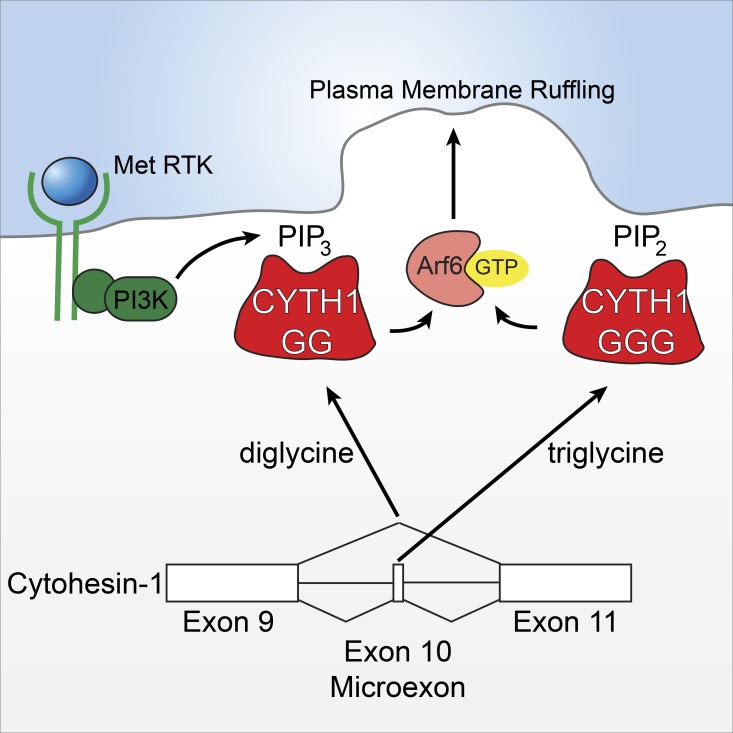
Splice variants of the Arf6 guanine exchange factor cytohesin-1 display differential affinity for PI(4,5)P2 and PI(3,4,5)P3. Ratcliffe et al. show that the specific lipid binding of the diglycine variant of cytohesin-1 is needed for HGF-dependent cell migration and establishment of the leading edge, thereby regulating cancer cell migration following activation of the proto-oncogenic receptor tyrosine kinase Met.
Abstract
Differential inclusion or skipping of microexons is an increasingly recognized class of alternative splicing events. However, the functional significance of microexons and their contribution to signaling diversity is poorly understood. The Met receptor tyrosine kinase (RTK) modulates invasive growth and migration in development and cancer. Here, we show that microexon switching in the Arf6 guanine nucleotide exchange factor cytohesin-1 controls Met-dependent cell migration. Cytohesin-1 isoforms, differing by the inclusion of an evolutionarily conserved three-nucleotide microexon in the pleckstrin homology domain, display differential affinity for PI(4,5)P2 (triglycine) and PI(3,4,5)P3 (diglycine). We show that selective phosphoinositide recognition by cytohesin-1 isoforms promotes distinct subcellular localizations, whereby the triglycine isoform localizes to the plasma membrane and the diglycine to the leading edge. These data highlight microexon skipping as a mechanism to spatially restrict signaling and provide a mechanistic link between RTK-initiated phosphoinositide microdomains and Arf6 during signal transduction and cancer cell migration.
Graphical Abstract
Introduction
The Met receptor tyrosine kinase (RTK) coordinates invasive growth in response to its ligand hepatocyte growth factor (HGF). This is tightly regulated during development to promote a morphogenic program that is essential for liver development and migration of muscle precursors into the limb bud as well as wound healing in the adult (Gherardi et al., 2012). Under pathophysiological conditions, deregulated signaling by the Met RTK leads to enhanced cell migration and metastatic spread of cancer cells (Gherardi et al., 2012; Parachoniak and Park, 2012; Knight et al., 2013).
The proinvasive properties of Met are tightly regulated by spatial localization of signaling complexes on subcellular compartments, including dorsal ruffles, invadopodia, lamellipodia, and endosomes (Maroun et al., 1999a,b; Palamidessi et al., 2008; Abella et al., 2010; Rajadurai et al., 2012; Ménard et al., 2014). Each of these compartments possesses distinct morphological and molecular features. While Met recruits many different effectors, not all complexes are assembled at each subcellular location where Met is active and additional determinants must define the localization of different signaling complexes. For example, the plasma membrane is the predominant source of phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) in the cell, however this lipid may be modified by phosphoinositide 3-kinase (PI3K) to locally generate phosphatidylinositol 3,4,5-trisphosphate (PI(3,4,5)P3; Whitman et al., 1988), thereby affecting local recruitment and activity of proteins possessing PI(3,4,5)P3-binding domains.
The small GTPase Arf6 is critical for Met-dependent invasive growth, although the molecular mechanisms that link Met to Arf6 activation in cancer cells are unknown (Tushir and D’Souza-Schorey, 2007). Arf small GTPases are members of a superfamily of molecular switches that mediate changes in cell morphology, endomembrane traffic, and cell signaling (Gillingham and Munro, 2007; Simanshu et al., 2017). Arf6 is unique among Arf proteins in that it localizes primarily to the plasma membrane and endosomes, as opposed to Arf1 and Arf3, which localize predominantly to the Golgi apparatus (Donaldson and Jackson, 2011). Active, GTP-bound Arf6 modulates processes that are critical for cell migration and tumor metastasis (Muralidharan-Chari et al., 2009; Yoo et al., 2016). These include endosomal recycling of the Met RTK or integrin receptors, clathrin-independent endocytosis, and rearrangement of the actin cytoskeleton (Powelka et al., 2004; Eyster et al., 2009; Parachoniak et al., 2011; Ratcliffe et al., 2016). Like the majority of small GTPases, Arf6 cycles between an “off” GDP-bound state and an “on” GTP-bound state. Cycling between these states is enhanced by guanine nucleotide exchange factors (GEFs), which promote GDP release, and GTPase activation proteins, which promote hydrolysis of GTP (Donaldson and Jackson, 2011; Simanshu et al., 2017). Subcellular localization of GEF and GTPase activation proteins imposes spatiotemporal regulation on small GTPase signaling, restricting activity to specific subcellular locations. However, the in vivo subcellular determinants for Arf6 activation remain to be fully defined.
Arf GEFs fall into seven families, three of which encompass putative Arf6 GEFs, including cytohesin (1–4), IQSEC (1–3), and PSD (1–4; Casanova, 2007; Donaldson and Jackson, 2011). These families are defined by the presence of a Sec7 domain that enhances the release of GDP from Arf proteins. In addition, there are multiple splice variants of Arf GEFs. Some differ by an entire domain, whereas others involve microexons (Ogasawara et al., 2000; Fukaya et al., 2016). The best-characterized microexon splice variants are two isoforms of cytohesin-2. These isoforms differ by a 3-nt microexon, whose splicing leads to an additional glycine residue within the cytohesin-2 pleckstrin homology (PH) domain (Cronin et al., 2004), yet a functional difference for these has not been determined.
Microexons are a recently described class of exons that are ≤27-nt long and predominantly found in structured regions of proteins. Microexons are frequently identified in brain-derived transcripts, including cytohesin-2, and are alternatively regulated in individuals with autism spectrum disorder (Irimia et al., 2014). However, their functional roles in other diseases, particularly cancer, are unknown. Furthermore, despite decades of research on signaling from RTKs, such as Met, the role of alternative splicing in generating multiple isoforms of downstream effectors is also largely unexplored. Here, we demonstrate differential functions for cytohesin-1 isoforms whereby a splice variant of cytohesin-1 that lacks a 3-nt microexon has distinct phospholipid binding and is uniquely required for HGF-dependent cell migration. Thus, we provide a mechanistic understanding into the regulation of HGF-dependent cell migration by microexon skipping in a specific effector acting downstream of Met.
Results
Cytohesin-1 regulates HGF-dependent cell migration
Stimulation of epithelial and many cancer cells with HGF promotes activation of the Met RTK and subsequent cellular morphological changes leading to enhanced cell migration. The migratory and invasive program induced by the Met RTK specifically requires the small GTPase Arf6 (Tushir and D’Souza-Schorey, 2007). Using previously described siRNAs targeting Arf family GTPases (Ratcliffe et al., 2016), we confirmed the role of Arf6 in Met-dependent rearrangement of the actin cytoskeleton (Fig. S1). However, the regulation of Arf6-dependent actin remodeling by Met is not fully understood. To identify Arf GEFs required for HGF-dependent cell migration, we measured the expression of putative Arf GEFs, which are defined by the presence of a Sec7 domain (Donaldson and Jackson, 2011). We used HeLa cells, which have been extensively studied for both Met-dependent migration and Arf6-dependent cell migration (Palamidessi et al., 2008; Parachoniak et al., 2011; Frittoli et al., 2014). Out of 10 putative Arf GEFs, six were detectably expressed in HeLa cells, as assessed by quantitative RT-PCR (Fig. S2 A). These include cytohesin-1, cytohesin-2, cytohesin-3, IQSEC1, IQSEC2, and PSD3.
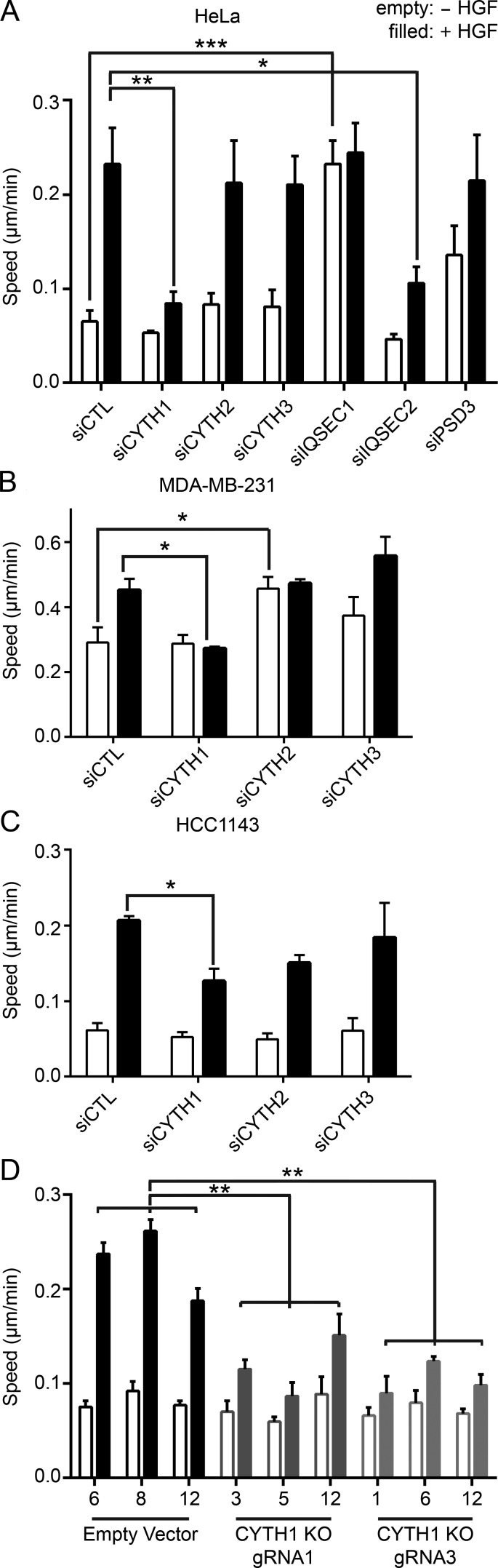
To assess the effect of Arf GEFs on cell migration, each Arf GEF was independently reduced by siRNA-mediated silencing to ≤30% (Fig. S2 B), and cells were imaged every 15 min for 24 h in the presence and absence of HGF. Quantification of cell speed between 16 and 24 h after stimulation revealed that silencing of cytohesin-1 and IQSEC2 reduced HGF-dependent cell migration to 36% and 46% of control cells, respectively (Fig. 1 A). In contrast, silencing of IQSEC1 (also known as BRAG2) enhanced HGF-independent cell migration, with no further increase of cell speed observed following HGF treatment (Fig. 1 A). This enhancement in cell speed is likely mediated by an increase in the cell surface levels of integrins caused by IQSEC1 silencing (Moravec et al., 2012).
