Neuronal morphology is maintained over long animal lifespans. Noblett et al. show that in Caenorhabditis elegans, DIP-2 acts to inhibit neurite outgrowth during aging and after axotomy, indicating that the maintenance of neuronal morphology and inhibition of axon regeneration may share a common mechanism.
Abstract
Neuronal morphology and circuitry established during early development must often be maintained over the entirety of animal lifespans. Compared with neuronal development, the mechanisms that maintain mature neuronal structures and architecture are little understood. The conserved disco-interacting protein 2 (DIP2) consists of a DMAP1-binding domain and two adenylate-forming domains (AFDs). We show that the Caenorhabditis elegans DIP-2 maintains morphology of mature neurons. dip-2 loss-of-function mutants display a progressive increase in ectopic neurite sprouting and branching during late larval and adult life. In adults, dip-2 also inhibits initial stages of axon regeneration cell autonomously and acts in parallel to DLK-1 MAP kinase and EFA-6 pathways. The function of DIP-2 in maintenance of neuron morphology and in axon regrowth requires its AFD domains and is independent of its DMAP1-binding domain. Our findings reveal a new conserved regulator of neuronal morphology maintenance and axon regrowth after injury.
Introduction
Neurons exhibit diverse morphologies that are important for proper connectivity and information processing. Morphogenesis of neurons involves specification of axon and dendritic processes, elaboration of dendritic arbors, and wiring and refinement of neuronal connections (Jan and Jan, 2010). Adult neurons are long lived and, although capable of plasticity-promoting remodeling of dendritic and synaptic structures, maintain relatively stable morphologies over the adult lifespan. Both intrinsic and extrinsically acting molecules have distinct roles in the maintenance of neuronal morphology. However, despite extensive studies, the mechanisms that maintain neuronal morphology and circuitry over long lifespans remain poorly understood. Moreover, regulation of such mechanisms may underlie adaptive responses that control neurite outgrowth or regeneration in response to axon damage.
The disco-interacting protein 2 (DIP2) family is conserved from Caenorhabditis elegans to mammals and contains a DNA methyltransferase-associated protein 1 (DMAP1) binding domain and two class I superfamily adenylate-forming domains (AFDs; Mukhopadhyay et al., 2002). AFDs, most commonly found in acyl-CoA synthetases in higher eukaryotes, activate fatty acids as acyl-adenylates during fatty acid metabolism (Schmelz and Naismith, 2009). DIP2 was first identified as a protein binding to the nuclear factor Disconnected (Disco) in Drosophila melanogaster (Mukhopadhyay et al., 2002), but the in vivo roles of the DIP2 family remain little understood. Humans and other mammals have three DIP2 genes (DIP2A–C). Human DIP2A was reported to be a candidate cell surface receptor for the secreted glycoprotein follistatin-like 1 (FSTL1) in endothelial cells (Ouchi et al., 2010). Mouse DIP2B is associated with methylated DNA in mitotic fetal epithelial progenitor cells during organogenesis (Hayashi et al., 2017). Drosophila has one DIP2 gene, recently shown to regulate stereotypical axon bifurcation of mushroom body neurons (Nitta et al., 2017).
In this study, we report that C. elegans DIP-2 functions postdevelopmentally to maintain neuronal morphology. Loss of dip-2 function results in progressive age-dependent increase in ectopic neurite formation. Overexpression of DIP-2 suppresses the normal progressive neuronal sprouting observed in aging C. elegans. Moreover, DIP-2 acts in mature neurons to inhibit axon regeneration after laser injury. Our findings reveal a common mechanism that restrains neurite outgrowth in mature neurons during aging and after axon injury.
Results and discussion
C. elegans DIP-2 regulates neuronal morphology
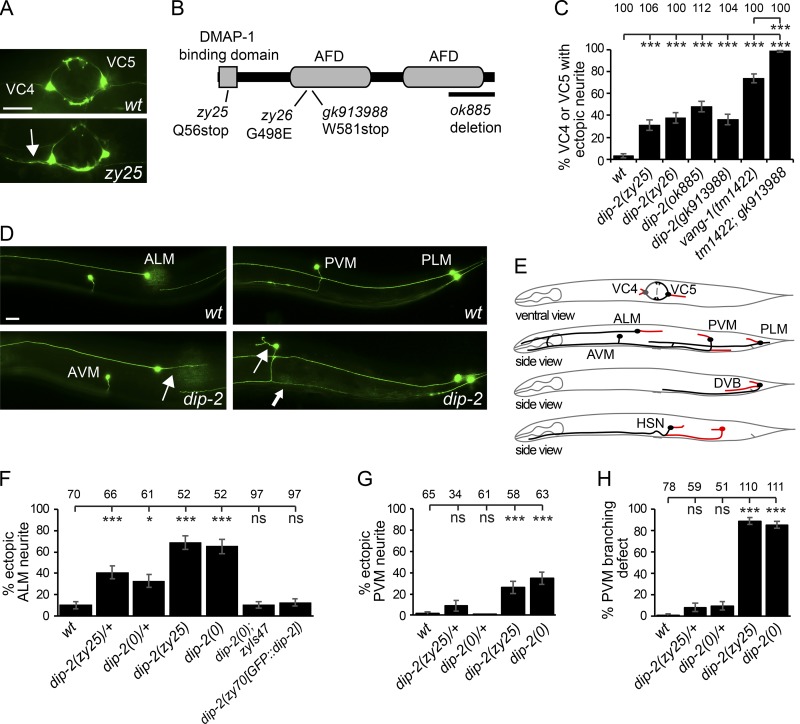
We previously reported the isolation of nde-5 mutants based on aberrant morphology of ventral cord (VC) motor neurons (Carr et al., 2016). In these mutants, the normally bipolar VC4 and VC5 neurons display additional neurite-like outgrowth from their cell bodies, resulting in distinct tripolar morphologies (Fig. 1 A). We performed whole-genome sequencing and found that nde-5 alleles affected the C. elegans orthologue of DIP2 (Fig. 1 B), hence we renamed the gene to dip-2. Transcriptomic data reveal several mRNAs produced from the dip-2 locus, encoding several protein isoforms with or without the DMAP1-binding domain (Lee et al., 2018). We confirmed one of these transcripts to encode an ORF of 1,681 amino acids including all conserved domains (Fig. 1 B). Two independently derived mutants, gk913988 and ok885, displayed similar VC neuronal morphology defects (Fig. 1 C). The neuronal defects of dip-2 mutants resemble those in planar cell polarity (PCP) pathway mutants such as vang-1/Van Gogh (Sanchez-Alvarez et al., 2011). We made double mutants of dip-2 and vang-1 and found the ectopic neurite defects to be further enhanced (Fig. 1 C), suggesting that dip-2 acts in a pathway distinct from PCP to block ectopic neurite formation. Because gk913988 is predicted to disrupt most if not all DIP-2 isoforms, we designate it as a null allele, dip-2(0). Apart from a defect in egg-laying (below), dip-2 mutants show normal growth and movement. In this study, we focus on the role of dip-2 in neurons.
