Maass et al. review interchromosomal interactions, which, like intrachromosomal interactions, are emerging as important regulators of genome organization and gene expression.
Abstract
Nuclei require a precise three- and four-dimensional organization of DNA to establish cell-specific gene-expression programs. Underscoring the importance of DNA topology, alterations to the nuclear architecture can perturb gene expression and result in disease states. More recently, it has become clear that not only intrachromosomal interactions, but also interchromosomal interactions, a less studied feature of chromosomes, are required for proper physiological gene-expression programs. Here, we review recent studies with emerging insights into where and why cross-chromosomal communication is relevant. Specifically, we discuss how long noncoding RNAs (lncRNAs) and three-dimensional gene positioning are involved in genome organization and how low-throughput (live-cell imaging) and high-throughput (Hi-C and SPRITE) techniques contribute to understand the fundamental properties of interchromosomal interactions.
Introduction
How DNA is folded and organized in the nucleus is a critical aspect of gene regulation and in turn of cell-fate determinations. Recent technical advances, such as molecular (e.g., chromosome conformation capture) and microscopic imaging approaches have provided important insight into where and how DNA is differentially packaged in normal and disease states. The nucleus is organized into chromosomal territories: each chromosome occupies its own distinct territory in the nuclear space (Cremer and Cremer, 2010). Many recent studies have resolved the fundamental properties of how chromosomes “fold” intra-molecularly into highly organized structures, such as topologically associated domains (TADs) and gene-enhancer looping in cis, which are readily detectable with molecular and microscopic approaches (Barutcu et al., 2018; Maass et al., 2018b). On a molecular level, these topological features are guided by a molecular toolkit consisting of lncRNAs, CTCF, cohesin, and other chromatin-associated protein complexes (Parelho et al., 2008; Rinn and Chang, 2012; Rinn and Guttman, 2014; Vance and Ponting, 2014; Dekker and Mirny, 2016; Engreitz et al., 2016). Despite the progress in understanding and mapping of intrachromosomal interactions, the fundamentals of interchromosomal organization remain poorly characterized. Therefore, elucidating the underlying mechanisms that form interchromosomal interactions is critical to understand the formation of nuclear bodies, as well as the crosstalk between chromosomal territories and DNA elements that regulate gene expression and are part of the 3D genome organization.
More than a decade ago, “intermingling” or “kissing” chromosomes were observed by microscopy approaches as overlapping regions between chromosomal territories (Branco and Pombo, 2006; Cremer and Cremer, 2010). We will refer to these interchromosomal interactions as non-homologous chromosomal contacts (NHCCs). The best known NHCC formation is the preassembly of the ribosomes by the coalescence of ribosomal RNA genes, encoded across five different human chromosomes, to form the nucleolus, the largest subnuclear compartment (McStay, 2016). Similarly, olfactory receptor (OR) genes are located across several different chromosomes and undergo a complex choreography to conglomerate in the same nuclear space (“olfactosome”) to regulate their expression (Lomvardas et al., 2006; Monahan et al., 2017, 2018).
In addition to the NHCCs forming the nucleolus and clusters of OR genes, NHCCs have been identified between defined genomic regions. For example, gene regulatory regions from one chromosome can activate a gene on another chromosome via NHCCs (Spilianakis et al., 2005). Moreover, lncRNA loci form NHCCs that affect gene-regulatory mechanisms in healthy and disease states (Maass et al., 2012; Hacisuleyman et al., 2014). In this paper, we will review studies identifying NHCCs in the context of a fundamental biological question: why and how do nonhomologous chromosomes communicate? In particular, we will provide a historical background and discuss the most recent findings on how NHCCs of coding and noncoding loci add information to the genome organization and to the control of gene regulation.
Principles of chromosomal structure and nuclear organization
Chromosomes are nonrandomly organized in defined territories within the nucleus, and these territories exist across different taxonomic orders: yeast, plants, and mammals (Fig. 1 A; Abranches et al., 2000; Parada et al., 2002, 2004; Bolzer et al., 2005; Branco and Pombo, 2006; Noma et al., 2006; Cremer and Cremer, 2010; Fritz et al., 2014; Sehgal et al., 2016; Maass et al., 2018c). For example, FISH experiments in various cells revealed that chromosomal territory arrangements are conserved in different primates (Old and New World monkeys) and in humans, suggesting a functional relevance for the spatial organization of the higher-order chromosomal architecture (Tanabe et al., 2002, 2005; Bolzer et al., 2005). However, chromosome and locus positioning can vary within the same organism, as has been described for different mouse cell types (Mayer et al., 2005; Hepperger et al., 2008).
Figure 1.
Interchromosomal interactions are a substantial part of genome organization and biological processes. (A) The interphase genome (1; Hoechst staining) consists of chromosomal territories (2) that share intermingling chromosomal regions in the 3D space of the nucleus. (3) Chromatin strands can loop out of their chromosomal territory and facilitate the formation of transcription factories. (B) The nucleus is organized in subnuclear compartments (e.g., speckles and nucleolus). In humans, five chromosomes are positioned around the nucleolus. (C) Intermingling chromosomal territories with highly specific NHCCs form the olfactosome to drive exclusive monogenic and mono-allelic OR gene expression. (D) Left: Heterochromatic foci appear as dense regions in the interphase genome, while dark (less dense) spots indicate less condensed chromatin. (1) Heterochromatic foci with H3K9me3 histone marks and silent gene loci can colocalize in the nucleus. (2) Transcription factors (TFs) facilitate the regulation of gene expression in tissue-specific transcription factories, where intermingling loci are in close spatial proximity.
Chromosomes are further organized into A- and B-type genomic compartments that represent active and inactive chromatin domains, respectively. The A-type compartment is associated with higher gene density and early replication—vice versa for the B-type compartment (Lieberman-Aiden et al., 2009; Pope et al., 2014). Each of these compartment types can consist of multiple subcompartments defined by their differential histone marks (Rao et al., 2014), and the changes in genomic compartmentalization are associated with changes in transcriptional activity (Barutcu et al., 2015; Dixon et al., 2015). Interactions between the compartments may be the driving force for the large-scale genomic organization, such as the nuclear speckles (Chen et al., 2018) or NHCCs.
A well-studied aspect of the nuclear architecture includes TADs and the intrachromosomal interactions (i.e., between enhancers and promoters) within them (Dixon et al., 2012; Nora et al., 2012; Sanyal et al., 2012; Terakawa et al., 2017; Ganji et al., 2018). TADs are clusters of genomic interactions 100 kb to 1 Mb in size in mammalian genomes (Dixon et al., 2012; Nora et al., 2012). The intra-TAD interactions are highly variable between cell types, but previous studies have shown that TADs are highly invariant between cell types and species (Dixon et al., 2012, 2015). More recently, single-cell studies determined that these structures are highly variable within individual nuclei (Nagano et al., 2013, 2017; Ramani et al., 2017; Stevens et al., 2017). Studying structural genomic variations, such as the deletion of boundaries between two TADs, revealed that this can result in disease phenotypes in some cases (Lupiáñez et al., 2015; Franke et al., 2016; Taberlay et al., 2016). However, in other cases, these perturbations do not lead to phenotypic changes with biological significance (Barutcu et al., 2018), unless larger regions between 200 and 400 kb of DNA are deleted (Nora et al., 2012; Rodríguez-Carballo et al., 2017). The genomic organization and its interactions within the TAD structure are explained by a model called “loop-extrusion” that proposes how intrachromosomal interactions bring otherwise distal, regulatory regions (e.g., enhancers) into 3D proximity to target genes (Fudenberg et al., 2016). This model suggests that two cohesin/condensin molecules slide toward each other while extruding the intervening DNA until two convergent CTCF sites are reached. The loop extrusion model has been shown to mediate intrachromosomal looping interactions and to form the majority of TADs (Fudenberg et al., 2016; Goloborodko et al., 2016; Ganji et al., 2018). A recent study has highlighted the possibility that large-scale genomic compartments can also be affected by the loop extrusion mechanism (Nuebler et al., 2018).
More recently, an emerging theme that explains the formation of cellular substructures via liquid-liquid phase separation has been proposed. Both DNA and RNA interact with proteins that harbor low-complexity regions (van der Lee et al., 2014; Protter et al., 2018) and form liquid, gel, or solid aggregates that may shape and compartmentalize the genome (Erdel and Rippe, 2018; Langdon et al., 2018; Maharana et al., 2018).
While great progress in understanding the intramolecular dynamics of chromosomal structure on a genomic scale have been made, there are several outstanding questions. What are the mechanisms of intrachromosomal interactions and, more importantly, of NHCCs? Are there common or distinctive molecular features between these two different classes of genomic interactions? We are beginning to understand the molecular mechanisms governing the principles of genome architecture, especially of intrachromosomal interactions, at an unprecedented pace. However, our understanding about the formation and the function of NHCCs is still in its infancy compared with other aspects in chromatin biology. Indeed, several studies have shown examples of NHCCs, as they are detectable in many contexts (Bolzer et al., 2005; Spilianakis et al., 2005; Branco and Pombo, 2006; Lomvardas et al., 2006; Maass et al., 2012, 2018b,c; Hacisuleyman et al., 2014; Horta et al., 2018; Quinodoz et al., 2018), yet their function remains elusive.
Kissing chromosomes: NHCCs
The first notion of NHCCs in the nucleus may have come from Carl Rabl, who, based on studies in sea urchins, proposed in 1885 that chromosomes occupy defined volumes at defined positions, and that they interact with neighboring chromosomes. Between 1902 and 1904, the married biologists Theodor Boveri and Marcella O’Grady Boveri studied the equine round worms Parascaris univalens and Parascaris equorum and postulated that chromosomal territories are stably arranged during interphase (Satzinger, 2008; Strickfaden et al., 2010). Their findings were validated by laser-UV-microbeam experiments in the 1980s (Cremer et al., 1982) and by radiosensitive and fluorescent replication labeling of neighboring chromosomal subdomains (Zink et al., 1998; Visser and Aten, 1999). Furthermore, it was shown that chromatin regions with high gene density and expression levels can extend from their chromosomal territory to colocalize at the interchromosomal space (Mahy et al., 2002); likewise, a mathematical model describes the probability that individual chromatin strands can pass through one another, thereby validating that direct interactions between different chromosomes are mathematically possible (Blackstone et al., 2011).