Figure 1.
Cytohesin-1 depletion reduces HGF-dependent cell migration. (A–C) Random cell migration of HeLa cells (A), MDA-MB-231 cells (B), or HCC1143 cells (C) treated with the indicated siRNA smartpool without (−, unfilled) or with (+, filled) HGF. (D) Random cell migration of CYTH1 KO or lentiCRISPR v2 empty vector HeLa clones treated with or without HGF. All quantified data indicate mean values ± SEM from three or four independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.00.1.
Whereas unstimulated IQSEC2-depleted cells were more spread and could readily be distinguished from control cells based on their morphology, cytohesin-1–depleted cells appeared morphologically indistinguishable from control cells but had diminished HGF-induced cell migration. This implies that IQSEC2 depletion may have a more general effect on the actin cytoskeleton, while the effects of cytohesin-1 silencing in these cells are required for HGF/Met signaling and cell migration.
Met and Arf6 activity have each been implicated in breast cancer cell migration (Hashimoto et al., 2004; Knight et al., 2013; Li et al., 2015). To establish if cytohesin-1 played a role in Met-dependent migration, the basal-like breast cancer cell lines MDA-MB-231 and HCC1143, which express cytohesin-1, cytohesin-2, and cytohesin-3, were investigated (Fig. S2 C). Migration speed of both cell lines is increased following HGF stimulation (Fig. 1, B and C), consistent with previous observations for MDA-MB-231 cells (Rajadurai et al., 2012). Silencing cytohesin-2 in MDA-MB-231 cells increased cell migration, and there was a trend toward reduced HGF-dependent cell migration in HCC1143 cells, supporting a role for cell-type–specific effects for cytohesin-2. Silencing cytohesin-1 reduced HGF-dependent cell migration in both MDA-MB-231 and HCC1143 cell lines (Fig. 1, B and C; and Fig. S2 D). Hence, we focused on cytohesin-1. The decreased migratory phenotype observed by siRNA-mediated depletion of cytohesin-1 was validated by generating HeLa cells with stable knockout (KO) of cytohesin-1 using the lentiCRISPR v2 system, with two independent guide RNAs targeting exon 2 of CYTH1 (Fig. S2 D). When HGF-dependent cell migration was compared in three control and CYTH1 KO clones, CYTH1 KO phenocopied siRNA-mediated silencing of cytohesin-1, whereby all clones displayed reduced HGF-dependent migration speed when compared with control clones (Fig. 1 D). We did not observe a significant difference in Met stability or Met, Akt, or Erk1/2phosphorylation between control and CYTH1 KO cells upon HGF treatment (Fig. S3). These data support a model in which cytohesin-1 regulates HGF-dependent cell migration independently of known downstream signaling targets and may represent a novel pathway to Arf6 activation.
Diglycine cytohesin-1 regulates HGF-dependent cell migration and invasion
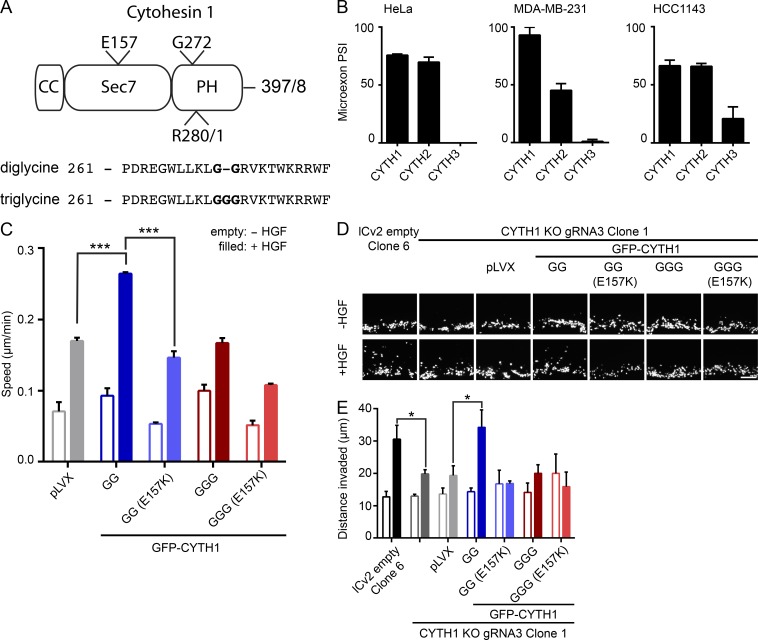
Cytohesin-1 belongs to a family of four proteins (cytohesin-1, cytohesin-2 [also known as ARNO], cytohesin-3 [also known as Grp1], and cytohesin-4). These proteins consist of a coiled-coiled domain, a Sec7 domain with Arf GEF activity, and a PH domain that selectively recognizes phosphoinositides (Chardin et al., 1996). Two isoforms of cytohesin family members are expressed that differ by the inclusion of a evolutionarily conserved 3-nt microexon resulting in an additional glycine residue in the PH domain (Ogasawara et al., 2000; Irimia et al., 2014). We refer to these isoforms as diglycine and triglycine variants (Fig. 2 A). The microexons of cytohesin-1 and cytohesin-2 show similar inclusion (or percent spliced in [PSI]) values in HeLa cells (76% and 69%) and HCC1143 cells (66% and 66%) but different PSI values in MDA-MB-231 cells (93% and 45%; Fig. 2 B), whereas cytohesin-3 shows a reduced PSI of 5% in HeLa and MDA-MB-231 cells and 21% in HCC1143 cells. To identify the splice variant of cytohesin-1 that mediates HGF-dependent cell migration, we generated a panel of stable cell lines expressing GFP-tagged isoforms or mutants of cytohesin-1 in the CYTH1 KO cell background (Fig. S4 A). Under these conditions the diglycine GFP-CYTH1, but not triglycine GFP-CYTH1, rescued HGF-dependent cell migration when compared with empty vector control (Fig. 2 C). Rescue was dependent on the nucleotide exchange activity of the diglycine isoform, as expression of a GFP-CYTH1 construct with a mutation of a glutamic acid essential for exchange activity (E157K) was unable to rescue HGF-dependent cell migration (Fig. 2 C). In addition to a decrease in cell migration in 2D, CYTH1 KO reduced HGF dependent invasion through a 3D collagen matrix, which could be rescued by expressing the diglycine, but not triglycine, isoform of cytohesin-1 (Fig. 2, D and E). Rescue of HGF-induced cell invasion is similarly dependent on the exchange activity of diglycine cytohesin-1, since the E157K cytohesin-1 mutant failed to rescue cell invasion in response to HGF (Fig. 2, D and E). Given these observations, we propose that the diglycine isoform of cytohesin-1 acts downstream of the Met RTK and is required for cancer cell HGF-dependent migration and invasion.
Figure 2.
Diglycine cytohesin-1 regulates HGF-dependent cell migration and invasion. (A) Domain organization of CYTH1. (B) PSI values of cytohesin-1, cytohesin-2, or cytohesin-3 microexons in HeLa, MDA-MB-231, and HCC1143 cell lines. (C) Random cell migration of pLVX empty vector or eGFP-CYTH1 variant expressing cells treated with or without HGF. (D) Representative images of invasion of DAPI stained cell lines. (E) Quantification of experiments shown in D as distance from seeded area. Scale bar, 100 µm. All quantified data indicate mean values ± SEM from three independent experiments. *, P < 0.05; ***, P < 0.001.
Cytohesin-1 splice variants differentially mediate membrane ruffling
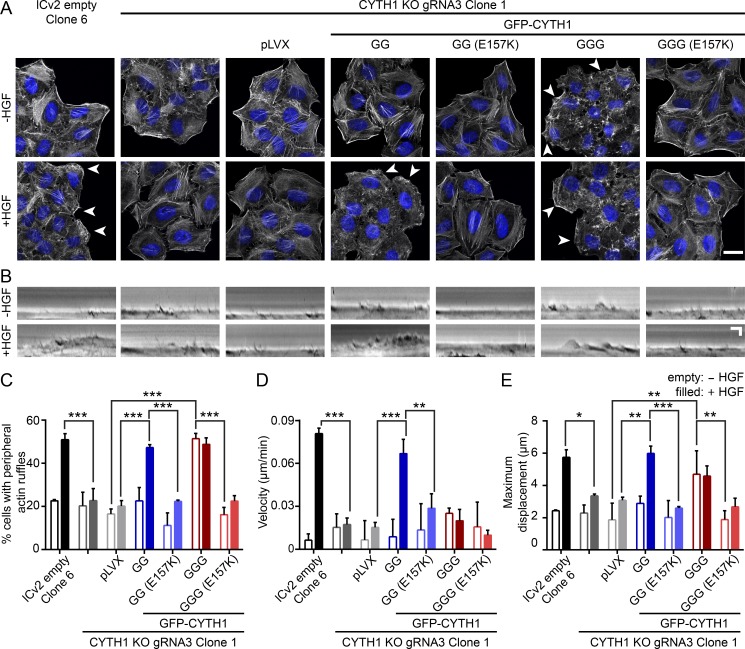
Cell migration requires the spatial coordination of multiple signals. Upon HGF stimulation, Met is rapidly internalized and a fraction of these receptors are recycled to the leading edge, where Rac1 is active and induces rearrangement of the actin cytoskeleton (Royal et al., 2000; Palamidessi et al., 2008; Parachoniak et al., 2011; Ménard et al., 2014). Consistent with our data and previous reports, HGF induces a rearrangement in the actin cytoskeleton with the formation of peripheral actin ruffles (Fig. 3 A). Consistently, in CYTH1 KO cells, the percentage of cells with HGF-induced peripheral actin ruffles was reduced (51% in CTL versus 23% in KO). This could be rescued by expression of diglycine GFP-CYTH1 (47%), but not the GEF exchange E157K mutant (22%; Fig. 3, A and C). Intriguingly, the majority of cells overexpressing the triglycine GFP-CYTH1 variant displayed peripheral actin ruffles (51%) in the absence of HGF stimulation that were not increased following HGF stimulation (Fig. 3, A and C). This suggests that while the triglycine variant was unable to rescue HGF-dependent cell migration, overexpression of this isoform is sufficient to promote downstream signals that enhance membrane ruffling. Consistent with this, the GEF inactive E157K triglycine mutant showed decreased ability to promote peripheral actin ruffles (16%; Fig. 3, A and C). These observations demonstrate a significant difference in cytohesin-1 isoform function and sensitivity to Met RTK stimulation.