Figure 1.
dip-2 regulates neuronal morphology and migration. (A) VC4 and VC5 neurons in WT and a zy25 mutant. Arrow points to ectopic neurite from VC4. (B) DIP-2 protein schematic showing domain organization and mutations. (C) Quantification of VC morphology defects in Dip-2 mutants. (D) Mechanosensory neuron images. (E) Worm schematics summarizing dip-2 neuronal morphology and migration defects (red). (F–H) Quantification of Dip-2 neuronal morphology defects in ALM (F) and PVM (G and H). dip-2 mutants display ectopic neurites (arrows) from cell bodies and axon branching (thick arrow in D) defects. Bars, 20 µm. Error bars indicate SEM of proportion (n = 51–112). Significance compared with WT using one-way ANOVA with Tukey post hoc test. *, P < 0.05; ***, P < 0.001.
To determine the extent of neuronal morphological defects in dip-2 mutants, we examined additional types of neurons including mechanosensory neurons (ALML/R, AVM, PVM, and PLML/R) and the DVB and HSN(L/R) motor neurons (see Materials and methods). In WT animals, these neurons have simple and mostly unipolar morphologies. In young adult dip-2(0) mutants, most of these neurons displayed ectopic neurites from cell bodies, like those in VC4 and VC5 (Fig. 1, D–G; and Fig. S1, A–D). PVM, which sends a single axon ventrally to the ventral nerve cord (VNC) and then anteriorly, was an exception as it also showed highly penetrant axon branching such that axons bifurcated at the VNC and extended a short ectopic posterior branch (Fig. 1, D and H). dip-2(0) mutations displayed partial dominance for ectopic neurites in ALM but not in other neurons (Fig. 1, F–H), suggesting that some neurons may be more sensitive to DIP-2 levels.
In dip-2(0) mutants, HSN neurons displayed incomplete migration (Fig. S1, C and E). It is possible that the egg-laying defect in dip-2(0) mutants is a result of defects in VC morphology and HSN migration. Despite penetrant neurite sprouting and cell migration defects (summarized in Fig. 1 E), dip-2 mutants displayed mostly normal axon pathfinding. Similarly, loss of function in Drosophila DIP2 does not generally affect axon guidance but results in ectopic axon branching (Nitta et al., 2017). Combined, these findings suggest that DIP2 proteins play evolutionarily conserved roles in neuronal development, particularly in ensuring stereotypic morphology by blocking ectopic protrusions from cell bodies or axons.
DIP-2 maintains the morphology of mature neurons
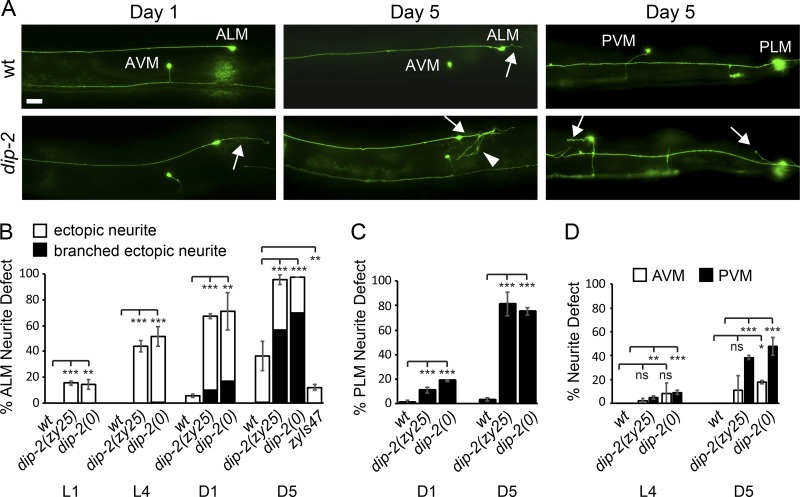
Neuronal morphology defects of dip-2 mutants were both more frequent and more severe in older adults compared with younger adults, and were more penetrant in animals cultured at 25°C compared with 20°C. As zy25 and gk913988 result in premature stop codons, leading to strong loss-of-function or null mutations, this temperature sensitivity is unlikely to be an effect of temperature-labile proteins. To quantify these effects, we examined mechanosensory neuron morphology from newly hatched first larval stage (L1) to day 5 (D5) adults in animals cultured at 20°C (Fig. S1 F) and 25°C (Fig. 2). ALM and PLM neurons are born embryonically and have undergone primary axon outgrowth and guidance before the L1 stage. AVM and PVM neurons arise postembryonically and attain their final positions and morphologies by L2 stage (Chalfie and Sulston, 1981). In dip-2(0) mutants, the percentage of ALMs displaying ectopic neurites increased from ∼14% in L1 to 71% and 98% in D1 and D5 adults, respectively (Fig. 2, A and B) compared with 36% ectopic neurites in ALMs of WT D5 adults. A similar progressive increase in axon morphology defects was seen in PLM, AVM, and PVM neurons (Fig. 2, C and D). Strikingly, ectopic ALM neurites in dip-2(0) mutants underwent secondary branching or developed arbor-like morphologies (Fig. 2 A) with increasing age (Fig. 2 B). This contrasts with the simple unbranched ectopic neurites associated with aging in the WT (Fig. 2 B; Pan et al., 2011; Tank et al., 2011; Toth et al., 2012). Notably, transgenic overexpression of DIP-2 (zyIs47) suppressed age-dependent sprouting in a WT background such that only 12% of ALMs displayed ectopic neurites in D5 adults (Fig. 2 B), suggesting that the ectopic neurites in aging WT adults might reflect a decline in dip-2 function.
