GC peripheral domain for the purpose of controlling veil protrusion (Mongiu et al., 2007; Tahirovic et al., 2010; Yang et al., 2012). Interestingly, HMM analysis of the multi-variate GC morphology trajectories of the unperturbed population revealed spontaneous, albeit rare, morphological transitions (Fig. 9, D [ii], E, and F) along the gradient of Rac1 activation (Fig. 10, B, C, and E–G) that resembled the transitions induced by acute Arp2/3 inhibition (Fig. 9 D, iii). Combined, these results (1) suggest that fluctuations in Rac1 signaling may occur under these experimental conditions, generating some of the natural variation in morphology and neurite outgrowth of the unperturbed population; and (2) highlight the complexity of Rac1 regulation by multiple GEFs/GAPs.
Changes in morphology/motility relations upon perturbation of Cdc42/RhoA signaling
Quantification at the GC scale revealed under-appreciated phenotypic nuances among the three selected perturbations enhancing neurite outgrowth (Cdc42, RhoA, and srGAP2). While srGAP2 KD predominately modified veil velocity, not morphology, on the GC scale, Cdc42 and RhoA KD induced distinct effects on GC morphology without changes in veil velocity (Fig. 7 A). GCs of RhoA KD cells showed markedly reduced veil/stems (Fig. 7 A) and sometimes shared composite feature profiles with the poorly elongating Trio KDs (Fig. 10, D and F), an intriguing finding as Trio is known to activate both RhoA and Rac1 (Debant et al., 1996). Cdc42 KD, in stark contradiction to the classic view of Cdc42 as a positive regulator of both neurite outgrowth (Chandran et al., 2016) and filopodia formation (Nobes and Hall, 1995), induced robustly elongating neurites (Fusco et al., 2016; Fig. 7 A) with corresponding GC morphologies characterized by enhanced filopodia lengths and decreased actin bundle veil-embedment (Fig. 7 C, i)—features correlated with poor neurite outgrowth in the unperturbed population (Fig. 6 D). Importantly, no decrease in the GC veil/stem thickness was observed for the Cdc42 KDs (Fig. 7, A and C, i), indicating the enhanced filopodia lengths were not simply a consequence of defects in the veil. While our observations are consistent with more recent studies suggesting that Cdc42 knockout fibroblasts retain filopodia (Czuchra et al., 2005), to our knowledge this is the first time an increase in filopodia length has been associated with a Cdc42 KD.
In our system, Cdc42 and RhoA KDs appear to induce long neurites due to suppression of GC collapse events (Fusco et al., 2016). This would imply a dual role for Cdc42/RhoA signaling—one regulating GC scale architecture, the other switching between neurite outgrowth/retraction states. Alternatively, altered relationships between morphology and neurite outgrowth may flag larger changes in the mechanism by which outgrowth occurs under these system perturbations. For instance, unlike in unperturbed GCs, the long filopodia associated with the Cdc42 KD may facilitate elongation in the context of the perturbed signaling. To better understand these more complex relations between signaling and morphology, Rho GTPase activity must be imaged directly within veil and individual filopodia. A combination of GCA segmentation, Förster resonance energy transfer biosensor technology (Fritz et al., 2013), and filopodia tracking will be the key to distinguishing the corresponding signaling of long filopodia associated with different outgrowth states.
The future of GC analysis
GCA provides the technical foundation for a wide array of future GC studies. The enhanced filopodia detections, combined with the extracted veil protrusion vectors (Machacek and Danuser, 2006; Ma et al., 2018), may be incorporated into previously developed tracking frameworks (Jaqaman et al., 2008) for the extraction of filopodia dynamics metrics from elongating/retracting GCs. Furthermore, GCA will expedite the extraction of high-confidence fluorescent signals with respect to a variety of GC morphological fiduciaries, including the tip/base of the filopodia, the veil edge, or the site of the GCs leading protrusion. HMM of multi-variate morphological GC features, as tested in Fig. 9, may prove useful for the identification of other, more subtle, correlations between GC morphology and neurite outgrowth, the quantification of the kinetics associated with specific morphology-state switches, and deconvolution of heterogeneous drug responses.
In summary, it is clear that the future challenge will be efficiently integrating information gleaned from the combinatorically expansive number of cell intrinsic/extrinsic scenarios to pinpoint physiologically relevant mechanistic commonalities and divergences governing neurite outgrowth and guidance. Automated GC analysis in vitro and in vivo allows for rapid, comprehensive cataloging of GC features, and thus will be fundamental for the synthesis of image data corresponding to different GC systems from independent laboratories.
Software availability
The MATLAB code and help files for the entire GCA pipeline, including segmentation, feature extraction, analytical tools, and visualization modules, are available at https://github.com/DanuserLab/GrowthConeAnalyzer. Example images, segmentation parameter files, and instructions for reproducing Fig. 3 C and Fig. 4, A–F, are included.
Materials and methods
Cell culture and transfection
N1E-115 neuroblastoma cells (American Tissue Culture Collection) were cultured in DMEM supplemented with 10% FBS, 1% l-glutamine, and 1% penicillin/streptomycin. For differentiation, N1E-115 cells were starved for 24 h in serum-free Neurobasal medium (Invitrogen) supplemented with 1% l-glutamine and 1% penicillin/streptomycin. Both siRNA and LifeAct GFP reporter were transfected simultaneously as previously described (Chong et al., 2006). Specifically, 400 ng of the plasmid pLenti-LifeAct-GFP, 20 pmol of the specific siRNA (Invitrogen Stealth Select), and 1 µl of TransFectin (Bio-Rad) were used in one transfection reaction. 48 h after transfection, cells were starved in Neurobasal medium. 72 h after transfection, cells were detached with Puck’s saline and replated on a glass-bottom 24-multiwell plate (MatTek), coated with 10 µg/ml laminin overnight at 4°C (Millipore). 24 h after plating, cells were imaged in Neurobasal medium (Invitrogen) in a heated closed chamber. Plasmid transfection rates were measured at ∼70%. 90% KD efficiency for select siRNAs used in this study was previously confirmed by Western blot or quantitative PCR as shown previously (Fusco et al., 2016). LifeAct GFP–transfected cells were therefore assumed to be also siRNA-transfected. Based on our previous siRNA screen on N1E-115 whole-cell scale neurite outgrowth (Fusco et al., 2016), we use specific siRNA sequences to knock down with the lowest possible amount of off-target effect (Table S2). Specifically, we used the siRNA that yielded the most similar whole-cell scale MS to the average MS of three distinct siRNAs.
Image acquisition of N1E-115 morphodynamics
All experiments were performed on an inverted Eclipse Ti microscope (Nikon) equipped with a motorized stage, hardware-based autofocus, and CoolLED light source, and controlled using Metamorph imaging software (Universal Imaging) using a PlanApo 60×/NA 1.4 objective. The images were acquired every 5 s for a total of 10 min. A Coolsnap HQ2 camera (Photometrics) was used for image acquisition. In the CK666 experiments, the cells were first imaged for 5 min without drug, and then for an additional 5 min with DMSO, or DMSO plus CK666 (corresponding to a final concentration of 25 µM CK666).
GCA: Overview
The GCA pipeline is illustrated in Fig. 1, and the full algorithmic scheme is outlined in Fig. S2.
Veil/Stem reconstruction
To detect the veil/stem of the neurite (Fig. 1 A, iii–v; and Fig. S2, Steps I–III) the algorithm combines a broadband ridge filter (Jacob and Unser, 2004; Fig. S2, Steps I and II) with an initial intensity-based thresholding (Niblack, 1985)/morphological opening (Maitre, 2008; Fig. S2, Step III). While the former detects the thick, ridge-like, consolidated regions of the neurite stem and its entrance into the image (Fig. 1 A, iii), the second step facilitates detection of more amorphous actin veils, which fail to conform to a ridge detector (Fig. 1 A, iv). When the stem is relatively thick, and of high signal to background (e.g., Fig. 1 A, iii–v), the ridge information mainly serves to identify the neurite entrance point in the image (Fig. 1 A, iii, black star). However, images of GCs positioned within more complicated local environments (Fig. 1 B, i) and/or exhibiting more complex veil/stem morphologies (Fig. 1 B, ii and iii) require the integration of the ridge information for successful segmentation (Fig. S2, Steps I–III; and Video 2).
Filopodia/Branch network reconstruction
A similar ridge localization scheme as employed for stem detection (Jacob and Unser, 2004) is used to detect filopodia with high sensitivity (Fig. 1 A, ii). However, this detection scheme has two limitations: (1) since the width of filopodia approaches the diffraction limit of the microscope (∼200 nm), high frequency noise is often also detected; and (2) regions where filopodia cross, branch, or connect to the veil give low signal, introducing breaks in the GC segmentation.
We solve both problems by first combining the veil/stem with the thin ridge information to create a “high confidence seed” (Fig. 1 A, vii), i.e., only filopodia segments attached to, and residing outside of, the veil/stem are maintained. These seed pixels serve as an anchor for (1) iterative reattachment of filopodia segments that may have been disconnected due to weak ridge detector responses at junctions (Fig. 1 A, viii, Veil), (2) resolution of crossing filopodia (Fig. 1 D and Video 3), (3) grouping thin filopodia/actin branched structures (Fig. 1, A [viii, Branch] and D; and Video 3), and (4) detection of actin bundles embedded in the veil (Fig. 1 C). Final segmentation is obtained via three different, but analogously formulated, graph matching steps: (1) a filopodia segment–building step that associates curvilinear segments of detected filopodia broken due to filopodia crossing/branching (Fig. 1 A, vi), (2) an iterative filopodia/branch network reconstruction step to reattach filopodia/branch candidates to seed pixels (Fig. 1 A, vii; and Fig. S3), and (3) an optional step to associate embedded actin bundle detections with their corresponding filopodia (Fig. 1 C).
Filopodia endpoint localization
To enhance the filopodia tip localization accuracy, we modified the intensity fitting approach used previously to localize microtubule tips (Demchouk et al., 2011). In this approach, the tip location was defined as the midpoint of a Gaussian survival function fitted along the microtubule axis in a manually defined region of interest. However, in contrast to the tip of a single microtubule, filopodia tips are not always best described by a single decay model. Rather, filopodia taper off as a result of spatial gradients in actin polymerization, possibly due to steric restriction of G-actin diffusion (Dobramysl et al., 2016). Thus, f-actin–labeled filopodia bundles may exhibit complex, multiple-decay intensity profiles. To localize filopodia endpoints with sufficient robustness, in a completely automated fashion, we use information regarding the local background intensity distribution (purple crosses) and the local minima (red) and maxima (green) in the slope of the intensity profile to isolate an optimal region of the intensity profile for data fitting. Fig. 1 D (iii) shows two such complex decay profiles, while Fig. 1 C (ii) illustrates that a similar approach can be applied to identify the endpoint of a veil-embedded actin bundle.
GCA: Algorithmic details and segmentation parameters
Segmentation parameter overview
Table S1 defines the default segmentation parameters used for the N1E-115 data and provides recommendations for users. Block colors in both Fig. S2 and Table S1 indicate a specific segmentation step within the pipeline. Note that while all segmentation parameters are made accessible to the user, many required no adjustment to achieve quality segmentation of our diverse image set of 72 N1E-115 neurite movies. The one exception was the local threshold patch size, which needed a course adjustment to the GC thickness. Importantly, the default segmentation parameter settings for the filopodia reconstructions also produced good results when applying the pipeline to other GC models (Fig. 3, A–D; Fig. 4, C and D; Fig. S6; and Videos 7 and 8) and even to non-GC images (Fig. 3, E–G; and Fig. 4, E–G) with only minimal, intuitive adjustments, primarily to account for the differences in pixel size among the datasets. Full documentation of each step in the algorithm and discussion of specific segmentation parameters are provided below. In addition, segmentation parameter files and corresponding images for reproducing Fig. 3 C and Fig. 4 (A–F) are included with the software release.