Figure 3.
Cytohesin-1 regulates membrane ruffling and actin cytoskeleton rearrangement. (A) Confocal images of cells counterstained with phalloidin (F-actin) and DAPI treated (−, unfilled; +, filled). Arrowheads indicate peripheral membrane ruffles. Scale bar, 20 µm. (B) Kymographs were generated from linescans of the cells’ leading edge imaged between 15 and 60 min after HGF treatment. (C) Quantification of experiments shown in A. (D and E) Quantification of experiments shown in B. All quantified data indicate mean values ± SEM from three or four independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To quantitatively assess the effect of HGF on plasma membrane dynamics, we performed live-cell imaging in response to HGF stimulation and assessed the relative position of the plasma membrane every 15 s between 15 min and 60 min after HGF stimulation. In control cells, lamellipodia are observed in response to HGF, with the formation of a leading edge that extends forward with a velocity of 0.081 µm/min and maximum displacement of 5.74 µm, compared with 0.006 µm/min and 2.43 µm in the absence of HGF (Fig. 3, B, D, and E). In contrast, in CYTH1 KO cells, both the velocity and maximum displacement of membrane protrusions in response to HGF were reduced to 21.4% and 58% of control cells, respectively (Fig. 3, B, D, and E). Consistent with its ability to rescue cell migration, diglycine GFP-CYTH1 rescued membrane protrusion velocity (82% of control) and maximum displacement (104% of control). Rescue depended on the Arf GEF activity of diglycine cytohesin-1, since cells expressing diglycine GFP-CYTH1 E157K failed to increase membrane velocity (35% of control) or maximum displacement (45% of control) relative to CYTH1 KO cells.
While there was no significant effect of overexpressing the triglycine isoform on net membrane velocity, we observed an HGF-independent increase in the maximum displacement (193% of control), indicating that these cells were actively ruffling but failed to produce a stable leading edge (Fig. 3, B, D, and E). This effect was also dependent on the Arf GEF activity of cytohesin-1, as the E157K triglycine mutant failed to increase the maximum displacement (Fig. 3, B and E). Together, this demonstrates that the diglycine PH domain mediates HGF-dependent cell migration and establishment of a leading edge in a migrating cell and suggests that phosphoinositide recognition regulates these processes.
Cytohesin-1 variants differentially recognize phosphoinositide headgroups
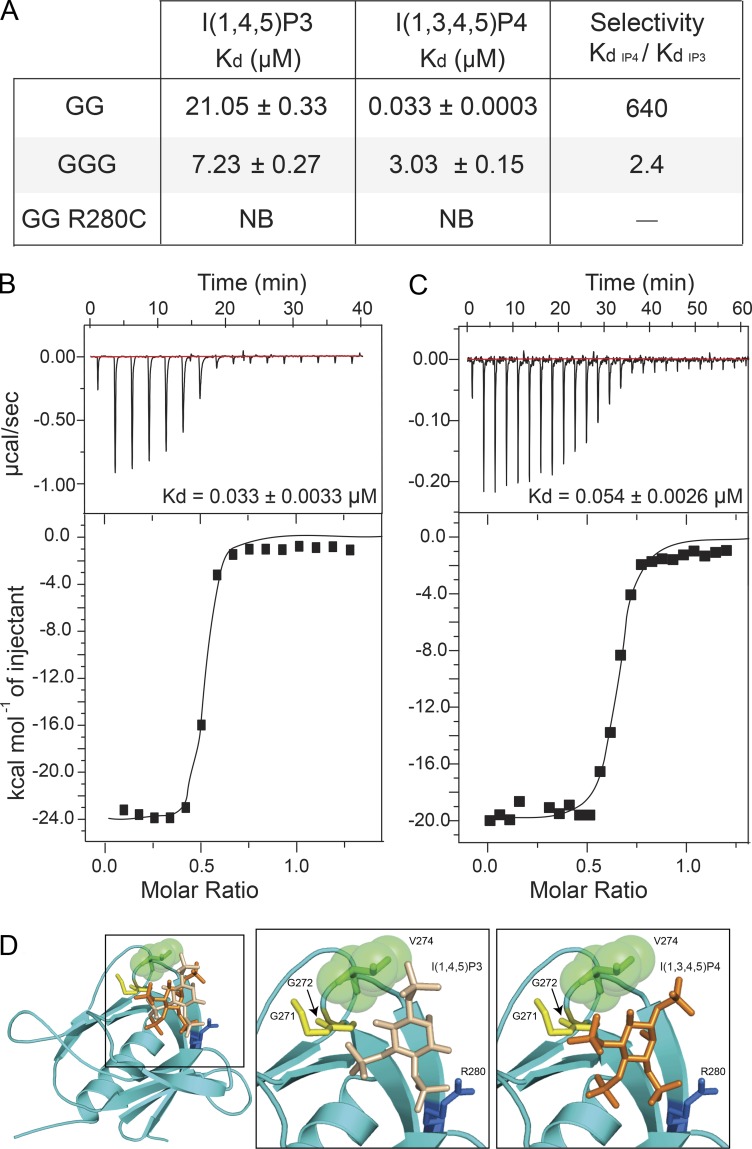
The binding affinities of the PH domain of cytohesin-2 and cytohesin-3 for different inositol phosphate headgroups have been extensively characterized (Klarlund et al., 2000; Cronin et al., 2004). These studies show that the diglycine variant of cytohesin-2 has a significantly stronger affinity (14-fold) for I(1,3,4,5)P4, which is the headgroup of PI(3,4,5)P3, relative to I(1,4,5)P3, the headgroup of PI(4,5,)P2 (Cronin et al., 2004), whereas the triglycine variant is less selective, binding to both with similar affinities. To characterize the specificity of cytohesin-1 for I(1,3,4,5)P4 and I(1,4,5)P3, we performed isothermal titration calorimetry (ITC) using recombinant cytohesin-1 PH domain variants (Fig. S4 B). We determined that the diglycine cytohesin-1 PH domain bound to I(1,3,4,5)P4 with a Kd of 0.033 µM and I(1,4,5)P3 with a Kd of 21.05 µM (Fig. 4, A and B). This indicates that cythohesin-1 has a 640-fold greater affinity for I(1,3,4,5)P4 over I(1,4,5)P3. The triglycine variant of cytohesin-1 PH domain binds to I(1,3,4,5)P4 with an affinity of 3.03 µM (∼100-fold lower than diglycine) and I(1,4,5)P3 with an affinity of 7.23 µM (∼3-fold higher than diglcyine; Fig. 4 A). These results, similar to those observed for cytohesin-2 by Cronin et al. (2004), support that the diglycine variant of cytohesin-1 preferentially interacts with PI(3,4,5)P3 on membranes. To test this, we titrated PI(3,4,5)P3-containing liposomes with the diglycine cytohesin-1 PH domain containing the C-terminal polybasic region and found that it bound with a comparable affinity (Kd = 0.054 µM) to the headgroup alone (Fig. 4 C), confirming the ability of the diglycine PH domain to recognize PI(3,4,5)P3 in the context of a lipid membrane.
Figure 4.
G272 defines the phosphoinositide binding selectivity of cytohesin-1. (A) Kd values measured by ITC. NB, no binding. (B) ITC trace of I(1,3,4,5)P4 titrated into diglycine CYTH1 PH domain. (C) ITC trace of diglycine CYTH1 PH domain (aa 243–397) titrated into PI(3,4,5)P3-containing liposomes. (D) Molecular model of the diglycine CYTH1 PH domain bound to I(1,3,4,5)P4 or I(1,4,5)P3. Error estimates shown are from fitting the data using a single site-binding model.
To gain further insight into the binding properties of the diglycine variant of cytohesin-1, we generated a homology model of its PH domain based on the structure of the PH domain of cytohesin-3 (Fig. 4 D). Since there is an ∼90% sequence identity between the PH domains of cytohesin family members, we anticipate that the phosphoinositide-binding pocket would be conserved. Based on previous studies, we predicted that R280 forms contacts with the 3′ phosphate of I(1,3,4,5)P4 or 4′ phosphate of I(1,4,5)P3, and this site is required for a detectable interaction. Consistent with this model, an R280C mutation when introduced into diglycine cytohesin-1 PH domain abrogated any interaction with I(1,3,4,5)P4 or I(1,4,5)P3 (Fig. 4 A). Together, these data indicate that the diglycine variant of cytohesin-1 specifically recognizes PI(3,4,5)P3, whereas the triglycine variant may bind both PI(4,5)P2 and PI(3,4,5)P3, albeit with ∼3-fold higher and ∼100-fold lower affinities, respectively, compared with the diglycine variant.
Phosphoinositide binding of CYTH1 regulates membrane ruffling and cell migration
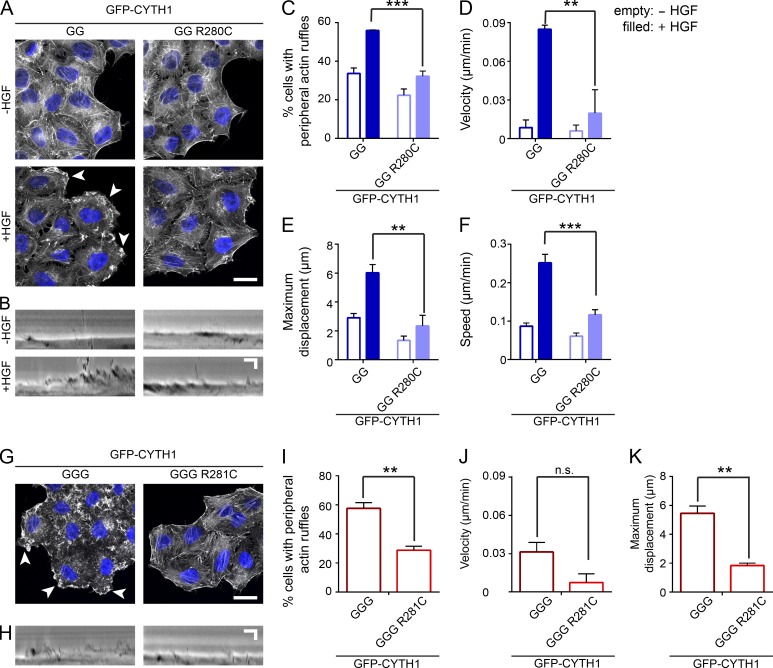
To test directly whether phosphoinositide recognition by cytohesin-1 is required for the formation of cell protrusions and HGF-dependent cell migration, we compared CYTH1 KO cells expressing WT diglycine GFP-CYTH1 or the R280C mutant using F-actin staining and live-cell imaging. In response to HGF, cells expressing the diglycine GFP-CYTH1 (R280C) had a reduced capacity to form peripheral actin ruffles compared with cells expressing a WT diglycine GFP-CYTH1 (Fig. 5, A and C). The velocity and maximum displacement of the leading edge were also reduced in cells expressing the R280C diglycine mutant compared with WT GFP-CYTH1 (23% and 39% of WT, respectively; Fig. 5, B, D, and E). Consistent with this, HGF-dependent cell migration was reduced in cells expressing the diglycine GFP-CYTH1 (R280C) compared with WT (47% of control; Fig. 5 F), supporting that phosphoinositide engagement by diglycine cytohesin-1 is required for HGF-dependent cell migration.