Figure 2.
dip-2 mutants display an age-dependent increase in neuronal morphology defects. (A) Representative images of mechanosensory neuron morphology in WT and dip-2 mutants at D1 and D5. Arrows indicate ectopic neurites. Bar, 20 µm. (B) ALM neurons in dip-2 mutants display a cumulative increase in ectopic neurites with increasing age compared with a lesser increase in WT animals. Some ectopic ALM neurites in dip-2 mutants show additional age-dependent secondary branching (arrowhead in A). (C and D) PLM, AVM, and PVM also display age-dependent increases in ectopic neurites. Error bars indicate SD for data collected from three independent counts of 40–75 ALM or PLM and 15–40 AVM or PVM neurons. Significance compared with WT at each life stage using one-way ANOVA with Tukey post hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
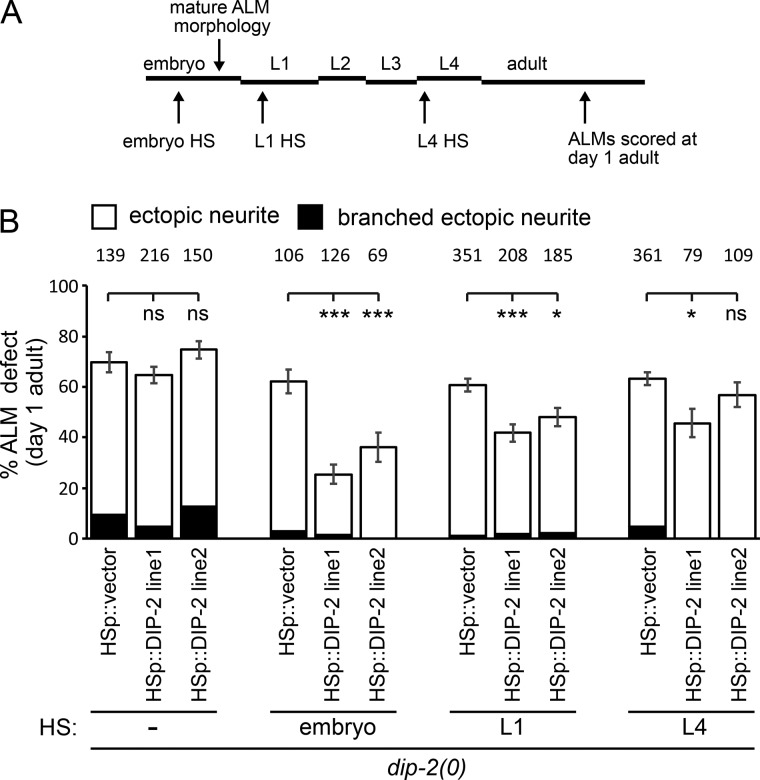
To determine whether DIP-2 acts postdevelopmentally to maintain neuronal morphology, we asked whether restoration of DIP-2 expression in dip-2(0) larvae mitigated the ALM ectopic neurite phenotype in adult animals. We used a heat shock (HS)-inducible promoter to drive DIP-2 expression with temporal control (see Materials and methods). We found that HS-induced (2 h at 35°C) DIP-2 expression in dip-2 mutants at the L1, and to a lesser extent, the L4 stage resulted in a partial but significant reduction in ectopic neurites in D1 adults (Fig. 3). We conclude that DIP-2 can act postdevelopmentally to maintain neuronal morphology.
Figure 3.
dip-2 maintains neuronal morphology in postembryonic stages. (A) Schematic indicating HS at embryonic, L1, or L4 stages in dip-2 mutants to trigger HS promoter–inducible DIP-2 expression and the quantification of ALM morphology defects in D1 adults. (B) In dip-2 mutants, HS-induced DIP-2 expression at embryonic and larval stages resulted in significantly less severe ALM defects in D1 adults compared with vector-only animals. Error bars indicate SEM of proportion (n = 69–361). Significance compared with vector-only worms using one-way ANOVA with Tukey post hoc test. *, P < 0.05; ***, P < 0.001.
DIP-2 localizes to the cytosol of neurons and acts cell autonomously
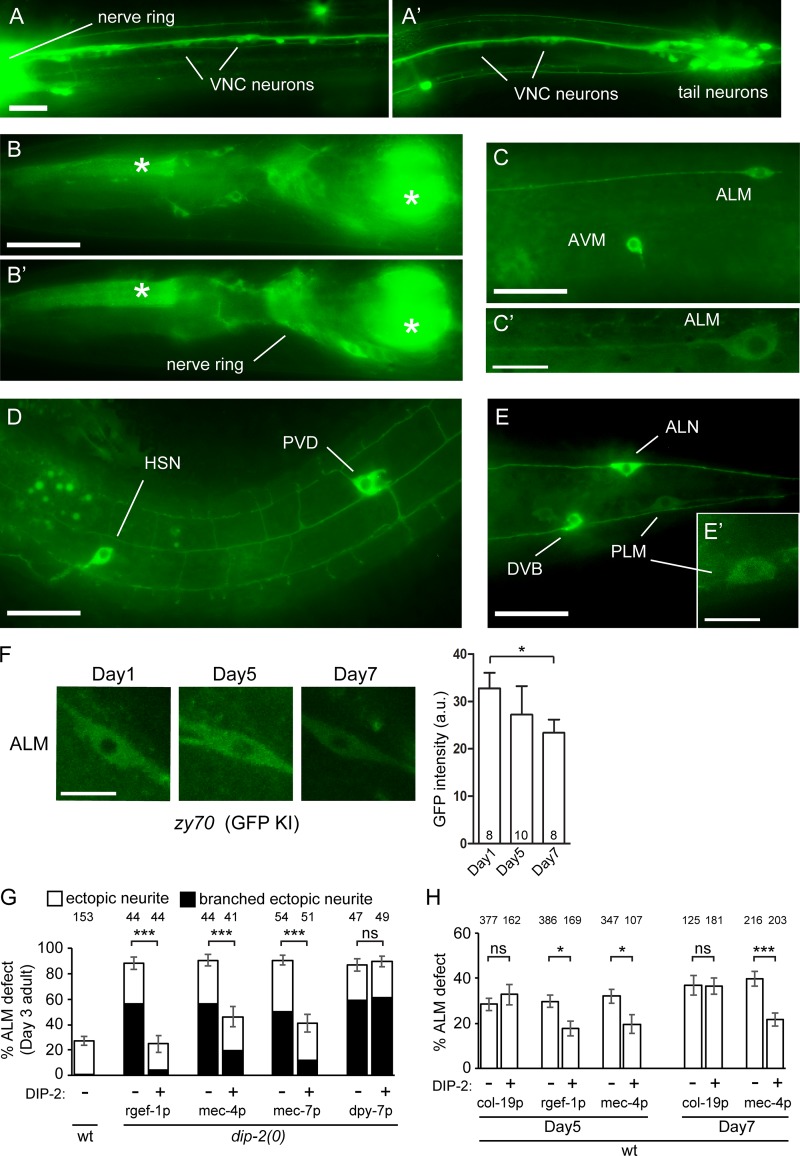
To determine where DIP-2 is expressed and localized, we generated transgenic lines expressing a transcriptional reporter (zyEx44) or a translational fusion protein with GFP at the C terminus (zyIs47) driven by ∼2.5-kb upstream regulatory sequences (see Materials and methods). We also used genome editing (Dickinson et al., 2015) to insert GFP at the start of the endogenous dip-2 gene (zy70). DIP-2::GFP expression from zyIs47 rescued Dip-2 defects, and zy70[GFP::DIP-2] displayed normal neuronal morphology (Fig. 1 F), indicating that GFP fusions to either the N or C terminus do not disrupt DIP-2.