Detection of neurite stem and determination of neurite entrance point
Currently GCA is designed to segment the GC and a small portion of the neurite stem, as this was the primary biological region of interest (Fig. 1). A small cropping tool to select this region of interest is included in the package. It is optional, and the only step requiring manual user input. The algorithm is then initiated by a procedure that locates the neurite entrance point (black stars in Fig. 1 B). For this to be successful, the cropping needs to ensure that some part of the neurite stem is included in the image sequence.
To identify stem-like regions of the neurite, GCA first filters the image using a multi-scale, fourth-order, steerable ridge filter (Jacob and Unser, 2004; Fig. S2, Step I). Ranges of sigma (σ) values defining the SDs of the Gaussian kernels on which the filters are based are set by the user via the segmentation parameter BBScale (Table S1, Step I, i). The ridge filtering is performed on the entire image for each sigma value assigned in the BBScale vector. For all movies in this study, multi-scale ridge filtering was performed using a sigma range of 5–10 pixels (∼1–2 µm) sampled at 1-pixel intervals, which is tuned to find relatively large line structures such as the stem without response to finer structures such as filopodia. A multi-scale approach is required to accommodate stems of variable thickness. The final multi-scale ridge response map is computed by finding for each pixel the maximum filter response over all scales. A nonmaximum suppression (NMS; Canny, 1986; Jacob and Unser, 2004) is performed on this multi-scale ridge response to identify the local maximum in the direction of the response gradient. The NMS map traces connected paths along the ridge-like portions of the veil/stem (orange lines, Fig. 1 B). Two ridge-cleaning steps are subsequently performed (Table S1, Step I, ii). First, an additional per-pixel thresholding of the NMS response is performed to remove very weak local-maximum ridge signals. The strictness of this threshold is dictated by a user adaptable parameter threshNMSResponse, set in this study to the 25th percentile of the total population of the NMS response values. Second, connected component objects from the final NMS ridge response less than MinCCRidgeBeforeConnect (set in this study to 3 pixels) are filtered, as it is difficult to extract high-confidence local orientation information, required for the subsequent connection step, in these cases.
If the veil/stem were a simple ridge with high signal to noise and invariant thickness, the NMS response map should trace a continuous path along the neurite. However, gaps along this path are expected in larger and/or more amorphous regions of the veil/stem where the ridge model is a poor assumption. (Detection of these amorphous regions will be addressed in the Veil/Stem reconstruction section.) However, gaps can arise even in relatively thin ridge-like regions of the neurite due to inherent limitations of the ridge filter. Small, geometrically noncontinuous gaps are often observed at sites where the neurite veil/stem abruptly changes thickness or the stem of the veil has very asymmetric filopodia density along either side. Larger, typically more geometrically aligned gaps in the large-scale ridge response may be observed when the response is weak due to poor signal to noise of the original image or from subtle deviations of the image object from the underlying ridge model. Therefore, we implement a linking step to bridge endpoints of connected components in the NMS map (Table S1, Step I, iii). The main goal of this step is to find rough connected component veil/stem paths leading from the neurite entrance point to each larger amorphous veil/stem structure. Accordingly, we allow relatively discontinuous linking geometries. If the gap distance is less than a user-defined cut-off radius, MaxRadiusNoGeoTerm, set in this study to 3 pixels (∼0.6 µm), connected components are linked regardless of the attachment geometry. Gaps with distances greater than MaxRadiusNoGeoTerm, but below a user adaptable cut-off, MaxRadiusLargeScaleLink, are bridged only if the respective connected components fulfill the constraint, that the angle (θRL) between the linking vector (L) and the local orientation vector at the connected component endpoint (R) does not exceed the value GeoThresh, in this study set to a lenient 90°. Potential links within a MaxRadiusLargeScaleLink (set to 10 pixels [∼2 µm] in this study) are found using a K-dimensional tree and linear linker path interpolated between candidate endpoints. To ensure only a single linkage for each ridge endpoint, a maximum weighted graph matching step (Avis, 1983; Kolmogorov, 2009) is performed using weights based upon distance only if the distance is below the MaxRadiusNoGeoTerm, or based on distance and geometry if the linkage distance is greater than MaxRadiusNoGeoTerm and less than MaxRadiusLargeScaleLink. After this initial linking step, detected endpoints of thick ridge candidates within MaxDistBorderFirstTry (10 pixels [∼2 µm] in our study) from the image edge are chosen as potential neurite stem entrance ridge candidates (Table S1, Step I, iv). The software assumes a reasonable entrance candidate for each frame of the movie exists; therefore, while MaxDistBorderFirstTry is technically a user-defined parameter, the value is widened automatically if no ridge candidate in a given frame meets this initial criterion. If more than one ridge candidate is found, such as is shown in (Fig. 1 B, i), the algorithm first filters candidate ridges based on length alone, as defined by the user-adaptable parameter MinCCEntranceRidgeFirstTry (set in our study to 5 pixels), in order to filter out any potential high-intensity noise. Using a summation of the mean fluorescence intensity (FI) and the length of the ridge candidate as a score, the program then determines on a per-frame basis which candidate ridge path most likely corresponds to the entering neurite stem. Occasionally, an incorrect ridge candidate is chosen as the neurite entrance ridge. As the orientation of the entering neurite stem within the image does not move significantly during the time resolution of interest in this study (total movie time ∼10 min), the correct orientation of the neurite within all images is the most frequent orientation of the automatically determined seed ridge for the entire movie. Frames in which the selected seed ridge fails to correspond with the majority neurite orientation for the movie—for example, due to a transient distracter object entering the image such as in Fig. 1 B, i—are corrected in a subsequent step so that the seed ridges for all frames are consistent (Fig. S2 and Table S1, Step II). This is accomplished by simply performing a crude spatial clustering via dilation of a logical mask marking all the estimated neurite entrance points by an input radius SizeOfConsistencyRestraint (default is 5 pixels). This dilation is then separated into connected components, and the connected component with the majority of frames is chosen as the correct neurite orientation cluster. The program then combines all the ridge neurite stem candidates from the majority cluster that are found in more than five frames as a base neurite orientation path. The best ridge candidate from the outlier frame that aligns to this path is then chosen as the new neurite entrance stem from which to build the veil/stem estimate in that frame. GCA has an optional visual quality control step, allowing the user to check the final neurite orientation selected for the movie (the checkOrient option; Table S1, Step II). If users see the wrong neurite entrance region is selected from the majority vote, they can select the correct entrance ridge to which the movie is aligned by the process described above. This feature also can help choose the correct SizeOfConsistencyRestraint. However, we found this manual refinement was typically not necessary as long as a distractor element, such as another GC, was absent or relatively transient. For example, incorrect entrance ridges in the movie corresponding to Fig. 1 B (i) were corrected automatically by the algorithm without any need for manual refinement. Notably, none of the datasets analyzed in this study, including the data of Fig. 3 (A–D), Fig. 4 (C and D), or Fig. S6, required this manual refinement.
Veil/Stem reconstruction
Gaps along the ridge path are expected in larger and/or more amorphous regions of the veil/stem where the ridge model is a poor assumption. A practical solution is to simply detect these more amorphous regions using local intensity Otsu thresholding (Niblack, 1985), a variant of traditional Otsu thresholding (Otsu, 1979), which allows for differences in the value of the intensity threshold per pixel throughout the image, depending on the intensity profile of the local surrounding region (Fig. S2, Step III). The size of the local region is controlled by the segmentation parameter LocalThresholdPatchSize (Table S1, Step III, i), which was set between 30 and 100 pixels in this study for the N1E-115 dataset, depending on the size of the GC (larger GCs required larger patch sizes). The default value is set to 75 pixels. Notably, global thresholding rather than local thresholding (Table S1, Step III, i) helped avoid over-emphasizing intracellular inhomogeneity associated with some datasets. Ultimately the method most appropriate here will depend upon the characteristic intensity distribution of a given imaging modality. Therefore, several different initial thresholding methods are made available for this step, as well as an option to import an externally generated mask (Table S1, Step III, i). Notably, this initial thresholding step is indeed the most likely segmentation parameter that may need modification/troubleshooting when applying GCA to a novel dataset. Importantly, however, as in GCA the main goal of this initial thresholding step is to find a reasonable detection of the veil and not the lower intensity filopodia, the final segmentation becomes less sensitive to this step as compared with other methods (Fig. 4, A–C, ii; Tsygankov et al., 2014; Urbančič et al., 2017). This vastly facilitates the segmentation parameter optimization. Low-fidelity information corresponding to filopodia from the binarization achieved via the initial thresholding step is removed via morphological opening (Maitre, 2008). Here morphological opening was performed using the imopen function included in MATLAB’s image processing toolbox, using the strel function to create a disk structuring element of a user-defined radius dictated by the veil/stem segmentation parameter DiskSizeLarge, set to 6 pixels (∼1.3 µm) for segmentation of all the GFP LifeAct images in this study (Table S1, Step III, ii). Note that in the case of the LifeAct images we found it worked best to keep the structuring disk for this morphological operation slightly larger than the true scale of the filopodia objects we aimed to remove, as residual fluorescence and overlapping filopodia are often merged/under-thresholded, leading to low-fidelity noise in the binary image thicker than a single filopodium. This was less of a problem in the case of the membrane-labeled images (recommended DiskSizeLarge is 4 pixels [∼0.86 µm] for the N1E-115 images), as filopodia exhibited lower signal to noise and thus were typically not detected by the local thresholding. However, morphological operations using larger disk sizes begin to remove thin pieces of the neurite stem/veil path, breaking connectivity. To maintain maximal connectivity along the veil stem path, we use geometric information to adapt the local disk size of the morphological operation employed in select regions of the neurite. We reasoned that ill-detected, thick filopodia/branch bundles, which we wish to remove in this step, and thinner portions of the veil/stem (Fig. S1 C, iii), which we wish to preserve, can be deciphered based on their connectivity with respect to the thicker regions of the neurite. Therefore, the algorithm scans for simple connected component paths of the neurite removed by morphological openings that span two larger-size veil/stem “pieces” in the binary threshold image. In these regions, the size of the structuring element is reduced to the user-defined radius DiskSizeSmall (Table S2, Step III, ii) set to 3 pixels (∼0.65 µm) for the N1E-115 dataset.
The neurite entrance ridge is then combined with the veil pieces detected via the initial thresholding/morphological opening step. All veil pieces overlapping the neurite entrance ridge are maintained. Subsequently, all ridge pieces overlapping with these selected veil pieces are added to the veil/stem detection. Any ridge detections within MaxRadiusBridgeRidges (set in this study to 5 pixels, ∼1 µm; Table S1, Step III, iii) of all veil/stem detection endpoints are linked to the current veil/stem detection. The process iterates until no more veil pieces overlap with the final veil/stem detection. When the reconstruction is complete, any endpoints, other than the neurite entrance point (black star in Fig. 1 B), of the veil/stem detection are eroded to create the final veil/stem mask (Fig. 1 B, far right panels, green outlines). The veil pieces are treated as nodes on a graph whose edges are defined by the detected thick ridges (potential stems). Only ridge detections spanning two separate veil nodes are maintained. Occasionally cycles are formed in this veil/stem graph (Fig. 1 B, iii), and these are resolved by solving for a minimal spanning tree (Prim, 1957) where the weights of the edges are dictated by the scale (i.e., thickness) and filter response strength of the connecting large-scale ridge, as well as its underlying original image intensity (Fig. S2, Step III). As the NMS only marks the central line of the stem, without providing information of the thickness/localization of the stem edges, remaining stem detections are dilated using MATLAB’s imdilate function included in MATLAB’s image processing toolbox, using the strel function to create a disk-structuring element with a radius set to 4 pixels (∼864 nm). (See Video 2 for a visual summary of these iterative steps for four example GCs with diverse morphology.)