Figure 5.
Cytohesin-1 phosphoinositide binding is required for membrane ruffling and HGF-dependent cell migration. (A) Confocal images of cells counterstained with phalloidin (F-actin) and DAPI, with or without HGF. (B) Kymographs were generated from linescans of the cells’ leading edge imaged between 15 and 60 min after HGF treatment. (C) Quantification of experiments shown in A. (D and E) Quantification of experiments shown in B. (F) Random cell migration of diglycine EGFP-CYTH1 or R280C-ovexpressing cells treated with or without HGF. (G) Confocal images of cells counterstained with phalloidin (F-actin) and DAPI and treated with or without HGF (H) Kymographs were generated from linescans of the cells’ leading edge imaged between 15 and 60 min after HGF treatment. (I) Quantification of experiments shown in G. (J and K) Quantification of experiments shown in H. Scale bars, 20 µm. Arrowheads indicate peripheral membrane ruffles. All quantified data indicate mean values ± SEM from three independent experiments. **, P < 0.01; ***, P < 0.001; n.s., not significant.
To establish if phosphoinositide binding was required for the constitutive membrane ruffling induced by overexpression of the triglycine variant, CYTH1 KO cells expressing WT triglycine GFP-CYTH1 or R281C (equivalent to diglycine R280C) were examined. Cells expressing the R281C mutant showed significantly fewer peripheral actin ruffles than WT (Fig. 5, G and I). When comparing the membrane dynamics of these cells, cells expressing triglycine GFP-CYTH1 R281C demonstrated reduced maximum displacement compared with WT (33.5% of WT triglycine; Fig. 5, H, J, and K). Hence, phosphoinositide recognition is a required step for both diglycine cytohesin-1–dependent membrane ruffling in response to HGF and constitutive membrane ruffling promoted by triglycine cytohesin-1.
Selective membrane recruitment of cytohesin-1 splice variants
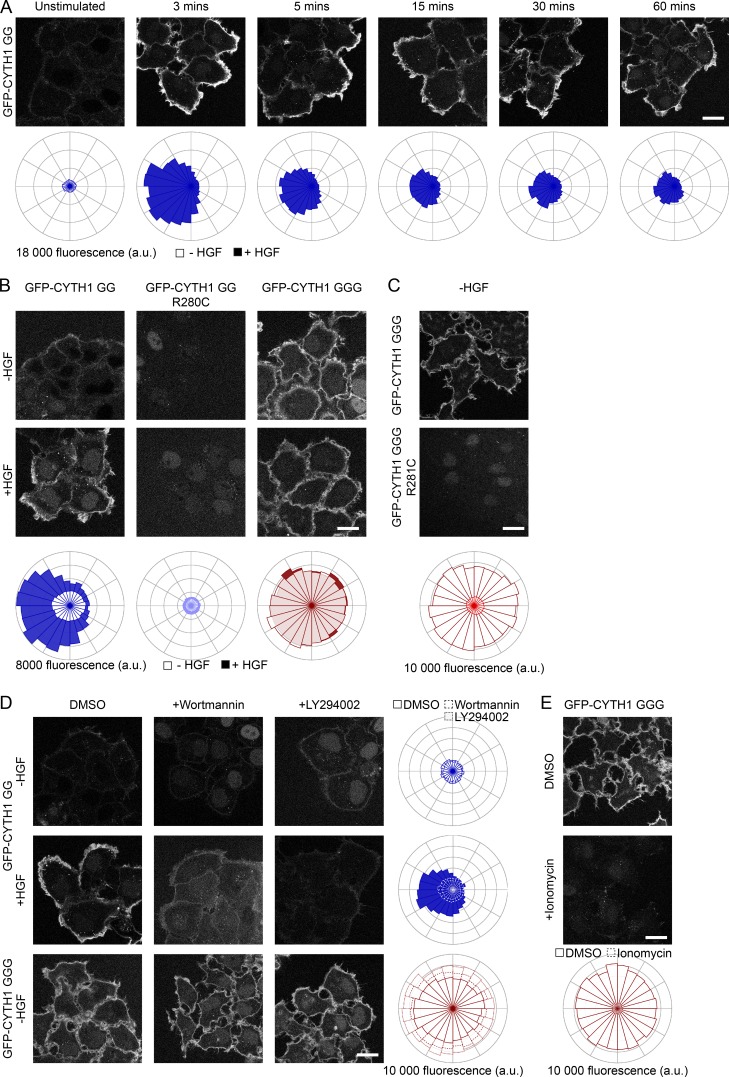
The abundance of PI(4,5)P2 at the plasma membrane is approximately two orders of magnitude higher than that of PI(3,4,5)P3 (Stephens et al., 1991; Malek et al., 2017). However, HGF stimulation activates PI3K, increasing PI(3,4,5)P3 and recruitment of PI(3,4,5)P3 binding proteins to PI(3,4,5)P3 microdomains (Maroun et al., 1999b). Hence, to test if the diglycine variant of cytohesin-1 is specifically recruited to the plasma membrane upon PI3K activation in response to HGF, cells expressing GFP-CYTH1 splice variants were stimulated with HGF for the indicated times, and membrane-bound cytohesin-1 was imaged by partially permeabilizing cells allowing for cytosolic GFP-CYTH1 to dissipate (Fig. S4 A and Video 1). Diglycine GFP-CYTH1 localized to the plasma membrane within 3 min after HGF stimulation, and recruitment was maintained for up to 60 min (Fig. 6 A). Notably, diglycine GFP-CYTH1 membrane localization was polarized toward the leading edge of the cell upon HGF stimulation. Importantly, mutation of the phosphoinositide-binding pocket (R280C) abrogated recruitment of diglycine GFP-CYTH1 to the leading edge (Fig. 6 B). In contrast, the triglycine GFP-CYTH1 variant is constitutively associated with the plasma membrane and is observed throughout the cell perimeter (Fig. 6 B). Recruitment was not further enhanced by HGF treatment, supporting a distinct mechanism of membrane recruitment that is distinct but also dependent on phosphoinositide binding, since a mutation in the phosphoinositide-binding pocket (R281C) abrogated recruitment of triglycine cytohesin-1 to the plasma membrane (Fig. 6 C).
Figure 6.
Cytohesin-1 localization is defined by G272. (A) HeLa cells stably expressing EGFP-tagged diglycine CYTH1 were either not treated (−, unfilled) or treated (+, filled) with HGF for the indicated time points, permeabilized with ice-cold 0.05% saponin in Pipes buffer, and imaged by confocal microscopy. (B) HeLa cells stably expressing EGFP-tagged variants of CYTH1 were either not treated (−, unfilled) or treated (+, filled) with HGF for 15 min, permeabilized with ice-cold 0.05% saponin in Pipes buffer, and imaged by confocal microscopy. (C and D) Cells were prepared as in A, except they were pretreated with DMSO, 0.2 µM Wortmannin, or 20 µM LY294002 for 30 min before HGF stimulation. (E) HeLa cells stably expressing triglcyine EGFP-CYTH1 were pretreated with 10 µM DMSO or ionomycin for 10 min, permeabilized with ice-cold 0.05% saponin in Pipes buffer, and imaged by confocal microscopy. Scale bars, 20 µm.
To test whether plasma membrane recruitment of cytohesin-1 was dependent on PI3K activity, cells expressing the diglycine or triglycine GFP-CYTH1 were pretreated with pan-PI3K inhibitors Wortmannin and LY294002 and localization of GFP-CYTH1 assessed. Pretreatment with either inhibitor abrogated HGF-dependent membrane recruitment of diglycine GFP-CYTH1, whereas localization of the triglycine GFP-CYTH1 to the plasma membrane was not significantly altered (Fig. 6 D). By comparing cells overexpressing the WT or catalytically dead PI(3,4,5)P3 lipid phosphatase (PTEN), we found that increased PTEN activity attenuated diglycine GFP-CYTH1 membrane recruitment (Fig. S5). These data support a requirement for PIP3 in diglycine GFP-CYTH1 localization. Consistent with the increased affinity of triglycine GFP-CYTH1 for PI(4,5)P2, pretreatment of cells with ionomycin to reduce plasma membrane levels of PI(4,5)P2 (Botelho et al., 2000) abolished peripheral localization of triglycine GFP-CYTH1 (Fig. 6 E). Together, these data show that cytohesin-1 splice variants are differentially recruited to the plasma membrane in vivo. Whereas the diglycine variant binds PI(3,4,5)P3 generated downstream from Met and growth factor signaling, the triglycine variant is constitutively recruited to the plasma membrane in a PI(4,5)P2-dependent manner. Therefore, the phosphoinositide-binding specificity of microexon-containing splice variants of CYTH1 defines the context for membrane ruffling, cell migration, and invasion.
Discussion
Initiation of cellular signaling through activation of RTKs is well recognized as a key event in cellular mitogenic or morphogenic response to growth factors. However, the cooperating molecular determinants of signal localization for these processes are still poorly understood. Arf proteins have been implicated in multiple biological processes that involve membrane ruffling, including cancer cell invasion (Morishige et al., 2008), Met-dependent migration (Parachoniak et al., 2011), and bacterial invasion (Humphreys et al., 2016). Activation of Arf proteins is modulated by a family of Arf GEFs, yet specificity for distinct Arf GEF functions is still poorly understood. In our screen of Arf GEFs that regulate cell migration in response to the proto-oncogenic RTK Met, we observed among six Arf GEFs expressed in HeLa cells, a specific requirement for cytohesin-1. Cytohesin proteins regulate phagocytosis in Dictyostelium discodium (Müller et al., 2013), adhesion of lymphoid cells (El azreq and Bourgoin, 2011), kidney repair following acute injury (Reviriego-Mendoza and Santy, 2015), and processes associated with cell migration.
Previous evidence suggests specific roles for cytohesins in the contexts of RTK signaling and integrin trafficking. For example, cytohesin-3, but not cytohesin-2, acts downstream of Met to promote integrin recycling and angiogenesis in endothelial cells (Hongu et al., 2015), whereas silencing cytohesin-2 or cytohesin-3 differentially affects β1 integrin trafficking in HeLa cells (Oh and Santy, 2010). The localization of cytohesin-1 at the periphery of the cell and known roles for Arf6 in endocytosis suggest that cytohesin-1 isoforms may differentially regulate Met endocytosis and trafficking (Mayor et al., 2014). While the different functions attributed to cytohesin proteins may be due to their differential expression (Ogasawara et al., 2000; Oh and Santy, 2012), their cognate mRNAs are also alternatively spliced, and the specific roles of the resulting isoforms have not been examined in the context of RTK signaling. Using multiple approaches, including structure function, live cell imaging, and CRISPR/Cas9-mediated genetic deletion, we find that a splice variant of cytohesin-1 lacking a 3-nt microexon is specifically required for HGF activation of Met RTK-dependent cytoskeletal dynamics and cell migration.