Expression from these three DIP-2 reporters was observed in multiple tissues in different developmental stages. In larvae and adults, dip-2 was expressed in most neurons including VNC motor neurons, HSN, PVD, and mechanosensory neurons (Fig. 4, A–E; and Fig. S2 A) as well as a subset of epidermal cells (Fig. S2 B). In neurons of larvae and adults, both transgenic DIP-2::GFP and endogenous GFP::DIP-2 proteins showed cytoplasmic localization in axons and cell somas and were mostly excluded from nuclei (Fig. 4, B–E; and Fig. S2 A). Cytoplasmic levels of GFP::DIP-2 (zy70) in neurons showed a decreasing trend with age (Fig. 4 F), consistent with our findings that DIP-2 overexpression can suppress age-dependent neuronal morphology defects in WT animals. Interestingly, however, in epidermal cells, transgenic and endogenous DIP-2 GFP fusions were localized to the membrane from embryo to late larvae (Fig. S2 B). Such differential subcellular localization of DIP-2 suggests that DIP-2 may function via divergent molecular mechanisms in different cell types.
Figure 4.
dip-2 is expressed in neurons and acts cell autonomously. (A) Ventral view of an L4 worm showing transcriptional dip-2p::GFP (zyEx44) expression in VNC neurons (anterior [A] and posterior [A′] segments from same worm). (B–E) Representative images of full-length DIP-2::GFP (zyIs47) expression in L4, showing cytoplasmic DIP-2::GFP localization in head (lateral [B] and medial [B′] focal planes from same worm) as well as neurons in midbody (C and D) and tail (E). Asterisks in B and B′ indicate myo-2p::mCherry coinjection marker bleedthrough in pharyngeal muscle. (C′ and E′) Representative images of endogenous GFP::DIP-2 (zy70) expression showing cytoplasmic localization in mechanosensory neurons. (F) Endogenous GFP::DIP-2 (zy70) in mechanosensory neurons remains cytoplasmic with increasing age. Left: Representative confocal images of ALM in D1, D5, and D7 animals. Right: Quantification of DIP-2 levels in ALM. Arbitrary units represent GFP intensity levels (mean ± SEM). Bars: 20 µm (A–E); 10 µm (C′, E′, and F). (G and H) Neuron-specific expression of DIP-2 rescues neuronal morphology defects in dip2(0) (G) and aged WT (H) animals. Error bars (G and H) indicate SEM of proportion (n = 41–386). All statistics, two-tailed t tests. *, P < 0.05; ***, P < 0.001.
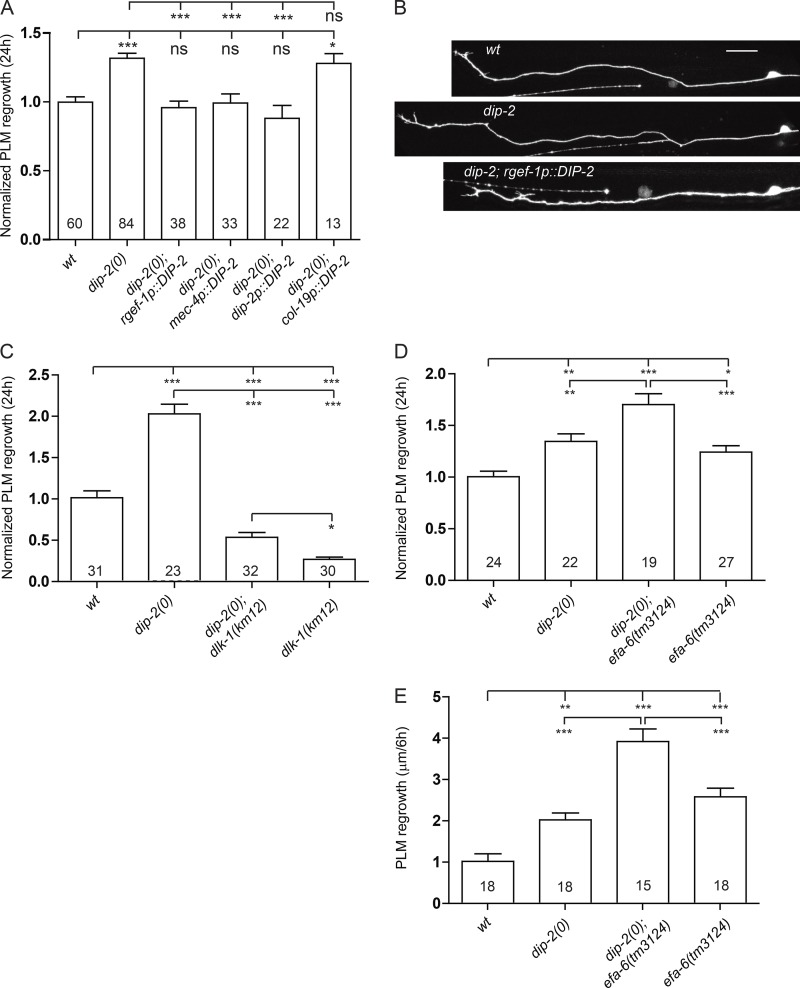
We next asked whether DIP-2 was required cell autonomously to regulate mechanosensory neuron morphology and HSN migration. We generated transgenic lines in which dip-2 was expressed from panneuronal (Prgef-1 or Punc-33), mechanosensory neuron (Pmec-4 or Pmec-7), or panepithelial (Pdpy-7 or Pcol-19) promoters and assessed them for their ability to rescue neuronal defects in dip-2(0) mutants and aged WT animals. dip-2 expression in neurons but not in the epidermis rescued dip-2 ALM morphology defects (Fig. 4 G), suppressed ectopic sprouting in WT D5 and D7 adults (Fig. 4 H), and rescued dip-2 HSN migration defects (Fig. S1 G), consistent with dip-2 acting in a cell autonomous manner.
