Ladstätter and Tachibana discuss changes in DNA methylation, chromatin accessibility, and topological architecture occurring during the reprogramming to totipotency in the early embryo.
Abstract
The early embryo is the natural prototype for the acquisition of totipotency, which is the potential of a cell to produce a whole organism. Generation of a totipotent embryo involves chromatin reorganization and epigenetic reprogramming that alter DNA and histone modifications. Understanding embryonic chromatin architecture and how this is related to the epigenome and transcriptome will provide invaluable insights into cell fate decisions. Recently emerging low-input genomic assays allow the exploration of regulatory networks in the sparsely available mammalian embryo. Thus, the field of developmental biology is transitioning from microscopy to genome-wide chromatin descriptions. Ultimately, the prototype becomes a unique model for studying fundamental principles of development, epigenetic reprogramming, and cellular plasticity. In this review, we discuss chromatin reprogramming in the early mouse embryo, focusing on DNA methylation, chromatin accessibility, and higher-order chromatin structure.
Introduction
One of the greatest feats of nature is that a single cell is able to activate an orchestrated developmental program that culminates in a complete multicellular organism. This property is the strictest definition of totipotency (Condic, 2014) and can be attributed to cells of the early embryo. A one-cell embryo (zygote) is generated by the fusion of terminally differentiated haploid gametes, namely, egg (oocyte) and sperm (Fig. 1). Epigenetic reprogramming, zygotic genome activation (ZGA), and several cleavage divisions give rise to the cells of the mammalian embryo (blastomeres) that specialize into the embryo proper and extraembryonic tissues to form an organism. Totipotency of the blastomeres is gradually lost during cleavage divisions to allow cell specification and differentiation (Fig. 1 and Table 1).
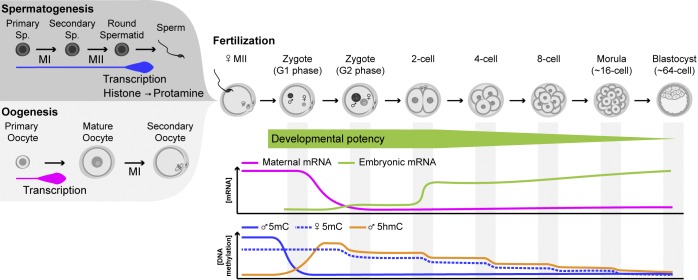
Figure 1.
Overview of molecular events during mouse gametogenesis and preimplantation development. Male and female gametogenesis entail distinct timings of meiotic divisions (MI and MII) in relation to transcriptional activity and differentiation. The sperm genome is packaged around protamines that are rapidly exchanged to maternal histones at fertilization. Fertilization triggers resumption of oocyte meiosis (MII), and the totipotent zygote forms, containing two parental pronuclei. Totipotency declines with subsequent cleavage divisions. Maternal transcripts are rapidly degraded before ZGA, which occurs in two waves: minor ZGA in the late zygote and major ZGA in the two-cell embryo. DNA methylation dynamically changes in the zygote: paternal 5mC is lost, de novo paternal 5hmC forms, and passive dilution of DNA methylation occurs during cleavage divisions. Sp., spermatocyte.
Table 1. Comparison of timings in human and mouse preimplantation development.
| Human | Mouse | |
|---|---|---|
| First cleavage division | 27 to 30 hpf (Mio and Maeda, 2008; Wong et al., 2010; Kirkegaard et al., 2012) | 16 to 20 hpf (Arav et al., 2008; Kirkegaard et al., 2012) |
| Blastocyst implantation | 6 to 8 dpf (Hertig et al., 1956) | 4 to 4.5 dpf (Finn and McLaren, 1967) |
| Totipotent embryo | Up to four-cell (Van de Velde et al., 2008) | Up to two-cell (Tarkowski, 1959; Rossant, 1976) |
| Major ZGA | Four- to eight-cell (Tesarík et al., 1987; Braude et al., 1988; Dobson et al., 2004; Vassena et al., 2011; Yan et al., 2013) | Two-cell (Flach et al., 1982; Aoki et al., 1997; Hamatani et al., 2004) |
| DNA demethylation | Before two-cell (Guo et al., 2014b) | In early one-cell (Mayer et al., 2000; Oswald et al., 2000) |
| Chromatin accessibility | Increases during cleavage stages; detectable promoter accessibility correlates with timing of ZGA (Gao et al., 2018; Li et al., 2018) | Increases during cleavage stages; detectable promoter accessibility correlates with timing of ZGA (Lu et al., 2016; Wu et al., 2016) |
| Higher-order chromatin structure | No data available | Gradual establishment during cleavage stages (Du et al., 2017; Flyamer et al., 2017; Gassler et al., 2017; Ke et al., 2017) |
dpf, days post-fertilization; hpf, hours post-fertilization.
One intriguing question in developmental biology is the following: What defines the chromatin state of a totipotent cell, and how does this determine the acquisition of totipotency? A comprehensive understanding of the nuclear and chromatin organization of the early embryo will provide crucial insights into the fundamental process of embryonic development and the complex network regulating cell identity and plasticity. The latter is of substantial interest considering the current limitations of cellular reprogramming, such as the generation of induced pluripotent stem cells, which is envisioned as a therapeutic tool for regenerative medicine (Robinton and Daley, 2012; Ohnuki and Takahashi, 2015; Srivastava and DeWitt, 2016).
The rising era of low-input genomic assays sets the stage for exploring the regulatory networks for acquisition of totipotency in sparsely available early embryos. This review will summarize aspects of chromatin reprogramming occurring in the totipotent mouse embryo with a focus on insights gained from recent technical advances relating to zygotic DNA methylation dynamics, chromatin conformations, and chromatin accessibility status. Of note, available data from early human embryos indicate that chromatin reprogramming events occur with different timings compared to mouse embryos (Table 1). Furthermore, the lineage-specific transcriptional program responsible for patterning the embryo differs substantially between mouse and human (Fougerousse et al., 2000; Niakan and Eggan, 2013; Fogarty et al., 2017). This suggests distinct fine-tuning of epigenetic and chromatin profiles that awaits further analysis in the human embryo.
Zygotic reprogramming: The first hours after fertilization
Dynamic changes in epigenetic marks that occur throughout zygotic interphase are thought to promote the acquisition of totipotency. These involve histone incorporation, establishment of histone modifications, timely changes in the DNA methylation status, changes in chromatin accessibility, and changes in three-dimensional chromatin conformations. The complex cellular events occurring after fertilization can be summarized as the “oocyte-to-zygote transition.” It is thought that maternal factors stored in the oocyte are important for this transition due to limited transcription in the first cell cycle after fertilization in mammals (Tadros and Lipshitz, 2009; Jukam et al., 2017). Vertebrate ZGA occurs in waves, with the major activation occurring at the two-cell stage in mouse (Flach et al., 1982; Aoki et al., 1997; Hamatani et al., 2004).
After fertilization, the parental genomes form two spatially separate entities—the maternal and paternal pronucleus. The cellular histories of the parental genomes are quite different (Fig. 1). Maternal chromosomes of the meiosis II oocyte are condensed, and fertilization triggers resumption of the second meiotic division, followed by decondensation and formation of interphase chromatin comprising the maternal pronucleus. Maternal chromatin is packaged with predeposited maternal histones and is segregated during the meiotic divisions in the absence of transcription (Bachvarova, 1985; Bouniol-Baly et al., 1999). On the other hand, meiotic divisions during spermatogenesis are followed by active transcription (Kierszenbaum and Tres, 1975), with subsequent protamine packaging of the paternal chromatin (Meistrich et al., 2003; Oliva, 2006; Rathke et al., 2014). In mice, ∼2–4% of the histones are retained in sperm (Bench et al., 1996; Carone et al., 2014). Shortly after fertilization, the protamines are exchanged to maternal histones containing the histone variant H3.3 (van der Heijden et al., 2005; Torres-Padilla et al., 2006), and the paternal chromatin decompacts to an interphase state (Rodman et al., 1981).
Additionally, the epigenetic signatures differ in the parental genomes. Histones of the paternal genome are initially mainly hypomethylated and hyperacetylated, while the maternal genome is enriched with repressive histone lysine methylation marks (reviewed in Burton and Torres-Padilla, 2010; Filipescu et al., 2014; Xu and Xie, 2018). Furthermore, DNA methylation underlies dynamic and parental specific events that will be discussed in detail. In brief, paternal DNA becomes demethylated, which is characterized by reduction of 5-methylcytosine (5mC; Mayer et al., 2000; Oswald et al., 2000) and acquires 5-hydroxymethylcytosine (5hmC; Gu et al., 2011; Inoue and Zhang, 2011; Iqbal et al., 2011; Wossidlo et al., 2011). In contrast, maternal DNA retains a higher 5mC level and does not gain notable de novo 5hmC. The functional significance of these epigenetic asymmetries remains to be determined.
Epigenetics of the early embryo: The complexity of zygotic DNA demethylation
In many eukaryotic clades, DNA methylation occurs at cytosine (5mC) of cytosine-guanine dinucleotides (CpG). This epigenetic modification correlates with gene repression in mammals, such as stable silencing of repetitive DNA, imprinting, and X chromosome inactivation (Jones, 2012; Schübeler, 2015). De novo establishment of 5mC is catalyzed by DNA methyltransferases (DNMTs) 3a, 3b, and 3c (Okano et al., 1999; Barau et al., 2016; Jain et al., 2017), while maintenance of 5mC throughout cell divisions is mediated by DNMT1 (Li et al., 1992). Loss of DNA methylation can proceed either by passive or active mechanisms. Passive dilution of 5mC occurs over consecutive cell cycles during DNA replication in the absence of maintenance DNMT, DNMT1 (Rougier et al., 1998). Active DNA demethylation mechanisms, which refer to enzyme-mediated changes in 5mC to other cytosine modifications or reversal to unmodified cytosine, can involve Ten-eleven translocation (Tet) methylcytosine dioxygenases (Wu and Zhang, 2017). Tet family proteins iteratively convert 5mC to 5hmC, 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC; Tahiliani et al., 2009; He et al., 2011; Ito et al., 2011; Fig. 2). Studies in mouse embryonic stem cells (mESCs) demonstrated that oxidized cytosines can be passively diluted but are also substrates for thymine-DNA glycosylase, which recognizes oxidized cytosines and generates an abasic site that is perceived by the base excision repair (BER) machinery, resulting in active DNA demethylation (Cortellino et al., 2011; He et al., 2011).