Alternative splicing diversifies the number of possible transcripts from a single gene. Indeed, it is believed that ∼95% of multiexon genes undergo alternative splicing (Pan et al., 2008). While significant effort has been put into establishing the regulatory mechanisms of alternative splicing, the functional significance of many of these events remains unknown. Microexons have been reported in both plants and metazoan (Beachy et al., 1985; Guo and Liu, 2015), and their role in generating alternative transcripts is becoming increasingly appreciated (Ustianenko et al., 2017). Many human diseases are characterized in part by defects or alterations in alternative splicing patterns (Baralle and Giudice, 2017), with emerging evidence suggesting the involvement of microexon splicing in their etiology. For example, microexons have been implicated in autism spectrum disorders, which is in keeping with their abundance in neuron-specific transcripts (Irimia et al., 2014). In a small cohort of symptomatic neuroblastoma patients, a highly phosphorylated neuronal microexon–containing splice variant of the non-RTK Src is lost (Brugge et al., 1985; Pyper and Bolen, 1990; Matsunaga et al., 1993; Keenan et al., 2015). In general, however, information on the roles of microexon splicing in cancer is very limited. This work defines a functional role for microexon alternative splice variants in cancer cell migration, suggesting the potential importance of microexons in processes involved in metastatic behavior, and emphasizes the importance of developing a functional understanding of the roles of microexons in health and disease.
Previous studies have shown that isoforms of cytohesin family members interact selectively with PI(4,5)P2 or PI(3,4,5)P3 (Klarlund et al., 2000; Cronin et al., 2004). However, a functional distinction between microexon splice variants has not been addressed. Here, we show by ITC and confocal microscopy that microexon splicing is a novel phosphoinositide switch whereby the shorter cytohesin-1 isoform lacking a single glycine residue (diglycine) binds PI(3,4,5)P3 in vivo and the longer isoform (triglycine) binds PI(4,5)P2. Thus, the diglycine cytohesin-1 variant, but not triglycine cytohesin-1, is a molecular link between Met activation, PI3K signaling, Arf6, and HGF-dependent biology. Initial data from brain tissue indicated that triglycine PI(4,5)P2-binding cytohesin-1 was the predominant isoform (Ogasawara et al., 2000). However, recent analysis of RNA-sequencing data from a variety of tissues indicates that PSI values for the cytohesin-1 microexon can vary from 25% in liver and epithelial cells (predominantly diglycine) to >95% in muscle and white blood cells (predominantly triglycine; Irimia et al., 2014). These values may reflect a need for constitutive versus growth factor dependent activation of Arf proteins in different tissues. To our knowledge, our study is the first to characterize a functional difference between cytohesin-1 splice variants. This points to a key role for these proteins and their splicing in normal development and disease.
HGF stimulation of Met promotes PI3K activation and recruitment of diglycine cytohesin-1 to the leading edge for prolonged periods (60 min). These data are consistent with an HGF-dependent rapid and prolonged recruitment of PI3K to a Met signaling complex at the plasma membrane and generation of PI(3,4,5)P3 at the leading edge of migrating cells (Maroun et al., 1999a,b; Frigault et al., 2008; Abella et al., 2010). The Gab1 scaffold is the major determinant for recruitment of the p85 adapter protein and PI3K activation following HGF stimulation of Met (Maroun et al., 1999b). Notably, in contrast to HGF, EGF stimulation promotes a transient increase in PI3K activity associated with Gab1 (Maroun et al., 1999b), and in response to EGF, cytohesin-1 is transiently recruited to the plasma membrane, supporting our model that diglycine cytohesin-1 is an important PI3K effector (Venkateswarlu et al., 1999). While the role for PI3K in cell migration has been examined extensively, many studies have focused on activation of the serine/threonine kinase Akt downstream of PI(3,4,5)P3 in cell proliferation and survival (Fruman et al., 2017). It is now understood that the modulation of cell migration by PI3K may predominantly involve Akt-independent pathways that are not well understood (Lien et al., 2017). PI(3,4,5)P3 is recognized by multiple GEFs, including DOCK180, Vav2, and P-REX1, that activate Rac to promote cell migration (Côté et al., 2005; Ménard et al., 2014; Graziano et al., 2017). Arf6 also acts upstream of the Rac1 GEF DOCK 180 to regulate cell migration (Santy et al., 2005; Koubek and Santy, 2018).
Within this context, our findings establish a function for the evolutionarily conserved alternatively spliced microexon in cytohesin-1 in phosphoinositide signaling, membrane dynamics and cancer cell migration. The in vivo relevance of microexons is only beginning to be understood and may have wide ranging implications from normal development, neurological disease, and cancer.
Materials and methods
Cell culture and transfection
HeLa and 293T cell lines were cultured under standard conditions at 37°C and 5% CO2 in 10% FBS. CYTH1 KO clonal lines were generated using the lentiCRISPR v2 system (Sanjana et al., 2014). Briefly, phosphorylated and annealed CYTH1-specific guide RNAs were cloned into the lentiCRISPRv2 vector using BsmBI restriction sites. Lentiviral particles were produced by Lipo2000 transfection of 293T cells with EGFP-CYTH1, psPAX2, and pMD2.G vectors according to the manufacturer’s protocol. Filtered supernatant was then used to infect HeLa cells, and 24 h after infection, cells were selected in puromycin (P7255; Sigma Aldrich) for 2 d. Clonal populations were established by limiting dilution and screened for cytohesin-1 expression by Western blot. To establish stable cell lines expressing EGFP-CYTH1 diglycine and triglycine isoforms, as well as mutants, triglycine EGFP-CYTH1 was first PCR amplified and subcloned into a pLVX-IRES-Hyg vector. NEB Q5 site-directed mutagenesis was used to generate protospacer adjacent motif silent mutations, the diglycine isoform, as well as cytohesin-1 mutants. Lentiviral particles were produced by Lipo2000 transfection of EGFP-CYTH1, psPAX2, and pMD2.G vectors into 293T lines. Filtered supernatant containing lentiviral particles was then concentrated by adding 1 vol PEG8000 to 3 vol supernatant, incubating overnight at 4°C, centrifuging at 2,750 g for 30 min at 4°C, and resuspending the pellet in DMEM. HeLa clonal population 1 expressing lentiCRISPR v2 gRNA3 was infected, and 2 d later, stable cell lines were selected and cultured in 10% FBS in DMEM and 600 µg/ml Hygromycin (10687010; Thermo Fisher). mCherry2-PTEN was generated by PCR amplifying mCherry2 (plasmid 54563; Addgene) and subcloning into pcDNA3 GFP-PTEN (plasmid 10759; Addgene) between BamHI and HindIII restriction sites. NEB Q5 site-directed mutagenesis was used to generate the C124S mutant. Stable cell lines expressing mCherry2-PTEN WT or C124S were generated by transfecting pcDNA3 vectors and, after 48 h, culturing in standard media with 1.2 mg/ml G418. After 1 wk, mCherry2-positive cells were sorted by FACS, and the mCherry2+ population was expanded.
Antibodies and reagents
Commercial antibodies to cytohesin-1 (2E11; mouse MA1-060) were obtained from Thermo Fisher; GAPDH (rabbit sc-25778) from Santa Cruz; GFP (rabbit A6455) from Life Technologies; and Akt (mouse 2920S), p-Akt1 (rabbit 9018S), p44/42 MAPK (mouse 9107S), p-p44/42 MAPK (T202/Y204; rabbit 9101L), Met (mouse 3127L), p-Met (rabbit 3077S), and PTEN (rabbit 9188S) from Cell Signaling. I(1,4,5)P3 (Q-0145), I(1,3,4,5)P4 (Q1345), and PI(3,4,5)P3 PolyPIPosomes (Y-P039) were obtained from Echelon Biosciences. Wortmannin (W-2990) was obtained from LC Laboratories, LY294002 (S1105) from Selleckchem, and ionomycin calcium salt from Streptomyces conglobatus (I0634) from Sigma Aldrich. AllStars Neg. Control siRNA (1027281) was obtained from QIAGEN. siRNA targeting Arf1 (5′-ACGUGGAAACCGUGGAGUA-3′), Arf3 (5′-UAUGAACGCUGCUGAGAUC-3′), and Arf6 (5′-GAUGAGGGACGCCAUAAUC-3′) were obtained from Dharmacon. SMARTpool siRNA against human CYTH1 (9267: 5′-GGAAUCAUCCCUUUAGAGA-3′, 5′-AAGAGACGCUGGUUCAUUC-3′, 5′-GGGAGAGAGAUGAGUUUAA-3′, 5′-GCAAUAAUGGCGUGUUCCA-3′), CYTH2 (9266: 5′-GCAAUGGGCAGGAAGAAGU-3′, 5′-AAACCGAACUGCUUUGAAC-3′, 5′-UGGCAGUGCUCCAUGCUUU-3′, 5′-GCCAAUGAGGGCAGUAAGA-3′), CYTH3 (9265: 5′-GGAGAAGGCCUAAAUAAGA-3′, 5′-CAGCAGAGAUCCCUUCUAU-3′, 5′-GAAGACCUCUCAUUAGAAG-3′, 5′-GGAAUCAUCCCGUUGGAAA-3′), IQSEC1 (9922: 5′-GGAAGAAAUUCACCGAUGA-3′, 5′-CGAGAAAUCUUCCUGUUCA-3′, 5′-GGACGAUGGUGAGGACAUU-3′, GACAGUCCUUCUCCUUGUA-3′), IQSEC2 (23096: 5′-GAAAUACCGUGUGGAGUCG-3′, 5′-GUACAGUCCCAGCGUCAAA, 5′-CCCAGAAAGUGGAGCGACU-3′, 5′-UCUGUAGAGUUGUGGCCAA-3′) and PSD3 (23362: 5′-CAACGAAUUUAGCAAACUA, 5′-GGACGUCGAUGAGUACAAA-3′, 5′-GACCAUCAGUCGUAUUGGA-3′, 5′-UGUCAGGAGUUCAUUGCAA-3′) were obtained from Dharmacon. All the following oligonucleotides were obtained from Integrated DNA Technologies. DNA oligonucleotides used for quantitative RT-PCR were CYTH1_FWD(5′-CAGTGACCTGACAGCAGAGG-3′) and REV(5′-TCTGAATGTCAGCCAGCAGC-3′); CYTH2_FWD(5′-AACCTGTACGACAGCATCCG-3′′) and REV(5′-CCGGTCCGGGTTGAAGAAG-3′); CYTH3_FWD(5′-CGTGCCTGAAGACCTCTCAT-3′) and REV(5′-TCGTTTTGCTCTCCTCTACGG-3′); IQSEC1_FWD(5′-GCACAGGATAGAGTCGGAGC-3′) and REV(5′-CCCGACTCCTTTTTGAGGCT-3′); IQSEC2_FWD(5′-TCCAGTCCCATATCCGGGTT-3′) and REV(5′-GAGGGCTGGGTTACAGACAC-3′); IQSEC3_FWD(5′-CTACCACTGCGAGAACCCAG-3′) and REV(5′-GATGCCCTTGTCGGGGTTTA-3′); PSD_FWD(5′-GGCTGTACCGACTAGATGGC-3′) and REV(5′-AGCTTGGTCCAGAGTCATGC-3′); PSD2_FWD(5′-ACCCTGATGACAGCACTTCG-3′) and REV(5′-TTTTGCCAATGTTGTGGCCG-3′); PSD3_FWD(5′-AGCGTGGCACATGAACAAAC-3′) and REV(5′-CCTCGACCCTTCCCCTAGAA-3′); and PSD4_FWD(5′-TTGGAGGCCATGTTTGGGTC-3′) and REV(5′-CACACTGACACACCTCCCTC-3′). DNA