DIP-2 function in neurons depends on AFD, not on its DMAP1-binding domain
AFD domain–containing enzymes such as acyl-CoA synthetases are present in many compartments of the cytoplasm and can also associate with membranes (Coleman et al., 2002). Drosophila DIP2 mutants have decreased acyl-CoA levels, suggesting a role in fatty acid metabolism (Nitta et al., 2017). However, none of the human DIP2 homologues have been shown to have acyl-CoA synthetase activity. To characterize the contribution of C. elegans DIP-2 AFD domains in its function and subcellular localization, we generated transgenes expressing DIP-2 lacking specific domains (Fig. S2 C). Deletion of either AFD domain but not of the DMAP1-binding domain completely abolished the ability of DIP-2 to rescue neuronal morphology and migration defects (Fig. S2, D and E). In mature neurons, DIP-2::GFP, expressed from these in-frame deletion constructs, showed cytoplasmic localization resembling that of full-length (FL) protein (Fig. S2 F). However, in embryonic epidermal cells, DIP-2::GFP proteins lacking AFD1 or AFD2 but not those lacking the DMAP1-binding domain showed partial to complete loss of membrane localization compared with FL (Fig. S2 G). These findings suggest a cytosolic site of action and potentially an AFD-dependent acyl-CoA synthetase role for DIP-2 in neurons. Human DIP2A was identified as a putative cell surface receptor in cultured endothelial cells (Ouchi et al., 2010). We found that DIP-2 shows membrane localization in epidermal cells. Such differential localization observed across species implies a membrane site of action for DIP2 proteins in nonneuronal cells.
DIP2 was first identified in Drosophila from its binding to Disco, a transcription factor implicated in axon guidance (Mukhopadhyay et al., 2002). However, fly dip2 mutant phenotypes differ from those of Disco, suggesting that the protein-binding interaction may not be relevant in vivo (Nitta et al., 2017). Drosophila and vertebrate DIP2 proteins are broadly expressed in the central nervous system (Mukhopadhyay et al., 2002; Zhang et al., 2015), and their subcellular localization in neurons had not been fully characterized (Nitta et al., 2017). We find that functional GFP::DIP-2 or DIP-2::GFP does not accumulate in the nucleus and that the DMAP1-binding domain is not required for DIP-2 function in neurons (Fig. S2, D and E). Moreover, Disco orthologues are detected only in insects. These results suggest that roles of C. elegans DIP-2 in neuronal maintenance are unlikely to be through interaction with transcription factors in epigenetic gene regulation.
DIP-2 inhibits the early phase of axon regeneration via its AFDs
The persistent expression of DIP-2 in the adult nervous system and the role of dip-2 in maintaining mature neuron morphology led us to ask whether it may mediate other forms of axon growth. We examined adult axon regeneration using femtosecond laser axotomy (Wu et al., 2007). In WT adults, axotomy of PLM axons triggers growth cone reformation and regrowth from the severed proximal end. dip-2(0) adults displayed enhanced regrowth compared with WT controls (Fig. 5, A and B; and Fig. S3 A). Expression of FL DIP-2 driven by pan-neural (Prgef-1) or mechanosensory (Pmec-4) neuron promoters but not by an adult epidermal promoter (Pcol-19) rescued the enhanced regrowth phenotype in dip-2(0) mutants (Fig. 5, A and B). These data indicate that dip-2 acts cell autonomously to inhibit the regenerative capacity of axons. GABAergic motor neurons also displayed enhanced regeneration (Fig. S3 B), suggesting broad effects of dip-2 in axon regeneration. DIP-2 lacking its DMAP1-binding domain was fully able to rescue enhanced regrowth phenotypes, whereas DIP-2 lacking either AFD1 or AFD2 lacked rescuing ability (Fig. S3, D and E).
Figure 5.
dip-2inhibits the early phase of axon regeneration in parallel to dlk-1 and efa-6. (A) PLM regrowth 24 h after axotomy (normalized to WT control). Increased regrowth length of dip2 mutants was rescued by expressing dip-2 pan-neuronally and in mechanosensory neurons but not in the epidermis. (B) Representative images of PLM axon regrowth at 24 h after axotomy in WT, dip-2, and rescued dip-2 transgenic animals. Bar, 20 µm. (C) Loss of dip-2 partially bypasses axon regeneration failure in dlk-1(0) mutants, suggesting that dip-2 acts in parallel to dlk-1 in PLM regrowth. Regrowth data are shown as normalized value >24 h after axotomy. (D) Loss of dip-2 shows additive enhanced regrowth in efa-6(0) >24 h after axotomy. (E) PLM regrowth 6 h after axotomy shows that dip-2 acts early, in parallel to efa-6. Data are absolute value of regrowth length. Statistics in A and C–E: one-way ANOVA. ***, P < 0.001; **, P < 0.01; *, P < 0.05. Error bars are SEM.
DIP-2 acts early in the axon regenerative response. WT PLM axons initiate regrowth ∼6 h after axotomy (Chen et al., 2011). dip-2(0) mutants showed significantly enhanced regrowth at 6 h after axotomy (Fig. 5 E), suggesting that DIP-2 inhibits the initial response to damage and/or growth cone reformation. Previous studies identified multiple pathways that regulate the early regrowth response including the MAPKKK DLK-1, which is essential for initiation of regrowth (Hammarlund et al., 2009; Yan et al., 2009), and the microtubule regulator EFA-6, which restrains regrowth (Chen et al., 2011, 2015). Loss of function in dip-2 partially suppressed the regeneration defects in dlk-1(0) (Fig. 5 C) as well as those of its downstream components, pmk-3 and cebp-1 mutants (Fig. S3 C). In contrast, loss of efa-6 leads to increased regrowth, and double mutants of efa-6 and dip-2 showed further enhanced regrowth (Fig. 5 D). These data indicate that DIP-2 likely acts in parallel to both DLK-1 and EFA-6.