Neurite length measurements
Given the morphological heterogeneity between GCs as well as the morphological variation over time, it is prohibitive to use the GC centroid for tracking neurite length changes, as was done previously (Tsygankov et al., 2014). We instead use the properties of veil/stem segmentation to obtain more robust neurite length measurements over time. We assume neurite length to extend from the entrance point to the tip of the leading veil protrusion (Fig. 5, A and B). This measurement can be achieved through skeletonization of the veil/stem (Fig. S2, Step IV). We used the “skel” operation of MATLAB’s bwmorph function. The skeleton is converted to a graph representation, where each node represents a pixel in the skeleton with a nearest neighbor number either equal to 1 (endpoint vertices) or greater than 2 (branch vertices). The shortest path through the skeleton graph from the neurite entrance point to each endpoint vertex of the skeleton (marking approximately the convex endpoint of a veil/stem protrusion) is calculated, and the longest of these paths is then chosen as the neurite length measurement for the given frame (Fig. 5 A, orange line). Note in a system of high symmetry there can be fluctuations in the orientation of the longest path from one frame to another as the neurite makes a directional decision. However, the overall neurite length outgrowth metric velocity can remain constant.
Optional veil/stem refinement step
Once the neurite length measurements are extracted (as described in the previous section), identification of outlier data points within an individual neurite outgrowth time series can serve as an automated indicator of segmentation error. Therefore, GCA includes an optional step (Fig. S2, Step V) to flag probable segmentation error frames, as indicated by an outlier veil/stem length measurement and the presence of a high-intensity floating “veil piece,” initially excluded from the final veil/stem segmentation due to failure to find a viable stem attachment path during reconstruction. While the exclusion of these detected “veil pieces” is desirable in certain cases—for instance, when there is surrounding high FI debris, or when the GC of interest is in the presence of a neighboring neurite (Fig. 1 B, i)—a coincident length outlier flag indicates a potential veil/stem truncation, which may be caused by limitations of the ridge filter at larger sigma values. For these identified frames, the longest path of the veil/stem skeleton in the frames toutlier + 1 and toutlier − 1 are combined, thinned, and then used in an attempt to bridge the previously excluded veil/stem piece and correct the segmentation for the outlier frame. All veil/stem masks calculated in this study were visually verified. Such truncations were infrequent and typically caused by dense, asymmetric filopodia distributions along one side of the stem to be detected. Notably, only two movies in this study required veil/stem refinement via this step, and therefore by default it is an optional step in the GCA pipeline.
Filopodia candidate detection
Filopodia-like structures were identified using the same fourth-order steerable ridge detector (Jacob and Unser, 2004) as for ridge identification, but applied using a single, much smaller Gaussian sigma value, set by the segmentation parameter FiloScale (Table S1, Step VI, i, in this study 1.5 pixels [324 nm]). The filter step was followed by an NMS step (Canny, 1986; Jacob and Unser, 2004) to isolate the center line of the filopodia images.
With smaller sigma Gaussians underlying the steerable filter, the responses tend to be noisier, requiring stringent filtering of false positives in the NMS map. Therefore, GCA takes a number of practical measures to limit the number of false-positive filopodia segments that are entered into the subsequent filopodia/branch optimization steps. First, a permissive threshold of the image intensity histogram estimates a rough background mask of the image, leaving a permissive filopodia search area surrounding each object, threshold: (μInt_Noise + 2σInt_Noise), where μInt_Noise and σInt_Noise are the mean and the SD, respectively, of the fit to the first Gaussian. These object masks can be cleaned and dilated if necessary if the filopodia search region surrounding the object is too small (i.e., the background mask is overestimated). This is an optional step (filterBackEst) in the pipeline (Table S1, Step VI, ii), but it is recommended to save computational time and to quickly avoid possible false-positive branch structures. Second, we applied a threshold directly upon the NMS values. We found a threshold of (μNoiseNMSResponse + 3σNoiseNMSResponse) performed well, where μNoiseNMSResponse and σNoiseNMSResponse are the mean and the SD, respectively, of the fit to the first Gaussian of the probability density function corresponding to the nonzero response values from the NMS. This value was sufficient for a majority of our needs in this study when combined with the above object-masking step. However, this value can be adjusted by the parameter multSTDNMSResponse (Table S1, Step VI, ii), and several quick automated checks are included in the software to check the integrity of these initial thresholds on the response values. If flagged, the software attempts a different thresholding method automatically, which may perform better given the shape of the intensity histogram. After thresholding, the NMS image is binarized, and any remaining branch/higher-order junctions are broken to obtain curvilinear ridge pieces that can be grouped in subsequent steps. Note that junctions typically show weak steerable ridge filter responses since they represent points in the image where the orientation of the ridge structure is ambiguous, and hence they are not typically well detected by the steerable filter. The subsequent filopodia/branch reconstruction steps resolve this problem (see below). In addition, ridge pieces ≤3 pixels (0.65 µm; segmentation parameter minCCRidge; Table S1, Step VI, ii) are removed from the NMS map, as the geometric measurements of these pieces, required for subsequent linking steps, are too uncertain. To classify this pool of segments into veil-exposed filopodia (Fig. 1 A, vi and vii) versus veil-embedded actin bundle candidates (orange lines, Fig. 1 C, i), we used the veil/stem mask and considered veil-exposed candidates connected directly to the mask as high-confidence filopodia detections. The combination of veil/stem edge pixels and their connected filopodia detections were subsequently used as a “high-confidence seed“ (Fig. 1 A, vii) for further filopodia candidate reattachment in the Filopodia/Branch Reconstruction and Optional Veil-Embedded Actin Bundle Detection modules documented below.
We also considered veil-exposed filopodia candidates not directly attached to the veil/stem mask. We reasoned that the population of connected candidates provided a distribution of filter response values representative for bona fide filopodia detections. Therefore, to remove weak filopodia candidates from the unattached candidate pool, we removed segments of less than 10 pixels in length or with a mean ridge filter response less than the fifth percentile of the veil/stem-attached filopodia distribution. Note this filtering step can be skipped by the user if the number of veil-attached filopodia is too small to obtain an estimate of the distribution. For example, this filtering step is not applicable in the case of a single dendritic filopodium (Fig. 4 G). The compilation of these initial selection steps produced the final unattached filopodia candidates as shown as orange lines in Fig. 1 A (vi).
Filopodia/Branch reconstruction
The filopodia segments were entered into the filopodia building step in which neighboring, linearly continuous fragments were merged (Fig. 1 A, vi). Candidates for merging were selected by finding matching endpoints within the distance defined by maxRadiusLink (set in this study to 5 pixels, ∼1 µm) using a K-dimensional tree search. Between potential matching endpoints, linear paths were then interpolated, and the link orientation represented by a unit-length vector. To determine the continuity between the filopodia segments and their corresponding link, the angle, θFL, between the link vector and the local orientation at the filopodia segment endpoint (black and orange arrows, respectively, in Fig. 1 A, vi) was calculated for each endpoint. While two θFL values, θFLij and θFLji, were calculated for each possible link, a single θFLij calculation is illustrated in Fig. 1 A (vi) for clarity. The tolerance with which links are allowed to deviate from perfect continuation (θFLij = 0°) is controlled by the parameter geoThresh (set in this study to θFL = ∼25° [cosθFL = 0.9]; Table S1, Step IV, iii). Hence, any endpoint whose local orientation vector and corresponding link vector formed an angle θFL > 25° was not considered for merging. Typically, only a single possible link between two endpoints fulfilled the stringent criteria for link selection. However, occasionally multiple links competed for the same endpoint. To ensure that each endpoint was associated with no more than one link, we formulated the link assignment as a maximum weighted graph matching problem (Avis, 1983; Kolmogorov, 2009). Each endpoint under consideration defines a node on the graph, and each interpolated link a graph edge. To calculate the weights, distances dij between candidate endpoints were converted to distance scores , where Dij represents the full set of dij values for a given frame. This conversion ensured that larger dij values result in smaller weights.
The scores Dij were then normalized as
where scoreDij′ is the final normalized distance score.
Weights of edges between two endpoints, i and j, were derived by simple summation of the normalized distance score Dij and the dot products between link orientation and local filopodia orientation in at each candidate endpoint:
Application of a maximum weighted matching to this graph determines which filopodia candidates are merged into a single candidate.
The final filopodia/branch reconstruction was then achieved by combining information from this new set of merged filopodia candidates (Fig. 1 A, vi) and the “high-confidence seed” (Fig. 1 A, vii), comprised of both the veil/stem edge pixels and its connected filopodia detections. Akin to the preceding filopodia candidate building step, we formulated a graph matching problem (Fig. 1 A, viii; Fig. S3; and Video 3): all pixels of the “seed” within a user-defined search radius maxRadiusConnectFiloBranch, set to 15 pixels, ∼3.2 µm in our study (Table S1, Step VI, iv), surrounding each filopodia endpoint were selected (Fig. S3 A, ii and iii). Linear paths are interpolated between candidate endpoints and all seed pixels using the Bresenham algorithm. These paths become the edges, and each graph node is either a single filopodia candidate or a pixel on the “seed” (Fig. S3 A, iii). In the second step (Fig. S3 B), weights are calculated for each path. These weights were designed to reflect that a path is more likely if (1) the Euclidean distance of the path is low, (2) the mean intensity of pixels along the path is high, and (3) the linkage results in a geometrically reasonable structure given the biophysical properties of actin filaments.
Specifically, geometric continuity between each filopodia candidate and each putative linkage was assessed by calculating the dot product of the unit vectors representing the the local orientation of the candidate filopodia in its endpoint and the direction of the linkage path. Given the relatively linear geometry of actin bundles of interest, dot products corresponding to an angle greater than the geoThreshFiloBranch segmentation parameter, set in this study to θFL = 60° (Table S1, Step VI, iv), were filtered before the final graph matching step (Fig. S3 B, i and ii). Any paths that crossed the veil/stem mask and/or crossed another ridge/candidate were likewise removed.
Similar to the preceding filopodia/ridge candidate building step, all individual distances, dij, between potential graph nodes were converted to individual distance scores , where D represents the full population of dij values for a given frame, for a given reconstruction iteration, before geometry-based path filtering (Fig. S3 B, i and ii). This conversion ensures that larger dij values result in a smaller scoreDij. scoreDijs were then normalized such that the maximum scoreDij is equal to 1,
where scoreDij′ is the final normalized distance score (Fig. S3 B, iii).
Unlike in the filopodia candidate building step, the graph weights corresponding to the final filopodia/branch reconstruction steps also accounted for the mean FI of each path. To this end, we calculated the mean FI ImeanPerPathij of a single path from graph node i, a ridge candidate endpoint, to graph node j, a pixel on the seed, and normalized the value such that the full population of potential paths maintained a value between 0 and 1:
Here, ImeanPerPath denotes the full distribution of path intensity mean values in a frame for a given filopodia/branch reconstruction iteration, and min(ImeanPerPath) and max(ImeanPerPath) denote the minimum and maximum values of this distribution, respectively.
Finally, the absolute value of the unit dot product corresponding to the local orientation vectors at the site of the filopodia segment endpoint (F) and the site of attachment to the seed (S), |cosθFSij|, was calculated.
The final weight (Fig. S3 B, vi), Wij, for each path was compiled by weighted summation of the above scores (Fig. S3 B, ii–v), such that
Weight values typically approached a Gaussian distribution (Fig. 3 B, vi, bottom). The maximum weighted graph matching step (Avis, 1983; Kolmogorov, 2009) solves the graph by choosing a subset of edges (black paths in Fig. S3 C) that maximizes the sum of the weights of the edges subject to each node being connected to another node not more than once. As each ridge candidate is considered a single node in the graph (Fig. S3 A, i) this formulation forces only a single optimized link to be chosen between filopodia segment and seed (Fig. S3 C). The attached fragments are then assigned labels based on their attachment site (Fig. 1 A, viii) and become part of the next iteration’s seed. The above steps iterate until no pixels in the current seed set fall within maxRadiusConnectFiloBranch of the remaining candidate filopodia segment endpoints (Video 3).