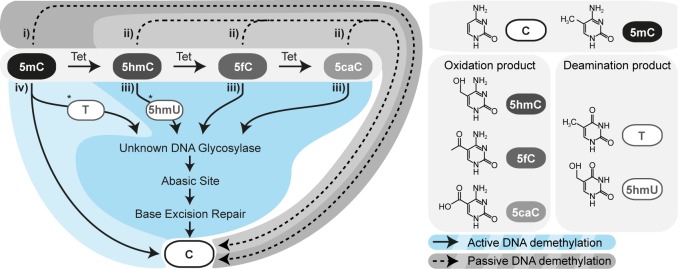
Figure 2.
Proposed mechanisms of zygotic DNA demethylation. Passive DNA demethylation mechanisms include (i) replication-dependent passive dilution of 5mC or (ii) its Tet-catalyzed oxidation products (5hmC, 5fC, and 5caC) in the absence of maintenance DNA methylase DNMT1 or Tet hydroxylase, respectively. Active DNA demethylation mechanisms comprise (iii) active removal of 5mC via Tet3-catalyzed oxidation products involving activity of yet unidentified deaminase (*) and DNA glycosylases feeding into the BER pathway or (iv) direct removal of 5mC independent of Tet action involving an unknown deaminase (*) or demethylase. C, cytosine; T, thymidine; 5hmU, 5-hydroxymethyluracil.
Our understanding of the regulation and function of DNA methylation during early embryonic development is still limited despite years of intensive research. Extensive and increasing studies show confounding aspects of zygotic DNA methylation dynamics. In 2000, immunofluorescence analysis showed epigenetic asymmetry between the two parental genomes in mouse zygotes: The paternal genome undergoes considerable loss of 5mC before DNA replication commences, while the maternal genome retains a high 5mC level (Mayer et al., 2000; Oswald et al., 2000). The timing of paternal DNA demethylation—occurring before DNA replication—led to the conclusion that an active mechanism is mediating this event, while the maternal genome is protected from active DNA demethylation (Nakamura et al., 2007; Hajkova et al., 2010). The maternal genome undergoes passive, replication-dependent DNA demethylation during subsequent embryonic cleavage divisions (Rougier et al., 1998), presumably due to limited availability of DNMT1 (Hirasawa et al., 2008).
The discovery of Tet family proteins stimulated the hypothesis that zygotic active paternal DNA demethylation involves 5mC oxidation. Indeed, the conversion of 5mC to 5hmC is observed on paternal DNA concurrently with 5mC loss in G1 phase (Gu et al., 2011; Inoue and Zhang, 2011; Iqbal et al., 2011; Wossidlo et al., 2011) and is accompanied by the timely appearance of 5fC and 5caC (Inoue et al., 2011). Among the Tet protein family members, Tet3 was identified to act predominantly in the mouse zygote. Depletion of Tet3 by siRNA or Tet3 conditional maternal knockout showed reduced paternal 5mC loss and abolished 5hmC accumulation in S/G2 phase zygotes (Gu et al., 2011; Wossidlo et al., 2011; Inoue et al., 2012, 2015; Tsukada et al., 2015). Immunofluorescence-based detection of cytosine modifications reflects global levels and does not allow discrimination between loci-specific 5mC loss by its oxidation followed by active excision and replacement by unmodified cytosine, further modifications, or passive dilution by DNA replication.
Recent progress in the generation of low-input genomic DNA methylation profiles is creating a distinct picture of zygotic DNA demethylation. Reduced representation bisulfite sequencing (RRBS) data revealed that both parental genomes are subjects of DNA demethylation, which is mediated primarily by passive replication-dependent dilution and to a lesser extent by active Tet3-dependent mechanisms (Guo et al., 2014a; Shen et al., 2014). This conclusion is supported by RRBS data from zygotes that were chemically inhibited to undergo DNA replication and postreplicative maternal Tet3 knockout zygotes. The RRBS results are apparently in contrast to the previous immunofluorescence-based observations, which led to the conclusion that 5mC loss occurs predominantly in the paternal genome with strong involvement of Tet3 activity (Mayer et al., 2000; Oswald et al., 2000; Gu et al., 2011; Wossidlo et al., 2011; Inoue et al., 2012, 2015; Tsukada et al., 2015). The discrepancy could in part be attributed to the enrichment for CpG-rich regions (bias to CpG islands) and the low genome coverage of 2–5% of all CpG sites in the RRBS data (Guo et al., 2014a; Shen et al., 2014). Whole-genome bisulfite sequencing can overcome a selective view on CpG methylation and was applied to wild-type and maternal Tet3 knockout zygotes (Peat et al., 2014). The majority (90%) of paternal intergenic regions and gene bodies are demethylated, and Tet3 does play a noteworthy, if only moderate, role in DNA demethylation at all genomic features (Peat et al., 2014). Interestingly, the whole-genome bisulfite sequencing data indicate an involvement of Tet3 in maintenance of the hypomethylated state of some CpG islands (Peat et al., 2014). This is further supported by qualitative (immunofluorescence) and impressive quantitative (liquid chromatography–mass spectrometry) data (Amouroux et al., 2016) showing DNMT3a- and DNMT1-driven accumulation of paternal 5hmC on de novo–generated 5mC in postreplicative zygotes. This may partially explain why some paternal 5mC is detected by immunofluorescence staining in late Tet3-depleted zygotes, which was previously interpreted as remaining 5mC due to impaired DNA demethylation (Gu et al., 2011; Wossidlo et al., 2011; Inoue et al., 2012, 2015; Tsukada et al., 2015).
Nevertheless, only a fraction of the paternal genome appears to undergo Tet3-independent DNA demethylation (Guo et al., 2014a; Peat et al., 2014; Shen et al., 2014). Whether this is solely mediated by DNA replication–dependent dilution of 5mC is challenged by recent observations that global 5mC loss occurs (a) temporally uncoupled from DNA replication in G1 phase and (b) before, as well as independently of, Tet3-mediated 5hmC accumulation (Amouroux et al., 2016; Ladstätter and Tachibana-Konwalski, 2016). The mechanism underlying this active and potential Tet3-independent DNA demethylation remains to be identified. Further work is necessary to understand zygotic Tet3-dependent active DNA demethylation since mouse knockout studies demonstrated that thymine-DNA glycosylase is not essential in zygotic active DNA demethylation (Santos et al., 2013; Guo et al., 2014a). Interestingly, mutations of several known DNA glycosylases result in viable mice that lack an obvious phenotype, suggesting redundant mechanisms (Friedberg et al., 1997). A contribution of a BER-coupled pathway in zygotic active DNA demethylation is supported by preferential localization of BER proteins to the paternal pronucleus (Hajkova et al., 2010; Wossidlo et al., 2010). Maternal knockout of the essential BER component Xrcc1 leads to accumulation of paternal DNA lesions during active DNA demethylation, demonstrating that Xrcc1 is required for their timely repair (Ladstätter and Tachibana-Konwalski, 2016). Furthermore, cohesin is necessary for the repair of endogenous paternal DNA lesions, which are generated by a mechanism requiring Tet3 activity (Ladstätter and Tachibana-Konwalski, 2016).
Currently, four mechanisms of zygotic DNA demethylation that might act redundantly and potentially possess specificity for certain genomic regions have been proposed (Fig. 2): (i) replication-dependent passive dilution of 5mC, (ii) replication-dependent passive dilution of oxidized 5mC (5hmC, 5fC, and 5caC), (iii) active removal of 5mC via Tet3-catalyzed oxidation products 5hmC, 5fC, and 5caC, and (iv) active removal of 5mC that is independent of Tet3 activity. The generation of extensive genomic profiles for zygotic 5mC and its oxidation products from staged zygotes (pre- versus post-replication) including separate analysis of parental genomes under conditions of Tet3 perturbation will help to further elucidate the intricate picture of DNA methylation dynamics and Tet3 function in early embryonic development. Of certain interest will be whether active DNA demethylation or 5hmC formation occurs at specific genomic regions to potentially establish a permissive chromatin state that may facilitate the acquisition of totipotency.
Biological relevance of zygotic DNA methylation dynamics
It remains to be determined whether zygotic DNA demethylation plays an essential role in mammalian development and contributes to the acquisition of totipotency. Intriguingly, DNA demethylation is observed in mammalian embryos from a range of species including mouse, rat, goat, bovine, and human (Mayer et al., 2000; Oswald et al., 2000; Dean et al., 2001; Fulka et al., 2004; Park et al., 2007; Lepikhov et al., 2008; Wossidlo et al., 2011; Efimova et al., 2015). Some species have been reported to not display zygotic DNA demethylation, like pig and sheep (Beaujean et al., 2004; Jeong et al., 2007). An extensive analysis of zygotic DNA methylation dynamics using different methods of detection in a multitude of mammalian species is still missing and might support either conserved or species-specific roles of DNA methylation during development.
Increasing evidence supports a role of 5mC in gene suppression (Jones, 2012; Schübeler, 2015), but the function of its oxidized forms remains elusive (Song et al., 2012). Some studies attempted to correlate ZGA with DNA demethylation and Tet3-mediated 5mC oxidation (Inoue et al., 2012; Peat et al., 2014; Shen et al., 2014; Tsukada et al., 2015; Zhu et al., 2017). Global analysis of zygotic transcription using semi-quantitative detection of incorporated UTP analogue or transcriptome analysis by RNA-seq in Tet3 knockdown or knockout zygotes, respectively (Inoue et al., 2012; Peat et al., 2014; Shen et al., 2014), suggests that Tet3 is dispensable for zygotic transcription. Additionally, a positive correlation of genes within demethylated paternal loci in the zygote to increased expression in the two-cell embryo does not change remarkably upon maternal Tet3 depletion (Peat et al., 2014), indicating that the majority of genes undergoing demethylation, and thus potential priming for ZGA, appear to do this in a Tet3-independent manner.
Nonetheless, it needs to be taken into account that bisulfite-based techniques cannot distinguish 5mC from 5hmC and unmodified cytosine from 5fC or 5caC (Huang et al., 2010; Jin et al., 2010; Nestor et al., 2010; Yu et al., 2012; Booth et al., 2015). Therefore, a subset of genes that might be marked with 5hmC or its further oxidized forms would not be identified. Furthermore, the detection of oxidized 5mC is challenging due to their low abundance (Ito et al., 2011) suggesting that these either are transient in nature or have a function at specific CpG islands. A recent methodological advance using chemical labeling–enabled C-to-T conversion sequencing that covers 15–27% of CpGs (Zhu et al., 2017) provided insights into the genomic distribution of 5fC in gametes and zygotes. Interestingly, 5fC is enriched at promoters, while its distribution among other genomic features is heterogeneous in the zygotic genome. Gene ontology analysis of promoters enriched for 5fC identified clusters of genes that are important for preimplantation development (Zhu et al., 2017), suggesting a regulatory role for 5fC in ZGA. Whether 5fC sites represent an intermediate stage toward cytosine demethylation or are directly involved in gene activation needs further investigation. Both scenarios are in principle possible since 5fC is associated with recruitment of DNA repair factors as well as transcriptional regulators (Iurlaro et al., 2013; Spruijt et al., 2013). A role of Tet3 in ZGA may be conceivable since evidence emerged that Tet3 partially restricts global transcription in late zygotes (Tsukada et al., 2015). Nevertheless, currently there is no clear evidence for a specific role of Tet3 in regulating ZGA.