In summary, we have shown that C. elegans DIP-2 maintains neuronal morphology. Loss of DIP-2 function leads to ectopic axon branching and neurite sprouting through adult life. Conversely, DIP-2 overexpression suppresses the progressive neuronal sprouting seen in the aging WT, suggesting that DIP-2 levels in normal aging might affect the stability of neuronal morphology. DIP-2 maintains neuronal morphology by suppressing nascent neurite outgrowth in mature neurons, unlike trophic factors that regulate the complexity of existing neuronal structures in adult animals (Emoto et al., 2006; Matter et al., 2009; Chen et al., 2017). We also show that DIP-2 negatively regulates regenerative regrowth after injury. Repression of neurite growth and inhibition of injury-triggered regrowth both depend on the AFDs of DIP-2 but not on its DMAP1-binding domain, providing the first evidence that maintenance of neurite morphology and axon regrowth might involve a common mechanism. It will be of interest to determine whether mammalian DIP2 family members play comparable roles in the mature nervous system.
Materials and methods
C. elegans strains and genetics
C. elegans strains were cultured using standard methods. The Bristol N2 strain was used as WT along with the following alleles and transgenes: LGI: dip-2(zy25, zy26, ok885, gk913988), dlk-1(km12), zdIs5[mec-4p::GFP]. LGII: juIs76[unc-25p::GFP]. LGIV: pmk-3(tm745, ok169), efa-6(tm3124), zyIs47 [DIP-2::GFP + myo-2p::mCherry], zdIs4[mec-4p::GFP], zdIs13[tph-1p::GFP], juSi329[mec4p::mKate]. LGX: vang-1(tm1422), cebp-1(tm2807). zyIs3[flp-10p::GFP + odr-1p::mCherry] (linkage unknown). Extrachromosomal transgenes: zyEx44[dip2p::gfp + myo-2p::mCherry], zyEx51[unc-33p::DIP-2::SL2::tagRFP + myo-2p::mCherry], zyEx53[rgef-1p::DIP-2::SL2::tagRFP + myo-2p::mCherry], zyEx55[dpy-7p::DIP-2::SL2::tagRFP + myo-2p::mCherry], zyEx57[mec-4p::DIP-2::GFP + myo-2p::mCherry], zyEx65[dip-2p::DIP-2::SL2::tagRFP + myo-2::mCherry], zyEx67[dip-2p::DIP-2 DMAP1Δ::SL2::tagRFP + myo-2::mCherry], zyEx69-70[dip-2p::DIP-2 AFD1Δ::SL2::tagRFP + myo-2::mCherry], zyEx72-73[dip-2p::DIP-2 AFD2Δ::SL2::tagRFP + myo-2::mCherry], zyEx75-77[col-19p::DIP-2::GFP + myo-2::mCherry], zyEx78[dip-2p::DIP-2::GFP + myo-2::mCherry], zyEx79[dip-2p::DIP-2 DMAP1Δ::GFP + myo-2::mCherry], zyEx81[dip-2p::DIP-2 AFD1Δ::GFP + myo-2::mCherry], zyEx82[dip-2p::DIP-2 AFD2Δ::GFP + myo-2::mCherry], zyEx83-84[hsp-16.2p::DIP-2::SL2::tagRFP + myo-2p::mCherry], zyEx85[hsp-16.2p (pPD49.78) vector], zyEx86[mec-7p::DIP-2].
Molecular biology and transgenic strains
Whole-genome paired sequence reads were mapped to the C. elegans reference genome version WS220 using the short-read aligner Burrows-Wheeler Aligner (Li and Durbin, 2009). Single-nucleotide variants (SNVs) were identified and filtered using the SAMtools toolbox (Li et al., 2009). Each SNV was annotated with a custom-made Perl script and gene information from WormBase (WS220). The read alignments in the regions of candidate SNVs were visually inspected with the IGV viewer (Thorvaldsdóttir et al., 2013).
A 5,046-bp cDNA corresponding with the DIP-2b transcript (F28B3.4b; WormBase) was isolated from mixed-stage total RNA by RT-PCR using SL1 trans-spliced lead (5′-GGTTTAATTACCCAAGTTTGAG-3′) and dip-2 C-terminal end (5′-TTACATGTGATACGCTACGTATAT-3′) primers and cloned into pBluescript. The dip-2p::GFP transcriptional reporter was made by PCR amplification of the 2,458-bp upstream regulatory sequences and inserted into the pPD95.77 vector. This construct was modified using Gibson Assembly to insert the dip-2 cDNA (excluding stop codon) upstream and in-frame with GFP to generate the dip-2p::DIP-2::GFP translational reporter.
Expression constructs containing promoters of dip-2 (2,458 bp), neuronal unc-33 (1,961 bp), neuronal rgef-1 (3,443 bp), and epidermal dpy-7 (434 bp) were made using three-fragment Gibson Assembly in which PCR-amplified promoters and the 5,046-bp dip-2 cDNA (including stop codon) were ligated upstream of SL2::tagRFP from a linearized pSM-SL2::tagRFP vector (a gift from C.-F. Chuang, University of Illinois, Chicago, IL). Overlap PCR extension was used to fuse the mechanosensory neuron mec-4p (1,020 bp) and epidermal col-19p (759 bp) promoters to DIP-2::GFP. PCR-amplified dip-2 cDNA was cloned into the KpnI site of the mec-7 expression plasmid pPD96.41. DIP-2 domains were identified using HMMER (Finn et al., 2011), and domain deletion constructs were made using Gibson Assembly in which either dip-2p::dip-2(cDNA)::SL2tagRFP or dip-2p::DIP-2::GFP plasmid was the backbone and overlap PCR was used to delete dip-2 cDNA nucleotides 7–345, 1,279–2,736, and 3,256–4,641 to generate in-frame deletion of DMAP1-binding domain (amino acids 3–115), AFD1 (amino acids 428–912), and AFD2 (amino acids 1,086–1,547) constructs, respectively. Gibson Assembly was used to insert a dip-2(cDNA)::SL2tagRFP PCR fragment into the EcoRV site of the hsp-16.2p vector pPD49.78 to generate hsp-16.2p::DIP-2::SL2::tagRFP. dip-2 cDNA-containing constructs were sequenced to verify PCR fidelity.