Among various subnuclear structures identified in the nucleus (Misteli et al., 1997; Lamond and Earnshaw, 1998; Pederson, 2002, 2011; Cremer and Cremer, 2010; Spector and Lamond, 2011), one of the most well-known and large-scale phenomena of NHCCs is the formation of the nucleolus. In human nuclei, ∼300 ribosomal genes located on five different acrocentric chromosomes (six in mouse) come into physical proximity to build the ribosomal preassembly in the nucleus (Fig. 1 B; Németh et al., 2010; Pliss et al., 2015; McStay, 2016). This spatial formation of the nucleolus is a conserved phenomenon and validates that nonhomologous chromosomes can intermingle in a nonrandom manner in all nuclei.
A structure equally as fascinating is the OR gene cluster, in which individual NHCCs allow the expression of single ORs in each cell to create a diverse repertoire of OR expression at the tissue level. At any given time, only a few of the ∼1,400 OR genes located on 18 different chromosomes converge in the same interchromosomal space (Horta et al., 2018). The regulation of OR genes is orchestrated by binding of Ldb1, Lhx2, and Ebf transcription factors to highly similar transcription factor motifs of multiple enhancers on different chromosomes, thereby leading to nondeterministic mono-allelic OR gene expression (Lomvardas et al., 2006; Markenscoff-Papadimitriou et al., 2014; Monahan and Lomvardas, 2015; Monahan et al., 2017, 2018). Remarkably, the monogenic and mono-allelic gene expression of OR genes is explained by the spatial clustering of inactive genes to the same heterochromatic foci in the olfactosome (Fig. 1, C and D; Clowney et al., 2012). Recent in situ Hi-C experiments of FACS-sorted, differentiated olfactory sensory neurons determined that, at very large scales (i.e., 500-kb resolution), NHCCs between OR genes are highly specific and frequent, and that they consist of multiple different chromosomes to regulate selectively and specifically the transcription of each individual OR gene (Horta et al., 2018).
NHCCs affect distinct transcriptional programs of biological pathways
At higher resolution, NHCCs have been observed between specific enhancer and gene targets. For example, the formation of an NHCC results in expression of cytokines in T cell types and also IFNγ expression in response to viral infection (Spilianakis et al., 2005; Apostolou and Thanos, 2008). Specifically, the promoter region of IFNγ on chromosome 10 is in physical proximity to regulatory regions of the Th2 cytokine locus on chromosome 11 to coordinate gene expression in a cell type–specific dynamic manner (Spilianakis et al., 2005). Taken together, these and other findings that we will discuss indicate that nonrandom NHCCs occur in homogenous cell populations to contribute to transcriptional regulation. For example, the formation of interchromosomal gene-specific regulatory events leads to the ability to smell or results in cytokine expression, underscoring the importance of NHCCs in diverse biological pathways. One common feature of these biological pathways, where NHCCs are involved, is that they occur in systems of variegated gene expression and seem to occur mostly in a cell type–specific fashion.
Furthermore, genomic interactions appear to be influenced by chromosomal location and transcription (Gandhi et al., 2012; Krueger et al., 2012). Human chromosomes share similar positions in interphase and prophase of the cell cycle (Chen et al., 2017), and the spatial positioning of genes and NHCCs in the 3D nucleus can be important for their transcriptional regulation (Kosak and Groudine, 2004; Maass et al., 2012). For instance, genes that are either actively transcribed or silent, are spatially recruited to subnuclear hubs, such as transcription factories and splicing speckles, consistent with the observation that experimental repositioning of chromosomes leads to gene expression changes (Finlan et al., 2008).
Gene positioning and NHCCs are also important during larval development in Drosophila melanogaster, where heterochromatin-mediated transcriptional silencing is due to discrete spatial proximity in the nucleus (Dernburg et al., 1996). In addition, mitotic processes, such as chromatin decondensation, and transcriptional activation can influence the genomic architecture and cause gene repositioning (Therizols et al., 2014). Indeed, the formation of NHCCs between early histone genes was accompanied by repositioning of gene loci toward the interior nucleus during the highest transcriptional activity in sea urchins (Matsushita et al., 2017). Similarly, a correlation between transcriptional activity and chromosomal intermingling occurs during differentiation of mouse embryonic stem cells (mESCs; Maharana et al., 2016). Evidence that stochasticity is involved in NHCC formation was described in the Drosophila eye, where the photoreceptor choice is driven by DNA elements that control allelic expression via NHCCs (Johnston and Desplan, 2014). Also, during myogenic differentiation, cell type–specific genes on different chromosomes have been shown to cluster in the nuclear space (Harada et al., 2015), and recently, a sequential FISH approach showed that sites of active transcription tend to interact more with other chromosomes than with sites along the same chromosome, supporting the idea that NHCCs are more frequent than previously appreciated (Shah et al., 2018). Collectively, these examples demonstrate a broad repertoire of interplay across chromosomes to establish cell-specific expression programs.
NHCCs and nuclear bodies
It has been proposed that a possible common feature of NHCCs can be the formation of subnuclear structures, such as the nucleolus. These subnuclear compartments can present transcriptional factories where many active genes from multiple chromosomes are brought into proximity to maintain their activated states (Sutherland and Bickmore, 2009; Ferrai et al., 2010; Edelman and Fraser, 2012; Papantonis and Cook, 2013). These transcription factories can take place between expression-dependent loci in cis (intrachromosomal; Tolhuis et al., 2002) or between NHCCs where transcriptionally active loci preferentially contact active rather than silent loci (Fig. 1; Brown et al., 1997; Osborne et al., 2004, 2007; Spilianakis et al., 2005; Ling et al., 2006; Lomvardas et al., 2006; Simonis et al., 2006; Zhao et al., 2006; Maass et al., 2012, 2018b; Hacisuleyman et al., 2014). Another well-studied example of a transcription factory is when active globin genes cooperate with hundreds of other transcribed genes both intrachromosomally and interchromosomally for efficient and coordinated transcriptional control (Schoenfelder et al., 2010). Molecular mechanisms are actively involved in forming NHCCs: for example, the genome organizer CTCF can generate a mono-allelic NHCC between the imprinted H19 locus/Igf2 and Wsb1/Nf1 (Ling et al., 2006).
Therefore, from transcription factories of related gene types to finer resolution of specific gene loci that are in physical proximity within the 3D space of the nucleus, it is becoming clear that NHCCs represent important regulatory interactions.
Interchromosomal contacts between homologous chromosomes (transvection)
Similar to contacts between nonhomologous chromosomes, nonmeiotic transvection between homologous chromosomes is another layer of epigenetic regulation to activate or repress genes. Transvection was microscopically observed in 1908 (Stevens, 1908), and the well-established example of interacting bithorax complexes in Drosophila (Lewis, 1954) is studied in the current era by live imaging (Lim et al., 2018). These homologous chromosomal contacts occur at the sites of DNA double-strand breaks (Gandhi et al., 2012). It was also shown in mESCs that DNA elements at the Oct4 promoter/enhancer locus mediate pairing of the Oct4 alleles, and perturbation of the Oct4/Sox2 binding sites at these elements leads to the disappearance of allele pairing (Hogan et al., 2015). Also, X-chromosome inactivation is accompanied by a transient physical interaction of both of the X-inactivation centers (Bacher et al., 2006), and the well-studied lncRNA XIST participates in the homologous chromosome pairing during X-chromosome inactivation (Marahrens, 1999).
However, the principle mechanisms of transvection and their impact on gene regulation and genome organization are not well understood and are out of the scope of this review about nonhomologous chromosomal contacts.
Toward identifying NHCCs with molecular techniques
The current understanding of nuclear organization derives from many technologies that have been developed to investigate the organizational features of chromatin and DNA packaging on a genomic scale. Multiple methods in both imaging (e.g., FISH and CRISPR live cell imaging [CLING]) and molecular (e.g., Hi-C and SPRITE) approaches probed the properties of chromosomal structure (Lieberman-Aiden et al., 2009; Maass et al., 2018a,b; Quinodoz et al., 2018). The advantage of imaging approaches is that they reveal cell-to-cell chromatin conformations, while the molecular methods assess the genome structure across a cell population.
The advent of chromatin conformation capture (3C) methods and their next-generation sequencing approaches (Dekker et al., 2002; Dekker, 2006; Ea et al., 2015; Barutcu et al., 2016), single-molecule imaging, and polymer simulations (Barbieri et al., 2013; Fudenberg and Imakaev, 2017) has rapidly expanded research in the genome organization field and resulted in a deeper understanding of genomic interactions. A plethora of advanced 3C-based methods, such as 4C (Simonis et al., 2006; Würtele and Chartrand, 2006; Zhao et al., 2006), 5C (Dostie et al., 2006), Hi-C (Lieberman-Aiden et al., 2009), ChIA-PET (Fullwood et al., 2009), capture Hi-C (Ma et al., 2015b), and the more recent genome architecture mapping, a ligation-free method to probe genomic interactions (Beagrie et al., 2017), resolve DNA–DNA interactions on a genome scale to unseal the entire intrachromosomal folding properties of each chromosome (Bickmore, 2013; Dekker and Misteli, 2015; Fraser et al., 2015; Barutcu et al., 2016; Bonev and Cavalli, 2016). More recently, improved Hi-C techniques measured interaction dynamics across individual nuclei within single nuclei (Nagano et al., 2013; Ramani et al., 2017; Stevens et al., 2017).