Considering that Tet3 is the major enzyme catalyzing 5mC oxidation in zygotes, together with its intricate role in DNA demethylation, it seems surprising that maternal Tet3 depletion results in no obvious defects in the preimplantation blastocyst, but rather in neonatal sublethality (Gu et al., 2011; Shen et al., 2014; Inoue et al., 2015). Elegant nuclear transfer experiments showed that Tet3-mediated paternal 5mC oxidation is not required for embryonic development to term and that Tet3 haploinsufficency likely causes sublethality (Inoue et al., 2015). However, it was not addressed whether Tet3-dependent changes on the maternal genome are crucial for full-term development, nor whether redundant mechanisms compensate for Tet3 loss in the early zygote. The other two known Tet protein family members (Tet1 and Tet2)—despite being poorly expressed in the zygote (Gu et al., 2011; Iqbal et al., 2011; Wossidlo et al., 2011)—might act redundantly in the absence of Tet3. Interestingly, CpG islands are hypermethylated and gene expression is perturbed in Tet1/2/3 knockout embryos, which develop morphologically normally until gastrulation (Dai et al., 2016). This is in line with the role of Tet3 in protecting against 5mC accumulation in the zygote (Peat et al., 2014; Amouroux et al., 2016), but does not exclude a function of Tet1/2 in maintenance of a hypomethylated state. The scarcity of early mammalian embryos as an in vivo model to study the role of dynamic DNA methylation has thus far prevented the generation of high-coverage genome-wide DNA methylation profiles. Further improvement of low-input techniques might be able to reveal whether Tet3-dependent gene regulation is set at distinct loci already in the zygote or later in development.
Integrating nuclear architecture with zygotic reprogramming
Revealing higher-order chromatin structure in the early embryo
The scarcity of material provides a challenge for analyzing genome architecture in totipotent early embryos. Studies of nuclear architecture in zygotes have hitherto been focused on nuclear subcompartments, which can be visualized in single cells by microscopy such as immunofluorescent detection of candidate proteins or FISH of genomic loci. Well-studied examples of subnuclear compartments include the nuclear envelope and lamina, nucleoli, promyelocytic leukemia and Cajal bodies, euchromatin and heterochromatin, and chromosome territories (reviewed in Borsos and Torres-Padilla, 2016).
The advances of genome-wide profiling of chromatin interactions (see text box) are revolutionizing the study of chromatin reprogramming in the early embryo. In 2017, four studies used high-throughput sequencing-based chromatin conformation capture methods (Hi-C) to detect higher-order chromatin structures in early mouse embryos (Du et al., 2017; Flyamer et al., 2017; Gassler et al., 2017; Ke et al., 2017). Topologically associating domains (TADs) can be detected from the zygote stage onward (Flyamer et al., 2017; Gassler et al., 2017) and become more pronounced, displaying increased intra-domain interactions between distal regions and progressive insulation around TAD boundaries during the course of embryonic cleavage divisions (Du et al., 2017; Gassler et al., 2017; Ke et al., 2017). This suggests a gradual shift toward a somatic cell-like chromatin state. It is not known whether this difference in TAD boundary insulation functionally correlates to the transition of totipotency toward lineage specification or might reflect an initial shortage of factors required for TAD formation that steadily increases with the onset of ZGA. Since TADs can be detected before minor ZGA in unperturbed G1 phase zygotes (Flyamer et al., 2017; Gassler et al., 2017), TAD formation does not strictly depend on transcription. Chemical inhibition of transcription in mouse zygotes as well as in the fly embryo is consistent with this (Du et al., 2017; Hug et al., 2017; Ke et al., 2017).
Importance and description of higher-order chromatin structure
Cellular identity and plasticity is mediated by differential gene expression that orchestrates the interplay of transcription factors, DNA sequence, and epigenetic modifications (Roy and Kundu, 2014; Sorrells and Johnson, 2015; Dong and Liu, 2017). This regulatory network is becoming conceptually extended by incorporating the spatial and temporal organization of the genome. Accommodation of the 2-m-long DNA into the three-dimensional nucleus of mammalian cells is non-random and cell type specific, implying a functional relevance of chromatin organization (Bickmore, 2013; Dekker and Misteli, 2015). Indeed, the three-dimensional arrangement of linear DNA can bring distal cis-regulatory elements, like enhancers or insulators, into close proximity with their target genes (Gaszner and Felsenfeld, 2006; Shlyueva et al., 2014).
The elucidation of higher-order nuclear architecture became experimentally amenable with the development of biochemical approaches that map physical genome interactions by DNA–DNA proximity ligation. These chromosome conformation capture assays, so called “C” techniques (3C, 4C, 5C, Hi-C, and derivatives; reviewed in Denker and de Laat, 2016) revealed multi-hierarchical structuring of chromatin. Thus, interphase chromatin is organized into loops, TADs, and compartments (Lieberman-Aiden et al., 2009; Rao et al., 2014; Nagano et al., 2017):
• Loops are formed by close association of two genomic regions that can be far apart in linear sequence and are characterized by closely associated CTCF-based anchor sites.
• TADs are large stretches of DNA that display high contact frequency and loci within these are less likely to interact with loci in other TADs. TADs may represent an assembly of dynamic loops, but the detailed relationship between loops and TADs awaits experimental verification.
• Compartments are spatially segregated regions that delineate transcriptionally active (A) and repressed (B) regions, which correlate with the chromatin state that is defined by epigenetic modifications.
The zygote stage displays the least defined higher-order chromatin structure during preimplantation development. While the detection of TADs and loops was reported in the parental genomes of the zygote (Flyamer et al., 2017), two subsequent studies reported weak to nearly undetectable TADs in zygotes (Du et al., 2017; Ke et al., 2017). The discrepancy was resolved by reanalysis of the bulk Hi-C data (Gassler et al., 2017) using algorithms with higher statistical power, which confirmed the presence of TADs and loops in these datasets (Du et al., 2017; Ke et al., 2017). It is worthwhile to point out that, despite epigenetic differences, TADs and loop strengths are similar between maternal and paternal genomes (Du et al., 2017; Flyamer et al., 2017; Gassler et al., 2017; Ke et al., 2017). There is the tendency of fewer distal interactions in the paternal genome (Du et al., 2017), suggesting a more relaxed chromatin state, in line with global hypomethylated DNA and histone modifications associated with transcriptionally active chromatin (Mayer et al., 2000; Oswald et al., 2000; Burton and Torres-Padilla, 2010). The allelic differences in TAD organization become evident upon interfering with one essential protein complex required for TAD formation or maintenance, namely cohesin (Gassler et al., 2017; Haarhuis et al., 2017; Hansen et al., 2017; Rao et al., 2017; Schwarzer et al., 2017; Wutz et al., 2017). Cohesin or its associated proteins are strong candidates for the loop extrusion factor, which has been proposed to progressively facilitate loop growth until encountering a boundary element (Nasmyth, 2001; Alipour and Marko, 2012; Sanborn et al., 2015; Fudenberg et al., 2016). Depletion of the cohesin-release factor Wapl affects the processivity of cohesin, which has different effects on parental chromatin structure: The maternal genome displays stronger compaction with longer loops, while loops and TAD insulation are stronger in the paternal genome (Gassler et al., 2017). This might indicate that loop extrusion dynamics in the parental genomes are different due to the underlying asymmetry of their epigenetic landscapes.
Interestingly, maternal and paternal chromatin structure are most distinct with respect to compartmentalization, which is weak or absent in zygotic maternal chromatin (Du et al., 2017; Flyamer et al., 2017; Gassler et al., 2017; Ke et al., 2017). This is of general interest because it suggests that establishment of compartments follows a different mechanism than TAD and loop formation. Indeed, the mechanism of TAD formation involves cohesin and thus loop extrusion, which are not required for and even antagonize compartmentalization (Gassler et al., 2017; Haarhuis et al., 2017; Hansen et al., 2017; Rao et al., 2017; Schwarzer et al., 2017; Wutz et al., 2017). In addition, the dynamics of TAD and compartment formation differ during the cell cycle of mESCs (Nagano et al., 2017). TAD insulation strength increases in G1 phase and reaches a plateau in S phase, while compartmentalization is weak in G1 and steadily increases from S phase onward until it reaches its maximum in G2 phase (Nagano et al., 2017). Assuming that higher-order chromatin structure is reestablished after every mitotic phase, one might expect that the maternal zygotic genome reestablishes chromatin conformation after completion of the second meiotic division, while the paternal genome might require de novo establishment of chromatin architecture after protamine–histone exchange (Fig. 1). Surprisingly, the maternal genome does not rapidly establish compartments after chromosome decondensation, while the paternal genome displays compartmentalization in G1-phase zygotes (Flyamer et al., 2017). This raises the question whether compartmentalization is inherited and/or simply established faster in the paternal genome. Potentially, a predetermined architecture might facilitate formation of paternal compartments after fertilization. Unexpectedly, bulk Hi-C data show that sperm chromatin conformation does not drastically differ from other mammalian cells apart from displaying increased long-range interactions and inter-chromosomal contacts, which are consistent with a compact chromatin state (Battulin et al., 2015; Jung et al., 2017). Single-cell sperm Hi-C data is needed to confirm these observations. A driving force for fast compartmentalization could be transcription since it is assumed that the paternal genome is more permissive for early transcription (Worrad et al., 1994; Aoki et al., 1997). Precise future experiments might shed light on whether epigenetic modifications and early transcription are required for paternal compartmentalization or vice versa.
Then again, a puzzling question is: What is special about the maternal chromatin and the lack of compartmentalization? Or, what are the prerequisites for de novo compartmentalization? Little is known about the actual mechanism behind formation of compartments. One hypothesis suggests that phase separation based on direct protein–protein interactions in the framework of epigenetic signatures drives segregation of chromatin types (Brackley et al., 2016; Di Pierro et al., 2016, 2017). Thus, the epigenetic asymmetry of histone modifications and DNA methylation in the parental genomes might account for distinct compartmentalization. The absence of maternal compartments might therefore be caused by the presence of factors specifically interacting with the maternal epigenome to prevent segregation of chromatin types. Alternatively, loop extrusion dynamics in the maternal genome might antagonize compartmentalization. Further investigations are required to delineate the determinants of de novo compartmentalization. An exciting topic of future research will be to determine whether there is a causal relationship between the epigenome and higher-order chromatin structure.