Optional veil-embedded actin bundle detection
Veil-embedded actin bundle candidates, as defined in the Filopodia Candidate Detection Step, were first cleaned using MATLAB’s bwmorph “spur” option (2 pixels). Any candidates without two well-defined endpoints were discarded. The endpoint closest to the veil/stem edge for both the exposed and embedded actin bundle candidate was chosen for potential matching. Any embedded actin bundles with endpoints equidistant from the veil/stem edge were discarded, as it was typically an indication that these ridge candidates corresponded to signal generated from the veil/stem edge itself. To further solve the problem of false-positive actin bundle detection, we also included a step that breaks the embedded ridge candidates at sites surpassing a user-defined curvature, curvBreakCandEmbed (Table S1, Step VI, v; set in this study to 0.05 1/pixels). This enforces the assumption that true embedded candidate ridges of interest are linear. If one wishes to detect more noncanonical highly curved embedded structures, one can simply set this value to 0, though at the potential risk of an increased false-positive rate.
All embedded actin-bundle, ridge candidate endpoints (orange lines in Fig. 1 C, i) within a 10-pixel (2.16-µm) radius of the high-confidence exposed filopodia seed points (green lines in Fig. 1 C, i) were found using a K-dimensional tree search. Although this study kept the radius fixed for all datasets, the software allows user adjustments via the segmentation parameter maxRadiusLinkEmbedded (Table S1, Step VI, v). Similar to the filopodia candidate building step (Fig. 1 A, vi), and the geometry filtering step of the main filopodia/branch reconstruction (Fig. S3 B, i and v), only connections exhibiting a linking geometry conforming to the specifications defined by the geoThreshEmbedded segmentation parameter (θFL = 60° in this study; Table S1, Step VI, v) were considered for linking. Again, to ensure only one linkage is maintained between two ridge candidate endpoints, the assignment problem was formulated as a maximum weighted graph matching problem (Avis, 1983; Kolmogorov, 2009), such that each endpoint under consideration corresponds to a node on the graph, and each interpolated linkage a graph edge path. In this case, weights, Wij, of individual edge paths were based completely on linkage geometry, such that
Of note, distance was not included in the weight. We empirically observed that confounding ridge signal corresponding to intracellular noise and the veil/stem edge frequently caused breaks along the ridge NMS extracted for the exposed filopodia ridge population. This, combined with generally lower ridge responses inside the image of the cytoplasm, often produced truncated ridge candidates with relatively large distance gaps between correct pairings. Hence, we reasoned in this case that geometry alone was the most reliable indicator of a high-fidelity link. Note that this framework allows only those embedded actin bundles with a veil-exposed filopodium counterpart to be detected (Fig. S5 C).
Filopodia endpoint localization
The thresholded steerable filter response of a filopodium provides only an inaccurate estimate of its endpoint, both because of the often weak signal in these regions, and because any signal at the ridge end inherently deviates from the geometric model underlying the detector, which assumes the ridge to be of infinite length. This may result in either an over- or underestimation of the true filopodia length. To improve on the tip localization, the local directional vector, identified from the steerable filter response, was employed to extrapolate each putative filopodia region by 10 pixels (∼2 µm). Examples of these extrapolated regions are shown in purple in Fig. 1 A (ix) and Fig. 1 D (iii) and are combined with the original filament fragment detection (green) to create a mask localizing the approximate region of each filament. For visualization, and to estimate the local intensity background surrounding a filopodium, each filament mask included ±2 pixels (∼0.4 µm) perpendicular to the filament’s centerline (achieved via the NMS of the steerable filter response or via the extrapolation step above). Image extraction from these regional masks confirmed good localization of filopodia centerline by the ridge detection followed by NMS thresholding (Fig. 1, A [ix] and D [iii]).
A weighted average of the pixel intensity values (not limited by the above mask), based upon the point spread function (PSF) of the microscope, was extracted for each pixel on the filament fragment’s backbone (Fig. 1, A [ix] and D [iii]). As the fluorescence decay of actin bundles can be complex (Fig. 1 D, iii), a series of steps was then employed to localize the region of each linescan corresponding to the first significant fluorescence signal decay. Specifically, we subjected a smoothened version of the signal’s derivative (obtained via cspas in MATLAB, P = 0.1) to MATLAB’s findpeaks function to locate the corresponding local minimum and maximum, denoted as green and red dots, respectively, in Fig. 1, A (ix) and D (iii).
The local background surrounding the filopodium was estimated using the extrapolated region of the filopodium mask (purple in Fig. 1, A [ix] and D [iii]). The center line of the projection mask was not included in the filament’s intensity background estimation. The first maximally decreasing region with signal above the mean of this background estimate was isolated, and signal within ±10 pixels (approximately ±2.14 µm) of this value was isolated for the final least-squares regression fitting of the intensity values. The maximum window size for least-squares regression can be modulated by the maxWindForLSQFit segmentation parameter (Table S1, Step VII). However, this window is currently partially self-adaptive. If a second maximally decreasing region is found within the initial window size, the values used for the least-squares regression are truncated to exclude these portions (Fig. 1 D, iii), as this typically indicates a second confounding intensity decay. The final window of the intensity profile is then fit to a Gaussian survival function (below equation) using the lsqcurvefit function in MATLAB:
Here I(x″) denotes the portion of the filament intensity profile, x″ is the distance along the filament bundle, IFil is the amplitude of the filament intensity above the mean background IBG, μFil corresponds to the mean value for the filament endpoint measurement, and σ(Fil + PSF) corresponds to the combined SD in the filament endpoint localization due to both the filament’s physical structure (Fil) and the PSF of the microscope.
GCA: Feature extraction
Integrated with GCA are a number of functions that extract from the segmented veil and filopodia morphological features for classification of GC behaviors. Importantly, GCA users are encouraged to define their own features. In the following sections, we define the morphological and morphodynamic features employed for the GC analysis in Figs. 6, 7, 8, 9, and 10.
Neurite length
We use neurite length as our primary feature describing GC function. It is defined as the longest path through the veil/stem skeleton as shown in Fig. 5 A. Further details are provided in the Neurite length measurements subsection under the GCA: Algorithmic details and segmentation parameters section.
Veil/Stem thickness
An illustration of the veil/stem thickness metric is shown in Fig. 5 B. The veil/stem thickness is defined as the mean of the Euclidian distance transform values along the longest path of the veil/stem skeleton, from the tip of the leading protrusion to a user-selected length (default 20 µm). The distance transform of the veil/stem binary mask was calculated using bwdist in MATLAB.
Veil/Stem dynamic features
Using the veil/stem detection mask (Fig. 1 A, iii), protrusion vector calculations and edge sector averaging of vectors were performed as described previously (Ma et al., 2018).
Specifically, cell edge velocities were derived from pixel-by-pixel matching of cell contours between consecutive time points. A B-form spline was fitted to the edge pixel positions of the segmented cell area, with nodes corresponding to each edge pixel. The spline representations of two consecutive frames were then divided into segments between their intersections. To map a correspondence between the edge splines on consecutive frames, the following objective function was iteratively minimized as:
| (1) |
with the topological constraints
| (2) |
The variable n denotes the number of nodes, which in the absence of down-sampling (see below) is equal to the number of edge pixels in that segment. p1,2,...n(t) are the parameters of the spline at time t defining equally spaced edge nodes x(t,pi), one at each edge pixel. The goal of Eqs. 1 and 2 is to identify n spline parameters o1,2...n(t+1) in between the intersection points e1 and en that define nonequally spaced nodes x(t+1,oi) at t+1 such that the overall displacement (SUMA) and strain, i.e., changes in spacing of nodes (SUMB), is minimized. Eq. 2 imposes the constraint to the minimization that displacement vectors must not cross. The two sums in Eq. 1 have different physical units. To balance them correctly, a factor ω is introduced as follows:
The factor ω is calculated only in the first iteration of the minimization, as the unit conversion by the ratio SUMA = SUMB changes insubstantially thereafter. The parameter w is a free user-control that allows the definition of the trade-off between minimal edge displacement and minimal lateral strain.
The minimization of Eq. 1 can be computationally costly when the number of edge pixels in a segment exceeds 100. To circumvent this problem, a control parameter 10 < Nmax < 100 is introduced. When the number of edge pixels in a segment is greater than Nmax, the number of nodes to Nmax is downsampled, and the boundary displacement for this number of nodes is calculated and then upsampled to the original number of edge pixels by interpolation. Once the boundary displacements are identified, the projections of these displacements onto the boundary normal vector are calculated to obtain a signed local measurement of the instantaneous normal edge velocity.
Edge sector numbering was initiated in each frame at the detected neurite entrance point (Fig. 1 B, black star). Propagation of this origin was set to false. Edge sectors were propagated using the “ConstantNumber” method, whereby the number of sampling edge sectors are held constant, allowing the width of each sector to vary as the length of the cell edge changes. The initial size of each edge sector, parallel to the cell edge, was set to ∼5 pixels (∼1.0 µm for the N1E-115 GC images). Edge sectors were reinitiated midway through the movie (frame 61) to help limit sector tracking errors. As extreme changes in the local veil velocity are predominately associated with rare veil/stem edge segmentation errors, to limit veil velocity analysis artifacts, outlier data points approximately greater than six times the mean were detected (Rousseeuw and Leroy, 1987) and removed for each veil velocity time series before further analysis (black crosses in Fig. 5 B, iii, indicate outlier data points). The velocity traces for each segment (Fig. 5 B, iii) were then processed as described previously (Mendoza et al., 2015), using a signal detection algorithm on the basis of empirical mode decomposition (Huang and Shen, 2014). Velocities within the noise level were classified as insignificant and the respective edge segment as inactive (Fig. 5 B, iv and v). Once a significant event was detected, it was classified as either protrusion or retraction, and the mean values were extracted. The event duration was also measured.
Design of filopodia/branch segment filters for feature extraction
Before any feature extraction, GCA’s workflow allows the user to design a filopodia/branch filter based on segment length (defined as μFilo, Fig. 1 A, ix), the quality of the intensity profile regression, as measured by IFilo compared with the residuals of the fitted data points (Fig. 1 A, ix), and segment branching order, such that order = 0 (veil-attached, no branch detected), order = 1 (veil-attached segment, with branch), order = 2 (branch segment attached to veil-attached segment type 1), order = 3 (branch attached to branch segment type 2), etc. (Fig. 5 D, top). Unless otherwise noted, segments with an μFilo length less than 0.3 µm and an IFilo amplitude less than the 95th percentile of the residuals from the intensity fit were excluded. For select features, these filter criteria were relaxed slightly as explained in the respective feature descriptions below.
Filopodia length
Filopodia lengths can be potentially calculated in two different ways using the intensity profile (Fig. 1 A, ix) fit information. One option, used in this study, is to use the value of μFil as the termination site of the filament and not consider variation in potential filament tapering that may be reflected in the SD of the fit. The second option is to use μFil + σ(Fil+PSF) as the sub-pixel filament termination point. While both metrics are reasonable, using μFil + σ(Fil+PSF) may yield filopodia lengths that are visually more satisfying. As filaments exhibiting a large σ, potentially due to excessive tapering of the actin bundle, may look visually underestimated if only the μ of the fit is displayed.
Unless otherwise noted, filopodia lengths were calculated from both branching and nonbranching veil/attached segments (branch order equal to either 0 or 1). Filopodia length was defined as the distance from μFil of the fitted segment outside the veil, to the detected veil/stem, as traced along the NMS of the ridge filter response (Fig. 5 C).