Surprisingly, many of the known NHCCs such as the nucleolus are not readily identified in many of the currently used 3C-based technical approaches. Apart from cell-to-cell variability, one reason for the limited evidence of functional NHCCs is most likely a bias of capturing predominantly intrachromosomal interactions rather than interchromosomal contacts in genome-wide 3C-based techniques (e.g., Hi-C; Maass et al., 2018b). This bias has been further compounded by modifying the existing 3C methodology. As the majority of genomic interactions arise from the insoluble fraction of the 3C material (Gavrilov et al., 2013), two independent groups have devised the in situ ligation protocol for 3C/Hi-C, which further enriches for intrachromosomal interactions over interchromosomal interactions (Rao et al., 2014; Nagano et al., 2015; Allahyar et al., 2018).
Some of these limitations have been mitigated by a more recent technique termed engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) in combination with next-generation sequencing (enChIP-Seq; Fujita et al., 2016). enChIP leverages locus-specific CRISPR targeting to probe for nearby DNA interactions. For example, applying enChIP-Seq to target the globin genes during erythroid differentiation revealed multiple NHCCs occurring in close physical proximity. Interestingly, a majority of these NHCCs contained transcriptionally active genes (Fujita et al., 2017). Another recent advancement for high-throughput identification of NHCCs is the split-pool recognition of interactions by tag extension (SPRITE) approach. SPRITE facilitates the detection of higher-order interactions occurring within the same nucleus at a new dimension (Quinodoz et al., 2018). Importantly, SPRITE robustly detects DNA–DNA as well as DNA–RNA NHCCs that form the nucleolus. Furthermore, it demonstrates that the nuclear bodies (splicing speckles, nucleolus, etc.) act as organizational hubs and scaffolds, where especially NHCCs are critical to shape the 3D chromatin folding and transcriptional programs. Furthermore, SPRITE also offers the opportunity to probe loci-specific interactions relative to different nuclear bodies inside the nucleus. Together, these recent technological advances are beginning to provide a genome-wide map of NHCCs.
Location matters for NHCCs in health and in disease
In addition to a role for NHCCs in coordinating nuclear organization in healthy states, where genomic loci and their interactions (intrachromosomal and NHCCs) reside in the nucleus has important implications for disease; for example, the ability to confidently phenotype different cancer subtypes based on changes in genome organization (Meaburn et al., 2009). Moreover, proper locus positioning and the occurrence of frequent functional NHCCs can be drawn from the detection of recurrent balanced or unbalanced chromosomal translocations that occur often in rare disease and in ∼40% of all cancers (Shaffer and Pandolfi, 2006). These translocations shed light on the genomic organization and NHCCs, since spatial proximity of chromosomal territories is associated with tissue-specific translocation prevalence (Parada et al., 2004; Branco and Pombo, 2006).
Deciphering where and when intrachromosomal and interchromosomal rearrangements appear can be tracked by induced DNA double-strand breaks in cell populations with high-throughput approaches. Although intrachromosomal translocations were found to be enriched in these studies, several hotspots for NHCCs suggest that 3D proximity of NHCCs in the nucleus can cause interchromosomal rearrangements, such as translocations, between nonhomologous chromosomes (Chiarle et al., 2011; Klein et al., 2011). Thus, the preexisting spatial proximity between chromosomal territories in the 3D nucleus is also related to recurrent translocations (Zhang et al., 2012; Sklyar et al., 2016). Finally, the direct correlation of cancer-associated translocations with Hi-C contacts (Engreitz et al., 2012), the reduction of NHCCs of gene-rich chromosomes in breast cancer cells compared with normal cells (Barutcu et al., 2015), and altered NHCCs due to a disease-associated deletion of HDAC4 (Maass et al., 2018c) indicate that rearrangements of the 3D genome architecture directly affect biological and pathogenic pathways. Moreover, anchors of long-range DNA loops have been associated with increased torsional stress that can lead to chromosomal breaks (Canela et al., 2017). As a whole, these examples suggest that the subnuclear positioning of chromosomal subdomains is critical to support specific gene expression programs for developmental and disease processes.
LncRNAs are involved in the 3D organization of NHCCs
While increasing evidence points to a pivotal role of noncoding RNAs (ncRNAs) in nuclear organization (Rinn and Chang, 2012; Rinn and Guttman, 2014; Vance and Ponting, 2014), the functional contribution of different species of ncRNAs in the assembly and regulation of long-range chromatin contacts, both intrachromosomal and of NHCCs, still remains poorly understood. So far, several lncRNAs have been determined to actively establish the nuclear architecture. Specifically, splicing speckle formation is associated with functional lncRNAs as mediators of nuclear organization. For example, the lncRNA NEAT1, in conjunction with the lncRNA MALAT1, can assemble a splicing speckle, which itself does not contain DNA, rather several DNA loci come in close proximity to process newly transcribed RNA (Ferrai et al., 2010; Spector and Lamond, 2011; Vera and Singer, 2014). ncRNAs can bind to multiple molecules and act as molecular scaffolds to shape the genome (Zappulla and Cech, 2006). Additionally, they can regulate looping interactions by binding to several RNA-binding proteins (RBPs) and/or chromatin modifiers (Hendrickson et al., 2016), thereby providing scaffolds for RNA–protein complexes that interact with and shape DNA organization (Santos-Pereira and Aguilera, 2015).
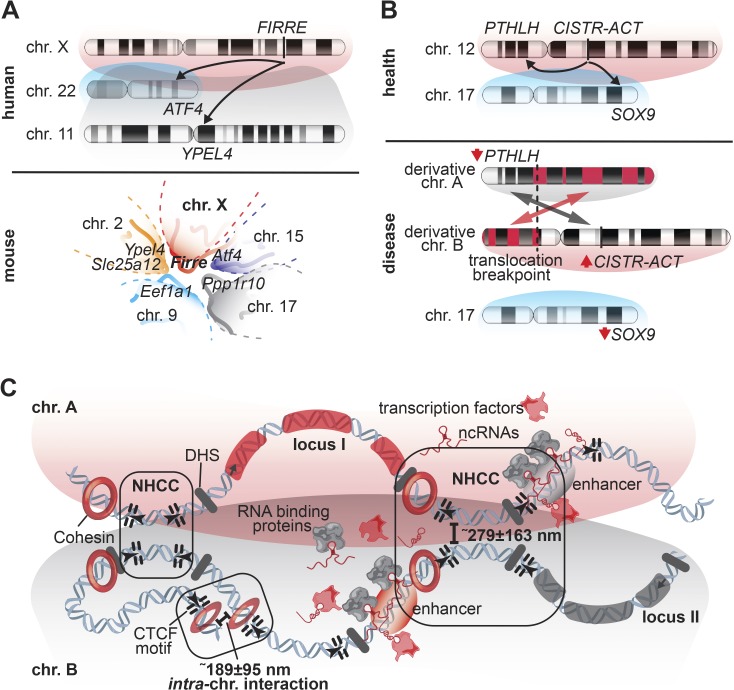
Dissecting the NHCCs of the lncRNA loci Firre and CISTR-ACT (Maass et al., 2012; Hacisuleyman et al., 2014), has provided insights into the involvement of lncRNAs in 3D chromosomal intermingling. The functional intergenic repeating RNA element (Firre, also known as linc-RAP-1) is a lncRNA locus that was detected in a loss-of-function screen as being required for proper adipogenesis (Sun et al., 2013). Firre is encoded on chromosome X, but it escapes X chromosome inactivation and interacts with the nuclear matrix factor hnRNPU (also known as SAF-A), a known mediator of genome organization (Nozawa et al., 2017; Fan et al., 2018). The Firre locus has an interesting property of forming NHCCs with several loci on nonhomologous mouse chromosomes; moreover, Firre RNA is bound at different chromosomes, identified by RNA affinity purification and RNA-FISH in mESCs (Hacisuleyman et al., 2014). More recently, the human FIRRE locus was found to make conserved NHCCs, with at least two of these NHCCs in human and mouse (Fig. 2 A; Maass et al., 2018b). The Firre locus is also implicated in shaping nuclear architecture, by anchoring the inactive X chromosome to the perinucleolar space (Yang et al., 2015). In addition to a possible role of FIRRE in forming NHCCs and tethering them to the nucleolar periphery is the interesting notion that the FIRRE locus harbors one of the highest densities of conserved CTCF motifs at its locus (Barutcu et al., 2018). Deleting this CTCF-rich Firre locus preserves its TAD boundary structure. Nevertheless, neither the Firre DNA sequence, promoter, nor CTCF motifs are required for this TAD boundary structure, as these elements may comprise a molecular toolkit of RNA and protein to establish and/or maintain NHCCs (Barutcu et al., 2018).
Figure 2.
LncRNA loci and their NHCCs, and mechanistic principles of NHCCs. (A) The lncRNA locus FIRRE interacts with ATF4 and YPEL4 in human cells. This 3D organization is conserved in the mouse genome, where Firre also interacts with Slc25a12, Eef1a1, and Ppp1r10. (B) The regulatory lncRNA locus CISTR-ACT facilitates 3D proximity to PTHLH, and the NHCC with SOX9, in normal cells. When balanced translocations disrupt this interaction and misplace CISTR-ACT onto a derivative chromosome, the positional effect leads to down-regulated PTHLH and SOX9 and up-regulated CISTR-ACT. (C) CLING determined that NHCCs have an average proximity of ∼279 ± 163 nm, while intrachromosomal interactions were 189 ± 95 nm apart. DNase I hypersensitivity sites (DHS) and convergent CTCF motifs are features of gene regulatory regions (loop-extrusion model) that are in spatial proximity to cooperate with tissue-specific transcription factors, ncRNAs, and RBPs to regulate the expression of genes that are located on different chromosomes (see transcription factory in Fig. 1 A).