The role of chromatin accessibility in preparing for ZGA
A more direct relationship of epigenetic modifications, transcription factor occupancy, and gene expression can be inferred when moving from global higher-order chromatin organization toward primary nucleosomal chromatin structure. The nucleoprotein structure of chromatin is the smallest unit guiding differential gene expression. Nucleosome positioning defines general chromatin accessibility (Radman-Livaja and Rando, 2010; Sadeh and Allis, 2011), while the four core histones of the octamer comprising the nucleosome carry post-translational modifications that can affect DNA binding affinity and serve as a binding platform for regulatory factors (Musselman et al., 2012).
The chromatin accessibility landscape of the early embryo is of great interest to identify pioneer transcription factors that may be important for the acquisition of totipotency (Zaret and Carroll, 2011; Iwafuchi-Doi and Zaret, 2014; Zaret, 2018). Pioneer factors are hypothesized to be critical for ZGA by initiating or priming regulatory events at closed chromatin regions to promote transcription. There are different modes of action. In its simplest form, a pioneer factor binds to closed chromatin, alters chromatin accessibility, and either directly initiates transcription or primes the genomic locus for other transcription factors. Pioneer factors have essential roles in cell fate transitions, such as hepatic differentiation of embryonic gut endoderm or B cell development from hematopoietic progenitors (Zaret and Carroll, 2011; Iwafuchi-Doi and Zaret, 2014; Zaret, 2018). The best studied pioneer factor in early embryonic development is the fly protein Zelda (Staudt et al., 2006; Liang et al., 2008). Zelda can facilitate transcription directly or indirectly (Harrison et al., 2011; Schulz et al., 2015; Sun et al., 2015) and was shown to affect chromatin conformation (Hug et al., 2017). However, a mammalian counterpart has not been identified. Interestingly, Oct4 and Sox2 homologues are important for ZGA in zebrafish (Lee et al., 2013; Leichsenring et al., 2013) and have been shown to bind to closed chromatin to induce cellular reprogramming during mammalian induced pluripotent stem cell generation (Soufi et al., 2012, 2015; King and Klose, 2017; Donaghey et al., 2018). However, Oct4 functions as a lineage specifier in mammalian embryos (Niakan and Eggan, 2013; Fogarty et al., 2017) that contributes to increased chromatin accessibility after ZGA in the eight-cell mouse embryo (Lu et al., 2016). Recently, the double homeobox transcription factor Dux emerged as a novel regulator of mammalian ZGA (De Iaco et al., 2017; Hendrickson et al., 2017; Whiddon et al., 2017). Dux expression coincides with ZGA. Chromatin immunoprecipitation (ChIP) data indicate that Dux preferentially binds to genes that are specific for early cleavage-stage embryos. The binding profile of Dux overlaps with chromatin accessibility profiles, further supporting its transcriptional role. Overexpression of Dux in mESCs induces transition to two-cell–like cells, which have a gene expression signature that to some extent resembles two-cell embryo blastomeres (Macfarlan et al., 2012; Ishiuchi et al., 2015; Eckersley-Maslin et al., 2016). A comparison of chromatin accessibility profiles of two-cell embryos and Dux-induced two-cell–like cells showed a correlation of accessible sites gained and lost upon mESC transition (Hendrickson et al., 2017). Interestingly, half of the gained accessible sites are bound by Dux, suggesting that Dux might act as a pioneer factor to open chromatin. It remains to be shown whether Dux is able to directly bind to closed chromatin and mediate its opening. Additionally, it is not known what mechanisms regulate Dux transcription in the early embryo. LINE1 RNA was recently shown to contribute to Dux silencing in mESCs and early embryos (Percharde et al., 2018). The existence of a yet unidentified upstream pioneer factor remains a likely possibility.
The identification of pioneer factors is thus far based on approaches aimed at identifying early binding factors in cellular transitions by ChIP or in vivo footprinting. ChIP-based approaches can generate genome-wide binding profiles, but are limited by assaying known candidate factors. Genome-wide in vivo chromatin accessibility assays, like micrococcal nuclease sensitive sites sequencing (MNase-seq), DNase I hypersensitive sites sequencing (DNase-seq), formaldehyde-assisted isolation of regulatory elements sequencing (FAIRE-seq), and assay for transposase-accessible chromatin sequencing (ATAC-seq; reviewed in Tsompana and Buck, 2014), can reveal sequence motifs for factors that bind to open chromatin. Zelda in Drosophila melanogaster and Dux in mammals were identified by motif analysis in a similar fashion (Staudt et al., 2006; Liang et al., 2008; De Iaco et al., 2017; Hendrickson et al., 2017; Whiddon et al., 2017).
Recent advances in low-input chromatin accessibility assays (Buenrostro et al., 2015; Cusanovich et al., 2015; Jin et al., 2015) make it feasible to analyze accessible chromatin in early embryos. Data have been gathered from cleavage stage embryos (Lu et al., 2016; Wu et al., 2016). Low-input DNase-seq and ATAC-seq techniques led to the discovery that parental genomes are similar in their chromatin accessibility profiles from genome activation in the two-cell mouse embryo onward (Lu et al., 2016; Wu et al., 2016), despite epigenetic asymmetry up to the eight-cell stage. Interestingly, open chromatin sites are increasing during cleavage stages in mouse and human embryos (Lu et al., 2016; Wu et al., 2016; Gao et al., 2018; Li et al., 2018), suggesting a gradual establishment during development. On the contrary, microscopy-based and Hi-C techniques suggest a rather permissive and open chromatin state in one-cell and two-cell embryos (Ahmed et al., 2010; Du et al., 2017; Flyamer et al., 2017; Gassler et al., 2017; Ke et al., 2017). However, microscopy and Hi-C reveal different levels of chromatin organization and can be used to shed light on higher-order structure, while DNase-seq and ATAC-seq reflect the primary chromatin structure of nucleosome and transcription factor occupancy. Thus, a rather susceptible and open higher-order chromatin structure does not necessarily imply an underlying general loose nucleosome array. DNase hypersensitive sites (DHSs) are mainly detected at promoters and distal regulatory regions. Detection at distal elements predominates at two-cell– and eight-cell–stage embryos (Lu et al., 2016), correlating to major ZGA and priming for lineage specification around the morula stage, respectively. Of note, the presence of DHS does not automatically indicate expression. Transcriptomic data have also revealed DHS sites at silent genes that are primed for expression at a subsequent developmental stage (Lu et al., 2016). Chromatin accessibility profiles identified putative regulatory elements that are part of the transcriptional networks accompanying early embryonic development. For example, the nuclear transcription factor Y α was shown to be involved in ZGA (Lu et al., 2016), and Nr5a2 (nuclear receptor subfamily 5, group A, member 2) regulates lineage specification (Wu et al., 2016).
Nevertheless, the questions of which genomic loci open up first after fertilization and how this is mediated remain to be answered. The challenge of experimental timing during embryo development and separation of data originating from the two parental genomes will be possible to overcome. In vitro fertilization can provide control of fertilization timing, while extraction of zygotic pronuclei or use of parental mice strains with distinct single-nucleotide polymorphisms can be used to separate the parental genomes. An interesting dataset was generated from late zygotes (S/G2 phase) applying pronuclear extraction and low-input DNase-seq (Inoue et al., 2017). Most accessible chromatin sites are similar between parental genomes, but a fraction displays allelic specificity that correlates with allelic gene expression profiles. Whether these sites might resemble loci primed for ZGA and potentially harbor information about putative pioneer factors of early embryonic development needs to be determined. However, it might be important to study zygotes at an earlier cell cycle stage in which putative pioneer factors might take first action and to test their ability to initiate a regulatory response that facilitates a totipotent state.
Perspectives
Recent technological advances in genome-wide low-input assays are starting to describe the chromatin state of the totipotent embryo. These will expand to obtain a comprehensive and detailed picture of embryonic histone modifications, DNA modifications, chromatin accessibility, and higher-order chromatin structure. Ultimately, functional assays will have to be developed to test the importance of the in vivo chromatin state for totipotency and developmental potential. Genetic perturbation experiments using maternal knockout strategies of candidate factors have great power to shed light on the essential regulators of early embryonic development. Thus, maternal-effect genes can be identified that are maternal factors nonessential for oogenesis but critical for embryonic development (Table 2). Genetic engineering is becoming easier with the advance of the CRISPR-Cas system (Cong et al., 2013; Wang et al., 2013). This may facilitate screening for pioneer factors that initiate a totipotent state by allowing cooperative action of a set of factors important for ZGA. The identification of such factors in Drosophila and zebrafish embryos (Staudt et al., 2006; Lee et al., 2013; Leichsenring et al., 2013) led to the assumption that mammalian counterparts exist. Pioneer factors might function not only by promoting transcription but also by inhibiting other transcriptional programs. This is exemplified through the role of Myt1l (myelin transcription factor 1-like) in protecting neuronal identity by repressing somatic lineage programs and allowing neuronal-specific transcription (Mall et al., 2017). It is possible that such transcriptional repressors also act during acquisition of totipotency to inhibit a multitude of differentiation programs.
Table 2. Maternal-effect genes found by genetic perturbation experiments with predominant developmental arrest at the zygote stage.
| Gene/aliases | KD/KO | Developmental role | References |
|---|---|---|---|
| Gas6/growth arrest specific 6 | KD by RNAi | Maternal cytoplasmic maturation, sperm chromatin decondensation, pronuclear formation | Kim et al., 2011, 2018 |
| Hira/histone cell cycle regulation defective homolog A | cKO using (Tg)Zp3-Cre and (Tg)Gdf9-Cre | Transcription, replication, paternal nucleosome assembly | Lin et al., 2014; Nashun et al., 2015 |
| Npm2/nucleoplasmin 2 | KO | Nuclear and nucleolar organization, chromatin remodeling | Burns et al., 2003 |
| Scc1 (Rad21)/double-strand-break repair protein rad21 homolog | cKO using (Tg)Zp3-Cre | Mitotic sister chromatid cohesion, repair of Tet3-dependent paternal DNA lesions, higher-order chromatin structure | Ladstätter and Tachibana-Konwalski, 2016; Gassler et al., 2017 |
| Zar1/zygote arrest 1 | KO | RNA processing, pronuclear fusion | Wu et al., 2003; Hu et al., 2010 |
| Ago2/argonaute 2, RISC catalytic component | cKO using (Tg)Zp3-Cre | miRNA homeostasis, post-transcriptional gene silencing of maternal factors | Kaneda et al., 2009; conflicting arrest after two-cell (Morita et al., 2007; Lykke-Andersen et al., 2008) |
| Hsf1/heat shock factor 1 | KO | Transcription, redox-homeostasis | Christians et al., 2000; Bierkamp et al., 2010 |
cKO, conditional KO; KD, knockdown.