Extrachromosomal transgenes were made by injecting each construct at 10 ng/µl with 5 ng/µl pCFJ90 (myo-2p::mCherry) coinjection marker and 115 ng/µl pBluescript. A dip-2p::DIP-2::GFP-containing extrachromosomal transgene was integrated to generate zyIs47. For ALM and HSN cell-specific rescue experiments, rgef-1 and unc-33 promoters, respectively, were used to drive panneural expression. For neuronal morphology and axon regeneration cell-specific rescue experiments, dpy-7 and col-19 promoters, respectively, were used to drive panepidermal expression.
zy70 CRISPR/Cas9-mediated GFP knock-in
The CRISPR/Cas9-mediated approach described in Dickinson et al. (2015) was used to insert an in-frame GFP cassette at the N terminus of endogenous dip-2 to generate dip-2(zy70). The CRISPR design tool (http://crispr.mit.edu) was used to identify the dip-2 N-terminal target site 5′-GTTGCAGATAATGAATGATCCGG-3′. Overlap fusion PCR was used to generate a plasmid containing C. elegans germline promoter driven Cas9 and dip-2-sgRNA (5′-GTTGCAGATAATGAATGATCGTTTTAGAGCTAGAAATAGCAAGT-3′). The GFP::DIP-2 homology-directed repair (HDR) plasmid was made using three-fragment Gibson Assembly in which dip-2 homology arms containing 508 bp upstream and 800 bp downstream of the dip-2 ATG start codon were PCR amplified from N2 genomic DNA and fused such that they flanked the GFP in the linearized GFP cassette–containing plasmid described in Dickinson et al. (2015). zy70 was verified by sequencing.
Quantification of neuronal morphology and migration defects
Neuronal morphology phenotypes were analyzed in worms grown at 25°C. Worms were mounted on a 2% agarose pad in 200 mM levamisole hydrochloride (31742; Sigma-Aldrich). Fluorescent imaging was performed by wide-field microscopy (Zeiss AxioImager M2) using either 20×/0.75-NA air or 63×/1.4-NA oil objectives. Images were acquired using a Zeiss Axiocam 506 mono camera and processed using Zeiss Axiovision (v.4.8) software. Ectopic neurites and axon branches (in PVM) were scored in young adults when extension length was >10 µm. For analysis of age dependence on neuronal morphology, worms were synchronized by bleaching and then grown on standard medium supplemented with 16 µM fluorodeoxyuridine (F0503; Sigma-Aldrich) to induce sterility and prevent premature death due to retention of eggs in aging dip-2 adults. In L4 and adult stages, neurites were scored as ectopic if length was >10 µm and as branched if they also showed secondary or complex branching. In L1, any ectopic ALM cell body protrusion >0 µm was scored as an ectopic neurite. HSN migration phenotypes were analyzed in young adults grown at 20°C. HSN positions were scored along the anterior–posterior (AP) axis between the vulva (0% AP position) and the tip of the tail (100% AP position). HSN was scored as undermigrated if at least one HSN per animal was located within bins defined as 12.5–37.5% (light gray), 37.5–75% (dark gray), and 75–100% (black) along the AP axis.
Heat shock–induced expression
Newly laid eggs (0–3 h after egg laying) or newly hatched L1 (0–3 h after hatching) from dip-2(0); Ex[hsp-16.2p::vector] or dip-2(0); Ex[hsp-16.2p::DIP-2] that were grown at 20°C were heat shocked, allowed to recover, and then scored for ALM neurite defects as described above as D1 adults. For embryonic heat shock, from worms grown at 20°C, newly laid eggs were heat shocked at 35°C for 2 h and then allowed to recover at 20°C. Newly hatched L1 were then transferred to 25°C for 48 h. For L1 heat shock, from worms grown at 20°C, newly hatched L1 were heat shocked at 35°C for 2 h and then allowed to recover at 25°C for 46 h. For L4 heat shock, from worms grown at 20°C, newly hatched L1 were transferred to 25°C for 24 h, heat shocked at 35°C for 2 h (early L4), and then transferred to 25°C for 22 h. Two independent lines containing hsp-16.2p::DIP-2 transgenic arrays were examined.
Age-dependent DIP-2 expression
Worms were mounted with levamisole in M9 buffer described above, and Z stack images of a cell of interest were acquired using the Plan Apochromat 63×/1.4 oil differential interference contrast objective lens of a Zeiss LSM710 confocal microscope at room temperature. GFP and mKate2 images were collected using the Zen 2011 SP3 (black edition) software and processed for maximum-intensity Z projection using Fiji (ImageJ; National Institutes of Health). In the final projected image, a circle fitting in the nucleus was drawn as a region of interest (ROI) in which GFP intensity was measured. Same-size ROIs were selected in the cytoplasm, one above the nucleus and the other below the nucleus. GFP intensity measured in these two cytoplasmic ROIs was averaged. The nuclear intensity considered as background was subtracted from the average cytoplasmic intensity. The size of ROI was identical in all the images analyzed using ImageJ.
Laser axotomy
Axons of PLM or GABAergic motor neurons were axotomized in anesthetized L4 larvae as described (Wu et al., 2007).
Statistical analysis
All statistical analysis used Prism (GraphPad Software). Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 shows that dip-2 mutants display neuronal morphology and migration defects. Fig. S2 shows requirements for the DMAP1 binding and AFD domains for DIP-2 neuronal development and protein localization. Fig. S3 shows supporting evidence for dip-2 in PLM axon regeneration.
Supplementary Material
Acknowledgments
We thank Cristina Slatculescu and Justin Evans for assistance with molecular biology and microscopy, Chiou-Fen Chuang for plasmids, Matt Andrusiak and Y. Jin for juSi329, and our laboratory members for comments and discussion.
Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work was supported by grants from the Canadian Institutes of Health Research (123513) and the Natural Sciences and Engineering Research Council of Canada (312460-2012) to A. Colavita and the National Institutes of Health (R01 NS093588) to A.D. Chisholm and Y. Jin.
The authors declare no competing financial interests.
Author contributions: N. Noblett and Z.H. Ding performed and analyzed neuronal morphology and migration assays. Z. Wu performed and analyzed axotomy. S. Park performed all confocal microscopy and image analysis. Z.H. Ding and T. Roenspies generated constructs. S. Flibotte analyzed whole-genome sequencing data. A.D. Chisholm, Y. Jin, and A. Colavita supervised the research and wrote the manuscript.