The cis- and trans-acting lncRNA locus CISTR-ACT is encoded on human chromosome 12 and forms a specific NHCC with the loci of the chondrogenic morphogenesis gene PTHLH on the same chromosome (cis) and with the transcription factor SOX9 on chromosome 17 (trans) to regulate chondrogenic gene expression (Fig. 2 B; Maass et al., 2012, 2018c). The physical disruption of this regulatory landscape by balanced translocations misplaces the CISTR-ACT locus to a derivative translocation chromosome, causing dysregulation of CISTR-ACT, PTHLH, and SOX9, thereby leading to the congenital cartilage malformation chondrodysplasia brachydactyly type E (shortened fingers and extremities; Maass et al., 2012). Labeling the entire chromosomes 12 and 17, as well as the individual CISTR-ACT and SOX9 loci, revealed that these two chromosomal territories frequently interact in a recurrent pattern, and although these chromosomal patterns are stable across different cell types, tissue-specific NHCCs occurred at the level of the individual gene loci (Maass et al., 2018c). These results support the concept that defined genomic loci come into 3D proximity to drive gene regulation in a highly specific manner. Any physical disruption of a tissue-specific transcription factory (Melnik et al., 2011) causes local reorganization of the genome, resulting in altered transcriptional programs that affect developmental programs and may cause disease (Fig. 2 B; Maass et al., 2018c). These findings indicate that the positioning of individual lncRNA loci may be a specific nonrandom feature in the 3D nuclear space, required to fulfill important functions.
Together, the FIRRE and CISTR-ACT lncRNA loci exemplify the emerging concept that noncoding loci with distinct features—multispecies conservation, DNase I hypersensitivity sites, open chromatin marks, enrichment of CTCF motifs and transcription factor binding sites, and noncoding transcription—can shape nuclear organization by facilitating the colocalization of euchromatic features between multiple nonhomologous chromosomes (Fig. 2 C). FIRRE and CISTR-ACT form specific NHCCs, and the fact that the interchromosomal contacts of FIRRE are CTCF/cohesin-independent suggests that gene regulation by intra- versus interchromosomal interactions operates by distinct, yet potentially overlapping mechanisms. Interestingly, albeit different gene order and content of linear orthologous sequences in different species, a spatial conservation of some NHCCs seems to exist (Chambers et al., 2013). It remains to be determined how many other lncRNA loci are involved in NHCCs and contribute to the 3D organization of the genome. More studies on NHCCs, compared with intrachromosomal regulatory processes, will decipher if similar or different regulators (transcription factors, chromatin modifying complexes, and CTCF and cohesin, etc.) provide the platform for interchromosomal communication.
Watching kissing chromosomes in real time: live-cell imaging of NHCCs
Fixation-based in situ methods, such as FISH-related and 3C-based approaches, are limited in their capabilities (Hoffman et al., 2015), since they cannot address the spatiotemporal dynamics of intra- or interchromosomal interactions. Therefore, live-cell imaging techniques are highly advantageous to understand the spatiotemporal chromatin dynamics in non–cross-linked living cells and to explore the spatial dimensions of NHCCs. Toward this goal, numerous studies have proven the principle of live-cell imaging of DNA using CRISPR-Cas technologies (Chen et al., 2013; Deng et al., 2015; Ma et al., 2015a, 2016b,a; Shechner et al., 2015; Fu et al., 2016; Shao et al., 2016; Wang et al., 2016; Guan et al., 2017; Qin et al., 2017; Takei et al., 2017; Zhou et al., 2017; Maass et al., 2018a,b; Wu et al., 2018). Collectively, these studies have established the principle that modification to guide sequences can target and label specific DNA loci that can be monitored in living cells.
One recent study leveraged CLING to measure the fundamental NHCC properties of two lncRNA loci. Specifically, CLING was applied to the previously determined NHCCs mediated by the FIRRE and CISTR-ACT loci to explore NHCCs and mechanisms of genome organization in living cells (Dekker, 2016; Fudenberg and Imakaev, 2017; Maass et al., 2018b). This revealed several fundamental properties of NHCCs. (a) NHCCs occur in a majority of the cell populations (>50%); and (b) NHCCs are stable over time. One possible way NHCCs could be missed is if they were in constant flux of movement. However, this is not the case, as they remain in close proximity over a substantial time period of the cell cycle. (c) Consistent with their stable proximity, NHCCs exhibit less tortuosity or tumble more slowly in the nucleus. (d) A substantial difference between intra- and interchromosomal interactions is the distance at which they occur. Specifically, the spatial distances for intrachromosomal interactions were found in the range of 189 ± 95 nm, and for NHCCs (FIRRE and CISTR-ACT), these interactions occurred at a larger distance (279 ± 163 nm; Maass et al., 2018b). Thus, in living cells, the NHCCs are frequent, stable, less mobile, and similar to intra-molecular interactions.
A fundamental feature of NHCCs is that one allele interacts with another one (Johnston and Desplan, 2014; Monahan and Lomvardas, 2015). However, this does not necessarily mean that NHCCs always represent interactions between both alleles. More often, in CLING experiments, only one allele of two different chromosomes interacts (∼40%), rather than both alleles forming NHCCs (∼17%; Maass et al., 2018b). This raises the interesting question whether NHCCs are nonrandom with respect to parental origin. For example, are NHCCs formed between paternal–paternal, maternal–maternal, or paternal–maternal chromosomes? Or do they have a random combination, that is, based on a determined order of the genomic architecture (Nagele et al., 1995; Weise et al., 2016)? Addressing this question would require either a molecular or imaging technique that distinguishes between the contacts of parental alleles in an allele-specific manner.
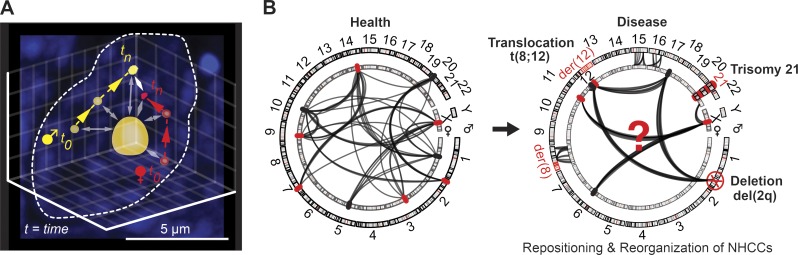
A very recent study developed and proved the principle of an allele-specific CRISPR-Cas9–based imaging approach, termed single nucleotide polymorphism (SNP)–CLING. In short, this approach leverages SNPs that can be used to target CRISPR-Cas9 constructs specifically to either the maternal or the paternal allele (Fig. 3 A). Applying SNP-CLING to the NHCC of Firre and Ypel4 revealed a slightly shifted NHCC distribution toward the maternal–paternal combination (Maass et al., 2018a). Thus, the possibility that 3D imprinting exists and contributes to cell type–specific gene expression still remains unresolved. Overall, SNP-CLING is a powerful approach to understand how specific alleles of chromosomes are positioned relative to each other and with respect to nuclear subcompartments, such as the nucleolus. Furthermore, for the first time, SNP-CLING allows the study of heterozygous states of genetic disease in living cells by distinguishing between healthy and affected alleles.
Figure 3.
Exploring NHCCs toward understanding their contribution to gene regulation and genome organization in health and disease. (A) SNP-CLING enables the study of allele-specific locus positioning and spatiotemporal dynamics in living cells (depicted example: mESC with exemplified time-lapse imaging of a maternal and paternal allele and their distances to the nucleolus). (B) Left: Maternal and paternal alleles of the interphase genome are in physical proximity and intermingle to control tissue-specific gene regulation in the 3D space of the nucleus. LncRNAs, proteins (chromatin modifiers, transcription factors, etc.), biophysical properties of the chromatin, genome organization, and stochastics cooperate to contribute to variable, but specific, biological processes. Right: Structural (i.e., deletions, translocations, etc.) and numerical chromosomal aberrations (e.g., trisomies) can disrupt and reorganize the intricate network of NHCCs. These aberrations cause altered transcriptional programs, repositioning of genomic loci, and reorganization of tissue-specific gene regulation that can influence genetic and developmental processes. They comprise a variable layer of genome organization and are often associated with pathogenic pathways and disease.
Collectively, these results show that genomic loci of the noncoding genome are actively involved in the 3D formation of NHCCs. Several different loci on nonhomologous chromosomes may share the same spatial hub, which can be conserved across species to regulate transcriptional programs. Therefore, studying genomic noncoding regions (enhancer and ncRNAs) by molecular high-throughput methods and imaging approaches provides immense potential to understand the formation and significance of NHCCs.
Perspective
The coordinated regulation of multiple genes within specific transcriptional programs requires physical proximity between genomic loci, either on the same chromosome or across different chromosomes. To accomplish defined transcriptional regulation, transcriptional hubs or factories can be formed around NHCCs in distinct nuclear locations. Specifically, subnuclear domains of NHCCs could emerge from phase transitions. This is seemingly the case for two of the larger subnuclear structures, the nucleolus and the olfactosome. Perhaps lncRNA loci and other NHCCs are also phase-state transitions, comprised of lncRNAs, RBPs, CTCF, and many other factors coalescing in physical proximity. For example, cohesin, CTCF, and transcription factors that are bound to loci on different chromosomes through RNA–protein interactions are the bridging factors that keep these loci in physical proximity (Fig. 2 C). In this scenario, the loop extrusion complexes on two different chromosomes would extend the DNA in the same spatial hub, similarly to an intrachromosomal interaction, to retain the chromatin strands of two different chromosomes in proximity (Fig. 2 C).
As presented in this review, increasing evidence supports a role for NHCCs in the establishment of proper gene-regulatory networks. Yet our understanding of NHCC’s impact on the nuclear architecture and of how repositioning of genomic loci and reorganization of tissue-specific gene regulation influences genetic processes and disease progression is still in its infancy (Fig. 3 B). Structural and numerical genomic aberrations add a variable dimension of genomic organization to both normal and disease states.
The reviewed results suggest that NHCCs could favor diverse, but recurrent and important, stochastic interactions that add an important informational layer to tissue-specific gene regulation. NHCCs, the biophysical properties of chromatin, and stochastics contribute to variable, but specific, gene regulation that supports monogenic and also mono-allelic expression of genes in a coordinated manner. Collectively, the examples covered here demonstrate that NHCCs account for a multitude of biological processes.