Next to in vivo functional assays addressing the interplay of chromatin state and developmental potential, insights may be gained from in vitro modeling. Considering the hypothesis of phase separation–guided genome compartmentalization, one can speculate that the epigenetic state of nucleosomes can be used to predict higher-order chromatin structure. Indeed, such a prediction has been computationally tested by integrating an array of several epigenetic markers in neuronal cells (Di Pierro et al., 2017). Nevertheless, how chromatin is organized in the embryonic interphase nucleus remains a fascinating subject for ongoing work. Advances in low-input ChIP (Dahl et al., 2016; Zhang et al., 2016) will help to define the epigenome of early embryos and might allow computational de novo modeling of higher-order chromatin structure.
We are now entering a new dimension to study fundamental biological processes such as cellular plasticity in sparse cells of the early embryo. How chromatin is reprogrammed to totipotency within hours after fertilization remains a central question in biology. A combination of mechanistic cell biology with genetics and genomics will shed light on how chromatin reorganization promotes totipotency and the essential regulators for this dramatic cell fate transition.
Acknowledgments
We thank B.J.H. Dequeker, J. Gassler, and T.S. Powell for critical reading of the manuscript.
Research in the Tachibana laboratory is funded by the Austrian Academy of Sciences, the European Research Council (ERC-StG-336460 ChromHeritance), the Human Frontier Science Program (RGP0057-2018), and the Austrian Science Fund in the form of the Doctoral Program (DK) Chromosome Dynamics grant (W1238-B20), Special Research Program (SFB) Chromosome Dynamics grant (F3419-B19), and Herzfelder Foundation grant (P 30613-B21).
The authors declare no competing financial interests.
Author contributions: S. Ladstätter wrote the manuscript and K. Tachibana edited the manuscript.
References
- Ahmed K., Dehghani H., Rugg-Gunn P., Fussner E., Rossant J., and Bazett-Jones D.P.. 2010. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One. 5:e10531 10.1371/journal.pone.0010531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipour E., and Marko J.F.. 2012. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 40:11202–11212. 10.1093/nar/gks925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amouroux R., Nashun B., Shirane K., Nakagawa S., Hill P.W.S., D’Souza Z., Nakayama M., Matsuda M., Turp A., Ndjetehe E., et al. . 2016. De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat. Cell Biol. 18:225–233. 10.1038/ncb3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki F., Worrad D.M., and Schultz R.M.. 1997. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev. Biol. 181:296–307. 10.1006/dbio.1996.8466 [DOI] [PubMed] [Google Scholar]
- Arav A., Aroyo A., Yavin S., and Roth Z.. 2008. Prediction of embryonic developmental competence by time-lapse observation and ‘shortest-half’ analysis. Reprod. Biomed. Online. 17:669–675. 10.1016/S1472-6483(10)60314-8 [DOI] [PubMed] [Google Scholar]
- Bachvarova R. 1985. Gene Expression During Oogenesis and Oocyte Development in Mammals. In Oogenesis. Browder L.W., editor. Springer US, Boston, MA: 453–524. 10.1007/978-1-4615-6814-8_11 [DOI] [PubMed] [Google Scholar]
- Barau J., Teissandier A., Zamudio N., Roy S., Nalesso V., Hérault Y., Guillou F., and Bourc’his D.. 2016. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science. 354:909–912. 10.1126/science.aah5143 [DOI] [PubMed] [Google Scholar]
- Battulin N., Fishman V.S., Mazur A.M., Pomaznoy M., Khabarova A.A., Afonnikov D.A., Prokhortchouk E.B., and Serov O.L.. 2015. Comparison of the three-dimensional organization of sperm and fibroblast genomes using the Hi-C approach. Genome Biol. 16:77 10.1186/s13059-015-0642-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaujean N., Hartshorne G., Cavilla J., Taylor J., Gardner J., Wilmut I., Meehan R., and Young L.. 2004. Non-conservation of mammalian preimplantation methylation dynamics. Curr. Biol. 14:R266–R267. 10.1016/j.cub.2004.03.019 [DOI] [PubMed] [Google Scholar]
- Bench G.S., Friz A.M., Corzett M.H., Morse D.H., and Balhorn R.. 1996. DNA and total protamine masses in individual sperm from fertile mammalian subjects. Cytometry. 23:263–271. [DOI] [PubMed] [Google Scholar]
- Bickmore W.A. 2013. The spatial organization of the human genome. Annu. Rev. Genomics Hum. Genet. 14:67–84. 10.1146/annurev-genom-091212-153515 [DOI] [PubMed] [Google Scholar]
- Bierkamp C., Luxey M., Metchat A., Audouard C., Dumollard R., and Christians E.. 2010. Lack of maternal Heat Shock Factor 1 results in multiple cellular and developmental defects, including mitochondrial damage and altered redox homeostasis, and leads to reduced survival of mammalian oocytes and embryos. Dev. Biol. 339:338–353. 10.1016/j.ydbio.2009.12.037 [DOI] [PubMed] [Google Scholar]
- Booth M.J., Raiber E.A., and Balasubramanian S.. 2015. Chemical methods for decoding cytosine modifications in DNA. Chem. Rev. 115:2240–2254. 10.1021/cr5002904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsos M., and Torres-Padilla M.-E.. 2016. Building up the nucleus: nuclear organization in the establishment of totipotency and pluripotency during mammalian development. Genes Dev. 30:611–621. 10.1101/gad.273805.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouniol-Baly C., Hamraoui L., Guibert J., Beaujean N., Szöllösi M.S., and Debey P.. 1999. Differential transcriptional activity associated with chromatin configuration in fully grown mouse germinal vesicle oocytes. Biol. Reprod. 60:580–587. 10.1095/biolreprod60.3.580 [DOI] [PubMed] [Google Scholar]
- Brackley C.A., Johnson J., Kelly S., Cook P.R., and Marenduzzo D.. 2016. Simulated binding of transcription factors to active and inactive regions folds human chromosomes into loops, rosettes and topological domains. Nucleic Acids Res. 44:3503–3512. 10.1093/nar/gkw135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braude P., Bolton V., and Moore S.. 1988. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature. 332:459–461. 10.1038/332459a0 [DOI] [PubMed] [Google Scholar]
- Buenrostro J.D., Wu B., Litzenburger U.M., Ruff D., Gonzales M.L., Snyder M.P., Chang H.Y., and Greenleaf W.J.. 2015. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 523:486–490. 10.1038/nature14590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns K.H., Viveiros M.M., Ren Y., Wang P., DeMayo F.J., Frail D.E., Eppig J.J., and Matzuk M.M.. 2003. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science. 300:633–636. 10.1126/science.1081813 [DOI] [PubMed] [Google Scholar]
- Burton A., and Torres-Padilla M.-E.. 2010. Epigenetic reprogramming and development: a unique heterochromatin organization in the preimplantation mouse embryo. Brief. Funct. Genomics. 9:444–454. 10.1093/bfgp/elq027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone B.R., Hung J.H., Hainer S.J., Chou M.T., Carone D.M., Weng Z., Fazzio T.G., and Rando O.J.. 2014. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev. Cell. 30:11–22. 10.1016/j.devcel.2014.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians E., Davis A.A., Thomas S.D., and Benjamin I.J.. 2000. Maternal effect of Hsf1 on reproductive success. Nature. 407:693–694. 10.1038/35037669 [DOI] [PubMed] [Google Scholar]
- Condic M.L. 2014. Totipotency: what it is and what it is not. Stem Cells Dev. 23:796–812. 10.1089/scd.2013.0364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., and Zhang F.. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science. 339:819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellino S., Xu J., Sannai M., Moore R., Caretti E., Cigliano A., Le Coz M., Devarajan K., Wessels A., Soprano D., et al. . 2011. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 146:67–79. 10.1016/j.cell.2011.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich D.A., Daza R., Adey A., Pliner H.A., Christiansen L., Gunderson K.L., Steemers F.J., Trapnell C., and Shendure J.. 2015. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 348:910–914. 10.1126/science.aab1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl J.A., Jung I., Aanes H., Greggains G.D., Manaf A., Lerdrup M., Li G., Kuan S., Li B., Lee A.Y., et al. . 2016. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 537:548–552. 10.1038/nature19360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H.Q., Wang B.A., Yang L., Chen J.J., Zhu G.C., Sun M.L., Ge H., Wang R., Chapman D.L., Tang F., et al. . 2016. TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling. Nature. 538:528–532. 10.1038/nature20095 [DOI] [PubMed] [Google Scholar]
- Dean W., Santos F., Stojkovic M., Zakhartchenko V., Walter J., Wolf E., and Reik W.. 2001. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc. Natl. Acad. Sci. USA. 98:13734–13738. 10.1073/pnas.241522698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A., Planet E., Coluccio A., Verp S., Duc J., and Trono D.. 2017. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 49:941–945. 10.1038/ng.3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., and Misteli T.. 2015. Long-Range Chromatin Interactions. Cold Spring Harb. Perspect. Biol. 7:a019356 10.1101/cshperspect.a019356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denker A., and de Laat W.. 2016. The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev. 30:1357–1382. 10.1101/gad.281964.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pierro M., Zhang B., Aiden E.L., Wolynes P.G., and Onuchic J.N.. 2016. Transferable model for chromosome architecture. Proc. Natl. Acad. Sci. USA. 113:12168–12173. 10.1073/pnas.1613607113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pierro M., Cheng R.R., Lieberman Aiden E., Wolynes P.G., and Onuchic J.N.. 2017. De novo prediction of human chromosome structures: Epigenetic marking patterns encode genome architecture. Proc. Natl. Acad. Sci. USA. 114:12126–12131. 10.1073/pnas.1714980114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A.T., Raja R., Abeyta M.J., Taylor T., Shen S., Haqq C., and Pera R.A.. 2004. The unique transcriptome through day 3 of human preimplantation development. Hum. Mol. Genet. 13:1461–1470. 10.1093/hmg/ddh157 [DOI] [PubMed] [Google Scholar]