References
- Carr D., Sanchez-Alvarez L., Imai J.H., Slatculescu C., Noblett N., Mao L., Beese L., and Colavita A.. 2016. A Farnesyltransferase Acts to Inhibit Ectopic Neurite Formation in C. elegans. PLoS One. 11:e0157537 10.1371/journal.pone.0157537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M., and Sulston J.. 1981. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev. Biol. 82:358–370. 10.1016/0012-1606(81)90459-0 [DOI] [PubMed] [Google Scholar]
- Chen C.-M., Orefice L.L., Chiu S.-L., LeGates T.A., Hattar S., Huganir R.L., Zhao H., Xu B., and Kuruvilla R.. 2017. Wnt5a is essential for hippocampal dendritic maintenance and spatial learning and memory in adult mice. Proc. Natl. Acad. Sci. USA. 114:E619–E628. 10.1073/pnas.1615792114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Wang Z., Ghosh-Roy A., Hubert T., Yan D., O’Rourke S., Bowerman B., Wu Z., Jin Y., and Chisholm A.D.. 2011. Axon regeneration pathways identified by systematic genetic screening in C. elegans. Neuron. 71:1043–1057. 10.1016/j.neuron.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chuang M, Koorman T, Boxem M, Jin Y, and Chisholm AD. 2015. Axon injury triggers EFA-6 mediated destabilization of axonal microtubules via TACC and doublecortin like kinase. eLife. 10.7554/eLife.08695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman R.A., Lewin T.M., Van Horn C.G., and Gonzalez-Baró M.R.. 2002. Do long-chain acyl-CoA synthetases regulate fatty acid entry into synthetic versus degradative pathways? J. Nutr. 132:2123–2126. 10.1093/jn/132.8.2123 [DOI] [PubMed] [Google Scholar]
- Dickinson D.J., Pani A.M., Heppert J.K., Higgins C.D., and Goldstein B.. 2015. Streamlined genome engineering with a self-excising drug selection cassette. Genetics. 200:1035–1049. 10.1534/genetics.115.178335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K., Parrish J.Z., Jan L.Y., and Jan Y.-N.. 2006. The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature. 443:210–213. 10.1038/nature05090 [DOI] [PubMed] [Google Scholar]
- Finn R.D., Clements J., and Eddy S.R.. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39(suppl):W29–W37. 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund M., Nix P., Hauth L., Jorgensen E.M., and Bastiani M.. 2009. Axon regeneration requires a conserved MAP kinase pathway. Science. 323:802–806. 10.1126/science.1165527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Lombaert I.M.A., Hauser B.R., Patel V.N., and Hoffman M.P.. 2017. Exosomal MicroRNA Transport from Salivary Mesenchyme Regulates Epithelial Progenitor Expansion during Organogenesis. Dev. Cell. 40:95–103. 10.1016/j.devcel.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan Y.N., and Jan L.Y.. 2010. Branching out: mechanisms of dendritic arborization. Nat. Rev. Neurosci. 11:316–328. 10.1038/nrn2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.Y.N., Howe K.L., Harris T.W., Arnaboldi V., Cain S., Chan J., Chen W.J., Davis P., Gao S., Grove C., et al. 2018. WormBase 2017: molting into a new stage. Nucleic Acids Res. 46(D1):D869–D874. 10.1093/nar/gkx998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter C., Pribadi M., Liu X., and Trachtenberg J.T.. 2009. δ-catenin is required for the maintenance of neural structure and function in mature cortex in vivo. Neuron. 64:320–327. 10.1016/j.neuron.2009.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay M., Pelka P., DeSousa D., Kablar B., Schindler A., Rudnicki M.A., and Campos A.R.. 2002. Cloning, genomic organization and expression pattern of a novel Drosophila gene, the disco-interacting protein 2 (dip2), and its murine homolog. Gene. 293:59–65. 10.1016/S0378-1119(02)00694-7 [DOI] [PubMed] [Google Scholar]
- Nitta Y., Yamazaki D., Sugie A., Hiroi M., and Tabata T.. 2017. DISCO Interacting Protein 2 regulates axonal bifurcation and guidance of Drosophila mushroom body neurons. Dev. Biol. 421:233–244. 10.1016/j.ydbio.2016.11.015 [DOI] [PubMed] [Google Scholar]
- Ouchi N., Asaumi Y., Ohashi K., Higuchi A., Sono-Romanelli S., Oshima Y., and Walsh K.. 2010. DIP2A functions as a FSTL1 receptor. J. Biol. Chem. 285:7127–7134. 10.1074/jbc.M109.069468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C.-L., Peng C.-Y., Chen C.-H., and McIntire S.. 2011. Genetic analysis of age-dependent defects of the Caenorhabditis elegans touch receptor neurons. Proc. Natl. Acad. Sci. USA. 108:9274–9279. 10.1073/pnas.1011711108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Alvarez L., Visanuvimol J., McEwan A., Su A., Imai J.H., and Colavita A.. 2011. VANG-1 and PRKL-1 cooperate to negatively regulate neurite formation in Caenorhabditis elegans. PLoS Genet. 7:e1002257 10.1371/journal.pgen.1002257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz S., and Naismith J.H.. 2009. Adenylate-forming enzymes. Curr. Opin. Struct. Biol. 19:666–671. 10.1016/j.sbi.2009.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank E.M., Rodgers K.E., and Kenyon C.. 2011. Spontaneous age-related neurite branching in Caenorhabditis elegans. J. Neurosci. 31:9279–9288. 10.1523/JNEUROSCI.6606-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., and Mesirov J.P.. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14:178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth M.L., Melentijevic I., Shah L., Bhatia A., Lu K., Talwar A., Naji H., Ibanez-Ventoso C., Ghose P., Jevince A., et al. 2012. Neurite sprouting and synapse deterioration in the aging Caenorhabditis elegans nervous system. J. Neurosci. 32:8778–8790. 10.1523/JNEUROSCI.1494-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Ghosh-Roy A., Yanik M.F., Zhang J.Z., Jin Y., and Chisholm A.D.. 2007. Caenorhabditis elegans neuronal regeneration is influenced by life stage, ephrin signaling, and synaptic branching. Proc. Natl. Acad. Sci. USA. 104:15132–15137. 10.1073/pnas.0707001104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Wu Z., Chisholm A.D., and Jin Y.. 2009. The DLK-1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell. 138:1005–1018. 10.1016/j.cell.2009.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Mabwi H.A., Palange N.J., Jia R., Ma J., Bah F.B., Sah R.K., Li D., Wang D., Bah F.B.M., et al. 2015. Expression Patterns and Potential Biological Roles of Dip2a. PLoS One. 10:e0143284 10.1371/journal.pone.0143284 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.