Moving forward, it is critical to elucidate the molecular mechanisms underlying NHCCs. Specifically, the role of regulatory noncoding DNA and lncRNA loci and the biophysical features underlying the regulation and formation of NHCCs will facilitate the identification of tissue-specific interchromosomal spatial hubs and broaden our current knowledge of gene regulation and of NHCCs. Exploring loci positioning and the spatial proximities of those loci interacting with regulatory DNA and RNA sequences will provide crucial insights into nuclear organization and etiologies therein.
Acknowledgments
We thank the Rinn laboratory, especially Chiara Gerhardinger, for intellectual input.
This work was supported by National Institutes of Health (U01 DA040612-01 and P01 GM09911) and the Howard Hughes Medical Institute Faculty Scholars Program (J.L. Rinn).
The authors declare no competing financial interests.
Author contributions: P.G. Maass, A.R. Barutcu, and J.L. Rinn wrote the manuscript.
References
- Abranches R., Santos A.P., Wegel E., Williams S., Castilho A., Christou P., Shaw P., and Stoger E.. 2000. Widely separated multiple transgene integration sites in wheat chromosomes are brought together at interphase. Plant J. 24:713–723. 10.1046/j.1365-313x.2000.00908.x [DOI] [PubMed] [Google Scholar]
- Allahyar A., Vermeulen C., Bouwman B.A.M., Krijger P.H.L., Verstegen M.J.A.M., Geeven G., van Kranenburg M., Pieterse M., Straver R., Haarhuis J.H.I., et al. 2018. Enhancer hubs and loop collisions identified from single-allele topologies. Nat. Genet. 50:1151–1160. 10.1038/s41588-018-0161-5 [DOI] [PubMed] [Google Scholar]
- Apostolou E., and Thanos D.. 2008. Virus Infection Induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell. 134:85–96. 10.1016/j.cell.2008.05.052 [DOI] [PubMed] [Google Scholar]
- Bacher C.P., Guggiari M., Brors B., Augui S., Clerc P., Avner P., Eils R., and Heard E.. 2006. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat. Cell Biol. 8:293–299. 10.1038/ncb1365 [DOI] [PubMed] [Google Scholar]
- Barbieri M., Chotalia M., Fraser J., Lavitas L.M., Dostie J., Pombo A., and Nicodemi M.. 2013. A model of the large-scale organization of chromatin. Biochem. Soc. Trans. 41:508–512. 10.1042/BST20120238 [DOI] [PubMed] [Google Scholar]
- Barutcu A.R., Lajoie B.R., McCord R.P., Tye C.E., Hong D., Messier T.L., Browne G., van Wijnen A.J., Lian J.B., Stein J.L., et al. 2015. Chromatin interaction analysis reveals changes in small chromosome and telomere clustering between epithelial and breast cancer cells. Genome Biol. 16:214 10.1186/s13059-015-0768-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barutcu A.R., Fritz A.J., Zaidi S.K., van Wijnen A.J., Lian J.B., Stein J.L., Nickerson J.A., Imbalzano A.N., and Stein G.S.. 2016. C-ing the Genome: A Compendium of Chromosome Conformation Capture Methods to Study Higher-Order Chromatin Organization. J. Cell. Physiol. 231:31–35. 10.1002/jcp.25062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barutcu A.R., Maass P.G., Lewandowski J.P., Weiner C.L., and Rinn J.L.. 2018. A TAD boundary is preserved upon deletion of the CTCF-rich Firre locus. Nat. Commun. 9:1444 10.1038/s41467-018-03614-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagrie R.A., Scialdone A., Schueler M., Kraemer D.C., Chotalia M., Xie S.Q., Barbieri M., de Santiago I., Lavitas L.M., Branco M.R., et al. 2017. Complex multi-enhancer contacts captured by genome architecture mapping. Nature. 543:519–524. 10.1038/nature21411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore W.A. 2013. The spatial organization of the human genome. Annu. Rev. Genomics Hum. Genet. 14:67–84. 10.1146/annurev-genom-091212-153515 [DOI] [PubMed] [Google Scholar]
- Blackstone T., Scharein R., Borgo B., Varela R., Diao Y., and Arsuaga J.. 2011. Modeling of chromosome intermingling by partially overlapping uniform random polygons. J. Math. Biol. 62:371–389. 10.1007/s00285-010-0338-8 [DOI] [PubMed] [Google Scholar]
- Bolzer A., Kreth G., Solovei I., Koehler D., Saracoglu K., Fauth C., Müller S., Eils R., Cremer C., Speicher M.R., and Cremer T.. 2005. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 3:e157 10.1371/journal.pbio.0030157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B., and Cavalli G.. 2016. Organization and function of the 3D genome. Nat. Rev. Genet. 17:661–678. 10.1038/nrg.2016.112 [DOI] [PubMed] [Google Scholar]
- Branco M.R., and Pombo A.. 2006. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 4:e138 10.1371/journal.pbio.0040138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.E., Guest S.S., Smale S.T., Hahm K., Merkenschlager M., and Fisher A.G.. 1997. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 91:845–854. 10.1016/S0092-8674(00)80472-9 [DOI] [PubMed] [Google Scholar]
- Canela A., Maman Y., Jung S., Wong N., Callen E., Day A., Kieffer-Kwon K.R., Pekowska A., Zhang H., Rao S.S.P., et al. 2017. Genome organization drives chromosome fragility. Cell. 170:507–521.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers E.V., Bickmore W.A., and Semple C.A.. 2013. Divergence of mammalian higher order chromatin structure is associated with developmental loci. PLOS Comput. Biol. 9:e1003017 10.1371/journal.pcbi.1003017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Gilbert L.A., Cimini B.A., Schnitzbauer J., Zhang W., Li G.W., Park J., Blackburn E.H., Weissman J.S., Qi L.S., and Huang B.. 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 155:1479–1491. 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Yusuf M., Hashimoto T., Estandarte A.K., Thompson G., and Robinson I.. 2017. Three-dimensional positioning and structure of chromosomes in a human prophase nucleus. Sci. Adv. 3:e1602231 10.1126/sciadv.1602231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhang Y., Wang Y., Zhang L., Brinkman E.K., Adam S.A., Goldman R., Steensel B.v., Ma J., and Belmont A.S.. 2018. TSA-seq mapping of nuclear genome organization. bioRxiv. Preprint posted April 25, 2018. [Google Scholar]
- Chiarle R., Zhang Y., Frock R.L., Lewis S.M., Molinie B., Ho Y.J., Myers D.R., Choi V.W., Compagno M., Malkin D.J., et al. 2011. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 147:107–119. 10.1016/j.cell.2011.07.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney E.J., LeGros M.A., Mosley C.P., Clowney F.G., Markenskoff-Papadimitriou E.C., Myllys M., Barnea G., Larabell C.A., and Lomvardas S.. 2012. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 151:724–737. 10.1016/j.cell.2012.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T., and Cremer M.. 2010. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2:a003889 10.1101/cshperspect.a003889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T., Cremer C., Baumann H., Luedtke E.K., Sperling K., Teuber V., and Zorn C.. 1982. Rabl’s model of the interphase chromosome arrangement tested in Chinese hamster cells by premature chromosome condensation and laser-UV-microbeam experiments. Hum. Genet. 60:46–56. 10.1007/BF00281263 [DOI] [PubMed] [Google Scholar]
- Dekker J. 2006. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat. Methods. 3:17–21. 10.1038/nmeth823 [DOI] [PubMed] [Google Scholar]
- Dekker J. 2016. Mapping the 3D genome: Aiming for consilience. Nat. Rev. Mol. Cell Biol. 17:741–742. 10.1038/nrm.2016.151 [DOI] [PubMed] [Google Scholar]
- Dekker J., and Mirny L.. 2016. The 3D Genome as Moderator of Chromosomal Communication. Cell. 164:1110–1121. 10.1016/j.cell.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., and Misteli T.. 2015. Long-Range Chromatin Interactions. Cold Spring Harb. Perspect. Biol. 7:a019356 10.1101/cshperspect.a019356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., and Kleckner N.. 2002. Capturing chromosome conformation. Science. 295:1306–1311. 10.1126/science.1067799 [DOI] [PubMed] [Google Scholar]
- Deng W., Shi X., Tjian R., Lionnet T., and Singer R.H.. 2015. CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc. Natl. Acad. Sci. USA. 112:11870–11875. 10.1073/pnas.1515692112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A.F., Broman K.W., Fung J.C., Marshall W.F., Philips J., Agard D.A., and Sedat J.W.. 1996. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell. 85:745–759. 10.1016/S0092-8674(00)81240-4 [DOI] [PubMed] [Google Scholar]
- Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., and Ren B.. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 485:376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J.R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J.E., Lee A.Y., Ye Z., Kim A., Rajagopal N., Xie W., et al. 2015. Chromatin architecture reorganization during stem cell differentiation. Nature. 518:331–336. 10.1038/nature14222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J., Richmond T.A., Arnaout R.A., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C., et al. 2006. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 16:1299–1309. 10.1101/gr.5571506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ea V., Baudement M.O., Lesne A., and Forné T.. 2015. Contribution of Topological Domains and Loop Formation to 3D Chromatin Organization. Genes (Basel). 6:734–750. 10.3390/genes6030734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman L.B., and Fraser P.. 2012. Transcription factories: genetic programming in three dimensions. Curr. Opin. Genet. Dev. 22:110–114. 10.1016/j.gde.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Engreitz J.M., Agarwala V., and Mirny L.A.. 2012. Three-dimensional genome architecture influences partner selection for chromosomal translocations in human disease. PLoS One. 7:e44196 10.1371/journal.pone.0044196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz J.M., Ollikainen N., and Guttman M.. 2016. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 17:756–770. 10.1038/nrm.2016.126 [DOI] [PubMed] [Google Scholar]