Embedded actin bundle length
An additional filter step removed embedded actin bundle segments from the feature distribution if their corresponding extra-veil filopodia segment failed to meet the confidence criteria of the filter. Embedded actin bundle length was calculated as the distance from μFil of the fitted segment inside the veil to the detected veil/stem, as traced along the NMS of the ridge filter response (Fig. 5 C). If no embedded segment of the filopodia was detected, or the detected segment did not meet the filter criteria, the embedded actin bundle length is set to 0. Only nonzero embedded actin bundle lengths were pooled for the distribution calculations in Fig. 6 (C and D), Fig. 7 A, and Fig. 8 B. Note, a fluorescent actin label, such as LifeAct GFP, is required for the calculation of this feature.
Full actin bundle length
Full actin bundle lengths were measured from both branching and nonbranching veil/attached segments (branch order equal to either 0 or 1), and calculated as the summation of the filopodia length and the embedded actin bundle length as defined in the above sections (Fig. 5 C). If no embedded segment of the filopodia was detected, or the detected segment did not meet the filter criteria, the embedded actin bundle length is set to 0.
Percent length each actin bundle veil embedded
The percentage each actin bundle veil embedded was calculated by dividing the embedded actin bundle length by the length of the full actin bundle (as defined in the above sections). Actin bundles with no significant veil-embedment (0 values) were excluded from this feature distribution, as a majority of detected filopodia do not have an embedded actin bundle counterpart and inclusion of these values skew the feature distribution to 0.
Branch length
Branch lengths were measured from second order branch segments only, and defined as the distance from μFil of the second order fitted branch segment, to its site of attachment along its respective first order branch, as traced along the NMS of the ridge filter response (Fig. 5 D).
Filopodia/Branch curvature
Detected filopodia/branch backbone coordinates obtained via the NMS of the steerable filter response were first smoothed using the spaps function in MATLAB, setting the tolerance parameter equal to 2. The local curvature was calculated at each pixel by fitting polygons to each of the filopodia backbone coordinates and solving for the curvature analytically from the polygons. The maximum curvature value along the length of the filopodia/branch (as defined in the Filopodia length and Branch length sections above) was recorded (Fig. 5 C, black arrows).
Filopodia/Branch LifeAct FI
For each detected segment of the filopodium/branch network, the background-subtracted FI profile along the filament’s backbone was averaged (Fig. 5, C and D), using pixels associated with the respective length calculations, where the end of the filopodium is defined as μFilo (Fig. 1 A, ix). These values were then normalized for LifeAct expression level by dividing by the mean of the veil/stem intensity distribution (excluding any high-intensity embedded actin bundles) calculated per frame.
The background mask used above was estimated via a coarse localization of the object regions in the image was first achieved as specified in the initial steps of the filopodia reconstruction (Table S1, Step VI, ii). To further ensure that none of the final detected object was considered in the background mask, the veil/stem and filopodia detections, plus the forward filopodia search projections, were combined and the mask dilated using imdilate in MATLAB. Inversion of these combined object masks were used to calculate the mean intensity of the background, and this value was subtracted from the original image. Any negative background-subtracted values were set to 0.
Filopodia/Branch orientation
Orientation for each filopodium is calculated as the angle between the filopodium’s directional vector at the site of veil/stem attachment and the local direction of the veil/stem boundary. Local veil/stem boundary vectors are defined as proceeding from the site of filopodia attachment, toward the neurite’s leading protrusion, in the direction along the veil/stem boundary. Given this definition, the orientation, θ, of the filopodia can assume a value between 0° and 180°, and for a relatively canonical GC architecture, filopodia with a θ value between 0° and 90° (Fig. 5 C, blue arrows) effectively point toward the “front” of the neurite, whereas filopodia with a θ value between 90° and 180° (Fig. 5 C, orange arrows) effectively point toward the “back” of the neurite. Note that a filopodium with a θ of 0° and filopodium with a θ of 180° are both considered fully collapsed onto the veil/stem, and a θ value of 90° indicates a filopodium positioned orthogonally (Fig. 5 C, green arrows) with respect to the local veil/stem. Both 0 and first-order segments were included in this feature distribution.
Likewise, branch filament orientations (Fig. 5 D) are defined as the angle between the two local vectors formed between branch and the root filament at the site of attachment. Root vectors are defined as proceeding from the site of branch attachment toward the tip of the root filament. Given these definitions, a branch lying along its filament stem pointing in the direction of the filament stem’s endpoint maintains a branch orientation, θ, of 0° and a branch lying along the filament stem and away from the filament stem’s endpoint maintains a branch orientation, θ, of 180°. Only second-order branch segments were included in this feature distribution.
Filopodia density
A filopodia/branch segment filter, as detailed in the Materials and methods section Design of filopodia/branch segment filters for feature extraction, was initially applied. However, as an accurate count of filopodia is of primary importance for the density calculation, and some intensity fits for long filopodia can be misleadingly poor due to crossing filopodia/complex decay profiles, this initial filter was relaxed such that any filopodium with a length greater than 5 µm and a mean intensity greater than the 50th percentile of the total distribution of segments was maintained for the density calculation, regardless of its corresponding IFilo value. Filopodia density (Fig. 5 C) was then calculated by dividing the number of filtered filopodia segments directly attached to the veil (branch order = 0 or 1) by the total length of the veil/stem boundary.
Branch density and complexity
Filopodia/branch segment filtering was performed as described in the Filopodia density section. Second-order branch densities (Fig. 5 D) were calculated as the number of second-order branches divided by the first-order segment length. 0-order segments, where branch density by definition equals 0, were excluded from the branch density distribution, as most filopodia segments attached to veil do not branch and hence, when included, these values skew the distribution toward 0. Branch complexity (Fig. 5 D) was defined as the total number of segments after filtering with a branch order greater than 1, divided by the total length of the complete filopodia/branch network.
GCA: Validation
Manual filopodia endpoint localization: Comparison among multiple annotators
Eight individual annotators were given both the image shown in Fig. 2 B (left) and the corresponding raw image. Using the “point” tool in ImageJ, they were asked to identify the endpoint coordinates (i.e., tip) of each filopodia/branch segment indicated by the 22 green dots (manually placed at the center/base of each filopodium). Annotators were allowed to modify the contrast of the raw image in ImageJ as they desired; however, they were not explicitly instructed to do so. The Euclidian distance between each filopodium endpoint annotation and the μFilo coordinate of GCA (Fig. 2 B, right) was calculated (Fig. 2 C). Each filopodium annotation was mapped to the closest point along the automated filopodium linescan (Fig. 2 D). Annotations mapped to linescan coordinates greater than μFilo (along the respective filopodium coordinate system) were assigned a positive value, while annotations mapped to linescan coordinates less than μFilo were assigned a negative value (Fig. 2 C).
While in general we observed good agreement between GCA-detected filopodia endpoint coordinates and manual filopodia endpoint annotations (Fig. 2 C), for select filopodia (Fig. 2 C, i and ii) we observed substantial variation among annotators, particularly for those filopodia exhibiting multiple FI decays along their length (Fig. 2 D, i and ii). In such cases, GCA automated detection often coincided well with the more subtle intensity decay identified by a subset of the eight annotators. Finally, while the endpoint localization appeared to be underestimated in select cases by GCA detection as compared with manual results (Fig. 2 C, iii), such apparent discrepancies typically were again due to semantics of the filopodia endpoint definition (Fig. 2 D, iii). We found these filopodia typically exhibited relatively slow fluorescent decay along their length and hence larger estimated σ values (Fig. 2 D, iii).
Further examination of the automated filopodia linescans marked by the manual annotator as a false positive often revealed ambiguous weak signals within complex environments (Fig. S4 A, vi and vii). Similarly, many false-negative filopodia were associated with complex intensity decays, whereby it became more difficult to quickly isolate an optimal fit region (Fig. S4 B).
Semi-automated validation of GCA filopodia length measurements
260 veil-attached filopodia segments from the N1E-115 dataset were randomly selected (n = 88 filopodia sampled from images of unperturbed GCs and n = 14–32 filopodia sampled per siRNA condition). The filopodia filter was set to the defaults used for the filopodia length feature extractions (default filter: ConnectToVeil_LengthInt: where filoTypes = [0,1]; filterByBundleLength= [0.3,inf]; filterByFit = true; saveFiloByLengthAndSig = [];.embedFitCriteria = 95; filoFitCriteria = 95). Only filopodia segments that passed these criteria were considered in the random sampling.
For each sampled filopodium, the annotator was provided with the intensity decay along the respective filopodium and a zoom corresponding to the raw image and segmentation overlay as in Fig. S4. The annotator could refine the length of the filopodium by moving the detected filopodium endpoint (Fig. S4 A, i, iv, vi, and vii) or the base of the veil/stem (Fig. S4 A, ii and v) along the detected linescan and iteratively evaluating the updated zoomed segmentation overlays until satisfied. Low confidence fits, resulting in false negatives (e.g., Fig. S4 B), were not included in Fig. 2 F, as these failed detections are not included in the final filopodia length distributions collected. An analogous method was used to evaluate the embedded actin bundle length errors in Fig. S5 D, where n = 100 embedded actin bundles were randomly selected from the unperturbed N1E-115 dataset. The semi-automatic filopodia refinement tool was custom-coded in MATLAB. Data corresponding to the automated versus manually refined lengths were fit to a simple linear model with no intercept, using fitlm in MATLAB.
Based on this semi-automatic, curated data, we confirmed the accuracy of the automated length metrics for a majority of the filopodia sampled, even for perturbation conditions, resulting in complex filopodia crossings (Fig. 2 E and Fig. S4 A, iii). Errors in endpoint detection typically arose in cases where the localization of the region for the best fit along the sigmoidal was ambiguous due to the shape of the intensity decay profile (Fig. S4 A, i). Notably, while filopodia length refinements were occasionally necessary due to errors in the veil/stem edge detection, they were often easily manually refined by using the automated information corresponding to the veil-embedded actin structure (Fig. S4, A, ii).
The complex and ambiguous intracellular decay profiles resulted in larger discrepancies in the automated versus manually refined embedded actin bundle lengths (Fig. S5 D) as compared with extra-veil filopodia detection (Fig. S4 A and Fig. S5 D, i, iii, and v).
Post-segmentation calculation of false-positive and false-negative errors
One frame for each movie in the N1E-115 dataset (66 movies total, excluding the acute perturbation movies of Fig. 8) was randomly selected. Filopodia filters were set to the defaults used for the filopodia density feature extraction (default filter: ConnectToVeil_Density: where; filoTypes = [0,1]; filterByBundleLength = [0.3,inf]; filterByFit = true; saveFiloByLengthAndSig = [5 inf; 50 100];.embedFitCriteria = 95; filoFitCriteria = 95). Overlays as in Fig. 2 F along with the raw image were provided to the annotator. The annotator was then asked to click on (1) false-positive and (2) false-negative filopodia in each image, and the corresponding coordinates were recorded. The numbers of GCA filopodia detections, false positives, and false negatives were pooled for all images sampled for a given perturbation condition, and the percentages of false positives and false negatives were calculated such that
and
The post-segmentation validation tool was custom-coded in MATLAB.
The above validation method confirmed a consistent false-positive rate 6–13% and a false-negative rate 6–16.5% for all perturbation images sampled. Further examination of the automated filopodia linescans marked by the manual annotator as a false positive often revealed ambiguous weak signal within complex environments (Fig. S4 A, vi and vii). Similarly, many false-negative filopodia were associated with complex intensity decays, whereby it became more difficult to quickly isolate an optimal fit region (Fig. S4 B).
The false-positive and false-negative error rate for the detection of the embedded actin bundles was slightly higher than for the traditional filopodia detections (18.1% and 17.2%; Fig. S5 A), as it can be more difficult to obtain robust endpoint fits that pass confidence criteria in the final filtering step due to the inhomogeneity/complexity in the intracellular intensity (Fig. S5 B). We note GCA is not designed to detect fully veil-embedded actin bundles (i.e., actin bundles without a corresponding extra-veil filopodia component) as part of the criteria for detection employs one-to-one geometric matching of each filopodium to its potential embedded counterpart (Fig. S5 C). Notably, many of the false-positive actin bundle counts were simply due to an orthogonal problem of incorrect veil/localization in very dense filopodia regions. However, in these cases, the automated full actin bundle length measurement typically remains robust (Fig. S4 A, ii; and Fig. S5 D, iv).