- Donaghey J., Thakurela S., Charlton J., Chen J.S., Smith Z.D., Gu H., Pop R., Clement K., Stamenova E.K., Karnik R., et al. . 2018. Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat. Genet. 50:250–258. 10.1038/s41588-017-0034-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P., and Liu Z.. 2017. Shaping development by stochasticity and dynamics in gene regulation. Open Biol. 7:170030 10.1098/rsob.170030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zheng H., Huang B., Ma R., Wu J., Zhang X., He J., Xiang Y., Wang Q., Li Y., et al. . 2017. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature. 547:232–235. 10.1038/nature23263 [DOI] [PubMed] [Google Scholar]
- Eckersley-Maslin M.A., Svensson V., Krueger C., Stubbs T.M., Giehr P., Krueger F., Miragaia R.J., Kyriakopoulos C., Berrens R.V., Milagre I., et al. . 2016. MERVL/Zscan4 Network Activation Results in Transient Genome-wide DNA Demethylation of mESCs. Cell Reports. 17:179–192. 10.1016/j.celrep.2016.08.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimova O.A., Pendina A.A., Tikhonov A.V., Fedorova I.D., Krapivin M.I., Chiryaeva O.G., Shilnikova E.M., Bogdanova M.A., Kogan I.Y., Kuznetzova T.V., et al. . 2015. Chromosome hydroxymethylation patterns in human zygotes and cleavage-stage embryos. Reproduction. 149:223–233. 10.1530/REP-14-0343 [DOI] [PubMed] [Google Scholar]
- Filipescu D., Müller S., and Almouzni G.. 2014. Histone H3 variants and their chaperones during development and disease: contributing to epigenetic control. Annu. Rev. Cell Dev. Biol. 30:615–646. 10.1146/annurev-cellbio-100913-013311 [DOI] [PubMed] [Google Scholar]
- Finn C.A., and McLaren A.. 1967. A study of the early stages of implantation in mice. J. Reprod. Fertil. 13:259–267. 10.1530/jrf.0.0130259 [DOI] [PubMed] [Google Scholar]
- Flach G., Johnson M.H., Braude P.R., Taylor R.A., and Bolton V.N.. 1982. The transition from maternal to embryonic control in the 2-cell mouse embryo. EMBO J. 1:681–686. 10.1002/j.1460-2075.1982.tb01230.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyamer I.M., Gassler J., Imakaev M., Brandão H.B., Ulianov S.V., Abdennur N., Razin S.V., Mirny L.A., and Tachibana-Konwalski K.. 2017. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature. 544:110–114. 10.1038/nature21711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty N.M.E., McCarthy A., Snijders K.E., Powell B.E., Kubikova N., Blakeley P., Lea R., Elder K., Wamaitha S.E., Kim D., et al. . 2017. Genome editing reveals a role for OCT4 in human embryogenesis. Nature. 550:67–73. 10.1038/nature24033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fougerousse F., Bullen P., Herasse M., Lindsay S., Richard I., Wilson D., Suel L., Durand M., Robson S., Abitbol M., et al. . 2000. Human-mouse differences in the embryonic expression patterns of developmental control genes and disease genes. Hum. Mol. Genet. 9:165–173. 10.1093/hmg/9.2.165 [DOI] [PubMed] [Google Scholar]
- Friedberg E.C., Meira L.B., and Cheo D.L.. 1997. Database of mouse strains carrying targeted mutations in genes affecting cellular responses to DNA damage. Mutat. Res. 383:183–188. 10.1016/S0921-8777(96)00057-2 [DOI] [PubMed] [Google Scholar]
- Fudenberg G., Imakaev M., Lu C., Goloborodko A., Abdennur N., and Mirny L.A.. 2016. Formation of Chromosomal Domains by Loop Extrusion. Cell Reports. 15:2038–2049. 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulka H., Mrazek M., Tepla O., and Fulka J. Jr. 2004. DNA methylation pattern in human zygotes and developing embryos. Reproduction. 128:703–708. 10.1530/rep.1.00217 [DOI] [PubMed] [Google Scholar]
- Gao L., Wu K., Liu Z., Yao X., Yuan S., Tao W., Yi L., Yu G., Hou Z., Fan D., et al. . 2018. Chromatin Accessibility Landscape in Human Early Embryos and Its Association with Evolution. Cell. 173:248–259.e15. 10.1016/j.cell.2018.02.028 [DOI] [PubMed] [Google Scholar]
- Gassler J., Brandão H.B., Imakaev M., Flyamer I.M., Ladstätter S., Bickmore W.A., Peters J.M., Mirny L.A., and Tachibana K.. 2017. A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J. 36:3600–3618. 10.15252/embj.201798083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M., and Felsenfeld G.. 2006. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 7:703–713. 10.1038/nrg1925 [DOI] [PubMed] [Google Scholar]
- Gu T.-P., Guo F., Yang H., Wu H.-P., Xu G.-F., Liu W., Xie Z.-G., Shi L., He X., Jin S.G., et al. . 2011. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 477:606–610. 10.1038/nature10443 [DOI] [PubMed] [Google Scholar]
- Guo F., Li X., Liang D., Li T., Zhu P., Guo H., Wu X., Wen L., Gu T.-P., Hu B., et al. . 2014a Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 15:447–459. 10.1016/j.stem.2014.08.003 [DOI] [PubMed] [Google Scholar]
- Guo H., Zhu P., Yan L., Li R., Hu B., Lian Y., Yan J., Ren X., Lin S., Li J., et al. . 2014b The DNA methylation landscape of human early embryos. Nature. 511:606–610. 10.1038/nature13544 [DOI] [PubMed] [Google Scholar]
- Haarhuis J.H.I., van der Weide R.H., Blomen V.A., Yáñez-Cuna J.O., Amendola M., van Ruiten M.S., Krijger P.H.L., Teunissen H., Medema R.H., van Steensel B., et al. . 2017. The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell. 169:693–707.e14. 10.1016/j.cell.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P., Jeffries S.J., Lee C., Miller N., Jackson S.P., and Surani M.A.. 2010. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 329:78–82. 10.1126/science.1187945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatani T., Carter M.G., Sharov A.A., and Ko M.S.. 2004. Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell. 6:117–131. 10.1016/S1534-5807(03)00373-3 [DOI] [PubMed] [Google Scholar]
- Hansen A.S., Pustova I., Cattoglio C., Tjian R., and Darzacq X.. 2017. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. eLife. 6:e25776 10.7554/eLife.25776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M.M., Li X.Y., Kaplan T., Botchan M.R., and Eisen M.B.. 2011. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 7:e1002266 10.1371/journal.pgen.1002266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y.-F., Li B.-Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., et al. . 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 333:1303–1307. 10.1126/science.1210944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson P.G., Doráis J.A., Grow E.J., Whiddon J.L., Lim J.W., Wike C.L., Weaver B.D., Pflueger C., Emery B.R., Wilcox A.L., et al. . 2017. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 49:925–934. 10.1038/ng.3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertig A.T., Rock J., and Adams E.C.. 1956. A description of 34 human ova within the first 17 days of development. Am. J. Anat. 98:435–493. 10.1002/aja.1000980306 [DOI] [PubMed] [Google Scholar]
- Hirasawa R., Chiba H., Kaneda M., Tajima S., Li E., Jaenisch R., and Sasaki H.. 2008. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 22:1607–1616. 10.1101/gad.1667008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Wang F., Zhu X., Yuan Y., Ding M., and Gao S.. 2010. Mouse ZAR1-like (XM_359149) colocalizes with mRNA processing components and its dominant-negative mutant caused two-cell-stage embryonic arrest. Dev. Dyn. 239:407–424. 10.1002/dvdy.22170 [DOI] [PubMed] [Google Scholar]
- Huang Y., Pastor W.A., Shen Y., Tahiliani M., Liu D.R., and Rao A.. 2010. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 5:e8888 10.1371/journal.pone.0008888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug C.B., Grimaldi A.G., Kruse K., and Vaquerizas J.M.. 2017. Chromatin Architecture Emerges during Zygotic Genome Activation Independent of Transcription. Cell. 169:216–228.e19. 10.1016/j.cell.2017.03.024 [DOI] [PubMed] [Google Scholar]
- Inoue A., and Zhang Y.. 2011. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 334:194 10.1126/science.1212483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A., Shen L., Dai Q., He C., and Zhang Y.. 2011. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 21:1670–1676. 10.1038/cr.2011.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A., Matoba S., and Zhang Y.. 2012. Transcriptional activation of transposable elements in mouse zygotes is independent of Tet3-mediated 5-methylcytosine oxidation. Cell Res. 22:1640–1649. 10.1038/cr.2012.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A., Shen L., Matoba S., and Zhang Y.. 2015. Haploinsufficiency, but not defective paternal 5mC oxidation, accounts for the developmental defects of maternal Tet3 knockouts. Cell Reports. 10:463–470. 10.1016/j.celrep.2014.12.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A., Jiang L., Lu F., Suzuki T., and Zhang Y.. 2017. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature. 547:419–424. 10.1038/nature23262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K., Jin S.-G., Pfeifer G.P., and Szabó P.E.. 2011. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. USA. 108:3642–3647. 10.1073/pnas.1014033108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiuchi T., Enriquez-Gasca R., Mizutani E., Bošković A., Ziegler-Birling C., Rodriguez-Terrones D., Wakayama T., Vaquerizas J.M., and Torres-Padilla M.-E.. 2015. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat. Struct. Mol. Biol. 22:662–671. 10.1038/nsmb.3066 [DOI] [PubMed] [Google Scholar]
- Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., and Zhang Y.. 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 333:1300–1303. 10.1126/science.1210597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iurlaro M., Ficz G., Oxley D., Raiber E.A., Bachman M., Booth M.J., Andrews S., Balasubramanian S., and Reik W.. 2013. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 14:R119 10.1186/gb-2013-14-10-r119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M., and Zaret K.S.. 2014. Pioneer transcription factors in cell reprogramming. Genes Dev. 28:2679–2692. 10.1101/gad.253443.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D., Meydan C., Lange J., Claeys Bouuaert C., Lailler N., Mason C.E., Anderson K.V., and Keeney S.. 2017. rahu is a mutant allele of Dnmt3c, encoding a DNA methyltransferase homolog required for meiosis and transposon repression in the mouse male germline. PLoS Genet. 13:e1006964 10.1371/journal.pgen.1006964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y.S., Yeo S., Park J.S., Koo D.B., Chang W.K., Lee K.K., and Kang Y.K.. 2007. DNA methylation state is preserved in the sperm-derived pronucleus of the pig zygote. Int. J. Dev. Biol. 51:707–714. 10.1387/ijdb.072450yj [DOI] [PubMed] [Google Scholar]