- Erdel F., and Rippe K.. 2018. Formation of Chromatin Subcompartments by Phase Separation. Biophys. J. 114:2262–2270. 10.1016/j.bpj.2018.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Lv P., Huo X., Wu J., Wang Q., Cheng L., Liu Y., Tang Q.Q., Zhang L., Zhang F., et al. 2018. The nuclear matrix protein HNRNPU maintains 3D genome architecture globally in mouse hepatocytes. Genome Res. 28:192–202. 10.1101/gr.224576.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrai C., de Castro I.J., Lavitas L., Chotalia M., and Pombo A.. 2010. Gene positioning. Cold Spring Harb. Perspect. Biol. 2:a000588 10.1101/cshperspect.a000588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan L.E., Sproul D., Thomson I., Boyle S., Kerr E., Perry P., Ylstra B., Chubb J.R., and Bickmore W.A.. 2008. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 4:e1000039 10.1371/journal.pgen.1000039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke M., Ibrahim D.M., Andrey G., Schwarzer W., Heinrich V., Schöpflin R., Kraft K., Kempfer R., Jerković I., Chan W.L., et al. 2016. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature. 538:265–269. 10.1038/nature19800 [DOI] [PubMed] [Google Scholar]
- Fraser J., Williamson I., Bickmore W.A., and Dostie J.. 2015. An Overview of Genome Organization and How We Got There: from FISH to Hi-C. Microbiol. Mol. Biol. Rev. 79:347–372. 10.1128/MMBR.00006-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz A.J., Stojkovic B., Ding H., Xu J., Bhattacharya S., and Berezney R.. 2014. Cell type specific alterations in interchromosomal networks across the cell cycle. PLOS Comput. Biol. 10:e1003857 10.1371/journal.pcbi.1003857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Rocha P.P., Luo V.M., Raviram R., Deng Y., Mazzoni E.O., and Skok J.A.. 2016. CRISPR-dCas9 and sgRNA scaffolds enable dual-colour live imaging of satellite sequences and repeat-enriched individual loci. Nat. Commun. 7:11707 10.1038/ncomms11707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G., and Imakaev M.. 2017. FISH-ing for captured contacts: towards reconciling FISH and 3C. Nat. Methods. 14:673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G., Imakaev M., Lu C., Goloborodko A., Abdennur N., and Mirny L.A.. 2016. Formation of Chromosomal Domains by Loop Extrusion. Cell Reports. 15:2038–2049. 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T., Yuno M., and Fujii H.. 2016. Efficient sequence-specific isolation of DNA fragments and chromatin by in vitro enChIP technology using recombinant CRISPR ribonucleoproteins. Genes Cells. 21:370–377. 10.1111/gtc.12341 [DOI] [PubMed] [Google Scholar]
- Fujita T., Yuno M., Suzuki Y., Sugano S., and Fujii H.. 2017. Identification of physical interactions between genomic regions by enChIP-Seq. Genes Cells. 22:506–520. 10.1111/gtc.12492 [DOI] [PubMed] [Google Scholar]
- Fullwood M.J., Liu M.H., Pan Y.F., Liu J., Xu H., Mohamed Y.B., Orlov Y.L., Velkov S., Ho A., Mei P.H., et al. 2009. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 462:58–64. 10.1038/nature08497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi M., Evdokimova V.N., T Cuenco K., Nikiforova M.N., Kelly L.M., Stringer J.R., Bakkenist C.J., and Nikiforov Y.E.. 2012. Homologous chromosomes make contact at the sites of double-strand breaks in genes in somatic G0/G1-phase human cells. Proc. Natl. Acad. Sci. USA. 109:9454–9459. 10.1073/pnas.1205759109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganji M., Shaltiel I.A., Bisht S., Kim E., Kalichava A., Haering C.H., and Dekker C.. 2018. Real-time imaging of DNA loop extrusion by condensin. Science. 360:102–105. 10.1126/science.aar7831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilov A.A., Gushchanskaya E.S., Strelkova O., Zhironkina O., Kireev I.I., Iarovaia O.V., and Razin S.V.. 2013. Disclosure of a structural milieu for the proximity ligation reveals the elusive nature of an active chromatin hub. Nucleic Acids Res. 41:3563–3575. 10.1093/nar/gkt067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloborodko A., Imakaev M.V., Marko J.F., and Mirny L.. 2016. Compaction and segregation of sister chromatids via active loop extrusion. eLife. 5:e14864 10.7554/eLife.14864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J., Liu H., Shi X., Feng S., and Huang B.. 2017. Tracking Multiple Genomic Elements Using Correlative CRISPR Imaging and Sequential DNA FISH. Biophys. J. 112:1077–1084. 10.1016/j.bpj.2017.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E., Goff L.A., Trapnell C., Williams A., Henao-Mejia J., Sun L., McClanahan P., Hendrickson D.G., Sauvageau M., Kelley D.R., et al. 2014. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 21:198–206. 10.1038/nsmb.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A., Mallappa C., Okada S., Butler J.T., Baker S.P., Lawrence J.B., Ohkawa Y., and Imbalzano A.N.. 2015. Spatial re-organization of myogenic regulatory sequences temporally controls gene expression. Nucleic Acids Res. 43:2008–2021. 10.1093/nar/gkv046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson D.G., Kelley D.R., Tenen D., Bernstein B., and Rinn J.L.. 2016. Widespread RNA binding by chromatin-associated proteins. Genome Biol. 17:28 10.1186/s13059-016-0878-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepperger C., Mannes A., Merz J., Peters J., and Dietzel S.. 2008. Three-dimensional positioning of genes in mouse cell nuclei. Chromosoma. 117:535–551. 10.1007/s00412-008-0168-2 [DOI] [PubMed] [Google Scholar]
- Hoffman E.A., Frey B.L., Smith L.M., and Auble D.T.. 2015. Formaldehyde crosslinking: a tool for the study of chromatin complexes. J. Biol. Chem. 290:26404–26411. 10.1074/jbc.R115.651679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan M.S., Parfitt D.E., Zepeda-Mendoza C.J., Shen M.M., and Spector D.L.. 2015. Transient pairing of homologous Oct4 alleles accompanies the onset of embryonic stem cell differentiation. Cell Stem Cell. 16:275–288. 10.1016/j.stem.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horta A., Monahan K., Bashkirova E., and Lomvardas S.. 2018. Cell type-specific interchromosomal interactions as a mechanism for transcriptional diversity. bioRxiv. Preprint posted March 23, 2018. [Google Scholar]
- Johnston R.J. Jr., and Desplan C.. 2014. Interchromosomal communication coordinates intrinsically stochastic expression between alleles. Science. 343:661–665. 10.1126/science.1243039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein I.A., Resch W., Jankovic M., Oliveira T., Yamane A., Nakahashi H., Di Virgilio M., Bothmer A., Nussenzweig A., Robbiani D.F., et al. 2011. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 147:95–106. 10.1016/j.cell.2011.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak S.T., and Groudine M.. 2004. Gene order and dynamic domains. Science. 306:644–647. 10.1126/science.1103864 [DOI] [PubMed] [Google Scholar]
- Krueger C., King M.R., Krueger F., Branco M.R., Osborne C.S., Niakan K.K., Higgins M.J., and Reik W.. 2012. Pairing of homologous regions in the mouse genome is associated with transcription but not imprinting status. PLoS One. 7:e38983 10.1371/journal.pone.0038983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A.I., and Earnshaw W.C.. 1998. Structure and function in the nucleus. Science. 280:547–553. 10.1126/science.280.5363.547 [DOI] [PubMed] [Google Scholar]
- Langdon E.M., Qiu Y., Ghanbari Niaki A., McLaughlin G.A., Weidmann C.A., Gerbich T.M., Smith J.A., Crutchley J.M., Termini C.M., Weeks K.M., et al. 2018. mRNA structure determines specificity of a polyQ-driven phase separation. Science. 360:922–927. 10.1126/science.aar7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E.B. 1954. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am. Nat. 88:225–239. 10.1086/281833 [DOI] [Google Scholar]
- Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O., et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 326:289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B., Heist T., Levine M., and Fukaya T.. 2018. Visualization of transvection in living Drosophila embryos. Mol. Cell. 70:287–296.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J.Q., Li T., Hu J.F., Vu T.H., Chen H.L., Qiu X.W., Cherry A.M., and Hoffman A.R.. 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 312:269–272. 10.1126/science.1123191 [DOI] [PubMed] [Google Scholar]
- Lomvardas S., Barnea G., Pisapia D.J., Mendelsohn M., Kirkland J., and Axel R.. 2006. Interchromosomal interactions and olfactory receptor choice. Cell. 126:403–413. 10.1016/j.cell.2006.06.035 [DOI] [PubMed] [Google Scholar]