Comparison of GCA to contemporary software packages
We compared the performance of GCA on our N1E-115 and in vivo zebrafish images to two recently released filopodia detection software packages, FiloQuant (Jacquemet et al., 2017) and Filopodyan (Urbančič et al., 2017; Fig. 4, A–C). Notably, in these packages, it was nearly impossible to establish segmentation parameter settings that would successfully translate to other frames/perturbation conditions in the same dataset (Fig. 4 B and Fig. S7). While the filter offered in FiloQuant (Fig. 4, A–C, iii) indeed appeared more sensitive than the thresholding methods offered in Filopodyan (Fig. 4, A–C, ii), the final threshold for this filter needed to be set manually in the package, making its utility quite limited in practice, and it was very difficult to use this enhanced sensitivity without introducing false positives (Fig. S7 E).
After tedious segmentation parameter optimization (Fig. S7, A–E), we found filopodia lengths were frequently underestimated in both packages (Fig. 4 B). Unlike GCA, neither contemporary algorithm resolves branching (Fig. 4 A) or crossing filopodia (Fig. 4 C), or ensures thin regions of the veil/stem are excluded from the filopodia detections (Fig. 4 C). Furthermore, using these images, we show GCA is less prone to filopodia length underestimation due to fragmentation, crossings, and high filopodia density (Fig. 4 E); extracts novel features, such as veil-embedded actin bundles (Fig. 4 F); and generalizes to dendritic filopodia (Fig. 4 G).
Statistical analysis
Correlation analysis: Unperturbed dataset
Net neurite outgrowth rate for each trajectory, as shown in Fig. 6 A, inset, was calculated by recording the difference between the neurite length (Fig. 5 A) at the start and the end of the neurite outgrowth trajectory. This value was then divided by the 10-min total time interval, for which each neurite was imaged. All other neurite movie features were extracted as illustrated in Fig. 5 and as described in the GCA: Feature extraction portion of the Materials and methods. Feature values were pooled for the entire neurite trajectory (Fig. 6 B). The mean value of each pooled distribution was calculated and used for the subsequent Spearman correlation analysis (Fig. 6, C and D). Spearman correlation coefficients and corresponding P values between each set of features were calculated using the corr function in MATLAB. Benjamini and Hochberg–adjusted P values were calculated using the original linear step-up procedure (Benjamini and Hochberg, 1995) via the mafdr function in MATLAB (input, “BHFDR” = true). The full distribution of 253 pairwise P values was used to perform the adjustments.
Hierarchical clustering of MSs
z-Score feature vectors comprising the perturbed condition MSs relative to control were calculated as described in Fig. 7 A. Hierarchical clustering, using unweighted average linkage and a Euclidean distance metric, was performed via the clustergram function in MATLAB.
Clustering of individual GC movies by neurite outgrowth rate
The neurite outgrowth rate in 10 min was calculated as in Figs. 5, 6, and 7 A for n = 66 N1E-115 GC movies, including seven different siRNA perturbation conditions. Using these data, movies were separated into two groups, high and low net outgrowth, via the kmeans function in MATLAB (K = 2, replicates = 20; Fig. 7 B), and for each cluster of movies, selected morphological features were directly compared between siRNA perturbation conditions and the control (Fig. 7 C).
Per filopodia length versus maximum curvature analysis
The length of each filopodium was calculated from its veil attachment point to the filopodium endpoint (defined in this study as μFilo; Fig. 1 A, ix) and the maximum curvature along this length extracted. To reduce potential curvature/length relationships due to branching and/or segmentation errors, only nonbranching filopodia, connected to the veil (branch order = 0), were considered for these analyses. Spearman correlation coefficients and corresponding P values were calculated using the corr function in MATLAB. Benjamini and Hochberg–adjusted correlation coefficient P values were calculated using the original linear step-up procedure (Benjamini and Hochberg, 1995) via the mafdr function in MATLAB (input, “BHFDR” = true). The full distribution of 69 P values was used to perform the adjustments.
Permutation t tests: siRNA data
Each data point in Fig. 7 A, left; Fig. 7 C; and Fig. 7 E represents the distribution mean for the respective GCA feature value pooled over all frames of a single N1E-115 GC movie (5-s intervals; 10-min duration). These values were plotted in MATLAB using notBoxplot.m written by R. Campbell (https://www.mathworks.com/matlabcentral/fileexchange/26508-notboxplot), which plots the individual data points, the mean (bar), the 95% CIs for the mean (dark patch), and ±SD (light patch) for a distribution of values. In Fig. 7 (A, C, and E), the SD estimates the GC-to-GC variation for the respective GC feature calculated per GC movie. A two-tailed permutation t test of the means (1,900 repetitions) implemented in MATLAB was performed between the control and each siRNA treatment distribution. The n values for Fig. 7 A (left) are as follows: control: 20 GC movies, 3,047–8,213 filopodia per movie; Cdc42: 6 GC movies, 2,872–5,607 filopodia per movie; RhoA: 5 GC movies, 2,743–6,298 filopodia per movie; srGAP2: 5 GC movies, 4,052–10,216 filopodia per movie; Rac1: 9 GC movies, 3,794–8,104 filopodia per movie; β-Pix: 9 GC movies, 2,498–6,911 filopodia per movie; Dock7: 6 GC movies, 2,146–5,634 filopodia per movie; and Trio: 3 GC movies, 3,386–4,329 filopodia per movie. (Note the full N1E-115 dataset, corresponding code, and instructions on how to reproduce Fig. 7 A are included with the GCA software release.)
Partitioning of neurite outgrowth trajectory into velocity states
The smoothed derivative of the neurite outgrowth trajectory was found using the csaps function in MATLAB where p, the smoothing parameter, was set to 0.01. Each frame of the trajectory was labeled by its instantaneous velocity such that frames corresponding to positive velocities greater than 0.5 µm/min were defined as growth, frames corresponding to negative velocities less than −0.5 µm/min were labeled as retraction, and velocities between 0.5 and −0.5 µm/min were labeled as pausing. The growth states were further partitioned into accelerating and decelerating by calculating the smoothed second derivative, again using the csaps function.
Acute drug perturbation: Permutation t tests
GCA features, filopodia length or the percent length each actin bundle veil embedded, were extracted using the GCA pipeline for each acutely treated N1E-115 GC movie (Fig. 8 B). For each movie, these values were pooled before and after treatment for each trajectory (Fig. 8 B, ii) and plotted using the boxplot.m function in MATLAB. Two-sided permutation t tests of the medians (10,000 repetitions) were performed comparing the before and after treatment feature distributions for each GC movie (Fig. 8 B, ii).
Per-frame feature dimensionality reduction
The median value for each frame for each of the seven features noted in Fig. 8 D, i, was calculated from the GCA output. Features are as defined in Fig. 5 and the GCA: Feature extraction section of Materials and methods. For the initial time series analysis, any feature selection based on the correlation analysis of the time-averaged data (Fig. 6) was consciously avoided to not exclude more subtle morphology/motility relationships that might be more difficult to detect via the pooled analysis. GCA features that did not maintain an adequate sample size per frame to be calculated robustly (e.g., branch orientation) were not included, nor were features that might be considered trivially correlated (e.g., the correlation between full actin bundle length and filopodia length in Fig. 6 B partially arises due to redundancy of the two metrics). Note, the percentage of each actin bundle veil-embedded feature, as calculated in Fig. 6 (C and D), Fig. 7 A, and Fig. 8 B, yielded an inadequate sample size per frame, as only actin bundles with significant veil embedment were considered. Hence this feature was excluded from the multi-dimensional per-frame analysis. Values for each distribution of features were normalized for scale such that the pooled values over all experimental conditions had a mean of 0 and a SD of 1.
Initially principal component analysis, using the pca function in MATLAB, was used to reduce the dimensionality of the seven features. However, the first two principal components of these data explained only ∼50% of the variability of the original seven features. Therefore, the dimensionality of the per-frame dataset was finally reduced to two dimensions via MDS using the MATLAB mdscale function (Borg, 1997). Kruskal’s stress-1 metric (total error in similarity representation) was computed to be 0.026.
Time series modeling of trajectories in reduced morphological space
HMM selection was performed as described previously (Monnier et al., 2015; Gordonov et al., 2016) using the MATLAB function hmm_process_cell_trajectory.m from the SAPHIRE package (http://saphire-hcs.org). Specifically, 2D shape space MDS coordinates for each GC movie were modeled as emissions et = (xt,yt) from K number of “hidden” shape states, , K = {1,2,3…}. Therefore, for each model, Mk, the full set of parameters, θk, that must be inferred from the data are defined as
where for each shape state (bivariate Gaussian distribution), μi and σi correspond to the state mean and the SD in shape space, respectively; is the probability of transitioning from state si to state sj within the state transition probability matrix; and is the probability of the GC starting state, si at the first time point.
The total marginalized likelihood of each model Mk is defined as
Summation over the hidden state sequences was performed using the forward algorithm, while Metropolis Markov Chain Monte Carlo with importance sampling integration was used to calculate the total marginalized likelihood of each model and corresponding maximum likelihood parameters. The model with the highest marginalized likelihood was chosen, and the most likely hidden shape state sequence was calculated by the Viterbi algorithm using its maximum likelihood parameters. Importantly, this method attempts to find the model that optimizes the number of morphological states based on each GC’s specific reduced-space morphological coordinates while penalizing increasing model complexity (i.e., increasing number of states). The maximum number of hidden shape states, K, tested for each GC movie time series was set to five in this study.
Morphological states were identified for 6 acute perturbation movies (for validation; Fig. 9, A and B) and 20 unperturbed movies (for screening; Fig. 9 D). The unperturbed GC trajectory in Fig. 9 D (ii) was identified computationally by screening among all well-separated state transitions identified by the HMM analysis (i.e., the distance between the mean of the first and the second Gaussian states of the transition is larger than two times the SD of the first state). Specifically, all HMM-identified morphological transitions were screened for switches into a persistent morphological state (longer than 90 s), where the neurite outgrowth state was classified as pausing/retracting for a majority of the state time points (greater than 85%), and accompanied by an increase in the percent time the neurite was pausing/retracting between the two morphology states (at least a 30% delta). Fig. 9 D (ii) shows the only trajectory, of the 20 unperturbed GC movies analyzed, exhibiting a morphology state transition meeting the above criteria.
After HMM, each frame of a respective trajectory is assigned to a given HMM state. Fig. 9 (C and F) was generated by pooling scale-normalized feature data (see Materials and methods, Per-frame feature dimensionality reduction) associated with each HMM state discovered along each respective trajectory. Heat maps show the median corresponding to the feature value distribution for each respective HMM state. The kruskalwallis P values were calculated via the kruskalwallis function in MATLAB. The resulting kruskalwallis stat structure for the filopodia length distributions was then input into MATLAB’s multcompare function to obtain the respective Tukey–Kramer P values.
Bagplot (2D boxplot) overlays
To visualize the localization of multiple individual GC trajectories within the reduced morphological MDS space and to remove potential outlier data points, 2D boxplots (i.e., bagplots) were constructed as described previously (Rousseeuw et al., 1999) using MATLAB code available for download at https://physionet.org/physiotools/ecg-kit/common/LIBRA/bagplot.m (Goldberger et al., 2000). These plots (Fig. 8 D, iii and iv; Fig. 9 D, iii; and Fig. 10, A–G) are 2D versions of the more frequently used one-dimensional boxplot; the inner contour, referred to as the “bag,” encloses 50% of the trajectory’s data points, while the outer contour, referred to as the “fence,” is the convex hull of the trajectory data points in 2D with outliers removed.