oligonucleotides used for determination of cytohesin family microexon splicing patterns were CYTH1_TIDE_FWD(5′-GCAGAGGAGCGTCAAGAACT-3′), CYTH1_TIDE_REV(5′-CGTAGAAAGGGTCCCTGCTG-3′), CYTH1_TIDE_seq(5′-GATAAGCCCACTGTGGAGAGGTT-3′); CYTH2_TIDE_FWD(5′-GAGGACGGCGTCTATGAACC-3′), CYTH2_TIDE_REV(5′-TGCTCCTGCTTCTTCTTGACT-3′), CYTH2_TIDE_seq(5′-GACAAGCCGGGCCTGGAGCGCTT3′); CYTH3_TIDE_FWD(5′-CGTGCCTGAAGACCTCTCAT-3′), CYTH3_TIDE_REV(5′-TCCTCGTTGCCAACATGTCA-3′) and CYTH3_TIDE_seq(5′-GACAAGCCCACGGCAGAACGGTT-3′). DNA oligonucleotides used to generate CYTH1 gRNA1_FWD(5′-CACCGGAACATCCGACGGAGAAAAC-3′) and REV(5′-AAACGTTTTCTCCGTCGGATGTTCC-3′); and CYTH1 gRNA3_FWD(5′-CACCGGCAGCTCCTGTTTTCTCCGT-3′) and _REV(5′-AAACACGGAGAAAACAGGAGCTGCC-3′). DNA oligonucleotides used to subclone cyothesin-1 were CYTH1_Xho1_EGFP FWD(5′-AAAACTCGAGATGGTGAGCAAGGGCGAGGAGC-3′), CYTH1_Xba1 Rev(AAAATCTAGATCAGTGTCGCTTCGTGGAGG); CYTH1_BamH1-PH_257_FWD(5′- AAAAGGATCCACGTTCTTCAACCCAGACCGAG-3′) and CYTH1_EcoR1-PH_381_REV (5′-TTTTGAATTCCTAGAAAGGGTCCCTGCTGATGGCTGCTTTAATGC-3′); and CYTH1_BamH1-PH_243 _FWD(5′-AAAAGGATCCCCCTTTAAAATCCCAGAAGAC-3′) and CYTH1_EcoR1-PH_397_REV(5′-TTTTGAATTCTCAGTGTCGCTTCGTGGAGGA-3′). DNA oligonucleotides used to subclone mCherry2 were HindIII-mCherry2_FWD(5′-AAAAAAGCTTGCCACCATGGTGAGCAAGGGCGAGGAGG-3′) and BamHI-mCherry2_REV(5′-TTTTGGATCCCTGAAGCTGCTGCTTGTACAGCTCGTCCATGCC-3′). DNA oligonucleotides used for site directed mutagenesis were CYTH1_gRNA3_PAM_SDM_FWD (5′-TGGAGAACATTCGACGGAGAAA-3′) and REV(5′-GTTCTTGACGCTCCTCTGCTG-3′); CYTH1_3Gto2G_SDM_FWD(5′-TGGCAGGGTAAAGACTTGGAAGAG-3′) and REV(5′-CCGAGTTTCAATAGCCAGCCTT-3′); CYTH1_E157K_SDM_FWD(5′-GCTACCCGGAAAGGCCCAGAA-3′) and REV(5′- CGGAAGCTCCACAGGAACTG-3′); CYTH1_R281C_SDM_FWD(5′-TTGGAAGAGATGCTGGTTCATTC-3′), GGG_R281C_REV(5′-GTCTTTACCCTGCCACCTC-3′) and GG_R280C_REV(5′-GTCTTTACCCTGCCACCG-3′); PTEN_C124S_FWD(5′-AGCAATTCACGCTAAAGCTGGAAAGGG-3′) and PTEN_C124S_REV(5′-GCAACATGATTGTCATCTTC-3′). All vectors were sequenced verified before use.
Microexon splicing patterns
Microexon PSI values were determined by adapting a protocol designed for estimating the frequency of genome editing events (Brinkman et al., 2014). cDNA from HeLa, MDA-MB-231, and HCC1143 cells was generated using the transcriptor first-strand cDNA synthesis kit (Roche). PCR reactions using cDNA as a template and primers flanking the microexon were performed using Dreamtaq green PCR master mix (Thermo Fisher). PCR conditions were 1 min at 95°C (once); 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C (35 times); and 5 min at 72°C (once). Amplicons were purified and sent for Sanger sequencing. The resulting chromatograms were analyzed using TIDE software (https://tide.deskgen.com) to determine PSI values.
Cell migration
Wells of a 24-well dish were coated with 25 µg/ml collagen for 1 h at 37°C and washed twice with PBS. Cells were plated at 7,500 cells per well in collagen-coated wells and, where indicated, immediately transfected with 20 nM siRNA using HiPerfect per the manufacturer’s instructions. Assays were performed 24–48 h after plating. Media was aspirated and replaced with growth media or growth media containing 0.5 nM HGF. The dish was then transferred to and images captured with an Axiovert 200 M inverted microscope (Carl Zeiss), LD A-Plan 20×/0.3 Ph1 objective lens, and AxioCam HRM (Carl Zeiss); all were contained within a transparent environment chamber Climabox (Carl Zeiss) maintained at 5% (vol/vol) CO2 at 37°C and driven by AxioVision LE software (Carl Zeiss). The motorized stage was preprogrammed to advance to defined locations, and images were captured every 15 min for 24 h. Three independent fields of view were then captured per condition, and 10 cells were tracked per field of view. Tracks between 16 and 24 h after stimulation were used for quantification. Cells were tracked using the track points application in MetaMorph (Molecular Devices).
Collagen invasion assays
Invasion assays were performed as previously described, with minor modifications (Knight et al., 2018). Collagen gels (3 mg/ml in PBS, pH 7.0–7.5) were polymerized in chambers of an eight-well chamber slide. 5 × 104 cells were seeded, and these were overlayed with an additional layer of collagen. Media without or without HGF was then added, and cells were grown for 7 d before fixation. Media was changed every 2 d. Fixed collagen gels were embedded in optimal cutting temperature compound and immediately frozen in liquid nitrogen. 8-µm sections were mounted on poly-L-lysine–coated microscope slides and stained with DAPI before mounting with a glass coverslip. Sections were imaged at 10× magnification using the DAPI light cube on a EVOS FL Cell Imaging System.
Kymograph analysis
24 h after plating 7,500 cells per well in a collagen-coated 24-well dish, cells were rinsed twice with 0.02% FBS in DMEM, and 0.9 ml 0.02% FBS in DMEM was added. Cells were replaced in the incubator overnight. The following morning, 0.1 ml 0.02% FBS in DMEM or 0.5 nM HGF in 0.02% FBS in DMEM was added to each well. To analyze membrane dynamics, the dish was transferred to and imaged with a Axiovert 200 M inverted microscope, LD A-Plan 20×/0.3 Ph1 objective lens, and AxioCam HRM; all were contained within a transparent environment chamber Climabox maintained at 5% (vol/vol) CO2 at 37°C and driven by AxioVision LE software. The motorized stage was preprogrammed to advance to defined locations, and images were captured every 15 s, beginning 15 min after HGF addition for 1 h. Images generated between 15 and 60 min after HGF stimulation were used to generated kymographs using the kymograph function in MetaMorph.
Analysis of actin rearrangement
For analysis of the actin cytoskeleton, cells were prepared as for kymograph analysis, except 1 h after HGF addition, the cells were fixed with 4% PFA. Coverslips were then processed and counterstained with Alexa Fluor 546–labeled Phalloidin and DAPI. Images were acquired using a 63× Plan Apochromat, 1.40 NA oil immersion objective on a LSM 800 laser scanning confocal microscope (Carl Zeiss) driven by ZEN Blue software (Carl Zeiss). 10 independent fields of view were chosen per condition, and cells at the periphery of colonies were manually scored for actin rich ruffles at the periphery of the cell. More than 100 cells per condition were scored.
PH domain purification
All recombinant proteins were expressed using Rosetta-2(DE3) Escherichia coli cells (EMD Biosciences) and purified using Glutathione Sepharose 4B resin (GE Healthcare). Bacterial cultures were grown at 37°C to an OD600 of 0.7–08 and then induced using 1 mM IPTG and incubated overnight at 25°C. The cells were harvested using a JLA-10.1 rotor at 6,000 relative centrifugal force. The cell pellets were resuspended in buffer A (50 mM Na/K phosphate buffer, pH 8.0, 150 mM NaCl, 0.1% β-mercaptoethanol, and 1× protease inhibitor cocktail; Roche). 200 µg/ml Lysozyme and 10 U/ml DNase1 was added to the cells and, after a 30-min incubation on ice, samples were lysed by sonication, followed by addition of 0.5% Triton X-100. Lysates were then cleared by centrifugation at 4°C (JA-20; 35 min at 35,000 relative centrifugal force) and filtered using a 0.45-μM filter. Glutathione Sepharose resin was added to the soluble fraction followed by incubation at 4°C for 1 h and then three washes with buffer A. GST-tagged PH domains were then eluted with 50 mM reduced glutathione in buffer A, and 50 U Precission Protease (GE Healthcare) was added to the supernatant and the sample was dialyzed using a 3,500–molecular weight cut-off membrane (Spectrum Laboratories) against buffer B (25 mM Tris, 100 mM NaCl, 1 mM DTT, and 1 mM EDTA, pH 7.3) overnight at 4°C. The sample was then incubated with Glutathione Sepharose resin for 1 h to remove cleaved GST, followed by separation of the resin and concentration of the sample using an Amicon 10K unit (Millipore). The concentrated sample was then applied to a Sephadex S75 column (GE Healthcare) using MOPS (100 mM NaCl, pH 7.5) as the running buffer. Sample purity was assessed by SDS-PAGE, and the purest fractions were pooled for further experiments (Fig. S4 B). The concentration of the sample was calculated using the Bio-Rad Bradford Protein Assay.
ITC
All ITC experiments were performed on a MicroCal microcalorimetry system (GE Healthcare). PH domains, phosphoinositide liposomes (Echelon Bioscience), and inositol phosphates (Echelon Biosciences) were prepared in 1× MOPS. To measure binding kinetics to lipososomes, the reaction cell contained 17.5 µM of PI(3,4,5)P3-containing liposomes and was titrated with 155 µM of recombinant CYTH1. To measure binding kinetics to inositol phosphates, the reaction cell was filled with 25 µM CYTH recombinant protein and the sample was titrated with either I(1,3,4,5)P4 or I(1,4,5)P3 (Echelon Biosciences). All experiments were performed at 20°C, the reaction cell contained 320 µl of sample and 70 µl of the titrant in the syringe, which was set between 19 and 38 injections at 2.5–1.25 µl per injection. The binding isotherm was fitted with a model that uses a single set of independent sites to determine the stoichiometry and thermodynamic binding constant.
Modeling of the CYTH1 PH domain
The 3D protein structural model of the diglycine CYTH1 PH domain was generated using the methods described in Swiss-Model (https://swissmodel.expasy.org/; Arnold et al., 2006). After target and template selection using the software, the final CYTH1 model was built against the structure of the PH domain from cytohesin-3 (Protein Data Bank accession numbers 2R09 and 2R0D). The ligand present in the template structure was transferred by homology to the model. Additional analysis of the binding pocket with the I(1,4,5)P3 and I(1,3,4,5)P4 ligands was performed by overlaying the model structure to the PH domains from ARNO (Protein Data Bank accession numbers 1U27 and 1U29).