- Jin S.G., Kadam S., and Pfeifer G.P.. 2010. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 38:e125 10.1093/nar/gkq223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Tang Q., Wan M., Cui K., Zhang Y., Ren G., Ni B., Sklar J., Przytycka T.M., Childs R., et al. . 2015. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature. 528:142–146. 10.1038/nature15740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.A. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13:484–492. 10.1038/nrg3230 [DOI] [PubMed] [Google Scholar]
- Jukam D., Shariati S.A.M., and Skotheim J.M.. 2017. Zygotic Genome Activation in Vertebrates. Dev. Cell. 42:316–332. 10.1016/j.devcel.2017.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y.H., Sauria M.E.G., Lyu X., Cheema M.S., Ausio J., Taylor J., and Corces V.G.. 2017. Chromatin States in Mouse Sperm Correlate with Embryonic and Adult Regulatory Landscapes. Cell Reports. 18:1366–1382. 10.1016/j.celrep.2017.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M., Tang F., O’Carroll D., Lao K., and Surani M.A.. 2009. Essential role for Argonaute2 protein in mouse oogenesis. Epigenetics Chromatin. 2:9 10.1186/1756-8935-2-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y., Xu Y., Chen X., Feng S., Liu Z., Sun Y., Yao X., Li F., Zhu W., Gao L., et al. . 2017. 3D Chromatin Structures of Mature Gametes and Structural Reprogramming during Mammalian Embryogenesis. Cell. 170:367–381.e20. 10.1016/j.cell.2017.06.029 [DOI] [PubMed] [Google Scholar]
- Kierszenbaum A.L., and Tres L.L.. 1975. Structural and transcriptional features of the mouse spermatid genome. J. Cell Biol. 65:258–270. 10.1083/jcb.65.2.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.H., Kim E.Y., Kim Y., Kim E., Lee H.S., Yoon S.Y., and Lee K.A.. 2011. Gas6 downregulation impaired cytoplasmic maturation and pronuclear formation independent to the MPF activity. PLoS One. 6:e23304 10.1371/journal.pone.0023304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.H., Kim E.Y., Lee S.Y., Ko J.J., and Lee K.A.. 2018. Oocyte Cytoplasmic Gas6 and Heparan Sulfate (HS) are Required to Establish the Open Chromatin State in Nuclei During Remodeling and Reprogramming. Cell. Physiol. Biochem. 45:37–53. 10.1159/000486221 [DOI] [PubMed] [Google Scholar]
- King H.W., and Klose R.J.. 2017. The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. eLife. 6:e22631 10.7554/eLife.22631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Agerholm I.E., and Ingerslev H.J.. 2012. Time-lapse monitoring as a tool for clinical embryo assessment. Hum. Reprod. 27:1277–1285. 10.1093/humrep/des079 [DOI] [PubMed] [Google Scholar]
- Ladstätter S., and Tachibana-Konwalski K.. 2016. A Surveillance Mechanism Ensures Repair of DNA Lesions during Zygotic Reprogramming. Cell. 167:1774–1787.e13. 10.1016/j.cell.2016.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.T., Bonneau A.R., Takacs C.M., Bazzini A.A., DiVito K.R., Fleming E.S., and Giraldez A.J.. 2013. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 503:360–364. 10.1038/nature12632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichsenring M., Maes J., Mössner R., Driever W., and Onichtchouk D.. 2013. Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science. 341:1005–1009. 10.1126/science.1242527 [DOI] [PubMed] [Google Scholar]
- Lepikhov K., Zakhartchenko V., Hao R., Yang F., Wrenzycki C., Niemann H., Wolf E., and Walter J.. 2008. Evidence for conserved DNA and histone H3 methylation reprogramming in mouse, bovine and rabbit zygotes. Epigenetics Chromatin. 1:8 10.1186/1756-8935-1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E., Bestor T.H., and Jaenisch R.. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 69:915–926. 10.1016/0092-8674(92)90611-F [DOI] [PubMed] [Google Scholar]
- Li L., Guo F., Gao Y., Ren Y., Yuan P., Yan L., Li R., Lian Y., Li J., Hu B., et al. . 2018. Single-cell multi-omics sequencing of human early embryos. Nat. Cell Biol. 10.1038/s41556-018-0123-2 [DOI] [PubMed] [Google Scholar]
- Liang H.L., Nien C.Y., Liu H.Y., Metzstein M.M., Kirov N., and Rushlow C.. 2008. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 456:400–403. 10.1038/nature07388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O., et al. . 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 326:289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.J., Koh F.M., Wong P., Conti M., and Ramalho-Santos M.. 2014. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev. Cell. 30:268–279. 10.1016/j.devcel.2014.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F., Liu Y., Inoue A., Suzuki T., Zhao K., and Zhang Y.. 2016. Establishing chromatin regulatory landscape during mouse preimplantation development. Cell. 165:1375–1388. 10.1016/j.cell.2016.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen K., Gilchrist M.J., Grabarek J.B., Das P., Miska E., and Zernicka-Goetz M.. 2008. Maternal Argonaute 2 is essential for early mouse development at the maternal-zygotic transition. Mol. Biol. Cell. 19:4383–4392. 10.1091/mbc.e08-02-0219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., Firth A., Singer O., Trono D., and Pfaff S.L.. 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 487:57–63. 10.1038/nature11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mall M., Kareta M.S., Chanda S., Ahlenius H., Perotti N., Zhou B., Grieder S.D., Ge X., Drake S., Euong Ang C., et al. . 2017. Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates. Nature. 544:245–249. 10.1038/nature21722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W., Niveleau A., Walter J., Fundele R., and Haaf T.. 2000. Demethylation of the zygotic paternal genome. Nature. 403:501–502. 10.1038/35000656 [DOI] [PubMed] [Google Scholar]
- Meistrich M.L., Mohapatra B., Shirley C.R., and Zhao M.. 2003. Roles of transition nuclear proteins in spermiogenesis. Chromosoma. 111:483–488. 10.1007/s00412-002-0227-z [DOI] [PubMed] [Google Scholar]
- Mio Y., and Maeda K.. 2008. Time-lapse cinematography of dynamic changes occurring during in vitro development of human embryos. Am. J. Obstet. Gynecol. 199:660.e1–660.e5. 10.1016/j.ajog.2008.07.023 [DOI] [PubMed] [Google Scholar]
- Morita S., Horii T., Kimura M., Goto Y., Ochiya T., and Hatada I.. 2007. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics. 89:687–696. 10.1016/j.ygeno.2007.01.004 [DOI] [PubMed] [Google Scholar]
- Musselman C.A., Lalonde M.E., Côté J., and Kutateladze T.G.. 2012. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 19:1218–1227. 10.1038/nsmb.2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T., Lubling Y., Várnai C., Dudley C., Leung W., Baran Y., Mendelson Cohen N., Wingett S., Fraser P., and Tanay A.. 2017. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature. 547:61–67. 10.1038/nature23001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Arai Y., Umehara H., Masuhara M., Kimura T., Taniguchi H., Sekimoto T., Ikawa M., Yoneda Y., Okabe M., et al. . 2007. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 9:64–71. 10.1038/ncb1519 [DOI] [PubMed] [Google Scholar]
- Nashun B., Hill P.W.S., Smallwood S.A., Dharmalingam G., Amouroux R., Clark S.J., Sharma V., Ndjetehe E., Pelczar P., Festenstein R.J., et al. . 2015. Continuous Histone Replacement by Hira Is Essential for Normal Transcriptional Regulation and De Novo DNA Methylation during Mouse Oogenesis. Mol. Cell. 60:611–625. 10.1016/j.molcel.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. 2001. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35:673–745. 10.1146/annurev.genet.35.102401.091334 [DOI] [PubMed] [Google Scholar]
- Nestor C., Ruzov A., Meehan R., and Dunican D.. 2010. Enzymatic approaches and bisulfite sequencing cannot distinguish between 5-methylcytosine and 5-hydroxymethylcytosine in DNA. Biotechniques. 48:317–319. 10.2144/000113403 [DOI] [PubMed] [Google Scholar]
- Niakan K.K., and Eggan K.. 2013. Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse. Dev. Biol. 375:54–64. 10.1016/j.ydbio.2012.12.008 [DOI] [PubMed] [Google Scholar]
- Ohnuki M., and Takahashi K. Present and future challenges of induced pluripotent stem cells. . Philos. Trans. R. Soc. B Biol. Sci. 2015;370 doi: 10.1098/rstb.2014.0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M., Bell D.W., Haber D.A., and Li E.. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 99:247–257. 10.1016/S0092-8674(00)81656-6 [DOI] [PubMed] [Google Scholar]
- Oliva R. 2006. Protamines and male infertility. Hum. Reprod. Update. 12:417–435. 10.1093/humupd/dml009 [DOI] [PubMed] [Google Scholar]
- Oswald J., Engemann S., Lane N., Mayer W., Olek A., Fundele R., Dean W., Reik W., and Walter J.. 2000. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 10:475–478. 10.1016/S0960-9822(00)00448-6 [DOI] [PubMed] [Google Scholar]
- Park J.S., Jeong Y.S., Shin S.T., Lee K.K., and Kang Y.K.. 2007. Dynamic DNA methylation reprogramming: active demethylation and immediate remethylation in the male pronucleus of bovine zygotes. Dev. Dyn. 236:2523–2533. 10.1002/dvdy.21278 [DOI] [PubMed] [Google Scholar]
- Peat J.R., Dean W., Clark S.J., Krueger F., Smallwood S.A., Ficz G., Kim J.K., Marioni J.C., Hore T.A., and Reik W.. 2014. Genome-wide bisulfite sequencing in zygotes identifies demethylation targets and maps the contribution of TET3 oxidation. Cell Reports. 9:1990–2000. 10.1016/j.celrep.2014.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M., Lin C.J., Yin Y., Guan J., Peixoto G.A., Bulut-Karslioglu A., Biechele S., Huang B., Shen X., and Ramalho-Santos M.. 2018. A LINE1-Nucleolin Partnership Regulates Early Development and ESC Identity. Cell. 174:391–405.e19. 10.1016/j.cell.2018.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman-Livaja M., and Rando O.J.. 2010. Nucleosome positioning: how is it established, and why does it matter? Dev. Biol. 339:258–266. 10.1016/j.ydbio.2009.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., and Aiden E.L.. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 159:1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.S.P., Huang S.C., Glenn St Hilaire B., Engreitz J.M., Perez E.M., Kieffer-Kwon K.R., Sanborn A.L., Johnstone S.E., Bascom G.D., Bochkov I.D., et al. . 2017. Cohesin Loss Eliminates All Loop Domains. Cell. 171:305–320.e24. 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathke C., Baarends W.M., Awe S., and Renkawitz-Pohl R.. 2014. Chromatin dynamics during spermiogenesis. Biochim. Biophys. Acta. 1839:155–168. 10.1016/j.bbagrm.2013.08.004 [DOI] [PubMed] [Google Scholar]