Separation statistic
The separation statistic, SS, in Fig. 10 H calculated between the control population and each respective siRNA treatment condition was formulated as an Inverse Davies–Bouldin index (Davies and Bouldin, 1979) such that
Here, ClustDist is the Euclidian distance between the centroid of the control and the centroid of the perturbation. Centroids of the respective populations were calculated from the pooled perturbation (control) per-frame data points. σiPert(Cont) is the Euclidean distance between the centroid of the perturbation (control) and the feature vector corresponding to the ith per-frame data point of the perturbation (control) population. NPert(Cont) is the total number of per-frame data points comprising the respective pooled perturbation/control population. Separation statistics were calculated using the full seven-dimensional feature vector as defined in Fig. 8 D (i). Values for each distribution of features were normalized for feature scale such that a pooled feature distribution over all conditions maintained a mean of 0 and a SD of 1. CIs for each separation calculation were constructed via bootstrapping, i.e., each pair of data was randomly resampled with replacement for 10,000 iterations maintaining the original labels. P value estimates were calculated by a permutation test, i.e., each pair of data was pooled and randomly reassigned group labels for 10,000 iterations to estimate the null distribution.
Online supplemental material
Fig. S1 shows raw image data segmented in Fig. 2 A, definitions of GC parts, and problems associated with global thresholding as a primary image processing method for GCs. Fig. S2 shows an extended flow chart of the GCA algorithm for segmentation, complementing Fig. 1. Fig. S3 details how GCA formulates the filopodia/branch reconstruction as a graph matching problem. Fig. S4 plots the GCA automated linescans corresponding to select veil/stem filopodia from Fig. 2 E as well as a false-negative filopodia resulting from a poor sigmoidal fit. Fig. S5 shows the results from the semi-automated validation of GCA’s embedded actin bundle detection and corresponding length measurements. Fig. S6 shows GCA segmentation and veil velocity analysis applied to previously published, primary mouse and differentiated induced pluripotent stem cell motor neurons. Fig. S7 shows how the segmentation parameters in Fig. 4 (A and B) for previously published software Filopodyan (Urbančič et al., 2017; ii) and FiloQuant (Jacquemet et al., 2017; iii) were chosen using systematic segmentation parameter scanning. Videos 1, 2, and 3 show steps in the segmentation algorithm for a canonical unperturbed GC (Video 1), several noncanonical GC veil/stems (Video 2), and two noncanonical GC filopodia/branch reconstructions (Video 3), respectively. Videos 4, 5, and 6 show GCA filopodia segmentation and local veil velocity overlays for example control (Video 4), Rho GTPase siRNA (Video 5), or select Rac1 GEF/GAP siRNA (Video 6)–treated N1E-115 GCs labeled with GFP LifeAct. Videos 7 and 8 compile movies of GCA filopodia segmentation and either veil/stem thickness (Video 7) or local veil velocity (Video 8) for GCs of varying size, type, and fluorescent label, imaged in vitro, ex vivo, and in vivo. Videos 9 and 10 show dynamic visualizations of how a GC morphology changes over time in reduced morphology space for an N1E-115 GC acutely treated with DMSO (Video 9) or 25 µM CK666 (Video 10). Data points in Videos 9 and 10 are colored by instantaneous neurite outgrowth (top; Fig. 8 E) or HMM morphology state (bottom; Fig. 9 A) classification. Table S1 lists GCA segmentation parameters, default settings, and recommendations. Table S2 provides the siRNA sequences used in this study.
Supplementary Material
Acknowledgments
We are grateful to the following colleagues for providing published and unpublished raw images: Paula Slater (Lowery laboratory); Nick Boyer (Gupton laboratory); James St. John (Griffith University); Rajeeve Sivadasan (Sendtner laboratory); Neset Ozel and Egemen Agi (Hiesinger laboratory); Tadamoto Isogai (Innocenti laboratory); and Kevin Dean (Danuser laboratory).
The project was funded by the Human Frontier Science Program (HFSP RGP0037_2010 to O. Pertz and G. Danuser) and grants from the International Foundation for Research in Paraplegia (to O. Pertz) and the National Institutes of Health (R01 GM067230 to G. Danuser).
The authors declare no competing financial interests.
Author contributions: Conceptualization: O. Pertz, G. Danuser, M.M. Bagonis, and L. Fusco. Data curation: M.M. Bagonis and L. Fusco. Formal analysis: M.M. Bagonis. Funding acquisition: O. Pertz and G. Danuser. Investigation (experimental work): L. Fusco. Methodology: M.M. Bagonis and G. Danuser. Project administration: O. Pertz and G. Danuser. Resources: O. Pertz and G. Danuser. Software: M.M. Bagonis. Validation: M.M. Bagonis. Visualization: M.M. Bagonis. Writing: M.M. Bagonis, G. Danuser, and O. Pertz.
References
- Amano M., Chihara K., Nakamura N., Fukata Y., Yano T., Shibata M., Ikebe M., and Kaibuchi K.. 1998. Myosin II activation promotes neurite retraction during the action of Rho and Rho-kinase. Genes Cells. 3:177–188. 10.1046/j.1365-2443.1998.00181.x [DOI] [PubMed] [Google Scholar]
- Avis D. 1983. A survey of heuristics for the weighted matching problem. Networks. 13:475–493. 10.1002/net.3230130404 [DOI] [Google Scholar]
- Barry D.J., Durkin C.H., Abella J.V., and Way M.. 2015. Open source software for quantification of cell migration, protrusions, and fluorescence intensities. J. Cell Biol. 209:163–180 10.1083/jcb.201501081. 10.1083/jcb.201501081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., and Hochberg Y.. 1995. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Series B Stat. Methodol. 57:289–300. [Google Scholar]
- Borg I. 1997. Modern Multidimensional Scaling: Theory and Applications. Springer-Verlag, New York: 614 pp. [Google Scholar]
- Bray D., and Chapman K.. 1985. Analysis of microspike movements on the neuronal growth cone. J. Neurosci. 5:3204–3213 10.1523/JNEUROSCI.05-12-03204.1985. 10.1523/JNEUROSCI.05-12-03204.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briançon-Marjollet A., Ghogha A., Nawabi H., Triki I., Auziol C., Fromont S., Piché C., Enslen H., Chebli K., Cloutier J.F., et al. 2008. Trio mediates netrin-1-induced Rac1 activation in axon outgrowth and guidance. Mol. Cell. Biol. 28:2314–2323. 10.1128/MCB.00998-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canny J. 1986. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 8:679–698. 10.1109/TPAMI.1986.4767851 [DOI] [PubMed] [Google Scholar]
- Chandran V., Coppola G., Nawabi H., Omura T., Versano R., Huebner E.A., Zhang A., Costigan M., Yekkirala A., Barrett L., et al. 2016. A Systems-Level Analysis of the Peripheral Nerve Intrinsic Axonal Growth Program. Neuron. 89:956–970. 10.1016/j.neuron.2016.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong K.W., Lee A.Y., Koay E.S., Seet S.J., and Cheung N.S.. 2006. pH dependent high transfection efficiency of mouse neuroblastomas using TransFectin. J. Neurosci. Methods. 158:56–63. 10.1016/j.jneumeth.2006.05.017 [DOI] [PubMed] [Google Scholar]
- Costantino S., Kent C.B., Godin A.G., Kennedy T.E., Wiseman P.W., and Fournier A.E.. 2008. Semi-automated quantification of filopodial dynamics. J. Neurosci. Methods. 171:165–173. 10.1016/j.jneumeth.2008.02.009 [DOI] [PubMed] [Google Scholar]
- Czuchra A., Wu X., Meyer H., van Hengel J., Schroeder T., Geffers R., Rottner K., and Brakebusch C.. 2005. Cdc42 is not essential for filopodium formation, directed migration, cell polarization, and mitosis in fibroblastoid cells. Mol. Biol. Cell. 16:4473–4484. 10.1091/mbc.e05-01-0061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D.L., and Bouldin D.W.. 1979. A cluster separation measure. IEEE Trans. Pattern Anal. Mach. Intell. 1:224–227. 10.1109/TPAMI.1979.4766909 [DOI] [PubMed] [Google Scholar]
- Debant A., Serra-Pagès C., Seipel K., O’Brien S., Tang M., Park S.H., and Streuli M.. 1996. The multidomain protein Trio binds the LAR transmembrane tyrosine phosphatase, contains a protein kinase domain, and has separate rac-specific and rho-specific guanine nucleotide exchange factor domains. Proc. Natl. Acad. Sci. USA. 93:5466–5471. 10.1073/pnas.93.11.5466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demchouk A.O., Gardner M.K., and Odde D.J.. 2011. Microtubule Tip Tracking and Tip Structures at the Nanometer Scale Using Digital Fluorescence Microscopy. Cell. Mol. Bioeng. 4:192–204. 10.1007/s12195-010-0155-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent E.W., Gupton S.L., and Gertler F.B.. 2011. The growth cone cytoskeleton in axon outgrowth and guidance. Cold Spring Harb. Perspect. Biol. 3:a001800 10.1101/cshperspect.a001800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobramysl U., Papoian G.A., and Erban R.. 2016. Steric Effects Induce Geometric Remodeling of Actin Bundles in Filopodia. Biophys. J. 110:2066–2075. 10.1016/j.bpj.2016.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangi A., Niessen W., Vincken K., and Viergever M.. 1998. Multiscale vessel enhancement. Lecture Notes in Computer Science. Springer-Verlag, Berlin, Germany: 130–137. [Google Scholar]
- Fritz R.D., Letzelter M., Reimann A., Martin K., Fusco L., Ritsma L., Ponsioen B., Fluri E., Schulte-Merker S., van Rheenen J., and Pertz O.. 2013. A versatile toolkit to produce sensitive FRET biosensors to visualize signaling in time and space. Sci. Signal. 6:rs12 10.1126/scisignal.2004135 [DOI] [PubMed] [Google Scholar]
- Fusco L., Lefort R., Smith K., Benmansour F., Gonzalez G., Barillari C., Rinn B., Fleuret F., Fua P., and Pertz O.. 2016. Computer vision profiling of neurite outgrowth dynamics reveals spatiotemporal modularity of Rho GTPase signaling. J. Cell Biol. 212:91–111. 10.1083/jcb.201506018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberger A.L., Amaral L.A., Glass L., Hausdorff J.M., Ivanov P.C., Mark R.G., Mietus J.E., Moody G.B., Peng C.K., and Stanley H.E.. 2000. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation. 101:E215–E220. 10.1161/01.CIR.101.23.e215 [DOI] [PubMed] [Google Scholar]
- Goodhill G.J., Faville R.A., Sutherland D.J., Bicknell B.A., Thompson A.W., Pujic Z., Sun B., Kita E.M., and Scott E.K.. 2015. The dynamics of growth cone morphology. BMC Biol. 13:10 10.1186/s12915-015-0115-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordonov S., Hwang M.K., Wells A., Gertler F.B., Lauffenburger D.A., and Bathe M.. 2016. Time series modeling of live-cell shape dynamics for image-based phenotypic profiling. Integr. Biol. 8:73–90. 10.1039/C5IB00283D [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier S., Coutinho-Budd J., Sassa T., Gresset A., Jordan N.V., Chen K., Jin W.L., Frost A., and Polleux F.. 2009. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 138:990–1004. 10.1016/j.cell.2009.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., and Lalli G.. 2010. Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb. Perspect. Biol. 2:a001818 10.1101/cshperspect.a001818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N.E., and Shen S.S.P.. 2014. Hilbert–Huang Transform and Its Applications. World Scientific Publishing Company, Singapore: 400 pp. [Google Scholar]