- Lupiáñez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R., et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 161:1012–1025. 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Naseri A., Reyes-Gutierrez P., Wolfe S.A., Zhang S., and Pederson T.. 2015a Multicolor CRISPR labeling of chromosomal loci in human cells. Proc. Natl. Acad. Sci. USA. 112:3002–3007. 10.1073/pnas.1420024112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Tu L.C., Naseri A., Huisman M., Zhang S., Grunwald D., and Pederson T.. 2016a CRISPR-Cas9 nuclear dynamics and target recognition in living cells. J. Cell Biol. 214:529–537. 10.1083/jcb.201604115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Tu L.C., Naseri A., Huisman M., Zhang S., Grunwald D., and Pederson T.. 2016b Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat. Biotechnol. 34:528–530. 10.1038/nbt.3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Ay F., Lee C., Gulsoy G., Deng X., Cook S., Hesson J., Cavanaugh C., Ware C.B., Krumm A., et al. 2015b Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat. Methods. 12:71–78. 10.1038/nmeth.3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass P.G., Rump A., Schulz H., Stricker S., Schulze L., Platzer K., Aydin A., Tinschert S., Goldring M.B., Luft F.C., and Bähring S.. 2012. A misplaced lncRNA causes brachydactyly in humans. J. Clin. Invest. 122:3990–4002. 10.1172/JCI65508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass P.G., Barutcu A.R., Shechner D.M., Weiner C.L., Melé M., and Rinn J.L.. 2018a Spatiotemporal allele organization by allele-specific CRISPR live-cell imaging (SNP-CLING). Nat. Struct. Mol. Biol. 25:176–184. 10.1038/s41594-017-0015-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass P.G., Barutcu A.R., Weiner C.L., and Rinn J.L.. 2018b Inter-chromosomal contact properties in live-cell imaging and in Hi-C. Mol. Cell. 69:1039–1045.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass P.G., Weise A., Rittscher K., Lichtenwald J., Barutcu A.R., Liehr T., Aydin A., Wefeld-Neuenfeld Y., Pölsler L., Tinschert S., et al. 2018c Reorganization of inter-chromosomal interactions in the 2q37-deletion syndrome. EMBO J. 37:e96257 10.15252/embj.201696257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharana S., Iyer K.V., Jain N., Nagarajan M., Wang Y., and Shivashankar G.V.. 2016. Chromosome intermingling-the physical basis of chromosome organization in differentiated cells. Nucleic Acids Res. 44:5148–5160. 10.1093/nar/gkw131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharana S., Wang J., Papadopoulos D.K., Richter D., Pozniakovsky A., Poser I., Bickle M., Rizk S., Guillén-Boixet J., Franzmann T.M., et al. 2018. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 360:918–921. 10.1126/science.aar7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahy N.L., Perry P.E., and Bickmore W.A.. 2002. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol. 159:753–763. 10.1083/jcb.200207115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y. 1999. X-inactivation by chromosomal pairing events. Genes Dev. 13:2624–2632. 10.1101/gad.13.20.2624 [DOI] [PubMed] [Google Scholar]
- Markenscoff-Papadimitriou E., Allen W.E., Colquitt B.M., Goh T., Murphy K.K., Monahan K., Mosley C.P., Ahituv N., and Lomvardas S.. 2014. Enhancer interaction networks as a means for singular olfactory receptor expression. Cell. 159:543–557. 10.1016/j.cell.2014.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita M., Ochiai H., Suzuki K.T., Hayashi S., Yamamoto T., Awazu A., and Sakamoto N.. 2017. Dynamic changes in the interchromosomal interaction of early histone gene loci during development of sea urchin. J. Cell Sci. 130:4097–4107. 10.1242/jcs.206862 [DOI] [PubMed] [Google Scholar]
- Mayer R., Brero A., von Hase J., Schroeder T., Cremer T., and Dietzel S.. 2005. Common themes and cell type specific variations of higher order chromatin arrangements in the mouse. BMC Cell Biol. 6:44 10.1186/1471-2121-6-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B. 2016. Nucleolar organizer regions: genomic ‘dark matter’ requiring illumination. Genes Dev. 30:1598–1610. 10.1101/gad.283838.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn K.J., Gudla P.R., Khan S., Lockett S.J., and Misteli T.. 2009. Disease-specific gene repositioning in breast cancer. J. Cell Biol. 187:801–812. 10.1083/jcb.200909127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnik S., Deng B., Papantonis A., Baboo S., Carr I.M., and Cook P.R.. 2011. The proteomes of transcription factories containing RNA polymerases I, II or III. Nat. Methods. 8:963–968. 10.1038/nmeth.1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T., Cáceres J.F., and Spector D.L.. 1997. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 387:523–527. 10.1038/387523a0 [DOI] [PubMed] [Google Scholar]
- Monahan K., and Lomvardas S.. 2015. Monoallelic expression of olfactory receptors. Annu. Rev. Cell Dev. Biol. 31:721–740. 10.1146/annurev-cellbio-100814-125308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan K., Schieren I., Cheung J., Mumbey-Wafula A., Monuki E.S., and Lomvardas S.. 2017. Cooperative interactions enable singular olfactory receptor expression in mouse olfactory neurons. eLife. 6:e28620 10.7554/eLife.28620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan K., Horta A., Mumbay-Wafula A., Li L., Zhao Y., Love P., and Lomvardas S.. 2018. Ldb1 mediates trans enhancement in mammals. bioRxiv. Preprint posted March 23, 2018. [Google Scholar]
- Nagano T., Lubling Y., Stevens T.J., Schoenfelder S., Yaffe E., Dean W., Laue E.D., Tanay A., and Fraser P.. 2013. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 502:59–64. 10.1038/nature12593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T., Várnai C., Schoenfelder S., Javierre B.M., Wingett S.W., and Fraser P.. 2015. Comparison of Hi-C results using in-solution versus in-nucleus ligation. Genome Biol. 16:175 10.1186/s13059-015-0753-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T., Lubling Y., Várnai C., Dudley C., Leung W., Baran Y., Mendelson Cohen N., Wingett S., Fraser P., and Tanay A.. 2017. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature. 547:61–67. 10.1038/nature23001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagele R., Freeman T., McMorrow L., and Lee H.Y.. 1995. Precise spatial positioning of chromosomes during prometaphase: evidence for chromosomal order. Science. 270:1831–1835. 10.1126/science.270.5243.1831 [DOI] [PubMed] [Google Scholar]
- Németh A., Conesa A., Santoyo-Lopez J., Medina I., Montaner D., Péterfia B., Solovei I., Cremer T., Dopazo J., and Längst G.. 2010. Initial genomics of the human nucleolus. PLoS Genet. 6:e1000889 10.1371/journal.pgen.1000889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K., Cam H.P., Maraia R.J., and Grewal S.I.. 2006. A role for TFIIIC transcription factor complex in genome organization. Cell. 125:859–872. 10.1016/j.cell.2006.04.028 [DOI] [PubMed] [Google Scholar]
- Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J., et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 485:381–385. 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa R.S., Boteva L., Soares D.C., Naughton C., Dun A.R., Buckle A., Ramsahoye B., Bruton P.C., Saleeb R.S., Arnedo M., et al. 2017. SAF-A regulates interphase chromosome structure through oligomerization with chromatin-associated RNAs. Cell. 169:1214–1227.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuebler J., Fudenberg G., Imakaev M., Abdennur N., and Mirny L.A.. 2018. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc. Natl. Acad. Sci. USA. 115:E6697–E6706. 10.1073/pnas.1717730115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne C.S., Chakalova L., Brown K.E., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J.A., Lopes S., Reik W., and Fraser P.. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36:1065–1071. 10.1038/ng1423 [DOI] [PubMed] [Google Scholar]
- Osborne C.S., Chakalova L., Mitchell J.A., Horton A., Wood A.L., Bolland D.J., Corcoran A.E., and Fraser P.. 2007. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 5:e192 10.1371/journal.pbio.0050192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papantonis A., and Cook P.R.. 2013. Transcription factories: genome organization and gene regulation. Chem. Rev. 113:8683–8705. 10.1021/cr300513p [DOI] [PubMed] [Google Scholar]
- Parada L.A., McQueen P.G., Munson P.J., and Misteli T.. 2002. Conservation of relative chromosome positioning in normal and cancer cells. Curr. Biol. 12:1692–1697. 10.1016/S0960-9822(02)01166-1 [DOI] [PubMed] [Google Scholar]
- Parada L.A., McQueen P.G., and Misteli T.. 2004. Tissue-specific spatial organization of genomes. Genome Biol. 5:R44 10.1186/gb-2004-5-7-r44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H.C., Jarmuz A., Canzonetta C., Webster Z., Nesterova T., et al. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 132:422–433. 10.1016/j.cell.2008.01.011 [DOI] [PubMed] [Google Scholar]
- Pederson T. 2002. Dynamics and genome-centricity of interchromatin domains in the nucleus. Nat. Cell Biol. 4:E287–E291. 10.1038/ncb1202-e287 [DOI] [PubMed] [Google Scholar]
- Pederson T. 2011. The nucleus introduced. Cold Spring Harb. Perspect. Biol. 3:a000521 10.1101/cshperspect.a000521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliss A., Fritz A.J., Stojkovic B., Ding H., Mukherjee L., Bhattacharya S., Xu J., and Berezney R.. 2015. Non-Random Patterns in the Distribution of NOR-Bearing Chromosome Territories in Human Fibroblasts: A Network Model of Interactions. J. Cell. Physiol. 230:427–439. 10.1002/jcp.24726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope B.D., Ryba T., Dileep V., Yue F., Wu W., Denas O., Vera D.L., Wang Y., Hansen R.S., Canfield T.K., et al. 2014. Topologically associating domains are stable units of replication-timing regulation. Nature. 515:402–405. 10.1038/nature13986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter D.S.W., Rao B.S., Van Treeck B., Lin Y., Mizoue L., Rosen M.K., and Parker R.. 2018. Intrinsically Disordered Regions Can Contribute Promiscuous Interactions to RNP Granule Assembly. Cell Reports. 22:1401–1412. 10.1016/j.celrep.2018.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P., Parlak M., Kuscu C., Bandaria J., Mir M., Szlachta K., Singh R., Darzacq X., Yildiz A., and Adli M.. 2017. Live cell imaging of low- and non-repetitive chromosome loci using CRISPR-Cas9. Nat. Commun. 8:14725 10.1038/ncomms14725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz S.A., Ollikainen N., Tabak B., Palla A., Schmidt J.M., Detmar E., Lai M.M., Shishkin A.A., Bhat P., Takei Y., et al. 2018. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell. 174:744–757.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani V., Deng X., Qiu R., Gunderson K.L., Steemers F.J., Disteche C.M., Noble W.S., Duan Z., and Shendure J.. 2017. Massively multiplex single-cell Hi-C. Nat. Methods. 14:263–266. 10.1038/nmeth.4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., and Aiden E.L.. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 159:1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J.L., and Chang H.Y.. 2012. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81:145–166. 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J., and Guttman M.. 2014. RNA Function. RNA and dynamic nuclear organization. Science. 345:1240–1241. 10.1126/science.1252966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Carballo E., Lopez-Delisle L., Zhan Y., Fabre P.J., Beccari L., El-Idrissi I., Huynh T.H.N., Ozadam H., Dekker J., and Duboule D.. 2017. The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes Dev. 31:2264–2281. 10.1101/gad.307769.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Pereira J.M., and Aguilera A.. 2015. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 16:583–597. 10.1038/nrg3961 [DOI] [PubMed] [Google Scholar]