Imaging subcellular localization of EGFP-cytohesin-1
Assays were performed 48 h after plating 7.5 × 104 cells in an ibidi glass-bottom dish (81158). Media was aspirated, and cells were rinsed twice with 0.02% FBS in DMEM, and 1.35 ml 0.02% FBS in DMEM was added. Cells were replaced in the incubator for 2–3 h. A bolus of 5 nM HGF was added to each plate (Cf = 0.5 nM). After the indicated time points, media was aspirated and ice-cold 0.05% saponin in Pipes buffer (80 mM Pipes KOH, pH 7.0, 5 mM EGTA, and 1 mM MgCl2) was added. Cells were imaged immediately after the cytosolic fraction had dissipated (∼2–5 min). When cells were treated with DMSO or inhibitor, this was added 20 min before HGF stimulation (Wortmannin and LY294002) or 10 min before permeabilization (ionomycin). Images were acquired using a 63× Plan Apochromat, 1.40 NA oil immersion objective on a LSM 800 laser scanning confocal microscope driven by ZEN Blue software. Using MetaMorph, a linescan with a width of 10 pixels was then manually drawn along the perimeter of the cell, starting from the innermost point of the cell relative to the colony. Linescan length was normalized and divided into 24 equal sections. The mean fluorescence of each section was averaged across cells and plotted using the polar histogram function in MATLAB.
SDS-PAGE and immunoblotting
HeLa cells were lysed in Triton X-100-glycerol-Hepes lysis buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X-100, 10% glycerol, 1 mM PMSF, 1 mM sodium vanadate, 10 µg/ml aprotinin, and 10 µg/ml leupeptin). Equal amounts of protein were aliquoted, and SDS sample buffer was added and boiled for 5 min. Samples were then resolved by SDS-PAGE and transferred to Immobilon-FL polyvinylidene fluoride transfer membranes. Membranes were blocked with blocking buffer (LI-COR Biosciences) and incubated with primary antibodies, followed by incubation with infrared conjugated secondary antibodies before detection on the Odyssey IR Imaging System (LI-COR Biosciences).
Quantitative RT-PCR
Total RNA was isolated using QIAGEN AllPrep DNA/RNA mini kit as per the manufacturer’s instructions. 1,000 ng of total RNA was used for QIAGEN OneStep RT-PCR kit as per the manufacturer’s instructions, and data were collected and analyzed using a Roche LightCycler 480. Data were normalized to GAPDH.
Quantification and statistical analysis
Quantitative data are presented as the means ± SEM. Statistical significance was assessed using a two-tailed Student’s t test or two-way ANOVA where indicated in the figure legend. P values and the number of experiments used for quantification and statistical analysis are indicated in the corresponding figure legends.
Online supplemental material
Fig. S1 shows the impact of Arf1, Arf3, or Arf6 depletion on HGF-dependent remodeling of the actin cytoskeleton. Fig. S2 shows the expression of Arf GEFs in HeLa cells, CYTH family member expression in MDA-MB-231 and HCC1143 cells, and the efficiency of siRNA-mediated silencing. It also shows the protein levels of cytohesin-1 in control and CYTH1 KO lines. Fig. S3 shows Met protein stability and downstream signaling in response to HGF. Fig. S4 shows expression levels of overexpressed EGFP-tagged cytohesin-1 isoforms and mutants as well as the purified PH cytohesin-1 PH domains. Fig. S5 shows the localization of diglycine GFP-CYTH1 in PTEN-overexpressing cells. Video 1 shows the localization of diglycine EGFP-CYTH1 upon saponin permeabilization in HGF-treated cells.
Supplementary Material
Acknowledgments
We thank members of the Park laboratory and Harvey W. Smith for their helpful comments on the manuscript, Genentech for HGF, Dr. Audrey Claing and Dr. Pierre-Luc Boulay for triglycine EGFP-CYTH1 cDNA, and Dongmei Zuo and the McGill Advanced Bioimaging Facility for technical assistance.
This research was supported by the Fonds de Recherche du Québec – Santé (doctoral studentships to P.P. Coelho and C.D.H. Ratcliffe) and the Rosalind Goodman Commemorative Scholarship (C.D.H. Ratcliffe) and Canadian Institutes of Health Research foundation operating grants 148423 (N. Sonenberg) and 242529 (M. Park). M. Park holds the Diane and Sal Guerrera Chair in Cancer Genetics.
The authors declare no competing financial interests.
Author contributions: Conceptualization, C.D.H. Ratcliffe and M. Park; Methodology, C.D.H. Ratcliffe and N. Siiddiqui; Formal Analysis, C.R., N. Siiddiqui, P.P Coelho, and T.N. Cookey; Investigation, C.D.H. Ratcliffe, N. Siddiqui, P.P Coelho, and N. Laterreur; Resources, N. Sonenberg and M. Park; Writing – Original Draft, C.D.H. Ratcliffe; Writing – Review & Editing, C.D.H. Ratcliffe, N. Siddiqui, and M.P; Visualization, C.D.H. Ratcliffe and N. Siddiqui; Supervision, N. Sonenberg and M. Park.
References
- Abella J.V., Parachoniak C.A., Sangwan V., and Park M.. 2010. Dorsal ruffle microdomains potentiate Met receptor tyrosine kinase signaling and down-regulation. J. Biol. Chem. 285:24956–24967. 10.1074/jbc.M110.127985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J., and Schwede T.. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 22:195–201. 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- Baralle F.E., and Giudice J.. 2017. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 18:437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachy P.A., Helfand S.L., and Hogness D.S.. 1985. Segmental distribution of bithorax complex proteins during Drosophila development. Nature. 313:545–551. 10.1038/313545a0 [DOI] [PubMed] [Google Scholar]
- Botelho R.J., Teruel M., Dierckman R., Anderson R., Wells A., York J.D., Meyer T., and Grinstein S.. 2000. Localized biphasic changes in phosphatidylinositol-4,5-bisphosphate at sites of phagocytosis. J. Cell Biol. 151:1353–1368. 10.1083/jcb.151.7.1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman E.K., Chen T., Amendola M., and van Steensel B.. 2014. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42:e168 10.1093/nar/gku936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugge J.S., Cotton P.C., Queral A.E., Barrett J.N., Nonner D., and Keane R.W.. 1985. Neurones express high levels of a structurally modified, activated form of pp60c-src. Nature. 316:554–557. 10.1038/316554a0 [DOI] [PubMed] [Google Scholar]
- Casanova J.E. 2007. Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic. 8:1476–1485. [DOI] [PubMed] [Google Scholar]
- Chardin P., Paris S., Antonny B., Robineau S., Béraud-Dufour S., Jackson C.L., and Chabre M.. 1996. A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature. 384:481–484. 10.1038/384481a0 [DOI] [PubMed] [Google Scholar]
- Côté J.-F., Motoyama A.B., Bush J.A., and Vuori K.. 2005. A novel and evolutionarily conserved PtdIns(3,4,5)P3-binding domain is necessary for DOCK180 signalling. Nat. Cell Biol. 7:797–807. 10.1038/ncb1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin T.C., DiNitto J.P., Czech M.P., and Lambright D.G.. 2004. Structural determinants of phosphoinositide selectivity in splice variants of Grp1 family PH domains. EMBO J. 23:3711–3720. 10.1038/sj.emboj.7600388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J.G., and Jackson C.L.. 2011. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 12:362–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El azreq M.-A., and Bourgoin S.G.. 2011. Cytohesin-1 regulates human blood neutrophil adhesion to endothelial cells through β2 integrin activation. Mol. Immunol. 48:1408–1416. [DOI] [PubMed] [Google Scholar]
- Eyster C.A., Higginson J.D., Huebner R., Porat-Shliom N., Weigert R., Wu W.W., Shen R.-F., and Donaldson J.G.. 2009. Discovery of new cargo proteins that enter cells through clathrin-independent endocytosis. Traffic. 10:590–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigault M.M., Naujokas M.A., and Park M.. 2008. Gab2 requires membrane targeting and the Met binding motif to promote lamellipodia, cell scatter, and epithelial morphogenesis downstream from the Met receptor. J. Cell. Physiol. 214:694–705. [DOI] [PubMed] [Google Scholar]
- Frittoli E., Palamidessi A., Marighetti P., Confalonieri S., Bianchi F., Malinverno C., Mazzarol G., Viale G., Martin-Padura I., Garré M., et al. . 2014. A RAB5/RAB4 recycling circuitry induces a proteolytic invasive program and promotes tumor dissemination. J. Cell Biol. 206:307–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruman D.A., Chiu H., Hopkins B.D., Bagrodia S., Cantley L.C., and Abraham R.T.. 2017. The PI3K Pathway in Human Disease. Cell. 170:605–635. 10.1016/j.cell.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya M., Ohta S., Hara Y., Tamaki H., and Sakagami H.. 2016. Distinct subcellular localization of alternative splicing variants of EFA6D, a guanine nucleotide exchange factor for Arf6, in the mouse brain. J. Comp. Neurol. 524:2531–2552. 10.1002/cne.24048 [DOI] [PubMed] [Google Scholar]
- Gherardi E., Birchmeier W., Birchmeier C., and Vande Woude G.. 2012. Targeting MET in cancer: rationale and progress. Nat. Rev. Cancer. 12:89–103. 10.1038/nrc3205 [DOI] [PubMed] [Google Scholar]
- Gillingham A.K., and Munro S.. 2007. The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 23:579–611. 10.1146/annurev.cellbio.23.090506.123209 [DOI] [PubMed] [Google Scholar]
- Graziano B.R., Gong D., Anderson K.E., Pipathsouk A., Goldberg A.R., and Weiner O.D.. 2017. A module for Rac temporal signal integration revealed with optogenetics. J. Cell Biol. 216:2515–2531. 10.1083/jcb.201604113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., and Liu C.-M.. 2015. A single-nucleotide exon found in Arabidopsis. Sci. Rep. 5:18087 10.1038/srep18087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto S., Onodera Y., Hashimoto A., Tanaka M., Hamaguchi M., Yamada A., and Sabe H.. 2004. Requirement for Arf6 in breast cancer invasive activities. Proc. Natl. Acad. Sci. USA. 101:6647–6652. 10.1073/pnas.0401753101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongu T., Funakoshi Y., Fukuhara S., Suzuki T., Sakimoto S., Takakura N., Ema M., Takahashi S., Itoh S., Kato M., et al. . 2015. Arf6 regulates tumour angiogenesis and growth through HGF-induced endothelial β1 integrin recycling. Nat. Commun. 6:7925 10.1038/ncomms8925 [DOI] [PubMed] [Google Scholar]