- Hyland C., Mertz A.F., Forscher P., and Dufresne E.. 2014. Dynamic peripheral traction forces balance stable neurite tension in regenerating Aplysia bag cell neurons. Sci. Rep. 4:4961 10.1038/srep04961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob M., and Unser M.. 2004. Design of steerable filters for feature detection using canny-like criteria. IEEE Trans. Pattern Anal. Mach. Intell. 26:1007–1019. 10.1109/TPAMI.2004.44 [DOI] [PubMed] [Google Scholar]
- Jacquemet G., Paatero I., Carisey A.F., Padzik A., Orange J.S., Hamidi H., and Ivaska J.. 2017. FiloQuant reveals increased filopodia density during breast cancer progression. J. Cell Biol. 216:3387–3403 10.1083/jcb.201704045. 10.1083/jcb.201704045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K., Loerke D., Mettlen M., Kuwata H., Grinstein S., Schmid S.L., and Danuser G.. 2008. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 5:695–702. 10.1038/nmeth.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleitman N., and Johnson M.I.. 1989. Rapid growth cone translocation on laminin is supported by lamellipodial not filopodial structures. Cell Motil. Cytoskeleton. 13:288–300. 10.1002/cm.970130407 [DOI] [PubMed] [Google Scholar]
- Kolmogorov V. 2009. Blossom V: a new implementation of a minimum cost perfect matching algorithm. Math. Program. Comput. 1:43–67. 10.1007/s12532-009-0002-8 [DOI] [Google Scholar]
- Kozma R., Sarner S., Ahmed S., and Lim L.. 1997. Rho family GTPases and neuronal growth cone remodelling: relationship between increased complexity induced by Cdc42Hs, Rac1, and acetylcholine and collapse induced by RhoA and lysophosphatidic acid. Mol. Cell. Biol. 17:1201–1211. 10.1128/MCB.17.3.1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubek M. 2016. Challenges and benchmarks in bioimage analysis. In Focus on Bio-Image Informatics. Vol. 219.De Vos W.H., Munck S., and Timmermans J., editors. Springer, Cham, Switzerland: 231–262. [DOI] [PubMed] [Google Scholar]
- Langen M., Agi E., Altschuler D.J., Wu L.F., Altschuler S.J., and Hiesinger P.R.. 2015. The Developmental Rules of Neural Superposition in Drosophila. Cell. 162:120–133. 10.1016/j.cell.2015.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery L.A., and Van Vactor D.. 2009. The trip of the tip: Understanding the growth cone machinery. Nat. Rev. Mol. Cell Biol. 10:332–343. 10.1038/nrm2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Dagliyan O., Hahn K.M., and Danuser G.. 2018. Profiling cellular morphodynamics by spatiotemporal spectrum decomposition. PLOS Comput. Biol. 14:e1006321 10.1371/journal.pcbi.1006321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machacek M., and Danuser G.. 2006. Morphodynamic profiling of protrusion phenotypes. Biophys. J. 90:1439–1452. 10.1529/biophysj.105.070383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitre H. 2008. Image Processing. ISTE Ltd and John Wiley & Sons Inc., London: 352 pp. [Google Scholar]
- Mason C.A., and Wang L.C.. 1997. Growth cone form is behavior-specific and, consequently, position-specific along the retinal axon pathway. J. Neurosci. 17:1086–1100. 10.1523/JNEUROSCI.17-03-01086.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza M.C., Vilela M., Juarez J.E., Blenis J., and Danuser G.. 2015. ERK reinforces actin polymerization to power persistent edge protrusion during motility. Sci. Signal. 8:ra47 10.1126/scisignal.aaa8859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H., Suetsugu S., and Takenawa T.. 1998. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 17:6932–6941. 10.1093/emboj/17.23.6932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misiak D., Posch S., Lederer M., Reinke C., Hüttelmaier S., and Möller B.. 2014. Extraction of protein profiles from primary neurons using active contour models and wavelets. J. Neurosci. Methods. 225:1–12 10.1016/j.jneumeth.2013.12.009. 10.1016/j.jneumeth.2013.12.009 [DOI] [PubMed] [Google Scholar]
- Mogilner A., and Rubinstein B.. 2005. The physics of filopodial protrusion. Biophys. J. 89:782–795. 10.1529/biophysj.104.056515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongiu A.K., Weitzke E.L., Chaga O.Y., and Borisy G.G.. 2007. Kinetic-structural analysis of neuronal growth cone veil motility. J. Cell Sci. 120:1113–1125. 10.1242/jcs.03384 [DOI] [PubMed] [Google Scholar]
- Monnier N., Barry Z., Park H.Y., Su K.C., Katz Z., English B.P., Dey A., Pan K., Cheeseman I.M., Singer R.H., and Bathe M.. 2015. Inferring transient particle transport dynamics in live cells. Nat. Methods. 12:838–840. 10.1038/nmeth.3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niblack W. 1985. An introduction to digital image processing. Strandberg Publishing Company, Birkerod, Denmark: 215 pp. [Google Scholar]
- Nilufar S., Morrow A.A., Lee J.M., and Perkins T.J.. 2013. FiloDetect: Automatic detection of filopodia from fluorescence microscopy images. BMC Syst. Biol. 7:66 10.1186/1752-0509-7-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes C.D., and Hall A.. 1995. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 81:53–62. 10.1016/0092-8674(95)90370-4 [DOI] [PubMed] [Google Scholar]
- Nolen B.J., Tomasevic N., Russell A., Pierce D.W., Jia Z., McCormick C.D., Hartman J., Sakowicz R., and Pollard T.D.. 2009. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 460:1031–1034. 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu N. 1979. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man Cybern. 9:62–66. 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- Özel M.N., Langen M., Hassan B.A., and Hiesinger P.R.. 2015. Filopodial dynamics and growth cone stabilization in Drosophila visual circuit development. eLife. 4:e10721 10.7554/eLife.10721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertz O.C., Wang Y., Yang F., Wang W., Gay L.J., Gristenko M.A., Clauss T.R., Anderson D.J., Liu T., Auberry K.J., et al. 2008. Spatial mapping of the neurite and soma proteomes reveals a functional Cdc42/Rac regulatory network. Proc. Natl. Acad. Sci. USA. 105:1931–1936. 10.1073/pnas.0706545105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., and Buckley T.R.. 2004. Model selection and model averaging in phylogenetics: advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst. Biol. 53:793–808. 10.1080/10635150490522304 [DOI] [PubMed] [Google Scholar]
- Prim R.C. 1957. Shortest Connection Networks And Some Generalizations. Bell Syst. Tech. J. 36:1389–1401. 10.1002/j.1538-7305.1957.tb01515.x [DOI] [Google Scholar]
- Ren Y., and Suter D.M.. 2016. Increase in Growth Cone Size Correlates with Decrease in Neurite Growth Rate. Neural Plast. 2016:3497901 10.1155/2016/3497901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseeuw P.J., and Leroy A.M.. 1987. Robust regression and outlier detection. John Wiley & Sons, Inc, New York: 329 pp. [Google Scholar]
- Rousseeuw P.J., Ruts I., and Tukey J.W.. 1999. The Bagplot: A Bivariate Boxplot. Am. Stat. 53:382–387. 10.1080/00031305.1999.10474494 [DOI] [Google Scholar]
- Saha T., Rathmann I., Viplav A., Panzade S., Begemann I., Rasch C., Klingauf J., Matis M., and Galic M.. 2016. Automated analysis of filopodial length and spatially resolved protein concentration via adaptive shape tracking. Mol. Biol. Cell. 27:3616–3626. 10.1091/mbc.e16-06-0406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarner S., Kozma R., Ahmed S., and Lim L.. 2000. Phosphatidylinositol 3-kinase, Cdc42, and Rac1 act downstream of Ras in integrin-dependent neurite outgrowth in N1E-115 neuroblastoma cells. Mol. Cell. Biol. 20:158–172. 10.1128/MCB.20.1.158-172.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivadasan R., Hornburg D., Drepper C., Frank N., Jablonka S., Hansel A., Lojewski X., Sterneckert J., Hermann A., Shaw P.J., et al. 2016. C9ORF72 interaction with cofilin modulates actin dynamics in motor neurons. Nat. Neurosci. 19:1610–1618. 10.1038/nn.4407 [DOI] [PubMed] [Google Scholar]
- Steketee M.B., Oboudiyat C., Daneman R., Trakhtenberg E., Lamoureux P., Weinstein J.E., Heidemann S., Barres B.A., and Goldberg J.L.. 2014. Regulation of intrinsic axon growth ability at retinal ganglion cell growth cones. Invest. Ophthalmol. Vis. Sci. 55:4369–4377. 10.1167/iovs.14-13882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John J.A., and Key B.. 2012. HuC-eGFP mosaic labelling of neurons in zebrafish enables in vivo live cell imaging of growth cones. J. Mol. Histol. 43:615–623. 10.1007/s10735-012-9462-7 [DOI] [PubMed] [Google Scholar]
- Suo D., Park J., Young S., Makita T., and Deppmann C.D.. 2015. Coronin-1 and calcium signaling governs sympathetic final target innervation. J. Neurosci. 35:3893–3902. 10.1523/JNEUROSCI.4402-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirovic S., Hellal F., Neukirchen D., Hindges R., Garvalov B.K., Flynn K.C., Stradal T.E., Chrostek-Grashoff A., Brakebusch C., and Bradke F.. 2010. Rac1 regulates neuronal polarization through the WAVE complex. J. Neurosci. 30:6930–6943. 10.1523/JNEUROSCI.5395-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa T., and Miki H.. 2001. WASP and WAVE family proteins: Key molecules for rapid rearrangement of cortical actin filaments and cell movement. J. Cell Sci. 114:1801–1809. [DOI] [PubMed] [Google Scholar]
- Takenawa T., and Suetsugu S.. 2007. The WASP-WAVE protein network: Connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8:37–48. 10.1038/nrm2069 [DOI] [PubMed] [Google Scholar]
- Tárnok K., Gulyás M., Bencsik N., Ferenc K., Pfizenmaier K., Hausser A., and Schlett K.. 2015. A new tool for the quantitative analysis of dendritic filopodial motility. Cytometry A. 87:89–96. 10.1002/cyto.a.22569 [DOI] [PubMed] [Google Scholar]
- ten Klooster J.P., Jaffer Z.M., Chernoff J., and Hordijk P.L.. 2006. Targeting and activation of Rac1 are mediated by the exchange factor beta-Pix. J. Cell Biol. 172:759–769. 10.1083/jcb.200509096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsygankov D., Bilancia C.G., Vitriol E.A., Hahn K.M., Peifer M., and Elston T.C.. 2014. CellGeo: A computational platform for the analysis of shape changes in cells with complex geometries. J. Cell Biol. 204:443–460. 10.1083/jcb.201306067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbančič V., Butler R., Richier B., Peter M., Mason J., Livesey F.J., Holt C.E., and Gallop J.L.. 2017. Filopodyan: An open-source pipeline for the analysis of filopodia. J. Cell Biol. 216:3405–3422 10.1083/jcb.201705113. 10.1083/jcb.201705113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M., John K.A., Janas J.A., Newey S.E., and Van Aelst L.. 2006. The Rac activator DOCK7 regulates neuronal polarity through local phosphorylation of stathmin/Op18. Neuron. 51:727–739. 10.1016/j.neuron.2006.07.020 [DOI] [PubMed] [Google Scholar]
- Welf E.S., Driscoll M.K., Dean K.M., Schäfer C., Chu J., Davidson M.W., Lin M.Z., Danuser G., and Fiolka R.. 2016. Quantitative Multiscale Cell Imaging in Controlled 3D Microenvironments. Dev. Cell. 36:462–475. 10.1016/j.devcel.2016.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Zhang X.F., Pollard T.D., and Forscher P.. 2012. Arp2/3 complex-dependent actin networks constrain myosin II function in driving retrograde actin flow. J. Cell Biol. 197:939–956. 10.1083/jcb.201111052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-F., Schaefer A.W., Burnette D.T., Schoonderwoert V.T., and Forscher P.. 2003. Rho-dependent contractile responses in the neuronal growth cone are independent of classical peripheral retrograde actin flow. Neuron. 40:931–944. 10.1016/S0896-6273(03)00754-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.