- Robinton D.A., and Daley G.Q.. 2012. The promise of induced pluripotent stem cells in research and therapy. Nature. 481:295–305. 10.1038/nature10761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodman T.C., Pruslin F.H., Hoffmann H.P., and Allfrey V.G.. 1981. Turnover of basic chromosomal proteins in fertilized eggs: a cytoimmunochemical study of events in vivo. J. Cell Biol. 90:351–361. 10.1083/jcb.90.2.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J. 1976. Postimplantation development of blastomeres isolated from 4- and 8-cell mouse eggs. J. Embryol. Exp. Morphol. 36:283–290. [PubMed] [Google Scholar]
- Rougier N., Bourc’his D., Gomes D.M., Niveleau A., Plachot M., Pàldi A., and Viegas-Péquignot E.. 1998. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 12:2108–2113. 10.1101/gad.12.14.2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., and Kundu T.K.. 2014. Gene regulatory networks and epigenetic modifications in cell differentiation. IUBMB Life. 66:100–109. 10.1002/iub.1249 [DOI] [PubMed] [Google Scholar]
- Sadeh R., and Allis C.D.. 2011. Genome-wide “re”-modeling of nucleosome positions. Cell. 147:263–266. 10.1016/j.cell.2011.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn A.L., Rao S.S.P., Huang S.C., Durand N.C., Huntley M.H., Jewett A.I., Bochkov I.D., Chinnappan D., Cutkosky A., Li J., et al. . 2015. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. USA. 112:E6456–E6465. 10.1073/pnas.1518552112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F., Peat J., Burgess H., Rada C., Reik W., and Dean W.. 2013. Active demethylation in mouse zygotes involves cytosine deamination and base excision repair. Epigenetics Chromatin. 6:39 10.1186/1756-8935-6-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D. 2015. Function and information content of DNA methylation. Nature. 517:321–326. 10.1038/nature14192 [DOI] [PubMed] [Google Scholar]
- Schulz K.N., Bondra E.R., Moshe A., Villalta J.E., Lieb J.D., Kaplan T., Mckay D.J., and Harrison M.M.. 2015. Zelda is differentially required for chromatin accessibility, transcription factor binding, and gene expression in the early Drosophila embryo. Genome Res. 25:1715–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W., Abdennur N., Goloborodko A., Pekowska A., Fudenberg G., Loe-Mie Y., Fonseca N.A., Huber W., H Haering C., Mirny L., and Spitz F.. 2017. Two independent modes of chromatin organization revealed by cohesin removal. Nature. 551:51–56. 10.1038/nature24281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Inoue A., He J., Liu Y., Lu F., and Zhang Y.. 2014. Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell. 15:459–471. 10.1016/j.stem.2014.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D., Stampfel G., and Stark A.. 2014. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 15:272–286. 10.1038/nrg3682 [DOI] [PubMed] [Google Scholar]
- Song C.X., Yi C., and He C.. 2012. Mapping recently identified nucleotide variants in the genome and transcriptome. Nat. Biotechnol. 30:1107–1116. 10.1038/nbt.2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrells T.R., and Johnson A.D.. 2015. Making sense of transcription networks. Cell. 161:714–723. 10.1016/j.cell.2015.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A., Donahue G., and Zaret K.S.. 2012. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 151:994–1004. 10.1016/j.cell.2012.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A., Garcia M.F., Jaroszewicz A., Osman N., Pellegrini M., and Zaret K.S.. 2015. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 161:555–568. 10.1016/j.cell.2015.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt C.G., Gnerlich F., Smits A.H., Pfaffeneder T., Jansen P.W.T.C., Bauer C., Münzel M., Wagner M., Müller M., Khan F., et al. . 2013. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 152:1146–1159. 10.1016/j.cell.2013.02.004 [DOI] [PubMed] [Google Scholar]
- Srivastava D., and DeWitt N.. 2016. In Vivo Cellular Reprogramming: The Next Generation. Cell. 166:1386–1396. 10.1016/j.cell.2016.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt N., Fellert S., Chung H.R., Jäckle H., and Vorbrüggen G.. 2006. Mutations of the Drosophila zinc finger-encoding gene vielfältig impair mitotic cell divisions and cause improper chromosome segregation. Mol. Biol. Cell. 17:2356–2365. 10.1091/mbc.e05-11-1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Nien C.Y., Chen K., Liu H.Y., Johnston J., Zeitlinger J., and Rushlow C.. 2015. Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Res. 25:1703–1714. 10.1101/gr.192542.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W., and Lipshitz H.D.. 2009. The maternal-to-zygotic transition: a play in two acts. Development. 136:3033–3042. 10.1242/dev.033183 [DOI] [PubMed] [Google Scholar]
- Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., and Rao A.. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 324:930–935. 10.1126/science.1170116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarkowski A.K. 1959. Experiments on the development of isolated blastomers of mouse eggs. Nature. 184:1286–1287. 10.1038/1841286a0 [DOI] [PubMed] [Google Scholar]
- Tesarík J., Kopecný V., Plachot M., and Mandelbaum J.. 1987. High-resolution autoradiographic localization of DNA-containing sites and RNA synthesis in developing nucleoli of human preimplantation embryos: a new concept of embryonic nucleologenesis. Development. 101:777–791. [DOI] [PubMed] [Google Scholar]
- Torres-Padilla M.-E., Bannister A.J., Hurd P.J., Kouzarides T., and Zernicka-Goetz M.. 2006. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int. J. Dev. Biol. 50(Next):455–461. 10.1387/ijdb.052073mt [DOI] [PubMed] [Google Scholar]
- Tsompana M., and Buck M.J.. 2014. Chromatin accessibility: a window into the genome. Epigenetics Chromatin. 7:33 10.1186/1756-8935-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y., Akiyama T., and Nakayama K.I.. 2015. Maternal TET3 is dispensable for embryonic development but is required for neonatal growth. Sci. Rep. 5:15876 10.1038/srep15876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden G.W., Dieker J.W., Derijck A.A.H.A., Muller S., Berden J.H.M., Braat D.D.M., van der Vlag J., and de Boer P.. 2005. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech. Dev. 122:1008–1022. 10.1016/j.mod.2005.04.009 [DOI] [PubMed] [Google Scholar]
- Van de Velde H., Cauffman G., Tournaye H., Devroey P., and Liebaers I.. 2008. The four blastomeres of a 4-cell stage human embryo are able to develop individually into blastocysts with inner cell mass and trophectoderm. Hum. Reprod. 23:1742–1747. 10.1093/humrep/den190 [DOI] [PubMed] [Google Scholar]
- Vassena R., Boué S., González-Roca E., Aran B., Auer H., Veiga A., and Izpisua Belmonte J.C.. 2011. Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development. 138:3699–3709. 10.1242/dev.064741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., and Jaenisch R.. 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 153:910–918. 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiddon J.L., Langford A.T., Wong C.-J., Zhong J.W., and Tapscott S.J.. 2017. Conservation and innovation in the DUX4-family gene network. Nat. Genet. 49:935–940. 10.1038/ng.3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C.C., Loewke K.E., Bossert N.L., Behr B., De Jonge C.J., Baer T.M., and Reijo Pera R.A.. 2010. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat. Biotechnol. 28:1115–1121. 10.1038/nbt.1686 [DOI] [PubMed] [Google Scholar]
- Worrad D.M., Ram P.T., and Schultz R.M.. 1994. Regulation of gene expression in the mouse oocyte and early preimplantation embryo: developmental changes in Sp1 and TATA box-binding protein, TBP. Development. 120:2347–2357. [DOI] [PubMed] [Google Scholar]
- Wossidlo M., Arand J., Sebastiano V., Lepikhov K., Boiani M., Reinhardt R., Schöler H., and Walter J.. 2010. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J. 29:1877–1888. 10.1038/emboj.2010.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M., Nakamura T., Lepikhov K., Marques C.J., Zakhartchenko V., Boiani M., Arand J., Nakano T., Reik W., and Walter J.. 2011. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2:241 10.1038/ncomms1240 [DOI] [PubMed] [Google Scholar]
- Wu X., and Zhang Y.. 2017. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 18:517–534. 10.1038/nrg.2017.33 [DOI] [PubMed] [Google Scholar]
- Wu J., Huang B., Chen H., Yin Q., Liu Y., Xiang Y., Zhang B., Liu B., Wang Q., Xia W., et al. . 2016. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 534:652–657. 10.1038/nature18606 [DOI] [PubMed] [Google Scholar]
- Wu X., Viveiros M.M., Eppig J.J., Bai Y., Fitzpatrick S.L., and Matzuk M.M.. 2003. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat. Genet. 33:187–191. 10.1038/ng1079 [DOI] [PubMed] [Google Scholar]
- Wutz G., Várnai C., Nagasaka K., Cisneros D.A., Stocsits R.R., Tang W., Schoenfelder S., Jessberger G., Muhar M., Hossain M.J., et al. . 2017. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36:3573–3599. 10.15252/embj.201798004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., and Xie W.. 2018. Epigenome in Early Mammalian Development: Inheritance, Reprogramming and Establishment. Trends Cell Biol. 28:237–253. 10.1016/j.tcb.2017.10.008 [DOI] [PubMed] [Google Scholar]
- Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J., et al. . 2013. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20:1131–1139. 10.1038/nsmb.2660 [DOI] [PubMed] [Google Scholar]
- Yu M., Hon G.C., Szulwach K.E., Song C.X., Zhang L., Kim A., Li X., Dai Q., Shen Y., Park B., et al. . 2012. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 149:1368–1380. 10.1016/j.cell.2012.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K.S. 2018. Pioneering the chromatin landscape. Nat. Genet. 50:167–169. 10.1038/s41588-017-0038-z [DOI] [PubMed] [Google Scholar]
- Zaret K.S., and Carroll J.S.. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25:2227–2241. 10.1101/gad.176826.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Zheng H., Huang B., Li W., Xiang Y., Peng X., Ming J., Wu X., Zhang Y., Xu Q., et al. . 2016. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature. 537:553–557. 10.1038/nature19361 [DOI] [PubMed] [Google Scholar]
- Zhu C., Gao Y., Guo H., Xia B., Song J., Wu X., Zeng H., Kee K., Tang F., and Yi C.. 2017. Single-Cell 5-Formylcytosine Landscapes of Mammalian Early Embryos and ESCs at Single-Base Resolution. Cell Stem Cell. 20:720–731.e5. 10.1016/j.stem.2017.02.013 [DOI] [PubMed] [Google Scholar]