- Humphreys D., Singh V., and Koronakis V.. 2016. Inhibition of WAVE Regulatory Complex Activation by a Bacterial Virulence Effector Counteracts Pathogen Phagocytosis. Cell Reports. 17:697–707. 10.1016/j.celrep.2016.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M., Weatheritt R.J., Ellis J.D., Parikshak N.N., Gonatopoulos-Pournatzis T., Babor M., Quesnel-Vallières M., Tapial J., Raj B., O’Hanlon D., et al. . 2014. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 159:1511–1523. 10.1016/j.cell.2014.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan S., Lewis P.A., Wetherill S.J., Dunning C.J., and Evans G.J.. 2015. The N2-Src neuronal splice variant of C-Src has altered SH3 domain ligand specificity and a higher constitutive activity than N1-Src. FEBS Lett. 589:1995–2000. 10.1016/j.febslet.2015.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarlund J.K., Tsiaras W., Holik J.J., Chawla A., and Czech M.P.. 2000. Distinct polyphosphoinositide binding selectivities for pleckstrin homology domains of GRP1-like proteins based on diglycine versus triglycine motifs. J. Biol. Chem. 275:32816–32821. 10.1074/jbc.M002435200 [DOI] [PubMed] [Google Scholar]
- Knight J.F., Lesurf R., Zhao H., Pinnaduwage D., Davis R.R., Saleh S.M.I., Zuo D., Naujokas M.A., Chughtai N., Herschkowitz J.I., et al. . 2013. Met synergizes with p53 loss to induce mammary tumors that possess features of claudin-low breast cancer. Proc. Natl. Acad. Sci. USA. 110:E1301–E1310. 10.1073/pnas.1210353110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight J.F., Sung V.Y.C., Kuzmin E., Couzens A.L., de Verteuil D.A., Ratcliffe C.D.H., Coelho P.P., Johnson R.M., Samavarchi-Tehrani P., Gruosso T., et al. . 2018. KIBRA (WWC1) Is a Metastasis Suppressor Gene Affected by Chromosome 5q Loss in Triple-Negative Breast Cancer. Cell Reports. 22:3191–3205. 10.1016/j.celrep.2018.02.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koubek E.J., and Santy L.C.. 2018. ARF1 and ARF6 regulate recycling of GRASP/Tamalin and the Rac1-GEF Dock180 during HGF-induced Rac1 activation. Small GTPases. 9:242–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Knight J.F., Park M., and Pendergast A.M.. 2015. Abl Kinases Regulate HGF/Met Signaling Required for Epithelial Cell Scattering, Tubulogenesis and Motility. PLoS One. 10:e0124960 10.1371/journal.pone.0124960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien E.C., Dibble C.C., and Toker A.. 2017. PI3K signaling in cancer: beyond AKT. Curr. Opin. Cell Biol. 45:62–71. 10.1016/j.ceb.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek M., Kielkowska A., Chessa T., Anderson K.E., Barneda D., Pir P., Nakanishi H., Eguchi S., Koizumi A., Sasaki J., et al. . 2017. PTEN Regulates PI(3,4)P2 Signaling Downstream of Class I PI3K. Mol. Cell. 68:566–580.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroun C.R., Holgado-Madruga M., Royal I., Naujokas M.A., Fournier T.M., Wong A.J., and Park M.. 1999b The Gab1 PH domain is required for localization of Gab1 at sites of cell-cell contact and epithelial morphogenesis downstream from the met receptor tyrosine kinase. Mol. Cell. Biol. 19:1784–1799. 10.1128/MCB.19.3.1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroun C.R., Moscatello D.K., Naujokas M.A., Holgado-Madruga M., Wong A.J., and Park M.. 1999a A conserved inositol phospholipid binding site within the pleckstrin homology domain of the Gab1 docking protein is required for epithelial morphogenesis. J. Biol. Chem. 274:31719–31726. [DOI] [PubMed] [Google Scholar]
- Matsunaga T., Shirasawa H., Tanabe M., Ohnuma N., Takahashi H., and Simizu B.. 1993. Expression of alternatively spliced src messenger RNAs related to neuronal differentiation in human neuroblastomas. Cancer Res. 53:3179–3185. [PubMed] [Google Scholar]
- Mayor S., Parton R.G., and Donaldson J.G.. 2014. Clathrin-independent pathways of endocytosis. Cold Spring Harb. Perspect. Biol. 6:a016758 10.1101/cshperspect.a016758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard L., Parker P.J., and Kermorgant S.. 2014. Receptor tyrosine kinase c-Met controls the cytoskeleton from different endosomes via different pathways. Nat. Commun. 5:3907 10.1038/ncomms4907 [DOI] [PubMed] [Google Scholar]
- Moravec R., Conger K.K., D’Souza R., Allison A.B., and Casanova J.E.. 2012. BRAG2/GEP100/IQSec1 interacts with clathrin and regulates α5β1 integrin endocytosis through activation of ADP ribosylation factor 5 (Arf5). J. Biol. Chem. 287:31138–31147. 10.1074/jbc.M112.383117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishige M., Hashimoto S., Ogawa E., Toda Y., Kotani H., Hirose M., Wei S., Hashimoto A., Yamada A., Yano H., et al. . 2008. GEP100 links epidermal growth factor receptor signalling to Arf6 activation to induce breast cancer invasion. Nat. Cell Biol. 10:85–92. 10.1038/ncb1672 [DOI] [PubMed] [Google Scholar]
- Müller R., Herr C., Sukumaran S.K., Omosigho N.N., Plomann M., Riyahi T.Y., Stumpf M., Swaminathan K., Tsangarides M., Yiannakou K., et al. . 2013. The cytohesin paralog Sec7 of Dictyostelium discoideum is required for phagocytosis and cell motility. Cell Commun. Signal. 11:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan-Chari V., Hoover H., Clancy J., Schweitzer J., Suckow M.A., Schroeder V., Castellino F.J., Schorey J.S., and D’Souza-Schorey C.. 2009. ADP-ribosylation factor 6 regulates tumorigenic and invasive properties in vivo. Cancer Res. 69:2201–2209. 10.1158/0008-5472.CAN-08-1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara M., Kim S.-C., Adamik R., Togawa A., Ferrans V.J., Takeda K., Kirby M., Moss J., and Vaughan M.. 2000. Similarities in function and gene structure of cytohesin-4 and cytohesin-1, guanine nucleotide-exchange proteins for ADP-ribosylation factors. J. Biol. Chem. 275:3221–3230. 10.1074/jbc.275.5.3221 [DOI] [PubMed] [Google Scholar]
- Oh S.J., and Santy L.C.. 2010. Differential effects of cytohesins 2 and 3 on β1 integrin recycling. J. Biol. Chem. 285:14610–14616. 10.1074/jbc.M109.043935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.J., and Santy L.C.. 2012. Phosphoinositide specificity determines which cytohesins regulate β1 integrin recycling. J. Cell Sci. 125:3195–3201. 10.1242/jcs.101683 [DOI] [PubMed] [Google Scholar]
- Palamidessi A., Frittoli E., Garré M., Faretta M., Mione M., Testa I., Diaspro A., Lanzetti L., Scita G., and Di Fiore P.P.. 2008. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell. 134:135–147. 10.1016/j.cell.2008.05.034 [DOI] [PubMed] [Google Scholar]
- Pan Q., Shai O., Lee L.J., Frey B.J., and Blencowe B.J.. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40:1413–1415. 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- Parachoniak C.A., and Park M.. 2012. Dynamics of receptor trafficking in tumorigenicity. Trends Cell Biol. 22:231–240. 10.1016/j.tcb.2012.02.002 [DOI] [PubMed] [Google Scholar]
- Parachoniak C.A., Luo Y., Abella J.V., Keen J.H., and Park M.. 2011. GGA3 functions as a switch to promote Met receptor recycling, essential for sustained ERK and cell migration. Dev. Cell. 20:751–763. 10.1016/j.devcel.2011.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powelka A.M., Sun J., Li J., Gao M., Shaw L.M., Sonnenberg A., and Hsu V.W.. 2004. Stimulation-dependent recycling of integrin β1 regulated by ARF6 and Rab11. Traffic. 5:20–36. 10.1111/j.1600-0854.2004.00150.x [DOI] [PubMed] [Google Scholar]
- Pyper J.M., and Bolen J.B.. 1990. Identification of a novel neuronal C-SRC exon expressed in human brain. Mol. Cell. Biol. 10:2035–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajadurai C.V., Havrylov S., Zaoui K., Vaillancourt R., Stuible M., Naujokas M., Zuo D., Tremblay M.L., and Park M.. 2012. Met receptor tyrosine kinase signals through a cortactin-Gab1 scaffold complex, to mediate invadopodia. J. Cell Sci. 125:2940–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe C.D.H., Sahgal P., Parachoniak C.A., Ivaska J., and Park M.. 2016. Regulation of Cell Migration and β1 Integrin Trafficking by the Endosomal Adaptor GGA3. Traffic. 17:670–688. [DOI] [PubMed] [Google Scholar]
- Reviriego-Mendoza M.M., and Santy L.C.. 2015. The cytohesin guanosine exchange factors (GEFs) are required to promote HGF-mediated renal recovery after acute kidney injury (AKI) in mice. Physiol. Rep. 3:e12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royal I., Lamarche-Vane N., Lamorte L., Kaibuchi K., and Park M.. 2000. Activation of cdc42, rac, PAK, and rho-kinase in response to hepatocyte growth factor differentially regulates epithelial cell colony spreading and dissociation. Mol. Biol. Cell. 11:1709–1725. 10.1091/mbc.11.5.1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santy L.C., Ravichandran K.S., and Casanova J.E.. 2005. The DOCK180/Elmo complex couples ARNO-mediated Arf6 activation to the downstream activation of Rac1. Curr. Biol. 15:1749–1754. 10.1016/j.cub.2005.08.052 [DOI] [PubMed] [Google Scholar]
- Simanshu D.K., Nissley D.V., and McCormick F.. 2017. RAS Proteins and Their Regulators in Human Disease. Cell. 170:17–33. 10.1016/j.cell.2017.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens L.R., Hughes K.T., and Irvine R.F.. 1991. Pathway of phosphatidylinositol(3,4,5)-trisphosphate synthesis in activated neutrophils. Nature. 351:33–39. 10.1038/351033a0 [DOI] [PubMed] [Google Scholar]
- Tushir J.S., and D’Souza-Schorey C.. 2007. ARF6-dependent activation of ERK and Rac1 modulates epithelial tubule development. EMBO J. 26:1806–1819. 10.1038/sj.emboj.7601644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustianenko D., Weyn-Vanhentenryck S.M., and Zhang C.. 2017. Microexons: discovery, regulation, and function. Wiley Interdiscip. Rev. RNA. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswarlu K., Gunn-Moore F., Tavaré J.M., and Cullen P.J.. 1999. EGF-and NGF-stimulated translocation of cytohesin-1 to the plasma membrane of PC12 cells requires PI 3-kinase activation and a functional cytohesin-1 PH domain. J. Cell Sci. 112:1957–1965. [DOI] [PubMed] [Google Scholar]
- Whitman M., Downes C.P., Keeler M., Keller T., and Cantley L.. 1988. Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature. 332:644–646. 10.1038/332644a0 [DOI] [PubMed] [Google Scholar]
- Yoo J.H., Shi D.S., Grossmann A.H., Sorensen L.K., Tong Z., Mleynek T.M., Rogers A., Zhu W., Richards J.R., Winter J.M., et al. . 2016. ARF6 is an actionable node that orchestrates oncogenic GNAQ signaling in uveal melanoma. Cancer Cell. 29:889–904. 10.1016/j.ccell.2016.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.