- Sanyal A., Lajoie B.R., Jain G., and Dekker J.. 2012. The long-range interaction landscape of gene promoters. Nature. 489:109–113. 10.1038/nature11279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satzinger H. 2008. Theodor and Marcella Boveri: chromosomes and cytoplasm in heredity and development. Nat. Rev. Genet. 9:231–238. 10.1038/nrg2311 [DOI] [PubMed] [Google Scholar]
- Schoenfelder S., Sexton T., Chakalova L., Cope N.F., Horton A., Andrews S., Kurukuti S., Mitchell J.A., Umlauf D., Dimitrova D.S., et al. 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 42:53–61. 10.1038/ng.496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal N., Fritz A.J., Vecerova J., Ding H., Chen Z., Stojkovic B., Bhattacharya S., Xu J., and Berezney R.. 2016. Large-scale probabilistic 3D organization of human chromosome territories. Hum. Mol. Genet. 25:419–436. 10.1093/hmg/ddv479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer D.R., and Pandolfi P.P.. 2006. Breaking the rules of cancer. Nat. Med. 12:14–15. 10.1038/nm0106-14 [DOI] [PubMed] [Google Scholar]
- Shah S., Takei Y., Zhou W., Lubeck E., Yun J., Eng C.L., Koulena N., Cronin C., Karp C., Liaw E.J., et al. 2018. Dynamics and spatial genomics of the nascent transcriptome by intron seqFISH. Cell. 174:363–376.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S., Zhang W., Hu H., Xue B., Qin J., Sun C., Sun Y., Wei W., and Sun Y.. 2016. Long-term dual-color tracking of genomic loci by modified sgRNAs of the CRISPR/Cas9 system. Nucleic Acids Res. 44:e86 10.1093/nar/gkw066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechner D.M., Hacisuleyman E., Younger S.T., and Rinn J.L.. 2015. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat. Methods. 12:664–670. 10.1038/nmeth.3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M., Klous P., Splinter E., Moshkin Y., Willemsen R., de Wit E., van Steensel B., and de Laat W.. 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 38:1348–1354. 10.1038/ng1896 [DOI] [PubMed] [Google Scholar]
- Sklyar I., Iarovaia O.V., Gavrilov A.A., Pichugin A., Germini D., Tsfasman T., Caron G., Fest T., Lipinski M., Razin S.V., and Vassetzky Y.S.. 2016. Distinct Patterns of Colocalization of the CCND1 and CMYC Genes With Their Potential Translocation Partner IGH at Successive Stages of B-Cell Differentiation. J. Cell. Biochem. 117:1506–1510. 10.1002/jcb.25516 [DOI] [PubMed] [Google Scholar]
- Spector D.L., and Lamond A.I.. 2011. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 3:000646 10.1101/cshperspect.a000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis C.G., Lalioti M.D., Town T., Lee G.R., and Flavell R.A.. 2005. Interchromosomal associations between alternatively expressed loci. Nature. 435:637–645. 10.1038/nature03574 [DOI] [PubMed] [Google Scholar]
- Stevens N.M. 1908. A study of the germ cells of certain Diptera with reference to the heterochromosomes and the phenomena of synapsis. J. Exp. Zool. 5:359–374. 10.1002/jez.1400050304 [DOI] [Google Scholar]
- Stevens T.J., Lando D., Basu S., Atkinson L.P., Cao Y., Lee S.F., Leeb M., Wohlfahrt K.J., Boucher W., O’Shaughnessy-Kirwan A., et al. 2017. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature. 544:59–64. 10.1038/nature21429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden H., Zunhammer A., van Koningsbruggen S., Köhler D., and Cremer T.. 2010. 4D chromatin dynamics in cycling cells: Theodor Boveri’s hypotheses revisited. Nucleus. 1:284–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Goff L.A., Trapnell C., Alexander R., Lo K.A., Hacisuleyman E., Sauvageau M., Tazon-Vega B., Kelley D.R., Hendrickson D.G., et al. 2013. Long noncoding RNAs regulate adipogenesis. Proc. Natl. Acad. Sci. USA. 110:3387–3392. 10.1073/pnas.1222643110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland H., and Bickmore W.A.. 2009. Transcription factories: gene expression in unions? Nat. Rev. Genet. 10:457–466. 10.1038/nrg2592 [DOI] [PubMed] [Google Scholar]
- Taberlay P.C., Achinger-Kawecka J., Lun A.T., Buske F.A., Sabir K., Gould C.M., Zotenko E., Bert S.A., Giles K.A., Bauer D.C., et al. 2016. Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations. Genome Res. 26:719–731. 10.1101/gr.201517.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei Y., Shah S., Harvey S., Qi L.S., and Cai L.. 2017. Multiplexed Dynamic Imaging of Genomic Loci by Combined CRISPR Imaging and DNA Sequential FISH. Biophys. J. 112:1773–1776. 10.1016/j.bpj.2017.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe H., Müller S., Neusser M., von Hase J., Calcagno E., Cremer M., Solovei I., Cremer C., and Cremer T.. 2002. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc. Natl. Acad. Sci. USA. 99:4424–4429. 10.1073/pnas.072618599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe H., Küpper K., Ishida T., Neusser M., and Mizusawa H.. 2005. Inter- and intra-specific gene-density-correlated radial chromosome territory arrangements are conserved in Old World monkeys. Cytogenet. Genome Res. 108:255–261. 10.1159/000080824 [DOI] [PubMed] [Google Scholar]
- Terakawa T., Bisht S., Eeftens J.M., Dekker C., Haering C.H., and Greene E.C.. 2017. The condensin complex is a mechanochemical motor that translocates along DNA. Science. 358:672–676. 10.1126/science.aan6516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P., Illingworth R.S., Courilleau C., Boyle S., Wood A.J., and Bickmore W.A.. 2014. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science. 346:1238–1242. 10.1126/science.1259587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B., Palstra R.J., Splinter E., Grosveld F., and de Laat W.. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 10:1453–1465. 10.1016/S1097-2765(02)00781-5 [DOI] [PubMed] [Google Scholar]
- Vance K.W., and Ponting C.P.. 2014. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 30:348–355. 10.1016/j.tig.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee R., Buljan M., Lang B., Weatheritt R.J., Daughdrill G.W., Dunker A.K., Fuxreiter M., Gough J., Gsponer J., Jones D.T., et al. 2014. Classification of intrinsically disordered regions and proteins. Chem. Rev. 114:6589–6631. 10.1021/cr400525m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera M., and Singer R.H.. 2014. Gene regulation: the HSP70 gene jumps when shocked. Curr. Biol. 24:R396–R398. 10.1016/j.cub.2014.03.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser A.E., and Aten J.A.. 1999. Chromosomes as well as chromosomal subdomains constitute distinct units in interphase nuclei. J. Cell Sci. 112:3353–3360. [DOI] [PubMed] [Google Scholar]
- Wang S., Su J.H., Zhang F., and Zhuang X.. 2016. An RNA-aptamer-based two-color CRISPR labeling system. Sci. Rep. 6:26857 10.1038/srep26857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise A., Bhatt S., Piaszinski K., Kosyakova N., Fan X., Altendorf-Hofmann A., Tanomtong A., Chaveerach A., de Cioffi M.B., de Oliveira E., et al. 2016. Chromosomes in a genome-wise order: evidence for metaphase architecture. Mol. Cytogenet. 9:36 10.1186/s13039-016-0243-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Mao S., Yang Y., Rushdi M.N., Krueger C.J., and Chen A.K.. 2018. A CRISPR/molecular beacon hybrid system for live-cell genomic imaging. Nucleic Acids Res. 46:e80 10.1093/nar/gky304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würtele H., and Chartrand P.. 2006. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended Chromosome Conformation Capture methodology. Chromosome Res. 14:477–495. 10.1007/s10577-006-1075-0 [DOI] [PubMed] [Google Scholar]
- Yang F., Deng X., Ma W., Berletch J.B., Rabaia N., Wei G., Moore J.M., Filippova G.N., Xu J., Liu Y., et al. 2015. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 16:52 10.1186/s13059-015-0618-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla D.C., and Cech T.R.. 2006. RNA as a flexible scaffold for proteins: yeast telomerase and beyond. Cold Spring Harb. Symp. Quant. Biol. 71:217–224. 10.1101/sqb.2006.71.011 [DOI] [PubMed] [Google Scholar]
- Zhang Y., McCord R.P., Ho Y.J., Lajoie B.R., Hildebrand D.G., Simon A.C., Becker M.S., Alt F.W., and Dekker J.. 2012. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 148:908–921. 10.1016/j.cell.2012.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Tavoosidana G., Sjölinder M., Göndör A., Mariano P., Wang S., Kanduri C., Lezcano M., Sandhu K.S., Singh U., et al. 2006. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 38:1341–1347. 10.1038/ng1891 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Wang P., Tian F., Gao G., Huang L., Wei W., and Xie X.S.. 2017. Painting a specific chromosome with CRISPR/Cas9 for live-cell imaging. Cell Res. 27:298–301. 10.1038/cr.2017.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D., Cremer T., Saffrich R., Fischer R., Trendelenburg M.F., Ansorge W., and Stelzer E.H.. 1998. Structure and dynamics of human interphase chromosome territories in vivo. Hum. Genet. 102:241–251. 10.1007/s004390050686 [DOI] [PubMed] [Google Scholar]